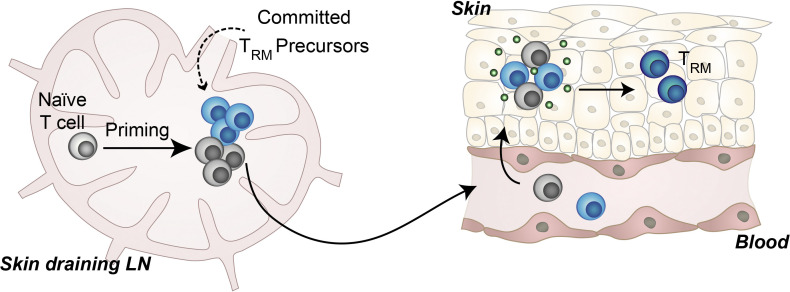
Following local inflammation, CD8+ T cells develop into tissue-resident memory T cells (TRM). Through combined lineage tracing and single-cell transcriptome analysis, the authors show that the circulating T cell pool harbors TRM precursors that commit to the TRM fate before tissue entry.
Abstract
An increasing body of evidence emphasizes the role of tissue-resident memory T cells (TRM) in the defense against recurring pathogens and malignant neoplasms. However, little is known with regard to the origin of these cells and their kinship to other CD8+ T cell compartments. To address this issue, we followed the antigen-specific progeny of individual naive CD8+ T cells to the T effector (TEFF), T circulating memory (TCIRCM), and TRM pools by lineage-tracing and single-cell transcriptome analysis. We demonstrate that a subset of T cell clones possesses a heightened capacity to form TRM, and that enriched expression of TRM–fate-associated genes is already apparent in the circulating TEFF offspring of such clones. In addition, we demonstrate that the capacity to generate TRM is permanently imprinted at the clonal level, before skin entry. Collectively, these data provide compelling evidence for early stage TRM fate decisions and the existence of committed TRM precursor cells in the circulatory TEFF compartment.
Graphical Abstract
Introduction
Upon local infection, antigen-specific naive CD8+ T cells undergo rapid clonal expansion to generate a large pool of effector T cells (TEFF) that are present in the circulation and at the affected peripheral site. Following pathogen clearance, this effector cell population contracts to form a small pool of memory T cells in the blood and secondary lymphoid organs (circulating memory T cells [TCIRCM]), and also at the site of pathogen entry (Steinert et al., 2015). The latter population, commonly refered to as tissue-resident memory T cells (TRM), has been shown to be important for local control of reinfection in tissues such as skin, intestine, and lung (Gebhardt et al., 2009; Masopust et al., 2010; Ariotti et al., 2012; Turner et al., 2014; Mueller and Mackay, 2016) and can be distinguished from its circulating counterpart by increased expression of markers such as CD103 and CD69 (Mackay et al., 2013, Mueller and Mackay, 2016).
A number of studies have provided evidence that certain subsets of TEFF possess an enhanced capacity to differentiate into TRM. Specifically, TEFF located in inflamed tissues that express CD69, CD103, or CD127, but lack killer cell lectin-like receptor G1 (KLRG1) expression, are considered to have a superior capacity to give rise to TRM (Sheridan et al., 2014; Mackay et al., 2013; Herndler-Brandstetter et al., 2018). Furthermore, those TEFF in peripheral tissues that are prone to differentiate into TRM display a unique transcriptome that differs from the transcriptional profile associated with TCIRCM formation (Milner et al., 2017). While these studies have established that the propensity to generate TRM is unequally distributed over the effector pool, prior work has also demonstrated that TRM and TCIRCM share a common clonal origin (Gaide et al., 2015). Thus, differences in TRM-forming capacity do not appear imprinted in naive CD8+ T cells, but a diversification in TRM generation potential is evident in the TEFF pool. A recent study has suggested that naive T cells can be poised for a TRM fate in steady-state conditions, through TGFβ signaling induced by migratory dendritic cells (Mani et al., 2019). However, it has not been elucidated whether such poising-signals result in variations in TRM generating potential between individual naive clones. Furthermore, at present, it has not been established at which point during an antigen-specific T cell response the progeny of naive T cells commits to the TRM lineage.
To address these issues, we tracked the offspring of individual naive CD8+ T cells responding to local skin vaccination by means of genetic barcoding. Using this lineage-tracing tool, we provide evidence that, whereas independent T cell clones possess an equal capacity to enter inflamed tissue during the effector phase, a subset of T cell clones possesses a heightened capacity to subsequently form tissue-resident T cell memory. Moreover, by combining lineage tracing with single-cell RNA sequencing (scRNA-seq), we report the existence of a circulatory TEFF subset that bears a strong transcriptional resemblance to TRM. Importantly, individual T cell clones contribute differentially to this population, and production of this TRM-poised TEFF subset by individual T cell clones is associated with their capacity to form TRM. Further support for the existence of a circulating TRM precursor comes from the observation that TRM-forming propensity is clonally acquired before tissue entry and is fixed upon secondary antigen encounter. Jointly, these experiments provide definitive evidence for the existence of a circulating TRM precursor population that commits to the TRM-fate before tissue entrance.
Results
Individual T cell clones contribute proportionally to the systemic and skin TEFF response
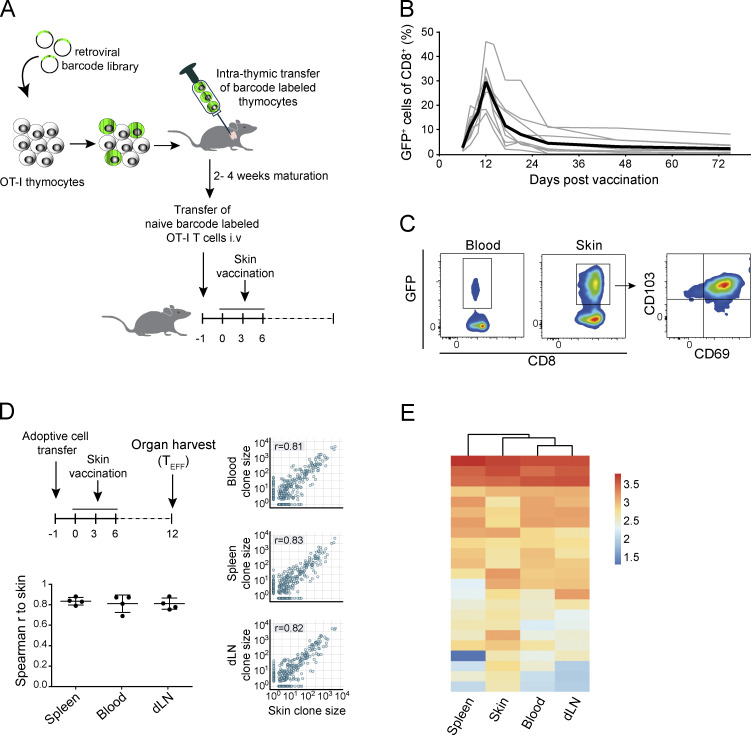
To evaluate how individual naive T cells contribute to the TRM lineage, and how the TRM population is developmentally related to the systemic CD8+ T cell subsets, we set out to track the progeny of individual naive CD8+ T cells within the TEFF, TCIRCM, and TRM compartment in vivo by genetic barcoding. To this purpose, we first generated a high-diversity retroviral barcode library (BC2.0) that comprises ∼200,000 unique cellular identifiers, thereby enabling the tracking of many individual cells in parallel. Using this BC2.0 genetic labeling system, we subsequently generated naive CD8+ T cells that each carry a unique DNA barcode (Gerlach et al., 2010, 2013). Specifically, thymocytes were transduced with the BC2.0 library and injected intrathymically into recipient mice to allow maturation into barcode-labeled naive T cells. This experimental approach allows for the genetic labeling of naturally cycling T cell precursors, thereby avoiding a requirement for in vitro activation of naive T cells. As shown previously, barcode-labeled T cells that are generated in this manner behave identically to unmanipulated naive OT-I T cells, in terms of both T cell response kinetics and effector differentiation potential (Gerlach et al., 2010). To be able to examine T cell fate and T cell development into the TRM lineage without TCR affinity as a confounder (Zehn et al., 2009), thymocytes were obtained from OT-I TCR transgenic mice, in which all CD8+ T cells carry the OT-I TCR specific for the OVA257–264-H2-Kb complex (Fig. 1 A).
Figure 1.
Proportional contribution of individual T cell clones to the systemic and skin effector response. OT-I thymocytes were transduced with the barcode library and intrathymically transferred into recipient mice. After maturation, barcode-labeled GFP+ OT-I T cells were transferred into secondary recipients that were subsequently exposed to skin vaccination. (A) Schematic overview of experimental setup. (B) Barcode-labeled GFP+ OT-I T cell response to DNA vaccination, measured in blood (n = 11 mice, gray lines). Black line represents group average. (C) Representative flow cytometry plots showing the presence of GFP+ memory T cells within CD8+ cells in blood and skin >60 d after vaccination. (D and E) Spleen, skin, draining LNs, and whole blood were collected from vaccinated recipient mice 12 d after start of vaccination. (D) Analysis of the contribution of individual T cell clones to the spleen, blood, and draining LN effector stage T cell compartment, relative to the skin effector–stage T cell compartment. Spearman correlation r was calculated over clones detected in both samples. Left: Spearman correlations for individual mice (n = 4), mean with whiskers representing SD. Right: Dots represent individual clones; P values were <0.0005. (E) Clonal output in all examined tissues of the 5% of largest clones detected in skin tissue. Heat map depicts log10-transformed clone sizes (read counts). D and E are representative data of two independent experiments.
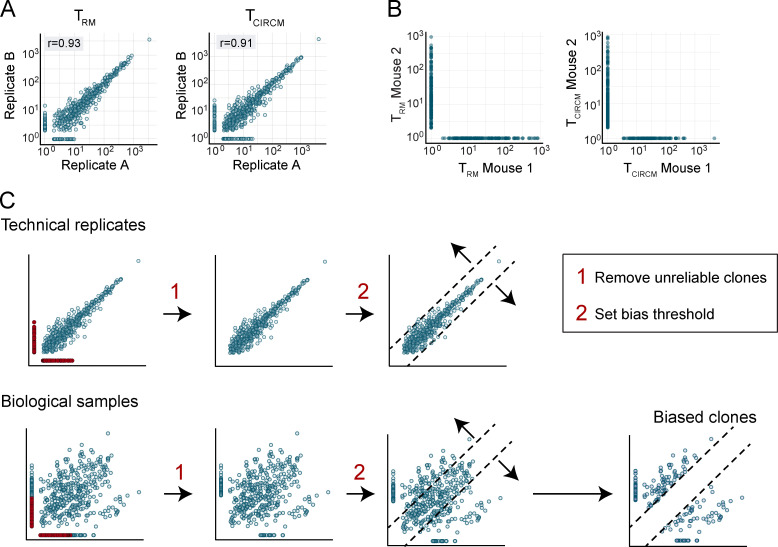
After in vivo development of barcode-labeled thymocytes into mature naive GFP+ OT-I T cell clones, cells were harvested, and physiologically relevant numbers (i.e., 500–1,000; Obar et al., 2008) of cells were transferred into wild-type recipient mice. Subsequently, a local immune response was induced by vaccination of hind leg skin of recipient mice with plasmid DNA encoding the OVA257–264 epitope (Fig. 1 A; Bins et al., 2005; Oosterhuis et al., 2012; Ahrends et al., 2016). Local vaccination induced clonal expansion and subsequent contraction of the barcode-labeled OT-I T cell pool (Fig. 1 B). At late time points (>60 d) after vaccination, GFP+ OT-I T cells remained detectable at low frequencies in both the circulation (Fig. 1 C, left) and at the site of skin vaccination. Consistent with prior work, the large majority of the barcode-labeled TRM harvested from the tissue site expressed the canonical tissue-residency markers CD69 and CD103 (Fig. 1 C, middle and right).
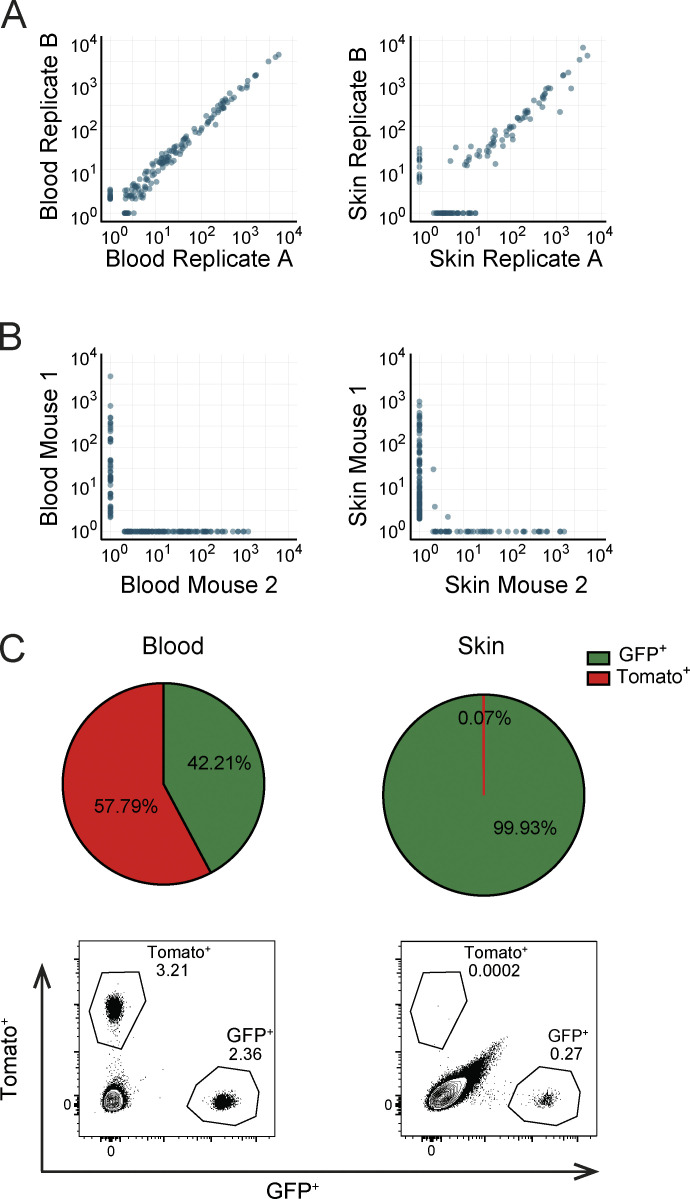
Having validated that skin vaccination induces clonal expansion of naive barcode-labeled T cells and their differentiation into TEFF, TCIRCM, and TRM, we aimed to assess whether individual naive T cells differ in their capacity to yield TEFF at distinct body sites. To this end, vaccinated recipient mice were sacrificed at the peak of the TEFF expansion phase (day 12); blood, spleen, draining lymph nodes, and affected skin tissue were collected; and clonal output was quantified by DNA barcode sequencing (Fig. 1 D, top). Barcode analysis of GFP+ OT-I T cells present in the blood compartment at the peak of the response showed that, similar to prior lineage-tracing studies involving Listeria monocytogenes–OVA257–264 infection (Buchholz et al., 2013; Gerlach et al., 2013), the capacity of individual naive T cells to expand in response to DNA vaccination was highly variable, with ∼7% of the clones producing ∼50% of the total TEFF pool. Comparison of clonal output in the sampled tissue sites showed that at the peak of the T cell response, the vast majority of clones contributed to the TEFF pool at all four examined locations (Fig. 1, D and E; controls in Fig. S1, A and B). Furthermore, the relative sizes of individual T cell clones at these different sites were highly correlated (r > 0.8), indicating that the progeny of different naive T cells possess a similar capacity to disseminate throughout the body during the TEFF phase (Fig. 1 D). As a control, the high clonal overlap between T cell compartments in skin and at other body sites was shown not to be explained by a potential contamination of skin samples with blood-borne T cells (Fig. S1 C). Thus, the ability to enter inflamed peripheral tissues is equally distributed over the progeny of responding naive T cell clones.
Figure S1.
Quality of barcode quantification and analysis of blood-borne T cell contamination in effector-phase skin samples. (A and B) Recipients of barcode-labeled T cells were vaccinated, and whole blood and organs were harvested 12 d after first vaccination. (A) Measured clone sizes detected in representative technical replicates of blood (left) and skin (right) samples. (B) Measured clone sizes detected in blood (left) and skin (right) of independent mice. Dots represent individual clones. (C) Analysis of the presence of blood-borne T cells in skin preparations. Recipients of GFP+ OT-I T cells were DNA vaccinated. 10 d after vaccination, mice received 1.5 × 106 Tomato+ OT-I T cells and were sacrificed 5 min later. Top: Pie charts depicting the relative percentage of GFP+ and Tomato+ cells in blood (left) and skin (right) preparations. Bottom: Representative flow cytometry plots. Cells are gated on live lymphocytes. Data are representative of four mice.
Clonal bias in TRM generation
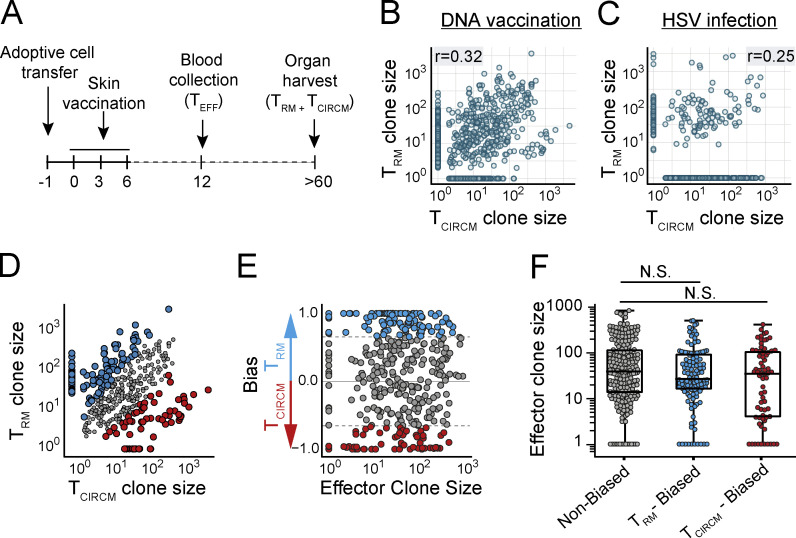
Having established that individual T cell clones display a similar capacity to disseminate to the skin and lymphoid compartments during the effector phase, we next evaluated whether this equal distribution of clones persisted into memory. To quantify the output of individual clones in the two memory compartments, recipient mice received a local skin vaccination, TEFF blood samples were drawn on day 12, and the skin TRM and TCIRCM populations from the same mice were isolated after memory formation (day >60; Fig. 2 A). In line with prior work (Gaide et al., 2015), comparison of clone sizes in the two memory pools revealed that the vast majority of naive T cells contributed to both the TCIRCM and TRM cell lineages (84.8%). Strikingly, however, the contribution of individual T cell clones to the TCIRCM or TRM pool was highly disparate (r = 0.32; Fig. 2 B, quality controls in Fig. S2, A and B). Importantly, this low degree of similarity was not due to suboptimal sampling, as shown by the high correlation (r > 0.9; Fig. S2 A) of technical replicates of either the skin TRM or the TCIRCM pool. Thus, although during the effector phase individual T cell clones contribute essentially equally to the T cell pool at different body sites, many clones preferentially contribute to either the TCIRCM or the TRM pool after contraction. This disparity in memory clone distribution is also present upon natural infection, as shown by DNA barcode analysis of the TCIRCM and TRM compartments after localized HSV-OVA257–264 infection (r = 0.25; Fig. 2 C). Specifically, after HSV-OVA257–264 infection, the average T cell clone preferentially contributed to either the TRM or the TCIRCM compartment by a factor of 11.34-fold (mean ratio of contributions to the two memory compartments, taking the lowest contribution as denominator and excluding nonshared clones). As a control, the average ratio between technical replicates was 2.19 (r = 0.86). By the same token, in response to DNA vaccination, T cell clones showed a preferential contribution toward either the circulating or skin-resident memory T cell compartment by a factor of 11.98 (factor of 1.66 between technical replicates).
Figure 2.
Clonal bias in TRM generation. (A) Representation of experimental timeline. Barcode-labeled TRM and TCIRCM were isolated from the skin and circulatory compartment (spleen, LN, and blood) of DNA-vaccinated mice (or HSV-OVA257–264–infected mice; C), and clonal output was quantified. (B) Comparison of clonal contribution to the skin TRM and TCIRCM compartment after DNA vaccination. (C) Comparison of clonal contribution to the skin TRM and TCIRCM compartment after HSV-OVA257–264 infection. (D) Clones responding to DNA vaccination were defined as TRM biased, TCIRCM biased, or nonbiased, based on their relative contribution to either memory compartment. Scatterplot similar to B highlighting TRM-biased (blue), TCIRCM-biased (red), and nonbiased (gray) T cell clones. Small clones for which clone size measurements were less reliable were excluded from analysis and are not depicted. (E and F) Comparison of effector stage burst size of nonbiased (gray), TRM-biased (blue), and TCIRCM-biased (red) T cell clones. In E, values on y axis depict (clone size TRM − clone size TCIRCM)/(clone size TRM + clone size TCIRCM) and represent the degree of preferential contribution to TRM or TCIRCM. Dashed lines indicate bias threshold of 4.8-fold. In F, median with whiskers representing minimum/maximum, Kruskal–Wallis test with Dunn’s multiple comparisons test; N.S., not significant. In B and C, Spearman correlation r was calculated over all clones that contributed to both samples, P < 0.0005 (B) and P = 0.01 (C). Dots represent individual clones. Data from four mice, representative of two individual experiments.
Figure S2.
Quality control and analyses of the barcode-labeled TRM and TCIRCM compartment. (A) Measured clone sizes detected in technical replicates of TRM (left) and TCIRCM (right) samples derived from the mice described in Fig. 2 B. Spearman correlation r was calculated over clones that were detected in both technical replicates: P < 0.0005 (left); P < 0.0005 (right). (B) Measured clone sizes detected in TRM (left) and TCIRCM (right) of independent mice described in Fig. 2 B. (C) Step-by-step description of the strategy used to filter biological data and define biased clones, as depicted in Fig. 2 D. First, unreliably detected clones (indicated in red) are removed. Second, a bias threshold (dashed lines) is set, such that 98% of the clones in technical replicates fall below this threshold. This threshold is subsequently applied to the biological data to identify clones with a bias in output that goes beyond the variation that occurs because of technical noise. For the data presented in Fig. 2 B, the threshold identified only clones contributing >4.8 times to one sample than to the other to be considered biased. Dots represent individual clones.
Next, we examined whether the bias in TRM and TCIRCM generation in response to DNA vaccination could be explained by differences in clonal TEFF expansion. First, to exclude clones that could show clonal bias because of random sampling variation, clones that were exclusively observed in one of the two memory T cell compartments and that represented <0.25% of that pool were removed (retaining 58.5% of barcodes and 97.2% of reads; before filtering, Fig. 2 B; after filtering, Fig. 2 D; filtering strategy, Fig. S2 C). Subsequently, to be able to identify biased clones, we defined a “bias threshold” based on comparison of technical replicates, a setting in which clonal bias can by definition not occur (Fig. S2 C). Application of the resulting threshold (fold difference of >4.8) to the lineage-tracing data revealed that close to 50% of T cell clones preferentially contributed to either memory T cell compartment, with 29.7% of clones being biased toward TRM formation and 16.9% toward TCIRCM formation (Fig. 2 D). Notably, analysis of effector phase burst sizes of TRM-biased, TCIRCM-biased, and nonbiased T cell clones showed that biased memory cell generation was also observed for TEFF-stage clones that had undergone massive or little expansion (Fig. 2, E and F). These results demonstrate that, independently of clonal burst size, a large fraction of T cell clones preferentially produces TRM or TCIRCM, indicating that TRM and TCIRCM are separated not only by location and phenotype but also by descent.
Nonstochastic formation of tissue-resident and systemic T cell memory
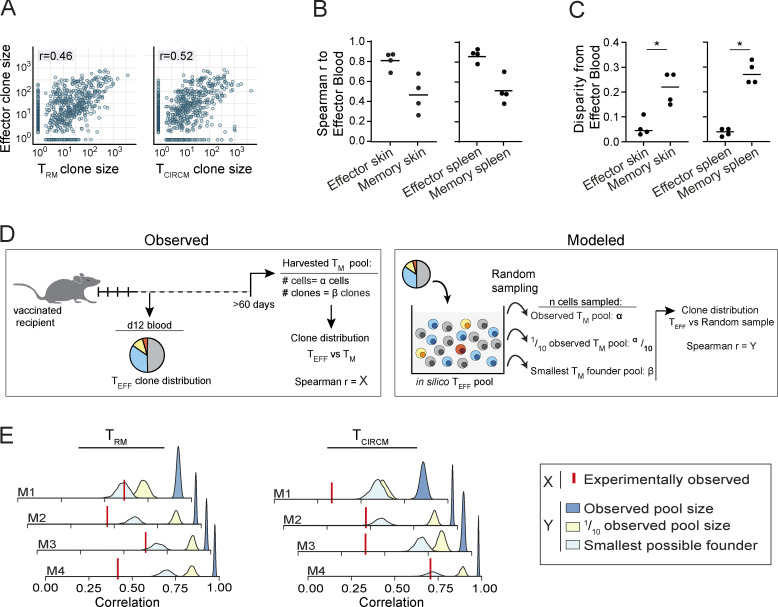
Next, we wanted to understand whether the clonal bias observed in memory (Fig. 2, B and D) was due to remodeling of either the circulatory or the skin-resident compartment during T cell contraction. As clonal hierarchy is highly similar at different body sites during the effector phase (Fig. 1, D and E), we reasoned that the TEFF pool in blood could be used as a “historical snapshot” of clonal distribution in all immune compartments before memory formation. Comparison of clone sizes of day-12 effector blood to the skin and spleen compartment in effector and memory phase demonstrated that both compartments in memory phase were substantially more disparate from TEFF blood than during effector phase (Fig. 3, A–C; and Fig. S3). Thus, during memory formation, both the skin-resident and the circulating T cell compartment undergo a substantial change in clonal hierarchy (Fig. 3, A–C; and Fig. S3), resulting in differential contributions of individual T cell clones to the two memory compartments (Fig. 2 D).
Figure 3.
Nonstochastic formation of tissue-resident and systemic T cell memory. (A) Contribution of T cell clones to the TRM (left) or TCIRCM (right) pool, relative to the effector stage blood compartment. Spearman correlation r was calculated over T cell clones that were detected in both samples; n = 4 mice. (B) Spearman correlations of clone sizes in skin (left) and spleen (right) samples collected during effector (n = 4 mice) and memory (n = 4 mice) phase to day-12 effector blood. (C) Clone size disparity of skin (left) and spleen (right) T cell pools in the effector and memory phase from the day-12 effector blood T cell pool. See Fig. S3 A for the definition of disparity. (D) Illustration depicting the strategy used to assess whether stochasticity can explain the observed clonal skewing during memory formation. Based on observed clone distribution in the TEFF pool, a virtual pool of TEFF cells is generated in silico, from which cells are sampled to form a randomly selected TRM or TCIRCM memory pool. The number of randomly sampled cells is equal to the number of observed cells in the biological memory (TM) pool (α), to 10% of the observed TM pool (α/10), or to the number of observed clones in the biological TM pool (β), which represents the smallest theoretically possible TM founder pool. The Spearman correlation coefficient between the randomly sampled cell pool and the experimentally observed TEFF pool is calculated (Y) and compared with the Spearman correlation coefficient between the experimentally observed TM pools and the experimentally observed TEFF pool (X). Only if Y approaches X, stochastic engraftment can explain the observed skewing in clonal output in the TM pool. (E) Stochastically formed TRM (left) and TCIRCM (right) pools were modeled 10,000 times in silico, as described in D, and the Spearman correlation between the modeled memory pools and the observed TEFF pool was calculated (Y). Graphs indicate individual mice (n = 4); histograms represent the distribution of Spearman r. Red vertical line indicates the correlation between the clonal distribution of the TEFF pool and the experimentally observed memory pool (X). Spearman correlations r were calculated over all clones detected in either the effector pool or the (modeled or experimental) memory pool. In A, dots represent output of individual clones. In B and C, dots represent individual mice. Spearman correlation r was calculated over clones that were detected in both samples: P < 0.0005 (A, left) and P < 0.0005 (A, right); Mann–Whitney U test, *, P < 0.05 (C). Representative data of two independent experiments.
Figure S3.
Remodeling of the skin-resident and circulating memory compartment. (A) Example plots depicting the strategy to determine the disparity between two cellular compartments, as applied in Figs. 3 C and 7 F. Disparity of compartments B and C to compartment A can be assessed by plotting the fraction of cumulative reads of clones in compartments B and C, which are ordered based on their size (largest to smallest) in compartment A (y axis), to the cumulative reads of the ordered clones in compartment A (x axis). Area between the compartment A reference curve and compartment B (left) and C (right) curves is calculated to generate a measure of disparity. (B) Left: Illustration of the subdivision of ordered effector-stage T cell clones (large to small) into four bins, with each bin containing 25% of all observed clones. Middle and right: Quantitative contribution of binned clones detected in effector blood to the TRM and TCIRCM compartment. Median with whiskers representing minimum/maximum; ***, P < 0.0005, Mann–Whitney U test. (C) Relative contribution of TEFF clones in bins 1–4 (highlighted in blue) to the TRM and TCIRCM compartments. In B and C, data are representative of two independent experiments; dots represent individual clones.
The observed divergence in clonal composition of T cell populations at the two sites could arise either through an intrinsic difference in cell fitness to survive in particular microenvironments or through the stochastic engraftment of cells at the individual sites. To test the latter hypothesis, we simulated the generation of TRM and TCIRCM pools that were derived from a founder population with a size that equaled either the experimentally observed T memory pool (indicated as α; Fig. 3 D), 10% of the observed T memory pool, or the smallest possible founder pool (i.e., the number of individual clones observed in the memory pool, indicated as β; Fig. 3 D). Subsequently, the correlation in clone sizes between the simulated T memory pools and the experimentally observed TEFF pool (indicated as Y; Fig. 3 D) were calculated and compared with the correlation between the experimentally observed T memory and TEFF pool (indicated as X; Fig. 3 D). Note that only when Y approaches X, stochastic engraftment of T cells can explain the observed clonal bias in memory phase. Interestingly, this analysis demonstrated that stochastic engraftment of a founder population with the size of the observed T memory pool (α) or one tenth of this size could not explain the observed skewing during T memory formation in any of the mice (Fig. 3 E). Furthermore, stochastic engraftment by the smallest possible founder pool was also insufficient to explain the skewing in the observed T cell memory pool in the majority of mice (Fig. 3 E). Collectively, these data indicate that the skewed composition of both the TRM and the TCIRCM pool is unlikely to be explained by stochastic survival or engraftment, thereby suggesting the existence of intrinsic differences between T cell clones in their capacity to form systemic and tissue-resident T cell memory.
The circulating TEFF pool harbors cells with a TRM-like transcriptional signature
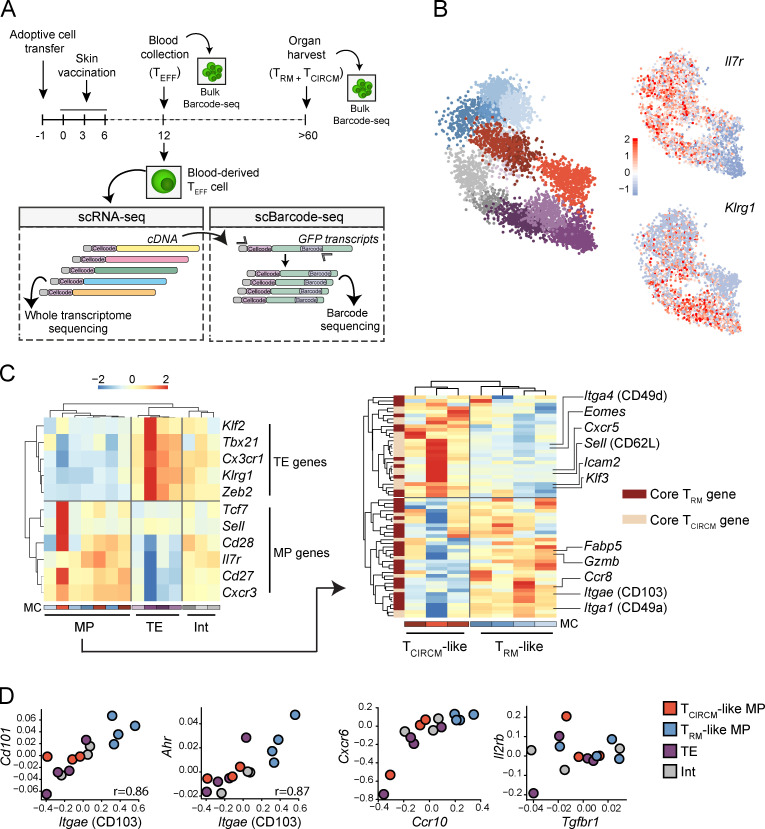
The pool of circulating TEFF is phenotypically and transcriptionally diverse and, next to the commonly recognized subsets of terminal effector (TE) cells and memory precursor (MP) cells, additional heterogeneity has been reported (Gerlach et al., 2016; Arsenio et al., 2014). To understand whether such heterogeneity could explain preferential TRM formation by individual T cell clones, we performed scRNA-seq on blood-derived barcode-labeled TEFF (day 12) and subsequently determined clonal output in the TRM and TCIRCM populations of the same mice at day >60. Importantly, as barcode sequences are contained within the 3′ untranslated region of GFP transcripts, scRNA-seq allowed for the parallel analysis of transcriptional state and clonal origin of individual vaccine-specific TEFF (see experimental setup in Fig. 4 A).
Figure 4.
scRNA-seq reveals a transcriptional TRM-like MP state in the circulating TEFF pool. Barcode-labeled TEFF were isolated from blood of recipient mice at day 12 after skin vaccination, and scRNA-seq was performed to map transcriptional profiles of circulating effector cells. In addition, barcode sequences were specifically amplified from single cell–derived cDNA and subsequently sequenced (single-cell barcode sequencing). Matching of cellcode sequences (sequences marking all transcripts derived from a single cell) in scRNA-seq and single-cell barcode sequencing datasets allows for the coupling of transcriptional profile and clone of origin of individual TEFF cells. (A) Schematic overview of the experimental procedure. (B) Left: 2D projection of 5,383 TEFF cells that, based on transcriptional profile, are grouped into 14 distinct MCs. MC colors indicate assigned TEFF state, as defined in C (red/brown, TCIRCM-like; blue, TRM-like MP; purple, TE; gray, Int). Right: 2D projections with superimposed expression of Il7r and Klrg1. Legend indicates gene expression Z-score. (C) Left: Hierarchical clustering of 14 transcriptionally distinct MCs based on the expression of TE- and MP-associated genes. Legend indicates log2 enrichment Z-score. Right: Hierarchical clustering, based on the expression of core TRM and TCIRCM genes, of the seven MCs that are classified as MP. (D) Gene expression comparison of genes associated with skin TRM biology between the four defined TEFF transcriptional states (i.e., TCIRCM-like, TRM-like MP, TE, and Int). Values on axis represent log2 enrichment Z-score. Spearman correlation r, P < 0.005. In B–D, data were obtained from three mice.
To delineate the transcriptional heterogeneity within the pool of sequenced TEFF, the MetaCell (MC) algorithm (Baran et al., 2019) was applied, resulting in the grouping of 5,383 T cells into 14 reproducibly detected MCs (Fig. 4 B and Fig. S4, A and B). Expression analysis of Ilr7a and Klrg1 (genes commonly used to identify the MP and TE populations, respectively) demonstrated substantial variation in expression over the MCs, underlining the variability in cell states within the TEFF pool. Next, to distinguish MCs that correspond to TE and MP TEFF cell states, expression of a multitude of genes associated with MP (Sell, Cd28, Il7r, Cd27, and Cxcr3) and TE (Klf2, Tbx21, Cx3cr1, Klrg1, and Zeb2; Chen et al., 2018) were analyzed at the MC level. Hierarchal clustering of MCs based on this gene set segregated the 14 MCs into three distinct classes: MP (7 MCs), TE (4 MCs), and intermediate (Int; 3 MCs) T cells (Fig. 4 C, left). To subsequently reveal possible heterogeneity within the seven MP MCs in expression of gene sets associated with TRM formation, we selected genes that have previously been described as differentially expressed between mature skin TRM and TCIRCM (Table S1; Mackay et al., 2013; Pan et al., 2017). Strikingly, clustering of the MP MCs based on core TRM and TCIRCM genes separated the MP population into two main clusters; one (three MCs) that displayed prominent expression of TCIRCM-related genes, such as the lymphoid homing markers Sell (CD62L) and Cxcr5, and also the transcription factors Klf3 and Eomes; and a second cluster (four MCs) that was strongly enriched for core TRM signature genes, such as Itgae (CD103), Itga1 (CD49a), and Fabp5 (Fig. 4 C, right; and Table S1). Based on this enrichment and depletion of TRM- and TCIRCM-associated genes, we classified these two MP clusters as TRM-like and TCIRCM-like MPs. In summary, based on gene expression profiles, we divided the high-complexity TEFF pool into four distinct transcriptional states: TE, Int, TCIRCM-like MP, and TRM-like MP (Fig. S4 C).
Figure S4.
Transcriptional TEFF profile of T cell clones is linked to memory generation capacity. scRNA-seq and single-cell barcode sequencing were performed on barcode-labeled TEFF cells responding to skin vaccination. The TEFF cells were grouped into 14 transcriptionally distinct MCs. (A) Relative contribution of each mouse to the 14 MCs. (B) Number of cells assigned to each MC. Colors indicate the transcriptional TEFF subset individual MCs were assigned to; see Fig. 4 C for the definition of the identified TEFF subsets. (C) Pie chart depicting the relative frequency of the four identified TEFF states. (D) Number of TEFF cells observed for each reliably detected clone. (E) Comparison of TCIRCM clone size to the relative contribution of clones to the indicated TEFF states (i.e., TCIRCM-like, TRM-like MP, TE, or Int). Spearman correlation r (when significant) and Spearman correlation P value are depicted. Dots represent individual clones. Clones that were not detected in the TCIRCM compartment were excluded. Red line represents the linear regression line. (F) Analysis as depicted in Fig. 6 B, but here depicting mean effector phase TE and Int output of large and small TCIRCM and TRM clones. (G) Comparison of the absolute production of TRM-like MP (top) and TCIRCM-like MP (bottom) of clones to their production of mature TRM. Black line indicates linear regression line with 95% confidence interval in gray.
To determine the resemblance of the TRM-like MP population observed in blood to bona fide skin TRM in more detail, we also evaluated expression of additional genes involved in TRM biology that are not included in the previously used gene set. Notably, genes encoding the surface molecule CD101 (Cd101; Kumar et al., 2017) and the nuclear aryl hydrocarbon receptor AhR (Ahr; Zaid et al., 2014), both considered signature skin TRM genes, were pronouncedly expressed in TRM-like MP cells (Fig. 4 D). In addition, a strong relation between the expression of these genes and Itgae (Cd101: r = 0.86, P < 0.0005; Ahr: r = 0.87, P < 0.0005) was observed across all 14 MCs (Fig. 4 D). Furthermore, TRM-like MP cells showed marked expression of the skin-migratory genes Ccr10 and Cxcr6 (Xia et al., 2014; Zaid et al., 2017) and displayed moderate to high expression of the IL-15 (Il2rb) and TGFβ (Tgfbr1) receptors, of which the ligands have been described to be essential for skin TRM differentiation and maintenance (Fig. 4 D; Mackay et al., 2015). Collectively, these data demonstrate the existence of a group of cells that transcriptionally mimic TRM, within the pool of circulating vaccine-specific TEFF.
T cell clones differ in their contribution to the TEFF states
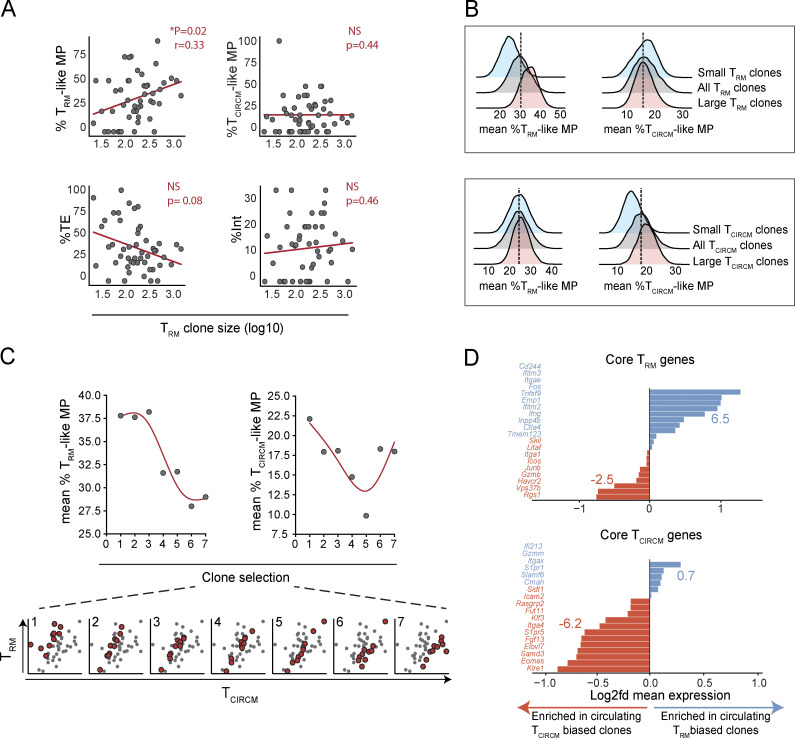
Next, we set out to test whether individual T cell clones differed in their contribution to the four TEFF states. To this end, mRNA-derived barcode sequences were mapped to their associated transcriptome by matching the cell code sequences that mark all transcripts derived from an individual cell. For 28% of the TEFF from which we had retrieved gene expression data (1,527 of the 5,383 cells), we could reliably determine barcode sequences, and thus, infer clonal origin. These 1,527 cells were distributed over 151 clones, ranging from 1 to 189 sampled cells per clone, with a mean and median count of 10 and 4 cells, respectively (Fig. S4 D). Analysis of the distribution of clonally related cells over the four effector subsets revealed that clones differed significantly in their TEFF output toward the different T cell states, as indicated by the deviation from the expected distribution in case of stochastic TEFF differentiation (χ square test, P < 0.0005). For example, while some clones almost exclusively produced TE (i.e., ∼12% of TEFF-stage clones consisted of >80% TE, versus a median of 37.5%), other clones were strongly skewed toward the production of TRM-like MP (i.e., ∼5.5% of TEFF-stage clones consisted of >70% of TRM-like MP, versus a median of 26.1%; Fig. 5 A). To evaluate whether the adoption of transcriptional biases in the TEFF pool could be driven by variations in clonal expansion, we assessed the relation between TEFF clone size, determined by bulk DNA barcode sequencing, and the relative contribution of each clone to the different TEFF subsets. No direct association between TEFF clone size and TEFF cell state was detectable in response to skin vaccination, as TEFF subset bias was observed for small and large clones (Fig. 5 B). Thus, clones responding to local skin vaccination differentially generate the subpopulations that jointly make up the TEFF pool, and this bias cannot be explained by level of clonal expansion.
Figure 5.
Differential contribution of antigen-specific T cell clones to distinct TEFF states, independent of clone size. The relative contribution of individual antigen-specific T cell clones to the four TEFF states was assessed. (A) Heat map depicting the contribution of 91 clones to the 4 identified TEFF states. The sum of each row equals 100%. (B) Comparison of the relative contribution to a transcriptional state (i.e., TCIRCM-like, TRM-like MP, TE, or Int) and clone size in day-12 effector blood. Spearman correlation P values are depicted. Dots represent individual clones. Red line represents the linear regression line. Note that biased output toward the four TEFF states is observed for both small and large clones and is not explained by stochasticity. Data obtained from three mice.
TRM-like transcriptional signature in effector phase predicts TRM-forming potential at the clonal level
The above data reveal the existence of a subgroup of circulating TRM-like cells in the effector phase and demonstrate that individual clones vary in their contribution to this subgroup of effector-phase T cells. To determine whether the observed TRM-like cells could be considered circulating TRM precursors, we analyzed the relationship between skin TRM clone size 75 d after vaccination and transcriptional state of the matched clone in the circulating effector phase compartment 12 d after vaccination. Notably, relative output of individual T cell clones to the TRM-like MP pool in the effector phase showed a significant correlation with TRM clone size in skin during memory, whereas such a correlation was not observed for the three other TEFF states (Fig. 6 A). To further understand the relationship between contribution to the skin TRM pool and TEFF states, we selected clones either randomly (n = 15, 10,000×) or with a proportional bias toward clones that dominated the skin TRM pool (i.e., in case clone A generated 2× more TRM than clone B; clone A was 2× more likely to be selected than clone B), or with a proportional bias toward small TRM clones (i.e., in case clone A generated 2× more TRM than clone B; clone A was 2× less likely to be selected than clone B). Analysis of mean TEFF state output by large (Fig. 6 B, red histogram) and by small (Fig. 6 B, blue histogram) TRM clones demonstrated that the propensity of clones to form TRM is predicted by the production of TRM-like MP by such clones in the effector phase. In contrast, production of TCIRCM-like MP during the effector phase was not predictive of TRM formation capacity. As a control, a similar analysis of the TCIRCM pool revealed that TCIRCM formation was predicted by the production of TCIRCM-like MP, but not TRM-like MP (Fig. 6 B, bottom), and this observation was corroborated by correlation analysis (Fig. S4 E). In line with expectations, a skewing of clonal output toward the TE state during the effector phase was associated with a diminished capacity to yield both TRM and TCIRCM (Fig. S4 F). Additional analysis of the relationship between TRM formation and the absolute quantities of circulating TRM-like and TCIRCM-like MPs produced by individual clones during the effector phase furthermore suggested that total TRM-like MP production better predicts mature TRM formation at the clonal level (R2 = 0.37, P < 0.0005; and r = 0.59, P < 0.0005), than the quantities of TCIRCM-like MP (R2 = 0.20, P = 0.012; and r = 0.47, P = 0.01; Fig. S4 G). Jointly, these data demonstrate that T cell clones that preferentially yield circulating TRM-like MP cells in the effector phase are endowed with a superior TRM forming capacity.
Figure 6.
Skewed production of circulating TRM-like MP cells by T cell clones in TEFF phase is associated with enhanced TRM generation. Clonal output in the TRM and TCIRCM compartments was assessed and compared with the transcriptional profiles of matched clones in the circulating TEFF compartment. (A) Comparison of TRM clone size in memory with the relative output of individual clones to the distinct transcriptional TEFF states (i.e., TCIRCM-like, TRM-like MP, TE, or Int) during the effector phase. Spearman correlation r (when significant) and Spearman correlation P value are depicted. Dots represent individual clones. Clones that were not detected in the TRM compartment were excluded. Red line represents the linear regression line. (B) Top: Analysis of relative production of TCIRCM-like and TRM-like MPs during the effector phase by either large or small TRM clones in memory phase. 15 clones (of a total of 49) were randomly selected 10,000 times (gray histogram, middle), selected with a bias toward large TRM clones (red histogram, front), or selected with a bias toward small TRM clones (blue histogram, back). The distribution of mean TCIRCM-like and TRM-like MP production of the sampled clones is plotted. Dotted line represents the most frequently observed mean production of randomly selected clones. Bottom: Similar analysis as in top, but performed for large and small TCIRCM clones, using random sampling of 69 clones. (C) Comparison of bias in memory generation of individual clones to their production of TCIRCM-like and TRM-like MP cells in the effector phase. Clones that contributed to both the TRM and TCIRCM pool (n = 40) were selected according to their bias in memory production: 10 clones per selection window, moving five clones with each step in the direction of TCIRCM-biased clones, depicted as red dots in scatterplots. The first selection window represents clones with the most prominent bias to TRM generation; the last selection window represents clones with the most prominent bias toward TCIRCM generation. Mean production of TRM-like (left) and TCIRCM-like MP (right) is plotted per window. Red lines represent smoothing spline curves. (D) Difference in expression of core TRM (top) and TCIRCM (bottom) genes between the most TRM-biased TEFF-stage clones (n = 10) and most TCIRCM-biased TEFF-stage clones (n = 10). X axis represents log2 fold difference of mean expression of TRM-biased clones over TCIRCM-biased clones (fold difference calculated as mean expression TRM-biased clones/mean expression TCIRCM-biased clones). Blue and red numbers indicate the sum of the log2 fold differences of genes enriched in TCIRCM- (red) or TRM- (blue) biased clones. Data obtained from three mice.
To test whether skewing toward TRM-like MP during the effector phase could explain not only TRM clone size in memory but also the preferential production of TRM over TCIRCM, as described in Fig. 2 D, clones with various degrees of memory bias (i.e., ratio clone size in TRM pool/clone size in TCIRCM pool) were selected in silico (Fig. 6 C, bottom), and relative production of TRM-like and TCIRCM-like MP by these clones during the effector phase was analyzed. Notably, production of TRM-like MP cells during the effector phase was positively associated with the subsequent preferential production of TRM over TCIRCM (Fig. 6 C). Moreover, gene-expression analysis of the 10 most TRM-biased and 10 most TCIRCM-biased clones demonstrated that TEFF-stage clones that preferentially produce TRM express elevated levels of core TRM genes, while being depleted of core TCIRCM genes (Fig. 6 D). In conclusion, the nonstochastic capacity of clones to preferentially form TRM is preceded by the acquisition of a TRM-fate poised transcriptional profile by these clones in the circulating TEFF compartment.
TRM differentiation is a clone-imprinted attribute that is preserved upon secondary antigen encounter
Based on the observed relationship between the capacity of clones to form TRM and the transcriptional profile of these clones during the effector phase, we hypothesized that the circulating TEFF pool harbors cells that are already committed to the TRM fate. If TRM fate decisions are indeed made before entry of the inflamed tissue site, a pool of responding T cell clones would be expected to reproducibly show the same TRM-forming capacity at different immunized skin sites. To test this, we generated two anatomically separated pools of skin TRM, by parallel vaccination of the right and left hind leg skin of mice (Fig. 7 A). If the development of TRM would be determined solely by stochastic encounter of inflamed skin-derived microenvironmental signals, clone size distributions in the two anatomically separate skin sites would be expected to be disparate. Conversely, if TRM fate commitment were to be imprinted in circulating TEFF-stage clones, the two skin sites would be expected to show a similar clonal distribution. Comparison of the clonal composition of either the left or the right leg skin TRM compartment with that of the TCIRCM compartment at day >60 after vaccination recapitulated the prior observation that a large fraction of naive T cells yield progeny that either preferentially form TRM or TCIRCM (TRM-LEFT − TCIRCM: r = 0.37, P < 0.0005; TRM-RIGHT − TCIRCM: r = 0.30, P < 0.0005), with the average T cell clone differing >10-fold in contribution to the skin and the systemic memory compartment (average ratio TRM-LEFT − TCIRCM: 10.14, average ratio TRM-RIGHT − TCIRCM: 11.67, Fig. 7, B and C). Strikingly, comparison of the TRM populations at the two spatially separated skin sites revealed a substantially higher degree of similarity (r = 0.78, P < 0.0005), with an average clone size ratio of 3.17 (Fig. 7, B and C). To compare the magnitude of this clone-intrinsic bias in TRM formation relative to a bias of individual T cell clones to yield either systemic central memory (TCM) or effector memory (TEM) T cells, we subsequently performed barcode lineage tracing of TRM from the two anatomically separate skin compartments, of TCM (defined as CD62L+) from LN and spleen, and of TEM (defined as CD62L–) from spleen. Complete-linkage clustering analysis again showed the highly similar clonal composition of the memory T cells at the two spatially separated skin compartments (Fig. 7 D). In addition, this analysis revealed that these two TRM compartments differ more strongly in clonal composition from all the three systemic memory T cell compartments than, for instance, splenic TEM and LN TCM differ from each other (Fig. 7 D). Thus, relative to differences in capacity to produce central memory or effector memory T cells, clonal imprinting of the capacity to yield tissue-resident T cell memory versus systemic T cell memory is profound.
Figure 7.
TRM generation capacity is a clone-imprinted attribute that is preserved upon secondary antigen encounter. (A) Schematic timeline used in B–D. (B) Contribution of T cell clones to the TRM pool present at two separate sites of primary vaccination (TRM-LEFT, TRM-RIGHT) relative to the TCIRCM pool (left) and relative to each other (right). Dots represent individual clones. (C) Spearman correlations (left) and ratios (right) of nine individual mice, comparing the clonal composition of the TRM-LEFT compartment to the TCIRCM and to the TRM-RIGHT compartment. Left: Mean with whiskers representing SD. Right: **, P < 0.005, Wilcoxon signed-ranked test. Data from nine mice from two independent experiments. (D) Output of individual OT-I T cells to different TRM and TCIRCM pools, as indicated in the columns. Heat map depicts log10-transformed clone sizes (read counts), clustered using Euclidian distance. Data from six mice from two independent experiments. (E and F) Recipient mice were vaccinated on the right hind leg (primary site) and >60 d later on the left hind leg (secondary site), and clonal composition at both sites was assessed >60 d after secondary vaccination. Top: Schematic time line used in E and F. Bottom left: Contribution of T cell clones to the TRM-SEC pool relative to the TRM-PRIM pool. Dots represent individual clones. Bottom right: Spearman correlations of six individual mice, mean with whiskers representing SD. (F) Disparity between TRM-LEFT and TCIRCM pool (red) and between the TRM-LEFT and TRM-right pool (cyan) in case of simultaneous or staggered vaccination. prim/prim, simultaneous vaccination; prim/sec, primary and secondary vaccination separated by >60 d. N.S., not significant; *, P < 0.05; ***, P < 0.0005; Mann–Whitney U test. Mean with whiskers representing SD. See Fig. S3 A for the definition of disparity. Dots represent individual mice. prim/prim and prim/sec groups each consisted of nine mice. Data from three independent experiments. (G) Illustration of proposed TRM differentiation model. After priming in the skin-draining lymph node, naive T cells undergo clonal expansion and a selection of activated T cells commit to the TRM fate. During the effector phase, these transcriptionally distinct TRM precursor cells migrate, along with non-TRM precursor cells, to the inflamed skin tissue. At the inflamed site, TRM precursors display a heightened capacity to mature into long-term persisting TRM in response to tissue-derived external signals, such as TGFβ, IL-15, and antigen. Note that formation of TRM precursor cells may occur early during clonal expansion, as depicted here, or may reflect heterogeneity in T cell potential that already exists before priming.
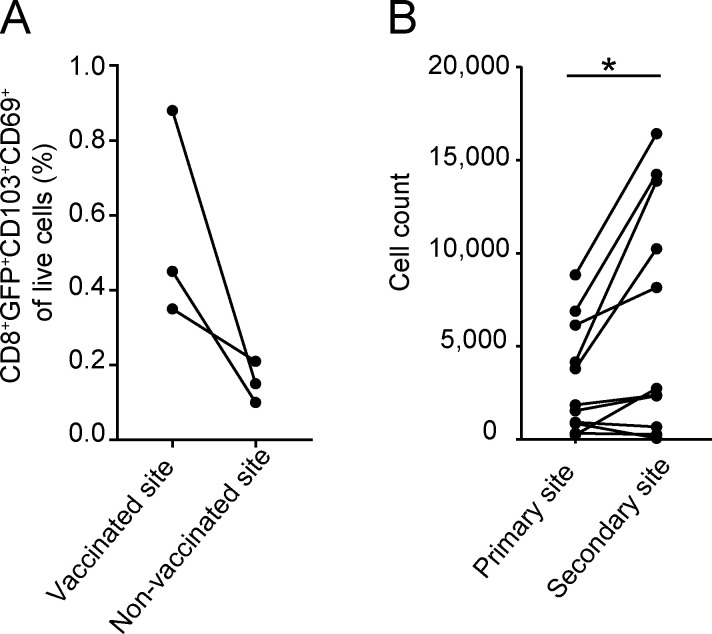
Finally, to test whether the acquisition of TRM generation potential is a stable property of CD8+ T cells, recipients of barcode-labeled naive OT-I T cells were subjected to a primary vaccination on the right hind leg, followed by a secondary vaccination on the left hind leg >60 d later (Fig. 7 E, top). In line with prior work (Casey et al., 2012; Jiang et al., 2012), low frequencies (on average fourfold less than at the vaccinated site) of TRM were detected at the initially unperturbed tissue site upon primary vaccination (Fig. S5 A). Following secondary vaccination at this site, local memory T cell numbers increased to exceed those seen at the primary vaccination site, indicative of de novo TRM formation induced by the secondary vaccine (Fig. S5 B). Subsequently, barcode abundance was separately assessed at the primary and secondary vaccination site >60 d after secondary vaccination, and was compared with clone abundance in the TCIRCM pool at the same time point. This analysis revealed that the secondary TRM pool was dissimilar to the TCIRCM compartment in terms of clonal hierarchy (average r = 0.5), but greatly resembled the TRM pool generated at the primary site of vaccination (average r = 0.73; Fig. 7 E). Furthermore, disparity analysis (Fig. 7 F; explained in Fig. S3 A) revealed that the clonal composition of these two TRM pools that were separated in time was equally similar as when two distinct TRM pools were generated simultaneously, indicating that the capacity of individual T cell clones to yield TRM is stable over time. Thus, these data reveal that, before skin entry, the ability of TEFF to form TRM is differentially and permanently imprinted at a clonal level.
Figure S5.
De novo TRM generation upon secondary vaccination in previously unperturbed sites. (A and B) Mice received GFP+ OT-I T cells (A) or barcode-labeled OT-I T cells (B) and were subjected to DNA vaccination on the right hind leg, whereas the other hind leg remained unperturbed. (A) Analysis of TRM frequencies in vaccinated (right leg) and nonvaccinated (left leg) skin sites >60 d after vaccination. (B) More than 60 d after primary vaccination, the nonvaccinated (left leg) skin site was subjected to DNA vaccination. More than 60 d after secondary vaccination, the primary and secondary vaccinated skin sites were harvested, and GFP+ TRM at the two sites were quantified. Number of barcode-labeled TRM detected at the primary and secondary skin vaccination site of nine mice. *, P < 0.05, Wilcoxon signed-rank test. Dots represent individual mice.
Discussion
The current data demonstrate that, while all naive T cells yield progeny that disseminate equally well to inflamed skin and the systemic lymphoid compartments, a subset of T cell clones yields offspring with a heightened capacity to persist long-term in peripheral tissues. The observation that tissue entry is equal between progeny derived from distinct clones implies that the selection of the TRM privileged clones is not driven by an enhanced capacity of a subset of circulating effector-stage clones to migrate into the inflamed tissue. Rather, the propensity of clones to generate TRM was linked to the transcriptional state of their circulating TEFF offspring, and in particular the production of MP cells that transcriptionally resemble skin TRM was associated with superior TRM formation. The observed link between transcriptional state during the effector phase and contribution to the TRM compartment following memory formation provides compelling evidence that the identified TEFF subgroup can be considered circulating TRM precursors. Furthermore, the notion of a committed TRM precursor pool in the circulation is also supported by the observation that the clonal composition of TRM pools that form at anatomically separate sites is highly similar, indicating that the propensity to efficiently produce TRM is imprinted into T cells in the circulatory compartment, before tissue entry. Previous reports have suggested that TEFF cells that develop into TRM enter the peripheral tissue early after immunization (Masopust et al., 2010; Milner et al., 2017). As the current data demonstrate that TEFF commit to TRM fate before tissue entry, fate decisions of circulating TRM precursors should then also occur early after immunization. In line with this, we observe that the capacity to generate TRM is unequally distributed over T cell clones, which implies that this property must be instilled before substantial clonal expansion. Collectively, our observations argue in favor of early stage TRM fate commitment by a subset of circulating TEFF. Although TRM fate decisions are, at least partially, made in the circulatory compartment, earlier work has established that skin microenvironmental cues, such as TGFβ, IL-15, and cognate antigen (Mackay et al., 2015; Muschaweckh et al., 2016), are essential in driving TRM formation. Jointly, these observations argue in favor of a model in which a subset of circulating TEFF transcriptionally diverge and subsequently develop a heightened capacity to respond to local cues, thereby selectively promoting the differentiation of their progeny into long-term persisting TRM (Fig. 7 G).
Through the combination of lineage-tracing and single-cell transcriptome analysis, we uncovered a transcriptional dichotomy within the pool of circulating MP TEFF cells that precedes the divergence in TRM and TCIRCM formation at a clonal level; however, the mechanisms that drive this dichotomy remain to be elucidated. Several studies have shown a link between TCR affinity and TRM generation potential (Frost et al., 2015; Fiege et al., 2019; Wang et al., 2019; Maru et al., 2017) and it would be of interest to determine whether variation in TCR affinity may influence the capacity of individual T cell clones to yield circulating TRM precursor cells. However, the current data indicate that differential production of circulating TRM-like MP and TRM generation potential can occur independently of variation in TCR affinity, suggesting that other T cell internal and/or external factors are involved. Indeed, an extensive body of work has demonstrated that external signals, such as cytokines and ligands of costimulatory receptors at the T cell priming site, can influence the production of functional memory T cells (Hendriks et al., 2005; Parameswaran et al., 2005; Mousavi et al., 2008; Scholer et al., 2008; Agarwal et al., 2009; Cui and Kaech, 2010; Ahrends et al., 2017). In addition, cross-priming by Batf3+ cDC1s (Iborra et al., 2016) and inhibition of mTOR activity (Araki et al., 2009; Sowell et al., 2014) have opposing roles in promoting TRM over TCIRCM fate commitment. Conceivably, differential exposure of individual T cell clones to these cues during the priming process forms the mechanistic basis for the observed formation of circulating TRM precursors. Furthermore, steady-state heterogeneity in naive T cell-intrinsic properties, such as developmental origin (Smith et al., 2018), prior TGFβ exposure (Mani et al., 2019), and stochastic variation in gene expression (Feinerman et al., 2008; Marchingo et al., 2016), could differentially precondition naive T cells for TRM fate. Evaluation of the role of these T cell external and internal factors should be of value to delineate the mechanistic processes that leads to the generation of the circulating TRM precursor population that we here identify.
Materials and methods
Mice
C57BL/6J-Ly5.1, C57BL/6J, OT-I, mTmG, and UCB-GFP mice were obtained from Jackson Laboratory, and strains were maintained in the animal department of the Netherlands Cancer Institute. The mTmG and UCB-GFP mice were crossed with OT-I mice to obtain mTmG-OT-I and GFP-OT-I strains, respectively. All animal experiments were approved by the Animal Welfare Committee of the Netherlands Cancer Institute, in accordance with national guidelines.
Generation of the BC2.0 high-diversity retroviral barcode library
The BC1DS_lib oligonucleotide (Table S2) containing a 21-nt random barcode sequence was PCR amplified (10 cycles: 10 s at 98°C, 30 s at 55°C, and 1 min at 72°C) with Phusion polymerase (New England Biolabs). The resulting PCR-amplified product was column purified (MinElute PCR cleanup kit; Qiagen) and digested with XhoI and EcoRI, followed by ligation into the 3′ untranslated region of the GFP cDNA sequence within the pMX retroviral vector, using the Electroligase kit (New England Biolabs). Electrocompetent DH10b bacteria (Invitrogen) were then electroporated with 16-ng ligation product, and a small fraction of the transformed bacteria were plated on Luria-Bertani agar plates to determine transformation efficiency; the remaining bacteria were grown overnight in 400 ml Luria-Bertani medium (VWR Life Science) supplemented with ampicillin (Sigma-Aldrich). DNA was extracted from the bacterial culture using the Maxiprep kit (Invitrogen).
Establishment of the barcode reference list
To be able to match barcode sequences observed in biological samples to a reference list of barcodes present in the BC2.0 library, barcode sequences in the library were PCR amplified in duplicate (repA and repB) and sequenced as independent samples. In brief, barcodes were amplified from 10 ng of retroviral library DNA using a combination of native Taq DNA polymerase (Invitrogen) and Deep Vent polymerase (New England Biolabs) at a 2:1 ratio, in three consecutive rounds of PCR. First-round PCR was performed using the Top_lib and Bot_lib primers (15 cycles: 5 s at 94°C, 5 s at 57.2°C, and 10 s at 72°C); second-round PCR was performed using the BC1v2DS_For and BC1v2DS_Rev primers (15 cycles: 5 s at 95°C, 5 s at 58°C, and 10 s at 72°C); third-round PCR was performed using the P5_For and P7_Index_Rev primers (7 cycles: 5 s at 94°C, 10 s at 58°C, and 10 s at 72°C). Resulting PCR products were sequenced on an Illumina hiSeq2500 lane. For primer sequences, see Table S2.
In the sequencing data of repA and repB, 349,439 and 333,422 unique barcode sequences were detected, respectively, with 64.32% of all detected sequences being shared between the two replicates. Many of these sequences are likely to be spurious, resulting from PCR and sequencing errors. Such spurious sequences derive from true “mother barcodes” that have a much higher abundance than the “child” sequences, with child sequences differing by up to several nucleotides from the mother sequence and having a reproducible frequency of occurrence of up to ∼5% of the abundance of the mother barcode (Beltman et al., 2016). To remove those spurious barcode variants, we removed all sequences that had a Levenshtein distance of ≤4 nucleotides (Levenshtein, 1966) from a potential mother barcode and that also had read count of ≤5% of that potential mother barcode. Additional spurious barcodes that occur at a very low abundance are likely to escape this cleaning procedure, for instance because they contain >4 nucleotide differences from their mother. For this reason, only barcodes that were detected ≥3 times in the two replicates combined were retained in the barcode reference list. After this filtering, a list of 263,582 unique barcodes was obtained, of which only 1.27% was not shared between technical replicates.
Generation of barcode-labeled T cells
Retrovirus of the barcode library was produced by transfection of Phoenix-E packaging cells using FuGene6 (Roche). Retroviral supernatant was harvested 48 h after transfection and stored at −80°C. To generate naive barcode-labeled OT-I T cells, thymocytes were harvested from 5–7-wk-old OT-I mice and transduced with the barcode library virus by spinfection (90 min, 400 g) in IMDM (Gibco Life Technologies) supplemented with 8% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 ng/ml recombinant murine IL-7 (PeproTech). To limit the fraction of T cells with multiple barcode integrations, barcode library virus was diluted before transduction to obtain a transduction efficiency of 8–10%. After 24 h of culture, cells were harvested, and viable thymocytes were enriched using Lympholite-M Cell Separation Medium (Cedarlane) followed by purification of GFP+ cells by FACS (FACSAria II [BD Biosciences] and MoFLo Astrios [Beckman Coulter]). Subsequently, ∼1 million sorted GFP+ thymocytes were intrathymically injected into 5–7-wk-old C57BL/6 or C57BL/6-Ly5.1 primary recipient mice, as described previously (Gerlach et al., 2010; 2013). After a maturation period of 2–4 wk, whole blood, spleen, and LNs (cervical, axillary, brachial, mesenteric, inguinal, and lumbar) were harvested and pooled, followed by enrichment of CD8+ T cells using the Mouse CD8 T Lymphocyte Enrichment Set (BD Biosciences). The fraction of GFP+ cells in the CD8+ T cell pool was determined by flow cytometry (Fortessa; BD Biosciences), and 500–1,000 GFP+ cells were adoptively transferred into 8–14-wk-old secondary C57BL/6 or C57BL/6-Ly5.1 recipient mice.
Immunization by DNA vaccination and HSV1 infection
1 d before vaccination with the HELP-OVA vector that encodes the OVA257–264 epitope (SIINFEKL), the shuffled HPV E7 sequence, and MHC-II class restricted helper epitopes (Oosterhuis et al., 2012; Ahrends et al., 2016), fur was removed from hind legs with Veet depilation cream (Reckitt Benckiser). Primary DNA vaccination was performed on days 0, 3, and 6 by tattooing (Bins et al., 2005) a 15-μl droplet of 2 μg/μl DNA solution in 10 mmol/l Tris, pH 8.0, and 1 mmol/l EDTA, pH 8.0, per leg, by means of a sterile disposable 9-needle bar mounted on a rotary tattoo device oscillating at a frequency of 100 Hz for 1 min with a needle depth of 1 mm (MT.DERM). For secondary vaccinations, mice received a single DNA tattoo with 20 μl of the 2 μg/μl plasmid solution on the inside and outside of the leg, >60 d after start of primary vaccination.
The HSVTOM-OVA virus, containing a CMV immediate-early promoter tomato-OVA257–264 gene cassette in the intergenic region between the UL26 and UL27 genes of the HSV-1 strain KOS (Halford et al., 2004), was grown in Vero cells, as described previously (Weeks et al., 2000). 1 d before infection, fur was removed from hind legs with Veet depilation cream (Reckitt Benckiser). On day 0, a 7-μl droplet containing ∼3.125 × 105 PFU HSVTOM-OVA in DMEM (Gibco Life Technologies) per area was given once to both legs of anesthetized mice by means of a tattoo, using a sterile disposable nine-needle bar mounted on a rotary tattoo device oscillating at a frequency of 100 Hz for 1 min with a needle depth of 0.5 mm (MT.DERM). The first macroscopic skin lesions became visible on treated areas on approximately day 3 after infection (not depicted).
Recovery of barcode-labeled T cells from vaccinated and HSV-infected recipient mice
To sample the TEFF pool without sacrificing the animal, a 300-μl blood sample was drawn from the tail vein. Erythrocytes were lysed using NH4Cl, and samples were stored as cell pellets at −80°C. To recover GFP+ T cells from skin and secondary lymphoid organs, in either the effector or memory phase, mice were sacrificed, whole blood was collected by heart puncture, and spleen and LNs (cervical, axillary, brachial, mesenteric, inguinal, and lumbar) were harvested. Blood, spleen, and LN samples derived from one mouse were processed as one sample, unless indicated otherwise. In addition, skin tissue from the hind legs was collected and processed separately. For isolation of barcode-labeled cells from skin tissue, Veet-depilated (Reckitt Benckiser) full-thickness skin was collected using scissors and forceps and minced into small pieces. Subsequently, skin fragments were taken up in DMEM (Gibco Life Technologies) supplemented with collagenase IV (Gibco) and II (Worthington Biochemical Corp.; both 1.25 mg/ml final), DNase type I (0.25 mg/ml final; Sigma-Aldrich), 4% FCS (Sigma-Aldrich), 0.25% BSA fraction IV (Thermo Fisher Scientific), and HBSS (Gibco Life Technologies) and rotated at 37°C overnight. After digestion, skin preparations were diluted with DMEM containing 8% FCS, filtered over 100- and 70-μm strainers (Falcon), washed twice, and taken up in HBSS supplemented with 0.5% BSA, pulmozyme (40 μg/ml final; Roche), and the indicated antibodies (Table S3). After staining for 30 min at 4°C, samples were washed and filtered through a 30-μm strainer (Miltenyi Biotec). To exclude dead cells, samples were stained with DAPI (Sigma-Aldrich). Barcode-labeled CD69+CD103+ skin-resident CD8+ memory T cells were sorted on a FACSAria II (BD Biosciences) or FACSAria Fusion (BD Biosciences). Typical yields were 1,000–10,000 GFP+ CD8+ cells per leg.
Harvested spleen and LN tissue of individual mice was mashed through a 70-μm strainer into single-cell suspensions and pooled with matched blood samples. This pooled cell pool, referred to as the circulatory compartment, was treated with NH4Cl to remove erythrocytes and stained with the indicated antibodies (Table S2). GFP+ CD8+ cells were then isolated by cell sorting on a MoFLo Astrios (Beckman Coulter), with typical yields of 1,000–10,000 GFP+ CD8+ cells per mouse. After isolation, sorted cells derived from either the skin or circulatory compartment were lysed in DirectPCR Lysis Reagent (Viagen Biotech) supplemented with 0.4 mg/ml Proteinase K (Sigma-Aldrich), and resulting samples were stored at −20°C.
Analysis of the presence of blood-borne T cells in the skin TEFF pool
To determine the fraction of blood-borne T cells in skin preparations of the vaccination site obtained during the effector phase, splenocytes of GFP-OT-I transgenic mice were first negatively enriched with the Mouse CD8 T Lymphocyte Enrichment Set (BD Biosciences). Subsequently, C57BL/6J-Ly5.1 animals received ∼700 naive GFP-OT-I splenocytes i.v., followed by primary DNA vaccination on Veet-depilated hind legs as described above. On day 10 after vaccination, mice received a one-time injection of 1.5 × 106 CD8+ negatively enriched mTmG-OT-I splenocytes as a reference for blood-borne T cells, 5 min before sacrificing the animals. Subsequently, blood and skin tissue was harvested, and cells were isolated from the two compartments, as described above. Single-cell suspensions were then stained with IR-dye (Thermo Fisher Scientific) and analyzed on an LSR II SORP (BD Biosciences).
Barcode DNA amplification and next-generation sequencing
Genomic DNA was isolated from frozen pellets of effector blood samples using DNeasy Blood and Tissue (Qiagen) for downstream PCR. Sorted samples of lymphoid tissues and from skin were lysed in DirectPCR Lysis Reagent (Viagen Biotech). Products of samples in experiments in which all samples contained more than ∼3,000 barcode-labeled T cells were used for PCR amplification without intermediate steps. To enhance barcode recovery in experiments with samples with a lower GFP+ cell count, barcode sequences were first captured from the obtained genomic DNA (gDNA) preparations, using biotinylated DNA capture oligonucleotides that anneal either 5′ or 3′ of the barcode sequence in the GFP gDNA (for oligonucleotide sequences, see Table S2). If at least one sample in an experiment contained <3,000 GFP+ cells, all samples in that experiment (independent of their GFP+ cell count) were subjected to the barcode gDNA capture protocol, to avoid the possible generation of bias by this procedure. In brief, gDNA was sheared on the ME220 Focused-ultrasonicator (Covaris) under the following conditions: time, 20 s; peak power, 70; duty%, 20; cycles/burst, 1,000. Next, sheared gDNA was denatured and mixed 1:1 with hybridization buffer (1 ml composition: 667.6 μl of 20× SSPE [Gibco]; 267.6 μl of 50× Denhardt’s solution [Sigma-Aldrich]; 13.2 μl of 20% SDS [Sigma-Aldrich]; 26.8 μl of 0.5 M EDTA, pH 8.0; and 26.8 μl water supplemented with the biotinylated Capt_For_BClibv2 [50 fmol] and Capt_Rev_BClibv2 [50 fmol] oligonucleotides). Hybridization with biotinylated capture oligonucleotides was performed overnight at 65°C. The next day, Streptavidin beads (Dynabeads MyOne streptavidin T1; Invitrogen) were washed with 2× B&W buffer (2 M NaCl in TE buffer, pH 8.0) in low-retention microtubes (Axygen) that were prerinsed with 400 ml of 10 mM Tris, pH 8.0, solution, and the hybridized gDNA was mixed with the streptavidin beads for 30 min at room temperature. Subsequently, bead-bound gDNA was isolated by magnetic pulldown using the Dynamag-2 magnet (Invitrogen). The isolated biotinylated gDNA beads were sequentially washed once with 500 μl of 1× B&W buffer (diluted in TE buffer, pH 8.0), 200 μl of 0.5× B&W buffer (diluted in Tris buffer, pH 8.0), 200 μl of 0.25× B&W buffer (diluted in Tris buffer, pH 8.0), and twice with 200 μl of 10 mM Tris buffer, pH 8.0. The bead-bound gDNA was directly used for downstream PCR amplification.
All samples were split into two separate technical replicates before the first PCR amplification. Genomic barcodes were amplified by nested PCRs using Taq polymerase (Invitrogen). First, the barcode sequence was amplified using the Top_Lib and Bot_Lib primers (30 cycles: 15 s at 95°C, 30 s at 57.2°C, and 30 s at 72°C). Subsequently, PCR products were subjected to a second amplification (30 cycles: 15 s at 95°C, 30 s at 57.2°C, and 30 s at 72°C) using the BC1v2_DS_For and BC1v2_DS_Rev primers that share the annealing sites of the Top_lib and Bot_lib primer, respectively, but are tailed with sequences representing the Illumina primer annealing sites. Finally, the resulting PCR products were subjected to a third amplification (15 cycles: 15 s at 95°C, 30 s at 57.2°C, and 30 s at 72°C) using the P5_For and P7_index_Rev primers that are tailed with the P5 or P7 adaptors, respectively. The P7_index_Rev primer harbors a unique 7-bp index sequence that allows multiplexed analysis of ≤144 samples on one sequencing lane. The 7-bp indexes had a Levenshtein distance of ≥3 bp from each other to avoid incorrect assignment of reads due to PCR or sequence errors (Faircloth and Glenn, 2012). The final PCR products of individual samples were pooled, 322-bp fragments were purified using E-gel extraction (Invitrogen), and PCR products were sequenced on a HiSeq2500 Illumina platform with a read length of 65 bp. For primer sequences, see Table S2.
Filtering of bulk lineage-tracing sequencing data
The reads obtained after sequencing were mapped to the barcode reference library, and reads that showed a 100% match to the barcode constant region, an index sequence that corresponded to one of the indices used during the PCR amplification, and a full match to one of the 21-bp barcode sequences listed in the reference library were retained. Using these filtering steps, ∼150–190 million reads (75–95% of total reads) were considered of appropriate quality for downstream analysis.
To determine barcode sampling efficiency in biological samples, reproducibility between technical replicates was analyzed, and biological samples were excluded from further analysis when the Spearman correlation coefficient between technical replicates was <0.7. Next, barcodes that were not detected in both technical replicates were excluded, removing on average 0.66% of the total reads (and hence inferred cell fraction) per biological sample. After removal of nonreproducibly detected barcodes, the normalized read counts of the barcodes detected in the two technical replicates were averaged. As an additional noise-filtering step, all barcodes that represented <0.01% of reads per sample were excluded. Finally, read counts were renormalized to 10,000, yielding values that represent relative T cell clone sizes in the biological samples. Data filtering and downstream analysis were performed in R version 3.6.0 (https://www.r-project.org/).
Bulk lineage-tracing data analysis after filtering
To allow the visualization of clones with a read count of 0 on a log scale, read counts of all clones were plotted as read count + 1, but original read count values were used for all calculations. Correlations between samples were calculated over the barcodes that were shared between the two compared samples, using Spearman rank correlation. For data visualization, R (ggplot2 and pheatmap) and GraphPad Prism 7.03 were used.
All ratios were calculated as: Clone SizeSampleA/Clone SizeSampleB, taking the inverse of this ratio if Clone SizeSampleA was lower than Clone SizeSampleB, ensuring all outcomes were ≥1. Nonshared barcodes were excluded from the ratio calculations.
To determine the clonal bias threshold described in Fig. 2 D, technical replicate samples of all biological samples used in Fig. 2 were used, with barcodes having a normalized read count of <0.5 excluded from the analysis. For all remaining barcodes, the ratio in read counts between technical replicates A and B was calculated, and a threshold was established such that 98% of barcodes detected in all technical replicates would have a ratio lower than this threshold (Fig. S2 C). This resulted in a clonal bias threshold of 4.8, indicating that a clone had to contribute ≥4.8 times more to one of the normalized cell compartments than to the other cell compartment to be considered biased. Biased clones that were detected only in either the TCIRCM or TRM compartment cannot be ascribed a read count ratio. To allow for the visualization of these clones in Fig. 2 E, we applied the formula (Clone SizeTRM − Clone SizeTCIRCM)/(Clone SizeTRM + Clone SizeTCIRCM), resulting in values that range from −1 to 1, with −1 being completely biased toward TCIRCM formation and 1 being completely biased toward TRM formation.
To allow statistical analysis of the magnitude of clonal disparity between different combinations of cell compartments, an additional measurement of disparity was established (applied in Figs. 3 C and 7 F). Specifically, to compare the magnitude of the differences between sample A and two other samples (i.e., A−B versus A–C), all barcodes observed in samples A, B, and C were ranked in descending order based on the normalized read counts observed in sample A (reference sample), taking along shared and nonshared barcodes detected in the biological samples. Next, the cumulative read count of the ordered barcodes in sample A was plotted against the cumulative read counts in sample A (providing a reference curve) and against the cumulative read counts in samples B and C (Fig. S3 A). The level of disparity was then determined by calculating the area between the reference curve and the curves obtained for samples B and C. In this analysis, a value of 0 signifies that samples are fully identical with respect to clonal composition, and a value of 0.5 signifies a complete lack of overlap between samples.
Modeling stochastic survival of memory T cells
To model the composition of a memory T cell pool that is purely formed by the stochastic survival of TEFF cells, random in silico sampling of barcodes detected in the effector cell pool present in peripheral blood was conducted (Fig. 3, D and E). Specifically, to mimic stochastic memory formation, the probability of a clone surviving was considered to be directly proportional to its relative contribution to the effector pool (i.e., if a clone represented 50% of the total TEFF pool, the probability of its offspring to be sampled per draw would be 0.5). In silico modeling of the memory pool of four mice was performed using the following conditions: (1) by drawing a number of cells that was equal to the number of experimentally observed TRM and TCIRCM cells; (2) by drawing a number of cells that was equal to a fraction 0.1 of the number of experimentally observed TRM and TCIRCM cells; and (3) by drawing a number of cells that was equal to the number of experimentally observed barcodes in the TRM and TCIRCM pool. The first setting models a situation in which the memory compartment is derived from the effector compartment without any further proliferation. The second setting models a situation in which the memory compartment is formed by a combination of cell death and expansion. The third scenario represents the most extreme bottleneck scenario in which each barcode observed in a memory compartment would be derived from a single cell that survived after the effector phase. Notably, for the second and third setting, we assumed that the final TRM pool is formed by proliferation of the drawn founder pool, and that during this expansion the hierarchy between founder clones does not alter. For the three settings, sampling was performed 1,000 times with replacement. To measure the resemblance of the modeled memory pool with the experimentally observed effector pool, Spearman correlations were calculated over the relative sizes of all clones and compared with the correlation between the experimentally observed effector pool and experimentally observed memory pool.
Single-cell barcode amplification from cDNA
From three vaccinated recipient mice, day-12 effector phase blood (300 μl) was collected and subsequently stained with fluorochrome-conjugated antibodies directed against CD8, CD45.2, and Vβ5 (Table S3). In addition, each individual blood sample was stained with distinct anti-mouse TotalSeq Hashtag antibodies (TotalSeq-A0301, TotalSeq-A0302, TotalSeq-A0303; BioLegend). After isolation of CD8+CD45.2+Vβ5+GFP+ cells (considered barcode-labeled OT-I T cells) by FACS sorting on a FACSAria Fusion (BD Biosciences), cells derived from the three mice were mixed 1:1:1 and taken up in PBS supplemented with 0.04% BSA. Next, single-cell RNA isolation and cDNA generation were performed according to the manufacturer’s protocol of the 10X Genomics Chromium Single Cell 3′ kit. Before the fragmentation step described in the manufacturer’s protocol, 20% of the amplified cDNA was taken out and used to specifically PCR amplify barcode sequences. Primers for barcode amplification were designed such that the cellcode and unique molecular identifier (UMI) sequence are both preserved in the amplified product. Barcodes were amplified using Q5 high-fidelity DNA polymerase (New England Biolabs) in two consecutive amplification rounds to make the product compatible with the Illumina sequencing platform. First, the barcode sequence was amplified using cDNA_r1_for and cDNA_r1_rev primers (Table S2); both primers were tailed with the Illumina sequencing primer annealing sites. Second, the resulting PCR products were amplified using the cDNA_r2_for and cDNA_r2_rev primers to add the P5 and P7 adaptors to the products. Products were subsequently sequenced on the Illumina MiSeq Micro platform. After sequencing, the transcripts with a read count of >4 were mapped to the barcode reference library, and transcripts that showed a 100% match to the barcode constant region and fully matched one of the 21-bp barcode sequences listed in the reference library were retained for downstream analysis.
scRNA-seq analysis
For analysis of single-cell transcriptomes, 25% of the single-cell cDNA library, generated according to the manufacturer’s protocol of the 10X Genomics Chromium Single Cell 3′ kit, was sequenced on a NextSeq550 Sequencing System (Illumina), resulting in the acquisition of a total of ∼400 million reads, and the detection of 6,173 cells, with a median of 2,400 genes detected per cell. Feature-barcode matrices were generated using the Cell Ranger software of the 10X Genomics Chromium pipeline. Cells that could be assigned to multiple mice or to no mouse (inferred from the detection of multiple or no Hashtags), cells with a transcript (UMI) count <2,000 or a mitochondrial gene fraction >0.075 were excluded from downstream analysis. Furthermore, cells that transcriptionally resembled B cells (n = 79, as determined by CD19 expression), erythrocytes (n = 11, as determined by expression of Hbb-bs/Hba-a1/Hbb-bt/Hbba-a2), or thrombocytes (n = 96, as determined by Ppbp/Pf4 expression) were excluded from the analysis. For further transcriptional profiling of the remaining 5,383 cells, the Seurat (Butler et al., 2018) and MC (Baran et al., 2019) algorithms were used.
Data from single-cell barcode sequencing and scRNAseq were merged based on matching of cell code sequences, and bulk DNA barcode sequencing data (of TEFF, TRM, and TCIRCM samples) was combined with single-cell data by matching barcode sequences to scRNA-derived barcode sequences. Clones were included for downstream analysis when transcriptional profiles were determined of three or more cells and when clones were also detected in the bulk DNA TEFF, TRM, or TCIRCM barcode sequencing data.
For analysis of differential expression of TRM- and TCIRCM-associated genes in the circulating effector phase compartment, as described in Fig. 6 D, the top 10 most TRM- and TCIRCM-biased clones were selected, and mean gene expression per clone was calculated. Next, the average mean expression of the 10 TRM- and the 10 TCIRCM- biased clones was calculated for the core TRM and TCIRCM genes (Table S1). To exclude lowly expressed genes that are inherently prone to show more noise, only genes with a mean expression level >0.1 normalized UMI count (calculated over all individual clones) are depicted in Fig. 6 D.
Statistics
Statistical analyses were performed using the two-tailed Mann-Whitney U test, Wilcoxon signed-rank test, Kruskal-Wallis test with Dunn’s multiple comparisons test, and Spearman correlation test, using R and GraphPad Prism 7.03. Results were regarded as statistically significant at a P value of <0.05, with *, P < 0.05; **, P < 0.005; and ***, P < 0.0005.
Data and code availability
Single-cell transcriptome data have been deposited in GEO under accession no. GSE152282. All other source data and codes are available upon reasonable request.
Online supplemental material
Fig. S1 shows quality controls of barcode quantification in skin and circulation during the effector phase. Fig. S2 describes the barcode filtering strategy and bias threshold determination. Fig. S3 shows the contribution of T cell clones to the TRM and TCIRCM pool in relation to their clonal burst size. Fig. S4 presents quality controls of the scRNA-seq analysis and the relation between TEFF subset production and memory generation potential of individual clones. Fig. S5 demonstrates the occurrence of de novo TRM generation upon secondary vaccination at previously unperturbed skin sites. Table S1 describes the core TRM and TCIRCM genes depicted in Fig. 4 C. Table S2 describes the primer and DNA-oligonucleotide sequences used to generate the barcode library and to amplify barcode sequences from biological samples. Table S3 provides details on the fluorochrome-conjugated antibodies used for flow-cytometric analysis.
Supplementary Material
shows core TRM and TCIRCM genes.
presents oligo-DNA and primer sequences used to generate the barcode library and to PCR-amplify barcode sequences from biological samples.
lists fluorochrome-conjugated antibodies used for flow-cytometric analysis.
Acknowledgments
We thank the Genomics Core facility, in particular Arno Velds and Marja Nieuwland, the Flow Cytometry facility, and the Animal Research facility of the Netherlands Cancer Institute for technical support; J. Rohr for providing training in specialized methodologies; M. Hoekstra for illustrations; and J. Borst (Leiden University Medical Center, Leiden, The Netherlands) for providing the OVA-vaccination plasmid. We are grateful to all members of the Schumacher group for scientific input.
This work was supported by European Research Council Advanced Grant grant Life-His-T (to T.N. Schumacher).
Author contributions: L. Kok and F.E. Dijkgraaf designed and performed mouse experiments. L. Kok performed lineage-tracing and scRNA-sequencing data analysis. J. Urbanus designed and produced the barcode library and performed DNA barcode capture experiments. K. Bresser assisted in the design of analysis methods. R.F. Cardoso contributed to design and execution of HSVTOM-OVA experiments. D.W. Vredevoogd and F.E. Dijkgraaf developed the TRM isolation protocol. L. Perié wrote the DNA barcode filtering script. J.B. Beltman generated the barcode reference list. L. Kok, F.E. Dijkgraaf, and T.N. Schumacher contributed to experimental design and prepared the manuscript with input of all co-authors.
References
- Agarwal P., Raghavan A., Nandiwada S.L., Curtsinger J.M., Bohjanen P.R., Mueller D.L., and Mescher M.F.. 2009. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J. Immunol. 183:1695–1704. 10.4049/jimmunol.0900592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrends T., Bąbała N., Xiao Y., Yagita H., van Eenennaam H., and Borst J.. 2016. CD27 Agonism Plus PD-1 Blockade Recapitulates CD4+ T-cell Help in Therapeutic Anticancer Vaccination. Cancer Res. 76:2921–2931. 10.1158/0008-5472.CAN-15-3130 [DOI] [PubMed] [Google Scholar]
- Ahrends T., Spanjaard A., Pilzecker B., Bąbała N., Bovens A., Xiao Y., Jacobs H., and Borst J.. 2017. CD4+ T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity. 47:848–861.e5. 10.1016/j.immuni.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Araki K., Turner A.P., Shaffer V.O., Gangappa S., Keller S.A., Bachmann M.F., Larsen C.P., and Ahmed R.. 2009. mTOR regulates memory CD8 T-cell differentiation. Nature. 460:108–112. 10.1038/nature08155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti S., Beltman J.B., Chodaczek G., Hoekstra M.E., van Beek A.E., Gomez-Eerland R., Ritsma L., van Rheenen J., Marée A.F., Zal T., et al. . 2012. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc. Natl. Acad. Sci. USA. 109:19739–19744. 10.1073/pnas.1208927109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenio J., Kakaradov B., Metz P.J., Kim S.H., Yeo G.W., and Chang J.T.. 2014. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat. Immunol. 15:365–372. 10.1038/ni.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran Y., Bercovich A., Sebe-Pedros A., Lubling Y., Giladi A., Chomsky E., Meir Z., Hoichman M., Lifshitz A., and Tanay A.. 2019. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 20:206 10.1186/s13059-019-1812-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltman J.B., Urbanus J., Velds A., van Rooij N., Rohr J.C., Naik S.H., and Schumacher T.N.. 2016. Reproducibility of Illumina platform deep sequencing errors allows accurate determination of DNA barcodes in cells. BMC Bioinformatics. 17:151 10.1186/s12859-016-0999-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bins A.D., Jorritsma A., Wolkers M.C., Hung C.F., Wu T.C., Schumacher T.N., and Haanen J.B.. 2005. A rapid and potent DNA vaccination strategy defined by in vivo monitoring of antigen expression. Nat. Med. 11:899–904. 10.1038/nm1264 [DOI] [PubMed] [Google Scholar]
- Buchholz V.R., Flossdorf M., Hensel I., Kretschmer L., Weissbrich B., Gräf P., Verschoor A., Schiemann M., Höfer T., and Busch D.H.. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science. 340:630–635. 10.1126/science.1235454 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. . 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zander R., Khatun A., Schauder D.M., and Cui W.. 2018. Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front. Immunol. 9:2826 10.3389/fimmu.2018.02826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., and Kaech S.M.. 2010. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol. Rev. 236:151–166. 10.1111/j.1600-065X.2010.00926.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth B.C., and Glenn T.C.. 2012. Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels. PLoS One. 7 e42543 10.1371/journal.pone.0042543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinerman O., Veiga J., Dorfman J.R., Germain R.N., and Altan-Bonnet G.. 2008. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 321:1081–1084. 10.1126/science.1158013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiege J.K., Stone I.A., Fay E.J., Markman M.W., Wijeyesinghe S., Macchietto M.G., Shen S., Masopust D., and Langlois R.A.. 2019. The Impact of TCR Signal Strength on Resident Memory T Cell Formation during Influenza Virus Infection. J. Immunol. 203:936–945. 10.4049/jimmunol.1900093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost E.L., Kersh A.E., Evavold B.D., and Lukacher A.E.. 2015. Cutting Edge: Resident Memory CD8 T Cells Express High-Affinity TCRs. J. Immunol. 195:3520–3524. 10.4049/jimmunol.1501521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaide O., Emerson R.O., Jiang X., Gulati N., Nizza S., Desmarais C., Robins H., Krueger J.G., Clark R.A., and Kupper T.S.. 2015. Common clonal origin of central and resident memory T cells following skin immunization. Nat. Med. 21:647–653. 10.1038/nm.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T., Wakim L.M., Eidsmo L., Reading P.C., Heath W.R., and Carbone F.R.. 2009. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10:524–530. 10.1038/ni.1718 [DOI] [PubMed] [Google Scholar]
- Gerlach C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., and von Andrian U.H.. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C., Rohr J.C., Perié L., van Rooij N., van Heijst J.W., Velds A., Urbanus J., Naik S.H., Jacobs H., Beltman J.B., et al. . 2013. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 340:635–639. 10.1126/science.1235487 [DOI] [PubMed] [Google Scholar]
- Gerlach C., van Heijst J.W., Swart E., Sie D., Armstrong N., Kerkhoven R.M., Zehn D., Bevan M.J., Schepers K., and Schumacher T.N.. 2010. One naive T cell, multiple fates in CD8+ T cell differentiation. J. Exp. Med. 207:1235–1246. 10.1084/jem.20091175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford W.P., Balliet J.W., and Gebhardt B.M.. 2004. Re-evaluating natural resistance to herpes simplex virus type 1. J. Virol. 78:10086–10095. 10.1128/JVI.78.18.10086-10095.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks J., Xiao Y., Rossen J.W., van der Sluijs K.F., Sugamura K., Ishii N., and Borst J.. 2005. During viral infection of the respiratory tract, CD27, 4-1BB, and OX40 collectively determine formation of CD8+ memory T cells and their capacity for secondary expansion. J. Immunol. 175:1665–1676. 10.4049/jimmunol.175.3.1665 [DOI] [PubMed] [Google Scholar]
- Herndler-Brandstetter D., Ishigame H., Shinnakasu R., Plajer V., Stecher C., Zhao J., Lietzenmayer M., Kroehling L., Takumi A., Kometani K., et al. . 2018. KLRG1+ Effector CD8+ T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity. 48:716–729.e8. 10.1016/j.immuni.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra S., Martínez-López M., Khouili S.C., Enamorado M., Cueto F.J., Conde-Garrosa R., Del Fresno C., and Sancho D.. 2016. Optimal generation of tissue-resident but not circulating memory T cells during viral infection requires crosspriming by DNGR-1+ dendritic cells. Immunity. 45:847–860. 10.1016/j.immuni.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Clark R.A., Liu L., Wagers A.J., Fuhlbrigge R.C., and Kupper T.S.. 2012. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 483:227–231. 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., et al. . 2017. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenshtein V. 1966. Binary codes capable of correcting deletions, insertions and reversals. Sov. Phys. Dokl. 10:707–710. [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. . 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Wynne-Jones E., Freestone D., Pellicci D.G., Mielke L.A., Newman D.M., Braun A., Masson F., Kallies A., Belz G.T., et al. . 2015. T-box Transcription Factors Combine with the Cytokines TGF-β and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity. 43:1101–1111. 10.1016/j.immuni.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Mani V., Bromley S.K., Äijö T., Mora-Buch R., Carrizosa E., Warner R.D., Hamze M., Sen D.R., Chasse A.Y., Lorant A., et al. . 2019. Migratory DCs activate TGF-β to precondition naïve CD8+ T cells for tissue-resident memory fate. Science. 366 eaav5728 10.1126/science.aav5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchingo J.M., Prevedello G., Kan A., Heinzel S., Hodgkin P.D., and Duffy K.R.. 2016. T-cell stimuli independently sum to regulate an inherited clonal division fate. Nat. Commun. 7:13540 10.1038/ncomms13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru S., Jin G., Schell T.D., and Lukacher A.E.. 2017. TCR stimulation strength is inversely associated with establishment of functional brain-resident memory CD8 T cells during persistent viral infection. PLoS Pathog. 13 e1006318 10.1371/journal.ppat.1006318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Choo D., Vezys V., Wherry E.J., Duraiswamy J., Akondy R., Wang J., Casey K.A., Barber D.L., Kawamura K.S., et al. . 2010. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207:553–564. 10.1084/jem.20090858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. . 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S.F., Soroosh P., Takahashi T., Yoshikai Y., Shen H., Lefrançois L., Borst J., Sugamura K., and Ishii N.. 2008. OX40 costimulatory signals potentiate the memory commitment of effector CD8+ T cells. J. Immunol. 181:5990–6001. 10.4049/jimmunol.181.9.5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., and Mackay L.K.. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Muschaweckh A., Buchholz V.R., Fellenzer A., Hessel C., König P.A., Tao S., Tao R., Heikenwälder M., Busch D.H., Korn T., et al. . 2016. Antigen-dependent competition shapes the local repertoire of tissue-resident memory CD8+ T cells. J. Exp. Med. 213:3075–3086. 10.1084/jem.20160888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar J.J., Khanna K.M., and Lefrançois L.. 2008. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 28:859–869. 10.1016/j.immuni.2008.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhuis K., Aleyd E., Vrijland K., Schumacher T.N., and Haanen J.B.. 2012. Rational design of DNA vaccines for the induction of human papillomavirus type 16 E6- and E7-specific cytotoxic T-cell responses. Hum. Gene Ther. 23:1301–1312. 10.1089/hum.2012.101 [DOI] [PubMed] [Google Scholar]
- Pan Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O’Malley J.T., Gehad A., Teague J.E., et al. . 2017. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 543:252–256. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran N., Suresh R., Bal V., Rath S., and George A.. 2005. Lack of ICAM-1 on APCs during T cell priming leads to poor generation of central memory cells. J. Immunol. 175:2201–2211. 10.4049/jimmunol.175.4.2201 [DOI] [PubMed] [Google Scholar]
- Scholer A., Hugues S., Boissonnas A., Fetler L., and Amigorena S.. 2008. Intercellular adhesion molecule-1-dependent stable interactions between T cells and dendritic cells determine CD8+ T cell memory. Immunity. 28:258–270. 10.1016/j.immuni.2007.12.016 [DOI] [PubMed] [Google Scholar]
- Sheridan B.S., Pham Q.M., Lee Y.T., Cauley L.S., Puddington L., and Lefrançois L.. 2014. Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity. 40:747–757. 10.1016/j.immuni.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N.L., Patel R.K., Reynaldi A., Grenier J.K., Wang J., Watson N.B., Nzingha K., Yee Mon K.J., Peng S.A., Grimson A., et al. . 2018. Developmental Origin Governs CD8+ T Cell Fate Decisions during Infection. Cell. 174:117–130.e14. 10.1016/j.cell.2018.05.029 [DOI] [PubMed] [Google Scholar]
- Sowell R.T., Rogozinska M., Nelson C.E., Vezys V., and Marzo A.L.. 2014. Cutting edge: generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J. Immunol. 193:2067–2071. 10.4049/jimmunol.1400074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert E.M., Schenkel J.M., Fraser K.A., Beura L.K., Manlove L.S., Igyártó B.Z., Southern P.J., and Masopust D.. 2015. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 161:737–749. 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L., Bickham K.L., Thome J.J., Kim C.Y., D’Ovidio F., Wherry E.J., and Farber D.L.. 2014. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7:501–510. 10.1038/mi.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang S., Goplen N.P., Li C., Cheon I.S., Dai Q., Huang S., Shan J., Ma C., Ye Z., et al. . 2019. PD-1hi CD8+ resident memory T cells balance immunity and fibrotic sequelae. Sci. Immunol. 4 eaaw1217 10.1126/sciimmunol.aaw1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks B.S., Ramchandran R.S., Hopkins J.J., and Friedman H.M.. 2000. Herpes simplex virus type-1 and -2 pathogenesis is restricted by the epidermal basement membrane. Arch. Virol. 145:385–396. 10.1007/s007050050030 [DOI] [PubMed] [Google Scholar]
- Xia M., Hu S., Fu Y., Jin W., Yi Q., Matsui Y., Yang J., McDowell M.A., Sarkar S., Kalia V., et al. . 2014. CCR10 regulates balanced maintenance and function of resident regulatory and effector T cells to promote immune homeostasis in the skin. J. Allergy Clin. Immunol. 134:634–644.e10. 10.1016/j.jaci.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid A., Hor J.L., Christo S.N., Groom J.R., Heath W.R., Mackay L.K., and Mueller S.N.. 2017. Chemokine Receptor-Dependent Control of Skin Tissue-Resident Memory T Cell Formation. J. Immunol. 199:2451–2459. 10.4049/jimmunol.1700571 [DOI] [PubMed] [Google Scholar]
- Zaid A., Mackay L.K., Rahimpour A., Braun A., Veldhoen M., Carbone F.R., Manton J.H., Heath W.R., and Mueller S.N.. 2014. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl. Acad. Sci. USA. 111:5307–5312. 10.1073/pnas.1322292111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D., Lee S.Y., and Bevan M.J.. 2009. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 458:211–214. 10.1038/nature07657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows core TRM and TCIRCM genes.
presents oligo-DNA and primer sequences used to generate the barcode library and to PCR-amplify barcode sequences from biological samples.
lists fluorochrome-conjugated antibodies used for flow-cytometric analysis.
Data Availability Statement
Single-cell transcriptome data have been deposited in GEO under accession no. GSE152282. All other source data and codes are available upon reasonable request.