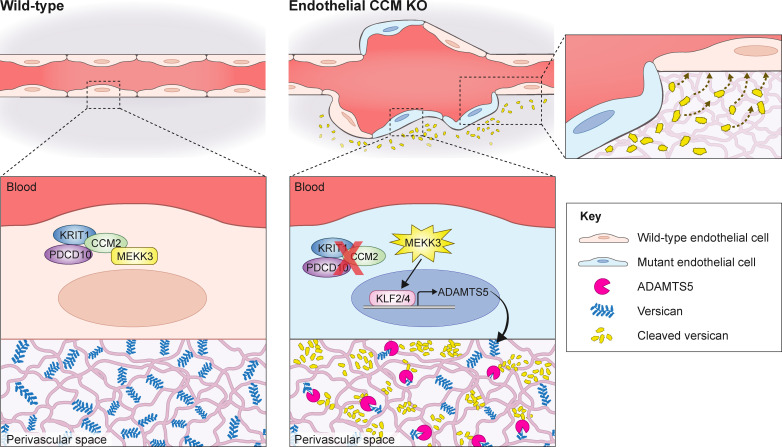
CCMs arise due to increased MEKK3-KLF2/4 signaling in brain endothelial cells, but the downstream mechanisms of CCM formation remain unclear. Hong et al. show that expression of the ADAMTS5 protease and proteolytic cleavage of the matrix proteoglycan versican by CCM-deficient endothelial cells contribute to CCM formation.
Abstract
Cerebral cavernous malformations (CCMs) form following loss of the CCM protein complex in brain endothelial cells due to increased endothelial MEKK3 signaling and KLF2/4 transcription factor expression, but the downstream events that drive lesion formation remain undefined. Recent studies have revealed that CCM lesions expand by incorporating neighboring wild-type endothelial cells, indicative of a cell nonautonomous mechanism. Here we find that endothelial loss of ADAMTS5 reduced CCM formation in the neonatal mouse model. Conversely, endothelial gain of ADAMTS5 conferred early lesion genesis in the absence of increased KLF2/4 expression and synergized with KRIT1 loss of function to create large malformations. Lowering versican expression reduced CCM burden, indicating that versican is the relevant ADAMTS5 substrate and that lesion formation requires proteolysis but not loss of this extracellular matrix protein. These findings identify endothelial secretion of ADAMTS5 and cleavage of versican as downstream mechanisms of CCM pathogenesis and provide a basis for the participation of wild-type endothelial cells in lesion formation.
Graphical Abstract
Introduction
Cerebral cavernous malformations (CCMs) are common vascular abnormalities that form predominantly in the central nervous system (CNS) and are a cause of hemorrhagic stroke and seizure (Al-Shahi Salman et al., 2008, 2012; Spiegler et al., 2018). CCMs occur due to loss of function mutations in one of three disease genes, KRIT1, CCM2, or PDCD10, in brain endothelial cells (BECs; Akers et al., 2017; Boulday et al., 2011; Chan et al., 2011; Fisher and Boggon, 2014; Plummer et al., 2005; Zhou et al., 2016b). Despite considerable molecular and genetic insight obtained into CCM disease over the past 10 yr, no effective medical therapies have been identified to treat it.
Human, mouse, and zebrafish genetic studies and biochemical studies have provided significant insight into the molecular basis of CCM disease. The CCM proteins form a ternary complex that binds and negatively regulates MEKK3, a MAPK that is selectively expressed in endothelial cells (ECs; Cullere et al., 2015; Fisher et al., 2015; Wang et al., 2015; Zhou et al., 2015, 2016b). MEKK3 mediates endothelial responses to both fluid shear forces and inflammatory cytokines, many of which are driven by downstream expression of the related transcription factors KLF2 and KLF4 (Huang et al., 2004; Tang et al., 2017; Zhou et al., 2015). Genetic and pharmacologic investigation of CCM disease mechanisms has been facilitated by the development of a faithful neonatal mouse model in which endothelial loss of CCM gene function, shortly after birth, confers CCM formation in the hindbrain and retina by postnatal day (P) 10 (Boulday et al., 2011; Zhou et al., 2016b). Using this model, endothelial gain of MEKK3 function and elevated KLF2/4 expression have been shown to be required for CCM formation (Cuttano et al., 2016; Zhou et al., 2016b). Recent mouse and human studies have demonstrated that signaling by the innate immune receptor TLR4, in response to circulating bacterial lipopolysaccharide derived from the gut microbiome, drives brain endothelial MEKK3-KLF2/4 signaling to induce CCMs (Tang et al., 2017, 2019). These findings have established a core molecular pathway for CCM pathogenesis and identified a major upstream input to that pathway; however, the effectors downstream of KLF2 and KLF4 that result in cavernoma formation remain unknown.
CCMs form selectively in the CNS and exhibit a natural history characterized by lesion genesis in the postcapillary venule and expansion to large, thin-walled vascular structures that are prone to hemorrhage (Frischer et al., 2008; Tanriover et al., 2013). Lineage tracing studies in the mouse neonatal model have recently demonstrated that lesion genesis is driven by CCM-deficient ECs, while lesion growth is largely the result of incorporation of neighboring wild-type ECs (Detter et al., 2018; Malinverno et al., 2019). These observations identify a cell nonautonomous mechanism (Akers et al., 2009) by which CCM-deficient ECs alter the behavior of neighboring wild-type ECs to create vascular malformations, but the molecular and cellular underpinnings remain undefined.
Studies of endothelial loss of CCM function in the developing heart have revealed a cell nonautonomous effect on myocardial cell growth mediated by elevated endothelial expression of ADAMTS5, a protease that cleaves the proteoglycan versican which is a major component of cardiac jelly (Zhou et al., 2015). Increased endothelial expression of ADAMTS5 and versican proteolysis in the developing heart require gain of MEKK3 signaling, identifying these events as bona fide downstream effectors of the pathway in that context (Zhou et al., 2015). In the present study, we have tested whether similar downstream events also underlie CCM formation in the postnatal brain. We find that brain endothelial loss of ADAMTS5 increases versican in the white matter, where CCMs preferentially form, and significantly rescues CCM formation. Using a tetracycline-inducible transgenic approach, we further show that gain of ADAMTS5 function is sufficient to confer vascular changes like those observed in early CCM lesions in the absence of increased MEKK3-KLF2/4 signaling, and strongly synergizes with KRIT1 deficiency to increase CCM formation. To determine if ADAMTS5-associated changes are due to cleavage of versican, we tested whether partial loss of versican alters CCM formation in the neonatal mouse model. Loss of versican reduced CCM formation, while versican cleavage products generated around developing CCM lesions increase endothelial sprouting and branching in vitro, consistent with a mechanism in which CCMs form due to the generation of versican proteolytic cleavage products rather than the loss of versican protein in the perivascular matrix. These studies identify ADAMTS5-mediated cleavage of versican as a downstream mechanism by which gain of MEKK3-KLF2/4 signaling may alter the function of both CCM-deficient and neighboring wild-type ECs in the brain.
Results
Global loss of ADAMTS5, but not ADAMTS1 or ADAMTS4, prevents CCM formation
Previous work by our group and others has demonstrated that CCM lesions arise from an endothelial gain of MEKK3-KLF2/4 signaling, but the downstream mechanisms by which this signaling pathway results in vascular malformations remain unidentified (Cuttano et al., 2016; Tang et al., 2017; Zhou et al., 2016b). We have shown that EC loss of CCM function in the embryonic heart and postnatal brain results in increased ADAMTS activity (Zhou et al., 2015, 2016b). We have also observed that the white matter of the brain, where most CCMs originate (Boulday et al., 2011; Golden et al., 2015; Hart et al., 2013; Zhou et al., 2016b), is rich in versican and that elevated levels of ADAMTS-cleaved versican appear immediately adjacent to postcapillary venules early in CCM pathogenesis in the neonatal mouse model (Zhou et al., 2016b). These findings suggested that ADAMTS-mediated proteolysis of versican may also be a causal downstream mechanism in CCM pathogenesis. To investigate whether versican-cleaving ADAMTS proteases (also known as versicanases) play a role in CCM pathogenesis, we adopted a genetic approach in which we generated neonatal mice with endothelial-specific deletion of Krit1 or Ccm2 (VE-Cadherin-CreERT2;Krit1fl/fl and VE-Cadherin-CreERT2;Ccm2fl/fl; hereafter denoted “Krit1ECKO” and “Ccm2ECKO,” respectively; see Table 1 for a summary of genotypes and associated phenotypes) in combination with genetic loss of ADAMTS function to determine if reduced ADAMTS activity could rescue CCM lesion formation. Similar genetic strategies have been used to establish the roles of endothelial TLR4, MEKK3, KLF2, and KLF4 in CCM pathogenesis (Tang et al., 2017; Zhou et al., 2016b). We have also previously demonstrated that endothelial loss of any of the three CCM genes (i.e., Krit1, Ccm2, or Pdcd10) confers CCM formation that can be reversed to a similar extent with partial loss of MEKK3 activity (Tang et al., 2019; Zhou et al., 2016b), and as such the genes are functionally interchangeable for endothelial-specific studies.
Table 1. Summary of described phenotypes arising from inactivation/activation of ADAMTS genes or versican in the CCM mouse model.
| Abbreviated name | Genotype | Phenotype | Vivarium | Strain |
|---|---|---|---|---|
| Krit1ECKO, Krit1BECKO | VE-Cadherin-CreERT2; Krit1fl/fl or Slco1c1(BAC)-Cre; Krit1fl/fl | Numerous lesions in the hindbrain | University of Pennsylvania | Mixed |
| Ccm2ECKO | VE-Cadherin-CreERT2; Ccm2fl/fl | Same as Krit1(B)ECKO | Centenary Institute | Mixed |
| N/A | PDGFb-CreERT2; Pdcd10fl/− | Same as Krit1(B)ECKO | Duke University | C57BL/6J |
| Krit1ECKO; Adamts4−/− | VE-Cadherin-CreERT2; Krit1fl/fl; Adamts4−/− | No reduction in lesion formation | University of Pennsylvania | Mixed |
| Krit1ECKO; Adamts1fl/− | VE-Cadherin-CreERT2; Krit1fl/fl; Adamts1fl/− | No reduction in lesion formation | University of Pennsylvania | Mixed |
| Krit1ECKO; Adamts1fl/fl; Adamts4−/− | VE-Cadherin-CreERT2; Krit1fl/fl; Adamts1fl/fl; Adamts4−/− | No reduction in lesion formation | University of Pennsylvania | Mixed |
| Ccm2ECKO; Adamts5−/− | VE-Cadherin-CreERT2; Ccm2fl/fl; Adamts5−/− | Significant reduction in lesion formation | Centenary Institute | Mixed |
| Krit1ECKO; Adamts5fl/fl; | VE-Cadherin-CreERT2; Krit1fl/fl; Adamts5fl/fl; | Significant reduction in lesion formation | University of Pennsylvania | Mixed |
| ADAMTS5BEC-GOF + doxycycline | Slco1c1(BAC)-Cre; LSL-rtTA; tetO-Adamts5 | Nascent CCM-like vascular changes | University of Pennsylvania | Mixed |
| Krit1BECKO + doxycycline | Slco1c1(BAC)-Cre; Krit1fl/fl | Significant reduction in lesion formation | University of Pennsylvania | Mixed |
| Krit1BECKO; ADAMTS5BEC-GOF + doxycycline | Slco1c1(BAC)-Cre; Krit1fl/fl; LSL-rtTA; tetO-Adamts5 | Increase in lesion formation | University of Pennsylvania | Mixed |
| Krit1ECKO; VcanRC/+ | VE-Cadherin-CreERT2; Krit1fl/fl; VcanRC/+ | Significant reduction in lesion formation | University of Pennsylvania | Mixed |
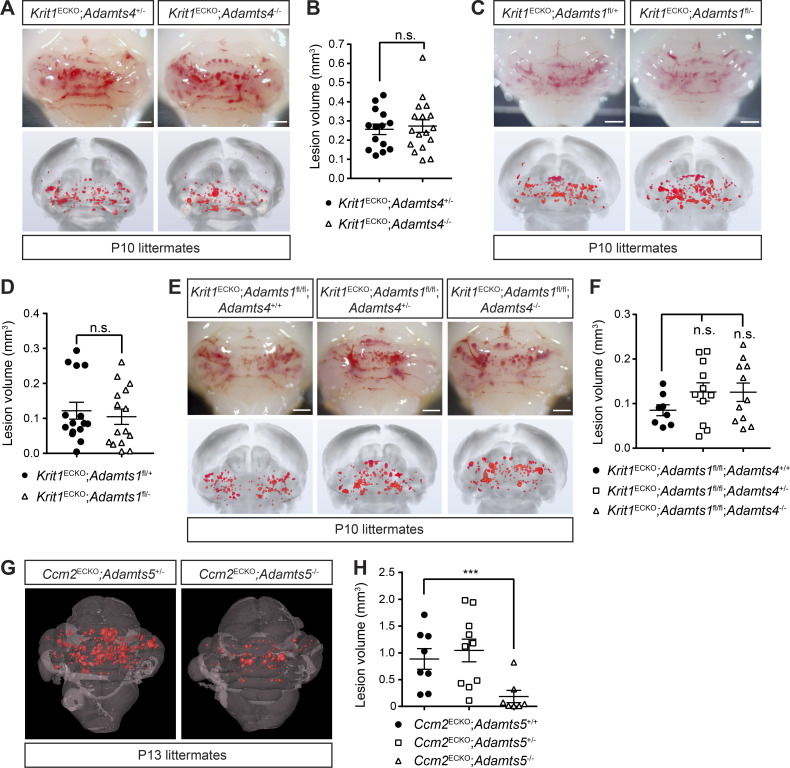
We first tested the role of ADAMTS4 because prior studies had revealed that Adamts4 is up-regulated in early CCM lesions (Zhou et al., 2016b). To do this, we generated Krit1ECKO mice on an ADAMTS4-deficient background. Due to the complexity of the breeding scheme required to generate animals with both inducible loss of CCM function and constitutive loss of ADAMTS4 function, most of these studies compared CCM loss of function on ADAMTS4 haplo-insufficient and fully deficient backgrounds. Visual appearance of lesion density in the hindbrain and blinded micro-computed tomography (microCT)–based quantitation of CCM lesion volumes were indistinguishable among Krit1ECKO;Adamts4+/− and Krit1ECKO;Adamts4−/− animals (Fig. 1, A and B).
Figure 1.
Loss of ADAMTS5, but not ADAMTS4 and ADAMTS1, rescues CCM formation. (A) Visual detection of CCM lesions in the hindbrains of P10 Krit1ECKO;Adamts4+/− (top left) and Krit1ECKO;Adamts4−/− (top right) littermates and corresponding composite microCT images (bottom). (B) Quantification of CCM lesion volumes (n = 14–17 from five litters, Student’s t test). (C) Visual detection of CCM lesions in the hindbrains of P10 Krit1ECKO;Adamts1fl/+ (top left) and Krit1ECKO;Adamts1fl/− (top right) littermates and corresponding composite microCT images (bottom). (D) Quantification of CCM lesion volumes (n = 15 from five litters, Student’s t test). (E and F) Combinatorial endothelial loss of ADAMTS1 and global loss of ADAMTS4 do not reduce CCM lesion burden. Visual appearance of CCM lesions (E, top), corresponding composite microCT images (E, bottom), and quantification of CCM lesion volumes (F; n = 8–11 from six litters, ANOVA + Tukey test). (G and H) Genetic rescue of CCM formation with global loss of Adamts5. MicroCT images of CCM lesions in Ccm2ECKO;Adamts5+/− and Ccm2ECKO;Adamts5−/− littermates (G) and quantification of CCM lesion volumes (H; n = 7–10 from five litters, ANOVA + Tukey test). Scale bars, 1 mm. Error bars represent ± SEM. Not significant (n.s.), P > 0.05; ***, P < 0.001.
Prior studies have shown that the metalloproteases ADAMTS1 and ADAMTS5 are also expressed by ECs and play key roles in versican proteolysis in the developing heart (Dupuis et al., 2011; Stankunas et al., 2008; Zhou et al., 2016b). Similar to Krit1ECKO;Adamts4−/− animals, however, we found that loss of ADAMTS1 (in Krit1ECKO;Adamts1fl/− animals, Fig. 1, C and D) as well as combinatorial loss of endothelial ADAMTS1 and global ADAMTS4 (in Krit1ECKO;Adamts1fl/fl; Adamts4−/− animals) had no significant effect on lesion burden (Fig. 1, E and F).
Several lines of evidence suggested that ADAMTS5 may play a role: (i) comparative studies of ADAMTS enzymatic activity have demonstrated that ADAMTS5 is a more potent versicanase than ADAMTS4 or ADAMTS1 (Gendron et al., 2007; Santamaria et al., 2019); (ii) our previous studies of the developing heart demonstrated that gain of ADAMTS5 activity underlies the loss of cardiac jelly conferred by endocardial loss of CCM function (Zhou et al., 2015); and (iii) we recently demonstrated a significant up-regulation of ADAMTS5 (log2[FC] = 4.2; P = 0.0003, false discovery rate–corrected) in the transcriptome of human lesional CCM neurovascular units (Koskimäki et al., 2019). We therefore hypothesized that gain of ADAMTS5 activity may play a causal role in CCM pathogenesis. To test the role of ADAMTS5, we measured lesion formation in Ccm2ECKO;Adamts5+/− and Ccm2ECKO;Adamts5−/− animals compared with littermate Ccm2ECKO;Adamts5+/+ controls. Global loss of ADAMTS5 resulted in a marked (80%) reduction in lesion burden (Fig. 1, G and H), consistent with a requirement for ADAMTS5 protease function during CCM formation.
Endothelial ADAMTS5 contributes to CCM formation
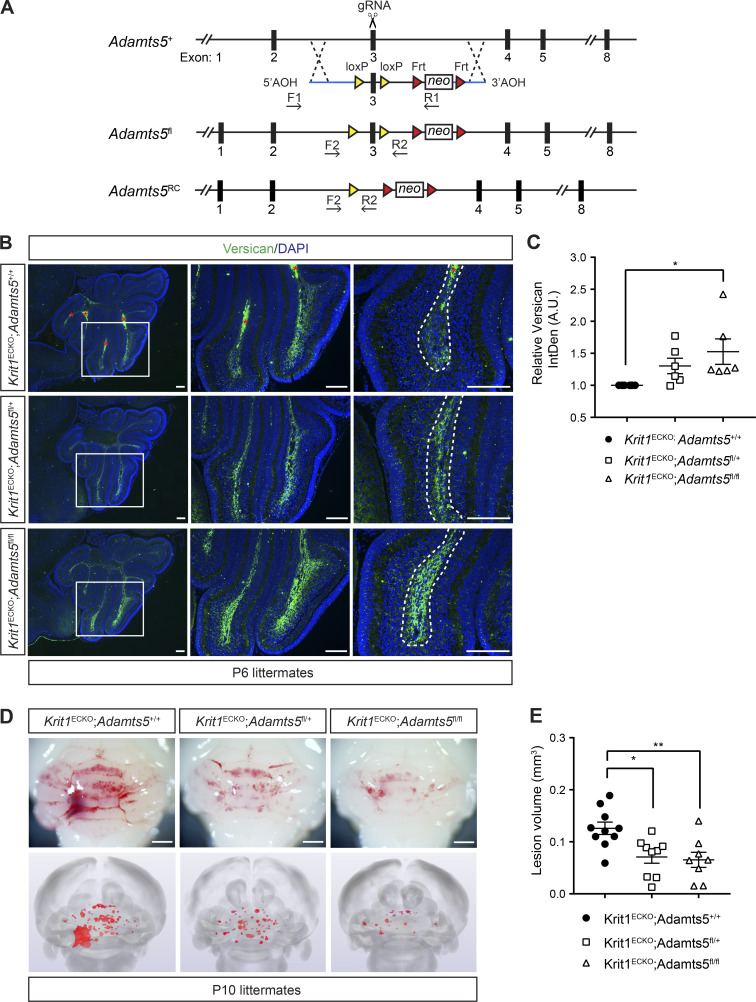
To address whether ADAMTS5 expression by ECs is required for CCM formation, and therefore whether ADAMTS5 might be likely to participate downstream of MEKK3-KLF2/4 signaling in CCM pathogenesis, we generated a conditional Adamts5 allele by flanking exon 3, which encodes the catalytic domain of the protease, with loxP sites (Fig. 2 A and Fig. S1). After crossing Adamts5fl/fl animals onto the Krit1ECKO background, immunostaining of P6 cerebellar sections revealed an increase (53%) in versican protein with loss of both Adamts5 alleles compared with Krit1ECKO;Adamts5+/+ littermates (Fig. 2, B and C; in contrast to combinatorial deletion of Adamts1 and Adamts4, Fig. S2), demonstrating that endothelial-derived ADAMTS5 is required for much of the versican proteolysis in the Krit1ECKO brain. Visual inspection of CCM formation in the intact hindbrain and blinded microCT quantitation of lesion volume revealed significant rescue of CCM formation (50%) with simultaneous endothelial loss of ADAMTS5 (Fig. 2, D and E). We have previously shown that the gut microbiome plays a critical role in CCM formation, and that differences in the microbiome may result in dramatic differences in lesion formation after genetic loss of CCM function in BECs (Tang et al., 2017). The differences in the absolute extent of lesion formation between the global and conditional genetic studies of ADAMTS5 deficiency described above are therefore likely to reflect the greater and lesser sensitivity to CCM formation associated with mouse microbiomes at the two institutions at which these studies were performed. Nonetheless, these findings identify gain of endothelial ADAMTS5 expression as causal for CCM formation, and suggest that ADAMTS5 is a critical downstream mechanism of MEKK3-KLF2/4 signaling in this disease. The observation that global loss of ADAMTS5 conferred a greater degree of rescue than endothelial-specific loss suggests that other cell types may also contribute to ADAMTS5 expression during CCM lesion formation.
Figure 2.
Endothelial loss of ADAMTS5 reduces CCM formation. (A) Schematic representation of Adamts5 targeting construct and strategy to create Adamts5fl and Adamts5RC alleles. RC denotes recombined allele following Cre activity. F1-R1 and F2-R2 represent primers used for screening and detecting recombination. (B) Immunostaining of versican in P6 cerebellums from Krit1ECKO;Adamts5+/+, Krit1ECKO;Adamts5fl/+, and Krit1ECKO;Adamts5fl/fl mice. Boxed regions are shown at higher magnification to the right. Dotted lines delineate white matter. Asterisks indicate erythrocyte autofluorescence. Scale bars, 150 µm. (C) Quantitation of versican IntDen along the white matter tracts relative to Krit1ECKO;Adamts5+/+ (n = 6 from three litters, ANOVA + Tukey test). A.U., arbitrary units. (D and E) Genetic rescue of CCM formation with endothelial-specific loss of ADAMTS5. Visual detection of CCM lesions in the hindbrains of P10 littermates (D, top), corresponding composite microCT images (D, bottom), and quantification of CCM lesion volumes (E; n = 8–10 from six litters, ANOVA + Tukey test). Scale bars, 1 mm. Error bars represent ± SEM; *, P < 0.05; **, P < 0.01.
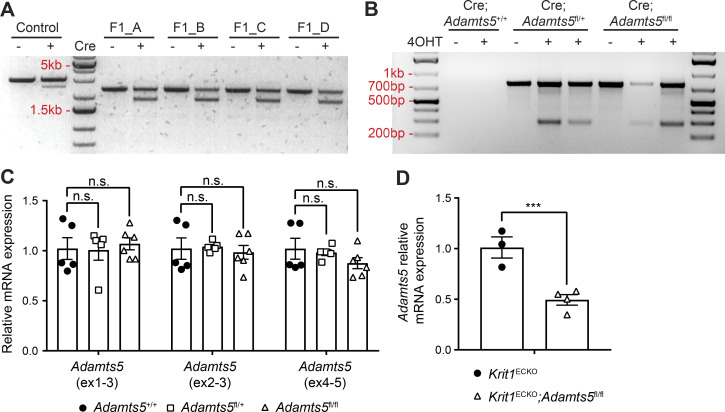
Figure S1.
Characterization of Adamts5fl mice. (A) In vitro Cre recombination of the Adamts5fl allele performed on PCR products from four F1 germline Adamts5fl animals. Note the generation of a smaller band (∼2 kb) with Cre exposure indicative of recombination. Primers outside of the arm of homology and in the NeoR cassette (F1-R1, Fig. 2 A) were used. (B) In vivo Cre recombination assessed in 4OHT-induced and noninduced Krit1ECKO;Adamts5+/+, Krit1ECKO;Adamts5fl/+, and Krit1ECKO;Adamts5fl/fl animals by PCR of tail DNA. Primers flanking the floxed region (F2-R2, Fig. 2 A) of the Adamts5fl allele were used to detect Cre recombination (at 288 bp). (C) qPCR analysis of Adamts5 mRNA levels in the hindbrain of P10 animals in the absence of Cre recombination (by three distinct primer sets; n = 5 or 6 from at least three litters, ANOVA + Tukey test). Results are presented relative to Adamts5+/+ mice. (D) qPCR analysis of Adamts5 mRNA levels in lung ECs isolated from 4OHT-induced animals at P10 using the exon1-3 primer set (n = 3 or 4 from two litters, Student’s t test). Error bars represent ± SEM. Not significant (n.s.), P > 0.05; ***, P < 0.001.
Figure S2.
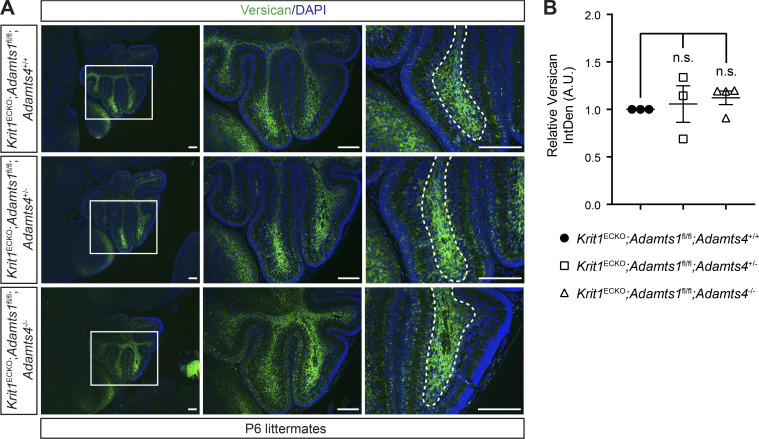
Combinatorial loss of ADAMTS1 and ADAMTS4 does not alter versican levels. (A) Versican immunostaining of cerebellums from P6 littermate animals. Boxed regions are shown at higher magnification to the right. Dotted lines delineate white matter. Scale bars, 150 µm. (B) Quantitation of versican IntDen along white matter tracts relative to Krit1ECKO;Adamts1fl/fl;Adamts4+/+ mice (n = 3 or 4 from three litters, ANOVA + Tukey test). Error bars represent ± SEM. Not significant (n.s.), P > 0.05.
Gain of brain endothelial ADAMTS5 expression results in white matter versican proteolysis
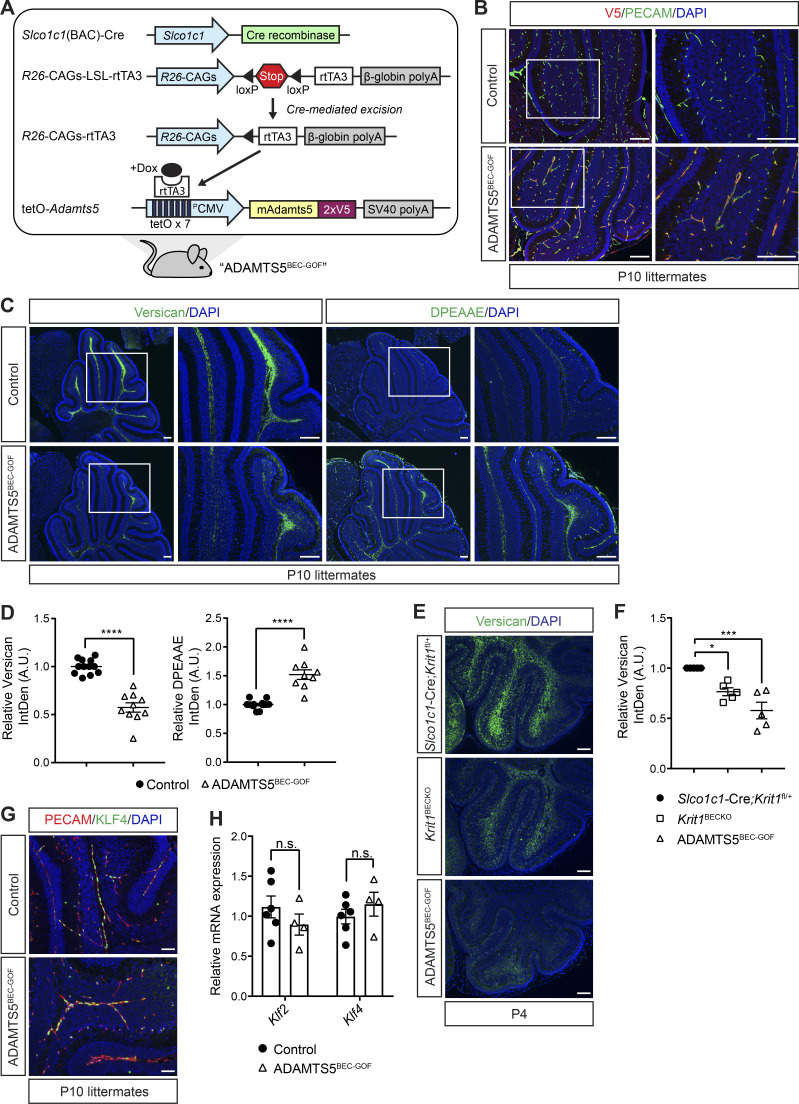
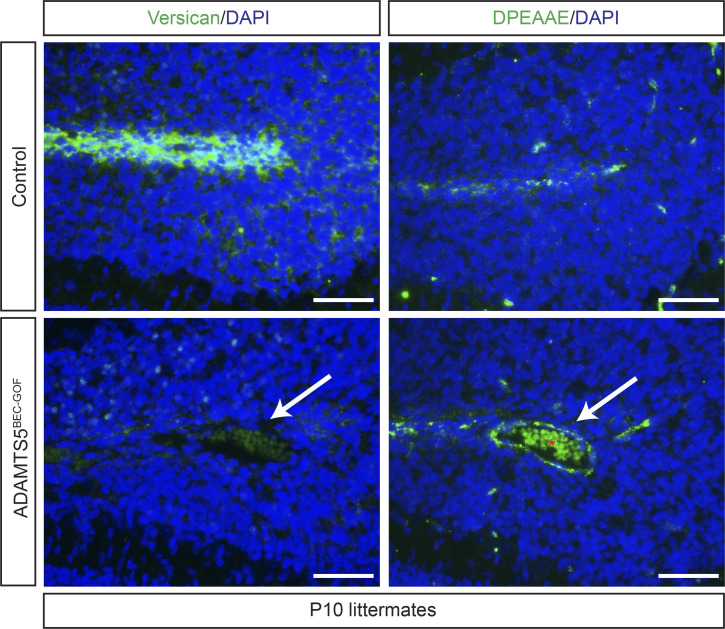
To test whether gain of ADAMTS5 is sufficient for CCM formation, we generated mice carrying a tetracycline operator (tetO)–Adamts5 allele to enable tetracycline-inducible expression of a C-terminally V5-tagged ADAMTS5 protein. The tetO-Adamts5-V5 transgene was crossed to constitutive brain endothelial-specific Cre and Cre-activated reverse tetracycline-controlled transactivator (rtTA) transgenes to generate animals in which ADAMTS5-V5 could be inducibly expressed in BECs of mice (Slco1c1(BAC)-Cre;LSL-rtTA;tetO-Adamts5; hereafter denoted “ADAMTS5BEC-GOF”; Fig. 3 A), independent of changes in CCM function or the MEKK3-KLF2/4 pathway. Doxycycline administration to neonatal ADAMTS5BEC-GOF mice resulted in endothelial-specific expression of V5-tagged ADAMTS5 in the cerebellum (Fig. 3 B). Gain of endothelial ADAMTS5 expression was associated with significantly reduced levels of total versican (43%) and increased levels of cleaved versican (anti-DPEAAE, 52%) in the white matter of the hindbrain (Fig. 3, C and D), the major site of CCM genesis. Comparison of versican immunostaining revealed that the level of ADAMTS activity observed in ADAMTS5BEC-GOF mice at P4 was similar to that observed in Krit1BECKO mice (Fig. 3, E and F). Importantly, immunostaining for KLF4 protein (Fig. 3 G) and quantitative PCR (qPCR) analysis of Klf2 and Klf4 mRNA in cerebellar ECs (Fig. 3 H) revealed no change in the expression of KLF2 and KLF4 with gain of ADAMTS5 expression. These studies identify a genetic approach by which functional ADAMTS5 protease can be inducibly expressed in BECs and reveal that ADAMTS5 does not induce KLF2 and/or KLF4 expression.
Figure 3.
Characterization of tetO-Adamts5 mice. (A) Schematic representation of the genetic strategy used to generate ADAMTS5BEC-GOF mice in which doxycycline drives inducible expression of V5-tagged ADAMTS5 specifically in BECs. (B) Co-immunostaining of V5 and PECAM (EC marker) in P10 cerebellums reveals expression of endothelial ADAMTS5-V5 in ADAMTS5BEC-GOF mice compared with littermate controls. Results are representative of n > 3 litters. (C) Immunostaining of versican (C, left) and DPEAAE (ADAMTS-cleaved versican; C, right) in serial cerebellar sections of P10 mice. Boxed regions are shown at higher magnification to the right. Scale bars, 150 µm. (D) Quantitation of versican (left) and DPEEAE (right) IntDen along the white matter tracts in ADAMTS5BEC-GOF mice relative to littermate controls (versican: n = 10–12 from eight litters; DPEEAE: n = 9–12 from seven litters, Student’s t test). (E and F) Versican immunostaining is reduced in ADAMTS5BEC-GOF mice to an extent comparable to Krit1BECKO animals at P4 (E). Scale bars, 100 µm. Quantitation of versican IntDen along cerebellar white matter tracts relative to Slco1c1-Cre;Krit1fl/+ mice (F; n = 5 from at least four litters, ANOVA + Tukey test). (G) Immunostaining for KLF4 in the cerebellar vessels of the white matter at P10. Scale bars: 50 µm. Results are representative of n > 3 litters. (H) qPCR analysis of mRNA levels of Klf2 and Klf4 in ECs isolated from control and ADAMTS5BEC-GOF P10 cerebellums (n = 4–6 from two litters, Student’s t test). Controls (B–D, G, and H) were littermate animals carrying one or two of the following alleles: the Cre transgene, the rtTA transgene, or the tetO-Adamts5-V5 allele. Error bars represent ± SEM. Not significant (n.s.), P > 0.05; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Gain of ADAMTS5 expression in BECs confers vascular changes like those in nascent CCM lesions
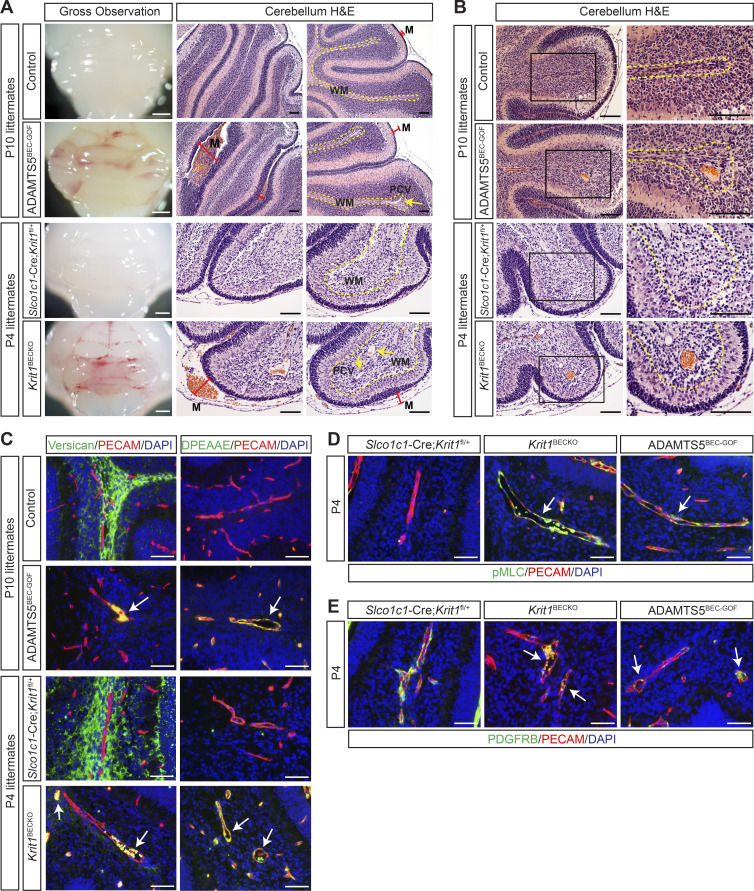
To test whether ADAMTS5 activity is sufficient for CCM lesion formation, we next characterized the brain vasculature of P4 and P10 ADAMTS5BEC-GOF mice with respect to changes that have been identified as characteristic of CCM lesion formation. In Slco1c1(BAC)-Cre mice in which the Krit1 gene is constitutively deleted in BECs (Slco1c1(BAC)-Cre;Krit1fl/fl; hereafter denoted “Krit1BECKO”), CCM lesions arose throughout the brain, but predominantly localized to the hindbrain. Beginning at P3–P4, the earliest lesions appeared in the postcapillary venule in the white matter as vascular dilatations that expanded to become large blood-filled caverns over the course of several days (Fig. S3). A second cardinal feature of CCM pathogenesis in the neonatal mouse model is meningeal venous dilatation (Boulday et al., 2011), detectable visually and histologically (Fig. S3). Visual analysis of P10 ADAMTS5BEC-GOF mice revealed a bloody appearance of the hindbrain, similar to that seen in the P4 Krit1BECKO mice (Fig. 4 A, left), but lacking darker, punctate lesions indicative of mature CCMs (Fig. S3). Histological analysis identified the cause of this appearance as dilated blood-filled meningeal vessels in both models (Fig. 4 A, middle). P10 ADAMTS5BEC-GOF mice also exhibited postcapillary venule dilatation (Fig. 4 A, right) and blood-filled malformations in the cerebellum identical to the nascent CCM lesions seen in the early Krit1ECKO and Krit1BECKO models (Fig. 4 B). These changes, like those observed in Krit1ECKO (Zhou et al., 2016b) and Krit1BECKO animals, were associated with decreased perivascular versican and increased abluminal, ADAMTS-cleaved versican (anti-DPEAAE; Fig. 4 C and Fig. S4). CCM lesions in humans as well as in the neonatal mouse model have been associated with a marked increase in EC phosphorylated-myosin light chain (pMLC; Zhou et al., 2016b) and a loss of associated pericytes (Tanriover et al., 2013; Zhou et al., 2016a). ADAMTS5BEC-GOF animals also exhibited increased endothelial pMLC (Fig. 4 D) and reduced pericyte coverage (anti-PDGFRB; Fig. 4 E). In contrast to the neonatal Krit1ECKO and Krit1BECKO models at later time points (e.g., P10 in Fig. S3), however, the vascular phenotype observed in ADAMTS5BEC-GOF mice failed to progress to large dilated caverns. These results demonstrate that ADAMTS5 overexpression is sufficient to recapitulate numerous key aspects of early-stage CCM lesions, but not for development of fully mature cavernomas.
Figure S3.
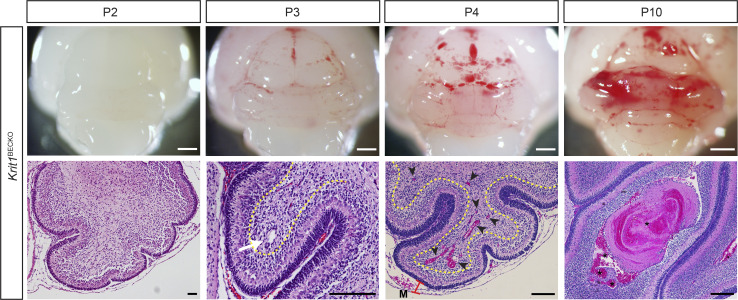
CCM lesion progression in the Krit1BECKO model. Key characteristics of lesions formed in the constitutive BEC-specific CCM knockout model. Vascular malformations are first detected at P3 as dilated venules (white arrow) in the cerebellar white matter (yellow dotted line) and grow to become large, blood-filled caverns by P10. Black arrowheads indicate multiple, small lesions formed at P4. CCM-deficient animals also exhibit characteristic dilatation of meningeal vessels (M, red bar). Asterisks mark trapped intravascular erythrocytes. Scale bars, 1 mm (white) and 150 µm (black).
Figure 4.
Forced expression of ADAMTS5 in BECs confers a vascular phenotype closely resembling early CCMs. (A) Hindbrains from P10 ADAMTS5BEC-GOF and P4 Krit1BECKO mice exhibit a similar bloody appearance visually (left). H&E staining reveals dilated, blood-filled meningeal vessels (M; red bars). Dilated postcapillary venules (PCV; yellow arrows, right) are also detected in the white matter tract of ADAMTS5BEC-GOF and Krit1BECKO animals. Scale bars, 1 mm (white) and 100 µm (black). Results are representative of n > 4 litters. WM, white matter. (B) A nascent CCM-like vascular phenotype characterized by dilated, blood-filled postcapillary venules in the white matter of the hindbrain is detected by H&E staining in ADAMTS5BEC-GOF but not control animals. Higher magnification images are shown on the right. Scale bars, 100 µm. Results are representative of n > 4 litters. (C) Co-immunostaining for versican and DPEEAE (ADAMTS-cleaved versican) with PECAM (EC marker) show similar changes around ADAMTS5BEC-GOF and Krit1BECKO lesions (white arrows). Asterisks indicate erythrocyte autofluorescence. Scale bars, 50 µm. (D) Anti-pMLC immunostaining in white matter venules (white arrows) of P4 ADAMTS5BEC-GOF and Krit1BECKO animals. Scale bars, 50 µm. (E) Anti-PDGFRB (pericyte marker) immunostaining in white matter venules (white arrows) of P4 ADAMTS5BEC-GOF and Krit1BECKO animals. Scale bars, 50 µm. Controls (A–E) were littermate animals carrying one or two of the following alleles: the Cre transgene, the rtTA transgene, or the tetO-Adamts5-V5 allele. Dotted lines in A and B delineate cerebellar white matter. Results shown in C–E are representative of three litters.
Figure S4.
Additional immunohistochemical characterization of ADAMTS5BEC-GOF animals. Versican and DPEAAE immunostaining in adjacent serial sections of P10 cerebellums. Controls were littermate animals carrying one or two of the following alleles: the Cre transgene, the rtTA transgene, or the tetO-Adamts5-V5 allele. White arrows indicate vascular changes within the white matter tract closely resembling nascent CCM lesions. Asterisks indicate erythrocyte autofluorescence. Scale bars, 50 µm.
Gain of ADAMTS5 synergizes with loss of KRIT1 during CCM formation
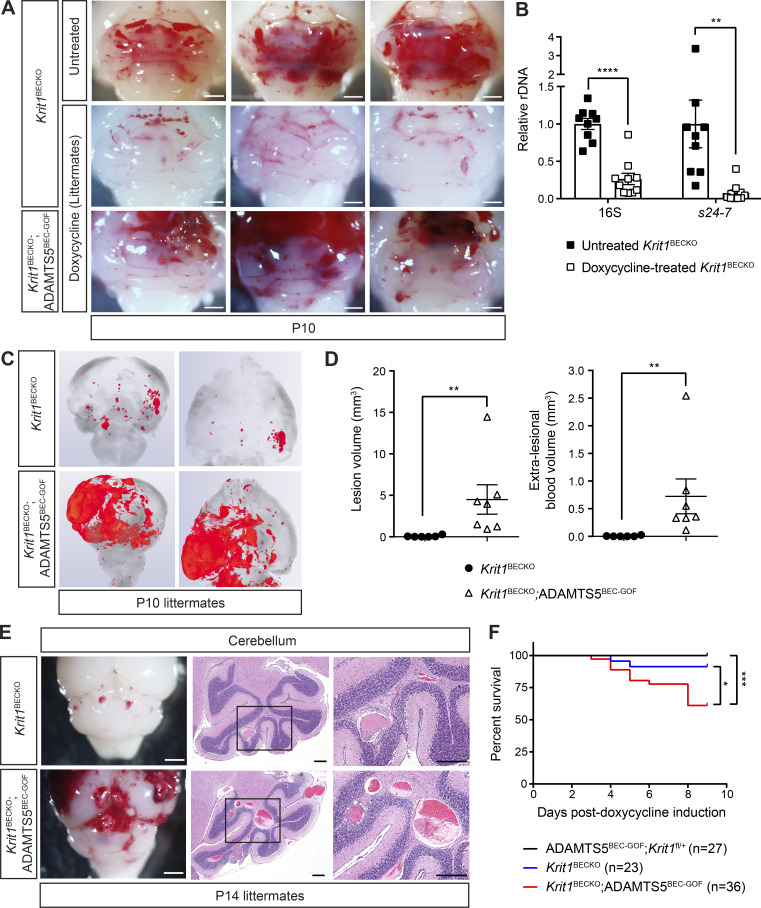
The studies described above revealed that ADAMTS5 plays an important role in CCM formation, but that gain of ADAMTS5 activity alone is not sufficient for full CCM lesion formation. One explanation for these findings is that other targets of MEKK3-KLF2/4 signaling are also required for full lesion progression. We have recently demonstrated that the gut microbiome is essential to drive MEKK3-KLF2/4 signaling in BECs during CCM pathogenesis, and that broad-spectrum antibiotic treatment prevents CCM formation in Krit1ECKO animals (Tang et al., 2017). Thus, administration of doxycycline, the antibiotic required to activate ADAMTS5 expression in ADAMTS5BEC-GOF animals, is predicted to also significantly reduce the level of MEKK3-KLF2/4 signaling in BECs and thereby inhibit CCM formation independent of any effect on the tetO-Adamts5 transgene. To determine whether the effect of gain of ADAMTS5 expression is modulated by the level of MEKK3-KLF2/4 signaling, we compared CCM lesion formation in doxycycline-treated Krit1BECKO and Krit1BECKO;ADAMTS5BEC-GOF littermates in which loss of KRIT1 augments MEKK3-KLF2/4 signaling. Compared with conventional, susceptible Krit1BECKO animals (Fig. 5 A, top row), doxycycline treatment resulted in a very low CCM lesion burden in Krit1BECKO mice (Fig. 5, A, C, and D), in association with a reduction in total gut bacterial load as measured by reduced 16S (74%) and gram-negative Bacteroidetes s24-7 levels (92%; Fig. 5 B). In contrast, Krit1BECKO;ADAMTS5BEC-GOF littermates exhibited a markedly more severe phenotype, with large hemorrhages and CCM lesions observed visually (Fig. 5 A) and a significant increase in lesional (63-fold) and extra-lesional blood (121-fold) volume determined by quantification of microCT images and histology (Fig. 5, C and D). Examination of H&E-stained sections confirmed a significant increase in the number and size of vascular lesions in Krit1BECKO;ADAMTS5BEC-GOF mice compared with Krit1BECKO control littermates (Fig. 5 E). Finally, brain endothelial gain of ADAMTS5 combined with KRIT1 loss resulted in the death of 39% of animals by P10, compared with only 9% lethality in Krit1BECKO littermates (Fig. 5 F). These studies provide evidence of strong synergy between KRIT1 loss and ADAMTS5 gain in BECs during CCM formation. The ability of ADAMTS5 gain of function to amplify CCM formation in antibiotic-treated animals is consistent with ADAMTS5 functioning downstream of MEKK3-KLF2/4 signaling.
Figure 5.
Endothelial gain of ADAMTS5 and loss of KRIT1 synergize during CCM formation. (A) Hindbrains from untreated Krit1BECKO animals or doxycycline-treated Krit1BECKO and Krit1BECKO;ADAMTS5BEC-GOF littermate animals at P10. Images from three different doxycycline-treated litters are shown and representative of seven litters. Scale bars, 1 mm. (B) Relative quantification of neonatal gut bacterial load measured by qPCR of bacterial 16S and Bacteroidetes s24-7 rRNA gene copies (n = 9 or 10 from at least three litters, Student’s t test). (C) Composite microCT images from the hindbrain (left) and forebrain (right) of Krit1BECKO and Krit1BECKO;ADAMTS5BEC-GOF littermates. (D) Volumetric analysis of lesional and extra-lesional blood. The distinction between lesional and extra-lesional blood is shown due to the high rate of peri-lesional hemorrhage in Krit1BECKO; ADAMTS5BEC-GOF animals compared with Krit1BECKO animals (n = 6 or 7 from six litters, Mann–Whitney U test). (E) Visual assessment (left) and corresponding H&E staining (center and right) of the hindbrain demonstrate an increase in both the size and number of CCM lesions in Krit1BECKO;ADAMTS5BEC-GOF mice compared with Krit1BECKO mice. Higher magnification images are shown on the right. Scale bars, 1.5 mm (white) and 250 µm (black). Results are representative of three litters. (F) Kaplan–Meier survival curves of ADAMTS5BEC-GOF;Krit1fl/+, Krit1BECKO, and Krit1BECKO;ADAMTS5BEC-GOF mice during the first 10 d of life following doxycycline induction at P1 (n = 23–36 per group from 19 litters, log-rank test). Error bars represent ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Deficiency of the ADAMTS5 substrate versican reduces CCM formation
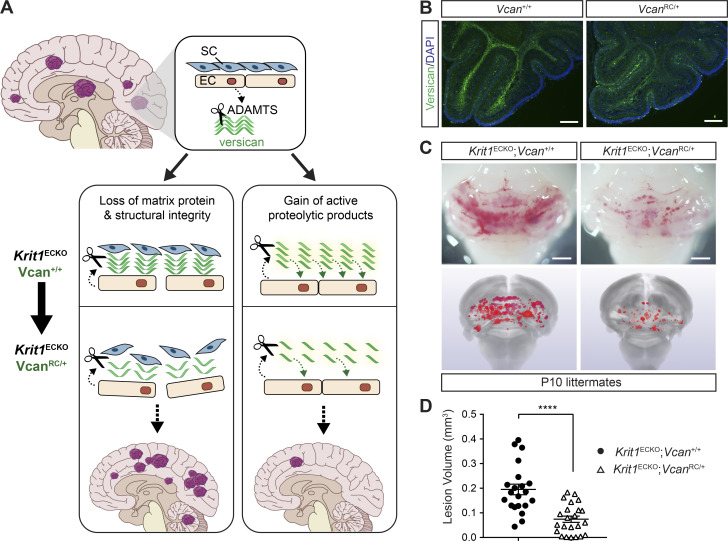
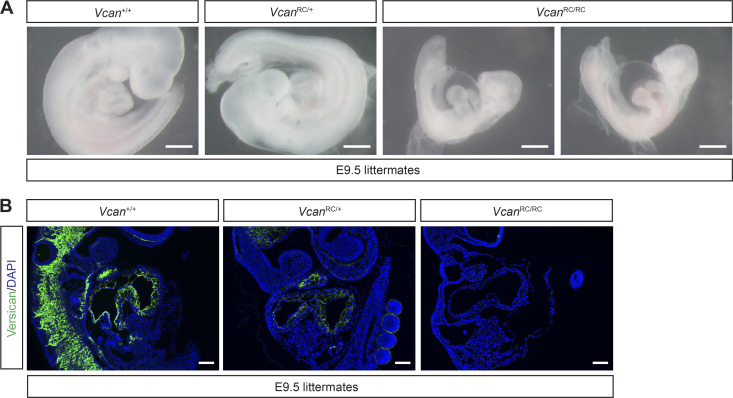
The ADAMTS5 protease cleaves proteoglycans including versican, aggrecan, brevican, and neurocan (Stanton et al., 2011). Our previous studies have shown that CCM lesions arise in the versican-rich white matter of the brain, and that increased staining for DPEAAE, a versican neoepitope that is exposed upon ADAMTS-mediated proteolysis, is detected very early during CCM lesion formation in Krit1ECKO mice (Zhou et al., 2016b; Fig. 4 C). These observations, and prior studies implicating ADAMTS5 in the degradation of versican in the developing heart following endocardial KRIT1 loss (Zhou et al., 2015), suggested that the role of ADAMTS5 in CCM lesion formation might be related to proteolytic cleavage of perivascular versican in the white matter of the brain. Previous studies have identified two distinct mechanisms by which ADAMTS-mediated cleavage of versican may alter the behavior of neighboring cells, loss of a structural matrix protein (Del Monte-Nieto et al., 2018) and gain of biologically active versican proteolytic fragments called versikines (Enomoto et al., 2010; McCulloch et al., 2009b). ADAMTS5-mediated proteolysis of versican could therefore support CCM lesion formation either by eliminating a required structural matrix protein or by generating new peptides with pathogenic effects (Fig. 6 A). To test the role of versican proteolysis in CCM formation, and potentially distinguish between these two possible mechanisms, we compared CCM formation in Krit1ECKO mice with normal (Vcan+/+) or reduced (VcanRC/+; RC, recombined) levels of versican. The VcanRC allele was generated by recombination of the Vcanfl allele. VcanRC/RC mice exhibit embryonic lethality at embryonic day (E) 9.5 associated with pericardial edema (Fig. S5 A) as similarly reported for versican-deficient animals (Mjaatvedt et al., 1998), consistent with a complete loss of versican function. If ADAMTS5 proteolysis of versican confers CCM formation by eliminating a required structural matrix protein, reduced versican levels in VcanRC/+ animals would be predicted to increase CCM lesion formation. Conversely, if ADAMTS5 proteolysis of versican generates biologically active peptides, VcanRC/+ animals would be predicted to exhibit less CCM lesion formation. We evaluated the effect of versican loss using constitutive, global VcanRC/+ animals because (i) complete versican deficiency results in early embryonic lethality (Mjaatvedt et al., 1998), (ii) single-cell RNA sequencing data indicate that versican is synthesized by non-ECs in the brain (Vanlandewijck et al., 2018), and (iii) versican protein is long-lived and slowly depleted following conditional gene deletion after birth (Kang et al., 2017). Immunostaining of postnatal Vcan+/+ and VcanRC/+ brains confirmed that loss of one allele of Vcan results in a significant reduction in versican protein in the white matter (Fig. 6 B), a result consistent with a similar level of versican loss in the E9.5 VcanRC/+ embryo (Fig. S5 B). Visual analysis of CCM formation in Krit1ECKO;Vcan+/+ and Krit1ECKO;VcanRC/+ neonates revealed a marked reduction in CCM lesion number and size in Krit1ECKO;VcanRC/+ animals (Fig. 6 C). Blinded microCT quantitation of CCM lesion volume confirmed a significant (62%) reduction in total lesion volume (Fig. 6 D). These findings closely mirror those reported in a prior study of ADAMTS proteolysis of versican during web regression in the developing mouse paw (McCulloch et al., 2009b), and support a mechanism in which ADAMTS5 secreted by CCM-deficient ECs drives CCM lesion formation by generating bioactive versican cleavage products rather than simply degrading versican protein.
Figure 6.
Loss of the ADAMTS5 substrate versican reduces CCM lesion formation. (A) Possible mechanisms by which versican proteolysis may contribute to CCM formation are diagrammed. Versican proteolysis may affect CCM formation through matrix degradation and/or generation of bioactive products. SC, stromal cells. (B) Versican immunostaining of P10 cerebellums from Vcan+/+ and VcanRC/+ mice. Scale bar, 150 µm. Images are representative of two independent experiments. (C) Visual assessment of hindbrains (top) and microCT images (bottom) of CCM lesions in P10 Krit1ECKO;Vcan+/+ and littermate Krit1ECKO;VcanRC/+ animals. Scale bars, 1 mm. (D) Blinded microCT quantitation of CCM lesion volume (n = 21–23 from eight litters, Student’s t test). Error bars represent ± SEM; ****, P < 0.0001.
Figure S5.
Characterization of the VcanRC allele. (A) Visual appearance of E9.5 control, VcanRC/+, and VcanRC/RC embryos. Scale bars, 500 µm. Note VcanRC/RC embryos are severely runted and exhibit pericardial edema compared with littermate controls. (B) Versican immunostaining in E9.5 VcanRC/RC and littermate control hearts. Scale bars, 100 µm. Images are representative of two independent experiments.
Versican cleavage products increase endothelial sprouting and are detected adjacent to wild-type ECs during CCM lesion growth
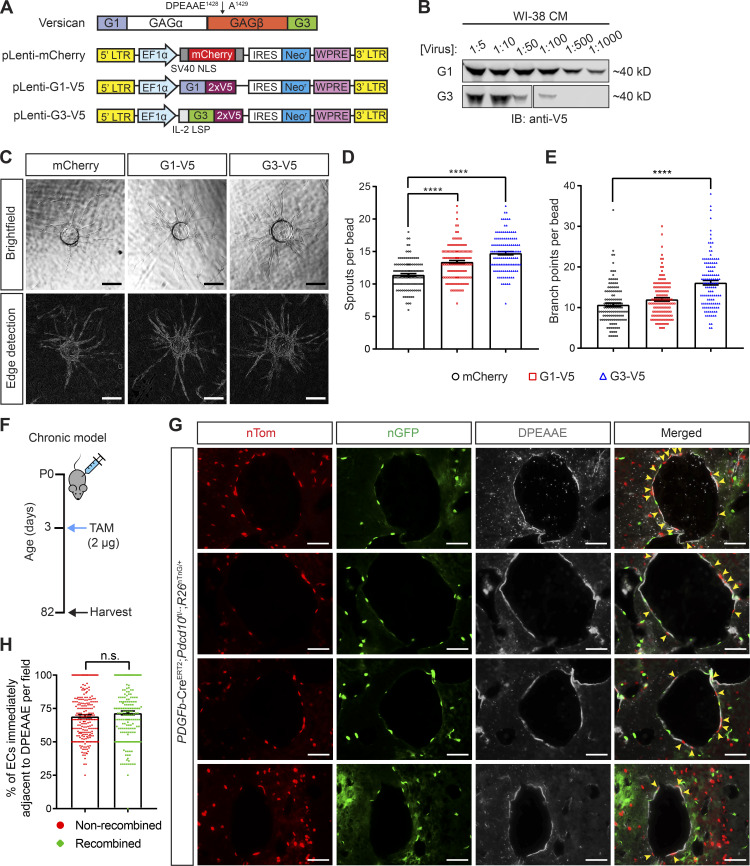
Prior studies have demonstrated biological roles for ADAMTS-generated versican fragments in several developmental and disease contexts (Enomoto et al., 2010; Gueye et al., 2017; Hope et al., 2016; McCulloch et al., 2009b). Additionally, in vitro studies have shown that the N- and C-terminal (i.e., G1 and G3; Fig. 7 A) domains of versican can modulate cell adhesion and proliferation (Yang et al., 1999; Yee et al., 2007; Zhang et al., 1998). To test whether versican fragments exert biological activity on ECs, we generated lentiviruses to exogenously express an mCherry vector control, V5-tagged G1 domain, or V5-tagged G3 domain (Fig. 7 A). To assess the biological function of these versican fragments, we used the lentiviruses shown in Fig. 7 A to generate a stable, neomycin-resistant fibroblast (WI-38) cell line. Analysis of conditioned medium from these cells using anti-V5 immunoblotting revealed extracellular expression of both the G1 and G3 peptides (Fig. 7 B). Since CCM disease arises specifically in highly angiogenic vascular beds such as the neonatal hindbrain and retina (Boulday et al., 2011; DiStefano et al., 2014; Wüstehube et al., 2010), we performed an in vitro three-dimensional sprouting assay to determine whether any of these versican fragments alter angiogenic endothelial behavior (Nakatsu et al., 2007). After 10 d of EC coculture with G1-V5 and G3-V5 expressing fibroblasts, both G1 and G3 exposures were associated with a greater number of sprouts (Fig. 7, C and D), and G3 also associated with increased branching (Fig. 7, C and E) compared with ECs that were cocultured with fibroblasts expressing mCherry vector. These studies are consistent with a mechanism in which ADAMTS5 cleavage of versican releases versikines that exert effects on surrounding ECs.
Figure 7.
Versican cleavage products increase endothelial sprouting in vitro and are detected adjacent to wild-type ECs in mature CCM lesions in vivo. (A) Schematic of the versican protein domain structure and the lentiviral constructs used to express the indicated recombinant versican peptides. The black arrow marks the canonical ADAMTS cleavage site within the GAGβ domain of versican by which the N-terminal “DPEEAE” fragment is generated. The lentiviral control construct encodes an mCherry fluorescent protein. pLenti-G1 and G3 encode the N- and C-terminal globular domains, respectively, of versican. All versican peptides are V5-tagged. LTR, long term repeat; NLS, nuclear localization signal; IRES, internal ribosomal entry site; WPRE, woodchuck posttranscriptional regulatory element; LSP, long signal peptide. (B) V5-immunoblot of conditioned medium collected from WI-38 fibroblasts 48 h post-lentiviral transduction indicates recombinant G1 and G3 are being secreted. Data are representative of two independent experiments. (C) HUVEC spheroids co-cultured with lentivirally infected WI-38 fibroblasts 10 d post-sprouting assay. Bright-field micrographs are shown on top. A Canny edge detection algorithm was applied to the image mask to identify vessel walls for visualization (bottom). Scale bars, 200 µm. Results are representative of at least two independent experiments. (D and E) Quantification of the mean number of sprouts and branch points per bead (n = 125 beads per group, ANOVA + Tukey test). (F) Schematic of the experimental design in which CCM lesions were generated in PDGFb-CreERT2;Pdcd10fl/−;R26nTnG/+ mice in a chronic model using low-dose tamoxifen (TAM). (G) Representative images of lesions from two PDGFb-CreERT2; Pdcd10fl/−;R26nTnG/+ mice in which cells expressing nTom (correlating with wild-type ECs) and nGFP (correlating with PDCD10-deficient ECs) were identified by endogenous fluorescent signals. Anti-DPEAAE immunostaining was used to identify ADAMTS-cleaved versican. Note the detection of DPEAAE staining adjacent to numerous nTom+ ECs (yellow arrows). Scale bars, 50 µm. (H) Quantitative analysis of the percentage of nonrecombined (nTom+) or recombined (nGFP+) lesional ECs identified adjacent to areas of positive DPEEAE staining (n = 233 random high-powered fields from two mice, Student’s t test). Error bars represent ± SEM. Not significant (n.s.), P > 0.05; ****, P < 0.0001.
Recent lineage tracing studies performed by two independent groups have demonstrated that growing CCM lesions incorporate large numbers of wild-type ECs (Detter et al., 2018; Malinverno et al., 2019). These studies provide support for a powerful cell nonautonomous mechanism in CCM pathogenesis (Akers et al., 2009), but how loss of CCM function and gain of MEKK3-KLF2/4 signaling in mutant BECs affects the behavior of neighboring wild-type ECs remains unexplained. The studies described above suggested that ADAMTS5 secretion and perivascular generation of versikines might explain how CCM-deficient ECs alter neighboring wild-type EC behavior during CCM lesion formation. To further test this hypothesis, we generated PDGFb-CreERT2;Pdcd10fl/−;R26nTnG/+ animals in which ECs express nuclear red fluorescence before, and nuclear green fluorescence following, Cre-mediated recombination at the ROSA26 locus. These animals were given a low dose of tamoxifen at P3 and harvested in late adulthood (P82), allowing for slow progression of CCM formation (Fig. 7 F). Since prior studies have demonstrated that the floxed Pdcd10 allele is efficiently recombined (Chan et al., 2011; Detter et al., 2018; Malinverno et al., 2019), nuclear red and green fluorescence is likely to label PDCD10 wild-type and deficient cells, respectively. To determine if versican proteolytic products are generated in close proximity to wild-type cells, immunostaining for DPEAAE was performed in sections containing mature lesions in PDGFb-CreERT2;Pdcd10fl/−;R26nTnG/+ animals. Anti-DPEEAE staining was detected immediately adjacent to both red, nonrecombined ECs and green, recombined ECs within CCM lesions (Fig. 7 G). Quantitative analysis revealed DPEAAE epitope adjacent to 69% of nonrecombined cells and 72% of recombined cells (Fig. 7 H). These findings suggest that wild-type ECs are exposed to ADAMTS-generated versican fragments to an extent similar to that of mutant ECs, and that extracellular, abluminal ADAMTS proteolysis of versican may therefore mediate the cell nonautonomous mechanism of CCM formation.
Discussion
Recent studies have identified the microbiome and gain of MEKK3 signaling as upstream molecular mechanisms of CCM pathogenesis in a model in which cavernoma formation results from elevated KLF2 and KLF4 transcription factor expression in BECs (Tang et al., 2017; Zhou et al., 2016b). However, the events downstream of KLF2/4 expression that drive CCM formation, how transcriptional changes in mutant ECs alter the behavior of neighboring wild-type ECs during CCM lesion growth, and why CCM lesions arise selectively in the white matter of the CNS remain open questions. Our studies begin to address these questions with the identification of gain of ADAMTS5 function and cleavage of its substrate versican as downstream mechanisms of CCM formation. Causal roles for a secreted protease and versican proteolysis explain both how wild-type EC behavior is altered by CCM-deficient ECs and why the versican-rich environment of the brain white matter is selectively affected by this disease. In addition to insights into CCM disease pathogenesis, our studies contribute to a growing body of evidence that ADAMTS cleavage of versican plays central roles in acquired cardiovascular diseases as well as in cardiovascular development.
The recent findings by two groups that small, early CCM lesions are composed primarily of CCM-deficient ECs while larger, more mature CCM lesions contain high numbers of wild-type ECs highlighted a cell nonautonomous effect in CCM pathogenesis (Akers et al., 2009; Detter et al., 2018; Malinverno et al., 2019). However, the molecular basis for such a cell nonautonomous mechanism was not clear. Prior studies in the developing mouse and zebrafish heart have demonstrated that endocardial loss of CCM function results in elevated expression of ADAMTS5 (Zhou et al., 2015), a metalloprotease that specifically cleaves versican and related proteoglycans. In the developing heart, increased ADAMTS5 expression results in loss of the versican-rich cardiac jelly that separates endothelial and myocardial cells and heart failure due to reduced cardiomyocyte proliferation (Zhou et al., 2015)—a cell nonautonomous effect. That the cell nonautonomous effects of endothelial CCM loss of function in the developing heart and postnatal brain share a common molecular mechanism is supported by the findings that endothelial-specific loss of ADAMTS5 rescues CCM formation, while endothelial-specific gain of ADAMTS5 confers hallmark CCM-deficient phenotypes, such as postcapillary venule and meningeal venous dilatation, and vastly augments CCM formation in the established neonatal model. Loss of pericyte coverage is a hallmark phenotype associated with both human and mouse CCMs (Tanriover et al., 2013; Zhou et al., 2016a), and we observe that ADAMTS5 gain of function alone confers pericyte loss in brain venules (Fig. 4 E). Thus, it is likely that pericyte loss is a second cell nonautonomous effect of ADAMTS5-mediated versican proteolysis during CCM pathogenesis. Together, these studies identify a conserved molecular mechanism that underlies a number of prenatal and postnatal CCM-deficient phenotypes. However, since gain of ADAMTS5 expression was not sufficient to confer a fully mature CCM phenotype when MEKK3-KLF2/4 signaling was suppressed by doxycycline administration, it is likely that other, yet to be identified, downstream mechanisms also contribute significantly to cavernoma formation.
ADAMTS5 may cleave proteoglycans other than versican, e.g., ADAMTS5 proteolysis of aggrecan has been implicated as an important mechanism of postnatal cardiovascular disease at other sites such as the aorta (Cikach et al., 2018; Dupuis et al., 2019; Fava et al., 2018), but several lines of evidence indicate that versican is a relevant ADAMTS substrate during CCM formation. First, immunostaining reveals that versican is highly expressed in the white matter of the brain where CCM lesions form in both the neonatal mouse model and in humans (Boulday et al., 2011; Golden et al., 2015; Hart et al., 2013; Zhou et al., 2016b), and temporal analysis of CCM formation reveals a close correlation between an increase in abluminal ADAMTS-cleaved versican and early lesion genesis (Zhou et al., 2016b). Second, aggrecan is expressed at much lower levels than versican in the CNS (Vanlandewijck et al., 2018), while combined loss of both brevican and neurocan, the other two known substrates of ADAMTS5, is well tolerated (Quaglia et al., 2008). Finally, our genetic studies demonstrate that reduced levels of versican significantly impact CCM formation. These findings and the observations that the phenotypes conferred by gain of ADAMTS5 expression are not associated with changes in KLF2/4 expression support the conclusion that ADAMTS-cleavage of versican is a key downstream mechanism of CCM formation. They further suggest that the abundance of perivascular versican in the white matter of the brain may explain why CCMs form selectively at that site.
The finding that loss of versican expression reduces, rather than augments, CCM formation provides unexpected insight into CCM disease pathogenesis specifically and the role of ADAMTS-mediated versican proteolysis in cardiovascular development and disease more generally. In the context of cardiovascular development, the role of versican-rich matrix has often been considered to be primarily mechanical. For example, the versican-rich cardiac jelly that separates the endocardial and myocardial cell layers in the developing heart is thought to control ventricular trabeculation by maintaining physical separation of these cell types, thereby modulating paracrine signals between them, such as those mediated by endothelial-derived neuregulin and cardiomyocyte ErbB receptors (Del Monte-Nieto et al., 2018). This straightforward, mechanical paradigm was recently challenged by genetic studies of ADAMTS protease and versican function during interdigital web regression that demonstrated that loss of versican worsened, rather than improved, defects in web regression caused by ADAMTS protease deficiency (McCulloch et al., 2009b). This study generated a new model in which ADAMTS-mediated versican cleavage is proposed to be required to release proteolytic products, termed versikines, with active biological properties. Our mouse genetic studies reproduce these seminal genetic findings in the context of a postnatal cerebrovascular disease, while our cell culture studies suggest that versikines may exert biological effects on endothelial function. The observation that wild-type ECs in mature CCM lesions are frequently in contact with versican cleavage products further supports a cell nonautonomous mechanism in which versican cleavage products may alter the behavior of wild-type as well as mutant ECs during CCM lesion growth. Future studies will be required to more precisely identify the mechanism(s) by which versican cleavage products affect endothelial function in order to clearly define their role in CCM lesion formation and perhaps other cardiovascular disease states.
Materials and methods
Mice
VE-Cadherin-CreERT2 (Wang et al., 2010), Krit1fl/fl (Mleynek et al., 2014), Ccm2fl/fl (Zheng et al., 2012), Pdcd10fl/fl (He et al., 2010), Adamts4−/− (Enomoto et al., 2010; McCulloch et al., 2009b), Adamts1fl/fl (Boerboom et al., 2011; Mittaz et al., 2004), Adamts1−/− (Lee et al., 2005), Adamts5−/− (McCulloch et al., 2009a), Slco1c1(BAC)-Cre (Lang et al., 2011), Vcanfl/fl (Choocheep et al., 2010), and PDGFb-CreERT2 (Claxton et al., 2008) animals have been previously described. C57BL/6J, DBA/2J, EIIaCre, and R26nTnG animals were obtained from The Jackson Laboratory. R26-CAGs-LSL-rtTA3 (LSL-rtTA) animals were obtained from L.E. Dow. Swiss Webster mice were obtained from Taconic Biosciences. Vcanfl/+ mice were crossed to EIIaCre mice to drive germline recombination (VcanRC). EIIaCre;VcanRC/+ animals were then crossed to wild-type mice to generate versican-deficient animals (VcanRC/+). All experiments were performed at least twice using different litters and used littermate controls on a mixed background unless otherwise indicated.
For neonatal CCM experiments using the VE-Cadherin-CreERT2;Krit1fl/fl (“Krit1ECKO”) or VE-Cadherin-CreERT2;Ccm2fl/fl (“Ccm2ECKO”) models, P1 pups were intragastrically injected by a 30-gauge needle with 40 µg of 4-hydroxytamoxifen (4OHT; Sigma-Aldrich, H7904) dissolved in 9% ethanol/corn oil vehicle (50 µl total volume per injection). The solution was freshly prepared from premeasured 4OHT powder for every injection. The P1 time point was defined by checking experimental breeding pairs every evening for new litters. The following morning (P1), pups were injected with 4OHT in a blinded fashion without knowledge of genotypes. Pups were then harvested, as previously described (Zhou et al., 2016b), at specified time points. For the R26-nTnG experiment, PDGFb-CreERT2;Pdcd10fl/−;R26nTnG/+ animals were induced with a single intragastric injection of 2 µg of tamoxifen (Sigma-Aldrich, T5648), dissolved in a 9:1 corn oil to ethanol solution, at P3 and harvested at P82 as previously described (Detter et al., 2018). All neonates were induced in a blinded fashion without knowledge of genotypes.
Generation and characterization of Adamts5 floxed allele
A targeting construct comprised of 1.5 kb flanking homology arms, floxed exon 3, and a flippase recombination target–flanked neomycin resistance cassette was synthesized by GenScript. Exon 3 encodes the catalytic domain of ADAMTS5 and has been previously targeted to generate a constitutively null allele (Stanton et al., 2005). A guide RNA sequence 5′-GGGCTTGGATGCATCGATGC-3′ targeting exon 3 was cloned into the PX330 plasmid (Addgene, 42230) and a corresponding protospacer adjacent motif site was mutated on the targeting construct. V6.5 embryonic stem (ES) cells were targeted by transfection of targeting construct and PX330 plasmids using XFect for mouse ES cells (Takara, 631320) followed by G418 selection. Individual clones were picked, expanded, and characterized by PCR reactions, Sanger sequencing, and in vitro Cre recombination. Chimeras were produced following microinjection of select ES cells into preimplantation embryos as described below.
To screen transgenic founders, DNA was isolated from F1 germline animals and amplified by PCR using a forward primer outside of the 5′ homology arm in the Adamts5 genomic locus and a reverse primer in the NeoR cassette (F1-R1 primers, Fig. 2 A). In vitro Cre recombination reaction was then performed (NEB, M0298). To verify Cre-mediated recombination in vivo, DNA samples from 4OHT-induced animals and noninduced Krit1ECKO;Adamts5+/+, Krit1ECKO;Adamts5fl/+, and Krit1ECKO;Adamts5fl/fl animals were amplified using primers flanking the loxP sites around exon 3 of Adamts5 (F2-R2 primers, Fig. 2 A). All PCR products were separated on an agarose gel and visualized by ethidium bromide staining.
Generation, characterization, and use of tetO-Adamts5 random transgenic allele
A mouse Adamts5 open reading frame with C-terminal double V5 epitope tags was synthesized by GenScript and cloned into the pTRE3G plasmid (Takara, 631168). The plasmid was then linearized, ethanol-precipitated, and used for pronuclear DNA injection to generate randomly integrated founders as described below.
Founders were initially screened by crossing to R26-rtTA lines, and the degree of rtTA-mediated transcription was examined by immunostaining for the V5 epitope tag. tetO-Adamts5 animals were crossed to the Cre-conditional reverse tetracycline-regulated transactivator (LSL-rtTA) and a constitutive brain endothelial-specific Cre (Slco1c1(BAC)-Cre) to enable inducible expression of V5-tagged ADAMTS5 from BECs of mice. To avoid daily manipulations of the neonatal pups, ADAMTS5 expression was induced by maintaining the nursing dams on a doxycycline diet (6 g/kg; Bio-Serv) from P0 until the specified time of harvest. Pups were also supplemented with a single intragastric injection of 200 µg of doxycycline (Sigma-Aldrich, D9891) in 20 µl total volume, administered at P1. Littermate animals carrying one or two of the alleles the Cre transgene, the rtTA transgene, or the tetO-Adamts5 transgene were used as controls for the gain-of-function (GOF) experiments unless otherwise stated.
Microinjection and manipulation of mouse embryos
To collect the embryos used to generate the transgenic founders, 6–8-wk-old B6D2 females were super-ovulated via intraperitoneal injection using 5 IU of pregnant mare serum gonadotropin, followed 48 h later by 5 IU of human chorionic gonadotropin (hCG), after which they were mated to B6D2 studs. B6D2 mice were generated from C57BL/6J females mated to DBA/2J males. For embryo transfers, Swiss Webster females were synchronized by using vasectomized males. After collection, embryos were incubated at 37°C with 5% CO2 using potassium-supplemented SOM media (Millipore, MR-202P-5F) covered with mineral oil (Millipore, ES-005-C). All the microinjections, embryo collection, and embryo transfers were performed at room temperature using CZB-Hepes media.
The zygotes used for DNA injections were collected 13–16 h after hCG from the ampulla of the oviduct. To remove the cumulus cells from the zygotes, the zygote/cumulus cell complexes were cultured for 2–3 min at room temperature in CZB-Hepes media supplemented with 3 mg/ml hyaluronidase. The blastocysts used for ES cell injection were flushed from the uterine horns 90 h after hCG. Using a glass capillary tubing with filament (GC100TF-15), the DNA solution was microinjected into the male pronuclei ∼2–3 h after collection. The injected embryos were further incubated for 2–3 h before they were transferred into the oviducts of E0.5 pseudo-pregnant recipient females.
For ES cells injections, 10–15 individual ES cells were injected into the blastocysts using a spiked beveled injection needle prepared by using a glass capillary tubing without a filament (GC100T-15). The blastocysts were injected ∼1 h after collection and were transferred into the uterine horns of E2.5 pseudo-pregnant recipient females 4–5 h after injection.
All the glass capillaries were pulled using a Micropipette puller model P-97. The injecting pressure was applied using an Eppendorf Cell Tram Air syringe, and the entire procedure was performed using an inverted Nikon Eclipse microscope.
Immunohistochemistry and quantification
Tissue samples were fixed in 4% paraformaldehyde (PFA)/PBS overnight at 4°C, dehydrated to 100% ethanol, and embedded in paraffin. Sections of 6 µm were used for H&E and immunohistochemistry staining. For frozen sections, tissue samples were fixed in 4% PFA/PBS overnight and washed through a graded series of 30% sucrose/PBS solutions before freezing in a 1:1 mixture of 30% sucrose/optimal cutting temperature compound. Sections of 10 µm were collected, dried overnight, and stored at −80°C. Cryosections were rehydrated with Tris-buffered saline/0.1% Tween 20, then subjected to blocking with PBS/1% BSA/0.3% Triton X-100 for 2 h at room temperature. The following primary antibodies were used for immunostaining: rabbit anti-versican (1:200, Millipore, AB1033), rabbit anti-DPEAAE (1:500, Pierce-Antibodies, PA1-1748A), rabbit anti-V5 (1:1,000, Abcam, AB9116), rat anti-platelet endothelial cell adhesion molecule (PECAM; 1:50; HistoBioTec, DIA-310), mouse anti-KLF4 (1:100, R&D, AF3158), rabbit anti-pMLC2 (1:200, Cell Signaling, 3674S), and goat anti-platelet-derived growth factor receptor beta (PDGFRB; 1:100, R&D, AF1042). Littermate control and experimental animal sections were placed on the same slide and stained at the same time under identical conditions. Images were acquired with ×4, ×10, ×20, and ×40 objectives on a Nikon 80i Eclipse or an Olympus DP80 microscope at the same exposure times using NIS Elements Digital Imaging software or Olympus CellSens acquisition software, respectively. ImageJ was used for image processing after data acquisition.
Versican protein and cleaved versican (DPEEAE) were quantified by measuring the average integrated fluorescence density (IntDen) of versican or DPEEAE-positive area along the cerebellar white matter tract. To minimize sampling bias, a minimum of 10 serial 6-µm-thick tissue sections were analyzed per hindbrain. Quantification for the R26-nTnG experiment was done by measuring the number of nTom-positive or nGFP-positive ECs immediately adjacent to DPEAAE-positive staining over total number of nTom or nGFP cells present within a lesion. Images taken from randomly selected high-powered fields, containing at least one CCM lesion, were assessed.
EC isolation, RNA isolation, and gene expression analysis
At the specified time points, cerebellar or lung ECs were isolated by enzymatic digestion followed by separation using magnetic-activated cell sorting by anti-CD31–conjugated magnetic beads (MACS MS system, Miltenyi Biotec), as previously described (Zhou et al., 2016b). The cerebella or a lobe of lung from neonatal mice were dissected and digested by a mixture of 10 mg/ml collagenase (Sigma-Aldrich, C0130), 5 mg/ml dispase (Sigma-Aldrich, D4693), and 1 mg/ml DNase I (Sigma-Aldrich, 10104159001) in DMEM for 30 min at 37°C with gentle shaking. The digestion was then passed thru a 70-µm strainer (Miltenyi Biotec), centrifuged, resuspended, and incubated with anti-CD31–conjugated magnetic beads for 30 min at 4°C. Microbead-bound cells were then washed, separated, and eluted on MACS MS columns (Miltenyi Biotec) according to the manufacturer’s protocol.
Total RNA was extracted from isolated ECs or tissue using TRIzol Reagent (Life Technologies) and the RNEasy Micro Kit (Qiagen, 74004) and reverse-transcribed using the SuperScript IV VILO Master Mix (Thermo Fisher Scientific, 11756050). To quantify transcript expression levels, real-time PCR was performed using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, 4368577). Relative gene expression was normalized to Gapdh levels and calculated using the ddCt method. The following mouse primers were used: Klf2 forward: 5′-CGCCTCGGGTTCATTTC-3′, Klf2 reverse: 5′-AGCCTATCTTGCCGTCCTTT-3′; Klf4 forward: 5′-GTGCCCCGACTAACCGTTG-3′, Klf4 reverse: 5′-GTCGTTGAACTCCTCGGTCT-3′; Adamts5 (exon1-3) forward: 5′-CGACCCTCAAGAACTTTTGC-3′, Adamts5 (exon1-3) reverse: 5′-CGTCATGAGAAAGGCCAAGT-3′; Adamts5 (exon2-3) forward: 5′-AATTGGGCATCTACTTGGCCT-3′, Adamts5 (exon2-3) reverse: 5′-TGGCTGACGTGCATTTGGA-3′; Adamts5 (exon4-5) forward: 5′-TGTCTGACCAAGAAGCTGCC-3′, Adamts5 (exon4-5) reverse: 5′-GGTAGGCAAACTGCACTCCT-3′; and Gapdh forward: 5′-AAATGGTGAAGGTCGGTGTGAACG-3′, Gapdh reverse: 5′-ATCTCCACTTTGCCACTGC-3′.
Bacterial DNA extraction and bacterial ribosomal DNA qPCR
Neonatal guts were dissected, snap-frozen, and stored at −80°C. As previously described (Tang et al., 2017), the QIAmp DNA Stool Mini Kit (Qiagen, 51504 or 51604) was used to extract bacterial DNA from the gut samples. Prior to following the manufacturer’s protocol, samples were mixed with stool lysis buffer provided by the Qiagen kit and homogenized with a TissueLyser LT (Qiagen, 69980) at 50 Hz for 10 min at 4°C. The concentration of isolated DNA was equalized, and qPCR was performed with 16 ng of DNA per reaction. The following primers were used: Universal 16S rRNA forward: 5′-ACTGAGAYACGGYCCA-3′, Universal 16S rRNA reverse: 5′-TTACCGCGGCTGCTGGC-3′; and Bacteroidetes s24-7 rRNA forward: 5′-GGAGAGTACCCGGAGAAAAAGC-3′, Bacteroidetes s24-7 rRNA reverse: 5′-TTCCGCATACTTCTCGCCCA-3′.
Quantification of CCM lesion burden and extra-lesional bleeding
For all experiments using microCT quantification of CCM lesion volume, brains were harvested and placed in 4% PFA/PBS. Brains remained in fixative until staining with nondestructive iodine contrast and subsequent microCT imaging performed as previously described (Girard et al., 2016).
Experiments where brains had observable hemorrhage (e.g., Krit1BECKO; ADAMTS5BEC-GOF) were subject to further analysis. For each mouse, the total blood volume (lesional + extra-lesional) was determine by microCT. Since extra-lesional bleeding cannot be distinguished from blood within caverns on microCT, histological analysis was used. With PECAM immunostaining, ECs were delineated on the largest sections of index lesions identified on microCT. Bleeding was identified on the respective histological images, and the area of extra-lesional blood (not confined by endothelium lining) and the lesional area (including any blood confined within endothelium lined caverns) were measured using the polygon area function of an Olympus DP2-SAL standalone connection kit attached to an Olympus DP22 digital camera mounted on top of an Olympus CX41 microscope.
Tissue processing, imaging, and volumetric quantification were performed in a blinded manner by investigators at the University of Chicago without prior knowledge of genotype or experimental details.
Cloning and generation of recombinant lentiviruses
To generate recombinant versican G1 or G3 domain-containing peptides, sequences for mouse Vcan exons 2–6 were used (which includes the endogenous signal peptide) for the G1 domain (N-terminal), and exons 9–14 preceded by an IL-2 signal peptide were used for the G3 domain (C-terminal). A 2× V5 epitope tag sequence was added immediately before the termination codon. A control mCherry fusion protein construct bearing 2× SV40 NLS was also generated. The described constructs were ordered as gene blocks from Integrative DNA Technologies, double-digested with BamHI and MluI restriction enzymes, cloned into the pLV-EIF1a-IRES-Neo lentiviral plasmid (Addgene), and verified by Sanger sequencing. Viral particles were generated by triple transfection of Lenti-X 293T cell line (Takara, 632180) using FuGENE 6 (Promega, E2691) with the transfer vector carrying the gene of interest, the psPAX2 packaging vector, and the pMD2.G envelope vector. This mixture was removed 24 h later and replaced with complete medium. 48, 72, and 96 h after initial transfection, the supernatant with viral particles was collected, pooled, and filtered through a 0.45-µm filter (Millipore). Viral supernatant was then concentrated using Lenti-X Concentrator (Takara, 631232) and titered using Lenti-X qRT-PCR Titration Kit (Takara, 631235). The virus was stored at −80°C until use.
Generation of cell lines and 3D sprouting assay
To generate WI-38 lung fibroblasts (American Type Culture Collection, CCL-75) expressing either mCherry, G1-V5, or G3-V5, WI-38 cells were transduced with the recombinant lentiviruses described above. The lentiviruses were resuspended in antibiotic-free Eagle’s Minimum Essential Medium supplemented with 10% FBS and 10 μg/ml polybrene and added to target cells. This mixture was removed 24 h later and replaced with complete medium. Conditioned media was collected 48 h post-transduction, and samples were analyzed for secretion by Western blotting using anti-V5 (1:1,000, Cell Signaling, 13202) and IRDye 800CW goat anti-rabbit (Licor) secondary antibody. 72 h after transduction, lentivirally transduced cells were selected in 500 µg/ml of G418 for 10 d with medium containing selection antibiotic replaced every 2 d.
Stable WI-38 cells that expressed mCherry, G1-V5, or G3-V5 were co-cultured with human umbilical vein ECs (HUVECs; Lonza, C2519A) in a three-dimensional sprouting assay as previously described (Nakatsu et al., 2007). Briefly, HUVECs were coated onto Cytodex 3 microcarrier beads (GE Healthcare, 17–0485-01; 106 HUVECs/2,500 beads) overnight and subsequently embedded into a fibrin gel (generated by the addition of thrombin to a fibrinogen/bead suspension). The lentivirally transduced WI-38 cells were then seeded on top of the fibrin gel at a concentration of 20,000 cells per well. The fibrin gel co-cultures were grown in endothelial basal medium supplemented with EGM-2 SingleQuot Kit (Lonza) and maintained for 10 d. Bright-field images of HUVEC spheroids were taken using a Nikon Eclipse Ti inverted microscope (×10 objective, NIS Elements Digital Imaging software), and images were analyzed by quantifying the number of sprouts and branch points per bead on ImageJ. A Canny–Deriche edge detection filter was applied to images using an ImageJ plugin for visualization purposes (seen in Fig. 7 C).
Statistics
All experimental and control animals were littermates, unless otherwise specified, and none were excluded from analysis at the time of harvest. Experimental animals were lost or excluded at two predefined points: (i) failure to properly inject the inducing agent (4OHT, tamoxifen, or doxycycline) and observation of significant leakage; and (ii) death before time of harvest due to injection. Sample sizes were estimated based on our prior experience with the neonatal CCM model and microCT quantification of lesion volume, as previously described (Tang et al., 2017; Zhou et al., 2016b). All data were analyzed with GraphPad Prism (version 7) and represented as mean ± SEM. P values were calculated using an unpaired two-tailed Student’s t test or one-way ANOVA plus Tukey post hoc analysis as indicated. Mann–Whitney nonparametric U tests were used for experiments that did not satisfy normality tests. A log-rank test was used to assess the Kaplan–Meier survival curves. P values <0.05 were considered statistically significant and are denoted as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Animal study approval
The global Adamts5 genetic rescue experiment was conducted with the approval of The Sydney Local Health District Animal Welfare Committee and according to the guidelines/regulations of Centenary Institute and the University of Sydney. The R26-nTnG experiment was performed in accordance with the Duke University Institutional Animal Care and Use Committee. All remaining animal experiments described were performed in accordance and approval of the University of Pennsylvania Institutional Animal Care and Use Committee.
Online supplemental material
Fig. S1 shows data regarding the characterization of the Adamts5 floxed allele, including recombination both in vitro and in vivo, unaffected cerebellar Adamts5 mRNA levels with retention of the neo-cassette in noninduced animals, and deletion efficiency in isolated lung ECs. Fig. S2 shows versican levels are not significantly changed with combinatorial loss of ADAMTS1 and ADAMTS4 in Krit1ECKO mice. Fig. S3 shows the progression of CCM formation in the Krit1BECKO model from P3-P10. Fig. S4 shows additional images of versican and DPEEAE staining in adjacent sections taken from ADAMTS5BEC-GOF animals. Fig. S5 shows the characterization of the VcanRC allele.
Acknowledgments
We thank the members of the Kahn laboratory and our colleagues E. Puré, S. Ryeom, R.K. Assoian, and E.E. Morrisey for their thoughtful comments and advice during the course of this work. We would also like to thank R.H. Adams (Max Planck Institute for Molecular Biomedicine, Münster, Germany) for the generous gift of the VE-Cadherin-CreERT2 mice, M.L. Iruela-Arispe (Northwestern University Feinberg School of Medicine, Chicago, IL) for the Adamts1 null mice, and J.S. Richards (Baylor College of Medicine, Houston, TX) for the Adamts1fl/+ ES cells used in this study.
This research was supported by National Institutes of Health grants R01HL094326 to M.L. Kahn; P01NS092521 to M.L. Kahn, D.A. Marchuk, and I.A. Awad; T32GM008076 and F31NS115256 to C.C. Hong; F30NS100252 to A.T. Tang; T32GM007171, F30HL140871, and American Heart Association 18PRE34060061 to M.R. Detter; and National Institutes of Health grant R01HL136507 to W. Min; and National Natural Science Foundation of China grant 81771240 and Australian National Health and Medical Research Council project grant APP1124011 to X. Zheng.
Author contributions: C.C. Hong designed and performed most of the experiments and wrote the manuscript. A.T. Tang also designed and performed experiments and helped write the manuscript. A.T. Tang designed and generated the Adamts5 floxed allele and tetO-Adamts5 random transgenic allele with the assistance of A.A. Guerrero, C.F. Wittig, and P. Mericko-Ishizuka. N.A. Leu and S. Sterling performed ES cell and pronuclear microinjections as part of the Penn Vet Transgenic Core. M.R. Detter, J.P. Choi, R. Wang, X. Yang, L.M. Goddard, A.A. Ren, M. Chen, J. Yang, and L. Li performed experiments. N. Hobson, R. Girard, R. Lightle, T. Moore, R. Shenkar, S.P. Polster, and I.A. Awad performed microCT CCM lesion imaging and quantification in a blinded manner. L.E. Dow, H. Watanabe, M. Schwaninger, and W. Min provided essential reagents. D.A. Marchuk and X. Zheng helped design the experiments. M.L. Kahn helped design the experiments and wrote the manuscript.
References
- Akers A.L., Johnson E., Steinberg G.K., Zabramski J.M., and Marchuk D.A.. 2009. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 18:919–930. 10.1093/hmg/ddn430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers A., Al-Shahi Salman R., A Awad I., Dahlem K., Flemming K., Hart B., Kim H., Jusue-Torres I., Kondziolka D., Lee C., et al. 2017. Synopsis of Guidelines for the Clinical Management of Cerebral Cavernous Malformations: Consensus Recommendations Based on Systematic Literature Review by the Angioma Alliance Scientific Advisory Board Clinical Experts Panel. Neurosurgery. 80:665–680. 10.1093/neuros/nyx091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahi Salman R., Berg M.J., Morrison L., and Awad I.A.; Angioma Alliance Scientific Advisory Board . 2008. Hemorrhage from cavernous malformations of the brain: definition and reporting standards. Stroke. 39:3222–3230. 10.1161/STROKEAHA.108.515544 [DOI] [PubMed] [Google Scholar]
- Al-Shahi Salman R., Hall J.M., Horne M.A., Moultrie F., Josephson C.B., Bhattacharya J.J., Counsell C.E., Murray G.D., Papanastassiou V., Ritchie V., et al. ; Scottish Audit of Intracranial Vascular Malformations (SAIVMs) collaborators . 2012. Untreated clinical course of cerebral cavernous malformations: a prospective, population-based cohort study. Lancet Neurol. 11:217–224. 10.1016/S1474-4422(12)70004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerboom D., Lafond J.F., Zheng X., Lapointe E., Mittaz L., Boyer A., Pritchard M.A., DeMayo F.J., Mort J.S., Drolet R., et al. 2011. Partially redundant functions of Adamts1 and Adamts4 in the perinatal development of the renal medulla. Dev. Dyn. 240:1806–1814. 10.1002/dvdy.22662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulday G., Rudini N., Maddaluno L., Blécon A., Arnould M., Gaudric A., Chapon F., Adams R.H., Dejana E., and Tournier-Lasserve E.. 2011. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J. Exp. Med. 208:1835–1847. 10.1084/jem.20110571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.C., Drakos S.G., Ruiz O.E., Smith A.C., Gibson C.C., Ling J., Passi S.F., Stratman A.N., Sacharidou A., Revelo M.P., et al. 2011. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J. Clin. Invest. 121:1871–1881. 10.1172/JCI44393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choocheep K., Hatano S., Takagi H., Watanabe H., Kimata K., Kongtawelert P., and Watanabe H.. 2010. Versican facilitates chondrocyte differentiation and regulates joint morphogenesis. J. Biol. Chem. 285:21114–21125. 10.1074/jbc.M109.096479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikach F.S., Koch C.D., Mead T.J., Galatioto J., Willard B.B., Emerton K.B., Eagleton M.J., Blackstone E.H., Ramirez F., Roselli E.E., et al. 2018. Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight. 3 e97167 10.1172/jci.insight.97167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claxton S., Kostourou V., Jadeja S., Chambon P., Hodivala-Dilke K., and Fruttiger M.. 2008. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 46:74–80. 10.1002/dvg.20367 [DOI] [PubMed] [Google Scholar]
- Cullere X., Plovie E., Bennett P.M., MacRae C.A., and Mayadas T.N.. 2015. The cerebral cavernous malformation proteins CCM2L and CCM2 prevent the activation of the MAP kinase MEKK3. Proc. Natl. Acad. Sci. USA. 112:14284–14289. 10.1073/pnas.1510495112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttano R., Rudini N., Bravi L., Corada M., Giampietro C., Papa E., Morini M.F., Maddaluno L., Baeyens N., Adams R.H., et al. 2016. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol. Med. 8:6–24. 10.15252/emmm.201505433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Monte-Nieto G., Ramialison M., Adam A.A.S., Wu B., Aharonov A., D’Uva G., Bourke L.M., Pitulescu M.E., Chen H., de la Pompa J.L., et al. 2018. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature. 557:439–445. 10.1038/s41586-018-0110-6 [DOI] [PubMed] [Google Scholar]
- Detter M.R., Snellings D.A., and Marchuk D.A.. 2018. Cerebral Cavernous Malformations Develop Through Clonal Expansion of Mutant Endothelial Cells. Circ. Res. 123:1143–1151. 10.1161/CIRCRESAHA.118.313970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiStefano P.V., Kuebel J.M., Sarelius I.H., and Glading A.J.. 2014. KRIT1 protein depletion modifies endothelial cell behavior via increased vascular endothelial growth factor (VEGF) signaling. J. Biol. Chem. 289:33054–33065. 10.1074/jbc.M114.582304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis L.E., McCulloch D.R., McGarity J.D., Bahan A., Wessels A., Weber D., Diminich A.M., Nelson C.M., Apte S.S., and Kern C.B.. 2011. Altered versican cleavage in ADAMTS5 deficient mice; a novel etiology of myxomatous valve disease. Dev. Biol. 357:152–164. 10.1016/j.ydbio.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis L.E., Nelson E.L., Hozik B., Porto S.C., Rogers-DeCotes A., Fosang A., and Kern C.B.. 2019. Adamts5-/- Mice Exhibit Altered Aggrecan Proteolytic Profiles That Correlate With Ascending Aortic Anomalies. Arterioscler. Thromb. Vasc. Biol. 39:2067–2081. 10.1161/ATVBAHA.119.313077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H., Nelson C.M., Somerville R.P.T., Mielke K., Dixon L.J., Powell K., and Apte S.S.. 2010. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 137:4029–4038. 10.1242/dev.050591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava M., Barallobre-Barreiro J., Mayr U., Lu R., Didangelos A., Baig F., Lynch M., Catibog N., Joshi A., Barwari T., et al. 2018. Role of ADAMTS-5 in Aortic Dilatation and Extracellular Matrix Remodeling. Arterioscler. Thromb. Vasc. Biol. 38:1537–1548. 10.1161/ATVBAHA.117.310562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher O.S., and Boggon T.J.. 2014. Signaling pathways and the cerebral cavernous malformations proteins: lessons from structural biology. Cell. Mol. Life Sci. 71:1881–1892. 10.1007/s00018-013-1532-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher O.S., Deng H., Liu D., Zhang Y., Wei R., Deng Y., Zhang F., Louvi A., Turk B.E., Boggon T.J., et al. 2015. Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex. Nat. Commun. 6:7937 10.1038/ncomms8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischer J.M., Pipp I., Stavrou I., Trattnig S., Hainfellner J.A., and Knosp E.. 2008. Cerebral cavernous malformations: congruency of histopathological features with the current clinical definition. J. Neurol. Neurosurg. Psychiatry. 79:783–788. 10.1136/jnnp.2007.132316 [DOI] [PubMed] [Google Scholar]
- Gendron C., Kashiwagi M., Lim N.H., Enghild J.J., Thøgersen I.B., Hughes C., Caterson B., and Nagase H.. 2007. Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J. Biol. Chem. 282:18294–18306. 10.1074/jbc.M701523200 [DOI] [PubMed] [Google Scholar]
- Girard R., Zeineddine H.A., Orsbon C., Tan H., Moore T., Hobson N., Shenkar R., Lightle R., Shi C., Fam M.D., et al. 2016. Micro-computed tomography in murine models of cerebral cavernous malformations as a paradigm for brain disease. J. Neurosci. Methods. 271:14–24. 10.1016/j.jneumeth.2016.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden M.J., Morrison L.A., Kim H., and Hart B.L.. 2015. Increased number of white matter lesions in patients with familial cerebral cavernous malformations. AJNR Am. J. Neuroradiol. 36:899–903. 10.3174/ajnr.A4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueye N.A., Mead T.J., Koch C.D., Biscotti C.V., Falcone T., and Apte S.S.. 2017. Versican proteolysis by ADAMTS proteases and its influence on sex steroid receptor expression in uterine leiomyoma. J. Clin. Endocrinol. Metab. 102:1631–1641. 10.1210/jc.2016-3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.L., Taheri S., Rosenberg G.A., and Morrison L.A.. 2013. Dynamic contrast-enhanced MRI evaluation of cerebral cavernous malformations. Transl. Stroke Res. 4:500–506. 10.1007/s12975-013-0285-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhang H., Yu L., Gunel M., Boggon T.J., Chen H., and Min W.. 2010. Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci. Signal. 3:ra26 10.1126/scisignal.2000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope C., Foulcer S., Jagodinsky J., Chen S.X., Jensen J.L., Patel S., Leith C., Maroulakou I., Callander N., Miyamoto S., et al. 2016. Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood. 128:680–685. 10.1182/blood-2016-03-705780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yang J., Lin Y., Walker C., Cheng J., Liu Z.G., and Su B.. 2004. Differential regulation of interleukin 1 receptor and Toll-like receptor signaling by MEKK3. Nat. Immunol. 5:98–103. 10.1038/ni1014 [DOI] [PubMed] [Google Scholar]
- Kang I., Harten I.A., Chang M.Y., Braun K.R., Sheih A., Nivison M.P., Johnson P.Y., Workman G., Kaber G., Evanko S.P., et al. 2017. Versican Deficiency Significantly Reduces Lung Inflammatory Response Induced by Polyinosine-Polycytidylic Acid Stimulation. J. Biol. Chem. 292:51–63. 10.1074/jbc.M116.753186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskimäki J., Girard R., Li Y., Saadat L., Zeineddine H.A., Lightle R., Moore T., Lyne S., Avner K., Shenkar R., et al. 2019. Comprehensive transcriptome analysis of cerebral cavernous malformation across multiple species and genotypes. JCI Insight. 4 e126167 10.1172/jci.insight.126167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M.F., Salinin S., Ridder D.A., Kleesiek J., Hroudova J., Berger S., Schütz G., and Schwaninger M.. 2011. A transgenic approach to identify thyroxine transporter-expressing structures in brain development. J. Neuroendocrinol. 23:1194–1203. 10.1111/j.1365-2826.2011.02216.x [DOI] [PubMed] [Google Scholar]
- Lee N.V., Rodriguez-Manzaneque J.C., Thai S.N., Twal W.O., Luque A., Lyons K.M., Argraves W.S., and Iruela-Arispe M.L.. 2005. Fibulin-1 acts as a cofactor for the matrix metalloprotease ADAMTS-1. J. Biol. Chem. 280:34796–34804. 10.1074/jbc.M506980200 [DOI] [PubMed] [Google Scholar]
- Malinverno M., Maderna C., Abu Taha A., Corada M., Orsenigo F., Valentino M., Pisati F., Fusco C., Graziano P., Giannotta M., et al. 2019. Endothelial cell clonal expansion in the development of cerebral cavernous malformations. Nat. Commun. 10:2761 10.1038/s41467-019-10707-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch D.R., Le Goff C., Bhatt S., Dixon L.J., Sandy J.D., and Apte S.S.. 2009a Adamts5, the gene encoding a proteoglycan-degrading metalloprotease, is expressed by specific cell lineages during mouse embryonic development and in adult tissues. Gene Expr. Patterns. 9:314–323. 10.1016/j.gep.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch D.R., Nelson C.M., Dixon L.J., Silver D.L., Wylie J.D., Lindner V., Sasaki T., Cooley M.A., Argraves W.S., and Apte S.S.. 2009b ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell. 17:687–698. 10.1016/j.devcel.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittaz L., Russell D.L., Wilson T., Brasted M., Tkalcevic J., Salamonsen L.A., Hertzog P.J., and Pritchard M.A.. 2004. Adamts-1 is essential for the development and function of the urogenital system. Biol. Reprod. 70:1096–1105. 10.1095/biolreprod.103.023911 [DOI] [PubMed] [Google Scholar]
- Mjaatvedt C.H., Yamamura H., Capehart A.A., Turner D., and Markwald R.R.. 1998. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev. Biol. 202:56–66. 10.1006/dbio.1998.9001 [DOI] [PubMed] [Google Scholar]
- Mleynek T.M., Chan A.C., Redd M., Gibson C.C., Davis C.T., Shi D.S., Chen T., Carter K.L., Ling J., Blanco R., et al. 2014. Lack of CCM1 induces hypersprouting and impairs response to flow. Hum. Mol. Genet. 23:6223–6234. 10.1093/hmg/ddu342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu M.N., Davis J., and Hughes C.C.W.. 2007. Optimized fibrin gel bead assay for the study of angiogenesis. J. Vis. Exp. (3):186 10.3791/186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer N.W., Zawistowski J.S., and Marchuk D.A.. 2005. Genetics of cerebral cavernous malformations. Curr. Neurol. Neurosci. Rep. 5:391–396. 10.1007/s11910-005-0063-7 [DOI] [PubMed] [Google Scholar]
- Quaglia X., Beggah A.T., Seidenbecher C., and Zurn A.D.. 2008. Delayed priming promotes CNS regeneration post-rhizotomy in Neurocan and Brevican-deficient mice. Brain. 131:240–249. 10.1093/brain/awm279 [DOI] [PubMed] [Google Scholar]
- Santamaria S., Yamamoto K., Teraz-Orosz A., Koch C., Apte S.S., de Groot R., Lane D.A., and Ahnström J.. 2019. Exosites in Hypervariable Loops of ADAMTS Spacer Domains control Substrate Recognition and Proteolysis. Sci. Rep. 9:10914 10.1038/s41598-019-47494-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegler S., Rath M., Paperlein C., and Felbor U.. 2018. Cerebral Cavernous Malformations: An Update on Prevalence, Molecular Genetic Analyses, and Genetic Counselling. Mol. Syndromol. 9:60–69. 10.1159/000486292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K., Hang C.T., Tsun Z.Y., Chen H., Lee N.V., Wu J.I., Shang C., Bayle J.H., Shou W., Iruela-Arispe M.L., et al. 2008. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell. 14:298–311. 10.1016/j.devcel.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton H., Rogerson F.M., East C.J., Golub S.B., Lawlor K.E., Meeker C.T., Little C.B., Last K., Farmer P.J., Campbell I.K., et al. 2005. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 434:648–652. 10.1038/nature03417 [DOI] [PubMed] [Google Scholar]
- Stanton H., Melrose J., Little C.B., and Fosang A.J.. 2011. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim. Biophys. Acta. 1812:1616–1629. 10.1016/j.bbadis.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Tang A.T., Choi J.P., Kotzin J.J., Yang Y., Hong C.C., Hobson N., Girard R., Zeineddine H.A., Lightle R., Moore T., et al. 2017. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature. 545:305–310. 10.1038/nature22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A.T., Sullivan K.R., Hong C.C., Goddard L.M., Mahadevan A., Ren A., Pardo H., Peiper A., Griffin E., Tanes C., et al. 2019. Distinct cellular roles for PDCD10 define a gut-brain axis in cerebral cavernous malformation. Sci. Transl. Med. 11 eaaw3521 10.1126/scitranslmed.aaw3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanriover G., Sozen B., Seker A., Kilic T., Gunel M., and Demir N.. 2013. Ultrastructural analysis of vascular features in cerebral cavernous malformations. Clin. Neurol. Neurosurg. 115:438–444. 10.1016/j.clineuro.2012.06.023 [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M., He L., Mäe M.A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Laviña B., Gouveia L., et al. 2018. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 554:475–480. 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- Wang Y., Nakayama M., Pitulescu M.E., Schmidt T.S., Bochenek M.L., Sakakibara A., Adams S., Davy A., Deutsch U., Lüthi U., et al. 2010. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 465:483–486. 10.1038/nature09002 [DOI] [PubMed] [Google Scholar]
- Wang X., Hou Y., Deng K., Zhang Y., Wang D.C., and Ding J.. 2015. Structural Insights into the Molecular Recognition between Cerebral Cavernous Malformation 2 and Mitogen-Activated Protein Kinase Kinase Kinase 3. Structure. 23:1087–1096. 10.1016/j.str.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Wüstehube J., Bartol A., Liebler S.S., Brütsch R., Zhu Y., Felbor U., Sure U., Augustin H.G., and Fischer A.. 2010. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc. Natl. Acad. Sci. USA. 107:12640–12645. 10.1073/pnas.1000132107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.L., Zhang Y., Cao L., and Yang B.B.. 1999. Cell adhesion and proliferation mediated through the G1 domain of versican. J. Cell. Biochem. 72:210–220. [DOI] [PubMed] [Google Scholar]
- Yee A.J., Akens M., Yang B.L., Finkelstein J., Zheng P.S., Deng Z., and Yang B.. 2007. The effect of versican G3 domain on local breast cancer invasiveness and bony metastasis. Breast Cancer Res. 9:R47 10.1186/bcr1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cao L., Yang B.L., and Yang B.B.. 1998. The G3 domain of versican enhances cell proliferation via epidermial growth factor-like motifs. J. Biol. Chem. 273:21342–21351. 10.1074/jbc.273.33.21342 [DOI] [PubMed] [Google Scholar]
- Zheng X., Xu C., Smith A.O., Stratman A.N., Zou Z., Kleaveland B., Yuan L., Didiku C., Sen A., Liu X., et al. 2012. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev. Cell. 23:342–355. 10.1016/j.devcel.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Rawnsley D.R., Goddard L.M., Pan W., Cao X.J., Jakus Z., Zheng H., Yang J., Arthur J.S.C., Whitehead K.J., et al. 2015. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell. 32:168–180. 10.1016/j.devcel.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.J., Qin L., Zhang H., Tang W., Ji W., He Y., Liang X., Wang Z., Yuan Q., Vortmeyer A., et al. 2016a Erratum: Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nat. Med. 22:1502–1502. 10.1038/nm1216-1502c [DOI] [PubMed] [Google Scholar]
- Zhou Z., Tang A.T., Wong W.Y., Bamezai S., Goddard L.M., Shenkar R., Zhou S., Yang J., Wright A.C., Foley M., et al. 2016b Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 532:122–126. 10.1038/nature17178 [DOI] [PMC free article] [PubMed] [Google Scholar]