Abstract
Background
Small nucleolar RNAs (snoRNAs) have been proved to play important roles in various cellular physiological process. Recently, dysregulation of snoRNA SNORA71A has been found involved in tumorigenesis of various malignant cancers. However, the emerging effects of SNORA71A in hepatocellular carcinoma (HCC) remain largely unclear. In this study, we aimed to explore the SNORA71A expression and its underlying significance in HCC.
Methods
Expression of SNORA71A in cell lines and clinical specimens was measured by quantitative real-time PCR. Then, all enrolled HCC patients were divided into low and high SNORA71A expression subgroups and then they were compared in the aspects of clinical features as well as survival outcome by respective statistical analysis methods.
Results
SNORA71A was significantly downexpressed in SK-HEP-1 (P = 0.001), Huh-7 (P < 0.001), Hep3B (P < 0.001), and clinical HCC specimens (P = 0.006). Comparing the clinical features between SNORA71A expression subgroups, it showed that low SNORA71A expression was significantly associated with large tumor diameter, multiple lesions, capsular invasion, bad tumor differentiation, and TNM stage (P < 0.05). Furthermore, it was found that HCC patients with lower SNORA71A expression had higher risk in postoperative tumor relapse (median time: 9.5 vs. 35.2 months; low vs. high; P < 0.001) and poor overall survival (median time: 36.8 vs. 52.9 months; low vs. high; P < 0.001). Besides, SNORA71A expression served as independent risk factors for tumor-free (HR = 0.450; 95% CI [0.263-0.770]; P = 0.004) and long-term survival (HR = 0.289; 95% CI [0.127-0.657]; P = 0.003).
Conclusions
Our study for the first time demonstrated that downregulation of SNORA71A could serve as a novel biomarker for clinical assessment and prognostic prediction of HCC patients.
1. Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and causes the fourth most cancer-related deaths in the world [1]. There are a series of risk factors to induce the tumorigenesis of HCC, mainly including chronic hepatitis virus infection, fatty diabetes, excessive alcohol intake, and nonalcoholic fatty liver disease [2]. Despite great achievements in the treatment of hepatocellular carcinoma with respect to liver resection and chemotherapy, the survival outcomes of HCC patients remain dissatisfactory for its high incidence of relapse, metastasis, and mortality [3]. Even as the most commonly and important means for HCC diagnosis, serum AFP monitoring is not sufficient to predict the postoperative survival for HCC patients effectively [4]. Therefore, it is vital and significant to identify a novel reliable clinical marker for HCC prognostication.
Small nucleolar RNA (snoRNA) is a type of noncoding RNA widely found in the nucleoli of eukaryotic cells [5]. It has a length of 0-300 nt and has conserved structural elements. Although these snoRNAs were once considered with single function and limited effects in pre-rRNA processing, recent accumulating evidence showed that some snoRNAs also participated in various cellular physiological processes, including cell proliferation, differentiation, epigenetic, regulation. And some dysregulation of snoRNA was proved to serve as tumor suppressor genes or oncogenes in various cancers [6–9]. A review from Han et al. clearly stated that SNHG1, SNHG6, SNHG15, SNHG16, and SNHG20 can play varied roles in HCC through different regulatory mechanisms. These SNHGs can promote and inhibit tumorigenesis [10]. For instance, snoRNA SNHG1 promoted tumorigenesis and progression of colorectal cancer via negative regulation of miR-137 [11]. Besides, snoRNA SNORA18 acted as a tumor suppressor gene in hepatocellular carcinoma [12].
SNORA71A, encoded by the third intron of snoRNA host gene 17 (SNHG17) cloning, commonly guided the pseudouridine of U406 in 18S rRNA [13]. In recent, more and more studies reported that SNORA71A was dysregulated in various types of cancers, such as lung cancer [14] and breast cancer [15], and played crucial roles in tumor progression. However, the emerging effects and potential mechanisms of SNORA71A in hepatocellular carcinoma remain largely unclear. In this study, we aimed to identify the SNORA71A expression features in HCC and its underlying clinical significance.
2. Materials and Methods
2.1. Patients and Tissue Samples
To reduce the potential confounding factors, none of the enrolled patients received any preoperative radiotherapy, chemotherapy, or endocrine therapy. All enrolled patients were diagnosed with HCC by histopathological examination. All patients involved in the study were written informed consent and approval. This study was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Zhejiang University (Hangzhou, China).
HCC tissues and adjacent liver tissues were received from 132 consecutive HCC patients, who underwent liver resection between January 2013 and December 2014. HCC tissue specimens underwent 2 pathological examinations by pathologists for TNM stage. Tissues were immediately snap frozen into liquid nitrogen postoperation and then stored at -80°C for RNA isolation. Patient data was collected from the database of the hospital electronic system, including individual information, liver function indexes, alpha fetal protein value, HBV infection, lesion features, pathological differentiation, and TNM classification.
2.2. Follow-Up of Tumor-Free and Long-Term Survival
After hepatectomy, all patients were followed up for 5 years. After the initial surgery, the time terminal of tumor-free survival included the recurrence of HCC, HCC distant metastasis, or death from any cause without cancer-related events, while long-term survival was calculated until death or the last follow-up time in 5 years. Death of patients was ascertained by the family members and the review of public hospital records. All staff who collected all the following-up data of enrolled patients were blind to participant status.
2.3. Cell Culture
Human normal hepatocyte cell line (QSG-7701) and HCC cell lines (SK-HEP-1, Huh-7, and Hep3B) obtained from the laboratory were maintained in the DMEM medium (BI, Israel) with 10% FBS (BI, Israel) as well as 100 U/ml penicillin and 100 mg/ml streptomycin (Sangon, China) at 37°C in a humidified incubator (Thermo Fisher, USA) containing 5% CO2.
2.4. RNA Extraction and Quantitative Real-Time PCR
Total RNA from cell lines and tissue specimens was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. RNA was reversely transcribed into cDNA using cDNA Reverse Transcription Kit (Vazyme, Nanjing, China). To determine the expression level of SNORA71A, cDNA sample was tested with quantitative real-time polymerase chain reaction (RT-PCR) (Roche, Basel, Switzerland). All samples were tested in triplicate. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control, and the relative expression of SNORA71A was calculated by comparative Ct method formula 2−ΔΔCt. The sequences of all PCR primers used were as follows (5′-3′): GAPDH: CAGGAGGCATTGCTGATGAT (forward), GAAGGCTGGGGCTCATTT (reverse); SNORA71A: AGGTCATTGATAGTGCAGGGAG (forward), GGTTCGGATGGGATAGGGT (reverse).
2.5. Statistical Analysis
All statistical analyses were performed using the SPSS (Statistical Package for the Social Sciences) 19.0 (SPSS, Chicago, IL). The differences between two independent groups were analyzed using Student's t-test. The correlations between SNORA71A expression and different clinical characteristics were performed with chi-squared test. The differences of tumor-free and long-term survival in different subgroups were assessed by Kaplan–Meier survival plots and log-rank tests. Associations between HCC prognosis and clinical parameters were first performed with univariate analysis and then gradually analyzed using multiple logistic regression for those significantly differing factors. All data were expressed with mean ± standard deviation (SD), and P < 0.05 was recognized as statistically significant difference.
3. Results
3.1. The Expression Levels of SNORA71A in HCC
To determine whether SNORA71A was differentially expressed in HCC, firstly human normal hepatocyte (QSG-7701) and HCC cell lines (SK-HEP-1, Huh-7, and Hep3B) were used to analyze SNORA71A expression by quantitative RT-PCR. As shown in Figure 1, the SNORA71A expression in SK-HEP-1 (P = 0.001), Huh-7 (P < 0.001), and Hep3B (P < 0.001) were remarkably lower than QSG-7701, respectively. Then, as shown in Figure 2, the similar results were also found in clinical tissues' examination that SNORA71A expression was mostly downexpressed in HCC tissues compared with adjacent liver tissues (P = 0.006). Therefore, these results suggested that SNORA71A might serve as a tumor suppressive role in HCC.
Figure 1.
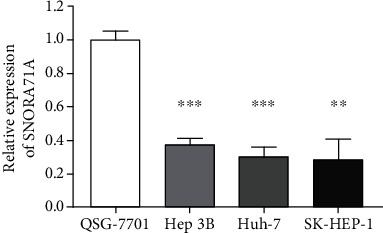
Comparison of SNORA71A expression in cell lines. Compared to normal liver cell line QSG-7701, SNORA71A was significantly suppressed in HCC cell lines (SK-HEP-1, P = 0.001; Huh-7, P < 0.001; Hep3B, P < 0.001). ∗∗P < 0.01 and ∗∗∗P < 0.001.
Figure 2.
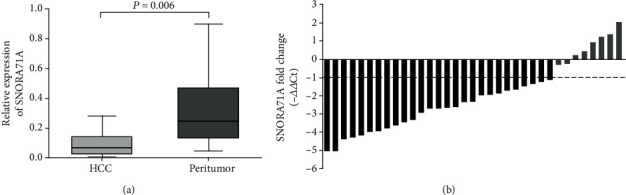
Expression patterns of SNORA71A in HCC tissue specimens. (a) Compared to corresponding adjacent liver tissues, SNORA71A expression was significantly lower in HCC tissues (P < 0.01). (b) Waterfall plot showed that SNORA71A was downregulated by at least twofold in 77.2% of paired HCC tissues. △△Ct = (Ct SNORA71A–Ct GAPDH) of HCC lesions − (Ct SNORA71A–Ct GAPDH) of adjacent liver tissue.
3.2. Relationships of SNORA71A Expression with Clinicopathologic Characteristics of HCC Patients
According to individual SNORA71A expression, all 132 HCC patients were divided into high and low expression subgroups. The relationships of SNORA71A expression with clinicopathologic characteristics of HCC patients were statistically analyzed. In Table 1, low SNORA71A expression was significantly associated with large tumor diameter (P = 0.016), multiple lesions (P = 0.017), capsular invasion (P = 0.015), bad tumor differentiation (P = 0.027), and TNM stage (P = 0.004). However, there was no statistical significance between SNORA71A expression with other clinicopathologic characteristics, such as hepatitis B, AFP, or cirrhosis.
Table 1.
Correlation between SNORA71A expression and clinicopathological features in HCC patients.
| Characteristics | Total | SNORA71A expression | P value | |
|---|---|---|---|---|
| Low (n = 66) | High (n = 66) | |||
| Age, ys | 0.138 | |||
| <60 | 85 | 46 (0.697) | 39 (0.591) | |
| ≥60 | 47 | 20 (0.303) | 27 (0.409) | |
| Gender | 0.212 | |||
| Female | 16 | 6 (0.091) | 10 (0.152) | |
| Male | 116 | 60 (0.909) | 56 (0.848) | |
| Hepatitis B | 0.310 | |||
| Positive | 113 | 58 (0.879) | 11 (0.167) | |
| Negative | 19 | 8 (0.121) | 55 (0.833) | |
| Cirrhosis | 0.361 | |||
| Present | 79 | 38 (0.576) | 41 (0.621) | |
| Absent | 53 | 28 (0.424) | 25 (0.379) | |
| AFP (ng/l) | 0.429 | |||
| <400 | 82 | 42 (0.636) | 40 (0.606) | |
| ≥400 | 50 | 24 (0.364) | 26 (0.394) | |
| Tumor diameter | 0.005∗ | |||
| <5 cm | 34 | 10 (0.152) | 24 (0.364) | |
| ≥5 cm | 98 | 56 (0.848) | 42 (0.636) | |
| Multiple lesions | 0.017∗ | |||
| Present | 22 | 16 (0.242) | 6 (0.091) | |
| Absent | 110 | 50 (0.758) | 60 (0.909) | |
| Vessel carcinoma embolus | 0.160 | |||
| Present | 34 | 20 (0.303) | 14 (0.212) | |
| Absent | 98 | 46 (0.697) | 52 (0.788) | |
| Microvascular invasion | 0.500 | |||
| Present | 7 | 4 (0.061) | 3 (0.045) | |
| Absent | 125 | 62 (0.939) | 63 (0.955) | |
| Capsular invasion | 0.015∗ | |||
| Present | 49 | 31 (0.470) | 18 (0.273) | |
| Absent | 83 | 35 (0.530) | 48 (0.727) | |
| Differentiation | 0.027∗ | |||
| Low | 70 | 41 (0.621) | 29 (0.439) | |
| High/moderate | 62 | 25 (0.379) | 37 (0.561) | |
| TNM stage | 0.004∗ | |||
| I~II | 91 | 38 (0.576) | 53 (0.803) | |
| III~IV | 41 | 28 (0.424) | 13 (0.197) | |
AFP = alpha fetal protein; TNM = tumor-node-metastasis; ∗P < 0.05; values are mean ± standard deviation or n (%).
3.3. Downregulation of SNORA71A Predicts Poor Prognosis in HCC Patients
To identify whether SNORA71A expression could serve as a novel biomarker to predict prognosis of HCC patients, the tumor-free and long-term survival data of two SNORD31 expression subgroups were compared using Kaplan–Meier method. According to the results from Figures 3 and 4, we found that HCC patients with high SNORA71A expression tended to present with lower tumor recurrence rate after hepatectomy and better overall survival condition. Between the subgroups with different SNORA71A expression levels, the median tumor-free survival time (9.5 vs. 35.2 months; low vs. high; P <0.001) and median long-term survival time (36.8 vs. 52.9 months; low vs high; P <0.001) were significantly different.
Figure 3.
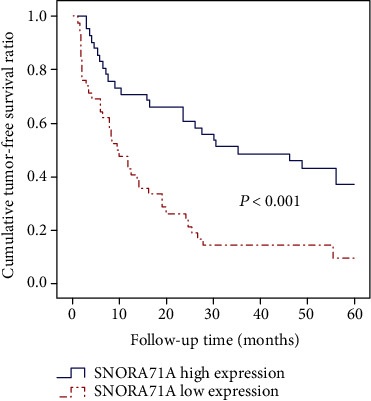
Cumulative tumor-free survival curves of patients in low and high SNORA71A expression subgroups.
Figure 4.
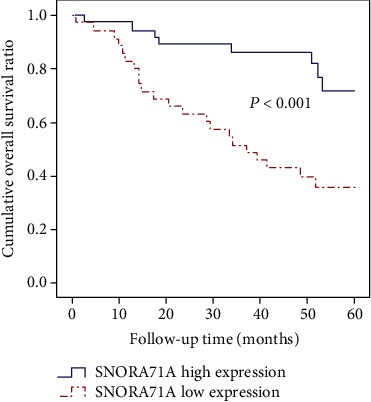
Cumulative overall survival curves of patients in low and high SNORA71A expression subgroups.
3.4. Identification of Independent Prognostic Factors for HCC Prognostication
To determine the independent risk factors for HCC patient's survival, various clinicopathological factors and SNORA71A expression were analyzed in univariate and multivariate Cox regression analysis. In Tables 2 and 3, SNORA71A expression was proved to independently relate with tumor-free (HR [95% CI]: 0.450 [0.263-0.770]; P = 0.004) as well as long-term survival (HR [95% CI]: 0.289 [0.127-0.657]; P = 0.003) of HCC patients. These results supported that SNORA71A expression was capable of serving as the biomarker for prognosis assessment in HCC patients.
Table 2.
Univariate and multivariate analysis of tumor-free survival in HCC patients.
| Clinicopathologic parameters | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (<60 vs. ≥60) | 0.734 (0.432-1.246) | 0.252 | ||
| Gender (female vs. male) | 1.397 (0.559-3.490) | 0.474 | ||
| Hepatitis B (negative vs. positive) | 1.608 (0.763-3.386) | 0.212 | ||
| AFP (<400 vs. ≥400) | 1.033 (0.618-1.725) | 0.901 | ||
| Cirrhosis (present vs. absent) | 1.172 (0.698-1.968) | 0.549 | ||
| Microvascular invasion (present vs. absent) | 1.272 (0.460-3.520) | 0.643 | ||
| Tumor differentiation (low vs. high/moderate) | 0.760 (0.457-1.262) | 0.289 | ||
| Capsular invasion (present vs. absent) | 1.521 (0.904-2.560) | 0.114 | ||
| Multiple lesions (present vs. absent) | 1.783 (0.980-3.242) | 0.058 | 1.189 (0.546-2.593) | 0.663 |
| Vessel carcinoma embolus (present vs. absent) | 1.699 (0.957-3.016) | 0.071 | 1.384 (0.771-2.485) | 0.276 |
| TNM stage (I~II vs. III~IV) | 2.006 (1.195-3.369) | 0.008 | 1.577 (0.922-2.699) | 0.097 |
| Tumor diameter (<5 cm vs. ≥5 cm) | 2.604 (1.402-4.839) | 0.002 | 2.154 (1.140-4.070) | 0.018∗ |
| SNORA71A expression (low vs. high) | 0.385 (0.228-0.650) | <0.001 | 0.450 (0.263-0.770) | 0.004∗ |
AFP = alpha fetal protein; TNM = tumor-node-metastasis; HR = hazard ratio; ∗P < 0.05 was considered statistically significant.
Table 3.
Univariate and multivariate analysis of overall survival in HCC patients.
| Clinicopathologic parameters | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (<60 vs. ≥60) | 1.067 (0.514-2.216) | 0.861 | ||
| Gender (female vs. male) | 0.541 (0.231-1.266) | 0.157 | ||
| Hepatitis B (negative vs. positive) | 0.895 (0.365-2.191) | 0.808 | ||
| AFP (<400 vs. ≥400) | 1.398 (0.682-2.866) | 0.360 | ||
| Cirrhosis (present vs. absent) | 1.208 (0.575-2.540) | 0.618 | ||
| Microvascular invasion (present vs. absent) | 1.791 (0.540-5.938) | 0.341 | ||
| Tumor differentiation (low vs. high/moderate) | 0.876 (0.424-1.809) | 0.720 | ||
| Capsular invasion (present vs. absent) | 2.175 (1.063-4.452) | 0.033 | 1.436 (0.662-3.116) | 0.360 |
| Tumor diameter (<5 cm vs. ≥5 cm) | 3.480 (1.210-10.005) | 0.021 | 1.952 (0.620-6.143) | 0.253 |
| Multiple lesions (present vs. absent) | 2.410 (1.140-5.095) | 0.021 | 1.125 (0.429-2.952) | 0.810 |
| Vessel carcinoma embolus (present vs. absent) | 2.075 (0.949-4.538) | 0.068 | 2.047 (0.898-4.665) | 0.088 |
| TNM stage (I~II vs. III~IV) | 3.061 (1.480-6.331) | 0.003 | 2.637 (1.263-5.507) | 0.010∗ |
| SNORA71A expression (low vs. high) | 0.257 (0.114-0.580) | 0.001 | 0.289 (0.127-0.657) | 0.003∗ |
AFP = alpha fetal protein; TNM = tumor-node-metastasis; HR = hazard ratio; ∗P < 0.05 was considered statistically significant.
4. Discussion
Hepatocellular carcinoma is the most common type of liver cancer and solid tumors with fast progress and poor prognosis [16]. For HCC commonly presented with occult progression in vivo, multiple HCC patients are diagnosed at middle or even late stages [17]. Although several clinical characteristics have listed into the criteria for predicting the prognosis of HCC patients, such as tumor diameters, TNM stage, and AFP, these classification schemes are limited to become common predictors for HCC patients due to dissatisfactory sensitivity and specificity [18]. In the past 40 years, AFP has been used for monitoring the HCC, however, with poor sensitivity and specificity: 39% to 65% and 76% to 94% [19, 20]. Moreover, Zhang and Yang's study indicated that AFP level rarely rose when HCC tumor mass was less than 2 cm in diameter [21]. Therefore, it is important and necessary to explore a novel biomarker in HCC patients.
As one kind of noncoding RNA, small nucleolar RNA (snoRNA) is originally found to make function in regulating the pseudouridine of rRNA [22]. Nowadays, emerging evidence showed that snoRNA also played important regulatory roles in tumorigenesis of various cancers, including liver cancer and breast cancer [23–25]. Among that, Cui et al. indicated that upregulation of snoRNA SNORA23 promoted invasion and metastasis of xenograft tumors in mice by modulating the spectrum repeat-containing nuclear envelope 2 [26]. Baral et al. reported that SNORD126, SNORD78, ACA11, SNORA47, and SNORD76 may also serve as an important prognostic marker related to clinical features. Abnormal expression of snoRNA promotes cell proliferation and leads to the development of liver cancer [27]. Besides, Cao's team reported for the first time that dysregulation of snoRNA SNORA18L5 could increase the risk of HBV-associated hepatocellular carcinoma for abnormity of ribosomal RNA maturation [28]. In HCC, moreover, snoRNA SNORD113-1 was also found to be significantly downregulated and functions as a tumor suppressor role [23]. SNHG16, SNORD76, and SnoU2_19 can regulate the development of HCC through Wnt/β-catenin signaling pathway [29]. Therefore, these indicated that some specific snoRNAs had underlying abilities to serve as biomarker for clinical evaluation of HCC patients.
In our study, we for the first time analyzed the expression level of SNORA71A in both HCC cell lines and tissues and elucidated that SNORA71A was universally downregulated in HCC. Moreover, a total of 132 patients' analyses proved that SNORA71A expression was correlated with tumor diameter, multiple lesions, capsular invasion, differentiation, and TNM stage. And our results showed that SNORA71A expression was an independent risk factors for both tumor-free survival and long-term survival of HCC patients.
In conclusion, our research for the first time demonstrated that SNORA71A could serve as a novel biomarker for prognostication and therapeutic monitoring for HCC patients. And more studies are needed to explore the specific mechanisms and verify our findings in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81572307 and 81773096), the Major Project of Medical and Health Technology Development Program in Zhejiang Province (No. 7211902), the Key Research and Development Project of Zhejiang Province (No. 2018C03085), the Public Welfare Technology Research Project of Zhejiang Province (No. LGD19C040006), and the General Research Project of the Zhejiang Provincial Education Department (No. Y201840044).
Abbreviations
- HCC:
Hepatocellular carcinoma
- snoRNA:
Small nucleolar RNA
- AFP:
Alpha fetal protein
- TNM:
Tumor-node-metastasis
- HR:
Hazard ratio.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
This retrospective study was conducted according to the ethical guidelines of the 1975 Declaration of Helsinki. All participants have signed written informed consent form. Ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University.
Conflicts of Interest
The authors report no conflicts of interest in this work.
Authors' Contributions
W.W. and Y.D. conceived and designed the study. Y.D., Z.S., S.Z., L.Z., and Q.X. conducted the study. D.Z., H.X., Y.B., and H.D. contributed to the acquisition of data. Z.S., S.Z., C.X., and Y.G. analyzed the data. W.W., Y.D., Z.S., and S.Z. interpreted the data. W.W., Y.D., Z.S., S.Z., and L.Z. reviewed and edited the manuscript. All authors read and approved the manuscript. Yuan Ding, Zhongquan Sun, and Sitong Zhang contributed equally to this paper.
References
- 1.el–Serag H. B., Rudolph K. L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Singal A. G., El-Serag H. B. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clinical Gastroenterology and Hepatology. 2015;13(12):2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum H. E. Hepatocellular carcinoma: therapy and prevention. World Journal of Gastroenterology. 2005;11(47):7391–7400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon A. M., Weiss N. S., Beste L. A., et al. No association between screening for hepatocellular carcinoma and reduced cancer-related mortality in patients with cirrhosis. Gastroenterology. 2018;155(4):1128–1139.e6. doi: 10.1053/j.gastro.2018.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiss T. NEW EMBO MEMBER'S REVIEW: Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. The EMBO Journal. 2001;20(14):3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mei Y. P., Liao J. P., Shen J., et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31(22):2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y., Chen E., Li Y., et al. H/ACA box small nucleolar RNA 7B acts as an oncogene and a potential prognostic biomarker in breast cancer. Cancer Cell International. 2019;19(1) doi: 10.1186/s12935-019-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong X.-Y., Rodriguez C., Guo P., et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Human molecular genetics. 2008;17(7):1031–1042. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Han L., Wei J., et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Scientific Reports. 2015;5(1, article 8588) doi: 10.1038/srep08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuwen H., Xi Y., Quan Q., Yin J., Miao D. Can small nucleolar RNA be a novel molecular target for hepatocellular carcinoma? Gene. 2020;733, article 144384 doi: 10.1016/j.gene.2020.144384. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y., Yin Y., Peng S., et al. Small nucleolar RNA host gene 1 promotes development and progression of colorectal cancer through negative regulation of miR-137. Molecular Carcinogenesis. 2019;58(11):2104–2117. doi: 10.1002/mc.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X. F., Thin K. Z., Ming X. L., et al. Small nucleolar RNA host gene 18 acts as a tumor suppressor and a diagnostic indicator in hepatocellular carcinoma. Technology in Cancer Research & Treatment. 2018;17 doi: 10.1177/1533033818794494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganot P., Bortolin M. L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89(5):799–809. doi: 10.1016/S0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 14.Tang G., Zeng Z., Sun W., et al. Small nucleolar RNA 71A promotes lung cancer cell proliferation, migration and invasion via MAPK/ERK pathway. Journal of Cancer. 2019;10(10):2261–2275. doi: 10.7150/jca.31077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulten H.-J., Bangash M., Karim S., et al. Comprehensive molecular biomarker identification in breast cancer brain metastases. Journal of Translational Medicine. 2017;15(1) doi: 10.1186/s12967-017-1370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J. D., Hainaut P., Gores G. J., Amadou A., Plymoth A., Roberts L. R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nature Reviews. Gastroenterology & Hepatology. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing W., Luo P., Zhu M., Ai Q., Chai H., Tu J. Prognostic and diagnostic significance of sdpr-cavin-2 in hepatocellular carcinoma. Cellular Physiology and Biochemistry. 2016;39(3):950–960. doi: 10.1159/000447803. [DOI] [PubMed] [Google Scholar]
- 18.Gish R. G. Hepatocellular carcinoma: overcoming challenges in disease management. Clinical Gastroenterology and Hepatology. 2006;4(3):252–261. doi: 10.1016/j.cgh.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Trevisani F., D'Intino P. E., Morselli-Labate A. M., et al. Serum α-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. Journal of Hepatology. 2001;34(4):570–575. doi: 10.1016/S0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 20.Alpert M. E., Uriel J., de Nechaud B. Alpha1 fetoglobulin in the diagnosis of human hepatoma. New England Journal of Medicine. 1968;278(18):984–986. doi: 10.1056/NEJM196805022781804. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B., Yang B. Combined α fetoprotein testing and ultrasonography as a screening test for primary liver cancer. Journal of Medical Screening. 1999;6(2):108–110. doi: 10.1136/jms.6.2.108. [DOI] [PubMed] [Google Scholar]
- 22.Ganot P., Caizergues-Ferrer M., Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Development. 1997;11(7):941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 23.Xu G., Yang F., Ding C. L., et al. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Molecular Cancer. 2014;13(1) doi: 10.1186/1476-4598-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H., Xu T., Ganapathy S., et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014;33(11):1348–1358. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 25.Siprashvili Z., Webster D. E., Johnston D., et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nature Genetics. 2015;48(1):53–58. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui L., Nakano K., Obchoei S., et al. Small nucleolar noncoding RNA SNORA23, up-regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat-containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. 2017;153(1):292–306.e2. doi: 10.1053/j.gastro.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 27.Baral D., Wu L., Katwal G., Yan X., Wang Y., Ye Q. Clinical significance and biological roles of small nucleolar RNAs in hepatocellular carcinoma (Review) Biomedical reports. 2018;8(4):319–324. doi: 10.3892/br.2018.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao P., Yang A., Wang R., et al. Germline duplication of SNORA18L5 increases risk for HBV-related hepatocellular carcinoma by altering localization of ribosomal proteins and decreasing levels of p53. Gastroenterology. 2018;155(2):542–556. doi: 10.1053/j.gastro.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Shuai Y., Ma Z., Lu J., Feng J. LncRNA SNHG15: a new budding star in human cancers. Cell Proliferation. 2020;53(1, article e12716) doi: 10.1111/cpr.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


