Abstract
The R2R3-MYB genes comprise one of the largest transcription factor gene families in plants, playing regulatory roles in plant-specific developmental processes, defense responses and metabolite accumulation. To date MYB family genes have not yet been comprehensively identified in the major staple fruit crop banana. In this study, we present a comprehensive, genome-wide analysis of the MYB genes from Musa acuminata DH-Pahang (A genome). A total of 285 R2R3-MYB genes as well as genes encoding three other classes of MYB proteins containing multiple MYB repeats were identified and characterised with respect to structure and chromosomal organisation. Organ- and development-specific expression patterns were determined from RNA-Seq data. For 280 M. acuminata MYB genes for which expression was found in at least one of the analysed samples, a variety of expression patterns were detected. The M. acuminata R2R3-MYB genes were functionally categorised, leading to the identification of seven clades containing only M. acuminata R2R3-MYBs. The encoded proteins may have specialised functions that were acquired or expanded in Musa during genome evolution. This functional classification and expression analysis of the MYB gene family in banana establishes a solid foundation for future comprehensive functional analysis of MaMYBs and can be utilized in banana improvement programmes.
Introduction
Banana (Musa spp.), including dessert and cooking types, is a staple fruit crop for a major world population, especially in developing countries. The crop is grown in more than 100 countries throughout the tropics and sub-tropics, mainly in the African, Asia-Pacific, and Latin American and Caribbean regions [1]. Bananas provide an excellent source of energy and are rich in certain minerals and in vitamins A, C and B6. Furthermore, this perennial, monocotyledonous plant provides an important source of fibre, sugar, starch and cellulose (used for paper, textiles). Bananas have also been considered as a useful tool to deliver edible vaccines [2]. Certain agronomic traits, such as stress and pest resistance as well as fruit quality, are thus of considerable interest. Banana improvement through breeding exercises has been challenging for various reasons. Therefore, genetic engineering-based optimisations hold great promise for crop improvement. For this purpose, candidate gene targets need to be identified. The release of a high quality banana genome sequence [3] provides an useful resource to understand functional genomics of important agronomic traits and to identify candidate genes to be utilized in banana improvement programmes.
Almost all biological processes in eukaryotic cells or organisms are influenced by transcriptional control of gene expression. Thus, the regulatory level is a good starting point for genetic engineering. Regulatory proteins are involved in transcriptional control, alone or complexed with other proteins, by activating or repressing (or both) the recruitment of RNA polymerase to promoters of specific genes [4]. These proteins are called transcription factors. As expected from their substantial regulatory complexity, transcription factors are numerous and diverse [5]. By binding to specific DNA sequence motifs and regulating gene expression, transcription factors control various regulatory and signaling networks involved in the development, growth and stress response in an organism.
One of the widest distributed transcription factor families in all eukaryotes is the MYB (myeloblastosis) protein family. In the plant kingdom, MYB proteins constitute one of the largest transcription factor families. MYB proteins are defined by a highly conserved MYB DNA-binding domain, mostly located at the N-terminus of the protein. The MYB domain generally consists of up to four imperfect amino acid sequence repeats (R) of about 50–53 amino acids, each forming three alpha–helices [summarised in 6]. The second and third helices of each repeat build a helix–turn–helix (HTH) structure with three regularly spaced tryptophan (or hydrophobic) residues, forming a hydrophobic core [7]. The third helix of each repeat was identified as the DNA recognition helix that makes direct contact with DNA [8]. During DNA contact, two MYB repeats are closely packed in the major groove, so that the two recognition helices bind cooperatively to the specific DNA recognition sequence motif. In contrast to vertebrates genomes, which only encode MYB transcription factors with three repeats, plants have different MYB domain organisations, comprising one to four repeats [6, 9]. R2R3-MYBs, which are MYB proteins with two repeats (named according to repeat numbering in vertebrate MYBs), are particularly expanded in plant genomes. Copy numbers range from 45 unique R2R3-MYBs in Ginkgo biloba [10] to 360 in Mexican cotton (Gossypium hirsutum) [11]. The expansion of the R2R3-MYB family was coupled with widening in the functional diversity of R2R3-MYBs, considered to regulate mainly plant-specific processes including secondary metabolism, stress responses and development [6]. As expected, R2R3-MYBs are involved in regulating several biological traits, for example wine quality, fruit color, cotton fibre length, pollinator preferences and nodulation in legumes.
In this study, we have used genomic resources to systematically identify members of the M. acuminata (A genome) R2R3-MYB gene family. We used knowledge from other plant species, including the model plant A. thaliana, leading to a functional classification of the banana R2R3-MYB genes based on the MYB phylogeny. Furthermore, RNA-Seq data was used to analyse expression in different M. acuminata organs and developmental stages and to compare expression patterns of closely grouped co-orthologs. The identification and functional characterization of the R2R3-MYB gene family from banana will provide an insight into the regulatory aspects of different biochemical and physiological processes, as those operating during fruit ripening as well as response to various environmental stresses. Our findings offer the first step towards further investigations on the biological and molecular functions of MYB transcription factors with the selection of genes responsible for economically important traits in Musa, which can be utilized in banana improvement programmes.
Material and methods
Search for MYB protein coding genes in the M. acuminata genome
A consensus R2R3-MYB DNA binding domain sequence [12] (S1 Table) was used as protein query in tBLASTn [13] searches on the M. acuminata DH-Pahang genome sequence (version 2) (https://banana-genome-hub.southgreen.fr/sites/banana-genome-hub.southgreen.fr/files/data/fasta/version2/musa_acuminata_v2_pseudochromosome.fna) in an initial search for MYB protein coding genes. To confirm the obtained amino acid sequences, the putative MYB sequences were manually analysed for the presence of an intact MYB domain. All M. acuminata MYB candidates from the initial BLAST were inspected to ensure that the putative gene models encode two or more (multiple) MYB repeats. The identified gene models were analysed to map them individually to unique loci in the genome and redundant sequences were discarded from the data set to obtain unique MaMYB genes. The identified MaMYB genes were matched to the automatically annotated genes from the Banana Genome Hub database [14]. The open reading frames of the identified MaMYB were manually inspected and, if available, verified by mapping of RNA-Seq data to the genomic sequence. Resulting MaMYB annotation is provided in the supporting information: Multi-FASTA files with MaMYB CDS sequences (S1 File) and MaMYB peptide sequences (S2 File), general feature format (GFF) file (S3 File) describing the MaMYB genes for use in genome viewers/browers.
Genomic distribution of MaMYB genes
The MaMYB genes were located on the corresponding chromosomes by the MapGene2chromosome web v2 (MG2C) software tool (http://mg2c.iask.in/mg2c_v2.0/) according to their position information of a physical map, available from the DH-Pahang genome annotation (V2).
Phylogenetic analyses
Protein sequences of 133 A. thaliana MYBs were obtained from TAIR (http://www.arabidopsis.org/). We also considered other multiple MYB-repeat proteins from A. thaliana in the phylogenetic analysis to determine orthologs in the M. acuminata genome: five AtMYB3R, AtMYB4R1 and AtCDC5. Additionally, 43 well-known, functionally characterised landmark plant R2R3-MYB protein sequences were collected from GenBank at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/).
Phylogenetic trees were constructed from ClustalΩ [15] aligned MYB domain sequences (293 MaMYBs, 132 AtMYBs and 43 plant landmark MYBs) using MEGA7 [16] with default settings. A mojority rule Maximum Likelihood (ML) consensus tree inferred from 1000 bootstrap replications was calculated. M. acuminata MYB proteins were classified according to their relationships with corresponding A. thaliana and landmark MYB proteins.
Expression analysis from RNA-Seq data
RNA-Seq data sets were retrieved from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) via fastq-dump v2.9.6 (https://github.com/ncbi/sra-tools) (S2 Table). STAR v2.5.1b [17] was applied for the mapping of reads to the Pahang v2 reference genome sequence [18] using previously described parameters [19]. featureCounts [20] was applied for quantification of the mapped reads per gene based on the Pahang v2 annotation. Previously developed Python scripts were deployed for the calculation of normalized expression values (https://github.com/bpucker/bananaMYB) [19].
Results and discussion
The first annotated reference genome sequence of M. acuminata (A genome) became available in 2012 [3]. It was obtained from a double haploid (DH) plant of the Pahang cultivar, derived through haploid pollen and spontaneous chromosome doubling from the wild subspecies Musa acuminata ssp. malaccensis [3]. This wild subspecies was involved in the domestication of the vast majority of cultivated bananas and its genetic signature is commonly found in dessert and cooking bananas (ProMusa, http://www.promusa.org). An improved version (DH-Pahang version 2) of the genome assembly and annotation was presented in 2016 comprising 450.7 Mb (86% of the estimated size) from which 89.5% are assigned to one of the 11 chromosomes, predicted to contain 35,276 protein encoding genes [18]. This M. acuminata DH-Pahang version 2 genome sequence provides the platform of this study.
Identification and genomic distribution of M. acuminata MYB genes
A consensus R2R3-MYB DNA binding domain sequence (deduced from Arabidopsis, grape and sugarbeet R2R3-MYBs, S1 Table) was used as protein query in tBLASTn searches on the DH-Pahang version 2 genome sequence to comprehensively identify MYB protein coding genes in M. acuminata. The resulting putative MYB sequences were proven to map to unique loci in the genome and were confirmed to contain an intact MYB domain. This ensured that the gene models contained two or more (multiple) MYB repeats. We identified a set of 285 R2R3-MYB proteins and nine multiple repeat MYB proteins distantly related to the typical R2R3-MYB proteins: six R1R2R3-MYB (MYB3R) proteins, two MYB4R proteins and one CDC5-like protein from the M. acuminata genome sequence (Table 1).
Table 1. List of annotated MYB genes in the Musa acuminata (DH Pahang) genome sequence.
| pseudochr. position | BHLH | coding | peptide | MYB | |||||
|---|---|---|---|---|---|---|---|---|---|
| gene ID | synonym | from | to | str. | motif | exons | length | type | functional assignment |
| Ma01_g00440 | 314253 | 315377 | - | 4 | 297 | R2R3 | stress response, hormone signaling | ||
| Ma01_g02850 | MusaMYB31 [21] | 1866501 | 1867245 | + | + | 3 | 200 | R2R3 | repressors PP, sinapate, lignin |
| Ma01_g04470 | 3005032 | 3006551 | + | 4 | 230 | R2R3 | ASR, flower morphogenesis, stilbene | ||
| Ma01_g10440 | 7520585 | 7526901 | + | 3 | 556 | R2R3 | anther development, stress response | ||
| Ma01_g10750 | 7722708 | 7723839 | - | 3 | 273 | R2R3 | photomorphogenesis | ||
| Ma01_g11890 | 8618982 | 8620082 | + | 2 | 234 | R2R3 | secondary cell wall, lignin | ||
| Ma01_g12250 | 8870030 | 8871190 | - | 3 | 334 | R2R3 | |||
| Ma01_g14370 | 10499811 | 10500874 | + | 4 | 195 | R2R3 | defense, stress response | ||
| Ma01_g15800 | 11477487 | 11478908 | + | 4 | 197 | R2R3 | secondary cell wall, lignin | ||
| Ma01_g16960 | 12409268 | 12410289 | + | 3 | 293 | R2R3 | |||
| Ma01_g17260 | 12621891 | 12623358 | - | 5 | 360 | R2R3 | |||
| Ma01_g17450 | 12782371 | 12783358 | + | 3 | 270 | R2R3 | defense, stress response | ||
| Ma01_g18470 | 13704465 | 13705557 | - | 2 | 330 | R2R3 | suberin | ||
| Ma01_g19610 | MYB32 [22] | 15271319 | 15272110 | - | + | 2 | 241 | R2R3 | repressors PP, sinapate, lignin |
| Ma01_g19960 | 15935305 | 15936912 | - | 5 | 282 | R2R3 | stress response, hormone signaling | ||
| Ma01_g21340 | 20977298 | 20978421 | - | 3 | 196 | R2R3 | stamen development | ||
| Ma02_g00280 | 2702046 | 2703056 | + | 3 | 281 | R2R3 | defense, stress response | ||
| Ma02_g00290 | 4086487 | 4089065 | + | 3 | 320 | R2R3 | flavonols, phlobaphene | ||
| Ma02_g03780 | 15236983 | 15238054 | + | 3 | 307 | R2R3 | mucillage, lignin, stomatal closure | ||
| Ma02_g04860 | 16268865 | 16270029 | + | 3 | 325 | R2R3 | |||
| Ma02_g05880 | 17024294 | 17029382 | - | 7 | 489 | 3R | cell cycle control | ||
| Ma02_g06190 | 17225149 | 17226252 | + | 3 | 301 | R2R3 | axillary meristem, root growth | ||
| Ma02_g06670 | 17595551 | 17596584 | + | 2 | 317 | R2R3 | suberin | ||
| Ma02_g09720 | MYB46 [22] | 19581503 | 19582518 | + | 2 | 297 | R2R3 | secondary wall, lignin | |
| Ma02_g09870 | 19645550 | 19646834 | - | 3 | 381 | R2R3 | |||
| Ma02_g13370 | 21804465 | 21806215 | + | 3 | 280 | R2R3 | photomorphogenesis | ||
| Ma02_g15770 | 23338515 | 23340804 | - | 3 | 444 | R2R3 | anther development, stress response | ||
| Ma02_g16570 | 23869343 | 23870389 | + | 3 | 297 | R2R3 | root development | ||
| Ma02_g17950 | MYB48 [22] | 24668376 | 24669235 | - | 4 | 205 | R2R3 | ||
| Ma02_g19650 | 25862380 | 25864514 | + | + | 3 | 258 | R2R3 | ||
| Ma02_g19770 | MYB63 [22] | 25962707 | 25963911 | - | 4 | 303 | R2R3 | PP, lignin | |
| Ma02_g20270 | 26305481 | 26306910 | + | 4 | 365 | R2R3 | |||
| Ma02_g21230 | 26925219 | 26926055 | + | 2 | 244 | R2R3 | general flavonoid, trichome | ||
| Ma02_g21760 | 27304216 | 27317415 | - | 11 | 1076 | 3R | cell cycle control | ||
| Ma02_g22540 | 27850957 | 27852253 | + | 3 | 229 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma02_g23870 | 28706512 | 28707545 | + | 4 | 253 | R2R3 | defense, stress response | ||
| Ma02_g24520 | 29095026 | 29096192 | - | 3 | 292 | R2R3 | defense, stress response | ||
| Ma03_g01260 | 944774 | 947014 | + | 4 | 124 | R2R3 | defense, stress response | ||
| Ma03_g06410 | 4438262 | 4439441 | - | 4 | 287 | R2R3 | root development | ||
| Ma03_g07620 | 5357989 | 5358809 | - | 3 | 205 | R2R3 | |||
| Ma03_g07840 * | 5566137 | 5567139 | - | + | 3 | 282 | R2R3 | proanthocyanidins | |
| Ma03_g07850 * | 5573753 | 5574630 | + | + | 3 | 238 | R2R3 | proanthocyanidins | |
| Ma03_g08300 | 5982225 | 5985925 | - | 5 | 287 | R2R3 | defense, stress response | ||
| Ma03_g08930 | 6570843 | 6573019 | + | 3 | 360 | R2R3 | embryogenesis, seed maturation | ||
| Ma03_g09310 | 6861304 | 6862367 | + | 3 | 289 | R2R3 | defense, stress response | ||
| Ma03_g09340 | 6900833 | 6902218 | + | 4 | 319 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma03_g09840 | 7294462 | 7296093 | + | 3 | 320 | R2R3 | anther-, tapetum development | ||
| Ma03_g11910 | 9246422 | 9247790 | + | 4 | 360 | R2R3 | |||
| Ma03_g12480 | 9621017 | 9623699 | - | 4 | 175 | R2R3 | defense, stress response | ||
| Ma03_g12720 | 9782781 | 9783893 | - | 4 | 290 | R2R3 | stress response, hormone signaling | ||
| Ma03_g14020 | 11197863 | 11198880 | - | 2 | 313 | R2R3 | suberin | ||
| Ma03_g18410 | 23945840 | 23948315 | + | 3 | 168 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma03_g19810 | 25073787 | 25074642 | + | 2 | 259 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma03_g20390 | 25556498 | 25558664 | - | 3 | 382 | R2R3 | embryogenesis, seed maturation | ||
| Ma03_g21920 | 26789598 | 26790783 | + | + | 5 | 254 | R2R3 | flavonoid repressor | |
| Ma03_g21970 | 26827699 | 26828817 | + | 3 | 303 | R2R3 | |||
| Ma03_g23170 | 27799667 | 27800542 | - | + | 4 | 217 | R2R3 | repressors PP, sinapate, lignin | |
| Ma03_g25780 | 29742629 | 29743879 | + | 4 | 344 | R2R3 | mucillage, lignin, stomatal closure | ||
| Ma03_g28720 | 31825604 | 31827664 | + | + | 3 | 274 | R2R3 | proanthocyanidins | |
| Ma03_g29070 | 32109761 | 32110872 | - | 3 | 325 | R2R3 | |||
| Ma03_g29510 | 32402508 | 32403434 | - | 2 | 288 | R2R3 | stress response, hormone signaling | ||
| Ma03_g29770 | 32613605 | 32614662 | + | 3 | 307 | R2R3 | |||
| Ma04_g00460 | 405252 | 406293 | + | 4 | 265 | R2R3 | defense, stress response | ||
| Ma04_g01010 | 896369 | 897222 | - | 3 | 226 | R2R3 | |||
| Ma04_g05460 | 4086523 | 4089539 | + | 4 | 254 | R2R3 | photomorphogenesis | ||
| Ma04_g06410 | 4737396 | 4738484 | + | 3 | 306 | R2R3 | |||
| Ma04_g09430 | 6706438 | 6707529 | + | 3 | 302 | R2R3 | stress response, hormone signaling | ||
| Ma04_g11930 | 8517172 | 8518916 | + | 3 | 228 | R2R3 | photomorphogenesis | ||
| Ma04_g12940 | 9788899 | 9789804 | + | 2 | 266 | R2R3 | stress response, hormone signaling | ||
| Ma04_g13260 | 10044518 | 10045599 | + | 3 | 308 | R2R3 | |||
| Ma04_g16770 | 16592910 | 16594109 | - | 3 | 348 | R2R3 | suberin | ||
| Ma04_g18740 | 20887894 | 20888897 | + | 4 | 146 | R2R3 | ASR, flower morphogenesis, stilbene | ||
| Ma04_g19500 | 22140585 | 22141813 | + | 2 | 347 | R2R3 | suberin | ||
| Ma04_g20120 | 22833201 | 22834619 | + | 4 | 322 | R2R3 | anther development, stress response | ||
| Ma04_g22200 | 24575839 | 24576649 | - | 3 | 195 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma04_g22930 | 25120275 | 25121367 | - | 4 | 254 | R2R3 | axillary meristem, root growth | ||
| Ma04_g23220 | 25400512 | 25401385 | - | 3 | 243 | R2R3 | PP, lignin | ||
| Ma04_g24670 | 26639014 | 26639915 | + | 3 | 243 | R2R3 | |||
| Ma04_g26220 | 27753189 | 27754723 | + | 4 | 283 | R2R3 | |||
| Ma04_g26550 | MYB85 [22] | 27973322 | 27974181 | - | 2 | 249 | R2R3 | secondary cell wall, lignin | |
| Ma04_g26660 | 28040859 | 28041969 | - | 4 | 128 | R2R3 | defense, stress response | ||
| Ma04_g26810 | 28140628 | 28141740 | - | 4 | 269 | R2R3 | stress response, hormone signaling | ||
| Ma04_g28300 | 29374644 | 29376644 | - | + | 3 | 232 | R2R3 | ||
| Ma04_g28510 | 29563855 | 29564811 | + | 4 | 219 | R2R3 | ASR, flower morphogenesis, stilbene | ||
| Ma04_g30160 | 30890260 | 30891484 | - | 2 | 368 | R2R3 | anther development, stress response | ||
| Ma04_g31800 | 32024735 | 32025857 | + | 3 | 279 | R2R3 | axillary meristem, root growth | ||
| Ma04_g31880 | 32081718 | 32084107 | - | 4 | 142 | R2R3 | photomorphogenesis | ||
| Ma04_g32240 | 32306362 | 32307003 | + | 2 | 187 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma04_g33920 | MYB72 [22] | 33328252 | 33329340 | + | 3 | 300 | R2R3 | PP, lignin | |
| Ma04_g34300 | 33575039 | 33576369 | + | 3 | 382 | R2R3 | anther development, stress response | ||
| Ma04_g34660 | 33734735 | 33735587 | + | 3 | 226 | R2R3 | axillary meristem, root growth | ||
| Ma04_g35350 | 34164151 | 34165124 | - | 3 | 272 | R2R3 | PP, lignin | ||
| Ma04_g35730 | 34361423 | 34362164 | + | 3 | 204 | R2R3 | PP, lignin | ||
| Ma04_g35890 | 34456334 | 34457640 | - | 3 | 279 | R2R3 | photomorphogenesis | ||
| Ma04_g38740 | 36139124 | 36140139 | + | 3 | 279 | R2R3 | axillary meristem, root growth | ||
| Ma05_g01100 | 651244 | 652022 | + | 3 | 206 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma05_g01880 | 1153222 | 1154262 | - | 3 | 281 | R2R3 | axillary meristem, root growth | ||
| Ma05_g03340 | 2404480 | 2405576 | + | 3 | 303 | R2R3 | PP, lignin | ||
| Ma05_g03690 | 2718554 | 2719768 | - | 4 | 240 | R2R3 | ASR, flower morphogenesis, stilbene | ||
| Ma05_g05670 | 4316981 | 4317987 | - | 4 | 246 | R2R3 | secondary cell wall, lignin | ||
| Ma05_g06310 | 4705834 | 4707231 | - | + | 3 | 375 | R2R3 | general flavonoid, trichome | |
| Ma05_g07140 | 5206984 | 5207790 | - | 1 | 269 | R2R3 | stress tolerance | ||
| Ma05_g07450 | 5427233 | 5429855 | - | 5 | 295 | R2R3 | defense, stress response | ||
| Ma05_g08960 | 6598702 | 6600983 | + | 5 | 192 | R2R3 | |||
| Ma05_g10430 | 7526380 | 7527556 | + | 3 | 279 | R2R3 | axillary meristem, root growth | ||
| Ma05_g12030 | 8749432 | 8751067 | + | 5 | 255 | R2R3 | flower meristem identity | ||
| Ma05_g14510 | 10595192 | 10596181 | - | 4 | 229 | R2R3 | ASR, flower morphogenesis, stilbene | ||
| Ma05_g17720 | 21202263 | 21203407 | - | 3 | 234 | R2R3 | |||
| Ma05_g18420 | 23864552 | 23865432 | + | 3 | 241 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma05_g18710 | 24606320 | 24608917 | - | 5 | 288 | R2R3 | defense, stress response | ||
| Ma05_g19630 | 28142012 | 28143079 | - | 3 | 295 | R2R3 | root development | ||
| Ma05_g20320 | 31975647 | 31976748 | + | 3 | 307 | R2R3 | suberin | ||
| Ma05_g20740 | 32405906 | 32411144 | + | 6 | 375 | R2R3 | |||
| Ma05_g20940 | 32642222 | 32646244 | + | 3 | 268 | R2R3 | |||
| Ma05_g23480 | 35547417 | 35548386 | + | 4 | 182 | R2R3 | defense, stress response | ||
| Ma05_g23640 | 35794926 | 35797163 | - | 3 | 321 | R2R3 | flavonols, phlobaphene | ||
| Ma05_g24200 | 36482627 | 36483576 | + | 3 | 207 | R2R3 | axillary meristem, root growth | ||
| Ma05_g24840 | 36981509 | 36982626 | + | 3 | 323 | R2R3 | anther-, tapetum development | ||
| Ma05_g25150 | 37166050 | 37167522 | + | 3 | 316 | R2R3 | axillary meristem, root growth | ||
| Ma05_g25490 | 37423048 | 37423913 | - | 3 | 226 | R2R3 | secondary cell wall, lignin | ||
| Ma05_g25630 | 37499891 | 37501199 | - | 6 | 290 | R2R3 | trichome branching, petal morphogenesis | ||
| Ma05_g25680 | 37532411 | 37533682 | + | 3 | 302 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma05_g28370 | 39396363 | 39402499 | - | + | 5 | 210 | R2R3 | repressors PP, sinapate, lignin | |
| Ma05_g30120 | 40637530 | 40638485 | + | 3 | 252 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma05_g30720 | 40993457 | 40994353 | + | 3 | 237 | R2R3 | cell cycle regulation | ||
| Ma05_g31160 | 41195712 | 41202456 | - | 4 | 397 | R2R3 | stress tolerance | ||
| Ma05_g31440 | 41356727 | 41357758 | + | 1 | 344 | R2R3 | leaf-, shoot-, germ morphogenesis | ||
| Ma06_g00910 | 744618 | 747868 | + | 5 | 394 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma06_g03570 | 2604334 | 2605906 | + | 3 | 333 | R2R3 | axillary meristem, root growth | ||
| Ma06_g04210 | 3055555 | 3059353 | - | + | 4 | 361 | R2R3 | proanthocyanidins | |
| Ma06_g04240 | 3072400 | 3074637 | - | 7 | 345 | R2R3 | trichome branching, petal morphogenesis | ||
| Ma06_g04270 | 3095165 | 3096766 | + | 4 | 354 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma06_g04370 | 3152489 | 3153586 | + | 4 | 250 | R2R3 | defense, stress response | ||
| Ma06_g05680 | 4227458 | 4228553 | + | 3 | 305 | R2R3 | root development | ||
| Ma06_g05960 | 4396784 | 4397869 | - | + | 3 | 275 | R2R3 | anthocyanins | |
| Ma06_g06660 | 4801530 | 4808116 | - | 7 | 517 | 3R | cell cycle control | ||
| Ma06_g08100 | 5748553 | 5749986 | + | 3 | 276 | R2R3 | embryogenesis, seed maturation | ||
| Ma06_g08440 | 5976699 | 5977672 | - | 3 | 243 | R2R3 | |||
| Ma06_g08910 | 6239164 | 6240262 | + | + | 5 | 199 | R2R3 | repressors PP, sinapate, lignin | |
| Ma06_g11140 | MaMYB3 [23] | 7827980 | 7828834 | + | + | 4 | 200 | R2R3 | starch degradation, flavonoid repressor |
| Ma06_g11270 | 7905384 | 7906681 | + | 3 | 308 | R2R3 | defense, stress response | ||
| Ma06_g12110 | 8408418 | 8411546 | - | 12 | 469 | R2R3 | guard cell division, root gravitropism | ||
| Ma06_g12160 | 8449881 | 8450871 | - | 4 | 256 | R2R3 | defense, stress response | ||
| Ma06_g14470 | 9914817 | 9917018 | - | + | 5 | 286 | R2R3 | repressors PP, sinapate, lignin | |
| Ma06_g16350 | 11053657 | 11054699 | + | + | 3 | 290 | R2R3 | general flavonoid, trichome | |
| Ma06_g16920 | 11470816 | 11471624 | + | 3 | 220 | R2R3 | cell cycle regulation | ||
| Ma06_g17440 | 11851435 | 11852673 | - | 4 | 275 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma06_g19030 | 13020150 | 13021197 | + | 2 | 263 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma06_g27210 | 29243310 | 29244238 | - | 4 | 237 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma06_g29060 | 30553280 | 30554423 | + | 3 | 195 | R2R3 | stamen development | ||
| Ma06_g31020 | 32237208 | 32238426 | + | 3 | 351 | R2R3 | defense, stress response | ||
| Ma06_g32530 | 33447718 | 33449121 | + | 3 | 315 | R2R3 | axillary meristem, root growth | ||
| Ma06_g33100 | 33858326 | 33871293 | + | 12 | 838 | 4R | SNAP complex | ||
| Ma06_g33190 | 33914360 | 33915233 | + | 3 | 244 | R2R3 | |||
| Ma06_g33430 | 34073015 | 34074005 | + | 3 | 273 | R2R3 | axillary meristem, root growth | ||
| Ma06_g33920 | 34371289 | 34372730 | + | 3 | 346 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma06_g35430 | 35255099 | 35260321 | + | 3 | 225 | R2R3 | stress tolerance | ||
| Ma06_g35620 | 35399156 | 35400628 | + | 4 | 400 | R2R3 | embryogenesis, seed maturation | ||
| Ma06_g37660 | 36662931 | 36664093 | + | 3 | 299 | R2R3 | anther-, trichome development | ||
| Ma06_g38880 | 37507220 | 37508388 | - | 3 | 311 | R2R3 | anther-, trichome development | ||
| Ma07_g00270 | 249167 | 250371 | + | 2 | 361 | R2R3 | |||
| Ma07_g02470 | 1968784 | 1973645 | - | 3 | 600 | R2R3 | anther development, stress response | ||
| Ma07_g05660 | 4114936 | 4116029 | + | 3 | 272 | R2R3 | defense, stress response | ||
| Ma07_g05780 | 4200621 | 4201699 | + | 2 | 251 | R2R3 | secondary cell wall, lignin | ||
| Ma07_g08110 | 6059949 | 6061103 | + | 5 | 256 | R2R3 | axillary meristem, root growth | ||
| Ma07_g10330 | 7698721 | 7708970 | + | 7 | 568 | 3R | cell cycle control | ||
| Ma07_g10340 | 7710266 | 7711261 | - | 1 | 332 | R2R3 | leaf-, shoot-, germ morphogenesis | ||
| Ma07_g11110 | 8262980 | 8264078 | - | 3 | 187 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma07_g12330 | 9212653 | 9213850 | + | 4 | 272 | R2R3 | secondary cell wall, lignin | ||
| Ma07_g13590 | 10213901 | 10216977 | - | + | 3 | 307 | R2R3 | general flavonoid, trichome | |
| Ma07_g17600 | 20759044 | 20760488 | + | 5 | 343 | R2R3 | |||
| Ma07_g19350 | 27376443 | 27377703 | - | 5 | 270 | R2R3 | defense, stress response | ||
| Ma07_g19470 | 27474578 | 27475521 | + | 4 | 241 | R2R3 | cell cycle regulation | ||
| Ma07_g19700 | 27655399 | 27656957 | - | 2 | 470 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma07_g19720 | 27682692 | 27684725 | + | 6 | 357 | R2R3 | trichome branching, petal morphogenesis | ||
| Ma07_g19880 * | 27793556 | 27794919 | + | + | 4 | 264 | R2R3 | proanthocyanidins | |
| Ma07_g19890 * | 27798326 | 27799369 | - | + | 3 | 269 | R2R3 | proanthocyanidins | |
| Ma07_g20020 | 27925622 | 27926695 | + | 4 | 259 | R2R3 | secondary cell wall, lignin | ||
| Ma07_g20990 | 28973984 | 28975550 | - | 3 | 341 | R2R3 | axillary meristem, root growth | ||
| Ma07_g22540 | 30450574 | 30451644 | - | 4 | 269 | R2R3 | defense, stress response | ||
| Ma07_g23060 | 30809797 | 30823608 | + | 3 | 551 | R2R3 | anther development, stress response | ||
| Ma07_g23180 | 30932263 | 30933303 | + | 3 | 289 | R2R3 | defense, stress response | ||
| Ma07_g23230 * | 30951709 | 30952887 | + | 3 | 274 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma07_g23240 * | 30951829 | 30952875 | + | 3 | 230 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma07_g26530 | 33294011 | 33295448 | - | 4 | 292 | R2R3 | |||
| Ma08_g01300 | 1210795 | 1211973 | - | 5 | 263 | R2R3 | defense, stress response | ||
| Ma08_g01760 | 1466390 | 1467139 | + | 1 | 250 | R2R3 | stress tolerance | ||
| Ma08_g02100 | 1707379 | 1708990 | + | 3 | 421 | R2R3 | embryogenesis, seed maturation | ||
| Ma08_g02450 | 1904860 | 1905752 | + | 3 | 241 | R2R3 | |||
| Ma08_g03420 | 2489605 | 2490759 | + | 3 | 319 | R2R3 | anther-, trichome development | ||
| Ma08_g10260 | 7475942 | 7478109 | - | 3 | 371 | R2R3 | flavonols, phlobaphene | ||
| Ma08_g10600 | 7758649 | 7759595 | - | 3 | 256 | R2R3 | defense, stress response | ||
| Ma08_g11120 | 8204512 | 8205519 | + | 3 | 278 | R2R3 | photomorphogenesis | ||
| Ma08_g12510 | 9469693 | 9470558 | + | 3 | 231 | R2R3 | PP, lignin | ||
| Ma08_g13070 | 10389299 | 10390584 | - | 3 | 313 | R2R3 | axillary meristem, root growth | ||
| Ma08_g14720 | 14652484 | 14653886 | + | 4 | 370 | R2R3 | |||
| Ma08_g15820 | 16034645 | 16037684 | - | + | 3 | 260 | R2R3 | ||
| Ma08_g15960 | 16626283 | 16635800 | - | 3 | 557 | R2R3 | |||
| Ma08_g16760 | 20724484 | 20725190 | - | + | 3 | 194 | R2R3 | flavonoid repressor | |
| Ma08_g17860 | 27301472 | 27302427 | - | 3 | 249 | R2R3 | defense, stress response | ||
| Ma08_g18540 | 32070622 | 32071809 | - | 4 | 276 | R2R3 | defense, stress response | ||
| Ma08_g23390 | 36792861 | 36793894 | + | 3 | 290 | R2R3 | proanthocyanidins | ||
| Ma08_g25570 | 38350630 | 38352260 | + | 3 | 307 | R2R3 | axillary meristem, root growth | ||
| Ma08_g25960 | MYBS3 [24] | 38629624 | 38630727 | + | 4 | 296 | R2R3 | stress response, hormone signaling | |
| Ma08_g26720 | 39203311 | 39211401 | - | 6 | 541 | R2R3 | |||
| Ma08_g30360 | 41652620 | 41653601 | + | 3 | 269 | R2R3 | defense, stress response | ||
| Ma08_g31720 | MYB83 [22] | 42548860 | 42549849 | + | 3 | 279 | R2R3 | secondary wall, lignin | |
| Ma08_g32760 | 43364702 | 43366041 | + | 4 | 200 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma08_g34230 | 44310614 | 44311366 | - | + | 3 | 203 | R2R3 | repressors PP, sinapate, lignin | |
| Ma08_g34710 | 44706501 | 44707468 | + | 3 | 245 | R2R3 | |||
| Ma09_g03310 | 2240622 | 2241647 | - | 1 | 342 | R2R3 | leaf-, shoot-, germ morphogenesis | ||
| Ma09_g03740 | 2481273 | 2483877 | + | 2 | 322 | R2R3 | stress tolerance | ||
| Ma09_g04930 | 3159224 | 3160371 | - | 3 | 302 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma09_g06730 | 4303577 | 4304525 | + | 3 | 204 | R2R3 | stamen development | ||
| Ma09_g08140 | 5353783 | 5354623 | + | 2 | 256 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma09_g08260 | 5451934 | 5452919 | - | + | 3 | 272 | R2R3 | repressors PP, sinapate, lignin | |
| Ma09_g09400 | 6192942 | 6193897 | - | 3 | 263 | R2R3 | |||
| Ma09_g09720 | 6398143 | 6410415 | - | 13 | 798 | 4R | SNAP complex | ||
| Ma09_g10800 | 7342974 | 7344121 | - | 3 | 327 | R2R3 | anther-, trichome development | ||
| Ma09_g11770 | 8000924 | 8002091 | - | 4 | 262 | R2R3 | defense, stress response | ||
| Ma09_g13170 | 8908685 | 8910010 | - | 3 | 310 | R2R3 | axillary meristem, root growth | ||
| Ma09_g14260 | 9743683 | 9745180 | - | 4 | 187 | R2R3 | root development | ||
| Ma09_g15050 | 10356705 | 10357805 | - | 4 | 248 | R2R3 | secondary cell wall, lignin | ||
| Ma09_g15130 | 10456847 | 10458264 | + | 4 | 211 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma09_g15440 | 10764272 | 10765275 | - | 4 | 257 | R2R3 | proanthocyanidins | ||
| Ma09_g15940 | 11297314 | 11298296 | - | 3 | 260 | R2R3 | defense, stress response | ||
| Ma09_g16940 | 12438026 | 12439426 | + | 4 | 279 | R2R3 | defense, stress response | ||
| Ma09_g16980 | 12476657 | 12478055 | + | 4 | 304 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma09_g20280 | 28998021 | 28999222 | + | 3 | 273 | R2R3 | axillary meristem, root growth | ||
| Ma09_g22730 | 34631677 | 34632781 | - | 3 | 315 | R2R3 | |||
| Ma09_g23100 | 35024484 | 35029665 | + | 4 | 288 | R2R3 | stress response, hormone signaling | ||
| Ma09_g23770 | 35511400 | 35518711 | + | 5 | 1120 | 2R | CDC5 | ||
| Ma09_g24640 | MYB31 [21] | 36291996 | 36292655 | - | 3 | 173 | R2R3 | repressors PP, sinapate, lignin | |
| Ma09_g25010 | 36615326 | 36616400 | + | 3 | 301 | R2R3 | defense, stress response | ||
| Ma09_g25590 | 36999501 | 37000692 | - | 3 | 323 | R2R3 | anther-, trichome development | ||
| Ma09_g27990 | 38852584 | 38853631 | - | + | 3 | 260 | R2R3 | anthocyanins | |
| Ma09_g28970 | 39592459 | 39593487 | + | 1 | 343 | R2R3 | leaf-, shoot-, germ morphogenesis | ||
| Ma09_g29010 | 39617399 | 39618193 | - | 3 | 214 | R2R3 | |||
| Ma09_g29660 | MYB52 [22] | 40061204 | 40062015 | - | 3 | 210 | R2R3 | cell wall, lignin, seed oil, axillary meristem | |
| Ma10_g01730 | 5102446 | 5104125 | - | 5 | 286 | R2R3 | defense, stress response | ||
| Ma10_g01750 | 5159022 | 5160613 | + | 5 | 288 | R2R3 | defense, stress response | ||
| Ma10_g04420 | 15095718 | 15097543 | - | 5 | 284 | R2R3 | |||
| Ma10_g04920 | 15562156 | 15563431 | - | 3 | 321 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma10_g05260 | 15989100 | 15989828 | + | 1 | 243 | R2R3 | stress tolerance | ||
| Ma10_g05680 | 17016063 | 17017842 | + | 3 | 499 | R2R3 | embryogenesis, seed maturation | ||
| Ma10_g06140 | 17599985 | 17600773 | + | 3 | 212 | R2R3 | |||
| Ma10_g09100 | 23301527 | 23306607 | - | 4 | 297 | R2R3 | anther-, trichome development | ||
| Ma10_g09370 | 23565764 | 23566747 | - | 3 | 268 | R2R3 | axillary meristem, root growth | ||
| Ma10_g10820 | 24568790 | 24569501 | + | 3 | 176 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma10_g11100 | 24705755 | 24706780 | - | 3 | 260 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma10_g13000 | 25942170 | 25943424 | + | 4 | 304 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma10_g13640 | 26378393 | 26379280 | - | 3 | 236 | R2R3 | cell cycle regulation | ||
| Ma10_g14150 | 26690247 | 26691396 | - | + | 4 | 298 | R2R3 | general flavonoid, trichome | |
| Ma10_g14950 | 27211124 | 27223165 | + | 16 | 869 | 3R | cell cycle control | ||
| Ma10_g16050 | 27917472 | 27918339 | + | + | 3 | 217 | R2R3 | repressors PP, sinapate, lignin | |
| Ma10_g17650 | 28964249 | 28965405 | + | + | 3 | 278 | R2R3 | anthocyanins | |
| Ma10_g18840 | 29621056 | 29624189 | + | 12 | 467 | R2R3 | guard cell division, root gravitropism | ||
| Ma10_g19130 | 29806495 | 29807615 | - | 3 | 297 | R2R3 | root development | ||
| Ma10_g19820 | 30234061 | 30235019 | - | 4 | 175 | R2R3 | defense, stress response | ||
| Ma10_g19970 | 30309772 | 30312309 | - | + | 4 | 206 | R2R3 | flavonoid repressor | |
| Ma10_g24510 | 33076512 | 33077586 | - | 4 | 305 | R2R3 | root development | ||
| Ma10_g25660 | 33694709 | 33695874 | + | 3 | 314 | R2R3 | root development | ||
| Ma10_g26540 | 34187707 | 34195688 | + | 7 | 567 | 3R | cell cycle control | ||
| Ma10_g26660 | 34247924 | 34248785 | + | 3 | 226 | R2R3 | |||
| Ma10_g29230 | 35877509 | 35878859 | - | 5 | 315 | R2R3 | defense, stress response | ||
| Ma10_g29290 | 35933319 | 35936352 | - | 3 | 562 | R2R3 | anther development, stress response | ||
| Ma10_g29660 | 36170205 | 36171708 | - | 7 | 289 | R2R3 | trichome branching, petal morphogenesis | ||
| Ma10_g29900 | 36334814 | 36335832 | + | 3 | 285 | R2R3 | secondary cell wall, lignin | ||
| Ma11_g00330 | 235287 | 236166 | - | 3 | 235 | R2R3 | |||
| Ma11_g00350 | 255492 | 257081 | + | 6 | 375 | R2R3 | |||
| Ma11_g02310 | 1659794 | 1660627 | - | 3 | 213 | R2R3 | PP, lignin | ||
| Ma11_g03860 | 2954888 | 2956813 | - | 4 | 294 | R2R3 | flower meristem identity | ||
| Ma11_g04680 | 3650672 | 3651636 | + | 3 | 264 | R2R3 | defense, stress response | ||
| Ma11_g06880 | 5505145 | 5505826 | - | 3 | 167 | R2R3 | repressors PP, sinapate, lignin | ||
| Ma11_g07330 | 5826847 | 5827930 | + | 3 | 261 | R2R3 | axillary meristem, root growth | ||
| Ma11_g07530 | 6014287 | 6015836 | - | 3 | 357 | R2R3 | suberin | ||
| Ma11_g08730 | 6941094 | 6942543 | - | 4 | 275 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma11_g10680 | 10271192 | 10273848 | - | 6 | 275 | R2R3 | defense, stress response | ||
| Ma11_g10710 | 10304514 | 10305532 | + | 3 | 275 | R2R3 | |||
| Ma11_g11300 | 12744937 | 12748426 | + | 6 | 251 | R2R3 | defense, stress response | ||
| Ma11_g11940 | 15469544 | 15470676 | - | 3 | 309 | R2R3 | defense, stress response | ||
| Ma11_g14670 | 20385602 | 20386542 | - | 3 | 262 | R2R3 | defense, stress response | ||
| Ma11_g15740 | 21412108 | 21413062 | - | 4 | 230 | R2R3 | PP, lignin | ||
| Ma11_g16150 | 21711896 | 21713795 | - | 5 | 233 | R2R3 | |||
| Ma11_g16430 | 21944276 | 21945343 | + | 3 | 287 | R2R3 | PP, lignin | ||
| Ma11_g19220 | 24164576 | 24166138 | - | 4 | 344 | R2R3 | defense, stress response | ||
| Ma11_g21160 | 25411345 | 25412681 | - | 6 | 303 | R2R3 | |||
| Ma11_g21730 | 25754364 | 25755420 | - | 2 | 326 | R2R3 | suberin | ||
| Ma11_g21820 | 25814103 | 25816846 | - | + | 3 | 235 | R2R3 | ||
| Ma11_g23010 | 26544741 | 26545608 | - | 3 | 241 | R2R3 | cell cycle regulation | ||
| Ma11_g23420 | 26767696 | 26769125 | + | 3 | 183 | R2R3 | cell wall, lignin, seed oil, axillary meristem | ||
| Ma00_g01590 | 9744573 | 9745661 | + | 2 | 283 | R2R3 | stress response, hormone signaling | ||
| Ma00_g04340 | 36355056 | 36356428 | + | 2 | 283 | R2R3 | secondary wall, lignin | ||
| Ma00_g04960 | 43098834 | 43099818 | - | 3 | 267 | R2R3 | defense, stress response | ||
The genes are ordered by DH Pahang version 2 [18] pseudochromosomes, from north to south. The annotation-version specific gene code describing the chromosomal assignment and position on pseudochromosomes is given in the first column. "Ma00" indicates genes located in sequences without chromosomal assignment. Functional assignment is based on the Maximum Likelihood tree presented in Fig 2. Multi-FASTA files with MaMYB CDS and peptide sequences together with a general feature format (GFF) file describing the annotated MaMYB genes is provided in the supplementaries. str.: strand
*: paralogs, ASR: abiotic stress response, PP: phenylpropanoid.
The number of R2R3-MYB genes is one of the highest among the species that have been studied to date, ranging from 45 in Ginkgo biloba [10] over 157 in Zea mays [25] and 249 in Brassica napus [26] to 360 in Gossypium hirsutum [11]. This is probably due to three whole-genome duplications (γ 100 Myr ago and α, β 65 Myr ago) that occurred during Musa genome evolution [3, 27]. The number of atypical multiple repeat MYB genes identified in M. acuminata is in the same range as those reported for most other plant species, up to six MYB3R and up to two MYB4R and CDC5-like genes.
The 285 MaR2R3-MYB genes identified constitute approximately 0.81% of the 35,276 predicted protein-coding M. acuminata genes and 9.0% of the 3,155 putative M. acuminata transcription factor genes [18]. These were subjected to further analyses. The identified MaR2R3-MYB genes were named following the nomenclature of the locus tags provided in the DH-Pahang version 2 genome annotation (Table 1). A keyword search in the NCBI database (http://www.ncbi.nlm.nih.gov/) revealed no evidence for M. acuminata MYB genes not present in Table 1.
Six publications dealing with M. acuminata MYB genes were identified: one study describes the elevated expression of nine MaMYB genes in transgenic banana plants overexpressing the NAC domain transcription factor MusaVND1 (vascular related NAC domain) indicating a role of these MaMYBs in the regulation of secondary wall deposition [22]. MYBS3 (Ma08_25960) was found to be differentially expressed between cold-sensitive and cold-tolerant bananas [24]. Another publication described the M. acuminata R2R3-MYB gene Ma05_03690 being upregulated in the early response to the endoparasitic root-knot nematode Meloidogyne incognita in roots [28]. MusaMYB31 (Ma01_02850) was identified as a negative regulator of lignin biosynthesis and the general phenylpropanoid biosynthesis pathway [21]. MaMYB3 (Ma06_11140) was found to repress starch degradation in fruit ripening [23] and MaMYB4 (Ma01_19610) was recently described to control fatty acid desaturation [29].
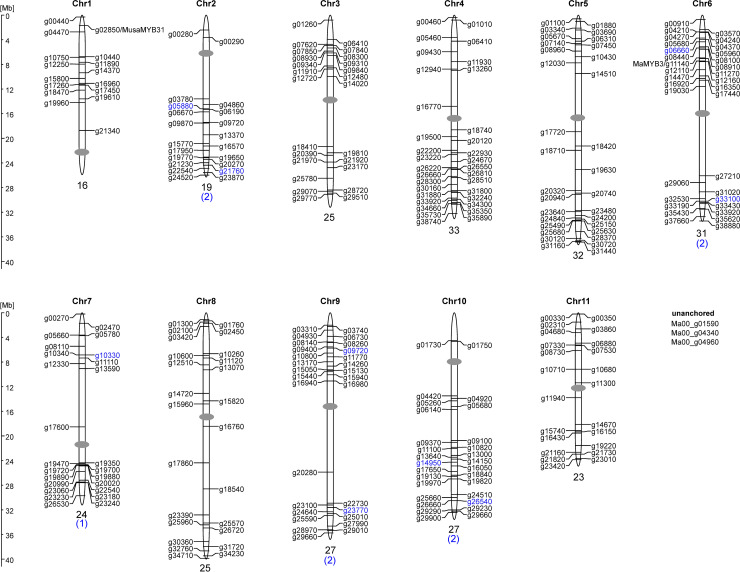
On the basis of the DH-Pahang version 2 annotation, 291 of the 294 MaMYB genes could be assigned to the eleven chromosomes. The chromosomal distribution of MaMYB genes on the pseudochromosomes is shown in Fig 1 and revealed that M. acuminata MYB genes are distributed across all chromosomes.
Fig 1. Distribution of MaMYB genes on the eleven M. acuminata chromosomes.
Gene structure analysis revealed that most MaR2R3-MYB genes (155 of 285; 54.4%) follow the previously reported rule of having two introns and three exons, and display the highly conserved splicing arrangement that has also been reported for other plant species [25, 30, 31]. A total of 23 (8.1%) MaR2R3-MYB genes have two exons and seven (2.5%) were single exon genes. 67 (23.5%) MaR2R3-MYB genes have four exons, 21 (7.4%) five exons, eight (2.8%) six exons and two of each with seven and twelve exons, respectively. The complex exon-intron structure of Ma06_12110 and Ma10_18840 is conserved in their A. thaliana orthologs AtMYB88 and AtMYB124/FOUR LIPS (FLP) containing ten and eleven exons, respectively. This supports their close evolutionary relationship, but also suggests the conservation of this intron pattern in evolution since the monocot-dicot split 140–150 Myr ago [32].
Chromosomes are drawn to scale. The positions of centromeres (grey ovals) are roughly estimated from repeat distribution data. The chromosomal positions of the MaMYB genes (given in DH Pahang version 2 annotation ID) are indicated. R2R3-MYB genes are given in black letters, MYB genes with more than two MYB repeats are given in blue letters. The number of MYB genes on each chromosome is given below the respective chromosome.
Phylogenetic analysis of the M. acuminata MYB family
With the aim to explore the putative function of the predicted M. acuminata MYBs, we assigned them to plant MYB proteins with known function. For this, we chose primarily data from A. thaliana, which is the source of most functional MYB characterisations. From comparable studies, MYB function appears conserved across MYB clades, suggesting that closely related MYBs recognise similar/same target genes and possess cooperative, overlapping or redundant functions.
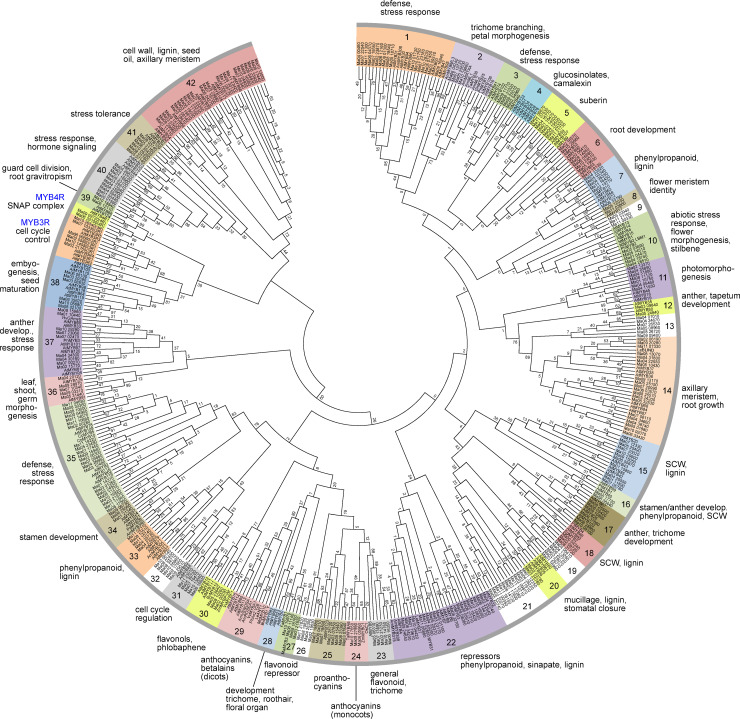
To unravel the relationships, we constructed a phylogenetic tree with 468 MYB domain amino acid sequences of MYB proteins. We used 293 MaMYBs (omitting the CDC5-like MaMYB), the complete A. thaliana MYB family (132 members, including 126 R2R3-MYB, five MYB3R and one MYB4R) and 43 functionally well characterised landmark R2R3-MYBs from other plant species. The phylogenetic tree topology allowed us to classify the analysed MYBs into one MYB3R clade, one MYB4R clade and 42 R2R3-MYB protein clades (Fig 2).
Fig 2. Phylogenetic Maximum Likelihood (ML) tree.
Most R2R3-MYB clades (31 of 42) include variable numbers of MYB proteins from Arabidopsis and banana, indicating that the appearance of most MYB genes in these two species predates the monocot-dicot split as observed in other studies [33]. Several of these clades also contain landmark MYBs from other plant species. The two clades 4 and 28 only contain Arabidopsis R2R3-MYB members, while seven clades (9, 13, 19, 21, 26, 29 and 32, displayed with white background in Fig 2) only contain banana R2R3-MYB members. Additionally, two clades, 24 with monocot anthocyanin biosynthesis regulators and 27 with the strawberry landmark flavonoid biosynthesis repressor FaMYB1 [34], do not contain any A. thaliana MYB (Fig 2).
ML consensus tree inferred from 1000 bootstraps with 468 MYB domain amino acid sequences of MYB proteins from Musa accuminata (Ma), Arabidopsis thaliana (At) and landmark MYBs from other plant species built with MEGA7. Clades are labeled with different colors and functional annotations are given. The numbers at the branches give bootstrap support values from 1000 replications. SCW, secondary cell wall.
The lineage specificity of some MYB clades could indicate that these clades may have been lost or gained in a single order or species during plant evolution, as indicated by other studies [35–37]. For example, clade 28 lacked M. acuminata orthologs, but includes the A. thaliana R2R3-MYBs AtMYB0/GLABRA1 and AtMYB66/WEREWOLF, which have been identified as being involved in the formation of trichomes and root hairs from epidermal cells [38, 39]. Similar observations have been made in maize (monocot) and sugarbeet (eudicot, caryophyllales) [30, 31], both not containing clade 28 orthologs, while grape (eudicot, rosid) and poplar (eudicot, rosid) do [12, 40]. It has been hypothesized that GLABRA1-like MYB genes have been acquired in rosids after the rosid-asterids split [41]. The absence of MaMYBs in clade 28 is consistent with this hypothesis, since monocots branched off before the separation of asterids and rosids in eudicots. Clade 4 also lacks banana R2R3-MYBs. This clade contains the glucosinolate biosynthesis regulators AtMYB28, AtMYB34 and AtMYB51 [42, 43]. The absence of MaMYBs in this clade is concordant with the fact that glucosinolates are only present in the Brassicaceae family. This clade is thought to have originated from a duplication event before the divergence of the genus Arabidopsis from Brassica [44].
The seven clades containing only MaMYBs were manually inspected by applying BLAST searches at the NCBI protein database in order to identify high similarity to functionally characterized landmark plant R2R3-MYBs. In no case could a landmark MYB be identified. Consequently, these clades could be described as a lineage-specific expansion in M. acuminata, reflecting a species-, genus- or order-specific evolutionary change. These MaMYB proteins may have specialised functions that were acquired or expanded in M. acuminata during genome evolution. Further research will be needed to decipher the biological roles of these MaMYB genes.
R2R3-MYBs may interact with basic helix-turn-helix (bHLH)-type transcription factors, together with WD-repeat (WDR) proteins, forming a trimeric MBW complex. These R2R3-MYBs are defined by a bHLH-binding consensus motif [D/E]Lx2[R/K]x3Lx6Lx3R [45] found in all bHLH-interacting R2R3-MYBs. A search in the MaMYB proteins for the mentioned bHLH-interaction motif identified 30 MaMYBs containing this motif (Table 1) and thus putatively interacting with bHLH proteins. 26 of these 30 MaMYBs were all functionally assigned to clades containing (potentially) known bHLH-interacting R2R3-MYBs: nine in clade 22 (repressors phenylpropanoid, sinapate-, lignin regulators), six in clade 25 (proanthocyanidin regulators), four in clade 23 (general flavonoid, trichome regulators), four in clade 27 (flavonoid repressors) and three in clade 24 (monocot anthocyanin regulators). Four MaMYBs, Ma02_19650, Ma04_28300, Ma08_15820 and Ma11_21820 were all found in the M. acuminata-specific clade 26. These MaMYBs may interact with bHLH-type transcription factors. The close relation to flavonoid biosynthesis regulators suggests that they may act similarly to R2R3-MYB proanthocyanidin regulators. Further research is needed to determine if they regulate a Musa-specific biosynthesis pathway.
Expression profiles for M. acuminata MYB genes in different organs and developmental stages
Since it is not unusual for large transcription factor families in higher organisms to have redundant functions, a particular transcription factor needs to be studied and characterised in the context of the whole family. In this regard, the gene expression pattern can provide a hint to gene function. In this study we used publicly-available RNA-Seq reads from the Sequence Read Archive (S2 Table) to analyse the expression of the 293 MaMYB genes in different organs and developmental stages: embryonic cell suspension, seedling, root and young, adult and old leaf, pulp stages S1-S4 and peel stages S1-S4. Filtered RNA-Seq reads were aligned to the reference genome sequence and the number of mapped reads per annotated transcript were quantified and compared across the analysed samples to calculate normalised RNA-Seq read values which are given in Table 2.
Table 2. The expression profiles of MaR2R3-MYB genes in different organs and developmental stages based on RNA-Seq data.
| gene | embryogenic | seedling | root | leaf | young_leaf | adult_leaf | old_leaf | pulp | pulpS1 | pulpS2 | pulpS3 | pulpS4 | peel | peelS1 | peelS2 | peelS3 | peelS4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ma00_g01590 | 50 | 35 | 6 | 151 | 77 | 175 | 348 | 6 | 5 | 5 | 8 | 5 | 8 | 2 | 19 | 10 | 2 |
| Ma00_g04340 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma00_g04960 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma01_g00440 | 181 | 49 | 38 | 129 | 53 | 175 | 276 | 32 | 31 | 58 | 34 | 7 | 30 | 9 | 63 | 35 | 11 |
| Ma01_g02850 | 9 | 2 | 14 | 27 | 21 | 31 | 38 | 20 | 24 | 23 | 23 | 10 | 11 | 7 | 9 | 17 | 12 |
| Ma01_g04470 | 1 | 0 | 12 | 4 | 1 | 6 | 2 | 9 | 13 | 9 | 7 | 8 | 6 | 6 | 3 | 7 | 7 |
| Ma01_g10440 | 3 | 16 | 4 | 5 | 7 | 5 | 5 | 4 | 5 | 5 | 3 | 3 | 3 | 3 | 2 | 3 | 4 |
| Ma01_g10750 | 0 | 0 | 2 | 2 | 0 | 0 | 4 | 1 | 1 | 1 | 1 | 2 | 4 | 5 | 4 | 4 | 5 |
| Ma01_g11890 | 0 | 0 | 3 | 3 | 10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma01_g12250 | 1 | 0 | 4 | 5 | 17 | 2 | 0 | 3 | 4 | 3 | 2 | 2 | 4 | 4 | 4 | 3 | 3 |
| Ma01_g14370 | 0 | 17 | 11 | 0 | 0 | 0 | 0 | 5 | 7 | 7 | 8 | 0 | 8 | 3 | 7 | 18 | 4 |
| Ma01_g15800 | 1 | 0 | 4 | 1 | 0 | 0 | 0 | 4 | 2 | 5 | 7 | 3 | 6 | 8 | 5 | 2 | 9 |
| Ma01_g16960 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma01_g17260 | 1 | 0 | 11 | 1 | 0 | 0 | 0 | 3 | 3 | 3 | 2 | 3 | 4 | 5 | 3 | 4 | 5 |
| Ma01_g17450 | 0 | 0 | 6 | 2 | 0 | 1 | 6 | 5 | 13 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 0 |
| Ma01_g18470 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 |
| Ma01_g19610 | 13 | 0 | 6 | 39 | 56 | 40 | 57 | 29 | 78 | 22 | 15 | 2 | 12 | 7 | 36 | 2 | 5 |
| Ma01_g19960 | 35 | 1 | 6 | 97 | 94 | 148 | 145 | 5 | 7 | 6 | 5 | 4 | 19 | 5 | 58 | 10 | 4 |
| Ma01_g21340 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma02_g00280 | 1 | 1 | 21 | 4 | 1 | 1 | 5 | 6 | 8 | 10 | 4 | 2 | 5 | 5 | 4 | 5 | 7 |
| Ma02_g00290 | 0 | 0 | 3 | 9 | 5 | 26 | 1 | 8 | 25 | 0 | 6 | 2 | 3 | 2 | 3 | 2 | 5 |
| Ma02_g03780 | 0 | 0 | 29 | 18 | 26 | 0 | 0 | 13 | 14 | 9 | 18 | 12 | 30 | 39 | 31 | 17 | 33 |
| Ma02_g04860 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma02_g05880 | 4 | 2 | 2 | 3 | 2 | 5 | 3 | 2 | 4 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 3 |
| Ma02_g06190 | 0 | 0 | 19 | 8 | 0 | 0 | 0 | 16 | 30 | 12 | 13 | 11 | 28 | 42 | 18 | 18 | 32 |
| Ma02_g06670 | 1 | 1 | 4 | 1 | 0 | 0 | 0 | 4 | 3 | 7 | 4 | 4 | 5 | 6 | 2 | 6 | 5 |
| Ma02_g09720 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Ma02_g09870 | 29 | 1 | 23 | 37 | 99 | 33 | 11 | 19 | 15 | 28 | 27 | 6 | 11 | 11 | 10 | 9 | 13 |
| Ma02_g13370 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma02_g15770 | 6 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| Ma02_g16570 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma02_g17950 | 8 | 0 | 46 | 14 | 4 | 6 | 13 | 33 | 53 | 35 | 16 | 26 | 19 | 19 | 14 | 20 | 22 |
| Ma02_g19650 | 0 | 0 | 3 | 3 | 5 | 1 | 0 | 7 | 10 | 5 | 3 | 11 | 9 | 15 | 8 | 6 | 5 |
| Ma02_g19770 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma02_g20270 | 3 | 0 | 3 | 16 | 57 | 4 | 0 | 1 | 1 | 2 | 2 | 1 | 4 | 3 | 4 | 3 | 7 |
| Ma02_g21230 | 2 | 2 | 1 | 5 | 14 | 3 | 1 | 3 | 8 | 3 | 0 | 0 | 1 | 0 | 3 | 2 | 0 |
| Ma02_g21760 | 0 | 3 | 7 | 2 | 2 | 0 | 0 | 8 | 11 | 7 | 8 | 7 | 8 | 11 | 5 | 5 | 12 |
| Ma02_g22540 | 1 | 0 | 7 | 4 | 2 | 6 | 7 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 |
| Ma02_g23870 | 3 | 0 | 0 | 7 | 4 | 20 | 3 | 2 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma02_g24520 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g01260 | 11 | 0 | 6 | 16 | 0 | 0 | 61 | 9 | 31 | 3 | 1 | 2 | 2 | 1 | 3 | 4 | 1 |
| Ma03_g06410 | 0 | 0 | 11 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 1 | 1 | 2 | 0 | 2 | 1 |
| Ma03_g07620 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g07840* | 0 | 0 | 2 | 1 | 2 | 0 | 0 | 10 | 24 | 9 | 3 | 5 | 2 | 1 | 3 | 1 | 2 |
| Ma03_g07850* | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 2 | 2 | 3 | 1 |
| Ma03_g08300 | 0 | 1 | 3 | 0 | 0 | 0 | 1 | 1 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 6 | 0 |
| Ma03_g08930 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g09310 | 0 | 0 | 0 | 12 | 42 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g09340 | 2 | 0 | 7 | 1 | 1 | 0 | 1 | 5 | 8 | 4 | 5 | 2 | 3 | 4 | 2 | 3 | 5 |
| Ma03_g09840 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g11910 | 5 | 8 | 25 | 35 | 76 | 25 | 6 | 12 | 10 | 13 | 12 | 13 | 24 | 26 | 30 | 18 | 22 |
| Ma03_g12480 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Ma03_g12720 | 16 | 10 | 4 | 45 | 59 | 49 | 70 | 2 | 2 | 2 | 2 | 3 | 18 | 2 | 32 | 35 | 3 |
| Ma03_g14020 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 4 | 1 | 3 | 2 | 3 | 1 | 3 | 2 |
| Ma03_g18410 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 4 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g19810 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g20390 | 5 | 1 | 1 | 2 | 2 | 0 | 4 | 2 | 1 | 3 | 2 | 2 | 0 | 1 | 0 | 0 | 1 |
| Ma03_g21920 | 5 | 0 | 2 | 17 | 13 | 32 | 22 | 4 | 12 | 2 | 2 | 0 | 2 | 0 | 6 | 3 | 0 |
| Ma03_g21970 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g23170 | 0 | 0 | 3 | 4 | 8 | 2 | 0 | 2 | 2 | 3 | 1 | 2 | 4 | 6 | 2 | 3 | 4 |
| Ma03_g25780 | 3 | 2 | 60 | 18 | 14 | 3 | 0 | 37 | 28 | 81 | 18 | 22 | 40 | 57 | 29 | 26 | 48 |
| Ma03_g28720 | 3 | 3 | 1 | 2 | 0 | 1 | 5 | 1 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Ma03_g29070 | 0 | 0 | 1 | 2 | 4 | 2 | 0 | 1 | 1 | 0 | 3 | 0 | 2 | 1 | 2 | 1 | 2 |
| Ma03_g29510 | 14 | 4 | 5 | 44 | 49 | 65 | 55 | 9 | 25 | 5 | 5 | 2 | 6 | 3 | 7 | 11 | 3 |
| Ma03_g29770 | 1 | 0 | 3 | 1 | 1 | 0 | 0 | 2 | 3 | 4 | 0 | 2 | 1 | 2 | 1 | 1 | 2 |
| Ma04_g00460 | 2 | 0 | 1 | 9 | 14 | 20 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 0 |
| Ma04_g01010 | 0 | 0 | 7 | 0 | 0 | 0 | 1 | 1 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| Ma04_g05460 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Ma04_g06410 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g09430 | 4 | 1 | 9 | 12 | 17 | 19 | 2 | 4 | 4 | 2 | 6 | 5 | 8 | 11 | 7 | 3 | 11 |
| Ma04_g11930 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g12940 | 4 | 0 | 2 | 3 | 6 | 2 | 4 | 2 | 3 | 1 | 2 | 0 | 1 | 0 | 1 | 0 | 0 |
| Ma04_g13260 | 0 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 3 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g16770 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 3 | 1 | 1 | 0 | 1 | 0 |
| Ma04_g18740 | 11 | 1 | 7 | 2 | 1 | 1 | 5 | 2 | 0 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 0 |
| Ma04_g19500 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 2 | 4 | 3 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Ma04_g20120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g22200 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 2 | 8 | 1 | 3 | 1 | 1 | 0 | 1 | 0 |
| Ma04_g22930 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 2 | 2 | 3 | 0 | 2 | 4 | 6 | 2 | 3 | 4 |
| Ma04_g23220 | 0 | 0 | 5 | 3 | 0 | 0 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 3 | 6 | 1 | 6 |
| Ma04_g24670 | 0 | 0 | 33 | 6 | 0 | 0 | 0 | 24 | 32 | 24 | 19 | 19 | 17 | 22 | 9 | 16 | 19 |
| Ma04_g26220 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 3 | 1 | 1 | 0 | 4 | 2 | 11 | 2 | 1 |
| Ma04_g26550 | 0 | 0 | 5 | 1 | 1 | 0 | 0 | 2 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| Ma04_g26660 | 0 | 2 | 22 | 1 | 0 | 1 | 3 | 12 | 27 | 11 | 5 | 4 | 1 | 0 | 2 | 1 | 2 |
| Ma04_g26810 | 41 | 78 | 15 | 153 | 258 | 145 | 185 | 39 | 35 | 74 | 32 | 17 | 33 | 13 | 54 | 50 | 14 |
| Ma04_g28300 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g28510 | 8 | 1 | 3 | 11 | 2 | 22 | 10 | 9 | 13 | 8 | 14 | 2 | 3 | 2 | 5 | 3 | 3 |
| Ma04_g30160 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g31800 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 1 | 6 | 0 |
| Ma04_g31880 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 2 | 0 | 1 |
| Ma04_g32240 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 4 | 2 | 0 | 0 | 1 | 0 | 0 | 1 |
| Ma04_g33920 | 3 | 7 | 2 | 2 | 4 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 3 | 1 |
| Ma04_g34300 | 1 | 0 | 0 | 1 | 2 | 1 | 0 | 2 | 4 | 1 | 1 | 2 | 1 | 1 | 0 | 0 | 1 |
| Ma04_g34660 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g35350 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g35730 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma04_g35890 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Ma04_g38740 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 |
| Ma05_g01100 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g01880 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 3 | 2 | 0 | 2 | 2 | 1 | 1 | 2 | 2 |
| Ma05_g03340 | 0 | 0 | 2 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g03690 | 10 | 0 | 1 | 14 | 2 | 25 | 28 | 1 | 0 | 1 | 1 | 1 | 2 | 0 | 8 | 1 | 0 |
| Ma05_g05670 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Ma05_g06310 | 4 | 1 | 1 | 4 | 4 | 4 | 8 | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ma05_g07140 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0 |
| Ma05_g07450 | 4 | 0 | 0 | 2 | 4 | 3 | 1 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g08960 | 27 | 1 | 9 | 34 | 47 | 53 | 23 | 17 | 38 | 11 | 8 | 10 | 9 | 10 | 3 | 9 | 12 |
| Ma05_g10430 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 2 | 3 | 3 | 6 | 2 | 1 | 4 |
| Ma05_g12030 | 4 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 8 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 1 |
| Ma05_g14510 | 1 | 0 | 4 | 48 | 0 | 1 | 190 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 3 | 0 |
| Ma05_g17720 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 6 | 4 | 2 | 4 |
| Ma05_g18420 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g18710 | 4 | 0 | 3 | 3 | 4 | 4 | 5 | 3 | 5 | 2 | 3 | 0 | 2 | 1 | 4 | 2 | 1 |
| Ma05_g19630 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 1 | 2 | 3 | 0 | 1 | 1 | 1 | 0 | 2 | 0 |
| Ma05_g20320 | 1 | 0 | 2 | 10 | 5 | 14 | 19 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| Ma05_g20740 | 0 | 4 | 1 | 2 | 9 | 0 | 0 | 2 | 5 | 1 | 2 | 0 | 1 | 0 | 2 | 1 | 0 |
| Ma05_g20940 | 1 | 0 | 5 | 11 | 0 | 0 | 44 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| Ma05_g23480 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ma05_g23640 | 0 | 0 | 1 | 1 | 0 | 3 | 0 | 2 | 7 | 0 | 2 | 0 | 2 | 1 | 3 | 0 | 2 |
| Ma05_g24200 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g24840 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g25150 | 0 | 0 | 6 | 2 | 0 | 0 | 0 | 3 | 4 | 4 | 5 | 1 | 3 | 3 | 2 | 2 | 4 |
| Ma05_g25490 | 0 | 1 | 2 | 2 | 1 | 3 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 |
| Ma05_g25630 | 0 | 0 | 0 | 4 | 15 | 0 | 1 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma05_g25680 | 1 | 0 | 1 | 2 | 2 | 1 | 3 | 7 | 24 | 2 | 3 | 1 | 0 | 0 | 1 | 1 | 0 |
| Ma05_g28370 | 0 | 0 | 4 | 4 | 9 | 4 | 1 | 1 | 0 | 1 | 0 | 1 | 2 | 2 | 1 | 1 | 2 |
| Ma05_g30120 | 0 | 3 | 16 | 1 | 0 | 1 | 0 | 4 | 7 | 4 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Ma05_g30720 | 0 | 0 | 19 | 1 | 0 | 0 | 0 | 7 | 12 | 10 | 2 | 4 | 3 | 3 | 3 | 1 | 6 |
| Ma05_g31160 | 9 | 3 | 5 | 6 | 7 | 7 | 7 | 5 | 7 | 4 | 4 | 3 | 2 | 2 | 3 | 2 | 4 |
| Ma05_g31440 | 0 | 3 | 2 | 68 | 266 | 2 | 0 | 43 | 12 | 88 | 71 | 2 | 26 | 2 | 89 | 9 | 2 |
| Ma06_g00910 | 0 | 7 | 1 | 5 | 1 | 1 | 15 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 3 | 0 |
| Ma06_g03570 | 0 | 0 | 4 | 2 | 0 | 0 | 0 | 3 | 5 | 3 | 3 | 4 | 5 | 7 | 4 | 3 | 7 |
| Ma06_g04210 | 0 | 0 | 5 | 2 | 1 | 0 | 0 | 5 | 5 | 2 | 7 | 4 | 3 | 4 | 3 | 2 | 5 |
| Ma06_g04240 | 1 | 0 | 1 | 1 | 4 | 0 | 0 | 7 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma06_g04270 | 11 | 1 | 1 | 1 | 0 | 1 | 2 | 3 | 6 | 3 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| Ma06_g04370 | 7 | 0 | 10 | 13 | 5 | 29 | 15 | 1 | 3 | 0 | 0 | 0 | 2 | 1 | 6 | 1 | 0 |
| Ma06_g05680 | 0 | 0 | 7 | 4 | 0 | 0 | 0 | 20 | 15 | 34 | 15 | 16 | 8 | 9 | 2 | 11 | 10 |
| Ma06_g05960 | 0 | 0 | 6 | 4 | 4 | 0 | 0 | 13 | 9 | 19 | 13 | 9 | 4 | 4 | 3 | 9 | 2 |
| Ma06_g06660 | 3 | 2 | 2 | 3 | 2 | 3 | 6 | 2 | 3 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| Ma06_g08100 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma06_g08440 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ma06_g08910 | 4 | 3 | 5 | 28 | 68 | 23 | 16 | 8 | 6 | 11 | 9 | 5 | 10 | 4 | 20 | 14 | 3 |
| Ma06_g11140 | 4 | 0 | 1 | 14 | 6 | 33 | 16 | 2 | 4 | 2 | 0 | 1 | 1 | 0 | 4 | 1 | 0 |
| Ma06_g11270 | 0 | 1 | 10 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 3 | 0 |
| Ma06_g12110 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 |
| Ma06_g12160 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 |
| Ma06_g14470 | 0 | 0 | 3 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | 0 | 1 | 3 | 0 |
| Ma06_g16350 | 0 | 1 | 0 | 3 | 4 | 2 | 5 | 1 | 2 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 |
| Ma06_g16920 | 3 | 1 | 40 | 38 | 30 | 13 | 13 | 60 | 108 | 51 | 34 | 46 | 54 | 72 | 36 | 45 | 61 |
| Ma06_g17440 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 2 |
| Ma06_g19030 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 | 8 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| Ma06_g27210 | 2 | 2 | 8 | 6 | 2 | 6 | 16 | 3 | 4 | 5 | 0 | 2 | 2 | 2 | 1 | 2 | 2 |
| Ma06_g29060 | 9 | 2 | 0 | 0 | 0 | 0 | 1 | 2 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma06_g31020 | 7 | 3 | 7 | 6 | 3 | 9 | 8 | 1 | 2 | 2 | 2 | 0 | 2 | 1 | 7 | 1 | 1 |
| Ma06_g32530 | 7 | 0 | 0 | 2 | 1 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ma06_g33100 | 4 | 6 | 3 | 3 | 2 | 3 | 3 | 4 | 5 | 3 | 2 | 6 | 3 | 4 | 2 | 2 | 5 |
| Ma06_g33190 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma06_g33430 | 0 | 1 | 11 | 2 | 1 | 1 | 0 | 8 | 20 | 7 | 2 | 4 | 3 | 5 | 3 | 4 | 3 |
| Ma06_g33920 | 0 | 7 | 10 | 4 | 3 | 0 | 1 | 5 | 5 | 4 | 3 | 6 | 10 | 13 | 8 | 9 | 9 |
| Ma06_g35430 | 6 | 15 | 28 | 31 | 49 | 14 | 31 | 20 | 37 | 22 | 13 | 8 | 33 | 24 | 63 | 25 | 17 |
| Ma06_g35620 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ma06_g37660 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma06_g38880 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma07_g00270 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ma07_g02470 | 5 | 1 | 4 | 5 | 5 | 5 | 7 | 5 | 6 | 7 | 2 | 3 | 3 | 3 | 2 | 2 | 4 |
| Ma07_g05660 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| Ma07_g05780 | 0 | 1 | 1 | 3 | 6 | 2 | 1 | 2 | 3 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 2 |
| Ma07_g08110 | 0 | 1 | 5 | 1 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Ma07_g10330 | 2 | 4 | 4 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 1 | 1 | 3 |
| Ma07_g10340 | 2 | 11 | 6 | 23 | 82 | 3 | 2 | 11 | 22 | 10 | 8 | 5 | 14 | 8 | 15 | 28 | 5 |
| Ma07_g11110 | 0 | 0 | 0 | 3 | 5 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma07_g12330 | 0 | 0 | 1 | 7 | 5 | 18 | 3 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ma07_g13590 | 0 | 0 | 1 | 6 | 4 | 16 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma07_g17600 | 1 | 1 | 8 | 2 | 4 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 2 | 2 | 2 | 2 |
| Ma07_g19350 | 2 | 0 | 1 | 22 | 18 | 44 | 23 | 4 | 15 | 0 | 0 | 1 | 9 | 1 | 26 | 11 | 0 |
| Ma07_g19470 | 3 | 0 | 15 | 7 | 5 | 6 | 6 | 54 | 80 | 62 | 33 | 42 | 10 | 14 | 6 | 9 | 10 |
| Ma07_g19700 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma07_g19720 | 0 | 0 | 7 | 7 | 26 | 1 | 0 | 5 | 11 | 6 | 3 | 1 | 4 | 2 | 12 | 1 | 1 |
| Ma07_g19880* | 1 | 1 | 5 | 4 | 0 | 0 | 2 | 10 | 14 | 7 | 11 | 8 | 9 | 12 | 3 | 8 | 12 |
| Ma07_g19890* | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma07_g20020 | 2 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Ma07_g20990 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 2 | 1 | 3 | 0 | 2 | 2 | 1 | 1 | 1 |
| Ma07_g22540 | 5 | 1 | 0 | 8 | 13 | 12 | 7 | 1 | 3 | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 0 |
| Ma07_g23060 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 2 | 4 | 2 | 3 | 1 | 1 | 2 | 1 | 1 | 1 |
| Ma07_g23180 | 3 | 1 | 1 | 3 | 8 | 1 | 0 | 4 | 14 | 1 | 1 | 0 | 3 | 4 | 2 | 1 | 5 |
| Ma07_g23230* | 0 | 171 | 9 | 2 | 2 | 1 | 3 | 34 | 6 | 10 | 12 | 106 | 9 | 1 | 2 | 33 | 2 |
| Ma07_g23240* | 16 | 23 | 7 | 9 | 9 | 12 | 11 | 9 | 7 | 10 | 12 | 6 | 5 | 6 | 3 | 3 | 7 |
| Ma07_g26530 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma08_g01300 | 6 | 0 | 3 | 6 | 12 | 3 | 7 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| Ma08_g01760 | 6 | 0 | 0 | 2 | 2 | 2 | 4 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 |
| Ma08_g02100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma08_g02450 | 1 | 3 | 5 | 2 | 2 | 1 | 3 | 7 | 7 | 10 | 9 | 1 | 5 | 3 | 13 | 2 | 3 |
| Ma08_g03420 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma08_g10260 | 1 | 0 | 3 | 7 | 16 | 3 | 1 | 4 | 12 | 1 | 1 | 3 | 13 | 18 | 6 | 7 | 19 |
| Ma08_g10600 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Ma08_g11120 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma08_g12510 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 2 | 3 | 2 | 2 | 0 | 2 | 2 | 1 | 1 | 5 |
| Ma08_g13070 | 0 | 0 | 10 | 4 | 0 | 0 | 0 | 17 | 12 | 30 | 18 | 8 | 15 | 19 | 7 | 12 | 21 |
| Ma08_g14720 | 0 | 1 | 5 | 5 | 14 | 1 | 0 | 2 | 4 | 1 | 3 | 2 | 6 | 8 | 6 | 2 | 7 |
| Ma08_g15820 | 0 | 1 | 4 | 2 | 1 | 1 | 2 | 6 | 7 | 6 | 6 | 4 | 4 | 5 | 4 | 5 | 2 |
| Ma08_g15960 | 2 | 6 | 3 | 1 | 2 | 1 | 1 | 4 | 5 | 1 | 2 | 5 | 2 | 2 | 1 | 1 | 2 |
| Ma08_g16760 | 2 | 0 | 2 | 10 | 5 | 13 | 21 | 3 | 5 | 3 | 0 | 3 | 3 | 1 | 4 | 7 | 0 |
| Ma08_g17860 | 0 | 0 | 1 | 2 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 7 | 0 |
| Ma08_g18540 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Ma08_g23390 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 6 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ma08_g25570 | 7 | 1 | 16 | 11 | 7 | 6 | 7 | 20 | 20 | 26 | 25 | 10 | 19 | 16 | 19 | 16 | 23 |
| Ma08_g25960 | 9 | 11 | 9 | 21 | 22 | 32 | 15 | 13 | 18 | 17 | 11 | 5 | 9 | 9 | 9 | 5 | 13 |
| Ma08_g26720 | 1 | 0 | 7 | 9 | 19 | 4 | 13 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Ma08_g30360 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 |
| Ma08_g31720 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma08_g32760 | 1 | 3 | 11 | 4 | 4 | 1 | 1 | 5 | 5 | 4 | 9 | 3 | 9 | 8 | 16 | 5 | 7 |
| Ma08_g34230 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 2 | 4 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 |
| Ma08_g34710 | 0 | 0 | 15 | 1 | 0 | 0 | 0 | 16 | 17 | 20 | 8 | 20 | 5 | 5 | 4 | 6 | 7 |
| Ma09_g03310 | 2 | 1 | 11 | 4 | 8 | 3 | 1 | 2 | 5 | 1 | 1 | 2 | 3 | 3 | 5 | 1 | 3 |
| Ma09_g03740 | 8 | 1 | 3 | 4 | 5 | 5 | 4 | 4 | 6 | 3 | 3 | 4 | 2 | 2 | 2 | 2 | 2 |
| Ma09_g04930 | 0 | 2 | 12 | 2 | 0 | 0 | 0 | 10 | 7 | 22 | 7 | 3 | 5 | 5 | 9 | 4 | 3 |
| Ma09_g06730 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma09_g08140 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 3 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma09_g08260 | 15 | 1 | 9 | 6 | 8 | 4 | 3 | 9 | 9 | 13 | 10 | 4 | 8 | 9 | 8 | 6 | 8 |
| Ma09_g09400 | 0 | 0 | 7 | 1 | 0 | 0 | 1 | 1 | 2 | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| Ma09_g09720 | 1 | 0 | 3 | 1 | 0 | 1 | 0 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 0 | 1 | 1 |
| Ma09_g10800 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 2 | 1 | 1 | 0 | 1 | 0 |
| Ma09_g11770 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma09_g13170 | 0 | 0 | 5 | 3 | 0 | 0 | 0 | 6 | 7 | 4 | 10 | 4 | 9 | 12 | 4 | 5 | 13 |
| Ma09_g14260 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 2 | 1 |
| Ma09_g15050 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| Ma09_g15130 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 1 | 1 |
| Ma09_g15440 | 0 | 0 | 1 | 2 | 5 | 1 | 0 | 5 | 9 | 3 | 2 | 4 | 1 | 1 | 2 | 2 | 1 |
| Ma09_g15940 | 2 | 0 | 2 | 3 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma09_g16940 | 3 | 0 | 0 | 4 | 5 | 8 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 1 |
| Ma09_g16980 | 0 | 1 | 7 | 0 | 0 | 0 | 0 | 1 | 4 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ma09_g20280 | 0 | 0 | 62 | 8 | 0 | 0 | 0 | 44 | 31 | 73 | 45 | 27 | 26 | 30 | 21 | 29 | 23 |
| Ma09_g22730 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ma09_g23100 | 32 | 2 | 3 | 36 | 45 | 65 | 32 | 10 | 9 | 21 | 9 | 1 | 5 | 2 | 12 | 4 | 3 |
| Ma09_g24640 | 0 | 0 | 8 | 4 | 3 | 0 | 8 | 7 | 12 | 6 | 3 | 6 | 4 | 5 | 2 | 6 | 4 |
| Ma09_g25010 | 4 | 0 | 2 | 4 | 3 | 4 | 7 | 1 | 1 | 1 | 2 | 0 | 2 | 1 | 2 | 1 | 2 |
| Ma09_g25590 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma09_g27990 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 2 | 1 | 1 | 3 |
| Ma09_g28970 | 1 | 0 | 0 | 7 | 26 | 2 | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma09_g29010 | 0 | 2 | 32 | 6 | 0 | 1 | 3 | 25 | 26 | 32 | 15 | 25 | 15 | 16 | 7 | 17 | 22 |
| Ma09_g29660 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g01730 | 0 | 0 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g01750 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g04420 | 0 | 0 | 1 | 4 | 10 | 2 | 2 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 3 | 0 | 0 |
| Ma10_g04920 | 3 | 0 | 0 | 1 | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ma10_g05260 | 0 | 0 | 0 | 1 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g05680 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g06140 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g09100 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g09370 | 0 | 0 | 4 | 3 | 1 | 0 | 5 | 2 | 2 | 3 | 3 | 1 | 3 | 3 | 2 | 4 | 2 |
| Ma10_g10820 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 4 | 14 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 |
| Ma10_g11100 | 0 | 1 | 6 | 2 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 7 | 0 |
| Ma10_g13000 | 3 | 0 | 5 | 10 | 2 | 21 | 16 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 |
| Ma10_g13640 | 2 | 1 | 40 | 31 | 30 | 34 | 12 | 22 | 25 | 19 | 20 | 25 | 35 | 48 | 21 | 28 | 44 |
| Ma10_g14150 | 0 | 1 | 1 | 2 | 2 | 3 | 4 | 1 | 3 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| Ma10_g14950 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 4 | 8 | 1 | 2 | 3 | 4 | 4 | 2 | 3 | 7 |
| Ma10_g16050 | 5 | 1 | 10 | 28 | 74 | 23 | 13 | 9 | 14 | 16 | 4 | 3 | 7 | 4 | 16 | 3 | 4 |
| Ma10_g17650 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 4 | 13 | 1 | 0 | 0 | 2 | 2 | 2 | 4 | 1 |
| Ma10_g18840 | 3 | 1 | 5 | 2 | 1 | 2 | 2 | 4 | 6 | 3 | 6 | 2 | 3 | 5 | 2 | 2 | 5 |
| Ma10_g19130 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| Ma10_g19820 | 1 | 0 | 2 | 6 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 12 | 9 | 0 |
| Ma10_g19970 | 1 | 0 | 0 | 2 | 6 | 3 | 1 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g24510 | 9 | 4 | 2 | 7 | 16 | 6 | 4 | 3 | 4 | 2 | 4 | 2 | 1 | 1 | 1 | 1 | 2 |
| Ma10_g25660 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g26540 | 5 | 7 | 6 | 4 | 3 | 4 | 4 | 6 | 8 | 4 | 4 | 6 | 4 | 5 | 2 | 3 | 6 |
| Ma10_g26660 | 2 | 0 | 6 | 3 | 1 | 2 | 2 | 10 | 14 | 11 | 8 | 5 | 4 | 3 | 3 | 5 | 6 |
| Ma10_g29230 | 6 | 0 | 1 | 4 | 6 | 9 | 3 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 |
| Ma10_g29290 | 4 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| Ma10_g29660 | 2 | 0 | 1 | 15 | 60 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma10_g29900 | 5 | 1 | 9 | 10 | 7 | 11 | 11 | 8 | 8 | 7 | 9 | 8 | 9 | 11 | 10 | 10 | 7 |
| Ma11_g00330 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Ma11_g00350 | 0 | 0 | 6 | 5 | 13 | 1 | 0 | 3 | 5 | 2 | 2 | 2 | 5 | 7 | 5 | 3 | 6 |
| Ma11_g02310 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 4 | 0 | 2 | 2 | 1 | 1 | 4 |
| Ma11_g03860 | 0 | 0 | 2 | 13 | 47 | 1 | 0 | 5 | 14 | 4 | 1 | 0 | 2 | 1 | 5 | 1 | 1 |
| Ma11_g04680 | 20 | 2 | 0 | 32 | 6 | 57 | 64 | 0 | 1 | 1 | 0 | 0 | 4 | 0 | 7 | 10 | 0 |
| Ma11_g06880 | 22 | 4 | 15 | 13 | 7 | 19 | 12 | 31 | 41 | 39 | 30 | 14 | 7 | 6 | 4 | 9 | 11 |
| Ma11_g07330 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma11_g07530 | 0 | 0 | 5 | 2 | 1 | 1 | 1 | 6 | 3 | 14 | 4 | 2 | 3 | 5 | 1 | 4 | 2 |
| Ma11_g08730 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| Ma11_g10680 | 1 | 2 | 5 | 2 | 1 | 0 | 3 | 12 | 21 | 20 | 6 | 3 | 6 | 7 | 5 | 8 | 6 |
| Ma11_g10710 | 0 | 1 | 2 | 1 | 4 | 0 | 0 | 2 | 5 | 2 | 1 | 2 | 3 | 1 | 6 | 2 | 1 |
| Ma11_g11300 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Ma11_g11940 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma11_g14670 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 5 | 0 |
| Ma11_g15740 | 4 | 0 | 2 | 5 | 7 | 4 | 6 | 6 | 9 | 4 | 6 | 5 | 2 | 1 | 3 | 2 | 1 |
| Ma11_g16150 | 1 | 2 | 3 | 10 | 37 | 2 | 1 | 4 | 7 | 4 | 6 | 1 | 3 | 3 | 3 | 2 | 4 |
| Ma11_g16430 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ma11_g19220 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 7 | 0 |
| Ma11_g21160 | 1 | 0 | 9 | 3 | 0 | 1 | 12 | 2 | 4 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ma11_g21730 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 2 | 2 | 3 | 0 | 1 | 2 | 4 | 0 | 2 | 2 |
| Ma11_g21820 | 0 | 0 | 0 | 2 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ma11_g23010 | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 3 | 3 | 5 | 2 | 2 | 2 | 3 | 1 | 2 | 3 |
| Ma11_g23420 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The intensity of green background is correlated to expression level given in FPKM.
* indicate paralogs.
Our expression analyses revealed that M. acuminata MYBs have diverse expression patterns in different organs. Many of the MaMYBs exhibited low transcript abundance levels with expression in only one or a few organs. This is consistent with other transcription factor genes, typically found to be expressed in this manner due to functional specificity and diversity. The highest number of expressed MaMYB genes (221; 75.4%) is observed in roots, followed by pulp (209; 71.3%), leaf (203; 69.3%), peel (196; 66.9%) and embryonic cell suspension (129; 44%). The fewest MaMYB genes are expressed in seedlings (98; 33.4%) in the considered dataset. 40 MaMYB genes (13.7%) were expressed in all samples analysed (albeit with varying expression levels), which suggested that these MaMYBs play regulatory roles at multiple developmental stages in multiple tissues. 13 MaMYB genes (4.4%) lacked expression information in any of the analysed samples, possibly indicating that these genes are expressed in other organs (e.g. pseudostem, flower, bract), specific cells, at specific developmental stages, under special conditions or are pseudogenes. 280 MaMYBs (95.6%) are expressed in at least one analysed sample, although the transcript abundance of some genes was very low. Some MaMYB genes were expressed in all analysed RNA-Seq samples at similar levels (e.g. R2R3-MYBs Ma06_g33440 and Ma11_g06880) while others show variance in transcript abundance with low (no) levels in one or several organs and high levels in others (or vice versa). For example, Ma02_g16570, Ma03_g09310, Ma07_g11110, Ma10_g01730, Ma10_g05260 and Ma10_g09100 show organ-specific expression, as their transcripts were exclusively detected in leaves, which hints to leaf-specific functionality. Ma03_g07840 and Ma07_g19470 were found to be predominantly expressed in pulp, showing expression also in other analysed organs, but not in the seedling. Overall, these results suggests that the corresponding MaMYB regulators are limited to distinct organs, tissues, cells or conditions.
Some paralogous MaMYB genes clustered in the genome (Tables 1 and 2) showed different expression profiles, while other clustered paralogous MaMYB genes did not. For example, the clustered R2R3-MYB genes Ma07_g19880 and Ma07_g19890 (both in the proanthocyanidin-related cluster 25): while Ma07_g19880 is expressed in pulps, peels, roots, and leaves, Ma07_g19890 is nearly not expressed in the analysed organs. The expression pattern of Ma03_g07840 and Ma03_g07850 (also both in the proanthocyanidin-related cluster 25) is, in contrast, very similar, with low expression in root, leaf, pulp and peel, but no expression in embyonic cells and roots. These results could point to functional redundancy of the genes Ma03_g07840 and Ma03_g07850, while Ma07_g19880 and Ma07_g19890 could be (partly) involved in distinct, tissue-specific aspects of specialised mebabolite biosynthesis. The functional categorisation and its expression domains make Ma07_g19880 a good candidate to encode a proanthocyanidin biosynthesis regulator in developing banana fruits (pulp and peel), and thus an excellent target for genetic manipulation of proanthocyanidin content, known to influence senescence of banana fruit [46], to maintain the freshness of harvested banana fruit.
Conclusions
The present genome-wide identification, chromosomal organisation, functional classification and expression analyses of M. acuminata MYB genes provide a first step towards cloning and functional dissection to decode the role of MYB genes in this economically interesting species. Further, knowledge of selected MYB genes can be biotechnologically utilized for improvement of fruit quality, yield, disease resistance, tolerance to biotic and abiotic stresses, and the biosynthesis of pharmaceutical compounds.
Supporting information
(DOCX)
(DOCX)
(FASTA)
(FASTA)
For use in genome viewers/browsers on the M. accuminata pseudochromosomes.
(GFF)
Acknowledgments
We are grateful to Melanie Kuhlmann for excellent technical assistance. We thank Nathanael Walker-Hale for language editing.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The research stay of AP at Weisshaar lab was supported by a grant from Alexander von Humbold Foundation (AvH) (AvH-IND/1184925). The publication costs were supported by Deutsche Forschungsgemeinschaft (DFG) and the Open Access Publication Fund of Bielefeld University. This is a general fund of Bielefeld University with DFG support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Frison EA, Sharrock SL. The economic, nutritional and social importance of bananas in the world In: Cea Picq, editor. Bananas and Food Security. Montpellier, France: INIBAP; 1999. p. 21–35. [Google Scholar]
- 2.Langridge WHR. Edible Vaccines. Scientific American Sp. 2006;16:46–53. [Google Scholar]
- 3.DHont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–7. 10.1038/nature11241 [DOI] [PubMed] [Google Scholar]
- 4.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–92. 10.1038/nature08449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–10. 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- 6.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends in Plant Science. 2010;15(10):573–81. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 7.Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, et al. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nature Structural Biology. 1996;3(2):178–87. 10.1038/nsb0296-178 [DOI] [PubMed] [Google Scholar]
- 8.Jia L, Clegg MT, Jiang T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiology. 2004;134(2):575–85. 10.1104/pp.103.027201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology. 2001;4:447–56. 10.1016/s1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Yu W, Zhang X, Wang G, Cao F, Cheng H. Identification and expression analysis under abiotic stress of the R2R3-MYB genes in Ginkgo biloba L. Physical and Molecular Biology of Plants. 2017;23(3):503–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salih H, Gong W, He S, Sun G, Sun J, Du X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genetics. 2016;17(1):129 10.1186/s12863-016-0436-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology. 2008;8:83 10.1186/1471-2229-8-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 14.Droc G, Larivière D, Guignon V, Yahiaoui N, This D, Garsmeur O, et al. The banana genome hub. Database (Oxford). 2013;2013(bat035). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution. 2016;33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin G, Baurens FC, Droc G, Rouard M, Cenci A, Kilian A, et al. Improvement of the banana "Musa acuminata" reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genetics. 2016;17(243). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haak M, Vinke S, Keller W, Droste J, Rückert C, Kalinowski J, et al. High Quality de Novo Transcriptome Assembly of Croton tiglium. Frontiers in Molecular Biosciences. 2018;5:62 10.3389/fmolb.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 21.Tak H, Negi S, Ganapathi TR. Overexpression of MusaMYB31, a R2R3 type MYB transcription factor gene indicate its role as a negative regulator of lignin biosynthesis in banana. PLoS ONE. 2017;12(2):e0172695 10.1371/journal.pone.0172695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negi S, Tak H, Ganapathi TR. Cloning and functional characterization of MusaVND1 using transgenic banana plants. Transgenic Research. 2015;24(3):571–85. 10.1007/s11248-014-9860-6 [DOI] [PubMed] [Google Scholar]
- 23.Fan ZQ, Ba LJ, Shan W, Yun-Yi X, Wang-Jin L, Jian-Fei K, et al. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. The Plant Journal. 2018;96(6):1191–1205. 10.1111/tpj.14099 [DOI] [PubMed] [Google Scholar]
- 24.Yang QS, Gao J, He WD, Dou TX, Ding LJ, Wu JH, et al. Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genomics. 2015;16:446 10.1186/s12864-015-1551-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du H, Feng BR, Yang SS, Huang YB, Tang YX. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7(6):e37463 10.1371/journal.pone.0037463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajiebrahimi A, Owji H, Hemmati S. Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome. 2017;60(10):797–814. 10.1139/gen-2017-0059 [DOI] [PubMed] [Google Scholar]
- 27.Wu W, Yang YL, He WM, Rouard M, Li WM, Xu M, et al. Whole genome sequencing of a banana wild relative Musa itinerans provides insights into lineage-specific diversification of the Musa genus. Scientific Reports. 2016;6(31586):srep31586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castañeda NEN, Alves GSC, Almeida RM, Amorim EP, Fortes Ferreira C, Togawa RC, et al. Gene expression analysis in Musa acuminata during compatible interactions with Meloidogyne incognita. Annals of Botany. 2017;119(5):915–30. 10.1093/aob/mcw272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song C, Yang Y, Yang T, Ba L, Zhang H, Han Y, et al. MaMYB4 recruits histone deacetylase MaHDA2 and modulates the expression of omega-3 fatty acid desaturase genes during cold stress response in banana fruit. Plant & cell physiology. 2019; 60(11):2410–2422. 10.1093/pcp/pcz142 [DOI] [PubMed] [Google Scholar]
- 30.Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biology. 2012;12(106):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stracke R, Holtgräwe D, Schneider J, Pucker B, Rosleff Sörensen T, Weisshaar B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biology. 2014;14(249). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. Journal of Molecular Evolution. 2004;58(4):424–41. 10.1007/s00239-003-2564-9 [DOI] [PubMed] [Google Scholar]
- 33.Du H, Liang Z, Zhao S, Nan MG, Phan Tran LS, Lu K, et al. The Evolutionary History of R2R3-MYB Proteins Across 50 Eukaryotes: New Insights Into Subfamily Classification and Expansion. Scientific Reports. 2015;5(11037). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal. 2001;28(3):319–32. [DOI] [PubMed] [Google Scholar]
- 35.Baum D. Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups. Nature Education. 2008;1(1):190. [Google Scholar]
- 36.Zhong Y, Yin H, Sargent DJ, Malnoy M, Cheng ZM. Species-specific duplications driving the recent expansion of NBS-LRR genes in five Rosaceae species. BMC Genomics. 2015;16(77):s12864-015-1291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta RS, Bryant D, Rosenberg NA. The probability of monophyly of a sample of gene lineages on a species tree. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(29):8002–9. 10.1073/pnas.1601074113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–93. 10.1016/0092-8674(91)90523-2 [DOI] [PubMed] [Google Scholar]
- 39.Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;24(5):473–83. [DOI] [PubMed] [Google Scholar]
- 40.Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiology. 2009;149(2):981–93. 10.1104/pp.108.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockington SF, Alvarez-Fernandez R, Landis JB, Alcorn K, Walker RH, Thomas MM, et al. Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Molecular Biology and Evolution. 2013;30(3):526–40. 10.1093/molbev/mss260 [DOI] [PubMed] [Google Scholar]
- 42.Sonderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One. 2007;2(12):e1322 10.1371/journal.pone.0001322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, Flugge UI. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. The Plant Journal. 2007;50(5):886–901. 10.1111/j.1365-313X.2007.03099.x [DOI] [PubMed] [Google Scholar]
- 44.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology. 2006;60(1):107–24. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- 45.Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. The Plant Journal. 2004;40:22–34. 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Li F, Li Y, Wang Y, Wang C, Yuan D, et al. Exogenous procyanidin treatment delays senescence of harvested banana fruit by enhancing antioxidant responses and in vivo procyanidin content. Postharvest Biology and Technology. 2019;158 10.1016/j.postharvbio.2019.110999 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(FASTA)
(FASTA)
For use in genome viewers/browsers on the M. accuminata pseudochromosomes.
(GFF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.