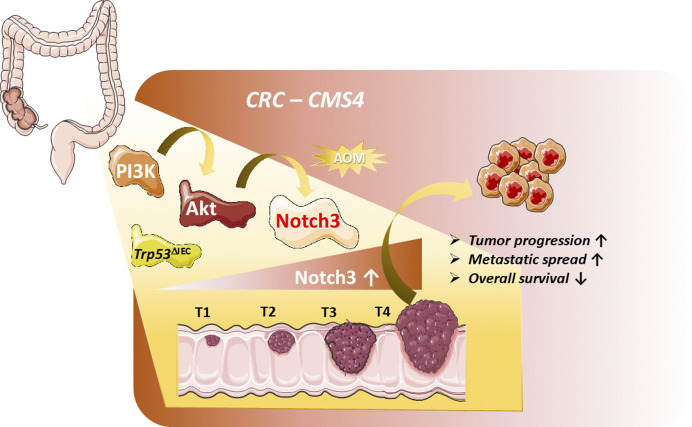
Varga et al. identified Notch3 as a promoter of tumor progression and metastasis in a mouse model of advanced colorectal cancer. In CRC patients, NOTCH3 expression was associated with poor overall survival. These findings feature NOTCH3 as putative therapeutic target for advanced CMS4 CRC patients.
Abstract
In this issue of JEM, Varga et al. (https://doi.org/10.1084/jem.20191515) describe a mouse model of invasive and metastatic colorectal cancer (CRC) closely resembling the human consensus molecular subtype (CMS) 4 associated with the poorest overall survival of the four CMSs. Transcriptomic and bioinformatic analysis combined with pharmacological and genetic studies identified Notch3 as a promoter of tumor progression and metastasis. NOTCH3 expression was up-regulated in CMS4 CRC patients and associated with tumor staging, lymph node and distant metastasis. These findings feature NOTCH3 as putative therapeutic target for advanced CMS4 CRC patients.
Colorectal cancer (CRC) is one of the major forms of cancer in adults and the second most common cause of cancer death in the western world (Siegel et al., 2020). Although advances in patient-screening procedures and adjuvant oncological therapies have improved patient outcome, mortality remains significant. CRC was one of the first solid tumors to be molecularly characterized, establishing a model of progressive and stepwise accumulation of genetic lesions that correlate with progression of disease from adenoma to metastatic carcinoma (Fearon and Vogelstein, 1990). However, genomic profiling of paired primary and metastatic human CRC tumors failed to identify causative metastasis-specific genetic alterations, suggesting that metastasis might instead be driven by transcriptional programs enabling tumor cells to invade, seed, and survive in foreign tissues (Brannon et al., 2014; Varga and Greten, 2017). More recently, an international consortium compared six CRC classification systems based on gene expression datasets, which resulted in a consensus molecular classification scheme for CRC. According to this gene expression–based classification scheme, most CRC tumors fall into four consensus molecular subtypes (CMSs) distinguishing mRNA expression together with distinct molecular and clinical features (Guinney et al., 2015).
Insights from Ute Koch and Freddy Radtke.
Genetically engineered mouse models (GEMMs) of cancer have been instrumental in understanding human disease biology and helped pave new paths in therapy; for example, immunotherapy. Although GEMMs for CRC have been generated based on the genetic alterations identified in human patients, only a few of them faithfully recapitulate human pathology. Major weaknesses of the current CRC GEMMs are that the tumors rarely progress from the adenoma to invasive carcinoma stage and they often lack penetrant metastasis in particular to distant organs and rarely resemble the human CRC CMS subtypes. Thus, most CRC GEMMs are suitable to study early-stage disease rather than tumor progression or metastasis (Jackstadt and Sansom, 2016).
The present study by Varga et al. (2020) is an exciting continuation of previous work by the Greten laboratory, in which they generated a mouse model of advanced CRC based on the intestinal specific gene inactivation of the tumor suppressor gene Trp53 (Trp53ΔIEC) in intestinal epithelia cells. Trp53 deficiency on its own is insufficient to drive intestinal tumorigenesis. However, when combined with the carcinogen azoxymethane (AOM), Trp53ΔIEC mice develop invasive colon tumors and lymph node metastasis with 20–30% penetrance (Schwitalla et al., 2013). Not only loss of TRP53, but also activation of the PI3K–AKT pathway has been associated with adenoma-to-carcinoma transitions and invasiveness in human CRC (Ishaque et al., 2018); thus, the authors correctly hypothesized that combining both traits might lead to a suitable mouse model in which to study advanced CRC. Although the number and size of primary colon tumors were comparable between AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice, compound animals revealed enhanced tumor progression with 100% of mice developing invasive colon tumors and 80% developing metastasis. The fact that the growth and number of primary tumors were comparable between the models suggests that PI3K–AKT activation does not alter tumor initiation or growth but rather promotes tumor progression, invasiveness, and aggressiveness of disease. Gene-expression profiling and phospho-proteomic analysis of tumor tissue derived from single- or double-mutant animals revealed that the Trp53ΔIECAktE17K tumors shared many pathways and characteristics typical for human CMS4, whereas Trp53ΔIEC tumors were more similar to CMS2. In addition, the authors identified Notch signaling as being differentially activated in Trp53ΔIECAktE17K tumors. The essential role of Notch signaling in maintenance of intestinal homoeostasis is well established. However, the function of Notch signaling in CRC, in particular during CRC progression, is less clear. Interestingly, the authors detected increased Notch3 transcripts and active Notch3 within tumor cells of the AOM-treated Trp53ΔIECAktE17K mice, while expression of other Notch receptors was unchanged. Although Notch3 has not been linked to any physiological function within the intestine, but rather to homeostasis of vascular smooth muscle cells and pericytes (Siebel and Lendahl, 2017), increased NOTCH3 expression in human CRC and CRC cell lines has been noted before (Aburjania et al., 2018). To confirm that increased Notch3 expression and signaling is indeed a cell-autonomous process mediated by Akt activation, the authors used sophisticated colon tumor–derived organoid models to tackle this question. The first model employed a knockdown of Pten in order to induce Akt activation in Trp53−/− tumor organoids, which indeed resulted in increased Notch3 expression. How Akt signaling mechanistically activates Notch3 expression has not been addressed and requires future investigation. Next, the authors generated organoids with defined mutation profiles containing deletions for the Apc, Trp53, and Tgfrb2 genes, as well as the oncogenic murine KrasG12D gene (APTK organoids). Introduction of a constitutive active myristoylated AKT gene in APTK organoids (APTKA) led to up-regulation of Notch3, similar to Pten knockdown in Trp53−/− organoids. Importantly, functional validation that Akt activation and, consequently, Notch 3 up-regulation was sufficient to enhance invasion and metastasis orthotopic transplantation of tumor organoids carrying these defined mutations was performed. Most importantly, expression of a dominant active form of Notch3 in APTK organoids lacking constitutive Akt activation also dramatically enhanced metastasis formation. Together, these results strongly suggest that Akt-dependent up-regulation of Notch3 indeed promotes metastasis formation. This notion was further confirmed and supported by a series of Notch loss-of-function approaches. The therapeutically most interesting proof-of-concept Notch loss-of-function assay was the treatment of AOM-induced Trp53ΔIECAktE17K mice with a Notch3-antagonistic antibody (α-NRR3; Yu et al., 2020), preventing activation of Notch3. α-NRR3 treatment of AOM-induced Trp53ΔIECAktE17K mice significantly reduced the extent of tumor invasion and markedly reduced the occurrence of lymph node metastasis, indicating that selective blockage of Notch3 in this mouse model is sufficient to interfere with tumor progression.
Akt-dependent up-regulation of Notch3 promotes tumor progression and metastasis formation. In a novel carcinogen-driven (AOM) mouse model of advanced CRC, constitutive activation of the PI3K–Akt pathway with combined loss of p53 leads to increased Notch3 signaling promoting tumor progression and metastasis. Phenotypic, gene expression profiling, and phospho-proteomic analysis of tumor tissue revealed characteristics typical of the human CMS4. Human NOTCH3 expression increased with tumor stages (T1–T4) and correlated positively with tumor invasion, metastasis, and reduced overall survival.
Analysis of publicly available CRC patient–derived expression datasets revealed that CRC patients with high NOTCH3 mRNA expression have a significantly worse overall survival compared with patients with unaltered or low NOTCH3 expression. Moreover, NOTCH3 expression increased with tumor stages (T1–T4) and correlated positively with tumor invasion and metastasis. In addition, increased NOTCH3 expression was specifically associated with the poor prognosis CRC CMS4 subtype. These correlative data imply that the observations made in this novel mouse model might bear clinical relevance for patients with CMS4 tumors.
This agrees with a recent report by the Sansom laboratory, who also identified a Notch gene signature score that is significantly associated with CMS4 and poor prognosis (Jackstadt et al., 2019). The same study reported activation of NOTCH1 in human CRC liver metastasis and the functional validation of the contribution of Notch signaling in CRC using a gut-specific murine model based on activation of K-Ras and Trp53-deficiency with concomitant overexpression of Notch1 (KPN). KPN mice developed intestinal adenocarcinoma and with a very high percentage (>80%) of metastasis to lymph nodes, lungs, and liver, while KP or PN mice developed only a few metastases, and very rarely to distant organs. The fact that both studies identified a Notch signature that specifically associates with CSM4 and that two independent Notch gain-of-function studies (N3ICD and N1ICD) in two different CRC mouse models enhances tumor progression and metastasis strongly supports a role for Notch signaling in tumor progression. This leads to the question of how Notch might promote tumor progression and metastasis. Jackstadt and colleagues suggest that forced Notch1 signaling drives metastasis through Tgf-β–dependent neutrophil recruitment, which creates an immune-suppressive and metastasis-permissive tumor microenvironment. Pharmacological inhibition of the recruitment of neutrophils into the tumor microenvironment attenuated metastasis and correlated with T cell activation (Jackstadt et al., 2019). The mechanistic aspect of how Notch3 might drive CRC tumor progression in the current study is not completely solved and awaits future investigations.
However, interestingly, Varga et al. (2020) show that, unlike NOTCH3, NOTCH1 mRNA expression did not correlate or associate with CRC patient survival, tumor stage, or metastasis, suggesting in human CMS4 patients NOTCH3 and not NOTCH1 might be promoting tumor progression. Which of the NOTCH receptors is involved in promoting tumor progression might at first glance not be of importance, but it could be highly relevant from a therapeutic point of view. Although pan-Notch inhibitors such as GSI, which block signaling mediated by all receptors, have shown beneficial effects in preclinical Notch-driven tumor models, none of these inhibitors have been clinically approved, largely due to on-target dose limiting toxicities (DLT) of the intestinal epithelium. To avoid this toxicity, clinical trials in Notch-driven cancers have relied on intermitting dosing of GSIs (Andersson and Lendahl, 2014). However, the question remains whether intermittent dosing strategies sustain Notch inhibition long enough to achieve therapeutic efficacy. Similarly, mAbs against NOTCH1 and/or antibodies that recognize both NOTCH2 and NOTCH3 also exhibited DLT in the intestine (Ferrarotto et al., 2018; Smith et al., 2019). Since NOTCH3-mediated signaling has no known physiological function in the intestine, a mAb targeting NOTCH3 with high specificity might have the greatest chance of being well tolerated and therapeutically relevant.
Although interfering with Notch signaling in two independent CRC mouse models has led to the attenuation of metastasis, an important clinically relevant question that needs to be addressed in the future is whether interfering with Notch signaling in a disease setting with already established metastasis is able to positively influence disease progression.
References
- Aburjania, Z., et al. 2018. Oncologist. 10.1634/theoncologist.2017-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, E.R., and Lendahl U.. 2014. Nat. Rev. Drug Discov. 10.1038/nrd4252 [DOI] [PubMed] [Google Scholar]
- Brannon, A.R., et al. 2014. Genome Biol. 10.1186/s13059-014-0454-7 [DOI] [Google Scholar]
- Fearon, E.R., and Vogelstein B.. 1990. Cell. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Ferrarotto, R., et al. 2018. Ann. Oncol. 10.1093/annonc/mdy171 [DOI] [PubMed] [Google Scholar]
- Guinney, J., et al. 2015. Nat. Med. 10.1038/nm.3967 [DOI] [Google Scholar]
- Ishaque, N., et al. 2018. Nat. Commun. 10.1038/s41467-018-07041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackstadt, R., and Sansom O.J.. 2016. J. Pathol. 10.1002/path.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackstadt, R., et al. 2019. Cancer Cell. 10.1016/j.ccell.2019.08.003 [DOI] [Google Scholar]
- Schwitalla, S., et al. 2013. Cancer Cell. 10.1016/j.ccr.2012.11.014 [DOI] [Google Scholar]
- Siebel, C., and Lendahl U.. 2017. Physiol. Rev. 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- Siegel, R.L., et al. 2020. CA Cancer J. Clin. 10.3322/caac.21601 [DOI] [Google Scholar]
- Smith, D.C., et al. 2019. Invest. New Drugs. 10.1007/s10637-018-0714-6 [DOI] [Google Scholar]
- Varga, J., and Greten F.R.. 2017. Nat. Cell Biol. 10.1038/ncb3611 [DOI] [PubMed] [Google Scholar]
- Varga, J., et al. 2020. J. Exp. Med. 10.1084/jem.20191515 [DOI] [Google Scholar]
- Yu, J., et al. 2020. J. Cell. Physiol. 10.1002/jcp.28960 [DOI] [Google Scholar]