Liver X receptors (LXRs) orchestrate cholesterol metabolism with diverse metabolic and immune functions. Chan et al. show that in the thymus, thymic epithelial cells protect against involution and promote regeneration, whereas T cells require LXRs to escape negative selection.
Abstract
The thymus is a primary lymphoid organ necessary for optimal T cell development. Here, we show that liver X receptors (LXRs)—a class of nuclear receptors and transcription factors with diverse functions in metabolism and immunity—critically contribute to thymic integrity and function. LXRαβ-deficient mice develop a fatty, rapidly involuting thymus and acquire a shrunken and prematurely immunoinhibitory peripheral T cell repertoire. LXRαβ’s functions are cell specific, and the resulting phenotypes are mutually independent. Although thymic macrophages require LXRαβ for cholesterol efflux, thymic epithelial cells (TECs) use LXRαβ for self-renewal and thymocytes for negative selection. Consequently, TEC-derived LXRαβ protects against homeostatic premature involution and orchestrates thymic regeneration following stress, while thymocyte-derived LXRαβ limits cell disposal during negative selection and confers heightened sensitivity to experimental autoimmune encephalomyelitis. These results identify three distinct but complementary mechanisms by which LXRαβ governs T lymphocyte education and illuminate LXRαβ’s indispensable roles in adaptive immunity.
Introduction
The main function of the thymus is to produce, via processes known as positive and negative selection, a repertoire of T lymphocytes that satisfies specific criteria for self-recognition and reactivity. Although cells that pass both selections can exit the thymus and enter the periphery, those that fail to pass even one selection die locally (Abramson and Anderson, 2017). Though necessary for proper T cell development, the selections are metabolically costly, as >90% of thymocytes that are generated in the thymus need to be eliminated (Sawicka et al., 2014, Wang et al., 2017). Exactly how the thymus disposes of the biomass that accrues as a consequence of thymocyte death is not entirely clear, but efficient elimination, which almost certainly involves efferocytosis (Elliott and Ravichandran, 2016), is likely critical for maintaining a healthy, unobstructed gland.
A somewhat unusual feature of the thymus is that, unlike many other organs, its size and cellular composition fluctuate dramatically. The thymus is largest in early life, slowly and progressively involutes as we age, and rapidly involutes after acute infection or injury (Palmer et al., 2018). Although involution reduces T cell output and compromises host defense, the thymus has a remarkable capacity to regenerate, which is crucial for restoring T cell production and immune competence, particularly during recovery after acute stress (Chaudhry et al., 2016). Such cellular dynamics suggest the existence of finely tuned mechanisms that control how the thymus adapts to thymocyte selection, how it involutes over time, and how it regenerates following stress.
Liver X receptors (LXRs) are nuclear receptors that sense intracellular oxysterols and intermediates of the cholesterol biosynthetic pathway such as desmosterol. Upon recognizing their ligands, LXRs regulate expression of genes involved in reverse cholesterol transport, fatty acid synthesis, and immune function (Hong and Tontonoz, 2014; Spann and Glass, 2013). As transcription factors with important roles in cell renewal and lipid shuttling, LXRs may be relevant contributors to thymic cell dynamics, and there are indirect clues this may indeed be the case. For example, age-related thymic involution has been associated with cholesterol accumulation within the thymic parenchyma (Youm et al., 2012) as well as with reduced proliferation of thymic epithelial cells (TECs; Aw and Palmer, 2011). However, notwithstanding a few studies documenting a link between LXRs and T cell biology (Bensinger et al., 2008; Cui et al., 2011), the role of these receptors in the thymus is unknown.
In this study, we first show that whole-body LXRαβ deficiency precipitates cholesterol accumulation in the thymus, accelerates thymic involution, and impairs T cell development. Using cell-specific LXRαβ deletion, we report that these phenotypes are causally independent. Specifically, macrophages use LXRs to regulate thymic lipid homeostasis, TECs use LXRs for self-renewal and thymic regeneration, and thymocytes rely on LXRs for calibrating negative selection strength. These findings identify LXRs as key regulators of thymic function and T cell output.
Results
LXRs maintain thymic function and T cell homeostasis
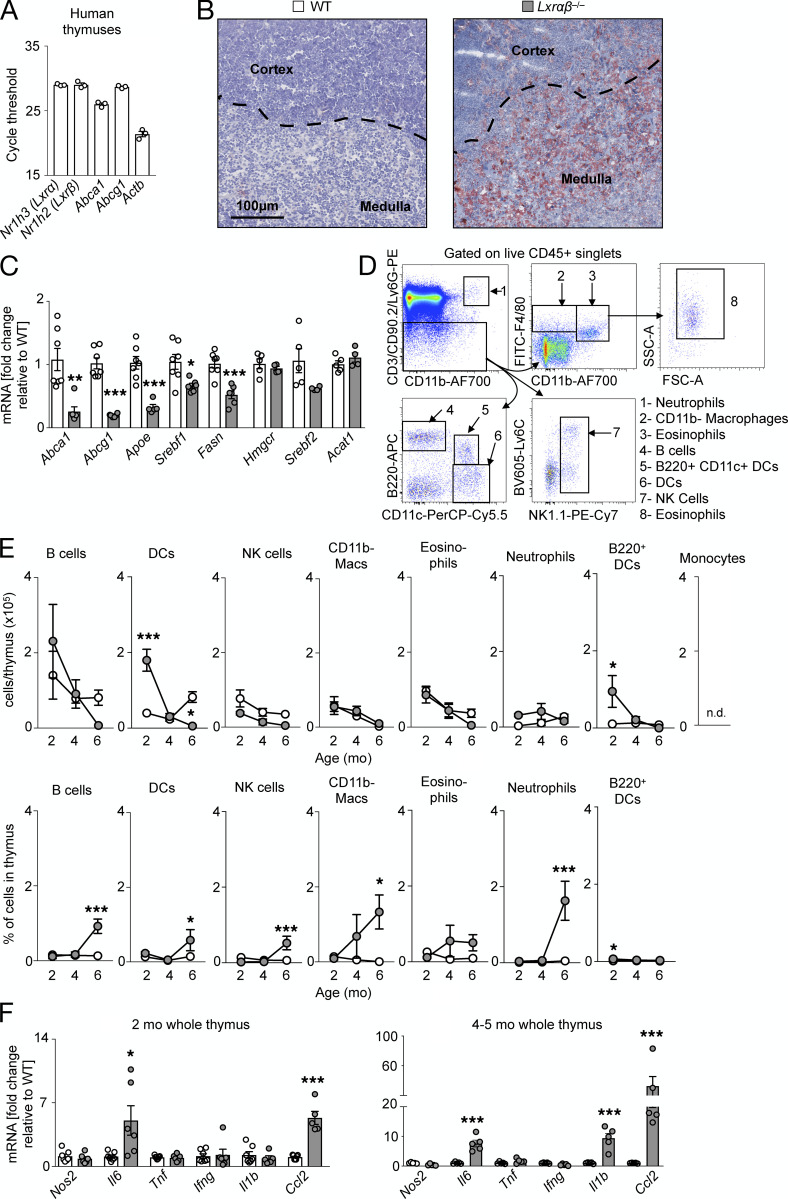
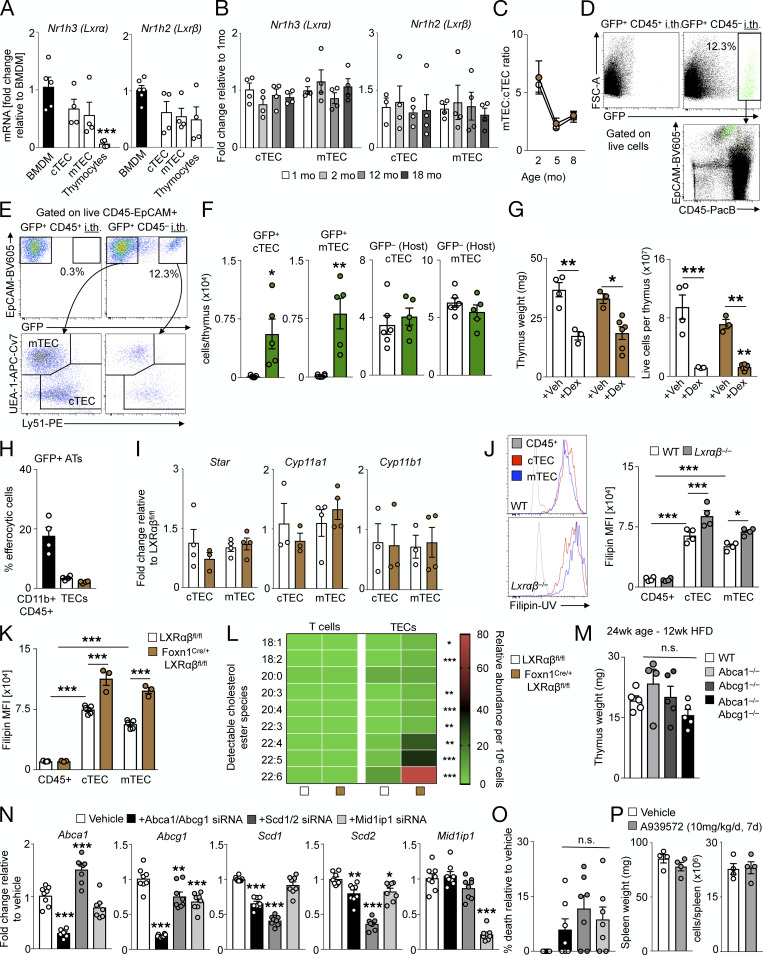
In healthy human thymuses, mRNA expression of Nr1h3 (Lxrα) and Nr1h2 (Lxrβ) and downstream target genes Abca1 and Abcg1 appeared comparable relative to Actb (Fig. S1 A), prompting us to investigate the role of LXRαβ in the mouse thymus. Compared with WT controls, thymus weight and cellularity in LXRα−/−β−/− (LXRαβ−/−) mice increased by 2–3 mo of age and then rapidly decreased, leading to complete involution and loss of cellularity by 6 mo of age (Fig. 1 A). Confirming that thymuses use LXRs, histological analysis of 2-mo-old LXRαβ−/− thymuses revealed neutral lipid enrichment (Fig. S1 B) and attenuated expression of LXR target genes Abca1, Abcg1, Apoe, Srebf1, and Fasn compared with WT, without alterations in genes associated with cholesterol synthesis (Hmgcr, Srebf2) and storage (Acat1; Fig. S1 C).
Figure S1.
LXR deficiency disrupts cholesterol homeostasis but does not induce inflammation. (A) Real-time PCR expression of Lxrα, Lxrβ, Abca1, Abcg1, and Actb in whole human thymuses from individuals aged 5 mo to 13 yr. n = 3 per group. (B) Representative histological images (n > 3 per group) from thymuses from 3-mo-old WT and LXRαβ−/− mice stained with Oil Red O and hematoxylin. Dashed lines indicate demarcation between cortex and medulla. (C) Real-time PCR expression of Abca1, Abcg1, Apoe, Srebf1, Fasn, Hmgcr, Srebf2, and Acat1 in whole thymuses from 2-mo-old WT and LXRαβ−/− mice. Data are n = 4–7 per group from two independent experiments. **P < 0.01, ***P < 0.001. (D) Flow cytometry gating for leukocyte subsets in the thymus. (E) Enumeration and frequency of nonthymocyte leukocytes in the thymuses of WT and LXRαβ−/− mice from 2 to 6 mo of age. Data are n = 4–6 per group from two independent experiments. *P < 0.05, ***P < 0.001. (F) Real-time PCR expression of Nos2, Il6, Tnf, Ifng, Il1b, and Ccl2 from whole thymuses from 2-mo-old and 4–5-mo-old WT and LXRαβ−/− mice. Data are n = 6 or 7 per group from two independent experiments. *P < 0.05, ***P < 0.001. Statistical analysis used was Student’s t test or two-way ANOVA followed by Bonferroni’s post-test. All data are mean ± SEM. SSC-A, side scatter area; FSC-A, forward scatter area; DC, dendritic cell; NK, natural killer; Mac, macrophage.
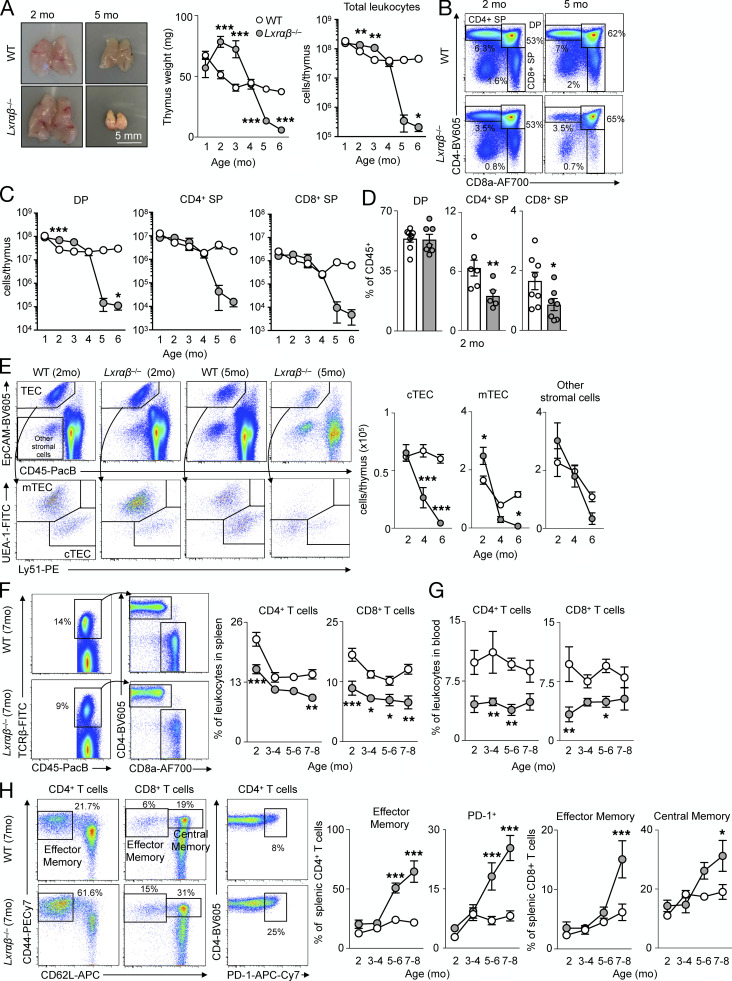
Figure 1.
LXR deficiency in mice disrupts thymic function and T cell homeostasis. (A) Representative images and quantification of thymus size and cellularity over time from C57BL/6J mice versus LXRαβ−/− mice. (B and C) Representative flow cytometric plots (B) and enumeration of major thymocyte subsets (C) in C57BL/6J mice versus LXRαβ−/− mice from 2–6 mo of age (n = 3–7 per group from at least three independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001. (D) Thymocyte subsets as a proportion of total leukocytes in the thymus in C57BL/6J mice versus LXRαβ−/− mice at 2 mo of age (n = 6 or 7 per group from at least two independent experiments). *P < 0.05, **P < 0.01. (E) Representative flow cytometry plots and quantification of TEC subsets and other stromal cells from C57BL/6J mice versus LXRαβ−/−mice from 2–6 mo of age (n = 4–7 per group from at least two independent experiments). *P < 0.05, ***P < 0.001. (F) Representative plots and quantification of CD4+ and CD8+ T cells from spleens of C57BL/6J mice versus LXRαβ−/− mice over time. (G) Proportion of circulating CD4+ and CD8+ T cells from C57BL/6J mice versus LXRαβ−/− mice over time (n = 5–9 per group). (H) Representative flow cytometry plots and quantification of effector memory or PD-1+ splenic CD4+ T cells and effector memory or central memory splenic CD8+ T cells. (F–H) n = 5–9 per group from at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis used was Student’s t test or two-way ANOVA followed by Sidak’s post-test. All data are mean ± SEM. EpCAM, epithelial cell adhesion molecule; PacB, Pacific blue.
In LXRαβ−/− thymuses, the major thymocyte subsets CD4+CD8+ double-positive (DP), CD4+ single-positive (SP), and CD8+ SP thymocytes disappeared over time compared with controls (Fig. 1, B and C). Interestingly, LXRαβ−/− thymuses had reduced CD4+ SP and CD8+ SP numbers but no changes in DP cell numbers at 2 mo of age (Fig. 1, C and D), suggesting a defect in T cell development downstream of DPs. An inventory of the resident thymic stromal cells revealed that cortical (c)TECs and medullary (m)TECs disappeared by 4 mo in LXRαβ−/− thymuses, whereas other stromal cells reduced similarly to WT (Fig. 1 E), indicating a potential TEC-specific defect in LXRαβ−/− thymuses. To consider if inflammation accompanied thymic involution, we quantified the number and frequency of other leukocytes (Fig. S1, D and E) and inflammatory gene expressions (Fig. S1 F), finding few changes between LXRαβ−/− and WT mice, suggesting that inflammation may be associated with thymic involution in this model.
In the periphery of LXRαβ−/− mice, we found fewer CD4+ and CD8+ T cells in the spleen (Fig. 1 F) and blood (Fig. 1 G) compared with WT controls. Over time, splenic CD4+ T cells progressively acquired an effector memory and immunoinhibitory PD-1+ phenotype, while splenic CD8+ T cells adopted an effector and central memory phenotype (Fig. 1 H), akin to previous observations (Bensinger et al., 2008). Together, these data show that LXRs limit thymic involution, reduce lipid accumulation, and sustain T cell development and output.
Cell-specific LXRαβ deletion separates thymic involution, lipid accumulation, and impaired T cell development
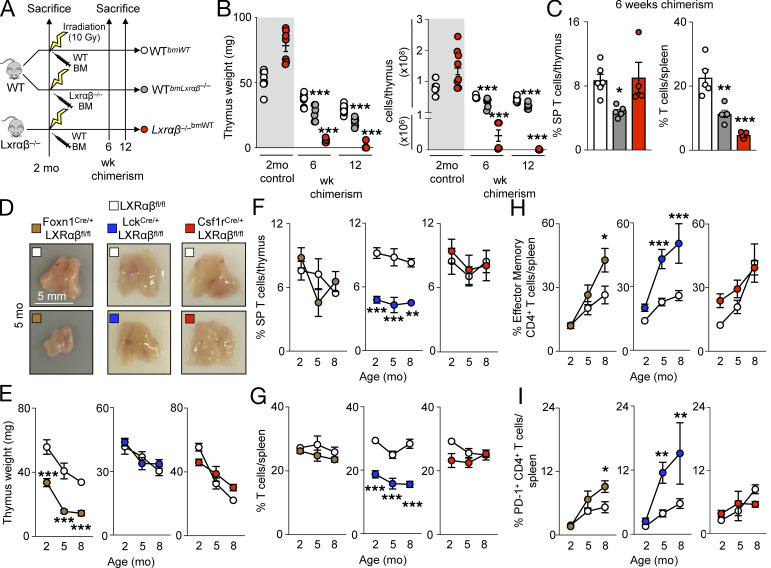
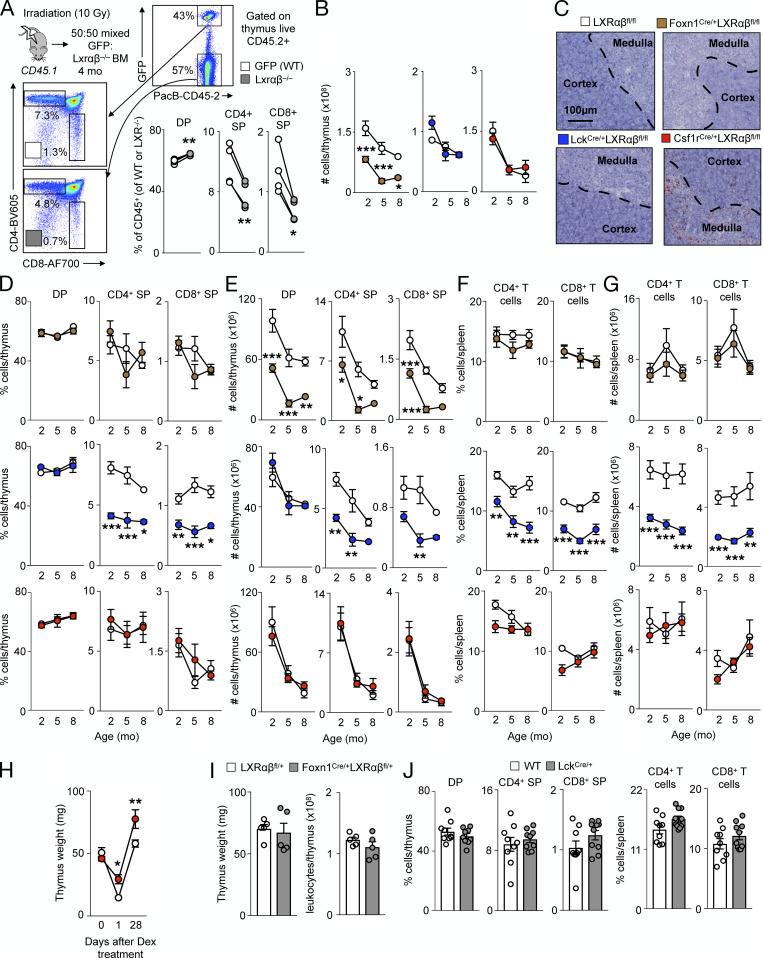
To determine if stromal or hematopoietic cells were responsible for the changes in the thymus, we reconstituted lethally irradiated WT mice with either WT or LXRαβ−/− bone marrow (BM) and reconstituted lethally irradiated LXRαβ−/− mice with WT BM (Fig. 2 A). We found that stromal and not hematopoietic LXR deficiency led to dramatic thymic involution (Fig. 2 B) and impaired thymocyte development (Fig. 2 C), but LXRαβ deficiency in either compartment gave rise to peripheral lymphopenia (Fig. 2 C). To exclude the possibility of cell-extrinsic effects of LXR deficiency impairing thymocyte development, we observed in mixed 50:50 GFP:LXRαβ−/− BM chimeras fewer LXRαβ−/− CD4+ and CD8+ SP cells developed in the thymus compared with equivalent GFP+ SP cells (Fig. S2 A). These results suggest that hematopoietic and stromal cells use LXRs distinctly.
Figure 2.
TEC LXRαβ deletion causes thymic involution, whereas LXRαβ deficiency from thymocytes impairs T cell homeostasis. (A) Schematic outlining the approach for BM chimera experiments. (B) Thymus weight and cellularity of nonirradiated and irradiated mouse groups. Gray bar indicates data that were not included in the statistical analyses. (C) Proportion of SP thymocytes and splenic T cells at 6 wk of age. (B and C) Data are n = 4–7 per group from at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (D) In FoxN1-LXRαβ−/−, Lck-LXRαβ−/−, and Csf1r-LXRαβ−/− versus respective littermate LXRαβfl/fl control mice, representative thymuses (of n > 5 per group) from 5-mo-old mice. Scale bar = 5 mm. (E–I) Quantification of thymus weight (E), proportion of SP thymocytes (F), proportion of splenic T cells (G), proportion of effector memory CD4+ T cells (H), and proportion of PD-1+ CD4+ T cells over time (I). Data are n = 4–8 per group from at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis used is one-way ANOVA followed by Bonferroni’s post-test or two-way ANOVA followed by Sidak’s post-test. All data are mean ± SEM.
Figure S2.
Cell-specific deletion of LXRαβ disrupts thymic homeostasis. (A) Schematic of mixed BM chimera experiment, representative flow cytometry gating, and quantification of DP, CD4+ SP, and CD8+ SP thymocyte percentages from GFP(WT) versus LXRαβ−/− fractions. Data are n = 4 and are representative of two independent experiments. *P < 0.05, **P < 0.01. (B) Enumeration of total thymic cells from Lck-LXRαβ−/− and Csf1r-LXRαβ−/− versus littermate LXRαβfl/fl control mice from 2–8 mo old. Data are n = 4–8 per group and are from more than two independent experiments. *P < 0.05, ***P < 0.001. (C) Representative histological images (n > 3 per group) from thymuses from 2-mo-old FoxN1-LXRαβ−/−, Lck-LXRαβ−/−, and Csf1r-LXRαβ−/− versus littermate LXRαβfl/fl control mice stained with Oil Red O and hematoxylin. Dashed lines indicate demarcation between cortex and medulla. (D and E) Frequency (D) and enumeration (E) of DP, CD4+ SP, and CD8+ SP thymocytes from FoxN1-LXRαβ−/−, Lck-LXRαβ−/−, and Csf1r-LXRαβ−/− versus littermate LXRαβfl/fl control mice over 2–8 mo of age. (F and G) Frequency (F) and enumeration (G) of splenic CD4+ and CD8+ T cells from FoxN1-LXRαβ−/−, Lck-LXRαβ−/−, and Csf1r-LXRαβ−/− versus littermate LXRαβfl/fl control mice over 2–8 mo of age. (D–G) Data are n = 4–8 per group and are from more than two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (H) Thymus weight of Csf1r-LXRαβ−/− versus littermate LXRαβfl/fl control mice over time after dexamethasone treatment (0.25 mg). Data are n = 3–5 per group from two independent experiments. *P < 0.05, **P < 0.01. (I) Thymus weight and cellularity of 2-mo-old LXRαβfl/+ versus FoxN1Cre/+LXRαβfl/+ mice. Data are n = 5 per group from two independent experiments. (J) Frequency of thymocytes and splenic T cells from 2-mo-old WT versus LckCre/+ mice. Data are n = 9 per group from two independent experiments. Statistical analysis used was paired Student’s t test (A) or two-way ANOVA followed by Bonferroni’s post-test (B and D–H). All data are mean ± SEM.
To identify the specific cell type responsible for the changes in the thymus, we deleted LXR from specific cells by breeding LXRαfl/flβfl/fl mice with FoxN1-Cre (FoxN1-LXRαβ−/−), Lck-Cre (Lck-LXRαβ−/−), and Csf1r-Cre (Csf1r-LXRαβ−/−) mice. We chose to test the TEC-specific FoxN1-Cre strain because TEC numbers were LXR dependent (Fig. 1 E) and because TECs regulate thymic size and output (Gray et al., 2006). To delete LXRs from T cells, the most numerous thymic inhabitants whose development appears LXR dependent (Fig. 1, C and D; and Fig. 2 C), we settled on the T cell–specific Lck-Cre. Finally, we deleted LXRs from macrophages by using Csf1r-Cre due to the importance of macrophages to efferocytosis in the thymus (A-Gonzalez et al., 2009; Tacke et al., 2015) .
We then evaluated thymic size and cellularity in the three groups and relevant controls at 2, 5, and 8 mo of age. First, we found that thymuses involuted only in FoxN1-LXRαβ−/− mice (Fig. 2, D and E; and Fig. S2 B), indicating that TECs used LXRs to maintain thymic integrity. Second, we observed that neutral lipids, as measured by Oil Red O staining, accumulated only in Csf1r-LXRαβ−/− mice (Fig. S2 C), suggesting that macrophages are the key cells participating in efferocytosis and cholesterol efflux. Third, we detected a lower frequency of SP thymocytes and peripheral T cells only in Lck-LXRαβ−/− mice (Fig. 2, F and G; and Fig. S2, D–G), which suggests that T cells somehow use LXR for their maturation. Finally, we observed augmented splenic CD4+ T cell activation and PD-1 acquisition among both FoxN1-LXRαβ−/− and Lck-LXRαβ−/− mice, but not Csf1r-LXRαβ−/− mice (Fig. 2, H and I).
Defective efferocytosis in LXRαβ−/− mice is associated with the inability of the thymus to involute after treatment with apoptosis-inducing agents such as dexamethasone (A-Gonzalez et al., 2009). Thymuses of Csf1r-LXRαβ−/− mice resisted dexamethasone-induced thymic involution and surprisingly remained larger even up to 28 d after injection (Fig. S2 H). These data are aligned with previous observations and likely explain why LXRαβ−/− thymuses are bigger at 2–3 mo of age before rapid involution (Fig. 1 A). Importantly, neither thymic involution nor changes in T cell development occurred in FoxN1-Cre–only and Lck-Cre–only mice, respectively (Fig. S2, I and J), ruling out potential effects of cell-specific Cre on thymic involution and T cell development. These data demonstrate that cell-specific LXRαβ deletion affects the thymus differentially: in macrophages, it causes lipid accumulation and resistance to acute thymic involution; in TECs, it induces thymic involution with modest effects on the activation state of T cells; and in thymocytes, it impairs T cell development.
TECs require LXRαβ for self-renewal
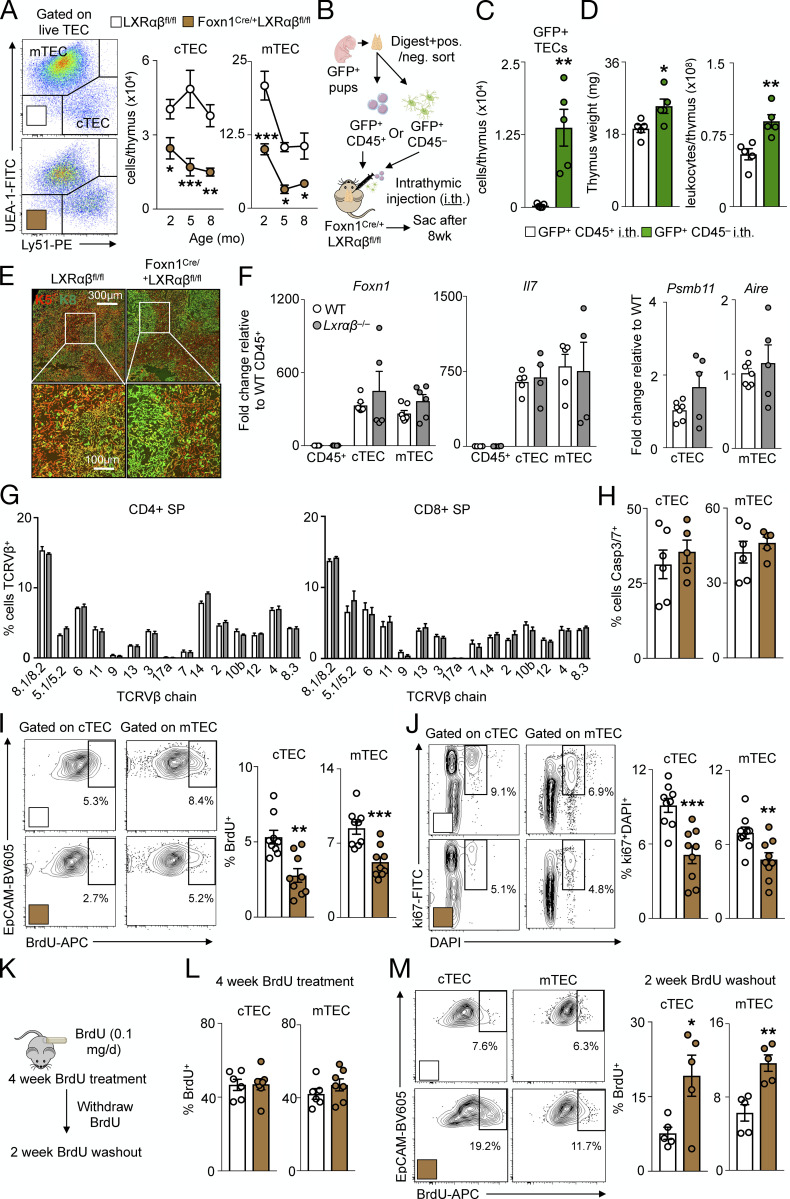
Having established that different cells use LXRs distinctly in the thymus, we next focused on the specific mechanisms that may help explain the phenomenon. We started by investigating the role of LXRs in TECs. Compared with BM-derived macrophages, cTECs and mTECs expressed comparable levels of Nr1h3 (Lxrα) and Nr1h2 (Lxrβ), whereas thymocytes expressed Lxrβ but not Lxrα, consistent with previous results (Fig. S3 A; Bensinger et al., 2008). Additionally, Lxrα and Lxrβ expression in TECs did not change over time (Fig. S3 B), suggesting that age-related thymic involution is not associated with altered LXR activity. In FoxN1-LXRαβ−/− mice, both cTEC and mTEC numbers waned over time (Fig. 3 A) at similar proportions (Fig. S3 C), suggesting that TEC loss due to LXR deficiency does not affect one subset over the other. After engrafting fetal thymic GFP+ CD45+ or CD45− cells (Fig. 3 B) directly into FoxN1-LXRαβ−/− thymuses and waiting for 8 wk, we noticed that only donor GFP+ TECs engrafted (Fig. 3 C; and Fig. S3, D–F) without affecting host cTEC and mTEC numbers (Fig. S3 F). Remarkably, the engraftment of donor GFP+ TECs into FoxN1-LXRαβ−/− thymuses boosted thymic weight and cellularity (Fig. 3 D). These data show that the loss of LXR-deficient TECs is cell intrinsic and that thymic involution can be reversed by LXR-sufficient TECs.
Figure S3.
LXRαβ-deficient TECs are functionally competent but display impaired proliferation. (A and B) Real-time PCR expression analysis of Nr1h3 and Nr1h2 from BM-derived macrophages (BMDM) versus sorted cTECs, mTECs, and total thymocytes from 2-mo-old WT mice (A; data are n = 4 or 5 per group from two independent experiments; ***P < 0.001) and sorted cTECs and mTECs from 1–18-mo-old WT mice (B; data are n = 3 or 4 per group representative of two independent experiments). (C) Ratio of mTECs to cTECs from FoxN1-LXRαβ−/− versus littermate LXRαβfl/fl control mice over 2–8 mo of age. Data are n = 5–8 per group from two independent experiments. (D) Representative flow cytometry plots from 3-mo-old FoxN1-LXRαβ−/− mice intrathymically injected with GFP+CD45+ cells or GFP+CD45− cells. Left: showing engrafted GFP cells and their identity in relation to endogenous (GFP−) cells. Right: example of flow cytometry plots gated on all EpCAM+CD45− (TECs) and engrafted GFP+ TECs. (E) Gating strategy further demonstrates the separation of GFP+ and GFP− into mTECs and cTECs. (F) Enumeration of engrafted GFP+ cTECs and mTECs and GFP− (Host) cTECs and mTECs from 3-mo-old FoxN1-LXRαβ−/− mice intrathymically injected with GFP+CD45+ cells or GFP+CD45− cells. Data are n = 5 or 6 per group from three independent experiments. *P < 0.05, **P < 0.01. (G) Thymus weight and cellularity from 2-mo-old FoxN1-LXRαβ−/− and littermate LXRαβfl/fl control mice treated with vehicle (10% EtOH in PBS) or dexamethasone (0.25 mg) and sacrificed 1 d later. Data are n = 4–6 per group from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (H) FACS-sorted WT CD11b+CD45+ cells, FoxN1-LXRαβ−/− TECs, and LXRαβfl/fl TECs were incubated with GFP+ apoptotic thymocytes for 4 h, and percentage of cells GFP+ after 4 h was measured. Data are n = 4 per group and are representative of two independent experiments. (I) Real-time PCR expression analysis of Star, Cyp11a1, and Cyp11b1 from cTECs and mTECs sorted from FoxN1-LXRαβ−/− versus littermate LXRαβfl/fl control mice. Data are n = 3 or 4 per group and are representative of two independent experiments. (J) Representative flow cytometry plots and quantification of mean fluorescence intensity (MFI) of filipin in CD45+, cTECs, and mTECs from 2-mo-old WT and LXRαβ−/− mice. Data are n = 4 per group and are representative of more than three independent experiments. *P < 0.05, ***P < 0.001. (K) Quantification of MFI of filipin in CD45+, cTECs, and mTECs from 2-mo-old FoxN1-LXRαβ−/− versus littermate LXRαβfl/fl control mice. Data are n = 3–5 per group and are representative of three independent experiments. ***P < 0.001. (L) Lipidomics analysis of cholesterol ester species from T cells and TECs FACS-sorted from 2-mo-old FoxN1-LXRαβ−/− versus littermate LXRαβfl/fl control mice. Data are n = 3 per group (of three pooled samples per n). *P < 0.05, **P < 0.01, ***P < 0.001. (M) Thymus weight from WT (C57BL6/J x DBA/1LacJ), Abca1−/−, Abcg1−/−, and Abca1−/−Abcg1−/− mice maintained on a high-fat diet (HFD) for 12 wk. Data are n = 4 or 5 per group from two independent experiments. (N) Real-time PCR expression of Abca1, Abcg1, Scd1, Scd2, and Mid1ip1 mRNA from MJC1-treated cells treated for 48 h with vehicle (lipofectamine only) or siRNA direct against target genes. Data are n = 8 per group of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (O) Cytotoxicity measured by MTT assay of MJC1 cells treated for 48 h with siRNA directed against Abca1/Abcg1, Scd1/Scd2, or Mid1ip1 versus vehicle. Data are n = 7 or 8 per group of three independent experiments. (P) Spleen weight and cellularity from WT mice treated with A939572 or vehicle for 7 d. Data are n = 4 per group, representative of two independent experiments. Statistical analysis used was Student’s t test or one-way ANOVA followed by Bonferroni’s post-test. All data are mean ± SEM. i.th., intrathymic injection; AT, apoptotic thymocytes.
Figure 3.
LXRαβ-deficient TECs are functionally competent but display impaired proliferation. (A) Representative flow cytometry plots and quantification over time from live FoxN1-LXRαβ−/− versus LXRαβfl/fl TECs (n = 5–8 per group from two independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001. (B–D) Experimental approach (B) and enumeration of engrafted GFP+CD45+ and GFP+CD45− cells (C) and thymus weight and cellularity (D) of FoxN1-LXRαβ−/− mice intrathymically injected GFP+CD45− or GFP+CD45− cells. Data are n = 4 or 5 per group from two independent experiments. *P < 0.05, **P < 0.01. (E) Histological staining of keratin-5 (K5) and keratin-8 (K8) from 2-mo-old Lxrαβfl/fl and FoxN1-Lxrαβ−/− mice (representative of three mice). White boxes indicate zoomed-in section represented in the lower panels. (F) In 2-mo-old C57BL6/J (WT) or LXRαβ−/− mice, Foxn1, Il7, Psmb11, and Aire levels by real-time qPCR from sorted CD45+, cTECs, and mTECs (n = 4–6 per group from two independent experiments). (G) Frequency of CD4+ SP or CD8+ SP thymocytes that express different TCRVβ chains (n = 3 or 4 per group, representative of two independent experiments). (H–J) From 2-mo-old Lxrαβfl/fl and FoxN1-Lxrαβ−/− mice, Caspase (Casp) 3/7+ cTECs and mTECs (H; n = 5 or 6 per group from two independent experiments), representative flow cytometry data and quantification of BrdU+ (I; injected 24 h prior), and ki67+DAPI+ cTECs and mTECs (J). (I and J) Data are n = 9 per group from two or three independent experiments. **P < 0.01, ***P < 0.001. (K) Experimental approach (n = 4 or 5 per group). (L and M) Proportion of BrdU+ cTECs and mTECs after 4-wk BrdU delivery (L) and representative flow cytometry images and quantification of BrdU+ cTECs and mTECs after the 2-wk washout BrdU withdrawal period (M). (K–M) Data are four or five per group from two independent experiments. *P < 0.05, **P < 0.01. Statistical analysis used was Student’s t test or two-way ANOVA followed by Sidak’s post-test. All data are mean ± SEM. Sac, sacrifice; i.th., intrathymic injection.
Next, we analyzed TEC distribution, identity, and function. On histology, the distribution of cTECs (K5+) and mTEC (K8+) in FoxN1-LXRαβ−/− thymuses appeared to be maintained and more discreetly demarcated compared with controls (Fig. 3 E), whereas expression of key TEC identity and function genes including FoxN1, Il7, Psmb11, and Aire did not differ between the groups (Fig. 3 F). Moreover, thymocytes from LXRαβ−/− thymuses showed an identical T cell receptor (TCR) Vβ chain expression pattern compared with controls (Fig. 3 G), suggesting no alteration in the quality of T cell selection by LXR deficiency. To consider efferocytosis, we noted that FoxN1-LXRαβ−/− mice did not resist thymic involution in response to dexamethasone (Fig. S3 G), and that FACS-sorted CD11b+ thymic leukocytes, but not TECs, incubated with apoptotic thymocytes ex vivo engulfed apoptotic cells (Fig. S3 H). Although LXRs can regulate glucocorticoid synthesis (Nader et al., 2012) and TEC-derived glucocorticoids are important in thymocyte selection (Mittelstadt et al., 2018), we observed no differences in steroidogenesis genes Star, Cyp11a1, and Cyp11b1 in cTECs and mTECs sorted from FoxN1-LXRαβ−/− mice versus controls (Fig. S3 I). Together, these data indicate that LXRs do not alter TEC identity and function.
We therefore turned our attention to TEC survival and self-renewal, reasoning that thymic involution due to TEC-specific LXR deficiency might be driven by either enhanced TEC apoptosis or impaired TEC proliferation. We found unchanged rates of apoptosis in both cTECs and mTECs of FoxN1-LXRαβ−/− and control mice, as indicated with an active Caspase 3/7 probe (Fig. 3 H). However, both cTECs and mTECs from FoxN1-LXRαβ−/− mice proliferated less versus controls according to 24-h BrdU+ incorporation and ki67+DAPI+ staining (Fig. 3, I and J). Although TEC apoptosis was far higher than proliferation in all groups and may not reflect the same cells incorporating BrdU or expressing ki67, to confirm these differences, we alternatively ascertained proliferation by delivering BrdU in osmotic mini pumps for 4 wk, followed by a washout period of 2 wk (Fig. 3 K). Though the percentages of BrdU+ cTECs and mTECs were equal after 4-wk labeling (Fig. 3 L) and despite fewer cTEC and mTEC numbers in FoxN1-LXRαβ−/− mice compared with controls (Fig. 3 A), a higher proportion of cTECs and mTECs remained BrdU+ in FoxN1-LXRαβ−/− mice than in controls after 2 wk of withdrawal (Fig. 3 M), indicating slower cell proliferation. These observations demonstrate that LXRs permit TEC proliferation but do not affect cell death.
TECs maintain self-renewal via stearoyl-CoA desaturase (Scd)1/2
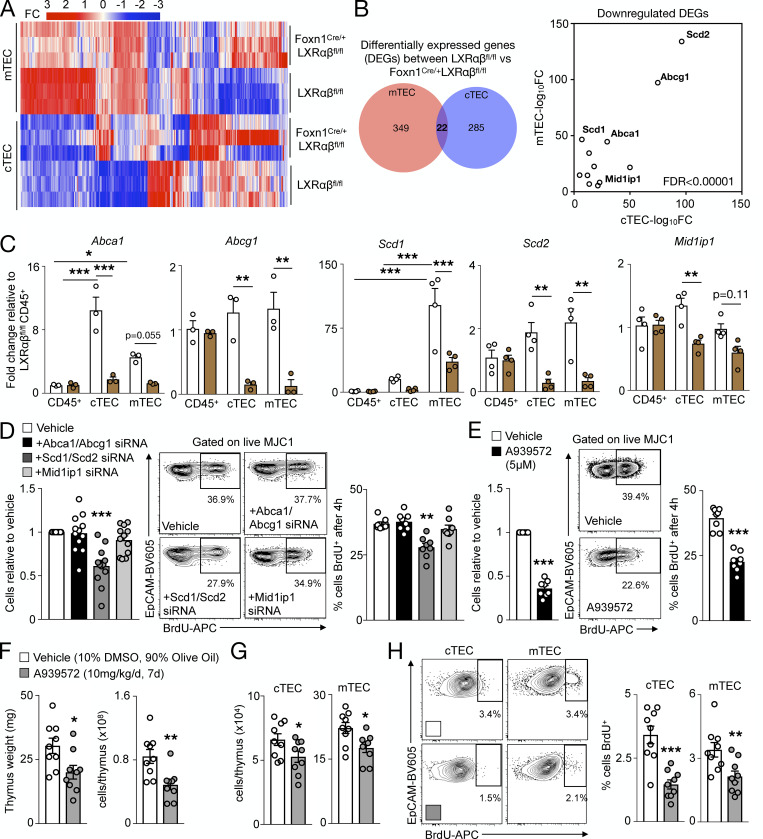
Our next goal was to determine how LXRs regulate TEC proliferation. To identify LXR target genes in TECs, we performed bulk RNA sequencing from FACS-sorted cTECs and mTECs of FoxN1-LXRαβ−/− and control mice. We observed broad transcriptional changes in FoxN1-LXRαβ−/− cTECs and mTECs relative to controls (Fig. 4 A). Of all the differentially expressed genes (DEGs) identified in FoxN1-LXRαβ−/− cTECs and mTECs, 22 DEGs were shared between cTECs and mTECs, with five targets identified as bona fide LXR target genes: Abca1, Abcg1, Mid1ip1, Scd1, and Scd2 (Fig. 4 B). We validated decreased expression of Abca1, Abcg1, Scd1, Scd2, and Mid1ip1 in FACS-sorted FoxN1-LXRαβ−/− cTECs and mTECs compared with CD45+ cells and control animals (Fig. 4 C), confirming that TECs express these LXR target genes.
Figure 4.
TECs maintain self-renewal via Scd1/2. (A and B) From 2-mo-old LXRαβfl/fl and FoxN1-LXRαβ−/− cTECs and mTECs (n = 3 per group of three pooled mice each), heatmap of DEGs (fold change in log2 scale) in bulk RNA-sequencing analysis (A) and Venn diagram and scatter plot of DEGs with normalized fold changes (log10 scale) of common DEGs between mTECs and cTECs (B; LXR target genes labeled; FDR < 0.00001). (C) LXR target gene expression in sorted CD45+ cells, cTECs, and mTECs from 2-mo-old mice (n = 3 or 4 per group, representative of two independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001. (D) In the MJC1 TEC cell line treated with siRNA for 48 h, proportion of MJC1 retrieved and representative plots and quantification of MJC1 BrdU+ (10 µM; 4 h). n = 12 per group. (E) In MJC1 TECs treated with A939572 (6.44 µM) or vehicle (DMSO) for 48 h, proportion of MJC1 retrieved and representative plots and quantification of MJC1 BrdU+ (10 µM; 4 h). n = 8 per group from at least two independent experiments. (D and E) **P < 0.01, ***P < 0.001. (F–H) In WT mice treated with A939572 (10 mg/kg/d, intraperitoneal) or vehicle (10% DMSO and 90% olive oil) for 7 d, thymus weight and cellularity (F), enumeration of cTECs and MTECs (G), and representative plots and quantification of BrdU+ cTECs and mTECs (H). n = 9 per group from at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis used was Student’s t test (unpaired or paired in E) or one-way ANOVA followed by Bonferroni’s post-test (nonrepeated or repeated measures in D). All data are mean ± SEM.
To identify which of these LXR targets were linked to thymic involution and TEC proliferation, we initially focused on Abca1 and Abcg1, the most well–studied LXR target genes that regulate cholesterol efflux (Yvan-Charvet et al., 2010b). As expected and consistent with the reduced Abca1 and Abcg1 expression in these cells, we detected greater membrane cholesterol levels by filipin staining in LXRαβ−/− and FoxN1-LXRαβ−/− TECs than in controls (Fig. S3, J and K) and higher cholesterol ester accumulation in FoxN1-LXRαβ−/− TECs than in T cells and controls using a lipidomics approach (Fig. S3 L). However, thymic involution did not occur in Abca1−/−, Abcg1−/−, and Abca1−/−Abcg1−/− mice, even under hypercholesterolemic conditions (Fig. S3 M), illustrating that neither Abca1 nor Abcg1 is responsible for limiting involution.
Using an immortalized cTEC cell line, MJC1 (Schuster et al., 2015), and siRNA knockdown against Abca1/Abcg1, Mid1ip1, or Scd1/Scd2 (Fig. S3 N), we sought to determine the LXR target genes that contribute to TEC proliferation in vitro. Knockdown of Scd1/Scd2 impaired MJC1 cell yield and proliferation, which did not occur with Abca1/Abcg1 or Mid1ip1 knockdown (Fig. 4 D). Importantly siRNA treatment of MJC1 did not modulate MJC1 apoptosis under any condition (Fig. S3 O). Moreover, an Scd inhibitor, A939572, likewise reduced MJC1 yield and proliferation (Fig. 4 E). Finally, in vivo treatment of WT mice with A939572 for 7 d led to thymic involution (Fig. 4 F), reduced cTEC and mTEC numbers (Fig. 4 G), and attenuated TEC proliferation (Fig. 4 H) and importantly did not change spleen weight or cellularity (Fig. S3 P). Collectively, these data demonstrate that LXR target genes Scd1/Scd2 drive TEC self-renewal to limit thymic involution.
LXRαβ protects thymocytes from negative selection and apoptosis via Abca1/Abcg1
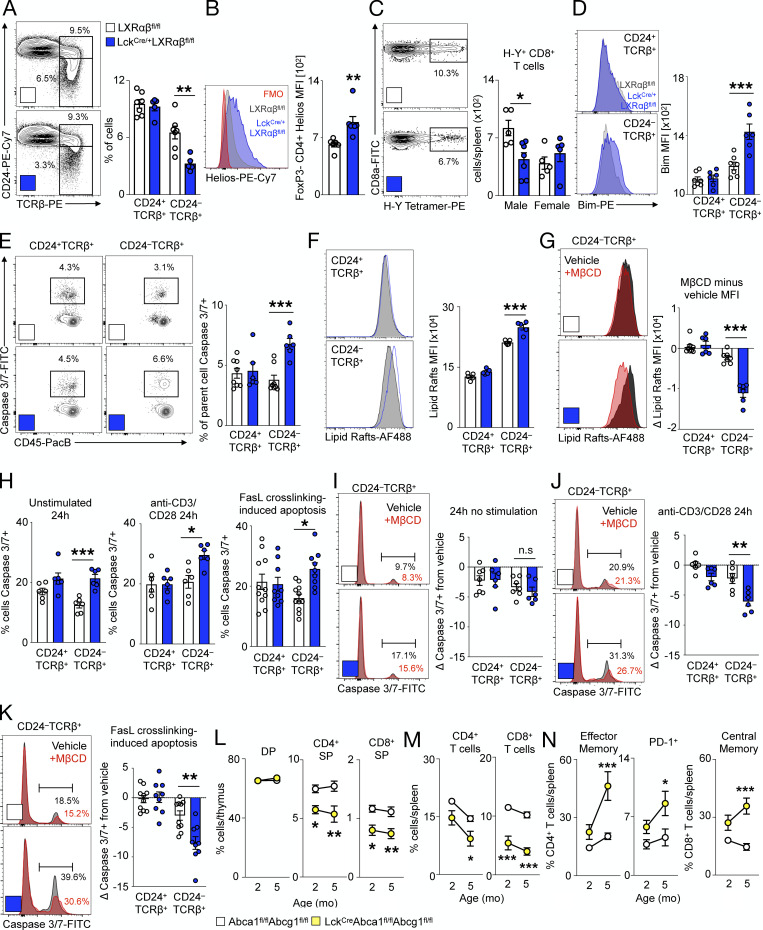
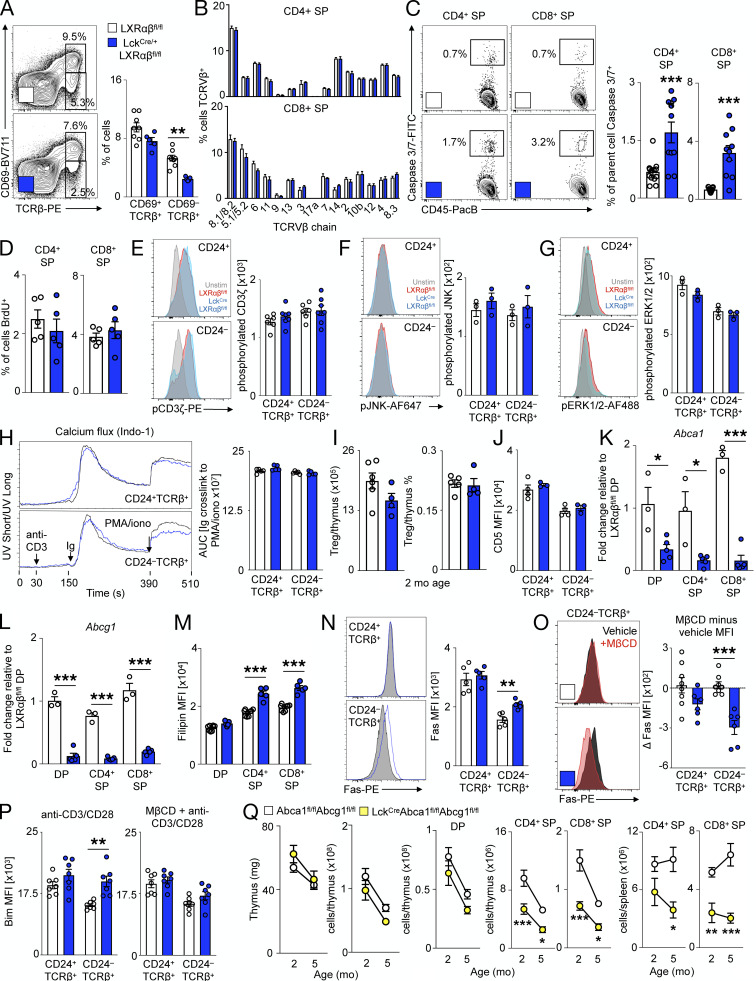
Having identified self-renewal as a mechanism by which TEC-specific LXRs prevent involution, we turned our attention to T cell–specific LXRs. Diminished SP thymocyte development may be attributed to defective positive selection or excessive negative selection. Thymocytes that pass positive selection receive survival signals from cTECs and begin to express TCRβ, CD69, and CD24 (Xu et al., 2013), whereas thymocytes that survive negative selection attenuate CD69 along with CD24. We noted similar frequencies of CD24+TCRβ+ and CD69+TCRβ+ cells between Lck-LXRαβ−/− and control mice (Fig. 5 A and Fig. S4 A). However, we enumerated substantially fewer CD24−TCRβ+ and CD69−TCRβ+ cells in Lck-LXRαβ−/− thymuses than in controls (Fig. 5 A and Fig. S4 A). We next examined surrogate measures of negative selection. Compared with Lck-LXRαβ−/− mice versus control, FoxP3−CD4+ SP thymocytes expressed more Helios (Fig. 5 B), a transcription factor reported to mark CD4+ SP cells undergoing negative selection (Daley et al., 2013). We also enumerated fewer splenic H-Y+ CD8+ T cells from male but not female Lck-LXRαβ−/− mice (Fig. 5 C). H-Y is male tissue specific, and H-Y+ CD8+ splenic T cells can be used to evaluate thymocyte negative selection efficiency (Wang et al., 2016); however, we cannot exclude the extrathymic effects of LXR deletion on reduced T cell numbers. Lck-LXRαβ−/− CD24−TCRβ+ but not CD24+TCRβ+ cells also expressed more Bim (Fig. 5 D), the pro-apoptotic Bcl-2 homologue induced by negative selection signals (Bouillet et al., 2002). We found no difference in the TCRVβ expression pattern of Lck-LXRαβ−/− thymocytes (Fig. S4 B), implying that the TCR repertoire selection of Lck-LXRαβ−/− T cells is unchanged.
Figure 5.
LXRαβ prevents negative selection of thymocytes via plasma membrane lipid rafts and Abca1/Abcg1. (A–J) From 2-mo-old LXRαβfl/fl and LckCre/+LXRαβfl/fl mice. (A) Representative plots and frequency/mean fluorescence intensity (MFI) of CD24+TCRβ+ and CD24−TCRβ+ thymocytes (n = 5–7 per group from at least two independent experiments. **P < 0.01). (B) MFI of Helios+CD4+FoxP3− SP thymocytes (n = 6 or 7 per group from at least two independent experiments. **P < 0.01). (C) Splenic H-Y+ CD8+ T cells (n = 5–7 per group from at least two independent experiments. *P < 0.05. (D) Bim expression of CD24+TCRβ+ and CD24−TCRβ+ thymocytes (n = 6 or 7 per group from at least two independent experiments, where ***P < 0.001). (E) Caspase 3/7+ CD24+TCRβ+ and CD24−TCRβ+ thymocytes (n = 6–8 per group from at least two independent experiments.) ***P < 0.001. (F and G) Representative plots and MFI of lipid raft staining (F; cholera toxin B) and change in lipid raft MFI after vehicle or MβCD ex vivo(G). Data are n = 6 or 7 per group from at least two independent experiments. ***P < 0.001. (H) Quantification of apoptotic thymocytes left unstimulated or treated with anti–CD3/CD28 for 24 h or Fas/FasL cross-linked thymocytes for 4 h. (I–K) Change in apoptosis after ex vivo MβCD compared with vehicle in thymocytes left unstimulated for 24 h (I), stimulated with anti–CD3/CD28 for 24 h (J), or stimulated with Fas/FasL cross-linking for 4 h (K). (H–K) Data are n = 5–9 per group from at least two independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001. (L–N) From 2–5-mo-old Abca1fl/flAbcg1fl/fl and Lck-A1G1−/− mice (n = 6–8 per group), frequency of thymocyte subsets (L), splenic CD4+ and CD8+ T cells (M), and splenic activated/PD-1+ CD4 and activated CD8 T cells (N). Data are n = 6–8 per group from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis used was Student’s t test or two-way ANOVA followed by Bonferroni’s post-test (repeated measures performed in G and H). All data are mean ± SEM.
Figure S4.
Thymocyte LXRαβ prevents excessive negative selection. From 2-mo-old LXRαβfl/fl and LckCre/+LXRαβfl/fl mice. (A) Representative plots and frequency of CD69+TCRβ+ and CD69−TCRβ+ thymocytes. Data are n = 5–7 per group from two independent experiments. **P < 0.01. (B) Frequency of CD4+ SP or CD8+ SP thymocytes expressing different TCRVβ chains. Data are n = 5 per group, representative of two independent experiments. (C) Frequency of Caspase3/7+ thymocytes. Data are n = 9 or 10 per group from three independent experiments. ***P < 0.001). (D) Frequency of BrdU+ thymocytes (2-h pulse). Data are n = 5 per group, representative of two independent experiments. (E–H) Thymocytes stimulated ex vivo with anti-CD3 (10 µg/ml) and anti-CD28 (2 µg/ml) for 5 min and representative plots and mean fluorescence intensity (MFI) of phosphorylated CD3ζ (E), phosphorylated JNK (F), and phosphorylated ERK/12 (G), and representative Indo-1 calcium flux from TCR cross-linking of CD24+TCRβ+ or CD24−TCRβ+ thymocytes and quantification of area under the curve (AUC) of Ig-induced cross-linking (H). (E–G) Data are n = 3–8 per group and are representative of two independent experiments. (H) Data are n = 5 per group representative of two independent experiments. (I) Enumeration and frequency of T reg cells. Data are n = 4 or 5 per group and are representative of two independent experiments. (J) CD5 MFI. Data are n = 3 or 4 per group and are representative of two independent experiments. (K and L) Real-time PCR expression of Abca1 (K) and Abcg1 (L) from FACS-sorted thymocytes. Data are n = 3 per group and are representative of two independent experiments. *P < 0.05, ***P < 0.001. (M) Quantification of thymocyte filipin MFI. Data are n = 5–8 per group from two independent experiments. ***P < 0.001. (N) Representative plots and thymocyte Fas MFI. Data are n = 5 per group and are representative of three independent experiments. **P < 0.01. (O) Change in thymocyte Fas MFI after MβCD or vehicle treatment ex vivo. Data are n = 6–8 per group from three independent experiments. ***P < 0.001. (P) Thymocyte Bim MFI after 4-h anti–CD3/CD28 stimulation with or without MβCD treatment. Data are n = 7 per group from two independent experiments. **P < 0.01. (Q) From 2–5-mo-old Abca1fl/flAbcg1fl/fl and Lck-A1G1−/− mice, thymus weight and cellularity, enumeration of splenic CD4 and CD8 T cells, and enumeration of splenic activated/immunoinhibitory CD4+ and activated CD8+ T cells. Data are n = 6–8 per group from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis used was Student’s t test or two-way ANOVA followed by Bonferroni’s post-test. All data are mean ± SEM.
These observations prompted us to explore the underlying mechanism by which T cell–specific LXRs regulate negative selection quantity. Lck-LXRαβ−/− CD24−TCRβ+ but not CD24+TCRβ+ cells was more apoptotic (Fig. 5 E). Though total apoptosis of both CD4+ SP and CD8+ SP thymocytes increased (Fig. S4 C), CD4+ SP and CD8+ SP proliferated similarly (Fig. S4 D). We found no differences in TCR-induced pCD3ζ, pERK1/2, pJNK expression or calcium flux between CD24+TCRβ+ and CD24−TCRβ+ thymocytes of Lck-LXRαβ−/− mice compared with those of controls (Fig. S4, E–H). Moreover, the proportion and number of T reg cells was unchanged in Lck-LXRαβ−/− mice and controls (Fig. S4 I), and we observed no differences in CD5 expression between Lck-LXRαβ−/− thymocytes versus controls (Fig. S4 J). These data suggest that LXRs do not alter the strength of T cell signaling.
We next surmised that LXR-induced changes in apoptosis and negative selection may be attributed to Abca1 and Abcg1, as these are the most highly differentially regulated LXR target genes in peripheral T cells (Bensinger et al., 2008). Indeed, FACS-sorted DP, CD4+ SP, and CD8+ SP thymocytes from Lck-LXRαβ−/− mice had decreased Abca1 and Abcg1 expression (Fig. S4, K and L) and increased membrane cholesterol in SP thymocytes (Fig. S4 M). As Abca1/Abcg1–dependent membrane cholesterol changes can enrich lipid raft microdomains (Yvan-Charvet et al., 2010a) and lipid rafts modulate T cell function (Robinson et al., 2017), we tested whether lipid rafts differ among thymocytes. We observed stronger lipid raft staining in CD24−TCRβ+ Lck-LXRαβ−/− thymocytes but not CD24+TCRβ+ Lck-LXRαβ−/− cells (Fig. 5 F). Using methyl-β-cyclodextrin (MβCD) to acutely deplete cholesterol and disrupt lipid rafts ex vivo (Kabouridis et al., 2000; Mahammad and Parmryd, 2015), we found selective attenuated lipid raft intensity in CD24−TCRβ+ Lck-LXRαβ−/− thymocytes but not CD24+TCRβ+ or control thymocytes (Fig. 5 G).
To determine causality between LXR, lipid rafts, and apoptosis, we next evaluated the sensitivity of Lck-LXRαβ−/− thymocytes to apoptosis by comparing cells left unstimulated for 24 h with cells stimulated with anti–CD3/CD28 for 24 h and Fas/Fas ligand (FasL) cross-linking–induced apoptosis. We examined Fas/FasL as an additional pathway of active apoptosis as (i) Fas is localized to lipid rafts on T cells (Legembre et al., 2006; Miyaji et al., 2005; Moretti et al., 2008), (ii) CD24−TCRβ+ thymocytes but not CD24+TCRβ+ cells or control cells of Lck-LXRαβ−/− mice displayed enriched cell surface Fas (Fig. S4 N), and (iii) only CD24−TCRβ+ Lck-LXRαβ−/− thymocytes exhibited reduced Fas cell surface expression after MβCD treatment (Fig. S4 O). Compared with controls, CD24−TCRβ+ thymocytes but not CD24+TCRβ+ cells of Lck-LXRαβ−/− mice displayed enhanced apoptosis in all conditions (Fig. 5 H). Although apoptosis in unstimulated thymocytes was not different after lipid raft disruption by MβCD (Fig. 5 I), apoptosis was reduced in CD24−TCRβ+ thymocytes from Lck-LXRαβ−/− mice and not control cells after anti–CD3/CD28 stimulation (Fig. 5 J) and Fas/FasL cross-linking (Fig. 5 K). Finally, Bim induction by anti–CD3/CD28 stimulation was enhanced in Lck-LXRαβ−/− CD24−TCRβ+ thymocytes, which was prevented after MβCD treatment (Fig. S4 P). These data collectively suggest that LXRs protect CD24−TCRβ+ thymocytes from activation-induced cell death by limiting Bim levels via lipid rafts.
To more firmly establish that these phenomena are Abca1 and Abcg1 dependent, we bred Lck-Cre mice to Abca1fl/flAbcg1fl/fl mice (Lck-A1G1−/−). Similar to Lck-LXRαβ−/− mice (Fig. 2, D–I), thymic involution did not occur in Lck-A1G1−/− mice (Fig. S4 Q), but T cell development was markedly impaired (Fig. 5 L and Fig. S4 Q), leading to peripheral T cell lymphopenia (Fig. 5 M and Fig. S4 Q) and premature T cell activation and immunoinhibitory phenotype (Fig. 5 N). Altogether, these data support the idea that thymocytes use LXRs to reduce lipid rafts in order to limit apoptosis and thus modulate negative selection.
LXRαβ is required for efficient thymic recovery and reconstitution of the T cell pool
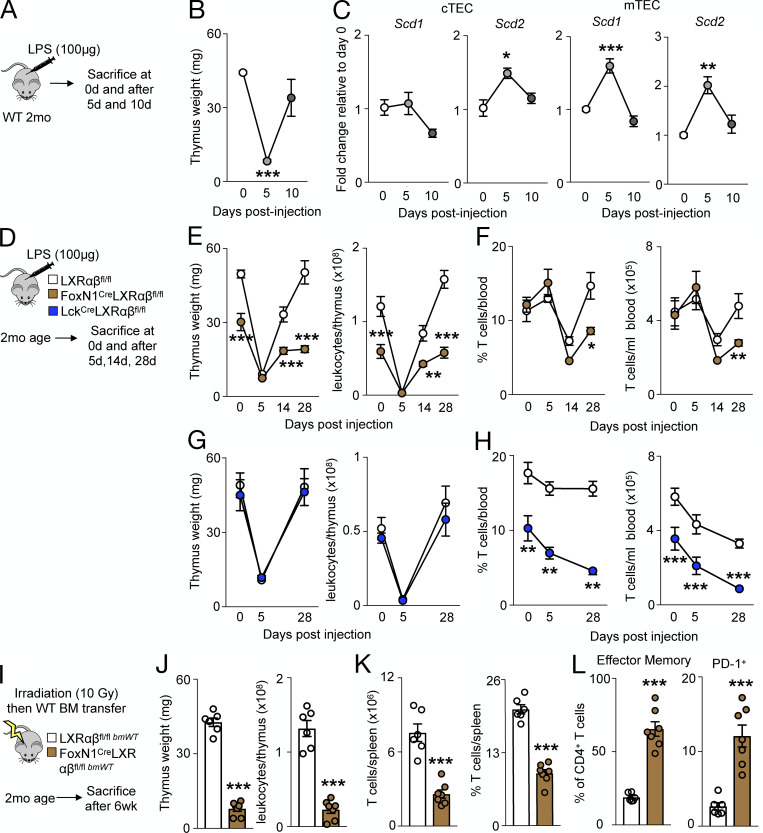
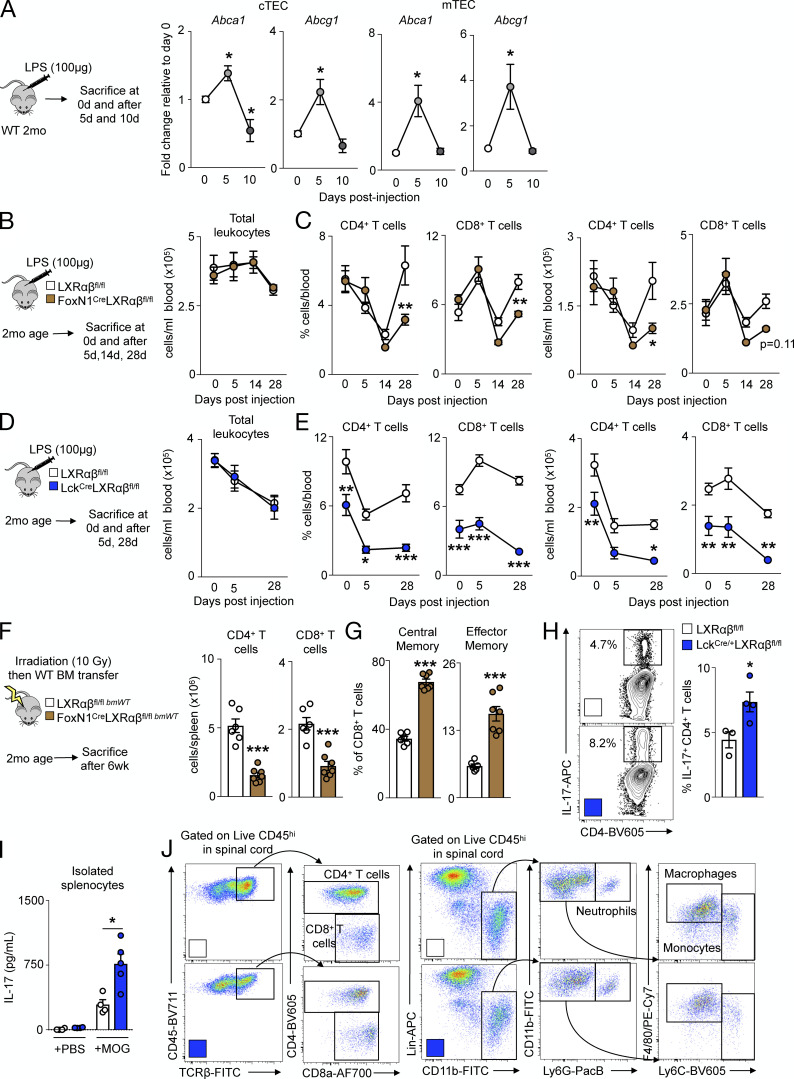
Thymic regeneration after acute stress relies on sustained TEC proliferation (Chaudhry et al., 2016). We therefore hypothesized that thymic injury mobilizes LXRs to mediate thymic recovery and to prevent T cell deficiency. Thymuses of WT mice receiving a single dose of LPS (Fig. 6 A) underwent thymic involution and quick regeneration within 10 d (Fig. 6 B). In FACS-sorted TECs, LPS-induced thymic involution associated with elevated, but transient expression of Scd1, Scd2, Abca1, and Abcg1 (Fig. 6 C and Fig. S5 A), suggesting that thymic regeneration after LPS-induced involution is LXR dependent. To test this directly, we injected LPS to FoxN1-LXRαβ−/− mice (Fig. 6 D) and found a similarly involuted thymus 5 d after injection (Fig. 6 E). However, thymuses of FoxN1-LXRαβ−/− mice failed to regenerate up to 28 d after LPS injection (Fig. 6 E), and these impairments were also reflected in the periphery (Fig. 6 F and Fig. S5, B and C). We did not observe impaired thymic recovery of Lck-LXRαβ−/− mice in response to LPS (Fig. 6 G) and no further changes on circulating leukocytes or T cell levels (Fig. 6 H and Fig. S5, D and E), showing that T cell LXRs do not contribute to thymic recovery.
Figure 6.
LXRαβ is required for efficient thymic recovery and reconstitution of the T cell pool. (A–C) 2-mo-old WT mice were injected with LPS (100 µg; A), and thymus weight over time after LPS injection (B) and Scd1 and Scd2 levels by real-time PCR from FACS-sorted WT cTECs and mTECs over time after LPS injection (C) are shown. (B and C) Data are n = 3 or 4 per group, representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (D–F) 2-mo-old LXRαβfl/fl mice, Lck-LXRαβ−/− mice, and FoxN1-LXRαβ−/− mice were injected with LPS (100 µg) intraperitoneally and sacrificed 5, 14, or 28 d after injection, with thymus weight over time quantified (E) and circulating T cell frequency and number enumerated (F). (G and H) Thymus weight over time after LPS injection in LXRαβfl/fl versus Lck-LXRαβ−/− mice (G) and circulating T cell frequency and number enumerated (H). (D–H) Data are n = 4–7 per group from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (I) Experimental approach of J–L: 2-mo-old LXRαβfl/fl and FoxN1-LXRαβ−/− mice were lethally irradiated and transplanted with WT BM. After 6 wk, mice were sacrificed. (J–L) Thymus weight and cellularity (J), frequency and enumeration of splenic T cells (K), and frequency of effector memory and immunoinhibitory splenic CD4+ T cells (L). (I–L) Data are n = 6–8 per group from two independent experiments. ***P < 0.001, by Student’s t test or two-way ANOVA followed by Bonferroni’s post-test. All data are mean ± SEM.
Figure S5.
LXRαβ is required for efficient thymic recovery, reconstitution of the T cell pool, and EAE. (A) Abca1 and Abcg1 levels by real-time PCR from FACS-sorted WT cTECs and mTECs over time after LPS injection. Data are n = 3 or 4 group and are representative of two independent experiments. *P < 0.05. (B) In 2-mo-old LXRαβfl/fl and FoxN1-LXRαβ−/− mice, mice were injected with LPS (100 µg) intraperitoneally and sacrificed 5, 14, or 28 d after injection, and circulating leukocytes were enumerated after LPS injection. (C) Frequency and enumeration of circulating CD4+ and CD8+ T cell levels after LPS injection are also shown. (D) In 2-mo-old LXRαβfl/fl and Lck-LXRαβ−/− mice, mice were injected with LPS (100 µg) intraperitoneally and sacrificed 5, 14, or 28 d after injection; circulating leukocytes were enumerated after LPS injection. (E) Frequency and enumeration of circulating CD4+ and CD8+ T cell levels after LPS injection are also shown. (B–E) Data are n = 4–7 per group from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (F) In 2-mo-old LXRαβfl/fl and FoxN1-LXRαβ−/− mice, mice were lethally irradiated and transplanted with WT BM and sacrificed after 6 wk, with circulating CD4+ and CD8+ T cell levels enumerated. (G) Quantification of frequency of effector memory and immunoinhibitory splenic CD8+ T cells is also shown. (F and G) Data are n = 6 or 7 per group from two independent experiments. ***P < 0.001. (H) Representative flow cytometry plots and quantification of IL-17+CD4+ T cells after 5-d Th17 polarization and 4-h PMA/ionomycin/GolgiPlug stimulation ex vivo from naive 2-mo-old LXRαβfl/fl and Lck-LXRαβ−/− mice. Data are n = 3 or 4 per group and are representative of two independent experiments. *P < 0.05. (I) IL-17 concentration (measured by ELISA) from supernatants of splenocytes stimulated with MOG35–55 peptide ex vivo for 3 d from EAE-treated 2-mo-old LXRαβfl/fl and Lck-LXRαβ−/− mice. Data are n = 4 or 5 per group and are representative of two independent experiments. *P < 0.05. (J) Representative flow cytometry plots of spinal cord gating of leukocytes from EAE-treated 2-mo-old LXRαβfl/fl and Lck-LXRαβ−/− mice. Lin, lineage (CD3, CD90.2 CD19, NK1.1). Statistical analysis used was Student’s t test or two-way ANOVA followed by Bonferroni’s post-test. All data are mean ± SEM.
To bolster the role of TEC LXR in thymic regeneration, we compared thymic recovery in response to lethal irradiation, which is a more severe form of acute stress. We reconstituted either WT or FoxN1-LXRαβ−/− mice with WT BM (Fig. 6 I). In FoxN1-LXRαβ−/− mice, thymic recovery was markedly attenuated (Fig. 6 J), which associated with T cell deficiency (Fig. 6 K and Fig. S5 F) and premature activation and PD-1 expression (Fig. 6 L and Fig. S5 G). These data collectively demonstrate that TEC LXRs are necessary to facilitate thymic regeneration and recovery of a depleted T cell pool.
Thymocyte LXRαβ enhances experimental autoimmune encephalomyelitis (EAE) in mice
As we observed fewer peripheral T cells in Lck-LXRαβ−/− animals (Fig. 2 G), we hypothesized there may be three potential outcomes following EAE, a T cell–mediated disease: (i) disease protection due to T cell deficiency; (ii) worsened disease due to enhanced Th17 sensitization in LXR-deficient T cells (Cui et al., 2011; Xu et al., 2009); and (iii) no effect.
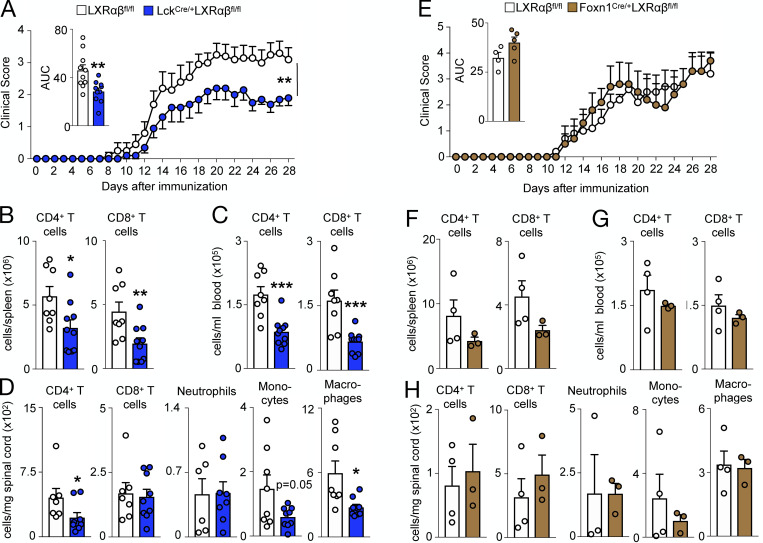
In accordance with previous observations (Cui et al., 2011; Xu et al., 2009), a higher frequency of T cells isolated from naive Lck-LXRαβ−/− mice expressed more IL-17 (Fig. S5 H), whereas splenocytes from Lck-LXRαβ−/− animals subjected to EAE produced more IL-17 upon myelin oligodendrocyte glycoprotein (MOG) stimulation ex vivo (Fig. S5 I). Despite this, Lck-LXRαβ−/− mice developed attenuated EAE (Fig. 7 A). This was associated with fewer splenic and circulating T cells (Fig. 7, B and C) and fewer CD4+ T cells but not CD8+ T cells in the spinal cord (Fig. 7 D and Fig. S5 J). Of other inflammatory leukocytes, the spinal cord of Lck-LXRαβ−/− mice contained fewer macrophages and monocytes but not neutrophils (Fig. 7 D). In contrast, induction of EAE in FoxN1-LXRαβ−/− mice did not alter disease progression (Fig. 7 E), peripheral T cell numbers (Fig. 7, F and G), or infiltrating leukocyte numbers in the spinal cord (Fig. 7 H), which highlights the divergent nature of the cell-specific LXR deletion. Thus, the protection against EAE in the absence of LXRαβ was cell specific. Altogether, the data indicate that LXRs contribute to thymic integrity and function via cell-specific mechanisms.
Figure 7.
T cell LXRαβ deficiency protects mice from autoimmune disease. (A–C) After EAE induction in 2-mo-old LXRαβfl/fl versus Lck-LXRαβ−/− mice, clinical score, with inset representing area under curve (AUC) quantified for each mouse (A) and frequency of splenic (B) and circulating (C) CD4+ T cells and CD8+ T cells at day 28 after EAE. (D) Frequency of spinal cord CD4+ T cells, CD8+ T cells, neutrophils, monocytes, and macrophages at day 28 after EAE. (A–D) Data are n = 8–10 per group from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (E–G) After EAE induction in 2-mo-old LXRαβfl/fl versus FoxN1-LXRαβ−/− mice, clinical score with inset representing AUC for each mouse (E; n = 5 per group) and frequency of splenic (F) and circulating (G) CD4+ T cells and CD8+ T cells at day 28 after EAE (n = 3 or 4 per group). (H) Frequency of spinal cord CD4+ T cells, CD8+ T cells, neutrophils, monocytes, and macrophages at day 28 after EAE. (E–H) Data are n = 3–5 per group and are representative of two independent experiments. Statistical analysis used was Student’s t test (two groups only) or two-way ANOVA followed by Bonferroni’s post-test (two groups over time). All data are mean ± SEM.
Discussion
In this study, we show that LXRs are important to lipid homeostasis, thymic integrity, and thymocyte negative selection. Although the global deletion to LXRs precipitates a series of phenotypes that may at first appear to be causally connected—and mechanistically bound to the capacity of LXRs to orchestrate reverse cholesterol transport—our data illuminate an unexpectedly complex and dynamic system that highlights the diverse molecular circuitry through which specific thymic denizens use the receptors.
Of the three cell types tested, deletion of LXRs from macrophages yielded the most expected and arguably straightforward results. Macrophages are professional efferocytes with well-defined roles in scavenging and eliminating failed thymocytes (Dzhagalov et al., 2013). As consumption of dying cells necessarily leads to accrual of various cell components, including lipids, the complete process of disposal requires efficient recycling of biomass, and macrophages are especially well equipped to do so, in part thanks to a coordinated program of lipid export (Remmerie and Scott, 2018). It is therefore unsurprising that deletion of LXRs from macrophages impaired their capacity to expel lipids, resulting in a fatty thymus.
Surprisingly, however, deletion of LXRs from macrophages neither involuted the thymus nor interfered with T cell production. Without insights from cell-specific approaches, it would stand to reason that lipid accrual catalyzes the remaining phenomena: a fatty thymus might be expected to be ineffectual at chaperoning thymocyte education, which could eventually spell its destruction and elicit lymphocyte impairment. However, thymic resilience in spite of lipid accumulation in macrophage-specific Lxrαβ−/− mice divulged the existence of additional macrophage-independent mechanisms, the elucidation of which constituted the bulk of our study.
TECs, of which there are two major kinds, are relatively rare thymic residents charged with the essential task of educating T cells via the sequential steps of positive and negative selection (Abramson and Anderson, 2017). Deleting LXRs from TECs did not have a substantial impact on T cell selection, but it accelerated thymic involution akin to the global knockout. Because involution was perhaps surprisingly Abca1/Abcg1 independent, we performed RNA sequencing, alongside other approaches to screen for additional LXR targets. We found that TEC-specific LXRs control expression of Scd1 and Scd2, which then regulate TEC proliferation. Hence, absence of TEC-specific LXRs impaired TEC self-renewal, leading to progressive TEC disappearance and thymic involution. Scds are enzymes that catalyze the production of monounsaturated fatty acids from saturated fatty acids, thus generating essential cell membrane components (Paton and Ntambi, 2009) that are in high demand during cell growth and mitosis (Coomans de Brachène et al., 2017; Igal, 2011; Yee et al., 2013). Our study therefore suggests that absence of LXRs in TECs impairs Scd-dependent proliferation not necessarily because Scds are obligate orchestrators of the cell cycle, but rather because Scds provide the building blocks without which a cell’s growth and reproduction is compromised.
Deficiency of LXRs in T cells led neither to lipid accumulation nor to thymic involution, but influenced T cell biology in a few important ways that confirmed, in some cases deviated from, and ultimately built on, previous work. LXRβ suppresses T cell proliferation in whole-body LXRβ−/− mice (Bensinger et al., 2008). Though we saw no such suppression, a key difference in approach was our study’s cell-specific deletion of both isoforms, precluding overcompensation that is known to occur, for example, in macrophages (Quinet et al., 2006). Consistent with previous studies (Cui et al., 2011; Xu et al., 2009), LXR deficiency increased the relative IL-17–producing pool of T cells. However, the most dramatic observations were peripheral lymphopenia, acquisition of effector memory, and increased production of the negative costimulator PD-1. In the thymus, these changes corresponded with exaggerated negative selection, which depended on heightened lipid raft–mediated activation-induced Bim induction and cell death of thymocytes. In other words, developing thymocytes produce LXRαβ, which induces Abca1/Abcg1 to reduce lipid rafts on the plasma membrane, leading to a reduction in Bim and diversion away from negative selection signals. LXRs participate in negative selection, we suggest, not by helping to distinguish self from nonself but by calibrating the strength of cells’ responsiveness to apoptosis-inducing signals, potentially limiting needless elimination of otherwise nonautoreactive lymphocytes. However, as LXRs did not modulate thymocyte TCR signaling strength, it remains to be determined what links LXRs and lipid rafts to Bim and negative selection.
The distinct and cell-specific LXR mechanisms we uncovered triggered different outcomes after acute stress and during chronic disease. That thymic regeneration following cytotoxic stress depended on TEC-specific LXRs was notable not only because thymic regeneration, which relies on multiple mechanisms including IL-22 (Dudakov et al., 2012), is important to immune fitness, but also because it highlighted the physiological imperative of sustaining TEC self-renewal. As for the contribution of T cell–produced LXRs on EAE, our findings draw attention to a quantity versus quality tension: whereas acquisition of IL-17 producers might aggravate disease, lymphopenia arising from overabundant thymocyte elimination can be expected to be protective. The results show that in this particular case, quantity may have dominated because absence of LXR, and the resultant lymphopenia, protected against EAE in spite of IL-17 skewing. Although these EAE data are consistent with negative selection data, our results do not exclude the possibility that extrathymic effects LXR deletion to fewer T cells contributing to this phenotype.
The nuanced and cell-specific mechanisms we describe in this study beg a fair if somewhat naive question: why should LXRs function differently in different cells? The answer probably requires appreciating that LXRs are pleiotropically integrated within the molecular networks of cells that express them. As multipotent transcription factors, their signature activity in a given cell reflects the particular needs of that cell: TECs do not rely on Abca1/Abcg1–dependent reverse cholesterol transport meaningfully maybe because TECs neither efferocytose nor phagocytose and are therefore unburdened by excess biomass accrual. Conversely, LXRs likely do not control macrophage proliferation and thus may be inconsequential to the thymus given redundancies unique to tissue macrophage renewal. Whether and how this line of argument helps explain the diversity of responses we document here should be investigated in the future.
Attenuated thymic output due to age-related involution, cytoreductive chemotherapy, chronic infections, and thymectomy associate with multiple complications including opportunistic infections, impaired vaccine responses, and cancer relapse (Chaudhry et al., 2016; Palmer et al., 2018; van den Brink et al., 2004). Pharmacological LXR activation suppresses inflammation and enhances cholesterol influx and is therefore therapeutically promising (Fessler, 2018). Our study reveals a dimension of LXR influence critical to T cell biology and cautions against using non–cell-specific approaches for manipulating factors endowed with diverse functional profiles. Overall, LXRs are critical to acquired immune responses by boosting thymic recovery and maintaining a healthy T cell pool. In concert with the suppressive effects of LXRs on innate immunity, our data show that LXR activation may be beneficial to treating diverse diseases such as atherosclerosis, infection, and cancer.
Materials and methods
Material availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by F.K. Swirski (fswirski@mgh.harvard.edu).
Experimental model and subject details
Mice
C57BL6/J, C57BL/6-Tg(UBC-GFP)30Scha/J (GFP), B6(Cg)-Foxn1tm3(cre)Nrm/J (FoxN1-Cre), B6.Cg-Tg(Lck-cre)548Jxm/J (Lck-Cre), and C57BL/6-Tg(Csf1r-cre)1Mnz/J (Csf1r-Cre) mice were purchased from The Jackson Laboratory. LXRαβ−/− and LXRαβfl/fl mice were provided from J.-A. Gusstafson and were >10 generations backcrossed to C57BL6/J. Lck-Cre, Csf1r-Cre, and FoxN1-Cre were bred to LXRαβfl/fl mice. Mice were maintained in a pathogen-free environment at the Massachusetts General Hospital animal facility, and all animal experiments were approved by the Subcommittee on Animal Research Care at Massachusetts General Hospital. Lck-Cre Abca1fl/flAbcg1fl/fl mice and Abcg1−/−, Abca1−/−, and Abca1−/−Abcg1−/− mice (Westerterp et al., 2017) were generated by L. Yvan-Charvet, M. Westerterp, and A. Tall and housed at Columbia University animal facility. Lck-Cre Abca1fl/flAbcg1fl/fl mice were >10 generations backcrossed to C57BL6/J. Abcg1−/−, Abca1−/−, and Abca1−/−Abcg1−/− mice were on a mixed C57BL6/J x DBA background with appropriate WT controls (mixed background). All experiments were performed on mice ranging from postnatal day 0 to 12 mo of age, using age- and sex-matched littermate controls.
Cell culture
For BM-derived macrophages, BM cells were incubated in RPMI 1640 containing macrophage-CSF (10 ng/ml) and 1× penicillin/streptomycin for 7 d. Adherent cells were collected for RNA analysis. The immortalized cTEC line MJC1 (Schuster et al., 2015) was maintained in MCDB-135 medium supplemented with 10% FBS and 1× penicillin/streptomycin.
Human samples
Human thymuses were obtained anonymously from three pediatric patients (5 mo old, 2 yr old, and 13 yr old) undergoing a median sternotomy because of elective cardiac or pulmonary surgery in the Amsterdam Universitair Medische Centra (UMC; Academic Medical Center), Amsterdam, Netherlands, following procedures for use of discarded tissue approved by the institutional review board of the Amsterdam UMC. Removal of the thymus is required to gain access to the mediastinum, and thus informed consent was not required for anonymously collected discarded tissue. No tissue was removed for research purposes only. Part of the thymus was directly snap frozen or in Tissue-Tek O.C.T. (optimal cutting temperature) compound and stored at −80°C until further use.
Method details
In vivo interventions
For BM chimeras, mice received a lethal radiation dose of 10 Gy followed by injection of purified BM cells (>5 × 106 cells) intravenously 8–24 h after irradiation. Mice were allowed at least 6 wk to recover before experiments were conducted.
Intrathymic injections were performed on anesthetized mice (95% oxygen mixed with 5% isoflurane for induction, then 1–2% isoflurane on maintenance). Mice were intubated and ventilated with a MiniVent Model 485 (Harvard Apparatus). After thoracotomy, purified cell populations were injected into the thymus with a 10-µl Hamilton syringe (5 µl per lobe). The thoracic cavity and skin were closed with sutures before mice were extubated, and mice were allowed to recover from surgery.
For proliferation experiments, mice received BrdU (1 mg) intraperitoneally 2 h or 24 h before sacrifice. In other experiments, mice were implanted with an osmotic mini pump (Model 2004) subcutaneously to infuse BrdU (0.1 mg/d) over 28 d. In a subset of these animals, the mini pump was removed, and mice were sacrificed 14 d after mini pump removal.
For Scd inhibition in vivo, A939572 (MedChem Express) was dissolved in DMSO (Sigma) and reconstituted in olive oil (Sigma) at a ratio of 1:10. Mice were injected intraperitoneally (10 mg/kg/d) daily for 7 d before sacrifice.
For thymic recovery or efferocytosis experiments, mice received either LPS (50 µg; Sigma) intraperitoneally once or dexamethasone (0.5 mg in 10% ethanol and 90% PBS; Sigma) and were sacrificed at various time points after injection (1 d, 5 d, 10 d, 14 d, or 28 d).
EAE was induced and disease severity scored according to manufacturer’s instructions (EK-2110; Hooke Laboratories). Briefly, mice were subcutaneously injected with MOG35-55/Complete Freud’s Adjuvant emulsion. 2 h and 24 h later, mice received an intraperitoneal injection of pertussis toxin (80 ng). Mice were monitored daily for 28 d before sacrifice.
Tissue processing
Blood was collected postmortem via the cardiac left ventricle, and red blood cells were lysed twice using 1× red blood cell lysis buffer (Biolegend). Spleens were passed through a 40-µm nylon mesh (BD Falcon) and washed with 1× PBS and centrifuged; then red blood cells were lysed to yield a single-cell suspension.
For thymocytes, thymuses were passed through a 40-µm nylon mesh and washed with 1× PBS, centrifuged, and resuspended to yield a single-cell suspension. For TECs, small incisions were made into the thymus and fragments triturated in PBS to release thymocytes. Thymuses were then transferred to a digestion cocktail of 2 ml Liberase TH (0.4 mg/ml) and DNase I (0.08 mg/ml) in RPMI 1640, incubated at 37°C for 6 min, and gently triturated. Every second period of digestion, supernatants were collected into FACS buffer and replaced with fresh digestion cocktail. Single-cell suspensions were stained with anti-CD90.2 (Biolegend) for 10 min and then incubated on an anti-IgG–coated panning plate for 20 min to enrich for TECs before being collected, washed, and resuspended in FACS buffer.
For spinal cord, tissues were harvested and finely minced in an enzymatic digestion cocktail containing 450 U/ml collagenase I, 125 U/ml collagenase XI, 60 U/ml DNase I, and 60 U/ml hyaluronidase (all Sigma) for 30–40 min. Tissues were then filtered through a 40-µm nylon mesh, washed with 1× PBS, centrifuged, and separated using a Percoll density gradient (30% upper, 70% lower). The mononuclear cell layer was collected and washed in PBS and then resuspended in FACS buffer before antibody staining.
Flow cytometry
Single-cell suspensions were stained in at 4°C in FACS buffer (0.5% BSA and 2 mM EDTA in PBS) with an antibody cocktail at a concentration of 1:700 unless otherwise specified. For cocktails containing biotin-conjugated antibodies, secondary streptavidin-fluorophore–conjugated antibodies were stained at 1:700 at 4°C. Live cells were identified using either Live/Dead Zombie Aqua (Biolegend) at a concentration of 1:1,000 and stained in 4°C or 7-aminoactinomycin D (7-AAD) before flow cytometric analysis.
For intracellular staining of FoxP3, ki67, Bim, and Helios, cells were fixed and permeabilized using the FoxP3/Transcription Factor Staining Buffer Kit according to manufacturer’s instructions (eBioscience), with DAPI/7-AAD added before analysis for ki67 staining. For BrdU staining, cells were processed using the BrdU Flow Kit according to the manufacturer’s instructions (BD Biosciences). For cholesterol quantification, antibody cocktail–labeled fixed cells (Fixation buffer; BD Biosciences) were stained with filipin (1:1,000; Sigma) for 1 h at room temperature immediately before flow cytometric analysis. Lipid rafts were stained in antibody-labeled nonfixed cells using the Vybrant Alexa Fluor 488 Lipid Raft Labeling Kit (cholera toxin B staining) according to the manufacturer’s instructions (Thermo Fisher Scientific). For membrane fluidity staining, antibody-labeled nonfixed cells were stained using the Membrane Fluidity Kit according to the manufacturer’s instructions (Abcam). Active Caspase-3/7 was measured according to the manufacturer’s instructions (Thermo Fisher Scientific). For analysis of TCRVβ chain analysis, thymocytes were stained using the Mouse Vβ TCR Screening Panel (BD Biosciences).
Data were acquired on an LSRII (BD Biosciences) and analyzed with FlowJo software.
Cell sorting
TECs and thymocytes underwent FACS using a FACSAria II cell sorter (BD Biosciences) with 7-AAD used for cell viability. For bulk TEC sorting, samples were stained with a primary antibody cocktail containing CD45, EpCAM, biotin-UEA-1, and Ly-51. Samples were then stained with a secondary antibody cocktail containing streptavidin-FITC. For thymocyte sorting, samples were stained with an antibody cocktail containing CD45, CD4, CD8, and CD5.
To isolate fetal thymic cells for intrathymic injection, thymuses from GFP mice were first digested in a Liberase/DNase I cocktail and stained with anti-CD45 microbeads (Miltenyi Biotec). Cell suspensions were passed through an LD column according to the manufacturer’s instructions (Miltenyi Biotec). Both the flow through (CD45−) and bound (CD45+) fractions were collected separately and verified to be >90% enriched by flow cytometry. Cells were then centrifuged and resuspended in sterile PBS before intrathymic injection.
For H-Y tetramer staining, single-cell suspensions of spleens from mice were first stained with PE anti–H-Y Tetramer (WMHHNMDLI:Db, 1:10; MBLI) for 1 h at 4°C, and then an antibody cocktail containing CD45, CD3, CD90.2, CD4, and CD8 was added in the last 15 min of staining. Samples were then stained with anti-PE microbeads (Miltenyi Biotec), washed, and passed through an LS column (Miltenyi Biotec). The bound fraction was collected and restained with the secondary antibody cocktail (CD45, CD3, CD90.2, CD4, and CD8) and Live/Dead Zombie Aqua viability dye before flow cytometric analysis.
Immunohistochemistry and histology
Thymuses were harvested, embedded in O.C.T. compound (Sakura Finetek), and snap frozen in 2-methylbutane (Sigma) chilled with dry ice. The tissues were sectioned (7 µm) and stored at −80°C until immunohistochemical staining. To detect lipid composition in thymuses, Oil Red O staining (Millipore-Sigma) with hematoxylin (Sigma) counterstaining was performed. For immunofluorescent double-staining, the tissue sections were blocked with 4% normal goat serum (Vector Laboratories) in PBS for 1 h at room temperature and then stained with rat Cytokeratin 8 antibody (Millipore) and rabbit Keratin 5 polyclonal antibody (BioLegend) overnight at 4°C. The next day, anti-rat IgG Alexa Fluor 488 and anti-rabbit IgG Alexa Fluor 594 were used as secondary antibodies to detect Cytokeratin 8 and Keratin 5 (Thermo Fisher Scientific), respectively. The nuclei were counterstained with DAPI (Thermo Fisher Scientific), and all the slides were scanned with the digital slide scanner NanoZoomer 2.0RS (Hamamatsu).
Real-time quantitative PCR (qPCR)
Total RNA from mouse and human thymuses was extracted using the RNEasy Mini kit (Qiagen) or E.Z.N.A. Total RNA Kit I (Omega Bio-Tek) according the manufacturer’s instructions. RNA from FACS-purified cell populations were extracted using the RNEasy Micro Kit (Qiagen) or E.Z.N.A. MicroElute Total RNA Kit (Omega Bio-Tek) according the manufacturer’s instructions. First-strand cDNA was synthesized using the High-Capacity RNA-to-cDNA kit (Applied Biosystems) according the manufacturer’s instructions. Taqman gene expression assays were used to quantify target genes (Thermo Fisher Scientific). Relative changes were normalized to GAPDH mRNA using the 2-ΔΔCt method.
Bulk RNA sequencing
cTECs and mTECs were sorted by FACS directly into lysis buffer, and RNA was extracted using the Nucleospin RNA XS kit according to the manufacturer’s protocol (Takara Bio). The RNA sequencing libraries were constructed using the SMARTer Low Input RNA kit v.4 (Takara Bio). Sequencing was performed on an Illumina HiSeq 2500 instrument, resulting in ∼30 million of 50 bp reads per sample. Sequencing reads were mapped in a splice-aware fashion to the Ensembl annotation of the mouse GRCm37/mm9 transcriptome using STAR (Dobin et al., 2013). Read counts over transcripts were calculated using HTSeq (Anders et al., 2015), followed by differential expression analysis using EdgeR (Robinson et al., 2010). Genes were classified as differentially expressed based on the cutoffs of twofold change in expression value and false discovery rates (FDRs) <0.05.
Lipidomics
cTECs and mTECs were sorted by FACS into FACS buffer. Pooled samples were centrifuged, and the cell pellet was snap frozen. Lipid analysis was performed at the University of California, San Diego Lipidomics Core (Quehenberger et al., 2010). Briefly, after spiking with internal standards, lipids were extracted by the Bligh-Dyer method, dried, and reconstituted in a buffer containing 59/40/1 (isopropyl alcohol, hexane, dH2O) with 10 mM NH4Oac. 10 µl of sample was injected and run on normal-phase ultra-high performance liquid chromatography (ACQUITY UPLC System; Waters) with data analyzed on a mass spectrometer (Sciex 6500; Qtrap).
Treatment of MJC1 cells
For siRNA treatment, 7.5 × 104 MJC1 cells per well were treated with 20 pmol siRNA for 48 h, with BrdU (10 µM) added to the medium in the final 4 h of treatment. For A939572 treatment, MJC1 cells were treated with A939572 (5 µM) or vehicle (DMSO) for 48 h, with BrdU (10 µM) added to the medium in the final 4 h of treatment. To quantify cell death, the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used according to manufacturer’s instructions (Abcam). Briefly, 104 cells were seeded into 96-well plates, treated with 20 pmol siRNA for 48 h, and then assayed.
Analysis of calcium flux
After 10 million thymocytes were labeled with cell surface antibodies against CD45, CD4, CD8, CD5, and TCRβ, cells were washed and resuspended in complete T cell media (RPMI 1640 + L-Glutamine, 10% FBS, 1× penicillin/streptomycin, 1× β-mercaptoethanol, and 25 mM Hepes) containing Indo-1 (2 µM) and incubated at 37°C for 30 min. Cells were washed and resuspended in complete T cell media and left to rest at room temperature for at least 30 min before analysis on a FACSAria II. For data acquisition, cells were run at 37°C, and baseline measurements were taken for 30 s. Anti-CD3 (3 µg/ml; Biolegend) was added, and measurements were taken for 120 s. Anti-hamster Ig (8 µg/ml; The Jackson Laboratory) was then added to cross-link anti-CD3, and data were acquired for 240 s. Finally, PMA (50 ng/ml; Sigma) and ionomycin (500 ng/ml; Sigma) were added, and data were acquired for another 120 s.
Thymocyte stimulation
After thymocytes were harvested, cells were allowed to rest for 2 h in complete T cell media at 37°C. For all conditions, cells were stimulated at 37°C with anti-CD3 (10 µg/ml) and anti-CD28 (2 µg/ml). For analysis of phosphorylated CD3ζ, JNK, and ERK1/2 (all BD Biosciences), cells were stimulated for 5 min, immediately fixed (1.5% paraformaldehyde) for 15 min at room temperature, and then fixed again with ice-cold methanol for 30 min at 4°C before staining for flow cytometry. For Bim induction, cells were stimulated for 4 h at 37°C, stained for cell surface markers and intracellular Bim (BD Biosciences), and analyzed by flow cytometry. For activation-induced cell death, cells were incubated for 24 h with or without anti-CD3/CD28.
Cyclodextrin treatment of cells ex vivo
Two to five million thymocytes were isolated and resuspended in complete T cell media containing either vehicle (dH2O) or MβCD (5 mM; Sigma) and incubated for 30 min at 37°C before further analysis.
FasL-induced apoptosis
Two to five million thymocytes were resuspended in complete T cell media containing recombinant FasL (2.5 µg/ml; R&D Systems) for 15 min at 37°C. After washing, cells were subsequently resuspended in complete T cell media containing HA Tag Antibody (2.5 µg/ml; R&D Systems) for 4 h at 37°C. Samples were then washed and stained with cell surface antibodies against TCRβ, CD24, CD45, CD4, CD5, and CD8, and active Caspase-3/7 was measured afterward.
Th17 cell polarization
Splenocytes were harvested, stained with anti-CD4 microbeads (Miltenyi Biotec), and positively sorted on an LS column (Miltenyi Biotec). Enriched CD4+ T cells were then resuspended in complete T cell media containing IL-6 (10 ng/ml), TGF-β (2 ng/ml), anti-IL4 (10 µg/ml), anti–IFN-γ (10 µg/ml), and anti-CD28 (2 µg/ml) and equally distributed into a 24-well plate 2–5 × 105 cells per well precoated with anti-CD3 (5 µg/ml). Cells were incubated at 37°C for 4 d, harvested, stimulated with PMA (50 ng/ml), ionomycin (500 ng/ml), and GolgiPlug (BD Biosciences) for 4 h, and then stained for cell surface TCRβ, CD45, and CD4 and intracellular IL-17A before flow cytometric analysis.
MOG stimulation of splenocytes
Two to five million splenocytes from EAE-treated animals were placed in a 48-well plate containing 20 µg/ml MOG35–55 peptide (Hooke Laboratories) in complete T cell medium for 3 d. After 3 d, the supernatant was taken, and IL-17 was measured using the Mouse IL-17 Quantikine Kit (R&D Systems) according to the manufacturer’s instructions.
Quantification and statistical analysis
No statistical methods were used to predetermine sample size. Data are presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism. As denoted, statistical significance of differences was determined by unpaired two-tailed Student’s t test (two groups), one-way ANOVA with Bonferroni’s post hoc analysis (more than two groups), or two-way ANOVA with Bonferroni’s post hoc analysis (analysis between two parameters). Where applicable, repeated measures or paired tests are defined. P values <0.05 were considered statistically significant. Asterisks denote statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001. Unless denoted, statistical significance denotes differences compared with the control animals or time or treatment control. The type of statistical tests performed and quantification of number (n) are indicated in the figure legends. For MJC1 cell culture, each data point corresponds to cells split from separate passages at different times. For RNA sequencing, DEGs are defined as a minimum of a twofold change with an FDR >0.01.
Data and code availability
Raw RNA sequencing data are available from Gene Expression Omnibus, accession no. GSE144342.
Online supplemental material
Fig. S1 shows human thymus LXRs and Abca1/Abcg1 gene expression and various readouts (Oil Red O histology, qPCR gene expression, flow cytometry) from WT and LXRαβ−/− mice. Fig. S2 shows flow cytometry data from mixed BM chimeras of WT/LXRαβ−/− mice; thymus size, Oil Red O histology, and leukocyte quantification from cell-specific LXR knockout mice; dexamethasone treatment of Csf1r-LXRαβ−/− mice; and control Lck-Cre and FoxN1-Cre data. Fig. S3 shows TEC data from LXR expression, engraftment, efferocytosis, glucocorticoid synthesis gene expression, cholesterol and lipidomics, siRNA knockdown and apoptosis, and extrathymic effects of Scd inhibition. Fig. S4 shows that in T cell LXR knockout mice, negative selection and apoptosis are enhanced but not proliferation, signaling, or T reg cell numbers; it also shows reduced Abca1/Abcg1 expression, cholesterol levels, Fas expression, and effects of MβCD on Fas and Bim expression and readouts from T cell Abca1/Abcg1 knockout mice. Fig. S5 shows readouts from WT, T cell LXR-deficient, and TEC LXR-deficient mice treated with LPS or irradiated and reconstituted with BM and data and gating strategy from EAE data using cell-specific LXR knockout animals.
Acknowledgments
The authors thank the Robert A. Welch Foundation. We thank Dr. Thomas Serwold at the Joslin Diabetes Center, Boston, MA, for providing MJC1 cells, the Harvard Stem Cell Institute-Center for Regnerative Medicine (HSCI-CRM) Flow Cytometry Core Facility at Massachusetts General Hospital (MGH), and the University of California, San Diego Lipidomics core for performing lipidomics analysis.
This work was funded by the National Institutes of Health (grants R35 HL135752, P01 HL131478, and P01 HL142494 to F.K. Swirski); the American Heart Association Established Investigator Award (grant 17EIA33410439 to F.K. Swirski); and the Patricia and Scott Eston MGH Research Scholar (to F.K. Swirski). C.T. Chan was supported by an American Heart Association Postdoctoral Fellowship (AHA 19POST34380057). N.K. Harder was supported by Boehringer Ingelheim Fonds.
Author contributions: C.T. Chan, A.M. Fenn, N.K. Harder, J.E. Mindur, C.S. McAlpine, J. Patel, C. Valet, S. Rattik, Y. Iwamoto, S. He, A. Anzai, F. Kahles, W.C. Poller, H. Janssen, and L.P. Wong conducted experiments and collected and analyzed data. C. Fernandez-Hernando, D.R. Koolbergen, A.M. van der Laan, L. Yvan-Charvet, R.I. Sadreyev, M. Nahrendorf, M. Westerterp, A.R. Tall, and J-A. Gustafsson provided reagents and discussed results and strategy. C.T. Chan and F.K. Swirski designed experiments, interpreted data, and wrote the manuscript. F.K. Swirski supervised, directed, and managed the study.
References
- A-Gonzalez N., Bensinger S.J., Hong C., Beceiro S., Bradley M.N., Zelcer N., Deniz J., Ramirez C., Díaz M., Gallardo G., et al. . 2009. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 31:245–258. 10.1016/j.immuni.2009.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramson J., and Anderson G.. 2017. Thymic Epithelial Cells. Annu. Rev. Immunol. 35:85–118. 10.1146/annurev-immunol-051116-052320 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw D., and Palmer D.B.. 2011. The origin and implication of thymic involution. Aging Dis. 2:437–443. [PMC free article] [PubMed] [Google Scholar]
- Bensinger S.J., Bradley M.N., Joseph S.B., Zelcer N., Janssen E.M., Hausner M.A., Shih R., Parks J.S., Edwards P.A., Jamieson B.D., et al. . 2008. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 134:97–111. 10.1016/j.cell.2008.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P., Purton J.F., Godfrey D.I., Zhang L.C., Coultas L., Puthalakath H., Pellegrini M., Cory S., Adams J.M., and Strasser A.. 2002. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 415:922–926. 10.1038/415922a [DOI] [PubMed] [Google Scholar]
- Chaudhry M.S., Velardi E., Dudakov J.A., and van den Brink M.R.. 2016. Thymus: the next (re)generation. Immunol. Rev. 271:56–71. 10.1111/imr.12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomans de Brachène A., Dif N., de Rocca Serra A., Bonnineau C., Velghe A.I., Larondelle Y., Tyteca D., and Demoulin J.B.. 2017. PDGF-induced fibroblast growth requires monounsaturated fatty acid production by stearoyl-CoA desaturase. FEBS Open Bio. 7:414–423. 10.1002/2211-5463.12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G., Qin X., Wu L., Zhang Y., Sheng X., Yu Q., Sheng H., Xi B., Zhang J.Z., and Zang Y.Q.. 2011. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J. Clin. Invest. 121:658–670. 10.1172/JCI42974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley S.R., Hu D.Y., and Goodnow C.C.. 2013. Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-κB. J. Exp. Med. 210:269–285. 10.1084/jem.20121458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakov J.A., Hanash A.M., Jenq R.R., Young L.F., Ghosh A., Singer N.V., West M.L., Smith O.M., Holland A.M., Tsai J.J., et al. . 2012. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 336:91–95. 10.1126/science.1218004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I.L., Chen K.G., Herzmark P., and Robey E.A.. 2013. Elimination of self-reactive T cells in the thymus: a timeline for negative selection. PLoS Biol. 11 e1001566 10.1371/journal.pbio.1001566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott M.R., and Ravichandran K.S.. 2016. The Dynamics of Apoptotic Cell Clearance. Dev. Cell. 38:147–160. 10.1016/j.devcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler M.B. 2018. The challenges and promise of targeting the Liver X Receptors for treatment of inflammatory disease. Pharmacol. Ther. 181:1–12. 10.1016/j.pharmthera.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D.H.D., Seach N., Ueno T., Milton M.K., Liston A., Lew A.M., Goodnow C.C., and Boyd R.L.. 2006. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 108:3777–3785. 10.1182/blood-2006-02-004531 [DOI] [PubMed] [Google Scholar]
- Hong C., and Tontonoz P.. 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug Discov. 13:433–444. 10.1038/nrd4280 [DOI] [PubMed] [Google Scholar]
- Igal R.A. 2011. Roles of StearoylCoA Desaturase-1 in the Regulation of Cancer Cell Growth, Survival and Tumorigenesis. Cancers (Basel). 3:2462–2477. 10.3390/cancers3022462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabouridis P.S., Janzen J., Magee A.L., and Ley S.C.. 2000. Cholesterol depletion disrupts lipid rafts and modulates the activity of multiple signaling pathways in T lymphocytes. Eur. J. Immunol. 30:954–963. [DOI] [PubMed] [Google Scholar]
- Legembre P., Daburon S., Moreau P., Moreau J.F., and Taupin J.L.. 2006. Modulation of Fas-mediated apoptosis by lipid rafts in T lymphocytes. J. Immunol. 176:716–720. 10.4049/jimmunol.176.2.716 [DOI] [PubMed] [Google Scholar]
- Mahammad S., and Parmryd I.. 2015. Cholesterol depletion using methyl-β-cyclodextrin. Methods Mol. Biol. 1232:91–102. 10.1007/978-1-4939-1752-5_8 [DOI] [PubMed] [Google Scholar]
- Mittelstadt P.R., Taves M.D., and Ashwell J.D.. 2018. Cutting edge: denovo glucocorticoid synthesis by thymic epithelial cells regulates antigen-specific thymocyte selection. J. Immunol. 200:1988–1994. 10.4049/jimmunol.1701328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji M., Jin Z.X., Yamaoka S., Amakawa R., Fukuhara S., Sato S.B., Kobayashi T., Domae N., Mimori T., Bloom E.T., et al. . 2005. Role of membrane sphingomyelin and ceramide in platform formation for Fas-mediated apoptosis. J. Exp. Med. 202:249–259. 10.1084/jem.20041685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S., Procopio A., Lazzarini R., Rippo M.R., Testa R., Marra M., Tamagnone L., and Catalano A.. 2008. Semaphorin3A signaling controls Fas (CD95)-mediated apoptosis by promoting Fas translocation into lipid rafts. Blood. 111:2290–2299. 10.1182/blood-2007-06-096529 [DOI] [PubMed] [Google Scholar]
- Nader N., Ng S.S., Wang Y., Abel B.S., Chrousos G.P., and Kino T.. 2012. Liver x receptors regulate the transcriptional activity of the glucocorticoid receptor: implications for the carbohydrate metabolism. PLoS One. 7 e26751 10.1371/journal.pone.0026751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S., Albergante L., Blackburn C.C., and Newman T.J.. 2018. Thymic involution and rising disease incidence with age. Proc. Natl. Acad. Sci. USA. 115:1883–1888. 10.1073/pnas.1714478115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton C.M., and Ntambi J.M.. 2009. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Endocrinol. Metab. 297:E28–E37. 10.1152/ajpendo.90897.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O., Armando A.M., Brown A.H., Milne S.B., Myers D.S., Merrill A.H., Bandyopadhyay S., Jones K.N., Kelly S., Shaner R.L., et al. . 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51:3299–3305. 10.1194/jlr.M009449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet E.M., Savio D.A., Halpern A.R., Chen L., Schuster G.U., Gustafsson J.A., Basso M.D., and Nambi P.. 2006. Liver X receptor (LXR)-beta regulation in LXRalpha-deficient mice: implications for therapeutic targeting. Mol. Pharmacol. 70:1340–1349. 10.1124/mol.106.022608 [DOI] [PubMed] [Google Scholar]
- Remmerie A., and Scott C.L.. 2018. Macrophages and lipid metabolism. Cell. Immunol. 330:27–42. 10.1016/j.cellimm.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G.A., Waddington K.E., Pineda-Torra I., and Jury E.C.. 2017. Transcriptional Regulation of T-Cell Lipid Metabolism: Implications for Plasma Membrane Lipid Rafts and T-Cell Function. Front. Immunol. 8:1636 10.3389/fimmu.2017.01636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka M., Stritesky G.L., Reynolds J., Abourashchi N., Lythe G., Molina-París C., and Hogquist K.A.. 2014. From pre-DP, post-DP, SP4, and SP8 Thymocyte Cell Counts to a Dynamical Model of Cortical and Medullary Selection. Front. Immunol. 5:19 10.3389/fimmu.2014.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C., Gerold K.D., Schober K., Probst L., Boerner K., Kim M.J., Ruckdeschel A., Serwold T., and Kissler S.. 2015. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity. 42:942–952. 10.1016/j.immuni.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann N.J., and Glass C.K.. 2013. Sterols and oxysterols in immune cell function. Nat. Immunol. 14:893–900. 10.1038/ni.2681 [DOI] [PubMed] [Google Scholar]
- van den Brink M.R., Alpdogan O., and Boyd R.L.. 2004. Strategies to enhance T-cell reconstitution in immunocompromised patients. Nat. Rev. Immunol. 4:856–867. 10.1038/nri1484 [DOI] [PubMed] [Google Scholar]
- Tacke R., Hilgendorf I., Garner H., Waterborg C., Park K., Nowyhed H., Hanna R.N., Wu R., Swirski F.K., Geissmann F., et al. . 2015. The transcription factor NR4A1 is essential for the development of a novel macrophage subset in the thymus. Sci Rep. 5:10055 10.1038/srep10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Beck-García K., Zorzin C., Schamel W.W., and Davis M.M.. 2016. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat. Immunol. 17:844–850. 10.1038/ni.3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Subramanian M., Yurdagul A. Jr., Barbosa-Lorenzi V.C., Cai B., de Juan-Sanz J., Ryan T.A., Nomura M., Maxfield F.R., and Tabas I.. 2017. Mitochondrial Fission Promotes the Continued Clearance of Apoptotic Cells by Macrophages. Cell. 171:331–345.E22. 10.1016/j.cell.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerterp M., Gautier E.L., Ganda A., Molusky M.M., Wang W., Fotakis P., Wang N., Randolph G.J., D’Agati V.D., Yvan-Charvet L., et al. . 2017. Cholesterol Accumulation in Dendritic Cells Links the Inflammasome to Acquired Immunity. Cell Metab. 25:1294–1304.E6. 10.1016/j.cmet.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wagoner G., Douglas J.C., and Drew P.D.. 2009. Liver X receptor agonist regulation of Th17 lymphocyte function in autoimmunity. J. Leukoc. Biol. 86:401–409. 10.1189/jlb.1008600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang S., Li P., Lu J., Xuan Q., and Ge Q.. 2013. Maturation and emigration of single-positive thymocytes. Clin. Dev. Immunol. 2013 282870 10.1155/2013/282870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J.K., Wahjudi P.N., Vega J., Lim S., Martin A., Patterson M.E., Cohen J.N., Mao C.S., and Lee W.N.. 2013. Stearoyl-CoA desaturase enzyme 1 inhibition reduces glucose utilization for de novo fatty acid synthesis and cell proliferation in 3T3-L1 adipocytes. Metabolomics. 9:809–816. 10.1007/s11306-013-0511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm Y.H., Kanneganti T.D., Vandanmagsar B., Zhu X., Ravussin A., Adijiang A., Owen J.S., Thomas M.J., Francis J., Parks J.S., et al. . 2012. The Nlrp3 inflammasome promotes age-related thymic demise and immunosenescence. Cell Rep. 1:56–68. 10.1016/j.celrep.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan-Charvet L., Pagler T., Gautier E.L., Avagyan S., Siry R.L., Han S., Welch C.L., Wang N., Randolph G.J., Snoeck H.W., et al. . 2010a ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 328:1689–1693. 10.1126/science.1189731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan-Charvet L., Wang N., and Tall A.R.. 2010b Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 30:139–143. 10.1161/ATVBAHA.108.179283 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw RNA sequencing data are available from Gene Expression Omnibus, accession no. GSE144342.