Abstract
Sex differences in overall recombination rates are well known, but little theoretical or empirical attention has been given to how and why sexes differ in their recombination landscapes: the patterns of recombination along chromosomes. In the first scientific review of this phenomenon, we find that recombination is biased towards telomeres in males and more uniformly distributed in females in most vertebrates and many other eukaryotes. Notable exceptions to this pattern exist, however. Fine scale recombination patterns also frequently differ between males and females. The molecular mechanisms responsible for sex-differences remain unclear, but chromatin landscapes play a role. Why these sex differences evolve also is unclear. Hypotheses suggest that they may result from sexually antagonistic selection acting on coding genes and their regulatory elements, meiotic drive in females, selection during the haploid phase of the life cycle, selection against aneuploidy, or mechanistic constraints. No single hypothesis, however, can adequately explain the evolution of sex differences in all cases. Sex-specific recombination landscapes have important consequences for population differentiation and sex chromosome evolution.
Keywords: Heterochiasmy, recombination, sex chromosomes, meiotic drive
Introduction
Rates and patterns of recombination differ strikingly between sexes in most species. These differences represent an evolutionary conundrum. A rich body of theory and data provides many insights into the evolution of overall recombination rates (Barton 1995; West et al. 1999; Otto 2009; Ritz et al. 2017; Stapley et al. 2017). But why recombination differs between males and females is far less well understood (Burt et al. 1991; Lenormand 2003; Lorch 2005; Hedrick 2007; Brandvain and Coop 2012; Lenormand et al. 2016).
Nearly all attention to this question has focused on overall map lengths. Achiasmy, in which recombination is lost in one sex, has evolved about 30 times, and the loss always occurs in the heterogametic sex (Haldane 1922; Huxley 1928; Burt et al. 1991). A much more common but less understood situation is heterochiasmy, in which both sexes recombine but at different rates. In these cases, sex differences in map lengths are not correlated with which sex is heterogametic (Burt et al. 1991; Lenormand 2003; Brandvain and Coop 2012), although overall recombination tends to be higher in females across animals and outcrossing plants (reviewed in Lenormand and Dutheil 2005; Brandvain and Coop 2012). Sex differences in map lengths are even found in species with non-genetic sex determination (Miles et al. 2009) and in hermaphrodites (Franch et al. 2006; Giraut et al. 2011; Jones et al. 2013; Theodosiou et al. 2016).
The historical focus on sex differences in overall map lengths overlooks a potentially more interesting and important phenomenon: sex differences in patterns of recombination along chromosomes. In humans, for example, crossovers are concentrated near the tips of chromosomes in males but not females (Broman et al. 1998). We call the distribution of crossovers along chromosomes the recombination landscape. These landscapes may have important implications for evolution, as variation in overall recombination along chromosomes influences adaptation, population differentiation and speciation (Birky and Walsh 1988; Butlin 2005; Presgraves 2005; Nachman and Payseur 2012; Burri et al. 2015; Ortiz-Barrientos et al. 2016).
This manuscript presents the first comprehensive review and synthesis of sex differences in recombination Landscapes. We begin with a meta-analysis that reveals several generalizations about these landscapes that extend across eukaryotes. Recombination rates near the ends of chromosomes are typically greater in males, regardless of which sex has the greater overall map length. Recombination rates in the middle of chromosomes, particularly near centromeres, are typically greater in females. Sex differences in recombination also occur at fine scales. These results indicate that sexual species do not have a single recombination landscape. Instead, they have two separate landscapes – one for each sex – which derive from a shared genome but can evolve independently. Several recent studies have provided key insight into the cellular and genetic mechanisms responsible for sex differences in recombination. We review these findings in the second section of this manuscript and discuss the evolutionary constraints or biases that they may contribute to the recombination landscape.
Few studies have asked how and why strikingly different landscapes have evolved in males and females. One possibility is that selection acts solely on the sex-averaged recombination rate, and that differences in the sexes result from genetic drift (Nei 1969; Burt et al. 1991). However, the consistent patterns of sex differences across several phyla renders this nuli hypothesis unlikely. Another possibility is that sex differences in the recombination landscape result from different mechanistic constraints imposed by oogenesis and spermatogenesis (Hunt and Hassold 2002; Petkov et al. 2007). A third possibility is that these differences are adaptive (Ritz et al. 2017). We review previously proposed adaptive hypotheses for sex differences in recombination, including potential roles for female meiotic drive or selection against aneuploidy. We also propose a new hypothesis in which sexually antagonistic selection (i.e., when an allele has positive fitness effects on one sex and negative effects on the other) acts on interactions between genes and their cis regulatory elements. All of the current adaptive hypotheses face theoretical problems, however, and no single hypothesis is likely to explain the origin of sex differences in recombination across all species.
Finally, we highlight potential consequences of sex differences in recombination for evolution. While the effects of recombination rates on adaptation, population differentiation, and speciation are well known (Ortlz-Barrientos et al. 2002; Butlin 2005; Presgraves 2005; Cooper 2007; Nachman and Payseur 2012; Burri et al. 2015; Ortiz-Barrientos et al. 2016; Ritz et al. 2017; Schumer et al. 2018), the effects of sex differences have received scant attention (Butlin 2005). Importantly, the genomic landscapes of adaptation are largely driven by the sex with the greatest variation in recombination rates across the genome, typically males. Recombination rates in just one sex, rather than the average across males and females, are also key to the evolution of sex chromosomes. We present six novel predictions regarding general patterns of sex chromosome evolution we expect to observe based on typical sex differences in recombination landscapes.
The patterns
Sex differences in the recombination landscape occur both at broad scales (megabases) and at fine scales (kilobases or less). The magnitude of these differences varies between species, but it is typically large. In humans, recombination rates are significantly less correlated between males and females (r = 0.66 over 10kb Windows) than between individuals of the same sex (r > 0.9) (Kong et al. 2010). The correlation between male and female recombination rates is similar in cattle (r = 0.64 between the sexes) (Ma et al. 2015), but much smaller in mice (r = 0.28 to 0.33 between the sexes; r = 0.50 to 0.66 between individuals of the same sex) (Shifman et al. 2006). Below, we discuss the broad and fine-scale patterns responsible for the relatively small correlations between male and female recombination rates.
Broad-scale patterns
We conducted a comprehensive literature review of sex differences in the recombination landscape across eukaryotes. We searched Web of Science and Google Scholar using relevant phrases such as “heterochiasmy”, “sex differences in recombination”, and “linkage map + sex” to identify studies that explicitly compared male and female recombination Ìandscapes. We also reviewed all studies cited in previous reviews of sex differences in overall map lengths (Lenormand 2003; Lenormand and Dutheil 2005; Brandvain and Coop 2012). In total, we identified 51 species for which data on recombination landscapes in both sexes are available. The results are shown in Figure 1 and Supp. Table 1.
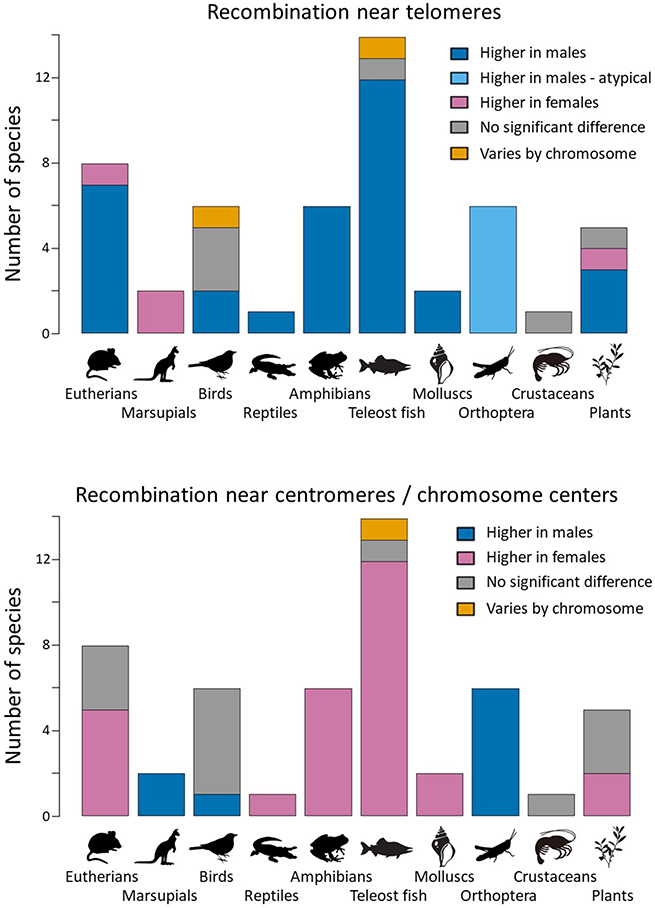
Figure 1:
Sex differences in the recombination landscape summarized across taxa. Top: Count of species with male-biased, female-biased, or no significant sex difference in recombination near telomeres. Grasshoppers have an unusual landscape in which recombination is high in males across the entire chromosome, but crossovers in females are strongly clustered towards telomeres. Bottom: Count of species with sex differences near centromeres (or chromosome middles when centromere position is unknown). Underlying data is summarized in Supp. Table 1.
Males and females typically differ in the distribution of crossovers (COs) along their chromosomes. Most often, COs cluster near telomeres in males, while they are more uniformly distributed or elevated near centromeres in females (see Figure 2 for examples). This results in a characteristic sex contrast: recombination rates near telomeres are higher in males, even when the overall recombination rate is higher in females, while rates in the middles of chromosomes are often higher in females (Figure 1). This pattern is widespread across eukaryotes. Male-biased recombination towards telomeres occurs in 33 of the 51 species we examined, including humans (Broman et al. 1998; Kong et al. 2002), nearly all other eutherian mammals, all but one teleost flsh, all frogs, some birds, both mollusks, and the majority of plants. We will refer to these as the typical landscapes in males and females.
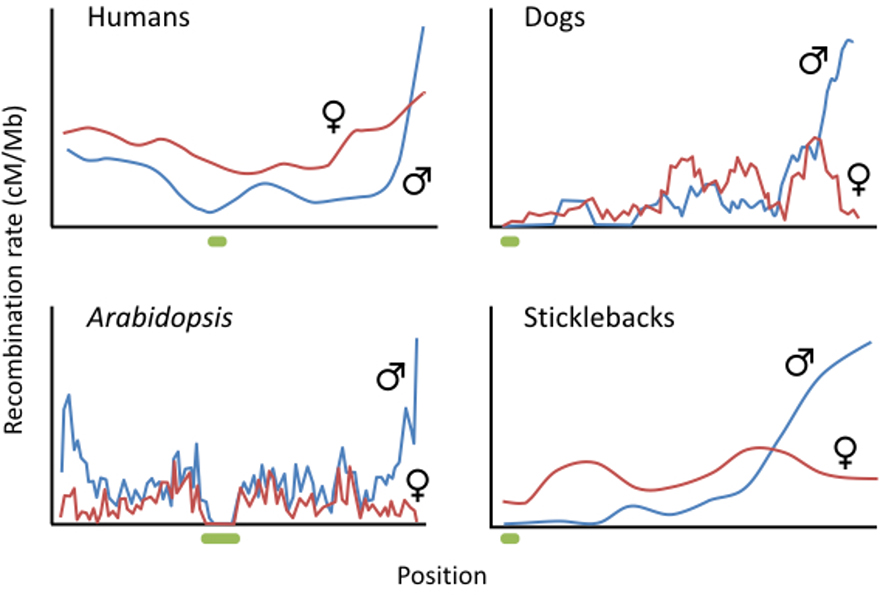
Figure 2:
Examples of the typical recombination landscapes. Local recombination rates are greater in males at chromosome ends. Local rates are often greater in females near the centromere (shown by the green bars). Top left: Recombination along human Chromosome 7 (Broman et al. 1998). Top right: Recombination along domestic dog Chromosome 19 (Wong et al. 2010). Bottom left: Recombination along Arabidopsis thaliana Chromosome 5 (Giraut et al. 2011). Bottom right: Distribution of crossovers as function of relative distance from centromere across long arms of all Gcisterosteus stickleback chromosomes (Sardell et al. 2018).
Although most of the species in our dataset exhibit male-biased recombination near telomeres, the degree of bias differs considerably between taxa. The most extreme sex differences occur in two frogs (Rana temporaria and Hyla arborea) and in guppies (Poecile reticulata). In these species, all COs ever observed in males have occurred very close to the telomeres (Brelsford et al. 2016a; Brelsford et al. 2016b; Bergero et al. 2019).
There are exceptions to the typical landscapes. In four species (both marsupials, the domestic pig, and tomatoes), recombination rates at the ends of chromosomes are higher in females than males. In all six species of grasshoppers, COs cluster more strongly towards telomeres in females, but recombination is higher along the entire chromosome in males (Cano and Santos 1990). Six species, including most birds and maize, do not exhibit pronounced sex differences. Landscapes can also vary between chromosomes within a species. In the gilthead sea bream (Sparus aurata), which is a sequential hermaphrodite, recombination is higher in females than males near the telomeres in some chromosomes, but the reverse in others (Franch et al. 2006). We note that the studies in our dataset are heavily biased towards vertebrates and that we have little or no data from some key clades (e.g., squamate reptiles, non-orthopteran insects, plants, and all minor animal phyla). More sex-specific linkage maps are needed to test for the generality of these patterns across all eukaryotes.
The typical broad-scale landscape may result from two primary effects. Several studies cited in Supp. Table 1 explicitly identified a sex-specific centromere effect, which reduces recombination in males and/or increases it in females. (In other cases, recombination is higher in chromosome centers in females, but the locations of centromeres are unknown.) Reduced recombination in chromosome centers can also result from mechanisms that favor clustering of COs near telomeres, such as telomere-guided initiation of recombination (Nachman 2002; Duret and Arndt 2008; Higgins et al. 2012; Nachman and Payseur 2012; Zickler and Kleckner 2016).
Acrocentric chromosomes, in which the centromere is located closer to one end of the chromosome, offer opportunities for disentangling the effects of centromeres and telomeres. Using a statistical approach, Sardell et al. (2018) showed that recombination landscapes in male stickleback fish (Gasterosteus) are governed both by the distances to the centromere and to the nearest telomere. Centromeres strongly suppress nearby recombination, while a weaker, independent telomere effect favors clustering of COs near chromosome tips and away from chromosome centers. Centromeres also suppress nearly all recombination in males on short arms of autosomes when one arm is much longer than the other. In females, however, centromeres and telomeres have little or no effect on recombination. Similar patterns are found in salmonid fish (Sutherland et al. 2017). The pattern is slightly different in humans: centromeres repress recombination in both sexes but do so much more strongly in males (Broman et al. 1998; Kong et al. 2002; Ottolini et al. 2015).
Yet another pattern is seen in red deer (Cervus elaphus), where female recombination is significantly elevated near the centromere on many chromosomes. Johnston et al. (2017) hypothesized that the lack of recombination suppression near their centromeres may be driven by several recent chromosome fissions. Young centromeres are small and expand by the accumulation of repeats, which can cause the suppression of recombination near centromeres to increase over evolutionary time (Liao et al. 2018). Elevated recombination near centromeres in females is also found in several other taxa, however, including Xenopus frogs (Furman and Evans 2018), the fìsh Notothenia coriiceps (Amores et al. 2017), and the plant Brassica nigra (Lagercrantz and Lydiate 1995). The latter two species have also undergone several recent chromosome fusions (Lagercrantz 1998).
Species with holocentric chromosomes, which lack centromeres, could provide additional insight to factors that govern broad-scale recombination landscapes. Unfortunately, little is known about recombination in those species. In the worm C. elegans, which is a hermaphrodite, overall recombination rates are lower during spermatogenesis, but the effects of telomeres differ between chromosomes (Zetka and Rose 1990; Meneely et al. 2002; Lim et al. 2008). Other holocentric taxa where recombination has been studied, including Lepidopterans and Hemipterans, are achiasmatic (Burt et al. 1991).
Fine-scale patterns
Recombination hotspots are typically defined as small regions of chromosomes where recombination is greater than the genomic mean. A hotspot is typically defined as “sex-specific” if recombination is significantly greater than the genome-wide average (Kong et al. 2010). We argue that this definition conflates two different situations. A motif might affect recombination in only one sex, directly causing fine-scale differences. Alternatively, a motif that increases fine-scale recombination in both sexes will be considered sex-specific if it lies in a region of the chromosome where broad-scale sex differences suppress recombination in one sex (see Supp.Fig. 1). In this second situation, the hotspot is identified as sex-specific by an artifact of the definition, rather than because of a true biological difference in fine-scale recombination between the sexes. Unfortunately, most studies published to date do not allow us to distinguish between these situations.
Several studies have identified sex-specific hotspots based on the standard definition. Approximately 15 percent of hotspots in humans (Kong et al. 2010) and 60 percent of hotspots in cattle (Ma et al. 2015) are reported to be unique to one sex. A large fraction of hotspots with sex-specific effects have also been reported in dogs (Wong et al. 2010), pigs (Tortereau et al. 2012), chickens (Elferink et al. 2010), mice (Shifman et al. 2006; de Boer et al. 2015), and maize (Kianian et al. 2018). Those findings, however, may be artifacts of under-powered experimental designs (Brick et al. 2018). Moreover, they did not control for the broad-scale sex differences in recombination discussed above.
A recent study of fine scale sex differences in recombination in mice by Brick et al. (2018) used a more refined approach to determine the sex-specific effects of hotspots. They first identified hotspots as regions with high rates of overall double-stranded break formation (the precursor to recombination). They then compared the recombination rate at each hotspot to rates in flanking regions in each sex. Under this approach, Brick et al. found that at least 97 percent of hotspots in mice increase local rates of recombination in both sexes. That is, almost no hotspots are sex-limited. The relative increase in recombination, however, differs significantly between males and females at 48 percent of autosomal hotspots, with a four-fold difference on average.
Many hotspots in mammals are assbciated with binding motifs for the zinc finger protein PRDM9 (Baudat et al. 2010; Berg et al. 2010; Parvanov et al. 2010). These hotspots are thought to be unaffected by sex (Kong et al. 2010; Lu et al. 2012), and the density of PRDM9 motifs is uncorrelated with sex differences in recombination rates in humans (Bhérer et al. 2017). Males and females differ, however, in the proportion of all hotspots that are associated with PRDM9. In mice, double-stranded breaks at non-PRDM9 sites occur more often in females than males (Brick et al. 2018). Likewise, a larger proportion of hotspots contain PRDM9 motifs in males than in females. PRDM9 is believed to localize COs away from gene regulatory regions. Thus, the increased prevalence of non-PRDM9 hotspots in females may explain why recombination is elevated in promoter regions in human females but not males (Bhérer et al. 2017). In maize, which lacks PRDM9, COs cluster at transcription start sites during female meiosis, but 400 bps upstream from transcription start sites on average during male meiosis (Kianian et al. 2018).
Mechanisms underlying sex differences in the landscape
Meiosis in males and females are fundamentally different processes (Hunt and Hassold 2002). The most conspicuous difference is that meiosis in males produces four gametes, whereas meiosis in females produces a single gamete and three inviable polar bodies. There are also many other differences, for example, how epigenetic marks are inherited, in the timing of the meiotic cell cycle, and in the cellular environment. Any or all of these factors could contribute to sex differences in the recombination landscape. One clear result is that sex chromosomes are not generally responsible for sex differences in recombination landscapes. Sex-reversal experiments in frogs (Rana temporaria) and pufferfish (Takifugu niphobles), and data from species with non-genetic sex determination indicate that differences in landscapes are instead determined by phenotypic sex (Miles et al. 2009; Giraut et al. 2011; Ieda et al. 2018; Rodrigues et al. 2018).
Several recent studies have provided new insight into the mechanisms responsible for sex differences in recombination. In this section, we first discuss cellular mechanisms that are thought to be responsible for sex differences in recombination. We then discuss the genetic basis of those differences.
Cellular, molecular, and physiological mechanisms
Sex differences in recombination rates can arise in two ways. First, males and females can differ in the rates that double-stranded breaks are initiated. This factor is responsible for many fine-scale sex differences in recombination in humans and mice (Gruhn et al. 2013; Gruhn et al. 2016; Brick et al. 2018). Second, the sexes can differ in the frequencies with which double-stranded breaks are resolved into either crossovers or gene conversion events (Duret and Arndt 2008; Kong et al. 2014; de Boer et al. 2015). This second factor is likely responsible for many broad-scale sex differences in the recombination landscape. In mice, recombination rates are higher near telomeres in males even though rates of double-stranded break formation near telomeres are higher in females (Brick et al. 2018).
Differences between the sexes in overall recombination rates may also result from crossover interference, which occurs when a crossover suppresses the formation of other COs nearby (Petkov et al. 2007). All else equal, recombination rates should be lower in the sex with stronger CO interference. Indeed, in humans (Hou et al. 2013) and the tree Prunus mume (Sun et al. 2017), CO interference is stronger and overall recombination rates are lower in males. The opposite sex bias is seen in cattle and Arabidopsis thaliana: CO interference is stronger and overall recombination rates are lower in females (Drouaud et al. 2007; Wang et al. 2016b). Crossover interference is unlikely, however, to explain sex differences in the distribution of COs along chromosomes. The typical male landscape, with elevated recombination near chromosome ends, occurs in all four of these species, despite their differences in the sex bias of CO interference.
The synaptonemal protein complex, which is responsible for chromosome pairing and synapsis during meiosis, may be important to sex differences in recombination (Petkov et al. 2007). The length of the synaptonemal complex is highly correlated with sex-specific map length in humans (Tease and Hultén 2004) and plants (Drouaud et al. 2007; Giraut et al. 2011; Phillips et al. 2015). Intriguingly, the structure of the synaptonemal complex in humans differs between males and females near the telomere and the centromere (Gruhn et al. 2016). This observation suggests that the synaptonemal complex may contribute to broad-scale sex differences in the recombination landscape in some taxa. Other evidence argues against its general importance as a driver of these sex differences. A genome wide-association study by Halldorsson et al. (2019) identified 47 variants at 35 loci associated with variation in recombination in humans, 11 of which are in genes that encode the synaptonemal complex. Only one of these 11 loci (in the gene SYCE3), however, is associated with sex differences in the localization of COs. Furthermore, the relationship between the synaptonemal complex and recombination does not seem to be universal, as synaptonemal complex length and recombination rates are not correlated in cichlid fish (Campos-Ramos et al. 2009).
Genomic imprinting, i.e. methylation that causes gene expression to differ between maternally and paternally inherited chromosomes, has been associated with increased recombination in humans (Lercher and Hurst 2003; Sandovici et al. 2006). Although imprinted genes are common in plants, they are rare in animals (about 0.1% in mammals, Burns et al. (2001)). Thus, they are unlikely to be responsible for much of the sex differences in the recombination landscapes of animals.
Other epigenetic modifications play a key role in determining fine-scale sex differences in recombination in mice, and likely other mammals. A fundamental difference between meiosis in male and female mammals is that the genome undergoes global cytosine demethylation during oogenesis, but not spermatogenesis (Seisenberger et al. 2012; Smith et al. 2012). Brick et al. (2018) found that the sex bias of recombination hotspots in mice is determined by the local epigenetic landscape. DNA methylation at a PRDM9 binding site increases local rates of double-stranded break formation. Because methylation is restricted to spermatogenesis, this results in a male-biased hotspot. DNA methylation in a region flanking a PRDM9 binding site decreases locai recombination in males, resulting in a female-biased hotspot. Methylation may also play a role in broad-scale sex differences in recombination. Methylation occurs primarily at CpG nucleotides (Bird 1986; Lister et al. 2009), and GC content is typically elevated near telomeres (Bernardi 2000; Arndt et al. 2005). We therefore propose that the typical landscapes in mammals may in large part be driven by broad-scale patterns of methylation in males. A different pattern is found in maize: male and female hotspots do not differ in their fine-scale methylation patterns, and the sexes exhibit no broad-scale differences in recombination (Kianian et al. 2018).
Physiological differences between males and females could also contribute to sex differences in recombination (Morelli and Cohen 2005). Recombination rates are often affected by temperature, which can differ considerably between male and female gonads (Plough 1917; Plough 1921; Phillips et al. 2015). In barley (Hordeum bulbosum), high temperatures increase the frequency of COs in the middle of chromosomes during male but not female meiosis (Phillips et al. 2015). In species with arrested female meiosis, telomeres in males and females spend different amounts of time clustered in the bouquet, which may affect patterns and rates of recombination (Paigen and Petkov 2010).
Finally, age also affects recombination differently in males and females. In humans, females have greater variation in map length (Broman et al. 1998), which is thought to result from increased recombination rates with age (Kong et al. 2004b; Coop et al. 2008). Similar effects of age occur in female Drosophila (Bridges 1927) and cattle (Wang et al. 2016b). The pattern, however, is not universal. The number of chiasma formed during meiosis, which is correlated with recombination rate, decreases with age in female hamsters (Sugawara and Mikamo 1983).
Genetic variation in the landscape
Some of the genetic variation for recombination rates is sex-specific. This observation implies that that sex differences in the recombination landscape are not the inevitable result of sex differences in meiosis. For example, artificial selection experiments on female recombination rate in Tribolium beetles caused correlated evolution of male recombination rates (Dewees 1975). The genetic correlation between male and female recombination rates, however, was not perfect: the differences in recombination between the high- and low-recombination selection lines was greater for females than males.
Polymorphism in several genes contributes to variation in sex differences in recombination. In humans, an allele of the gene RNF212 is associated with higher recombination in males but lower recombination in females (Kong et al. 2008; Chowdhury et al. 2009). The same locus has similar effects in Soay sheep (Johnston et al. 2016). A chromosomal inversion on human Chromosome 17 is associated with higher recombination in females but not males (Stefansson et al. 2005; Chowdhury et al. 2009). A genome wide association study by Halldorsson et al. (2019) identified 47 variants at 35 genes that are associated with variation in recombination rates or the locations of COs in humans. All of these variants have an effect that is restricted to one sex in at least one aspect of recombination, and 12 had opposite effects in males and females. Finally, at least three loci in cattle effect recombination in females but not in males (Ma et al. 2015).
The most detailed genetic characterization of sex differences in recombination rates comes from house mice (Mus musculus), which vary substantially between strains and subspecies (Dumont and Payseur 2011a). Males from one strain of M. m. musculus have 30% more COs than those from a strain of M. m. castaneus, but females have nearly identical CO frequencies (Dumont and Payseur 2011b). Surprisingly, a castaneus allele at an X-linked locus increases genome-wide recombination when introgressed into musculus (Dumont and Payseur 2011b; Dumont 2017). These authors suggest that lower recombination in both sexes evolved first in castaneus, and that later selection for higher rates in females favored the sex-linked modifier. The current sex differences in recombination rates result because females carry two copies of the allele, while males carry only one. Although this example involves overall recombination rates, it is plausible that differences in the distribution of COs along chromosomes could evolve in response to similar selection pressures.
Evolution of sex differences in the landscape
There are two large families of hypotheses that might explain the evolution of sex differences in recombination landscapes. First, they may arise as a by-product of mechanistic differences between meiosis in males and females. Alternatively, the differences may be adaptive.
We have already seen that meiosis in males and females differs in many ways, and these differences could impose constraints on recombination in each sex. Mechanistic constraints might, for example, explain why the typical landscapes are shared across distantly-related phyla. Such constraints are not absolute, however, as several observations suggest that sex differences in the landscapes can evolve readily. First, achiasmy and achiasmatic sex chromosomes have evolved repeatedly (Blackmon and Brandvain 2017). Second, the degree of sexual dimorphism in the landscape varies dramatically between taxa, and exceptions to the typical landscapes are not rare. Third, sex-specific genetic variation for recombination rates exists in many taxa. Finally, recombination landscapes also differ between mating types in the unicellular haploid fungus Cryptococcus deneoformans, suggesting that different recombination landscapes can evolve without differences in gametogenesis (Roth et al. 2018). Together, these observations suggest that sex differences in recombination may be adaptive rather than an artifact of mechanistic constraints.
Several adaptive hypotheses have been proposed for heterochiasmy. Trivers (1988) suggested that overall map lengths are generally smaller in males because sexual selection favors tighter linkage between loci important for male reproductive success. This hypothesis did not survive a formal population genetic model analyzed by Lenormand (2003), who showed that selection acting on autosomes during the diploid phase of the life cycle generally does not favor the evolution of sex differences in recombination. He did find exceptions to that generalization, however, and we now turn to them. While none of these hypotheses can explain the contrasting recombination landscapes in males and females by themselves, combinations of them might, and we return to that possibility in the Conclusions.
The SACE hypothesis
Lenormand (2003) found that one way that sex differences in recombination can evolve is when two conditions are met. The first is that the strength of epistasis between alleles at a pair of loci on a chromosome depends on whether they are in a cis or trans relationship (i.e., if they are on the same physical chromosome or on different homologues). Second, the relative strengths of the cis and trans epistasis must differ in males and females. Lenormand (2003) thought the first condition implausible, arguing that if two loci produce gene products, interactions between those products is unaffected by whether their alleles are in cis or trans.
We propose a hypothesis in which both of Lenormand’s conditions can in fact be met: when there are sex differences in epistasis between coding regions and their cis regulatory regions. We refer to this idea as “sexually antagonistic cis epistasis”, or SACE. Consider a coding locus C and a regulatory region R that are expressed in males but not females. Polymorphism is maintained at both loci by some form of frequency-dependent selection, and there is epistasis between them. To make the ideas concrete, imagine that the loci contribute to alternative mating strategies, as in the ruff, Phylomachus pugnax (Küpper et al. 2016). The alternate alleles at C code for lesser or greater intensity of a plumage color, and the alleles at R cause C to be expressed at higher or lower levels. Epistasis occurs because the alleles for higher expression and greater color intensity are favorable to one male morph that has high fitness, while the alleles for lower expression and lesser intensity are favorable to an alternate morph that also has high fitness. Those conditions favor the spread of a mutation at a linked modifier locus M that decreases recombination between R and C in males, but that has no effect (or even the opposite effect) in females (Lenormand 2003, Equation 32).
To verify Lenormand’s conclusions and gain further insight, we carried out simulations of a very simple model. Initially, loci R and C recombine at a rate rrc, while loci C and M recombine at rate rCM in both sexes. A neutral mutation at M is introduced that decreases the recombination rates rRC and rCM in males by the same proportion, as would happen if the modifier globally decreases the number of crossovers in males. We assume the mutation increases recombination in females by the same factor, so that the overall recombination remains unchanged. The proportional increase in the mutation’s frequency in each generation while it is stili rare gives the effective selection coefficient (or eigenvalue), which is the strength of selection on a mutation needed to produce the observed rate of spread. Further details are given in the Supplemental Materials.
The simulations show that decreased recombination in males does indeed evolve, but that the effective selection on the modifier is weak (see Supp. Materials). For example, take the case in which the epistatic selection coefficient is ε = 0.01, the initial recombination rates are rRC= 0.005 and rCM = 0.1, and allele frequencies at the R and C loci are 0.5. A mutation that decreases recombination between the three loci in males by one half experiences an effective selection coefficient of 3 × 10−7. As intuition suggests, simulations show that the effective selection coefficient is proportional to the strength of epistasis, the amount by which the modifier decreases male recombination, and the heterozygosity at R and C.
That the selection on the modifier generated by just one pair of loci is very weak even under relatively strong epistatic selection suggests that a single pair of loci is unlikely to drive fixation of a recombination modifier by itself. The force of selection will be increased, however, if the modifier affects multiple pairs of loci. That situation is plausible: a mutation that decreases male recombination along the center of a chromosome (for example, by moving crossovers towards the telomeres) will affect all the loci in that region. The cumulative effects on the modifier are expected to be additive (Kirkpatrick et al. 2002). So, if ten pairs of loci experience SACE selection like that described in the last paragraph, the effective selection coefficient would increase by a factor of ten (from 3 × 10−7 to 3 × 10−6). A recombination modifier only evolves in response to selection on linked loci, so SACE selection occurring on other chromosomes will not increase the effective strength of selection. There is, however, the possibility that recombination modifiers have global effects, that is, they modify recombination landscapes on all chromosomes as pleiotropic effects of modifying recombination on their own chromosome (e.g., by locating COs near telomeres). In that event, the evolution of decreased male recombination could result from the cumulative effect of several modifiers throughout the genome which each effect many pairs of epistatic loci. Stili, even under favorable conditions, the SACE mechanism is a weak evolutionary force. It may have been acting over long evolutionary timescales, however, in which case its cumulative effects might have contributed to the recombination landscapes we see today.
At present, the SACE hypothesis is a theoretical possibility without empirical support. A further difficulty for the hypothesis is that male meioses in hermaphrodites tend to show the typical male recombination landscape. The mechanism could stili operate in this situation if SACE selection is acting in loci expressed in the testes. Another challenge to the hypothesis is that gene density is typically lower ciose to the centromere than near telomeres (Bernardi 2000). This pattern suggests that SACE by itself should favor clustering of COs near centromeres, where they will be least likely to fall between a gene and its regulatory regions, rather than the typical clustering of COs near telomeres in males. On the other hand, SACE may explain the fine-scale pattern in mammals where COs occur near promoters less frequently in males compared to females (Bhérer et al. 2017), as predicted by this hypothesis.
Haploid selection and genomic imprinting
Lenormand (2003) also found that adaptive sex differences in recombination can evolve by a second (and very different) pathway: when selection acts on the haploid phase of the life cycle. Here selection favors sex differences if the strength and/or direction of epistatic selection is different in sperm and eggs (or pollen and ovules in the case of plants). Evidence for this hypothesis is mixed (Lenormand et al. 2016). Pollen is often subject to strong haploid selection, which may favor the evolution of heterochiasmy in plants. As predicted by the model, female-biased heterochiasmy is strongly correlated with outcrossing, which increases competition among pollen (Lenormand and Dutheil 2005). Opportunities for selection on the haploid phase, however, are much more limited in animals, since many fewer genes are expressed in sperm and eggs (Joseph and Kirkpatrick 2004). Furthermore, heterochiasmy is uncorrelated with the strength of sperm competition in mammals, in contrast to the prediction of the haploid selection model (Mank 2009).
Sex differences in recombination are also favored when the fitness effects of an allele in the diploid phase of the life cycle depend on whether it is inherited from the mother or the father – a situation very similar to selection in the haploid phase (Lenormand (2003). The most familiar way in which that can happen is by genomic imprinting. Since imprinting affects so few genes in animals (about 0.1% in mammals, Burns et al. (2001)), this seems unlikely to be the typical way that sex differences in recombination evolve. Lenormand noted, however, that it might explain the observation that imprinted regions of the genome can show large sex differences in recombination rates (Lercher and Hurst 2003; Sandovici et al. 2006).
Female meiotic drive
Meiotic drive can generate selection on haploid combinations of alleles in gametes, and so can favor heterochiasmy (Haig 2010; Brandvain and Coop 2012). Meiotic drive during oogenesis can lead to the evolution of recombination rates in females. Drive can occur during either Meiosis I or Meiosis Il, but the mechanisms are quite different (Brandvain and Coop 2012).
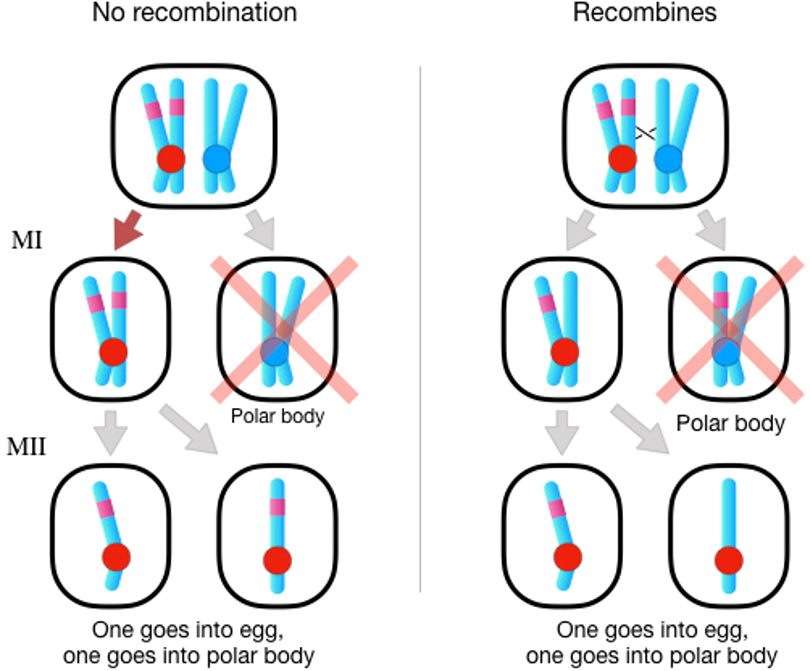
Female meiotic drive can happen during Meiosis I when a centromere biases the chance that it is passed to an egg rather than a polar body (Figure 3). An allele at a Locus linked to the centromere will increase in frequency if it causes (or enhances) the drive of the centromere on its chromosome. This allele’s advantage is disrupted, however, if there is a crossover between that locus and the centromere, because that decouples the allele from the centromere. Meiotic drive often has deleterious effects on fecundity (Haig and Grafen 1991; Fishman and Saunders 2008; Fishman and Kelly 2015), and unlinked mutations that increase female recombination near centromeres will spread because they disrupt these meiotic drive conspiracies. (Haig (2010) proposed an alternate model of female drive during Meiosis I in which an individual’s fitness depends on the genetic composition of its dyads, rather than its genotype at the drive Locus. This model predicts that drive during Meiosis I favors reduced recombination, which is opposite to the typical landscape in females.)
Figure 3:
Meiotic drive during Meiosis I in oogenesis favors the spread of unlinked modifiers that increase recombination in females, particularly near the centromere. When drive decreases fecundity, these modifiers are favored because they disrupt drive. Left: When there is no recombination, the drive allele (magenta) increases the likelihood that its centromere will be transmitted to Meiosis II, with the non-drive chromatids transmitted to inviable polar bodies (denoted by pink “X”). Right: When there is a crossover between the drive allele and its centromere, one copy of the drive allele is present on both pairs of sister chromatids, eliminating the opportunity to increase its chance of transmission since it has no effect on the outcome of Meiosis II.
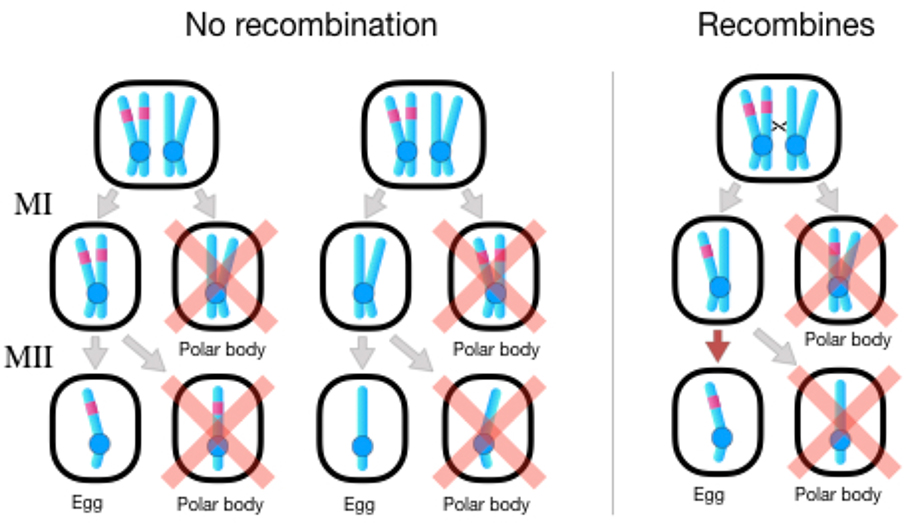
The situation is different for drive during Meiosis II (Figure 4). Here, drive requires a crossover between the driving locus and the centromere, rendering it heterozygous in both potential products of Meiosis I. Mutations that increase female recombination are favored when they are linked to the driving allele and hitchhike along with it. If drive incurs a fecundity cost, then rest of the genome again favors suppressing it. In this case, however, drive is suppressed by decreasing female recombination, contrary to the pattern seen in most taxa.
Figure 4:
Meiotic drive during Meiosis II in oogenesis favors the spread of unlinked modifiers that decrease recombination or coupled modifiers that increase recombination in females. Left: A Meiosis II drive allele has no effect without recombination. Whether or not the drive allele is transmitted to the egg is solely determined the outcome of Meiosis I, which is unaffected by the drive allele. As a result, there is an equal chance that the chromatids containing the allele are both transmitted to Meiosis Il (far left) or that they are deposited in inviable polar bodies (middle, denoted by pink “X”). Right: When there is a crossover between the drive allele and its centromere, the product of Meiosis I will always compose one chromatid with the drive allele and one without. Meiotic drive will then favor transmission of the drive allele to the egg during Meiosis II.
In sum, Brandvain and Coop (2012) predict higher recombination in females (speciflcally, near centromeres) can be favored in two ways. The first is when meiotic drive occurs during Meiosis I, drive incurs a fecundity cost, and unlinked modifiers act on recombination throughout the genome. The second occurs during Meiosis II when a driving mutation appears at a locus and a modifier link to it increases recombination between that locus and its centromere.
Several observations are consistent with this hypothesis. The rapid evolution of centromeres and their associated histones is strongly suggestive of an evolutionary arms race involving centromere drive and other loci that are selected to suppress it (Malik 2009; Malik and Henikoff 2009). Female drive during Meiosis I has been well characterized in several species (Dawe and Cande 1996; Fishman and Saunders 2008; Chmátal et al. 2014). Evidence of female drive during Meiosis II comes from humans, where recombinant chromatids are more likely to be transmitted to the egg during human oogenesis (Ottolini et al. 2015). On the other hand, there are theoretical reasons to question how important female meiotic drive may be to the evolution of recombination (Veller, pers. comm.). Mutations that increase Crossing over must occur in the small window of time while the driver is at very low frequency. Further, if drive occurs during Meiosis Il, the recombination modifier must be linked to the locus where drive is occurring. Thus, the status of this hypothesis is also in question.
Aneuploidy
Crossing over is criticai to proper chromatid segregation during chiasmatic meiosis, and failure to cross over properly often results in aneuploid gametes. Aneuploidy is surprisingly common and has strong fitness consequences: it is seen in 20% to 40% of human fertilizations, and is a primary cause of miscarriage (Nagaoka et al. 2012). The risk of aneuploidy is affected by the location of COs along chromosomes: chiasmata falling ciose to the centromere are frequently associated with improper segregation during meiosis (May et al. 1990; Lamb et al. 2005a; Rockmill et al. 2006). This suggests that the typical male recombination landscape (elevated recombination rates near chromosome ends and decreased recombination near centromeres) should minimize the fitness costs of meiotic errors. If true, then the challenge lies in explaining why female landscapes differ from this pattern, supporting the meiotic drive hypothesis (which predicts elevated recombination near centromeres in females) over the SACE hypothesis (which typically entails selection on recombination in males).
The necessity of crossovers could also contribute to the typical landscapes. Say that reduced recombination in males is favored (for example, by SACE) but selection against aneuploidy enforces a minimum of one crossover per chromosome and imposes a fitness cost of localizing COs near centromeres. These goals can be realized by localizing the chiasma at the chromosome tips, where Crossing over will minimize the number of pairs of loci that recombine (see also Veller et al. 2019).
Selection against aneuploidy may also favor the evolution of greater overall female map lengths in species that undergo extended meiotic arrest, such as humans. Chiasmata may lose cohesion during years of pachytene arrest prior to adulthood, increasing the risk of aneuploidy upon the completion of meiosis. A greater number of chiasmata may reduce the risk of such catastrophic failure (Brandvain and Coop 2012). There are several lines of support for this idea: a positive correlation between maternal recombination rate and fertility in humans (Kong et al. 2004a; Stefansson et al. 2005), an increased risk of aneuploidy with age in humans (Hassold and Hunt 2001; Lamb et al. 2005b; Nagaoka et al. 2012), and loss of sister chromosome cohesion with age in mice (Hodges et al. 2005; Chiang et al. 2010; Lister et al. 2010). When selection favors increased crossover number in females, interference may prevent multiple chiasma from clustering near telomeres, necessitating their more even distribution along chromosomes. This hypothesis cannot, however, explain why similar patterns are also observed in species that do not have meiotic arrest.
Evolutionary consequences
Although the importance of recombination to evolution is well understood, very little attention has been given to the consequences of sex-specific recombination patterns. This neglect is likely because the linkage disequilibrium between two loci often depends only on the sex average of the recombination rates between them. But ignoring sex differences in recombination can hinder our understanding of several features of evolution. In this section, we discuss how sex differences in the recombination landscape affect population differentiation and sex chromosome evolution.
Population differentiation, introgression, and speciation
Several general patterns link the recombination landscape to population divergence and speciation. Across the genome, differentiation (measured by Fst or dxy) is often negatively correlated with the local recombination rate (Van Doren et al. 2017; Vijay et al. 2017; Wolf and Ellegren 2017). Recombination landscapes also affect patterns of hybridization and speciation, as alleles are less likely to introgress between species in regions of low recombination, such as near centromeres (Wu 2001; Butlin 2005; Baack and Rieseberg 2007; Juric et al. 2016; Schumer et al. 2018). These relationships drive a characteristic pattern across eukaryotes, in which closely-related species show elevated Fst and dxy in chromosome centers relative to chromosome ends (Berner and Roesti 2017; Haenel et al. 2018).
Our meta-analysis shows that the typical pattern of differentiation seen along chromosomes reflects the typical landscape found in males. To understand why males should have the major effect, consider a scenario in which recombination rates are constant along the chromosome in females, but elevated at chromosome ends in males. In this case, the male landscape is responsible for all the variation in the sex-average of the local recombination rates along chromosomes, which in turn determines the genomic landscape of population differentiation.
We propose that sex differences in recombination can also have important consequences for introgression between species when hybrid fitness differs between males and females, as it commonly does (Haldane 1922; Coyne and Orr 1989). In these cases, using the sex-average recombination landscape in population genetics models can lead to erroneous conclusions. When hybrid females are infertile or inviable, for example, all recombination between the parental genomes in the F1 generation occurs in males. If sex differences in fitness carry over into later generations of hybrids, the effects of sex differences in recombination will be amplified. Sex-biased dispersal may further complicate this process. A formal model and data are needed to test the potential effects of asymmetric hybrid fitness and sex-biased dispersal on introgression when recombination varies between sexes.
Finally, strong sex differences in recombination may affect patterns of introgression between species when loci are subject to sexually antagonistic selection. Runemark et al. (2018) proposed that alleles that are beneficial to the sex with very little or no recombination will introgress less readily than alleles that are beneficial to the recombining sex. Modeling is again required to evaluate this idea.
Sex chromosomes
In many groups of eukaryotes, sex determination frequently changes from one pair of chromosomes to another, a process called sex chromosome “turnover”. These turnovers can be caused by the appearance of a new sex-determining factor or by the translocation of a sex-determining factor from one chromosome to another (van Doorn and Kirkpatrick 2007; Bachtrog et al. 2014; Beukeboom and Perrin 2014). Recombination plays a key role in two adaptive hypotheses for sex chromosome turnover. In the first, a new sex determination factor invades because it is linked to an autosomal locus that is under sexually antagonistic (“SA”) selection (Charlesworth and Charlesworth 1980; van Doorn and Kirkpatrick 2007; van Doorn and Kirkpatrick 2010). In the second hypothesis, Y and W chromosomes degenerate when they cease recombining with their homologues, which favors invasion of a new sex chromosome that has not yet degenerated (Blaser et al. 2013).
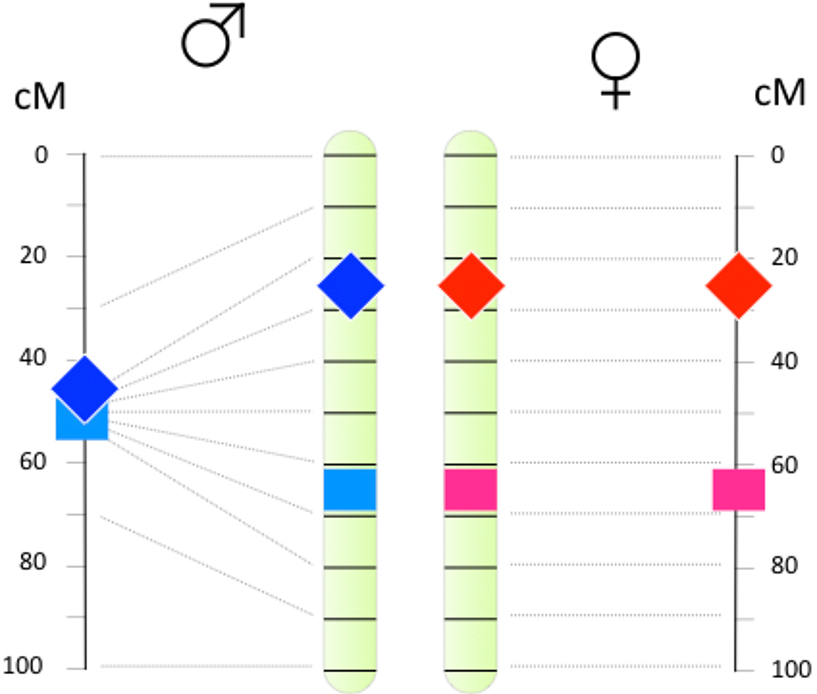
Sex differences in recombination landscapes have important implications for both hypotheses (Sardell et al. 2018). A turnover causes the recombination landscape of the new sex chromosome to immediately change from the sex-averaged landscape it experienced as an autosome to the sex-specific one it experiences as a sex chromosome. Conversely, a sex chromosome that reverts to an autosome sees its recombination landscape change in the reverse direction. As shown in Figure 5, these shifts can have implications for turnovers driven by SA selection. These turnovers are more likely when the recombination rate between the new sex determiner and the locus under SA selection is low (Charlesworth and Charlesworth 1980; van Doorn and Kirkpatrick 2007; van Doorn and Kirkpatrick 2010). Data suggests that divergence between young X and Y chromosomes at loci subject to SA selection in guppies (Poecilia reticulata) may indeed be fostered by extreme clustering of COs near chromosome ends in males (Bergero et al. 2019).
Figure 5:
The potential for sexually antagonistic selection to establish new sex chromosomes is strongly affected by sex differences in the recombination landscape. The chromosome’s physical map is at center, and the linkage maps in males at the far left and in females at the far right. Sex-determining mutations are shown as diamonds, and alleles with sexually antagonistic effects as squares. Left: With the typical male recombination landscape, a new male determining mutation and a male-beneficial allele can be tightly linked even if they are many megabases apart. This favors establishment of a new Y chromosome. Right: With the typical female landscape, a female determining mutation and a female beneficial allele that are many megabases apart will be only loosely linked. This inhibits establishment of a new W chromosome.
Sex differences in recombination also have potential consequences for turnovers driven by sex chromosome degeneration. In frogs (Brelsford et al. 2016b) and guppies (Bergero et al. 2019), crossovers in males occur only at the ends of chromosomes. These conditions may foster degeneration of Y chromosomes without need for the chromosomal inversions that are responsible for blocking recombination between the X and Y in many other taxa (Jeffries et al. 2018). Even very low rates of recombination, including gene conversion, are sufficient to prevent Y chromosome degeneration, however, and there is no evidence for Y chromosome degeneration in frogs or guppies (Jeffries et al. 2018; Bergero et al. 2019).
In light of these hypotheses, our observations about sex differences in the recombination landscape lead to three testable predictions regarding patterns of sex chromosome turnover:
-
1
Rates of sex chromosome turnover will be higher in taxa with XY sex determination.
-
2
Male determining factors are more likely to originate in the middles of chromosomes, and female determining factors more likely to originate near the telomeres.
-
3
XY sex determination will be more common than ZW sex determination.
Under the typical landscape, males have lower recombination along most of their chromosomes. Thus, a new male determining mutation is more likely to be tightly linked to a Locus under SA selection than is a new female determining mutation (see Figure 5). Further, when recombination is very strongly suppressed across most of the chromosome in males, Y chromosomes may degenerate more rapidly than W chromosomes. Both factors will favor more rapid turnover in species with XY sex determination. Indeed, the true frogs (family Ranidae), which have XY sex determination and extreme clustering of COs in males, show some of the highest observed rates of sex chromosome turnover yet observed (Jeffries et al. 2018).
Prediction (2) is difficult to test at present because many sex chromosomes have undergone extensive rearrangements since their sex determining genes were first established. Prediction (3) is consistent with the available data (Bachtrog et al. 2014), and several notable examples of ZW sex determination occur in species with atypical sex-specific recombination landscapes (e.g., most birds) or achiasmy (e.g., butterflies). Interestingly, Prediction (3) does not follow if turnovers are driven by sex chromosome degeneration: transitioning to ZW sex determination would provide an opportunity to escape the vicious cycle of sex chromosome degeneration that may be imposed by extreme male recombination landscapes in frogs.
Sex differences in the recombination landscape lead to two additional predictions about how sex chromosomes will evolve once they have been established. Sexually antagonistic selection acting on a locus in the recombining (or pseudoautosomal) region of a sex chromosome favors decreased recombination between that locus and the sex determining region (Rice 1987; Charlesworth et al. 2005). This change can occur by the fixation of inversions that block recombination between the X and Y or Z and W. Similarly, SA selection acting on an autosomal locus can favor a fusion between that chromosome and the sex chromosome (Charlesworth and Charlesworth 1980).
In the cases of both fusions and inversions, the strength of selection that favors the chromosome rearrangement to spread is proportional to how much it decreases recombination between the locus under SA selection and the sex determining region (Rice 1987). That leads to the following predictions under the typical landscapes where recombination is higher in females across much of the chromosome:
-
4
Sex chromosome-autosome fusions will be more common in XY species.
-
5
Inversions on sex chromosomes willfix more frequently in ZW taxa or when a male-determining locus is located near a telomere on a Y chromosome.
Prediction (4) is consistent with published data on frequency of sex chromosome-autosome fusions (Pennell et al. 2015). Prediction (5) is consistent with observations from Xenopus frogs, which have ZW sex determination and an atypical landscape of elevated recombination in chromosome middles in females (Furman and Evans 2018). The old sex chromosomes of X. laevis have retained a very small nonrecombining sex-linked region at one end of the chromosome (a region of low recombination in females). In contrast, the much younger sex chromosomes of X. borealis have rapidly evolved a large nonrecombining sex-linked region that covers much of the middle of the chromosome (a region of high recombination in females).
Prediction (5) could also explain an intriguing correlation found in reptiles. Most snakes and lizards with ZW determination have heteromorphic sex chromosomes, while those with XY sex determination have homomorphic sex chromosomes (Augstenová et al. 2018). We hypothesize that Z and W chromosomes are fixing inversions more frequently than X and Y chromosomes, which triggers the W to degenerate and be identified as heteromorphic.
A final prediction involves gene expression. Connallon and Clark (2010) developed a model showing how recombination differences between the sexes can affect the evolution of sex-biased expression. Their results lead to the following prediction in species where males do not recombine over most of their chromosomes:
-
6
Genes with sex-biased expression will accumulate more rapidly on sex chromosomes than autosomes if they are female-beneficial, and on autosomes if they are male beneficial.
Genes with female-biased expression are enriched on the sex chromosome in the frog Rana temporaria (Toups et al. 2018), in which all COs in males cluster at the extreme chromosome ends. Genes with male-biased expression are not significantly enriched on any chromosome. The first finding supports the predictions of Connallon and Clark’s model, but the second one does not.
These six predictions provide a framework for testing the sexual antagonism and sex chromosome degeneration hypotheses. Both are well understood theoretically but neither has yet been confronted by data. Sex-specific patterns of recombination are currently known only in a limited number of species, and additional studies are needed to gain key insights into sex chromosome evolution.
Conclusions
Sex differences in the recombination landscape are pervasive. In the typical Landscapes, recombination in males is higher near the telomeres and lower in the middle of chromosomes, but more uniform along chromosomes in females. Our meta-analysis found that pattern in the majority of the 51 species sampled from 8 animal phyla and 5 plant species, though there are exceptions. The full extent of this pattern across all eukaryotes is unclear, however, as the relevant data are not available from many clades.
Two families of hypotheses might explain sex differences in the recombination landscape. The first is that they are the mechanistic side-effects of the many fundamental differences between meiosis in males and females. This idea is a plausible explanation for some general patterns, for example higher overall recombination rates in females. It does not, however, explain the exceptions. Further, it is not consistent with evidence of sex-specific genetic variation for recombination that would allow adaptive differences in the male and female landscapes to evolve.
Another family of hypotheses proposes sex differences in the landscapes are adaptative. Several ideas have some support, but each has weaknesses. First, epistatic selection favoring combinations of alleles at coding loci and their cis-regulatory factors (the “SACE” hypothesis) can favor the evolution of decreased recombination in males. Modeling shows that this effect is very weak, but perhaps could be a factor if there are many of these epistatically interacting loci throughout the genome. SACE also should generally favor clustering of COs near centromeres more than telomeres, and only affects the recombination landscape in one sex, requiring additional explanations for why the sexes differ. It is uniquely able, however, to explain some fine-scale patterns. Second, meiotic drive can favor the evolution of increased recombination in females. There is growing evidence for female drive in diverse animals and plants. Models show, however, that drive can only favor changes in recombination under quite limited conditions. Further, meiotic drive is unaffected by male recombination, and so other factors are required to explain why recombination differs between males and females. Third, sexual selection during the haploid stage may drive sex differences in recombination in plants but is unlikely to affect landscapes in animals. Fourth, the necessity of crossovers to prevent aneuploidy can contribute to the typical landscapes. If decreased recombination in males is favored for any reason, localizing crossovers at the chromosome tips will minimize the recombination they cause. Finally, a variety of evolutionary forces may favor a certain sex-averaged recombination rate (which can vary across the genome) (Otto 2009). If one or more of the first three factors favor sex-specific changes in recombination, this last factor could inhibit recombination in the other sex following along as a correlated response, thus leading to sex differences in the recombination landscape.
In sum, none of the mechanistic or adaptive hypotheses by themselves offer a compelling explanation for sex differences in recombination that appear to be widespread across eukaryotes. At present, it seems that some combination of these hypotheses (or some as-yet unknown factor) is responsible.
Regardless of how and why sexual dimorphism in the recombination landscape has evolved, it has major consequences for the study of evolution that remain underappreciated. They likely play key roles in the adaptive evolution of recombination, can potentially influence the outcome of hybridization and population differentiation, and can have dramatic impacts on sex chromosome evolution and the resolution of sexually antagonistic selection. These considerations argüe that it may often be most useful to consider recombination in males and females as independent features of the genome, and to incorporate those differences into evolutionary theory.
Supplementary Material
Acknowledgments
We thank Maria Servedio, Thomas Lenormand, Yoel Stuart, members of the Kirkpatrick lab, and an anonymous reviewer for helpful feedback. This work was supported by National Institutes of Health grant R01-GM116853 to MK.
The authors wish to be identified to the reviewers
Literature Cited
- Amores A, Wilson CA, Allard CA, Detrich HW, and Postlethwait JH 2017. Cold fusion: massive karyotype evolution in the Antarctic bullhead notothen Notothenia coriiceps. G3: Genes, Genomes, Genetics 7:2195–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt PF, Hwa T, and Petrov DA 2005. Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects. Journal of Molecular Evolution 60:748–763. [DOI] [PubMed] [Google Scholar]
- Augstenová B, Johnson Pokorná M, Altmanová M, Frynta D, Rovatsos M, and Kratochvíl L 2018. ZW, XY, and yet ZW: Sex chromosome evolution in snakes even more complicated. Evolution 72:1701–1707. [DOI] [PubMed] [Google Scholar]
- Baack EJ, and Rieseberg LH 2007. A genomic view of introgression and hybrid speciation. Current Opinion in Genetics & Development 17:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, Hahn MW et al. 2014. Sex determination: why so many ways of doing it? PLoS Biology 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT et al. 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Research 20:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N 1995. A general model for the evolution of recombination. Genetics Research 65:123–144. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G et al. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J, Hayman D, and Hope R 1986. Novel sex differences in linkage values and meiotic chromosome behaviour in a marsupial. Nature 323:59–60. [DOI] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam K-WG, Sarbajna S, Odenthal-Hesse L, May CA, and Jeffreys AJ 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nature Genetics 42:859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Gardner J, Bader B, Yong L, and Charlesworth D 2019. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proceedings of the National Academy of Sciences 116:6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241:3–17. [DOI] [PubMed] [Google Scholar]
- Berner D, and Roesti M 2017. Genomics of adaptive divergence with chromosome-scale heterogeneity in crossover rate. Molecular Ecology 26:6351–6369. [DOI] [PubMed] [Google Scholar]
- Beukeboom LW, and Perrin N 2014, The evolution of sex determination. Oxford, UK, Oxford University Press, USA. [Google Scholar]
- Bhérer C, Campbell CL, and Auton A 2017. Refined genetic maps reveal sexual dimorphism in human meiotic recombination at multiple scales. Nature Communications 8:14994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP 1986. CpG-rich islands and the function of DNA methylation. Nature 321:209–213. [DOI] [PubMed] [Google Scholar]
- Birky CW, and Walsh JB 1988. Effects of linkage on rates of molecular evolution. Proceedings of the National Academy of Sciences 85:6414–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmon H, and Brandvain Y 2017. Long-term fragility of Y chromosomes is dominated by short-term resolution of sexual antagonism. Genetics 207:1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, and Perrin N 2013. Sex-chromosome turnovers: the hot-potato model. The American Naturalist 183:140–146. [DOI] [PubMed] [Google Scholar]
- Brandvain Y, and Coop G 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics 190:709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A, Dufresnes C, and Perrin N 2016a. High-density sex-specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 116:177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A, Rodrigues N, and Perrin N 2016b. High-density linkage maps fail to detect any genetic component to sex determination in a Rana temporaria family. Journal of Evolutionary Biology 29:220–225. [DOI] [PubMed] [Google Scholar]
- Brick K, Thibault-Sennett S, Smagulova F, Lam K-WG, Pu Y, Pratto F, Camerini-Otero RD et al. 2018. Extensive sex differences at the initiation of genetic recombination. Nature 561:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB 1927. The relation of the age of the female to crossing over in the third chromosome of Drosophila melanogaster. The Journal of General Physiology 8:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, and Weber JL 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. The American Journal of Human Genetics 63:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Jackson DA, and Hassan AB 2001. A view through the clouds of imprinting. The FASEB Journal 15:1694–1703. [DOI] [PubMed] [Google Scholar]
- Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Research 25:1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Bell G, and Harvey PH 1991. Sex differences in recombination. Journal of Evolutionary Biology 4:259–277. [Google Scholar]
- Butlin RK 2005. Recombination and speciation. Molecular Ecology 14:2621–2635. [DOI] [PubMed] [Google Scholar]
- Calderon P, and Pigozzi M 2006. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosome Research 14:605–612. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Bhérer C, Morrow BE, Boyko AR, and Auton A 2016. A pedigree-based map of recombination in the domestic dog genome. G3: Genes, Genomes, Genetics 6:3517–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ramos R, Harvey SC, and Penman DJ 2009. Sex-specific differences in the synaptonemal complex in the genus Oreochromis (Cichlidae). Genetica 135:325–332. [DOI] [PubMed] [Google Scholar]
- Cano M, and Santos J 1990. Chiasma frequencies and distributions in gomphocerine grasshoppers: a comparative study between sexes. Heredity 64:17. [Google Scholar]
- Castaño-Sánchez C, Fuji K, Ozaki A, Hasegawa O, Sakamoto T, Morishima K, Nakayama I et al. 2010. A second generation genetic linkage map of Japanese flounder (Paralichthys olivaceus). BMC Genomics 11:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, and Charlesworth B 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genetical Research 35:205–214. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, and Marais G 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118. [DOI] [PubMed] [Google Scholar]
- Chiang T, Duncan FE, Schindler K, Schultz RM, and Lampson MA 2010. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Current Biology 20:1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martlnez-Vargas J, Ventura J, Searle JB, Schultz RM et al. 2014. Centromere strength provides the celi biological basis for meiotic drive and karyotype evolution in mice. Current Biology 24:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Bois PR, Feingold E, Sherman SL, and Cheung VG 2009. Genetic analysis of variation in human meiotic recombination. PLoS Genetics 5:e1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, and Clark AG 2010. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64:3417–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MA, Joshi R, Moore EC, Nandamuri SP, Gammerdinger WJ, Roberts RB, Carleton KL et al. 2018. Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes. bioRxiv:383992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, and Przeworski M 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319:1395–1398. [DOI] [PubMed] [Google Scholar]
- Cooper TF 2007. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biology 5:e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE et al. 2009. A new standard genetic map for the laboratory mouse. Genetics 182:1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, and Orr HA 1989. Patterns of speciation in Drosophila. Evolution 43:362–381. [DOI] [PubMed] [Google Scholar]
- Dawe RK, and Cande WZ 1996. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proceedings of the National Academy of Sciences 93:8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Jasin M, and Keeney S 2015. Local and sex-specific biases in crossover vs. noncrossover outcomes at meiotic recombination hot spots in mice. Genes & Development 29:1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vicente M, and Tanksley S 1991. Genome-wide reduction in recombination of backcross progeny derived from male versus female gametes in an interspecific cross of tomato. Theoretical and Applied Genetics 83:173–178. [DOI] [PubMed] [Google Scholar]
- Devaux P, Kilian A, and Kleinhofs A 1995. Comparative mapping of the barley genome with male and female recombination-derived, doubled haploid populations. Molecular and General Genetics 249:600–608. [DOI] [PubMed] [Google Scholar]
- Dewees AA 1975. Genetic modification of recombination rate in Tribolium castaneum. Genetics 81:537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, Zanni V et al. 2007. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genetics 3:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL 2017. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics 205:155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, and Payseur BA 2011a. Evolution of the genomic recombination rate in murid rodents. Genetics 187:643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- —. 2011b. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genetics 7:e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, and Arndt PF 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genetics 4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink MG, van As P, Veenendaal T, Crooijmans RP, and Groenen MA 2010. Regional differences in recombination hotspots between two chicken populations. BMC Genetics 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, and Kelly JK 2015. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution 69:1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, and Saunders A 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322:1559–1562. [DOI] [PubMed] [Google Scholar]
- Franch R, Louro B, Tsalavouta M, Chatziplis D, Tsigenopoulos CS, Sarropoulou E, Antonello J et al. 2006. A genetic linkage map of the hermaphrodite teleost fish Sparus aurata L. Genetics 174:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman BL, and Evans BJ 2018. Divergent evolutionary trajectories of two young, homomorphic, and closely related sex chromosome systems. Genome Biology and Evolution 10:742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi K, Gautier A, Danzmann RG, Gharbi S, Sakamoto T, H0yheim B, Taggart JB et al. 2006. A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172:2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, and Mézard C 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genetics 7:e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn JR, Al-Asmar N, Fasnacht R, Maylor-Hagen H, Peinado V, Rubio C, Broman KW et al. 2016. Correlations between synaptic initiation and meiotic recombination: a study of humans and mice. The American Journal of Human Genetics 98:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn JR, Rubio C, Broman KW, Hunt PA, and Hassold T 2013. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PloS One 8:e85075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, and Berner D 2018. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Molecular Ecology 27:2477–2497. [DOI] [PubMed] [Google Scholar]
- Haig D 2010. Games in tetrads: segregation, recombination, and meiotic drive. The American Naturalist 176:404–413. [DOI] [PubMed] [Google Scholar]
- Haig D, and Grafen A 1991. Genetic scrambling as a defence against meiotic drive. Journal of Theoretical Biology 153:531–558. [DOI] [PubMed] [Google Scholar]
- Haldane JBS 1922. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics 12:101–109. [Google Scholar]
- Halldorsson BV, Palsson G, Stefansson OA, Jonsson H, Hardarson MT, Eggertsson HP, Gunnarsson B et al. 2019. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363:eaau1043. [DOI] [PubMed] [Google Scholar]
- Hassold T, and Hunt P 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nature Reviews Genetics 2:280–291. [DOI] [PubMed] [Google Scholar]
- Hayman D, Moore H, and Evans E 1988. Further evidence of novel sex differences in chiasma distribution in marsupials. Heredity 61:455–458. [Google Scholar]
- Hedrick PW 2007. Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution 61:2750–2771. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ et al. 2012. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. The Plant Cell 24:4096–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges CA, Revenkova E, Jessberger R, Hassold TJ, and Hunt PA 2005. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nature Genetics 37:1351–1355. [DOI] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J et al. 2013. Genome analyses of single human oocytes. Cell 155:1492–1506. [DOI] [PubMed] [Google Scholar]
- Hunt PA, and Hassold TJ 2002. Sex matters in meiosis. Science 296:2181–2183. [DOI] [PubMed] [Google Scholar]
- Huxley J 1928. Sexual difference of linkage in Gammarus chevreuxi. Journal of Genetics 20:145–156. [Google Scholar]
- Ieda R, Hosoya S, Tajima S, Atsumi K, Kamiya T, Nozawa A, Aoki Y et al. 2018. Identification of the sex-determining locus in grass puffer (Takifugu niphobles) provides evidence for sex-chromosome turnover in a subset of Takifugu species. PLOS One 13:e0190635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN et al. 2018. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nature Communications 9:4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Bérénos C, Slate J, and Pemberton JM 2016. Conserved genetic architecture underlying individual recombination rate variation in a wild population of Soay sheep (Ovis aries). Genetics 203:583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Huisman J, Ellis PA, and Pemberton JM 2017. A high-density linkage map reveals sexual dimorphism in recombination landscapes in red deer (Cervus elaphus). G3: Genes, Genomes, Genetics 7:2859–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DB, Jerry DR, Khatkar MS, Raadsma HW, van der Steen H, Prochaska J, Forêt S et al. 2017. A comparative integrated gene-based linkage and locus ordering by linkage disequilibrium map for the Pacific white shrimp, Litopenaeus vannamei. Scientific Reports 7:10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DB, Jerry DR, Khatkar MS, Raadsma HW, and Zenger KR 2013. A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genomics 14:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SB, and Kirkpatrick M 2004. Haploid selection in animals. Trends in Ecology & Evolution 19:592–597. [Google Scholar]
- Juric I, Aeschbacher S, and Coop G 2016. The strength of selection against Neanderthal introgression. PLoS Genetics 12:e1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian PM, Wang M, Simons K, Ghavami F, He Y, Dukowic-Schulze S, Sundararajan A et al. 2018. High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nature Communications 9:2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Johnson T, and Barton N 2002. General models of multilocus evolution. Genetics 161:1727–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Barnard J, Gudbjartsson DF, Thorleifsson G, Jonsdottir G, Sigurdardottir S, Richardsson B et al. 2004a. Recombination rate and reproductive success in humans. Nature Genetics 36:1203–1206. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S et al. 2002. A high-resolution recombination map of the human genome. Nature Genetics 31:241–247. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Masson G, Gudbjartsson DF, Villemoes R, Magnusdottir E et al. 2014. Common and low-frequency variants associated with genome-wide recombination rate. Nature Genetics 46:11. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467:1099–1103. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, Jonsdottir GM et al. 2008. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319:1398–1401. [DOI] [PubMed] [Google Scholar]
- Kong X, Murphy K, Raj T, He C, White P, and Matise T 2004b. A combined linkage-physical map of the human genome. The American Journal of Human Genetics 75:1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper C, Stocks M, Risse JE, dos Remedios N, Farrell LL, McRae SB, Morgan TC et al. 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nature Genetics 48:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150:1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U, and Lydiate DJ 1995. RFLP mapping in Brassica nigra indicates differing recombination rates in male and female meioses. Genome 38:255–264. [DOI] [PubMed] [Google Scholar]
- Lamb N, Sherman S, and Hassold T 2005a. Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenetic and Genome Research 111:250–255. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Yu K, Shaffer J, Feingold E, and Sherman SL 2005b. Association between maternal age and meiotic recombination for trisomy 21. The American Journal of Human Genetics 76:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-Y, Lee W-J, Streelman JT, Carleton KL, Howe AE, Hulata G, Slettan A et al. 2005. A second-generation genetic linkage map of tilapia (Oreochromis spp.). Genetics 170:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T 2003. The evolution of sex dimorphism in recombination. Genetics 163:811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, and Dutheil J 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biology 3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Engelstädter J, Johnston SE, Wijnker E, and Haag CR 2016. Evolutionary mysteries in meiosis. Philosophical Transactions of the Royal Society B: Biological Sciences 371:20160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, and Hurst LD 2003. Imprinted chromosomal regions of the human genome have unusually high recombination rates. Genetics 165:1629–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Zhang X, Li B, Liu T, Chen J, Bai Z, Wang M et al. 2018. Comparison of Oryza sativa and Oryza brachyantha genomes reveals selection-driven gene escape from the centromeric regions. The Plant Cell 30:1729–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Gidskehaug L, Moen T, Hayes BJ, Berg PR, Davidson WS, Omholt SW et al. 2011. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics 12:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JG, Stine RR, and Yanowitz JL 2008. Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex. Genetics 180:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A et al. 2010. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Current Biology 20:1511–1521. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch PD 2005. Sex differences in recombination and mapping adaptations, Pages 39–47 in Mauricio R, ed. Genetics of Adaptation, Springer. [DOI] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P et al. 2012. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338:1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, O’Connell JR, VanRaden PM, Shen B, Padhi A, Sun C, Bickhart DM et al. 2015. Cattle sex-specific recombination and genetic control from a large pedigree analysis. PLoS Genetics 11:e1005387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS 2009. The centromere-drive hypothesis: a simple basis for centromere complexity, Pages 33–52 Centromere, Springer. [DOI] [PubMed] [Google Scholar]
- Malik HS, and Henikoff S 2009. Major evolutionary transitions in centromere complexity. Cell 138:1067–1082. [DOI] [PubMed] [Google Scholar]
- Mank JE 2009. The evolution of heterochiasmy: the role of sexual selection and sperm competition in determining sex-specific recombination rates in eutherian mammals. Genetics Research 91:355–363. [DOI] [PubMed] [Google Scholar]
- May K, Jacobs P, Lee M, Ratcliffe S, Robinson A, Nielsen J, and Hassold T 1990. The parental origin of the extra X chromosome in 47 XXX females. The American Journal of Human Genetics 46:754. [PMC free article] [PubMed] [Google Scholar]
- Meneely PM, Farago AF, and Kauffman TM 2002. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162:1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles LG, Isberg SR, Glenn TC, Lance SL, Dalzell P, Thomson PC, and Moran C 2009. A genetic linkage map for the saltwater crocodile (Crocodylusporosus). BMC Genomics 10:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T, Hayes B, Baranski M, Berg PR, Kj0glum S, Koop BF, Davidson WS et al. 2008. A linkage map of the Atlantic salmon (Salmo salar) based on EST-derived SNP markers. BMC Genomics 9:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli MA, and Cohen PE 2005. Not all germ cells are created equal: aspects of sexual dimorphism in mammalian meiosis. Reproduction 130:761–781. [DOI] [PubMed] [Google Scholar]
- Nachman MW 2002. Variation in recombination rate across the genome: evidence and implications. Current Opinion in Genetics & Development 12:657–663. [DOI] [PubMed] [Google Scholar]
- Nachman MW, and Payseur BA 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philosophical Transactions of the Royal Society B: Biological Sciences 367:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, and Hunt PA 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nature Reviews Genetics 13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M 1969. Linkage modification and sex difference in recombination. Genetics 63:681–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Engelstadter J, and Rieseberg LH 2016. Recombination rate evolution and the origin of species. Trends in Ecology & Evolution 31:226–236. [DOI] [PubMed] [Google Scholar]
- Ortíz-Barrientos D, Reiland J, Hey J, and Noor MA 2002. Recombination and the divergence of hybridizing species, Pages 167–178 Genetics of Mate Choice: From Sexual Selection to Sexual Isolation, Springer. [PubMed] [Google Scholar]
- Otto SP 2009. The evolutionary enigma of sex. American Naturalist 174:S1–S14. [DOI] [PubMed] [Google Scholar]
- Ottolini CS, Newnham LJ, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK et al. 2015. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nature Genetics 47:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, and Petkov P 2010. Mammalian recombination hot spots: properties, control and evolution. Nature Reviews Genetics 11:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH et al. 2008. The recombinational anatomy of a mouse chromosome. PLoS Genetics 4:e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, and Paigen K 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327:835–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Kirkpatrick M, Otto SP, Vamosi JC, Peichel CL, Valenzuela N, and Kitano J 2015. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genetics 11::e1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Broman KW, Szatkiewicz JP, and Paigen K 2007. Crossover interference underlies sex differences in recombination rates. Trends in Genetics 23:539–542. [DOI] [PubMed] [Google Scholar]
- Phillips D, Jenkins G, Macaulay M, Nibau C, Wnetrzak J, Fallding D, Colas I et al. 2015. The effect of temperature on the male and female recombination landscape of barley. New Phytologist 208:421–429. [DOI] [PubMed] [Google Scholar]
- Pigozzi M, and Solari A 1999. Equal frequencies of recombination nodules in both sexes of the pigeon suggest a basic difference with eutherian mammals. Genome 42:315–321. [PubMed] [Google Scholar]
- Plough HH 1917. The effect of temperature on crossingover in Drosophila. Journal of Experimental Zoology Part A: Ecological Genetics and Physiology 24:147–209. [Google Scholar]
- —. 1921. Further studies on the effect of temperature on crossing over. Journal of Experimental Zoology Part A: Ecological Genetics and Physiology 32:187–202. [Google Scholar]
- Presgraves DC 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Current Biology 15:1651–1656. [DOI] [PubMed] [Google Scholar]
- Reid DP, Smith C-A, Rommens M, Blanchard B, Martin-Robichaud D, and Reith M 2007. A genetic linkage map of Atlantic halibut (Hippoglossus hippoglossus L.). Genetics 177:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41:911–914. [DOI] [PubMed] [Google Scholar]
- Ritz KR, Noor MA, and Singh ND 2017. Variation in recombination rate: adaptive or not? Trends in Genetics 33:364–374. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Voelkel-Meiman K, and Roeder GS 2006. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics 174:1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N, Studer T, Dufresnes C, and Perrin N 2018. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Molecular Biology and Evolution 35:942–948. [DOI] [PubMed] [Google Scholar]
- Roth C, Sun S, Billmyre RB, Heitman J, and Magwene PM 2018. A high resolution map of meiotic recombination in Cryptococcus deneoformans demonstrates decreased recombination in unisexual reproduction. Genetics 209:567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runemark A, Eroukhmanoff F, Nava-Bolaños A, Hermansen JS, and Meier JI 2018. Hybridization, sex-specific genomic architecture and local adaptation. Phil. Trans. R. Soc. B 373:20170419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Danzmann RG, Gharbi K, Howard P, Ozaki A, Khoo SK, Woram RA et al. 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155:1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandovici I, Kassovska-Bratinova S, Vaughan JE, Stewart R, Leppert M, and Sapienza C 2006. Human imprinted chromosomal regions are historical hot-spots of recombination. PLoS Genetics 2:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, Cheng C, Dagilis AJ, Ishikawa A, Kitano J, Peichel CL, and Kirkpatrick M 2018. Sex differences in recombination in sticklebacks. G3: Genes, Genomes, Genetics 8:1971–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C et al. 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular Cell 48:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, and Flint J 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biology 4:e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, Perlman H, Yan Y, Walker C, Corley-Smith G, Brandhorst B, and Postlethwait J 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Mugal CF, Qvarnström A, and Ellegren H 2016. High-resolution mapping of crossover and non-crossover recombination events by whole-genome re-sequencing of an avian pedigree. PLoS Genetics 12:e1006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, and Meissner A 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Feulner PG, Johnston SE, Santure AW, and Smadja CM 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philosophical Transactions of the Royal Society B: Biological Sciences 372:20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A et al. 2005. A common inversion under selection in Europeans. Nature Genetics 37:129–137. [DOI] [PubMed] [Google Scholar]
- Sugawara S, and Mikamo K 1983. Absence of correlation between univalent formation and meiotic nondisjunction in aged female Chlnese hamsters. Cytogenetic and Genome Research 35:34–40. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang J, Sang M, Jiang L, Zhao B, Cheng T, Zhang Q et al. 2017. Landscaping crossover interference across a genome. Trends in Plant Science 22:894–907. [DOI] [PubMed] [Google Scholar]
- Sutherland BJ, Rico C, Audet C, and Bernatchez L 2017. Sex chromosome evolution, heterochiasmy and physiological QTL in the salmonid Brook Charr Salvelinus fontinalis. G3: Genes, Genomes, Genetics 7:2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tease C, and Hultén MA 2004. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenetic and Genome Research 107:208–215. [DOI] [PubMed] [Google Scholar]
- Theodosiou L, McMillan W, and Puebla O 2016. Recombination in the eggs and sperm in a simultaneously hermaphroditic vertebrate . Proceedings of the Royal Society B: Biological Sciences 283:20161821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgasheva AA, and Borodin PM 2017. Immunocytological analysis of meiotic recombination in the gray goose (Anser anser). Cytogenetic and Genome Research 151:27–35. [DOI] [PubMed] [Google Scholar]
- Tortereau F, Servin B, Frantz L, Megens H-J, Milan D, Rohrer G, Wiedmann R et al. 2012. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics 13:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MA, Rodrigues N, Perrin N, and Kirkpatrick M 2018. A reciprocal translocation radically reshapes sex-linked inheritance in the common frog. Molecular Ecology:doi: 10.1111/mec.14990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers R 1988. Sex differences in rates of recombination and sexual selection, Pages 270–286 in Michod RE, and Levin BR, eds. The evolution of sex: an examination of current ideas. Sunderland, MA, Sinauer Press. [Google Scholar]
- van Doorn GS, and Kirkpatrick M 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449:909–912. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, and Kirkpatrick M 2010. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186:629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren BM, Campagna L, Helm B, Illera JC, Lovette IJ, and Liedvogel M 2017. Correlated patterns of genetic diversity and differentiation across an avian family. Molecular Ecology 26:3982–3997. [DOI] [PubMed] [Google Scholar]
- Van Oers K, Santure A, De Cauwer I, Van Bers NE, Crooijmans RP, Sheldon BC, Visser M et al. 2014. Replicated high-density genetic maps of two great tit populations reveal fine-scale genomic departures from sex-equal recombination rates. Heredity 112:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veller C, Kleckner N, and Nowak MA 2019. A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and Mendel’s second law. Proceedings of the National Academy of Sciences 116:1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn O, Turner I, Mathieson I, de Groot N, Bontrop R, and McVean G 2014. Strong male bias drives germline mutation in chimpanzees. Science 344:1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay N, Weissensteiner M, Burri R, Kawakami T, Ellegren H, and Wolf JB 2017. Genome-wide patterns of variation in genetic diversity are shared among populations, species and higher order taxa. Molecular Ecology 26:4284–4295. [DOI] [PubMed] [Google Scholar]
- Wang J, Li L, and Zhang G 2016a. A high-density SNP genetic linkage map and QTL analysis of growth-related traits in a hybrid family of oysters (Crassostrea gigas × Crassostrea angulata) using genotyping-by-sequencing. G3: Genes, Genomes, Genetics 6:1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Shen B, Jiang J, Li J, and Ma L 2016b. Effect of sex, age and genetics on crossover interference in cattle. Scientific Reports 6:37698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson I, and Callan H 1963. The form of bivalent chromosomes in newt oocytes at first metaphase of meiosis. Journal of Cell Science 3:281–295. [Google Scholar]
- West SA, Lively CM, and Read AF 1999. A pluralist approach to sex and recombination. Journal of Evolutionary Biology 12:1003–1012. [Google Scholar]
- Wolf JB, and Ellegren H 2017. Making sense of genomic islands of differentiation in light of speciation. Nature Reviews Genetics 18:87. [DOI] [PubMed] [Google Scholar]
- Wong AK, Ruhe AL, Dumont BL, Robertson KR, Guerrero G, Shull SM, Ziegle JS et al. 2010. A comprehensive linkage map of the dog genome. Genetics 184:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woram R, McGowan C, Stout J, Gharbi K, Ferguson M, Hoyheim B, Davidson E et al. 2004. A genetic linkage map for Arctic char (Salvelinus alpinus): evidence for higher recombination rates and segregation distortion in hybrid versus pure strain mapping parents. Genome 47:304–315. [DOI] [PubMed] [Google Scholar]
- Wu CI 2001. The genic view of the process of speciation. Journal of Evolutionary Biology 14:851–865. [Google Scholar]
- Zetka M-C, and Rose AM 1990. Sex-related differences in crossing over in Caenorhabditis elegans. Genetics 126:355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, and Kleckner N 2016. A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Seminars in Cell & Developmental Biology 54:135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.