An extensive crosstalk between host and intestinal microbiota contributes to the development and maturation of intestinal epithelium and immune system. This review details the interplay between nucleotide-binding domain leucine-rich repeat containing proteins (NLR, or NOD-like receptors) signaling to host-microbiota homeostasis.
Abstract
The gastrointestinal tract harbors a highly complex microbial community, which is referred to as gut microbiota. With increasing evidence suggesting that the imbalance of gut microbiota plays a significant role in the pathogenesis of multiple diseases, interactions between the host immune system and the gut microbiota are now attracting emerging interest. Nucleotide-binding and leucine-rich repeat–containing receptors (NLRs) encompass a large number of innate immune sensors and receptors, which mediate the activation of Caspase-1 and the subsequent release of mature interleukin-1β and interleukin-18. Several family members have been found to restrain rather than activate inflammatory cytokines and immune signaling. NLR family members are central regulators of pathogen recognition, host immunity, and inflammation with utmost importance in human diseases. In this review, we focus on the potential roles played by NLRs in controlling and shaping the microbiota community and discuss how the functional axes interconnecting gut microbiota with NLRs impact the modulation of colitis, inflammatory bowel diseases, and colorectal cancer.
Introduction
Intestinal microbiome
Microorganisms cover almost all host mucosal surfaces, residing in different body niches including the gastrointestinal tract, skin, oral cavity, urogenital tract, and respiratory tract. These microorganisms, collectively referred to as microbiota, form an ecological community of commensal, symbiotic, and pathogenic microorganisms and have received increasing attention over the last decade for their immense impact on human health (Cho and Blaser, 2012; Ley et al., 2008). Microbial communities are complex and dynamic, composed of bacteria, fungi, viruses, protozoa, and archaea. The gastrointestinal tract is inhabited by ∼100 trillion microorganisms and over 1,000 different bacterial species, making it the greatest microbial compartment in the body (Gareau et al., 2010; Hooper and Macpherson, 2010). Most microbial analyses rely on fecal samples, which are noninvasively and readily available. However, it is worth noting that fecal profiling does not represent a complete picture of what is occurring within the host and cannot account for the spatial heterogeneity of gut microbiota (Donaldson et al., 2016; Tropini et al., 2017). Within the gastrointestinal system, bacterial species and communities are not uniformly distributed throughout the lumen, with the density and diversity of bacteria increasing along the gut longitudinal axis. Usually, the small intestine is dominated by fast-growing facultative anaerobes, while the cecum and colon favor saccharolytic anaerobes. Meanwhile, the mucous layer also provides habitat for a significant fraction of bacteria in mice and humans (Rogala et al., 2020). Increasing evidence suggests that a highly diverse microbiota helps maintain a healthy mucosal barrier and vice versa (Jarret et al., 2020; Okumura et al., 2016; Propheter et al., 2017; Vaishnava et al., 2011). The interactions between the mucosal-associated microbiome and the host innate immune system deserves much attention as a complement to fecal community studies.
Undeniably, the exciting advances in high-throughput sequencing methods and biological assays during the last 15 yr have accelerated knowledge about the genetic composition of gut microbiota under physiological and pathological conditions, shedding light on the various functions provided by this “forgotten organ” (Durack and Lynch, 2019). The entire genome sequence of a microbial community is referred to as the microbiome. By using metagenomics, metatranscriptomics, metaproteomics, and metametabolomics, combined with the use of various in vivo models (such as germ-free mice), the bacterial diversity, composition, function, and metabolic capacity of the gut microbiota have been widely studied (Schloss and Handelsman, 2005; Turnbaugh and Gordon, 2008; Verberkmoes et al., 2009). It has been shown that gut microbes have essential roles in regulating host metabolism and immunity. The imbalance or dysbiosis of a microbial community is associated with potential diseases, risks, and even to the clear onset of clinical symptoms (Cho and Blaser, 2012; Durack and Lynch, 2019; Gareau et al., 2010). As the study of the microbiota has matured, it has become apparent that the microbiota is affected by multiple complex factors, including the host’s genetics, age, sex, diet, lifestyle, living environment, disease status, infections, and metabolism (Rogers et al., 2014; Stappenbeck and Virgin, 2016; Zaneveld et al., 2017). Studies in mouse models suggest that in addition to the genetics background, nongenetic confounding factors such as maternal inheritance and vivarium differences should also be taken into account in order to draw meaningful conclusions and to increase reproducibility in this intense research area (Stappenbeck and Virgin, 2016). Littermate controls and fecal microbiota transplants in germ-free mice are highlighted by many researchers as rigorous and critical experimental designs in this field (Elinav et al., 2018; Mamantopoulos et al., 2018; Wullaert et al., 2018).
Nucleotide-binding domain and leucine-rich repeat–containing receptors (NLRs)
Communication between the host immune system and gut microbiota occurs through the action of pattern recognition receptors (PRRs) expressed on or in immune cells. One family of PRRs which is essential for communication between commensal microbes and the host immune system is NLR proteins (Hooper and Macpherson, 2010). NLRs are highly conserved intracellular proteins composed of a central nucleotide-binding and oligomerization domain, a C-terminal leucine-rich repeat domain, and an N-terminal protein–protein interaction domain. There are over 20 identified NLR proteins in humans and over 30 in mice, divided into groups based on structures of the variable N-terminus. The NLRP group of NLRs, for example, contains a pyrin domain at the N-terminus, while the NLRC group contains a caspase activation and recruitment domain (Kanneganti et al., 2007; Ting and Davis, 2005).
As in other PRRs, the defining function of the NLR family is to sense pathogen- or damage-associated molecular patterns (Ting and Davis, 2005; Kanneganti et al., 2007). The NLR family is unique among PRRs in its immense diversity of ligands and downstream effector functions. NLRs act as signaling molecules and scaffolding proteins, and the earliest member, CIITA, is a master transcription factor (Gregory et al., 2011; Ting et al., 2010). A well-defined function of some NLRs, such as NLRP1, NLRP3, NLRC4, and NLRP6, is the activation of a multi-protein complex resulting in the release of mature IL-1 and IL-18, known as the inflammasome. Another major function of NLR proteins involves the modulation of inflammatory signaling pathways, including NF-κB and MAPK. While some NLR family members, such as NOD1 and NOD2, can cause activation of these pathways in response to stimuli, select NLRs, such as NLRX1, NLRC3, and NLRP12, act as negative regulators of inflammatory pathways, adding to the complexity of NLR biology (Allen et al., 2011; Lich et al., 2007; Schneider et al., 2012; Williams et al., 2005; Xia et al., 2011).
The NLR protein family is a diverse group of receptors with critical functions in the regulation of host immunity. In addition, it is becoming more and more evident that the microbiome plays a long-underappreciated role in shaping the host immune system. The current review highlights the growing body of literature characterizing the crosstalk between NLR proteins and the gut microbiome, as well as the immense impact of these complex interactions on intestinal homeostasis and inflammation.
NOD1/2 (NLRC1/2)
NOD1 and NOD2, which share similar structural compositions, are the first and among the best-studied members of the NLR family. NOD1/2 are expressed ubiquitously in a variety of cell types, such as epithelial cells, stromal cells, and endothelial cells (Bertin et al., 1999; Gutierrez et al., 2002; Inohara et al., 1999). These cytosolic proteins are composed of a C-terminal leucine-rich repeat region, a centrally located nucleotide-binding oligomerization domain, and an N-terminal caspase activation and recruitment domain (Chamaillard et al., 2003; Girardin et al., 2003b; Ogura et al., 2001b). As well-known PRRs, they can recognize cytosolic bacterial peptidoglycan fragments with high specificity. Upon ligand binding, NF-κB–ERK–MAPK signaling is activated, resulting in the expression of various pro-inflammatory cytokines and chemokines, as well as production of antimicrobial peptides (AMPs) and reactive oxygen species (Moreira and Zamboni, 2012). As NOD1/2 contribute to cytosolic surveillance, it is not surprising that these proteins act as important regulators of host–microbe interactions, which also control susceptibility to abnormal intestinal inflammation (Girardin et al., 2003b).
Early studies provide supporting evidence for the antimicrobial role of NOD1 in multiple bacterial infections (Allison et al., 2009; Berrington et al., 2010; Boneca et al., 2007; Frutuoso et al., 2010; Girardin et al., 2003a; Hasegawa et al., 2011; Kim et al., 2004, 2010; Travassos et al., 2005, 2004; Zilbauer et al., 2007). However, the influence of NOD1 in the whole gut microbial community is less evident (Table 1). Bouskra et al. (2008) evaluated the whole bacterial kingdoms in Nod1-deficient mice by quantitative PCR of 16S ribosomal RNA (rRNA) and showed an ∼100-fold increase in total bacteria compared with WT mice. Moreover, there were different relative amounts of bacteria in Nod1-deficient mice, including Clostridiales, Bacteroides, and Enterobacteriaceae. However, as these earlier experiments used nonlittermate controls, more work needs to be done to further verify this conclusion. Recently, Yu et al. (2020) found that WT mice (WT2), which were generated by crossing C57BL/6 mice to Nod1-deficient mice, developed significantly more colon tumors in the azoxymethane (AOM)–dextran sodium sulfate (DSS) colitis-associated colon cancer (CAC) model than did the “pure” WT mice (WT1), which were purchased from Jackson Laboratory. Although both WT1 and WT2 colonies originated from C57BL/6 mice from the same vendor and were housed in the same mouse room, they had distinct microbiome compositions. This microbiome difference was directly associated with differential tumor susceptibilities as confirmed by fecal material transplant, cohousing, and cross-fostering experiments. This study not only emphasizes that the gut microbiome can be typically determined by maternal transmission but also indirectly suggests Nod1-deficient mice may harbor a dysbiotic microbiome, which contributes to increased tumor susceptibility.
Table 1. Influences of NOD1/2 on intestinal microbial community structures.
| Genotype | Species | Sample tissue | Sequence method | Disease model | Microbiome changes | Littermates? | Reference |
|---|---|---|---|---|---|---|---|
| Nod2 mutation | Human | Ileum | 454 seq | CD, UC | Shifts in microbial composition | Frank et al., 2011 | |
| Nod2 mutation | Human | Ileum | qPCR | CD | ↑Bacteroidetes and Firmicutes | Rehman et al., 2011 | |
| Nod2 mutation | Human | Intestinal biopsies | 16S seq | ↑Enterobacteriaceae | Knights et al., 2014 | ||
| Nod2−/− | Mice | Ileum, feces | qPCR | Steady state | ↑Bacterial load; ↑Bacteroides, Bacillus, and Firmicutes | Yes | Petnicki-Ocwieja et al., 2009 |
| Nod2−/− | Mice | Ileum, feces | 454 seq | Steady state | ↑Bacterial load; ↑Bacteroidetes and Firmicutes | No | Rehman et al., 2011 |
| Nod2−/− | Mice | Feces | 454 seq | Steady state | ↓Bacterial diversity and richness; ↑Rikenellaceae, Bacteroidaceae, and Bacteroides acidifaciens; ↓Prevotellaceae | No | Mondot et al., 2012 |
| Nod2−/− | Mice | Colon | 454 seq | DSS | ↑Bacterial load | Yes | Smith et al., 2012 |
| Nod2−/− | Mice | Colon | 454 seq | DSS, DSS-AOM | ↑Bacteroidetes | No | Couturier-Maillard et al., 2013 |
| Nod2−/− | Mice | Feces | 16S seq | Steady state | ↑Bacteroides vulgatus | No | Ramanan et al., 2014 |
| Nod1−/− | Mice | Ileum | qPCR | Steady state | ↑Bacterial load; ↑Bacteroidetes, Clostridiales, and Enterobacteriaceae; ↓Lactobacillaceae | No | Bouskra et al., 2008 |
| Nod1/2 DKO | Mice | Ileum, cecum, colon | qPCR | Steady state | No change | Yes | Robertson et al., 2013 |
| Nod1/2 DKO | Mice | Cecum | qPCR | Salmonella DaroA infection | No change | Yes | Robertson et al., 2016 |
Summary of recent studies in both human and mice indicating the effect of NOD1 and NOD2 on the intestinal microbiome. DKO, double-knockout; qPCR, quantitative PCR; seq, sequencing.
It has been appreciated that NOD2 expression can be induced by bacterial components (lipopolysaccharides) or metabolites (short-chain fatty acids, e.g., butyrate; Inohara and Nuñez, 2003; Leung et al., 2009; Ogura et al., 2001a). Petnicki-Ocwieja et al. (2009) showed that Nod2-deficient mice harbored a larger load of bacteria in the ileum and feces than did littermate controls, with elevated abundances of Bacteroidetes and Firmicutes (Table 1). Nod2-deficient mice also exhibited decreased antibacterial activity and increased susceptibility to colonization by opportunistic pathogens in the terminal ileum due to impaired crypts function. The advent of next-generation sequencing has made it possible to take an in-depth snapshot of the intestinal bacterial ecosystem and to delineate microbial community structures and composition. A more direct study of NOD2’s influence on the gut microbiome using 16S rRNA gene–based clone library sequencing and high throughput pyrosequencing (Rehman et al., 2011) confirmed that Nod2-deficient mice displayed an elevated bacterial load in fecal and terminal ileal samples compared with WT counterparts. Another study demonstrated that Nod2 deficiency resulted in an expansion of Bacteroides vulgatus, which mediated small intestinal abnormalities and inflammation by affecting interferon-γ–expressing intraepithelial lymphocytes (Ramanan et al., 2014).
NOD2 and its interaction with gut microbiota have been implicated in inflammatory bowel disease (IBD), including both Crohn’s disease (CD) and ulcerative colitis (UC) in humans (Trindade and Chen, 2020). Mutations in Nod2 have been investigated as one of the strongest known genetic risk factors in the development of CD (Hugot et al., 2001; Ogura et al., 2001a). Three major single-nucleotide polymorphisms (SNPs) of the Nod2 gene (SNP8, SNP12, and SNP13) are reported to be associated with CD and to cause a loss-of-function phenotype toward muramyl dipeptide (Chen et al., 2017b). Rehman et al. (2011) investigated ileal biopsies and feces from CD patients with or without the Nod2 SNP13 mutation and found patients carrying Nod2 variants had increased loads of Bacteroidetes and Firmicutes. A recent work genotyped 178 intestinal samples collected from CD, UC, and control patients for Nod2 risk alleles and suggested that the Nod2 composite genotype was significantly associated with shifts in microbial compositions, especially with compositions of Faecalibacterium and Escherichia (Frank et al., 2011). Others indicated that NOD2 was associated with an increase of Enterobacteriaceae in subjects with higher Nod2 risk allele dosage (Knights et al., 2014).
Consistent with clinical findings, several mouse studies have also confirmed the interaction between NOD2 and microbiota (Table 1). In a 2,4,6-trinitrobenzene sulfonic acid (TNBS)–induced mice colitis model, the protective capacity of Lactobacillus salivarius Ls33 was shown to be NOD2 dependent and correlated with local IL-10 production (Macho Fernandez et al., 2011). Nod2-deficient mice also showed an increased sensitivity and susceptibility in a DSS–induced colitis model (Couturier-Maillard et al., 2013). Surprisingly, this effect was transmissible via a cohousing or cross-fostering strategy. The mortality and morbidity of WT mice were significantly enhanced after being cohoused with Nod2-deficient mice, concurrent with higher IL-6 production from dendritic cells (DCs). This effect was further confirmed by a fecal transplantation experiment showing germ-free WT mice suffered more severe disease after receiving feces from Nod2-deficient mice, while germ-free Nod2-deficient mice were rescued after being recolonized with fecal flora from WT mice. Although the majority of studies elucidated the interference of NOD2 with host–microbe interactions, it is worth noting that one group reported NOD2 was not associated with intestinal microbial composition and density (Robertson et al., 2016, 2013). This conflicting conclusion may be due to the use of nonlittermate controls.
NLRP3
NLRP3 is an inflammasome-forming NLR family member that plays a critical and well-defined role in the host innate immune response to a diverse array of stimuli (Davis et al., 2011). It is therefore not surprising that NLRP3 activity is implicated in diverse human diseases, including IBD (Davis et al., 2011; Menu and Vince, 2011). It is well documented that NLRP3 responds to various microbes, including commensals, as well as microbial products and metabolites (Camell et al., 2015; Macia et al., 2015; Seo et al., 2015; Singh et al., 2019). However, the consequences of NLRP3 activation in the context of IBD is controversial, as different laboratories have reported conflicting results when mice with genetic knockout or mutation of Nlrp3 undergo models of intestinal inflammation (Table 2). While some studies support a protective role for NLRP3 in maintaining intestinal homeostasis by keeping pathogenic bugs at bay, others report that NLRP3 activation by the microbiota contributes to pathogenesis.
Table 2. Comparison of methodologies and results from studies examining relationships between NLRP3, NLRP6, NLRP12, and the gut microbiota.
| Nlrp3 genotype | Disease model | NLRP3 function | Microbiome implicated | Germ-free (GF) strategy | Microbiome changes | Cellular mechanism | Littermates? | Reference |
|---|---|---|---|---|---|---|---|---|
| Nlrp3−/− | DSS, TNBS | Protective | Antibiotic treatment | N/A | Commensal overgrowth and bacteremia | ↓Non-hematopoietic cell IL-18 production | No | Zaki et al., 2010 |
| Nlrp3−/− | DSS, TNBS | Protective | TRFLP seq | N/A | ↑Enterobacteriaceae ↑Mycobacterium; ↑Clostridium; etc. | ↓Colonic IL-1β, IL-10, TGFβ, and antimicrobial secretions; ↓Neutrophil and macrophage responses | Yes | Hirota et al., 2011 |
| Nlrp3−/− | DSS, TNBS | Pathogenic | Antibiotic treatment, cohousing | N/A | N/A | ↑Lamina propria CD103+ tolerogenic DCs | No | Bauer et al., 2012 |
| Nlrp3R258W (hyper-active) | DSS, AOM-DSS | Protective | Antibiotic treatment, cohousing, fecal transplantation, 16S seq | Fecal transplantation to GF-WT recipients | ↓Actinobacteria; ↓Verrucomicrobia; ↓Akkermansia; ↑Lactobacillus | ↑IL-1β and AMP from lamina propria immune cells; ↑Regulatory T cells | Yes | Yao et al., 2017 |
| Nlrp6 genotype | Disease model | NLRP6 function | Microbiome implicated | Germ-free (GF) strategy | Microbiome changes | Cellular mechanism | Littermates? | Reference |
| Nlrp6−/− | Steady state, DSS | Protective | Antibiotic treatment, cohousing, 16S seq | N/A | ↑Prevotellaceae | ↓Non-hematopoietic IL-18 production; ↑CCL5 production; ↑Immune cell recruitment | No | Elinav et al., 2011 |
| Nlrp6−/− | Listeria, Salmonella, etc., infection | Pathogenic | Cohousing, 16S seq | N/A | ↑Bacteroidetes (Prevotellaceae family) | ↑Monocyte and neutrophil recruitment; ↑NF-κB and ERK activation | No | Anand et al., 2012 |
| Nlrp6−/− | AOM-DSS | Protective | Antibiotic treatment, cohousing | N/A | N/A | ↑Tumorigenesis mediated by IL-18, CCL5, and IL-6 | No | Hu et al., 2013 |
| Nlrp6−/− | Citrobacter rodentium infection | Protective | Cohousing | N/A | N/A | ↓Autophage in goblet cells; ↓Mucus secretion | No | Wlodarska et al., 2014 |
| Nlrp6−/− | Steady state, IL-18, LPS, DSS | Protective | Antibiotic treatment, cohousing, 16S seq | GF-WT and GF-Nlrp6−/−, cohousing in GF-WT | Different microbial compositions compared with WT | ↓IL-18 production; ↓AMPs | No | Levy et al., 2015 |
| Nlrp6−/−, Nlrp6−/−, Il10−/− DKO | Il10−/− spontaneous colitis | Protective | Cohousing, 16S seq | Fecal transplantation to GF-WT and GF-Nlrp6−/−recipients | ↓Bacterial richness and diversity; ↑Akkermansia; Bacteroides; Prevotella; etc. | ↓IL-18 production; ↑Significant colitis | Yes | Seregin et al., 2017 |
| Nlrp6−/− | Steady state, DSS | No impact on colitis | Cohousing, 16S seq | N/A | No impact on microbiota composition | N/A | Yes | Lemire et al., 2017 |
| Nlrp6−/− | Steady state, DSS | No impact on colitis | Cohousing, 16S seq | GF-Pycard−/− mice | No impact on microbiota composition | N/A | Yes | Mamantopoulos et al., 2017 |
| Nlrp6−/− | Steady state | N/A | Fecal transplantation, 16S seq | N/A | ↑Prevotellaceae; ↑Helicobacteraceae; etc. | N/A | No | Gálvez et al., 2017 |
| Nlrp6−/− | Apigenin treatment, DSS | Protective | Cohousing, 16S seq | N/A | ↓Bacterial diversity, compared with WT | Antiproliferative effect of apigenin dependents on NLRP6 pathway | No | Radulovic et al., 2018 |
| Nlrp6−/− | Graft-versus-host disease (GVHD) | Pathogenic | Antibiotic treatment, cohousing, fecal transplantation | Fecal transplantation to GF-WT and GF-Nlrp6−/− recipients | ↑Porphyromonadaceae; ↑Prevotellaceae | GVHD is independent of microbiome; ↑Gastrointestinal homeostasis after allo-BMT | Yes | Toubai et al., 2019 |
| Nlrp12−/− | DSS | Protective | Cohousing, fecal transplantation, 16S seq | Fecal transplantation to GF-WT and GF-Nlrp12−/− recipients | ↑Lachnospiraceae family; ↑Erysipelotrichaceae family; ↓Bacteroidales order; ↓Clostridiales order | ↑Inflammatory signaling; ↑inflammatory cytokine production from colonic DCs | Yes | Chen et al., 2017a |
Summary of recent studies regarding the correlations between NLRP3, NLRP6, and NLRP12 and the gut microbiome in murine models of intestinal infection and inflammation. AMP, antimicrobial peptide; BMT, bone marrow transplant; DKO, double-knockout; LPS, lipopolysaccharide; N/A, not applicable; seq, sequencing; TRFLP, terminal–restriction fragment length polymorphism.
Multiple groups have reported a protective role for NLRP3 in mouse models of colitis and colorectal cancer (CRC), which is one of the most common forms of cancer, with prolonged colitis as the major risk factor for its development (Hu et al., 2013). Zaki et al. (2010) reported an exacerbated inflammatory phenotype in Nlrp3−/− mice in both DSS- and TNBS-colitis models, and they attributed the protective role for NLRP3 to the production of IL-18 from nonhematopoietic cells, which promoted the integrity of the epithelial barrier between gut microbes and host immune cells. Indeed, Nlrp3−/− mice experienced bacteremia, and administration of antibiotics was able to ameliorate the exacerbated colitis phenotype, directly implicating the microbiota. We found an increased inflammatory phenotype accompanied by increased tumor burden in the AOM-DSS induced cancer model in Nlrp3-, Asc-, or Casp1-deficient mice compared with WT counterparts (Allen et al., 2010). Bone-marrow chimera experiments determined that NLRP3-mediated protection depended on the expression of the NLRP3 inflammasome components in hematopoietic cells. Although the mechanistic conclusions differ, both studies suggest that the interaction between NLRP3 and the commensal microbiota is critical for intestinal homeostasis and protection against inflammation.
Shortly following these initial studies, Hirota et al. (2011) directly investigated the impact of NLRP3 on the commensal microbial community and observed an exacerbated colitis phenotype in Nlrp3−/− mice associated with reduced IL-1β, IL-10, and TGFβ expression and linked the phenotype to impaired responses of macrophages and neutrophils to microbial products. They also reported reduced antimicrobial secretions and altered colonic β-defensin expression, resulting in a distinct microbiome in Nlrp3−/− mice compared with WT controls derived from littermates. Using terminal–restriction fragment length polymorphism analyses, they reported several bacterial candidates that were more abundant in either WT or Nlrp3−/− mice including members of the Enterobacteriaceae family and the Mycobacterium and Clostridium genera. This study provided the first characterization of the effect of NLRP3 on the makeup of intestinal microbiota.
More support for a critical role of NLRP3 in maintaining intestinal homeostasis came from more recent studies in the context of hyperactive NLRP3. Cryopyrin-associated periodic syndrome (CAPS) is an autoinflammatory disease in humans caused by hyperactive mutations in NLRP3 leading to inflammation in skin, joints, and eyes (Booshehri and Hoffman, 2019; Menu and Vince, 2011). Yao et al. (2017) used mice harboring the Nlrp3R258W mutation, a common mutation found in CAPS patients, to investigate the consequence of hyperactive NLRP3 in the intestine. They found that hyperactive NLRP3 led to decreased abundance of bacteria in the phyla of Actinobacteria, Verrucomicrobia, and Akkermansia, but increased abundance of Lactobacillus. Strikingly, mice with the mutation were highly resistant to both DSS-colitis and DSS-AOM CAC. Mechanistically, NLRP3 promoted IL-1β secretion from immune cells in the lamina propria and downstream AMP secretion to remodel the microbiota and induce regulatory T cells as the CAC phenotype was restored in Nlrp3R258W × Il1r−/− mice or with depletion of regulatory T cells via anti-CD25 (Yao et al., 2017). This provides further evidence that NLRP3 functions to maintain intestinal homeostasis through regulation of the microbiota.
Contrary to the previously discussed reports, the Schnurr group reported a proinflammatory and pathogenic role for NLRP3 in the context of colitis (Bauer et al., 2010). The authors reported an ameliorated colitis phenotype, with less severe weight loss and histological scores, in Nlrp3−/− animals and implicated IL-1β production from macrophages in disease pathogenesis. In an attempt to reconcile their results from the conflicting reports, this group published a follow-up study to directly test the impact of the gut microbiome on the severity of DSS-colitis in their Nlrp3-deficient mice (Bauer et al., 2012). They associated protection with an increase in tolerogenic CD103+ DCs in the lamina propria, a finding recently expanded upon in Mak’Anyengo et al. (2018), which demonstrated NLRP3-dependent inhibition of CD103+ DCs via IL-1β. A later study reported a detrimental role for NLRP3 and its interaction with gut microbiota in the context of intestinal inflammation (Seo et al., 2015). The authors reported that during DSS-colitis, the commensal pathobiont Proteus mirabilis induced robust NLRP3-dependent IL-1β production from infiltrating monocytes but not from resident macrophages present in the lamina propria. NLRP3-dependent IL-1β production in this case was pathogenic, as Il1b−/− mice were significantly protected from weight loss and pathology. Together, studies such as these support a detrimental role for NLRP3 in immune cells during intestinal inflammation.
In sum, these studies investigating the impact of NLRP3–microbiota interactions in the context of intestinal homeostasis and inflammation have provided a less-than-straightforward connection between commensal bacteria and the inflammasome pathway. Evidence supports that NLRP3 expression in resident immune cells and nonhematopoietic cells can promote barrier integrity via IL-18 and maintain a “healthy” balance of commensal bacteria via secretion of IL-1β and AMPs. Alternatively, studies also support that certain commensals may provide the first signal to prime NLRP3 inflammasome activation in circulating immune cells to promote detrimental inflammatory responses. Ultimately, some of these conflicting findings may be dependent on the environment, such as the preexisting microbiota load and composition, the housing environment, the control mice that were used (bred in the same facility or not, littermates or not), and the animal chow, as well as the fine specifics of the disease-induction process.
NLRP6
NLRP6 is another well-studied, inflammasome-forming NLR family member (Elinav et al., 2011; Grenier et al., 2002). NLRP6 is predominantly expressed in epithelial cells from the small intestine, colon, kidney, liver, and lung (Chen et al., 2011; Elinav et al., 2011; Normand et al., 2011). In the intestinal tract, NLRP6 has been shown to have a preferential expression in enterocytes and goblet cells, where it plays a critical role in regulating intestinal homeostasis and defending against infection, autoimmune responses, and tumorigenesis (Birchenough et al., 2016; Wlodarska et al., 2014).
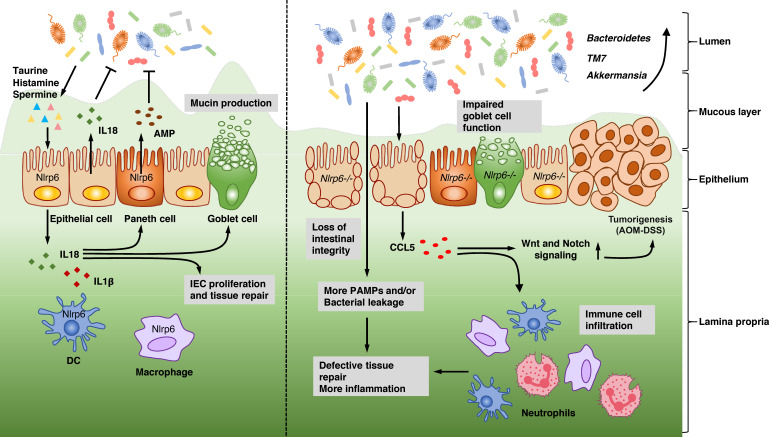
Over the past few years, there has been an explosive increase in interest in whether and how NLRP6 regulates intestinal microbiota and participates in the onset of intestinal inflammation and tumorigenesis (Fig. 1 and Table 2). It was first reported that NLRP6-ASC inflammasome signaling shapes the intestinal microbiota community and regulates host immunity against chemically induced colitis (Elinav et al., 2011). Their results showed that both Nlrp6-deficient and Asc-deficient mice were more susceptible to DSS-induced colitis and that the exacerbated phenotype could be transferred to cohoused WT mice. By using a bacterial 16S rRNA–based analysis, the authors demonstrated that Nlrp6-deficient mice harbored a distinct fecal microbiota with significant increases in the bacterial phyla Bacteroidetes (family Prevotellaceae) and TM7 compared with WT counterparts. Moreover, Il18-deficient mice could transfer a colitis-prone phenotype to cohoused WT mice, which suggested that the NLRP6-ASC–IL-18 axis played a striking role in maintaining a “healthy” microbiome.
Figure 1.
Mechanisms controlled by NLRP6 in regulating intestinal microbiota and the onset of intestinal inflammation as well as tumorigenesis. In normal physiological conditions, NLRP6 is expressed by both intestinal cells (such as epithelial cells, goblet cells, and Paneth cells) and hematopoietic cells (such as DCs and macrophages). The NLRP6 inflammasome regulates the secretion of IL-1β and IL-18, which play an important role in maintaining a homeostatic bacterial community and promoting epithelial cell proliferation, tissue repair, mucin production, and AMP secretion. Meanwhile, NLRP6 also inhibits canonical NF-κB and MAPK signaling in an inflammasome-independent manner. The metabolites produced by microbiota, such as taurine, histamine, and spermine, also modulate NLRP6-dependent IL-18 production and further help to maintain a healthy intestinal environment. In the absence of NLRP6, the intestinal microbiome is altered with an increase of potentially pathogenic species, such as Bacteroidetes, TM7, or Akkermansia. As IL-18 expression is abolished, goblet cell function is impaired, resulting in less mucin production along with defective epithelium repair, leading to the loss of intestinal integrity and the failure of defense against bacteria during acute inflammation induced by DSS challenge. CCL5 is induced by gut microbiota and increases immune cell infiltration together with promotion of epithelial cell proliferation, which is dependent on Wnt and Notch signaling, promoting cancer formation. IEC, intestine epithelial cell; PAMP, pathogen-associated molecular pattern.
The same laboratory published another study on the influence of NLRP6–microbiota interactions in the pathogenesis of the AOM-DSS model of CRC (Hu et al., 2013). Enhanced tumorigenesis was observed in Nlrp6- and Asc-deficient mice and could be transmitted to cohoused WT. In another paper, Anand et al. (2012) studied the function of NLRP6 in regulating host defense against bacterial pathogens. Although they confirmed that the microbiome in Nlrp6-deficient mice was different from that in WT mice, the resistance mediated by NLRP6 against Listeria monocytogenes infection was independent of its microflora composition. Enteric pathogens trigger multiple impairments in gastrointestinal physiology, including motility reduction and intrinsic enteric-associated neuron loss. A recent paper unveiled that neuronal-specific NLRP6 is the main effector of infection-induced intrinsic enteric-associated neuron death, which could be reversed by manipulation of gut microbiota. This work provides another clue as to the correlation between NLRP6 and the gut microbiome (Matheis et al., 2020).
To obtain more insights into how the inflammasome participates in maintaining a healthy host-microbial mutualism, Levy et al. (2015) performed a metabolomics analysis of fecal samples from germ-free recipients cohoused with either WT or Asc-deficient mice. The authors found that the metabolites taurine, histamine, and spermine were involved in NLRP6 inflammasome modulation by inducing epithelial IL-18 secretion and downstream AMP production. As a result, these microbial metabolites modulated the microbial community, host physiology, and disease susceptibility (Levy et al., 2015). Recently, another work uncovered that a flavonoid metabolite, apigenin, also conferred protection against DSS-induced colitis through NLRP6-dependent signaling (Radulovic et al., 2018). Because the DSS-induced colitis model is more suitable for investigating host responses during epithelial injury and repair processes but not appropriate for examining the whole pathogenesis (Brown et al., 2007; Kaser et al., 2010), Seregin et al., (2017) further confirmed NLRP6-microbiome interactions in a spontaneous colitis model using Il10-deficient mice, which have increased pathogenic Th1 responses and develop chronic enterocolitis (Davidson et al., 1996; Seregin et al., 2017). They demonstrated that Il10/Nlrp6 double knockout mice were more prone to spontaneous colitis and harbored an altered microbiota with increased abundance of the pathobiont Akkermansia muciniphila. This work underscored a new mechanistic underpinning of how NLRP6 maintains intestinal homeostasis by limiting the colonization of specific colitogenic bacteria. A recent paper suggests that NLRP6 recognizes lipoteichoic acid derived from gram-positive bacteria to cause IL-18 release, which paradoxically leads to exacerbated infection. Whether this impacts the gut microbiota remains to be studied (Hara et al., 2018).
However, not all groups illustrated NLRP6 as a hallmark host factor shaping the gut microbiome and modulating intestinal inflammation. In 2017, two independent laboratories published results that revealed no direct effect of Nlrp6 genetic background on gut microbiota composition using littermate controls (Lemire et al., 2017; Mamantopoulos et al., 2017). These results also emphasized that Nlrp6 deficiency could not predispose mice to higher susceptibility to DSS-induced colitis. Interestingly, another work was published together with the work of Lemire et al. (2017) and further supported NLRP6’s function in regulating the gut microbiome in a fecal material transfer experiment (Gálvez et al., 2017). An additional paper found a pathogenic role for Nlrp6 in gastrointestinal graft-versus-host disease after bone marrow transplant that is not affected by the microbiota (Toubai et al., 2019). Wlodarska et al. (2014) demonstrated that Nlrp6 deficiency caused an abrogation of autophagy in goblet cells, which directly affected their function by decreasing mucin granule exocytosis and resultant mucous layer formation. The impaired mucin production made Nlrp6-deficient mice more susceptible to infection by intestinal pathogens (e.g., Citrobacter rodentium) and less capable of maintaining microbial homeostasis. This work showed a mechanism by which the absence of NLRP6 inflammasome leads to changes in intestinal microbial community composition and biogeographical distribution. However, it is important to note that this finding is different from another work, which used littermate controlled Nlrp6-deficient mice (Volk et al., 2019). By performing a series of ex vivo and in vivo analyses that provided highly reproducible quantitative assessment of inner mucous layer function, Volk et al. (2019) showed that mice lacking Nlrp6 formed a mucous layer that was functionally indistinguishable from that of WT animals.
In response to these nonignorable contradictions, authors with opposite perspectives debated either for or against NLRP6-dependent regulation of gut microbiota and expressed their opinions regarding how to minimize experimental discrepancies and better investigate the cause and effect interplay between gut microbiota and the host immune system. Elinav et al. (2018) argued that the impact of NLRP6 on regulation of the microbiome community requires exposure to a sufficiently diversified bacterial group, and the effect of NLRP6 on eliminating intestinal inflammation and metabolic dysfunction is mainly mediated through its microbiome-modulatory activity. Wullaert et al. (2018), on the other hand, emphasized that NLRP6 does not perform a generalizable effect on the host microbiota based on several littermate-controlled studies (Mamantopoulos et al., 2018). Interestingly, both groups reached a consensus that to avoid partial and nongeneralizable conclusions, multiple complementary experimental modalities, not only littermate breeding strategies but also microbiota recolonization of germ-free mice, should be included in future microbiome research. Both approaches are needed to draw correct conclusions about host-microbiome correlations. A microbiome study should not solely rely on only one approach because different phenotypes may occur in littermate-controlled studies versus studies done in germ-free colonized mice for the following reasons. First, a large amount of bacterial influx during fecal transfer is not a physiological process and may cause colonization resistance or inflammatory reactions (Wullaert et al., 2018). Second, as we mentioned above, bacterial species and communities are not uniform within different organs. The microbiota in feces is distinct from that in the cecum or colon and may not represent the real microbial community in specific gastrointestinal regions. Besides, many anaerobes are likely to die immediately during fecal collection or processing time. In this case, although fecal microbiota transfers or recolonizations of germ-free mice are powerful approaches in microbiome studies, littermate breeding strategies should be used together to dissect the impact of host genetics.
NLRP12
The NLRP12 protein has been shown to cause caspase-1 processing and IL-1β release in response to Yersinia and Plasmodium chabaudi (Ataide et al., 2014; Vladimer et al., 2012); however, multiple evidence indicates that another dominant function of NLRP12 is independent of inflammasome formation and immunosuppressive in nature. Our group and others identified NLRP12, formerly known as Monarch-1, as a negative regulator of inflammatory immune activation in vitro using human monocytic cell lines (Lich et al., 2007; Wang et al., 2002; Williams et al., 2005). These studies showed that NLRP12 suppressed both noncanonical and canonical NF-κB signaling and subsequent inflammatory cytokine, chemokine, and surface protein expression. In humans, mutations in Nlrp12, which result in less efficient inhibition of NF-κB, are associated with periodic fever syndromes (Jéru et al., 2008). In mice, NLRP12 has been implicated in multiple disease models, including colitis and CAC, obesity, and multiple sclerosis (Allen et al., 2012; Chen et al., 2017a; Gharagozloo et al., 2015; Lukens et al., 2015; Truax et al., 2018; Zaki et al., 2011).
To define the relevance of NLRP12 in intestinal inflammation, two studies investigated the consequence of Nlrp12 deficiency on DSS-colitis and AOM-DSS CRC in mice. Both groups described a protective role for NLRP12 in colon inflammation and tumorigenesis, as Nlrp12−/− mice in both studies experienced exacerbated disease, increased NF-κB activation, and enhanced expression of inflammatory cytokines and chemokines (Allen et al., 2012; Zaki et al., 2011). However, the groups described different cellular mechanisms of protection. One group concluded from bone marrow chimera experiments that NLRP12 was required for the suppression of canonical NF-κB and ERK signaling in hematopoietic cells to attenuate tumorigenesis (Zaki et al., 2011). Alternatively, another group described functions for both hematopoietic cells and nonhematopoietic cells during initial inflammation and found that NLRP12 in nonhematopoietic cells was required for protection against tumor growth and implicated noncanonical NF-κB signaling in the exacerbated phenotype (Allen et al., 2012). Although both groups appreciated the impact of the commensal microbiota on colitis and CAC pathogenesis, these studies did not include direct testing of its involvement in the exacerbated inflammatory phenotypes or its potential interaction with NLRP12.
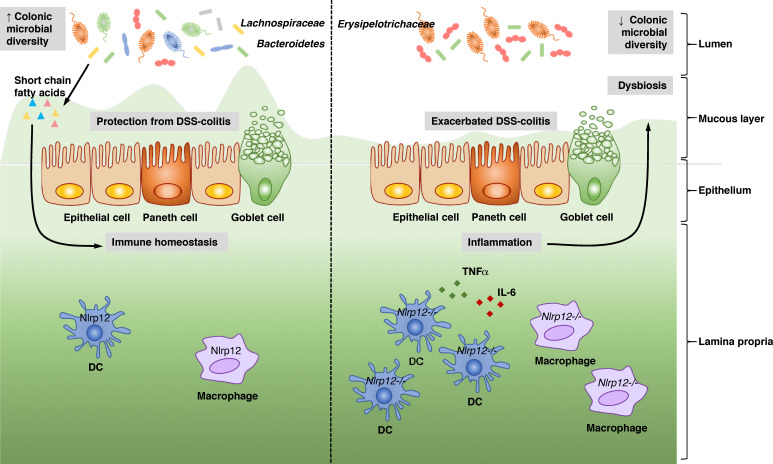
A recent study connected the colitis phenotype in Nlrp12−/− mice to the microbiome (Fig. 2 and Table 2; Chen et al., 2017a), where 16S sequencing demonstrated that Nlrp12−/− harbored a significantly altered microbiota, with significantly less diversity, compared with that of WT littermates. Specifically, Nlrp12−/− mice contained a significantly lower abundance of bacteria in the orders of Bacteriodales and Clostridiales, including bacteria in the family Lachnospiraceae and a reciprocal increase in the abundance of the family Erysipelotrichaceae. Cohousing experiments, fecal transplant experiments performed in germ-free mice, and littermate studies strongly implicated the dysbiosis observed in Nlrp12−/− mice as a major factor contributing to exacerbated DSS-colitis. Further, reconstitution with Lachnospiraceae was sufficient to attenuate the exacerbated colitis phenotype, inflammatory cytokines such as IL-6, TNFα, NF-κB, and MAPK inflammatory signaling in Nlrp12−/− mice. Significantly, these same beneficial bacteria also are promoted by NLRP12 to mitigate obesity, and the short-chain fatty acids, known to be secreted by Lachnospiraceae, mimicked the effect of Lachnospiraceae (Truax et al., 2018). These studies ultimately provide insight into the delicate balance of immune responses and the commensal microbiota and identify NLRP12 as a key mediator between them.
Figure 2.
NLRP12 and the microbiome in intestinal inflammation. Chen et al. (2017a) describe a role for NLRP12 in protecting against intestinal inflammation through regulation of the microbiome. The authors show that NLRP12 functions in hematopoietic cells to promote microbial diversity and the colonization of commensals in the Bacteroidetes and Lachnospiraceae families. Lachnospiraceae promotes homeostasis and protects against DSS-colitis through the production of short-chain fatty acids. In the absence of NLRP12, DCs express increased inflammatory cytokines including TNFα and IL-6, resulting in dysbiosis, which involves the loss of Lachnospiraceae and an increase in Erysipelotrichaceae. Mice deficient in Nlrp12 therefore experience exacerbated DSS-colitis, which can be ameliorated by reconstitution of Lachnospiraceae or treatment with SCFAs.
NLRC4
NLRC4 (formerly known as ICE-protease-activating factor; Poyet et al., 2001) is an inflammasome-forming NLR that, when coupled with its partner NAIP proteins, can be activated by flagellin or the type III secretory system from gram-negative bacterial pathogens (Broz et al., 2010; Franchi et al., 2006; Mariathasan et al., 2004; Miao et al., 2006; Sutterwala et al., 2007; Suzuki et al., 2007). Activation of the NLRC4 inflammasome results in the production of IL-1β and IL-18 and can induce caspase-mediated cell death. Interestingly, activation of the NLRC4 inflammasome does not require ASC, although NLRC4-induced caspase-1 activation is enhanced in the presence of ASC (Duncan and Canna, 2018). Gain-of-function NLRC4 mutations have been associated with both enteric and systemic autoinflammatory diseases in humans (Romberg et al., 2017). Heterozygous gain-of-function mutations in NLRC4 are the cause of autoinflammation with infantile enterocolitis, which is a chronic inflammatory disease characterized by episodes of infantile diarrhea and other systemic presentations linked to macrophage activation in the skin, central nervous system, and liver (Canna et al., 2014). In addition, other NLRC4 mutations have been linked to syndromes that are phenotypically similar to NLRP3-associated autoinflammatory diseases (Romberg et al., 2017). The potential role of the gut microbiome in human NLRC4 inflammasomopathies, however, has not been described.
Studies in mouse models have revealed a protective role of NLRC4 in intestinal inflammation and inflammation-induced tumorigenesis. A study by Carvalho et al. (2012) found that Nlrc4-deficient animals exhibited more severe DSS-induced colitis than WT littermates. The authors also demonstrated that Nlrc4-deficient mice were more susceptible to intestinal Salmonella infection. Increased mortality in these mice correlated with decreased production of IL-1β (Carvalho et al., 2012). Another study demonstrated a similar protective role for NLRC4 in the AOM-DSS model of CRC. This study found significantly increased tumor numbers and tumor load in Nlrc4-deficient mice compared with age- and sex-matched cohoused WT mice (Hu et al., 2010). However, it is worth noting that a previous work from our laboratory showed no significant difference in disease progression or outcome in Nlrc4−/− mice compared with similarly treated WT animals (Allen et al., 2010). One possible explanation is the difference in the microbiota makeup at different institutions. Further studies with rigorous and critical experimental designs are necessary to investigate the potential role of NLRC4 in regulating the components of the intestinal microbiota and potential consequences for intestinal disease.
Future directions and challenges
The interaction between host immunity and the gut microbiota remains an exciting area of study and potential therapeutic discovery in the context of intestinal homeostasis and inflammation. However, conflicting conclusions and concerns of reproducibility between individual laboratories prove increasingly challenging in the microbiome field. Many previous studies underappreciated the effect of nongenetic factors (e.g., housing conditions, diet, mouse facilities, disease models, and sequencing methods) that are now verified to have strong influences on microbiome composition, which may lead to discordant results. Host–microbiota interaction is still a young and growing field for scientists to explore. With new collective knowledge, it is critical to conduct standardized littermate-controlled experimental design and to include germ-free mice and fecal microbiome transplantations when investigating host effects on microbiome community structure to minimize any confounding effects caused by nongenetic factors.
Acknowledgments
The authors acknowledge many investigators in the field whose primary data could not be cited in this review because of space limitations.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (grant R01-AI029564), National Institutes of Health (grant R35CA232109), National Institute of Diabetes and Digestive and Kidney Diseases (grant P01-DK094779), and RadCCORE (grant AI067798 to J.P.Y. Ting) and ITCMS T32 (grant 5T32CA009156 to S.A. Gibson).
Author contributions: H. Guo wrote the initial draft, prepared and created the figures and tables, and revised the paper. S.A. Gibson wrote the initial draft and prepared and created the figures and Table 2. J.P.Y. Ting provided oversight and leadership responsibility for this paper and edited the manuscript.
References
- Allen I.C., TeKippe E.M., Woodford R.M., Uronis J.M., Holl E.K., Rogers A.B., Herfarth H.H., Jobin C., and Ting J.P.. 2010. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207:1045–1056. 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen I.C., Moore C.B., Schneider M., Lei Y., Davis B.K., Scull M.A., Gris D., Roney K.E., Zimmermann A.G., Bowzard J.B., et al. . 2011. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 34:854–865. 10.1016/j.immuni.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen I.C., Wilson J.E., Schneider M., Lich J.D., Roberts R.A., Arthur J.C., Woodford R.M., Davis B.K., Uronis J.M., Herfarth H.H., et al. . 2012. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity. 36:742–754. 10.1016/j.immuni.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison C.C., Kufer T.A., Kremmer E., Kaparakis M., and Ferrero R.L.. 2009. Helicobacter pylori induces MAPK phosphorylation and AP-1 activation via a NOD1-dependent mechanism. J. Immunol. 183:8099–8109. 10.4049/jimmunol.0900664 [DOI] [PubMed] [Google Scholar]
- Anand P.K., Malireddi R.K., Lukens J.R., Vogel P., Bertin J., Lamkanfi M., and Kanneganti T.D.. 2012. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 488:389–393. 10.1038/nature11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide M.A., Andrade W.A., Zamboni D.S., Wang D., Souza M.C., Franklin B.S., Elian S., Martins F.S., Pereira D., Reed G., et al. . 2014. Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS Pathog. 10 e1003885 10.1371/journal.ppat.1003885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C., Duewell P., Mayer C., Lehr H.A., Fitzgerald K.A., Dauer M., Tschopp J., Endres S., Latz E., and Schnurr M.. 2010. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 59:1192–1199. 10.1136/gut.2009.197822 [DOI] [PubMed] [Google Scholar]
- Bauer C., Duewell P., Lehr H.A., Endres S., and Schnurr M.. 2012. Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig. Dis. 30(s1, Suppl 1):82–90. 10.1159/000341681 [DOI] [PubMed] [Google Scholar]
- Berrington W.R., Iyer R., Wells R.D., Smith K.D., Skerrett S.J., and Hawn T.R.. 2010. NOD1 and NOD2 regulation of pulmonary innate immunity to Legionella pneumophila. Eur. J. Immunol. 40:3519–3527. 10.1002/eji.201040518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin J., Nir W.J., Fischer C.M., Tayber O.V., Errada P.R., Grant J.R., Keilty J.J., Gosselin M.L., Robison K.E., Wong G.H., et al. . 1999. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J. Biol. Chem. 274:12955–12958. 10.1074/jbc.274.19.12955 [DOI] [PubMed] [Google Scholar]
- Birchenough G.M., Nyström E.E., Johansson M.E., and Hansson G.C.. 2016. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 352:1535–1542. 10.1126/science.aaf7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca I.G., Dussurget O., Cabanes D., Nahori M.A., Sousa S., Lecuit M., Psylinakis E., Bouriotis V., Hugot J.P., Giovannini M., et al. . 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. USA. 104:997–1002. 10.1073/pnas.0609672104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booshehri L.M., and Hoffman H.M.. 2019. CAPS and NLRP3. J. Clin. Immunol. 39:277–286. 10.1007/s10875-019-00638-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D., Brézillon C., Bérard M., Werts C., Varona R., Boneca I.G., and Eberl G.. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 456:507–510. 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- Brown S.L., Riehl T.E., Walker M.R., Geske M.J., Doherty J.M., Stenson W.F., and Stappenbeck T.S.. 2007. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J. Clin. Invest. 117:258–269. 10.1172/JCI29159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., and Monack D.M.. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755. 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camell C., Goldberg E., and Dixit V.D.. 2015. Regulation of Nlrp3 inflammasome by dietary metabolites. Semin. Immunol. 27:334–342. 10.1016/j.smim.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canna S.W., de Jesus A.A., Gouni S., Brooks S.R., Marrero B., Liu Y., DiMattia M.A., Zaal K.J., Sanchez G.A., Kim H., et al. . 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140–1146. 10.1038/ng.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F.A., Nalbantoglu I., Aitken J.D., Uchiyama R., Su Y., Doho G.H., Vijay-Kumar M., and Gewirtz A.T.. 2012. Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges. Mucosal Immunol. 5:288–298. 10.1038/mi.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M., Girardin S.E., Viala J., and Philpott D.J.. 2003. Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell. Microbiol. 5:581–592. 10.1046/j.1462-5822.2003.00304.x [DOI] [PubMed] [Google Scholar]
- Chen G.Y., Liu M., Wang F., Bertin J., and Núñez G.. 2011. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186:7187–7194. 10.4049/jimmunol.1100412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wilson J.E., Koenigsknecht M.J., Chou W.C., Montgomery S.A., Truax A.D., Brickey W.J., Packey C.D., Maharshak N., Matsushima G.K., et al. . 2017a NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 18:541–551. 10.1038/ni.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Salem M., Boyd M., Bornholdt J., Li Y., Coskun M., Seidelin J.B., Sandelin A., and Nielsen O.H.. 2017b Relation between NOD2 genotype and changes in innate signaling in Crohn’s disease on mRNA and miRNA levels. NPJ Genom. Med. 2:3 10.1038/s41525-016-0001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I., and Blaser M.J.. 2012. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13:260–270. 10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier-Maillard A., Secher T., Rehman A., Normand S., De Arcangelis A., Haesler R., Huot L., Grandjean T., Bressenot A., Delanoye-Crespin A., et al. . 2013. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123:700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson N.J., Leach M.W., Fort M.M., Thompson-Snipes L., Kühn R., Müller W., Berg D.J., and Rennick D.M.. 1996. T helper cell 1-type CD4+ T cells, but not B cells, mediate colitis in interleukin 10-deficient mice. J. Exp. Med. 184:241–251. 10.1084/jem.184.1.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.K., Wen H., and Ting J.P.. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29:707–735. 10.1146/annurev-immunol-031210-101405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G.P., Lee S.M., and Mazmanian S.K.. 2016. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14:20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J.A., and Canna S.W.. 2018. The NLRC4 Inflammasome. Immunol. Rev. 281:115–123. 10.1111/imr.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J., and Lynch S.V.. 2019. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 216:20–40. 10.1084/jem.20180448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., et al. . 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Henao-Mejia J., Strowig T., and Flavell R.. 2018. NLRP6 and Dysbiosis: Avoiding the Luring Attraction of Over-Simplification. Immunity. 48:603–604. 10.1016/j.immuni.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. . 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Frank D.N., Robertson C.E., Hamm C.M., Kpadeh Z., Zhang T., Chen H., Zhu W., Sartor R.B., Boedeker E.C., Harpaz N., et al. . 2011. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17:179–184. 10.1002/ibd.21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutuoso M.S., Hori J.I., Pereira M.S., Junior D.S., Sônego F., Kobayashi K.S., Flavell R.A., Cunha F.Q., and Zamboni D.S.. 2010. The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes Infect. 12:819–827. 10.1016/j.micinf.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Gálvez E.J.C., Iljazovic A., Gronow A., Flavell R., and Strowig T.. 2017. Shaping of Intestinal Microbiota in Nlrp6- and Rag2-Deficient Mice Depends on Community Structure. Cell Rep. 21:3914–3926. 10.1016/j.celrep.2017.12.027 [DOI] [PubMed] [Google Scholar]
- Gareau M.G., Sherman P.M., and Walker W.A.. 2010. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7:503–514. 10.1038/nrgastro.2010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharagozloo M., Mahvelati T.M., Imbeault E., Gris P., Zerif E., Bobbala D., Ilangumaran S., Amrani A., and Gris D.. 2015. The nod-like receptor, Nlrp12, plays an anti-inflammatory role in experimental autoimmune encephalomyelitis. J. Neuroinflammation. 12:198 10.1186/s12974-015-0414-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin S.E., Boneca I.G., Carneiro L.A., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M.K., Labigne A., Zähringer U., et al. . 2003a Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 300:1584–1587. 10.1126/science.1084677 [DOI] [PubMed] [Google Scholar]
- Girardin S.E., Boneca I.G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D.J., and Sansonetti P.J.. 2003b Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869–8872. 10.1074/jbc.C200651200 [DOI] [PubMed] [Google Scholar]
- Gregory S.M., Davis B.K., West J.A., Taxman D.J., Matsuzawa S., Reed J.C., Ting J.P., and Damania B.. 2011. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 331:330–334. 10.1126/science.1199478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier J.M., Wang L., Manji G.A., Huang W.J., Al-Garawi A., Kelly R., Carlson A., Merriam S., Lora J.M., Briskin M., et al. . 2002. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 530:73–78. 10.1016/S0014-5793(02)03416-6 [DOI] [PubMed] [Google Scholar]
- Gutierrez O., Pipaon C., Inohara N., Fontalba A., Ogura Y., Prosper F., Nunez G., and Fernandez-Luna J.L.. 2002. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-kappa B activation. J. Biol. Chem. 277:41701–41705. 10.1074/jbc.M206473200 [DOI] [PubMed] [Google Scholar]
- Hara H., Seregin S.S., Yang D., Fukase K., Chamaillard M., Alnemri E.S., Inohara N., Chen G.Y., and Núñez G.. 2018. The NLRP6 Inflammasome Recognizes Lipoteichoic Acid and Regulates Gram-Positive Pathogen Infection. Cell. 175:1651–1664.e14. 10.1016/j.cell.2018.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Yamazaki T., Kamada N., Tawaratsumida K., Kim Y.G., Núñez G., and Inohara N.. 2011. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J. Immunol. 186:4872–4880. 10.4049/jimmunol.1003761 [DOI] [PubMed] [Google Scholar]
- Hirota S.A., Ng J., Lueng A., Khajah M., Parhar K., Li Y., Lam V., Potentier M.S., Ng K., Bawa M., et al. . 2011. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Dis. 17:1359–1372. 10.1002/ibd.21478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., and Macpherson A.J.. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10:159–169. 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- Hu B., Elinav E., Huber S., Booth C.J., Strowig T., Jin C., Eisenbarth S.C., and Flavell R.A.. 2010. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc. Natl. Acad. Sci. USA. 107:21635–21640. 10.1073/pnas.1016814108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Elinav E., Huber S., Strowig T., Hao L., Hafemann A., Jin C., Wunderlich C., Wunderlich T., Eisenbarth S.C., et al. . 2013. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc. Natl. Acad. Sci. USA. 110:9862–9867. 10.1073/pnas.1307575110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O’Morain C.A., Gassull M., et al. . 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 411:599–603. 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- Inohara N., and Nuñez G.. 2003. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3:371–382. 10.1038/nri1086 [DOI] [PubMed] [Google Scholar]
- Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., et al. . 1999. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 274:14560–14567. 10.1074/jbc.274.21.14560 [DOI] [PubMed] [Google Scholar]
- Jarret A., Jackson R., Duizer C., Healy M.E., Zhao J., Rone J.M., Bielecki P., Sefik E., Roulis M., Rice T., et al. . 2020. Enteric Nervous System-Derived IL-18 Orchestrates Mucosal Barrier Immunity. Cell. 180:50–63.e12. 10.1016/j.cell.2019.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jéru I., Duquesnoy P., Fernandes-Alnemri T., Cochet E., Yu J.W., Lackmy-Port-Lis M., Grimprel E., Landman-Parker J., Hentgen V., Marlin S., et al. . 2008. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc. Natl. Acad. Sci. USA. 105:1614–1619. 10.1073/pnas.0708616105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti T.D., Lamkanfi M., and Núñez G.. 2007. Intracellular NOD-like receptors in host defense and disease. Immunity. 27:549–559. 10.1016/j.immuni.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Kaser A., Zeissig S., and Blumberg R.S.. 2010. Inflammatory bowel disease. Annu. Rev. Immunol. 28:573–621. 10.1146/annurev-immunol-030409-101225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.G., Lee S.J., and Kagnoff M.F.. 2004. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by toll-like receptors. Infect. Immun. 72:1487–1495. 10.1128/IAI.72.3.1487-1495.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.L., Jeong H.G., Kasper C.A., and Arrieumerlou C.. 2010. IKKα contributes to canonical NF-κB activation downstream of Nod1-mediated peptidoglycan recognition. PLoS One. 5 e15371 10.1371/journal.pone.0015371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D., Silverberg M.S., Weersma R.K., Gevers D., Dijkstra G., Huang H., Tyler A.D., van Sommeren S., Imhann F., Stempak J.M., et al. . 2014. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6:107 10.1186/s13073-014-0107-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire P., Robertson S.J., Maughan H., Tattoli I., Streutker C.J., Platnich J.M., Muruve D.A., Philpott D.J., and Girardin S.E.. 2017. The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition. Cell Rep. 21:3653–3661. 10.1016/j.celrep.2017.12.026 [DOI] [PubMed] [Google Scholar]
- Leung C.H., Lam W., Ma D.L., Gullen E.A., and Cheng Y.C.. 2009. Butyrate mediates nucleotide-binding and oligomerisation domain (NOD) 2-dependent mucosal immune responses against peptidoglycan. Eur. J. Immunol. 39:3529–3537. 10.1002/eji.200939454 [DOI] [PubMed] [Google Scholar]
- Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y., et al. . 2015. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 163:1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Lozupone C.A., Hamady M., Knight R., and Gordon J.I.. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6:776–788. 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lich J.D., Williams K.L., Moore C.B., Arthur J.C., Davis B.K., Taxman D.J., and Ting J.P.. 2007. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J. Immunol. 178:1256–1260. 10.4049/jimmunol.178.3.1256 [DOI] [PubMed] [Google Scholar]
- Lukens J.R., Gurung P., Shaw P.J., Barr M.J., Zaki M.H., Brown S.A., Vogel P., Chi H., and Kanneganti T.D.. 2015. The NLRP12 Sensor Negatively Regulates Autoinflammatory Disease by Modulating Interleukin-4 Production in T Cells. Immunity. 42:654–664. 10.1016/j.immuni.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho Fernandez E., Valenti V., Rockel C., Hermann C., Pot B., Boneca I.G., and Grangette C.. 2011. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 60:1050–1059. 10.1136/gut.2010.232918 [DOI] [PubMed] [Google Scholar]
- Macia L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S., Maruya M., Ian McKenzie C., Hijikata A., Wong C., et al. . 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6:6734 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- Mak’Anyengo R., Duewell P., Reichl C., Hörth C., Lehr H.A., Fischer S., Clavel T., Denk G., Hohenester S., Kobold S., et al. . 2018. Nlrp3-dependent IL-1β inhibits CD103+ dendritic cell differentiation in the gut. JCI Insight. 3 e96322 10.1172/jci.insight.96322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamantopoulos M., Ronchi F., Van Hauwermeiren F., Vieira-Silva S., Yilmaz B., Martens L., Saeys Y., Drexler S.K., Yazdi A.S., Raes J., et al. . 2017. Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity. 47:339–348.e4. 10.1016/j.immuni.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Mamantopoulos M., Ronchi F., McCoy K.D., and Wullaert A.. 2018. Inflammasomes make the case for littermate-controlled experimental design in studying host-microbiota interactions. Gut Microbes. 9:374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Matheis F., Muller P.A., Graves C.L., Gabanyi I., Kerner Z.J., Costa-Borges D., Ahrends T., Rosenstiel P., and Mucida D.. 2020. Adrenergic Signaling in Muscularis Macrophages Limits Infection-Induced Neuronal Loss. Cell. 180:64–78.e16. 10.1016/j.cell.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menu P., and Vince J.E.. 2011. The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin. Exp. Immunol. 166:1–15. 10.1111/j.1365-2249.2011.04440.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Mondot S., Barreau F., Al Nabhani Z., Dussaillant M., Le Roux K., Doré J., Leclerc M., Hugot J.P., and Lepage P.. 2012. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut. 61:634–635. 10.1136/gutjnl-2011-300478 [DOI] [PubMed] [Google Scholar]
- Moreira L.O., and Zamboni D.S.. 2012. NOD1 and NOD2 Signaling in Infection and Inflammation. Front. Immunol. 3:328 10.3389/fimmu.2012.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand S., Delanoye-Crespin A., Bressenot A., Huot L., Grandjean T., Peyrin-Biroulet L., Lemoine Y., Hot D., and Chamaillard M.. 2011. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. USA. 108:9601–9606. 10.1073/pnas.1100981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., et al. . 2001a A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 411:603–606. 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- Ogura Y., Inohara N., Benito A., Chen F.F., Yamaoka S., and Nunez G.. 2001b Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 276:4812–4818. 10.1074/jbc.M008072200 [DOI] [PubMed] [Google Scholar]
- Okumura R., Kurakawa T., Nakano T., Kayama H., Kinoshita M., Motooka D., Gotoh K., Kimura T., Kamiyama N., Kusu T., et al. . 2016. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 532:117–121. 10.1038/nature17406 [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T., Hrncir T., Liu Y.J., Biswas A., Hudcovic T., Tlaskalova-Hogenova H., and Kobayashi K.S.. 2009. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA. 106:15813–15818. 10.1073/pnas.0907722106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet J.L., Srinivasula S.M., Tnani M., Razmara M., Fernandes-Alnemri T., and Alnemri E.S.. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309–28313. 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- Propheter D.C., Chara A.L., Harris T.A., Ruhn K.A., and Hooper L.V.. 2017. Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc. Natl. Acad. Sci. USA. 114:11027–11033. 10.1073/pnas.1711395114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic K., Normand S., Rehman A., Delanoye-Crespin A., Chatagnon J., Delacre M., Waldschmitt N., Poulin L.F., Iovanna J., Ryffel B., et al. . 2018. A dietary flavone confers communicable protection against colitis through NLRP6 signaling independently of inflammasome activation. Mucosal Immunol. 11:811–819. 10.1038/mi.2017.87 [DOI] [PubMed] [Google Scholar]
- Ramanan D., Tang M.S., Bowcutt R., Loke P., and Cadwell K.. 2014. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 41:311–324. 10.1016/j.immuni.2014.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A., Sina C., Gavrilova O., Häsler R., Ott S., Baines J.F., Schreiber S., and Rosenstiel P.. 2011. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 60:1354–1362. 10.1136/gut.2010.216259 [DOI] [PubMed] [Google Scholar]
- Robertson S.J., Zhou J.Y., Geddes K., Rubino S.J., Cho J.H., Girardin S.E., and Philpott D.J.. 2013. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 4:222–231. 10.4161/gmic.24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S.J., Geddes K., Maisonneuve C., Streutker C.J., and Philpott D.J.. 2016. Resilience of the intestinal microbiota following pathogenic bacterial infection is independent of innate immunity mediated by NOD1 or NOD2. Microbes Infect. 18:460–471. 10.1016/j.micinf.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Rogala A.R., Oka A., and Sartor R.B.. 2020. Strategies to Dissect Host-Microbial Immune Interactions That Determine Mucosal Homeostasis vs. Intestinal Inflammation in Gnotobiotic Mice. Front. Immunol. 11:214 10.3389/fimmu.2020.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.B., Kozlowska J., Keeble J., Metcalfe K., Fao M., Dowd S.E., Mason A.J., McGuckin M.A., and Bruce K.D.. 2014. Functional divergence in gastrointestinal microbiota in physically-separated genetically identical mice. Sci. Rep. 4:5437 10.1038/srep05437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg N., Vogel T.P., and Canna S.W.. 2017. NLRC4 inflammasomopathies. Curr. Opin. Allergy Clin. Immunol. 17:398–404. 10.1097/ACI.0000000000000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., and Handelsman J.. 2005. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 6:229 10.1186/gb-2005-6-8-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M., Zimmermann A.G., Roberts R.A., Zhang L., Swanson K.V., Wen H., Davis B.K., Allen I.C., Holl E.K., Ye Z., et al. . 2012. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat. Immunol. 13:823–831. 10.1038/ni.2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.U., Kamada N., Muñoz-Planillo R., Kim Y.G., Kim D., Koizumi Y., Hasegawa M., Himpsl S.D., Browne H.P., Lawley T.D., et al. . 2015. Distinct Commensals Induce Interleukin-1β via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity. 42:744–755. 10.1016/j.immuni.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seregin S.S., Golovchenko N., Schaf B., Chen J., Pudlo N.A., Mitchell J., Baxter N.T., Zhao L., Schloss P.D., Martens E.C., et al. . 2017. NLRP6 Protects Il10-/- Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep. 19:2174 10.1016/j.celrep.2017.05.074 [DOI] [PubMed] [Google Scholar]
- Singh V., Yeoh B.S., Walker R.E., Xiao X., Saha P., Golonka R.M., Cai J., Bretin A.C.A., Cheng X., Liu Q., et al. . 2019. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut. 68:1801–1812. 10.1136/gutjnl-2018-316250 [DOI] [PubMed] [Google Scholar]
- Smith P., Siddharth J., Pearson R., Holway N., Shaxted M., Butler M., Clark N., Jamontt J., Watson R.P., Sanmugalingam D., et al. . 2012. Host genetics and environmental factors regulate ecological succession of the mouse colon tissue-associated microbiota. PLoS One. 7 e30273 10.1371/journal.pone.0030273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck T.S., and Virgin H.W.. 2016. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 534:191–199. 10.1038/nature18285 [DOI] [PubMed] [Google Scholar]
- Sutterwala F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., and Flavell R.A.. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245. 10.1084/jem.20071239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Inohara N., Sasakawa C., and Nuñez G.. 2007. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3 e111 10.1371/journal.ppat.0030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J.P., and Davis B.K.. 2005. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annu. Rev. Immunol. 23:387–414. 10.1146/annurev.immunol.23.021704.115616 [DOI] [PubMed] [Google Scholar]
- Ting J.P., Duncan J.A., and Lei Y.. 2010. How the noninflammasome NLRs function in the innate immune system. Science. 327:286–290. 10.1126/science.1184004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubai T., Fujiwara H., Rossi C., Riwes M., Tamaki H., Zajac C., Liu C., Mathew A.V., Byun J., Oravecz-Wilson K., et al. . 2019. Host NLRP6 exacerbates graft-versus-host disease independent of gut microbial composition. Nat. Microbiol. 4:800–812. 10.1038/s41564-019-0373-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos L.H., Girardin S.E., Philpott D.J., Blanot D., Nahori M.A., Werts C., and Boneca I.G.. 2004. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 5:1000–1006. 10.1038/sj.embor.7400248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos L.H., Carneiro L.A., Girardin S.E., Boneca I.G., Lemos R., Bozza M.T., Domingues R.C., Coyle A.J., Bertin J., Philpott D.J., et al. . 2005. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J. Biol. Chem. 280:36714–36718. 10.1074/jbc.M501649200 [DOI] [PubMed] [Google Scholar]
- Trindade B.C., and Chen G.Y.. 2020. NOD1 and NOD2 in inflammatory and infectious diseases. Immunol. Rev. 297:139–161. 10.1111/imr.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C., Earle K.A., Huang K.C., and Sonnenburg J.L.. 2017. The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe. 21:433–442. 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truax A.D., Chen L., Tam J.W., Cheng N., Guo H., Koblansky A.A., Chou W.C., Wilson J.E., Brickey W.J., Petrucelli A., et al. . 2018. The Inhibitory Innate Immune Sensor NLRP12 Maintains a Threshold against Obesity by Regulating Gut Microbiota Homeostasis. Cell Host Microbe. 24:364–378.e6. 10.1016/j.chom.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., and Gordon J.I.. 2008. An invitation to the marriage of metagenomics and metabolomics. Cell. 134:708–713. 10.1016/j.cell.2008.08.025 [DOI] [PubMed] [Google Scholar]
- Vaishnava S., Yamamoto M., Severson K.M., Ruhn K.A., Yu X., Koren O., Ley R., Wakeland E.K., and Hooper L.V.. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 334:255–258. 10.1126/science.1209791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes N.C., Russell A.L., Shah M., Godzik A., Rosenquist M., Halfvarson J., Lefsrud M.G., Apajalahti J., Tysk C., Hettich R.L., et al. . 2009. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3:179–189. 10.1038/ismej.2008.108 [DOI] [PubMed] [Google Scholar]
- Vladimer G.I., Weng D., Paquette S.W., Vanaja S.K., Rathinam V.A., Aune M.H., Conlon J.E., Burbage J.J., Proulx M.K., Liu Q., et al. . 2012. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 37:96–107. 10.1016/j.immuni.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk J.K., Nyström E.E.L., van der Post S., Abad B.M., Schroeder B.O., Johansson Å., Svensson F., Jäverfelt S., Johansson M.E.V., Hansson G.C., et al. . 2019. The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function. J. Exp. Med. 216:2602–2618. 10.1084/jem.20190679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Manji G.A., Grenier J.M., Al-Garawi A., Merriam S., Lora J.M., Geddes B.J., Briskin M., DiStefano P.S., and Bertin J.. 2002. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J. Biol. Chem. 277:29874–29880. 10.1074/jbc.M203915200 [DOI] [PubMed] [Google Scholar]
- Williams K.L., Lich J.D., Duncan J.A., Reed W., Rallabhandi P., Moore C., Kurtz S., Coffield V.M., Accavitti-Loper M.A., Su L., et al. . 2005. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J. Biol. Chem. 280:39914–39924. 10.1074/jbc.M502820200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Thaiss C.A., Nowarski R., Henao-Mejia J., Zhang J.P., Brown E.M., Frankel G., Levy M., Katz M.N., Philbrick W.M., et al. . 2014. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 156:1045–1059. 10.1016/j.cell.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullaert A., Lamkanfi M., and McCoy K.D.. 2018. Defining the Impact of Host Genotypes on Microbiota Composition Requires Meticulous Control of Experimental Variables. Immunity. 48:605–607. 10.1016/j.immuni.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Xia X., Cui J., Wang H.Y., Zhu L., Matsueda S., Wang Q., Yang X., Hong J., Songyang Z., Chen Z.J., et al. . 2011. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 34:843–853. 10.1016/j.immuni.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Zhang C., Xing Y., Xue G., Zhang Q., Pan F., Wu G., Hu Y., Guo Q., Lu A., et al. . 2017. Remodelling of the gut microbiota by hyperactive NLRP3 induces regulatory T cells to maintain homeostasis. Nat. Commun. 8:1896 10.1038/s41467-017-01917-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.I., Zhao L., Eaton K.A., Ho S., Chen J., Poe S., Becker J., Gonzalez A., McKinstry D., Hasso M., et al. . 2020. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep. 31 107471 10.1016/j.celrep.2020.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Boyd K.L., Vogel P., Kastan M.B., Lamkanfi M., and Kanneganti T.D.. 2010. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 32:379–391. 10.1016/j.immuni.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Vogel P., Malireddi R.K., Body-Malapel M., Anand P.K., Bertin J., Green D.R., Lamkanfi M., and Kanneganti T.D.. 2011. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 20:649–660. 10.1016/j.ccr.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld J.R., McMinds R., and Vega Thurber R.. 2017. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121 10.1038/nmicrobiol.2017.121 [DOI] [PubMed] [Google Scholar]
- Zilbauer M., Dorrell N., Elmi A., Lindley K.J., Schüller S., Jones H.E., Klein N.J., Núnez G., Wren B.W., and Bajaj-Elliott M.. 2007. A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell. Microbiol. 9:2404–2416. 10.1111/j.1462-5822.2007.00969.x [DOI] [PubMed] [Google Scholar]