Abstract
Artificial intelligence (AI) is transforming healthcare delivery. The digital revolution in medicine and healthcare information is prompting a staggering growth of data intertwined with elements from many digital sources such as genomics, medical imaging and electronic health records. Such massive growth has sparked the development of an increasing number of AI-based applications that can be deployed in clinical practice. Pulmonary specialists who are familiar with the principles of AI and its applications will be empowered and prepared to seize future practice and research opportunities. The goal of this review is to provide pulmonary specialists and other readers with information pertinent to the use of AI in pulmonary medicine. First, we describe the concept of AI and some of the requisites of machine learning and deep learning. Next, we review some of the literature relevant to the use of computer vision in medical imaging, predictive modelling with machine learning, and the use of AI for battling the novel severe acute respiratory syndrome-coronavirus-2 pandemic. We close our review with a discussion of limitations and challenges pertaining to the further incorporation of AI into clinical pulmonary practice.
Short abstract
Artificial intelligence (AI) is changing the landscape in medicine. AI-based applications will empower pulmonary specialists to seize modern practice and research opportunities. Data-driven precision medicine is already here. https://bit.ly/324tl2m
Introduction
The sheer volume of healthcare-related data generated from digital sources such as high-resolution imaging, genomic studies, continuous biosensor monitoring, and electronic health records is staggering: 150 exabytes (an exabyte is one quintillion (1018) bytes or one billion gigabytes) in the United States alone, and it grows 48% annually [1]. Its analysis is overwhelming for physicians and healthcare scientists, who, like other humans, are limited by an ability to process only six or fewer data points simultaneously [2]. In contrast, computers can seamlessly analyse millions and even billions of data points, making artificial intelligence (AI), and its subfields such as machine learning and deep learning, a potential game changer for modern healthcare delivery.
Despite many obstacles, not the least of which is that AI is still in its infancy [3], AI shows great promise for changing the way we practice pulmonary medicine. A PubMed literature search performed on May 5, 2020 using the search terms “artificial intelligence OR machine learning OR deep learning AND pulmonary AND medicine” identified 930 articles published between 2015 and 2019. This is a five-fold increase compared to only 171 articles found in a similar search for the years 2010–2014. Furthermore, our query for full-text, English language systematic and narrative reviews published from 2018 onward using the search terms “machine learning in respiratory medicine” identified 32 scientific papers: 14 pertained to cardiac or critical care medicine, 14 were dedicated to specific lung disorders, and four described the role of machine learning in general pulmonary medicine [4–7].
However, incorporating AI into how we process data and make medical decisions takes time. In addition to a general lack of awareness about its potential applications, many healthcare professionals perceive AI as a threat to medical jobs [8]. Validation studies, particularly prospective work in clinical settings, are lacking, and some fear that over-reliance on AI might result in de-skilling our workforce as well as prompt unsatisfactory outcomes, especially if algorithms are poorly generalisable or built on data that are not well structured [9].
In this review, we describe several requisites of machine learning and deep learning. Using mostly publications from the last 4 years, we also provide examples of how AI is used in pulmonary medicine: for computer vision in medical imaging, predictive modelling with machine learning, and in the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) pandemic. Given the narrative nature of our work, articles were carefully selected to complement other reviews and to provide readers with a general understanding of these topics. We close our review with a discussion of limitations and challenges pertaining to the further incorporation of AI into clinical pulmonary practice.
A brief description of AI and machine learning
The field of AI was born at Dartmouth College (Hanover, NH, USA) in 1956 when a group of computer scientists gathered to discuss mathematical theorems, language processing, game theory and how computers learn from analysing training examples. At first, a rules-based system governed AI. By the 1980s, medical uses were noted, including applications in pulmonary medicine [10–12]. Rules could represent knowledge coded into the system [13], providing direction for different clinical scenarios. Essentially, the programmer defined what the computer had to do. This system could help guide decision-making for interpreting ECGs, evaluating the risk of myocardial infarction and diagnosing diseases [14]. Originally, the concept of probabilities was applied to represent uncertainties [15], and logic-based explicit expressions of decision-rules and human-authored updates were required. Performance was limited by low sensitivity, incomplete medical knowledge datasets and insufficient ability to integrate probabilistic reasoning.
With machine learning, modern systems are considered artificially intelligent because they use algorithms that enable computers (the machine) to learn functions from a specific and potentially ever-changing dataset (a dataset is a set of samples). A function is the deterministic mapping of output values from a set of input values such that the output value is always the same for any specific set of input values (for example, 3×3 is always 9). By at least one definition, an algorithm is a process the computer uses to analyse the dataset and identify patterns. Primarily, the computer programme can learn, and software can adapt its behaviour automatically to better match the requirements of the task [16].
Functions learned from the data can be represented as simple arithmetic operations or complex neural network architectures. In machine-learning models, training (derivation) and testing (validation) datasets are used to help alleviate algorithmic bias (systemic errors that result in unfair outcomes such as preferring one arbitrary group over others) [17–20]. To improve reproducibility, cross-validation is commonly performed using multiple splits within the training set to reduce the effects of randomness of the split, especially if datasets are small. While details of how a particular cross-validation scheme is performed depend on the data and research question, the goal remains the same: to create an accurate model that predicts outcomes in an externally validated dataset with minimal algorithmic bias [21].
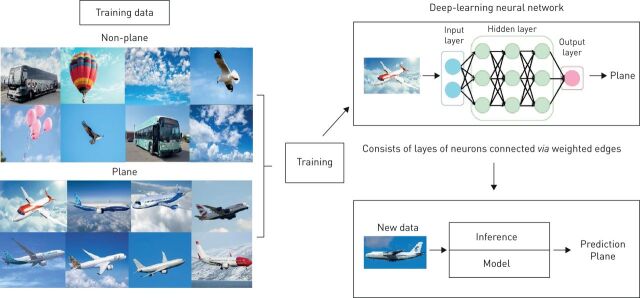
In medicine, several machine-learning models are commonly used (table 1). For example, an artificial neural network (ANN) is a subtype of machine learning that comprises a series of algorithms designed to recognise underlying relationships in a set of data using a process inspired by the way neurons communicate in the human brain. ANNs consist of multiple layers of “neurons” where each neuron in a given layer is connected to every other neuron in adjacent layers, and the weight attributed to each connection is optimised through an iterative process using linear algebra and calculus. The optimisation process is both science and art, where theory often follows heuristics (figure 1).
TABLE 1.
Examples of machine-learning algorithms
| Example | Description | |
| Supervised learning algorithm | ||
| Regularised regression | LASSO | An extension of classic regression algorithms in which a penalty is enforced to the fitted model to minimise its complexity and reduce the risk of overfitting |
| Tree-based model | Classification and regression trees, random forest, gradient boosted trees (XGBoost) | Based on decision trees (a decision support tool which is a sequence of “if-then-else” splits are derived by iteratively separating data into groups based on the relationship between attributes and outcomes) |
| Support vector machine | Linear, hinge loss, radial basis function kernel | Represents data in a multidimensional feature space and fits a “hyperplane” that best separates data based on outcomes of interest |
| KNN | KNN | Represents data in a multidimensional feature space and uses local information about observations closest to a new dataset to predict outcomes for the new dataset |
| Neural network | Deep neural networks, ANNs | Nonlinear algorithms built using multiple layers of nodes that extract features from the data and perform combinations that best predict outcomes |
| Unsupervised learning algorithm | ||
| Dimensionality reduction algorithms | Principal component analysis, linear discriminant analysis | Exploits inherent structure to transform data from high-dimensional space into a low-dimensional space which retains some meaningful attributes of the original data |
| LCA | LCA | Identifies hidden population subgroups (latent classes) in the data. Used in datasets with complex constructs that have multiple behaviours. The probability of class membership is indirectly estimated by measuring patterns in the data |
| Cluster analysis | K-means, hierarchical cluster analysis | Uses inherent structures in the data to best organise data into subgroups of maximum commonality based on some distance measure between features |
| Reinforcement learning algorithm | ||
| Reinforcement learning | Markov decision process and Q learning | Provides tools to optimise sequences of decision for the best outcomes or to maximise rewards. Learns by trial and error. An action is reinforced with the action that results in a positive outcome (reward), and vice versa. The algorithm can improve performance in scenarios where a learning system can choose to repeat known decisions (exploitation) or make novel decisions expecting to gain even greater rewards (exploration) |
KNN: K-nearest neighbour; LCA: latent class analysis; LASSO: least absolute shrinkage and selection operator; ANN: artificial neural network.
FIGURE 1.
An example of an artificial neural network. Simple image classification: the model is trained on a few thousand images of planes and non-planes, then is later able to predict if a given image is a plane or not.
Machine learning is challenging, in part, because of difficulties for conventional statistical analyses such as logistic regression to isolate relationships between predictors and outcomes, especially when relationships are nonlinear and when the number of variables is large. Furthermore, most datasets include a large amount of noise, and information provided by the data may not allow a single best solution. This prompts the computer to identify a series of possible solutions that match the data.
In 2014, only AliveCor's algorithm for detection of atrial fibrillation was approved for clinical use. Today, it appears that medical specialties such as radiology, cardiology, dermatology and pathology are most conducive to AI-based applications. To date, >75 AI algorithms are approved by the US Food and Drug Administration (FDA), and one report states that AI-based medical imaging investments have grown exponentially to USD 1.17 billion [22, 23]. To our knowledge, a number of algorithms related to pulmonary medicine have received 510(k) premarket approval (legal, regulatory recognition that a medical device is safe and effective) from the FDA and several more are CE-marked (Conformité Européenne is the mandatory regulatory marking for products sold within the European Economic Area) [22, 24] (table 2). Lagging growth in pulmonary and critical care medicine compared with some other medical fields [22] warrants a need for increased awareness among physicians of this specialty.
TABLE 2.
Examples of Conformité Européenne (CE)-marked and US Food and Drug Administration (FDA)-approved artificial intelligence (AI) algorithms
| Name of algorithm/parent company | Short description | CE-marked | US FDA-approved |
| Lung nodule (chest CT) | |||
| InferRead CT Lung (InferVision) | CT lung nodule detection, report generation, multi-time point analysis and aims to aid early-stage diagnosis | Yes | Yes |
| Lung AI (Arterys) | Automatic detection of solid, part solid and ground-glass nodules and supports Lung-RADS reporting and multi-time point analysis | Yes | Yes |
| Veolity (MeVis Medical Solutions) | CT lung nodule detection, segmentation, quantification, temporal registration and nodule comparison | Yes | Yes |
| ClearReadCT compare (Riverain) | Compares, tracks nodules and provides nodule volumetric changes over time for solid, part-solid and ground-glass nodules | Yes | Yes |
| JLD-01K (JLK Inc.) | CT lung nodule detection, measures the diameter and volume of the nodules, categorises LungRADS category | Yes | No |
| VUNO Med-LungCT (VUNO) | Quantifies pulmonary nodules and automatically categorises the Lung-RADS | Yes | No |
| Veye Chest (Aidence) | CT lung nodule detection, nodule classification, volume quantification and growth calculation | Yes | No |
| RevealAI Lung (Mindshare Medical) | Provides a malignancy similarity index from lung CT scans that aids risk assessment of lung nodules | Yes | No |
| COVID-19 pneumonia (chest CT) | |||
| Icolung (Icometrix) | Objective quantification of disease burden in COVID-19 patients | Yes | No |
| InferRead CT Pneumonia (InferVision) | An alert system that warns if there is a suspected positive case of COVID-19 | Yes | No |
| Pulmonary embolism (chest CT) | |||
| Aidoc (Aidoc) | Analysis of CT images and flags presence of pulmonary embolism | Yes | Yes |
| Emphysema/COPD/ILD (chest CT) | |||
| LungQ (Thirona) | Lung volume segmentation and quantification, volume density analysis, airway morphology, fissure completeness analysis | Yes | Yes |
| LungPrint Discovery (VIDA) | Provides visual and quantitative information relevant to COPD and ILD. Provides high-density tissue quantification by lobe, trachea analysis and quantification | Yes | Yes |
| Lung Density Analysis (Imbio) | Provides visualisation and quantification of lung regions with abnormal tissue density. Provides a mapping of normal lung, air-trapping and areas of persistent low density | Yes | Yes |
| Lung texture analysis (Imbio) | Transforms a chest CT into a map of the lung textures to identify ILDs and other fibrotic conditions | Yes | No |
| Lung densities (Quibim) | Provides quantification of imaging biomarkers: lung volumes, vessel volumes and emphysema volume ratios | Yes | No |
| Pneumothorax (chest radiography) | |||
| Red Dot (behold.ai) | Assessment of adult chest radiographs with features suggestive of pneumothorax | Yes | Yes |
| Triage (Zebra Medical Vision) | Identifies findings suggestive of pneumothorax based on chest radiography; outputs an alert | Yes | Yes |
| Pulmonary function tests | |||
| ArtiQ.PFT (ArtiQ) | Automated pulmonary function test interpretation | Yes | No |
CT: computed tomography; COVID-19: coronavirus disease 2019; ILD: interstitial lung disease; Lung-RADS: Lung Imaging Reporting and Data System.
Requisites of machine learning
Machine learning uses datasets, functions and algorithms to search through data and identify output values that best match the data. However, statistical analysis, human expertise, and trial and error modelling of potential neural networks are labour-intensive. Therefore, a precise determination of how well each function matches the data and leads to an acceptable output is also needed. In AI jargon, this measure of quality, of how well each function is used to match the data, is referred to as “fitness”. How the elements of machine learning (data, functions, algorithms and fitness) work together defines three different categories of machine learning, each of which plays a role in the elaboration of AI-based medical decision-making (table 1).
Supervised learning refers to an algorithm in which each input of the dataset is matched to a specific output value. The dataset must be labelled to predict known outcomes. For example, the input could be a clinical characteristic and the output an event of interest (e.g. mortality). Once the algorithm is successfully trained, it will be capable of making outcome predictions when applied to a new dataset. To be used successfully, each input value of the dataset must have a target output value. This can require a large amount of data and input can be very time-consuming. Selecting the appropriate output target value often requires human expertise. With supervised learning, the algorithm can subsequently use each output target value to help the learning process by comparing the outputs created by a specific function with known outputs specified in the dataset, and use a fitness measure to search for the best function [19, 25, 26].
Unsupervised learning refers to an algorithm that finds clusters within the input data. The algorithm identifies functions that map input datasets into clusters so that data points within each cluster are more similar than data points in other clusters [19, 20, 27]. An example of unsupervised learning is in data mining of electronic medical records [28], where goals are to reveal patterns for patients who share clinical, genetic or molecular characteristics that might theoretically respond to targeted therapies directed at a specific pathophysiology as in precision medicine.
Reinforcement learning is useful for control tasks such as those needed in robotics, or in constructing policies where medical decisions require sequenced procedures; for example, whether a robot should move forward or backward, or whether a treatment regimen requires changes in drug-dosing or clinical reassessment. Reinforcement learning has been used to elaborate dynamic treatment regimens for cardiovascular diseases, in mental illnesses where long-term treatment involves a sequence of medical interventions, and in radiation therapy where treatment is geared to destroying cancer cells while sparing as many normal cells as possible. Learning is often by trial and error: the function is viewed as an action for the agent to interact favourably with its environment. If the action results in a positive outcome (often referred to as a “reward”), that action is reinforced. An obvious challenge lies in the design of training mechanisms that distribute positive or negative rewards appropriately. Examples include algorithms designed for studying protocols in sepsis, sedation and mechanical ventilation [29, 30]. The technique is particularly helpful when outcomes are not clearly supported by high-quality evidence from randomised controlled trials or meta-analyses, or when input data include a wide selection of physiological data, clinical notes, vital sign time series and radiological images difficult to analyse using supervised or unsupervised learning algorithms [29].
What is deep learning?
Deep learning is a subfield of AI in which features needed for a particular task are automatically learned from the raw data. Deep learning requires costly and time-consuming human effort to accumulate raw data, provide statistical analysis and domain expertise, as well as build experimental models with specific feature sets and potentially complex algorithms.
Deep learning is already an integral part of our daily lives. It can be applied to all three machine-learning modalities, and with increasing computational power and the massive datasets that make up “big data”, it is especially promising for healthcare applications. Deep learning is used for common tasks such as internet data searches and speech recognition on our smart phones. As our ability to manipulate images [31], language [32] and speech [33] increases, and with the advent of new hardware such as neuromorphic chips and quantum computing, larger datasets are becoming analysable. A number of commercial companies and academic institutions are building massive datasets in the hope that deep learning derived algorithms will favourably impact disease prediction, prevention, diagnosis, treatment and healthcare-related economics [34].
Deep learning often outperforms other machine-learning modalities when these huge datasets are manipulated. This is how DeepMind's AlphaGo defeated the world's greatest Go player [35]. It is also how millions of pixels are analysed to assist with medical image processing or facial recognition [36]. It often relies on principles similar to the working of the human brain, using multiple layers of ANNs composed of input and output layers of data representations called neurons. There are also hidden layers (neurons that cannot be defined as either input or output layers). All layers are arranged sequentially such that a representation of one layer is fed into the following layer [37], the depth of the network being linked to the number of hidden and output layers (figure 1).
Inspired by the human brain with its billions of neurons interconnected via dendrites and axons, ANNs are networks of information processing units. Complex nonlinear mappings of input/output relationships start from simple interactions between a large number of data points. The deeper the network, the more powerful the model in regards to its ability to learn complex nonlinear mappings. Deep learning networks are trained by “iteration”, a process of running and re-running networks while optimising neuronal parameters to improve performance and minimise errors. Massive datasets drive technological innovation and a quest for better algorithms and faster computational hardware.
Applications in clinical pulmonary medicine
AI, particularly pattern recognition using deep and machine learning, has numerous potential applications in pulmonary medicine, whether in image analysis, decision-making or prognosis prediction [5–7]. In this section, we provide examples of how AI is also used for computer vision in medical imaging, predictive modelling with machine learning, and in battling the novel SARS-CoV-2 pandemic (table 3).
TABLE 3.
Model training, validation, algorithm type and data source for selected studies in pulmonary medicine
| First author [ref.] | Population/type of study | Source of data | Main findings | Reference standard/ground truth | Algorithm type | Datasets | Type of internal validation/availability of external validation |
| Solid pulmonary nodules/masses | |||||||
| Ardila [38] | CT chest of lung cancer screening patients/retrospective | NLST | Prediction of cancer risk based on CT findings For testing dataset, ROC of 94.4% (95% CI 91.1–97.3%) For validation set, AUC of 95.5% (95% CI 88.0–98.4%) |
Histology; follow-up | CNN | 42 290 CT images from 14 851 patients | Not reported/yes |
| Baldwin [39] | CT chest of lung cancer screening patients/retrospective | The IDEAL study (Artificial Intelligence and Big Data for Early Lung Cancer Diagnosis) | The AUC for CNN was 89.6% (95% CI 87.6–91.5%), compared with 86.8% (95% CI 84.3–89.1%) for the Brock model (p≤0.005) | Histology; follow-up | CNN | 1397 nodules in 1187 patients | Not reported/yes |
| Massion [40] | CT chest of lung cancer screening patients/retrospective | NLST for model derivation and internal validation/externally tested on cohorts from two academic institutions | The AUC for CNN was 83.5% (95% CI 75.4–90.7%) and 91.9% (95% CI 88.7–94.7%) on two different cohorts (Vanderbilt and Oxford University) | Histology; follow-up | CNN | 14 761 benign nodules from 5972 patients, and 932 malignant nodules from 575 patients | Not reported/yes |
| Ciompi [41] | CT chest of lung cancer screening patients/retrospective | Training dataset from the Multicentric Italian Lung Detection trial and validation dataset from the Danish Lung Cancer Screening Trial | CNN can achieve performance at classifying nodule type within the interobserver variability among human experts (Cohen κ-statistics ranging from 0.58 to 0.65) | Expert consensus | CNN | 1352 nodules for training set and 453 nodules for validation set | Random split sample validation/yes |
| Nam [42] | Chest radiographs to detect malignant nodule/retrospective | Analysis of data collected from Seoul National University Hospital, Boramae Hospital and National Cancer Center, University of California San Francisco Medical Center | Chest radiograph classification and nodule detection performances of deep learning-based automatic detection were a range of 0.92–0.99 (AUROC) | Expert consensus | CNN | 43 292 chest radiographs from 34 676 patients | Random split sample validation/yes |
| Wang [43] | Mediastinal lymph node metastasis of NSCLC from 18F-FDG PET/CT images/retrospective | Data collected at the Affiliated Tumor Hospital of Harbin Medical University | The performance of CNN is not significantly different from classic machine-learning methods and expert radiologists | Expert consensus | CNN | 1397 lymph node stations from 168 patients | Resampling method/no |
| Wang [44] | Solitary pulmonary nodule ≤3 cm, histologically confirmed adenocarcinoma/retrospective | Analysis of data collected from Fudan University Shanghai Cancer Center | Algorithm showed AUROC of 0.892, which was higher than three expert radiologists in classifying invasive adenocarcinoma from pre-invasive lesions | Histology | CNN | CT scan from 1545 patients | Random split sample validation/no |
| Zhao [45] | Thin-slice chest CT scan before surgical treatment; nodule diameter ≤10 mm/retrospective | Secondary analysis of data from Huadong Hospital affiliated to Fudan University | Based on classification of tumour invasiveness, deep-learning algorithm achieved better classification performance than the radiologists (63.3% versus 56.6%) | Histology | CNN | Pre-operative thin-slice CT; 523 nodules for training/128 nodules for testing | Not reported/no |
| Fibrotic lung diseases | |||||||
| Walsh [46] | HRCT showing diffuse fibrotic lung disease confirmed by at least two thoracic radiologists/retrospective | Secondary analysis of data from La Fondazione Policlinico, Universitario and University of Parma (Italy) | Interobserver agreement between the algorithm and the radiologists’ majority opinion (n=91) was good (κw=0·69) | Expert consensus | CNN | HRCT; 929 scans for training/89 scans for validation | Not reported/yes |
| Christe [47] | HRCT showing NSIP or UIP confirmed by two thoracic radiologists/retrospective | HRCT dataset from the Lung Tissue Research Consortium | Interobserver agreements between the algorithm and the radiologists’ opinion were fair to moderate (κw=0.33 and 0.47) | Expert consensus | CNN | HRCT 105 patients (54 of NSIP and 51 for UIP) | Not reported/no |
| Raghu [48] | The whole-transcriptome RNA sequencing data from transbronchial biopsy samples/prospective | Bronchial Sample Collection for a Novel Genomic Test (BRAVE) study in 29 US and European sites | The molecular signatures had high specificity (88%) and sensitivity (70%) against diagnostic reference pathology (ROC-AUC 0.87, 95% CI 0.76–0.98) | Histology | ML; type not reported | 94 patients in clinical utility analysis | Not reported/no |
| PH | |||||||
| Sweatt [49] | Peripheral blood biobank/prospective | Peripheral blood biobanked at Stanford University, USA and University of Sheffield, UK | Four distinct immunological clusters were identified. Cluster I had unique sets of upregulated proteins (TRAIL, CCL5, CCL7, CCL4, MIF), which was the cluster with the least favourable 5-year transplant-free survival rates (47.6%, 95% CI 35.4–64.1%) | N/A | Unsupervised ML | Blood biobanked; 281 patients for discovery cohort/104 patients for validation cohort | Resampling method/yes |
| Leha [50] | Echocardiographic parameters/retrospective | King's College Hospital (UK); University Medical Center Gottingen and University of Regensburg (Germany) | Among five ML algorithms, random forest of regression trees is the best method to identify PH patients (AUC 0.87, 95% CI 0.78–0.96) with accuracy of 0.83 | Right heart catheterisation | Five ML algorithms (random forest of classification trees, random forest of regression trees, lasso-penalised logistic regression, boosted classification trees, SVM) | 90 patients with invasively determined PAP with corresponding echocardiographic estimations of PAP | Resampling method/no |
| Asthma | |||||||
| Wu [51] | 100 clinical, physiological, inflammatory and demographic variables/prospective | Severe Asthma Research Program (SARP) cohort from National Heart, Lung, and Blood Institutes (USA) | Four asthma clusters with differing CS responses were identified. Those in CS-responsive cluster were older, more nasal polyps, and high blood eosinophils. After CS, there was the highest increase in lung function in this group | N/A | Unsupervised ML; MML-MKKC | 346 adult asthmatics with paired (before and after CS) sputum data | Random split sample validation/no |
| Pleural effusion | |||||||
| Khemasuwan [52] | 19 candidate clinical variables from retrospective cohort of patients with pleural infection | A tertiary care, university-affiliated hospital, Utah, USA | Candidate predictors of tPA/DNase failure were the presence of pleural thickening (48% relative importance) and presence of an abscess/necrotising pneumonia (24%) | N/A | Supervised ML (extreme gradient boosting and coupled with decision trees) | 84 patients with pleural infection and received intrapleural tPA/DNase | Random split sample validation/no |
| PFT interpretation and clinical diagnosis | |||||||
| Topalovic [53] | PFT tests and clinical diagnosis/prospective | University Hospital Leuven (Belgium) | Pulmonologists’ interpretation of PFTs matched guideline in 74.4±5.9% of cases and made correct diagnosis in 44.6±8.7% versus AI algorithm matched the PFT pattern interpretations in 100% and assigned correct diagnosis in 82% (p<0.0001) | ATS/ERS guideline and expert panel | ML; type not reported | Dataset based on 1430 historical cases/50 cases in prospective analysis | Not reported/yes |
| SARS-CoV-2 pandemic | |||||||
| Wang [54] | CT chest of patients with atypical pneumonia/retrospective | Xi'an Jiaotong University First Affiliated Hospital, Nanchang University First Hospital and Xi'An No.8 Hospital of Xi'An Medical College (China) | An internal validation achieved a total accuracy of 82.9% with specificity of 80.5% and sensitivity of 84%. The external testing dataset showed a total accuracy of 73.1% with specificity of 67% and sensitivity of 74% | Confirmed nucleic acid testing of SARS-CoV-2 | CNN | CT images from 99 patients, of which 44 were confirmed cases of SARS-CoV-2 | Random split sample validation/no |
| Li [55] | CT chest of patients with atypical pneumonia/retrospective | Six medical centres, China | AUC values for COVID-19 was 0.96 (95% CI 0.94–0.99). Sensitivity of 90% (95% CI 83–94%) and specificity 96% (95% CI 93–98%) | Confirmed nucleic acid testing of SARS-CoV-2 | CNN | 4356 chest CT examinations from 3322 patients | Not reported/yes |
PH: pulmonary hypertension; PFT: pulmonary function test; SARS-CoV-2: severe acute respiratory syndrome-coronavirus-2; CT: computed tomography; NLST: National Lung Screening Trial; ROC: receiver operating characteristic; AUC: area under the curve; CNN: convolutional neural network; AUROC: area under the ROC curve; NSCLC: nonsmall cell lung cancer; 18F-FDG PET: fluorine-18 2-fluoro-2-deoxy-d-glucose positron emission tomography; HRCT: high-resolution computed tomography; κw: weighted κ-coefficient; NSIP: nonspecific interstitial pneumonia; UIP: usual interstitial pneumonia; ML: machine learning; TRAIL: tumor necrosis factor-related apoptosis-inducing ligand; CCL: C-C motif chemokine ligand; MIF: macrophage migration inhibitory factor; N/A: not applicable; SVM: support vector machine; PAP: pulmonary arterial pressure; CS: corticosteroids; MML-MKKC: multiview learning-multiple Kernel k-means clustering; tPA: intrapleural tissue plasminogen activator; DNase: deoxyribonuclease; ATS: American Thoracic Society; ERS: European Respiratory Society; COVID-19: coronavirus disease 2019.
Computer vision for lung nodule detection and prediction risk for malignancy
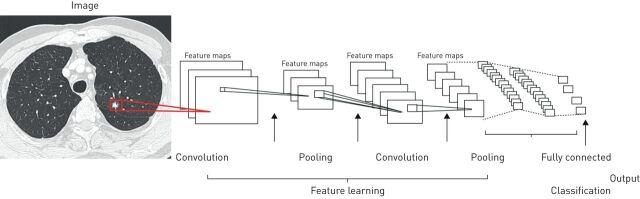
Computer vision is a form of deep learning where objects are identified directly from raw image pixels. Experts first label each image with the correct diagnosis, representing what is described as “ground truth”. Computer vision then focuses on image and video recognition that handles assigned tasks such as object classification, detection and interpretation in order to categorise predefined outcomes. Convolutional neural networks (CNNs) form deep learning algorithms designed to process input images, assigning importance to various aspects in order to differentiate one image from another. The structural architecture of CNNs is comparable to that of the connectivity pattern of neurons in the human brain, following a hierarchical model that creates a funnel-like framework to provide a fully connected layer where all the neurons are connected and output is processed (figure 2). Of historical interest, state-of-the-art computer vision accuracy was favourably compared with human accuracy in object-classification tasks. The study was based on the ImageNet Large Scale Visual Recognition Challenge dataset (an annual competition using a publicly available dataset of random objective images from a massive collection of human-annotated photographs) [31]. Expert-level performances were also achieved in studies of diabetic retinopathy [56], skin lesion classification [57] and metastatic lymph node detection [58].
FIGURE 2.
An example of convolutional neural network (CNN). A sequence of layers. Each layer transforms one volume of activations to another through a differentiable function. Three main types of layers build CNN architectures: convolutional layer, pooling layer and fully connected layer. The convolution layers merge two sets of information with the use of a filter to produce a feature map as an output. The pooling layers reduce the number of parameters and computation in the network. The fully connected output layer provides the final probabilities for each label as a final classification.
Lung cancer screening using low-dose computed tomography (CT) has been shown to reduce mortality by 20% in the National Cancer Institute's National Lung Screening Trial (NLST). It is currently included in North American screening guidelines. False-positive results that lead to an invasive procedure are high [59–61], but AI is being used to improve diagnostic accuracy. Studies compare performance between expert radiologists and deep-learning algorithms using chest imaging. Ardila et al. [38] proposed an end-to-end approach for detecting lung cancer using only input CT data from the NLST. A three-dimensional CNN model was created with an end-to-end analysis of whole-CT volumes using pathology-confirmed lung cancer as ground truth in training data (a “full-volume” model). Subsequently, CNN was trained to identify regions of interest (ROI) and to develop a CNN cancer risk prediction model that operated on outputs from the full-volume model and cancer ROI detection model. In the testing dataset, this model achieved an area under the receiver operating characteristic (ROC) curve of 94.4%. A comparison group of expert radiologists had performance at or below the algorithm's ROC. Given the perceived “black-box” nature of deep learning, investigators did not clearly understand if the model incorporates other features outside the ROI in its predictions. The term “black box” is used to describe complex models, including neural networks. Sometimes, even coders do not clearly understand how their algorithms work. Further examination using model attribution techniques may allow radiologists to take advantage of visual features used by the algorithm to predict the risk of malignancy.
Liu et al. [62] performed a meta-analysis of 69 studies to evaluate the diagnostic accuracy of deep-learning algorithms versus that of healthcare professionals. Deep-learning algorithms had better or equivalent accuracy compared with readings by expert radiologists. Nonetheless, there were limitations. Firstly, all of the referred studies were retrospective, in silico and based on previously assembled datasets. Secondly, the reporting on handling missing information in these datasets was poor. Most studies did not state whether portions of data were missing or how missing data were handled. Lastly, some studies [39–45] did not externally validate findings with external datasets, missing a crucial step to evaluate the model's performance with completely independent datasets.
Computer vision and molecular signatures for diagnosis of pulmonary fibrosis
Despite the 2018 Fleischner Society statement expanding diagnostic recommendations for radiographic evaluation of fibrotic lung disease, diagnosis remains challenging due to substantial interobserver variability, even between experienced radiologists [63–65]. Deep-learning algorithms proposed by several investigators classify fibrotic lung diseases using high-resolution computed tomography (HRCT) images. Walsh et al. [46] used deep learning for automated classification of fibrotic lung disease on HRCT based on the Fleischner Society and other international respiratory society guidelines. The study used a database of 1157 anonymous HRCTs of patients with diffuse fibrotic lung disease. The performance of the algorithm was compared with the majority vote of 91 expert thoracic radiologists. The median accuracy of thoracic radiologists was 70.7% compared with 73.3% accuracy of the algorithm. There was a good interobserver agreement (weighted κ-coefficient (κw) 0.69) between the algorithm and expert readings. Christe et al. [47] trained a deep-learning algorithm to classify HRCT scans of 105 cases of pulmonary fibrosis into four diagnostic categories according to Fleischner Society recommendations and compared performance with interpretations by two radiologists. Interobserver agreements between the algorithm and the radiologists' opinions were fair to moderate (κw 0.33 and 0.47).
In different studies, machine learning was used to develop an algorithm based on genomic data from surgical biopsy samples in order to detect molecular signatures for usual interstitial pneumonia (UIP). Signatures were concordant with the histopathological diagnosis from tissues obtained by transbronchial biopsy [48, 66, 67]. In one study, the whole-transcriptome ribonucleic acid sequencing data from transbronchial biopsy samples was used to train and validate the machine-learning algorithm. Molecular signatures had high specificity (88%) and acceptable sensitivity (70%) against diagnostic reference pathology (ROC area under the curve (AUC) 0.87, 95% CI 0.76–0.98) [48]. In addition, this study demonstrated how a molecular classifier helped make a definitive diagnosis in patients without a definitive UIP pattern on HRCT: positive predictive value was 81% (95% CI 54–96%) for underlying biopsy-proven UIP. The authors cautiously suggested that results need to be interpreted in the appropriate clinical setting and only after consideration by a multidisciplinary team according to standard-of-care guidelines [48].
Predictive models in pulmonary hypertension, asthma, pleural infections and pulmonary function tests
Machine learning improves prediction accuracy compared with conventional regression models. In pulmonary hypertension-related vascular injury, for example, the roles of inflammation and cytokines are not clearly understood. In one study, unsupervised machine learning was used to classify World Health Organization group I pulmonary arterial hypertension (PAH) patients into proteomic immune clusters (cytokines, chemokines and growth factors) [49]. Clinical characteristics and outcomes were subsequently compared across clusters. Four PAH clusters with distinct proteomic profiles were identified. These immune clusters had significant cluster-specific differences in clinical risk parameters and long-term prognosis. Compared with other clusters, patients in cluster 1 had unique sets of upregulated proteins (tumour necrosis factor-related apoptosis-inducing ligand, microphage migration inhibitory factor, chemokine ligand 4, 5 and 7) and had the least favourable 5-year transplant-free survival rates (30.8%, 95% CI 17.3–54.8%) [49]. The major limitation of this study was cross-section analysis from single time points, which may not reflect a patient's dynamic immune phenotype in clinical practice.
In asthma, systemic steroids are often used for rescue therapy, but have numerous side-effects. Cluster analysis has identified subphenotypes of steroid-responsive asthma using large datasets and regression analysis [68, 69]. An unsupervised learning approach was proposed to identify differential systemic corticosteroid response patterns. A multiview learning-multiple kernel k-means clustering (MML-MKKC) was developed to identify patient clustering by incorporating different types of variables and assigning them into different views based on clinical importance. MML-MKKC is a subset of cluster analysis in an unsupervised learning algorithm that combines different data modalities by capturing view-specific data patterns. Top relevant variables were selected from 100 clinical, physiological, inflammatory and demographic variables obtained from 346 patients in a Severe Asthma Research Program cohort categorised into four clusters using an MML-MKKC model. A multiclass support vector machine (SVM) algorithm with a 10-fold cross-validation strategy was deployed. SVM is a subset of a supervised learning algorithm to identify a hyperplane that separates a dataset based on outcomes of interest. This algorithm identified the top 12 variables that predicted a cluster of test samples with 81% overall accuracy. The clinical characteristics in the corticosteroid-responsive cluster were older, with the latest age at onset, more nasal polyps and high blood eosinophils. After corticosteroids, the highest increase in lung function was noted in this group [51]. In contrast, a cluster less responsive to corticosteroids included characteristics such as obesity, female sex and early-onset asthma. Approximately half were African American.
Patients with empyema and complex parapneumonic effusions are sometimes treated with intrapleural tissue plasminogen activator and deoxyribonuclease (tPA/DNase) [70]. Some require surgical intervention. Khemasuwan et al. [52] used extreme gradient boosting (XGBoost) to evaluate 19 candidate predictors of tPA/DNase failure in a retrospective cohort. XGBoost is a subset of a supervised learning algorithm deployed to assess the importance of candidate variables in terms of their ability to predict the outcomes. XGBoost identified pleural thickening and presence of abscess/necrotising pneumonia as risk factors for failure of combined intrapleural agents. This was confirmed using best-subset logistic regression, a technique that is fundamentally different from XGBoost. In the presence of these two variables (pleural thickening and presence of abscess/necrotising pneumonia), the logistic model demonstrated an AUC of 0.981 with a sensitivity of 96% and specificity of 78%.
In order to test the value of a clinical decision support system based on the interpretation of pulmonary function tests (PFTs), Topalovic and co-workers [71–73] developed a machine-learning model using 1430 cases structured from pre-defined, established diagnoses of eight categories of respiratory disease, coupled with software in line with gold-standard American Thoracic Society/European Respiratory Society guidelines for PFT pattern interpretation [74]. A random sample of 50 cases with PFT and clinical information was used to prospectively compare the accuracy in pattern recognition and diagnosis of the AI algorithm with the performance of 120 mostly senior pulmonologists from 16 European hospitals. PFT interpretation results from pulmonologists matched the guidelines in 74.4±5.9% of the cases (range 56–88%) with only moderate to substantial agreement (κ=0.67). Pulmonologists made a correct diagnosis in 44.6±8.7% of the cases (range 24–62%) with large inter-rater variability (κ=0.35). In contrast, the AI algorithm perfectly matched PFT pattern interpretations (100%), and assigned a correct diagnosis in 82% of all cases (p<0.0001) [53]. The superior performance demonstrated by this AI algorithm for both pattern recognition and clinical diagnosis is promising for future clinical decision-making.
AI in the SARS-CoV-2 pandemic
Novel coronavirus disease 2019 (COVID-19) has ravaged the world since its discovery as a flu-like outbreak in Wuhan, China, in January 2020. Rapid recognition of the outbreak was, in part, related to an AI epidemiology algorithm called BlueDot, which skimmed foreign-language news reports and official announcements to provide clients with news of potential epidemics [75]. The BlueDot algorithm predicted the early spread of COVID-19 outside of Wuhan based on travel data generated from the International Air Transport Association [75]. Previously, BlueDot algorithms successfully predicted the international spread of the Zika virus in South Florida in 2016 [76].
In addition to infectious disease-related tracking and prediction models, AI might help elaborate early diagnosis and treatment strategies. For example, deep learning can help diagnose COVID-19 using image analysis from chest radiographs [77] and CT scans [54, 55, 78]. Unsupervised learning algorithms are being developed to help predict immune response to influenza vaccine [79] and to predict an inhibitory potency of atazanavir and remdesivir against the SARS-CoV-2 3C-like proteinase [80]. A deep-learning algorithm called AlphaFold [81] was developed by Google DeepMind to improve protein structure predictions, which might otherwise take many months using traditional experimental approaches. The algorithm is used to predict the structural protein shape of COVID-19 [82], providing vital information for COVID-19 vaccine developers [83].
There are various efforts to predict infectious cases [84], coronavirus-related mortality [85] and burden on medical resources [86, 87]. Models use the latest data science and AI techniques to forecast outcomes. Proposed models help authorities form public policy and be more future-informed. The complex, dynamic and heterogeneous nature of AI-based models warrants that outcomes be continuously recalibrated using the newest data on a daily basis [88]. However, prediction models for COVID-19 may not always be accurate and reliable. One reason is the lack of sufficient historic data to build a model that can accurately track and forecast the virus's spread. Another is that many studies tend to use Chinese-based samples, and could thus be biased [89]. Because AI prediction models mostly rely on previous disease patterns, an outlier event with unprecedented data, such as occurred with COVID 19, can be disruptive to the model. For these reasons, many forecasting experts avoid using AI methods and prefer using epidemiological models such as susceptible-infected-recovered [90].
Limitations of using AI in pulmonary medicine
AI provides opportunities to improve quality of care and accelerate the evolution of precision medicine. However, its limitations have nurtured the field of AI ethics and the study of the impact of AI on technology, individual lives, economics and social transformation [91].
One illustration relates to how AI-based algorithms may cause iatrogenic risk to a large group of patients. For example, hundreds of hospitals around the world use IBM Watson for recommending cancer treatments. Algorithms based on a small number of synthetic fictional cases using limited input (real patient data) from oncologists [92, 93] resulted in output treatment recommendations that were eventually shown to be erroneous [94]. This supports systemic debugging, audits, extensive iteration and evidence from robust, prospective validation studies before algorithms are widely implemented in clinical practice [95].
Another example relates to the inequities of healthcare delivery. By inputting low socioeconomic status as a major risk factor for premature mortality [96], AI algorithms may be biased against patients of a particular ethnicity or socioeconomic status, which might widen the gap in health outcomes. Combined with concerns for exacerbating pre-existing inequities, the potential for embedding bias by excluding minorities from datasets is a real hazard. Embedded prejudice must be mitigated, and massive datasets should provide a true representative cross-section of all populations.
Another hurdle facing the use of AI in healthcare relates to a lack of prospective validation studies and difficulty improving an algorithm's performance. Many investigations are validated in silico by dividing a single pre-existing dataset into a training and testing dataset. However, external validation using an independent dataset is critical prior to implementation in a real-world environment, and inherently opaque machine-learning algorithm black-box models should be avoided as much as possible.
Finally, the future of AI-related medical applications depends on how well safety, confidentiality and data security can be assured. In light of hacking and data breaches, there will be little interest in using algorithms that risk revealing patient identity [93], and of course, adverse effects on clinician workloads may arise from an overdependence on automated machine-learning systems or increases in medical errors [97]. One mitigating strategy uses a novel process called federated learning, so that machine-learning models can analyse confidential, decentralised data without transfer to a central server [98]. Multiple organisations thus share data without compromising patient privacy. The effectiveness and safety of such techniques require prospective studies and large randomised controlled trials.
Conclusion
Data-driven decision-making in pulmonary medicine can be leveraged by incorporating machine-learning and deep-learning algorithms into daily practice. Massive quantities of clinical, physiological, epidemiological and genetic data are already being analysed using algorithms that serve clinicians in the form of manageable, interpretable and actionable knowledge that augments decision-making capacity. In addition, AI-based data analyses provide increasingly accurate predictive modelling and lay the foundation for genuinely data-driven precision medicine that will decompress our reliance on human resources. As computational power evolves, algorithms will ingest and meaningfully process massive sets of data even more quickly, more accurately and less onerously than human minds. With advances in technology and a new generation of computer-literate physicians, future practitioners will inevitably incorporate AI into clinical care. Considering how AI favourably affects other fields, the future is already here.
Footnotes
Provenace: Submitted article, peer reviewed.
Conflict of interest: D. Khemasuwan has nothing to disclose.
Conflict of interest: J.S. Sorensen has nothing to disclose.
Conflict of interest: H.G. Colt has nothing to disclose.
References
- 1.Stanford Medicine. 2017. Harnessing the Power of Data in Health. Stanford Medicine 2017 Health Trends Report. https://med.stanford.edu/content/dam/sm/sm-news/documents/StanfordMedicineHealthTrendsWhitePaper2017.pdf
- 2.Kahneman D. Thinking, Fast and Slow. New York, Farrar, Straus and Giroux, 2011. [Google Scholar]
- 3.Catherwood PA, Rafferty J, McLaughlin J. Artificial intelligence for long-term respiratory disease management. In: Proceedings of the 32nd International BCS Human Computer Interaction Conference. 2018; 32: pp. 1–5. [Google Scholar]
- 4.Mekov E, Miravitlles M, Petkov R. Artificial intelligence and machine learning in respiratory medicine. Expert Rev Respir Med 2020; 14: 559–564. doi: 10.1080/17476348.2020.1743181 [DOI] [PubMed] [Google Scholar]
- 5.Gonem S, Janssens W, Das N, et al. . Applications of artificial intelligence and machine learning in respiratory medicine. Thorax 2020; 75: 695–701. doi: 10.1136/thoraxjnl-2020-214556 [DOI] [PubMed] [Google Scholar]
- 6.Angelini E, Dahan S, Shah A. Unravelling machine learning: insights in respiratory medicine. Eur Respir J 2019; 54: 1901216. doi: 10.1183/13993003.01216-2019 [DOI] [PubMed] [Google Scholar]
- 7.Mlodzinski E, Stone DJ, Celi LA. Machine learning for pulmonary and critical care medicine: a narrative review. Pulm Ther 2020; 6: 67–77. doi: 10.1007/s41030-020-00110-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das N, Topalovic M, Janssens W. Artificial intelligence in diagnosis of obstructive lung disease: current status and future potential. Curr Opin Pulm Med 2018; 24: 117–123. doi: 10.1097/MCP.0000000000000459 [DOI] [PubMed] [Google Scholar]
- 9.Lovejoy CA, Phillips E, Maruthappu M. Application of artificial intelligence in respiratory medicine: has the time arrived? Respirology 2019; 24: 1136–1137. doi: 10.1111/resp.13676 [DOI] [PubMed] [Google Scholar]
- 10.Pan J, Tompkins WJ. A real-time QRS detection algorithm. IEEE Trans Biomed Eng 1985; 32: 230–236. doi: 10.1109/TBME.1985.325532 [DOI] [PubMed] [Google Scholar]
- 11.Aikins JS, Kunz JC, Shortliffe EH, et al. . PUFF: an expert system for interpretation of pulmonary function data. Comput Biomed Res 1983; 16: 199–208. doi: 10.1016/0010-4809(83)90021-6 [DOI] [PubMed] [Google Scholar]
- 12.Snow MG, Fallat RJ, Tyler WR, et al. . Pulmonary consult: concept to application of an expert system. J Clin Eng 1988; 13: 201–206. doi: 10.1097/00004669-198805000-00010 [DOI] [Google Scholar]
- 13.Grosan C, Abraham A. Rule-based expert systems. In: Intelligent Systems. Intelligent Systems Reference Library, Vol 17. Berlin, Heidelberg, Springer. 2011; pp. 149–185. [Google Scholar]
- 14.Garvey JL, Zegre-Hemsey J, Gregg R, et al. . Electrocardiographic diagnosis of ST segment elevation myocardial infarction: an evaluation of three automated interpretation algorithms. J Electrocardiol 2016; 49: 728–732. doi: 10.1016/j.jelectrocard.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szolovits P, Pauker SG. Categorical and probabilistic reasoning in medical diagnosis. Artif Intell 1978; 11: 115–144. doi: 10.1016/0004-3702(78)90014-0 [DOI] [Google Scholar]
- 16.Alpaydin E. Machine Learning. Cambridge, MA, MIT Essential Knowledge Series, 2016; p. IX. [Google Scholar]
- 17.Berner ES, Webster GD, Shugerman AA, et al. . Performance of four computer-based diagnostic systems. N Engl J Med 1994; 330: 1792–1796. doi: 10.1056/NEJM199406233302506 [DOI] [PubMed] [Google Scholar]
- 18.Szolovits P, Patil RS, Schwartz WB. Artificial intelligence in medical diagnosis. Ann Intern Med 1988; 108: 80–87. doi: 10.7326/0003-4819-108-1-80 [DOI] [PubMed] [Google Scholar]
- 19.Deo RC. Machine learning in medicine. Circulation 2015; 132: 1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol 2019; 19: 64. doi: 10.1186/s12874-019-0681-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JH, Asch SM. Machine learning and prediction in medicine – beyond the peak of inflated expectations. N Engl J Med 2017; 376: 2507–2509. doi: 10.1056/NEJMp1702071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Medical Futurist. FDA-approved A.I.-based Algorithms. www.medicalfuturist.com/fda-approved-ai-based-algorithms/ Date last accessed: 15 July 2020. Date last updated: 6 June 2019.
- 23.Alexander A, Jiang A, Ferreira C, et al. . An intelligent future for medical imaging: a market outlook on artificial intelligence for medical imaging. J Am Coll Radiol 2020; 17: 165–170. doi: 10.1016/j.jacr.2019.07.019 [DOI] [PubMed] [Google Scholar]
- 24.AI for Radiology. An Implementation Guide. www.grand-challenge.org/aiforradiology/?subspeciality=Chest&modality=All&search= Date last accessed: 10 August 2020.
- 25.James G, Witten D, Hastie T, et al. . An Introduction to Statistical Learning. New York, Springer. 2013; Vol. 112. [Google Scholar]
- 26.Gençay R, Qi M. Pricing and hedging derivative securities with neural networks: Bayesian regularization, early stopping, and bagging. IEEE Trans Neural Netw 2001; 12: 726–734. doi: 10.1109/72.935086 [DOI] [PubMed] [Google Scholar]
- 27.Seymour CW, Gomez H, Chang CH, et al. . Precision medicine for all? Challenges and opportunities for a precision medicine approach to critical illness. Crit Care 2017; 21: 257. doi: 10.1186/s13054-017-1836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Research Council (US) Committee on a Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, National Academies Press, 2011. [PubMed] [Google Scholar]
- 29.Gottesman O, Johansson F, Komorowski M, et al. . Guidelines for reinforcement learning in healthcare. Nat Med 2019; 25: 16–18. doi: 10.1038/s41591-018-0310-5 [DOI] [PubMed] [Google Scholar]
- 30.Komorowski M, Celi LA, Badawi O, et al. . The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018; 24: 1716–1720. doi: 10.1038/s41591-018-0213-5 [DOI] [PubMed] [Google Scholar]
- 31.Russakovsky O, Deng J, Su H, et al. . ImageNet large scale visual recognition challenge. Int J Comput Vis 2015; 115: 211–252. doi: 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 32.Hirschberg J, Manning CD. Advances in natural language processing. Science 2015; 349: 261–266. doi: 10.1126/science.aaa8685 [DOI] [PubMed] [Google Scholar]
- 33.Geoffrey H, Deng L, Yu D, et al. . Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag 2012; 29: 82–97. [Google Scholar]
- 34.Chen D, Liu S, Kingsbury P, et al. . Deep learning and alternative learning strategies for retrospective real-world clinical data. NPJ Digit Med 2019; 2: 43. doi: 10.1038/s41746-019-0122-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver D, Schrittwieser J, Simonyan K, et al. . Mastering the game of Go without human knowledge. Nature 2017; 550: 354–359. doi: 10.1038/nature24270 [DOI] [PubMed] [Google Scholar]
- 36.Hosny A, Parmar C, Quackenbush J, et al. . Artificial intelligence in radiology. Nat Rev Cancer 2018; 18: 500–510. doi: 10.1038/s41568-018-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 38.Ardila D, Kiraly AP, Bharadwaj S, et al. . End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019; 25: 954–961. 10.1038/s41591-019-0536-x. [DOI] [PubMed] [Google Scholar]
- 39.Baldwin DR, Gustafson J, Pickup L, et al. . External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules. Thorax 2020; 75: 306–312. doi: 10.1136/thoraxjnl-2019-214104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massion PP, Antic S, Ather S, et al. . Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. Am J Respir Crit Care Med 2020; 202: 241–249. doi: 10.1164/rccm.201903-0505OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciompi F, Chung K, van Riel SJ, et al. . Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017; 7: 46479. doi: 10.1038/srep46479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nam JG, Park S, Hwang EJ, et al. . Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 2019; 290: 218–228. doi: 10.1148/radiol.2018180237 [DOI] [PubMed] [Google Scholar]
- 43.Wang HK, Zhou ZW, Li YC, et al. . Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res 2017; 7: 11. doi: 10.1186/s13550-017-0260-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Wang R, Zhang S, et al. . 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤3 cm using HRCT. Quant Imaging Med Surg 2018; 8: 491–499. doi: 10.21037/qims.2018.06.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao W, Yang J, Sun Y, et al. . 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res 2018; 78: 6881–6889. doi: 10.1158/0008-5472.CAN-18-0696 [DOI] [PubMed] [Google Scholar]
- 46.Walsh SL, Calandriello L, Silva M, et al. . Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med 2018; 6: 837–845. [DOI] [PubMed] [Google Scholar]
- 47.Christe A, Peters AA, Drakopoulos D, et al. . Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Invest Radiol 2019; 54: 627–632. doi: 10.1097/RLI.0000000000000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raghu G, Flaherty KR, Lederer DJ, et al. . Use of a molecular classifier to identify usual interstitial pneumonia in conventional transbronchial lung biopsy samples: a prospective validation study. Lancet Respir Med 2019; 7: 487–496. doi: 10.1016/S2213-2600(19)30059-1 [DOI] [PubMed] [Google Scholar]
- 49.Sweatt AJ, Hedlin HK, Balasubramanian V, et al. . Discovery of distinct immune phenotypes using machine learning in pulmonary arterial hypertension. Circ Res 2019; 124: 904–919. doi: 10.1161/CIRCRESAHA.118.313911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leha A, Hellenkamp K, Unsöld B, et al. . A machine learning approach for the prediction of pulmonary hypertension. PLoS One 2019; 14: e0224453. doi: 10.1371/journal.pone.0224453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu W, Bang S, Bleecker ER, et al. . Multiview cluster analysis identifies variable corticosteroid response phenotypes in severe asthma. Am J Respir Crit Care Med 2019; 199: 1358–1367. doi: 10.1164/rccm.201808-1543OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khemasuwan D, Sorensen J, Griffin DC. Predictive variables for failure in administration of intrapleural tissue plasminogen activator/deoxyribonuclease in patients with complicated parapneumonic effusions/empyema. Chest 2018; 154: 550–556. doi: 10.1016/j.chest.2018.01.037 [DOI] [PubMed] [Google Scholar]
- 53.Topalovic M, Das N, Burgel PR, et al. . Artificial intelligence outperforms pulmonologists in the interpretation of pulmonary function tests. Eur Respir J 2019; 53: 1801660. doi: 10.1183/13993003.01660-2018 [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Kang B, Ma J, et al. . A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv 2020: doi:10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Qin L, Xu Z, et al. . Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020; 296: 200905. doi: 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gulshan V, Peng L, Coram M, et al. . Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316: 2402–2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 57.Esteva A, Kuprel B, Novoa RA, et al. . Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. . Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318: 2199–2210. doi: 10.1001/jama.2017.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.National Lung Screening Trial Research Team, Aberle DR, Berg CD, et al. . The National Lung Screening Trial: overview and study design. Radiology 2011; 258: 243–253. doi: 10.1148/radiol.10091808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinsky PF, Bellinger CR, Miller DP Jr. False-positive screens and lung cancer risk in the National Lung Screening Trial: implications for shared decision-making. J Med Screen 2018; 25: 110–112. doi: 10.1177/0969141317727771 [DOI] [PubMed] [Google Scholar]
- 61.Jemal A, Fedewa SA. Lung cancer screening with low-dose computed tomography in the United States – 2010 to 2015. JAMA Oncol 2017; 3: 1278–1281. doi: 10.1001/jamaoncol.2016.6416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, Faes L, Aditya U, et al. . A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health; 1: e271–e297. doi: 10.1016/S2589-7500(19)30123-2 [DOI] [PubMed] [Google Scholar]
- 63.Walsh SL, Calandriello L, Sverzellati N, et al. . Interobserver agreement for the ATS/ERS/JRS/ALAT criteria for a UIP pattern on CT. Thorax 2016; 71: 45–51. doi: 10.1136/thoraxjnl-2015-207252 [DOI] [PubMed] [Google Scholar]
- 64.Tominaga J, Sakai F, Johkoh T, et al. . Diagnostic certainty of idiopathic pulmonary fibrosis/usual interstitial pneumonia: the effect of the integrated clinico-radiological assessment. Eur J Radiol 2015; 84: 2640–2645. doi: 10.1016/j.ejrad.2015.08.016 [DOI] [PubMed] [Google Scholar]
- 65.Gruden JF. CT in idiopathic pulmonary fibrosis: diagnosis and beyond. AJR Am J Roentgenol 2016; 206: 495–507. doi: 10.2214/AJR.15.15674 [DOI] [PubMed] [Google Scholar]
- 66.Kim SY, Diggans J, Pankratz D, et al. . Classification of usual interstitial pneumonia in patients with interstitial lung disease: assessment of a machine learning approach using high-dimensional transcriptional data. Lancet Respir Med 2015; 3: 473–482. doi: 10.1016/S2213-2600(15)00140-X [DOI] [PubMed] [Google Scholar]
- 67.Pankratz DG, Choi Y, Imtiaz U, et al. . Usual interstital pneumonia can be detected in transbronchial biopsies using machine learning. Ann Am Thorac Soc 2017; 14: 1646–1654. doi: 10.1513/AnnalsATS.201612-947OC [DOI] [PubMed] [Google Scholar]
- 68.Haldar P, Pavord ID, Shaw DE, et al. . Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med 2008; 178: 218–224. doi: 10.1164/rccm.200711-1754OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore WC, Meyers DA, Wenzel SE, et al. . Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med 2010; 181: 315–323. doi: 10.1164/rccm.200906-0896OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rahman NM, Maskell NA, West A, et al. . Intrapleural use of tissue plasminogen activator and DNase in pleural infection. N Engl J Med 2011; 365: 518–526. doi: 10.1056/NEJMoa1012740 [DOI] [PubMed] [Google Scholar]
- 71.Topalovic M, Laval S, Aerts JM, et al. . Automated interpretation of pulmonary function tests in adults with respiratory complaints. Respiration 2017; 93: 170–178. doi: 10.1159/000454956 [DOI] [PubMed] [Google Scholar]
- 72.Decramer M, Janssens W, Derom E, et al. . Contribution of four common pulmonary function tests to diagnosis of patients with respiratory symptoms: a prospective cohort study. Lancet Respir Med 2013; 1: 705–713. doi: 10.1016/S2213-2600(13)70184-X [DOI] [PubMed] [Google Scholar]
- 73.Topalovic M, Das N, Troosters T, et al. . Applying artificial intelligence on pulmonary function tests improves the diagnostic accuracy. Eur Respir J 2017; 50: Suppl. 61, OA3434. [Google Scholar]
- 74.Pellegrino R, Viegi G, Brusasco V, et al. . Interpretative strategies for lung function tests. Eur Respir J 2005; 26: 948–968. doi: 10.1183/09031936.05.00035205 [DOI] [PubMed] [Google Scholar]
- 75.Bogoch II, Watts A, Thomas-Bachli A, et al. . Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med 2020; 27: taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bogoch II, Brady OJ, Kraemer MUG, et al. . Anticipating the international spread of Zika virus from Brazil. Lancet 2016; 387: 335–336. doi: 10.1016/S0140-6736(16)00080-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozturk T, Talo M, Yildirim EA, et al. . Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 2020; 121: 103792. doi: 10.1016/j.compbiomed.2020.103792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Majidi H, Niksolat F. Chest CT in patients suspected of COVID-19 infection: a reliable alternative for RT-PCR. Am J Emerg Med 2020; in press [ 10.1016/j.ajem.2020.04.016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tomic A, Tomic I, Rosenberg-Hasson Y, et al. . SIMON, an automated machine learning system, reveals immune signatures of influenza vaccine responses. J Immunol 2019; 203: 749–759. doi: 10.4049/jimmunol.1900033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beck BR, Shin B, Choi Y, et al. . Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug–target interaction deep learning model. Comput Struct Biotechnol J 2020; 18: 784–790. doi: 10.1016/j.csbj.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Senior AW, Evans R, Jumper J, et al. . Improved protein structure prediction using potentials from deep learning. Nature 2020; 577: 706–710. doi: 10.1038/s41586-019-1923-7 [DOI] [PubMed] [Google Scholar]
- 82.Jumper J, Tunyasuvunakool K, Kohli P, et al. . 2020. Computational Predictions of Protein Structures Associated with COVID-19. www.deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19 Date last accessed: 9 September 2020 Date last updated: 4 August 2020.
- 83.Alimadadi A, Aryal S, Manandhar I, et al. . Artificial intelligence and machine learning to fight COVID-19. Physiol Genomics 2020; 52: 200–202. doi: 10.1152/physiolgenomics.00029.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z, Zeng Z, Wang K, et al. . Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J Thorac Dis 2020; 12: 165–174. doi: 10.21037/jtd.2020.02.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Verity R, Okell LC, Dorigatti I, et al. . Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis 2020; 20: 669–677. doi: 10.1016/S1473-3099(20)30243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Institute of Health Metrics and Evaluation (IHME) at University of Washington. COVID-19 Projections. https://covid19.healthdata.org/united-states-of-america Date last accessed: 9 September 2020 Date last updated: 3 September 2020.
- 87.The MRC Centre for Global Infectious Disease Analysis at the Imperial College. Short-term Forecasts of COVID-19 Deaths in Multiple Countries. https://mrc-ide.github.io/covid19-short-term-forecasts/index.html Date last accessed: 9 September 2020. Date last updated: 31 August 2020.
- 88.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20: 533–534. doi: 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naudé W. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. AI Soc 2020; 35: 761–765. 10.1007/s00146-020-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song P, Wang L, Zhou Y, et al. . An epidemiological forecast model and software assessing interventions on COVID-19 epidemic in China. medRxiv 2020: doi:10.1101/2020.02.29.20029421. [Google Scholar]
- 91.Coeckelbergh M. AI Ethics. Cambridge, MIT Press Essential Knowledge Series. 2020; p. 7. [Google Scholar]
- 92.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25: 44–56. doi: 10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 93.Brundage, M, Avin S, Clark J, et al. . The malicious use of artificial intelligence: forecasting, prevention, and mitigation. 2018. www.arxiv.org/ftp/arxiv/papers/1802/1802.07228.pdf
- 94.Somashekhar SP, Sepúlveda MJ, Puglielli S, et al. . Watson for oncology and breast cancer treatment recommendations: agreement with an expert multidisciplinary tumor board. Ann Oncol 2018; 29: 418–423. doi: 10.1093/annonc/mdx781 [DOI] [PubMed] [Google Scholar]
- 95.Miliard M. 2018. As FDA Signals Wider AI Approval, Hospitals Have a Role to Play. Healthcare IT News. www.healthcareitnews.com/news/fda-signals-wider-ai-approval-hospitals-have-role-play Date last accessed: 9 September 2020. Date last updated: 31 May 2018.
- 96.Stringhini S, Carmeli C, Jokela M, et al. . Socioeconomic status and the 25×25 risk factors as determinants of premature mortality: a multicohort study and meta-analysis of 1.7 million men and women. Lancet 2017; 389: 1229–1237. doi: 10.1016/S0140-6736(16)32380-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y, Chen PC, Krause J, et al. . How to read articles that use machine learning: users’ guides to the medical literature. JAMA 2019; 322: 1806–1816. doi: 10.1001/jama.2019.16489 [DOI] [PubMed] [Google Scholar]
- 98.Najafabadi MM, Villanustre F, Khoshgoftaar TM, et al. . Deep learning applications and challenges in big data analytics. J Big Data 2015; 2: 1. doi: 10.1186/s40537-014-0007-7 [DOI] [Google Scholar]