Varga et al. describe a novel genetically modified mouse model of human, poor-prognosis mesenchymal colorectal cancer. Using this model, they identify NOTCH3 as a potential therapeutic target for this specific colorectal cancer subtype.
Abstract
Recently, a transcriptome-based consensus molecular subtype (CMS) classification of colorectal cancer (CRC) has been established, which may ultimately help to individualize CRC therapy. However, the lack of animal models that faithfully recapitulate the different molecular subtypes impedes adequate preclinical testing of stratified therapeutic concepts. Here, we demonstrate that constitutive AKT activation in intestinal epithelial cells markedly enhances tumor invasion and metastasis in Trp53ΔIEC mice (Trp53ΔIECAktE17K) upon challenge with the carcinogen azoxymethane. Gene-expression profiling indicates that Trp53ΔIECAktE17K tumors resemble the human mesenchymal colorectal cancer subtype (CMS4), which is characterized by the poorest survival rate among the four CMSs. Trp53ΔIECAktE17K tumor cells are characterized by Notch3 up-regulation, and treatment of Trp53ΔIECAktE17K mice with a NOTCH3-inhibiting antibody reduces invasion and metastasis. In CRC patients, NOTCH3 expression correlates positively with tumor grading and the presence of lymph node as well as distant metastases and is specifically up-regulated in CMS4 tumors. Therefore, we suggest NOTCH3 as a putative target for advanced CMS4 CRC patients.
Introduction
Although both colorectal cancer (CRC) incidence and mortality have been declining over the past decades, CRC still remains the third leading cancer type for estimated new cancer cases and deaths in both genders (Siegel et al., 2018). Most CRCs develop via the adenoma-carcinoma sequence and are driven by the stepwise accumulation of mutations in certain oncogenes and tumor suppressors (Fearon and Vogelstein, 1990). Even though CRC driver mutations are well known, recent large-scale genomic profiling of primary tumors and their matched metastasis failed to identify consistent metastasis-specific mutations (Brannon et al., 2014), suggesting that instead of genetic alterations, changes in transcriptional programs endow cells with metastatic traits (Varga and Greten, 2017). Recently, an international consortium developed an integrated and unbiased approach for the gene expression–based classification of CRC and defined four distinct consensus molecular subtypes (CMSs; Guinney et al., 2015). Importantly, CMS categories show correlation with patient survival, and patients with tumors, which fall into the CMS4 group, have the worst relapse-free and overall survival (Dienstmann et al., 2017; Guinney et al., 2015). Interestingly, CMS4 tumors are characterized by profound stromal infiltration, extracellular matrix remodeling, epithelial-to-mesenchymal transition (EMT), and TGF-β pathway activation, supporting the notion that signals deriving from the stromal tumor microenvironment rather than the underlying genetic profile of a colorectal tumor are particularly important.
The lack of appropriate genetically engineered mouse models (GEMMs) of advanced CRC is a limitation in the development of new therapeutic strategies (McIntyre et al., 2015). To date, only a few GEMMs have been generated that faithfully recapitulate advanced human CRC (Jackstadt and Sansom, 2016), and none of them has been shown to closely mimic the CMS categories. Previously, we developed a mouse model of advanced CRC in which genetic inactivation of Trp53 in combination with administration of the procarcinogen azoxymethane (AOM) leads to invasive tumors and, in ∼20 to 30% of the animals, lymph node metastasis (Schwitalla et al., 2013). Apart from TP53 loss, the activation of the phosphoinositide 3-kinase (PI3K)–AKT pathway is also associated with the adenoma-carcinoma transition and acquisition of an invasive phenotype in human CRC (Fearon, 2011; Markowitz and Bertagnolli, 2009). Therefore, we hypothesized that a mouse model with the combination of these two genetic alterations could provide us with a valuable tool for the understanding of the pathogenesis of advanced CRC.
The PI3K–AKT pathway mediates signals from a large number of growth factors and cytokines, as well as other extracellular mediators released into the tumor microenvironment, to regulate a plethora of key cellular processes, including proliferation, survival, motility, and metabolism (Thorpe et al., 2015). Many downstream targets of PI3K–AKT signaling have already been identified; however, the exact contribution of PI3K–AKT signaling to colorectal tumor progression is still not completely understood. Moreover, studies using various mouse models of PI3K–AKT activation-associated colorectal carcinogenesis have yielded conflicting results (Byun et al., 2011; Davies et al., 2014; He et al., 2007; Leystra et al., 2012; Marsh et al., 2008; Shao et al., 2007).
NOTCH signaling is an evolutionarily conserved signaling pathway that is crucial for proper embryonic development as well as homeostasis in the adult organism. NOTCH receptors (NOTCH1–4) are transmembrane heterodimers, composed of a ligand-binding extracellular and an intracellular signaling domain. Upon ligand binding, the receptor is proteolytically cleaved, and the intracellular domain translocates to the nucleus to regulate gene expression (Aster et al., 2017). In the intestine, NOTCH maintains intestinal stem cell function and, in concert with other signaling pathways, regulates cell fate decisions (Fre et al., 2005; Milano et al., 2004; Stanger et al., 2005; van Es et al., 2005). Although NOTCH signaling can confer both tumor-suppressive and -promoting properties depending on cancer type (Siebel and Lendahl, 2017), in CRC, NOTCH appears to be protumorigenic (Chanrion et al., 2014; Chu et al., 2010; Fre et al., 2009; Sonoshita et al., 2011; Ueo et al., 2012). Activating NOTCH mutations are very rare in CRC, yet overexpression of NOTCH receptors and ligands has been described (Noah and Shroyer, 2013). The exact mechanisms underlying NOTCH activation in CRC, as well as its consequences particularly during late stages of carcinogenesis, are still ill defined.
Here we show that constitutive AKT1 activation along with Trp53 loss enhances carcinogen-induced tumor progression. Furthermore, we demonstrate that these tumors resemble human tumors of the CMS4 subtype, and we propose that NOTCH3 may represent a promising target in this bad prognosis subtype.
Results
Combined loss of Trp53 and AKT activation enhances AOM-induced tumor progression
To examine whether constitutive AKT activation would affect tumor progression in AOM-challenged Trp53ΔIEC mice, we crossed LSL-AktE17K mice to Trp53ΔIEC animals to selectively induce AKT in Trp53-deficient intestinal epithelial cells (IECs; Fig. 1 a). The substitution of glutamic acid with a lysine at amino acid position 17 in AKT leads to its constitutive activation and enhanced downstream signaling even in the absence of activating stimuli (Carpten et al., 2007). Trp53 deletion combined with constitutive AKT activation was not sufficient to induce spontaneous colon tumors but led to tumor formation in the small intestine (SI) within 6 mo (not depicted). Although a proportion of the SI tumors were invasive, these tumors never gave rise to metastasis. To induce colonic tumorigenesis, we subjected Trp53ΔIECAktE17K and control Trp53ΔIEC mice to repetitive AOM treatment (Fig. 1 b). AOM is a DNA-alkylating procarcinogen that causes mutations in several well-known oncogenes and tumor suppressors that are particularly important for colorectal carcinogenesis and induces tumors specifically in the colon (Rosenberg et al., 2009).
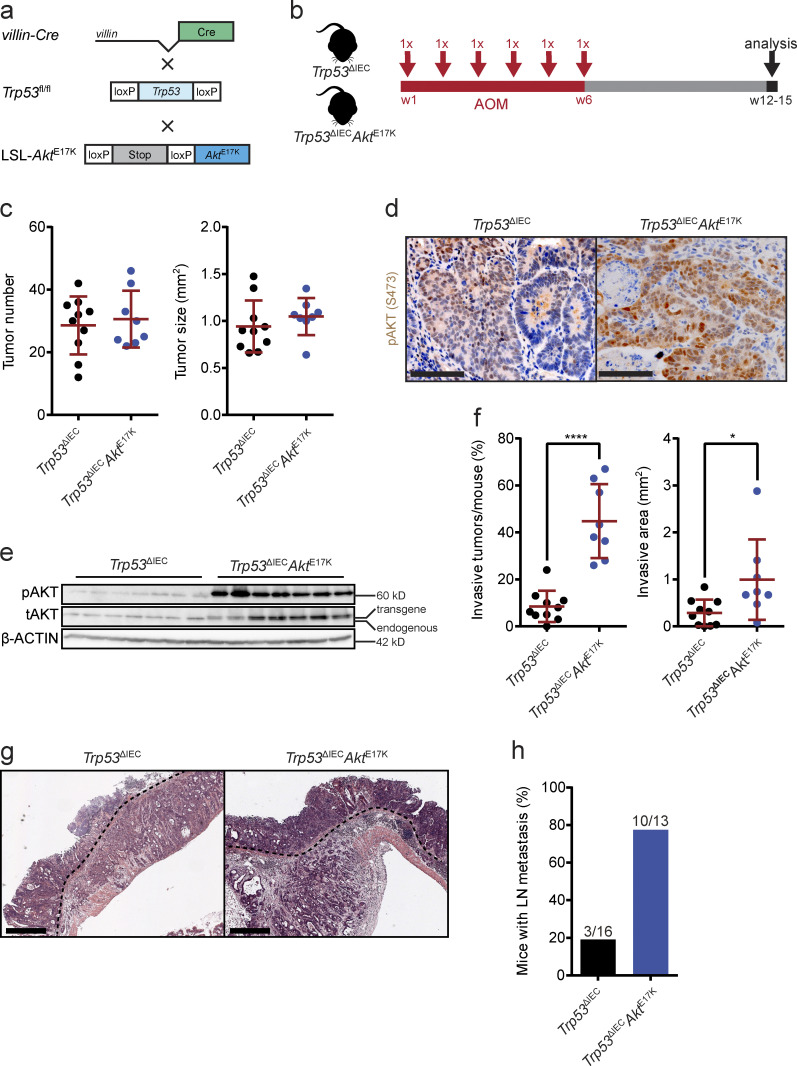
Figure 1.
AOM-treated Trp53ΔIECAktE17K mice develop highly invasive tumors with lymph node metastasis. (a) Schematic representation of the strategy used for the generation of Trp53ΔIECAktE17K mice (fl, floxed; wt, wild type). (b) Experimental design of the AOM treatment of Trp53ΔIECAktE17K mice. (c) Primary tumor number and primary tumor size in AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice. Data are mean ± SD, n ≥ 8 per genotype. Not significant by t test. (d) Representative images of pAKT immunohistochemistry (IHC) on sections from AOM-induced colon tumors in Trp53ΔIEC and Trp53ΔIECAktE17K mice. Scale bars = 100 µm. (e) Immunoblot analysis of phosphorylated (p) and total (t) AKT levels in whole-tumor lysates of AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice. Lower band corresponds to endogenous AKT; upper band represents transgenic AKT. β-ACTIN was used as loading control. n = 7 per genotype. (f) Frequency of invasive tumors per mouse and extent of invasive area in AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice. Data are mean ± SD, n ≥ 8 per genotype; *, P < 0.05; ****, P < 0.0001 by t test. (g) Representative images of H&E-stained tumor sections from AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice (dashed lines mark invasive fronts). Scale bars = 500 µm. (h) Frequency of mice with lymph node metastasis in AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice. n ≥ 13 per genotype. Numbers indicate the mice with metastasis/all mice.
In response to AOM, both control Trp53ΔIEC and Trp53ΔIECAktE17K mice developed colon tumors of comparable number and size (Fig. 1 c). Apart from colon tumors, we also observed AOM-induced tumor formation in the SI in both genotypes. Because tumorigenesis predominantly affects the colon and rectum in patients, we focused our further analysis on the colon tumors only.
Pronounced AKT1 activation in Trp53ΔIECAktE17K tumors was confirmed by immunohistochemistry and immunoblot analysis (Fig. 1, d and e). The similar number and size of the tumors indicated that PI3K–AKT pathway activation does not affect tumor initiation or tumor growth in this model. However, the number of invasive tumors was significantly higher and the invasive areas were larger in Trp53ΔIECAktE17K mice (Fig. 1, f and g). Moreover, Trp53ΔIECAktE17K mice developed lymph node metastases more frequently, in line with a more aggressive phenotype (Fig. 1 h).
Gene expression profiles of Trp53ΔIECAktE17 tumors closely resemble human CMS4 subtype profiles
To understand the molecular basis for the more aggressive phenotype of Trp53ΔIECAktE17K tumors, we subjected whole-tumor tissues from AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice to RNA sequencing and phosphoproteomic analysis. Because mice from both genotypes developed several tumors, we decided to combine all tumors from one mouse into a single sample. With this approach, we hoped to minimize the effect of intertumoral heterogeneity on the analysis. RNA sequencing of tumors of both genotypes identified 1,697 differentially expressed genes (Padj < 0.05). Based on these genes, Ingenuity Pathway Analysis (IPA) predicted differential activation of several signaling pathways related to invasion and metastasis, such as adherens and tight junction signaling, EMT signaling, signaling by Rho GTPases and Rho GDP-dissociation inhibitors, actin-cytoskeleton signaling, planar cell polarity signaling, and integrin-linked kinase signaling, as well as inflammatory pathways including agranulocyte and granulocyte adhesion and diapedesis, IL-6, IL-17, CXR4, and chemokine signaling pathways, all of which have been previously shown to have tumor-promoting effects (Bollrath et al., 2009; Cabrero-de Las Heras and Martínez-Balibrea, 2018; Grivennikov et al., 2012; Fig. S1, a and b). Taken together, these data suggest that Trp53ΔIECAktE17K tumors have proinvasive and prometastatic properties and possess a unique microenvironment.
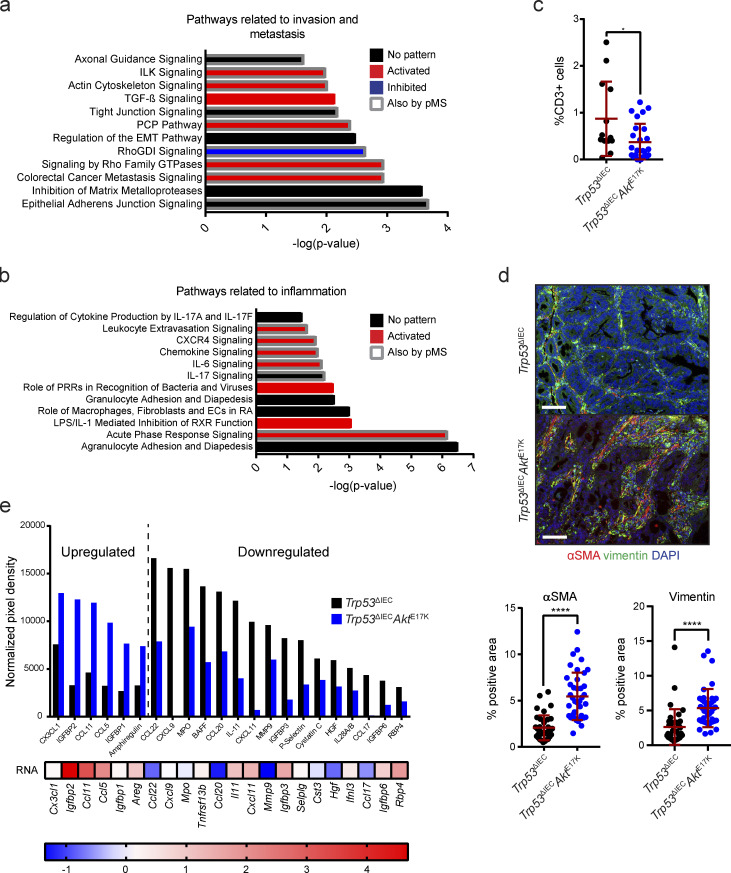
Figure S1.
Transcriptomic and phosphoproteomic analysis of Trp53ΔIECAktE17K tumors reveals regulation of pathways associated with invasion and metastasis as well as inflammation. (a and b) Signaling pathways associated with invasion and metastasis (a) as well as inflammation (b), which are significantly regulated in response to AKT hyperactivation. Predictions were made based on RNA sequencing of whole tumor tissues from AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice using IPA. P < 0.05 with right-tailed Fisher’s exact test, activation: z-score > 0, inhibition: z-score < 0, no pattern: pathway is differentially regulated, but z-score is not available. Regulated pathways which were also identified by phosphoprotein-specific MS of whole-tumor tissues of AOM-treated Trp53ΔIEC and Trp53ΔIECAktE17K mice using IPA are highlighted by a gray frame. P < 0.05. (c) Quantification of CD3 immunohistochemistry (IHC) in the invasive tumors of Trp53ΔIEC and Trp53ΔIECAktE17K mice. Cell numbers are shown as percentage of total cells, and at least one tumor and one invasive front was quantified per mouse. Data are mean ± SD; *, P ≤ 0.05 by t test. ROUT test with Q = 1% was used to remove outliers. n = 4 mice per genotype. (d) Representative images and quantification of immunofluorescent analysis of αSMA (red) and vimentin (green) expression in the tumors of Trp53ΔIEC and Trp53ΔIECAktE17K mice. Cell nuclei are stained with DAPI (blue). Data are mean ± SD; ****, P ≤ 0.0001 by t test. n ≥ 3 mice per genotype. Scale bars = 100 µm. (e) Cytokine array analysis of whole-tumor lysates from Trp53ΔIEC and Trp53ΔIECAktE17K tumors. A protein was considered to be up-regulated in Trp53ΔIECAktE17K tumors with a fold change >1.5 and was considered to be down-regulated with a fold change <0.75. n ≥ 6 per genotype.
To validate activation of these various signaling cascades on the protein expression level, we performed label-free phosphoproteomic analysis of whole-tumor tissues from Trp53ΔIEC and Trp53ΔIECAktE17K mice by mass spectrometry (MS). With this approach, we were able to identify >3,000 p-sites, of which 119 showed a greater than twofold significant up-regulation of phosphorylation and 84 showed a greater than twofold significant down-regulation of phosphorylation in the tumors of Trp53ΔIECAktE17K mice. IPA analysis of the 119 up- and 84 down-regulated p-sites demonstrated a great overlap with the pathways identified by RNA sequencing (Fig. S1, a and b).
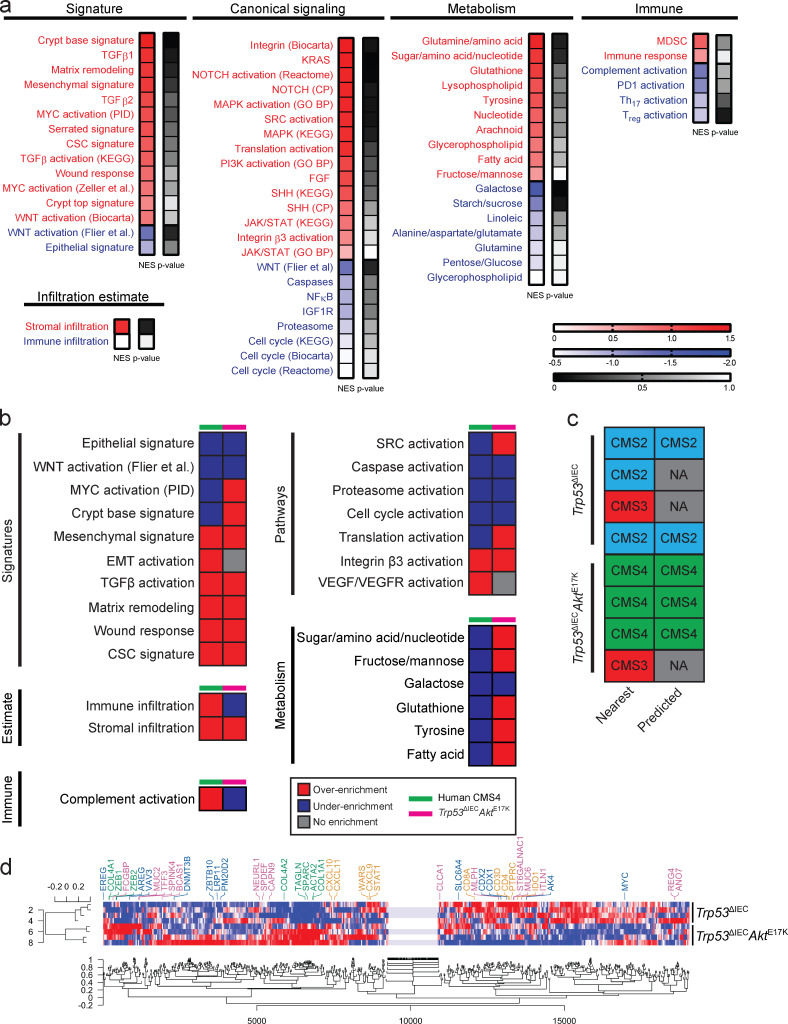
In the past years, extensive research has revealed that tumor characteristics are not only defined by their genetic make-up, but are also greatly influenced by their gene expression profile. Based on this finding, a classification system discriminating four distinct CMS categories of human CRC has been established (Guinney et al., 2015). To characterize Trp53ΔIECAktE17K tumors from this perspective, we performed gene set enrichment analysis (GSEA) using signatures for various signaling pathways, cellular, and other biological processes, which were used to describe the biological characteristics of the four distinct CMS categories (Fig. 2 a). Interestingly, differently activated signaling pathways in Trp53ΔIECAktE17K tumors seemed to substantially overlap with the gene sets typically found to be altered in CMS4 tumors. Human CMS4 tumors are characterized by stromal infiltration, increased mesenchymal and decreased epithelial signature, activation of EMT and TGFβ signaling, down-regulation of WNT signaling, angiogenesis, and matrix remodeling, and the CMS4 subtype is associated with worse overall and relapse-free survival (Guinney et al., 2015). Indeed, Trp53ΔIECAktE17K tumors showed high correlation with human CMS4 tumors in terms of the quantity and quality of stromal infiltration, including increased mesenchymal and decreased epithelial signature (Fig. 2 b). Furthermore, similar to human CMS4 tumors, Trp53ΔIECAktE17K tumors were characterized by enhanced matrix remodeling, wound response, and cancer stem cell signature as well as activation or inhibition of canonical pathways, including pathways such as WNT, caspase, integrin, proteasome, and cell cycle signaling. Despite substantial similarities between human CMS4 and Trp53ΔIECAktE17K tumors, there were also differences, especially in terms of metabolic pathways, but also in SRC and MYC signaling activation, translation activation, immune infiltration, and complement activation (Fig. 2 b). Regarding the immune microenvironment, so far human CMS4 tumors have not been described to possess a unique immune profile that would clearly distinguish them from the other subtypes. However, they are characterized by elevated TGFβ production and related immunosuppression (Dienstmann et al., 2017; Guinney et al., 2015). In line with this, we also found activated TGFβ signaling (Fig. 2, a and b) and significantly decreased numbers of CD3+ T cells in mesenchymal tumors of Trp53ΔIECAktE17K mice (Fig. S1 c). Applying the “single-sample CMS classifier” with the published CMS classifier R package further confirmed that murine Trp53ΔIECAktE17K tumors resembled the human CMS4 subtype, whereas the gene expression profile of control Trp53ΔIEC tumors appeared to be closer to canonical CMS2 tumors (Fig. 2 c). Furthermore, gene module–based coexpression analysis revealed a strong coexpression of stroma-related genes in the Trp53ΔIECAktE17K tumors that was absent in the control Trp53ΔIEC tumors (Fig. 2 d). Indeed, immunohistochemical analysis confirmed significantly enhanced expression of both α-smooth muscle actin (αSMA) and vimentin in Trp53ΔIECAktE17K tumors (Fig. S1 d). Moreover, proteome profiling validated differential expression of various cytokines and chemokines in Trp53ΔIECAktE17K tumors (Fig. S1 e). Thus, AOM-challenged Trp53ΔIECAktE17K mice may comprise a good model to study the pathogenesis of CMS4 tumors.
Figure 2.
Gene expression profile of Trp53ΔIECAktE17K tumors resembles the gene expression profile of human CMS4 tumors. (a) GSEA of the differentially expressed genes in in Trp53ΔIEC and Trp53ΔIECAktE17K tumors using signatures of various signaling pathways, cellular as well as other biological processes. A group of these gene sets was used to describe CMS categories (Guinney et al., 2015). Overenrichment was considered at normalized enrichment score (NES) >0.4 and underenrichment was considered at NES less than −0.4. n = 4 per genotype. (b) Comparison of the over- and underenriched gene sets in Trp53ΔIECAktE17K tumors and human CMS4 CRC, showing expression signatures of particular interest in human CMS4 tumors, especially those associated with stromal infiltration, canonical pathway activation, immune signature, and metabolism. GSEA for human CRC was performed previously, and overenrichment was defined as positive enrichment score and Padj < 0.05; underenrichment was defined as negative enrichment score and Padj < 0.05 (Guinney et al., 2015). For mouse CRC, overenrichment was considered at NES > 0.4 and underenrichment was considered at NES less than −0.4. n = 4 per genotype. (c) CMS subtyping of Trp53ΔIEC and Trp53ΔIECAktE17K tumors using the published single-sample CMS classifier R package (Guinney et al., 2015). Nearest and predicted CMS indicates different statistical stringency. n = 4 per genotype. (d) Verification of assigned subtypes by coexpression analysis. Genes associated with immune response (CMS1) are indicated in orange; colon epithelial differentiation (CMS2) in blue; goblet cell differentiation (CMS3) in pink; and stroma (CMS4) in green. n = 4 per genotype.
NOTCH signaling is activated in Trp53ΔIECAktE17K tumors
IPA suggested that one of the activated pathways in Trp53ΔIECAktE17K tumors consisted of NOTCH signaling (not depicted), which was confirmed by GSEA in a cross-species comparison using human NOTCH signaling–related genes (Figs. 2 a and 3 a). NOTCH signaling is indispensable for the maintenance of intestinal homeostasis and plays a sometimes controversial but essential role in various types of cancer. Although the importance of NOTCH signaling for intestinal homeostasis is undisputed, less is known about its precise role or regulation in CRC, and particularly during CRC progression.
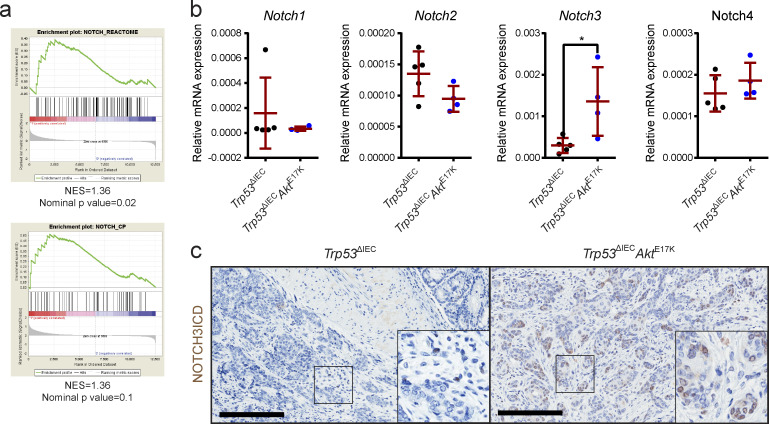
Figure 3.
NOTCH3 signaling is up-regulated in the tumors of Trp53ΔIECAktE17K mice. (a) Cross-species comparison of the differentially regulated genes identified by RNA sequencing to human gene sets related to NOTCH signaling (Guinney et al., 2015) by GSEA. Positive NES indicates a positive enrichment. n = 4 per genotype. (b) Relative gene expression levels of Notch1–4 in the tumors of Trp53ΔIEC and Trp53ΔIECAktE17K mice as determined by quantitative RT-PCR (qRT-PCR). Data are mean ± SD, n ≥ 4 per genotype; *, P < 0.05 by t test. (c) Representative images of N3ICD immunohistochemistry (IHC) on sections from AOM-induced colon tumors in Trp53ΔIEC and Trp53ΔIECAktE17K mice. Scale bars = 200 µm.
Interestingly, we detected a specific up-regulation of Notch3 receptor gene expression, whereas transcripts of the other three members of the NOTCH family (Notch1, Notch2, and Notch4) seemed unaffected by AKT activation (Fig. 3 b). Importantly, the functional activation of NOTCH3 signaling in tumor cells was confirmed by immunohistochemistry using an antibody specific for NOTCH3 intracellular domain (N3ICD; Choy et al., 2017; Fig. 3 c). Collectively, these data clearly demonstrated that NOTCH and specifically NOTCH3 signaling is up-regulated in AOM-induced Trp53ΔIECAktE17K tumors.
AKT signaling up-regulates Notch3 expression
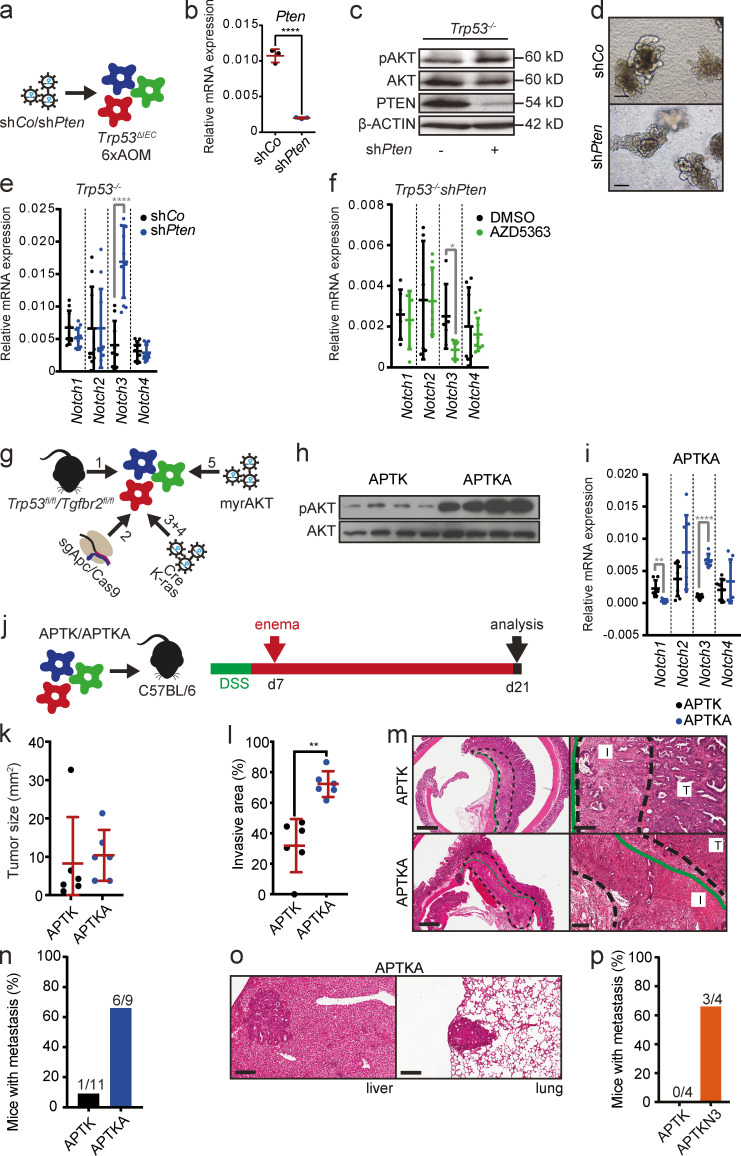
To confirm that AKT increased Notch3 expression directly in a cell-autonomous manner, we generated tumor organoids from AOM-induced tumors from Trp53ΔIEC mice (Trp53−/− organoids). Trp53−/− tumor organoids were transduced with either a control hairpin (shCo) or a hairpin targeting Pten (shPten) to induce AKT activation (Fig. 4 a), and Pten knockdown and consequent AKT activation was confirmed on gene as well as protein expression levels (Fig. 4, b and c). Although organoids with Pten knockdown did not show any obvious differences in morphology and growth (Fig. 4 d), Pten silencing significantly increased Notch3 mRNA levels but left the expression of other NOTCH receptors unaffected (Fig. 4 e). AKT-dependent Notch3 up-regulation was confirmed using the AKT1-3 inhibitor AZD5363 (Fig. 4 f). Comparable results were obtained with two additional independently generated tumor organoids (not depicted). Because Trp53−/− organoids still harbor unknown mutations due to AOM exposure, we aimed to generate organoids with a defined mutation profile (Fig. 4 g). Therefore, we first established colon organoids from unchallenged Trp53fl/flTgfbr2fl/fl mice. Next, Apc deletion was introduced via CRISPR/Cas9 as described previously (Schwank et al., 2013), and organoids were selected via removal of R-spondin from the growth medium (A organoids). To induce Trp53 and Tgfbr2 deletion, A organoids were transduced with a plasmid encoding Cre recombinase, and organoids with Trp53 and Tgfbr2 deletion were selected with the MDM2 inhibitor nutlin3 (APT organoids). We subsequently introduced oncogenic murine KrasG12D into APT organoids by retroviral infection and selected the correctly targeted organoids with puromycin (APTK organoids). Finally, constitutive AKT activation was achieved by introduction of a retroviral vector expressing human myristoylated AKT, and stably infected organoids were selected with hygromycin (APTKA organoids). Activation of AKT signaling in APTKA organoids was confirmed by immunoblot analysis (Fig. 4 h). Importantly, similarly to Pten knockdown in Trp53−/− organoids, introduction of myristoylated AKT into APTK organoids also led to up-regulated Notch3 expression (Fig. 4 i).
Figure 4.
AKT signaling up-regulates Notch3 expression and induces a more aggressive phenotype. (a) Schematic representation of the strategy used for the establishment of Trp53−/− mouse tumor organoids expressing a control hairpin (shCo) or a hairpin against Pten (shPten). (b) Relative mRNA expression of Pten in Trp53−/−shCo and Trp53−/−shPten mouse tumor organoids by qRT-PCR. Data are mean ± SD of n = 1 experiment performed in triplicate; ****, P < 0.0001 by t test. (c) Immunoblot analysis of phosphorylated (p) and total (t) AKT and PTEN in Trp53−/−shCo and Trp53−/−shPten mouse tumor organoids. β-ACTIN was used as loading control. (d) Representative morphology of the Trp53−/−mouse tumor organoids with or without Pten knockdown. Scale bars = 50 µm. (e) Relative mRNA expression levels of Notch1–4 genes in Trp53−/−shCo and Trp53−/−shPten mouse tumor organoids by qRT-PCR. Data are shown for a dataset of n = 3 experiments performed in triplicate. Data are mean ± SD; ****, P < 0.0001 by t test. (f) Relative mRNA expression levels of Notch1–4 genes in DMSO- and AZD5363-treated Trp53−/−shPten mouse tumor organoids by qRT-PCR. Organoids were treated with 3 µM AZD5363 or appropriate concentrations of DMSO for 3 d. Data are shown for a dataset of n = 2 experiments performed in triplicate. Data are mean ± SD; *, P < 0.05 by t test. (g) Schematic representation of the strategy used for the establishment of APTK and APTKA mouse tumor organoids. fl, floxed; myr, myristoylated. (h) Immunoblot analysis of phosphorylated (p) and total (t) AKT in APTK and APTKA mouse tumor organoids. (i) Relative mRNA expression levels of Notch1–4 genes in APTK and APTKA mouse tumor organoids by qRT-PCR. Data are shown for a dataset of n = 2 experiment performed in triplicate and quadruplicate. Data are mean ± SD; **, P < 0.01; ****, P < 0.0001 by t test. (j) Schematic timeline of the strategy used for orthotopic transplantation of APTK and APTKA mouse tumor organoids into C57BL/6 mice. (k) Tumor size upon orthotopic transplantation of APTK and APTKA mouse tumor organoids into C57BL/6 mice. Data are shown for a dataset of n = 2 experiments with n = 6 mice per group. Data are mean ± SD. (l) Extent of the invasive area relative to total tumor size upon orthotopic transplantation of APTK and APTKA mouse tumor organoids into C57BL/6 mice. Data are shown for a dataset of n = 2 experiments with n = 6 mice per group. Data are mean ± SD. **, P < 0.01 by t test. (m) H&E-stained sections of invasive primary tumors in C57BL/6 mice orthotopically transplanted with APTK and APTKA mouse tumor organoids. T, noninvasive tumor area; I, invasive area. Dashed line demarcates the invasive area; continuous green line marks the inner border of the external muscle layer of the colon wall. Scale bars = 900 µm (left); 200 µm (right). (n) Frequency of mice with metastasis upon orthotopic transplantation of APTK and APTKA mouse tumor organoids into C57BL/6 mice. Numbers indicate mice with metastasis/all mice. Data are shown for a dataset of n = 3 experiments with n ≥ 9 mice per group. (o) H&E-stained sections of liver and lung metastasis in C57BL/6 mice orthotopically transplanted with APTKA mouse tumor organoids. Scale bars = 200 µm. (p) Frequency of mice with metastasis upon orthotopic transplantation of APTK and APTKN3 mouse tumor organoids into Rag1 mice. Numbers indicate the number of mice with metastasis/total number of mice. Data are shown for a dataset of n = 1 experiments with n = 4 mice per group.
Next, we wanted to examine whether activation of AKT signaling and consequent Notch3 up-regulation was sufficient to enhance invasion and metastasis using these organoids in an orthotopic transplantation model. In this model, first, colonic epithelial damage is induced by administration of dextran sodium sulfate (DSS) for five consecutive days. This step is essential for organoid engraftment (Yui et al., 2012). 2 d after DSS treatment, APTK and APTKA organoids were dissociated into single-cell suspensions and instilled into the colon of C57BL/6 mice (Fig. 4 j). Tumor growth was monitored by endoscopy, and tumors were histologically analyzed 21 d after transplantation. As expected, both APTK and APTKA organoids formed a single primary colorectal tumor that spontaneously metastasized. Similar to the AOM model, primary organoid-derived tumor growth was comparable between APTK and APTKA tumors (Fig. 4 k). However, although both APTK and APTKA organoids were capable of forming invasive tumors in vivo, the invasive area of APTKA tumors was significantly larger compared with APTK tumors (Fig. 4, l and m). Most importantly, mice with APTKA tumors developed lung and liver metastases much more frequently than mice with APTK tumors (Fig. 4, n and o). Moreover, overexpression of N3ICD in APTK organoids (APTKN3) dramatically enhanced metastasis formation even in the absence of constitutively active AKT (Fig. 4 p), strongly supporting the notion that AKT-dependent Notch3 up-regulation supports formation of metastatic disease.
NOTCH3 signaling drives a gene expression program associated with tumor cell survival and motility, and NOTCH3 inhibition blocks tumor progression
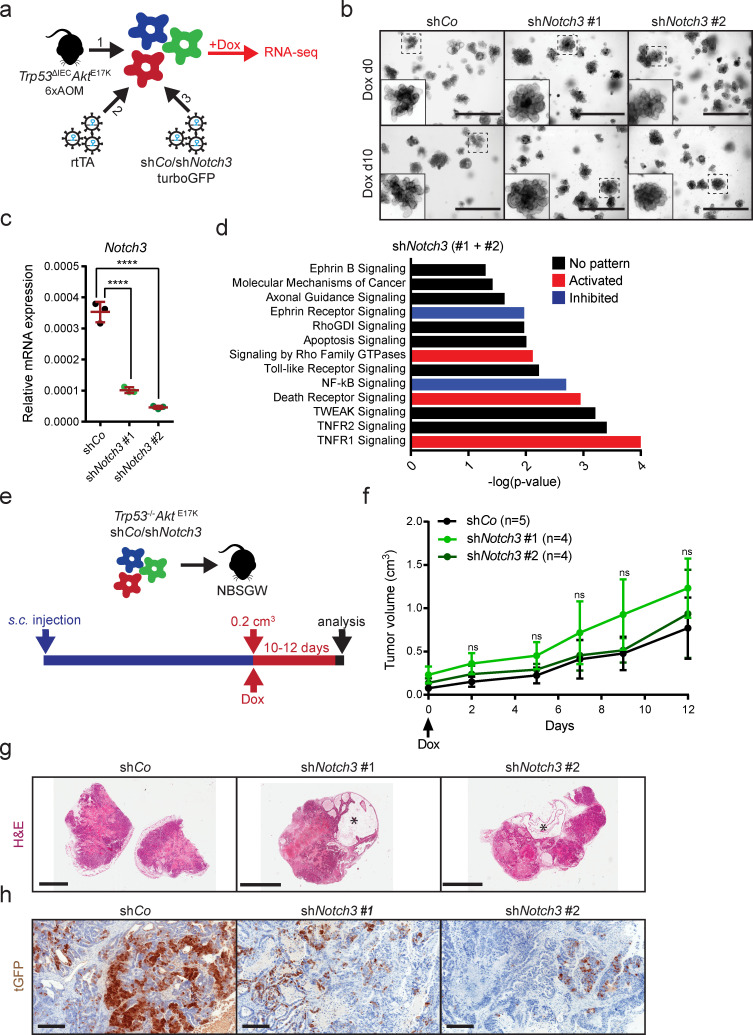
To investigate Notch3-mediated downstream effects, we first transduced AOM-induced tumor organoids from Trp53ΔIECAktE17K mice (Trp53−/−AktE17K organoids) with a plasmid encoding a reverse transactivator to allow doxycycline-inducible expression of shRNAs. Next, successfully targeted organoids were infected with a retrovirus encoding a control hairpin (shCo) or two different hairpins targeting Notch3. All constructs contained turboGFP (tGFP) as a reporter (Fig. 5 a). This approach enabled temporal control of Notch3 gene expression by the addition or withdrawal of doxycycline.
Figure 5.
NOTCH3 signaling regulates tumor cell invasion and survival. (a) Schematic depiction of the strategy used for the generation of Trp53−/−AktE17K organoids with doxycycline-inducible Notch3 knockdown. RNA was isolated and used for RNA sequencing after culture in doxycycline-containing medium for 10 d. (b) Representative morphology of Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids without doxycycline and after culture in doxycycline-containing medium for 10 d. Scale bars = 1,000 µm. (c) Relative Notch3 mRNA expression in Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids after culture in doxycycline-containing medium for 10 d by qRT-PCR. Data are mean ± SD; ****, P ≤ 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparison test. Data are shown for a dataset of n = 1 experiment performed in triplicate. (d) Differentially regulated pathways in response to Notch3 silencing. Predictions were made based on RNA-sequencing results of Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids after culture in doxycycline-containing medium for 10 d using IPA. P < 0.05 with right-tailed Fisher’s exact test; activation: z-score > 0, inhibition: z-score < 0, no pattern: pathway is differentially regulated, but z-score is not available. (e) Schematic depiction of the strategy used for subcutaneous injection of Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids and doxycycline administration in the drinking water. (f) Representative growth curves of subcutaneous tumors in NBSGW mice transplanted with Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids. Tumor size is shown starting from the first day of doxycycline administration (day 0). Doxycycline was administered for 12 d. Experiment was performed in three independent sets for 10 or 12 d, data are shown for one set of 12 d. Data are mean ± SEM. Not significant (ns) by one-way ANOVA followed by Dunnett’s multiple comparison test. n ≥ 4 per group. (g) Representative images of H&E-stained sections of subcutaneous tumors from NBSGW mice transplanted with Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids at day 10 after the start of doxycycline administration. Stars mark large cysts filled with liquid. Scale bars = 5 mm. (h) Representative images of tGFP immunohistochemistry (IHC) on sections from subcutaneous tumors of NBSGW mice transplanted with Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids at day 10 after the start of doxycycline administration. Scale bars = 200 µm.
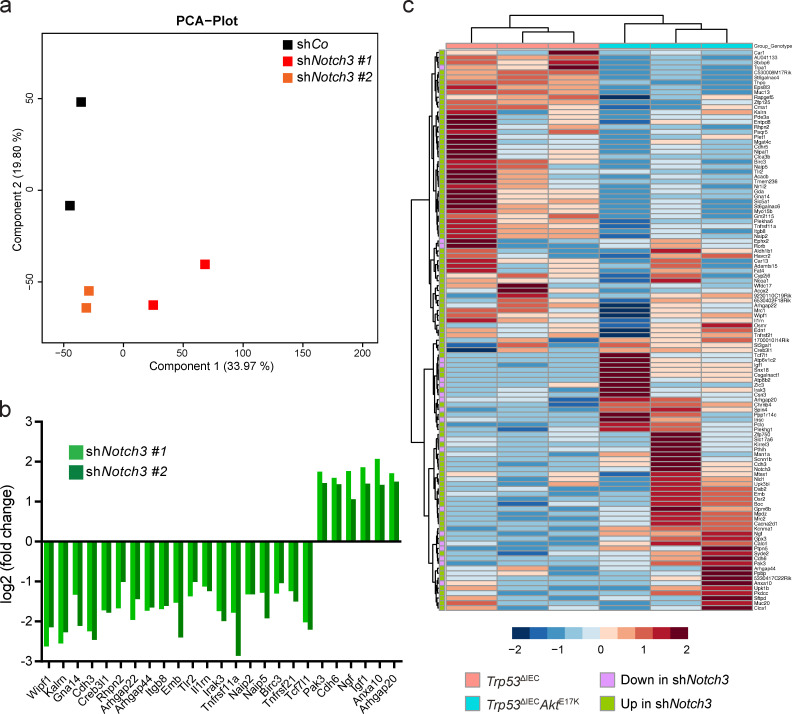
We then cultured Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids in the presence of doxycycline for 10 d without observing any obvious effect on organoid growth rate or morphology (Fig. 5 b), while successful knockdown of Notch3 was confirmed (Fig. 5 c). At day 10, RNA was collected from Trp53−/−AktE17K/shCo and Trp53−/−AktE17K/shNotch3 organoids and subjected to next-generation RNA sequencing, which revealed 301 (shNotch3 #1) and 244 (shNotch3 #2) differentially regulated genes, of which 113 genes were common. Principal component analysis showed that Trp53−/−AktE17K organoids expressing shNotch3 #1 and #2 clustered together, and they substantially differed from the Trpr53−/−AktE17K organoids expressing the control hairpin (Fig. S2 a). Notch3 knockdown decreased the expression of genes encoding proteins associated with the cytoskeleton or cytoskeletal rearrangements, such as Wipf, Cdh3, Rhpn2, Kalrn, and Arhgap22; apoptosis, such as Naip2, Birc3, and Tnfrsf21; and inflammation, such as Tlr2, Il1rn, and Irak3 (Fig. S2 b). Interestingly, only a few genes were up-regulated by Notch3 knock-down. These included genes encoding proteins involved in the assembly and regulation of the cytoskeleton, such as Pak3 and Cdh6, the proapoptotic gene Anxa10, and secreted growth factors, such as Igf and Ngf (Fig. S2 b). IPA predicted differentially regulated pathways in response to Notch3 silencing, including pathways with a potential role in invasion and metastasis, such as signaling by Rho family GTPases, Rho GDP-dissociation inhibitor signaling, and Ephrin signaling (Fig. 5 d). In addition, a significant proportion of differentially regulated pathways were related to cell death and survival, such as TNFR1 and 2 signaling, TWEAK signaling, death receptor signaling, and apoptosis signaling (Fig. 5 d). The regulation of the NF-κB and TLR pathways indicated that Notch3 might play a role in the formation of an inflammatory tumor microenvironment (Fig. 5 d). To demonstrate that NOTCH3 target genes are regulated in response to AKT activation, we investigated the expression of genes that were up- or down-regulated upon Notch3 knockdown in Trp53ΔIEC tumors and CMS4-like Trp53ΔIECAktE17K tumors. Although gene expression differed between organoid and tumors, heatmap and cluster analysis demonstrated that many of the Notch3-regulated genes were differentially expressed between the two genotypes (Fig. S2 c). More importantly, samples from Trp53ΔIEC and Trp53ΔIECAktE17K mice clustered together, indicating that in our model, AKT is upstream of NOTCH3 and its activity affects the expression of NOTCH3 target genes.
Figure S2.
Analysis of gene expression changes in response to Notch3 silencing in Trp53−/−AktE17K organoids. (a) Principal component analysis (PCA) of the RNA-sequencing results of Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids cultured in the presence of doxycycline for 10 d. (b) Significant gene expression changes induced by Notch3 knockdown for selected genes as determined by RNA sequencing. Changes in the gene expression in Trp53−/−AktE17KshNotch3 organoids were determined relative to control Trp53−/−AktE17KshCo organoids. Experiment was performed in duplicate; Padj < 0.05. (c) Heatmap and cluster analysis showing expression z-scores of NOTCH3 target genes in AOM-treated CMS2-like Trp53ΔIEC and CMS4-like Trp53ΔIECAktE17K tumors. n = 3 mice per genotype.
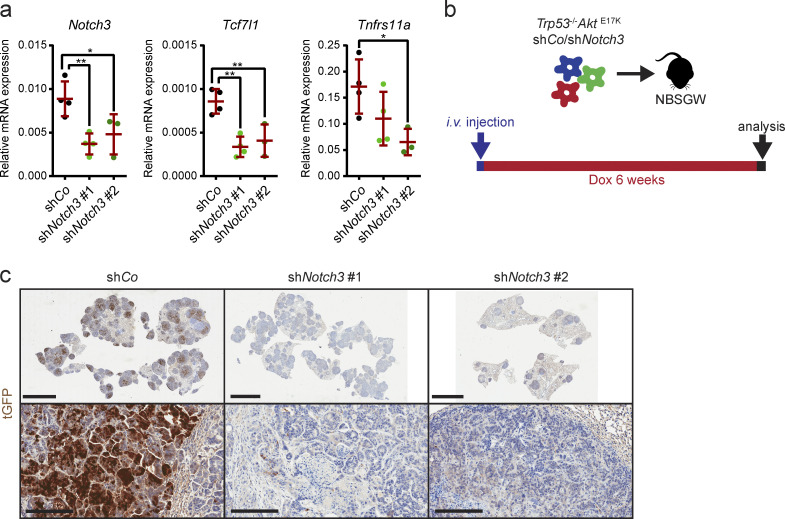
To examine the consequence of Notch3 knockdown in vivo, Trp53−/−AktE17K/shCo and Trp53−/−AktE17K/shNotch3 organoids were injected subcutaneously into immunodeficient NBSGW mice (Fig. 5 e). Although subcutaneous tumors grew relatively slowly, all mice developed noticeable tumors ∼5 wk after injection. When tumors reached a size of ∼0.2 cm3, mice were given doxycycline in drinking water for a period of 10–12 d to induce hairpin expression (Fig. 5 e). Surprisingly, Notch3 knockdown did not affect relative increase in tumor growth (Fig. 5 f), despite a substantial Notch3 knockdown and reduced expression of genes that were identified as Notch3 targets by RNA sequencing (Fig. S3 a). Histological examination revealed that tumors with Notch3 knockdown contained large necrotic areas and liquid-filled cysts (Fig. 5 g) and marked reduction of tGFP-expressing tumor cells, indicating hairpin loss (Fig. 5 h). Similar results were obtained when Trp53−/−AktE17K/shCo and Trp53−/−AktE17K/shNotch3 organoids were injected i.v. to induce lung metastasis (Fig. S3 b). Also in this model, tGFP+ shNotch3–expressing cells were eliminated, indicating that Notch3 knockdown in vivo is unfavorable and cells with diminished Notch3 expression are outcompeted (Fig. S3 c). Thus, shRNA-mediated Notch3 knockdown is not suitable for a detailed in vivo analysis of NOTCH3 function in tumor progression, and NOTCH3 seems essential for tumor cell survival.
Figure S3.
Notch3 silencing in s.c. and i.v. tumors. (a) Relative mRNA expression level of the indicated genes in s.c. tumors of NBSGW mice transplanted with Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids day 10 after the start of doxycycline administration by qRT-PCR. Data are mean ± SD; n ≥ 3 per group; *, P < 0.05; **, P < 0.01 by one-way ANOVA followed by Dunnett’s multiple comparison test. Data are shown for a dataset of n = 1 experiment. (b) Schematic depiction of the strategy used for i.v. injection of Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids and doxycycline administration. (c) Representative images of tGFP immunohistochemistry (IHC) on sections from lung tumors of NBSGW mice i.v. injected with Trp53−/−AktE17KshCo or Trp53−/−AktE17KshNotch3 organoids 6 wk after the start of doxycycline administration. Scale bars = 5 mm (upper row); 200 µm (lower row).
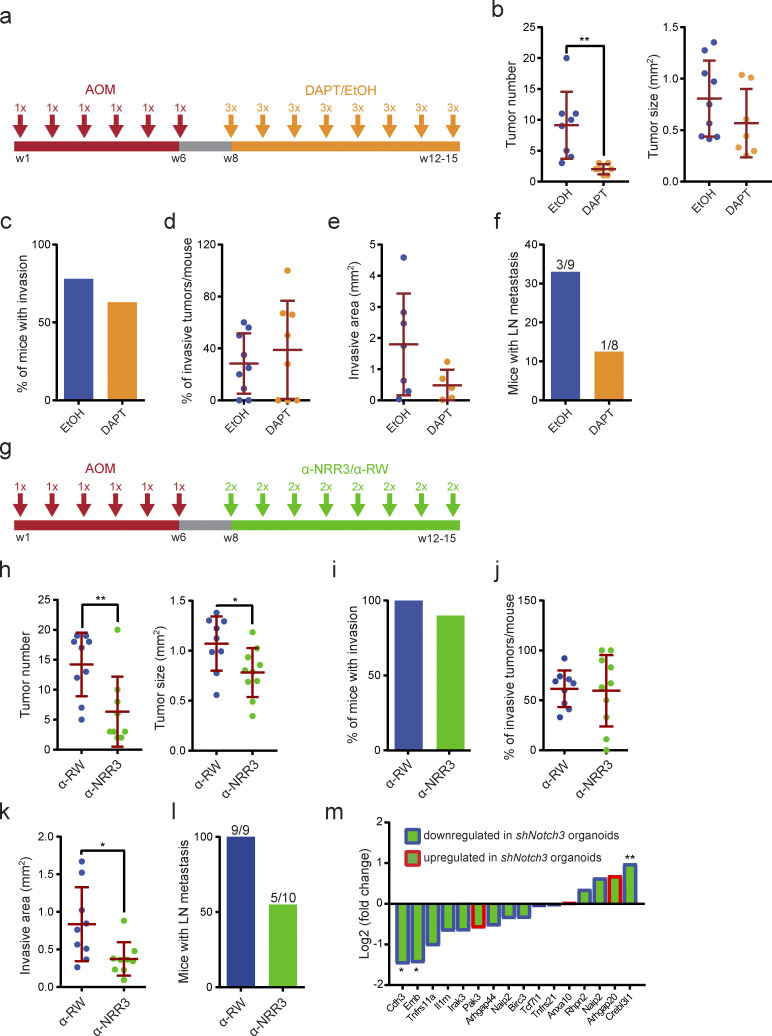
To circumvent the technical drawback of the knockdown approach yet examine whether targeting NOTCH3 could represent a therapeutic option of CMS4-like tumors, we treated AOM-induced Trp53ΔIECAktE17K mice with the γ-secretase inhibitor DAPT. γ-Secretase is a proteolytic enzyme complex required for the liberation of the intracellular domain of all NOTCH receptors; therefore, DAPT blocks the function of all four NOTCH receptors. AOM-treated Trp53ΔIECAktE17K mice were randomly divided into two groups and treated with either DAPT or control ethanol/oil mixture by oral gavage in a 3-d-on/4-d-off regimen starting from week 8 (Fig. 6 a). Application of γ-secretase inhibitors in a 3-d-on/4-d-off schedule was reported to minimize intestinal toxicity associated with γ-secretase inhibition and enable long-term administration of DAPT (Strosberg et al., 2012; Tammam et al., 2009). Mice were sacrificed when they showed signs of disease, such as weight loss, diarrhea, and rectal bleeding. DAPT treatment strongly interfered with tumor initiation, as reflected by the significantly lower number of tumors in the DAPT-treated group, but had little effect on tumor size (Fig. 6 b). Both DAPT-treated and control mice developed invasive tumors (Fig. 6 c), and the frequency of the invasive tumors was comparable between the two groups (Fig. 6 d). However, DAPT administration reduced the extent of the invasive area, although the difference did not reach statistical significance (Fig. 6 e). More importantly, DAPT treatment strongly reduced the number of mice with lymph node metastasis compared with the control group (Fig. 6 f).
Figure 6.
In vivo NOTCH3 inhibition reduces colorectal tumor development and progression. (a) Schematic depiction of the treatment strategy of AOM-induced Trp53ΔIECAktE17K mice with DAPT or ethanol/oil mixture. DAPT was administered in a 3-d-on/4-d-off regimen at 10 mg/kg. (b) AOM-induced primary tumor number and primary tumor size in Trp53ΔIECAktE17K mice treated with DAPT or ethanol/oil mixture. Data are shown for a dataset of n = 2 experiments with n ≥ 8 mice per group. Data are mean ± SD; **, P < 0.01 by t test. Grubbs’ test with α = 0.05 was used to remove outliers. (c) Frequency of mice with AOM-induced invasive tumors in Trp53ΔIECAktE17K mice treated with DAPT or ethanol/oil mixture. Data are shown for a dataset of n = 2 experiments with n ≥ 8 mice per group. (d) Frequency of AOM-induced invasive tumors per mouse in Trp53ΔIECAktE17K mice treated with DAPT or ethanol/oil mixture. Data are shown for a dataset of n = 2 experiments with n ≥ 8 mice per group. Data are mean ± SD; not significant by t test. Grubbs’ test with α = 0.05 was used to remove outliers. (e) Extent of the invasive area in AOM-induced Trp53ΔIECAktE17K mice treated with DAPT or ethanol/oil mixture. Data are shown for a dataset of n = 2 experiments with n ≥ 5 mice per group. Data are mean ± SD; not significant by t test. Grubbs’ test with α = 0.05 was used to remove outliers. (f) Frequency of mice with lymph node metastasis in AOM-induced Trp53ΔIECAktE17K mice treated with DAPT or ethanol/oil mixture. Data are shown for a dataset of n = 2 experiments with n ≥ 8 mice per group. Numbers indicate the number of mice with metastasis/total number of mice. (g) Schematic depiction of the treatment strategy of AOM-induced Trp53ΔIECAktE17K mice with α-NRR3 or α-RW. α-NRR3 or α-RW was administered twice per week at 30 mg/kg. (h) AOM-induced primary tumor number and primary tumor size in Trp53ΔIECAktE17K mice treated with α-NRR3 or α-RW. Data are shown for a dataset of n = 2 experiments with n ≥ 9 mice per group. Data are mean ± SD; *, P < 0.05 by t test; **, P < 0.01. Grubbs’ test with α = 0.05 was used to remove outliers. (i) Frequency of mice with AOM-induced invasive tumors in Trp53ΔIECAktE17K mice treated with α-NRR3 or α-RW. Data are shown for a dataset of n = 2 experiments with n ≥ 9 mice per group. (j) Frequency of AOM-induced invasive tumors per mouse in Trp53ΔIECAktE17K mice treated with α-NRR3 or α-RW. Data are shown for a dataset of n = 2 experiments with n ≥ 9 mice per group. Data are mean ± SD; not significant by t test. Grubbs’ test with α = 0.05 was used to remove outliers. (k) Extent of the invasive area in AOM-induced Trp53ΔIECAktE17K mice treated with α-NRR3 or α-RW. Data are shown for a dataset of n = 2 experiments with n ≥ 9 mice per group. Data are mean ± SD; *, P < 0.05 by t test. Grubbs’ test with α = 0.05 was used to remove outliers. (l) Frequency of mice with lymph node metastasis in AOM-induced Trp53ΔIECAktE17K mice treated with α-NRR3 or α-RW. Data are shown for a dataset of n = 2 experiments with n ≥ 9 mice per group. Numbers indicate mice with metastasis/all mice. (m) Log2 fold change of the expression of the indicated NOTCH3 target genes in AOM-induced Trp53ΔIECAktE17K mice treated with α-NRR3 relative to α-RW treated mice by qRT-PCR. Genes with a blue frame were found to be down-regulated in Trp53−/−AktE17KshNotch3 organoids compared with control Trp53−/−AktE17KshCo organoids. Genes with a red frame were found to be up-regulated in Trp53−/−AktE17KshNotch3 organoids compared with control Trp53−/−AktE17KshCo organoids. Data are shown for a dataset of n = 1 experiments with n = 5 mice per group. *, P < 0.05; **, P < 0.01 by t test.
DAPT inhibits all four NOTCH receptors, and its long-term administration might be associated with adverse side effects. Moreover, ubiquitous inhibition of NOTCH signaling might even be protumorigenic in some tissues (Nowell and Radtke, 2017). Therefore, we aimed to target NOTCH3 in a more direct manner and treated AOM-induced Trp53ΔIECAktE17K mice with a NOTCH3 antagonist antibody (α-NRR3; Yu et al., 2020). α-NRR3 targets the negative regulatory region (NRR) of the NOTCH3 receptor that prevents proteolytic cleavage and subsequent activation of the NOTCH3 receptor (Choy et al., 2017; Li et al., 2008). AOM-challenged Trp53ΔIECAktE17K mice were again randomly divided into two groups and received either α-NNR3 or α-RW control antibody i.p. twice a week starting from week 8 (Fig. 6 g). α-NRR3 treatment significantly reduced both multiplicity and size of colon tumors (Fig. 6 h). Similar numbers of α-NRR3– and α-RW–treated mice developed invasive tumors (Fig. 6 i), and the frequency of invasive tumors per mouse was comparable between the two groups (Fig. 6 j). However, α-NRR3 administration significantly reduced the extent of invasion (Fig. 6 k) and markedly blocked occurrence of lymph node metastases (Fig. 6 l). Gene expression analysis of α-NRR3–treated tumors confirmed down-regulation of several of the NOTCH3 target genes that we had identified by Notch3 knockdown compared with α-RW–treated control tumors (Fig. 6 m). Thus, selectively blocking NOTCH3 seems sufficient to block tumor progression in this model.
NOTCH3 up-regulation is associated with human CRC progression and the CMS4 subtype
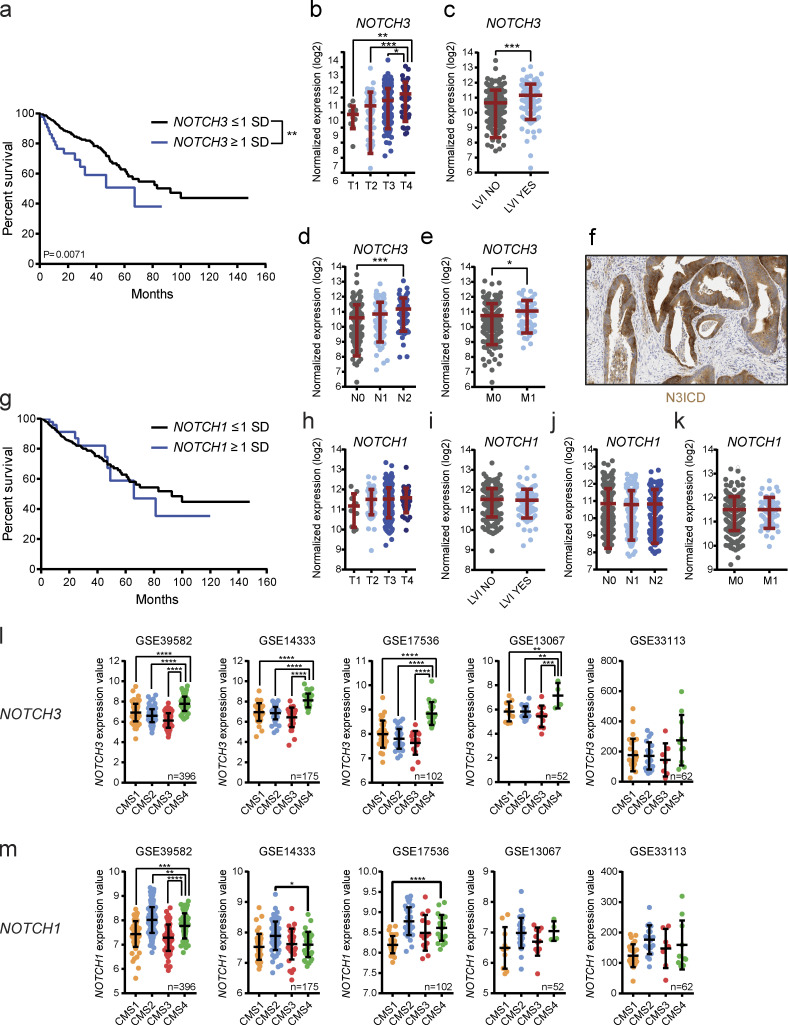
To investigate whether NOTCH3 may be up-regulated in a stage- or tumor subtype–specific manner in human CRC, genomic and clinical data from The Cancer Genome Atlas (TCGA) Research Network database were analyzed using the cBio Cancer Genomic Portal, an open-access resource for the analysis and visualization of large-scale multidimensional genomic, transcriptomic, and expression data (Cerami et al., 2012; Gao et al., 2013). As hypothesized, CRC patients with elevated NOTCH3 mRNA expression have significantly worse overall survival compared with patients with unaltered or low expression of NOTCH3 mRNA (Fig. 7 a). Furthermore, NOTCH3 mRNA gradually increased with the tumor stage (T) and showed a significant positive correlation with lymphovascular invasion (LVI), lymphatic metastatic status (N), and distant metastasis (M; Fig. 7, b–e). Immunohistochemistry using an antibody specific for human active NOTCH3 (N3ICD) in a separate cohort of 28 CRC patients confirmed epithelial NOTCH3 activation in 16 of 19 patient samples that stained positive (Fig. 7 f). In 9 of these 19 patient samples, we also detected stromal expression. However, unlike NOTCH3, NOTCH1 mRNA expression did not show any association with CRC patient survival (Fig. 7 g). Accordingly, NOTCH1 mRNA expression was not associated with tumor stage, LVI, lymphatic metastatic status (N), or distant metastasis (M; Fig. 7, h–k). Notably, analysis of CMS-classified GEO human CRC datasets revealed that increased NOTCH3 expression is specifically and significantly associated with the CMS4 subtype (Fig. 7 l), whereas NOTCH1 expression did not show any consistent correlation with any of the four CMS categories (Fig. 7 m). These data clearly show that NOTCH3 up-regulation is associated with human CRC progression and the poor-prognosis CMS4 subtype, thus suggesting that NOTCH3 may represent a valuable target for this CRC subtype.
Figure 7.
High NOTCH3 expression correlates with CRC progression and CMS4 subtype in CRC patients. (a) Kaplan–Meier survival curve of CRC patients with NOTCH3 under- or overexpression. Data were generated by using the cBioPortal and TCGA RNA sequencing V2 dataset with a z-score threshold of ±1. Underexpression was considered for expression values smaller than one SD below the mean (n = 327), overexpression was considered for expression values greater than one SD above the mean (n = 46). Curve comparison was performed with log-rank (Mantel–Cox) test. (b–e) NOTCH3 normalized expression in CRC patients according to the tumor stage (T1–T4; b), LVI (c), lymphatic metastasis (N0–N2; d), and distant metastasis (M0–M1; e). Data were generated by using the cBioPortal and TCGA RNA sequencing V2 dataset with a z-score threshold of ±1. Data are mean ± SD, n = 374. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test or one-way ANOVA followed by Tukey’s multiple comparison test. (f) Representative image of N3ICD immunohistochemistry (IHC) in human CRC. Scale bar = 100 µm. (g) Kaplan–Meier survival curve of CRC patients with NOTCH1 under- or overexpression. Data were generated by using the cBioPortal and TCGA RNA sequencing V2 dataset with a z-score threshold of ±1. Underexpression was considered for expression values smaller than one SD below the mean (n = 320), overexpression was considered for expression values greater than one SD above the mean (n = 52). Curve comparison was performed with log-rank (Mantel–Cox) test. (h–k) NOTCH1 normalized expression in CRC patients according to the tumor stage (T1–T4; h), LVI (i), lymphatic metastasis (N0–N2; j), and distant metastasis (M0–M1; k). Data were generated by using the cBioPortal and TCGA RNA sequencing V2 dataset with a z-score threshold of ±1. Data are mean ± SD; n = 374. Not significant by t test or one-way ANOVA followed by Tukey’s multiple comparison test. (l) NOTCH3 expression in the indicated human CRC datasets according to the CMS category. Data are mean ± SD; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparison test. (m) NOTCH1 expression in the indicated human CRC datasets according to the CMS category. Data are mean ± SD; *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparison test.
Discussion
The majority of anticancer compounds that show promising efficacy in preclinical testing fail often in early clinical trials and are only very rarely approved for cancer treatment (Nass et al., 2018). One reason for the extremely high failure rate of oncology drug candidates is the lack of appropriate preclinical models. For CRC, preclinical models include CRC cell lines and their xenografts, patient-derived organoids and xenografts, and GEMMs (McIntyre et al., 2015). Moreover, the fact that, for example, in CMS4 tumors, tumor progression is defined by the tumor stroma rather than a particular genetic profile within the actual cancer cells underlines the difficulties in creating adequate tumor models. Thus, to date only few immune-competent GEMMs exist that truly recapitulate all the steps of CRC progression and faithfully mimic advanced metastatic human CRC.
In this study, we analyzed a mouse model in which concomitant inactivation of TP53 signaling and activation of PI3K–AKT signaling in combination with chemical carcinogenesis induces invasive colon tumors in 100% of the mice. Importantly, these invasive tumors give rise to lymph node metastasis with a very high frequency (80%) within <13 wk. Of note, tumor cells from lymph node metastases can reenter the circulation and give rise to distant metastasis, and do not represent dead ends as previously believed (Brown et al., 2018; Pereira et al., 2018). Other metastatic mouse models of CRC include models with various combinations of inactivating genetic modifications in main tumor suppressors, such as Apc, Trp53, Pten, Tgfbr2, and Cdkn2a, and activating mutations in oncogenes, such as Kras, Braf, and Notch1. However, in contrast to AOM-treated Trp53ΔIECAktE17K mice, none of these mouse models develop invasive tumors with 100% penetrance, and the frequency of lymph node and distant metastasis is relatively low (<50%; Jackstadt and Sansom, 2016). Gene expression and phosphoproteomic profiling of AOM-induced Trp53ΔIEC and Trp53ΔIECAktE17K tumors demonstrated that AKT signaling activation triggered inflammatory and proinvasive features. Differential regulation of pathways such as adherens and tight junction signaling might indicate a loss of epithelial barrier integrity, which has been shown to contribute to the formation of an inflammatory tumor microenvironment and drive CRC growth and progression (Grivennikov et al., 2012). Further analysis of gene expression data also suggested altered recruitment of a wide range of immune cells (granulocyte and agranulocyte adhesion and diapedesis) in response to AKT activation, possibly leading to the substantially different inflammatory tumor microenvironment in Trp53ΔIEC and Trp53ΔIECAktE17K tumors.
Most importantly, gene expression profiling of AOM-induced Trp53ΔIEC and Trp53ΔIECAktE17K tumors also showed that Trp53ΔIECAktE17K mice mimic advanced human CRC not only phenotypically, but also on a transcriptional level. We were able to demonstrate by several different approaches that the gene expression profile of the Trp53ΔIECAktE17K tumors is highly similar to the gene expression profile of human CMS4 tumors. This is of particular importance, as patients with CMS4 tumors have the worst progression-free and overall survival (Guinney et al., 2015). The CMS4 subgroup is characterized by the high expression of stroma-related genes, decreased epithelial signature, and activation of EMT, integrin, and TGFβ signaling. Moreover, CMS4 tumors, similarly to Trp53ΔIECAktE17K tumors, possess a prometastatic, angiogenic, inflammatory, and immunosuppressive microenvironment (Becht et al., 2016). Importantly, our results further suggest that, similarly to the human CMS4 subtype, Trp53ΔIECAktE17K tumors also possess an immune-suppressed microenvironment characterized by elevated TGFβ and decreased T cell infiltration.
Based on the above characteristics, several therapeutic approaches have been suggested for the treatment of CMS4 tumors, including anti-integrin therapy (Élez et al., 2015), anti-inflammatory therapy, antiangiogenic therapy (Becht et al., 2016), and TGFβ inhibition (Calon et al., 2015; Tauriello et al., 2018). Among these treatment modalities, TGFβ inhibition shows the most promise, and it is already being tested in a phase II clinical trial in combination with chemotherapy in patients with CMS4 signature (MoTriColor, 2015). The observation that TGFβ signaling was activated in Trp53ΔIECAktE17K tumors suggests that, in addition to NOTCH3 signaling, TGFβ signaling could contribute to the CMS4 phenotype, and that blocking TGFβ may potentially impact tumor progression in our model as well. However, TGFβ signaling has a dual role in CRC, as it is a tumor promoter in the stroma and a tumor suppressor in the tumor cells at the same time (Calon et al., 2015; David et al., 2016), and therefore TGFβ inhibitors should be applied only after careful consideration. Hence, there is a clear need for more therapeutic options for the treatment of CMS4 tumors. In fact, CMS4 tumors are not characterized by a high frequency of PTEN, PI3K, or AKT mutations. Instead, we suggest that stroma-derived signals, including IL-6 family members such as IL-11, fuel gp130-dependent PI3K–AKT activation even in the absence of mutations (J. Varga and F.R. Greten, unpublished data) to induce Notch3 transcription. Undoubtedly, AKT activation will lead to a wide range of additional downstream effectors, yet we suggest that Notch3 represents one important target considering that overexpression of N3ICD suffices to induce distant metastases in an organoid-based orthotopic transplantation setting.
An interplay between PI3K–AKT signaling and NOTCH3 signaling has already started to appear. In stratified epithelia, PDK1, which controls both PKC and AKT, is important for asymmetric cell divisions in the basal keratinocytes. Keratinocytes that are deficient for PDK1 fail to undergo asymmetric cell divisions and are unable to activate the NOTCH-induced differentiation program. Reintroduction of active NOTCH3 rescued defective asymmetric cell divisions and restored proper keratinocyte differentiation (Dainichi et al., 2016). Data from human cancer samples also support the link between PI3K–AKT and NOTCH3. NOTCH3 overexpression and nuclear localization correlate with phospho-AKT levels in human pancreatic ductal adenocarcinoma (Doucas et al., 2008). NOTCH3 knockdown in cholangiocarcinoma decreases the transcription of several genes related to AKT signaling and diminishes the activation of AKT and mTOR signaling (Guest et al., 2016).
Recently, a high NOTCH score was suggested to be associated with CMS4 tumors, and NOTCH1 activation was detected in liver metastases (Jackstadt et al., 2019). Here we demonstrate that particularly increased NOTCH3 expression in primary CRC associates with worse overall survival and increases with tumor progression. Most importantly, NOTCH3 expression was highest in the CMS4 tumors. Although relatively few studies have focused specifically on the role of NOTCH3 in human CRC, the contribution of NOTCH3 to CRC progression is beginning to emerge. Increased NOTCH3 expression has been shown to characterize primary and metastatic CRC (Serafin et al., 2011), and down-regulation of NOTCH3 reduces the invasiveness of human CRC cells (Wang et al., 2015).
When we treated Trp53ΔIECAktE17K mice with the γ-secretase inhibitor DAPT or the NOTCH3 antagonistic antibody α-NRR3, we were able to significantly block tumor progression. The fact that NOTCH3 inhibition is equally as, if not more, effective in reducing invasion and metastasis compared with DAPT administration suggests that it is primarily signaling via NOTCH3 (but not other NOTCH receptors), which drives tumor progression. This finding is important, because broad inhibition of NOTCH signaling by γ-secretase inhibitors is associated with adverse side effects, and in certain tissues it can be even protumorigenic (Doody et al., 2013). Because of these considerations, antibodies that target individual NOTCH receptors have been developed and are being tested in preclinical models (Choy et al., 2017; Gurney and Hoey, 2011; Lafkas et al., 2015; Li et al., 2008; Wu et al., 2010). NOTCH3 is also highly expressed by vascular smooth muscle cells and pericytes and is important for their homeostasis (Fouillade et al., 2012; Siebel and Lendahl, 2017). We could also detect NOTCH3 activation in stromal cells of CRC patients. Hence, it is possible that NOTCH3 inhibition affects not only tumor cells but also stromal cells, which could further contribute to the metastasis-reducing effect of NOTCH3 inhibition. Collectively, our findings suggest that NOTCH3 inhibition might be an effective therapeutic approach in the management of advanced human CRC, particularly CMS4 tumors.
Materials and methods
Mouse strains and treatments
For the generation of LSL-AktE17K mice an Akt1E17K cDNA (codon optimized for expression in mouse and including an N-terminal Flag tag) was cloned after a loxP-flanked stop cassette (LSL) into a Rosa26 targeting vector (Pechloff et al., 2010). The targeting vector was electroporated into 129J/Ola embryonic stem (ES) cells, and successfully targeted ES cells were selected to generate chimeric mice which were bred for germline transmission (S. Wanninger and J. Ruland, unpublished data). LSL-AktE17K mice were crossed to Trp53ΔIEC mice (Schwitalla et al., 2013) to generate Trp53ΔIECAktE17K double mutants. Trp53ΔIECAktE17K mice used for initial characterization were backcrossed to FvB background for at least four generations, and Trp53ΔIEC mice on FvB background were used as controls. For the treatment with DAPT and α-NRR3, Trp53ΔIECAktE17K mice on mixed genetic background were also used. For the isolation of colon organoids, Trp53fl/fl mice (JAX; 008462) were crossed to Tgfbr2fl/fl mice (JAX; 012603) harboring loxP sites flanking exon 4 of the Tgfbr2 gene. Trp53fl/flTgfbr2fl/fl mice were on C57BL/6J background. To induce colon tumors, mice were injected with 10 mg/kg AOM i.p. once per week for a period of 6 wk. DAPT was applied in 5% EtOH/oil mixture at a dose of 10 mg/kg in a 3-d-on/4-d-off regimen by oral gavage and given to AOM-treated Trp53ΔIECAktE17K mice from week 8 until the time of sacrifice. NOTCH3 blocking α-NRR3 and the control α-RW antibodies (Genentech) were injected i.p. twice a week at 30 mg/kg. AOM-injected Trp53ΔIECAktE17K mice were treated from week 8 until the time of sacrifice. All mouse experiments were reviewed and approved by the Regierungspräsidium Darmstadt, Darmstadt, Germany.
Subcutaneous, lung metastasis, and orthotopic mouse models
To generate subcutaneous tumors, NBSGW (McIntosh et al., 2015) mice were injected s.c. with organoid single-cell suspension containing 300,000 cells in 20% Matrigel/PBS solution. To induce shRNA expression, doxycycline was given in sucrose-containing drinking water at a concentration of 0.2 mg/ml starting from when tumor size reached a volume of ∼0.2 cm3. Subcutaneous tumor volume was calculated according to the modified ellipsoid formula V = 0.5 (length × width2). To generate mice with lung tumors, mice were i.v. injected with organoid single-cell suspension containing 300,000 cells in PBS. For i.v. injected mice, doxycycline treatment was started 24 h after injection. Mice were sacrificed 6 wk after the i.v. injection. Orthotopic transplantation was essentially performed as described in Yui et al. (2012) with minor modifications. First, C57BL/6J or Rag1 mice received 4% or 2% DSS, respectively, for five consecutive days (days 0–5) in the drinking water. On day 7, organoids were dissociated by mechanical disruption using a narrowed glass pipette followed by incubation in Accutase solution (Sigma-Aldrich; A6964) at 37°C for 5 min. Single cells were resuspended in 20% Matrigel/PBS solution. Mice were anesthetized with ketamine/xylazine, and dissociated organoids (1–2 × 106 cells/mouse) in 150 μl of 20% Matrigel/PBS solution were flushed intrarectally using a flexible plastic catheter inserted 2 cm into the anus. After enema, the anus was covered with a thick layer of Vaseline to prevent leakage. All mouse experiments were reviewed and approved by the Regierungspräsidium Darmstadt.
Histology
To determine tumor number and size and the extent of invasion, entire mouse colons were dissected and embedded in paraffin as “Swiss rolls.” Complete Swiss rolls were serially sectioned in 200-μm steps. Sections were stained with H&E, scanned using an Aperio ScanScope CS2 (Leica) and analyzed by Aperio Image Scope software (Leica). Tumor size was determined for every individual tumor on each serial section, and the biggest area of a tumor was used for further quantification. An area was considered invasive when tumor cells were penetrating the basal membrane and infiltrating the submucosa or the external muscle layer. The invasive area for each invasive tumor was quantified on every individual serial section, and the biggest measured area was used for further quantification. For both tumor size and extent of the invasive area, a mean value is shown for each mouse. For the AOM-induced models, tumor size was determined without the invasive area. For determination of tumor size in the orthotopic model, both the noninvasive and invasive areas were considered. For detection of lymph node metastases, lymph nodes were either embedded and sectioned together with the colon or embedded separately and sectioned in 100-µm steps. In both cases, lymph nodes were stained with H&E, scanned using an Aperio ScanScope CS2 (Leica), and analyzed by Aperio Image Scope. Immunohistochemistry was performed using an automated Bond-MAX autostainer (Leica) with primary antibodies against phospho-AKT S473 (Cell Signaling Technologies; 3787) 1:100, mouse and human N3ICD (Genentech) 10 μg/ml, tGFP (Thermo Fisher Scientific; PA5-22688) 1:600, and CD3 (Dako; IS503) 1:2. For the detection of primary antibodies, the Bond Polymer Refine Detection kit (Leica) was used. For quantification of CD3+ cells, sections were scanned using an Aperio ScanScope CS2 (Leica), and positive and negative cells were counted using Aperio Image Scope. Immunofluorescence analysis was performed manually. Briefly, formalin-fixed and paraffin-embedded sections were deparaffinized and rehydrated, and heat-mediated antigen unmasking was performed. After blocking with 3% BSA/PBS at RT for 40 min, sections were incubated with primary antibodies against 1:200 αSMA (Sigma-Aldrich; A2547) and 1:300 vimentin (Abcam; ab92547) overnight. The next day, sections were incubated with the appropriate fluorescently labeled secondary antibodies for 1 h at RT and covered with ProLong Gold antifade mounting medium with DAPI (Thermo Fisher Scientific; P36931). 20× images were taken with an Axio Imager 2 (Zeiss), and the fluorescent signal was quantified using ImageJ (v1.51j8; National Institutes of Health) as a percentage of the total image area.
Nucleic acid extraction
To extract RNA from mouse tumor tissue and mouse tumor organoids, small pieces of snap-frozen tissue or organoids were incubated in RLT Buffer with 1% β-mercaptoethanol and homogenized using a Precellys 24 homogenizator (Bertin Technologies). For further steps, Qiagen RNeasy Mini kit (Qiagen) was used according to the manufacturer’s instructions. RNA quality and concentration were determined by Nanodrop spectrophotometer.
Gene expression analysis
For quantitative PCR, cDNA was synthesized using 200–1,000 ng RNA. cDNA was prepared by using Superscript II reverse transcription kit (Invitrogen). First, RNA was incubated with 1 μl oligo dT (50 μM) and 1 μl dNTP Mix (10 mM each) in a total volume of 13 μl at 65°C for 5 min. The mixture was cooled on ice, and 4 μl of 5× First Strand Buffer, 1 μl of dithiothreitol (DTT; 0.1 M), 1 μl of RNase OUT (40 U/μl), and 1 μl of SuperScript II reverse transcription (200 U/μl) was added. Reverse transcription was performed at 42°C for 60 min. cDNA was diluted 1:5 with RNase-free water. Gene expression analysis was performed with a 96-well StepOne real-time PCR machine (Applied Biosystems) using 2× FastStart Universal SYBR Green Master Mix (Roche). Gene expression levels were determined relative to the expression of the housekeeping gene Ppia (cyclophilin). A complete list of primers can be found in Table S1.
To prepare the library for RNA sequencing, 500 ng of total RNA was used. The library was generated with TruSeq mRNA Sample Preparation v2 Kit (Illumina; RS-122-2002). Quantification of cDNA libraries was performed with the QuantiFluor ds DNA System (Promega), and fragment size was determined using the Fragment Analyzer (Advanced Bioanalytical). 50-bp single-end sequencing was performed on the Illumina HiSeq 4000 platform. Read counts were analyzed in the R/Bioconductor environment (version 3.2) using the DESeq2 package (version 1.8.1). Differentially regulated genes were filtered to a minimum of twofold change and false discovery rate–corrected P value < 0.05. Candidate genes were used for further analysis by IPA software (Qiagen) to predict differentially regulated pathways and biological processes. Ingenuity predicts up- and down-regulation of the pathway from the z-score, which is calculated based on the match between expected relationship direction (based on Ingenuity Knowledge Database) and observed gene expression in the experimental dataset.
GSEA was performed after differential expression analysis using the GSEA tool (Subramanian et al., 2005) and compared with the Consensus Molecular Subtypes curated gene sets signatures (https://www.synapse.org/#!Synapse:syn2321865). CMS classification was performed after data normalization with a B-spline linear model using the single-sample CMS classifier (CMSclassifier::classifyCMS(t(sampleData), method=“SSP”) and the published CMS classifier R package (Guinney et al., 2015). Coexpression patterns of genes associated with immune response (CD4, STAT1, CXCL10, IDO1, WARS, CD3D, PTPRC, CD8A, CXCL9, and CXCL11), colon epithelial differentiation (CDX2, AREG, EREG, VAV3, CDX1, DNMT3B, SLC6A4, MYC, ZBTB10, PM20D2, LRP11, and AK4), goblet cell–like (MUC6, MUC2, SPINK4, REG4, TFF3, ANO7, NEURL1, CLCA1, ITLN1, LINC00261, FCGBP, ST6GALNAC1, CAPN9, SPDEF, MLPH, and BCAS1), and stroma (SPARC, ZEB1, ZEB2, COL4A1, COL4A2, ACTA2, TAGLN, and COL1A1), which represent CMS1, CMS2, CMS3, and CMS4, respectively, were inspected to support the CMS class assigned by the classifier. To map mouse genes to human genes, the BiomaRt R package was used. All RNA sequencing data have been deposited at the GEO database and are available under accession no. GSE136257.
Protein analysis
To extract proteins from tumor tissue, small snap-frozen tissue pieces were homogenized in 200 µl of complete protein lysis buffer (50 mM Tris, pH 7.5, 250 mM sodium chloride, 30 mM EDTA, 30 mM EGTA, 25 mM sodium pyrophosphate, 1% Triton X-100, 0.5% NP-40, 10% glycerol, and 1 mM DTT) containing protease and phosphatase inhibitors. To isolate proteins from organoids, organoids were collected and incubated on ice in cell recovery solution (Corning) to remove Matrigel. Organoids were then centrifuged and resuspended in 100 μl of complete protein lysis buffer containing protease and phosphatase inhibitors and lysed on ice for 15 min. Sample homogenization was performed using the Precellys 24 homogenizer (Bertin Technologies). For immunoblot analysis, protein lysates were mixed with Laemmli buffer supplemented with 5% β-mercaptoethanol and boiled at 95°C for 10 min. Proteins were separated via SDS-PAGE and transferred to polyvinylidene fluoride membrane using common immunoblot protocols. After blocking, membranes were incubated with the following primary antibodies: phospho-AKT S473 1:1,000 (Cell Signaling Technologies; 3787), total AKT 1:1,000 (Cell Signaling Technologies; 9272), PTEN 1:500 (Abcam; 32199), and β-actin 1:3,000 (Sigma-Aldrich; A4700). HRP-linked secondary antibodies were used at 1:5,000, and the chemiluminescent signal was imaged using the Fusion Fx7 System (Vilber). The Proteome Profiler Mouse XL Cytokine Array (R&D Systems; ARY028) was used according to the manufacturer’s instructions with 300 μg of total protein per genotype.
Label-free quantitative global phosphoproteomic analysis
Phosphoproteomic analysis was performed basically as described before (Häupl et al., 2019). In brief, after cell harvest and lysis in modified radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.5, 150 mM sodium chloride, 1 mM EDTA, 1% NP-40, 0.1% Na-deoxycholate, 1 mM DTT, and protease and phosphatase inhibitors) using a Precellys 24 homogenizer (Bertin Technologies), 0.5 mg of protein was subjected to acetone precipitation, dissolved in denaturation buffer, reduced, and alkylated. The samples were then sequentially digested with LysC and trypsin, and the resulting peptide mixtures were purified using SepPak C18 cartridges. After phosphopeptide enrichment by TiO2 metal oxide affinity chromatography, the samples were fractionated using strong cation-exchange chromatography. The fractions were purified with C18 Stage tips and subjected to liquid chromatography–tandem MS analysis on a Q Exactive HF Orbitrap mass spectrometer coupled online to an Ultimate 3000 RSLCnano HPLC system via an electrospray ionization interface (all Thermo Fisher Scientific). First, peptides were washed on a reversed phase C18 trap column (length 5 mm, diameter 0.3 mm) for 3 min. Then the trap column was switched online with the analytical column (length 30 cm, diameter 75 μm) packed with ReproSil-Pur C18 AQ 1.9 μm reversed phase material (Dr. Maisch GmbH). Solvent A consisted of 0.1% formic acid in water, and the peptides were eluted from the column with a linear solvent gradient from 5 to 35% solvent B (0.1% formic acid, 80% acetonitrile in water) over 88 min at a flow rate of 300 nl/min. MS data were acquired in a data-dependent top 15 scheme, in which the 15 precursor ions with the most intense signals within the mass range of 350–1,600 m/z were selected for high-energy collisional dissociation fragmentation with a normalized collision energy of 28 from an MS1 survey scan (AGC target 1E6, maximum injection time 50 ms, resolution 120,000). Precursors were selected for fragmentation with an isolation window of 1.6 Th and a dynamic exclusion of 30 s, and fragment ions were analyzed in MS2 scans at a resolution of 30,000 (AGC target 1E5, maximum injection time 110 s).
Raw MS data were processed with MaxQuant software (version 1.6.0.16; Cox and Mann, 2008). For protein identification, tandem MS spectra were searched with the integrated Andromeda search engine (Cox et al., 2011) against the UniProt reference database of murine proteins containing 54,211 entries (November 15, 2018) and an internal database of potential contaminants (246 entries) with the default settings. Acetylation at the protein N-terminus, oxidation of methionine, and phosphorylation on serine, threonine, or tyrosine were set as variable modifications, and carbamidomethylation of cysteine was defined as fixed modification. Tryptic digestion was considered with a peptide length of ≥7 aa, allowing up to two missed cleavage sites. On both the peptide and protein levels, a false discovery rate of 1% was tolerated by application of a forward-and-reverse concatenated decoy database search approach. Relative label-free quantification (LFQ) was conducted with the MaxLFQ module of MaxQuant (Cox et al., 2014), and the “match between runs” option was enabled.
Downstream data analysis was performed with Perseus software (version 1.6.0.7; Tyanova et al., 2016). First, identified phosphosites derived from potential contaminants and hits to the decoy database were removed. Furthermore, phosphosites were filtered for a localization probability of ≥75% (class I phosphosites). After filtering for at least two-thirds valid intensity values in each sample group, normalization of each sample to the median overall LFQ intensity was conducted. The normalized intensities were log2 transformed, and missing values were imputed using the default settings. After a two-sample t test between both groups, phosphosites with at least twofold change and P < 0.05 were considered regulated and subjected to IPA (Qiagen). The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD014778.
Cloning
97-mer oligos targeting the coding sequence of the mouse Pten and Notch3 and the control shRNA sequences were obtained from the previously published shRNA list (Fellmann et al., 2013) and can be found in Table S2. To achieve efficient and stable knockdown of Pten, a murine stem cell virus–based retroviral vector coexpressing a microRNA-30–based shRNA and GFP was used (kind gift from Lars Zender, University Hospital Tübingen, Tübingen, Germany; University of Tübingen, Tübingen, Germany; German Cancer Research Center, Heidelberg, Germany; Dickins et al., 2005). Briefly, the hairpin against Pten or the control hairpin was introduced into a murine stem cell virus retroviral vector, in which the expression of the shRNA and EGFP is driven by the LTR promoter using XhoI and EcoRI restriction enzymes. To generate organoids enabling the doxycycline-regulatable expression of Notch3-specific hairpins, retroviral vectors RT3GEP expressing tGFP (kind gift from Johanes Zuber, Research Institute of Molecular Pathology, Vienna BioCenter, Vienna, Austria; Addgene; 111164) and rtTA3-PGK-Hygro (kind gift from Lars Zender) were used. shRNAs targeting murine Notch3 or the control hairpin were cloned into RT3GEP recipient vector using XhoI and EcoRI restriction enzymes as previously described (Fellmann et al., 2013). To generate APTK and APTK-myrAKT organoids, the following constructs were used: (1) plasmid encoding the sgRNA targeting the Apc gene (sgRNA-Apc-T1; Schwank et al., 2013), (2) human myrAKT cloned downstream of IRES and hygromycin in rtTA3-PGK-Hygro, (3) murine KrasG12D cDNA cloned downstream of IRES and puromycin resistance gene in RT3GEP, (4) pPGK-Cre-bpA (kind gift from Klaus Rajewsky, Max Delbrück Center for Molecular Medicine, Berlin, Germany; Addgene; plasmid 11543). Lentiviral constructs encoding mouse N3ICD were designed and synthetized by VectorBuilder. Briefly, the sequence encoding mouse NOTCH3-ICD was cloned after an N-terminal FLAG sequence into a pLV backbone. To generate the control plasmid, a 300-bp “stuffer” sequence was cloned after the N-terminal FLAG sequence.
Organoid culture and treatments
Normal colon organoids were isolated from unchallenged Trp53fl/flTgfbr2fl/fl mice as previously described (Sato et al., 2009) and maintained in Matrigel (Corning; 356231) with medium containing Advanced DMEM F12 (Thermo Fisher Scientific; 12634-028), 1% penicillin/streptomycin (Thermo Fisher Scientific; 15140-22), 1× Glutamax (Thermo Fisher Scientific; 35050061), Hepes (Thermo Fisher Scientific; 15630056), in-house-made mouse noggin and R-spondin, 1× N2 (Thermo Fisher Scientific; 17502–048), 1× B27 (Thermo Fisher Scientific; 17504-044), 80 μM N-acetylcysteine (Sigma-Aldrich; A9165), 200 ng/ml mouse EGF (Thermo Fisher Scientific; PMG8043), 10 nM Y-27632 (Sigma-Aldrich; Y0503), 5 μM CHIR 99021 (Axon Medchem; 1386), 500 μM A83-01 (Sigma-Aldrich; SML0788). Mouse tumor organoids were isolated from AOM-treated Trp53ΔIEC and Trp53 ΔIECAktE17K mice. Single tumors were pooled and digested with 0.1 mg/ml Collagenase A (Sigma-Aldrich; C0130) and Dispase II (Sigma-Aldrich; D4693) in Advanced DMEM F12 containing 1% penicillin/streptomycin, 0.05 mg/ml DNase I (Roche; 4716728001), 0.03 ng/ml hydrocortisone (Sigma-Aldrich; H0888), 50 ng/ml murine EGF, and 1× insulin-transferrin-selenium-ethanolamine (Thermo Fisher Scientific; 51500056) for 2 h at 37°C. Mouse tumor organoids were maintained in Matrigel with medium containing Advanced DMEM F12, 1% penicillin/streptomycin, 1× Glutamax, 1× Hepes, 80 μM N-acetylcysteine, and 200 ng/ml mouse EGF. To induce the expression of the doxycycline-inducible hairpins and tGFP, organoids were maintained for the indicated times in the presence of 0.5 µg/ml doxycycline in the culture medium. Retroviral and lentiviral vectors were introduced into 293T cells by standard calcium phosphate coprecipitation. Virus-containing supernatant was collected after 24 h every 6 h at least four times. Virus-containing supernatant was then concentrated by centrifugation overnight at 10,000 g at 4°C, and the pellet was resuspended in mouse tumor organoid medium containing 8 μg/ml polybrene. Organoids were dissociated into single-cell suspensions and infected by spinoculation. Selection started 48 h after infection. To deliver sgRNA-Apc-T1, Lipofectamine–nucleic acid complexes were prepared according to the standard Lipofectamine protocol and were delivered by spinoculation.
TCGA data analysis
Transcriptomic and clinical data from TCGA Research Network database were retrieved with the help of the cBio Cancer Genomic Portal and were further processed with GraphPad Prism (Cerami et al., 2012; Gao et al., 2013). Tumor samples with RNA sequencing V2 data (n = 382) with a z-score threshold ±1 were analyzed. Overexpression was defined as expression more than one standard deviation (SD) from mean. To obtain CMS-classified human CRC samples, frozen robust multiarray analysis normalized expression data for the GEO datasets GSE13067 (Jorissen et al., 2008), GSE14333 (Jorissen et al., 2009), GSE17536 (Smith et al., 2010), GSE33113 (Kemper et al., 2012), and GSE39582 (Marisa et al., 2013) were obtained from the Colorectal Cancer Subtyping Consortium’s Synapse platform (https://www.synapse.org/#!Synapse:syn2634742). Annotation with Entrez IDs was performed in R (version 3.3.3) using the biomaRt package, and samples were CMS classified using the single-sample prediction provided by the R package CMS classifier (version 1.0.0; Guinney et al., 2015).
Statistical analysis
Appropriate statistical tests were used for the analysis of all experiments. Statistical analysis was performed using GraphPad Prism 7. Statistical significance was calculated with unpaired, two-tailed t test or one-way ANOVA followed by Tukey’s or Dunnett’s multiple comparison test. Where applicable, outliers were filtered out by applying ROUT (Q = 1%) or Grubbs’ test. CRC patient Kaplan–Meier survival curves were generated by Prism 7 using data retrieved from the cBio Cancer Genomic Portal, and significance was calculated by log-rank (Mantel–Cox) test.
Online supplemental material
Fig. S1 lists selected pathways that are significantly differentially regulated in response to AKT activation and gives information on the tumor microenvironment in Trp53ΔIEC and Trp53ΔIECAktE17K mice. Fig. S2 shows additional analysis of the RNA-sequencing results of Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids. Fig. S3 provides supporting information for Notch3 knockdown in the s.c. transplantation model and shows results of the i.v. metastasis model using Trp53−/−AktE17KshCo and Trp53−/−AktE17KshNotch3 organoids. Table S1 lists primers used for gene expression analysis in this study. Table S2 lists sequences of shRNAs.
Supplementary Material
lists PCR primers.
lists shRNAs.
Acknowledgments
We thank Hana Kunkel, Susanne Bösser, Kathleen Mohs, Preeti Gupta, Eva Rudolf, and Christin Danneil for expert technical assistance, as well as the staff at the Animal Facility and the Histology and Flow Cytometry Core Facilities at the Georg-Speyer-Haus. We are grateful to the Transcriptome and Genome Analysis Laboratory DNA Microarray and Deep Sequencing Core Facility at the University Hospital Göttingen for performing RNA sequencing.
Work in the laboratory of F.R. Greten is supported by institutional funds from the Georg-Speyer-Haus, by the LOEWE Center Frankfurt Cancer Institute funded by the Hessen State Ministry for Higher Education, Research and the Arts [III L 5 - 519/03/03.001 - (0015)], and Deutsche Forschungsgemeinschaft (FOR2438: Gr1916/11-1; SFB 815, 1177, and 1292, as well as GRK 2336). The Institute for Tumor Biology and Experimental Therapy, Georg-Speyer-Haus, is funded jointly by the German Federal Ministry of Health and the Hessen State Ministry for Higher Education, Research and the Arts.
Author contributions: J. Varga, A. Nicolas, and V. Petrocelli performed and analyzed experiments. F.R. Greten, J. Varga, and M. Pesic designed the experiments. GSEA was performed by A. Mahmoud and H. Medyouf. CMS classification of mouse tumors and gene-module analysis were designed and performed by E. Etlioglu and S. Tejpar. CMS classification of human CRC samples was performed by B.E. Michels and H.F. Farin. P.K. Ziegler, K. Bankov, and P.J. Wild provided human tissue sections. Phospho-MS analysis was designed, performed, and analyzed by T. Oellerich, D. Yepes, and B. Häupl. J. Ruland, and S. Wanninger generated LSL-AktE17K mice, and C.W. Siebel provided α-NRR3 and mouse and human α-N3ICD antibodies. J. Varga, V. Petrocelli, and F.R. Greten wrote the manuscript.
References
- Aster J.C., Pear W.S., and Blacklow S.C.. 2017. The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. 12:245–275. 10.1146/annurev-pathol-052016-100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., de Reyniès A., Giraldo N.A., Pilati C., Buttard B., Lacroix L., Selves J., Sautès-Fridman C., Laurent-Puig P., and Fridman W.H.. 2016. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 22:4057–4066. 10.1158/1078-0432.CCR-15-2879 [DOI] [PubMed] [Google Scholar]
- Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., et al. . 2009. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 15:91–102. 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Brannon A.R., Vakiani E., Sylvester B.E., Scott S.N., McDermott G., Shah R.H., Kania K., Viale A., Oschwald D.M., Vacic V., et al. . 2014. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 15:454 10.1186/s13059-014-0454-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M., Assen F.P., Leithner A., Abe J., Schachner H., Asfour G., Bago-Horvath Z., Stein J.V., Uhrin P., Sixt M., et al. . 2018. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science. 359:1408–1411. 10.1126/science.aal3662 [DOI] [PubMed] [Google Scholar]
- Byun D.-S., Ahmed N., Nasser S., Shin J., Al-Obaidi S., Goel S., Corner G.A., Wilson A.J., Flanagan D.J., Williams D.S., et al. . 2011. Intestinal epithelial-specific PTEN inactivation results in tumor formation. Am. J. Physiol. Gastrointest. Liver Physiol. 301:G856–G864. 10.1152/ajpgi.00178.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrero-de Las Heras S., and Martínez-Balibrea E.. 2018. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J. Gastroenterol. 24:4738–4749. 10.3748/wjg.v24.i42.4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M., Sevillano M., Palomo-Ponce S., Tauriello D.V.F., Byrom D., et al. . 2015. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47:320–329. 10.1038/ng.3225 [DOI] [PubMed] [Google Scholar]
- Carpten J.D., Faber A.L., Horn C., Donoho G.P., Briggs S.L., Robbins C.M., Hostetter G., Boguslawski S., Moses T.Y., Savage S., et al. . 2007. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 448:439–444. 10.1038/nature05933 [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. . 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanrion M., Kuperstein I., Barrière C., El Marjou F., Cohen D., Vignjevic D., Stimmer L., Paul-Gilloteaux P., Bièche I., Tavares S.R., et al. . 2014. Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. Nat. Commun. 5:5005 10.1038/ncomms6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy L., Hagenbeek T.J., Solon M., French D., Finkle D., Shelton A., Venook R., Brauer M.J., and Siebel C.W.. 2017. Constitutive NOTCH3 Signaling Promotes the Growth of Basal Breast Cancers. Cancer Res. 77:1439–1452. 10.1158/0008-5472.CAN-16-1022 [DOI] [PubMed] [Google Scholar]
- Chu D., Li Y., Wang W., Zhao Q., Li J., Lu Y., Li M., Dong G., Zhang H., Xie H., et al. . 2010. High level of Notch1 protein is associated with poor overall survival in colorectal cancer. Ann. Surg. Oncol. 17:1337–1342. 10.1245/s10434-009-0893-7 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M.. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13:2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Dainichi T., Hayden M.S., Park S.G., Oh H., Seeley J.J., Grinberg-Bleyer Y., Beck K.M., Miyachi Y., Kabashima K., Hashimoto T., et al. . 2016. PDK1 Is a Regulator of Epidermal Differentiation that Activates and Organizes Asymmetric Cell Division. Cell Rep. 15:1615–1623. 10.1016/j.celrep.2016.04.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.J., Huang Y.H., Chen M., Su J., Zou Y., Bardeesy N., Iacobuzio-Donahue C.A., and Massagué J.. 2016. TGF-β Tumor Suppression through a Lethal EMT. Cell. 164:1015–1030. 10.1016/j.cell.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E.J., Marsh Durban V., Meniel V., Williams G.T., and Clarke A.R.. 2014. PTEN loss and KRAS activation leads to the formation of serrated adenomas and metastatic carcinoma in the mouse intestine. J. Pathol. 233:27–38. 10.1002/path.4312 [DOI] [PubMed] [Google Scholar]
- Dickins R.A., Hemann M.T., Zilfou J.T., Simpson D.R., Ibarra I., Hannon G.J., and Lowe S.W.. 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37:1289–1295. 10.1038/ng1651 [DOI] [PubMed] [Google Scholar]
- Dienstmann R., Vermeulen L., Guinney J., Kopetz S., Tejpar S., and Tabernero J.. 2017. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 17:79–92. 10.1038/nrc.2016.126 [DOI] [PubMed] [Google Scholar]
- Doody R.S., Raman R., Farlow M., Iwatsubo T., Vellas B., Joffe S., Kieburtz K., He F., Sun X., Thomas R.G., et al. ; Semagacestat Study Group . 2013. A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 369:341–350. 10.1056/NEJMoa1210951 [DOI] [PubMed] [Google Scholar]
- Doucas H., Mann C.D., Sutton C.D., Garcea G., Neal C.P., Berry D.P., and Manson M.M.. 2008. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J. Surg. Oncol. 97:63–68. 10.1002/jso.20894 [DOI] [PubMed] [Google Scholar]
- Élez E., Kocáková I., Höhler T., Martens U.M., Bokemeyer C., Van Cutsem E., Melichar B., Smakal M., Csőszi T., Topuzov E., et al. . 2015. Abituzumab combined with cetuximab plus irinotecan versus cetuximab plus irinotecan alone for patients with KRAS wild-type metastatic colorectal cancer: the randomised phase I/II POSEIDON trial. Ann. Oncol. 26:132–140. 10.1093/annonc/mdu474 [DOI] [PubMed] [Google Scholar]
- Fearon E.R. 2011. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 6:479–507. 10.1146/annurev-pathol-011110-130235 [DOI] [PubMed] [Google Scholar]
- Fearon E.R., and Vogelstein B.. 1990. A genetic model for colorectal tumorigenesis. Cell. 61:759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y., et al. . 2013. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5:1704–1713. 10.1016/j.celrep.2013.11.020 [DOI] [PubMed] [Google Scholar]
- Fouillade C., Monet-Leprêtre M., Baron-Menguy C., and Joutel A.. 2012. Notch signalling in smooth muscle cells during development and disease. Cardiovasc. Res. 95:138–146. 10.1093/cvr/cvs019 [DOI] [PubMed] [Google Scholar]
- Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., and Artavanis-Tsakonas S.. 2005. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 435:964–968. 10.1038/nature03589 [DOI] [PubMed] [Google Scholar]
- Fre S., Pallavi S.K., Huyghe M., Laé M., Janssen K.-P., Robine S., Artavanis-Tsakonas S., and Louvard D.. 2009. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc. Natl. Acad. Sci. USA. 106:6309–6314. 10.1073/pnas.0900427106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. . 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Wang K., Mucida D., Stewart C.A., Schnabl B., Jauch D., Taniguchi K., Yu G.-Y., Osterreicher C.H., Hung K.E., et al. . 2012. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 491:254–258. 10.1038/nature11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest R.V., Boulter L., Dwyer B.J., Kendall T.J., Man T.-Y., Minnis-Lyons S.E., Lu W.-Y., Robson A.J., Gonzalez S.F., Raven A., et al. . 2016. Notch3 drives development and progression of cholangiocarcinoma. Proc. Natl. Acad. Sci. USA. 113:12250–12255. 10.1073/pnas.1600067113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J., Dienstmann R., Wang X., de Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. . 2015. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21:1350–1356. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney A., and Hoey T.. 2011. Anti-DLL4, a cancer therapeutic with multiple mechanisms of action. Vasc. Cell. 3:18 10.1186/2045-824X-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häupl B., Urlaub H., and Oellerich T.. 2019. Phosphoproteomic Analysis of Signaling Pathways in Lymphomas. Methods Mol. Biol. 1956:371–381. 10.1007/978-1-4939-9151-8_19 [DOI] [PubMed] [Google Scholar]
- He X.C., Yin T., Grindley J.C., Tian Q., Sato T., Tao W.A., Dirisina R., Porter-Westpfahl K.S., Hembree M., Johnson T., et al. . 2007. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet. 39:189–198. 10.1038/ng1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackstadt R., and Sansom O.J.. 2016. Mouse models of intestinal cancer. J. Pathol. 238:141–151. 10.1002/path.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackstadt R., van Hooff S.R., Leach J.D., Cortes-Lavaud X., Lohuis J.O., Ridgway R.A., Wouters V.M., Roper J., Kendall T.J., Roxburgh C.S., et al. . 2019. Epithelial NOTCH Signaling Rewires the Tumor Microenvironment of Colorectal Cancer to Drive Poor-Prognosis Subtypes and Metastasis. Cancer Cell. 36:319–336.e7. 10.1016/j.ccell.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen R.N., Gibbs P., Christie M., Prakash S., Lipton L., Desai J., Kerr D., Aaltonen L.A., Arango D., Kruhøffer M., et al. . 2009. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin. Cancer Res. 15:7642–7651. 10.1158/1078-0432.CCR-09-1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen R.N., Lipton L., Gibbs P., Chapman M., Desai J., Jones I.T., Yeatman T.J., East P., Tomlinson I.P.M., Verspaget H.W., et al. . 2008. DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin. Cancer Res. 14:8061–8069. 10.1158/1078-0432.CCR-08-1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K., Versloot M., Cameron K., Colak S., de Sousa e Melo F., de Jong J.H., Bleackley J., Vermeulen L., Versteeg R., Koster J., et al. . 2012. Mutations in the Ras-Raf Axis underlie the prognostic value of CD133 in colorectal cancer. Clin. Cancer Res. 18:3132–3141. 10.1158/1078-0432.CCR-11-3066 [DOI] [PubMed] [Google Scholar]
- Lafkas D., Shelton A., Chiu C., de Leon Boenig G., Chen Y., Stawicki S.S., Siltanen C., Reichelt M., Zhou M., Wu X., et al. . 2015. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 528:127–131. 10.1038/nature15715 [DOI] [PubMed] [Google Scholar]
- Leystra A.A., Deming D.A., Zahm C.D., Farhoud M., Olson T.J.P., Hadac J.N., Nettekoven L.A., Albrecht D.M., Clipson L., Sullivan R., et al. . 2012. Mice expressing activated PI3K rapidly develop advanced colon cancer. Cancer Res. 72:2931–2936. 10.1158/0008-5472.CAN-11-4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Li Y., Wu W., Gordon W.R., Chang D.W., Lu M., Scoggin S., Fu T., Vien L., Histen G., et al. . 2008. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 283:8046–8054. 10.1074/jbc.M800170200 [DOI] [PubMed] [Google Scholar]
- Marisa L., de Reyniès A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.-C., Schiappa R., Guenot D., Ayadi M., et al. . 2013. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 10 e1001453 10.1371/journal.pmed.1001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz S.D., and Bertagnolli M.M.. 2009. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 361:2449–2460. 10.1056/NEJMra0804588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh V., Winton D.J., Williams G.T., Dubois N., Trumpp A., Sansom O.J., and Clarke A.R.. 2008. Epithelial Pten is dispensable for intestinal homeostasis but suppresses adenoma development and progression after Apc mutation. Nat. Genet. 40:1436–1444. 10.1038/ng.256 [DOI] [PubMed] [Google Scholar]
- McIntosh B.E., Brown M.E., Duffin B.M., Maufort J.P., Vereide D.T., Slukvin I.I., and Thomson J.A.. 2015. Nonirradiated NOD,B6.SCID Il2rγ-/- Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports. 4:171–180. 10.1016/j.stemcr.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre R.E., Buczacki S.J.A., Arends M.J., and Adams D.J.. 2015. Mouse models of colorectal cancer as preclinical models. BioEssays. 37:909–920. 10.1002/bies.201500032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano J., McKay J., Dagenais C., Foster-Brown L., Pognan F., Gadient R., Jacobs R.T., Zacco A., Greenberg B., and Ciaccio P.J.. 2004. Modulation of notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82:341–358. 10.1093/toxsci/kfh254 [DOI] [PubMed] [Google Scholar]
- MoTriColor 2015. Molecularly guided trials with specific treatment strategies in patients with advanced newly molecular defined subtypes of colorectal cancer. http://www.motricolor.eu/project/ (Accessed Jan. 10, 2015).
- Nass S.J., Rothenberg M.L., Pentz R., Hricak H., Abernethy A., Anderson K., Gee A.W., Harvey R.D., Piantadosi S., Bertagnolli M.M., et al. . 2018. Accelerating anticancer drug development - opportunities and trade-offs. Nat. Rev. Clin. Oncol. 15:777–786. 10.1038/s41571-018-0102-3 [DOI] [PubMed] [Google Scholar]
- Noah T.K., and Shroyer N.F.. 2013. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu. Rev. Physiol. 75:263–288. 10.1146/annurev-physiol-030212-183741 [DOI] [PubMed] [Google Scholar]
- Nowell C.S., and Radtke F.. 2017. Notch as a tumour suppressor. Nat. Rev. Cancer. 17:145–159. 10.1038/nrc.2016.145 [DOI] [PubMed] [Google Scholar]
- Pechloff K., Holch J., Ferch U., Schweneker M., Brunner K., Kremer M., Sparwasser T., Quintanilla-Martinez L., Zimber-Strobl U., Streubel B., et al. . 2010. The fusion kinase ITK-SYK mimics a T cell receptor signal and drives oncogenesis in conditional mouse models of peripheral T cell lymphoma. J. Exp. Med. 207:1031–1044. 10.1084/jem.20092042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E.R., Kedrin D., Seano G., Gautier O., Meijer E.F.J., Jones D., Chin S.-M., Kitahara S., Bouta E.M., Chang J., et al. . 2018. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science. 359:1403–1407. 10.1126/science.aal3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. . 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47(D1):D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg D.W., Giardina C., and Tanaka T.. 2009. Mouse models for the study of colon carcinogenesis. Carcinogenesis. 30:183–196. 10.1093/carcin/bgn267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., et al. . 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K., et al. . 2013. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13:653–658. 10.1016/j.stem.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Schwitalla S., Ziegler P.K., Horst D., Becker V., Kerle I., Begus-Nahrmann Y., Lechel A., Rudolph K.L., Langer R., Slotta-Huspenina J., et al. . 2013. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 23:93–106. 10.1016/j.ccr.2012.11.014 [DOI] [PubMed] [Google Scholar]
- Serafin V., Persano L., Moserle L., Esposito G., Ghisi M., Curtarello M., Bonanno L., Masiero M., Ribatti D., Stürzl M., et al. . 2011. Notch3 signalling promotes tumour growth in colorectal cancer. J. Pathol. 224:448–460. 10.1002/path.2895 [DOI] [PubMed] [Google Scholar]
- Shao J., Washington M.K., Saxena R., and Sheng H.. 2007. Heterozygous disruption of the PTEN promotes intestinal neoplasia in APCmin/+ mouse: roles of osteopontin. Carcinogenesis. 28:2476–2483. 10.1093/carcin/bgm186 [DOI] [PubMed] [Google Scholar]
- Siebel C., and Lendahl U.. 2017. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 97:1235–1294. 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., and Jemal A.. 2018. Cancer statistics, 2018. CA Cancer J. Clin. 68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- Smith J.J., Deane N.G., Wu F., Merchant N.B., Zhang B., Jiang A., Lu P., Johnson J.C., Schmidt C., Bailey C.E., et al. . 2010. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 138:958–968. 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M., Aoki M., Fuwa H., Aoki K., Hosogi H., Sakai Y., Hashida H., Takabayashi A., Sasaki M., Robine S., et al. . 2011. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 19:125–137. 10.1016/j.ccr.2010.11.008 [DOI] [PubMed] [Google Scholar]
- Stanger B.Z., Datar R., Murtaugh L.C., and Melton D.A.. 2005. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA. 102:12443–12448. 10.1073/pnas.0505690102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strosberg J.R., Yeatman T., Weber J., Coppola D., Schell M.J., Han G., Almhanna K., Kim R., Valone T., Jump H., et al. . 2012. A phase II study of RO4929097 in metastatic colorectal cancer. Eur. J. Cancer. 48:997–1003. 10.1016/j.ejca.2012.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. . 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammam J., Ware C., Efferson C., O’Neil J., Rao S., Qu X., Gorenstein J., Angagaw M., Kim H., Kenific C., et al. . 2009. Down-regulation of the Notch pathway mediated by a gamma-secretase inhibitor induces anti-tumour effects in mouse models of T-cell leukaemia. Br. J. Pharmacol. 158:1183–1195. 10.1111/j.1476-5381.2009.00389.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauriello D.V.F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., Sevillano M., Ibiza S., Cañellas A., Hernando-Momblona X., et al. . 2018. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 554:538–543. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- Thorpe L.M., Yuzugullu H., and Zhao J.J.. 2015. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 15:7–24. 10.1038/nrc3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J.. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13:731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Ueo T., Imayoshi I., Kobayashi T., Ohtsuka T., Seno H., Nakase H., Chiba T., and Kageyama R.. 2012. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 139:1071–1082. 10.1242/dev.069070 [DOI] [PubMed] [Google Scholar]
- van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., et al. . 2005. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 435:959–963. 10.1038/nature03659 [DOI] [PubMed] [Google Scholar]
- Varga J., and Greten F.R.. 2017. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat. Cell Biol. 19:1133–1141. 10.1038/ncb3611 [DOI] [PubMed] [Google Scholar]
- Wang X.W., Xi X.Q., Wu J., Wan Y.Y., Hui H.X., and Cao X.F.. 2015. MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol. Rep. 33:1402–1410. 10.3892/or.2015.3731 [DOI] [PubMed] [Google Scholar]
- Wu Y., Cain-Hom C., Choy L., Hagenbeek T.J., de Leon G.P., Chen Y., Finkle D., Venook R., Wu X., Ridgway J., et al. . 2010. Therapeutic antibody targeting of individual Notch receptors. Nature. 464:1052–1057. 10.1038/nature08878 [DOI] [PubMed] [Google Scholar]
- Yu J., Siebel C.W., Schilling L., and Canalis E.. 2020. An antibody to Notch3 reverses the skeletal phenotype of lateral meningocele syndrome in male mice. J. Cell. Physiol. 235:210–220. 10.1002/jcp.28960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K., et al. . 2012. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 18:618–623. 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists PCR primers.
lists shRNAs.