Abstract
Malaria has been a global epidemic health threat since ancient times. It still claims roughly half a million lives every year in this century. Artemisinin and its derivatives, are frontline antimalarial drugs known for their efficacy and low toxicity. After decades of wide use, artemisinins remain our bulwark against malaria. Here, we review decades of efforts that aim to understand the mechanism of action (MOA) of artemisinins, which help explain the specificity and potency of this anti-malarial drug. We summarize the methods and approaches employed to unravel the MOA of artemisinin over the last three decades, showing how the development of advanced techniques can help provide mechanistic insights and resolve some long-standing questions in the field of artemisinin research. We also provide examples to illustrate how to better repurpose artemisinins for anti-cancer therapies by leveraging on MOA. These examples point out a practical direction to engineer artemisinin for broader applications beyond malaria.
Keywords: Artemisinin, Mechanism of action, Anti-malaria, Repurpose of artemisinin
Abbreviations: CQ, chloroquine; SP, sulfadoxine-pyrimethamine; TCM, traditional Chinese medicine; DHA, dihydroartemisinin; ACTs, artemisinin-based combination therapies; MOA, mechanism of action; ROS, reactive oxygen species; DFO, desferrioxamine; TCTP, translational controlled tumor protein; SERCA, sarco/endoplasmic reticulum membrane calcium; MS, mass spectrometry; IC50, 50% inhibiting concentration; ALA, aminolevulinic acid
1. Introduction
Malaria is a life-threatening global epidemic disease which has existed since ancient times and has claimed more than 400,000 lives in the year of 2019 according to the 2019 world malaria report. The cause of the disease remained unclear until the discovery of Plasmodium in the blood of malaria patients by Charles Louis Alphonse Laveran in 1880 and Anopheles mosquitoes as the vectors for transmission of malaria by Ronald Ross in 1897. In recognition of their seminal discoveries in understanding the causes of malaria, Laveran and Ross were awarded Nobel Prizes in Physiology or Medicine at 1907 and 1902, respectively. (Cox, 2010).
In the course of combat with malaria, scientists discovered several antimalarial drugs, such as chloroquine (CQ) and sulfadoxine-pyrimethamine (SP). The worldwide use of CQ led to great progress in the elimination of malaria. However, resistance to CQ emerged at a time when the World Health Organization launched the Global Malaria Eradication Campaign. This resistance is linked to multiple mutations in PfCRT (chloroquine resistance transporter), a protein that functions as a transporter in the parasite's digestive vacuole membrane. The mutated PfCRT gained the function to efflux CQ from the vacuole wherein CQ reacts to damage parasites. (Wellems & Plowe, 2001). CQ resistance made malaria control and treatment more difficult, especially in Africa, where few affordable alternatives were available. The resurgence of malaria and increased mortality posed a significant global challenge, especially in Southeast Asian countries. This severe circumstance stimulated drug discovery programs worldwide. In the 1970s, inspired by records in traditional Chinese medicine (TCM), Tu's group discovered the antimalarial drug artemisinin. Artemisinin is derived from the sweet wormwood plant, Artemisia annua L. Since its discovery, artemisinin and its derivatives (collectively called artemisinins in this review) have saved two hundred million malaria patients and become the most effective antimalarial drug.
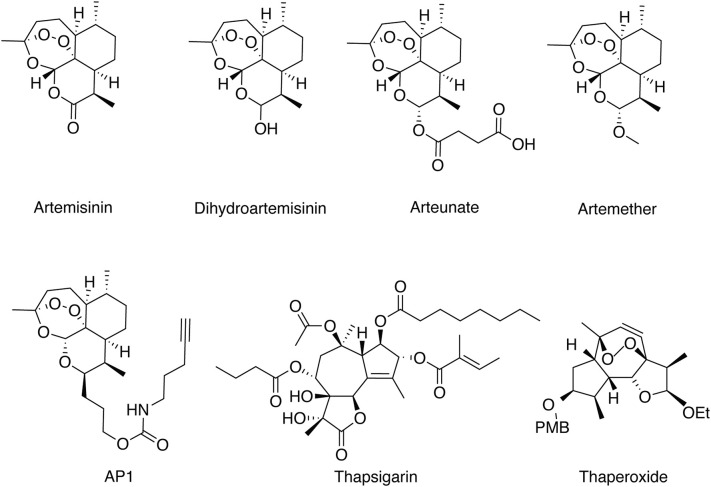
Artemisinin is a sesquiterpene lactone containing an unusual peroxide bridge with a formula of C15H22O5.(Fig. 1 ) Its unique peroxide bridge is essential for its anti-malaria activity. Because artemisinin itself is poorly soluble in water or oil, engineering efforts were made to remove the carbonyl group of artemisinin to obtain dihydroartemisinin (DHA). Additional modifications on DHA led to the synthesis of the water-soluble artesunate and oil-soluble artemether and arteether. All those four artemisinin derivatives have significantly improved antimalarial activity (Cui & Su, 2009; Miller & Su, 2011), and are included in the frontline antimalarial artemisinin-based combination therapies (ACTs).(Fig. 1) Furthermore, some fully synthetic peroxides have also been developed based on the key pharmacophore in artemisinins. For example, 1,2,4-trioxolane OZ439 and the trioxaquine SAR116242 have reach Phase II and preclinical stages (Jefford, 2012).
Fig. 1.
Chemical structures of artemisinin and its derivatives, artemisinin-based chemical engineered probe (AP1), thapsigargin and thaperoxide.
The ACT regimen is typically composed of one artemisinin derivative, such as artemether, artesunate, dihydroartemisinin, and another longer-lasting antimalarial, including lumefantrine, amodiaquine, mefloquine, piperaquine, pyronaridine and SP. A three-day course of ACT is recommended by the WHO to treat uncomplicated Plasmodium falciparum malaria. There are several reasons that underpinned this regimen design. Firstly, artemisinins are potent and safe anti-malaria drugs that rapidly kill parasites at nanomolar concentrations. However, the elimination half-lives of artemisinins are relatively short – less than 5 h. Thus, the recommended ACT regimens contain both artemisinins that would rapidly eliminate most of the malaria parasites, and a partner drug with a much longer half-life, such as mefloquine, which would eliminate the remaining parasites. Secondly, the artemisinin monotherapy requires longer treatment and it might increase selection pressure and promote the development of artemisinin resistance. Thirdly, considering drug adherence, a three-day course of ACT is also a better choice.
Since the discovery of artemisinins, considerable efforts have been made to elucidate the mechanism of action (MOA) of artemisinins. Understanding the MOA of artemisinins is of great interest for both optimizing the treatment regimens and aiding the targeted design of future antimalarial drugs. Despite some gaps and controversy, the principal MOA of artemisinin has come to be understood. At the heart of the question is the understanding of how artemisinin is activated and the identities of its direct targets (Li & Zhou, 2010; Robert, Dechy-Cabaret, Cazelles, & Meunier, 2002). Here we will review the recent understanding of the mechanism of anti-malaria actions of artemisinins, which includes drug activation and drug target identification. As the discovery of artemisinin drug targets is closely synchronized with the development of analytical techniques, we will also discuss the development of the methods used to identify drug targets. Finally, we would summarize the recent studies regarding the repurposing of artemisinin, which would broaden the application of artemisinins and benefit a wider range of patients.
2. The activation of artemisinins
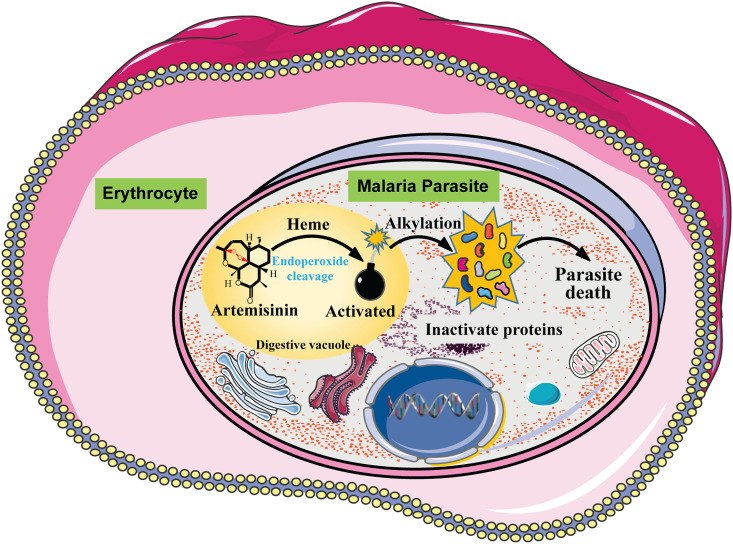
Owing to the great effort of researchers, the mechanism of activation of artemisinins has been gradually revealed. The function of artemisinins is directly linked to the special chemical structure of artemisinins(Meshnick et al., 1993a; Zhang, Gosser, & Meshnick, 1992). It was found that the characteristic endoperoxide bridge of artemisinins could be cleaved inside the infected erythrocytes (Haynes, Cheu, N'Da, Coghi, & Monti, 2014; O'Neill, Barton, & Ward, 2010). Analogues losing this chemical feature also lose the anti-malaria activity (Jefford et al., 1996; Klayman, 1985). The cleavage of the endoperoxide bridge produces highly reactive carbon-centered radicals that can alkylate susceptible proteins and cellular metabolites, and generate reactive oxygen species (ROS), which collectively lead to the death of parasites (Stocks et al., 2007). Generally, artemisinins are prodrugs that are specifically activated at intracellular and/or intraparasite environments (Haynes et al., 2014; O'Neill et al., 2010). Thus, the key question here is what triggers the specific cleavage of the endoperoxide bridge so that the resulting radicals only attack malaria parasites but not human cells. When parasites propagate inside the hosting erythrocytes, they rely on massive hemoglobin digestion to obtain nutrients that are essential for their growth and maturation. Hemoglobin digestion has been strongly linked with the susceptibility of parasites toward artemisinin treatment (Klonis et al., 2011; Xie et al., 2016). The digestion of hemoglobin releases abundant free redox-active heme and free ferrous iron (Klonis et al., 2011; Lew, Tiffert, & Ginsburg, 2003). Like the Fenton reaction, in which H2O2 is catalyzed by ferrous iron to generate free HO radicals, both free ferrous iron and heme could interact with artemisinin to produce carbon-centered radicals (Wu et al., 1998). Thus, free ferrous iron (Fe2+) and heme were proposed as the factors required for the specific activation of artemisinins (O'Neill et al., 2010). Even though there is still some debate over whether one or the other is the predominant activator (which will be discussed in detail in the following two sections), the data clearly links the unique metabolic feature of malarial parasites – hemoglobin digestion – with drug activation, explaining the specificity of artemisinin toward malarial parasites but not human cells.
2.1. Iron in artemisinin activation and activity
Free ferrous iron was proposed as the activator of artemisinin and supported by the following evidence. First, it was found that artemisinin is decomposed in an iron-dependent manner, which converts artemisinin into carbon-centered radicals through the generation of oxy radicals and subsequent electronic rearrangements (Meshnick et al., 1993b; Posner & Oh, 1992). Several models were proposed to elucidate the free radical generation pathways of artemisinin after iron-mediated decomposition (O'Neill et al., 2010; O'Neill & Posner, 2004). Second, iron chelators, such as pyridoxal benzoylhydrazone,1,2-dimethyl-3-hydroxypyrid-4-one and desferrioxamine (DFO)(selective for the non-heme source of iron), antagonize the efficacy of artemisinin (Eckstein-Ludwig et al., 2003; Meshnick et al., 1993b). Third, artemisinin derivatives carrying fluorescent tags were found to accumulate in parasites' cytoplasm and food vacuole, and this characteristic labeling and enrichment were abolished by the iron chelator DFO (Eckstein-Ludwig et al., 2003; Stocks et al., 2007). These data suggested an iron-dependent artemisinin activation mechanism, which seems to be important for the anti-malarial function of artemisinin. Intriguingly, Haynes and co-workers generated two highly active artemisinin derivatives that only had a relatively weak reaction with free ferrous iron (Haynes et al., 2004). This finding suggested that artemisinin activation might not require the presence of free ferrous iron. Furthermore, DFO could protect against oxidative stress through inhibition of cytotoxic heme and ferryl heme reduction, which is independent of its function of iron chelation (Reeder, Hider, & Wilson, 2008; Reeder & Wilson, 2005). Thus, the antagonism of iron chelator against artemisinin induced cell death might be simply because of its anti-oxidative activity that attenuates the oxidative stress resulting from artemisinin activation rather than the interference of the activation per se. Taken together, despite some supporting evidence, the iron-dependent artemisinin activation mechanism still requires further clarification.
2.2. Heme in artemisinin activation and activity
On the other hand, the evidence that supports heme as the activator of artemisinin seems to be more convincing. Meshnick and co-workers first isolated artemisinin-heme adducts from artemisinin-treated P. falciparum and confirmed hemin catalyzed decomposition of artemisinin by cyclic voltammetry experiment (Butler et al., 1998; Meshnick, Thomas, Ranz, Xu, & Pan, 1991; Zhang et al., 1992). Experiments comparing the reactions of artemisinin with different redox forms of heme, ferrous iron, and deoxygenated and oxygenated hemoglobin under similar in vitro conditions showed that redox active heme reacted with artemisinin much more efficiently than the other iron-containing species, suggesting its role as the primary activator of artemisinin (Zhang & Gerhard, 2008). Recently, a study using a live parasites system observed that a cysteine protease inhibitor that blocks the digestion of hemoglobin to release heme inhibited the activation of artemisinin, whereas the addition of iron chelator DFO made no difference. More direct evidence came from the in vitro experiment, in which every component of the reaction could be strictly controlled. It was found that artemisinin only reacts with its target protein in the presence of heme but not free ferrous iron. Thus, this again suggests that heme, rather than free ferrous iron, plays a predominant role in artemisinin activation (Meunier & Robert, 2010; Jigang Wang et al., 2015).
Stability assays of peroxide antimalarials (including artemisinin) in the presence of human oxyhemoglobin, the most abundant form of iron in healthy erythrocytes, showed that peroxide antimalarials did not react with intact human hemoglobin, consistent with the view that heme is trapped inside hemoglobin and is only released upon parasite-induced hemoglobin digestion (Creek et al., 2009). Stage-specific sensitivity assays showed that artemisinin acted effectively against parasites in both rings and schizonts stages (Skinner, Manning, Johnston, & Davis, 1996). This result seemed to contradict the heme-centric hypothesis for artemisinin activation because it was thought that parasites in the ring stage did not digest hemoglobin (Skinner et al., 1996). This apparent discrepancy was clarified by studies that demonstrated the existence of heme in malarial parasites at the ring stage. On the one hand, there is active heme biosynthesis in the malarial parasite at the ring stage (Wang et al., 2015). On the other hand, it was found that hemoglobin digestion starts at the ring stage, and parasites even accumulate hemozoin crystal at late-ring-stage (Bakar, Klonis, Hanssen, Chan, & Tilley, 2010). In summary, heme-centric activation might be the underpinning mechanism for the specific activation of artemisinin.
2.3. Other activation mechanisms
Interestingly, Zhou and co-workers proposed another scenario in which the activation of artemisinin was approached from a different angle. Using a yeast model and a rodent malarial model as the proxy, it was found that mitochondria play an important role in the activation and action of artemisinin. They suggested that some unknown factors in mitochondria, possibly the components of electron transport chains, activated artemisinin, which subsequently generates active radicals that damage mitochondria and lead to mitochondrial dysfunction. Meanwhile, the activated artemisinin also contributes ROS generation that further compromise cellular function (Li et al., 2005; Sun, Li, Cao, Long, & Zhou, 2015; Wang et al., 2010). However, the mitochondria-based model cannot exclude the influence of heme on artemisinin activation as heme is mainly synthesized in mitochondria. Furthermore, additional experiments are needed to explain why only the mitochondria of malarial parasites can specifically activate artemisinin but not mitochondria of the human cell or Artemisia annua cell.
Taken together, even though we cannot fully exclude the possibility that other factors also contribute to artemisinin activation, the existing evidence favors the hypothesis that heme is the predominant factor. Importantly, this working model fully explains the specificity of artemisinin.
3. Identifying the targets of artemisinin
Identifying the targets of artemisinin is the key to understanding the potency of artemisinin as an anti-malarial drug that not only quickly kills parasites but has not suffered any emergence of real drug resistance after decades of wide usage. In most cases, it is believed that one drug has one specific or major target that changes the related pathway and produces the corresponding biological effect. However, unlike most of the conventional chemical drugs, the activated artemisinin is converted into carbon-centered radicals, which makes it difficult to profile its target in an unbiased and systematic manner. Here, we will briefly summarize the methods employed – that gradually evolved from traditional laborious and low throughput approaches to unbiased and systemic methods – to identify artemisinin binding targets (Fig. 2 ). With the targets and related pathways uncovered, we can now start to understand why artemisinin is such a “miracle” for anti-malarial treatment.
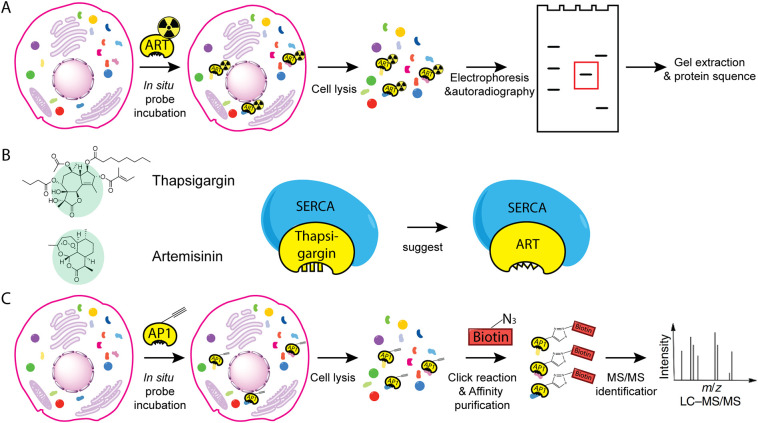
Fig. 2.
General workflow of some methods used to identify targets of artemisinin. (A) Radiolabeling and gel-based protein target identification. The radiolabeled artemisinin was synthesized and used to treat cells. The lysed parasite extracts were separated by electrophoresis. By autoradiography, protein targets of artemisinin were identified and subjected to protein sequencing. (B) Using structural similarity to identify artemisinin targets. Artemisinin and thapsigargin share a similar sesquiterpene lactone structure, thus the target of thapsigargin, SERCA, was proposed to be a target of artemisinin. (C) Using chemical proteomics to identify artemisinin targets. AP1, artemisinin derivative with a clickable alkyne moiety, was used to treat cells for in situ labeling. The lysed parasite extracts were labeled with a biotin moiety through click chemistry before affinity purification and MS-based protein identification.
3.1. Methods used to identify targets
3.1.1. Using radiolabeling and gel-based protein target identification to probe the targets of artemisinin
The first attempt to identify artemisinin binding proteins was based on the combination of radiolabeling and 2-dimensional electrophoresis. Briefly, the radiolabeled artemisinin was synthesized and used to treat P. falciparum-infected erythrocytes. The resulting parasite extracts were first separated by isoelectric focusing tube gel and followed by SDS-polyacrylamide gel electrophoresis. The protein bands that were still carrying radioactivity were the protein targets that covalently bond with radiolabeled artemisinin. It was found that several proteins were artemisinin targets, one of which was identified as the P. falciparum translational controlled tumor protein (TCTP) homolog (Asawamahasakda, Ittarat, Pu, Ziffer, & Meshnick, 1994; Bhisutthibhan et al., 1998).
3.1.2. Using structural similarity to identify artemisinin targets
In many cases, structurally similar drugs turn out to share the same drug targets. Guided by this principle, Krishna and co-workers proposed that thapsigargin (Fig. 1), a specific mammalian sarco/endoplasmic reticulum membrane calcium ATPase (SERCA) inhibitor that shares similar sesquiterpene lactone with artemisinin, might provide some clues to search for artemisinin target in malarial parasites. The only orthologue of SERCA in P. falciparum is PfATP6 that is indeed a bona fide artemisinin target validated by various biochemical experiments (Eckstein-Ludwig et al., 2003; Jung, Kim, Ki, & Kyoung, 2005; Uhlemann et al., 2005). In hindsight, even though it is ingenious to deduce artemisinin targets based on its structure information at a time when we knew little about the MOA of this drug, thapsigargin lacks the endoperoxide bridge – the pharmacophore of artemisinin, which already suggests the limitation of this approach. Building on this, a class of optimized chemical structure known as thaperoxides was synthesized, containing an endoperoxide bridge in the reduced thapsigargin-like scaffold (Fig. 1). Predictably, thaperoxide was found to be a more potent antimalarial and inhibitor of PfATP6 than thapsigargin (Pulcini et al., 2013).
3.1.3. Using yeast models to search for artemisinin targets
Compared with other well-established model systems, P. falciparum is refractory to genetic modification, limiting the options of using genetic approaches to screen for mutants of malaria parasites that are resistant to artemisinin treatment, which in turns provides information on its targets. Yeast, on the contrary, is a system where versatile genetic tools are available and indeed provides insights into many complex human diseases. Researchers developed a yeast system that was rendered sensitive to artemisinin treatment with a similar growth inhibition concentration [IC50] to P. falciparum. Taking advantage of the yeast system, researchers found that mitochondria are involved in the activation of artemisinin and that mitochondria themselves are a direct target of artemisinin (Li et al., 2005; Sun et al., 2015; Juan Wang et al., 2010). However, malaria parasites have a unique life cycle and digest hemoglobin as a source of amino acids and inevitably generate high concentrations of heme, all of which are hard to recapitulate in the yeast system. Thus, the information that can be extrapolated from the yeast studies is limited.
3.1.4. Using chemical proteomics to identify artemisinin targets
Chemical proteomics is a more recently developed approach in drug target discovery, which uses chemical methods to engineer the drug that enables subsequent affinity purification of its targets and identification by mass spectrometry (MS) (Böttcher, Pitscheider, & Sieber, 2010; Böttcher & Sieber, 2008; Evans & Cravatt, 2006; Fonović & Bogyo, 2008; Gersch, Kreuzer, & Sieber, 2012; Liu et al., 2012; Nomura, Dix, & Cravatt, 2010; Ovaa et al., 2003; Paulick & Bogyo, 2008; Speers, Adam, & Cravatt, 2003; Willems et al., 2011; Yang et al., 2010). This method provides an unbiased platform for drug target discovery and is validated by several studies wherein targets of the investigated drug are successfully identified (Böttcher et al., 2010; Su et al., 2013; Ziegler, Pries, Hedberg, & Waldmann, 2013). Special attention needs to be paid to make sure the chemically engineered drug is pharmacologically identical to its parent compound so that the incorporation of the additional tag does not disrupt authentic drug-protein interaction or bring in non-specific interaction. In the case of artemisinin, a small clickable alkyne tag was introduced to generate AP1 (Fig. 1). Drug activity analysis showed that AP1 was as potent as artemisinin, whereas the clickable alkyne tag enabled the labeling of a fluorescent dye or a biotin moiety through click chemistry after AP1-treated malarial parasites were lysed. Importantly, the addition of an excessive amount of artemisinin totally disrupted AP1 binding with its targets, suggesting AP1 and artemisinin are indeed pharmacologically identical to each other. Using this approach, the extent of in vivo artemisinin activation and targeting could be visualized and quantified by fluorescent scanning, and the details of the protein targets could be uncovered by MS analysis after affinity purification (Wang et al., 2015). Besides Wang's group, Ward's group independently applied a similar probe-based proteomics study and likewise identified multiple promiscuous drug targets of artemisinins (Ismail et al., 2016). It is evident that the chemical proteomics approach has great advantages for drug target discovery at physiological conditions.
3.2. Potential targets of artemisinins
3.2.1. Heme
Heme is a byproduct of hemoglobin digestion, which provides peptides and amino acids for the maturation and development of hematophagous malarial parasites. However, the high quantities of released free heme dimerize to hematin, a toxic molecule that causes lipid peroxidation and subsequent membrane lysis of parasites (Fitch et al., 1983). To circumvent the toxicity of hematin, malarial parasites developed a detoxification mechanism that converts hematin to non-toxic hemozoin (malaria pigment) through a biomineralization process (Egan, 2008). Since this hemozoin formation is essential to the survival of malarial parasites, drugs that inhibit this process, such as chloroquine and mefloquine, effectively kill the parasites (Kumar, Guha, Choubey, Maity, & Bandyopadhyay, 2007). Alkylation of heme by artemisinin was first reported by Meshnick and colleagues, in which they used MS to identify heme-artemisinin adducts (Meshnick et al., 1991; Ying-Zi, Little, & Meshnick, 1994). Subsequent experiments showed that activated artemisinins alkylates heme, inhibiting hemozoin formation both in vivo and in vitro and contributing to hematin-mediated malarial death (Cazelles, Robert, & Meunier, 2001; Loup, Lelièvre, Benoit-Vical, & Meunier, 2007; Meunier & Robert, 2010; Robert, Benoit-Vical, Claparols, & Meunier, 2005). Thus, heme can serve as both the activator and target of artemisinin (Meunier & Robert, 2010).
3.2.2. PfATP6
Inspired by artemisinin's structural similarity to thapsigargin, PfATP6 was identified as a potential target. On the one hand, artemisinin could specifically inhibit PfATP6 to an extent that is comparable to that of thapsigargin. On the other hand, thapsigargin antagonizes the parasiticidal activity of artemisinin. Besides, there is a correlation between the inhibitory effects of artemisinin on PfATP6 activity and killing potencies of artemisinin for parasites. The labeling of malarial parasites by a fluorescent derivative of thapsigargin (BODIPY-thapsigargin) could be abolished by an excess amount of either thapsigargin or artemisinin (Eckstein-Ludwig et al., 2003; Jung et al., 2005; Uhlemann et al., 2005). A field isolate with mutations in PfATP6 was somewhat resistant to artemether treatment (Jambou et al., 2005). However, another study with the same field isolate showed full sensitivity toward artemisinins (Cojean, Hubert, Le Bras, & Durand, 2006). Futhermore, the data associating PfATP6 with ART action was produced in Xenopus oocytes, which is known to be noisy with poor signal to background ratios (Eckstein-Ludwig et al., 2003). Although some experimental results have suggested PfATP6 as a drug target of artemisinins by different approaches (Arnou et al., 2011; Krishna, Pulcini, Fatih, & Staines, 2010; Krishna, Pulcini, Moore, Teo, & Staines, 2014), other results have contradicted this theory (Cojean et al., 2006; El Garah, Stigliani, Coslédan, Meunier, & Robert, 2009). The controversial outcomes so far suggest that PfATP6 might not be the main target that underlies the antimalarial effects of artemisinin.
3.2.3. TCTP
As previously described, the 25 kDa TCTP was identified through the radiolabeling method. The in vitro interaction between TCTP and artemisinin is heme-dependent, which is consistent with heme-centric activation mechanisms (Bhisutthibhan et al., 1998). Using mass spectroscopy and chemical-probe-based enrichment assay, it was found that activated artemisinin binds to the N-terminal region of TCTP and could alkylate multiple amino acids from Phe12 to Tyr22 of TCTP in the presence of heme (Eichhorn et al., 2013, Li et al., 2016). However, the molecular function of TCTP is obscure, and it is hard to explain why artemisinin could efficiently and quickly kill malaria parasites if TCTP is its major target.
3.2.4. Promiscuous protein alkylation
The promiscuous targeting of activated artemisinin was first reported when the chemical proteomics method described above (Section 3.1.4) was employed to study the MOA of artemisinin. In total, it was found that 124 proteins are covalently bound with heme-activated artemisinin in live parasites (Wang et al., 2015). Importantly, a similar promiscuous targeting mechanism was independently confirmed by another probe-based proteomics study (Ismail et al., 2016). Previously known targets of artemisinin, such as TCTP, PfCRT, Pfmdr1, and PfATG6, could also be observed by this high throughput chemical proteomics method. The protein targets involved in many essential pathways, including hemoglobin catabolic, antioxidant defence, glycolytic, nucleoside, and protein synthesis (Ismail et al., 2016; Wang et al., 2015). These results strongly suggest that artemisinin does not fit into the classical paradigm of “one-drug-one-target”. Instead, the heme-activated artemisinin can simultaneously target many malarial proteins that disrupt many essential pathways and lead to death of parasites, consistent with previous findings that activated artemisinin is a carbon-centered radical (Wang et al., 2015; Zhou, Li, & Xiao, 2016) Furthermore, several studies showed that artemisinins could synergize with parasite proteasome inhibitors, which might lead to a promising drug combination with great potential for future use (Bridgford et al., 2018; Dogovski et al., 2015; Kirkman et al., 2018; Li et al., 2016; Stokes et al., 2019).
With a deep understanding of the activation and targeting mechanisms of artemisinin (Fig. 3 ), we can now start to understand and appreciate why artemisinin is such an ideal anti-malaria drug from the molecular level. The key is the endoperoxide bridge of artemisinin that mainly requires heme for activation. Because of the metabolic feature of malarial parasites that rely on hemoglobin digestion to provide the essential nutrients for the progression of the cell cycle, an excessive amount of heme release is unavoidable – in other words, the activation of artemisinin is unavoidable. The activated artemisinin, converting into carbon-centered free radicals, alkylate and damage proximal proteins inside the parasite, resulting in disruption of cellular pathways and the death of parasites. Thus, artemisinin is like an elegantly “designed” drug, which is specifically activated by the byproduct of parasite metabolism, thus precisely targeting parasites and avoiding attacking healthy erythrocytes. The promiscuous targeting of artemisinin also confers a great advantage, as it is much more difficult to develop drug resistance to multiple targets and pathways than gain resistance to a single protein target. This also explains why, after decades of widespread use as front line anti-malarials, no conventional drug resistance has emerged.
Fig. 3.
A model of the mechanism of action of artemisinin. Artemisinin is activated by a heme-dependent endoperoxide bridge cleavage in the digestive vacuoles of malaria parasites. The activated artemisinin alkylates and damages proximal proteins inside the parasite, resulting in disruption of cellular pathways and the death of parasites.
On this topic, it is worth noting that the phenomenon of “artemisinin resistance” was first reported when P .falciparum displaying a delayed clearance rate after a 3-day ACT treatment first emerged in Southeast Asia. These parasites frequently bear mutations in the parasite kelch13 (K13) gene, which results in a reduction of Kelch13 at the protein level (Siddiqui, Srivastava, Russell, & Creek, 2017). Kelch13 protein localizes to the periphery of parasites, and has been shown to regulate the endocytosis of hemoglobin (Birnbaum et al., 2020; Yang et al., 2019). Thus, parasites with mutated Kelch13 showed reduced hemoglobin endocytosis (Birnbaum et al., 2020; Yang et al., 2019). As previously summarized, hemoglobin endocytosis is essential both for the activation of artemisinins and for parasite growth. The reduced hemoglobin endocytosis renders parasites partially resistant to artemisinin exposure but also results in a lengthened ring stage in their life cycle (Birnbaum et al., 2020; Yang et al., 2019). Although the “resistant” parasites could withstand a short exposure of artemisinins as in the 3-day ACT regimens, the parasites would inevitably have to reach the trophozoite stage, wherein massive hemoglobin endocytosis and activation of artemisinins occur simultaneously. The activated radical artemisinins would then damage cellular proteins, which results in the death of the parasite. Thus, unlike conventional drug resistance, which fails to eliminate the parasites after a full course of treatment, the full course 7-day treatment of ACT is still efficacious to eradicate these K13 mutant parasites (Krishna & Kremsner, 2013). Nevertheless, although the artemisinins are still efficacious to P. falciparum mutants, the mutational evolution of parasites should continue to be closely monitored so that ACTs can continue to be effectively deployed or modified as necessary (Wang et al., 2019).
4. Repurposing of artemisinin
Due to its efficacy, affordability, and clinical safety, considerable efforts are invested in applying artemisinin in non-malarial areas, including anti-cancer, anti-inflammatory, anti-parasite (other than Plasmodium) and anti-viral therapies (Crespo-Ortiz & Wei, 2012; Efferth et al., 2016; Ho, Peh, Chan, & Wong, 2014; Lai, Singh, & Sasaki, 2013; Willoughby et al., 2009). Here, we will only focus on the recent progress in the anti-cancer application of artemisinin as it serves as a great example showing how to leverage on the MOA of artemisinin to better engineer it to treat cancer.
Since first reported in 1993, the selective cytotoxicity of artemisinin against different cancer types, both in vitro and in vivo has been reported (Efferth et al., 2003; Lai & Singh, 1995; Woerdenbag et al., 1993). Even though a wide range of pathways and mechanisms were deliberately identified and their roles in anticancer effects of artemisinin have been explained, it is quite clear that the efficacy of artemisinin is rather limited, much lower than it as an anti-malarial drug (Crespo-Ortiz & Wei, 2012; Efferth, 2015; Lai et al., 2013; Willoughby et al., 2009). Since artemisinin relies on heme for activation as an anti-malarial drug, could we exploit its activation mechanism to improve its efficacy in anti-cancer treatment? Interestingly, it was found that the level of heme synthesis and availability are elevated in cancer cells (Hooda et al., 2013; Hooda, Alam, & Zhang, 2015; Hooda, Shah, & Zhang, 2014). Artemisinin is also activated by heme in the cancer cell and the anticancer activity correlates with the level of heme synthesis (Mercer, Copple, Maggs, O'Neill, & Park, 2011; Stockwin et al., 2009; Wang et al., 2017; Zhang, Chen, & Gerhard, 2010; Zhang & Gerhard, 2009). As mitochondria are the site where heme synthesis takes place in the mammalian cell, an artemisinin derivative with a specific mitochondria-targeting tag increases the anti-cancer activity of artemisinin by ten-fold (Zhang et al., 2015; Zhang et al., 2016). Consistently, the addition of aminolevulinic acid (ALA), the rate-limiting precursor used for heme biosynthesis, greatly enhances the anti-colorectal-cancer activity of artemisinin. Interestingly, ALA is a clinically approved chemical used in photodynamic therapy, suggesting the combination of artemisinin and ALA treatment should be safe. Currently, preliminary data from clinical trials using artemisinin derivatives to treat non-small cell lung cancer, cervical cancer, and colorectal cancer is safe, well-tolerated, and effective to a certain extent (Jansen et al., 2011; Krishna et al., 2015; Zhang et al., 2008). Future studies should test the safety and efficacy of the more optimized artemisinin-based anti-cancer therapy, such as the artemisinin plus ALA treatment.
Other applications of artemisinin have been continuously studied. There is no doubt that artemisinin has certain anti-inflammatory, anti-viral, and anti-obesity effects (Bunnag, Viravan, Looareesuwan, Karbwang, & Harinasuta, 1991; Efferth, 2018; Efferth et al., 2008; Ho et al., 2014; Keiser & Utzinger, 2007; Lam, Long, Su, Zhuan, & Lu, 2018; Saeed, Krishna, Greten, Kremsner, & Efferth, 2016). The important question for future work is how to optimize and improve the efficacy of artemisinin-based therapies from what we have already known about its MOA to treat those devastating diseases currently without cures. For instance, it is of interest to explore the potential application of artemisinins against the severe acute respiratory coronavirus 2 (SARS-CoV-2). Preliminary in vitro data has so far indicated that artemisinin could bind to the SARS-CoV-2 spike protein, which is essential for the host infection of SARS-CoV-2 (Cao et al., 2020; Sehailia & Chemat, 2020). However, much more cautious and in-depth research would be necessary before exploring any potential clinical use of artemisinins toward SARS-CoV-2.
5. Summary
Artemisinin and its derivatives are the first-line treatment for malaria cases and have saved millions of lives of patients suffering from malaria. As a novel antimalarial drug, artemisinins are effective, well-tolerated and affordable. These outstanding pharmacological characteristics result from the unique MOAs of artemisinins, whereby artemisinins are specifically activated and act inside parasites. Guided by the detailed MOA of artemisinins at molecular level, the applications of artemisinins in non-malarial areas have been considerably studied. Preliminary data present promising applications of artemisinins in anti-cancer, anti-inflammation, anti-parasites and anti-viral areas. While great strides have been made to reveal the mystery of artemisinins, there are some gaps in knowledge of artemisinins that needs to be investigated thoroughly. Firstly, the mechanism of activation and targets of artemisinins in artemisinin-resistant malaria needs to be further elucidated. This study would reveal the molecular mechanism underpinning artemisinin resistance, which would help to adjust the clinical ACTs regimens to kill artemisinin-resistant parasites more efficiently. Secondly, more efforts are needed to reveal the specific MOAs at different disease models. These studies are essential for broadening the application of artemisinins in non-malarial areas and eventually bring artemisinins to the bedsides of more patients.
Declaration of Competing Interest
All authors have no conflicts of interest to declare.
Acknowledgments
We gratefully acknowledge financial support from the National Natural Science Foundation of China (81903588, 81803456, 82074098, 81702580 and 81841001), TCM One Belt and One Road Project of CACMS (GH201807), the Major National Science and Technology Program of China for Innovative Drug (2017ZX09101002-001-001-05), the National Key R&D Program of China (2020YFA0908000, 2020YFA0908004).
References
- Arnou B., Montigny C., Morth J.P., Nissen P., Jaxel C., Møller J.V., Le Maire M. The Plasmodium falciparum Ca2+-ATPase PfATP6: Insensitive to artemisinin, but a potential drug target. Biochemical Society Transactions. 2011;39(3) doi: 10.1042/BST0390823. [DOI] [PubMed] [Google Scholar]
- Asawamahasakda W., Ittarat I., Pu Y.M., Ziffer H., Meshnick S.R. Reaction of antimalarial endoperoxides with specific parasite proteins. Antimicrobial Agents and Chemotherapy. 1994;38(8):1854–1858. doi: 10.1128/AAC.38.8.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakar N.A., Klonis N., Hanssen E., Chan C., Tilley L. Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of Plasmodium falciparum. Journal of Cell Science. 2010;123(3):441–450. doi: 10.1242/jcs.061499. [DOI] [PubMed] [Google Scholar]
- Bhisutthibhan J., Pan X.Q., Hossler P.A., Walker D.J., Yowell C.A., Carlton J.…Meshnick S.R. The Plasmodium falciparum translationally controlled tumor protein homolog and its reaction with the antimalarial drug artemisinin. Journal of Biological Chemistry. 1998;273(26) doi: 10.1074/jbc.273.26.16192. [DOI] [PubMed] [Google Scholar]
- Birnbaum J., Scharf S., Schmidt S., Jonscher E., Maria Hoeijmakers W.A., Flemming S.…Spielmann T. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367(6473):51–59. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- Böttcher T., Pitscheider M., Sieber S.A. Natural products and their biological targets: Proteomic and metabolomic labeling strategies. Angewandte Chemie - International Edition. 2010;49(15):2680–2698. doi: 10.1002/anie.200905352. [DOI] [PubMed] [Google Scholar]
- Böttcher T., Sieber S.A. β-Lactones as privileged structures for the active-site labeling of versatile bacterial enzyme classes. Angewandte Chemie - International Edition. 2008;47(24) doi: 10.1002/anie.200705768. [DOI] [PubMed] [Google Scholar]
- Bridgford J.L., Xie S.C., Cobbold S.A., Pasaje C.F.A., Herrmann S., Yang T.…Tilley L. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nature Communications. 2018;9(1):1–9. doi: 10.1038/s41467-018-06221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnag D., Viravan C., Looareesuwan S., Karbwang J., Harinasuta T. Clinical trial of artesunate and artemether on multidrug resistant falciparum malaria in Thailand. A preliminary report. Southeast Asian Journal of Tropical Medicine and Public Health. 1991;22(3) [PubMed] [Google Scholar]
- Butler A.R., Gilbert B.C., Hulme P., Irvine L.R., Renton L., Whitwood A.C. EPR evidence for the involvement of free radicals in the iron-catalysed decomposition of qinghaosu (artemisinin) and some derivatives; antimalarial action of some polycyclic endoperoxides. Free Radical Research. 1998;28(5) doi: 10.3109/10715769809066884. [DOI] [PubMed] [Google Scholar]
- Cao R., Hu H., Li Y., Wang X., Xu M., Liu J.…Zhong W. Anti-SARS-CoV-2 potential of artemisinins in vitro. ACS Infectious Diseases. 2020;6(9):2524–2531. doi: 10.1021/acsinfecdis.0c00522. [DOI] [PubMed] [Google Scholar]
- Cazelles J., Robert A., Meunier B. Alkylation of heme by artemisinin, an antimalarial drug. Comptes Rendus de l’Academie Des Sciences - Series IIC: Chemistry. 2001;4(2) doi: 10.1016/S1387-1609(00)01188-9. [DOI] [Google Scholar]
- Cojean S., Hubert V., Le Bras J., Durand R. Resistance to dihydroartemisinin [7] Emerging Infectious Diseases. 2006;12(11) doi: 10.3201/eid1211.060903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox F.E. History of the discovery of the malaria parasites and their vectors. Parasites and Vectors. 2010;3(1):5. doi: 10.1186/1756-3305-3-5. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D.J., Ryan E., Charman W.N., Chiu F.C.K., Prankerd R.J., Vennerstrom J.L., Charman S.A. Stability of peroxide antimalarials in the presence of human hemoglobin. Antimicrobial Agents and Chemotherapy. 2009;53(8) doi: 10.1128/AAC.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Ortiz M.P., Wei M.Q. Antitumor activity of artemisinin and its derivatives: From a well-known antimalarial agent to a potential anticancer drug. Journal of Biomedicine and Biotechnology. 2012;2012 doi: 10.1155/2012/247597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Su X.Z. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Review of Anti-Infective Therapy. 2009;7(8):999–1013. doi: 10.1586/ERI.09.68. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogovski C., Xie S.C., Burgio G., Bridgford J., Mok S., McCaw J.M.…Tilley L. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biology. 2015;13(4) doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein-Ludwig U., Webb R.J., Van Goethem I.D.A., East J.M., Lee A.G., Kimura M.…Krishna S. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 2003;424(6951):957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- Efferth T. Artemisinin–Second career as anticancer drug? World Journal of Traditional Chinese Medicine. 2015;1(4):2–25. doi: 10.15806/j.issn.2311-8571.2015.0036. [DOI] [Google Scholar]
- Efferth T. Beyond malaria: The inhibition of viruses by artemisinin-type compounds. Biotechnology Advances. 2018;36(6) doi: 10.1016/j.biotechadv.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Efferth T., Romero M.R., Wolf D.G., Stamminger T., Marin J.J.G., Marschall M. The antiviral activities of artemisinin and artesunate. Clinical Infectious Diseases. 2008;47(6):804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- Efferth T., Romero R., Rita Bilia A., Galal Osman A., ElSohly M., Wink M.…Marin J. Expanding the therapeutic spectrum of artemisinin: Activity against infectious diseases beyond malaria and novel pharmaceutical developments. World Journal of Traditional Chinese Medicine. 2016;2(2):1–23. doi: 10.15806/j.issn.2311-8571.2016.0002. [DOI] [Google Scholar]
- Efferth T., Sauerbrey A., Olbrich A., Gebhart E., Rauch P., Weber H.O.…Funk J.O. Molecular modes of action of artesunate in tumor cell lines. Molecular Pharmacology. 2003;64(2) doi: 10.1124/mol.64.2.382. [DOI] [PubMed] [Google Scholar]
- Egan T.J. Recent advances in understanding the mechanism of hemozoin (malaria pigment) formation. Journal of Inorganic Biochemistry. 2008;102(5–6):1288–1299. doi: 10.1016/j.jinorgbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Eichhorn T., Winter D., Büchele B., Dirdjaja N., Frank M., Lehmann W.D.…Efferth T. Molecular interaction of artemisinin with translationally controlled tumor protein (TCTP) of Plasmodium falciparum. Biochemical Pharmacology. 2013;85(1):38–45. doi: 10.1016/j.bcp.2012.10.006. [DOI] [PubMed] [Google Scholar]
- El Garah F.B., Stigliani J.L., Coslédan F., Meunier B., Robert A. Docking studies of structurally diverse antimalarial drugs targeting PfATP6: No correlation between in silico binding affinity and in vitro antimalarial activity. ChemMedChem. 2009;4(9) doi: 10.1002/cmdc.200900200. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Cravatt B.F. Mechanism-based profiling of enzyme families. Chemical Reviews. 2006;106(8):3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Fitch C.D., Chevli R., Kanjananggulpan P., Dutta P., Chevli K., Chou A.C. Intracellular ferriprotoporphyrin IX is a lytic agent. Blood. 1983;62(6) doi: 10.1182/blood.v62.6.1165.bloodjournal6261165. [DOI] [PubMed] [Google Scholar]
- Fonović M., Bogyo M. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Review of Proteomics. 2008;5(5):721–730. doi: 10.1586/14789450.5.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersch M., Kreuzer J., Sieber S.A. Electrophilic natural products and their biological targets. Natural Product Reports. 2012;29(6) doi: 10.1039/c2np20012k. [DOI] [PubMed] [Google Scholar]
- Haynes R., Cheu K.-W., N’Da D., Coghi P., Monti D. Considerations on the mechanism of action of artemisinin antimalarials: Part 1 - The carbon radical and heme hypotheses. Infectious Disorders - Drug Targets. 2014;13(4) doi: 10.2174/1871526513666131129155708. [DOI] [PubMed] [Google Scholar]
- Haynes R.K., Ho W.Y., Chan H.W., Fugmann B., Stetter J., Croft S.L.…Robinson B.L. Highly antimalaria-active artemisinin derivatives: Biological activity does not correlate with chemical reactivity. Angewandte Chemie - International Edition. 2004;43(11):1381–1385. doi: 10.1002/anie.200352343. [DOI] [PubMed] [Google Scholar]
- Ho W.E., Peh H.Y., Chan T.K., Wong W.S.F. Artemisinins: Pharmacological actions beyond anti-malarial. Pharmacology and Therapeutics. 2014;142(1):126–139. doi: 10.1016/j.pharmthera.2013.12.001. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Hooda J., Alam M., Zhang L. Measurement of heme synthesis levels in mammalian cells. Journal of Visualized Experiments. 2015;2015(101) doi: 10.3791/51579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda J., Cadinu D., Alam M.M., Shah A., Cao T.M., Sullivan L.A.…Zhang L. Enhanced heme function and mitochondrial respiration promote the progression of lung cancer cells. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0063402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda J., Shah A., Zhang L. Heme, an essential nutrient from dietary proteins, critically impacts diverse physiological and pathological processes. Nutrients. 2014;6(3):1080–1102. doi: 10.3390/nu6031080. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail H.M., Barton V., Phanchana M., Charoensutthivarakul S., Wong M.H.L., Hemingway J.…Ward S.A. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):2080–2085. doi: 10.1073/pnas.1600459113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambou R., Legrand E., Niang M., Khim N., Lim P., Volney B.…Mercereau-Puijalon O. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366(9501):1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- Jansen F.H., Adoubi I., Comoe J.C., De Cnodder T., Jansen N., Tschulakow A., Efferth T. First study of oral artenimol-R in advanced cervical cancer: Clinical benefit, tolerability and tumor markers. Anticancer Research. 2011;31(12) [PubMed] [Google Scholar]
- Jefford C.W., Vicente M.G.H., Jacquier Y., Favarger F., Mareda J., Millasson-Schmidt P.…Burger U. 124. The deoxygenation and isomerization of artemisinin and artemether and their relevance to antimalarial action. Helvetica Chimica Acta. 1996;79(5) doi: 10.1002/hlca.19960790520. [DOI] [Google Scholar]
- Jefford W. Synthetic peroxides as potent antimalarials. News and views. Current Topics in Medicinal Chemistry. 2012;12(5):373–399. doi: 10.2174/156802612799362940. [DOI] [PubMed] [Google Scholar]
- Jung M., Kim H., Ki Y.N., Kyoung T.N. Three-dimensional structure of Plasmodium falciparum Ca2+- ATPase(PfATP6) and docking of artemisinin derivatives to PfATP6. Bioorganic and Medicinal Chemistry Letters. 2005;15(12):2994–2997. doi: 10.1016/j.bmcl.2005.04.041. [DOI] [PubMed] [Google Scholar]
- Keiser J., Utzinger J. Artemisinins and synthetic trioxolanes in the treatment of helminth infections. Current Opinion in Infectious Diseases. 2007;20(6) doi: 10.1097/QCO.0b013e3282f19ec4. [DOI] [PubMed] [Google Scholar]
- Kirkman L.A., Zhan W., Visone J., Dziedziech A., Singh P.K., Fan H.…Lin G. Antimalarial proteasome inhibitor reveals collateral sensitivity from intersubunit interactions and fitness cost of resistance. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(29):E6863–E6870. doi: 10.1073/pnas.1806109115. National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klayman D.L. Qinghaosu (artemisinin): An antimalarial drug from China. Science. 1985;228(4703) doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- Klonis N., Crespo-Ortiz M.P., Bottova I., Abu-Bakar N., Kenny S., Rosenthal P.J., Tilley L. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Ganapathi S., Ster I.C., Saeed M.E.M., Cowan M., Finlayson C.…Kumar D. A randomised, double blind, placebo-controlled pilot study of oral artesunate therapy for colorectal cancer. EBioMedicine. 2015;2(1) doi: 10.1016/j.ebiom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Kremsner P.G. Antidogmatic approaches to artemisinin resistance: Reappraisal as treatment failure with artemisinin combination therapy. Trends in Parasitology. 2013;29(7):313–317. doi: 10.1016/j.pt.2013.04.001. Elsevier Current Trends. [DOI] [PubMed] [Google Scholar]
- Krishna S., Pulcini S., Fatih F., Staines H. Artemisinins and the biological basis for the PfATP6/SERCA hypothesis. Trends in Parasitology. 2010;26(11) doi: 10.1016/j.pt.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Krishna S., Pulcini S., Moore C.M., Teo B.H.Y., Staines H.M. Pumped up: Reflections on PfATP6 as the target for artemisinins. Trends in Pharmacological Sciences. 2014;35(1):4–11. doi: 10.1016/j.tips.2013.10.007. Elsevier. [DOI] [PubMed] [Google Scholar]
- Kumar S., Guha M., Choubey V., Maity P., Bandyopadhyay U. Antimalarial drugs inhibiting hemozoin (β-hematin) formation: A mechanistic update. Life Sciences. 2007;80(9):813–828. doi: 10.1016/j.lfs.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Lai H., Singh N.P. Selective cancer cell cytotoxicity from exposure to dihydroartemisinin and holotransferrin. Cancer Letters. 1995;91(1):41–46. doi: 10.1016/0304-3835(94)03716-V. [DOI] [PubMed] [Google Scholar]
- Lai H.C., Singh N.P., Sasaki T. Development of artemisinin compounds for cancer treatment. Investigational New Drugs. 2013;31(1):230–246. doi: 10.1007/s10637-012-9873-z. [DOI] [PubMed] [Google Scholar]
- Lam N.S., Long X., Su X., Zhuan, Lu F. Artemisinin and its derivatives in treating helminthic infections beyond schistosomiasis. Pharmacological Research. 2018;133 doi: 10.1016/j.phrs.2018.04.025. [DOI] [PubMed] [Google Scholar]
- Lew V.L., Tiffert T., Ginsburg H. Excess hemoglobin digestion and the osmotic stability of Plasmodium falciparum - Infected red blood cells. Blood. 2003;101(10) doi: 10.1182/blood-2002-08-2654. [DOI] [PubMed] [Google Scholar]
- Li H., O’Donoghue A.J., Van Der Linden W.A., Xie S.C., Yoo E., Foe I.T.…Bogyo M. Structure-and function-based design of Plasmodium-selective proteasome inhibitors. Nature. 2016;530(7589):233–236. doi: 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhou B. Biological actions of artemisinin: Insights from medicinal chemistry studies. Molecules. 2010;15(3):1378–1397. doi: 10.3390/molecules15031378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhou Y., Tang G., Xiao Y. Characterization of the artemisinin binding site for translationally controlled tumor protein (TCTP) by bioorthogonal click chemistry. Bioconjugate Chemistry. 2016;27(12):2828–2833. doi: 10.1021/acs.bioconjchem.6b00556. [DOI] [PubMed] [Google Scholar]
- Li W., Mo W., Shen D., Sun L., Wang J., Lu S.…Zhou B. Yeast model uncovers dual of mitochondria in the action of artemisinin. PLoS Genetics. 2005;1(3):329–334. doi: 10.1371/journal.pgen.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.X., Yin Q.Q., Zhou H.C., Wu Y.L., Pu J.X., Xia L.…Chen G.Q. Adenanthin targets peroxiredoxin I and II to induce differentiation of leukemic cells. Nature Chemical Biology. 2012;8(5):486–493. doi: 10.1038/nchembio.935. [DOI] [PubMed] [Google Scholar]
- Loup C., Lelièvre J., Benoit-Vical F., Meunier B. Trioxaquines and heme-artemisinin adducts inhibit the in vitro formation of hemozoin better than chloroquine. Antimicrobial Agents and Chemotherapy. 2007;51(10) doi: 10.1128/AAC.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer A.E., Copple I.M., Maggs J.L., O’Neill P.M., Park B.K. The role of heme and the mitochondrion in the chemical and molecular mechanisms of mammalian cell death induced by the artemisinin antimalarials. Journal of Biological Chemistry. 2011;286(2):987–996. doi: 10.1074/jbc.M110.144188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshnick S.R., Yang Y.Z., Lima V., Kuypers F., Kamchonwongpaisan S., Yuthavong Y. Iron-dependent free radical generation from the antimalarial agent artemisinin (qinghaosu) Antimicrobial Agents and Chemotherapy. 1993;37(5):1108–1114. doi: 10.1128/AAC.37.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshnick S.R., Yang Y.Z., Lima V., Kuypers F., Kamchonwongpaisan S., Yuthavong Y. Iron-dependent free radical generation from the antimalarial agent artemisinin (qinghaosu) Antimicrobial Agents and Chemotherapy. 1993;37(5) doi: 10.1128/AAC.37.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshnick S.R., Thomas A., Ranz A., Xu C.M., Pan H.Z. Artemisinin (qinghaosu): The role of intracellular hemin in its mechanism of antimalarial action. Molecular and Biochemical Parasitology. 1991;49(2):181–189. doi: 10.1016/0166-6851(91)90062-B. [DOI] [PubMed] [Google Scholar]
- Meunier B., Robert A. Heme as trigger and target for trioxane-containing antimalarial drugs. Accounts of Chemical Research. 2010;43(11):1444–1451. doi: 10.1021/ar100070k. [DOI] [PubMed] [Google Scholar]
- Miller L.H., Su X. Artemisinin: Discovery from the Chinese herbal garden. Cell. 2011;146(6):855–858. doi: 10.1016/j.cell.2011.08.024. Cell Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura D.K., Dix M.M., Cravatt B.F. Activity-based protein profiling for biochemical pathway discovery in cancer. Nature Reviews Cancer. 2010;10(9):630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill P.M., Barton V.E., Ward S.A. The molecular mechanism of action of artemisinin - The debate continues. Molecules. 2010;15(3):1705–1721. doi: 10.3390/molecules15031705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill P.M., Posner G.H. A medicinal chemistry perspective on artemisinin and related endoperoxides. Journal of Medicinal Chemistry. 2004;47(12) doi: 10.1021/jm030571c. [DOI] [PubMed] [Google Scholar]
- Ovaa H., van Swieten P.F., Kessler B.M., Leeuwenburgh M.A., Fiebiger E., van den Nieuwendijk A.M.C.H.…Overkleeft H.S. Chemistry in living cells: Detection of active proteasomes by a two-step labeling strategy. Angewandte Chemie. 2003;115(31):3754–3757. doi: 10.1002/ange.200351314. [DOI] [PubMed] [Google Scholar]
- Paulick M.G., Bogyo M. Application of activity-based probes to the study of enzymes involved in cancer progression. Current Opinion in Genetics and Development. 2008;18(1):97–106. doi: 10.1016/j.gde.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner G.H., Oh C.H. A regiospecifically oxygen-18 labeled 1,2,4-trioxane: A simple chemical model system to probe the mechanism(s) for the antimalarial activity of artemisinin (Qinghaosu) Journal of the American Chemical Society. 1992;114(21) doi: 10.1021/ja00047a076. [DOI] [Google Scholar]
- Pulcini S., Staines H.M., Pittman J.K., Slavic K., Doerig C., Halbert J.…Krishna S. Expression in yeast links field polymorphisms in PfATP6 to in vitro artemisinin resistance and identifies new inhibitor classes. Journal of Infectious Diseases. 2013;208(3):468–478. doi: 10.1093/infdis/jit171. [DOI] [PubMed] [Google Scholar]
- Reeder B.J., Hider R.C., Wilson M.T. Iron chelators can protect against oxidative stress through ferryl heme reduction. Free Radical Biology and Medicine. 2008;44(3):264–273. doi: 10.1016/j.freeradbiomed.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Reeder B.J., Wilson M.T. Desferrioxamine inhibits production of cytotoxic heme to protein cross-linked myoglobin: A mechanism to protect against oxidative stress without iron chelation. Chemical Research in Toxicology. 2005;18(6):1004–1011. doi: 10.1021/tx049660y. [DOI] [PubMed] [Google Scholar]
- Robert A., Benoit-Vical F., Claparols C., Meunier B. The antimalarial drug artemisinin alkylates heme in infected mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(38):13676–13680. doi: 10.1073/pnas.0500972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A., Dechy-Cabaret O., Cazelles J., Meunier B. From mechanistic studies on artemisinin derivatives to new modular antimalarial drugs. Accounts of Chemical Research. 2002;35(3) doi: 10.1021/ar990164o. [DOI] [PubMed] [Google Scholar]
- Saeed M.E.M., Krishna S., Greten H.J., Kremsner P.G., Efferth T. Antischistosomal activity of artemisinin derivatives in vivo and in patients. Pharmacological Research. 2016;110 doi: 10.1016/j.phrs.2016.02.017. [DOI] [PubMed] [Google Scholar]
- Sehailia M., Chemat S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 spike protein than hydroxychloroquine: Potential repurposing of artenimol for COVID-19. Journal of Biomolecular Structure and Dynamics. 2020:1–11. doi: 10.1080/07391102.2020.1796809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui G., Srivastava A., Russell A.S., Creek D.J. Multi-omics based identification of specific biochemical changes associated with PfKelch13-mutant artemisinin-resistant Plasmodium falciparum. Journal of Infectious Diseases. 2017;215(9):1435–1444. doi: 10.1093/infdis/jix156. [DOI] [PubMed] [Google Scholar]
- Skinner T.S., Manning L.S., Johnston W.A., Davis T.M.E. In vitro stage-specific sensitivity of Plasmodium falciparum to quinine and artemisinin drugs. International Journal for Parasitology. 1996;26(5) doi: 10.1016/0020-7519(96)89380-5. [DOI] [PubMed] [Google Scholar]
- Speers A.E., Adam G.C., Cravatt B.F. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. Journal of the American Chemical Society. 2003;125(16):4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- Stocks P.A., Bray P.G., Barton V.E., Al-Helal M., Jones M., Araujo N.C.…O’Neill P.M. Evidence for a common non-heme chelatable-iron-dependent activation mechanism for semisynthetic and synthetic endoperoxide antimalarial drugs. Angewandte Chemie - International Edition. 2007;46(33):6278–6283. doi: 10.1002/anie.200604697. [DOI] [PubMed] [Google Scholar]
- Stockwin L.H., Han B., Yu S.X., Hollingshead M.G., Elsohly M.A., Gul W.…Newton D.L. Artemisinin dimer anticancer activity correlates with heme-catalyzed reactive oxygen species generation and endoplasmic reticulum stress induction. International Journal of Cancer. 2009;125(6):1266–1275. doi: 10.1002/ijc.24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes B.H., Yoo E., Murithi J.M., Luth M.R., Afanasyev P., da Fonseca P.C.A.…Fidock D.A. Covalent Plasmodium falciparum-selective proteasome inhibitors exhibit a low propensity for generating resistance in vitro and synergize with multiple antimalarial agents. PLoS Pathogens. 2019;15(6) doi: 10.1371/journal.ppat.1007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Ge J., Zhu B., Zheng Y.G., Zhu Q., Yao S.Q. Target identification of biologically active small molecules via in situ methods. Current Opinion in Chemical Biology. 2013;17(5) doi: 10.1016/j.cbpa.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Sun C., Li J., Cao Y., Long G., Zhou B. Two distinct and competitive pathways confer the cellcidal actions of artemisinins. Microbial Cell. 2015;2(1):14–25. doi: 10.15698/mic2015.01.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann A.C., Cameron A., Eckstein-Ludwig U., Fischbarg J., Iserovich P., Zuniga F.A.…Krishna S. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nature Structural and Molecular Biology. 2005;12(7) doi: 10.1038/nsmb947. [DOI] [PubMed] [Google Scholar]
- Wang J., Xu C., Liao F.L., Jiang T., Krishna S., Tu Y. A temporizing solution to “artemisinin resistance.”. New England Journal of Medicine. 2019;380(22):2087–2089. doi: 10.1056/NEJMp1901233. Massachussetts Medical Society. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang C.J., Chia W.N., Loh C.C.Y., Li Z., Lee Y.M.…Lin Q. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nature Communications. 2015;6 doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang J., Shi Y., Xu C., Zhang C., Wong Y.K.…Lin Q. Mechanistic investigation of the specific anticancer property of artemisinin and its combination with aminolevulinic acid for enhanced anticolorectal cancer activity. ACS Central Science. 2017;3(7):743–750. doi: 10.1021/acscentsci.7b00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Huang L., Li J., Fan Q., Long Y., Li Y., Zhou B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS One. 2010;5(3) doi: 10.1371/journal.pone.0009582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems T.E., Plowe C.V. Chloroquine-resistant malaria. The Journal of Infectious Diseases. 2001;184(6):770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- Willems L.I., Van Der Linden W.A., Li N., Li K.Y., Liu N., Hoogendoorn S.…Overkleeft H.S. Bioorthogonal chemistry: Applications in activity-based protein profiling. Accounts of Chemical Research. 2011;44(9):718–729. doi: 10.1021/ar200125k. [DOI] [PubMed] [Google Scholar]
- Willoughby J.A., Sundar S.N., Cheung M., Tin A.S., Modiano J., Firestone G.L. Artemisinin blocks prostate cancer growth and cell cycle progression by disrupting Sp1 interactions with the cyclin-dependent kinase-4 (CDK4) promoter and inhibiting CDK4 gene expression. Journal of Biological Chemistry. 2009;284(4):2203–2213. doi: 10.1074/jbc.M804491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerdenbag H.J., Moskal T.A., Pras N., Malingré T.M., El-Feraly F.S., Kampinga H.H., Konings A.W.T. Cytotoxicity of artemisinin-related endoperoxides to Ehrlich ascites tumor cells. Journal of Natural Products. 1993;56(6):849–856. doi: 10.1021/np50096a007. [DOI] [PubMed] [Google Scholar]
- Wu Wen-Min, Wu Yikang, Wu Yu-Lin, Yao Zhu-Jun, Zhou Cheng-Ming, Li Ying, Shan Feng. Unified Mechanistic Framework for the Fe(II)-Induced Cleavage of Qinghaosu and Derivatives/Analogues. The First Spin-Trapping Evidence for the Previously Postulated Secondary C-4 Radical. Journal of the American Chemical Society. 1998;120(14):3316–3325. doi: 10.1021/ja973080o. [DOI] [Google Scholar]
- Xie S.C., Dogovski C., Hanssen E., Chiu F., Yang T., Crespo M.P.…Tilley L. Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. Journal of Cell Science. 2016;129(2):406–416. doi: 10.1242/jcs.178830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.Y., Liu K., Ngai M.H., Lear M.J., Wenk M.R., Yao S.Q. Activity-based proteome profiling of potential cellular targets of orlistat - An FDA-approved drug with anti-tumor activities. Journal of the American Chemical Society. 2010;132(2):656–666. doi: 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- Yang T., Yeoh L.M., Tutor M.V., Dixon M.W., McMillan P.J., Xie S.C.…Cobbold S.A. Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Reports. 2019;29(9):2917–2928. doi: 10.1016/j.celrep.2019.10.095. e5. [DOI] [PubMed] [Google Scholar]
- Ying-Zi Y., Little B., Meshnick S.R. Alkylation of proteins by artemisinin. Effects of heme, pH, and drug structure. Biochemical Pharmacology. 1994;48(3) doi: 10.1016/0006-2952(94)90287-9. [DOI] [PubMed] [Google Scholar]
- Zhang C.J., Wang J., Zhang J., Lee Y.M., Feng G., Lim T.K.…Liu B. Mechanism-guided design and synthesis of a mitochondria-targeting artemisinin analogue with enhanced anticancer activity. Angewandte Chemie - International Edition. 2016;55(44):13770–13774. doi: 10.1002/anie.201607303. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gosser D.K., Meshnick S.R. Hemin-catalyzed decomposition of artemisinin (qinghaosu) Biochemical Pharmacology. 1992;43(8) doi: 10.1016/0006-2952(92)90713-S. [DOI] [PubMed] [Google Scholar]
- Zhang S., Chen H., Gerhard G.S. Heme synthesis increases artemisinin-induced radical formation and cytotoxicity that can be suppressed by superoxide scavengers. Chemico-Biological Interactions. 2010;186(1):30–35. doi: 10.1016/j.cbi.2010.03.021. [DOI] [PubMed] [Google Scholar]
- Zhang S., Gerhard G.S. Heme activates artemisinin more efficiently than hemin, inorganic iron, or hemoglobin. Bioorganic and Medicinal Chemistry. 2008;16(16):7853–7861. doi: 10.1016/j.bmc.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Zhang S., Gerhard G.S. Heme mediates cytotoxicity from artemisinin and serves as a general anti-proliferation target. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ba Q., Gu Z., Guo D., Zhou Y., Xu Y.…Liu H. Fluorescent coumarin-artemisinin conjugates as mitochondria-targeting theranostic probes for enhanced anticancer activities. Chemistry - A European Journal. 2015;21(48):17415–17421. doi: 10.1002/chem.201502543. [DOI] [PubMed] [Google Scholar]
- Zhang Z.Y., Yu S.Q., Miao L.Y., Huang X.Y., Zhang X.P., Zhu Y.P.…Li D.Q. Artesunate combined with vinorelbine plus cisplatin in treatment of advanced non-small cell lung cancer: A randomized controlled trial. Journal of Chinese Integrative Medicine. 2008;6(2) doi: 10.3736/jcim20080206. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Li W., Xiao Y. Profiling of multiple targets of artemisinin activated by hemin in cancer cell proteome. ACS Chemical Biology. 2016;11(4) doi: 10.1021/acschembio.5b01043. [DOI] [PubMed] [Google Scholar]
- Ziegler S., Pries V., Hedberg C., Waldmann H. Target identification for small bioactive molecules: Finding the needle in the haystack. Angewandte Chemie - International Edition. 2013;52(10) doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]