This study shows that IgG memory B cells developing in HBV vaccinees and in rare individuals able to naturally control chronic HBV infection express a panoply of neutralizing antibodies, some of which are potent viremia suppressors in vivo.
Abstract
Rare individuals can naturally clear chronic hepatitis B virus (HBV) infection and acquire protection from reinfection as conferred by vaccination. To examine the protective humoral response against HBV, we cloned and characterized human antibodies specific to the viral surface glycoproteins (HBsAg) from memory B cells of HBV vaccinees and controllers. We found that human HBV antibodies are encoded by a diverse set of immunoglobulin genes and recognize various conformational HBsAg epitopes. Strikingly, HBsAg-specific memory B cells from natural controllers mainly produced neutralizing antibodies able to cross-react with several viral genotypes. Furthermore, monotherapy with the potent broadly neutralizing antibody Bc1.187 suppressed viremia in vivo in HBV mouse models and led to post-therapy control of the infection in a fraction of animals. Thus, human neutralizing HBsAg antibodies appear to play a key role in the spontaneous control of HBV and represent promising immunotherapeutic tools for achieving HBV functional cure in chronically infected humans.
Introduction
Chronic hepatitis B virus (HBV) infection is a major health issue affecting more than 250 million people worldwide despite the availability of effective vaccines (World Health Organization, 2017). The infection accounts for ∼1 million deaths per year due to HBV-associated cirrhosis, liver failure, and hepatocellular carcinoma (World Health Organization, 2017). HBV belongs to the Hepadnaviridae family, and its replication cycle is characterized by the production of infectious virions, or Dane particles, and a large excess of noninfectious subviral particles (Seeger et al., 2013). These defective particles outnumbering infectious HBV virions are thought to act as immune decoys (Seeger et al., 2013). Virions and subviral particles display at their surface three forms of HBV envelope glycoproteins or HBV surface antigens (HBsAg): the large HBsAg (L-HBsAg), the middle HBsAg (M-HBsAg), and the small HBsAg (S-HBsAg). All HBsAg forms share a common C-terminal region called the extracellular antigenic loop or major hydrophilic region, comprising the immunodominant “a” determinant (~aa 124–147), which is the prime target of antibodies induced by vaccination and infection (Bertoletti and Ferrari, 2016). Current therapies for treating chronic HBV rarely achieve functional cure as defined by HBsAg loss and anti-HBs antibody seroconversion. Still, HBV infection in adults can be successfully controlled by natural immune responses during the acute phase, and for ∼1% of chronically infected patients, called HBV seroconverters or natural controllers, who spontaneously clear the infection (Bauer et al., 2011; Chu and Liaw, 2016; McMahon, 2009). Robust and multispecific HBV-specific CD4+ and CD8+ T cell responses are key immune effectors in controlling the infection (Bauer et al., 2011; Bertoletti and Ferrari, 2012, 2016). However, B cells and their secreted antibodies are also instrumental for the long-term clearance and protection from viral rebound after functional cure (Bertoletti and Ferrari, 2016; Corti et al., 2018; Gehring and Protzer, 2019; Maini and Burton, 2019). For instance, despite that HBsAg/anti-HBs seroconversion in patients is correlated with an undetectable level of HBV DNA (McMahon, 2009), functionally cured individuals undergoing B cell depletion therapy to treat non-Hodgkin lymphoma have a higher risk of HBV reactivation, which can quickly lead to severe liver dysfunction (Kusumoto et al., 2019; Perrillo et al., 2015). Hence, key immune components to control HBV infection likely include a broad and robust antigen-specific T cell response, as well as the development of neutralizing anti-HBs antibodies mediating HBsAg clearance and lifelong protective immunity (Bertoletti and Ferrari, 2016; Corti et al., 2018).
Neutralizing antibodies produced in response to HBV infection target either the antigenic loop of L-, M-, or S-HBsAg and interfere with the attachment to heparane sulfate proteoglycans on hepatocytes or the preS1 domain of L-HBsAg and block the binding to the host cellular receptor, the sodium taurocholate cotransporting polypeptide (Corti et al., 2018). IgG antibodies against the immunodominant “a” determinant of HBsAg induced by vaccination (based on recombinant S-HBsAg [rS-HBsAg] immunogens), or administrated to individuals at risk of exposure by polyclonal Ig infusions, confer protection against HBV infection (Samuel et al., 1993; West and Calandra, 1996). Anti-HBs neutralizing antibodies may contribute to the viral clearance and long-term suppression in HBV seroconverters as they do for protecting vaccinees from infection. Several neutralizing anti-preS and anti-HBs antibodies have been isolated from immunized mice and vaccinated humans (Corti et al., 2018). However, no study has yet been performed on the memory B cell response to HBV in functionally cured HBV-infected individuals by cloning and characterizing human HBsAg-specific antibodies. As seroconversion to anti-HBs antibodies and clearance of HBsAg is the hallmark of a functional cure in chronic HBV, the molecular dissection of the antibody repertoire and reactivity of virus-specific IgG memory B cells in natural HBV controllers should provide insights on the humoral parameters participating in the control and protection against HBV infection.
Here, we report on the antibody features of HBV-specific memory B cells circulating in vaccinees and natural controllers. We isolated HBsAg-specific memory B cells from both cohorts using single-cell capture and following antibody expression cloning, we characterized a panel of 72 human anti-HBs monoclonal antibodies at a molecular and functional level. Our analyses reveal the diversity of the memory B cell antibody response to HBV in terms of Ig gene repertoire, cross-genotypic reactivity, neutralizing activity, and targeted S-HBsAg epitopes. These studies also demonstrate that neutralizing anti-HBs memory antibodies dominate in HBV controllers. Finally, monotherapy with a potent controller-derived cross-neutralizer can suppress viremia in mouse models, opening the path toward a therapeutic evaluation of potent broadly neutralizing HBV antibodies in chronically infected individuals.
Results
Human anti-HBs monoclonal antibodies from HBV vaccinees and controllers
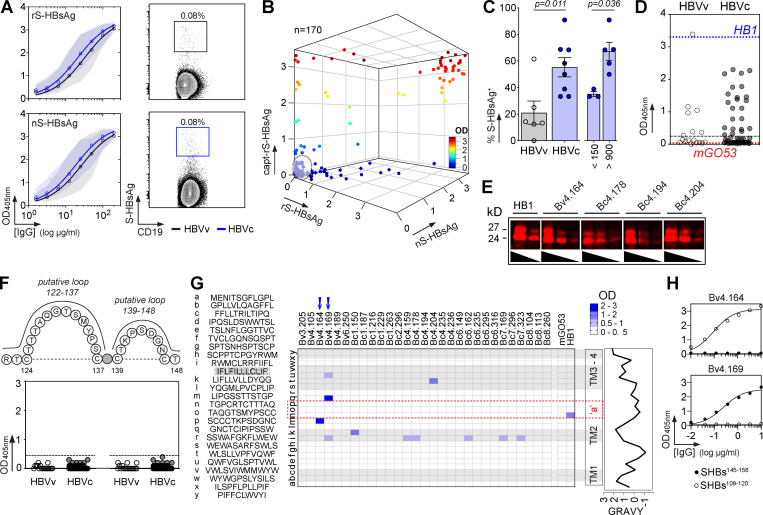
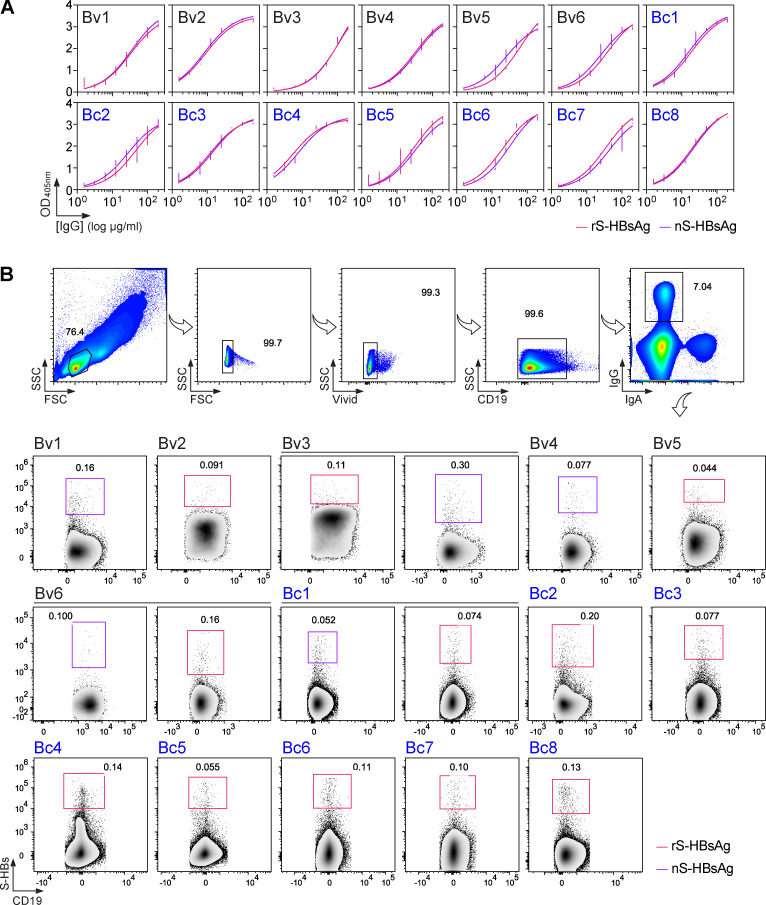
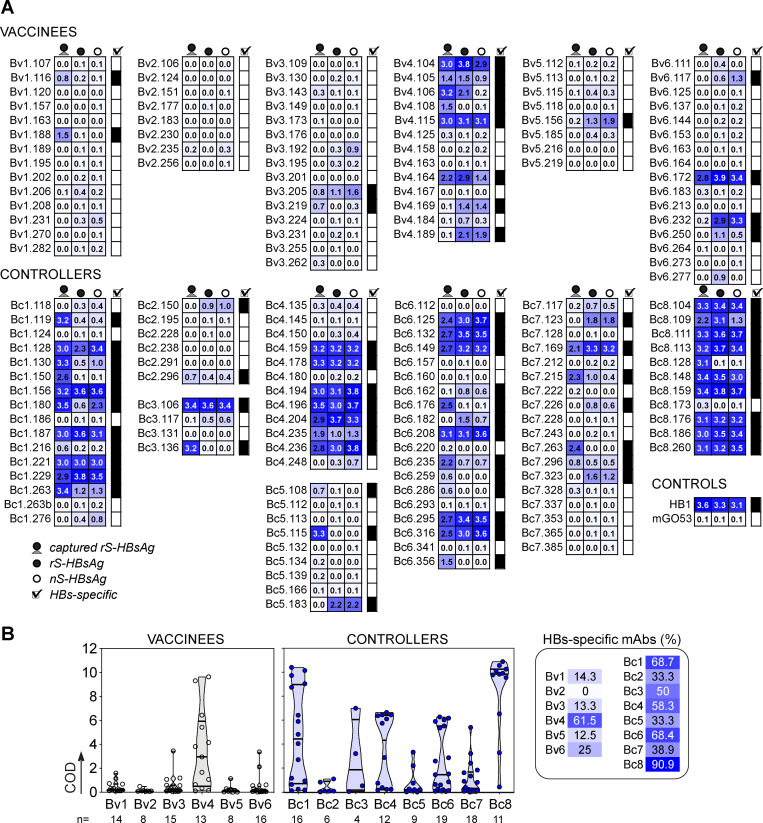
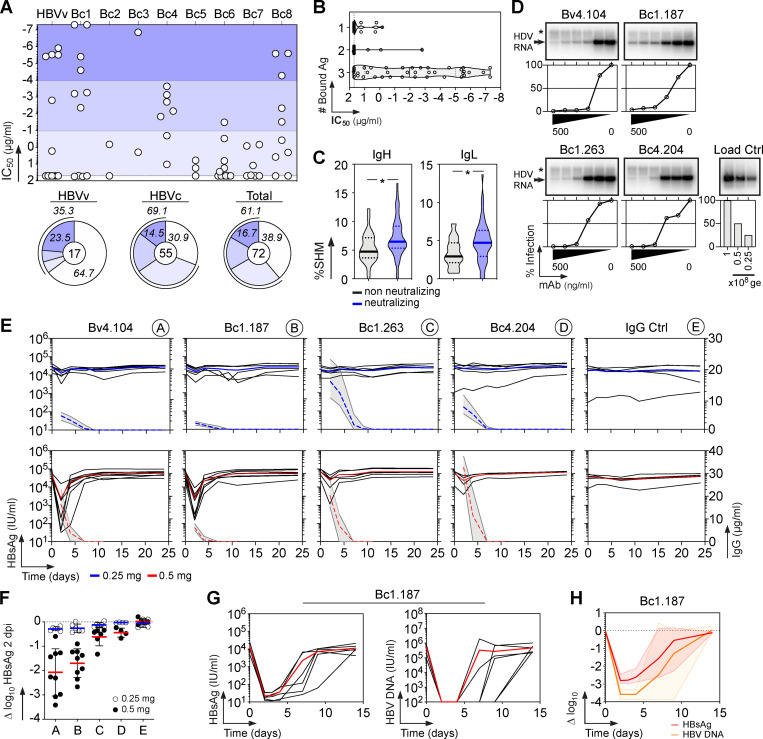
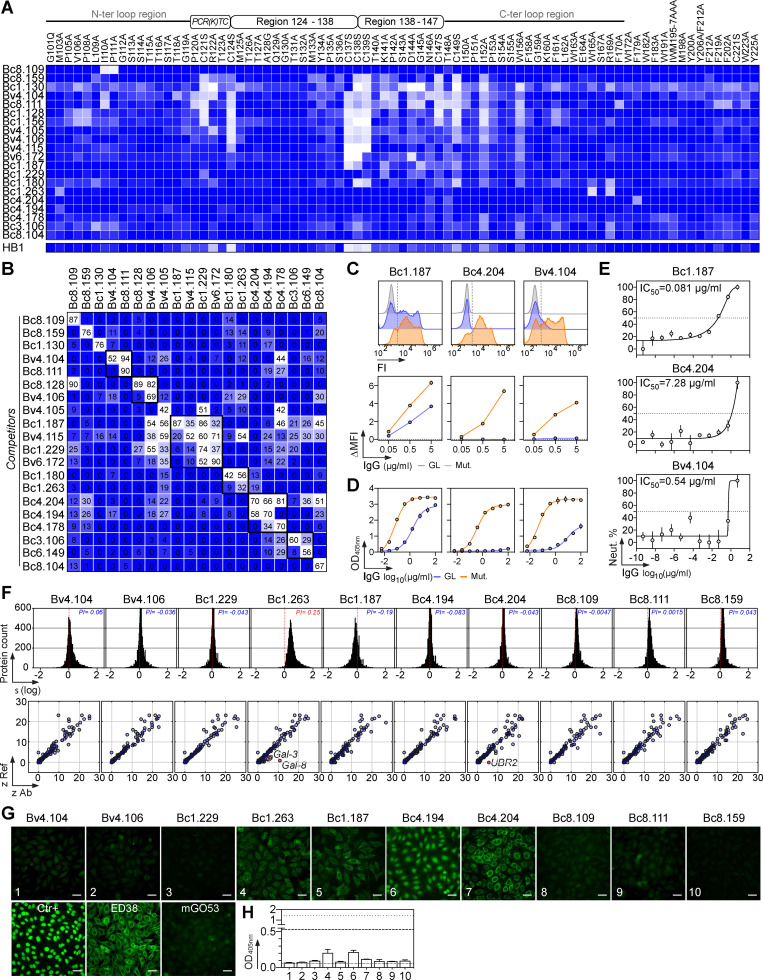
To characterize the memory B cell antibody response to the HBV surface glycoproteins, peripheral blood B cells from six vaccinees and eight controllers with high serum anti-HBs IgG antibody titers (Table S1) were stained with fluorescently labeled rS-HBsAg or native S-HBsAg (nS-HBsAg) particles (Fig. 1 A and Fig. S1). From the 3,452 S-HBsAg–binding IgG+ memory B cells captured by single-cell flow-cytometric sorting, we produced a total of 170 unique human monoclonal antibodies by recombinant expression cloning (Tiller et al., 2008). ELISA binding analyses of S-HBsAg+ memory B cell antibodies cloned from HBV vaccinees and controllers to S-HBsAg particles showed that only 21.1% (0–61.5%) and 55.2% (33.3–90.9%) had high affinity for HBsAg, respectively (P = 0.011; Fig. 1, B and C; and Fig. S2). In HBV controllers, high titers of circulating anti-HBs antibodies were associated to a greater recovery of HBV-specific B cells (35.2% vs. 67.3%; P = 0.036; Fig. 1 C). Bystander B cells lacking S-HBsAg reactivity by ELISA were almost all polyreactive (not shown) and thus may have been captured by interacting with nonenvelope components of the viral particles. 29 anti-HBs IgG antibodies (40.3%; n = 72) showed binding to a recombinant form of the S-HBsAg protein lacking transmembrane (TM) domains (ΔTM-rS-HBsAg; Fig. 1 D), but only four recognized denatured S-HBsAg by immunoblotting (Fig. 1 E). Moreover, none bound to the “a” determinant peptides (Fig. 1, F and G), whereas two reacted with linear epitopes flanking the “a” determinant (Fig. 1, G and H), indicating that most anti-HBs memory antibodies target conformational epitopes.
Figure 1.
S-HBsAg–specific memory antibodies cloned from HBV vaccinees and controllers. (A) Average S-HBsAg reactivity of serum IgGs from HBV vaccinees (HBVv, n = 6, black gate, top) and controllers (HBVc, n = 8, blue gate, bottom) as shown in Fig. S1 A. Shaded regions indicate value ranges. Representative flow-cytometry plots showing S-HBsAg-binding IgG+ memory B cells in HBV vaccinees and controllers as in Fig. S1 B; Bv4 and Bc3 are shown). nS-HBsAg and rS-HBsAg are human-derived nS-HBsAg and rS-HBsAg particles, respectively. (B) S-HBsAg-ELISA reactivities of S-HBsAg–captured IgG+ memory B cell antibodies. capt-rS-HBsAg, rS-HBsAg capture ELISA. Means of triplicate values are shown as measured in Fig. S2 A. (C) Percentage of S-HBsAg–specific monoclonal antibodies (% S-HBsAg+) isolated from HBVv and HBVc (right). % S-HBsAg+ according to anti-HBs antibody titers in HBVc (<150 and >900 IU/ml) are indicated. Groups were compared using the Mann–Whitney test. (D) ELISA reactivity of anti-HBs antibodies against TM domains–deleted S-HBsAg protein (ΔTM-rS-HBsAg). HB1 and mGO53 are positive and negative controls, respectively. The dotted line indicates the cutoff OD405nm for positive reactivity. Means of assay triplicates from three independent experiments are shown. (E) Infrared immunoblot shows anti-HBs memory IgGs reactive against denatured S-HBsAg proteins. The immunoreactive bands correspond to HBV p24 and gp27 proteins. (F) Same as in D but for the cyclic peptides corresponding to putative S-HBsAg loops 122–137 and 139–148. (G) Heat-mapped reactivity of ΔTM-rS-HBsAg–binding antibodies against S-HBsAg overlapping linear peptides. Means of triplicate OD values from one representative experiment (n = 3) are shown. HB1 and mGO53 are positive and negative controls, respectively. Amino acid sequences (left) and values of grand average of hydropathy (GRAVY; right) of S-HBsAg peptides are indicated. Blue arrows indicate peptide-reactive human anti-HBs antibodies. (H) ELISA binding curves of peptide-reactive anti-HBs antibodies are shown (means of assay quadruplicates ± SD).
Figure S1.
Binding of purified serum IgGs and blood IgG+ memory B cells to S-HBsAg. Related to Fig. 1. (A) Representative ELISA graphs showing the reactivity of purified serum IgG antibodies from HBV vaccinees (HBVv) and controllers (HBVc) against rS-HBsAg and human-derived nS-HBsAg particles. Error bars indicate the SEM of duplicate values. (B) Flow-cytometric cytograms showing the gating strategy used to single-cell sort IgG+ memory B cells binding to fluorescently labeled rS-HBsAg and nS-HBsAg proteins used as baits. The S-HBsAg–reactive IgG+ memory B cell population is shown for all donors. SSC, side scatter; FSC, forward scatter.
Figure S2.
S-HBsAg reactivity of S-HBsAg-captured IgG+ memory B cell antibodies. Related to Fig. 1. (A) Heat map showing the ELISA reactivity against nS-HBsAg and rS-HBsAg (immobilized and captured) of S-HBsAg–binding memory antibodies cloned from HBV vaccinees and controllers. Means of triplicate OD values are shown. (B) Violin plots showing the cumulative ELISA OD (COD) values for the bindings shown in A. The proportion of HBs-specific antibodies cloned from S-HBsAg-captured IgG+ memory B cells is shown per donor (right).
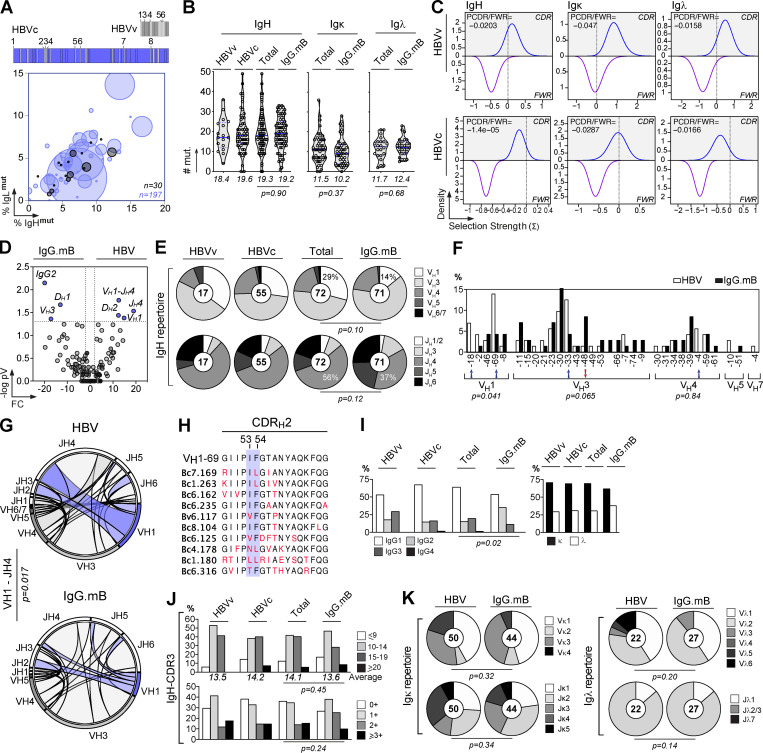
HBV-specific memory B cells were mainly part of clonal expansions and expressed somatically mutated Ig genes displaying antigen-driven maturation hallmarks (Fig. 2, A–C; and Table S2). Comparison of Ig gene features with IgG+ memory B cells from healthy controls revealed an increased usage in the HBV-specific B cell repertoire of VH1 (i.e., VH1-69 and VH1-18; P = 0.041), JH4 (P = 0.0017), and rearranged VH1(DH)JH4 (P = 0.017) genes, as well as IgG1/IgG3 subclass expression (P = 0.019; Fig. 2, D–G and I; and Table S2). Among the HBV antibodies using VH1-69 genes (14% of total; Fig. 2 F), 60% possessed the germline (GL)-encoded phenylalanine at position 54 in the hydrophobic complementary determining region (CDR) H2 tip (Fig. 2 H), which was shown to be essential for numerous human neutralizing antibodies against influenza virus, hepatitis C virus, and HIV-1 (Chen et al., 2019). No specific IgH CDRH3 loop and IgL chain gene features were significantly associated with S-HBsAg reactivity (Fig. 2, J and K). We conclude that the majority of human S-HBsAg–specific IgG+ memory B cells express conformation-dependent IgG1 and IgG3 antibodies enriched in VH1-coding Ig.
Figure 2.
Immunoglobulin gene repertoire of S-HBsAg-specific IgG+ memory B-cells. (A) Bubble plot showing the level of clonal expansions according to percentages of somatic mutations in the IgH and IgL chain variable domains of S-HBsAg–specific IgG antibodies. The size of the expansions for each donor is indicated in the bar graph below. (B) Violin plots comparing the number of mutations in VH, Vκ, and Vλ genes in HBs-specific and control IgG+ memory B cells (n = 72). The average number of mutations (# mut.) is indicated below each dot plot. Numbers of mutations were compared across groups of antibodies using the unpaired Student t test with Welch’s correction. (C) Graphs showing the Bayesian estimation of antigen-driven selection based on anti-S-HBs IgH and IgL sequences. FWR, framework regions. (D) Volcano plot analysis comparing the Ig gene repertoire of S-HBsAg–specific IgG+ B cells from HBV immune donors and IgG+ memory B cells from healthy individuals (IgG.mB; Prigent et al., 2016). Blue dots indicate statistically significant differences between both Ig gene repertoires. pV, P value; FC, fold changes. (E) Pie charts comparing the distribution of VH/JH gene usage of blood S-HBsAg–specific IgG+ memory B cells and IgG+ memory B cells from healthy individuals (IgG.mB). The number of antibody sequences analyzed is indicated in the center of each pie chart. (F) Bar graph comparing the distribution of single Ig VH genes expressed by S-HBsAg–specific and control IgG+ memory B cells. (G) Circos plots comparing the VH(DH)JH rearrangement frequencies between S-HBsAg–specific IgG+ memory B cells and IgG+ memory B cells from healthy individuals (IgG.mB). (H) Amino acid alignment of the CDRH2 region (defined by Kabat) of VH1-69–expressing anti-HBs antibodies. Residues in red indicate substitutions compared with the GL VH gene (on the top). (I) Bar graph comparing the distribution of IgG subtypes (left) and κ- vs. λ-Ig chain usage (right) as in F. (J) Same as in I but for the CDRH3 lengths and positive charge numbers. The average of CDRH3 lengths is indicated below each histogram. (K) Same as in E but for Vκ/Jκ and Vλ/Jλ gene usages. Groups were compared (in D–G and I–K) using 2 × 2 and 2 × 5 Fisher’s exact tests.
In vitro and in vivo HBV neutralization by human anti-HBs memory B cell antibodies
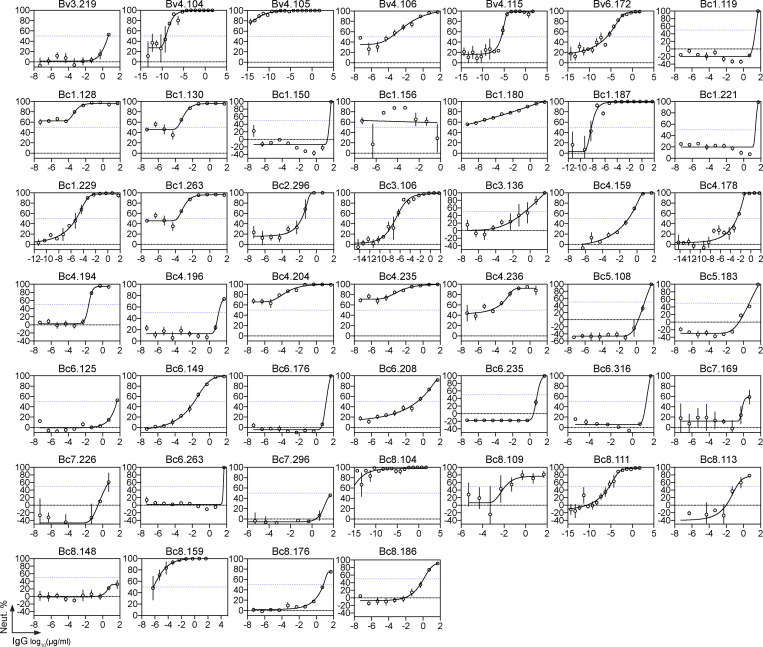
To determine whether the S-HBsAg memory B cell antibodies neutralize HBV, we measured their in vitro neutralizing activity against genotype D viruses in the HepaRG cell–based assay. Overall, 61% of the anti-HBs antibodies blocked HBV infection with 50% inhibitory concentrations (IC50) ranging from 50 µg/ml down to 0.05 pg/ml (Fig. 3 A, Fig. S3, and Table S2). In HBV controllers, 69% of the antibodies were neutralizing, including 35% of potent neutralizers with IC50 values below 50 ng/ml (Fig. 3 A and Table S2). Anti-HBs antibodies with broader reactivity to S-HBsAg particles and higher hypermutation loads were more prone to inhibit HBV infection (Fig. 3, B and C). HBV neutralizers equally expressed VH1-69 F54 and L54 alleles (Fig. 2 H and Table S2). Hepatitis delta virus (HDV) is a defective virus and an obligate satellite of HBV, which utilizes envelope proteins of its helper to assemble infectious HDV virions (Sureau and Negro, 2016). We thus asked whether HBV neutralizing antibodies could also prevent HDV infection in vitro. As expected, selected antibodies neutralized HDV in the Huh-106 cell assay as efficiently as HBV (Fig. 3 D).
Figure 3.
Neutralizing activity of human anti-HBs memory antibodies. (A) Dot plot showing the neutralizing activity of anti-HBs IgG antibodies (n = 72) against in vitro infection of HepaRG cells by genotype D HBV. Dots represent IC50 values for each antibody calculated from Fig. S3. Pie charts (bottom) show the distribution of nonactive (white) vs. neutralizing (shades of blue) antibodies according to neutralization potency. (B) Violin plots comparing the neutralization capacity of anti-HBs IgGs according to bound S-HBsAg proteins (nS-HBsAg, rS-HBsAg, and captured rS-HBsAg as measured in Fig. S2). (C) Violin plots comparing the percentage of somatic hypermutations (%SHM) between nonneutralizing and neutralizing antibody groups. Groups in B and C were compared using the Mann–Whitney test. (D) In vitro neutralizing activity of selected anti-HBs monoclonal IgG antibodies against HDV using HDV RNA quantification in Huh-106 cells by Northern blotting. ge, genome equivalents. Asterisk indicates ribosomal RNA. (E) In vivo neutralization activity of human anti-HBs antibodies in HBV-carrier mice. Circulating blood HBsAg levels were monitored in HBV-carrier mice treated once i.v. with 0.25 mg of anti-HBs antibodies Bv4.104 (n = 6), Bc1.187 (n = 6), Bc1.263 (n = 6), Bc4.204 (n = 6), or mGO53 isotype control (n = 5), and 0.5 mg of anti-HBs antibodies Bv4.104 (n = 9), Bc1.187 (n = 9), Bc1.263 (n = 6), Bc4.204 (n = 4), or mGO53 isotype control (n = 5). Blue and red lines represent averages. Graphs also show the evolution of human IgG titers of anti-HBs neutralizers over time in treated HBV-carrier mice. Dotted lines indicate the means and gray area the value ranges. (F) Log10 changes of HBsAg titers at nadir (2 dpi) upon antibody administration of 0.25 mg (white) and 0.5 mg (black) per mouse are shown. Means are indicated in red (0.5 mg) and blue (0.25 mg). (G) Circulating blood HBsAg and HBV DNA levels were monitored in HBV-carrier mice (n = 6) injected once i.v. with 1 mg of the anti-HBs antibody Bc1.187. (H) Average log10 changes over time of HBsAg (red) and HBV DNA (orange) levels.
Figure S3.
In vitro HBV neutralization by human anti-HBs antibodies. Related to Fig. 3. Graphs show neutralization curves of genotype D HBV viruses by selected human anti-HBs antibodies as measured in vitro using the HepaRG cell assay. The dotted horizontal line indicates 50% neutralization, from which the IC50 value can be derived from the antibody concentration on the x axis. Error bars indicate the SEM of triplicate measurements.
To evaluate the in vivo activity of neutralizing anti-HBs antibodies, we generated HBV-persistent mice based on the liver-targeted transduction of a recombinant HBV-encoding adeno-associated virus (AAV; Dion et al., 2013). Four potent HBV neutralizers were selected and passively transferred into HBV carrier mice with high levels of circulating HBsAg (>104 IU/ml). A single i.v. injection of antibodies at ∼20 mg/kg (0.5 mg per mouse) led to a marked drop in viremia 2 d post-injection (dpi), whereas treatments with a lower antibody dose (∼10 mg/kg, 0.25 mg per mouse) had milder effects due to the rapid exhaustion of antibodies by outnumbering HBV particles (Fig. 3 E). The two most potent antibodies, Bc1.187 and Bv4.104, induced an average decrease of 1.7 and 2.1 log10 at nadir (2 dpi), respectively (Fig. 3 F). A viral rebound 2 d after drop accompanied antibody decay with a return to baseline levels of circulating HBsAg at 7 dpi (Fig. 3 E). As the decline of viremia in vivo was dependent on the amount of antibody administrated, we next determined the effect of an increased dose of the HBV cross-neutralizing antibody Bc1.187. HBV carrier mice receiving a single injection of Bc1.187 at ∼40 mg/kg (1 mg i.v. per mouse) showed a maximum decrease of viremia of 2.8 log10 in average at 2 dpi (Fig. 3, G and H), with HBsAg levels sustained below baseline for at least 12 d (Fig. 3, G and H). In treated animals, HBV DNA loads followed the evolution of serum HBsAg titers and dropped an average 3.6 log10 to reach undetectable levels up to 1 wk after the last Bc1.187 injection in four out of six mice before viral rebound (Fig. 3, G and H).
Cross-genotypic activity of potent human HBV neutralizing antibodies
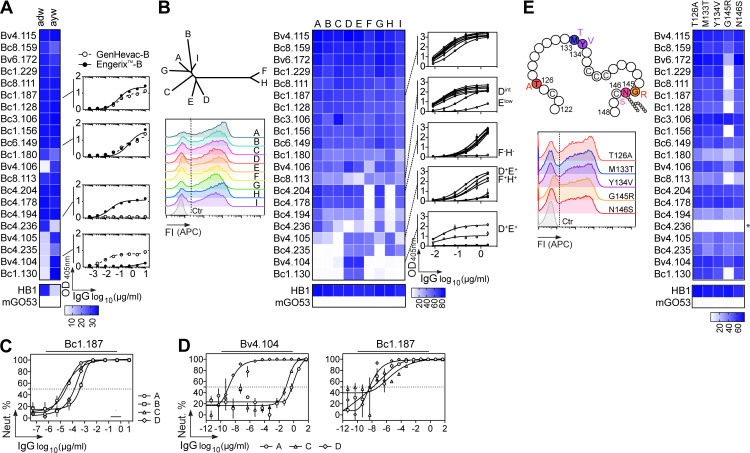
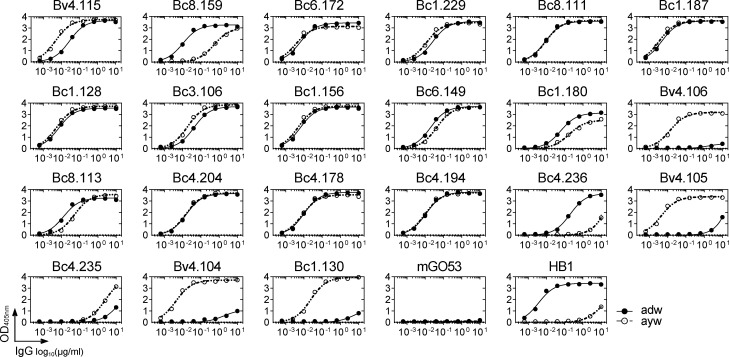
HBV is classified into four major serotypes based on specific amino acid variations in the “a” determinant (adr, adw, ayr, ayw), and 10 genotypes according to viral genome-based phylogeny (A to J; Kato et al., 2016). Binding analyses showed that most potent HBV neutralizing antibodies (71%) are able to recognize both ayw and adw HBsAg particles used in GenHevac-B and Engerix-B vaccines, respectively (Fig. 4 A and Fig. S4). Importantly, about half of the neutralizers cross-reacted equally with consensus S-HBsAg proteins from nine different genotypes (A to I; Fig. 4 B) and Fig. S5, A and B). The others displayed more heterogeneous binding profiles, with antibodies not reacting with F and H genotypes (19%, all from donor Bc4), or binding to D–F and H, or only to D and E (Fig. 4 B). To validate that cross-genotypic HBV binding reflects cross-neutralization potential, we measured the in vitro neutralizing activity of the HBV cross-reactive neutralizer Bc1.187 against infection of primary human hepatocytes (PHHs) with HBV virions from genotypes A, B, C, and D. As anticipated, Bc1.187 neutralized HBV viruses from all four genotypes, although more efficiently genotypes A and C than B and D (Fig. 4 C). In the HepaRG cell assay, genotype A and C virions were also highly sensitive to Bc1.187 as opposed to Bv4.104 that only neutralized potently genotype D HBV, in agreement with its reactivity profile (Fig. 4 D).
Figure 4.
Cross-reactivity of human HBV neutralizing antibodies. (A) Heat map comparing the ELISA reactivity of HBV neutralizing antibodies against adw and ayw genotype D S-HBsAg particles (measured as area under the curve values from Fig. S4). Representative ELISA graphs on the right show the reactivity of selected antibodies against recombinant HBV vaccines Engerix-B (adw) and GenHevac-B (ayw). Error bars indicate the SD of assay duplicates. (B) Heat map comparing the reactivity of HBV neutralizing antibodies against S-HBsAg antigens from genotypes depicted in the phylogenic tree (top left), as % of bound S-HBsAg-expressing cells determined by flow cytometry. Data represent one of two independent experiments as shown in Fig. S5 B. HB1 and mGO53 are positive and negative controls, respectively. Cytograms in the bottom left shows a representative reactivity profile of HB1 antibody. ELISA graphs on the right show the reactivity of selected antibodies against rS-HBsAg proteins from all genotypes but G. Error bars indicate the SD of assay duplicates. (C) Graph comparing the neutralizing (Neut.) activity of Bc1.187 against infection of PHHs by HBV viruses from A to D genotypes. Error bars indicate the SEM of assay triplicates. (D) Graphs show neutralization curves of HBV viruses from genotype A, C, and D by Bv4.104 and Bc1.187 antibodies as determined by the HepaRG neutralization assay. Error bars indicate the SEM of assay triplicates. (E) Same as in B but for S-HBsAg mutant proteins depicted on the diagram in the top left (Fig. S5 C). Asterisk indicates that this antibody was initially not reactive against genotype D S-HBsAg. FI, fluorescence intensity.
Figure S4.
Binding of HBV neutralizing antibodies to recombinant serotype-specific S-HBsAg. Related to Fig. 4. Representative ELISA graph showing the binding of selected HBV neutralizing antibodies to purified recombinant adw (straight lines) and ayw (dotted lines) S-HBsAg particles. HB1 and mGO53 are positive and negative control, respectively. Mean values ± SEM of assay duplicates from one or two independent experiments are shown.
Figure S5.
Cross-reactivity of HBV neutralizing antibodies against genotype-specific and mutant S-HBsAg proteins. Related to Fig. 4. (A) Amino acid alignment of the consensus S-HBsAg protein sequences from different HBV genotypes used in B. Residue variations are highlighted in blue. (B) Cytograms comparing the reactivity profiles of selected HBV neutralizing antibodies against genotype-specific S-HBsAg. Data represent one of two independent experiments. HB1 and mGO53 are positive and negative control, respectively. Ctr, nontransfected cell control (gray); FI, fluorescence intensity. (C) Cytograms comparing the reactivity of selected HBV neutralizing antibodies against genotype D S-HBsAg mutant proteins displaying naturally occurring escape mutations (T126A, M133T, Y134V, or G145R), or a mutation in the S-HBsAg N-glycosylation site (N146S). Data represent one of two independent experiments. HB1 and mGO53 are positive and negative control, respectively. Ctr, nontransfected cell control (gray).
In humans, several HBV escape mutations have been described, of which G145R is the most prevalent substitution in the S protein region (Chotiyaputta and Lok, 2009; Huang et al., 2012). We thus evaluated whether some of these mutations could affect HBV recognition by the potent neutralizers (Fig. 4 E and Fig. S5 C). Significant decrease or loss of S-HBsAg binding was only detected with the G145R mutant protein for about half of the antibodies, indicating that the G145R mutation known to destabilize the “a” determinant (Huang et al., 2012; Salisse and Sureau, 2009) can be detrimental to the neutralizing activity of anti-HBs antibodies. Mutant binding analyses also showed that none of the neutralizing antibodies interact with unique N-glycans on the S-HBsAg found at position N146 (Fig. 4 E and Fig. S5 C). Collectively, these data show that HBsAg-specific memory B cells in HBV controllers can produce potent cross-neutralizing antibodies able to dramatically decrease levels of circulating HBsAg and HBV DNA in vivo.
Binding features of potent human HBV neutralizing antibodies
To map the epitopes targeted by potent neutralizing HBV antibodies, we performed alanine-scanning ELISA experiments using a library of mutant HBsAg proteins with substitutions covering the major hydrophilic region of HBsAg. Although all tested antibodies presented a unique recognition pattern, common binding profiles could be identified based on mutation sensitivity (Fig. 5 A). Antigenic determinants were scattered throughout the antigenic loop (aa 101–170). Some epitopes could be mapped to a restricted domain in the N-terminal and C-terminal regions of S-HBsAg (Bc8.109 and Bc1.263, respectively). Others could not be unambiguously determined (Bc4.204, Bc4.194, Bc4.178, Bc3.106, and Bc8.104; Fig. 5 A). Neutralizing epitopes in the “a” determinant were quite complex, comprising major interacting residues in the (i) conserved peptide 120-PCR(K)TC-124 and amino acid sequence 138–149 (Bc8.159 and Bc1.130), (ii) two adjacent regions aa 124–138 and aa 138–147 (Bv4.105 and Bv4.106), (iii) all three aforementioned domains (Bv4.104, Bc8.111, Bc1.128, and Bc1.156), or (iv) peptide region 138–147 mainly (Bv4.115, Bv6.172, Bc1.187, Bc1.229, and Bc1.180; Fig. 5 A). Next, we performed competition S-HBsAg–binding ELISA analyses to evaluate the extent of overlap between neutralizing epitopes. In each aforementioned antibody class, cross-competition molecules were mostly found as independent tandems: Bv4.104/Bc8.111, Bc8.128/Bv4.106, Bc1.180/Bc1.263, and Bc3.106/Bc6.149 (Fig. 5 B). The trio of neutralizers Bc4.194/Bc4.204/Bc4.178 recognized a common nonconformational epitope not well defined by the mapping (Fig. 1 E and Fig. 5, A and B), but likely located in the most distal portion of the S-HBsAg N or C terminus. In this regard, all three antibodies were unable to bind F and H genotypes, which principally diverge from the others in a region predicted to be the TM domain 3 (Q178-Y225; Fig. S5 A). Moreover, Bc4.204 had its binding decreased by the F179A mutation and also weakly bound to a linear peptide in C-ter (191–200; Fig. 1 G and Fig. 5 A). The larger epitopic cluster contained neutralizing antibodies having epitopes more centered on the aa 138–149 region (Bv4.115, Bv6.172, Bc1.187, and Bc1.229), but also able to interfere with the S-HBsAg binding of others neutralizers (Fig. 5, A and B).
Figure 5.
Binding features of human HBV neutralizing antibodies. (A) Heat map shows the ELISA binding of selected HBV neutralizing antibodies to recombinant HBsAg mutant proteins. Color value is proportional to the reactivity level. (B) Heat map showing competition for S-HBsAg binding of the HBV neutralizing antibodies tested in duplicate. Lighter colors indicate stronger inhibition; dark blue indicates no competition. (C) S-HBsAg binding of the selected HBV neutralizing antibodies and their GL counterparts as measured in duplicate by flow cytometry. Representative cytograms are shown on the top. (D) Same as in C but as determined by ELISA. Means ± SD of triplicate assay values are shown. (E) In vitro neutralizing activity of the GL versions of Bc1.187, Bc4.204, and Bv4.104 against genotype D HBV viruses as determined by the HepaRG neutralization assay. Error bars indicate the SD of assay triplicates. (F) Reactivity profile of selected anti-HBs human antibodies on human protein microarrays. Frequency histograms showing the log10 protein displacement (σ) of the mean fluorescence intensity signals for anti-HBs antibodies compared with nonreactive antibody mGO53 (top). The PI corresponds to the Gaussian mean of all array protein displacements. Microarray reactivity plots showing anti-HBs antibody binding to human proteins (bottom). Each spot correspond to the z-scores given on a single protein by the reference (Ref: mGO53, y axis) and test antibody (x axis). Red dots indicate immunoreactive proteins (z > 5). (G) Binding of selected anti-HBs antibodies to HEp2-expressing self-antigens was assayed by indirect immunofluorescence assay. Ctr+, positive control of the kit. mGO53 and ED38 are negative and positive control antibodies, respectively. The scale bars represent 40 µM. (H) Bar graph shows the HEp-2 reactivity as measured by ELISA. Means ± SD of values from two independent experiments performed in duplicate are shown.
To examine whether somatic mutations contribute to the activity of HBV neutralizing antibodies, we reverted mutations in three representative potent neutralizers to produce their putative GL precursors. Representative GL versions showed heterogeneous HBV recognition profiles: Bc4.204-GL failed reacting with S-HBsAg, whereas Bc1.187-GL and Bv4.104-GL still bound but with much lower relative affinities compared with their matured counterparts (Fig. 5, C and D). GL IgGs displayed weak, as compared with the mutated antibodies, but significant inhibitory activities against in vitro HepaRG cell infection by genotype D HBV, particularly Bc1.187-GL with an IC50 of 0.08 µg/ml (Fig. 5 E). This implies that although certain GL antibodies expressed by B cell precursors can bind and neutralize HBV at a high concentration, somatic mutations are required for their high-affinity HBsAg binding and potent HBV neutralization.
Human class-switched memory B cells including high-affinity B cell clones against viral antigens can cross-react with self-antigens (Andrews et al., 2015; Prigent et al., 2016, 2018; Tiller et al., 2007). We thus evaluated the self-reactivity of 10 selected potent HBV neutralizers using clinical autoantibody assays (HEp-2 cells IFA and ELISA), and microarray immunoblotting (>9,000 human proteins). Apart from Bc1.263, none of the HBV neutralizers showed polyreactivity as measured by a global shift of the microarray fluorescence signals compared with the isotype control (Fig. 5 F). Only Bc1.263 and Bc4.204 antibodies displayed a significant cross-reactivity (Z-scores > 5) against galectin-3/-8 and E3 ubiquitin-protein ligase UBR2, respectively (Fig. 5 F). Low (Bc1.187 and Bc1.263) to moderate (Bc4.194 and Bc4.204) binding to HEp-2 cell antigens was detected by IFA using high antibody concentrations (Fig. 5 G), but no HBV neutralizers showed positive HEp-2 ELISA reactivity (Fig. 5 H).
In vivo HBV suppression by potent broadly neutralizing antibody Bc1.187
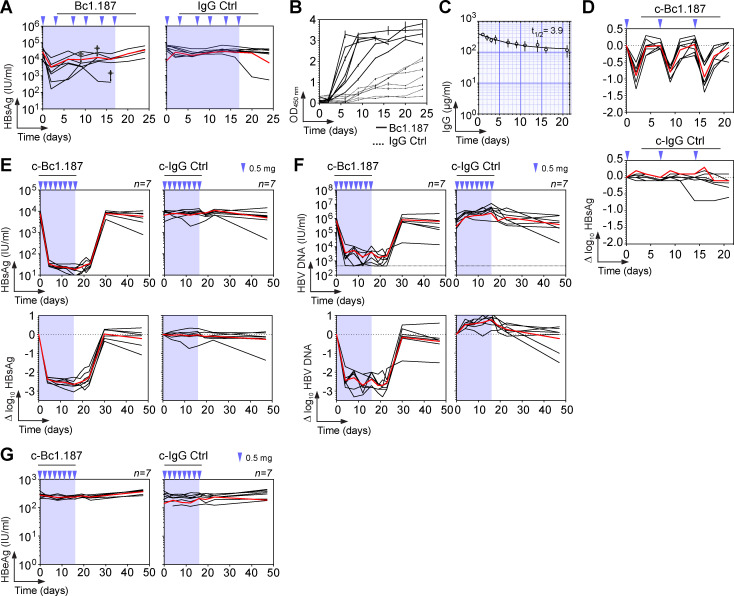
To determine whether potent HBV neutralizers from natural controllers can stably suppress HBV viremia in vivo, HBV-carrier mice were treated for 17 d with Bc1.187 antibody (0.5 mg i.v., ∼20 mg/kg, twice a week; Fig. 6 A). Viremic mice experienced a decrease in circulating HBsAg levels upon treatment with Bc1.187 but not with the isotype control (Fig. 6 A). However, the development of murine anti-human IgG antibodies (anti-drug antibodies [ADAs]) rapidly altered therapy effectiveness (Fig. 6 B). To overcome or limit ADA production, we generated a chimeric version of Bc1.187 by combining the antibody’s variable domains with the murine Igγ2a and IgK constant regions. The chimeric Bc1.187 antibody (c-Bc1.187) had a serum half-life of 3.9 d in nontransduced wild-type mice (Fig. 6 C), and led to reproducible viremia drops when administrated weekly in HBV-carrier mice (0.88 ± 0.07 log10 in average at 2 dpi during 3 consecutive weeks; Fig. 6 D). 16 d of treatment with 0.5 mg i.v. injections of c-Bc1.187 every 2 d but not control antibody led to a loss of circulating HBsAg in all mice from day 4 with an average 2.5 log10 fold decrease compared with the set-point (Fig. 6 E). During c-Bc1.187 therapy, serum HBV DNA content was also drastically diminished by an average 2.5 log10 fold and reached the detection limit by day 21 in all but one mouse (Fig. 6 F). HBsAg and HBV viremia were still suppressed for 2 wk after the last antibody injection before rising back to baseline levels (Fig. 6, E and F). As expected in this model, serum levels of HBe antigen, a surrogate marker for viral replication, remained unchanged during therapy (Fig. 6 G).
Figure 6.
Bc1.187 antibody therapy in HBV-carrier mice. (A) Circulating blood HBsAg levels over time in AAV-HBV–transduced mice (n = 7) treated every 3–4 d for 17 d with 0.5 mg i.v. of human anti-HBs antibody Bc1.187 or mGO53 isotype control (blue arrows). Blue-shaded area indicates the antibody therapy period. (B) Murine anti-human IgG antibody levels in the treated mice shown in A as measured in duplicate by ELISA. (C) IgG concentrations of passively administrated chimeric Bc1.187 antibody (0.5 mg i.v.) in B6 mice (n = 4). Error bars represent SD values. Half-life in days (t1/2) of chimeric antibody Bc1.187 is indicated in the upper-right corner. (D) Δlog10 HBsAg levels over time in C57BL/6J mice receiving weekly i.v. injections (0.5 mg) of chimeric antibody Bc1.187 (c-Bc1.187) and mGO53 isotype control (c-mGO53). Red lines indicate the averages. (E) Evolution of blood HBsAg levels over time in HBV-carrier mice (n = 7 per group) treated every 2 d for 16 d with 0.5 mg i.v. of anti-HBs Bc1.187 or isotype control mGO53 chimeric antibody (blue arrows). Red lines indicate the averages. Blue-shaded area indicates the antibody therapy period. Corresponding Δlog10 HBsAg levels compared with baseline are shown at the bottom. (F) Same as in E but for blood HBV DNA levels. (G) Same as in E but for blood HBeAg levels.
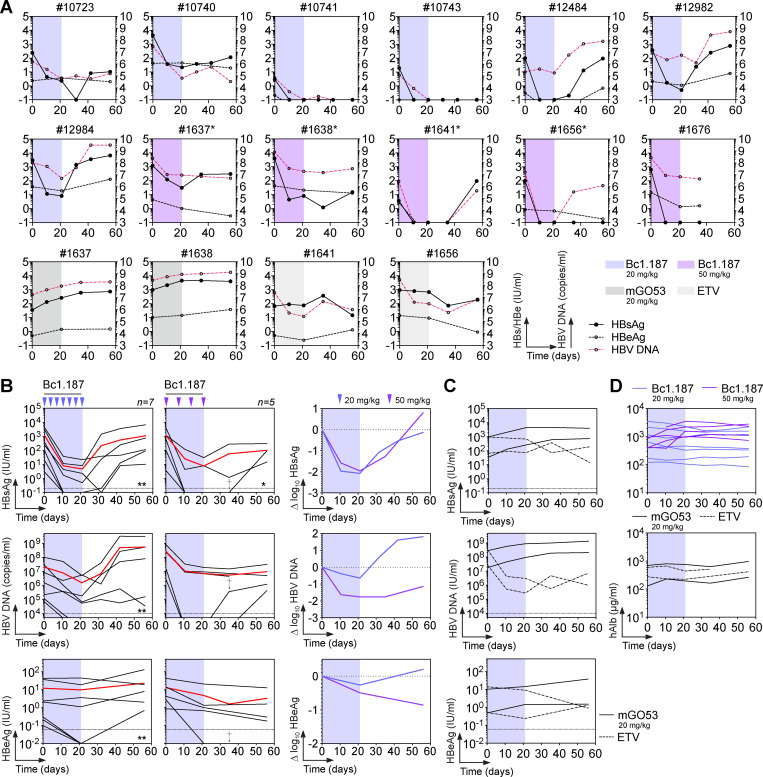
We next determined whether Bc1.187 could alter the natural course of HBV infection in vivo. BALB/c Rag2−/−SirpaNODAlb-uPAtg/tg mice stably engrafted with human hepatocytes (HUHEP; Dusseaux et al., 2017; Strick-Marchand et al., 2015) were infected with genotype D HBV. Once infection was established, mice received i.p. injections of Bc1.187 for 3 wk, either bi-weekly (20 mg/kg per mouse) or weekly (50 mg/kg per mouse; Fig. 7 A). Treatment with Bc1.187, but not isotype antibody and nucleoside analogue (Entecavir) controls, induced a decrease of circulating HBsAg levels in viremic HUHEP mice by an average 2.1 and 2 log10 fold at 21 dpi for the 20 mg/kg and 50 mg/kg dosing regimens, respectively (Fig. 7, A–C). However, HBV viremia declined more effectively in the group receiving weekly Bc1.187 injections of 50 mg/kg (average drop at 21 dpi of 1.76 log10 vs. 0.64 log10 for 20 mg/kg; Fig. 7 B). At the end of the follow-up, two mice with low pretreatment viremia showed complete viral suppression in response to the 20 mg/kg dosing regimen (28.6%, n = 7; Fig. 7, A and B). Circulating HBsAg levels dropped and remained undetectable for 2 wk after the last injection of 50 mg/kg Bc1.187 in 60% of the mice (n = 5). Of these three mice, one died, another experienced viral rebound, and the last still controlled the infection for more than a month after treatment before HBV DNA reappearance (Fig. 7, A and B). Circulating levels of HBeAg were also diminished upon Bc1.187 antibody treatment but to a lower extent compared with HBsAg (average fold changes at 21 dpi of 0.26 log10 for 20 mg/kg and 0.49 log10 for 50 mg/kg; Fig. 7 B). Serum titers of human albumin remained unchanged during the monitoring period (Fig. 7 D), indicating that engraftments were stable and not affected by treatments.
Figure 7.
Bc1.187 antibody therapy in HBV-infected HUHEP mice. (A) Blood HBsAg, HBeAg, and HBV DNA levels over time for each individual HUHEP mouse infected with genotype D HBV, and treated with anti-HBs antibody Bc1.187 for 3 wk with either 20 mg/kg or 50 mg/kg human antibody i.p. Asterisk indicates mice in control groups that were reused for a treatment regimen of Bc1.187 antibody at 50 mg/kg. (B) Evolution of infection over time in HBV-infected HUHEP mice receiving i.p. injections of human anti-HBs antibody Bc1.187 (20 mg/kg ∼0.4 mg every 3–4 d, n = 7; 50 mg/kg ∼1 mg, weekly, n = 5) for 3 wk. Red lines indicate the averages. Dagger (†) and asterisk indicate dead and controlled mice, respectively. The number of asterisks indicates the number of controlled mice. Circulating blood HBsAg, HBeAg, and HBV DNA levels (left) and average Δlog10 values compared with baseline (right, 20 mg/kg, blue lines; 50 mg/kg, purple lines) are shown. (C) Evolution of HBV infection over time in HBV-infected HUHEP mice receiving a treatment with the non-HBV isotype control mGO53 (20 mg/kg i.p.) or Entecavir (ETV) every 3–4 d for 3 wk. Blood levels of HBsAg (black lines), HBeAg (magenta dotted lines), and HBV DNA (black dotted lines) are shown. (D) Graphs showing the levels of human serum albumin over time in infected HUHEP mice and treated i.p. with 20 mg/kg and 50 mg/kg of Bc1.187 antibody as shown in B.
Discussion
In chronic HBV infection, most HBsAg-specific B cells are atypical memory lymphocytes presenting several B cell dysfunctions such as an altered capacity to differentiate into antibody-producing cells and thus, to potentially produce HBV neutralizing antibodies (Burton et al., 2018; Salimzadeh et al., 2018). In contrast, circulating B cells in individuals who resolved the infection, both during the acute and chronic phase, secrete HBsAg antibodies likely participating in the seroconversion (Salimzadeh et al., 2018; Xu et al., 2015). To characterize memory B cells directed against the HBsAgs in immune donors, we investigated the molecular and functional properties of recombinant human antibodies cloned from single S-HBsAg–specific IgG+ B cells circulating in the blood of HBV controllers and vaccinees. Our study reveals that despite a moderate enrichment in VH1-JH4–encoding clones, circulating S-HBsAg–specific memory B cells from HBV controllers, but also from vaccinees, express a diverse gene repertoire of Ig recognizing mostly conformational epitopes on the HBV surface glycoproteins. The majority of anti-HBs IgGs cloned from controllers could neutralize HBV in vitro, with some being active at very low concentrations, including against HDV infection. Potent neutralizers evaluated in vivo also showed antiviral effects by decreasing both serum HBsAg level and HBV viremia, suggesting that in infected humans, these antibodies can be functionally important. Most HBV neutralizing antibodies were broadly reactive against various viral genotypes. The majority recognized epitopes of the “a” determinant including a main region, spanning the cysteine 138 to 147, recognized by the potent cross-neutralizing antibody Bc1.187, albeit others were also mapped outside the antigenic loop. A single passive infusion of Bc1.187 was particularly effective in vivo in a mouse model of chronic HBV infection to completely suppress for a few days serum HBsAg and HBV viremia. Interestingly, the GL version of Bc1.187 could still bind and neutralize HBV with an IC50 <0.1 µg/ml. This suggests that B cell precursors expressing such Ig could rapidly affinity mature and thus be readily active in neutralizing HBV. Although neutralizing antibodies to the preS1 domain not investigated here might develop in HBV controllers, our data argue in favor of an essential role of anti-HBs antibodies in clearing the infection.
Current therapies for treating chronic HBV infection include potent direct antivirals and polyethylene glycol (PEG)–IFN-α. However, antiviral treatments have no significant impact on the serum HBsAg levels due principally to the production of subviral particles from HBV covalently closed circular DNA minichromosomes or HBV DNA integrated in the host cell genome. To bypass the immune tolerance observed in chronically infected individuals and induce effective anti-HBV antibody responses is therefore still a major challenge. One promising strategy is the development of immunotherapies based on the use of potent HBV neutralizing antibodies (Corti et al., 2018; Gao et al., 2017; Tu and Urban, 2018). Preclinical models in mice and monkeys, as well as several phase I clinical trials in chronically infected humans, have shown the in vivo efficacy of neutralizing HBV monoclonal antibodies, which when administrated in infected recipients altered the course of the infection by suppressing HBsAg and reducing HBV DNA content (Eren et al., 2000; Galun et al., 2002; Lee et al., 2019; Lever et al., 1990; Li et al., 2017; van Nunen et al., 2001; Zhang et al., 2016; Zhu et al., 2016). In this study, we showed using two different mouse models that treating viremic animals with a potent human broadly neutralizing antibody had profound effects on HBV infection. First, Bc1.187 therapy induced a rapid loss and/or substantial decrease in circulating HBsAg and HBV DNA that persisted in most mice for days to weeks after treatment. Furthermore, in vivo experiments demonstrated that weakly viremic mice could fully clear HBV infection upon Bc1.187 antibody treatment. Antibodies are versatile immune effectors. Besides neutralization, they can exert a variety of immunological effector functions by engaging innate immune cells to eliminate infected cells such as antibody-dependent cellular cytotoxicity, antibody-dependent cellular phagocytosis, and complement-dependent cytotoxicity. Complement-dependent cytotoxicity and antibody-dependent cellular phagocytosis were described in murine anti-HBs antibody-mediated hepatocyte killing using in vitro and in vivo systems (Shouval et al., 1982a, b; Tharinger et al., 2017; Zhang et al., 2016). Moreover, a human neutralizing HBV antibody against the preS1 region has been shown to exert therapeutic activity with sustained virological suppression in part by eliciting Fc-dependent effector functions including antibody-dependent cellular cytotoxicity (Li et al., 2017). Similarly, neutralizing anti-HBs antibodies produced by HBV controllers as those we describe very likely opsonize infected hepatocytes, and may therefore promote their elimination by effector cells such as Kupffer cells. Finally, antibody therapy by engaging the host immune system can lead to stimulation of immune responses, a property better known as vaccine-like effects (Pelegrin et al., 2015), which could be essential in breaking HBV-induced immune tolerance. Hence, we propose that in addition to blocking de novo infection and possibly eliminating infected hepatocytes via Fc-dependent mechanisms, some of the potent human broadly neutralizing HBV antibodies identified here could be used in chronically infected patients to greatly reduce serum HBsAg level. Such antibodies could indeed act as a powerful “antigenic sink,” which, when combined with therapies aiming at restoring the host’s innate and adaptive immune responses, i.e., INF-α, therapeutic vaccines, TLR agonists, and checkpoint inhibitors (Fanning et al., 2019; Gehring and Protzer, 2019; Maini and Burton, 2019) could facilitate viral clearance and eventually lead to long-term control of infection and a functional HBV cure. Besides their therapeutic potential, human HBV neutralizing antibodies could also serve as efficient alternatives for polyclonal hepatitis B Ig used in pre- and post-exposure prophylactic settings such as for preventing mother-to-child transmission (Corti et al., 2018).
Materials and methods
Human samples
Blood samples from HBV vaccinees and chronically HBV-infected individuals who experienced a spontaneous HBsAg/anti-HBs seroconversion (controllers) were obtained from the healthy donors cohort of the Investigation Clinique et Accès aux Ressources Biologiques biobanking platform (Institut Pasteur) under the CoSimmGen protocol approved by the French Agence Nationale de Sécurité du Médicament on May 24, 2012, and the Comité de Protection des Personnes on January 17, 2014. The main clinical–biological characteristics of the donors are summarized in Table S1. All samples from HBV vaccinees and seroconverters were obtained as part of the clinical research protocol Réponses Anticorps Protectrices contre l’Infection par le Virus de l’Hépatite B (RAPIVIB) performed in accordance with and after ethical approval from all the French legislation and regulation authorities. The RAPIVIB protocol received approval from the Comité de Recherche Clinique of the Institut Pasteur on July 30, 2015 (2014-058), the Agence Nationale de Sécurité du Médicament on April 29, 2015 (150457B-41), and the Comité de Protection des Personnes Ile-de-France III on June 10, 2015 (2015-100220-49/3267). The protocol was subjected to the MR-001 reference methodology of the Commission Nationale de l’Informatique et des Libertés. All donors gave written consent to participate in this study, and data were collected under pseudo-anonymized conditions using subject coding. Peripheral blood mononuclear cells (PBMCs) were isolated from donors’ blood using Ficoll Plaque Plus (GE Healthcare), and plasma or serum IgG antibodies were purified by batch/gravity-flow affinity chromatography using protein G sepharose 4 fast flow beads (GE Healthcare).
Antigens and antibody controls
nS-HBsAg purified from infected subjects and rS-HBsAg particles (ayw subtype) were prepared and biotinylated by Roche. Recombinant HBsAg particles (ayw) were produced in Chinese hamster ovary cells (Aventis Pasteur; Michel et al., 1984). Adw-subtype HBsAg particles (HBV genotype A) were produced in the yeast Pichia pastoris expression system, and purified (<95% purity) at the Center for Genetic Engineering and Biotechnology production facilities (Havana, Cuba; a kind gift from J. Aguilar). Consensus genotype-specific S-HBsAg fragments were constructed by multiple amino acid alignment of individual HBV genotypic sequences (A, n = 205; B, n = 495; C, n = 1,322; D, n = 382; E, n = 314; F, n = 271; G, n = 6; H, n = 40; I, n = 7) using CLC Main Workbench 7 software (v7.5.3, QIAGEN Aarhus A/S; see Fig. S5 A). Codon-optimized nucleotide fragments encoding the consensus S-HBsAg, a G3S linker, 10xHis-, and Avi-tags were cloned into a pcDNA 3.1/Zeo(+) expression vector (Thermo Fisher Scientific) using Anza 5 and 11 restriction enzymes (Thermo Fisher Scientific). For mutant proteins, single point mutations (T126A, M133T, Y134V, G145R, and N146S) were introduced into the construct using the QuickChange Site-Directed Mutagenesis kit (Agilent Technologies) following the manufacturer’s instructions. For the production of the TM domains-deleted S-HBsAg protein (ΔTM-S-HBsAg), the same construct but with the S-HBsAg DNA fragment lacking TM1, TM3, and TM4 domains, and having the TM2 replaced by a (G3S)5 linker, was generated. Disulfide-bridged S-HBs/D 122–137 (RTCTTTAQGTSMYPSC) and 139–148 (CTKPSDGNCT) loop peptides, and 3 amino acids–overlapping 12-mer peptides (n = 24) encompassing the entire ConsD S-HBsAg region but one TM domain 2 peptide with high hydrophobicity (IFLFILLLCLIF), were synthetized and desalted (GenScript HK Limited). Non-HBV antibody mGO53 (Wardemann et al., 2003) was used as isotype control. As positive control, neutralizing murine HB1 antibody specific to the 119-GPCRTCT-125 linear epitope of the “a” determinant of S-HBsAg (Kucinskaite-Kodze et al., 2016), was produced as a chimeric recombinant IgG1 with a human Fc using the expression-cloning system described below. Murine anti-preS2 antibody 1F6 (Küttner et al., 1999) was produced as chimeric human Fab fragments to be used for detection of bound subviral particles in the alanine scan mapping described below.
Single B cell FACS and expression-cloning of antibodies
Peripheral blood human B cells were isolated from donors’ PBMCs by CD19 MACS (Miltenyi Biotec), and stained with LIVE⁄DEAD fixable dead cell stain kit (Thermo Fisher Scientific). Purified B cells were then incubated for 30 min at 4°C with 1 µg of biotinylated rS-HBsAg or nS-HBsAg antigens, washed with 1% FBS-PBS (FACS buffer), and incubated for 30 min at 4°C with a cocktail of mouse anti-human antibodies (CD19 A700 [HIB19], IgG BV786 [G18-145], CD27 PE-CF594 [M-T271; BD Biosciences], IgA FITC [IS11-8E10; Miltenyi Biotec]), and streptavidin R-PE conjugate (Thermo Fisher Scientific). Stained cells were washed and resuspended in 1 mM EDTA FACS buffer. Single S-HBs+CD19+IgG+ B cells were sorted into 96-well PCR plates using a FACS Aria III sorter (Becton Dickinson) as previously described (Tiller et al., 2008). Single-cell cDNA synthesis using SuperScript IV reverse transcription (Thermo Fisher Scientific) followed by nested-PCR amplifications of IgH, Igκ, and Igλ genes, and sequences analyses for Ig gene features were performed as previously described (Tiller et al., 2008). For the reversion to GL of the selected antibodies, sequences were constructed by replacing the mutated VH-(DH)-JH and VL-JL gene segments with their GL counterparts as previously described (Mouquet et al., 2012). Purified digested PCR products were cloned into human Igγ1-, Igκ-, or Igλ-expressing vectors as previously described (Tiller et al., 2008). Murine Igγ2- and Igκ-expression vectors were generated from the original IgG1-expression vectors (Tiller et al., 2008) by substituting the DNA sequences coding for the human Igγ1/Igκ constant regions by the ones of the mouse Igγ2- and Igκ (synthetic DNA fragment, GeneArt, Thermo Fisher Scientific), and then used for the cloning of chimeric mGO53 and Bc1.187 antibodies. Recombinant antibodies were produced by transient cotransfection of Freestyle 293-F suspension cells (Thermo Fisher Scientific) using the polyethylenimine (PEI)-precipitation method as previously described (Lorin and Mouquet, 2015). Recombinant human IgG antibodies were purified by protein G affinity chromatography (Protein G Sepharose 4 Fast Flow, GE Healthcare). Purified antibodies were dialyzed against PBS. Preparations for in vivo infusions were micro-filtered (Ultrafree-CL devices – 0.1 µm polyvinylidene difluoride membrane, Merck-Millipore), and checked for endotoxins levels using the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (GenScript).
ELISAs
ELISAs were performed as previously described (Planchais et al., 2019). Briefly, high-binding 96-well ELISA plates (Costar, Corning) were coated overnight with purified rS-HBsAg and nS-HBsAg (125 ng/well in PBS). After washings with 0.05% Tween 20–PBS (PBST), plates were blocked 2 h with 2% BSA, 1 mM EDTA-PBST (Blocking solution), washed, and incubated with serially diluted purified serum IgG and recombinant monoclonal antibodies in PBS. For sandwich ELISAs, plates were coated overnight with purified IgG antibodies (250 ng/well in PBS) and treated as aforementioned before incubation with biotylinated-rS-HBsAg (100 ng/well in PBS). After washings, plates were revealed by addition of goat HRP-conjugated anti-human IgG or HRP-conjugated streptavidin (0.8 µg/ml final in blocking solution, Immunology Jackson ImmunoReseach) and HRP chromogenic substrate (ABTS solution, Euromedex). For competition ELISAs, purified antibodies were biotinylated using the EZ-Link Sulfo-NHS-Biotin kit (Thermo Fisher Scientific). rS-HBsAg–coated plates were blocked, washed, incubated for 2 h with biotinylated antibodies (at a concentration 0.33 nM) in 1:2 serially diluted solutions of antibody competitors in PBS (IgG concentration range from 0.83 to 106.7 nM), and revealed using HRP-conjugated streptavidin. Experiments were performed using a HydroSpeed microplate washer and Sunrise microplate absorbance reader (Tecan Männedorf), with OD405nm. Binding of anti-HBs antibodies to cyclic and overlapping linear peptides was tested using the same procedure as previously described (Mouquet et al., 2006). All antibodies were tested in duplicate or triplicate in at least two independent experiments, which included mGO53 negative and appropriate positive controls.
HBV neutralization assay
HepaRG and HepG2.2.15 cell lines were obtained, respectively, from Biopredic International and Dr. Michael Nassal (University Hospital Freiburg, Freiburg, Germany). HepaRG cells were cultured in Williams E medium (Gibco, Thermo Fisher Scientific) supplemented with 10% HepaRG growth supplement (Biopredic), and differentiated using 1.8% DMSO for at least 2 wk before infection. PHHs isolated from chimeric uPA/SCID mice with humanized livers by a collagenase perfusion method (Tateno et al., 2015) were obtained from PhoenixBio. PHHs were plated on type I collagen-coated 96-well plates at a concentration of 7 × 104 cells per well in culture media provided by Phoenix Bio. HBV genotype D viruses were produced in HepG2.2.15 cell culture supernatant and concentrated using PEG precipitation (Hantz et al., 2009). HBV viruses from genotypes A to C were purified from HBV-containing serum (American Red Cross) by gradient ultracentrifugation. Briefly, 1.5 ml of serum was applied on an OptiPrep gradient (10–50%, Sigma-Aldrich) and centrifuged at 32,000 rpm for 3 h at 4°C. Fractions (2 ml) were collected and analyzed for HBsAg antigen expression using the HBsAg CLIA Kit (Autobio) and quantitative PCR quantification. Whole genome sequences of all purified virus isolates were obtained by ultra-deep sequencing (DDL Diagnostic Laboratory), and HBV genotypes were determined using the Hepatitis B Virus database HBVdb (http://hbvdb.ibcp.fr; Hayer et al., 2013). The neutralizing activity of HBV antibodies was evaluated by incubating HBV virions (multiplicity of infection, 20–30) with serially diluted antibodies in HepaRG or PHH complete media for 1 h at room temperature. HBV-antibody mixtures were then added to the cells in 96-well plates with a final concentration of 4% PEG 8000 (Sigma-Aldrich). Infected cells were incubated for 20 h at 37°C, and then washed four times with PBS to remove the HBV inoculum and refilled with complete media. 6 dpi, in-supernatant HBsAg antigen content was quantified using the HBsAg CLIA Kit (Autobio) according to the manufacturer’s instructions.
HDV neutralization assay
In vitro neutralization assays were conducted using the HDV model as previously described (Sureau, 2010), except that sodium taurocholate cotransporting polypeptide–expressing Huh-106 cells substituted for HepaRG cells (Verrier et al., 2016). At day 1 after seeding, Huh-106 cells (105 cells/16.5-mm-diameter well) were exposed to 5 × 107 genome equivalents of HDV virions for 16 h, in the presence of 4% PEG 8000. To assay neutralization, the inoculums were mixed with 1:2 dilutions of 500 ng/ml of monoclonal antibodies, and the mixture was incubated for 1 h at 37°C before inoculation. Cells were harvested at 9 d after inoculation for measurement of intracellular HDV RNA that served as a marker of infection. HDV RNA signals were detected by Northern blot analysis using a 32P-labeled RNA probe and quantified using a phosphorimager instrument (BAS-1800 II; Fuji).
Flow cytometry binding assay
Freestyle 293-F were transfected with S-HBs-encoding vectors (0.65 µg plasmid DNA per 106 cells) using the PEI-precipitation method as previously described (Lorin and Mouquet, 2015). 48 h after transfection, transfected and nontransfected control cells were fixed and permeabilized using Cytofix/Cytoperm solution kit (BD Biosciences), and 0.5 × 106 cells were incubated with IgG antibodies for 45 min at 4°C (10 μg/ml, except if specified otherwise, in Perm/Wash solution; BD Biosciences). After washings, cells were incubated 20 min at 4°C with AF647-conjugated goat anti-human IgG antibodies (1:1,000 dilution; Thermo Fisher Scientific), washed, and resuspended in PBS. Data were acquired using a CytoFLEX flow cytometer (Beckman Coulter) and analyzed using FlowJo software (v10.3; FlowJo LLC).
Alanine scan mapping
HBV subviral particles bearing wild-type or alanine mutant L-, M-, and S-HBsAg proteins were produced by transfection of Huh-7 cells with pT7HB2.7 plasmid or mutant derivatives using FuGENE 6 reagent (Roche) as previously described (Salisse and Sureau, 2009). High-binding 96-well ELISA plates (Costar, Corning) were coated overnight with purified human anti-HBs antibodies (0.5 µg/well in PBS). After PBST washings and a 2 h-blocking step, plates were incubated 2 h with culture supernatants containing wild-type and mutant HBV envelope proteins. After washings, plates were incubated for 1 h with purified His-tagged anti-preS2 1F6 Fab fragments (125 ng/well). Wells were washed before addition of rabbit HRP-conjugated anti-6xHis-tag antibodies (1:4,000-diluted in blocking solution, ab1187, Abcam) and HRP chromogenic substrate (ABTS solution, Euromedex) as described above. Percentage of binding was calculated following the formula ([OD]mutant / [OD]wild-type) × 100.
Protein microarrays
All experiments were performed at 4°C using ProtoArray Human Protein Microarrays (Thermo Fisher Scientific). Microarrays were blocked for 1 h in blocking solution (Thermo Fisher Scientific), washed, and incubated for 1 h 30 min with IgG antibodies at 2.5 µg/ml as previously described (Planchais et al., 2019). After washings, arrays were incubated for 1 h 30 min with AF647-conjugated goat anti-human IgG antibodies (at 1 µg/ml in PBS; Thermo Fisher Scientific), and revealed using GenePix 4000B microarray scanner (Molecular Devices) and GenePix Pro 6.0 software (Molecular Devices) as previously described (Planchais et al., 2019). Fluorescence intensities were quantified using Spotxel software (SICASYS Software GmbH), and mean fluorescence intensity signals for each antibody (from duplicate protein spots) were plotted against the reference antibody mGO53 (non-polyreactive isotype control) using GraphPad Prism software (v8.1.2, GraphPad Prism Inc.). For each antibody, Z-scores were calculated using ProtoArray Prospector software (v5.2.3, Thermo Fisher Scientific), and deviation (σ) to the diagonal, and polyreactivity index (PI) values were calculated as previously described (Planchais et al., 2019). Antibodies were defined as polyreactive when PI >0.21.
HEp-2 cell binding assays
Binding of human anti-HBs and control monoclonal IgG antibodies (mGO53 and ED38; Meffre et al., 2004; Wardemann et al., 2003) to HEp-2 cell–expressing autoantigens was analyzed at 50 µg/ml by ELISA (AESKULISA ANA-HEp-2, Aesku.Diagnostics), and indirect immunofluorescence assay (IFA; ANA HEp-2 AeskuSlides, Aesku.Diagnostics) following the manufacturer’s instructions. IFA sections were examined using the fluorescence microscope Axio Imager 2 (Zeiss), and pictures were taken at magnification ×40 with 5,000 ms-acquisition using ZEN imaging software (Zen 2.0 blue version, Zeiss) at the Photonic BioImaging platform (Institut Pasteur).
HBV-AAV mouse model
HBV infection was established in 6–8-wk-old C57BL/6 mice (Janvier Labs) by a single i.v. injection (retro-orbital venous sinus) of 5 × 1010 viral genome of an adeno-associated virus serotype 2/8 carrying a replication competent HBV-DNA genome (Dion et al., 2013). Virus stocks were produced and titrated as virus genome equivalents by the Centre de Production de Vecteurs Viraux (Institut National de la Santé et de la Recherche Médicale [INSERM] U1089, Nantes, France). 6 wk after transduction, antibodies (0.25, 0.5, or 1 mg per injection per mouse) were administrated intravenously to HBV-carrier mice. Blood samples were collected and stored at −20°C. Serum HBsAg and HBeAg levels were determined using Monolisa S-HBsAg ULTRA (Bio-Rad) ELISA kits and ETI-EBK Plus NO140 (Diasorin SA), respectively. Concentrations were calculated by reference to standard curves established with known concentrations of HBsAg (Architect HBsAg Calibrators, Abbott) and of the Paul-Ehrlich-Institut standard and are expressed in IU per milliliter and PEI units per milliliter, respectively. HBV DNA was purified from mouse sera using QIAamp Blood Mini kits (Qiagen), and quantified by quantitative PCR as previously described (Cougot et al., 2012). Serial dilutions of the payw1.2 plasmid containing 1.2 copies of HBV genome were used as quantification standards. Results are expressed in IU per milliliter with a detection threshold at 1,000 IU/ml. Concentrations of human IgGs in mouse sera were determined by ELISA as previously described (Tiller et al., 2008). ADA titers were measured by ELISA as described above but using anti-HBs IgG-coated plates, and HRP-conjugated anti-mouse IgG antibodies (0.8 µg/ml final in blocking solution, Immunology Jackson ImmunoReseach). All experimental animal protocols have been reviewed and approved by the institutional animal care committee of Institut Pasteur for compliance with the French and European regulations on Animal Welfare and validated by the French Ministry of Education and Research (APAFIS no. 15408-2018060517005070 v1). All experiments with HBV infections were performed in an A3 animal facility (Institut Pasteur).
HBV-infected HUHEP mouse model
To generate the HUHEP model, BALB/c Rag2−/−IL-2Rγc−/−NOD.sirpa uPAtg/tg (BRGS-uPA) mice were intrasplenically injected with 7 × 105 freshly thawed HUHEP (BD Biosciences; Strick-Marchand et al., 2015). Liver chimerism was determined on plasma samples with a species-specific human albumin ELISA (Bethyl Laboratories), and HUHEP mice with ≥100 µg/ml human albumin were intraperitoneally infected with 107 HBV genome equivalents (Dusseaux et al., 2017). HBV-infected mice with >105 HBV DNA copies/ml were given an intraperitoneal injection of either Bc1.187 or an isotype control (mGO53) IgG every 3–4 d at 20 mg/kg mouse, or every week at 1 mg/mouse, or Entecavir 0.3 mg/kg/d (Baraclude, BMS) delivered in MediDrop Sucralose (Clear H2O) by mouth. For the rebound phase, mice were returned to drinking water. Plasma HBV DNA and HBsAg quantifications were determined as described above for the AAV-HBV model. Blood HbeAg was quantified with the Monolisa HBeAg ULTRA (Bio-Rad) ELISA kit. Animals were housed in isolators under pathogen-free conditions with human care. Experiments were approved by an institutional ethical committee at the Institut Pasteur (Paris, France), and validated by the French Ministry of Education and Research (MENESR no. 02162.02).
Statistics
For Ig gene repertoire analyses, groups were compared using two-sided 2 × 2 and 2 × 5 Fisher’s exact tests. The numbers of VH, Vκ, and Vλ mutations were compared across groups of antibodies using the unpaired Student t test with Welch’s correction. The volcano plot analysis was performed by comparing 206 individual antibody gene features between groups, and reporting for each parameter the Δmean (x axis) and the −log10 P values given by the two-sided 2 × 2 Fisher’s exact test (y axis). Statistical analyses were performed using GraphPad Prism software (v8.1.2, GraphPad Prism Inc.), and SISA online tools for the 2 × 5 Fisher test (http://www.quantitativeskills.com/sisa).
Online supplemental material
Fig. S1 shows the binding of purified serum IgGs and blood IgG+ memory B cells to S-HBsAg. Fig. S2 shows S-HBsAg the reactivity of S-HBsAg–captured IgG+ memory B cell antibodies. Fig. S3 shows the in vitro HBV neutralizing activity of human anti-HBs antibodies. Fig. S4 shows the binding of HBV neutralizing antibodies to recombinant serotype-specific S-HBsAg. Fig. S5 shows the cross-reactivity of HBV neutralizing antibodies against genotype-specific and mutant S-HBsAg proteins. Table S1 lists the clinical and immunovirological characteristics of the HBV immune donors. Table S2 lists the Ig gene features and HBV neutralization IC50 values of human anti-HBs antibodies.
Supplementary Material
shows clinical and immunovirological characteristics of the HBV immune donors.
shows Ig gene repertoire and neutralizing activity of human anti-HBs antibodies.
Acknowledgments
We are grateful to all participants who consented to be part of this study. We are grateful to members of the Center for Translational Sciences (Institut Pasteur), Cassandre Von Platen (Clinical core) for the preparation of the RAPIVIB protocol, and Gloria Morizot (Investigation Clinique et Accès aux Ressources Biologiques) for collecting biological samples and associated personal data. We also thank Sandrine Schmutz, Sophie Novault (UTechS CB, Center for Translational Sciences, Institut Pasteur), and Ayrin Kök for their technical help with single cell sorting, and Dr. J. Aguilar (Center for Genetic Engineering and Biotechnology, Havana, Cuba) for providing us with the purified adw HBsAg particles.
H. Mouquet received core grants from the G5 Institut Pasteur Program, the Milieu Intérieur Program (ANR-10-LABX-69-01) and INSERM. This work was funded by Institut Roche. V. Hehle and M. Beretta were supported by postdoctoral fellowships from Roche. V. Hehle and T. Hieu were also supported by the European Research Council–Seventh Framework Program (ERC-2013-StG 337146). H. Strick-Marchand was supported by the Institut Pasteur and Agence Nationale de Recherches sur le Sida et les Hépatites Virales (2016-16365, 2019-19241).
Author contributions: V. Hehle, M. Beretta, M. Bourgine, M. Ait-Goughoulte, C. Planchais, S. Morrise, B. Vesin, A. Stauffer, C. Sureau, V. Lorin, O. Fiquet, J.D. Dimitrov, C. Sureau, H. Strick-Marchand, and N. Pelletier performed and analyzed the experiments. T. Hieu carried out bioinformatics analyses. M.-N. Ungeheuer and S. Pol provided human samples and personal data. C. Sureau and M.-L. Michel contributed with key reagents and expertise. M. Bourgine, J.P. Di Santo, H. Strick-Marchand, N. Pelletier, and H. Mouquet supervised the experiments. H. Mouquet conceived and supervised the study, designed and analyzed experiments, and wrote the manuscript with contributions from all the authors.
References
- Andrews S.F., Huang Y., Kaur K., Popova L.I., Ho I.Y., Pauli N.T., Henry Dunand C.J., Taylor W.M., Lim S., Huang M., et al. 2015. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7 316ra192 10.1126/scitranslmed.aad0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer T., Sprinzl M., and Protzer U.. 2011. Immune control of hepatitis B virus. Dig. Dis. 29:423–433. 10.1159/000329809 [DOI] [PubMed] [Google Scholar]
- Bertoletti A., and Ferrari C.. 2012. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut. 61:1754–1764. 10.1136/gutjnl-2011-301073 [DOI] [PubMed] [Google Scholar]
- Bertoletti A., and Ferrari C.. 2016. Adaptive immunity in HBV infection. J. Hepatol. 64(1, Suppl):S71–S83. 10.1016/j.jhep.2016.01.026 [DOI] [PubMed] [Google Scholar]
- Burton A.R., Pallett L.J., McCoy L.E., Suveizdyte K., Amin O.E., Swadling L., Alberts E., Davidson B.R., Kennedy P.T., Gill U.S., et al. 2018. Circulating and intrahepatic antiviral B cells are defective in hepatitis B. J. Clin. Invest. 128:4588–4603. 10.1172/JCI121960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tzarum N., Wilson I.A., and Law M.. 2019. VH1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design. Curr. Opin. Virol. 34:149–159. 10.1016/j.coviro.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotiyaputta W., and Lok A.S.. 2009. Hepatitis B virus variants. Nat. Rev. Gastroenterol. Hepatol. 6:453–462. 10.1038/nrgastro.2009.107 [DOI] [PubMed] [Google Scholar]
- Chu C.-M., and Liaw Y.-F.. 2016. Natural History of Hepatitis B Virus Infection In Hepatitis B Virus in Human Diseases. Humana Press, Totowa, NJ: pp. 217–247. 10.1007/978-3-319-22330-8_11 [DOI] [Google Scholar]
- Corti D., Benigni F., and Shouval D.. 2018. Viral envelope-specific antibodies in chronic hepatitis B virus infection. Curr. Opin. Virol. 30:48–57. 10.1016/j.coviro.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Cougot D., Allemand E., Rivière L., Benhenda S., Duroure K., Levillayer F., Muchardt C., Buendia M.A., and Neuveut C.. 2012. Inhibition of PP1 phosphatase activity by HBx: a mechanism for the activation of hepatitis B virus transcription. Sci. Signal. 5:ra1 10.1126/scisignal.2001906 [DOI] [PubMed] [Google Scholar]
- Dion S., Bourgine M., Godon O., Levillayer F., and Michel M.L.. 2013. Adeno-associated virus-mediated gene transfer leads to persistent hepatitis B virus replication in mice expressing HLA-A2 and HLA-DR1 molecules. J. Virol. 87:5554–5563. 10.1128/JVI.03134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusséaux M., Masse-Ranson G., Darche S., Ahodantin J., Li Y., Fiquet O., Beaumont E., Moreau P., Rivière L., Neuveut C., et al. 2017. Viral Load Affects the Immune Response to HBV in Mice With Humanized Immune System and Liver. Gastroenterology. 153:1647–1661.e9. 10.1053/j.gastro.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren R., Ilan E., Nussbaum O., Lubin I., Terkieltaub D., Arazi Y., Ben-Moshe O., Kitchinzky A., Berr S., Gopher J., et al. 2000. Preclinical evaluation of two human anti-hepatitis B virus (HBV) monoclonal antibodies in the HBV-trimera mouse model and in HBV chronic carrier chimpanzees. Hepatology. 32:588–596. 10.1053/jhep.2000.9632 [DOI] [PubMed] [Google Scholar]
- Fanning G.C., Zoulim F., Hou J., and Bertoletti A.. 2019. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discov. 18:827–844. 10.1038/s41573-019-0037-0 [DOI] [PubMed] [Google Scholar]
- Galun E., Eren R., Safadi R., Ashour Y., Terrault N., Keeffe E.B., Matot E., Mizrachi S., Terkieltaub D., Zohar M., et al. 2002. Clinical evaluation (phase I) of a combination of two human monoclonal antibodies to HBV: safety and antiviral properties. Hepatology. 35:673–679. 10.1053/jhep.2002.31867 [DOI] [PubMed] [Google Scholar]
- Gao Y., Zhang T.Y., Yuan Q., and Xia N.S.. 2017. Antibody-mediated immunotherapy against chronic hepatitis B virus infection. Hum. Vaccin. Immunother. 13:1768–1773. 10.1080/21645515.2017.1319021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring A.J., and Protzer U.. 2019. Targeting Innate and Adaptive Immune Responses to Cure Chronic HBV Infection. Gastroenterology. 156:325–337. 10.1053/j.gastro.2018.10.032 [DOI] [PubMed] [Google Scholar]
- Hantz O., Parent R., Durantel D., Gripon P., Guguen-Guillouzo C., and Zoulim F.. 2009. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J. Gen. Virol. 90:127–135. 10.1099/vir.0.004861-0 [DOI] [PubMed] [Google Scholar]
- Hayer J., Jadeau F., Deléage G., Kay A., Zoulim F., and Combet C.. 2013. HBVdb: a knowledge database for Hepatitis B Virus. Nucleic Acids Res. 41(Database issue, D1):D566–D570. 10.1093/nar/gks1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.H., Yuan Q., Chen P.J., Zhang Y.L., Chen C.R., Zheng Q.B., Yeh S.H., Yu H., Xue Y., Chen Y.X., et al. 2012. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J. Hepatol. 57:720–729. 10.1016/j.jhep.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Kato H., Sugiyama M., and Mizokami M.. 2016. Hepatitis B Virus Genotypes In Hepatitis B Virus in Human Diseases. Humana Press, Totowa, NJ: pp. 63–78. 10.1007/978-3-319-22330-8_3 [DOI] [Google Scholar]
- Kucinskaite-Kodze I., Pleckaityte M., Bremer C.M., Seiz P.L., Zilnyte M., Bulavaite A., Mickiene G., Zvirblis G., Sasnauskas K., Glebe D., et al. 2016. New broadly reactive neutralizing antibodies against hepatitis B virus surface antigen. Virus Res. 211:209–221. 10.1016/j.virusres.2015.10.024 [DOI] [PubMed] [Google Scholar]
- Kusumoto S., Arcaini L., Hong X., Jin J., Kim W.S., Kwong Y.L., Peters M.G., Tanaka Y., Zelenetz A.D., Kuriki H., et al. 2019. Risk of HBV reactivation in patients with B-cell lymphomas receiving obinutuzumab or rituximab immunochemotherapy. Blood. 133:137–146. 10.1182/blood-2018-04-848044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küttner G., Kramer A., Schmidtke G., Giessmann E., Dong L., Roggenbuck D., Scholz C., Seifert M., Stigler R.D., Schneider-Mergener J., et al. 1999. Characterization of neutralizing anti-pre-S1 and anti-pre-S2 (HBV) monoclonal antibodies and their fragments. Mol. Immunol. 36:669–683. 10.1016/S0161-5890(99)00074-7 [DOI] [PubMed] [Google Scholar]
- Lee H.W., Park J.Y., Hong T., Park M.S., and Ahn S.H.. 2019. Efficacy of Lenvervimab, a Recombinant Human Immunoglobulin, in Treatment of Chronic Hepatitis B Virus Infection. Clin. Gastroenterol. Hepatol. S1542-3565(19)31089-4 10.1016/j.cgh.2019.09.038 [DOI] [PubMed] [Google Scholar]
- Lever A.M., Waters J., Brook M.G., Karayiannis P., and Thomas H.C.. 1990. Monoclonal antibody to HBsAg for chronic hepatitis B virus infection with hypogammaglobulinaemia. Lancet. 335:1529 10.1016/0140-6736(90)93069-2 [DOI] [PubMed] [Google Scholar]
- Li D., He W., Liu X., Zheng S., Qi Y., Li H., Mao F., Liu J., Sun Y., Pan L., et al. 2017. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. eLife. 6 e26738 10.7554/eLife.26738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorin V., and Mouquet H.. 2015. Efficient generation of human IgA monoclonal antibodies. J. Immunol. Methods. 422:102–110. 10.1016/j.jim.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Maini M.K., and Burton A.R.. 2019. Restoring, releasing or replacing adaptive immunity in chronic hepatitis B. Nat. Rev. Gastroenterol. Hepatol. 16:662–675. 10.1038/s41575-019-0196-9 [DOI] [PubMed] [Google Scholar]
- McMahon B.J. 2009. The natural history of chronic hepatitis B virus infection. Hepatology. 49(5, Suppl):S45–S55. 10.1002/hep.22898 [DOI] [PubMed] [Google Scholar]
- Meffre E., Schaefer A., Wardemann H., Wilson P., Davis E., and Nussenzweig M.C.. 2004. Surrogate light chain expressing human peripheral B cells produce self-reactive antibodies. J. Exp. Med. 199:145–150. 10.1084/jem.20031550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M.L., Pontisso P., Sobczak E., Malpièce Y., Streeck R.E., and Tiollais P.. 1984. Synthesis in animal cells of hepatitis B surface antigen particles carrying a receptor for polymerized human serum albumin. Proc. Natl. Acad. Sci. USA. 81:7708–7712. 10.1073/pnas.81.24.7708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Farci S., Joly P., Maillère B., Leblond J., Drouot L., Leprince J., Tonon M.C., Loiseau P., Charron D., et al. 2006. A truncated alternative spliced isoform of human desmoglein 1 contains a specific T cell epitope binding to the pemphigus foliaceus-associated HLA class II DRbeta1*0102 molecule. J. Immunol. 177:6517–6526. 10.4049/jimmunol.177.9.6517 [DOI] [PubMed] [Google Scholar]
- Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J.F., Halper-Stromberg A., Gnanapragasam P.N., Spencer D.I., Seaman M.S., et al. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 109:E3268–E3277. 10.1073/pnas.1217207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrin M., Naranjo-Gomez M., and Piechaczyk M.. 2015. Antiviral Monoclonal Antibodies: Can They Be More Than Simple Neutralizing Agents? Trends Microbiol. 23:653–665. 10.1016/j.tim.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrillo R.P., Gish R., and Falck-Ytter Y.T.. 2015. American Gastroenterological Association Institute technical review on prevention and treatment of hepatitis B virus reactivation during immunosuppressive drug therapy. Gastroenterology. 148:221–244.e3. 10.1053/j.gastro.2014.10.038 [DOI] [PubMed] [Google Scholar]
- Planchais C., Kök A., Kanyavuz A., Lorin V., Bruel T., Guivel-Benhassine F., Rollenske T., Prigent J., Hieu T., Prazuck T., et al. 2019. HIV-1 Envelope Recognition by Polyreactive and Cross-Reactive Intestinal B Cells. Cell Rep. 27:572–585.e7. 10.1016/j.celrep.2019.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent J., Lorin V., Kök A., Hieu T., Bourgeau S., and Mouquet H.. 2016. Scarcity of autoreactive human blood IgA+ memory B cells. Eur. J. Immunol. 46:2340–2351. 10.1002/eji.201646446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent J., Jarossay A., Planchais C., Eden C., Dufloo J., Kök A., Lorin V., Vratskikh O., Couderc T., Bruel T., et al. 2018. Conformational Plasticity in Broadly Neutralizing HIV-1 Antibodies Triggers Polyreactivity. Cell Rep. 23:2568–2581. 10.1016/j.celrep.2018.04.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimzadeh L., Le Bert N., Dutertre C.A., Gill U.S., Newell E.W., Frey C., Hung M., Novikov N., Fletcher S., Kennedy P.T., et al. 2018. PD-1 blockade partially recovers dysfunctional virus-specific B cells in chronic hepatitis B infection. J. Clin. Invest. 128:4573–4587. 10.1172/JCI121957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisse J., and Sureau C.. 2009. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J. Virol. 83:9321–9328. 10.1128/JVI.00678-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel D., Muller R., Alexander G., Fassati L., Ducot B., Benhamou J.P., and Bismuth H.. 1993. Liver transplantation in European patients with the hepatitis B surface antigen. N. Engl. J. Med. 329:1842–1847. 10.1056/NEJM199312163292503 [DOI] [PubMed] [Google Scholar]
- Seeger C., Zoulim F., and Mason W.S.. 2013. Hepadnaviruses. . Wolters Kluwer/Lippincott Williams & Wilkins Health, New York, Philadelphia, Baltimore. [Google Scholar]
- Shouval D., Shafritz D.A., Zurawski V.R. Jr., Isselbacher K.J., and Wands J.R.. 1982a Immunotherapy in nude mice of human hepatoma using monoclonal antibodies against hepatitis B virus. Nature. 298:567–569. 10.1038/298567a0 [DOI] [PubMed] [Google Scholar]
- Shouval D., Wands J.R., Zurawski V.R. Jr., Isselbacher K.J., and Shafritz D.A.. 1982b Selecting binding and complement-mediated lysis of human hepatoma cells (PLC/PRF/5) in culture by monoclonal antibodies to hepatitis B surface antigen. Proc. Natl. Acad. Sci. USA. 79:650–654. 10.1073/pnas.79.2.650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick-Marchand H., Dusséaux M., Darche S., Huntington N.D., Legrand N., Masse-Ranson G., Corcuff E., Ahodantin J., Weijer K., Spits H., et al. 2015. A novel mouse model for stable engraftment of a human immune system and human hepatocytes. PLoS One. 10 e0119820 10.1371/journal.pone.0119820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau C. 2010. The use of hepatocytes to investigate HDV infection: the HDV/HepaRG model. Methods Mol. Biol. 640:463–473. 10.1007/978-1-60761-688-7_25 [DOI] [PubMed] [Google Scholar]
- Sureau C., and Negro F.. 2016. The hepatitis delta virus: Replication and pathogenesis. J. Hepatol. 64(1, Suppl):S102–S116. 10.1016/j.jhep.2016.02.013 [DOI] [PubMed] [Google Scholar]
- Tateno C., Kawase Y., Tobita Y., Hamamura S., Ohshita H., Yokomichi H., Sanada H., Kakuni M., Shiota A., Kojima Y., et al. 2015. Generation of Novel Chimeric Mice with Humanized Livers by Using Hemizygous cDNA-uPA/SCID Mice. PLoS One. 10 e0142145 10.1371/journal.pone.0142145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharinger H., Rebbapragada I., Samuel D., Novikov N., Nguyen M.H., Jordan R., Frey C.R., and Pflanz S.. 2017. Antibody-dependent and antibody-independent uptake of HBsAg across human leucocyte subsets is similar between individuals with chronic hepatitis B virus infection and healthy donors. J. Viral Hepat. 24:506–513. 10.1111/jvh.12667 [DOI] [PubMed] [Google Scholar]
- Tiller T., Tsuiji M., Yurasov S., Velinzon K., Nussenzweig M.C., and Wardemann H.. 2007. Autoreactivity in human IgG+ memory B cells. Immunity. 26:205–213. 10.1016/j.immuni.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., and Wardemann H.. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 329:112–124. 10.1016/j.jim.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu T., and Urban S.. 2018. Virus entry and its inhibition to prevent and treat hepatitis B and hepatitis D virus infections. Curr. Opin. Virol. 30:68–79. 10.1016/j.coviro.2018.04.004 [DOI] [PubMed] [Google Scholar]
- van Nunen A.B., Baumann M., Manns M.P., Reichen J., Spengler U., Marschner J.P., and de Man R.A.; International Study Group . 2001. Efficacy and safety of an intravenous monoclonal anti-HBs in chronic hepatitis B patients. Liver. 21:207–212. 10.1034/j.1600-0676.2001.021003207.x [DOI] [PubMed] [Google Scholar]
- Verrier E.R., Colpitts C.C., Bach C., Heydmann L., Weiss A., Renaud M., Durand S.C., Habersetzer F., Durantel D., Abou-Jaoudé G., et al. 2016. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology. 63:35–48. 10.1002/hep.28013 [DOI] [PubMed] [Google Scholar]
- Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., and Nussenzweig M.C.. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. 10.1126/science.1086907 [DOI] [PubMed] [Google Scholar]
- West D.J., and Calandra G.B.. 1996. Vaccine induced immunologic memory for hepatitis B surface antigen: implications for policy on booster vaccination. Vaccine. 14:1019–1027. 10.1016/0264-410X(96)00062-X [DOI] [PubMed] [Google Scholar]
- World Health Organization 2017. Global hepatitis report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
- Xu X., Shang Q., Chen X., Nie W., Zou Z., Huang A., Meng M., Jin L., Xu R., Zhang J.Y., et al. 2015. Reversal of B-cell hyperactivation and functional impairment is associated with HBsAg seroconversion in chronic hepatitis B patients. Cell. Mol. Immunol. 12:309–316. 10.1038/cmi.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.Y., Yuan Q., Zhao J.H., Zhang Y.L., Yuan L.Z., Lan Y., Lo Y.C., Sun C.P., Wu C.R., Zhang J.F., et al. 2016. Prolonged suppression of HBV in mice by a novel antibody that targets a unique epitope on hepatitis B surface antigen. Gut. 65:658–671. 10.1136/gutjnl-2014-308964 [DOI] [PubMed] [Google Scholar]
- Zhu D., Liu L., Yang D., Fu S., Bian Y., Sun Z., He J., Su L., Zhang L., Peng H., et al. 2016. Clearing Persistent Extracellular Antigen of Hepatitis B Virus: An Immunomodulatory Strategy To Reverse Tolerance for an Effective Therapeutic Vaccination. J. Immunol. 196:3079–3087. 10.4049/jimmunol.1502061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows clinical and immunovirological characteristics of the HBV immune donors.
shows Ig gene repertoire and neutralizing activity of human anti-HBs antibodies.