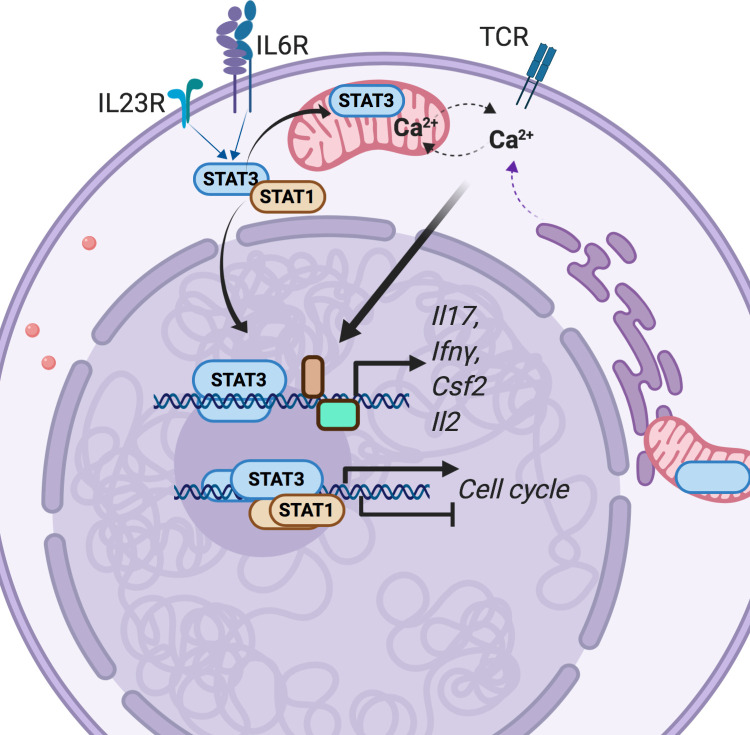
STAT3 transcription factor activity is critical for Th17 cell development. Poholek et al. report that STAT3 maintains proliferation and function of autoimmune effector Th17 cells by limiting STAT1 activation and sustaining mitochondrial membrane potential to allow cytokine production in response to TCR stimulation.
Abstract
The STAT3 signaling pathway is required for early Th17 cell development, and therapies targeting this pathway are used for autoimmune disease. However, the role of STAT3 in maintaining inflammatory effector Th17 cell function has been unexplored. Th17ΔSTAT3 mice, which delete STAT3 in effector Th17 cells, were resistant to experimental autoimmune encephalomyelitis (EAE), a murine model of MS. Th17 cell numbers declined after STAT3 deletion, corresponding to reduced cell cycle. Th17ΔSTAT3 cells had increased IL-6–mediated phosphorylation of STAT1, known to have antiproliferative functions. Th17ΔSTAT3 cells also had reduced mitochondrial membrane potential, which can regulate intracellular Ca2+. Accordingly, Th17ΔSTAT3 cells had reduced production of proinflammatory cytokines when stimulated with myelin antigen but normal production of cytokines when TCR-induced Ca2+ flux was bypassed with ionomycin. Thus, early transcriptional roles of STAT3 in developing Th17 cells are later complimented by noncanonical STAT3 functions that sustain pathogenic Th17 cell proliferation and cytokine production.
Graphical Abstract
Introduction
T helper 17 (Th17) cells promote pathology in a variety of autoimmune conditions, including psoriasis, ankylosing spondylitis, inflammatory bowel disease, and multiple sclerosis (Gaffen et al., 2014; Patel and Kuchroo, 2015). New therapies targeting Th17 cells via IL-23 blockade or by blocking IL-17 or the IL-17 receptor are proving highly effective in autoimmune therapy (Gaffen et al., 2014; Patel and Kuchroo, 2015). In healthy individuals, Th17 cells maintain microbial homeostasis in barrier sites such as the gut and skin and are important regulators of extracellular bacterial and fungal pathogens (McGeachy and McSorley, 2012). Humans with loss-of-function mutations in IL-23R have reduced susceptibility to inflammatory bowel disease (Di Meglio et al., 2011; Di Meglio et al., 2013; Rioux et al., 2007), while mutations that reduce the activation or function of Th17 cells result in increased susceptibility to mucocutaneous fungal infections (Okada et al., 2016). One such disease is hyper-IgE syndrome, in which STAT3 mutations cause reduced signaling downstream of Th17 cell–promoting cytokines, leading to reduced numbers of Th17 cells and increased rates of infection with Staphylococcus aureus and Candida albicans (de Beaucoudrey et al., 2008; Steward-Tharp et al., 2014).
Th17 cell differentiation is known to require signals from multiple cytokines in a step-wise manner, culminating in proinflammatory effector cells that induce inflammation in the target tissue through production of cytokines, including IL-17, GM-CSF, and IFN-γ (Zúñiga et al., 2013). Several Th17 cell–promoting cytokines, including IL-6, IL-21, and IL-23, activate STAT3, a critically required transcription factor during early Th17 cell differentiation (Yang et al., 2007; Zhou et al., 2007). In the absence of IL-6 and STAT3 signaling, expression of the master transcription factor RORγt is not sufficiently stabilized, and instead, Foxp3 is up-regulated, leading to induction of regulatory T cells (Durant et al., 2010; Geng et al., 2017; Korn et al., 2008). STAT3 also effects epigenetic modifications that promote the Th17 gene transcriptional program (Ciofani et al., 2012; Durant et al., 2010). In addition to directing cell phenotypic fate, STAT3 is required for efficient proliferation and survival of recently activated T cells (Durant et al., 2010).
Given the apparent importance of STAT3 in pathogenic Th17 cell development, it is no surprise that therapeutics targeting upstream STAT3 activation, via cytokines and JAK inhibitors, are being investigated as treatments for autoimmune disease (Schwartz et al., 2017). The importance of STAT3 has mostly been studied in systems in which STAT3 was deleted before initiation of the Th17 response, such as in mice with constitutive STAT3 deletion in T cells or humans with genetic mutations present from birth. However, patients with autoimmune disease have already developed effector and memory Th17 cells. We therefore sought to establish the roles and importance of STAT3 in Th17 effector cell function using mice in which STAT3 is deleted only after IL-17 is already expressed.
Results
Effector Th17 cells require STAT3 for pathogenic function in experimental autoimmune encephalomyelitis (EAE)
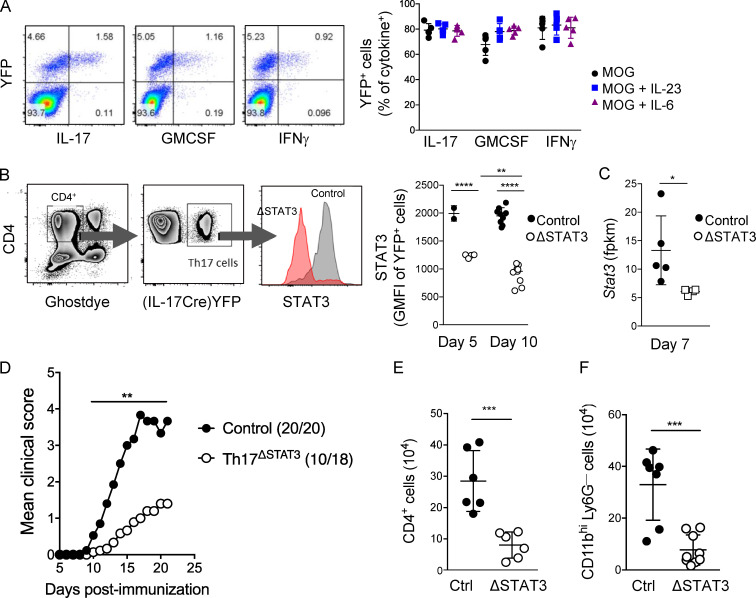
To examine the function of STAT3 in effector Th17 cells, we used IL-17creROSA26YFPfl/fl fate-tracker mice, in which Cre is induced by the IL-17A promoter to drive YFP expression (Hirota et al., 2011). Following immunization with myelin oligodendrocyte glycoprotein (MOG) peptide (35–55) in CFA to induce EAE, the T cells that produced cytokine in response to MOG stimulation were mostly YFP+ (Fig. 1 A). IL-17creROSA26YFPfl/fl mice were crossed with STAT3fl/fl mice, designated here Th17ΔSTAT3. Importantly, Th17 cells are still able to develop in Th17ΔSTAT3 mice, as only those cells that have already received sufficient signals to produce IL-17A (a process critically dependent on early STAT3 expression) express Cre and delete STAT3. We confirmed that STAT3 expression is significantly decreased in YFP+ cells from day 5 after immunization and further decreased by day 10 when effector Th17 cells are generated, although STAT3 did not appear to be completely ablated in this system (Fig. 1, B and C). Th17ΔSTAT3 mice were resistant to EAE, with reduced disease severity and incidence compared with IL-17creROSA26YFPfl/flSTAT3fl/+ littermate controls, hereafter termed Th17ctrl (Fig. 1 D). Consistent with reduced clinical severity, numbers of total CD4+ T cells and inflammatory macrophages infiltrating the central nervous system (CNS) of Th17ΔSTAT3 mice were significantly reduced on day 13 after immunization (Fig. 1, E and F). Hence, we conclude that continued STAT3 expression has an important role after initial Th17 cell activation to allow full effector Th17 cell function in autoimmune CNS inflammation.
Figure 1.
Effector Th17 cells require STAT3 for pathogenic function in EAE. Th17ΔSTAT3 and Th17ctrl mice were immunized with MOG(35–55) in CFA to induce EAE. (A) dLNs were harvested on day 10 and stimulated with MOG(35–55) for 18 h, with GolgiPlug added during the final 5 h. Representative FACS plots are shown to demonstrate that cytokine production is largely restricted to YFP+ cells after gating on live CD4+ cells, and the graph shows YFP expression in cytokine-producing populations after stimulation. (B) STAT3 expression analyzed in live CD4+YFP+ cells on days 5 and 10 after immunization, gated as indicated in representative plot. (C) Stat3 gene expression in live CD4+YFP+ cells sorted from dLNs of indicated mice on day 10 after immunization and analyzed by RNA sequencing. (D) EAE clinical scores, with number per group and incidence indicated next to group name. (E) Number of CD4+ cells in CNS on day 13 after immunization. (F) Number of inflammatory macrophages on day 13 after immunization analyzed by flow cytometry. Data in A–C, E, and F indicate mean ± SD of individual mice, pooled from two experiments; data in D are pooled from four experiments with 18–20 mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Student’s t test (except D, Mann–Whitney U test) for each time point.
STAT3 deletion in effector Th17 cells reduces their numbers in vivo
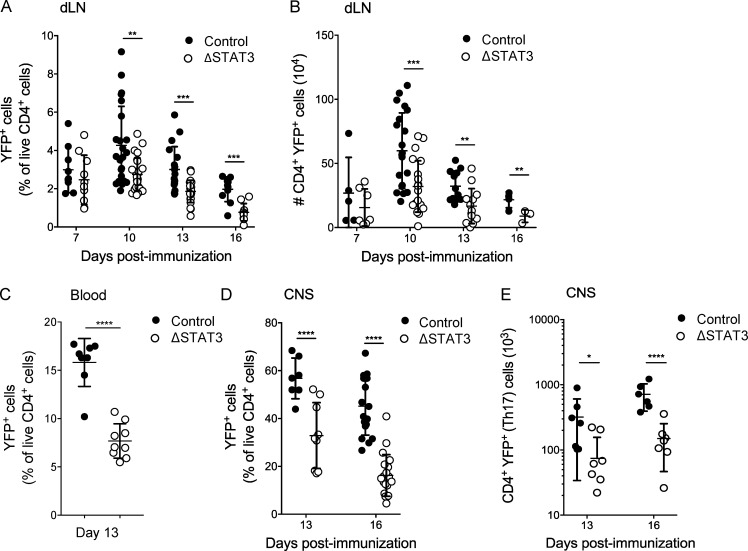
Analysis of YFP+Th17ΔSTAT3 cells after immunization for EAE induction revealed that early generation of Th17 cells was not impaired (Fig. 2, A and B). This was expected, since STAT3 is present for initial Th17 cell activation in these mice. However, the numbers of YFP+Th17ΔSTAT3 cells were reduced in the draining LN (dLN) beginning on day 10 after immunization, the peak of LN priming (Fig. 2, A and B). Reduced frequencies of STAT3-deleted Th17 cells in LNs at later time points resulted in strongly reduced frequencies of effector YFP+Th17ΔSTAT3 cells in blood (Fig. 2 C) and accordingly fewer YFP+Th17ΔSTAT3 cells in CNS tissue compared with controls (Fig. 2, D and E). Thus, reduced EAE disease severity corresponds with reduced numbers of effector Th17 cells generated in LNs, resulting in fewer effector Th17 cells present in blood and CNS tissue.
Figure 2.
STAT3 deletion reduces effector Th17 cell numbers in vivo. Th17ΔSTAT3 and Th17ctrl mice were immunized with MOG(35–55) in CFA to induce EAE. (A–E) At the indicated time points, live CD4+YFP+ cells were analyzed by flow cytometry in dLNs (A and B), blood (C), and the CNS (D and E) on indicated days after immunization. Data points show values for individual mice with mean ± SD, pooled from two to four experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA (except C, Student’s t test).
STAT3 regulates proliferation, but not apoptosis, in effector Th17 cells
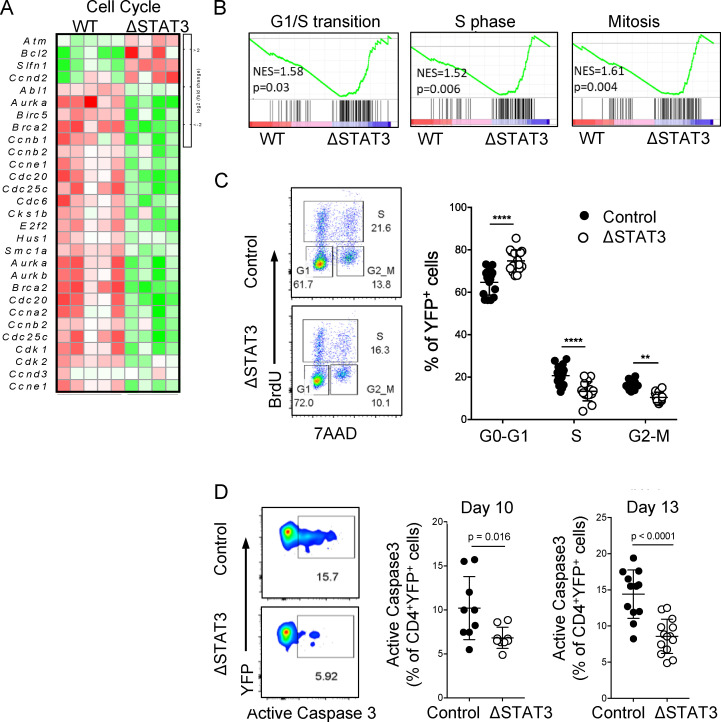
Cell population numbers in a body tissue can be controlled by the rates of proliferation, cell death, and migration into and out of that site. The reduced numbers of STAT3-deleted effector Th17 cells in LNs, blood, and CNS argued against a migration defect, since we did not see accumulation at any site. Gene expression analysis of YFP+ Th17ΔSTAT3 and Th17ctrl cells isolated from dLNs on day 10 after immunization indicated that STAT3 deletion resulted in perturbation of genes associated with cell cycle (Fig. 3 A). Gene set enrichment analysis (GSEA) further highlighted that Th17ΔSTAT3 cells had reduced cell cycle progression (Fig. 3 B). Correspondingly, flow cytometric analysis of YFP+Th17ΔSTAT3 cells after in vivo BrdU administration showed increased proportions of cells in G0/G1 phase and decreased proportions of cells in S phase and G2/M phase compared with Th17ctrl cells (Fig. 3 C). In contrast, GSEA indicated that apoptosis pathways were not significantly enriched after STAT3 deletion, and we did not find any increase in annexin V expression in YFP+ Th17ΔSTAT3 cells (data not shown). Intriguingly, caspase 3 activity was consistently reduced in STAT3-deleted cells, perhaps reflecting the reduced proliferation rate and hence reduced activation-induced cell death of this population (Fig. 3 D). Taken together, these data indicate that reduced proliferation underlies the defective expansion of Th17 effector cells after STAT3 deletion.
Figure 3.
STAT3 regulates proliferation, but not apoptosis, in Th17 cells. (A and B) Live CD4+YFP+ cells were sorted from dLNs on day 10 after immunization and analyzed by RNA sequencing. (A) Heatmap of cell cycle–associated genes differentially expressed in Th17ctrl versus Th17ΔSTAT3 cells. (B) GSEA of cell cycle stage. NES, normalized enrichment score. (C) Cell cycle stage analyzed by flow cytometry on day 10 after immunization, following BrdU injection in vivo. (D) Apoptosis analyzed by flow cytometry for activated caspase 3 on indicated days after immunization. Data points show values for individual mice with mean ± SD, pooled from two or three experiments. **, P < 0.01; ****, P < 0.0001, one-way ANOVA (C) and Student’s t test (D).
STAT3 deletion in effector Th17 cells does not phenocopy IL-23R blockade
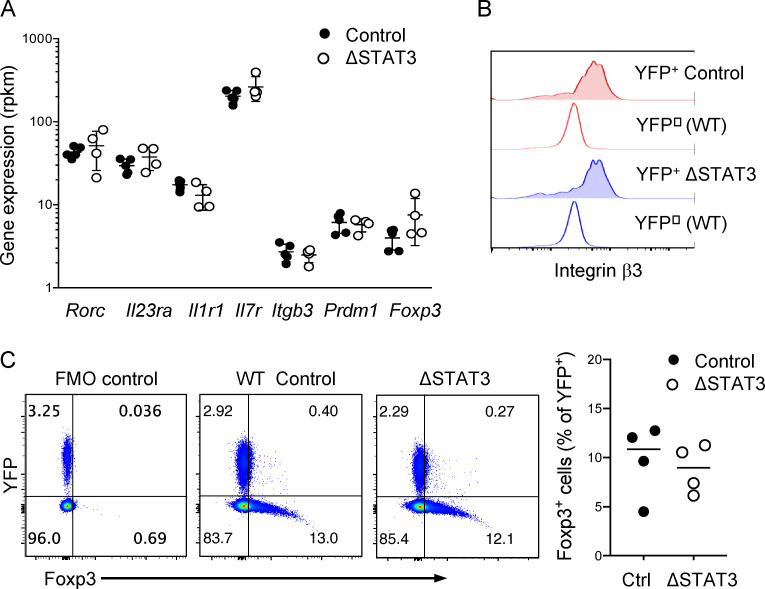
Proliferation defects leading to reduced STAT3-deleted Th17 cells in blood and CNS was reminiscent of Th17 cell defects observed following IL-23R blockade, and in fact, many of the same cell cycle genes were affected (Duhen et al., 2013; Haines et al., 2013). We therefore further investigated whether STAT3 deletion mimicked IL-23R blockade. Surprisingly, expression of factors previously identified to be regulated by IL-23 in effector Th17 cells was in fact unchanged following STAT3 deletion (Codarri et al., 2011; Du et al., 2016; El-Behi et al., 2011; Hirota et al., 2011; Jain et al., 2016; McGeachy et al., 2009). These included receptors for IL-23, IL-1β, and IL-7; the migration mediator integrin β3; and the transcription factor Blimp1 (encoded by Prdm1; Fig. 4 A). We confirmed normal expression of integrin β3 protein on STAT3-deleted effector Th17 cells (Fig. 4 B). IL-23 inhibits Foxp3+ T regulatory cell generation in the intestine, and STAT3 was shown to favor RORγt over Foxp3 expression for developing Th17 cells (Ahern et al., 2010; Geng et al., 2017). However, Rorc and Foxp3 gene expression was not different between YFP+ Th17ΔSTAT3 and YFP+ Th17ctrl cells, and we confirmed that Foxp3 protein expression was not increased after STAT3 deletion in Th17 cells (Fig. 4, A and C). We also confirmed that IL-10 was not increased in response to MOG stimulation either by flow cytometry or ELISA (data not shown). These data thus suggest that loss of IL-23R signaling through STAT3 does not fully explain the functional defects observed in YFP+ Th17ΔSTAT3 cells.
Figure 4.
STAT3 deletion in effector Th17 cells does not phenocopy IL-23R blockade. (A) Relative gene expression for select IL-23 targets, generated by RNA sequencing from indicated YFP+ cells sorted from dLNs on day 10 after immunization. (B) Integrin β3 expression analyzed by flow cytometry in YFP+ and YFP− CD4+ dLN cells from Th17ΔSTAT3 and Th17ctrl mice 10 d after immunization with MOG(35–55) in CFA. Plots are representative of three or four mice per group in three experiments. (C) Foxp3 expression in YFP+ cells analyzed by flow cytometry on day 10 after immunization. FMO, fluorescence minus one. Data points show values for individual mice with mean ± SD, pooled from two experiments. Differences between groups were not significant by one-way ANOVA (A) and Student’s t test (C).
STAT3 counterregulates STAT1 activation by IL-6 in effector Th17 cells
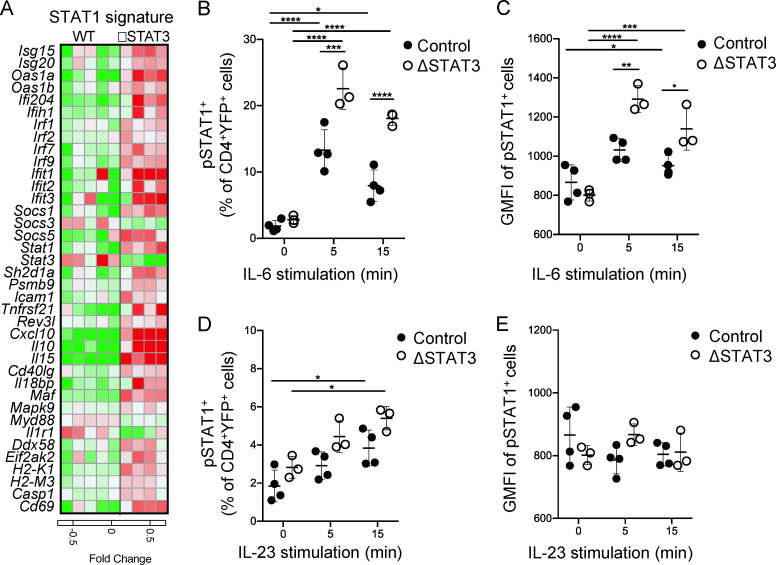
IL-6 signaling activates both STAT1 and STAT3. These transcription factors have opposing effects on proliferation; STAT3 promotes while STAT1 inhibits cell cycle progression in T cells (Bromberg et al., 1996; Durant et al., 2010; Gil et al., 2006). Gene expression analysis of YFP+ Th17ΔSTAT3 and YFP+Th17ctrl cells indicated an enrichment of type-I IFN– and IFN-γ–associated expression. Since IFNs strongly drive STAT1 activation, we generated a map of STAT1-associated gene expression that included many of the IFN-associated genes (Twohig et al., 2019). These data supported a pattern of enhanced STAT1 activation in STAT3-deleted cells (Fig. 5 A), including STAT1 itself as previously reported (Costa-Pereira et al., 2002; Der et al., 1998; Wan et al., 2015). Activation of naive T cells increases expression of protein tyrosine phosphatases that tune STAT1 activation (Twohig et al., 2019). However, we did not observe changes in expression of these genes in the absence of STAT3 (data not shown). We next tested whether STAT1 phosphorylation in response to IL-6 stimulation was enhanced after STAT3 deletion in Th17 effector cells. IL-6 induced phosphorylation of STAT1 in YFP+Th17ctrl cells, as expected (Fig. 5, B and C). YFP+ Th17ΔSTAT3 cells responded to IL-6 with increased STAT1 phosphorylation both as a proportion of responders and amount of pSTAT1 per cell (Fig. 5, B and C). IL-23 stimulation also activated STAT1, but, compared with IL-6, IL-23 induced overall weaker STAT activation and an attenuated trend toward increased STAT1 activity in absence of STAT3 (Fig. 5, D and E). Taken together, these data suggest that STAT3 competitively reduces STAT1 activation following Th17 cell–promoting cytokine stimulation.
Figure 5.
STAT3 counterregulates STAT1 activation by IL-6 in effector Th17 cells. (A) Heatmap of relative gene expression for known STAT1 targets generated by RNA sequencing from indicated YFP+ cells sorted from dLN on day 10 after immunization. (B–E) dLN cells were isolated from indicated mice on day 10 after immunization with MOG(35–55) in CFA and stimulated with IL-6 (B and C) or IL-23 (D and E) for indicated times. (B and D) Percentage of YFP+ cells expressing pSTAT1. (C and E) Geometric mean fluorescence intensity (GMFI) of pSTAT1-expressing YFP+ cells. Data points show values for individual mice with mean ± SD, pooled from two experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA.
STAT3 is required for TCR-stimulated cytokine production in Th17 cells
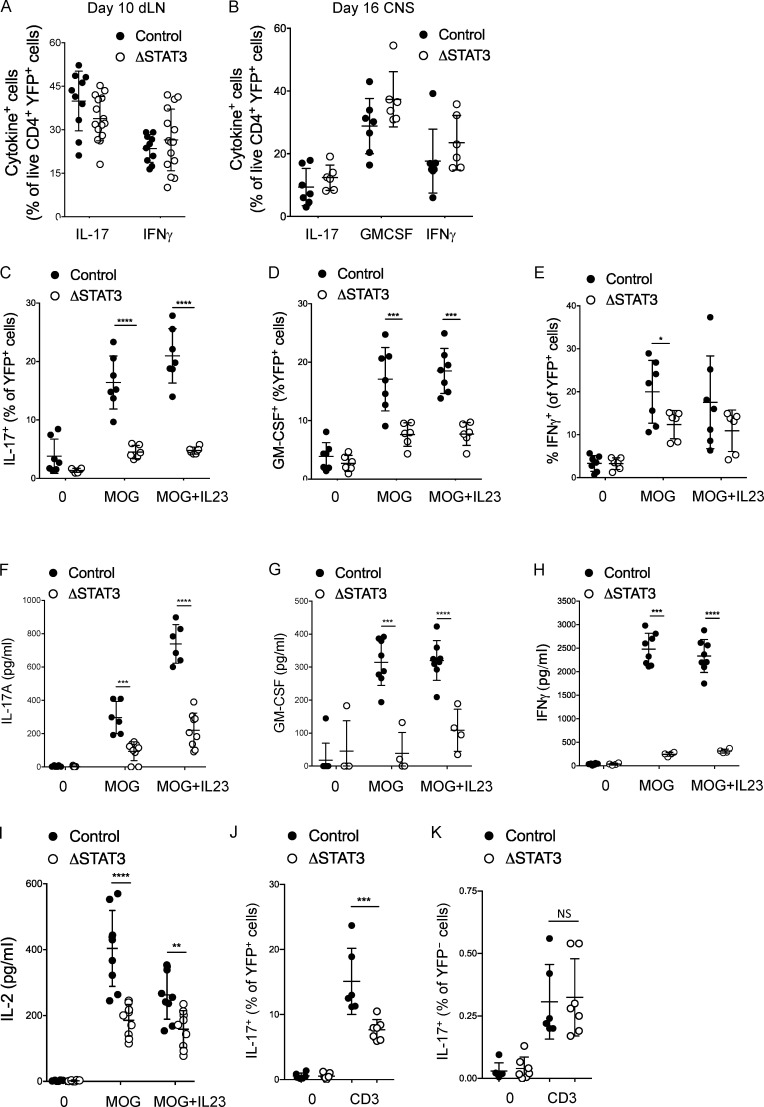
Stimulation with PMA and ionomycin bypasses the intracellular regulation of Ca2+ signaling pathways, and we queried whether this had masked a functional defect in STAT3-deleted Th17 cells (Fig. 6, A and B). YFP+Th17ΔSTAT3 cells (from dLNs of mice immunized 10 d prior) were therefore stimulated overnight with MOG(35–55) before the addition of Golgi inhibitor and assessment of intracellular cytokine by flow cytometry. In contrast to stimulation with PMA and ionomycin, antigen stimulation revealed a significant defect in production of IL-17, GM-CSF, and, to a lesser extent, IFN-γ in YFP+Th17ΔSTAT3 compared with YFP+Th17ctrl cells (Fig. 6, C–E). The geometric mean fluorescence intensity of cytokine-positive cells showed a trend toward decreased production per cell in YFP+Th17ΔSTAT3 compared with YFP+Th17ctrl cells, but this did not reach statistical significance. Cytokine secretion assessed by ELISA after only 10 h of restimulation was also clearly impaired in Th17ΔSTAT3 mice compared with Th17ctrl cells (Fig. 6, F–H). This second set of analyses was performed on day 7 when YFP+ cell frequencies were similar, and the short stimulation avoids the effects of altered proliferation rates. Strikingly, the defect in cytokine production by STAT3-deleted cells was apparent even without addition of IL-23 (Fig. 6, C–H). IL-2, the prototypical cytokine response to TCR triggering, was also reduced in STAT3-deleted cells (Fig. 6 I), further suggesting that TCR activation, rather than only cytokine signaling, could be regulated by STAT3. Finally, stimulation of immunized dLN cells with anti-CD3 demonstrated a similar defect in IL-17 production by YFP+Th17ΔSTAT3 cells, while YFP– cells from Th17ΔSTAT3 and Th17ctrl mice had no difference in IL-17 production, confirming that the defect was intrinsic to the Th17 cells rather than host effects (Fig. 6, J and K). Taken together, these data show an unexpected regulation of cytokine production in response to antigen that is dependent on STAT3 and can be overridden by PMA and ionomycin stimulation.
Figure 6.
STAT3 sustains TCR-stimulated cytokine production. (A and B) Cytokine expression in live CD4+ YFP+ cells from indicated mice analyzed by flow cytometry following stimulation with PMA and ionomycin in the presence of GolgiPlug in dLNs on day 10 and the CNS on day 16 after immunization with MOG(35–55) in CFA. (C–E) dLN cells were isolated from indicated mice on day 10 after immunization and stimulated with MOG(35–55) with or without IL-23 for 19 h, GolgiPlug was added for the final 5 h, and the percentage of YFP+ cells expressing the indicated cytokine was analyzed by flow cytometry. (F–H) dLN cells were isolated from indicated mice on day 7 after immunization and stimulated with MOG(35–55) with or without IL-23 for 10 h, and the indicated cytokines were analyzed by ELISA in supernatants. (I) dLN cells were isolated from indicated mice on day 7 after immunization and stimulated with MOG(35–55) with or without IL-23 for 19 h, and supernatant IL-2 was assessed by ELISA. (J and K) dLN cells were isolated from Th17ctrl mice on day 10 after immunization with MOG(35–55) in CFA and stimulated with 10 μg/ml anti-CD3 for 14 h, GolgiPlug was added for the final 5 h, and the percentage of YFP+ (J) and YFP− (K) cells expressing IL-17 was analyzed by flow cytometry. Data points show values for individual mice with mean ± SD, pooled from two experiments (except B and C, showing one experiment representative of three with similar results). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA.
STAT3 sustains mitochondrial membrane potential (Δψm) and Ca2+ signaling in effector Th17 cells
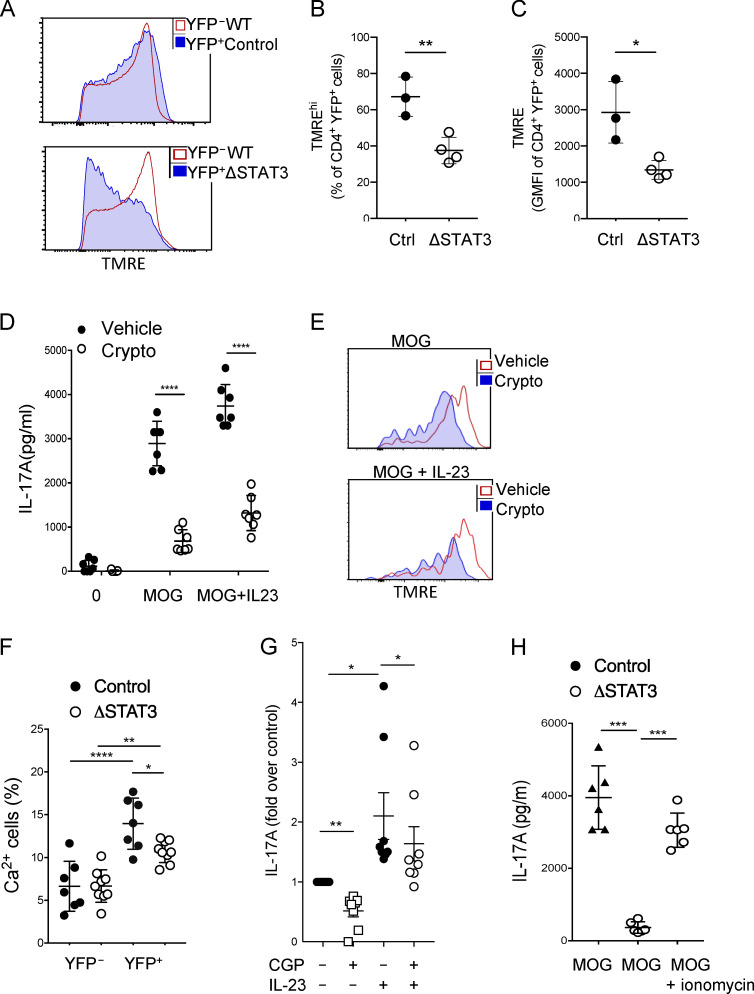
The prior data pointed to a role for STAT3 in promoting TCR signaling in addition to mediating transcription in response to classical cytokine signaling. We therefore examined the nontranscriptional role reported for STAT3 in maintaining the integrity of Δψm (Meier and Larner, 2014; Rincon and Pereira, 2018). Indeed, STAT3-depleted YFP+ cells had reduced Δψm on day 10 after immunization when compared with STAT3-sufficient YFP−CD4+ cells within the same host and compared with YFP+ cells from immunized Th17ctrl mice (Fig. 7, A–C). To confirm that Δψm was maintained by STAT3 in effector Th17 cells, we immunized Th17ctrl mice and stimulated the cells ex vivo in presence of STAT3 inhibitor cryptotanshinone, which inhibits both p727 and p703 sites (Brambilla et al., 2015; Genini et al., 2017). STAT3 inhibition during restimulation of effector Th17 cells resulted in significantly reduced IL-17 production, corresponding to reduced Δψm (Fig. 7, D and E). Reduced Δψm can disrupt mitochondrial Ca2+ storage, with consequences for intracellular Ca2+-regulated signaling (Yang et al., 2015). Accordingly, YFP+Th17ΔSTAT3 cells had a reduced intracellular Ca2+ response to stimulation with MOG(35–55) ex vivo compared with YFP+Th17ctrl cells from similarly immunized mice (Fig. 7 F). To further demonstrate that mitochondrial Ca2+ regulation contributes to MOG-induced IL-17 production in effector Th17 cells, we restimulated dLN cells from immunized mice in presence or absence of CGP-37157 to block mitochondrial Ca2+ export. IL-17 production in response to MOG and MOG + IL-23 was significantly reduced in presence of mitochondrial Ca2+ export blockade (Fig. 7 G). Interestingly, IL-23–induced IL-17 was clearly less affected than MOG alone in this assay, suggesting a separation between STAT3 transcriptional events downstream of cytokine signaling and STAT3 mitochondrial maintenance for TCR signaling. Finally, to determine whether overcoming intracellular Ca2+ signaling could override the defective IL-17 production in Th17ΔSTAT3 cells, we restimulated dLN cells from Th17ΔSTAT3 and Th17ctrl mice with MOG in the presence of ionomycin to bypass the TCR-induced store-operated calcium entry pathway of Ca2+ flux. Indeed, ionomycin was sufficient to increase MOG-induced IL-17 production by STAT3-depleted Th17 cells (Fig. 7 H). Hence, we conclude that sustained STAT3 regulation of Δψm and intracellular Ca2+ is required for effector cytokine production in response to antigen recognition through the TCR.
Figure 7.
STAT3 regulates Δψm. (A–C) dLN cells were isolated from indicated mice on day 10 after immunization with MOG(35–55) in CFA and labeled with TMRE for flow cytometric analysis of Δψm. (A) Representative TMRE labeling in YFP+ and YFP– cells from indicated mice. (B) Percentage of cells labeled TMREhi in indicated YFP+ cells. (C) Mean fluorescence intensity of TMRE labeling in YFP+ cells. Data are representative of three experiments with three or four mice per group. (D and E) dLN cells were isolated from Th17ctrl mice on day 10 after immunization with MOG(35–55) in CFA and stimulated with MOG(35–55) in the presence or absence of the STAT3 inhibitor cryptotanshinone for 19 h; IL-17A was assessed in the supernatant by ELISA (D), and TMRE was assessed by flow cytometry (E). Plots are representative of data from two experiments with three mice each. (F) dLN cells were isolated from indicated mice on day 10 after immunization, labeled with the intracellular Ca2+ detector Rhod3-AM, and stimulated with MOG(35–55) for 10 min before flow cytometric analysis. (G) On day 10 after immunization, dLN cells from Th17ctrl mice were stimulated in the presence of MOG ± IL-23 and mitochondrial Ca2+ inhibitor CGP-37157 or vehicle control overnight, and then IL-17 was assessed by ELISA. Data are shown as paired fold change pooled from two experiments. (H) dLN cells were isolated from indicated mice on day 10 after immunization and stimulated with MOG(35–55) with or without ionomycin for 19 h, and IL-17 was assessed by ELISA. Data are pooled from two experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Student’s t test (B and C) and one-way ANOVA (D–H).
Discussion
STAT3 transcriptional activity is critical for early Th17 development. Here, we have demonstrated that STAT3 also plays a critical role after initiation of the Th17 program and that this role extends beyond the expected transcription factor functions. In fact, we unexpectedly found that STAT3 was not directly required for transcriptional activation of effector cytokines, since there was no defect in cytokines elicited by PMA and ionomycin stimulation. Early STAT3 activity stabilizes Th17 differentiation both through RORγt expression and epigenetic modifications (Durant et al., 2010). Our data suggest that RORγt expression is unchanged and therefore sufficient to sustain Th17 cytokine response when ionomycin is present to override requirements for intracellular Ca2+ regulation. Indeed, a recent report using the same Il17aCre system to delete Rorc demonstrated that RORγt deletion in effector Th17 cells led to loss of IL-17–producing capacity (Brucklacher-Waldert et al., 2016). These findings do not preclude a role for STAT3 acting as a transcription factor downstream of cytokine signaling, but rather suggest that it is not absolutely required for this role in effector Th17 cells.
Our data point toward a critical role for STAT3 in maintaining the capacity of Th17 cells to produce cytokine in response to antigenic stimuli, and this was associated with reduced Δψm. In cancer cells and cardiac myocytes, STAT3-regulated Δψm is thought to be important for mitochondrial metabolic activity required to support cell proliferation, as well as counteracting damaging effects of reactive oxygen species generated under hypoxic conditions (Meier and Larner, 2014; Yang et al., 2015). Mitochondrial STAT3 is known to interact with components of the electron transport chain, including GRIM19, but the mechanisms by which this interaction regulates Δψm are as yet unclear (Rincon and Pereira, 2018). The role of STAT3 in mitochondria has not been extensively studied in T cells. However, mitochondrial STAT3 induction by IL-6 has been clearly shown to sustain IL-21 and IL-4 production in recently activated T cells (Yang et al., 2015). Unlike tumor cells, mitochondrial functions of STAT3 in recently activated T cells were uncoupled from energy generation (Yang et al., 2015). Similarly, we did not observe any changes in metabolic pathway or mitochondrial genes in STAT3-deleted Th17 effector cells.
Mitochondria play an often-underappreciated role in balancing intracellular Ca2+ stores. Mitochondrial and ER membranes are intimately associated in the cytoplasm, and Δψm regulation is important for ER store-operated Ca2+ flux (Duszyński et al., 2006; Marchi et al., 2017). TCR-induced Ca2+ flux leads to activation of the transcription factor nuclear factor of activated T cells (NFAT), a key driver of IL-2 production in recently activated T cells that has also been linked to IL-17 production in Th17 cells. Itk-deficient T cells have defective activation of the nuclear factor of activated T cells (NFAT) pathway downstream of TCR ligation, leading to impaired IL-17 production that can be overcome by ionomycin. Compared with naive T cells, effector T cells have greatly reduced store-operated Ca2+ entry in response to TCR stimulation (Bohineust et al., 2018; Manjunath et al., 1999) and so are likely to be especially susceptible to changes in mitochondrial-regulated Ca2+ concentrations. We propose that a major role of STAT3 in effector Th17 cell function is to regulate the responsiveness of Th17 cells to TCR engagement by maintaining Δψm and thus intracellular Ca2+ balance. These data also suggest that assessment of cytokine production by PMA and ionomycin stimulation may erroneously report cytokine production capability during therapeutic blockade of the STAT3 pathway and that Δψm may better reflect the functional capacity of effector Th17 cells.
There were interesting differences between IL-23R blockade and STAT3 deletion in effector Th17 cells, as might be expected, since STAT3 is also activated by IL-6 and IL-21 to support Th17 development (Heink et al., 2017; Wei et al., 2007). A key difference was in induction of effector molecules. IL-23R blockade results in reduced production of IL-17 and IFN-γ by day 10 after immunization for EAE, whether measured by PMA and ionomycin stimulation or antigen stimulation with MOG(35–55) (McGeachy et al., 2009). In contrast, STAT3 deletion after IL-17 production did not affect PMA- and ionomycin-elicited cytokine production or expression of other previously identified IL-23–dependent effector molecules (Codarri et al., 2011; Du et al., 2016; El-Behi et al., 2011; Hirota et al., 2011; Jain et al., 2016; McGeachy et al., 2009). This disparity could perhaps be explained by the fact that the absolute requirement for IL-23 signaling in Th17 cell effector function (at least in the EAE model) occurs within the first 5 d in vivo, despite the functional outcomes of IL-23 blockade appearing during the effector stages starting on day 7 (Chen et al., 2006; McGeachy et al., 2009).
Both Il23ra−/− and STAT3-deleted Th17 cells both show defective proliferation associated with reduced numbers of effector Th17 cells in blood and CNS (Haines et al., 2013; McGeachy et al., 2009). Our data suggest that enhanced STAT1 activation, which has well-established antiproliferative functions (Bromberg et al., 1996; Durant et al., 2010; Gil et al., 2006), further contributes to reduced proliferation following STAT3 deletion, although we have not directly tested the contribution of STAT1 to proliferative defects in these cells. IL-6 and IL-23 signaling are typically associated with driving STAT3 phosphorylation, but like most cytokines, they activate multiple STAT proteins. The interplay between STAT3 and STAT1 can determine specificity of response to cytokine signaling, as has been characterized for IL-6 and IL-21 signaling (Hirahara et al., 2015; Wan et al., 2015). STAT3 deletion in murine naive T cells or fibroblasts, and dysfunctional STAT3 in patients with hyper-IgE syndrome, result in enhanced STAT1 activity and gene expression signature (Costa-Pereira et al., 2002; Hirahara et al., 2015; Wan et al., 2015). Gain-of-function mutations in STAT1 have been shown to inhibit Th17 responses and cause chronic mucocutaneous candidiasis that is clinically similar to Th17 cell–associated immunodeficiency (Eren Akarcan et al., 2017). Thus, these data and our results suggest that negative regulation of STAT1 activation is an important function of STAT3 in maintaining healthy Th17 cell populations at barrier surfaces.
In summary, our data suggest a new model in which lack of STAT3 activation temporarily “stuns” effector Th17 cells by impairing their ability to respond to antigen through the TCR. In vivo, this would be an efficient safeguard against inappropriate activation of a proinflammatory Th17 response in absence of infection, when local concentrations of STAT3-activating cytokines (such as IL-6 and IL-23) remain low. However, we speculate that tissue damage or bystander infection could overcome this safeguard by inducing increased inflammatory cytokines that allow potent activation of tissue-resident memory cells to induce local inflammation, thus providing a potential mechanism to link recent infection or tissue injury with flare of autoimmune disease.
Materials and methods
Mice
C57BL/6, STOPfl/flROSAYFP and Il17aCre (Hirota et al., 2011) mice were from the Jackson Laboratory, and STAT3fl/fl mice were provided by Dr. Levy (New York University, New York, NY). All experiments included age- and sex-matched littermate controls, and both males and females aged between 7 and 18 wk were used. Mice were housed under specific pathogen–free conditions in an Association for Assessment and Accreditation of Laboratory Animal Care–approved facility at the University of Pittsburgh. Protocols were approved by the University of Pittsburgh institutional animal care and use committee and adhered to guidelines in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
EAE
Mice were immunized subcutaneously in four sites on the back with 100 µg MOG(35–55) peptide (Biosynthesis) emulsified with CFA with M. tuberculosis strain H37Ra (DIFCO Laboratories). Mice also received 100 ng pertussis toxin (List Biological Laboratories) intraperitoneally on days 0 and 2. Mice were assessed daily and scored as follows: 1, flaccid tail; 2, impaired righting reflex and hindlimb weakness; 3, partial hindlimb paralysis; 4, complete hindlimb paralysis; 5, hindlimb paralysis with partial forelimb paralysis; or 6, moribund.
Flow cytometry
Single-cell suspensions were obtained from LNs and CNS (brain and spinal cord) tissues by mechanical isolation. The single-cell suspensions were filtered before staining. CNS tissue homogenates were incubated in digestion medium containing 0.5 mg/ml Collagenase Type I (Worthington Biochemical) and 1,000 U/ml DNase I (Sigma-Aldrich) for 45 min, following myelin debris removal using a Percoll gradient. Blood was collected in PBS containing EDTA following red blood cell lysis using ACK lysis buffer (Gibco). For flow cytometry and FACS sorting, cells were first stained with Ghost Dye Violet 510 or Ghost Dye Violet 450 viability dyes (TONBO Biosciences) in PBS to allow exclusion of dead cells, followed by labeling with cell-surface and intracellular FACS antibodies or other reagents. For cytokine analysis, cells were cultured in complete medium with 50 ng/ml PMA and 500 ng/ml ionomycin (Sigma-Aldrich) in the presence of GolgiPlug (BD Biosciences) for 4 h before FACS staining. For intracellular cytokines, staining was performed using Cytofix-cytoperm kit from BD Biosciences. For Foxp3 costaining with YFP, cells were fixed with 2% formaldehyde for 30 min at 4°C and then permeabilized with permeabilization buffer (eBioscience) for 30 min followed by intracellular staining. Phosphoflow cytometry analysis was performed using BD Phosflow buffers (557870 and 558050). FACS antibodies and staining reagents were purchased from the following companies: eBioscience: Ly-6G/Gr-1 (1A8-Ly6g), GM-CSF (MP1-22E9), IL-17A (eBio17B7), IFN-γ (XMG1.2); BD: IL-2 (JES6-5H4), GM-CSF (MP1-22E9), STAT3 (M59-50), Ki-67 (B56), CD178 (MFL3), IL-17A (TC11-18H10), pSTAT1 (4a); Invitrogen: CD4 (RM4-5), Rhod-3 Calcium Imaging Kit (R10145), TMRE (tetramethylrhodamine, ethyl ester; T669); and BioVision: caspase 3 (K183) Staining Kit. Flow cytometry data were acquired on a FACS Fortessa (BD Biosciences) and analyzed with FlowJo software.
Cell cycle analysis using BrdU
BrdU Flow Kit was purchased from BD Biosciences (552598). 1 mg BrdU was injected into MOG(35–55)–immunized animals intraperitoneally 4 h before analysis. Following BrdU staining (according to the manufacturer’s instructions), cells were resuspended in PBS containing 7AAD for staining of total DNA. Data were acquired on a FACS Fortessa (BD Biosciences) and analyzed with FlowJo software.
RNA sequencing and GSEA
Th17Ctrl (five mice total, combined from two separate experiments) and Th17ΔSTAT3 (four mice total combined from two separate experiments) mice were immunized with MOG(35–55) in CFA to induce EAE. On day 10 after immunization, dLNs were harvested and CD4+YFP+ Th17 cells were sorted on a FACSAria II, directly into SmartSeq low-input RNA kit lysis buffer. Libraries were prepared using Nextera XT DNA library prep kits, and RNA sequencing was performed on Illumina NextSeq500 by the Health Sciences Sequencing Core at University of Pittsburgh. Reads were aligned to mouse reference genome mm10 using TopHat and Bowtie2 following gene identification using Cufflinks. Gene expression values (fragments per kilobase exon per million mapped reads) were calculated using Cuffdiff. Relative expression shown in heatmaps was calculated as the fragments per kilobase exon per million mapped reads value for each sample divided by mean expression of that gene in all samples per each experiment. GSEA from the Broad Institute (http://www.broad.mit.edu/gsea) was used to calculate enrichment of genes in each set. Pathways were curated from Reactome Pathway Database (Fabregat et al., 2018).
MOG restimulation and ELISA assays
dLN cells from mice immunized with MOG(35–55) in CFA were cultured in flat bottom 96-well plates at a cell density of 1 million cells per well in complete medium (RPMI) with 100 μg/ml MOG(35–55) in the presence or absence of 20 ng/ml IL-23. STAT3 inhibitor cryptotanshinone (S2285; Selleck Chemicals) was added to cultures at a final concentration of 5–10 μM. The mitochondrial Ca2+ inhibitor CGP-37157 (Tocris) was added to cultures at final concentration of 10 μM. Cytokine production was analyzed from culture supernatants taken 14 h after MOG rechallenge using the following ELISA kits from eBioscience: IL-17A (88–7371-76), GM-CSF (88–7334-22), IFN-γ (88–7314-76), and IL-2 (88–7024-22). For intracellular cytokine analysis by flow cytometry, GolgiPlug (BD Biosciences) was added to the cell cultures at 14 h for a further 5 h before intracellular FACS staining.
Calcium flux assay
dLNs from mice immunized with MOG(35–55) were labeled with red fluorescent Rhod-3 AM dye according to the manufacturer’s guidelines (R10145; Invitrogen). Total LN cells were stimulated with 100 μg/ml MOG(35–55) in HBSS medium containing 5% FCS for 10 min immediately before flow cytometry analysis. Positive cells were assessed as percentage cells above baseline set with unstimulated cells.
Statistics
Experimental results were analyzed for significance using one-way ANOVA (for multiple groups) or Student’s t test, except EAE clinical data, which were analyzed by Mann–Whitney test, on each day. Statistical analyses were performed using GraphPad Prism. P values are shown as *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 where statistical significance was found, and data are represented as mean ± SD. Individual points in graphs represent biological replicates (i.e., individual mice).
Data availability
RNA-sequencing datasets from this study are available in the Gene Expression Omnibus under accession number GSE151447.
Acknowledgments
The authors thank the Division of Laboratory Animal and Research support staff for animal husbandry and the United Flow Core of University of Pittsburgh and Flow Core of Children’s Hospital for assistance with flow cytometry and sorting. D. Wu acknowledges the encouragement of Dr. Fen Wang. The graphical abstract was generated in BioRender.
Funding for this study was provided by the National Institutes of Health (grants AI110822, AI148356, and DE023815 to M.J. McGeachy and AI089443 T32 training grant to I. Raphael). D. Wu was sponsored by the Third Xiangya Hospital, Central South University. S. Majumder was supported by a trainee fellowship from the American Association of Immunologists. This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided.
Author contributions: C.H. Poholek, I. Raphael, D. Wu, S. Revu, S. Majumder, N. Rittenhouse, and M.J. McGeachy performed experiments and data analysis. A.C. Poholek, U.U. Uche, and L.P. Kane assisted with experimental design and analysis. M.J. McGeachy designed and directed the study and wrote the manuscript with assistance from I. Raphael and C.H. Poholek.
References
- Ahern P.P., Schiering C., Buonocore S., McGeachy M.J., Cua D.J., Maloy K.J., and Powrie F.. 2010. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 33:279–288. 10.1016/j.immuni.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohineust A., Garcia Z., Beuneu H., Lemaître F., and Bousso P.. 2018. Termination of T cell priming relies on a phase of unresponsiveness promoting disengagement from APCs and T cell division. J. Exp. Med. 215:1481–1492. 10.1084/jem.20171708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla L., Genini D., Laurini E., Merulla J., Perez L., Fermeglia M., Carbone G.M., Pricl S., and Catapano C.V.. 2015. Hitting the right spot: Mechanism of action of OPB-31121, a novel and potent inhibitor of the Signal Transducer and Activator of Transcription 3 (STAT3). Mol. Oncol. 9:1194–1206. 10.1016/j.molonc.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Horvath C.M., Wen Z., Schreiber R.D., and Darnell J.E. Jr.. 1996. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. USA. 93:7673–7678. 10.1073/pnas.93.15.7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucklacher-Waldert V., Ferreira C., Innocentin S., Kamdar S., Withers D.R., Kullberg M.C., and Veldhoen M.. 2016. Tbet or Continued RORγt Expression Is Not Required for Th17-Associated Immunopathology. J. Immunol. 196:4893–4904. 10.4049/jimmunol.1600137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Langrish C.L., McKenzie B., Joyce-Shaikh B., Stumhofer J.S., McClanahan T., Blumenschein W., Churakovsa T., Low J., Presta L., et al. . 2006. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J. Clin. Invest. 116:1317–1326. 10.1172/JCI25308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. . 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., and Becher B.. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12:560–567. 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]
- Costa-Pereira A.P., Tininini S., Strobl B., Alonzi T., Schlaak J.F., Is’harc H., Gesualdo I., Newman S.J., Kerr I.M., and Poli V.. 2002. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc. Natl. Acad. Sci. USA. 99:8043–8047. 10.1073/pnas.122236099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beaucoudrey L., Puel A., Filipe-Santos O., Cobat A., Ghandil P., Chrabieh M., Feinberg J., von Bernuth H., Samarina A., Jannière L., et al. . 2008. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J. Exp. Med. 205:1543–1550. 10.1084/jem.20080321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der S.D., Zhou A., Williams B.R., and Silverman R.H.. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 95:15623–15628. 10.1073/pnas.95.26.15623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio P., Di Cesare A., Laggner U., Chu C.C., Napolitano L., Villanova F., Tosi I., Capon F., Trembath R.C., Peris K., et al. . 2011. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS One. 6 e17160 10.1371/journal.pone.0017160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio P., Villanova F., Napolitano L., Tosi I., Terranova Barberio M., Mak R.K., Nutland S., Smith C.H., Barker J.N.W.N., Todd J.A., et al. . 2013. The IL23R A/Gln381 allele promotes IL-23 unresponsiveness in human memory T-helper 17 cells and impairs Th17 responses in psoriasis patients. J. Invest. Dermatol. 133:2381–2389. 10.1038/jid.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Garg A.V., Kosar K., Majumder S., Kugler D.G., Mir G.H., Maggio M., Henkel M., Lacy-Hulbert A., and McGeachy M.J.. 2016. Inflammatory Th17 Cells Express Integrin αvβ3 for Pathogenic Function. Cell Rep. 16:1339–1351. 10.1016/j.celrep.2016.06.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen R., Glatigny S., Arbelaez C.A., Blair T.C., Oukka M., and Bettelli E.. 2013. Cutting edge: the pathogenicity of IFN-γ-producing Th17 cells is independent of T-bet. J. Immunol. 190:4478–4482. 10.4049/jimmunol.1203172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L., Watford W.T., Ramos H.L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H.W., Kanno Y., Powrie F., et al. . 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 32:605–615. 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszyński J., Kozieł R., Brutkowski W., Szczepanowska J., and Zabłocki K.. 2006. The regulatory role of mitochondria in capacitative calcium entry. Biochim. Biophys. Acta. 1757:380–387. 10.1016/j.bbabio.2006.04.017 [DOI] [PubMed] [Google Scholar]
- El-Behi M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G.X., Dittel B.N., and Rostami A.. 2011. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12:568–575. 10.1038/ni.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren Akarcan S., Ulusoy Severcan E., Edeer Karaca N., Isik E., Aksu G., Migaud M., Evin Gurkan F., Azarsiz E., Puel A., Casanova J.L., et al. . 2017. Gain-of-Function Mutations in STAT1: A Recently Defined Cause for Chronic Mucocutaneous Candidiasis Disease Mimicking Combined Immunodeficiencies. Case Reports Immunol. 2017 2846928 10.1155/2017/2846928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. . 2018. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46(D1):D649–D655. 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen S.L., Jain R., Garg A.V., and Cua D.J.. 2014. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14:585–600. 10.1038/nri3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Yu S., Zhao H., Sun X., Li X., Wang P., Xiong X., Hong L., Xie C., Gao J., et al. . 2017. The transcriptional coactivator TAZ regulates reciprocal differentiation of TH17 cells and Treg cells. Nat. Immunol. 18:800–812. 10.1038/ni.3748 [DOI] [PubMed] [Google Scholar]
- Genini D., Brambilla L., Laurini E., Merulla J., Civenni G., Pandit S., D’Antuono R., Perez L., Levy D.E., Pricl S., et al. . 2017. Mitochondrial dysfunction induced by a SH2 domain-targeting STAT3 inhibitor leads to metabolic synthetic lethality in cancer cells. Proc. Natl. Acad. Sci. USA. 114:E4924–E4933. 10.1073/pnas.1615730114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil M.P., Salomon R., Louten J., and Biron C.A.. 2006. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood. 107:987–993. 10.1182/blood-2005-07-2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines C.J., Chen Y., Blumenschein W.M., Jain R., Chang C., Joyce-Shaikh B., Porth K., Boniface K., Mattson J., Basham B., et al. . 2013. Autoimmune memory T helper 17 cell function and expansion are dependent on interleukin-23. Cell Rep. 3:1378–1388. 10.1016/j.celrep.2013.03.035 [DOI] [PubMed] [Google Scholar]
- Heink S., Yogev N., Garbers C., Herwerth M., Aly L., Gasperi C., Husterer V., Croxford A.L., Möller-Hackbarth K., Bartsch H.S., et al. . 2017. Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat. Immunol. 18:74–85. 10.1038/ni.3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K., Onodera A., Villarino A.V., Bonelli M., Sciumè G., Laurence A., Sun H.W., Brooks S.R., Vahedi G., Shih H.Y., et al. . 2015. Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity. Immunity. 42:877–889. 10.1016/j.immuni.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Duarte J.H., Veldhoen M., Hornsby E., Li Y., Cua D.J., Ahlfors H., Wilhelm C., Tolaini M., Menzel U., et al. . 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12:255–263. 10.1038/ni.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Chen Y., Kanno Y., Joyce-Shaikh B., Vahedi G., Hirahara K., Blumenschein W.M., Sukumar S., Haines C.J., Sadekova S., et al. . 2016. Interleukin-23-Induced Transcription Factor Blimp-1 Promotes Pathogenicity of T Helper 17 Cells. Immunity. 44:131–142. 10.1016/j.immuni.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Korn T., Mitsdoerffer M., Croxford A.L., Awasthi A., Dardalhon V.A., Galileos G., Vollmar P., Stritesky G.L., Kaplan M.H., Waisman A., et al. . 2008. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. USA. 105:18460–18465. 10.1073/pnas.0809850105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N., Shankar P., Stockton B., Dubey P.D., Lieberman J., and von Andrian U.H.. 1999. A transgenic mouse model to analyze CD8(+) effector T cell differentiation in vivo. Proc. Natl. Acad. Sci. USA. 96:13932–13937. 10.1073/pnas.96.24.13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S., Bittremieux M., Missiroli S., Morganti C., Patergnani S., Sbano L., Rimessi A., Kerkhofs M., Parys J.B., Bultynck G., et al. . 2017. Endoplasmic Reticulum-Mitochondria Communication Through Ca2+ Signaling: The Importance of Mitochondria-Associated Membranes (MAMs). Adv. Exp. Med. Biol. 997:49–67. 10.1007/978-981-10-4567-7_4 [DOI] [PubMed] [Google Scholar]
- McGeachy M.J., and McSorley S.J.. 2012. Microbial-induced Th17: superhero or supervillain? J. Immunol. 189:3285–3291. 10.4049/jimmunol.1201834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O’Shea J.J., and Cua D.J.. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10:314–324. 10.1038/ni.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J.A., and Larner A.C.. 2014. Toward a new STATe: the role of STATs in mitochondrial function. Semin. Immunol. 26:20–28. 10.1016/j.smim.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Puel A., Casanova J.L., and Kobayashi M.. 2016. Chronic mucocutaneous candidiasis disease associated with inborn errors of IL-17 immunity. Clin. Transl. Immunology. 5 e114 10.1038/cti.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.D., and Kuchroo V.K.. 2015. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity. 43:1040–1051. 10.1016/j.immuni.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Rincon M., and Pereira F.V.. 2018. A New Perspective: Mitochondrial Stat3 as a Regulator for Lymphocyte Function. Int. J. Mol. Sci. 19:1656 10.3390/ijms19061656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. . 2007. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39:596–604. 10.1038/ng2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.M., Kanno Y., Villarino A., Ward M., Gadina M., and O’Shea J.J.. 2017. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 17:78 10.1038/nrd.2017.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward-Tharp S.M., Laurence A., Kanno Y., Kotlyar A., Villarino A.V., Sciume G., Kuchen S., Resch W., Wohlfert E.A., Jiang K., et al. . 2014. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood. 123:2978–2987. 10.1182/blood-2013-09-523167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twohig J.P., Cardus Figueras A., Andrews R., Wiede F., Cossins B.C., Derrac Soria A., Lewis M.J., Townsend M.J., Millrine D., Li J., et al. . 2019. Activation of naïve CD4+ T cells re-tunes STAT1 signaling to deliver unique cytokine responses in memory CD4+ T cells. Nat. Immunol. 20:458–470. 10.1038/s41590-019-0350-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C.K., Andraski A.B., Spolski R., Li P., Kazemian M., Oh J., Samsel L., Swanson P.A. II, McGavern D.B., Sampaio E.P., et al. . 2015. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc. Natl. Acad. Sci. USA. 112:9394–9399. 10.1073/pnas.1511711112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Laurence A., Elias K.M., and O’Shea J.J.. 2007. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 282:34605–34610. 10.1074/jbc.M705100200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., and Dong C.. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363. 10.1074/jbc.C600321200 [DOI] [PubMed] [Google Scholar]
- Yang R., Lirussi D., Thornton T.M., Jelley-Gibbs D.M., Diehl S.A., Case L.K., Madesh M., Taatjes D.J., Teuscher C., Haynes L., and Rincón M.. 2015. Mitochondrial Ca2+ and membrane potential, an alternative pathway for Interleukin 6 to regulate CD4 cell effector function. eLife. 4:e06376. 10.7554/eLife.06376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Ivanov I.I., Spolski R., Min R., Shenderov K., Egawa T., Levy D.E., Leonard W.J., and Littman D.R.. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8:967–974. 10.1038/ni1488 [DOI] [PubMed] [Google Scholar]
- Zúñiga L.A., Jain R., Haines C., and Cua D.J.. 2013. Th17 cell development: from the cradle to the grave. Immunol. Rev. 252:78–88. 10.1111/imr.12036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-sequencing datasets from this study are available in the Gene Expression Omnibus under accession number GSE151447.