SUMMARY
The developmental journey of cortical interneurons encounters several activity-dependent milestones. During the early postnatal period in developing mice, GABAergic neurons are transient preferential recipients of thalamic inputs and undergo activity-dependent migration arrest, wiring, and programmed cell-death. Despite their importance for the emergence of sensory experience and the role of activity in their integration into cortical networks, the collective dynamics of GABAergic neurons during that neonatal period remain unknown. Here, we study coordinated activity in GABAergic cells of the mouse barrel cortex using in vivo calcium imaging. We uncover a transient structure in GABAergic population dynamics that disappears in a sensory-dependent process. Its building blocks are anatomically clustered GABAergic assemblies mostly composed by prospective parvalbumin-expressing cells. These progressively widen their territories until forming a uniform perisomatic GABAergic network. Such transient patterning of GABAergic activity is a functional scaffold that links the cortex to the external world prior to active exploration.
In Brief
Using in vivo imaging in the barrel cortex, Modol et al. show that prospective parvalbumin interneurons form patches of correlated activity that merge during postnatal development. These assemblies are regulated by whisker inputs and altered by sensory deprivation.
INTRODUCTION
Spontaneous neuronal activity shifts from highly coordinated to sparse before the start of active exploration in most, if not all, cortical sensory regions (Golshani et al., 2009; Rochefort et al., 2009; Wolfe et al., 2010). This process is “internally mediated” as it can occur under sensory deprivation, at least in upper cortical layers (Golshani et al., 2009; Rochefort et al., 2009; Che et al., 2018). The circuit mechanisms supporting the “sparsification” of activity are not completely understood but certainly involve the emergence of a powerful inhibitory brake, particularly through canonical feedforward perisomatic inhibition (Butt et al., 2017; Wolfe et al., 2010). The emergence of perisomatic inhibition is also the signal for the opening of the so-called critical period of development (Toyoizumi et al., 2013). Interestingly, in contrast to the finding that sparsification is internally mediated, several studies indicate that the maturation of GABAergic perisomatic circuits depends on electrical activity and sensory drive (Butt et al., 2017; Chattopadhyaya et al., 2007; De Marco García et al., 2015; De Marco García et al., 2011; Deng et al., 2017; Micheva and Beaulieu, 1995a; Micheva and Beaulieu, 1995b; Oh et al., 2016; Tuncdemir et al., 2016; Wamsley and Fishell, 2017). More generally, the firing of interneurons critically controls several key steps in the postnatal development of GABAergic neurons themselves, including migration, wiring, and programmed cell death (Bortone and Polleux, 2009; Butt et al., 2017; Chattopadhyaya et al., 2004; De Marco García et al., 2015; Marques-Smith et al., 2016; Takesian et al., 2018; Tuncdemir et al., 2016; Wong et al., 2018). Accordingly, increasing evidence indicates that GABAergic neurons may form transiently interconnected schemes prior to communicating with their local glutamatergic partners (Allene et al., 2012; Anastasiades et al., 2016; Butt et al., 2017; De Marco García et al., 2015; Tuncdemir et al., 2016). Therefore, GABAergic circuits seem to have an important function in closing the period of recurring population bursts by inducing the activity-dependent development of perisomatic inhibition and coordinating the emergence of normal sensory experience.
Despite their critical positioning at the interface between intracortical circuits and externally driven developmental processes, as well as their profound activity-dependent remodeling prior to active exploration, the structure of GABAergic population dynamics throughout the neonatal period of cortical development remains unexplored territory. Indeed, prior studies of population dynamics have focused on mapping the activity of glutamatergic cells (for example, through the genetic restriction of GCaMP expression [Antón-Bolaños et al., 2019; Kirmse et al., 2010; Kummer et al., 2016] or the recording of extracellular electrophysiological signals (Khazipov and Luhmann, 2006]). In addition, imaging perisomatic interneurons is technically challenging due to the protracted expression of parvalbumin, their main neurochemical marker.
We therefore examined the coordination of activity among GABAergic cells in the barrel cortex. We focused on the period of intracortical circuit development that spans between the establishment of barrels and the start of active whisking, a time when most interneurons undergo activity-dependent programmed cell death (Lim et al., 2018). We used two-photon calcium imaging in non-anaesthetized mouse pups for which expression of the genetically encoded calcium indicator (GCaMP6s) was restricted to GABAergic neurons. For this, we took advantage of several driver mouse lines: (1) the GAD67-Cre mouse line (Melzer et al., 2012), which allows both the mapping of interneuron activity without any a priori focus on a particular subtype as well as imaging the ring of axonal activity provided by GABAergic cells around the somatic compartment of non-GABA containing cells (Muldoon et al., 2015); (2) the Lhx6i-Cre mouse line (Fogarty et al., 2007), which allows imaging the activity from the two main types of interneurons derived from the medial ganglionic eminences (MGE), i.e., the somatostatin (SST)- and parvalbumin (PV)-expressing cells; (3) the 5HT3aR-Cre mouse line (Lee et al., 2010), which targets neurons originating from the caudal ganglionic eminence (CGE); and (4) the SST-Cre mouse line to image from SST-expressing interneurons.
We observed a transient patterning of global GABAergic population dynamics in the form of spontaneously recurring synchronous events composed of activated, anatomically clustered but functionally orthogonal (Malvache et al., 2016) GABA assemblies spanning both superficial and deep cortical layers. Prospective PV neurons, with their perisomatic axonal domains, were the main contributors to these anatomically and functionally segregated assemblies, whereas SST neurons synchronized across larger territories. In contrast to the internally mediated desynchronization of global population dynamics reported in supragranular layers (Golshani et al., 2009), here the GABA assembly size and combined recruitment was a sensory-dependent process that was modulated–positively initially, negatively at later developmental stages–by whisker stimulation and disrupted in deep cortical layers upon whisker plucking. Therefore, perisomatic GABAergic circuits, despite the protracted emergence of the parvalbumin marker, display early structured population dynamics. A disruption of this functional template by genetic or environmental factors impacts interneuron developmental fate (see also companion paper by Duan et al., 2019) and may well serve as a substrate for sensory perception disorders.
RESULTS
Spontaneous Correlated Activity of Diverse GABAergic Components In Vivo
To record spontaneous activity of large ensembles of GABAergic neurons in vivo, we generated mice expressing the calcium reporter protein GCaMP6s exclusively in GABA neurons by crossing GAD67Cre mice (Melzer et al., 2012) with the reporter line Ai96 (RCL-GCaMP6s) (The Jackson Laboratory, Figures 1A and 1A1). For simplification, pups born from these crossings will be referred to as GAD67-GCaMP6. GCaMP6s expression was stable and constrained to GABAergic cells across development in all cortical layers (Figure 1B). GCaMP6 was equally expressed in all GABAergic cells, independent from their embryonic origin (Figure S1). Pups were then used to monitor neuronal calcium dynamics in vivo, following window implantation (see STAR Methods). Experiments were performed during the neonatal period that spans from the end of anatomical barrel formation in deep layers (P4), to the onset of active whisking (P11) (Figures 1A and 1A1; Video S1–S3). A field of 600×600pm was imaged at different cortical depths up to 500pm below the surface (Figures 1A and 1A2). Data points were pooled in three successive groups (P4-P6, P7-P9, and P10-P11) based on similarity in terms of dynamics revealed by post hoc analysis. After movement correction, cell centers were manually detected. Contours surrounding co-active pixels were automatically detected by filtering the data with a Gaussian (see STAR Methods) and then extracting the correlated pixels that accounted for the maximum variance (Pnevmatikakis et al., 2016) (Figures 1C and 1C1). To estimate neuronal firing rates from changes in the fluorescent signal, we used a Markov Chain Monte Carlo approach initialized by a constrained deconvolution method (Pnevmatikakis et al., 2013) (see STAR Methods).
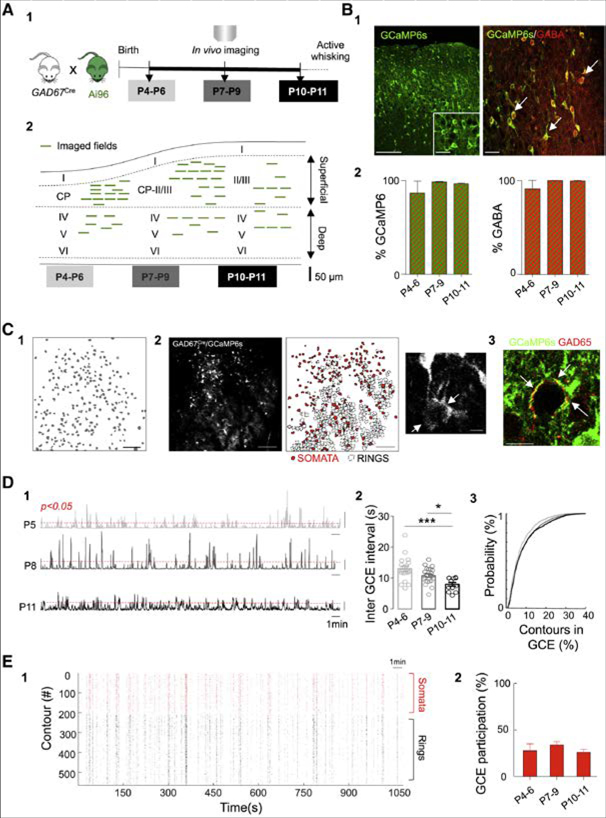
Figure 1. GABAergic Calcium Events (GCE) Recurring during Postnatal Development.
(A) Schematic representation of the experimental timeline (A1). Experiments were performed in non-anesthetized GAD67-GCaMP6s pups between postnatal days 4 (P4) and 11 (P11). Location of the fields of view (FOV, green lines) at P4-P6 (superficial layers = 13, and deep = 6), P7-P9 (superficial = 17, deep = 7) and P10-P11 (superficial = 7, deep = 5) (A2). Each line represents one imaged field of view.
(B) Immunohistochemistry and quantification of GCaMP6s expression and GABA co-labeling to test the GABAergic identity of imaged cells (B1 and B2). White arrows indicate co-expression of GCaMP6s (green) and GABA (red). Scale bars, 100 μm and 20 μm, respectively. Bar plots (B2) indicate the fraction of GCaMP6s positive contours expressing GABA (left) as a function of developmental time (P4-P6: 87% ± 12%; P7-P9: 99% ± 0.5%; P10-P11: 97% ± 0.8%, n = 2, 3, 2 FOV for each age, 2 animals per age) as well as the fraction of GABA contours expressing GcaMP6s (right) (P4-P6: 90% ± 9%; P7-P9: 99% ± 0.1%; P10-P11: 99% ± 0.5%, n = 2, 3, 2 FOV for each age, 2 animals per age).
(C) Representative contour plot of the active SOMATA detected in a FOV (scale bar 100 μm) (C1). Maximum intensity projection of a representative field of view (600 × 600 μm; scale bar, 100 μm) and detection of GABAergic SOMATA (red filled) and RINGS (black contours) (C2). Magnification of a Ring detail (scale bar 10 μm). Immunohistochemical analysis of GCaMP6s (green) expression and GAD65 (red) shows that RINGS are often co-labeled with GAD65 (red) (see arrows, scale bar 20 μm) (C3).
(D) Representative histograms showing the number of active contours as a function of time within imaging sessions at P5 (top), P8 (middle), and P11 (bottom) (D1). Scale bar, 50 contours. Red dotted line indicates statistical threshold above which the number of co-active contours exceeds chance levels (p < 0.05). Bar plots of the average time interval between GCEs as a function of age (open circles indicate one FOV) (D2). Two-way ANOVA shows no interaction (F(2,49) = 2.23, p = 0.11) or depth effect (F(1,49) = 0.65, p = 0.42) but an age main effect (2, F(2,49) = 13.69, p < 0.0001). Post hoc test (Tukey) indicates a decrease in the inter-GCE interval overtime: P10-P11 versus P4-P6 (p < 0.001) and P10-P11 versus P7-P9 (p < 0.05). Cumulative probability distribution of contours recruitment into GCEs at P4-P6, P7-P9 and P10-P11. Kolgomorov-Smirnov test, adjusted α = 0.05/3. P4-P6 versus P7-P9 and P10-P11, p < 0.01 (D3).
(E) Representative rasterplot indicating calcium events onsets detected in SOMATA (red) and RINGS (black) in a given imaging session as a function of time (E1). Fraction of SOMATA recruited in GCEs at different developmental stages (E2). One-way ANOVA indicates no differences between the fraction of SOMATA recruited within GCEs over time; F(2,53) = 0.66; p = 0.51. Data in the plot are represented by mean ± SEM.
As previously reported, using the same driver mouse line in the CA1 pyramidal layer of the hippocampus (Muldoon et al., 2015), calcium fluorescence transients could be detected not only in the soma of GABAergic neurons, but also in putative perisomatic axonal terminals forming a thin ring of fluorescence around unlabeled cell bodies (Figures 1C and 2). These fluorescent rings were often co-labeled with GAD65 antibody staining, further supporting their axonal identity (Figures 1C and 3). Contours reflecting “somatic” versus “perisomatic” activity were then manually classified as SOMATA or RINGS.
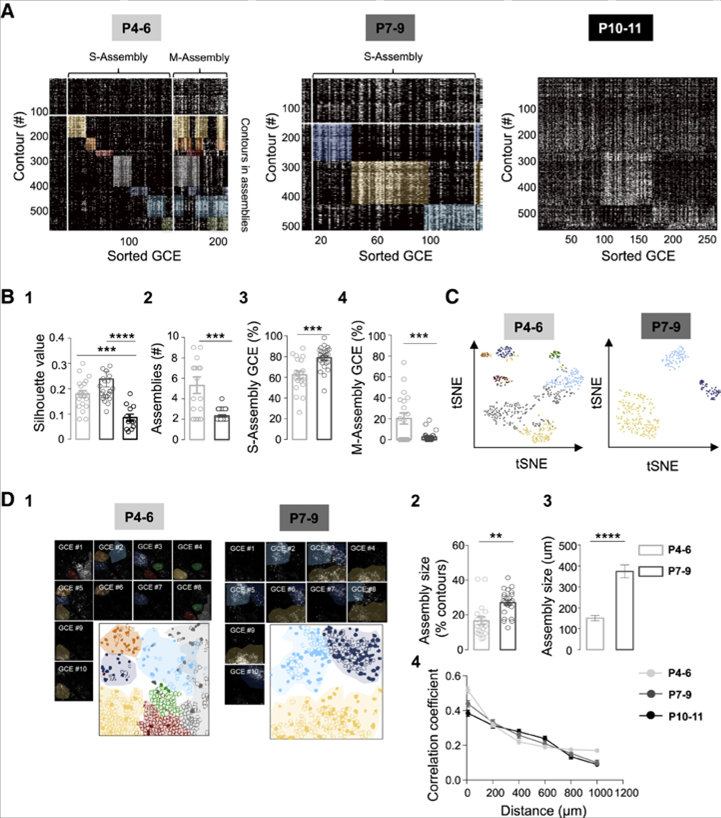
Figure 2. GABA Assemblies during the Early Postnatal Period.
(A) Representative rasterplots of GCEs for one imaging session at P5, P8, and P11, sorted according to our functional clustering algorithm. Contours are comprised of SOMATA and RINGS. P4–P6: 5 mice, 13 superficial and 6 deep FOVs; P7–P9: 5 mice, 17 superficial and 7 deep FOVs; P10–P11: 3 mice, 7 superficial and 5 deep FOVs.
(B) Two-way ANOVA was performed to analyze changes across development (age factor) in superficial and deep cortical layers (depth factor). Unpaired t test was applied when there was no interaction effect but the two-way analysis reported simple main effects. S or M assembly: GCEs recruiting a single or multiple assemblies; Functional coherence of assemblies (silhouette value) decreases at P10–P11 (B1). Two-way ANOVA report no interaction (F(2,49) = 1.29, p = 0.28) or depth effect (F(1,49) = 0.80, p = 0.37) but main simple effect in the age factor (F(2,49) = 17.57, p < 0.0001), Post hoc (Tukey) P10–P11 versus P4–P6, p < 0.001 and P10– P11 versusP7–P9, p < 0.0001.Number of functional assemblies decreases across development (B2). Two-way ANOVA report no interaction (F(1,39) = 0.01, p = 0.91) or depth effect (F(1,39) = 0.27, p = 0.59) but main simple effect in the Age factor (F(1,39) = 14.04, p = 0.0006). Unpaired t test: t(41) = 4.12, p = 0.0002. Fraction of GCEs per imaging session recruiting a single assembly (S-Assembly) as a function of age (B3). Two-way ANOVA report no interaction (F(1,39) = 0.20, p = 0.65) or depth effect (F(1,39) = 3.32, p = 0.07) but main simple effect in the age factor (F(1,39) = 12, p = 0.001). Unpaired t test: t(41) = 3.92, p= 0.0003. FractionofGCEsper imaging session recruiting multiple assemblies (M-Assembly) as a function of age (B4). Two-way ANOVA report no interaction (F(1,39)= 0.02, p = 0.87) or depth effect (F(1,39) = 0.06, p = 0.80) butmain simple effect in the age factor (F(1,39) = 12.60, p = 0.001). Unpaired t test: t(41) = 3.62, p = 0.0008. Data in the plots are mean ± SEM.
(C) Stochastic neighbor embedding (tSNE) showing a two-dimensional representation of the functional correlation between the local structure (contours) and the global structure (assemblies) color-coded according to their belonging to functional assemblies displayed in (A).
(D) Representative movie frames showing GCEs imaged at P5 and P8 and the contour map of SOMATA (filled) and RINGS (open), color-coded according to their assembly membership (D1). Mean fraction of active contours involved in one assembly at P4–P6 and P7–P9 (D2). Two-way ANOVA, no interaction effect (F(1,39) = 0.05, p = 0.819) or depth effect (F(1,39) = 0.59, p = 0.44), but main simple effect in the Age factor: F(1,39) = 16.83, p = 0.0002. Subsequent unpaired t test, t(41) = 4.05, p = 0.0002. Mean size of assemblies in μm at P4–P6 (n = 21 assemblies, 3 mice) and P7–P9 (n = 6 assemblies, 3 mice) (D3). Unpaired t test: t(25) = 7.94, p < 0.0001. Each circle represents a FOV. Data in the plots are represented by mean ± SEM 4, pairwise correlation as a function of distance for all possible cell pairs as a function of age. Each dot represents the mean value for the pooled data set.
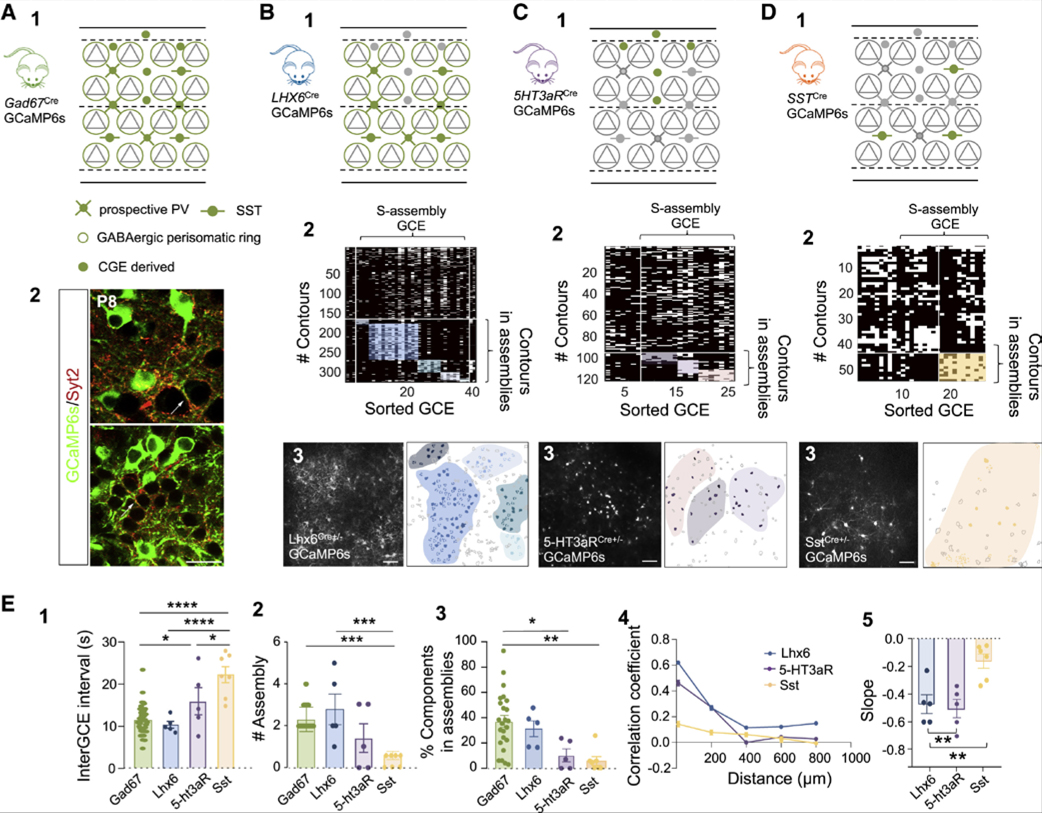
Figure 3. Specific Involvement of Different Subtypes of Interneurons into Assemblies.
(A) Schematic representation of the cells (GABAergic neurons) targeted with the GAD67-GCaMP mouse line (A1). Gray triangles represent glutamatergic neurons and green indicates cells expressing GCaMP6s; perisomatic RINGS are large open circles. Confocal images indicating co-labeling (arrows) between GCaMP (green) and synaptotagmin-2 (Syt2, red) at P8 around the somata of putative glutamatergic neurons in the deep (top) and superficial (bottom) layers of the barrel cortex (A2). Scale bar, 20 μm.
(B-D) Same as (A1) for the different interneuron subtypes targeted with the following different mouse lines: (B) Lhx6-GCaMP6 (3 mice, 5 FOVs); (C) 5-HT3aR-GCaMP6 (3 mice, 5 FOVs); and (D) SST.GCaMP6 (3 mice, 6 FOVs) (B1, C1, and D1). Representative rasterplots of GCEs re-ordered following functional clustering occurring within one representative imaging session in the mouse lines indicated in 1 (B2, C2, and D2). Maximum intensity projection of a representative field of view (scale bar, 50 μm) and corresponding contour map of active SOMATA (filled) and RINGS (doted contours), color-coded according to their assembly membership following to 2 (B3, C3, and D3).
(E) Bar plots of the average time interval between GCEs in different interneuron subtypes (E1). One-way ANOVA indicates significant differences between groups: F(3, 37) = 18.80; p < 0.0001. Post hoc (Tukey’s) shows differences when comparing GAD67-GCaMP6 versus SST-GCaMP6 (p < 0.0001), Lhx6-GCaMP6 versus SST-GCaMP6 (p < 0.0001), SST-GCaMP6 versus 5-HT3aR-GCaMP6 (p < 0.05), and GAD67-GCaMP6 versus 5-HT3aR-GCaMP6 (p < 0.05). Comparison between numbers of functional assemblies observed in GAD67-GCaMP6 (green), Lhx6-GCaMP6 (blue), 5-HT3aR-GCaMP6 (purple), and SST-GCaMP6 (yellow) pups (E2). One-way ANOVA indicates significant differences between groups: F(3, 37) = 9.31; p = 0.0001. Post hoc (Tukey’s) shows that SST-GCaMP6mice display a significantly lower number of assemblies when compared to GAD67-GCaMP6 (p < 0.001) and Lhx6-GCaMP6 (p < 0.001). Comparison between the percentage of contours that are involved in assemblies in GAD67-GCaMP6 (green), Lhx6-GCaMP6 (blue), 5-HT3aR-GCaMP6 (purple), and SST-GCaMP6 (yellow) mice (E3). One-way ANOVA indicates differences between groups (F(3, 37) = 6.68; p = 0.001), particularly when comparing GAD67-GCaMP6 with SST-GCaMP6 (p < 0.01) and 5-HT3aR-GCaMP6 (p < 0.05). Pairwise correlation of activity onsets as a function of distance recorded in Lhx6-GCaMP6 (blue), 5-HT3aR-GCaMP6 purple) and SST-GCaMP6 (yellow) mice and (E4), regression coefficient (slope) of the distributions of pairwise correlation as a function of distance (E5). One-way ANOVA indicates differences between interneuron subtypes, particularly when comparing SST-GCaMP6 to Lhx6-GCaMP6 (p < 0.01, post-hoc Tukey’s) and SST-GCaMP6 to 5HT3aR-GCaMP6 (p < 0.01, post-hoc Tukey’s). Each dot is a FOV. Data in the plots are mean ± SEM.
Significant peaks of synchronous activity originating from GABAergic contours (GABAergic calcium events [GCEs]) were detected (Figures 1D and 1). GCEs recurred on average about once every ten seconds and their frequency slightly increased as a function of age in both superficial and deep cortical layers (13 ± 1 s (P4-P6, n = 19 movies from 5 mice), 11 ± 0.5 s (P7-P9, n = 24 movies from 5 mice) and 8 ± 1 s (P10-P11, n = 12 movies from 3 mice, Figures 1D and 2). Interestingly, GCEs occurred significantly more often, at least at P4-P6, than global network events involving both glutamatergic and GABAergic cells (Figure S1, methods, p < 0.0001). The fraction of active contours involved per GCE was variable (interquartile range: 3%−10%, log-normal distribution, 55 movies from 13 mice) and it significantly increased from P4-P6 to P7 onward (Figures 1D and 3). On average, SOMATA represented the smallest fraction of the contours recruited in GCEs across ages (16% ± 1% at P4-P6, 16% ± 1% at P7-P9, and 18% ± 2% at P10-P11, n = 55 movies, 13 mice, Figures 1E, 1E1, and 1E2), with RINGS being the major contributors. We observed that the activation of RINGS significantly preceded that of SOMATA in superficial layers at all ages (difference in the mean onset [At] between RINGS and SOMATA at P4-P6, p < 0.01; P7-P9, p < 0.001, Figure S1; and P10-P11, p < 0.01). This delayed SOMATA activation was not observed in deeper layers until P10–11 (Figure S1). These differential dynamics of recruitment suggest that GCEs coordinate functionally diverse sources of GABAergic activity.
Our results indicate that, even at a stage when GABAergic input onto principal cells is thought to be limited in vivo, particularly the one provided by soma-targeting cells, GABAergic cells already operate as a functional network displaying recurring coordinated activity in the form of GCEs with sub-compartment-and layer-specific temporal dynamics.
Functionally Orthogonal and Spatially Clustered GABA Assemblies
Given that GCEs variably synchronized GABA contours, we next asked whether GCEs could be preferentially recruiting specific assemblies. To detect and sort the different types of GCEs, we used a previously designed k-means clustering algorithm (Malvache et al., 2016) based on pairwise correlation between contour (SOMATA and RINGS) recruitment in GCEs (Figure 2). Single contours (both SOMATA and RINGS) were assigned to the assembly they mostly contributed to (Figure 2A). In this way, functional cell assemblies were uncovered from P4 to P9. Earlier than that, at P3 (n = 2 mice, FOV = 9, Figure S2), local patches of coordinated GABAergic activity could be observed, but the cellular origin of the calcium signals could not be distinguished (Figure S2), which precluded further analysis. After that, at P10-P11, a significant drop in the silhouette value was observed (Figures 2B and 1), indicating that GCEs did not display any significant cluster. Hence, only the time periods displaying an assembly type of organization (P4-P6 and P7-P9) were further examined.
The patterning of GCEs was significantly different between P4-P6 and P7-P9. Indeed, GCEs segregated into three main categories: (1) “single assembly GCEs” (S-Assembly GCE) that accounted for the majority of events (62% ± 4% at P4-P6 versus 79% ± 4% at P7-P9, Figure 2); these repeatedly recruited one assembly, with almost three times more assemblies per imaging session at P4-P6 than at P7-P9 (p < 0.001, Figure 2B). (2) “Multiple assembly GCEs” (M-Assembly GCE): a significant fraction of GCEs at P4-P6, but not later, recruited more than one assembly (20% ± 5% at P4-P6, p < 0.01, Figure 2). (3) The remaining GCEs (17% ± 3% at P4-P6 and 18% ± 2% at P7-P9) did not involve any defined set of neurons (Figure S2). The change in network functional structure observed between P4-P6 and P7-P9 can readily be captured by computing the t-distributed stochastic neighbor embedding (t-SNE). This machine learning algorithm for visualization is a dimensionality reduction technique that segregates data points (here spike trains of active contours) onto a two-dimensional space (see STAR Methods) according to how similar they are. This visualization further allows representing the global network structure (here assemblies), which shows that contours belonging to the same cluster are more segregated in that two-dimensional space at P7-P9 than P4-P6, as expected from the disappearance of multiple assembly activation (Figure 2C). Importantly, despite the smaller contribution of SOMATA to GCEs than RINGS, the developmental patterning of GCEs into separate assemblies described above also holds when only somatic activity is considered (Figure S3). Lastly, segregation of GCEs into distinct categories did not differ between superficial and deep layers, including layer 4 (n = 8, fields of view [FOVs], Figure S2).
Therefore, our results show that GABAergic contours displayed stereotyped dynamics across cortical layers in the form of functional assemblies becoming independent from each other at the end of the first postnatal week. We next investigated the anatomical structure of these assemblies.
In all imaged layers, assemblies were not only functionally segregated, but also anatomically clustered and non-overlapping in physical space both at P4-P6 and P7-P9 (Figures 2D and 1). Indeed, the mean distance between assembly contour pairs was significantly smaller than the average distance between all active contour pairs (Figures 2D and 2). The spatial extent of assemblies doubled between P4–P6 (150 ± 12 μm, n = 3 movies, 3 mice) and P7–P9 (374 ± 30 μm, n = 3 movies, 3 mice, Figure 2D), whereas the average barrel size (estimated using immunoreactivity for the vesicular glutamate transporter, VGluT2, Figure S4) only slightly increased from 104 ± 5 μm at P4–P6 (n = 9) to 138 ± 6.5 μm at P7–P9 (n = 9). This indicates that functional GABA assemblies first matched the size of single barrels and then broadened their spatial domains across multiple barrels. Interestingly, contours involved in GCEs at P10–P11 were no longer spatially clustered (Figure S5). In addition, pairwise correlations distributed similarly as a function of distance across different age groups (Figures 2D and 4), indicating that whereas distance is the main predictor of whether two cells are correlated, it cannot account for the population dynamics described above.
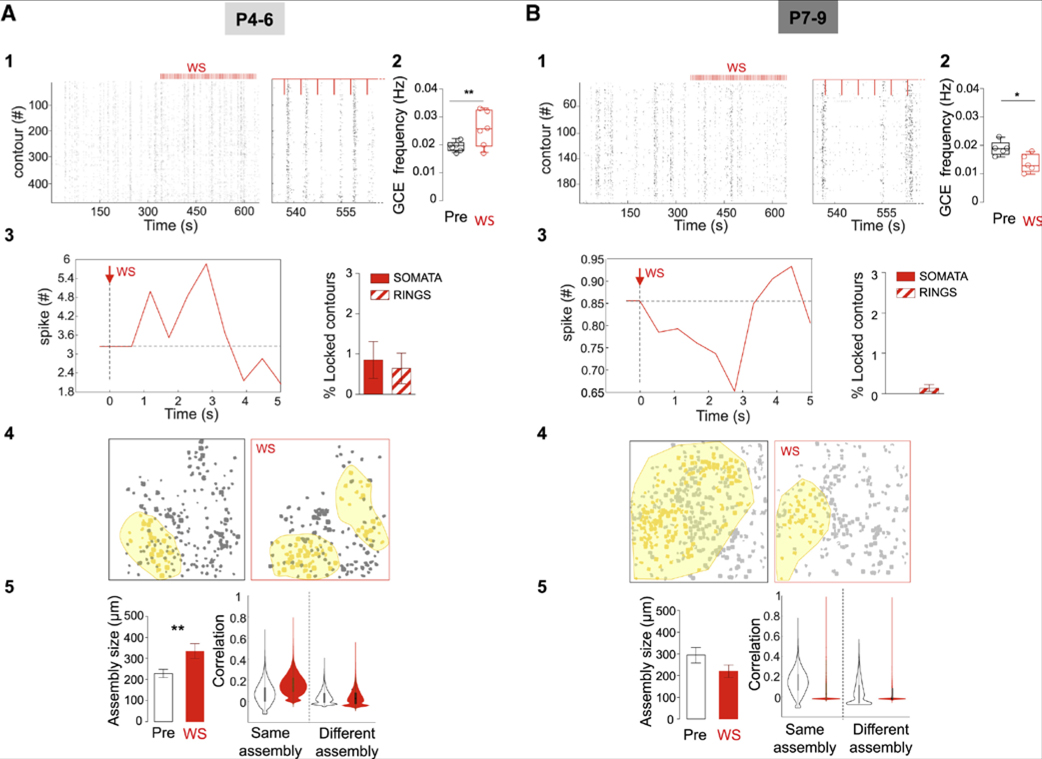
Figure 4. Impact of Whisker Stimulation on GABAergic Dynamics as a Function of Age.
(A and B) Rasterplots of calcium events onsets as a function of time in all active contours within an imaging session (right rasterplot at an expanded timescale) that included whisker stimulations (WS) period in a representative P5 (A1) and P7 (B1) pup (P4–P6: 3 superficial and 3 deep FOVs, 2 mice; P7–P9: 3 superficial and 2 deep FOV, 2 mice); Red vertical lines indicate stimulation pulses (every 4 s). (A2 and B2) Boxplots indicating the average GCE frequency before and during WS for all FOVs at P4–P6 (A2) and P7–P9 (B2). GCE frequency significantly increases during WS at P4–P6 (unpaired t test, t(10) = 2.95, p = 0.01), but decreases at P7–P9 (unpaired t test, t(8) = 2.84, p = 0.02). (A3 and B3) Averaged number of activation onsets per contours during WS (red line) as compared to the pre-stimulation period (baseline, dotted line) at P4–P6 (A3) and P7–P9 (B3). Boxplots indicate the fraction of active contours (SOMATA and RINGS) that display a time-locked response to single WS. (A4 and B4) Contour map of active cells during pre-stimulation and WS periods; yellow filled contours are members of the same assembly for a representative case at P4–P6 (A4) and P7–P9 (B4). (A5 and B5) Mean size of assemblies in micrometers at P4–P6 (A5) (15 spontaneous and 13 stimulation registered assemblies, n = 2 mice) (A5) and P7–P9 (B5) (14 spontaneous and 10 stimulation registered assemblies, n = 2 mice) (B5) during baseline (left) and WS. Unpaired t test: t(26) = 2.70, p = 0.01 and t(22) = 1.51, p = 0.14. Data in the plots are represented by mean ± SEM. Left boxplots indicate the average assembly size before stimulation (PRE) and WS periods (red). Violin plots represent the pairwise correlation between cells belonging to the same or different assemblies during the pre-stimulation and WS periods (red) at P4–P6 (A5) and P7–P9 (B5).
Taken together, our data delineate two critical developmental time points in the organization of collective GABAergic dynamics in the barrel cortex, the first at the beginning of the second postnatal week (P7-P9), where local assemblies broaden but sharpen their borders as evidenced by the disappearance of multiple-assembly events and as observable using t-SNE visualization, and the second just before the onset of active whisking (P10-P11) when GABAergic microcircuits change from recurring assembly activation to global network events involving spatially intermingled GABA contours. Our data also indicate that perisomatic GABA RINGS are active components of population dynamics throughout that neonatal period. We next investigated the identity of the GABA neurons involved in the assemblies.
Perisomatic MGE-Derived GABAergic Neurons Are the Main Components of GABA Assemblies
In order to determine whether GABA assemblies indiscriminately involved all interneurons or specific subsets, we restricted GCaMP6s expression in defined subfamilies using the following Cre lines: (1) Lhx6i-Cre (n = 5 movies, 3 pups) to select MGE-derived interneurons, i.e., mainly the PV- and SST-expressing subtypes (Fogarty et al., 2007); (2) 5HT3aR-Cre (n = 5 movies, 3 pups) that targets neurons originating from the CGE (Lee et al., 2010); and (3) SST-Cre (n = 7 movies, 3 mice). For simplification, the pups born from these crossings will be designated as Lhx6-GCaMP, 5HT3aR-GCaMP, or SST-GCaMP. Imaging was performed at P7-P9, the time when assemblies become functionally orthogonal (Figures 3 and S6). Recurring synchronous calcium events were observed in all three cases and occurred at a similar rate as GCEs in Lhx6-GCaMP mice, but at a lower rate in both 5-HT3aR-GCaMP6 (p < 0.05) and SST-GCaMP6 pups (p < 0.0001). The latter displayed a twice slower frequency (Figures 3E and 3E1). In addition, about one third of MGE- and a small minority of CGE-derived interneurons (31% ± 6% active contours and 10.2% ± 5% respectively) segregated into functionally and spatially clustered assemblies as previously described in Gad67-GCaMP6 mice (Figures 3E, 3E2, and 3E3). This was not observed in SST-GCaMP6 pups, for which contour pairs were similarly correlated across the entire FOV irrespective of anatomical distance (Figures 3D, 3D1, 3E4, 3E5; Figure S6). We conclude that assemblies were more likely comprised of MGE-derived neurons excluding the SST-expressing subtype, leaving PV-expressing interneurons as the main component of GABA assemblies. Accordingly, RINGS were mainly detected in Lhx6-GCaMP (Figures 3B, 3B3, 3E, 3E3) where they represented more than half of the detected active contours (56% ± 3%, Figure S6), indicating they likely originated from future PV-expressing cells. In order to further establish their future PV identity, we performed immunohistochemistry against synaptotagmin-2 (Syt2), a marker that mimics and precedes PV expression in synaptic boutons (Sommeijer and Levelt, 2012). With this proxy, we confirmed the identity of fluorescent RINGS on Gad67-GCaMP mice as future PV cell terminals (Figures 3A and 3A2).
We conclude that before the expression of PV, prospective PV cells and their perisomatic arborization (with a minor contribution of CGE cells) form local patches of GABAergic activity, while SST cells coordinate their activation at a slower pace in a distance-independent manner.
Because the development of GABAergic circuits was previously shown to depend on sensory drive (Butt et al., 2017; Chattopadhyaya et al., 2007; Cheet al., 2018; De Marco García et al., 2015; Deng et al., 2017; Micheva and Beaulieu, 1995b; Oh et al., 2016; Tuncdemir et al., 2016; Wamsley and Fishell, 2017) in contrast to the sparsification of neuronal activity in upper cortical layers (Golshani et al., 2009), we last analyzed the modulation of GABA assemblies by sensory inputs.
GABA Assemblies Can Be Evoked by Whisker Stimulation and Altered in Sensory-Deprived Pups
We first asked whether collective GABA dynamics could be modulated by sensory stimulation. To do so, the activity of GABA contours was monitored while stimulating the whiskers periodically (Figure 4). Whisker stimulation (WS) induced opposite effects on GABA contours when comparing P4-P6 (Figure 4A) to P7-P9 (Figure 4B). Indeed, while WS at P4-P6 triggered a time-locked increase in the activation onsets of a small subset of SOMATA and RINGS (typically about eight in a FOV), a silencing of activity was observed at P7-P9 (Figures 4A and 4B). Accordingly, this resulted in a significant increase in the frequency of GCE occurrence during WS at P4-P6 as compared to the pre-stimulus period (p < 0.05, Figures 4A1, 4A2 and 4A3), and a significant decrease at P7-P9 (Figures 4B1,4B2 and 4B3). We next compared the spatial and functional patterning of GCEs occurring before and during the WS period (Figures 4A, 4B, 4A4, and 4A5). At P4-P6, we found that the assemblies detected during WS were covering an almost twice broader territory (comparable to the P7-P9 situation) than those occurring spontaneously (Figures 4A and 4A4). Interestingly, WS selectively strengthened the functional links between contours belonging to the same assembly because a significant increase in pairwise correlation during WS only occurred in contour pairs belonging to the same assembly prior to WS (Figure 4A). In contrast, at P7-P9, WS induced a loss of correlation between cells independent from their assembly membership (Figure 4B). These results suggest that thalamic inputs may contribute to the consolidation and expansion of GABA assemblies at P4-P6, whereas they favor their disappearance from P7-P9.
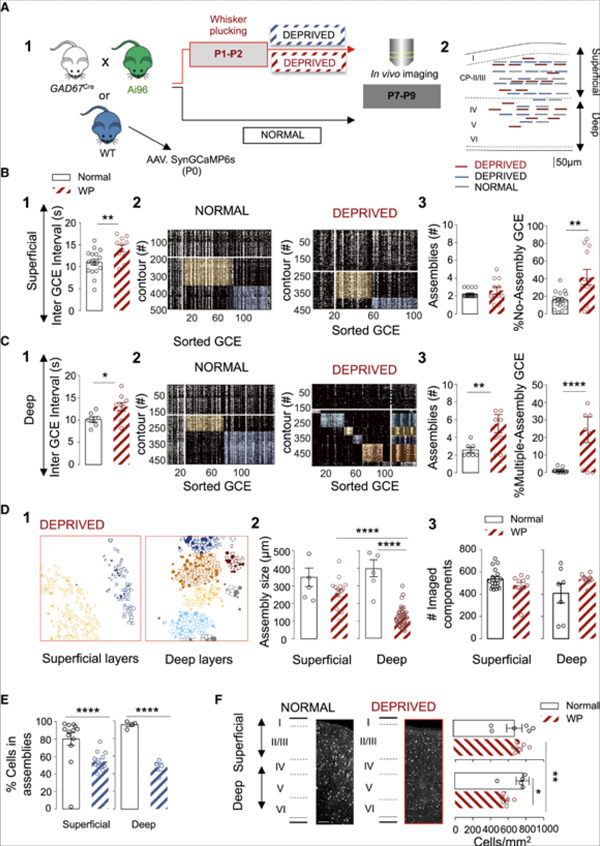
Last, we asked whether a chronic early sensory deprivation could modify the developmental patterning of GABAergic activity. To this aim, we plucked all principal whiskers bilaterally from P1 to P2 while preserving the follicles. Imaging was performed at P7-P9 (Figures 5A, 5A1, and 5A2, n = 20 movies; 4 mice). Such early sensory deprivation was previously shown to induce changes in the intracortical circuitry and functional response to stimulation of adult rodents (Micheva and Beaulieu, 1995b; Fox, 1992; Simons and Land, 1987) without disrupting the cytoarchitectural and development of the barrel field (Figure S4 and (Simons and Land, 1987)) or the sparsening of global cortical dynamics (Golshani and Portera-Cailliau, 2008). We confirmed here using VGluT2 immunoreactivity that the barrel architecture was preserved (Figure S4). This deprivation, however, had remarkable impacts on the functional organization of neonatal GABAergic dynamics. First, the frequency of GCEs was globally decreased by about one-third in sensory-deprived animals in both superficial and deep layers (superficial p < 0.01, Figures 5B and 1, and deep layers, p < 0.05, Figures 5C and 1). Second, sensory deprivation also affected GABA assemblies (Figures 5B and 5C, 2 and 3). The strongest alterations were observed in deep layers (including layer IV, 4 FOVs, Video S4). There, the patterning of activity at P7-P9 was comparable to the one observed at earlier stages (P4-P6, Figure 5 and Video S5) in normal pups with three times more assemblies (p < 0.01, Figures 5C and 3) and an increased incidence of “M-assembly” GCEs (p < 0.0001, Figures 5C and 3). The spatial organization of GABAergic assemblies in deep layers also remained halted to a previous developmental time point (Figures 5D and 1). The averaged physical distance between contours recruited in the same assembly divided by the average contour distance showed differences in deep layers of sensory-deprived animals (Figure S7; p < 0.05). Additionally, we observed a significant decrease in the fraction of contours recruited in assemblies when comparing deep layers of deprived and control animals (p < 0.0001), but also when comparing to superficial layers of sensory deprived pups (p < 0.0001), Figure 5D). Milder changes were observed in superficial layers, where the proportion of GCEs with “No-Assembly” structure was increased when compared to control pups (p < 0.01, Figures 5B and 3). This last result further indicates that sensory-deprivation affected differentially superficial and deep cortical layers. These changes could not be explained by a difference in the total amount of imaged contours (Figures 5D and 3) SOMATA or RINGS (Figure S7), nor the percentage of contours in assemblies (Figure S7).
Figure 5. Differential Impact of Sensory Deprivation on Global Population Dynamics and GABAergic Assemblies.
(A) Schematic representation of the experimental paradigm (A1) and location of the imaged fields in superficial and deep cortical layers at P7-P9 (A2). Red lines indicate sensory deprived GAD67-GCaMP6 (superficial: 11 FOVs, 4 mice; deep 9 FOVs, 4 mice); blue lines indicate sensory deprived wild types (WT, 13 superficial and 4 deep FOVs, 3 mice); and gray lines control WT pups (13 superficial and 4 deep FOVs, 6 mice).
(B and C) Impact of whisker plucking (WP) on GCEs and assemblies at P7-P9 (normal = black; WP = red;) in superficial (B) and deep (C) cortical layers. Two-way ANOVA test used with two independent factors: sensory deprivation and depth. Unpaired t test was applied when there was no interaction effect but the two-way analysis reported simple main effects. Boxplots indicating that WP significantly increases GCE interval in superficial (B1) and deep cortical layers (C1). Two-way ANOVA shows no interaction effect (F(1,40) = 0.04, p < 0.834), but simple main effect on the deprivation factor (F(1,40) = 13.20, p = 0.0008). Unpaired t test superficial (t(26) = 3.007, p = 0.005) and deep layers (t(14) = 2.39, p = 0.031). (B2 and C2) Representative rasterplots of GCEs within one imaging session at P9, sorted according to their similarity in normal and WP pups. (B3 and C3) Boxplots indicate the effect of WP on the number of functional assemblies, the number of GCEs that did not display assembly organization (B3), and the fraction of GCEs recruiting multiple assemblies (M-Assembly, C3). Sensory deprivation increases the number of assemblies only in deep cortical layers (C3). Interaction effect: Two-way ANOVA F(1,40) = 6.64, p = 0.001. Post-hoc (Tukey): main effect in deep layers WP versus normal, p < 0.01. WP increases the ratio of GCEs that did not display assembly organization only in superficial layers (B3): interaction effect: two-way ANOVA F(1,40) = 15.03, p = 0.004. Post-hoc comparisons (Tukey): main effect in superficial layers, WP versus normal, p < 0.01; the ratio of GCE recruiting multiple assemblies is exclusively increased in deep layers: interaction effect, two-way ANOVA F(1,40) = 13.39, p = 0.0007. Post-hoc comparisons (Tukey): Main effect in deep layers, WP versus normal, p < 0.0001.
(D) Representative contour maps of SOMATA (filled) and RINGS (open), in WP conditions at P9 in superficial and deep cortical layers color-coded according to their assembly membership in (B) and (C) (D1). Mean assembly size in μm at P7-P9, in WP and normal conditions in superficial and deep cortical layers (Normal: superficial n = 6 and deep n = 6, 3 mice; WP: superficial n = 15 and deep n = 40 assemblies, 3 mice) and P7-P9 (n = 6 assemblies, 3 mice) (D2). WP prevents spatial territory broadening of GABAergic assemblies in deep layers. Interaction effect: Two-way ANOVA F(1,63) = 14.3, p = 0.0003. Post-hoc (Tukey): main effect in deep layers WP versus normal (deep) and superficial versus deep in WP conditions, p < 0.0001 for both. Shows that effects are not due to a change in the total number of imaged contours. Two-way ANOVA no interaction effect (F(1,40) = 3.38, p = 0.07), or WP effect (F(1,40) = 0.43, p = 0.51) (D3).
(E) Boxplots show that WP (blue) induces a decrease in the percentage of contours recruited in assemblies in superficial and deep cortical layers of WT pups. Interaction effect, two-way ANOVA F(1,30) = 4.09, p = 0.05. Post hoc (Tukey): Superficial normal versus deprived, p < 0.0001, and deep normal versus deprived, p < 0.0001.
(F) Density of GAD67-positive cells per square millimeter in superficial (layers I/II/III) and deep layers in the barrel cortex of normal and WP animals aged P9 (n = 3 animals normal and n = 3 animals WP); scale bar 100 μm. Sensory deprivation reduces the density of GAD67-positive cells in deep layers (Two-way ANOVA, Interaction effect F(1,20) = 4.39, p = 0.049). Post-hoc (Tukey’s), indicates a decrease in the density of GABA cells per mm2 when comparing to WP superficial layers (p < 0.01), but also with deep layers of non-sensory deprived animals (p < 0.05). Each dot represents a FOV. Data are mean ± SEM.
Therefore, our results indicate that sensory inputs provided by the whiskers contribute to the normal coordination of perisomatic GABA circuits, including prospective PV-expressing cells. This is in striking contrast with previous results in which similar whisker plucking did not affect activity correlation at the global population level, i.e., when analyzing together the activity of both glutamatergic and GABAergic neurons. To determine if the impact of sensory deprivation was specific to GABAergic networks or could be observed at the whole population level, we next performed the same experiments in pups injected with AAV-Syn-GCaMP6s at P0 (Figure S7; normal: n = 17 movies, 6 mice; sensory-deprived: n = 17 movies, 3 mice) to label all cortical neurons and applied a similar clustering analysis. We found no significant difference in the frequency of synchronous calcium events (SCEs) between deprived and control mice (Figure S7) except for a decreased proportion of contours recruited in functional clusters in sensory-deprived pups (p < 0.0001 for superficial and deep layer, Figure 5E). This shows that sensory deprivation has a stronger impact on GABAergic components than on their glutamatergic counterparts.
The developmental period addressed here matches a developmental episode characterized by interneuron cell death (Wong et al., 2018, see also Duan et al., 2019). We last asked whether sensory deprivation could specifically affect interneuron survival and thus induce a change in the total amount of GABAergic cells. To do so, we counted GABAergic interneurons using the GAD67-green fluorescent protein (GFP) knockin (KI) (GAD67-GFP) mouse (Tamamaki et al., 2003). The number of GFP-expressing cells per mm2 in superficial and deep layers of the somatosensory cortex from deprived (n = 3) and control pups (n = 3) at P9 was computed (Figure 5F). We observed a significant loss in the number of GFP-positive cells only in deep layers of deprived pups as compared to control and superficial layers (p < 0.05 and p < 0.01 respectively, Figure 5F). Altogether this further indicates that interneurons located in deep layers are particularly sensitive to sensory inputs during that early developmental stage, most likely in an activity-dependent manner.
DISCUSSION
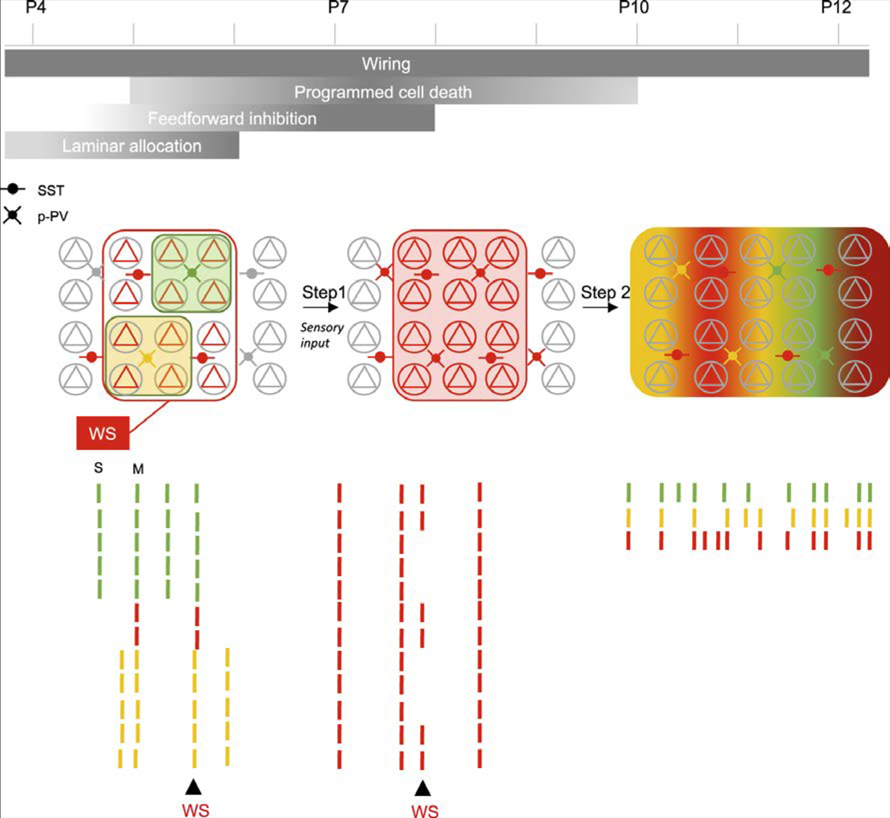
Here, we studied how cortical GABAergic cells work as a population during neonatal development without any a priori assumption on the role of a specific subtype. We show that prospective PV-containing perisomatic interneurons segregate within distinct assemblies critically modulated by sensory inputs. Two fast developmental transitions in their functional organization are uncovered: the first occurs at the end of the postnatal week with the emergence of anatomically and functionally clustered GABA assemblies that switch from positively to negatively modulated by whisker stimulation and the second two days later is the merging of GABA assemblies into a fully functionally connected network.
The main contributor to the coordinated GABA dynamics observed here are the somata and terminals of perisomatic cells, most likely belonging to the prospective-PV expressing basket subtype (Chattopadhyaya et al., 2004). This conclusion is supported by several results. First, our fate-mapping analysis using three different mouse lines shows that distinct GABA assemblies are mainly observed among MGE-derived GABA neurons that did not express SST, leaving prospective PV neurons as the main candidate subtype. In addition, the fluorescent RINGS that contributed to the majority of coactive contours in assemblies most likely originated from co-active somatic GABAergic terminals: (1) they were frequently found to express GAD65, a marker of GABAergic synapses (Chattopadhyaya et al., 2004), as well as Syt2, an early marker of PV-containing terminals (Sommeijer and Levelt, 2012); (2) they were automatically detected as co-active pixels around non-labeled somata (Muldoon et al., 2015); (3) their activation dynamics were temporally distinct from that of somata. Importantly, assemblies segregated even when only somatic contribution was taken into account. The contribution of perisomatic GABAergic synapses to early correlated activity (probably as early as P3, Figure S2) may seem at odds with the delayed maturation of PV cells but it is compatible with early reports indicating that the distribution of GABA synapses along the somato-dendritic domain of principal cells is the same at P5 and P60 (Micheva and Beaulieu, 1995a). Interestingly, in contrast to prospective PV neurons or CGE-derived interneurons, SST cells were correlated across spatial extents comprising several barrels (over 500 μm). In addition, spontaneous co-activations of SST interneurons occurred at a similar rate as multiple-assembly events, therefore less often than single-assembly events at P5-P7. This suggests that SST cells may be mainly recruited when at least two assemblies of prospective PV neurons are co-activated and as such be involved in the binding of segregated assemblies into larger segregated functional domains. This is in agreement with the demonstrated hub function in vitro of some of these cells with an early birthdate (Mòdol et al., 2017; Wang et al., 2019), their importance in the development and coordination of GABAergic networks in vitro, their role in the development of perisomatic GABAergic inputs, and their early preferential wiring to thalamic inputs (Mòdol et al., 2017; Picardo et al., 2011; Tuncdemir et al., 2016). One appealing possibility is that thalamic inputs would recruit deep layer SST cells at P4-P6, which in turn would synchronize segregated clusters of perisomatic inhibition, most likely through an excitatory GABA action (Kirmse et al., 2015), while a few days later, feedforward inhibition following thalamic stimulation is already mature, (Daw et al., 2007), even onto GABA assemblies, as confirmed by the whisker stimulation experiments performed here.
If single assemblies of prospective PV neurons may be co-activated into multiple-assembly events through a circuit involving SST neurons, the synchronization of components within single assemblies may instead be triggered by nearby excitatory glutamatergic neurons directly driven by patterned, sensory-independent thalamic inputs from barrelettes (Antón-Bolaños et al., 2019; Minlebaev et al., 2011; Mizuno et al., 2018). This would concur with the fact that single assemblies match the size of single barrels. Other mechanisms cannot be excluded, including the coordination through electrical synapses (Allene et al., 2012; Dupont et al., 2006 or by neuromodulatory inputs, such as cholinergic ones (Bruel-Jungerman et al., 2011).
The developmental function of these assemblies is an important issue. An appealing possibility would be that the patterning of GABA collective dynamics (rather than single-cell activation rates) may act as a selective mechanism to prevent interneuron cell death (see Duan et al., 2019). The first postnatal week is indeed known to exhibit a major wave of apoptosis with the peak of neuronal loss occurring at P5 for pyramidal cells (Blanquie et al., 2017; Nikolic et al., 2013; Southwell et al., 2012) and P7-P8 for interneurons (Wong et al., 2018), thus coinciding with the enlargement of functional GABAergic assemblies reported here. In that respect, delaying the merging of assemblies into wider and functionally segregated domains as reported here following sensory deprivation correlates with an increased GABA cell death (see also Micheva and Beaulieu, 1995b; Narducci et al., 2018), whereas accelerating such broadening increases GABA survival (see companion paper by Duan et al., 2019 and Figure 6). Coordinated activation of perisomatic GABA neurons within assemblies may coordinate their loss or survival, but it may as well contribute to perisomatic GABAergic synaptogenesis, in agreement with previous reports (Chattopadhyaya et al., 2004; Jiao et al., 2011; Marques-Smith et al., 2016; Tuncdemir et al., 2016).
Figure 6. Schematic Representation of the Developmental Evolution of GABAergic Assemblies.
Developmental timeline representing the major steps in the maturation of GABAergic circuits for the period studied here. The functional structure of the active circuits corresponding to each period are schematized below with glutamatergic cells (pyramids), SST-expressing interneurons (SST) and prospective interneurons (p-PV). Assemblies are schematized by colored domains. Schematic rasterplots of the activity color-coded according to assembly membership, and effects of Whisker Stimulation (WS) is indicated. S and M indicate examples of a single or multiple assembly event, respectively. Step 1 depends on sensory input.
Importantly, the present data also show that developing perisomatic GABAergic networks can be modulated externally by environmental influences, whereas the development of the majority of cells, including glutamatergic ones, may be following an internally set process (Golshani et al., 2009; Che et al., 2018). Indeed, sensory deprivation significantly affected the coordination of activity between GABA cells without any major impact on their glutamatergic partners. When analyzing the activity of all cells, we did not observe any sizeable effect, except for a significant change in the percentage of contours recruited in assemblies. This mild impact of an impaired functional organization of GABAergic circuits at population level agrees with previous whole-cell recordings, indicating that glutamatergic neurons in the granular layer of the rat barrel cortex receive few, if any, spontaneous IPSCs at early postnatal stages until P6 (Minlebaev et al., 2011).
In sum, this study shows that GABAergic neurons are ideally suited to coordinate the development of local cortical networks with sensory signals during a narrow postnatal critical period. GABA perisomatic circuits would actively tailor sensory cortical circuits to the external world before active exploration and the opening of the critical period. As such, understanding the mechanisms governing the coordination of GABAergic assemblies should provide major insights into the pathogenesis of neuropsychiatric diseases.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Rosa Cossart at rosa.cossart@inserm.fr
This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal use protocols were performed under the guidelines of the French National Ethics Committee for Sciences and Health report on “Ethical Principles for Animal Experimentation” in agreement with the European Community Directive 86/609/EEC under agreement APAFIS#18185-2018122110204650. A total number of 42 mice were used in the study (24 female and 18 males). Data from all animals was included in the analysis. No effects related to the animals sex were observed. We used RCL-GCaMP6s (Ai96) (The Jackson laboratories) crossed with a double heterozygous GAD67Cre-RCL-GCaMP6s (Melzer et al., 2012) for in vivo experiments. For simplification, pups issued from such crosses will be termed GAD67-GCaMP6. Lhx6.iCre (The Jackson laboratories), SST-Cre and 5HT3aR.Cre (a gift from N. Heintz, Rockefeller University) were also crossed with RCL-GCaMP6s. GAD67-GFP-KI (Tamamaki et al., 2003) crossed with Swiss (C.E Janvier, France) were used for the ex vivo experiments and cell counting (n = 6 mice with 3 sensory deprived); 5-HT3aR-GFP-KI (a gift from G. Fishell, Harvard Medical School) were used for cell counting (n = 5 mice). Additional Swiss pups (n = 6 sensory deprived) (C.E Janvier, France) were used for the viral injections (AAV1.syn.GCaMP6s.WPRE.SV40, UPenn viral core). All crossings were performed at 7-to 8-weeks-old. All efforts were made to minimize both the suffering and the number of animals used. In agreement with a recent study (Steinmetz et al., 2017) no developmental abnormalities or aberrant activity was observed in GAD67Cre-RCL-GCaMP6s.
METHOD DETAILS
Viral Injections
In order to perform large-scale calcium imaging, pups were injected at birth with a viral solution (titer, 1012 genomes copy/mL; Penn Vector Core) of AAV1.syn.GCaMP6s.WPRE.SV40 (UPenn viral core). Virus stock was diluted 1:2. Mice were separated from the dam and anesthetized by hypothermia. The procedure lasted 10min approx. in order to minimize separation from the dam. Glass micropipettes with a tip of 30–45 μm diameter attached to a Nanoject (Drummond) injector were used for injections. A total amount of 200nl of virus was injected in the cortex 200 μm below the surface (RC: 0.5mm; ML:1.85mm). Location of injections sites was confirmed post-mortem and after the imaging. Pups were allowed to recover on a heat pad at 37Co and then returned to the dam. We did not observe any neglecting or consuming behavior after this procedure.
Cranial Window Surgery
All procedures were performed following (Golshani and Portera-Cailliau, 2008). Briefly, Betadineand lidocaine (cream) was applied to the skin tissue adjacent to the intended incision, 20 min before starting the surgical procedures. Mice aged between P3-P11 (male and female) were anaesthetized with isoflurane (1.5% via a nose cone) and placed in a stereotaxic frame containing a heating pad with bedding. All surgical instruments were sterilized. After skin removal, a custom-made head plate containing two 3.5mm diameter holes was fixed to the skull using the veterinary adhesive Vetabond (3M Vetbond Tissue Adhesive). Once the head plate was fixed, stabilized and the pup comfortably placed in a warm cotton bed, a ~3 mm diameter craniotomy was performed over the primary somatosensory barrel cortex and covered with a glass coverslip. Care was taken not to damage the dura or the underlying vessels during the procedure. A glass window was then sealed with uncured kwik-sil (WPI) and fixed to the head plate using Vetbond to reinsure the stability of the window. The head plate was then fixed to the skull using Super-bond (DSM Dentaire). Body temperature was continuously monitored and maintained at close to physiological values (34–37C°) by means of a heating pad during the whole surgical process.
In Vivo Two-Photon Calcium Imaging
After surgical procedure, pups were allowed to recover for 90 min in the same stereotaxic frame where the craniotomy was done. It contained cotton bedding and a heat pad. Throughout this period and the following experiments, pups were constantly monitored and fed with veterinary milk ad libitum. They typically remained calm while head-fixed allowing calcium imaging experiments that lasted 2–3 h. Mice did not exhibit any sign of pain or distress during the in vivo imaging. Movies were 1800 or 3600 frames at 512×512 pixels. Each movie lasted approximately 20 min. Imaging was performed with a single beam multiphoton-pulsed laser scanning system coupled to a microscope (TriM Scope II, LaVision Biotech). The Ti: sapphire excitation laser (Chameleon Ultra II, Coherent) was operated at 920 nm. GCaMP fluorescence was isolated using a bandpass filter (510nm/25nm). Images were acquired through a GaSP PMT (H7422–40, Hamamatsu) using a 16X immersion objective (NIKON, NA0.8). Using Imspector software (LaVision Biotech), the fluorescence signal from a 600 μm2 field of view was acquired at 1.5 or 3.3Hz with a 5.7 μs or 2.8 μs dwell time per pixel (1.2 μm/pixel), respectively. Movies at a faster frame rate (5, 7 and 8Hz) were also acquired in order to probe our temporal resolution. Because spontaneous activity during development is typically on the low range kinetics (Golshani et al., 2009; Kummer et al., 2016), we found that dynamics were comparable in low and high scanning rates. Movies from non-overlapping fields of view were performed in the same pup and used for the analysis (FoV are indicated in their respective figure legends).
Whisker Stimulation
Whisker stimulation was obtained by brief air puffs delivered by a picospritzer unit (Picospritzer III, Parker Hannifin) via 1 mm diameter plastic tubes placed perpendicularly ~20 mm in front of the whiskers contralateral to the imaging side. Stimulation was directed to the snout. Air puffs (100 ms duration) were given every 4 s. The air puff pressure was adjusted to avoid a startle response.
Sensory Deprivation
At P1-P2, mice were briefly removed from the dam and anesthetized by hypothermia (10 min); all whiskers were plucked using sterile forceps. Pups were allowed to recover on a heating pad before returning to their dam.
Immunohistochemistry
After anesthesia with isoflurane, Gad67Cre/GCaMP6s pups were perfused with paraformaldehyde before brain extraction. Brains were post-fixed overnight, washed in PBS, and 70 μm thick coronal sections were obtained. Coronal brain sections from fixed brains (70 μm thick sections, n = 9 pups: 3 at P5, 3 at P9 and 3 at P11) were processed for immunohistochemistry as described previously (Modol et al., 2017). Briefly, sections were blocked with PBS-Triton 0.3% and normal donkey serum (at 5%; Merk Millipore), and incubated overnight at 4° C with the following primary antibodies: rabbit anti-GABA (1:1000; Sigma_A2052; RRID:AB_477652), guinea pig anti-GAD2 (1:100; Synaptic Systems_198104; RRID:AB_10557995), guinea pig anti-VGluT2 (1:1000, Merk Millipore_AB2251-I; RRID:AB_2665454) and mouse anti-Syt2 (1:1000; Developmental Studies Hybridoma Bank_AB_2315626; RRID:AB_2315226). After several washes, sections were incubated for 2 h at room temperature with conjugated secondary antibodies Alexa Fluor 594 (1:200; Life Technologies). GABA co-labeling with GCaMP6s expression was used to quantify the constrained expression of GCaMP6s in interneurons (Figures 1 and S1). Perisomatic identity of GABAergic terminals was determined with GAD65 and GCaMP6s co-labeling (Chattopadhyaya et al., 2004).
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of Calcium Dynamics
Cell and Calcium Transients Detection
Movies were first motion-corrected using a rigid registration method (Pnevmatikakis and Giovannucci, 2017). The field of view (FOV) was split into overlapping patches of 1/4 and then 1/8th of the spatial dimension. During registration, a rigid translation was estimated for each frame of each patch by matching it to a template (initially the median image of the first 250 frames). Each registered frame was then used to update the running mean of previously registered frames. For each of the conditions two iterations were run decreasing the maximum shift allowed for each patch. Next, using the same patch dividing scheme and method as above, a FFT-based algorithm was used (Guizar-Sicairos et al., 2008), which up-sampled the signal after the initial registration, and allowed for a smooth motion field that was applied to account for non-rigid movement between patches.
Contours and calcium transients were detected using the constrained non-negative matrix factorization framework (Pnevmatikakis et al., 2016). Last, a Markov Chain Monte Carlo approach was used (Pnevmatikakis etal., 2013) initialized by the fast OOPSI algorithm (Vogelstein et al., 2010) to infer spikes from the deconvolved fluorescence trace after detrending and extracting the 20% Df/f. We then mapped selected pixels onto the ROIs detected using the non-negative matrix factorization. The remaining ROIs were then classified the as either RINGS or noise depending on their correlation image and processed time-series. RINGS were included in the analysis of GABAergic calcium events (GCEs). Pixels belonging to SOMATA were then manually sampled by going through each movie frame.
GCE and SCE Detection
To detect population events we randomly circularly reshuffled each spike vector, hence maintaining within contour spike dependencies while randomizing the population and correcting for differences in baseline firing for different animals and ages. One thousand surrogate distributions were created. The spikes of each frame for these distributions were computed and the 99th percentile of the resulting “sum of spikes” vector was used as a statistical threshold. Peaks above the threshold that were at least separated by 7 frames were considered as synchronous calcium events. A population vector is a binary representation of the spiking activity of the event (peak +− 3 frames), where a 1 is assigned when a contour has a spike within the interval and 0 otherwise.
Spike Distributions
To detect the calcium onsets of contours (SOMATA and RINGS) and quantify their individual contribution to synchronous network events, the averaged onset time of the first spike within an event was considered for all contours as well as the ratio of events that recruited a given contour and then divided the contours into the previously assigned subgroups either SOMATA and RINGS. We then computed the probability density function of onsets and scaled them to the maximum participation rate per group. Statistical analyses were then applied to measure differences in the onset dynamics (see below).
Identification of Cell Assemblies in GCE and SCE
Analyses were performed as described in (Malvache et al., 2016). Cell assemblies were identified using a clustering algorithm based on GCE or SCE similarity for cell participation followed by a statistical test for cell participation in each GCE/SCE cluster. The GCE/SCE similarity metric was the squared Euclidian distance between columns of the normalized covariance matrix. This similarity metric allowed for a more efficient clustering. Unsupervised clustering of GCE/SCE was obtained by running the k-means algorithm on this metric with cluster numbers ranging from 2 to 19. Hundred iterations of k-means were run for each cluster number and the iteration that resulted in the best-averaged silhouette value was kept. The silhouette value was computed as described previously (Malvache et al., 2016). A random distribution of average silhouette values for each cluster was calculated byreshuffling cell participation across different GCE/SCE and applying the same algorithm. Clusters with average silhouette values exceeding the 95th percentile of the random case were considered as statistically significant. Each cluster was then associated to a cell assembly that comprised those cells that significantly participated to the GCE/SCE events within that particular cluster. Cell participation to a given cluster was considered statistically significant if the fraction of synchronous events in that cluster that activated the cell exceeded the 95th percentile of reshuffled data. If a cell was significantly active in more than one GCE/SCE cluster, it was associated to the one in which it participated the most (percentage wise). The overlap between assemblies was quantified by calculating the silhouette value of each cell (with the normalized hamming distance between each cell pair as a dissimilarity metric). A cell was significantly involved in a single assembly if its silhouette value was higher than expected by chance (95th percentile after reshuffling). GCE/SCE were finally sorted with respect to their projection onto cell assemblies. A GCE/SCE was activating a given neuronal assembly if the number of cells recruited in that assembly was higher than expected by chance (95th percentile after reshuffling).
Topological Distribution of Cell Assemblies
For each cell assembly, the spatial silhouette value was computed using the pairwise anatomical distance between cells as a dissimilarity metric. To assess the statistical significance of spatial clustering for each cell assembly, its silhouette value was compared to the ones obtained after cell reshuffling (1000 iterations). Spatial clustering was considered significant if the silhouette value of the empirical assembly exceeded a certain percentile of the reshuffled silhouette value distribution. For each session, the percentile was set at (1 – 0.05/(number of clusters) to account for the inter-dependence of the comparisons (Bonferroni correction).
T-distributed Stochastic Neighbor Embedding (t-SNE)
Due to commonly associated shortcomings of the k-means algorithm, we used the t-distributed stochastic neighbor embedding method (Van Der Maaten and Hinton, 2008), to visualize the data and also verify the concurrence with the k-means analysis. The t-SNE technique converts correlations between contour vectors to joint probabilities and tries to minimize the Kullback-Leibler divergence (KL divergence) between the joint probabilities of the low-dimensional embedding and the high-dimensional data. We first reduced the dimensionality of the data by considering the first 20 Principal Components of contours participation in GCEs. These components were then non-linearly mapped onto a two-dimensional space in a way that preserved both local and global structure of the data. The perplexity parameter (the number of nearest neighbors) was chosen in an iterative procedure that minimized the KL divergence. These values ranged between 15 and 25. We then color-coded each component according to the previous k-means analysis.
Cell Counting
The density of GAD67+ cells per mm2 was calculated based on confocal images at 20x magnification obtained from normal and sensory deprived pups. 4 sections containing superficial and deep cortical layers of the somatosensory barrel fields were used to obtain an average count for each animal (n = 6 pups in total). Cell counting was performed using an ImageJ cell counting plug-in for single color fluorescence image. Density was calculated as counts/area.
Statistical Analysis
Data are given as mean and Standard Error of the Mean (SEM). Statistics and graphs were performed using Prism 7 (Graphpad) or MATLAB. Statistical analysis of changes in GCE during development in superficial and deep cortical layers was performed using two-way ANOVA in order to see the interaction between the two factors (“age factor” and “depth factor”). In order to correct for multiple comparisons we performed Tukey’s post hoc test when necessary. One-way ANOVA or t test was applied where there was no significant interaction but we observed a simple main effect (Figures 1, 2, 3, 4, 5, 6 and Figures S1, S2, S3, S5, S6, S7). The level of statistical significance is indicated with asterisks, *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001; ns, p > 0.05). All degrees of freedom for the ANOVA and the t test are indicated in their respective legends. Kolmogorov-Smirnov was used to analyze cumulative probability distributions (Figure 1).
DATA AND CODE AVAILABILITY
All the datasets generated in this study are available upon request. The codes can be downloaded from: http://gitlab.com/cossartlab.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GABA | Sigma-Aldrich | Cat#: A2052; RRID:AB_477652 |
| Guinea pig polyclonal anti-GAD2 | Synaptic Systems | Cat#: 198104; RRID: AB_10557995 |
| Guinea pig polyclonal anti-VGluT2 | Merck Millipore | Cat#: AB2251-I; RRID:AB_2665454 |
| Mouse monoclonal anti-Syt2 | Developmental Studies Hybridoma Bank | Cat#: A2315626; RRID: AB_2315226 |
| Bacterial and Virus Strains | ||
| AAV1.syn.GCaMP6s.WPRE.SV40 | UPenn viral core | Cat#: 100843 |
| Experimental Models: Organisms/Strains | ||
| Swiss Webster (CFW) | C.E Janvier | SW-F/ SW-M |
| Gad67.Cre | a gift from H. Monyer Heidelberg University | N/A |
| RCL-GCaMP6s (Ai96) | Jackson Laboratories | 02 4106 |
| SST.Cre | a gift from N. Heintz, Rockefeller University | N/A |
| Lhx6.Cre | Jackson Laboratories | 026555 |
| 5HT3aR.Cre | a gift from N. Heintz, Rockefeller University | N/A |
| GAD67-GFP-KI | a gift from K. Obata Department of Morphological Brain Science, Kyoto | N/A |
| 5-HT3aR-GFP-KI | a gift from G. Fishell Harvard University | N/A |
| Software and Algorithms | ||
| CaImAn | Pnevmatikakis et al., 2016; Giovannucci et al., 2018 | https://github.com/flatironinstitute/CaImAn-MATLAB |
| NoRMCorre | Pnevmatikakis and Giovannucci, 2017 | https://github.com/flatironinstitute/NoRMCorre |
| Iterative K-means clustering | Malvache et al., 2016 | http://gitlab.com/cossartlab |
| t-SNE | Van Der Maaten and Hinton, 2008 | https://lvdmaaten.github.io/tsne/#implementations |
Highlights.
In the developing barrel cortex, GABAergic neurons form functional assemblies
Prospective parvalbumin interneurons segregate into spatially clustered assemblies
Somatostatin interneurons display broadly correlated firing
GABA assemblies are regulated by sensory inputs
ACKNOWLEDGMENTS
This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (646925). L.M. was supported by Horizon 2020 MSCA-IF-2014 project 660795. T.T. was supported by The French National Research Agency (ANR-14-CE13-0016). Y.B. was supported by ERC (646925). A.B. and R.C. are supported by CNRS. N.V.D.: 1R01MH110553, NIMH; CURE 15-17, Citizens United for Research in Epilepsy; R.B.: 5F30MH117939, NIMH; R.B. and Z.S.D.: T32GM007739, NIGMS. We are grateful to Pr. H. Monyer for providing the GAD67-Cre mouse line. Wethank I. Bureau, R. Boyce, and R. Khazipov for critical comments on the manuscript; Dr. Pnevmatikakis for help with the analysis; and Dr. V. Villette for advice on the in vivo imaging. We thank L. Cagnacci for technical support as well as S. Pellegrino-Corby, M. Kurz, and F. Michel from the INMED animal and imaging facilities (InMagic).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.neuron.2019.10.007.
A video abstract is available at https://doi.org/10.1016/j.neuron.2019.10.007#mmc8.
REFERENCES
- Allene C, Picardo MA, Becq H, Miyoshi G, Fishell G, and Cossart R (2012). Dynamic changes in interneuron morphophysiological properties mark the maturation of hippocampal network activity. J. Neurosci 32, 6688–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiades PG, Marques-Smith A, Lyngholm D, Lickiss T, Raffiq S, Kätzel D, Miesenböck G, and Butt SJB (2016). GABAergic interneurons form transient layer-specific circuits in early postnatal neocortex. Nat. Commun 7, 10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón-Bolaños N, Sempere-Ferràndez A, Guillamón-Vivancos T, Martini FJ, Pérez-Saiz L, Gezelius H, Filipchuk A, Valdeolmillos M, and López-Bendito G (2019). Prenatal activity from thalamic neurons governs the emergence of functional cortical maps in mice. Science 364, 987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquie O, Yang J-W, Kilb W, Sharopov S, Sinning A, and Luhmann HJ (2017). Electrical activity controls area-specific expression of neuronal apoptosis in the mouse developing cerebral cortex. eLife 6, 2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortone D, and Polleux F (2009). KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron 62, 53–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel-Jungerman E, Lucassen PJ, and Francis F (2011). Cholinergic influences on cortical development and adult neurogenesis. Behav. Brain Res 221, 379–388. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Stacey JA, Teramoto Y, and Vagnoni C (2017). A role for GABAergic interneuron diversity in circuit development and plasticity of the neonatal cerebral cortex. Curr. Opin. Neurobiol 43, 149–155. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, and Huang ZJ (2004). Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J. Neurosci 24, 9598–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Wu CZ, Knott G, Kuhlman S, Fu Y, Palmiter RD, and Huang ZJ (2007). GAD67-mediated GABA synthesis and signaling regulate inhibitory synaptic innervation in the visual cortex. Neuron 54, 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che A, Babij R, lannone AF, Fetcho RN, Ferrer M, Liston C, Fishell G, and De Marco García NV (2018). Layer I Interneurons Sharpen Sensory Maps during Neonatal Development. Neuron 99, 98–116.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw MI, Ashby MC, and Isaac JTR (2007). Coordinated developmental recruitment of latent fast spiking interneurons in layer IV barrel cortex. Nat. Neurosci 10, 453–61. [DOI] [PubMed] [Google Scholar]
- De Marco García NV, Karayannis T, and Fishell G (2011). Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature 472, 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco García NV, Priya R, Tuncdemir SN, Karayannis T, and Fishell G (2015). Sensory inputs control the integration of neurogliaform interneurons into cortical circuits. Nat. Neurosci 18, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R, Kao JPY, and Kanold PO (2017). Distinct Translaminar Glutamatergic Circuits to GABAergic Interneurons in the Neonatal Auditory Cortex. Cell Rep. 19, 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan ZS, Che A, Chu P, Modol L, Bollman Y, Babij R, Fetcho RN, Otsuka T, Fuccillo MV, Liston C, et al. (2019). GABAergic Restriction of Network Dynamics Regulates Interneuron Survival in the Developing Cortex. Neuron 105 Published online November 25, 2019 https://doi.org/10.10167j.neuron.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont E, Hanganu IL, Kilb W, Hirsch S, and Luhmann HJ (2006). Rapid developmental switch in the mechanisms driving early cortical columnar networks. Nature 439, 79–83. [DOI] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, and Kessaris N (2007). Spatial genetic patterning ofthe embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci 27, 10935–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K (1992). A critical period for experience-dependent synaptic plasticity in rat barrel cortex. J. Neurosci 12, 1826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golshani P, and Portera-Cailliau C (2008). In vivo 2-photon calcium imaging in layer 2/3 of mice. J. Vis. Exp 13, e681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, Kalfon J, Koay SA, Taxidis J, Najafi F, Gauthier JL, Zhou P, Tank DW, et al. (2018). CalmAn: An open source tool for scalable calcium imaging data analysis. eLife. 10.7554/eLife.38173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golshani P, Goncalves JT, Khoshkhoo S, Mostany R, Smirnakis S, and Portera-Cailliau C (2009). Internally mediated developmental desynchronization of neocortical network activity. J. Neurosci 29, 10890–10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizar-Sicairos M, Thurman ST, and Fienup JR (2008). Efficient subpixel image registration algorithms. Opt. Lett 33, 156–158. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Zhang Z, Zhang C, Wang X, Sakata K, Lu B, and Sun Q-Q (2011). A key mechanism underlying sensory experience-dependent maturation of neocortical GABAergic circuits in vivo. Proc. Natl. Acad. Sci. USA 108, 12131–12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazipov R, and Luhmann HJ (2006). Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci. 29, 414–418. [DOI] [PubMed] [Google Scholar]
- Kirmse K, Witte OW, and Holthoff K (2010). GABA depolarizes immature neocortical neurons in the presence ofthe ketone body 3-hydroxybutyrate. J. Neurosci 30, 16002–16007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmse K, Kummer M, Kovalchuk Y, Witte OW, Garaschuk O, and Holthoff K (2015). GABA depolarizes immature neurons and inhibits network activity in the neonatal neocortex in vivo. Nat. Commun 6, 7750. [DOI] [PubMed] [Google Scholar]
- Kummer M, Kirmse K, Zhang C, Haueisen J, Witte OW, and Holthoff K (2016). Column-like Ca(2+) clusters in the mouse neonatal neocortex revealed by three-dimensional two-photon Ca(2+) imaging in vivo. Neuroimage 138, 64–75. [DOI] [PubMed] [Google Scholar]
- Lee S, Hjerling-Leffler J, Zagha E, Fishell G, and Rudy B (2010). The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J. Neurosci 30, 16796–16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Mi D, Llorca A, and Marin O (2018). Development and Functional Diversification of Cortical Interneurons. Neuron 100, 294–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvache A, Reichinnek S, Villette V, Haimerl C, and Cossart R (2016). Awake hippocampal reactivations project onto orthogonal neuronal assemblies. Science 353, 1280–1283. [DOI] [PubMed] [Google Scholar]
- Marques-Smith A, Lyngholm D, Kaufmann A-K, Stacey JA, Hoerder-Suabedissen A, Becker EBE, Wilson MC, Molnàr Z, and Butt SJB(2016). A Transient Translaminar GABAergic Interneuron Circuit Connects Thalamocortical Recipient Layers in Neonatal Somatosensory Cortex. Neuron 89, 536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Michael M, Caputi A, Eliava M, Fuchs EC, Whittington MA, and Monyer H (2012). Long-range-projecting GAB Aergic neurons modulate inhibition in hippocampus and entorhinal cortex. Science 335, 1506–1510 [DOI] [PubMed] [Google Scholar]
- Micheva KD, and Beaulieu C (1995a). Postnatal development ofGABAneurons in the rat somatosensory barrel cortex: a quantitative study. Eur. J. Neurosci 7, 419–30. [DOI] [PubMed] [Google Scholar]
- Micheva KD, and Beaulieu C (1995b). Neonatal sensory deprivation induces selective changes in the quantitative distribution of GABA-immunoreactive neurons in the rat barrel field cortex. J. Comp. Neurol 361, 574–584. [DOI] [PubMed] [Google Scholar]
- Minlebaev M, Colonnese M, Tsintsadze T, Sirota A, and Khazipov R (2011). Early g oscillations synchronize developing thalamus and cortex. Science 334, 226–229. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Ikezoe K, Nakazawa S, Sato T, Kitamura K, and Iwasato T (2018). Patchwork-Type Spontaneous Activity in Neonatal Barrel Cortex Layer 4 Transmitted via Thalamocortical Projections. Cell Rep. 22, 123–135 [DOI] [PubMed] [Google Scholar]
- Modol L, Sousa VH, Malvache A, Tressard T, Baude A, and Cossart R(2016). Spatial Embryonic Origin Delineates GABAergic Hub Neurons Driving Network Dynamics in the Developing Entorhinal Cortex. Cereb. Cortex 27, 4649–4661. [DOI] [PubMed] [Google Scholar]
- Muldoon SF, Villette V, Tressard T, Malvache A, Reichinnek S, Bartolomei F, and Cossart R (2015). GABAergic inhibition shapes interictal dynamics in awake epileptic mice. Brain 138, 2875–2890. [DOI] [PubMed] [Google Scholar]
- Narducci R, Baroncelli L, Sansevero G, Begenisic T, Prontera C, Sale A, Cenni MC, Berardi N, and Maffei L (2018). Early impoverished environment delays the maturation of cerebral cortex. Sci. Rep 8, 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M, Gardner HAR, and Tucker KL (2013). Postnatal neuronal apoptosis in the cerebral cortex: physiological and pathophysiological mechanisms. Neuroscience 254, 369–378. [DOI] [PubMed] [Google Scholar]
- Oh WC, Lutzu S, Castillo PE, and Kwon H-B (2016). De novo synaptogenesis induced by GABA in the developing mouse cortex. Science 353, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardo MA, Guigue P, Bonifazi P, Batista-Brito R, Allene C, Ribas A, Fishell G, Baude A, and Cossart R (2011). Pioneer GABA cells comprise a subpopulation of hub neurons in the developing hippocampus. Neuron 71, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, and Giovannucci A (2017). NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94. [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Merel J, Pakman A, and Paninski L (2013). Bayesian spike inference from calcium imaging data. https://arxiv.org/abs/1311.6864v1.
- Rochefort NL, Garaschuk O, Milos R-I, Narushima M, Marandi N, Pichler B, Kovalchuk Y, and Konnerth A (2009). Sparsification of neuronal activity in the visual cortex at eye-opening. Proc. Natl. Acad. Sci. USA 106, 15049–15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons DJ, and Land PW (1987). Early experience oftactile stimulation influences organization of somatic sensory cortex. Nature 326, 694–697. [DOI] [PubMed] [Google Scholar]
- Sommeijer J-P, and Levelt CN (2012). Synaptotagmin-2 is a reliable marker for parvalbumin positive inhibitory boutons in the mouse visual cortex. PLoS ONE 7, e35323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Paredes MF, Galvao RP, Jones DL, Froemke RC, Sebe JY, Alfaro-Cervello C, Tang Y, Garcia-Verdugo JM, Rubenstein JL, et al. (2012). Intrinsically determined cell death of developing cortical interneurons. Nature 491, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Buetfering C, Lecoq J, Lee CR, Peters AJ, Jacobs EAK, Coen P, Ollerenshaw DR, Valley MT, de Vries SEJ, et al. (2017). Aberrant Cortical Activity in Multiple GCaMP6-Expressing Transgenic Mouse Lines. eNeuro 4, 10.1523/ENEURO.0207-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takesian AE, Bogart LJ, Lichtman JW, and Hensch TK (2018). Inhibitory circuit gating of auditory critical-period plasticity. Nat. Neurosci. 21, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, and Kaneko T (2003). Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J. Comp. Neurol. 467, 60–79. [DOI] [PubMed] [Google Scholar]
- Toyoizumi T, Miyamoto H, Yazaki-Sugiyama Y, Atapour N, Hensch TK, and Miller KD (2013).Atheory ofthetransition to critical period plasticity: inhibition selectively suppresses spontaneous activity. Neuron 80, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncdemir SN, Wamsley B, Stam FJ, Osakada F, Goulding M, Callaway EM, Rudy B, and Fishell G (2016). Early Somatostatin Interneuron Connectivity Mediates the Maturation of Deep Layer Cortical Circuits. Neuron 89, 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605. [Google Scholar]
- Vogelstein JT, Packer AM, Machado TA, Sippy T, Babadi B, Yuste R, and Paninski L (2010). Fast nonnegative deconvolution for spike train inference from population calcium imaging. J. Neurophysiol. 104, 3691–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamsley B, and Fishell G (2017). Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat. Rev. Neurosci. 18, 299–309. [DOI] [PubMed] [Google Scholar]
- Wang C-Z, Ma J, Xu Y-Q, Jiang S-N, Chen T-Q, Yuan Z-L, Mao X-Y, Zhang S-Q, Liu L-Y, Fu Y, and Yu Y-C (2019). Early-generated interneurons regulate neuronal circuit formation during early postnatal development. eLife 8, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe J, Houweling AR, and Brecht M (2010). Sparse and powerful cortical spikes. Curr. Opin. Neurobiol. 20, 306–312. [DOI] [PubMed] [Google Scholar]
- Wong FK, Bercsenyi K, Sreenivasan V, Portales A, Fernandez-Otero M, and Marin O (2018). Pyramidal cell regulation of interneuron survival sculpts cortical networks. Nature 557, 668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets generated in this study are available upon request. The codes can be downloaded from: http://gitlab.com/cossartlab.