Abstract
Background. Severe acute respiratory syndrome coronavirus (SARS-CoV), which emerged with highly contagious and life-threatening characteristics in 2002, remains a potential risk for future outbreaks. Membrane (M) and envelope (E) proteins are major structural proteins of the SARS-CoV. The M protein has been determined as a protective antigen in humoral responses. However, its potential roles in stimulating cellular immunity remain elusive.
Methods. In this study, a panel of peptides derived from M and E proteins were tested by in vitro refolding, T2 cell-binding assays, and responses stimulated by cytotoxic T-lymphocyte (CTL) epitopes in HLA-A2.1/Kb transgenic mice and human peripheral blood mononuclear cells (PBMCs).
Results. A nonameric epitope Mn2 and a decameric epitope Md3 derived from the M protein were identified and used for the evaluation of M protein-specific immunity. Responses stimulated by M protein-specific CTL epitopes have been found in the PBMCs of donors who had recovered from SARS infection. Additionally, the transmembrane domain of the M protein may contain a T cell epitope cluster revealed by the immunogenic and structural analysis of a panel of truncated peptides overlapping with Mn2 and Md3.
Conclusions. The M protein of SARS-CoV holds dominant cellular immunogenicity. This, together with previous reports of a strong humoral response against the M protein, may help to further explain the immunogenicity of SARS and serves as potential targets for SARS-CoV vaccine design.
Severe acute respiratory syndrome (SARS) emerged in Foshan City, Guangdong Province, China, in November 2002 [1–3] and caused tremendous impact on the world health systems and economies because of its high transmissibility, rapid deteriorating process, and poor prognosis [4–10]. The etiological agent of SARS was iden- tified as a new member of human coronavirus (family Coronaviridae) named SARS-associated coronavirus (SARS-CoV) [2, 3]. It has been proven that SARS-CoV may be an interspecies-transmissible virus [11–13]. However, the true scenario of the crossspecies transmission, virus adaptation to humans, and sudden disappearance remains mysterious because current data are limited to genetic sequence and serology [14, 15]. This indicates that SARS-CoV is still a potential reemerging pathogen and a good experimental model system for research.
SARS-CoV causes by far the most severe disease among all human coronaviruses, a so-called cytokine storm characterized by serious lung injury (acute respiratory distress syndrome [ARDS]) [16, 17]. It is believed that the severe clinical outcome is due to the overreaction of the human immune system because the virus is relatively “new” to the host, evidenced by the positive effect of corticosteroid treatment of infected patients [8]. Immune protection against SARS-CoV infection is convincingly confirmed by some experimental data [18–21], although the pros and cons of the host immune response contributing to the pathogenicity are controversial [16, 22, 23]. It is believed that humoral immunity plays an important role in host immune protection [18, 19], but host cellular immunity for protection against SARS-CoV infection is also recognized [20, 21]. The immune response of cytotoxic T lymphocytes (CTLs) is the major specific defense against viral invasion, and a series of studies have demonstrated that human memory T cell responses to SARS-CoV persists for months to years in the absence of antigen [21, 24–27]. Thus, immunological monitoring of T cell immunity against SARS-CoV infection is crucial to evaluate the functions of CTLs in viral clearance and their status in immunopathogenesis.
The antigenicity of specific proteins within a viral proteome differs depending on the diversity of quantity and immunogenic antigenicity of the immune epitopes comprised in the proteins. It has been demonstrated that structural proteins of SARS-CoV such as spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins possess much higher immunogenicity for T cell responses than the nonstructural proteins [20]. It has been shown that the S and N proteins are characterized by high immunogenicity to engage the humoral and cellular response to SARS-CoV [19, 28]. The M and E proteins are the other 2 significant structural proteins anchoring on the envelope membrane surface of the SARS-CoV particles. The M protein is a typical transmembrane glycoprotein composed of a triplemembrane domain spanning 80 amino acids that account for about one-third of the entire protein (221 residues in total) [29]. Acting as the most abundant structural protein in the virion of SARS-CoV [29], M also plays a significant role in virus-specific humoral response and is able to elicit efficient neutralizing antibodies in SARS patients [30]. The E protein is a small integral membrane polypeptide that forms an ion channel [31]. Either the absence or inactivation of the E protein results in attenuated virulence because of the alterations in virion morphology or tropism [32, 33].
We previously reported an efficient strategy of screening HLA-A*0201 restricted CTL epitopes, which led to the identification of an immunodominant epitope from the S protein restricted by HLA-A*0201 [34]. Following the same strategy, in the present study we identified a CTL-related epitope clustering region containing 2 CTL epitopes restricted by HLAA* 0201 in the transmembrane domain of the M protein. The 2 epitopes have been further functionally and structurally characterized. We also found a strong response by both enzymelinked immunospot (ELISPOT) assay and human leukocyte antigen (HLA) tetramer staining in peripheral blood mononuclear cells (PBMCs) obtained from patients who had recovered from SARS-CoV infection. We conclude that the SARSCoV M protein acts as a dominant immunogen for both the humoral response (from previous reported data) and cellular immune response.
Materials and Methods
Prediction and synthesis of candidate peptides. The potential nonamer and decamer peptides in the M and E proteins of SARS-CoV (strain TJF; GenBank accession no. AY654624) were predicted by estimating the half-life of disassociation from the HLA-A*0201 molecule through BIMAS online analysis software [35]. Fourteen peptides with high scores were synthesized and purified with a purity of ∼90% (Table 1). HLA-A*0201-restricted epitope of influenza A matrix peptide p58–66 (Flu, GILGFVFTL) [36] and SARS-CoV S protein-derived peptide P15 (KLPDDFMGCV) [34] served as the positive control. The negative control was performed by simian immunodeficiency virus Mamu-A*01-restricted peptide CM9 (CTPYDINQM) [37]. These peptides were dissolved in dimethyl sulfoxide after synthesis and stored in aliquots at −80°C in a lyophilized environment.
Table 1.
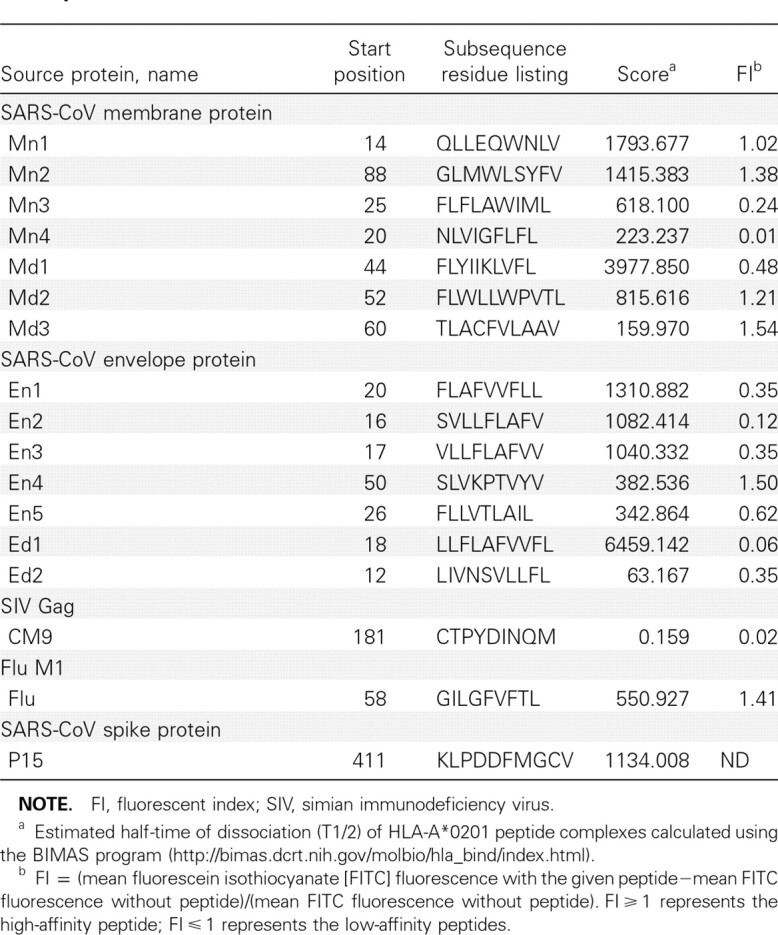
NOTE. FI, fluorescent index; SIV, simian immunodeficiency virus. a Estimated half-time of dissociation (T1/2) of HLA-A*0201 peptide complexes calculated using the BIMAS program (http://bimas.dcrt.nih.gov/molbio/hla_bind/index.html). b FI = (mean fluorescein isothiocyanate [FITC] fluorescence with the given peptide—mean FITC fluorescence without peptide)/(mean FITC fluorescence without peptide). Fl ⩾ 1 represents the high-affinity peptide; FI⩾1 represents the low-affinity peptides.
In vitro refolding of the synthesized peptides with HLAA* 0201 heavy chain and β2 microglobulin. HLA-A*0201 heavy chain and β2 microglobulin were overexpressed as recombinant proteins in Escherichia coli and subsequently refolded in vitro and assembled in the presence of a high concentration of specific peptides as described elsewhere [38]. The refolded protein was concentrated and analyzed by Superdex 200 10/300 GL gel-filtration chromatography (Amersham Pharmacia Biotech). The fractionated samples were collected and analyzed with sodium dodecyl sulfate polyacrylamide gel electrophoresis to check the refolding efficiency.
Stabilization assay of HLA-A*0201 in T2 cells incubated with synthetic peptides. To further determine the binding capability of synthetic peptides to HLA-A*0201 molecules present on the cell surface, the T2 cell-binding assay was performed as described elsewhere [21, 34]. HLA-A*0201 binding peptides increased the stable expression of HLA-A*0201 molecules on the T2 cells. Briefly, T2 cells suspended in serum-free Roswell Park Memorial Institute 1640 medium (RPMI) were incubated with the peptides (50 μmol/L), supplemented with 1 μmol/L human β2 microglobulin (Sigma) for 18 h at 37°C with 5% b2 CO2. The expression of HLA-A*0201 on T2 cells was determined by staining with an anti-human HLA-A2 antibody (Serotech) conjugated with fluorescein isothiocyanate (FITC) and then analyzed by flow cytometry (FACScan; BD Biosciences). Results were calculated as the fluorescent indexes (FI), which were determined as follows: FI = (mean FITC fluorescence with the given peptide)/(mean FITC fluorescence without peptide). Peptides with FI ⩾ 1 were arbitrarily regarded as high-affinity epitopes [34], which means that these peptides could increase the mean fluorescence of T2 cells by at least 1-fold.
ELISPOT assay for IFN-γ production. The CTL epitope- specific response was measured by performing the interferon g (IFN-γ) ELISPOT assays as described elsewhere [34, 39]. The IFN-γ production after the CTL epitope stimulation is defined as CTL epitope-specific response regardless of the IFN-γ production cells. The number of spots was determined using an automatic ELISPOT reader and image analysis software (Cellular Technology Limited).
Peptide-HLA tetramer staining. Two kinds of recombinant HLA-A*0201 heavy chain with biotin tag were used to produce major histocompatibility complex (MHC) tetramers as described elsewhere [34]: (1) wild type and (2) α3 region substitution with mouse H-Kd to enhance the binding to mouse CD8 cells. In vitro biotinylation of the peptide-MHC complexes was achieved by incubating the sample with the biotin protein ligase Bir A, D-biotin, and adenosine triphosphate. The samples were purified again through gel filtration to remove free biotin, and then the multimers were produced by using streptavidin conjugated with phycoerythrin (Sigma). Cells from the subjects were stained with the tetramer (0.05 μg/μL) and FITC-conjugated anti-CD8 antibody and then immediately analyzed with flow cytometry.
PBMCs obtained from donors previously infected with SARS. The frozen PBMCs collected from 3 HLA-A2+ patients who had been recovered from SARS for 20–22 months were stored in Peking Union Hospital with the donors' informed consent. The cells were thawed and incubated with 10 μmol/ L peptides and 20 U/mL recombinant human interleukin 2 in RPMI containing 10% fetal calf serum at 37°C with 5% CO2. The PBMCs of a healthy HLA-A2+ donor were obtained with informed consent. All the manipulations of human samples have been approved by an institutional ethics committee.
Inoculation of mice with peptides. The HLA-A*0201 transgenic mice with C57BL/6 background [40] were maintained in an animal care facility under pathogen-free conditions. Each experimental group contained 8 members with equivalent male and female mice and was inoculated with a different candidate peptide. The 6- to 8-week old transgenic mice were injected subcutaneously at multiple sites with candidate peptides and the N-terminal fragment N333 (23–355aa) of murine gp96 [41] emulsified in complete Freund's adjuvant. Mice were administered the mixture of peptides, gp96 fragment, and incomplete Freund's adjuvant twice in 2-week intervals. Mice were killed at day 7 after the final inoculation.
Inoculation of mice with SARS-CoV membrane DNA plasmid (pD3-M). The SARS-CoV membrane gene (derived from strain TJF) was cloned in the pcDNA3-FLAG vector (pD3-M) using forward primer 5'-GCGGATCCGCCACCATGGCAGACAACGGTACTATT- 3' with a BamHI restriction site followed by a Kozak sequence and reverse primer 5'-GCCTCGAGCTGTACTAGCAAAGCAATATTGTCG- 3'. The expression of M protein coded by pD3-M in 293 T cells was confirmed by Western blot using an anti-FLAG antibody. Each transgenic mouse was injected intramuscularly with 100 μg of plasmid. The injection was administered at weeks 0, 3, and 6, and the CTL epitope- specific responses were measured 10 days after the last boost.
X-ray crystallography. HLA-A*0201/peptide complexes were refolded and purified as described above. Crystals were grown by the hanging drop vapor diffusion method at 4°C. Single HLA- A*0201-Md3 crystals grew in a final concentration of 10 mg/mL in 0.1 mol/L BIS-TRIS pH 5.5 gel and 10% wt/ vol polyethylene glycol 3350. Single crystals of HLA- A*0201- Mn2 were grown over the course of 3 days by microseeding in a concentration of 20 mg/mL using 0.1 mol/L ammonium acetate, 0.1 mol/L BIS-TRIS, pH 6.0 gel, and 13%–17% polyethylene glycol 10,000. For cryoprotection, crystals were transferred to reservoir solutions containing 20% glycerol. Crystallographic data were collected at 100 K in-house on a Rigaku MicroMax007 rotating-anode X-ray generator operated at 40 kV and 20 mA (Cu Kαλ = 1.5418) equipped with an R-AXIS VII++ image-plate detector. Data were indexed and scaled using the DENZO program and the HKL2000 software package (HKL Research).
Structure determination, refinement, and analysis. The structures of HLA-A*0201/Mn2 and HLA-A*0201/Md3 were determined using molecular replacement with the program MolRep. The search model was Protein Data Bank entry 1JF1 [42] with water coordinates omitted. Extensive model building was performed manually using the COOT program [43] and restrained refinement using the REFMAC5 program. The additional rounds of refinement were performed by using the PHENIX.refine program implemented in the PHENIX software package with isotropic adenosine diphosphate refinement and bulk solvent modeling [44]. The stereochemical quality of the final model was assessed with the program PROCHECK [45]. The atomic coordinates of these 2 crystal structures have been deposited in the Protein Data Bank (accession nos. 3I6G and 3I6K).
Results
Identification of potential HLA-A*0201-binding peptides. The complete sequences of SARS-CoV membrane and envelope protein were analyzed with the online HLA Peptide Binding Prediction software to predict the potential HLA-A*0201-binding peptides. Fourteen candidate peptides with relatively high estimated half-time of dissociation from HLA-A*0201 were selected (Table 1). All the peptides were synthesized, with purity up to 90%. Eleven of the predicted peptides could refold into HLAA* 0201 complexes. Of these peptides, Mn2, Mn3, Md1, Md2, Md3, and En4 possess a higher affinity for binding to HLAA* 0201 molecules and the ability to assist the refolding of heavy chain with β2 microglobulin (Figure 1).
Figure 1.
Binding affinity of the synthesized peptides to HLA-A*0201 molecules. A, The refolding results were analyzed by fast-performance liquid chromatography Superdex 200 10/300 GL gel-filtration chromatography, which was calibrated with the standard protein marker, showing that the predicted molecular weight of the protein coming out with 15.2 mL eluting buffer would be 45 kDa, coincident with the molecular mass of the human leukocyte antigen (HLA) complex. A positive control (PC) was performed by the HLA-A*0201-restricted epitope Flu, and the negative control (NC) peptide was CM9 (Table 1). B, In the T2 cell-binding assay, the distribution histograms of cell populations are presented with different fluorescence intensity of fluorescein isothiocyanate (FITC)-anti-HLA-A2 staining of T2 cells cocultured with PC (Flu), NC (CM9), and high-affinity (Fl ⩾ 1) peptides Mn1, Mn2, Md2, Md3, and En4. The results are representative of 3 independent experiments.
T2 cell-peptide binding assay. The T2 cell-peptide binding assay was used to further investigate the binding affinity of the peptides to HLA-A*0201 molecules (Figure 1). Of the 14 synthesized candidate peptides, only Mn1, Mn2, Md2, Md3, and En4 could increase the surface expression of HLA-A*0201 by >2-fold, which was demonstrated as . The FI ⩾ of these 5 high-affinity peptides were 1.02, 1.38, 1.21, 1.54, and 1.50, respectively. The positive control Flu peptide had a high affinity, with FI = 1.41, whereas the negative control CM9 peptide had an affinity of FI = 0.02, indicating no binding ability with HLA-A*0201. The results of the HLA-A*0201 complex refolding assay and T2 cell-peptide binding assay were taken into account together, and 4 peptides-Mn2, Md2, Md3, and En4- were chosen for additional experiments in vivo.
In vivo generation of CTL epitope-specific CD8+ immune response in HLA-A*0201/Kb-transgenic mice inoculated with high-affinity peptides. After 3 rounds of fortnightly in vivo injections, the ELISPOT assay was performed using fresh splenocytes (Figure 2A). No specific reactivity of IFN-γ secretion could be detected in splenocytes separated from transgenic mice inoculated with either peptide Md2 or peptide En4. Thus, these 2 peptides were excluded from additional studies. CD8+ T cells from splenocytes of transgenic mice responding to peptide Md3 presented strong IFN-γ production, whereas peptide Mn2 elic ited a relative lower response. However, both of the groups inoculated with Md3 and Mn2 peptides possessed a statistically significant difference compared with the group inoculated with phosphate-buffered saline (PBS). After an in vitro proliferation of the splenocytes for an additional 10 days, HLA-A*0201 tetramers of peptide Mn2 and Md3 were prepared to stain peptide- specific CD8+ T lymphocytes (presumably CTLs). The splenocytes from Mn2- and Md3-inoculated mice contained 5.2% and 8.6% of peptide-specific CD8+ T cells, respectively, after 10 days of stimulation in vitro (Figure 3A). No peptidespecific CD8+ T cells were identified from splenocytes of mice inoculated with PBS stained with either HLA-A*0201/Mn2 or HLA-A*0201/Md3 tetramer. The positive controlHLA-A*0201/ Flu tetramer showed 8.5% of Flu-specific CD8+ T cells within splenocytes stimulated in vitro from transgenic mice inoculated with Flu peptide (data not shown). These results indicated that peptides Mn2 and Md3 may induce specific CD8+ T cell immune response in vivo in transgenic mice and furthermore possess the capability to significantly stimulate specific CD8+ T cell expansion in vitro.
Figure 2.
Detection of peptide-specific CD8+ T cells in HLA-A2.1/Kb transgenic mice. A, Fresh splenocytes from dead mice inoculated with peptides that had double-positive results with both refolding and T2 cellbinding assays were manipulated with the enzyme-linked immunospot (ELISPOT) assay. The bars represent the quantity of spot-forming cells (SFCs) in 105 splenocytes. The results of peptides Mn2 and Md3 consistent with the positive control Flu and the nonspecific stimulus phytohemagglutinin (PHA) have a statistically significant difference compared with the results from peptides Md2 and En4 (P < .05). B, Survey of the peptide-specific CD8+ T cells induced in HLA-A2.1/Kb transgenic mice inoculated with DNA plasmid. The mean numbers of the SFCs in 105 Mn2- and Md3- stimulated splenocytes from mice inoculated with pD3-M, represented by the gray bars, are significantly higher than the white bar, which represents the SFC numbers of Mn2- and Md3-stimulated splenocytes from mice inoculated with pcDNA3, according to the statistics. There is no statistically significant difference between the PHA-stimulated splenocytes of these 2 groups.
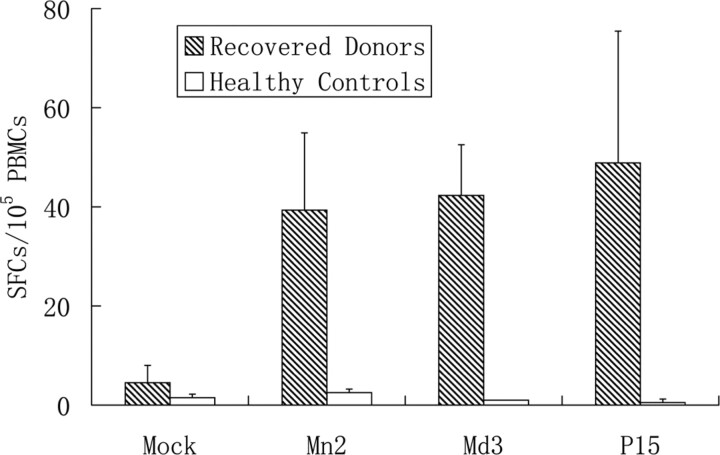
Figure 3.
Measurement of peptide-specific CD8+ T cells from peripheral blood mononuclear cells (PBMCs) of donors recovered from severe acute respiratory syndrome (SARS) by enzyme-linked immunospot (ELISPOT) assay. The thawed PBMCs were manipulated with the ELISPOT assay kit. Shaded bars represent the mean numbers of spot-forming cells (SFCs) in the PBMCs of HLA-A2+ recovered donors, whereas white bars represent the PBMCs from HLA-A2+ healthy donors. The former differs statistically from the latter when stimulated with the peptides. Nevertheless, no difference has been shown between them when stimulated with nonspecific stimulus phytohemagglutinin (PHA).
Detection of CTL epitope-specific CD8+ T cell immune response in HLA-A*0201/Kb-transgenic mice inoculated with DNA vaccine. To further demonstrate whether peptides Mn2 and Md3 were naturally processed and presented peptides, we constructed a DNA vaccine encoding SARS-CoV M protein: pD3-M. Splenocytes isolated from the dead mice after the third boost of the pD3-M DNA vaccine were analyzed with the ELISPOT assay. The results of the ELISPOT assay showed that Mn2 and Md3 could induce high IFN-γ production by specific CD8+ T cells in freshly isolated splenocytes of transgenic mice (Figure 2B). These results suggested that peptides Mn2 and Md3 could be naturally processed in vivo and induce specific CD8+ T cell immune responses (presumably mainly CTLs).
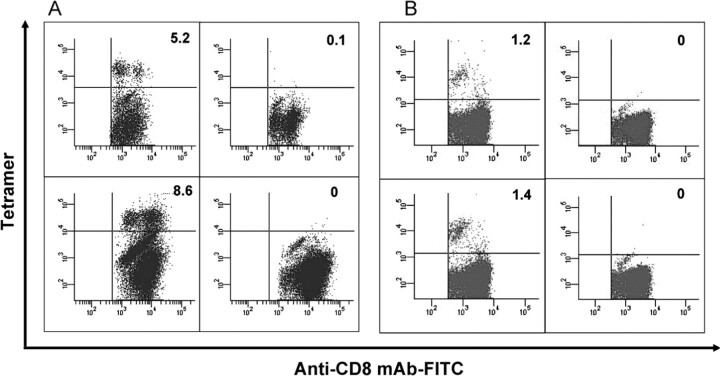
Presence of peptide-specific CD8+ T cells in PBMCs obtained from donors recovered from SARS infection. In the ELISPOT assays, peptides Mn2 and Md3 elicited high IFN-γ release from specific T cells in PBMCs obtained from recovered donors (Figure 4). Peptide Mn2 pulsed a specific T cell response by IFN-γ production, with a mean number of 34.8±19.7 spot-forming cells per 105 PBMCs. Likewise, for peptide Md3, the mean number is 44.1 ± 10.8 cells per 105 PBMCs. Peptide P15 (KLPDDFMGCV) from the SARS S protein, the positive control in parallel assays, provoked similar specific CD8+ T cell responses, with a statistical number of 38.0 ± 14.1 spot-forming cells per 105 PBMCs. Such T cell responses were not observed in the PBMCs donated from the control patients. Tetramers of peptides Mn2 and Md3 were also prepared to stain CD8+ T lymphocytes bearing the peptide-specific T cell receptors (TCRs). Through the peptide-specific tetramer staining and subsequent flow cytometry analysis, the PBMCs obtained from recovered donors possessed CD8+ T lymphocytes specific for peptides Mn2 and Md3 after 20–22 months from the onset of the disease. These 2 peptide tetramers showed 1.2% and 1.4% of specific T cells in the CD8+ T lymphocytes, respectively, from the PBMCs of recovered donors (Figure 3B). However, no peptide-specific CD8+ T cells were excavated from the PBMCs of the 3 healthy donors.
Figure 4.
Frequency of Mn2- and Md3-specific CD8+ T cells. A, In the tetramer staining, the splenocytes from mice inoculated with peptides were cultured in vitro for 10 days. Next, the cells were stained with phycoerythrin (PE)-labeled HLA-A*0201/peptide chimeric tetramer together with fluorescein isothiocyanate (FITC)-labeled anti-mouse CD8 antibody. Top left: Mn2-inoculated mice stained with Mn2 tetramer. Top right: Phosphatebuffered saline (PBS)-inoculated mice stained with Mn2 tetramer. Bottom left: Md3-inoculated mice stained with Md3 tetramer. Bottom right: PBS-inoculated mice stained with Md3 tetramer. The results are representatives of 3 independent experiments. B, Mn2-specific (top left) and Md3-specific (bottom left) CD8+ T cells were detected from the thawed peripheral blood mononuclear cells (PBMCs) of HLA-A2+ donors recovered from severe acute respiratory syndrome, by staining with fluorescein isothiocyanate-labeled anti-CD8 mAb along with PE-labeled HLA-A*0201/peptide tetramers. No specific CD8+ T cells were stained with Mn2 (top right) or Md3 tetramers among HLA-A2+ healthy donors (bottom right).
Structural evidence of peptide presentation of both Mn2 and Md3 by HLA-A*0201/ microglobulin. To further confirm that Mn2 and Md3 are typical HLA-A2-restricted epitopes, the crystal structures of peptides Mn2 and Md3 bound to HLA-A2 were solved at 2.2 and 2.8 Å resolutions, respectively. The statistics of the complex data are shown in Table 2.
Table 2.
This table is available in its entirety in the online version of Journal of Infectious Diseases.
For both complexes, unambiguous electron density is observed for the bound peptides, which are well defined inside the peptide-binding grooves of HLA-A*0201 (Figure 5A, 5B, 5C, and 5D). The residues in position 2 from the N-terminus and the C-terminal are similar for both peptides, with L2 deeply buried in the B pocket and V9 (for Mn2) or V10 (for Md3) in the F pocket, indicating typical anchor residues [42]. Residues in positions 3 and 6 are commonly secondary anchors in some HLA-A*0201 complexes. For position 3, M3 of peptide Mn2 inserts into the D pocket, and for position 6, S6 of peptide Mn2 and L7 of peptide Md3 point their side chains into pocket C. All of these anchor residues help the peptides tightly bind the HLA-A*0201 heavy chain and enhance the stability of the entire complex. Amino acids W4, L5, Y7, F8 of Mn2, and C4 and V6 of Md3 protrude their side chains away from the HLA-A* 0201 surface and might be involved in the TCR docking (Figure 5C, 5D, and 5F). Like other decameric HLA-A*0201 restricted epitopes, the bulge conformation of Md3 is also observed to be drastically different from the extended conformation of nonamer Mn2. This is especially apparent around the central residues of the peptides of the main chains. Md3 is raised up ∼2.25 Å (distance between carbonyl carbon atom of residues in position 5: L5 of Mn2 and V6 of Md3) compared with Mn2 (Figure 5E). These conformational differences between Mn2 and Md3 might partially contribute to the variances in the T cell recognition of these peptides, which lead to the immunogenic diversity of the peptides.
Figure 5.
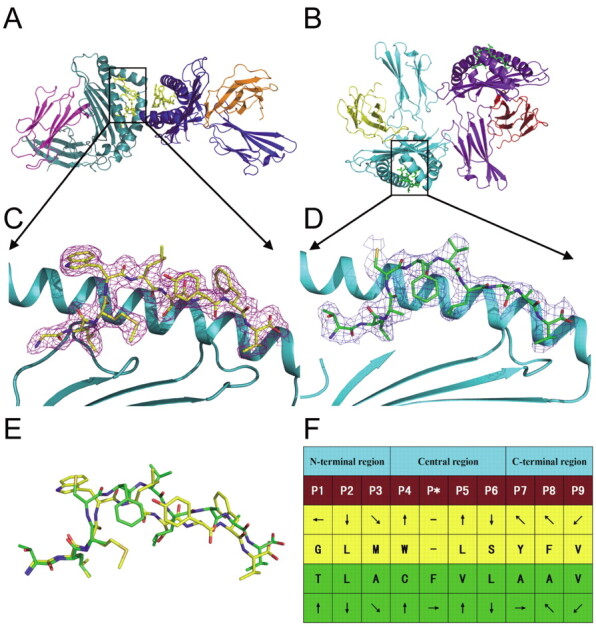
Structures of HLA-A*0201 complexed with Mn2 (GLMWLSYFV) and Md3 (TLACFVLAAV). Structural overviews of HLA-A2 with peptides Mn2 (A) and Md3 (B) demonstrate that both have 2 molecules in 1 asymmetric unit. 2Fo-Fc electron density for Mn2 (C) and Md3 (D) peptides are shown as violet and blue wire (contoured at 1.0 δ) viewed in profile through the HLA-A2 α2 helix. The carbon atoms are shown as yellow and green for Mn2 and Md3, respectively. E, Comparison of Mn2 (yellow) and Md3 (green) peptide structures by superimposition of the second residues from the N-terminal residues (Leu) and the C-terminal residues (Val). F, General side chain orientation for the 2 peptides as viewed in profile from the peptide N terminus toward the C terminus (up is toward the T cell receptor; down is toward the floor of the peptide binding groove; left is toward the a1 helix domain; right is toward the α2 helix domain). The figure was generated using the PyMOL program (http://www.pymol.org/).
Discussion
Clearance of viruses from infected patients depends not only on neutralizing antibodies but also on the optimal CTLs. The SARS-CoV M protein has been proven to induce strong neutralizing antibodies [30]. As most parts of the M protein are located inside the cell or virion and are triply embedded in the cellular or viral membrane, the M protein is most likely a good target for the CD8+ T cells because it is treated as an internal gene. Therefore, CTL epitopes might be found in theMprotein, as well as in the E protein (which may also be considered an internal gene). In this study, 2 CTL epitopes restricted by HLAA* 0201 have been identified from the M protein, using an optimized rational strategy covering functional and structural approaches. Epitope candidates were initially selected through online motif prediction on the basis of computer algorithms and then tested with in vitro refolding, T2 cell binding, and animal vaccination. Finally, these 2 defined Mn2 and Md3 epitopes, presented by HLA-A*0201 as a tetramer, can be recognized by specific CD8+ T cells in the PBMCs obtained from donors recovered from SARS infection 20 months after the initial onset of the disease.
In addition to all these functional studies, we determined the 3-dimensional structures of the peptide-HLA complexes containing Mn2 and Md3. As shown in the peptide-HLA struc tures, Mn2 and Md3 lie in the grooves formed by HLA heavy chains in a similar conformation, with 2 anchor residues inserting deep into the binding pockets, as seen in other typical HLA-A*0201-restricted epitopes. The analysis of the 2 structures not only confirms that Mn2 and Md3 possess structural properties typical of HLA-A*0201-restricted epitopes but also determines that they are minimal stimulatory peptides, because both use the C-terminal residue to anchor in pocket F of the heavy chain.
Furthermore, we have analyzed the immunogenic features of a panel of Mn2 and Md3 derived peptides (which means 1 amino acid deletion or addition around the defined epitopes) to see whether these regions are T cell epitope hot spots (unpublished data). Indeed, they could induce CD8+ T cell reaction from the transgenic mice inoculated with pD3-M. The crystal structure of the HLA-A2 complexed to peptide Md3-C9 (LACFVLAAV), equivalent to 9 residues in the C-terminal of Md3, revealed a distinct conformation of the peptide, which may demonstrate that this peptide acts as a new epitope in a different way with Md3 (nonamer versus decamer epitopes). All of these assays may reveal that the transmembrane domain of M protein contains a T cell epitope cluster that contributes significantly to the M protein-specific cellular immunity.
In general, protective memory immunity could be detected for a long period after the onset of viral diseases. Unfortunately, it has been reported that the geometric mean reciprocal titers of SARS-CoV-specific immunoglobulin G and neutralizing antibodies dropped significantly to a very low level in the 30th month after infection and were even undetectable in some proportion of the samples [46]. However, studies of the cellular immune response demonstrated that memory CD8+ T cells could be detectable from recovered donors years after the onset of the disease [21, 24, 47]. Long-term existence of cellular immunity was also observed in the studies of the SARS-CoV N protein-specific immunity [47]. In this study, CD8+ T cells against the M protein (as shown with peptides Mn2 and Md3) and against the S protein (as shown with peptide P15) [34] still existed in the PBMCs of recovered donors 20 months after infection.
In conclusion, this study has successfully identified a novel and defined CTL epitope clustering region containing 2 novel HLA-A*0201-restricted, immunogenic CTL epitopes from the M protein (Figure 6). Taking the earlier reports into account that the M protein induces strong neutralizing antibodies [18, 30, 48], we propose that the M protein could be a good candidate antigen for a prophylactic vaccine inducing both dominant cellular and humoral immunogenicity. Our current and previous studies [34, 49] also indicate that the combination of bioinformatics, cell biology, mouse model, and structural biology is a good method to evaluate the CTL epitope-specific immune response.
Figure 6.
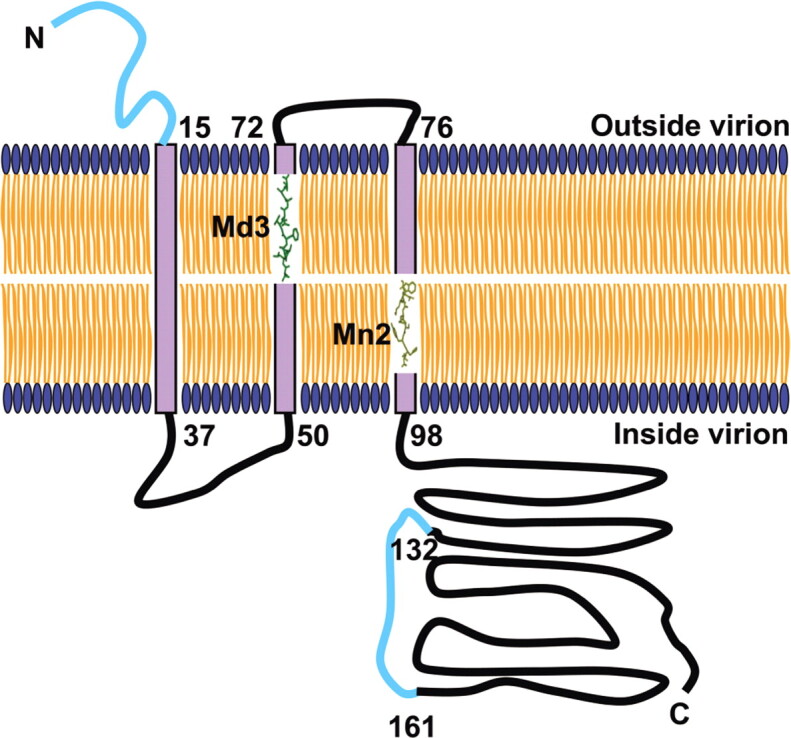
Schematic diagram of severe acute respiratory syndrome coronavirus (SARS-CoV) membrane (M) protein on the virion envelope, addressing the immune epitopes. The M protein comprises a domain spanning a triple membrane of ∼80 amino acid residues, a short Nterminus protruding out of the membrane, and a long cytoplasmic Cterminus. Md3 (green) and Mn2 (yellow) are completely included in the second and the third transmembrane domain, respectively. The identified B cell epitopes that could be recognized by antibodies are also represented in the diagram (cyan). The predicted structure of the M protein is based on previous studies [48].
Acknowledgements
We thank Dr Christopher J. Vavricka, Jr, and Ms Zhenying Liu from the Institute of Microbiology, Chinese Academy of Sciences, for their excellent suggestions and technical assistance.
Footnotes
Potential conflicts of interest: none reported.
Financial support: This study is supported by the China National Grand S and T Special Project (grants 2009ZX10004–201 and 2009ZX10004–305) and the Key International Science and Technology Cooperation Projects (grants 2006DFB32010 and 2007DFC30240). The China-Japan Joint Laboratory of Molecular Immunology and Molecular Microbiology is supported in part by the Japan Ministry of Education, Culture, Sports, Science, and Technology and the International Science and Technology Cooperation Projects of the Chinese Academy of Sciences (grant GJHZ0619). J.L. is partially supported for this project by the Doctoral Candidate Innovation Research Support Program by Science and Technology Review (grant kjdb20090102-4). G.F.G. is a Distinguished Young Investigator of the National Natural Science Foundation of China (award 30525010).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schlagenhauf P, Ashraf H. Severe acute respiratory syndrome spreads worldwide. Lancet. 2003;361:1017. doi: 10.1016/S0140-6736(03)12843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Chan KS, Zheng JP, Mok YW, et al. SARS: prognosis, outcome and sequelae. Respirology. 2003;8(Suppl):S36–S40. doi: 10.1046/j.1440-1843.2003.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly CA, Ghani AC, Leung GM, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riley S, Fraser C, Donnelly CA, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 7.Wong KT, Antonio GE, Hui DS, et al. Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology. 2003;228:401–406. doi: 10.1148/radiol.2282030593. [DOI] [PubMed] [Google Scholar]
- 8.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Gao GF. Towards our understanding of SARS-CoV, an emerging and devastating but quickly conquered virus. Comp Immunol Microbiol Infect Dis. 2007;30:309–327. doi: 10.1016/j.cimid.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Feng Y, Liu D, Gao GF. Avian influenza virus, Streptococcus suis serotype 2, severe acute respiratory syndrome-coronavirus and beyond: molecular epidemiology, ecology and the situation in China. Philos Trans R Soc Lond B Biol Sci. 2009;364:2725–2737. doi: 10.1098/rstb.2009.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 12.Ren W, Li W, Yu M, et al. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J Gen Virol. 2006;87:3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- 13.Hon CC, Lam TY, Shi ZL, et al. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu D, Tu C, Xin C, et al. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J Virol. 2005;79:2620–2625. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu RH, He JF, Evans MR, et al. Epidemiologic clues to SARS origin in China. Emerg Infect Dis. 2004;10:1030–1037. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Zhou Y, Siddiqui P, Niu J, Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J Clin Microbiol. 2005;43:3718–3726. doi: 10.1128/JCM.43.8.3718-3726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchholz UJ, Bukreyev A, Yang L, et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li CK, Wu H, Yan H, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YD, Sin WY, Xu GB, et al. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YH, Huang YH, Chuang YH, et al. Autoantibodies against human epithelial cells and endothelial cells after severe acute respiratory syndrome (SARS)-associated coronavirus infection. J Med Virol. 2005;77:1–7. doi: 10.1002/jmv.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Wo J, Shao J, et al. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J Clin Virol. 2003;28:239–244. doi: 10.1016/S1386-6532(03)00195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Hou J, Jiang X, et al. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol. 2005;175:591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- 25.Yang LT, Peng H, Zhu ZL, et al. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Peng H, Zhu Z, et al. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J Gen Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng H, Yang LT, Li J, et al. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8:2424–2431. doi: 10.1016/j.micinf.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Li J, Heck S, Lustigman S, Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J Virol. 2006;80:5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang H, Liu Y, Han X, et al. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J Gen Virol. 2004;85:3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- 31.Pervushin K, Tan E, Parthasarathy K, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeDiego ML, Alvarez E, Almazan F, et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeDiego ML, Pewe L, Alvarez E, Rejas MT, Perlman S, Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou M, Xu D, Li X, et al. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 35.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 36.Gotch F, McMichael A, Smith G, Moss B. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J Exp Med. 1987;165:408–416. doi: 10.1084/jem.165.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu F, Lou Z, Chen YW, et al. First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape. J Immunol. 2007;178:944–952. doi: 10.4049/jimmunol.178.2.944. [DOI] [PubMed] [Google Scholar]
- 38.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Zhou M, Han J, et al. Generation of murine CTL by a hepatitis B virus-specific peptide and evaluation of the adjuvant effect of heat shock protein glycoprotein 96 and its terminal fragments. J Immunol. 2005;174:195–204. doi: 10.4049/jimmunol.174.1.195. [DOI] [PubMed] [Google Scholar]
- 42.Sliz P, Michielin O, Cerottini JC, et al. Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes. J Immunol. 2001;167:3276–3284. doi: 10.4049/jimmunol.167.6.3276. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 46.Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 47.Li T, Xie J, He Y, et al. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLoS ONE. 2006;1:e24. doi: 10.1371/journal.pone.0000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y, Wen J, Tang L, et al. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteomics Bioinformatics. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Liu J, Yang M, et al. Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses. J Gen Virol. 2010;91(4):919–930. doi: 10.1099/vir.0.016766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]