Abstract
Coronavirus disease‐2019 (COVID‐19) is a global pandemic and caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which has resulted in millions of deaths worldwide. Reports denote SARS‐CoV‐2 uses angiotensin‐converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2) as its primary entry point into the host cell. However, understanding the biology behind this viral replication, disease mechanism and drug discovery efforts are limited due to the lack of a suitable experimental model. Here, we used single‐cell RNA sequencing data of human organoids to analyze expressions of ACE2 and TMPRSS2, in addition to an array of RNA receptors to examine their role in SARS‐CoV‐2 pathogenesis. ACE2 is abundant in all organoids, except the prostate and brain, and TMPRSS2 is omnipresent. Innate immune pathways are upregulated in ACE2(+) cells of all organoids, except the lungs. Besides this, the expression of low‐density lipoprotein receptor is highly enriched in ACE2(+) cells in intestinal, lung, and retinal organoids, with the highest expression in lung organoids. Collectively, this study demonstrates that the organoids can be used as an experimental platform to explore this novel virus disease mechanism and for drug development.
Keywords: ACE2, COVID‐19, human organoids, SARS‐CoV‐2, scRNA sequencing, TMPRSS2
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) receptors, angiotensin‐converting enzyme 2 (ACE2), and transmembrane serine protease 2 (TMPRSS2) are expressed in different cell clusters of various human organoids.
Human organoids will be a suitable model for rapid translation SARS‐CoV‐2 research.
1. INTRODUCTION
An important obstacle in the development of therapies against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the lack of viable in vivo models of disease that are needed to recapitulate the infectivity and clinical features of coronavirus disease 2019 (COVID‐19), the disease manifestation of SARS‐CoV‐2. A recent review has outlined the current animal models utilized in SARS‐CoV‐2 research (Cleary et al., 2020). At present, there are limited rodent models that replicate features of COVID‐19. The only experimental models available involve the mouse‐adapted SARS‐Co‐V virus strain that replicates features of SARS and is highly lethal (Day et al., 2009), and transgenic mice expressing the human angiotensin‐converting enzyme 2 (ACE2) that can be infected successfully with SARS‐CoV‐2 (Bao et al., 2020). However, the use of both models requires BSL‐3 or BSL‐4 animal husbandry environments, which are not readily available in many animal facilities. This underscores the need to develop novel approaches for drug testing/discovery in SARS‐CoV‐2.
Organoids are artificially developed three‐dimensional (3D) cultured structures or miniature organs (derived from stem cells or organ progenitors) encompassing several cells of specific organ systems (Eiraku et al., 2008; Sasai, 2013; Takahashi, 2019; Trinkaus & Groves, 1955). Organoids provide two distinct advantages over other experimental models, they develop and expand in their destined lineage, thereby mimicking the in vivo niche (Lancaster & Knoblich, 2014; Mahe et al., 2013), secondly, tissue‐derived human organoids maintain their in vivo genetic and physiological characteristics, without any significant alterations for many generations (DiMarco et al., 2014; Grabinger et al., 2014). Recently, the use of multi‐cell type, organoid coculture models with stromal and immune cells have been developed to recapitulate the complex in vivo niche for developing disease models, preclinical testing, and biomarker discoveries (Shamir & Ewald, 2014; Tsai et al., 2018). Indeed, organoids have outplayed conventional animal models and cell culture systems as experimental models (Jensen & Teng, 2020; Mead & Karp, 2019) and can serve greatly in the current COVID‐19 pandemic, as efficient experimental models for SARS‐CoV‐2 drugs. Moreover, organoids as experimental models will also serve to gain better insights into the molecular mechanisms and/or role of cellular targets like receptors or transporters by which the pathogenesis of life‐threatening SARS‐CoV‐2 is mediated (Clevers, 2020; Monteil et al., 2020).
The co‐morbid conditions of COVID‐19, including cardiac, renal, central nervous system (CNS), prostate, and gastrointestinal dysfunctions cause significant mortality globally (Zaim, Chong, Sankaranarayanan, & Harky, 2020). Amid the lack of effective preventive and remedial measures against the impending threat of COVID‐19; studies on its pathogenic mechanisms and drug target identification are greatly warranted (Harrison, 2020; Shi et al., 2020). SARS‐CoV‐2 uses angiotensin‐converting enzyme 2 (ACE2) receptor for host cell entry and the transmembrane serine protease 2 (TMPRSS2) for S protein priming (Hoffmann et al., 2020). In this study, we have analyzed the single‐cell RNA (scRNA) sequencing data to show the differential expression patterns of ACE2 and TMPRSS2 receptors in specific tissue organoids including the intestine, prostate, kidney, brain, retina, and lung, the major sites of COVID‐19 pathogenesis (Fujii et al., 2018; McCray, Moline, Baumann, Vander Griend, & Nonn, 2019; Miller et al., 2020; Sridhar et al., 2020; Velasco et al., 2019; Wu et al., 2018). We believe that the results obtained from the scRNA sequencing of various organoids will aid in understanding the spatiotemporal effect of new SARS‐CoV‐2 antiviral drugs (using organoids as experimental models) in regulating disease pathogenesis.
2. METHODS
2.1. Data sets
All the gene expression data either in the normalized count or raw counts were downloaded from the Gene expression Omnibus or Array Express database. There were eight organoids data sets used in this study: intestine conventional and improved condition (GSE119969), kidney Morizane and Takasato methods (GSE118184), prostate (GSE130318), lung (E‐MTAB‐8221), retina (GSM4231315), and brain (GSE129519).
2.2. Data analysis
The gene expression matrix or raw count matrix was analyzed using Seurat v3.0 (Stuart et al., 2019). The following criteria were used for filtering the cells for the clustering analysis of each sample separately; genes that were seen in at least three cells, cells should express 100 genes and the mitochondrial gene expression less than 20%. The normalized data function was used for the normalization with the default parameter. The variation across the cells was regressed out using the ScaleData function with default settings. For clustering analysis, 2000 highly variable genes were selected using the FindVariableGenes function and the expression matrix was centered and scaled. Next, the principal component analysis was applied to generate 100 principal components (PCs) and the JackStraw function was used to select the significant PCs to be used for further clustering and dimensionality reduction. To identify clusters of transcriptionally similar cells, unsupervised clustering was employed using the FindClusters function with the k. param set to 10 and the resolution set to 0.5. For dimensionality reduction, we used a Uniform Manifold Approximation and Projection (UMAP) method employed in Seurat. To identify the differential expression analysis between ACE2(+) and negative cells, the cells were grouped based on the expression of ACE2 and labeled as ACE2(+) cells when the read counts were greater than 0, similarly, the counts were equal to 0 and labeled as ACE2(−) cells. Then, differential expression analysis was performed in Seurat using a Wilcoxon test and Bonferroni p‐value correction. There were no ACE2 cells were observed in the prostate and brain organoids data set. The cell types in clustering for each sample were identified based on known and reported markers (Figure S1).
2.3. Enrichment analysis
The upregulation genes from ACE2(+) cells compared to the ACE2(−) cells were used as the input in the Enrichr (Chen et al., 2013) server for the enrichment analysis. From the gene ontology (GO) biological process, significant immune and viral related pathways were selected and plotted. For the Lung data set, there were no immune and viral related pathways observed.
3. RESULTS
3.1. Identification of cell types in various organoids
The scRNA sequencing raw count data or normalized gene expression data of intestine (conventional and improved methods), Kidney (Morizane and Takasato methods), prostate, lung, retina, and brain were analyzed by clustering and dimension reduction using the Seurat package. There were 71 cell clusters identified from these organoids and these cell clusters were annotated based on the respective published articles (Figure S1; Fujii et al., 2018; McCray et al., 2019; Miller et al., 2020; Sridhar et al., 2020; Velasco et al., 2019; Wu et al., 2018). In the intestinal organoid, five cell types were identified in the conventional organoid (goblet, transamplifying 1, enterocyte, stem cell, and transamplifying 2) while seven cell types were identified in the intestine organoid improved (stem cell, transamplifying 1, enterocyte, goblet, enteroendocrine, M, and transamplifying 2). Prostate organoids are androgen‐sensitive, and they possess basal and luminal cells arranged as a pseudostratified epithelium in their organ system. In the prostate organoid, we have identified six cell types (luminal, basal, dividing cells, arrested cells, stem cells, and unidentified cells). In the Morizane kidney organoid, we have identified 13 cell types (mesenchyme 1, proximal tubule 1, podocyte 1, mesenchyme 2, podocyte 2, podocyte 3, proximal tubule 2, mesenchyme 3, neural 1, neural 3, neural 2, muscle cells, and endothelial cells). In the Takasato kidney organoid, 13 cell types were identified including (proximal tubule 1, neural cells 3, mesenchyme 3, proximal tubule 3, mesenchyme 1, proximal tubule 2, neural progenitors, podocytes, neural 1, melanocytes (Me), mesenchyme 2, neural 2, and endothelial cells), and in the lung organoid, eight cell types (bud tip progenitor cells, bud tip adjacent cells, basal cells, multiciliated cells, secretory progenitor cells, neuroendocrine cells, submucosal gland basal cells) were identified. In the brain organoid, we have identified 9 cell types (immature PNs, corticofugal peripheral neurons [PNs], immature colossal PNs, and outer radial glia, callosal PNs, Radial glia, immature corticofugal PNs, IPCs/immature PNs, cycling progenitors). In the retinal organoid, we have identified 10 cell types (cone, T3, retinal ganglion, progenitor/mature glial cells, Müller glial cells, Amacrine cells, rod cells, bipolar cells [BP], progenitors, and horizontal cells [HCs]).
3.2. Expression pattern of ACE2 and TMPRSS2 in eight human organoids
The viral spike protein from SARS‐CoV‐2 binds to ACE2 as a host cell receptor for entry and it is triggered by TMPRSS2. Hence, we analyzed the expression patterns of ACE2 and TMPRSS2 in these organoids (Figure 1). In the intestinal organoid (conventional and improved), ACE2 expression is specifically enriched in the enterocytes, and to some extent, in the goblet and stem cells, whereas TMPRSS2 expression is observed in almost all the cell types. In the kidney organoids (Morizane and Takasato), both ACE2 and TMPRSS2 were observed specifically in the proximal tubule cells. In the retina, ACE2 expression is limited to very few cells of progenitor glia and rod; TMPRSS2 was seen in muller glia, rod, progenitor glia, progenitors, T3, cone, and bipolar cell clusters. In the lung organoids, a few cells of bud tip progenitor cells, multiciliated cells, and secretory progenitor cells showed the expression of ACE2 and almost all the clusters except submucosal gland basal cells showed TMPRSS2 expression. In the prostate and brain organoids, there was no ACE2 expression seen, and TMPRSS2 expression was observed in almost all the cell types. The expression of genes in scRNA sequencing is influenced by the organoid growing methods, cell dissociation, sequencing method, and sequencing depth. Therefore, the lack of ACE2 expression in the brain and prostate may be due to any one or a combination of those factors. A recent study reported on the role of TMPRSS4 in virus entry into the host cells (Zang et al., 2020), therefore we checked the expression profile of TMPRSS4 across organoids (Figure S2). All the ACE2 and TMPRSS2 positive cell types showed the expression of TMPRSS4 except the retinal organoid, where there was no expression observed.
Figure 1.
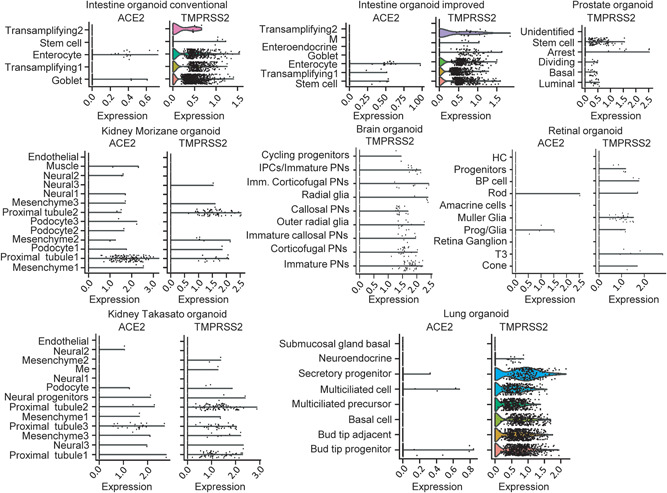
The expression profiles of ACE2 and TMPRSS2 in various organoids. The violin plot shows the ACE2 and TMPRSS2 expression in intestine, prostate, kidney, brain, retinal, and lung organoids. There is no ACE2 transcript observed in prostate and brain organoids. ACE2, angiotensin‐converting enzyme 2; TMPRSS2, transmembrane serine protease 2
3.3. The expression profile of curated single‐stranded RNA receptors (scRNA) in ACE2(+) positive and ACE2(−) negative cells in various organoids
A recent study showed that ACE2 expression is correlated with other scRNA receptors such as ANPEP, ENPEP, and DPP4 (Qi, Qian, Zhang, & Zhang, 2020). Therefore, we checked the curated scRNA receptors expression pattern from ACE2(+) positive and ACE2(−) negative cells in various organoids (Figure 2). The ACE2(+) cells of conventional and improved intestine organoids showed the expression of CXADR, PVR, CD55, ITGB6, EGFR, OCLN, LDLR, CEACAM1, ANPEP, and TFRC along with the ACE2. Similarly, in the kidney organoids (Morizane and Takasato), the expression of PHB, CXCADR, ITGB3, CD46, CLDN1, MERTK, ANPEP, DPP4, and AXL showed consistent expression along with ACE2. In the lung organoid, ACE2(+) cells showed the expression of PHB, NCAM1, CDHR3, ITGB8, ITGA5, ITGB1, CD55, ICAM1, SCARB2, ITGB6, EFNB2, EGFR, ITGAV, OCLN, LDLR, CD81, CEACAM1, LAMP1, TYRO3, and DAG1. The ACE2(+) cells of the retinal organoid showed the expression of PHB, VCAM1, ITGB1, SCARB2, ITGB6, EFNB2, ANXA5, LDLR, CD81, and LAMP1. The receptor LDLR is consistently expressed along with ACE2 in all the organoids except the kidney, suggesting that LDLR may also play a significant role in COVID‐19 transmission.
Figure 2.
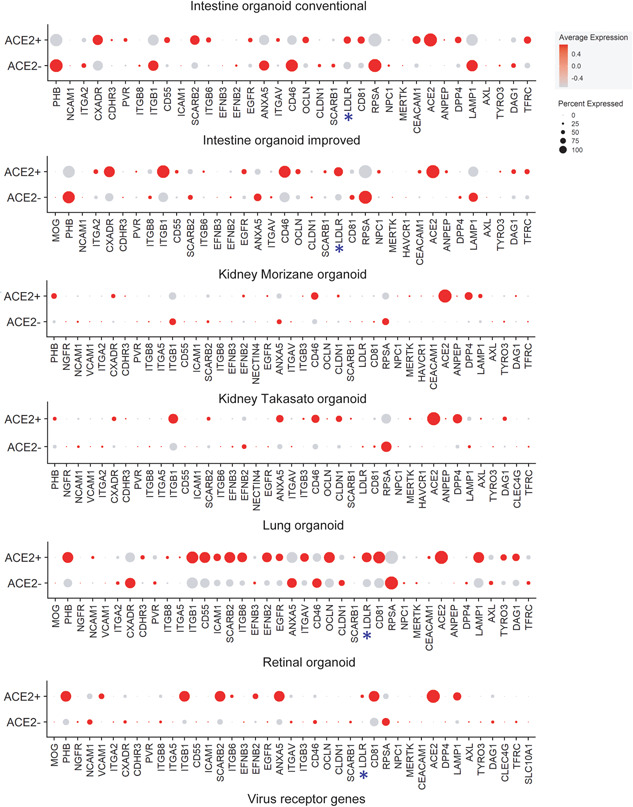
The expression profile of curated scRNA receptors in ACE2(+) and ACE2(−) negative cells in various organoids. The dot plot shows the expression of multiple scRNA receptors transcript in the intestine, kidney, lung, and retinal organoids. ACE2, angiotensin‐converting enzyme 2; TMPRSS2, transmembrane serine protease 2
3.4. Immune specific enrichment profile for ACE2(+) and ACE2(−) cells in various organoids
To further characterize the ACE2(+) cells, differential gene expression analysis was performed between ACE2(+) and ACE2(−) cells, and the pathway enrichment profiles specific to immune‐related pathways were identified for the ACE2(+) cell upregulated genes (Figure 3). Except for lung organoids, all showed that the cytokine and neutrophil related pathways are enriched in ACE2(+) cells, which suggests that these cells from organoids can trigger a strong immune response after the virus entry. The recent analysis of the RNA‐seq data from COVID‐19 patients showed the expression of neutrophil and cytokine/chemokines genes (Didangelos, 2020). Furthermore, the pathway analysis indicated neutrophil response and inflammation response are enriched. These suggest that the neutrophils could play a role in COVID‐19 inflammation.
Figure 3.
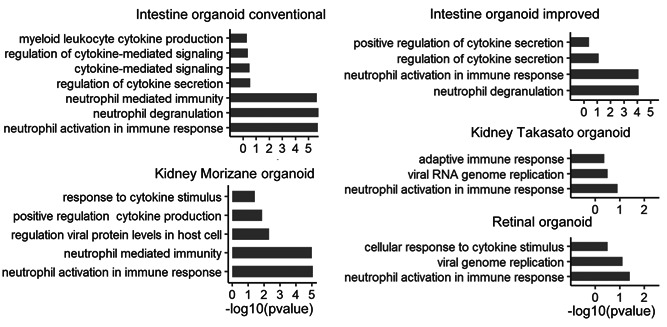
Immune specific enrichment profile for ACE2(+) and ACE2(−) negative cells in various organoids. The upregulation genes in the ACE2(+) cells are used for GO biological process enrichment analysis with specific immune‐related pathways in the intestine, kidney, and retinal organoids. ACE2, angiotensin‐converting enzyme 2; GO, gene ontology
4. DISCUSSION
Given the fact that SARS‐CoV‐2 mediates multiorgan dysfunction, the underlying pathogenic mechanisms might be orchestrated via diverse in‐cell mediators specific for each organ. In such instances, the requirement for experimental models exhibiting key features as in in vivo is crucial in understanding the pathogenic mechanism and developing suitable/personalized drug candidates. Organoids exhibiting key features of the tissue of origin, will greatly serve to delineate in‐cell mediators of SARS‐CoV‐2 pathologies, and testing efficacy of SARS‐CoV‐2 drugs (Maenhoudt et al., 2020), thereby enabling rapid translation research and clinical applications (Clevers, 2016). In this study, we analyzed the expression of ACE2, TMPRSS2, and other scRNA receptors in ACE2(+) and ACE2(−) cells in eight different organoids. To the best of our knowledge, this is the first study investigating ACE2 expression across a wide variety of organoids. Although there are several studies (Peng et al., 2020; Tai et al., 2020; Zhang et al., 2020) reporting about COVID‐19 virus transmission, tissue tropism, and pathogenic mechanism, many questions cannot be answered due to the lack of suitable research models. The organoids are a suitable model to explore the study of host‐microbe interaction (Dutta & Clevers, 2017) because they retain the biology of individual tissues. The 3D organoid structures can be developed from either induced pluripotent stem cells or multipotent adult stem cells (Lancaster & Knoblich, 2014). The advantage of organoid models is that they resemble the organ through self‐organizing multiple differentiated cell types that reflect the true morphology of the tissues. Our clustering results (Figure S1) indicate that the eight different organoids captured the respective tissue cell types including stem cells or basal cells.
The organoids have been shown before as a suitable system to explore virus replication (Ettayebi et al., 2016) as well as the mechanism of pathogenesis (Platt & Miner, 2017; Tang et al., 2016). The outbreak of COVID‐19 is causing significant mortality and morbidity across the globe. The virus transmits from person to person causing influenza‐like symptoms, mid to severe lung damage, affecting multiple organs including the gut, kidney, brain, eye, and liver (Huang et al., 2020; Wang et al., 2020; Zhu et al., 2020). Therefore, it is necessary to find the best model to study virus infection and mechanism. As the organoids resemble real tissue and organs in vitro, scientists have started to use this organoid model to examine the effect of the virus. Monteil et al. (2020) showed that the virus can infect kidney organoids derived from induced pluripotent stem cells, and another study (Lamers et al., 2020) has used the human adult stem cell‐derived intestinal organoids to study virus infection and replication. Furthermore, Lamers et al showed that the infection of SARS‐CoV‐2 triggered the broad expression of cytokines and interferon‐stimulated genes related to type I and III interferon responses. In our enrichment analysis, immune‐related genes are already present in the ACE2(+) cells in the organoids, which can be further triggered after the infection.
Here, we have demonstrated that the expression of ACE2 is seen in the enterocytes cell population in both conventional and improved intestine organoids, and in the proximal tubule cells in the kidney organoids. The other organoids such as retina and lung also show the expression of ACE2, suggesting the suitability of these organoids to study the virus infection. However, prostate and brain organoids do not show the expression of the ACE2. There are several scRNA receptors showing expression along with ACE2 in the organoids. Interestingly, the receptor LDLR shows consistent expression in ACE2(+) cells of all the organoids except the kidney, which suggests that LDLR may play a role in virus transmission and infection.
The organoids are developed by different methods, and it is important to ensure that those organoids depict multiple cell types of respective tissue. It is also necessary to examine the transcriptome profile to understand the suitability of these organoids for SARS‐CoV‐2 infection study. The organoids can be also used to identify the potential drug molecules, which may be used for COVID‐19 therapies. A recent study demonstrated that intestine (Krüger et al., 2020) and lung organoids (Suzuki et al., 2020) are suitable to explore the viral life cycle as well as the efficacy of antiviral drugs. This opens a new door for COVID‐19 drug discovery.
Despite the presence of multi‐factorial etiologies and an array of receptor involvements in SARS‐CoV‐2 pathologies, the requirement of personalized therapy might be warranted for protecting specific organ systems in SARS‐CoV‐2 patients. Hence, the need for efficient and rapid experimental models against diverse cellular targets in specific organ systems is highly warranted. Recently, organoid experimental models have been widely utilized in conditions like regenerative medicine, cancer personalized therapy in renal diseases (Grassi et al., 2019), in ovarian cancer preclinical models (Maenhoudt et al., 2020) and in host‐pathogen interaction studies (Duque‐Correa, Maizels, Grencis, & Berriman, 2020). There are several high‐throughput methods developed to screen the drug in the organoids such as organ‐on‐a‐chip (Domansky et al., 2010), cancer‐on‐a‐chip (Zervantonakis et al., 2012) and 3D gut‐liver chip (Lee, Ha, Choi, & Sung, 2017). These technologies have advanced the usage of the organoids for drug development. Similar technology or modified methods can be adapted as an in vitro platform to screen either the existing FDA approved drugs or new molecules to effectively kill the virus. To conclude, we believe that organoids can be the best suitable experimental model in pursuit of the rapid requirement to study viral replication, disease mechanisms, and personalized drugs to each organ affected in SARS‐CoV‐2 patients.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Supplementary Figure 1: The cell types for each sample were identified and annotated from the know cell markers from the respective publication and the top differentially expressed genes in each cluster.
Supplementary Figure 2: The expression profile of TMPRSS4 in various organoids.
ACKNOWLEDGMENTS
This study was supported by grants from the American Heart Association (19TPA34880039 and 18IPA34170497 to Rajarajan A. Thandavarayan) and the Roswell and Ann Vaughan Fund to Guha Ashrith.
Mahalingam R, Dharmalingam P, Santhanam A, et al. Single‐cell RNA sequencing analysis of SARS‐CoV‐2 entry receptors in human organoids. J Cell Physiol. 2021;236:2950–2958. 10.1002/jcp.30054
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available. All the gene expression data either in the normalized count or raw counts were downloaded from the Gene expression Omnibus or Array Express database. There were eight organoids data sets used in this study: Intestine conventional and improved condition (GSE119969), Kidney Morizane and Takasato methods (GSE118184), prostate (GSE130318), lung (E‐MTAB‐8221), retina (GSM4231315), and brain (GSE129519).
REFERENCES
- Bao, L. , Deng, W. , Huang, B. , Gao, H. , Liu, J. , Ren, L. , … Qin, C (2020). The pathogenicity of SARS‐CoV‐2 in hACE2 transgenic mice. Nature, 583(7818), 830–833. 10.1038/s41586-020-2312-y [DOI] [PubMed] [Google Scholar]
- Chen, E. Y. , Tan, C. M. , Kou, Y. , Duan, Q. , Wang, Z. , Meirelles, G. V. , … Ma'ayan, A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, S. J. , Pitchford, S. C. , Amison, R. T. , Carrington, R. , Robaina Cabrera, C. L. , Magnen, M. , … Page, C. P. (2020). Animal models of mechanisms of SARS‐CoV‐2 infection and COVID‐19 pathology. British Journal of Pharmacology, bph.15143. 10.1111/bph.15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H. (2016). Modeling development and disease with organoids. Cell, 165(7), 1586–1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Clevers, H. (2020). COVID‐19: Organoids go viral. Nature Reviews Molecular Cell Biology, 21(7), 355–356. 10.1038/s41580-020-0258-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, C. W. , Baric, R. , Cai, S. X. , Frieman, M. , Kumaki, Y. , Morrey, J. D. , … Barnard, D. (2009). A new mouse‐adapted strain of SARS‐CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology, 395(2), 210–222. 10.1016/j.virol.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didangelos, A. (2020). COVID‐19 hyperinflammation: What about neutrophils? mSphere, 5(3), 10.1128/mSphere.00367-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMarco, R. L. , Su, J. , Yan, K. S. , Dewi, R. , Kuo, C. J. , & Heilshorn, S. C. (2014). Engineering of three‐dimensional microenvironments to promote contractile behavior in primary intestinal organoids. Integrative Biology, 6(2), 127–142. 10.1039/c3ib40188j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domansky, K. , Inman, W. , Serdy, J. , Dash, A. , Lim, M. H. , & Griffith, L. G. (2010). Perfused multiwell plate for 3D liver tissue engineering. Lab on a Chip, 10(1), 51–58. 10.1039/b913221j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque‐Correa, M. A. , Maizels, R. M. , Grencis, R. K. , & Berriman, M. (2020). Organoids ‐ New Models for Host‐Helminth Interactions. Trends in Parasitology, 36(2), 170–181. 10.1016/j.pt.2019.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, D. , & Clevers, H. (2017). Organoid culture systems to study host–pathogen interactions. Current Opinion in Immunology, 48, 15–22. 10.1016/j.coi.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku, M. , Watanabe, K. , Matsuo‐Takasaki, M. , Kawada, M. , Yonemura, S. , Matsumura, M. , … Sasai, Y. (2008). Self‐organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell, 3(5), 519–532. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Ettayebi, K. , Crawford, S. E. , Murakami, K. , Broughman, J. R. , Karandikar, U. , Tenge, V. R. , … Estes, M. K. (2016). Replication of human noroviruses in stem cell‐derived human enteroids. Science, 353(6306), 1387–1393. 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, M. , Matano, M. , Toshimitsu, K. , Takano, A. , Mikami, Y. , Nishikori, S. , … Sato, T. (2018). Human intestinal organoids maintain self‐renewal capacity and cellular diversity in niche‐inspired culture condition. Cell Stem Cell, 23(6), 787‐793), e786. 10.1016/j.stem.2018.11.016 [DOI] [PubMed] [Google Scholar]
- Grabinger, T. , Luks, L. , Kostadinova, F. , Zimberlin, C. , Medema, J. P. , Leist, M. , & Brunner, T. (2014). Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death & Disease, 5, e1228. 10.1038/cddis.2014.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi, L. , Alfonsi, R. , Francescangeli, F. , Signore, M. , De Angelis, M. L. , Addario, A. , … Bonci, D. (2019). Organoids as a new model for improving regenerative medicine and cancer personalized therapy in renal diseases. Cell Death & Disease, 10(3), 201. 10.1038/s41419-019-1453-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, C. (2020). Drug researchers pursue new lines of attack against COVID‐19. Nature Biotechnology, 38(6), 659–662. 10.1038/d41587-020-00013-z [DOI] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Kruger, N. , Herrler, T. , Erichsen, S. , … Pohlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280 e278. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, C. , & Teng, Y. (2020). Is it time to start transitioning from 2D to 3D cell culture? Frontiers in Molecular Biosciences, 7, 33. 10.3389/fmolb.2020.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger, J. , Groß, R. , Conzelmann, C. , Müller, J. , Koepke, L. , Sparrer, K. , … Münch, J. (2020). Remdesivir but not famotidine inhibits SARS‐CoV‐2 replication in human pluripotent stem cell‐derived intestinal organoids. In: bioRxiv. [DOI] [PMC free article] [PubMed]
- Lamers, M. M. , Beumer, J. , van der Vaart, J. , Knoops, K. , Puschhof, J. , Breugem, T. I. , … Clevers, H. (2020). SARS‐CoV‐2 productively infects human gut enterocytes. Science, 369(6499), 50–54. 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, M. A. , & Knoblich, J. A. (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science, 345(6194), 1247125. 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- Lee, D. W. , Ha, S. K. , Choi, I. , & Sung, J. H. (2017). 3D gut‐liver chip with a PK model for prediction of first‐pass metabolism. Biomedical Microdevices, 19(4), 100. 10.1007/s10544-017-0242-8 [DOI] [PubMed] [Google Scholar]
- Maenhoudt, N. , Defraye, C. , Boretto, M. , Jan, Z. , Heremans, R. , Boeckx, B. , … Vankelecom, H. (2020). Developing organoids from ovarian cancer as experimental and preclinical models. Stem Cell Reports, 14(4), 717–729. 10.1016/j.stemcr.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe, M. M. , Aihara, E. , Schumacher, M. A. , Zavros, Y. , Montrose, M. H. , Helmrath, M. A. , … Shroyer, N. F. (2013). Establishment of gastrointestinal epithelial organoids. Current Protocols in Mouse Biology, 3(4), 217–240. 10.1002/9780470942390.mo130179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray, T. , Moline, D. , Baumann, B. , Vander Griend, D. J. , & Nonn, L. (2019). Single‐cell RNA‐Seq analysis identifies a putative epithelial stem cell population in human primary prostate cells in monolayer and organoid culture conditions. American Journal of Clinical and Experimental Urology, 7(3), 123–138. [PMC free article] [PubMed] [Google Scholar]
- Mead, B. E. , & Karp, J. M. (2019). All models are wrong, but some organoids may be useful. Genome Biology, 20(1), 66. 10.1186/s13059-019-1677-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. J. , Yu, Q. , Czerwinski, M. , Tsai, Y. H. , Conway, R. F. , Wu, A. , … Spence, J. R. (2020). In vitro and in vivo development of the human airway at single‐cell resolution. Developmental Cell, 53(1), 117–128 e116. 10.1016/j.devcel.2020.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil, V. , Kwon, H. , Prado, P. , Hagelkruys, A. , Wimmer, R. A. , Stahl, M. , … Penninger, J. M. (2020). Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell, 181(4), 905–913 e907. 10.1016/j.cell.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Wan, L. , Fan, C. , Zhang, P. , Wang, X. , Sun, J. , … Wei, C. (2020). Cholesterol metabolism—Impacts on SARS‐CoV‐2 infection prognosis. medRxiv, 2020.2004.2016.20068528, 10.1101/2020.04.16.20068528 [DOI] [Google Scholar]
- Platt, D. J. , & Miner, J. J. (2017). Consequences of congenital Zika virus infection. Current Opinion in Virology, 27, 1–7. 10.1016/j.coviro.2017.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, F. , Qian, S. , Zhang, S. , & Zhang, Z. (2020). Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochemical and Biophysical Research Communications, 526(1), 135–140. 10.1016/j.bbrc.2020.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai, Y. (2013). Cytosystems dynamics in self‐organization of tissue architecture. Nature, 493(7432), 318–326. 10.1038/nature11859 [DOI] [PubMed] [Google Scholar]
- Shamir, E. R. , & Ewald, A. J. (2014). Three‐dimensional organotypic culture: experimental models of mammalian biology and disease. Nature Reviews Molecular Cell Biology, 15(10), 647–664. 10.1038/nrm3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Zhang, X. , Mu, K. , Peng, C. , Zhu, Z. , Wang, X. , … Zhu, W. (2020). D3Targets‐2019‐nCoV: A webserver for predicting drug targets and for multi‐target and multi‐site based virtual screening against COVID‐19. Acta Pharmaceutica Sinica B, 10, 1239–1248. 10.1016/j.apsb.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar, A. , Hoshino, A. , Finkbeiner, C. R. , Chitsazan, A. , Dai, L. , Haugan, A. K. , … Reh, T. A. (2020). Single‐cell transcriptomic comparison of human fetal retina, hPSC‐derived retinal organoids, and long‐term retinal cultures. Cell Reports, 30(5), 1644–1659 e1644. 10.1016/j.celrep.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T. , Butler, A. , Hoffman, P. , Hafemeister, C. , Papalexi, E. , Mauck, W. M. , … Satija, R. (2019). Comprehensive integration of single‐cell data. Cell, 177(7), 1888–1902. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T. , Itoh, Y. , Sakai, Y. , Saito, A. , Okuzaki, D. , Motooka, D. , … Takayama, K. (2020). Generation of human bronchial organoids for SARS‐CoV‐2 research. bioRxiv, 2020.2005.2025.115600, 10.1101/2020.05.25.115600 [DOI] [Google Scholar]
- Tai, W. , He, L. , Zhang, X. , Pu, J. , Voronin, D. , Jiang, S. , … Du, L. (2020). Characterization of the receptor‐binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & Molecular Immunology, 17(6), 613–620. 10.1038/s41423-020-0400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T. (2019). Organoids for drug discovery and personalized medicine. Annual Review of Pharmacology and Toxicology, 59, 447–462. 10.1146/annurev-pharmtox-010818-021108 [DOI] [PubMed] [Google Scholar]
- Tang, H. , Hammack, C. , Ogden, S. C. , Wen, Z. , Qian, X. , Li, Y. , … Ming, G. L. (2016). Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell, 18(5), 587–590. 10.1016/j.stem.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkaus, J. P. , & Groves, P. W. (1955). Differentiation in culture of mixed aggregates of dissociated tissue cells. Proceedings of the National Academy of Sciences of the United States of America, 41(10), 787–795. 10.1073/pnas.41.10.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S. , McOlash, L. , Palen, K. , Johnson, B. , Duris, C. , Yang, Q. , … James, M. A. (2018). Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer, 18(1), 335. 10.1186/s12885-018-4238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco, S. , Kedaigle, A. J. , Simmons, S. K. , Nash, A. , Rocha, M. , Quadrato, G. , … Arlotta, P. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature, 570(7762), 523–527. 10.1038/s41586-019-1289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. , Du, Z. , Zhu, F. , Cao, Z. , An, Y. , Gao, Y. , & Jiang, B. (2020). Comorbidities and multi‐organ injuries in the treatment of COVID‐19. Lancet, 395(10228), e52. 10.1016/S0140-6736(20)30558-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H. , Uchimura, K. , Donnelly, E. L. , Kirita, Y. , Morris, S. A. , & Humphreys, B. D. (2018). Comparative analysis and refinement of human PSC‐derived kidney organoid differentiation with single‐cell transcriptomics. Cell Stem Cell, 23(6), 869–881, e868. 10.1016/j.stem.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaim, S. , Chong, J. H. , Sankaranarayanan, V. , & Harky, A. (2020). COVID‐19 and multiorgan response. Current Problems in Cardiology, 45(8), 100618. 10.1016/j.cpcardiol.2020.100618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, R. , Gomez Castro, M. F. , McCune, B. T. , Zeng, Q. , Rothlauf, P. W. , Sonnek, N. M. , … Ding, S. (2020). TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Science Immunology, 5(47), eabc3582. 10.1126/sciimmunol.abc3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervantonakis, I. K. , Hughes‐Alford, S. K. , Charest, J. L. , Condeelis, J. S. , Gertler, F. B. , & Kamm, R. D. (2012). Three‐dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proceedings of the National Academy of Sciences of the United States of America, 109(34), 13515–13520. 10.1073/pnas.1210182109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Honko, A. , Zhou, J. , Gong, H. , Downs, S. N. , Vasquez, J. H. , … Zhang, L. (2020). Cellular nanosponges inhibit SARS‐CoV‐2 infectivity. Nano Letters, 20(7), 5570–5574. 10.1021/acs.nanolett.0c02278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , … Research, T. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: The cell types for each sample were identified and annotated from the know cell markers from the respective publication and the top differentially expressed genes in each cluster.
Supplementary Figure 2: The expression profile of TMPRSS4 in various organoids.
Data Availability Statement
The data that support the findings of this study are openly available. All the gene expression data either in the normalized count or raw counts were downloaded from the Gene expression Omnibus or Array Express database. There were eight organoids data sets used in this study: Intestine conventional and improved condition (GSE119969), Kidney Morizane and Takasato methods (GSE118184), prostate (GSE130318), lung (E‐MTAB‐8221), retina (GSM4231315), and brain (GSE129519).


