Abstract
Human interleukin 15 (IL-15) circulates in blood as a stable molecular complex with the soluble IL-15 receptor alpha (sIL-15Rα). This heterodimeric IL-15:sIL-15Rα complex (hetIL-15) shows therapeutic potential by promoting the growth, mobilization and activation of lymphocytes and is currently evaluated in clinical trials. Favorable pharmacokinetic properties are associated with the heterodimeric formation and the glycosylation of hetIL-15, which, however, remains largely uncharacterized. We report the site-specific N-and O-glycosylation of two clinically relevant large-scale preparations of HEK293-derived recombinant human hetIL-15. Intact IL-15 and sIL-15Rα and derived glycans and glycopeptides were separately profiled using multiple LC-MS/MS strategies. IL-15 Asn79 and sIL-15Rα Asn107 carried the same repertoire of biosynthetically-related N-glycans covering mostly α1–6-core-fucosylated and β-GlcNAc-terminating complex-type structures. The two potential IL-15 N-glycosylation sites (Asn71 and Asn112) located at the IL-2 receptor interface were unoccupied. Mass analysis of intact IL-15 confirmed its N-glycosylation and suggested that Asn79-glycosylation partially prevents Asn77-deamidation. IL-15 contained no O-glycans, whereas sIL-15Rα was heavily O-glycosylated with partially sialylated core 1 and 2-type mono- to hexasaccharides on Thr2, Thr81, Thr86, Thr156, Ser158, and Ser160. The sialoglycans displayed α2–3- and α2–6-NeuAc-type sialylation. Non-human, potentially immunogenic glycoepitopes (e.g. N-glycolylneuraminic acid and α-galactosylation) were not displayed by hetIL-15. Highly reproducible glycosylation of IL-15 and sIL-15Rα of two batches of hetIL-15 demonstrated consistent manufacturing and purification. In conclusion, we document the heterogeneous and reproducible N- and O-glycosylation of large-scale preparations of the therapeutic candidate hetIL-15. Site-specific mapping of these molecular features is important to evaluate the consistent large-scale production and clinical efficacy of hetIL-15.
Keywords: N-glycosylation, O-glycosylation, LC-MS/MS, Glycoprotein, IL-15, IL-15Rα
Introduction
The γ chain cytokine interleukin 15 (IL-15) plays a key role in lymphocyte development, survival, proliferation, and activation [1–5]. These characteristics have been associated with the anti-cancer effects of IL-15 demonstrated in several preclinical cancer models [6–8]. Thus, IL-15 is considered as a promising therapeutic candidate in cancer immunotherapy [9–11].
IL-15 functions by binding to the beta and gamma subunits of the interleukin 2 receptor complex (IL-2Rβ:IL-2Rγ also called CD122/CD132) presented on responding cells [12]. Co-expression of the IL-15 receptor alpha (IL-15Rα) subunit is a requirement for the efficient production and secretion of IL-15 under physiological conditions [13–15]. Mechanistically, this requirement is explained by the intracellular association and the resulting stability enhancement of IL-15 and IL-15Rα. This bioactive heterodimeric complex is efficiently transported to the cell surface where it is rapidly cleaved and released into circulation as a stable bioactive cytokine composed of two polypeptide chains, IL-15 and soluble IL-15Rα (sIL-15Rα) [16–18]. This soluble complex was named heterodimeric IL-15 (hetIL-15). Due to its therapeutic potential, we recently developed robust methods for the recombinant production and purification of soluble bioactive hetIL-15 [19]. The heterodimer was synthesized, processed, and secreted by a stable, clonal human embryonic kidney 293 (HEK293) cell line. This optimized expression method allowed molecular characterization of hetIL-15 confirming Gly170-His171 of IL-15Rα as the single proteolytic cleavage site facilitating cell membrane release, and enabled evaluation of its pharmacokinetics and in vivo bioactivity [19]. Importantly, hetIL-15 was found to be heavily N- and O-glycosylated, and these post-translational modifications and the heterodimer formation were shown to affect the physicochemical properties and enhance the bioactivity of IL-15 [19]. Nonetheless, the exact glycan structures, their sites and distribution of the hetIL-15 complex remain to date uncharacterized.
Since a growing body of literature documents the modulatory effects of glycosylation on cytokine function [20–24], we performed a detailed site-specific characterization of the N-and O-linked glycosylation of HEK-derived IL-15 and IL-15Rα using multiple analytical LC-MS/MS-based approaches including glycoprofiling of released glycans, glycopeptides and intact glycoproteins of two clinically relevant large-scale preparations of hetIL-15. Our results contribute to the characterization of the structure and function of this heterodimeric cytokine, important for the homeostasis of the immune system.
Materials and methods
Production and purification of two large-scale preparations of human hetIL-15
Human hetIL-15 was expressed as previously described [19]. In brief, RNA/codon optimized genes encoding the two polypeptide chains of hetIL-15, IL-15 and full-length IL-15Rα (see Table 1 for sequences) were expressed in a stable clonal HEK293 cell line using endotoxin-free DNA plasmids (Qiagen Endofree Giga kit, Hilden, Germany). Cells were adapted to and grown in serum-free media.
Table 1.
Overview of the full length and mature amino acid sequences and numbering (subscript) and the identified N- and O-glycosylation sites (occupied sites in bold, red; unoccupied sites in bold, black) of human IL-15 and sIL-15Rα. The same amino acid sequences of IL-15 and sIL-15Rα were used for the two large-scale preparations of hetIL-15 (i.e. the cGMP and EN lots)
| Protein (UniProt) |
Engineered sequences and observed glycosylation sites of human IL-15 and sIL-15Rα (deleted/truncated sequences in the secreted/soluble hetIL-15 complex) |
|---|---|
|
Human IL-15 (P40933) - cGMP lot - EN lot |
(MRISKPHLRSISIQCYLCLLLNSHFLTEAGIHVFILGCFSAGLPKTE)N1WVNVISDLKKIE DLIQSMHIDATLYTESDVHPSCKVTAMKCFLLELQVISLESGDASIHDTVENLIILAN71 NSLSSNGN79VTESGCKECEELEEKNIKEFLQSFVHIVQMFIN112TS114 |
|
Human sIL-15Rα (Q13261) - cGMP lot - EN lot |
(MAPRRARGCRTLGLPALLLLLLLRPPATRG)I1T2CPPPMSVEHADIWVKSYSLYSRERYI CNSGFKRKAGTSSLTECVLNKATNVAHWTTPSLKCIRDPALVHQRPAPPSTVT81TAG VT86PQPESLSPSGKEPAASSPSSN107NTAATTAAIVPGSQLMPSKSPSTGTTEISSHESS HGTPSQTTAKNWELT156AS158AS160HQPPGVYPQG170(HSDTTVAISTSTVLLCGLSAVSLL ACYLKSRQTPPLASVEMEAMEALPVTWGTSSRDEDLENCSHHL) |
Large-scale production and purification of hetIL-15 was performed by Xcellerex, Inc. (Marlborough, MA). Samples of the final purified bulk drug substance from two large-scale production and purification lots were obtained for our analysis. The first lot (EN627–01-13–001) was the “engineering lot” (hereafter referred to as the “EN lot”) that was used in previous toxicology studies. The second lot (627–01-13–002) was manufactured by the same procedure under current good manufacturing practices (cGMP) (hereafter referred to as the “cGMP lot”) for the manufacture of biotechnology drug substances. The cGMP lot was approved for investigational human use. The facilities and equipment used for the cGMP lot are qualified with regular calibration and maintenance programs in place. The manufacturing process took place in the Xcellerex FlexFactory in which, as far as is practical, all product contact surfaces are disposable, and each unit operation within the FlexFactory is self-contained in its own controlled environment module.
The clarified raw harvest of the hetIL-15 lots were concentrated through ultrafiltration/diafiltration and subjected to ion exchange chromatography and viral inactivation in low pH buffers. After a second ultrafiltration/diafiltration step, the purified hetIL-15 complex was subjected to ceramic hydroxyapatite chromatography, viral filtration (Planova) and a final ultrafiltration/diafiltration step.
Separation and detection of human IL-15 and sIL-15Rα of hetIL-15 preparations
The two polypeptide chains of the hetIL-15 complex (IL-15 and sIL-15Rα) were separated by reversed-phase high performance liquid chromatography (RP-HPLC) under non-reducing conditions. Slightly different conditions were used to separate the two examined hetIL-15 preparations (i.e., EN and cGMP lots): The EN lot of hetIL-15 was separated at a flow rate of 5 ml/min on a preparative PLRP-S polymeric C18 RP-HPLC column (i.d., 50 mm, length, 150 mm, Agilent). Solvent A was 0.1 % (vol: vol) trifluoroacetic acid (TFA) in water. The gradient of solvent B (0.1 % TFA in acetonitrile (ACN)) was: 15–60 % (vol:vol), 280 min; and isocratic at 60 %, 10 min. The cGMP lot of hetIL-15 was separated at a flow rate of 5 ml/min on a preparative μBondapak C18 Radial-Pak RP-HPLC column (i.d., 40 mm, length, 100 mm, Waters). The gradient of solvent B (0.1 % TFA in ACN) was: 13.8 % − 70 %, 130 min; and 70 %, 10 min. For both samples a Dionex HPLC system equipped with Ultimate 3000 binary pumps model #HPG-3400A, Ultimate 3000 photodiode array detector model #PDA-3000, Ultimate 3000 column compartment model #TCC-3000, Ultimate 3000 solvent rack and degasser model #SRD-3400, Isco fraction collector model #Foxy 200 were used. Proteins eluted from the column were analyzed by SDS-PAGE, immunoblot analysis and N-terminal sequencing. The SDS-PAGE analyses of IL-15 and sIL-15Rα were performed under native and denaturing conditions on polyacrylamide gradient gels (4–20 %), and visualized by Coomassie Blue staining. The identity of IL-15 and sIL-15Rα was confirmed by Western immunoblotting using anti-human IL-15 or anti-human IL-15Rα polyclonal goat IgG antibodies (product #AF315 and #AF247, respectively, R&D Systems).
Release and handling of N- and O-glycans of human IL-15 and sIL-15Rα
The N- and O-glycans of purified and separated IL-15 and sIL-15Rα from the EN and cGMP lots of hetIL-15 were released and handled as previously described [25]. In brief, IL-15 and sIL-15Rα (approximately 60 pmol of each protein per preparation) were immobilized in separate spots on a primed 0.45 μm Immobilon-PSQ PVDF membrane (Millipore). The spots were stained with Direct Blue (Sigma-Aldrich), excised and washed in separate wells in a flat bottom polypropylene 96-well plate (Corning Incorporated, Corning, NY). N-glycans were exhaustively released using 2.5 U N-glycosidase F (Flavobacterium meningosepticum, Roche) per spot in 10 μl water by allowing incubation for 16 h at 37 °C. The released N-glycans were collected and reduced to alditols by treatment with 1 M NaBH4 in 50 mM potassium hydroxide (aq) (final concentration stated if not stated otherwise) for 3 h at 50 °C. The de-N-glycosylated spots were washed in water and O-glycans were subsequently quantitatively released by reductive β-elimination by incubation with 10 μl 0.5 M NaBH4 in 50 mM potassium hydroxide (aq) for 16 h at 50 °C. The N- and O-glycans were desalted as described [25], dried and redissolved in 10 μl water for LC-MS/MS glycome profiling.
Proteolysis of human IL-15 and sIL-15Rα
Purified and separated IL-15 and sIL-15Rα of the clinical-grade cGMP lot of hetIL-15 were reduced by incubation for 1 h at 50 °C in 15 mM dithiothreitol (aq) and carbamidomethylated by incubation for 30 min in the dark at 25 °C in 50 mM iodoacetamide (aq). The alkylation reaction was quenched by the addition of 50 mM dithiothreitol. The samples were adjusted to pH 8 with 50 mM ammonium bicarbonate (aq). The sIL-15Rα protein component was digested for 10 h at 37 °C using two additions (1 h interval) of 1 % (w/w) sequence-grade porcine trypsin (Promega) in 50 mM ammonium bicarbonate (aq). IL-15 was digested firstly with trypsin as described above and then for another 4 h at 37 °C using 2 % (w/w) of sequence-grade Staphylococcus aureus V8 endoproteinase GluC (Promega). The resulting peptide mixtures were aliquoted and dried for storage. For LC-MS/MS of the non-enriched peptide mixtures, aliquots were taken up into 0.1 % (vol/vol) formic acid (aq) and used for multiple LC-MS/MS injections. Separate aliquots were used for glycopeptide enrichment as described below.
Glycopeptide enrichment
In a separate set of experiments, glycopeptide enrichments of sIL-15Rα peptide mixtures of the clinical-grade cGMP lot of hetIL-15 were performed using hydrophilic interaction liquid chromatography (HILIC) in a micro-column solid phase extraction (SPE) format prior to LC-MS/MS analysis. The dried peptide mixtures were redissolved in 10 μl of HILIC loading solvent consisting of 80 % (vol/vol) ACN in 0.1 % (vol/vol) TFA (aq). The enrichments were carried out as described previously with minor modifications [26, 27]. In short, a stationary phase consisting of washed ZIC-HILIC resin (particle size, 10 μm; pore size, 200 Å) (Sequant/Merck) was used to pack a HILIC SPE column (approximately length, 10 mm) in a GeLoader tip (Eppendorf). The column was equilibrated in 20 μl loading solvent and the peptide mixtures were loaded manually at low flow. The column was washed twice with 10 μl loading solvent before the retained glycopeptides were eluted with 10 μl eluting solvent consisting of 0.1 % (vol/vol) TFA (aq). The eluate was dried and taken up in 20 μl 0.1 % (vol/vol) formic acid (aq) and used for multiple LC-MS/MS injections.
LC-ESI-MS/MS of N- and O-glycans released from human IL-15 and sIL-15Rα
Released N- and O-glycan alditols from the EN and cGMP lots of hetIL-15 (from ~5 pmol protein/injection) were analyzed in separate experiments on a Hypercarb porous graphitized carbon (PGC) column (particle size, 5 μm; i.d., 320 μm; length, 100 mm, Thermo Scientific) on an Agilent 1100 capillary LC (Agilent Technologies) connected to an Agilent MSD 3D ion-trap XCT Plus mass spectrometer. Separations were carried out at a constant flow rate of 2 μl/min using a multi-segmented increasing linear gradients of solvent B (10 mM ammonium bicarbonate in ACN) in solvent A (10 mM ammonium bicarbonate (aq)): 2–16 % (vol/vol) solvent B for 45 min, 16–45 % solvent B for 20 min, 45 % solvent B for 6 min and re-equilibration in solvent A for 10 min. ESI-MS/MS was performed in negative ion polarity mode with the following settings: drying gas temperature, 325 °C; drying gas flow, 7 l/min; nebulizer gas, 18 psi; skimmer, trap drive and capillary exit, −40 V, −99.1 V and −166 V, respectively; smart fragmentation was ramping in the interval 30–200 % amplitude with a maximum accumulation time of 200 ms and ICC ion target of 100,000 ions. Data acquisition was performed with two scan events: An MS full scan (m/z 100–2,200) with a scan speed of 8,100 m/z/s and data-dependent MS/MS scans after collision induced dissociation (CID) of the top two most intense precursor ions with a minimum threshold of 30,000 and a relative threshold of 5 % relative to the base peak. No dynamic exclusion was activated to enable MS/MS triggering of closely eluting yet separated N-glycan isoforms. Two separate LC-MS/MS injections and analyses were performed for all glycome profiles.
LC-ESI-MS/MS of human IL-15 and sIL-15Rα peptides, enriched glycopeptides and intact protein
Human IL-15 and sIL-15Rα peptides of the clinical-grade cGMP lot of hetIL-15 (1–5 pmol/injection) were analyzed with and without prior glycopeptide enrichment using LC-ESI-MS/MS in positive polarity mode on an HCT 3D ion trap (Bruker Daltonics) coupled to an Ultimate 3000 LC (Dionex). Samples were loaded onto a ProteCol C18aq RP-HPLC column (i.d., 300 μm; length, 100 mm; particle size, 3 μm and pore size, 300 Å) (SGE, Australia). The column was equilibrated in 100 % solvent A (0.1 % (vol/vol) formic acid (aq)), the sample loaded and a gradient up to 50 % (0.8 %/min slope) solvent B (0.1 % (vol/vol) formic acid in ACN) was applied before washing the column in 80 % solvent B for 10 min and re-equilibration in the starting LC solvent condition. A constant flow rate of 5 μl/min was used and the column was heated to 45 °C. Separate data acquisition strategies were used to cover two fragmentation modes: 1) LC-MS/MS analysis, where a MS full scan (m/z 400–2,200, scan speed: 8,100 m/z/s) was followed by data-dependent fragmentation of the four most abundant precursors in each full scan using CID (ICC target 200,000 ions, maximum accumulation time: 200 ms) and 2) the same LC-MS/MS setup as in 1), but with electron transfer dissociation (ETD) using the following settings of the top two most abundant precursor ions in each full scan: ICC reactant target: 600,000, reactant accumulation time: 7–20 ms (maximum 200 ms), reaction time: 100 ms.
Intact IL-15 of the clinical-grade cGMP lot of hetIL-15 (~1 μg protein/injection) was analyzed by LC-ESI-MS in positive ion polarity mode using a high resolution/high mass accuracy quadrupole time-of-flight (QTOF) 6538 mass spectrometer (Agilent Technologies) coupled to a capillary LC (Agilent 1260 Infinity) installed with a C4 RP-HPLC column (Proteocol C4Q, SGE; particle size, 3 μm; pore size, 300 Å; i.d., 300 μm; length, 100 mm). The column was equilibrated in identical mobile phases as for the C18 column (described above) with a gradient up to 60 % (2 %/min slope) of solvent B before washing the column in 99 % solvent B for 10 min and re-equilibration in the starting LC solvent condition. A constant flow rate of 5 μl/min and 1 μl injections were used. The following MS settings were used in high resolution (4 GHz) mode: MS full scan (m/z 400–2,500); fragmentor potentials, 200 V; drying gas temperature, 300 °C; drying gas flow rate, 8 l/min; nebulizer pressure, 10 psig; capillary potential, 4,300 V; skimmer potential, 65 V. The mass accuracy of the mass spectrometer was calibrated using a tune mix (Agilent) prior to data acquisition. A mass calibrant (Agilent) was infused continuously during the LC-MS run to allow accurate and automated in-spectrum mass calibration. Generally, mass accuracies better than 2 ppm were achieved.
Profiling of intact sIL-15Rα of the clinical-grade cGMP lot of hetIL-15 was performed using matrix assisted laser desorption ionization (MALDI)-TOF MS on a MicroFlex (Bruker Daltonics). In brief, 10 pmol reduced and alkylated human sIL-15Rα was mixed with 6 mg/ml sinapinic acid in a 1:1 (vol:vol) mixture of 0.1 % (vol/vol) TFA (aq):ACN on a MALDI target and dried. In total, 300–500 laser shots (20 Hz) were accumulated with the lowest possible laser energy. Data acquisition was performed in linear mode using positive ionization polarity mode after a multi-point mass axis calibration using a protein calibrant (Bruker). All mass spectra were viewed and analyzed using DataAnalysis v4.0 (Bruker Daltonics).
Determination of monosaccharide compositions, glycan structures, relative abundances, amino acid attachment sites and site occupancies
The monosaccharide compositions and specific N- and O-glycan structures of IL-15 and sIL-15Ra were manually annotated from the PGC-LC-MS/MS glycomics data using three analytical features including molecular mass, CID-MS/MS fragmentation data and PGC-LC retention time as recently described [28]. Only species containing interpretable MS/MS spectra with a qualitative match to previously recorded spectra of validated N-glycan structures found in UniCarbKB (http://unicarbkb.org) or in our recently published glycome datasets [28, 29] were included in the study. Structural ambiguity of the monosaccharide linkage types and branch points remained for a subset of the glycans following PGC-LC-ESI-MS/MS in particular for the O-glycan structures since no common core exist. Such structures were left without complete structural assignment. The N-glycans were partially assigned using the knowledge and established constraints of the human biosynthetic machinery. The relative molar abundances of the N- and O-glycans were determined based on the relative EIC areas of the observed charge states of all the identified glycan species. This has been shown to be an accurate method for profiling released glycans [30].
The site-specific glycosylation data of IL-15 and sIL-15Rα were manually annotated as described [31, 32]. For sIL-15Rα, the LC-MS/MS data of the non-enriched tryptic peptide mixtures were used to estimate the O-glycosylation site-occupancies and the HILIC-enriched glycopeptide data were used to determine the site-specific distribution of the O-glycans. The ratios of the EIC area of the glycosylated species to the sum of the EIC areas of both the non-glycosylated and glycosylated species were used to estimate the glycan occupancies. This has recently been shown to be a reasonably accurate estimate when evaluating the occupancy of small neutral glycans [33]. The site localization of the O-glycans was, in part, performed using CID fragmentation of the individual glycopeptides to validate the site-specific monosaccharide compositions and, in part, by ETD fragmentation to determine the exact glycan attachment sites. The assignment of c- and z-ions was aided by matching the fragment ions to in silico fragmentation products using GPMAW v.9.51 (Lighthouse, Odense, Denmark). For IL-15, the localization of the single N-glycosylation site was performed using non-enriched tryptic and GluC digested peptide mixtures. The site distribution of IL-15 N-glycans was assessed qualitatively on the glycoprotein and glycopeptide level and quantitatively using the glycomics data, which could be performed since only a single site was found to be occupied. The N-glycosylation site occupancy of IL-15 was assessed using the more quantitative SDS-PAGE and HPLC techniques.
Raw mass spectra of intact IL-15 were handled using MassHunter workstation v.B.06 (Agilent). BioConfirm was used to deconvolute the acquired spectra using default settings. GPMAW v.9.51 was used to generate theoretical isotopic distributions of intact IL-15 displaying different degrees of Asn77-deamidation i.e. 0 %, 25 %, 75 %, and 100 %. The best fit of the theoretical isotope distributions of IL-15 to the experimental data was manually assessed.
Mapping the spatial presentation of the N- and O-glycosylation sites of hetIL-15
The identified N- and O-glycosylation sites of human hetIL-15 were spatially mapped on a 3D structure of a quaternary complex of hetIL-15 with a heterodimeric surface receptor complex human IL-2Rβ:IL-2Rγ using Open-source Pymol v1.3. The crystal structure (PDB: 4GS7, resolution: 2.35 Å) covered the entire glycosylated region of IL-15, but only the lightly O-glycosylated N-terminal region of the sIL-15Rα polypeptide chain [34]. At present, no other 3D structure or homology model covering a larger region of sIL-15Rα is available. Water molecules and heterodimeric atoms were removed upon visual presentation of the IL-15 and sIL-15Rα glycosylation sites on the quaternary complex.
Results
Analysis of the purity of the EN and cGMP lots of hetIL-15 and isolation of IL-15 and sIL-15Rα
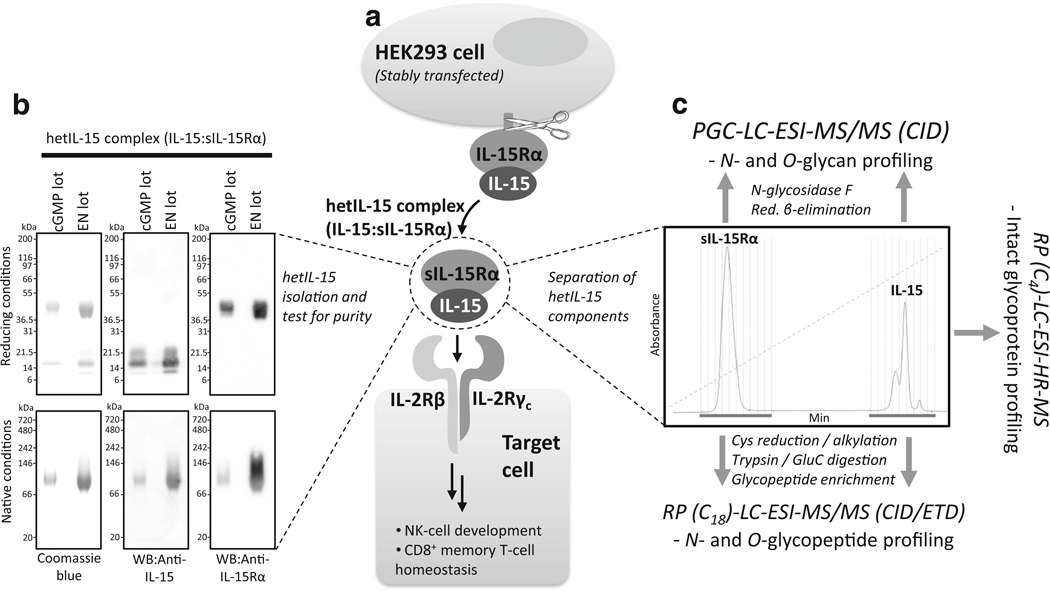
To facilitate a deep structural glycosylation-centric characterization of the hetIL-15, the engineered heterodimer was expressed by a stably-transfected HEK293 cell clone and the soluble complex purified from the culture medium, Fig. 1a. SDS-PAGE analyses followed by Coomassie staining and Western blotting were utilized to demonstrate that the two clinically relevant large-scale preparations of hetIL-15 (i.e., EN and cGMP lots) were purified to high purity, Fig. 1b. The EN lot of hetIL-15 was the “engineering lot”, which previously has been used for toxicology studies, and the cGMP lot of hetIL-15 was manufactured the same way, but under current good manufacturing practices (cGMP) to obtain clinical-grade hetIL-15 in qualities allowing human administration.
Fig. 1.
Overview of the expression, cellular presentation, function and analysis of the soluble human hetIL-15 complex. a. Engineered human IL-15 and IL-15Rα were co-expressed and secreted by HEK293 cells as a soluble heterodimeric complex (hetIL-15) after proteolytic cleavage from the cell surface. The complex binds to the IL-2Rβ/γ receptor complex located on target cells, where it initiates a cellular response. b. The hetIL-15 complex was isolated and its purity monitored using reducing (upper) and native (lower) SDS-PAGE with Coomassie blue staining (left gels) and Western blotting using anti-IL-15 and anti-IL-15Rα antibodies (right gels). The two clinically relevant preparations of hetIL-15 (i.e. the EN and cGMP lots) are shown. c. IL-15 and sIL-15Rα were separated from their heterodimeric complex by non-reductive RP-HPLC and individually subjected to LC-MS/MS-based glycan (top), glycopeptide (bottom) and glycoprotein (right) profiling in order to characterize their N- and O-glycosylation in a detailed and site-specific manner
Under reducing and denaturing conditions, hetIL-15 produced two major bands of ~15–17 kDa and ~45 kDa. Western blots confirmed that the two bands corresponded to IL-15 and sIL-15Rα, respectively. Under native conditions, hetIL-15 migrated as a single high molecular mass band, showing that all IL-15 protein components associate stably with the sIL-15Rα components in the soluble heterodimeric complex. No difference was observed between the EN and cGMP lots of the hetIL-15 preparations. The purity and identity of the two desired gene products making up the heterodimeric complex were validated using N-terminal Edman sequencing (data not shown). Subsequently, the hetIL-15 complex was separated into its individual polypeptide chains by non-reductive RP-HPLC, Fig. 1c. The N- and O-linked glycosylation of IL-15 and sIL-15Rα were then profiled separately using LC-MS/MS-based glycan, glycopeptide and intact glycoprotein analysis.
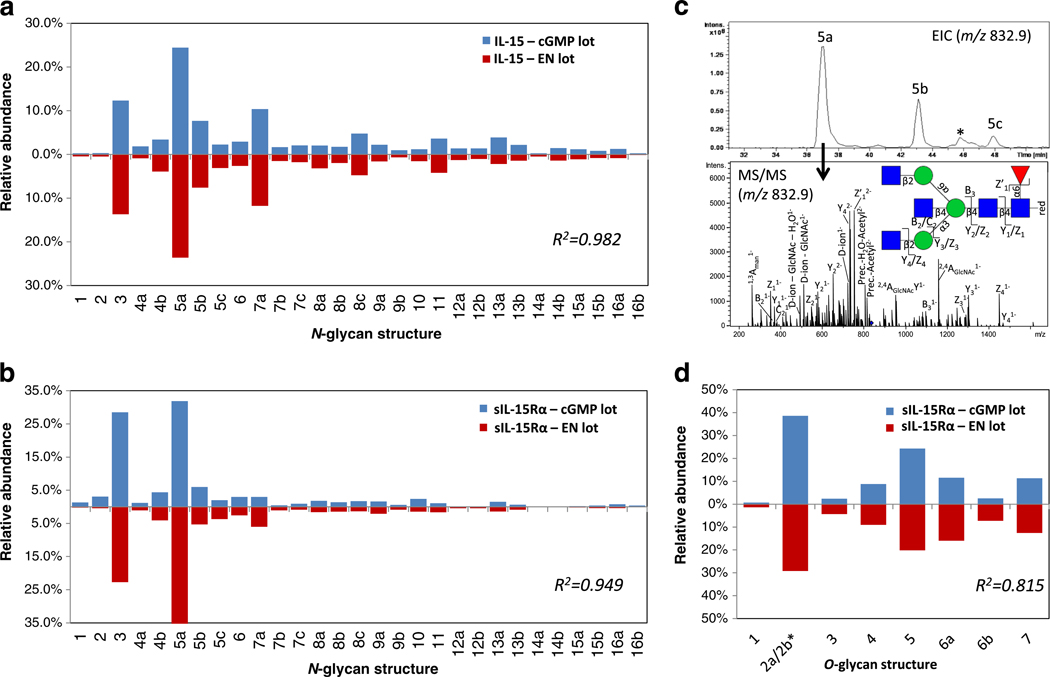
Glycomics profiling illustrates extensive and reproducible N- and O-glycan heterogeneity of IL-15 and sIL-15Rα of the EN and cGMP lots of hetIL-15
Porous graphitized carbon (PGC)-LC-ESI-MS/MS was utilized to profile and compare the N- and O-glycomes of the EN and cGMP lots of hetIL-15. Extensive N-glycan heterogeneity was observed for IL-15 and sIL-15Rα by the characterization of 29 individual glycan structures covering 16 monosaccharide compositions, Fig. 2a–b and Supplementary Table S1. The N-glycans, some of which were found to be isobaric variants, were characterized using the molecular mass of the glycan, its response to CID-MS/MS fragmentation and its relative PGC-LC retention time, see Fig. 2c and Supplementary Fig. S1 for examples of CID fragment spectra. The O-glycome profiles of sIL-15Rα showed comparably less micro-heterogeneity by the characterization of only eight glycan structures belonging to seven monosaccharide compositions, Fig. 2d and Supplementary Table S2. No O-glycans of IL-15 were detected for the two hetIL-15 preparations (data not shown). The glycan distributions of IL-15 and sIL-15Rα were established using label-free EIC-based relative quantitation, which was previously found to yield accurate estimates of the relative glycan abundances when evaluated against other glycan profiling techniques [30]. Comparison of the EN and cGMP lots of hetIL-15 demonstrated that IL-15 and sIL-15Rα were similarly N- and O-glycosylated across the two preparations (R2 = 0.815–0.982) indicating that reproducible glycosylation was achieved during the production and purification of the heterodimeric complex. The data also indicated that the IL-15 and sIL-15Rα protein components of the complex were similarly N-glycosylated (direct comparison not shown).
Fig. 2.
Glycome profiling demonstrates extensive and reproducible N-and O-glycosylation of IL-15 and sIL-15Rα in the large-scale preparations of hetIL-15 (cGMP and EN lots). N-glycans of IL-15 (a) and sIL-15Rα (b) were structurally characterized and quantitatively profiled using PGC-LC-ESI-negative ion-CID-MS/MS. c. Several isobaric N-glycan isomers were identified as exemplified by the extracted ion chromatogram (EIC) of the abundant Man3GlcNAc5Fuc1 composition (m/z 832.9 [M – 2 H]2−, upper panel) and the corresponding CID-MS/MS (bottom panel, see Fig. 3 for key to monosaccharide symbols) demonstrating three isobaric GlcNAc-terminating N-glycan isomers i.e. N-glycan structure 5a (shown), 5b and 5c (fragment spectra for the two latter N-glycans are presented in Supplementary Fig. S1). ‘*’ represents a non-glycan signal interference. d. The O-glycome profiling of sIL-15Rα showed less micro-heterogeneity. *Structure 2a/2b could not be consistently separated and were thus combined for quantitation purposes. No O-glycosylation was detected for IL-15 (data not shown). The N- and O-glycosylation profiles of IL-15 and sIL-15Rα of the EN (red bars) and cGMP (blue bars) lots of hetIL-15 were similar as evaluated by their high correlation coefficients (R2 = 0.815–0.982). The relative glycan quantities are averages of technical duplicates (see Supplementary Table S1 and S2 for exact values). The corresponding N- and O-glycan structures and their biosynthetic relationship are depicted in Fig. 3
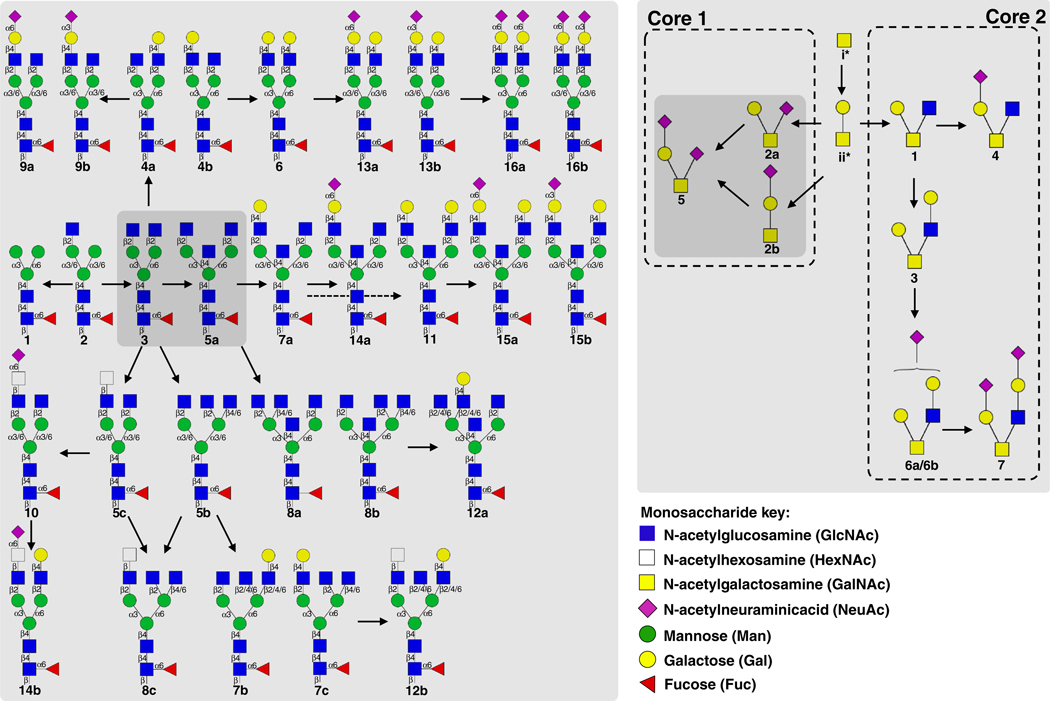
With the exception of a single truncated structure of the paucimannosidic type [35], the N-glycans, which all showed a remarkable biosynthetic relatedness, were of the complex type displaying α1–6-linked (core) fucosylation, Fig. 3. A significant number of β-GlcNAc-terminating structures were observed covering both β1–4-bisecting GlcNAcylated and β1–2/4/6-linked mono-, bi- and triantennary structures (e.g. the abundant N-glycans #3 and 5a). The remaining N-glycans predominantly displayed α2–3- and α2–6-sialylation as well as β1–4-galactosylation as terminal epitopes. The N-glycomes indicated lower levels of the more unusual N, N-diacetyllactosamine (LacdiNAc) (#5c, 8c and 12b) and sialyl-LacdiNAc (#10 and 14b) moieties. The most abundant O-glycans were core 1 type structures (#2a/b and 5), whereas core 2 type glycans (#1, 3, 4, 6 and 7) were of less abundance albeit more heterogeneous. The smaller biosynthetic precursors for the observed sIL-15Rα O-glycans i.e. Tn (GalNAcα) and T (Galβ1–3GalNAcα) glycoepitopes, which were not observed in the glycome profiling due to weak PGC retention, were identified in the O-glycopeptide analysis.
Fig. 3.
Structures and biosynthetic relationship of the observed IL-15 and sIL-15Rα N- and O-glycans. The designated numbers of the individual N-linked (left) and O-linked (right) glycans correspond to the numbering used in Fig. 2, Fig. 5 and Supplementary Tables S1–S2. Their biosynthetic interconnectivity is presented with arrows symbolizing single glycosylation enzyme reactions. The most abundant N- and O-glycans are shaded in dark grey. Monosaccharide symbols are presented according to the Essentials of Glycobiology/Consortium for Functional Glycomics nomenclature. Key: fucose (red triangle), mannose (green circle), GlcNAc (blue square), sialic acid (NeuAc) (purple diamond), galactose (yellow circle) and HexNAc (unspecified GlcNAc or GalNAc) (open square)
Site-specific glycoprofiling of sIL-15Rα reveals multiple O-glycosylation sites
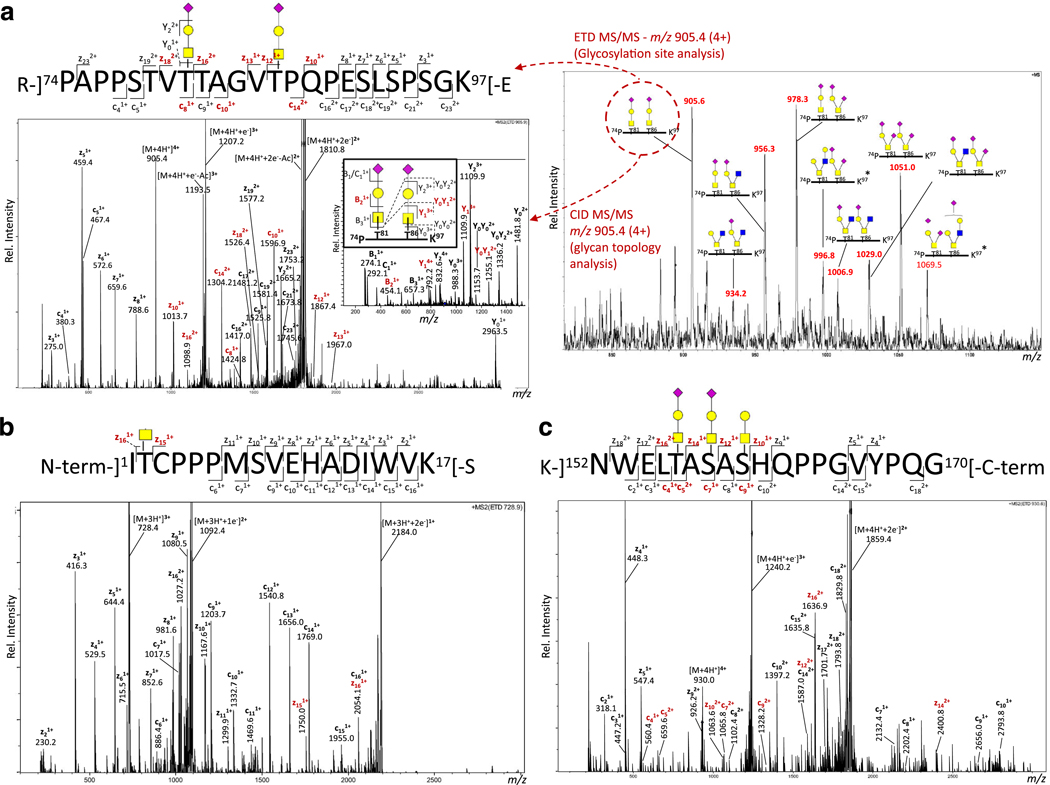
The site-specific positions of the O-glycosylation of sIL-15Rα isolated from the clinical-grade cGMP lot of hetIL-15 were investigated using RP (C18) LC-ESI-MS/MS analysis of HILIC-enriched and non-enriched tryptic peptides. Several glycoforms were identified of the doubly O-glycosylated 74PAPPSTVTTAGVTPQPESLSPSGK97 peptide; the two O-glycosylation sites (Thr81 and Thr86) and the structures of the conjugated O-glycans were validated using CID and ETD fragmentation, Fig. 4a. These orthogonal dissociation methods also validated the unusual N-terminal proline residue of this peptide formed by the cleavage of the usually trypsinresistant Arg-Pro peptide bond. In addition, O-glycosylation was identified in the sIL - 15Rα termini by the characterization of the singly O-glycosylated N-terminal peptide i.e. 1ITCPPPMSVEHADIWVK17 (Thr2) and the triply O-glycosylated C-terminal peptide i.e. 152NWELTASASHQPPGVYPQG170 (Thr156, Ser158 and Ser160), Fig. 4b–c. Characterization of these peptides simultaneously validated the termini of the mature sIL-15Rα polypeptide chain. Similar to the N-terminal peptide, the 50ATNVAHWTTPSLK62 peptide was identified to display a single GalNAc (Tn) epitope, but the site was not determined to amino acid resolution, see Table 2 for complete overview of the identified sIL-15Rα O-glycopeptides. The singly O-glycosylated peptides showed partial occupancy by the identification of the non-glycosylated counterparts in the non-enriched tryptic peptide mixture derived from sI L-15Rα. The relative abundance of the multiple O-glycosylated peptides showed limited heterogeneity of sialylated and asialylated core 1 and 2-type O-glycans in agreement with the structures identified in the O-glycome profile of sIL-15Rα. The polypeptide sequence of sIL-15Rα displayed only a single putative N-glycosylation site (sequon) at Asn107. The theoretical Asn107-containing tryptic glycopeptide (i.e. K-]98EPAASSPSSNNTAATTAAIVPGSQLMPSK126-[S) was not observed in any form in any LC-MS/MS analyses (with or without glycopeptide enrichment) possibly because of heavy O-glycosylation of this region. Thus, such modified tryptic peptides would be very large, acidic and heterogeneous, features that may suppress their ionization. Dense O-glycosylation of this region was supported by the absence of de-N-glycosylated and desialylated tryptic Asn107-peptides upon N-glycosidase F and sialidase treatments in separate LC-MS/MS experiments (data not shown). Future experiments combining the treatment of multiple endo- (N-glycosidase F) and exoglycosidases (sialidase and galactosidase) may be required to lower the charge and molecular heterogeneity of this region even further to increase the chances of observing this heavily modified region of the polypeptide chain. The peptide sequence coverage of sIL-15Rα is visualized in Supplementary Fig. S2a.
Fig. 4.
Site-specific O-glycoprofiling of sIL-15Rα of clinical-grade hetIL-15 (cGMP lot) using RP (C18)-LC-ESI-positive ion-CID/ETD-MS/MS. a. The MS1 level profile (right) indicated multiple Thr81- and Thr86 - glycoforms on the tryptic O - glycopeptide R - ] 74 PA P PAPPSTVTTAGVTPQPESLSPSGK97[−E. ETD and CID fragmentation (left spectra) confirmed the O-glycosylation sites and the structure of the two conjugated core 1-type O-glycansans (HexHexNAcNeuAc, corresponding to structure 2b, Fig. 3), m/z 905.4 (4+). Additional examples of ETD-MS/MS fragment spectra of two other sIL-15Rα tryptic O-glycopeptides i. e. b. The N-terminal-1ITCPPPMSVEHADIWVK17-[S peptide conjugated with a single HexNAc (corresponding to structure i, Fig. 3) m/z 728.9 (3+) and c. the C-terminal peptide K-]152NWELTASASHQPPGVYPQG170[−conjugated with two core 1-type O-sialoglycans (HexHexNAcNeuAc, corresponding to structure 2a or 2b, latter shown) and one core 1-type O-asialoglycan (HexHexNAc, structure ii, Fig. 3) m/z 930.8 (4+). Key fragment ions for exact site localization are presented in red. See Fig. 3 for monosaccharide key
Table 2.
Overview of the identified sIL-15Rα tryptic O-glycopeptides. The modified amino acid residues are underlined where known or listed as ND where unknown. The theoretical glycopeptide masses are based on carbamidomethylated cysteine residues. For peptides where the non-glycosylated variants were observed, the non-enriched LC-MS/MS data were used to establish the relative site-occupancy; otherwise glycopeptide-enriched LC-MS/MS data were used to establish the relative glycoform distribution. See Fig. 4 for examples of assigned ETD/CID-MS/MS O-glycopeptide spectra
| Peptide | Obs. m/z (Z) | Obs. mass (M, Da) | Theo. mass (M, Da) | Δmass (Da) | Hex | HexNAc | NeuAc | MS/MS | # O-glycanmoieties | Modified amino acid residue | Rel. abundance (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1ITCPPPMSVEHADIWVK17[−S | |||||||||||
| 660.7 (3+) | 1979.1 | 1979.0 | 0.1 | CID/ETD | 0 | None | 87.9 | ||||
| 728.4 (3+) | 2182.2 | 2182.0 | 0.2 | 1 | CID/ETD | 1 | 2Thr | 12.1 | |||
| K-]50ATNVAHWTTPSLK62[−C | |||||||||||
| 476.0 (3+) | 1425.0 | 1424.7 | 0.3 | CID/ETD | 0 | None | ~100.0 | ||||
| 543.6 (3+) | 1627.8 | 1627.8 | 0.0 | 1 | CID | 1 | ND | Trace | |||
| R-]74PAPPSTVTTAGVTPQPESLSPSGK97[−E | |||||||||||
| 905.4 (4+) | 3617.6 | 3617.6 | 0.0 | 2 | 2 | 2 | CID/ETD | 2 | 81Thr/86Thr | 12.0 | |
| 956.1 (4+) | 3820.4 | 3820.7 | 0.3 | 2 | 3 | 2 | CID/ETD | 2 | 81Thr/86Thr | 14.8 | |
| 978.2 (4+) | 3908.8 | 3908.7 | 0.1 | 2 | 2 | 3 | CID/ETD | 2 | 81Thr/86Thr | 24.5 | |
| 996.8 (4+) | 3983.2 | 3982.8 | 0.4 | 3 | 3 | 2 | CID | 2–3 | ND | 5.0 | |
| 1029.0 (4+) | 4112.0 | 4111.8 | 0.2 | 2 | 3 | 3 | CID | 2–3 | ND | 13.6 | |
| 1051.0 (4+) | 4200.0 | 4199.8 | 0.2 | 2 | 2 | 4 | CID/ETD | 2 | 81Thr/86Thr | 18.5 | |
| 1069.5 (4+) | 4274.0 | 4273.9 | 0.1 | 3 | 3 | 3 | CID | 2–3 | ND | 11.5 | |
| K-]152NWELTASASHQPPGVYPQG170 | |||||||||||
| 929.9 (4+) | 3715.6 | 3715.5 | 0.1 | 3 | 3 | 2 | CID/ETD | 3 | 156Thr/158Ser/160Ser | 57.7 | |
| 1002.7 (4+) | 4006.8 | 4006.6 | 0.2 | 3 | 3 | 3 | CID/ETD | 3 | 156Thr/158Ser/160Ser | 42.3 | |
Further support for regions of sIL-15Rα containing unidentified dense O-glycosylation was generated by low resolution MALDI-TOF MS of intact sIL-15Rα displaying a heterogeneous molecular mass of 30 ± 9 kDa (data not shown). This mass range was significantly higher than the theoretical mass interval of the mature unmodified sIL-15Rα polypeptide chain (~18 kDa) and indeed also the aggregate mass range of the mature sIL-15Rα polypeptide chain modified with the identified N- and O-glycosylation (~23–25 kDa). Thus, we predict that at least eight to ten O-glycans, perhaps even more glycan moieties, remain unaccounted for in the analysis of sIL-15Rα. Alternatively, non-glycan modifications may contribute to the large mass discrepancy observed here. Systematic MS infusion of relatively large amounts of a wide range of HPLC off-line separated tryptic sIL-15Rα peptides covering these putatively dense O-glycosylated regions and utilizing analytical strategies previously shown to be successful for extremely challenging O-glycopeptides [36] did not yield further insight into the site-specific O-glycan pattern of sIL-15Rα (data not shown). Multiple attempts of profiling intact sIL-15Rα using a high resolution Q-TOF platform with and without LC conjugation were also unsuccessful. This demonstrates the extreme analytical challenges still associated with the site-specific mapping of densely O-glycosylated peptide regions.
Site-specific glycoprofiling of IL-15 documents heterogeneous N-glycosylation of Asn79
The N-glycosylation of IL-15 from the clinical-grade cGMP lot of hetIL-15 was site-specifically profiled using RP (C18) LC-ESI-MS/MS analysis of non-enriched tryptic and GluC digested peptides. The E-]65NLIILANNSLSSNGNVTE82]-S glycopeptide showed multiple N-glycoforms; The conjugated N-glycans could be site-specifically localized to the Asn79 residue using CID and ETD fragmentation, Fig. 5a. In fact, neither Asn71 on the same peptide nor Asn112 on another trypsin/GluC-derived peptide, both forming potential N-glycosylation sites, were found to be occupied (data not shown for the latter). The peptide sequence coverage of IL-15 is visualized in Supplementary Fig. S2b. The compositions of the Asn79-glycans agreed well with the N-glycans initially mapped by the glycome profiling of IL-15. The tentative identification of LacdiNAc and sialyl-LacdiNAc containing structures were supported by the presence of CID fragments at m/z 407 (HexNAc2) and m/z 495/698 (NeuAcHexNAc/NeuAcHexNAc2), respectively, in the glycopeptide LC-MS/MS data as have been reported to be informative diagnostic ions for such glycoepitopes [37].
Fig. 5.
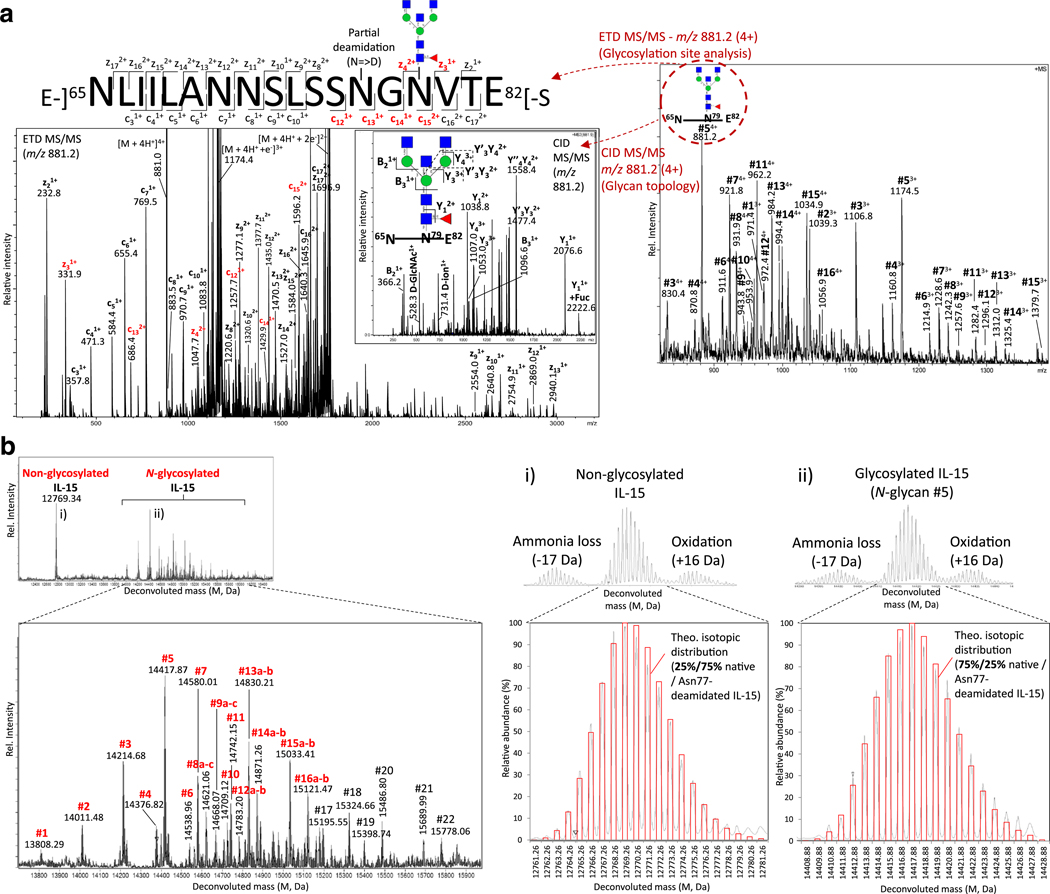
Site-specific N-glycoprofiling of IL-15 of clinical-grade hetIL-15 (cGMP lot) using RP (C18)-LC-ESI-positive ion-CID/ETD-MS/MS. a. The MS1 level profile (right, numbering corresponds to structures in Fig. 3) indicated multiple glycoforms of the N-glycopeptide 65NLIILANNSLSSNGNVTE82. ETD- (left large spectrum, key site-localizing c/z-ions shown in red) and CID- (inserted spectrum) based fragmentation confirmed the N-glycosylation site on the Asn79 sequon as opposed to the Asn71 sequon and confirmed the structure of the conjugated complex-type GlcNAc-terminating N-glycan (Hex3HexNAc5Fuc, corresponding to structure 5a (shown), 5b or 5c, Fig. 3), m/z 881.2 (4+). b. Accurate mass analysis of intact IL-15 using RP (C4)-LC-ESI-positive ion-QTOF-MS confirmed the N-glycosylation of IL-15 (glycoforms observed in the glycome profile shown in red, numbering corresponds to structures in Fig. 3) and indicated some under-glycosylation of IL-15. Zoom of deconvoluted MS spectra (right) of the isotopic distribution of i) non-glycosylated and ii) glycosylated (N-glycan #5) IL-15. Overlaid, a theoretical isotope distribution (red bars). The ‘best fit’ showed partial Asn77-deamidation of IL-15, which appeared to be dependent on the glycosylation status of Asn79, see also Supplementary Fig. S4. See Fig. 3 for key to monosaccharide symbols
Accurate mass analysis of intact IL-15 from the clinical-grade cGMP lot of hetIL-15 using RP (C4) LC-ESI-QTOF-MS confirmed that IL-15 carries a single N-glycan and indicated that a proportion of IL-15 is not occupied by N-glycans, Fig. 5b. Quantitative HPLC and densitometry after SDS-PAGE analysis established that ~95 % of the population of IL-15 molecules are N-glycosylated, Supplementary Fig. S3. Interestingly, several additional, mostly sialylated, IL-15 glycoforms, not observed in the glycan and glycopeptide analysis, were detected in the intact protein profile (structure #17–22, see Supplementary Table S1 for compositions). These glycoforms will need further documentation to validate their attachment to Asn79 of IL-15. In addition, by comparing the isotopic distribution of the deconvoluted spectra of isotopically-resolved intact IL-15 to the theoretical isotopic distributions of IL-15, partial deamidation (Δm = +0.984 Da) was indicated, Fig. 5b i–ii. The extent of the single deamidation reaction, which could be localized to Asn77 by peptide fragmentation analysis, appeared to be dependent on the glycosylation status of the proximal Asn79. More complete Asn77-deamidation (~75 %) was observed for non-glycosylated Asn79 in comparison to Asn79-glycosylated forms of IL-15, for which relative little deamidation (~25 %) was observed, Supplementary Fig. S4. This trend was site-specifically confirmed on the peptide level and no other asparagine residues of IL-15 other than Asn77 were identified as being deamidated (data not shown). The mass analysis also indicated minor oxidation (+16 Da) and ammonia loss (−17 Da) of intact IL-15; however, further studies are needed to confirm that these modifications are not artificially introduced in the sample preparation and/or during the MS analysis. Finally, the accurate mass analysis of intact IL-15 confirmed the lack of O-glycosylation and validated the polypeptide termini of this component of the heterodimeric complex.
Discussion
We previously optimized vectors for the coordinated recombinant expression of the native form of the circulating human IL-15 cytokine complex, hetIL-15 [11, 16, 17, 38], and demonstrated its efficient expression and purification from a stably transfected HEK293 human cell line [19]. IL-15 has been shown to have anti-tumoral activity in a variety of mouse cancer models [6–8, 39]. Clinical experience with IL-15 is limited to the administration of the monomeric, E.coli-produced non-glycosylated IL-15 [40–42]. Although administration of the monomeric IL-15 was overall well-tolerated, transient neutropenia and hypotension were observed [42, 43]. These toxic effects were mainly related to the transient spike in serum IL-15 levels. Several studies have demonstrated superior bioactivity (e.g. lymphocyte stimulation capacity) and favorable pharmacokinetic and pharmacodynamics properties (e.g. prolonged in vivo half-life) of hetIL-15 relative to E.coli-produced IL-15 and such beneficial features were, thus, related to the heterodimer formation and the glycosylation status [8, 17, 19, 44]. These favorable characteristics warrant an evaluation of the therapeutic potential of hetIL-15 in different clinical settings. At present, hetIL-15 is in clinical trial for the treatment of a range of metastatic tumors (i.e. skin, renal cell, non-small cell lung, and head and neck squamous cell carcinoma) in adult patients (trial number NCT02452268). Despite its extensive functional and pharmacological characterization, the exact glycan structures and their glycosylation sites and occupancies of hetIL-15 remain until now undetermined. To shed light on the structural aspects of these functionally important modifications, we performed a detailed LC-MS/MS-based site-specific glycosylation analysis of two clinically relevant large-scale batch preparations (i.e. EN and cGMP lots) of the bioactive HEK293-derived hetIL-15 complex.
The combined glycan, glycopeptide and intact glycoprotein analysis of hetIL-15 were in general in excellent agreement unlike our recent experience when using these multiple analytical strategies to analyze unconventional (truncated) N-glycoproteins [45] and, thus, confirmed the N- and O-glycosylation of hetIL-15. The sIL-15Rα component carried a single occupied N-glycosylation site at Asn107 and multiple O-glycans at Thr2, Thr81, Thr86, Thr156, Ser158, and Ser160. In contrast, IL-15 was only relatively lightly glycosylated with only a single occupied N-glycosylation site at Asn79. Interestingly, Asn79 of IL-15 and Asn107 of sIL-15Rα were similarly N-glycosylated by the same repertoire of biosynthetically related complex type core fucosylated structures. The similar N-glycosylation pattern is most likely explained by the simultaneous processing of the two protein components of the complex under exactly the same cellular conditions (protein trafficking rate and availability of nucleotide donors and glycosylation enzymes) in the HEK293 cells thus eliminating variability in glycosylation arising from cellular factors [46]. We have previously shown close correlation between N-glycan processing and glycosylation site solvent accessibility on maturely folded proteins and that this relationship can explain the commonly observed feature of subcellular-specific N-glycosylation [47, 48]. Thus, the similar N-glycosylation of sIL-15Rα and IL-15 indicated that their two glycosylation sites are equally accessible for the processing glycosylation enzymes in the biosynthetic machinery and supports their co-trafficking in the secretory pathway. Extensive β-GlcNAc-branching and non-reducing-end termination of complex N-glycans including the abundance of β1–4-bisecting GlcNAcylation and β1–2/4/6-GlcNAc branching (bi- and triantenna formation) and core fucosylation are commonly reported cell-specific features of the HEK293 cell line [37, 49–51]. Lower levels of α2–3- and α2–6-sialylation, as observed here for hetIL-15, have also been reported for other glycoproteins produced by HEK293 [50, 52]. In addition, both IL-15 and sIL-15Rα carried interesting diHexNAc features presumably GalNAcβ1–4GlcNAc-R (LacdiNAc) epitopes that were reported to be characteristic features of HEK293 cells, and not expressed in the widely used CHO cells due to lower β4-GalNAc transferase and higher β4-Gal transferase activities in CHO cell cultures [37, 49, 50, 53, 54].
The interplay between IL-15 Asn77-deamidation and Asn79-glycosylation demonstrated here is interesting and to the best of our knowledge not previously reported. Glycosylation of Asn79 clearly protected the proximal Asn77, albeit not entirely, from deamidation. Asparagine deamidation is a spontaneous and highly sequence-dependent non-enzymatic reaction formed under physiological conditions and is associated with regulatory functions [55, 56]. However, deamidation is usually unwanted in protein therapeutics due to its association with protein degradation [57] and cellular apoptosis [58]. Asn77 has been reported as the main deamidation site of human IL-15 and efforts involving optimization of the protein processing, formulation and targeted amino acid changes have been undertaken to minimize this chemical reaction [59]. It is possible that some of the deamidation of Asn77, which is flanked by an N-terminal serine and a C-terminal glycine residue, a sequence arrangement that dramatically increase the deamidation rate [60], occurred post-protein expression due to the experimental conditions (e.g. low pH buffers) during the HPLC separation and molecular characterization of hetIL-15. Further structure-function studies are needed to validate the deamidation-glycosylation interplay of the two proximal asparagine residues of IL-15, which may also reveal in more detail, protective functions of specific volume enhancing features of the observed Asn79-glycans. For example, it may be speculated that features such as core fucosylation and bisecting β1–4-GlcNAcylation, which are known to dramatically alter the conformation and the function of N-glycans [61], or simply the sheer size/volume of the relatively large N-glycans identified on Asn79 of IL-15, are mechanisms that serve the purpose of regulating the rate of the spontaneous deamidation of the proximal Asn77.
It is of interest to note the rather dramatic differences in O-glycosylation between the two components of the heterodimer. The sIL-15Rα component is heavily O-glycosylated by core 1 and 2-type sialo- and asialoglycans, whereas IL-15 completely lacks O-glycosylation. Mechanistically, this difference may relate to the generally higher presence of proline, serine and threonine residues in sIL-15Rα (10.1 %, 13.9 % and 10.9 %) relative to IL-15 (1.9 %, 11.1 % and 4.9 %, respectively) and the multiple ‘SS’ (5×) and ‘TT’ (5×) motifs presented uniquely by sIL-15Rα, which are favorable substrates for the 20 GalNAc-transferases that can add the first mucin-type O-GalNAc residue to the polypeptide backbone [62, 63]. In support, the neural network-based mucin-type O-GalNAc site predictor NetOGlyc v4.0 (http://www.cbs.dtu.dk/services/NetOGlyc/) predicted no O-glycosylation sites for IL-15 (all scores were less than the 0.5 score thresh-old), but predicted no less than 39 mucin-type O-glycosylation sites for sIL-15Rα (mean score of 0.84, all six identified O-glycosylation sites reported here were correctly predicted) [64]. In addition to hosting the favorable Ser/Thr/Pro-rich motifs, intact IL-15Rα may also benefit from being membrane-tethered and thereby in relative closer proximity to the membrane-bound GalNAc-transferases during the ER/Golgi transport than the IL-15 chain [62]. The presentation of core 1 and core 2-type O-glycans as observed on sIL-15Rα is a previously reported feature of HEK293 cells [49, 53].
It is also worth mentioning that no non-human glycoepitopes were carried by the identified N- or O-glycans of IL-15 and sIL-15Rα including the immunogenic N-glycolylneuraminic acid [65] and α-galactose [66]. Such glycoepitopes, which may be produced by the rodent CHO, SP2 and NS0 cells commonly used as expression systems for recombinant human proteins [67–69], are generally unwanted in therapeutic glycoproteins for human use due to their xenoantigenic character, which may provoke an undesired immune response upon administration in patients.
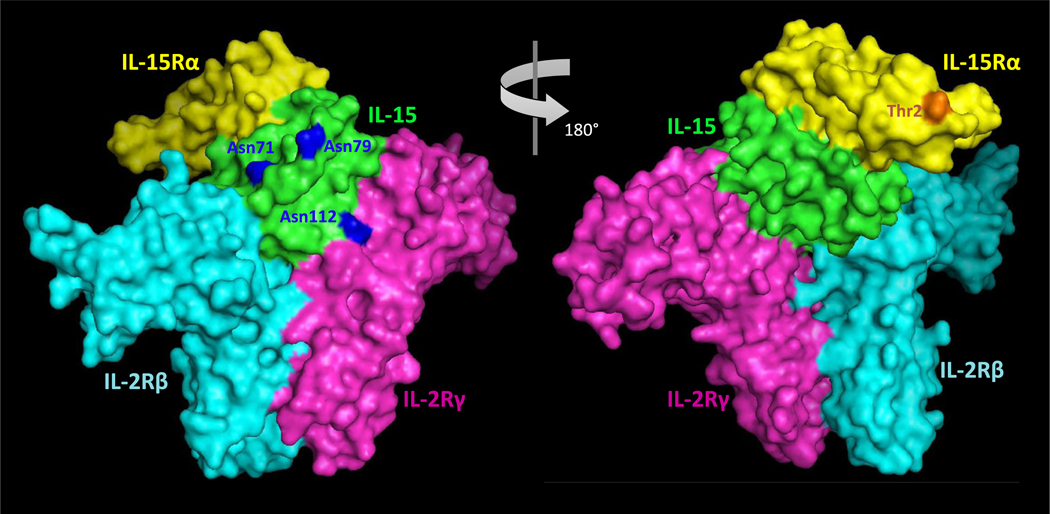
Functionally, the soluble hetIL-15 as well as the membrane-bound IL-15:IL-15Rα complex are known to engage with the IL-2Rβ:IL-2Rγ (also known as CD122/CD132) complex presented on neighboring target cells to initiate a ‘trans’-cellular response [12] (see Fig. 1a for schematic illustration). IL-15 is the protein component that directly interacts with the IL-2Rβ: IL-2Rγ receptor complex, whereas IL-15Rα appears to be distal to the quaternary complex-interface, Fig. 6. Spatial mapping of the identified glycosylation sites onto the quaternary complex showed that the two potential N-glycosylation sites (Asn71 and Asn112) of IL-15 are found directly on the interface to IL-2Rβ and IL-2Rγ, respectively, which may explain their unoccupied status in the bioactive hetIL-15 complex. Voluminous complex-type N-glycans, in particular structures rich in core fucosylation, LacdiNAcylation and bisected GlcNAcylation characteristic of HEK293-derived hetIL-15, would likely impede this interaction if conjugated to Asn71 and Asn112. The occupied Asn79, in contrast, appears to be located in a surface-accessible position on IL-15 in sufficient distance from the interface to IL-2Rβ:IL-2Rγ to avoid interference when occupied. The mechanism creating the differential occupancy of the three IL-15 N-glycosylation sequons i.e. Asn71 (NNS), Asn79 (NVT) and Asn112 (NTS) may relate to the nature of the two amino acid residues C-terminal to the occupied asparagine residues, the location of the sequon in the polypeptide chain or other protein or cellular factors [46]. It is interesting to note that the non-N-glycosylated IL-2 is known to bind to IL-2Rβ:IL-2Rγ in a nearly indistinguishable manner, but with different receptor affinities, relative to IL-15 [34]. This suggest that the glycosylation status of IL-15 may play a role in defining the receptor affinities towards IL-2Rβ: IL-2Rγ and perhaps even the interaction to sIL-15Rα as well as its circulatory half-life [70]. Unfortunately, the heavily N-and O-glycosylated region of IL-15Rα is not covered by the available crystal structure. The biological relevance of the N-and O-glycosylation of sIL-15Rα may be illuminated upon further structure-function studies and when a crystal structure covering the entire complex becomes available.
Fig. 6.
Spatial map of the N- and O-glycosylation sites of hetIL-15 in its quaternary complex with IL-2Rβ and IL-2Rγ. The three putative N-glycosylation sites of IL-15 (green) are shown in blue. Only Asn79 was found to be occupied; Asn71 and Asn112 found on the interface to IL-2Rβ (cyan) and IL-2Rγ (magenta), respectively, were not utilized as N-glycosylation sites when expressed in HEK293. The available crystal structure (PDB: 4GS7) covered only the lightly O-glycosylated N-terminal region of the sIL-15Rα polypeptide chain (yellow) [34]. The occupied O-glycosylation site at Thr2 covered by this region is shown in orange. See also Fig. 1a. for schematic illustration of the quaternary complex
In summary, we report on the detailed and site-specific glycosylation of the engineered hetIL-15 expressed in HEK293 cells. We conclude that this soluble bioactive heterodimeric complex carries heterogeneous N- and O-glycosylation that can be reproducibly expressed and purified to clinical-grade quality in large quantities. Considering the important modulatory role of glycosylation affecting the physicochemical properties and therapeutic efficacy of (glyco)protein therapeutics, it is likely that the glycosylation of hetIL-15 has multiple and diverse in vivo functions, including influencing the pharmacokinetic properties and the strength of interactions with the binding partners in intricate ways. The site-specific N- and O-glycosylation map provided here enable more advanced structural-functional studies that may allow us to understand the molecular design of this heterodimeric complex and eventually enable us to manipulate and further improve the therapeutic properties of hetIL-15.
Supplementary Material
Acknowledgments
This research was facilitated through access to the Australian Proteomics Analysis Facility (APAF). ESXM was supported by an international Macquarie University Research Scholarship (iMQRES). M.T.-A. was supported by an Early Career Fellowship from the Cancer Institute, NSW, Australia. This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E and by the Intramural Research Program of the National Cancer Institute, National Institutes of Health (NCI/NIH). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We thank Admune Therapeutics for supplying hetIL-15 preparations.
Abbreviations
- ACN
Acetonitrile
- cGMP
Current good manufacturing practices
- CID
Collision induced dissociation
- EIC
Extracted ion chromatogram
- ESI
Electrospray ionization
- ETD
Electron transfer dissociation
- GlcNAc
N-acetylglucosamine
- HexNAc
N-acetylhexosamine
- HILIC
Hydrophilic interaction liquid chromatography
- HEK293
Human embryonic kidney 293
- hetIL-15
Heterodimeric IL-15:sIL-15Rα complex
- IL-2R
Interleukin 2 receptor
- IL-15
Interleukin 15
- IL-15Rα
Interleukin 15 receptor alpha
- sIL-15Rα
Soluble interleukin 15 receptor alpha
- LacdiNAc
N,N-diacetyllactosamine
- LC
Liquid chromatography
- MALDI
Matrix assisted laser desorption ionization
- MS/MS
Tandem mass spectrometry
- NeuAc
N-acetylneuraminic acid
- PGC
Porous graphitized carbon
- QTOF
Quadrupole time-of-flight
- RP-HPLC
Reversed-phase high performance liquid chromatography
- SPE
Solid phase extraction
- TFA
Trifluoroacetic acid
- TOF
Time-of-flight
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10719-015-9627-1) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Disclosure of potential conflicts of interest CB, BKF and GNP are inventors on US Government-owned patents and patent applications related to hetIL-15 and gene expression optimization.
References
- 1.Ma A, Koka R, Burkett P: Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 24, 657–679 (2006). doi: 10.1146/annurev.immunol.24.021605.090727 [DOI] [PubMed] [Google Scholar]
- 2.Berard M, Brandt K, Bulfone-Paus S, Tough DF: IL-15 promotes the survival of naive and memory phenotype CD8+ T cells. J. Immunol. 170(10), 5018–5026 (2003) [DOI] [PubMed] [Google Scholar]
- 3.Burton JD, Bamford RN, Peters C, Grant AJ, Kurys G, Goldman CK, Brennan J, Roessler E, Waldmann TA: A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc. Natl. Acad. Sci. U. S. A. 91(11), 4935–4939 (1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson WE, Giri JG, Lindemann MJ, Linett ML, Ahdieh M, Paxton R, Anderson D, Eisenmann J, Grabstein K, Caligiuri MA: Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J. Exp. Med. 180(4), 1395–1403 (1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Sun S, Hwang I, Tough DF, Sprent J: Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 8(5), 591–599 (1998) [DOI] [PubMed] [Google Scholar]
- 6.Dubois S, Patel HJ, Zhang M, Waldmann TA, Muller JR: Preassociation of IL-15 with IL-15R alpha-IgG1-Fc enhances its activity on proliferation of NK and CD8+/CD44high T cells and its antitumor action. J. Immunol. 180(4), 2099–2106 (2008) [DOI] [PubMed] [Google Scholar]
- 7.Epardaud M, Elpek KG, Rubinstein MP, Yonekura AR, Bellemare-Pelletier A, Bronson R, Hamerman JA, Goldrath AW, Turley SJ: Interleukin-15/interleukin-15R alpha complexes promote destruction of established tumors by reviving tumor-resident CD8+ T cells. Cancer Res. 68(8), 2972–2983 (2008). doi: 10.1158/0008-5472.CAN-08-0045 [DOI] [PubMed] [Google Scholar]
- 8.Stoklasek TA, Schluns KS, Lefrancois L: Combined IL-15/IL-15Ralpha immunotherapy maximizes IL-15 activity in vivo. J. Immunol. 177(9), 6072–6080 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheever MA: Twelve immunotherapy drugs that could cure cancers. Immunol. Rev. 222, 357–368 (2008). doi: 10.1111/j.1600-065X.2008.00604.x [DOI] [PubMed] [Google Scholar]
- 10.Croce M, Orengo AM, Azzarone B, Ferrini S: Immunotherapeutic applications of IL-15. Immunotherapy. 4(9), 957–969 (2012). doi: 10.2217/imt.12.92 [DOI] [PubMed] [Google Scholar]
- 11.Bergamaschi C, Kulkarni V, Rosati M, Alicea C, Jalah R, Chen S, Bear J, Sardesai NY, Valentin A, Felber BK, Pavlakis GN: Intramuscular delivery of heterodimeric IL-15 DNA in macaques produces systemic levels of bioactive cytokine inducing proliferation of NK and T cells. Gene Ther. 22(1), 76–86 (2015). doi: 10.1038/gt.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois S, Mariner J, Waldmann TA, Tagaya Y: IL-15Ralpha recycles and presents IL-15 in trans to neighboring cells. Immunity. 17(5), 537–547 (2002) [DOI] [PubMed] [Google Scholar]
- 13.Sandau MM, Schluns KS, Lefrancois L, Jameson SC: Cutting edge: transpresentation of IL-15 by bone marrow-derived cells necessitates expression of IL-15 and IL-15R alpha by the same cells. J. Immunol. 173(11), 6537–6541 (2004) [DOI] [PubMed] [Google Scholar]
- 14.Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A: Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8 + T cell homeostasis. J. Exp. Med. 200(7), 825–834 (2004). doi: 10.1084/jem.20041389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koka R, Burkett PR, Chien M, Chai S, Chan F, Lodolce JP, Boone DL, Ma A: Interleukin (IL)-15R[alpha]-deficient natural killer cells survive in normal but not IL-15R[alpha]-deficient mice. J. Exp. Med. 197(8), 977–984 (2003). doi: 10.1084/jem.20021836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergamaschi C, Jalah R, Kulkarni V, Rosati M, Zhang GM, Alicea C, Zolotukhin AS, Felber BK, Pavlakis GN: Secretion and biological activity of short signal peptide IL-15 is chaperoned by IL-15 receptor alpha in vivo. J. Immunol. 183(5), 3064–3072 (2009). doi: 10.4049/jimmunol.0900693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergamaschi C, Rosati M, Jalah R, Valentin A, Kulkarni V, Alicea C, Zhang GM, Patel V, Felber BK, Pavlakis GN: Intracellular interaction of interleukin-15 with its receptor alpha during production leads to mutual stabilization and increased bioactivity. J. Biol. Chem. 283(7), 4189–4199 (2008). doi: 10.1074/jbc.M705725200 [DOI] [PubMed] [Google Scholar]
- 18.Bergamaschi C, Bear J, Rosati M, Beach RK, Alicea C, Sowder R, Chertova E, Rosenberg SA, Felber BK, Pavlakis GN: Circulating IL-15 exists as heterodimeric complex with soluble IL-15Ralpha in human and mouse serum. Blood. 120(1), e1–e8 (2012). doi: 10.1182/blood-2011-10-384362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chertova E, Bergamaschi C, Chertov O, Sowder R, Bear J, Roser JD, Beach RK, Lifson JD, Felber BK, Pavlakis GN: Characterization and favorable in vivo properties of heterodimeric soluble IL-15 IL-15Ralpha cytokine compared to IL-15 monomer. J. Biol. Chem. 288(25), 18093–18103 (2013). doi: 10.1074/jbc.M113.461756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamorey AL, Magne N, Pivot X, Milano G: Impact of glycosylation on the effect of cytokines A special focus on oncology. Eur. Cytokine Netw. 13(2), 154–160 (2002) [PubMed] [Google Scholar]
- 21.Sola RJ, Griebenow K: Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 98(4), 1223–1245 (2009). doi: 10.1002/jps.21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dissing-Olesen L, Thaysen-Andersen M, Meldgaard M, Hojrup P, Finsen B: The function of the human interferon-beta 1a glycan determined in vivo. J. Pharmacol. Exp. Ther. 326(1), 338–347 (2008). doi: 10.1124/jpet.108.138263 [DOI] [PubMed] [Google Scholar]
- 23.Beq S, Rozlan S, Gautier D, Parker R, Mersseman V, Schilte C, Assouline B, Rance I, Lavedan P, Morre M, Cheynier R: Injection of glycosylated recombinant simian IL-7 provokes rapid and massive T-cell homing in rhesus macaques. Blood. 114(4), 816–825 (2009). doi: 10.1182/blood-2008-11-191288 [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi M, Kobata A: Structures and functional roles of the sugar chains of human erythropoietins. Glycobiology. 1(4), 337–346 (1991) [DOI] [PubMed] [Google Scholar]
- 25.Jensen PH, Karlsson NG, Kolarich D, Packer NH: Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7(7), 1299–1310 (2012). doi: 10.1038/nprot.2012.063 [DOI] [PubMed] [Google Scholar]
- 26.Thaysen-Andersen M, Thogersen IB, Lademann U, Offenberg H, Giessing AM, Enghild JJ, Nielsen HJ, Brunner N, Hojrup P: Investigating the biomarker potential of glycoproteins using comparative glycoprofiling - application to tissue inhibitor of metalloproteinases-1. Biochim. Biophys. Acta. 1784(3), 455–463 (2008). doi: 10.1016/j.bbapap.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 27.Mysling S, Palmisano G, Hojrup P, Thaysen-Andersen M: Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82(13), 5598–5609 (2010). doi: 10.1021/ac100530w [DOI] [PubMed] [Google Scholar]
- 28.Sethi MK, Kim H, Park CK, Baker MS, Paik YK, Packer NH, Hancock WS, Fanayan S, Thaysen-Andersen M: In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology. (2015). doi: 10.1093/glycob/cwv042 [DOI] [PubMed] [Google Scholar]
- 29.Venkatakrishnan V, Thaysen-Andersen M, Chen SC, Nevalainen H, Packer NH: Cystic fibrosis and bacterial colonization define the sputum N-glycosylation phenotype. Glycobiology. 25(1), 88–100 (2015). doi: 10.1093/glycob/cwu092 [DOI] [PubMed] [Google Scholar]
- 30.Leymarie N, Griffin PJ, Jonscher K, Kolarich D, Orlando R, McComb M, Zaia J, Aguilan J, Alley WR, Altmann F, Ball LE, Basumallick L, Bazemore-Walker CR, Behnken H, Blank MA, Brown KJ, Bunz SC, Cairo CW, Cipollo JF, Daneshfar R, Desaire H, Drake RR, Go EP, Goldman R, Gruber C, Halim A, Hathout Y, Hensbergen PJ, Horn DM, Hurum D, Jabs W, Larson G, Ly M, Mann BF, Marx K, Mechref Y, Meyer B, Moginger U, Neusubeta C, Nilsson J, Novotny MV, Nyalwidhe JO, Packer NH, Pompach P, Reiz B, Resemann A, Rohrer JS, Ruthenbeck A, Sanda M, Schulz JM, Schweiger-Hufnagel U, Sihlbom C, Song E, Staples GO, Suckau D, Tang H, Thaysen-Andersen M, Viner RI, An Y, Valmu L, Wada Y, Watson M, Windwarder M, Whittal R, Wuhrer M, Zhu Y, Zou C: Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: the ABRF glycoprotein research multi-institutional study 2012. Mol. Cell. Proteomics. 12(10), 2935–2951 (2013). doi: 10.1074/mcp.M113.030643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumer-Bayraktar Z, Nguyen-Khuong T, Jayo R, Chen DD, Ali S, Packer NH, Thaysen-Andersen M: Micro- and macroheterogeneity of N-glycosylation yields size and charge isoforms of human sex hormone binding globulin circulating in serum. Proteomics. 12(22), 3315–3327 (2012). doi: 10.1002/pmic.201200354 [DOI] [PubMed] [Google Scholar]
- 32.Sumer-Bayraktar Z, Kolarich D, Campbell MP, Ali S, Packer NH, Thaysen-Andersen M: N-glycans modulate the function of human corticosteroid-binding globulin. Mol. Cell. Proteomics. 10(8), M111 009100 (2011). doi: 10.1074/mcp.M111.009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Varon Silva D, Fuchser J, Kaspar S, Rapp E, Seeberger PH, Kolarich D: Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 48(6), 627–639 (2013). doi: 10.1002/jms.3210 [DOI] [PubMed] [Google Scholar]
- 34.Ring AM, Lin JX, Feng D, Mitra S, Rickert M, Bowman GR, Pande VS, Li P, Moraga I, Spolski R, Ozkan E, Leonard WJ, Garcia KC: Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol. 13(12), 1187–1195 (2012). doi: 10.1038/ni.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thaysen-Andersen M, Venkatakrishnan V, Loke I, Laurini C, Diestel S, Parker BL, Packer NH: Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 290(14), 8789–8802 (2015). doi: 10.1074/jbc.M114.631622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thaysen-Andersen M, Wilkinson BL, Payne RJ, Packer NH: Site-specific characterisation of densely O-glycosylated mucin-type peptides using electron transfer dissociation ESI-MS/MS. Electrophoresis. 32(24), 3536–3545 (2011). doi: 10.1002/elps.201100294 [DOI] [PubMed] [Google Scholar]
- 37.Zeck A, Pohlentz G, Schlothauer T, Peter-Katalinic J, Regula JT: Cell type-specific and site directed N-glycosylation pattern of FcgammaRIIIa. J. Proteome Res. 10(7), 3031–3039 (2011). doi: 10.1021/pr1012653 [DOI] [PubMed] [Google Scholar]
- 38.Jalah R, Rosati M, Kulkarni V, Patel V, Bergamaschi C, Valentin A, Zhang GM, Sidhu MK, Eldridge JH, Weiner DB, Pavlakis GN, Felber BK: Efficient systemic expression of bioactive IL-15 in mice upon delivery of optimized DNA expression plasmids. DNA Cell Biol. 26(12), 827–840 (2007). doi: 10.1089/dna.2007.0645 [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi H, Dubois S, Sato N, Sabzevari H, Sakai Y, Waldmann TA, Tagaya Y: Role of trans-cellular IL-15 presentation in the activation of NK cell-mediated killing, which leads to enhanced tumor immunosurveillance. Blood. 105(2), 721–727 (2005). doi: 10.1182/blood-2003-12-4187 [DOI] [PubMed] [Google Scholar]
- 40.Conlon KC, Lugli E, Welles HC, Rosenberg SA, Fojo AT, Morris JC, Fleisher TA, Dubois SP, Perera LP, Stewart DM, Goldman CK, Bryant BR, Decker JM, Chen J, Worthy TA, Figg WD Sr., Peer CJ, Sneller MC, Lane HC, Yovandich JL, Creekmore SP, Roederer M, Waldmann TA: Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J. Clin. Oncol. 33(1), 74–82 (2015). doi: 10.1200/JCO.2014.57.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lugli E, Goldman CK, Perera LP, Smedley J, Pung R, Yovandich JL, Creekmore SP, Waldmann TA, Roederer M: Transient and persistent effects of IL-15 on lymphocyte homeostasis in nonhuman primates. Blood. 116(17), 3238–3248 (2010). doi: 10.1182/blood-2010-03-275438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldmann TA, Lugli E, Roederer M, Perera LP, Smedley JV, Macallister RP, Goldman CK, Bryant BR, Decker JM, Fleisher TA, Lane HC, Sneller MC, Kurlander RJ, Kleiner DE, Pletcher JM, Figg WD, Yovandich JL, Creekmore SP: Safety (toxicity), pharmacokinetics, immunogenicity, and impact on elements of the normal immune system of recombinant human IL-15 in rhesus macaques. Blood. 117(18), 4787–4795 (2011). doi: 10.1182/blood-2010-10-311456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger C, Berger M, Hackman RC, Gough M, Elliott C, Jensen MC, Riddell SR: Safety and immunologic effects of IL-15 administration in nonhuman primates. Blood. 114(12), 2417–2426 (2009). doi: 10.1182/blood-2008-12-189266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubinstein MP, Kovar M, Purton JF, Cho JH, Boyman O, Surh CD, Sprent J: Converting IL-15 to a superagonist by binding to soluble IL-15R{alpha}. Proc. Natl. Acad. Sci. U. S. A. 103(24), 9166–9171 (2006). doi: 10.1073/pnas.0600240103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loke I, Packer NH, Thaysen-Andersen M: Complementary LC-MS/MS-based N-glycan, N-glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil cathepsin G. Biomolecules. 5(3), 1832–1854 (2015). doi: 10.3390/biom5031832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudd PM, Dwek RA: Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 32(1), 1–100 (1997). doi: 10.3109/10409239709085144 [DOI] [PubMed] [Google Scholar]
- 47.Thaysen-Andersen M, Packer NH: Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology. 22(11), 1440–1452 (2012). doi: 10.1093/glycob/cws110 [DOI] [PubMed] [Google Scholar]
- 48.Lee LY, Lin CH, Fanayan S, Packer NH, Thaysen-Andersen M: Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 5, 404 (2014). doi: 10.3389/fimmu.2014.00404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Tao S, Orlando R, Brockhausen I, Kan FW: Structures and biosynthesis of the N- and O-glycans of recombinant human oviduct-specific glycoprotein expressed in human embryonic kidney cells. Carbohydr. Res. 358, 47–55 (2012). doi: 10.1016/j.carres.2012.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harvey DJ, Baruah K, Scanlan CN: Application of negative ion MS/MS to the identification of N-glycans released from carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1). J. Mass Spectrom. 44(1), 50–60 (2009). doi: 10.1002/jms.1470 [DOI] [PubMed] [Google Scholar]
- 51.Chitlaru T, Kronman C, Velan B, Shafferman A: Overloading and removal of N-glycosylation targets on human acetylcholinesterase: effects on glycan composition and circulatory residence time. Biochem. J. 363(Pt 3), 619–631 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chitlaru T, Kronman C, Zeevi M, Kam M, Harel A, Ordentlich A, Velan B, Shafferman A: Modulation of circulatory residence of recombinant acetylcholinesterase through biochemical or genetic manipulation of sialylation levels. Biochem. J. 336(Pt 3), 647–658 (1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geoghegan KF, Song X, Hoth LR, Feng X, Shanker S, Quazi A, Luxenberg DP, Wright JF, Griffor MC: Unexpected mucin-type O-glycosylation and host-specific N-glycosylation of human recombinant interleukin-17A expressed in a human kidney cell line. Protein Expr. Purif. 87(1), 27–34 (2013). doi: 10.1016/j.pep.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 54.Van den Nieuwenhof IM, Koistinen H, Easton RL, Koistinen R, Kamarainen M, Morris HR, Van Die I, Seppala M, Dell A, Van den Eijnden DH: Recombinant glycodelin carrying the same type of glycan structures as contraceptive glycodelin-a can be produced in human kidney 293 cells but not in Chinese hamster ovary cells. Eur. J. Biochem. 267(15), 4753–4762 (2000) [DOI] [PubMed] [Google Scholar]
- 55.Eggleton P, Haigh R, Winyard PG: Consequence of neoantigenicity of the ‘altered self’. Rheumatology (Oxford). 47(5), 567–571 (2008). doi: 10.1093/rheumatology/ken014 [DOI] [PubMed] [Google Scholar]
- 56.Weintraub SJ, Deverman BE: Chronoregulation by asparagine deamidation. Sci. STKE. 2007(409), re7 (2007). doi: 10.1126/stke.4092007re7 [DOI] [PubMed] [Google Scholar]
- 57.Vlasak J, Bussat MC, Wang S, Wagner-Rousset E, Schaefer M, Klinguer-Hamour C, Kirchmeier M, Corvaia N, Ionescu R, Beck A: Identification and characterization of asparagine deamidation in the light chain CDR1 of a humanized IgG1 antibody. Anal. Biochem. 392(2), 145–154 (2009). doi: 10.1016/j.ab.2009.05.043 [DOI] [PubMed] [Google Scholar]
- 58.Zhao R, Oxley D, Smith TS, Follows GA, Green AR, Alexander DR: DNA damage-induced Bcl-xL deamidation is mediated by NHE-1 antiport regulated intracellular pH. PLoS Biol. 5(1), e1 (2007). doi: 10.1371/journal.pbio.0050001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nellis DF, Michiel DF, Jiang MS, Esposito D, Davis R, Jiang H, Korrell A, Knapp GC, Lucernoni LE, Nelson RE, Pritt EM, Procter LV, Rogers M, Sumpter TL, Vyas VV, Waybright TJ, Yang X, Zheng AM, Yovandich JL, Gilly JA, Mitra G, Zhu J: Characterization of recombinant human IL-15 deamidation and its practical elimination through substitution of asparagine 77. Pharm. Res. 29(3), 722–738 (2012). doi: 10.1007/s11095-011-0597-0 [DOI] [PubMed] [Google Scholar]
- 60.Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, Robinson AB: Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J. Pept. Res. 63(5), 426–436 (2004). doi: 10.1111/j.1399-3011.2004.00151.x [DOI] [PubMed] [Google Scholar]
- 61.Takahashi M, Kuroki Y, Ohtsubo K, Taniguchi N: Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydr. Res. 344(12), 1387–1390 (2009). doi: 10.1016/j.carres.2009.04.031 [DOI] [PubMed] [Google Scholar]
- 62.Tran DT, Ten Hagen KG: Mucin-type O-glycosylation during development. J. Biol. Chem. 288(10), 6921–6929 (2013). doi: 10.1074/jbc.R112.418558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA: Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 22(6), 736–756 (2012). doi: 10.1093/glycob/cwr182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H: Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32(10), 1478–1488 (2013). doi: 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diswall M, Angstrom J, Karlsson H, Phelps CJ, Ayares D, Teneberg S, Breimer ME: Structural characterization of alpha1, 3-galactosyltransferase knockout pig heart and kidney glycolipids and their reactivity with human and baboon antibodies. Xenotransplantation. 17(1), 48–60 (2010). doi: 10.1111/j.1399-3089.2009.00564.x [DOI] [PubMed] [Google Scholar]
- 66.Galili U: The alpha-gal epitope and the anti-gal antibody in xenotransplantation and in cancer immunotherapy. Immunol. Cell Biol. 83(6), 674–686 (2005). doi: 10.1111/j.1440-1711.2005.01366.x [DOI] [PubMed] [Google Scholar]
- 67.Bosques CJ, Collins BE, Meador JW 3rd, Sarvaiya H, Murphy JL, Dellorusso G, Bulik DA, Hsu IH, Washburn N, Sipsey SF, Myette JR, Raman R, Shriver Z, Sasisekharan R, Venkataraman G: Chinese hamster ovary cells can produce galactose-alpha-1,3-galactose antigens on proteins. Nat. Biotechnol. 28(11), 1153–1156 (2010). doi: 10.1038/nbt1110-1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoo EM, Chintalacharuvu KR, Penichet ML, Morrison SL: Myeloma expression systems. J. Immunol. Methods. 261(1–2), 1–20 (2002) [DOI] [PubMed] [Google Scholar]
- 69.Baker KN, Rendall MH, Hills AE, Hoare M, Freedman RB, James DC: Metabolic control of recombinant protein N-glycan processing in NS0 and CHO cells. Biotechnol. Bioeng. 73(3), 188–202 (2001) [DOI] [PubMed] [Google Scholar]
- 70.Eisenman J, Ahdieh M, Beers C, Brasel K, Kennedy MK, Le T, Bonnert TP, Paxton RJ, Park LS: Interleukin-15 interactions with interleukin-15 receptor complexes: characterization and species specificity. Cytokine. 20(3), 121–129 (2002) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.