Abstract
Background
Cervical cancer (CC) is a common gynecological tumor that affects women’s health. Circular RNA hsa_circ_0084927 (hsa_circ_0084927) has been reported to be upregulated in CC. However, the role and regulatory mechanism of hsa_circ_0084927 in CC are unclear.
Methods
Expression of hsa_circ_0084927, microRNA (miR)-634, and tumor protein D52 (TPD52) mRNA in CC tissues and cells was examined by quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation, colony formation, cell cycle progression, apoptosis, migration, and invasion of CC cells were determined with cell counting kit-8 (CCK-8), plate clone, flow cytometry, or transwell assays. The levels of cyclin D1, cleaved-caspase-3 (c-caspase 3), matrix metalloproteinase (MMP)-2, MMP-9, and TPD52 protein were evaluated with Western blotting. The targeting relationship between hsa_circ_0084927 or TPD52 and miR-634 was verified via dual-luciferase reporter and/or RNA immunoprecipitation (RIP) assays. Xenograft assay was conducted to confirm the role of hsa_circ_0084927 in vivo.
Results
Hsa_circ_0084927 and TPD52 were upregulated while miR-634 was downregulated in CC tissues and cells. Hsa_circ_0084927 silencing reduced tumor growth in vivo and induced cell cycle arrest, apoptosis, and curbed proliferation, colony formation, migration, and invasion of CC cells in vitro. Hsa_circ_0084927 regulated TPD52 expression through sponging miR-634. MiR-634 inhibitor reversed hsa_circ_0084927 knockdown-mediated impact on the malignancy of CC cells. TPD52 elevation abolished the repressive influence of miR-634 mimics on the malignancy of CC cells.
Conclusion
Hsa_circ_0084927 accelerated CC advancement via upregulating TPD52 via sponging miR-634, offering a new evidence to support hsa_circ_0084927 as a promising target for CC treatment.
Keywords: CC, hsa_circ_0084927, miR-634, TPD52
Introduction
Cervical cancer (CC) is a common gynecological tumor, which ranks fourth among female cancer death causes worldwide.1 In recent years, the age of CC patients tends to younger.2 Despite encouraging progress in the treatment of CC, the 5-year survival rate of patients is still only 40%-50%.3,4 Therefore, exploring the mechanism of CC progression is indispensable for finding new strategies to treat CC.
Circular RNAs (circRNAs) are a type of non-coding RNAs, which are produced by reverse splicing of precursor mRNA and expression in tissue-specific or cell-specific manner.5 CircRNAs have been revealed to take part in tumor advancement by acting as sponges for microRNAs (miRs).6 For example, circRNA circ_SLC8A1 impeded bladder cancer development through modulating PTEN expression via sponging miR-130b and/or miR-494.7 Circular RNA hsa_circ_0084927 (hsa_circ_0084927) also termed as circRNA epithelial splicing regulatory protein 1 (ESRP1), is located at chromosome 8 (q22.1) with 287 bp in length (http://www.circbase.org/cgi-bin/simplesearch.cgi). Hsa_circ_0084927 has been reported to sensitize cell chemotherapy in small cell lung cancer.8 One report pointed out that hsa_circ_0084927 expression was elevated in CC tissues.9 At present, the biological function of hsa_circ_0084927 in CC is unclear.
MiRs are short non-coding RNAs that directly modulate the expression of mRNAs in different biological functions.10 Increasing researches have demonstrated that miRs exert a vital role in tumor progression via serving as tumor promoters or suppressors.11 For instance, miR-423-5p exerted a tumor repressive role in ovarian cancer12 and nasopharyngeal cancer,13 but miR-423-5p acted as a tumor promoter in breast cancer14 and glioblastomas.15 It was reported that miR-634 acted as a suppressor in some tumors, such as gastric cancer16 and pancreatic cancer.17 In CC, miR-634 could induce cancer cell apoptosis and reduce cancer cell proliferation through targeting the mTOR pathway.18 However, the regulator mechanism of miR-634 in CC remains indistinct.
Tumor protein D52 (TPD52) is considered to be an oncogene in human cancers, which is isolated from the amplification region of human chromosome 8q21.19 Previous researches demonstrated that TPD52 acted as an oncogene in lung squamous cell cancer,20 nasopharyngeal cancer,21 CC,22 and prostate cancer.23 However, the mechanism by which TPD52 participates in CC progression is unclear.
In the current study, we demonstrated that hsa_circ_0084927 exerted a cancerogenic role in CC. Also, hsa_circ_0084927 accelerated CC progression via upregulating TPD52 via sponging miR-634, which provided a promising target for CC treatment.
Materials and Methods
Patient-Derived Samples
Samples of CC tissues and paired adjacent normal tissues (n = 46) were provided by CC patients who had undergone surgical resection at The Third Hospital of Ji’nan. All CC patients had complete clinicopathological data and did not receive any medical intervention, radiotherapy, or chemotherapy before surgery. Participants had signed written informed consent prior to the analysis of tissues samples. The research was approved by the Ethics Committee of The Third Hospital of Ji’nan. The clinicopathological features of CC patients were displayed in Table 1.
Table 1.
Association Between the Clinicopathologic Characteristics and Hsa_circ_0084927 Expression in Patients with Cervical Cancer
| Factor | Number | hsa_circ_0084927 | P value | |
|---|---|---|---|---|
| Low Expression (n=23) |
High Expression (n=23) |
|||
| Age | 0.640 | |||
| <55 | 22 | 10 | 12 | |
| ≥55 | 24 | 13 | 11 | |
| Tumor size | 0.242 | |||
| <4 | 28 | 12 | 16 | |
| ≥4 | 18 | 11 | 7 | |
| Histological grade | 0.324 | |||
| Well-differentiated | 18 | 10 | 8 | |
| Moderately or poor differentiated | 28 | 13 | 15 | |
| FIGO stage | 0.031* | |||
| I+II | 21 | 13 | 8 | |
| III | 25 | 10 | 15 | |
| Lymph node metastasis | 0.014* | |||
| No | 26 | 16 | 10 | |
| Yes | 20 | 7 | 13 | |
| Depth of cervical invasion | ||||
| <2/3 | 29 | 17 | 12 | 0.025* |
| ≥2/3 | 17 | 6 | 11 | |
Note: Chi-square test, *P < 0.05.
Cell Culture
Human cervical epithelial cells End1/E6E7 and CC cell lines (Hela, SiHa, CaSki, and C-33A) were purchased from BeNa Culture Collection (Suzhou, Jiangsu, China). These cells were maintained at 37°C in a moist atmosphere with 5% CO2. End1/E6E7 cells were cultured in Keratinocyte Serum-Free Medium (K-SFM) (Life Technologies, Grand Island, NY, USA). CC cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, St Louis, MO, USA) (for Hela, SiHa, and C-33A cells) or Roswell Park Memorial Institute (RPMI)-1640 medium (Sigma) (for CaSki cells) supplemented with fetal bovine serum (FBS, 10%, Solarbio, Beijing, China) and streptomycin/penicillin (1%, Solarbio).
Cell Transfection
When the confluence reached 70%-90%, the oligonucleotides or vectors were transfected into CC cells by Lipofectamine 3000 reagent (Life technologies). Small interference (si) RNA targeting hsa_circ_0084927 (si-circRNA#1, si-circRNA#2, and si-circRNA#3) and matched negative control (si-NC) were synthesized from GenePharma (Shanghai, China). Lentivirus carrying short hairpin (sh) RNA targeting hsa_circ_0084927 (sh-circRNA) and its corresponding negative control (sh-NC) were bought from GenePharma. MiR-634 mimics and negative control (NC mimics), as well as miR-634 inhibitor and negative control (NC inhibitor), were purchased from RiboBio (Guangzhou, China). For pCD-ciR-hsa_circ_0084927 (hsa_circ_0084927) construction, the full-length of hsa_circ_0084927 was synthesized and inserted into the EcoRI and BamHI sites of the pCD-ciR vector (Geneseed, Guangzhou, China). For pcDNA3.1-TPD52 (TPD52) generation, the full-length sequence of TPD52 (NM_005079) was synthesized and subcloned into the Smal and EcoRV sites of the pcDNA3.1 vector (pcDNA) (Life Technologies).
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNAiso Plus (TaKaRa, Tokyo, Japan) was utilized to isolate total RNA from tissue samples and cells. For RNase R treatment, total RNA of CC cells was incubated with 3 U/μg RNase R (Geneseed) for 15 min at 37°C. For complementary DNA generation, total RNA was reversely transcribed with the HiScript Q RT SuperMix for qPCR Kit (Vazyme, Nanjing, China) or commercial miR reverse transcription PCR kit (RiboBio). The synthesized complementary DNA was used for qRT-PCR in the CFX96 Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the SYBR Green PCR Master Mix (Bio-Rad) based on previous studies.24,25. The following primers were used: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (F:5ʹ-GACTCCACTCACGGCAAATTCA-3ʹ; R:5ʹ-TCGCTCCTGGAAGATGGTGAT-3ʹ), hsa_circ_0084927 (F:5ʹ-TTGTAAGTGAGGAGCACCGAGAC-3ʹ; R:5ʹ-CGTGCCCTGACTACGGTGTTAT-3ʹ), ESRP1 (F:5ʹ-TGCGTTGAGGAAGCATAAAG-3ʹ; R:5ʹ-GGGTTGGAAGTGGAATGAGA-3ʹ), TPD52 (F:5ʹ-AGCATCTAGCAGAGATCAAGCG-3ʹ; R:5ʹ-AGCCAACAGACGAAAAAGCAG-3ʹ), miR-634 (F:5ʹ-CAGTCTCAAACCAGCACC-3ʹ; R:5ʹ-TATGGTTGTTCACGACTCCTTCAC-3ʹ), and U6 small nuclear RNA (U6) (F:5ʹ-GCTCGCTTCGGCAGCACA-3ʹ; R:5ʹ-GAGGTATTCGCACCAGAGGA-3ʹ). Relative expression levels were figured with the 2−ΔΔCt method, and GAPDH or U6 was used as an internal control for hsa_circ_0084927, ESRP1, TPD52, and miR-634.
Cell Proliferation Assay
The proliferation of CC cells was analyzed with cell counting kit-8 (CCK-8) assay according to previous studies.26,27 CC cells (1×103 cells/well) were seeded into 96-well plates and maintained for 24 h, 48 h, 72 h, or 96 h. Next, the CCK-8 solution (20 μL, Beyotime, Shanghai, China) was added to each well. Thereafter, the Microplate Reader (Bio-Rad) was used to measure the absorbance at 450 nm.
Plate Clone Assay
For cell colony formation analysis, CC cells (1×102 cells/well) were seeded into 6-well plates and cultured for 2 weeks. After washing with phosphate buffer solution, the cells were fixed with paraformaldehyde (4%, Sigma) and stained with crystal violet (0.5%, Beyotime). Thereafter, the number of colonies (> 50 cells) were counted and photographed with a light microscope (Olympus, Tokyo, Japan).
Flow Cytometry Assay
CC cells were harvested and treated with trypsin, and then washed with phosphate buffer solution. For cell cycle distribution analysis, the cells were fixed with ethanol (70%) at 4°C overnight. Thereafter, the cells were incubated with propidium iodide (PI) (50 μg/mL, Solarbio) and RNase A (100 μg/mL, Solarbio). For cell apoptosis evaluation, the cells were stained with the Annexin V-fluorescein isothiocyanate (FITC)/PI Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). The distribution of the cells was analyzed by a FACScan flow cytometry (BD Biosciences) with FACS Diva Software (BD Biosciences).
Transwell Assay
Cell migration and invasion were evaluated with an 8 μm pore membrane filter (Costar, Cambridge, MA, USA). The upper chamber of the invasion assay was pre-coated with Matrigel (Sigma), but the migration assay was not added. In short, the lower chamber was supplemented with DMEM (600 μL) containing FBS (10%), and the upper chamber was supplemented with serum-free DMEM (200 μL) containing CC cells (1×105 cells). After 24 h incubation, the migrated or invasive cells on the lower surface were fixed with paraformaldehyde (4%, Sigma) and then stained with crystal violet (0.5%, Beyotime). The light microscope (Olympus) (magnification, 100×) was applied to count the migrated or invasive cells.
Western Blotting
The RIPA lysis buffer (Beyotime) was applied to extract total protein form tissue samples and cells. Western blotting was executed as previously described.24,28 The primary antibodies including: anti-GAPDH (#5174, 1:1000), anti-cyclin D1 (#2922, 1:1000), anti-cleaved-caspase-3 (c-caspase 3) (#9661, 1:1000), anti-matrix metalloproteinase (MMP)-2 (#4022, 1:1000), anti-MMP-9 (#3852, 1:1000), anti-TPD52 (ab182578, 1:10,000). GAPDH was deemed as a loading control. Goat anti-rabbit IgG (#7074, 1:2000) was used as a secondary antibody. Antibody against TPD52 was purchased from Abcam (Cambridge, MA, USA), and other antibodies were purchased from Cell Signaling Technology (Boston, MA, USA).
Dual-Luciferase Reporter Assay
The binding sites of hsa_circ_0084927 or TPD52 in miR-634 were predicted with the CircInteractome or Starbase databases. The fragments of wild type hsa_circ_0084927, mutant hsa_circ_0084927 (mut-hsa_circ_0084927), wild type 3ʹ untranslated regions (UTR) of TPD52 (TPD52 3ʹUTR), and mutant 3ʹUTR of TPD52 (mut-TPD52 3ʹUTR) containing miR-634 binding sites were synthesized and inserted into the psiCHECK-2 vectors (Promega, Madison, WI, USA) to generate the luciferase reporter vectors, respectively. Next, CC cells were cotransfected with the luciferase reporter vectors together with NC mimics or miR-634 mimics. After transfection for 48 h, the firefly and Renilla luciferase activities were quantified with the luciferase reporter assay system (Promega). Relative luciferase intensity was determined by normalizing the firefly luminescence to Renilla luminescence.
RNA Immunoprecipitation (RIP) Assay
The RIP assay was executed used the Magna RIP Kit (Millipore, Billerica, MA, USA). In brief, CC cells (80%-90% confluence) were lysed in RIP lysis buffer. Next, cell extract (100 μL) was incubated with RIP buffer containing magnetic beads conjugated to Anti-Ago2 (ab32318, Abcam) or Anti-IgG (Millipore). After RNA isolation, the levels of hsa_circ_0084927 and miR-634 were analyzed with qRT-PCR.
Xenograft Assay
For stable knockdown of hsa_circ_0084927, the lentivirus carrying sh-circRNA was transfected into 293T cells and then infected with SiHa cells. For xenograft assay, the SiHa cells (5×106 cells/200 μL) with sh-circRNA or sh-NC were subcutaneously injected into the right flank of female BALB/c nude mice (4–6 weeks old, Vital River, Beijing, China) (5 mice/group). Tumor volume was measured every 4 d with a digital caliper, and tumor volume was calculated in accordance with the following equation: Volume = (length ×width2)/2. 28 days after injection, all were sacrificed by cervical dislocation under isoflurane (5%) to obtain their tumor tissues. The protocols of xenograft assay were authorized by the Animal Ethics Committee of The Third Hospital of Ji’nan. The guidelines for the care and use of laboratory animals strictly followed the “Guidelines for the Ethical Review of Laboratory Animal Welfare” (GB/T 35892–2018).
Statistical Analysis
The experiments in vitro were repeated at last 3 times. Statistical analysis was implemented with SPSS 20.0 software (SPSS, Chicago, IL, USA). The Kolmogorov–Smirnov test was applied to evaluate whether the data follow a normal distribution. The homogeneity of variance between 2 groups was analyzed by F-test. The difference between 2 groups was evaluated with a paired or unpaired Student’s t test. For 3 groups or above, the differences were assessed by one-way variance analysis (ANOVA) with Turkey’s post hoc test. The correlation among hsa_circ_0084927, miR-634, and TPD52 mRNA were analyzed with Pearson’s correlation analysis. Data exhibited as the mean ± standard deviation. Differences were deemed significant if P < 0.05.
Results
Characteristic and Expression of hsa_circ_0084927 in CC
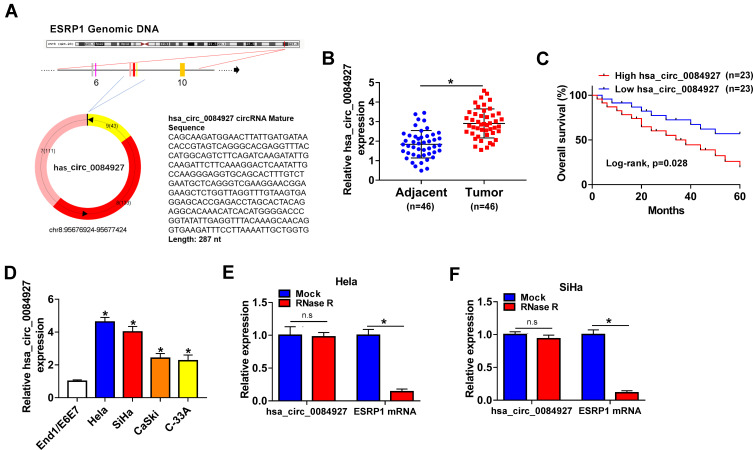
Hsa_circ_0084927 arises from the ESRP1 gene, which is located at chromosome 8 and consists of the head-to-tail splicing of exons 7 to 9 (Figure 1A). Previous study reported that hsa_circ_0084927 was upregulated in CC tissues.9 To explore the biological role of hsa_circ_0084927 in CC, we examined the expression of hsa_circ_0084927 in 46 paired CC tissues and adjacent non-tumor tissues with qRT-PCR. In contrast to the adjacent non-tumor tissues, hsa_circ_0084927 expression was observably elevated in CC tissues (Figure 1B). Next, we divided the patients into two groups (high hsa_circ_0084927 expression group and low hsa_circ_0084927 expression group) according the median expression. High hsa_circ_0084927 expression was associated with International Federation of Gynecology and Obstetrics (FIGO) stage, lymph node metastasis, and depth of cervical invasion (Table 1). Kaplan-Meier analysis and log rank test exhibited that CC patients with high hsa_circ_0084927 expression had a shorter 5-year survival rate (Figure 1C). Subsequently, we detected the expression of hsa_circ_0084927 in CC cells. Compared to the End1/E6E7 cells, hsa_circ_0084927 expression was overtly increased in CC cells (Hela, SiHa, CaSki, and C-33A) (Figure 1D). Then, we used RNase R to pretreat the RNA of Hela and SiHa cells. QRT-PCR exhibited that hsa_circ_0084927 was resistant to RNase R, but the linear ESRP1 mRNA was apparently digested (Figure 1E and F). These results indicated that hsa_circ_0084927 was stable and highly expressed in CC.
Figure 1.
Characteristic and expression of hsa_circ_0084927 in CC. (A) Schematic illustration exhibiting the circularization of ESRP1 exons 7 to 9 forming hsa_circ_0084927. (B) QRT-PCR was used to analyze hsa_circ_0084927 expression in 46 paired CC tissues and adjacent non-tumor tissues. (C) The 5-year survival rate of CC patients with high and low hsa_circ_0084927 expression was evaluated with Kaplan-Meier analysis and log rank test. (D) QRT-PCR was utilized to detect hsa_circ_0084927 expression in CC cells (Hela, SiHa, CaSki, and C-33A) and End1/E6E7 cells. (E and F) After RNase R treatment, the levels of hsa_circ_0084927 and ESRP1 mRNA were analyzed with qRT-PCR. *P < 0.05 and n.s.: no significance.
Hsa_circ_0084927 Silencing Reduced the Malignancy of CC Cells
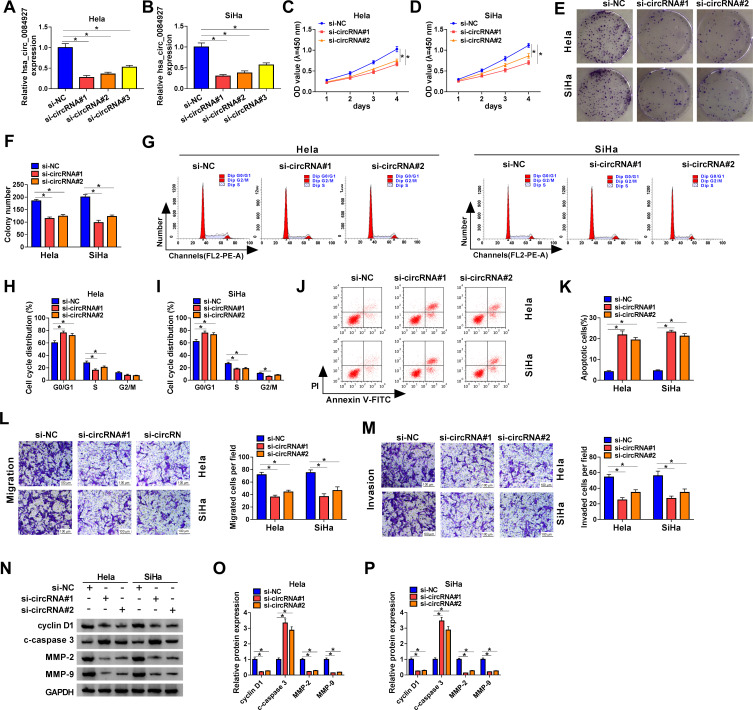
Next, we explored the role of hsa_circ_0084927 in CC through loss-of-function experiments. We observed that hsa_circ_0084927 expression was signally reduced in Hela and SiHa cells after si-circRNA#1, si-circRNA#2, or si-circRNA#3 compared to the control si-NC (Figure 2A and B). Then, we explored the influence of hsa_circ_0084927 inhibition on the malignancy of CC cells. CCK-8 assay presented that hsa_circ_0084927 inhibition decreased the proliferation of Hela and SiHa cells relative to the control group (Figure 2C and D). Plate clone assay indicated that silenced hsa_circ_0084927 expression reduced the colony number of Hela and SiHa cells (Figure 2E and F). Flow cytometry assay displayed that hsa_circ_0084927 knockdown elevated cell number in G0/G1 stage and reduced cell number in S stage in Hela and SiHa cells, indicating that hsa_circ_0084927 inhibited induced cell cycle arrest (Figure 2G-I). Flow cytometry assay also exhibited that hsa_circ_0084927 downregulation could increase the apoptotic rate of Hela and SiHa cells (Figure 2J and K). Furthermore, transwell assay exhibited that the migration and invasion abilities of hsa_circ_0084927-silenced Hela and SiHa cells were reduced compared to the control group (Figure 2L and M). In addition, we measured the levels of cyclin D1, c-caspase 3, MMP-2, and MMP-9 through using Western blotting. The results exhibited that the levels of cyclin D1, MMP-2, and MMP-9 were reduced in hsa_circ_0084927-silenced Hela and SiHa cells relative to the control group, while the level of c-caspase-3 was elevated (Figure 2N-P). Collectively, these data manifested that hsa_circ_0084927 knockdown could decrease the malignancy of CC cells.
Figure 2.
Inhibition of hsa_circ_0084927 decreased the malignancy of CC cells. (A and B) The expression of hsa_circ_0084927 in Hela and SiHa cells after si-circRNA#1, si-circRNA#2, si-circRNA#3, or si-NC transfection was assessed by qRT-PCR. (C-P) Hela and SiHa cells were transfected with si-circRNA#1, si-circRNA#2, or si-NC. (C-M) The proliferation, colony formation, cell cycle progression, apoptosis, migration, and invasion of Hela and SiHa cells were determined with CCK-8, plate clone, flow cytometry, or transwell assays. (N-P) The levels of cyclin D1, c-caspase 3, MMP-2, and MMP-9 in Hela and SiHa cells were measured by Western blotting. *P < 0.05.
Hsa_circ_0084927 Acted as a Sponge for miR-634, Which Targeted TPD52 in CC Cells
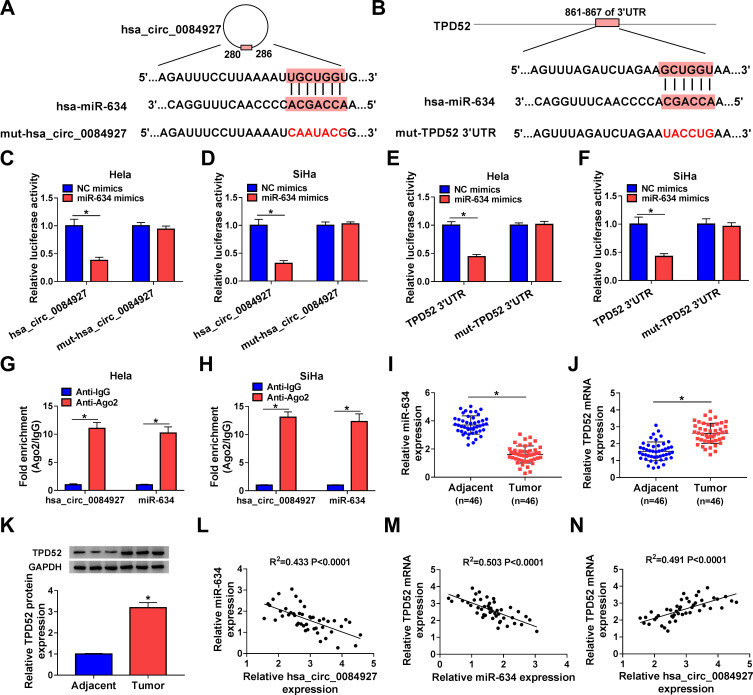
To investigate the underlying molecular mechanism of hsa_circ_0084927 in CC, we predicted miRs with complementary sites to hsa_circ_0084927 through the CircInteractome database. The results exhibited that miR-634 had latent binding sites complementary to hsa_circ_0084927 (Figure 3A). Moreover, we discovered that TPD52 was a latent target for miR-634 through using the Starbase database (Figure 3B). Dual-luciferase reporter assay exhibited that miR-634 mimics could reduce the luciferase activity of luciferase vectors with hsa_circ_0084927 or TPD52 3ʹUTR in Hela and SiHa cells in contrast to the control NC mimics, while the luciferase activity of luciferase vectors with mut-hsa_circ_0084927 or mut-TPD52 3ʹUTR did not change (Figure 3C-F). Then, we executed an RIP assay to observe whether hsa_circ_0084927 could bind miR-634. The results presented that hsa_circ_0084927 and miR-634 were enriched in the Anti-Ago2 group compared to the control Anti-IgG (Figure 3G and H). We also conducted qRT-PCR on 46 paired CC tissues and adjacent non-tumor tissues and found that miR-634 expression was decreased while TPD52 mRNA expression was elevated in CC tissues in contrast to the adjacent non-tumor tissues (Figure 3I and J). In the next step, we observed that TPD52 protein level was higher in CC tissues in comparison to the adjacent non-tumor tissues (Figure 3K). Additionally, we found that miR-634 and hsa_circ_0084927 or TPD52 mRNA had a negative correlation in CC tissues, but hsa_circ_0084927 and TPD52 mRNA had a positive correlation (Figure 3L-N). In summary, these data indicated that hsa_circ_0084927 served as a sponge for miR-634, which targeted TPD52 in CC cells.
Figure 3.
Hsa_circ_0084927 served as a sponge for miR-634, which targeted TPD52 in CC cells. (A and B) Schematic diagrams exhibiting the potential binding sites of hsa_circ_0084927 or TPD52 and miR-634. (C-F) Dual-luciferase reporter assay was performed to analyze the luciferase activity in Hela and SiHa cells cotransfected with NC mimics or miR-634 mimics and luciferase vectors containing hsa_circ_0084927, mut-hsa_circ_0084927, TPD52 3ʹUTR, or mut-TPD52 3ʹUTR. (G and H) RIP assay was performed to determine whether hsa_circ_0084927 could bind miR-634 in Hela and SiHa cells. (I-K) The levels of miR-634, TPD52 mRNA, and TPD52 protein in 46 paired CC tissues and adjacent non-tumor tissues were analyzed by qRT-PCR or Western blotting. (L-N) The correlation among hsa_circ_0084927, miR-634, and TPD52 mRNA were assessed with Pearson’s correlation analysis. *P < 0.05.
Hsa_circ_0084927 Regulated TPD52 Expression via Sponging miR-634 in CC Cells
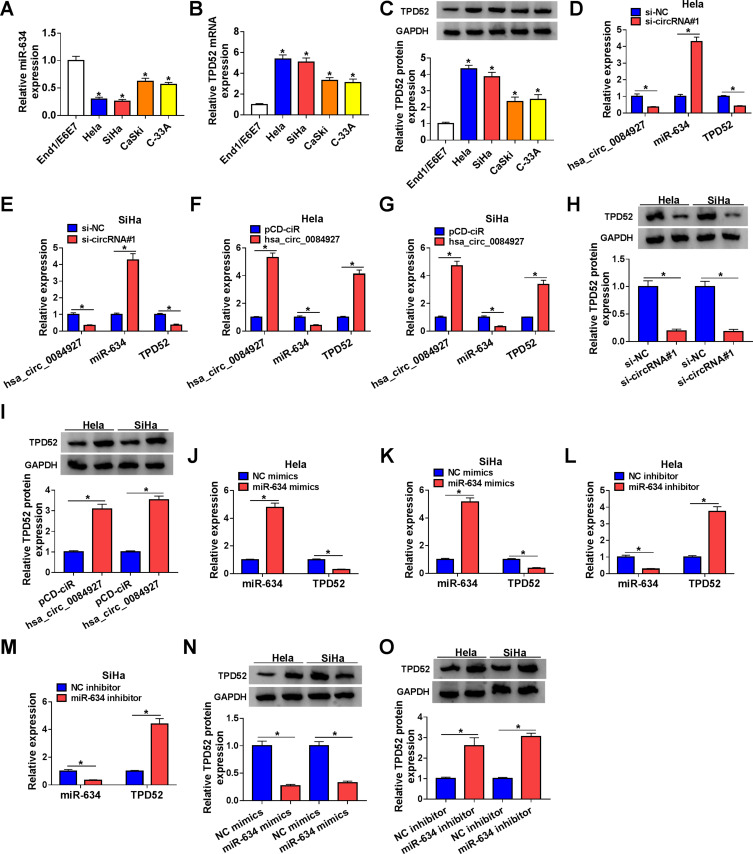
Considering that hsa_circ_0084927 served as a sponge for miR-634 and miR-634 targeted TPD52 in CC cells, we further verified whether hsa_circ_0084927 regulated TPD52 expression via miR-634 in CC. QRT-PCR and Western blotting exhibited that miR-634 expression was reduced while TPD52 mRNA and protein levels were increased in CC cells (Hela, SiHa, CaSki, and C-33A) than that in End1/E6E7 cells (Figure 4A-C). Moreover, we discovered that inhibition of hsa_circ_0084927 reduced hsa_circ_0084927 and TPD52 mRNA expression in Hela and SiHa cells, but elevated the expression of miR-634 (Figure 4D and E). Also, hsa_circ_0084927 and TPD52 mRNA expression were elevated while miR-634 expression was decreased in Hela and SiHa cells after hsa_circ_0084927 transfection compared to the control pCD-ciR (Figure 4F and G). Western blotting displayed that TPD52 protein levels were decreased in hsa_circ_0084927-inhibited Hela and SiHa cells and elevated in hsa_circ_0084927-overexpressed Hela and SiHa cells relative to their matched negative controls (Figure 4H and I). Furthermore, miR-634 elevation decreased TPD52 mRNA and protein levels in Hela and SiHa cells, but miR-634 inhibition elevated TPD52 mRNA and protein levels in Hela and SiHa cells (Figure 4J-O). Together, these data indicated that hsa_circ_0084927 could regulate TPD52 expression through sponging miR-634 in CC cells.
Figure 4.
Hsa_circ_0084927 regulated TPD52 expression via miR-634 in CC cells. (A-C) QRT-PCR and Western blotting were performed to examine the levels of miR-634, TPD52 mRNA, and TPD52 protein in CC cells (Hela, SiHa, CaSki, and C-33A) and End1/E6E7 cells. (D-I) QRT-PCR and Western blotting were employed to examine the levels of hsa_circ_0084927, miR-634, TPD52 mRNA, and TPD52 protein in Hela and SiHa cells transfected with si-NC, si-circRNA#1, pCD-ciR, or hsa_circ_0084927. (J-O) QRT-PCR and Western blotting were executed to analyze the levels of miR-634, TPD52 mRNA, and TPD52 protein in Hela and SiHa cells transfected with NC mimics, miR-634 mimics, NC inhibitor, or miR-634 inhibitor. *P < 0.05.
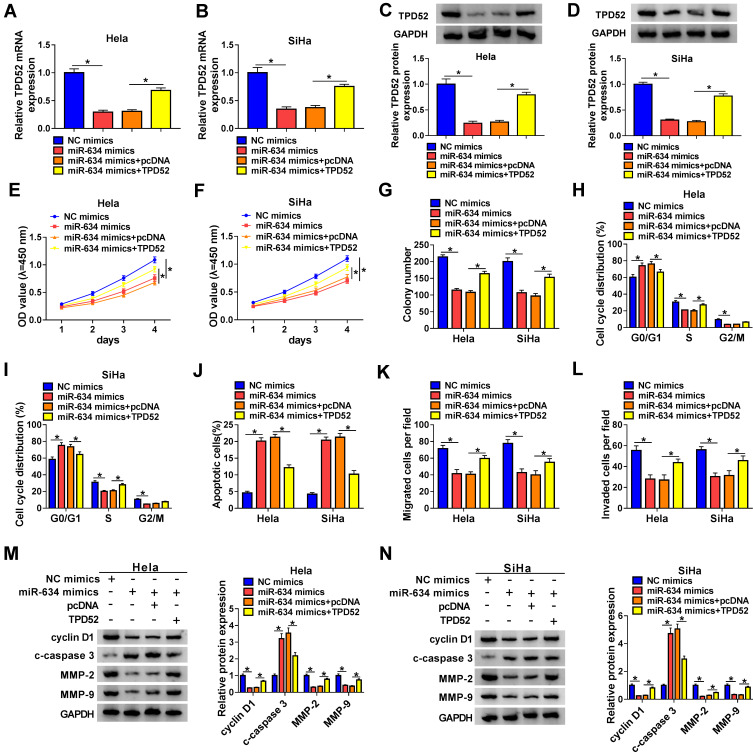
TPD52 Overexpression Reversed miR-634 Mimics-Mediated Influence on the Malignancy of CC Cells
Based on the above results, we verified whether miR-634 regulated the malignancy of CC cells via TPD52. The results exhibited that miR-634 mimics reduced the levels of TPD52 mRNA and protein in Hela and SiHa cells, but this influence was partly reversed after TPD52 transfection (Figure 5A-D). CCK-8 and plate clone assays exhibited that miR-634 mimics reduced the proliferation and colony formation abilities of Hela and SiHa cells, while this trend was overturned by TPD52 overexpression (Figure 5E-G). Flow cytometry assay manifested that elevated miR-634 expression induced cell cycle arrest and apoptosis in Hela and SiHa cells, but this impact was abolished by forcing TPD52 expression (Figure 5H-J). Transwell assay exhibited that forced TPD52 expression overturned the repressive influence of miR-634 mimics on migration and invasion of Hela and SiHa cells (Figure 5K and L). Furthermore, miR-634 overexpression reduced the levels of cyclin D1, MMP-2, and MMP-9 while elevated the level of c-caspase 3 in Hela and SiHa cells, but these tendency were reversed by TPD52 elevation (Figure 5M and N). These results manifested that miR-634 exerted its role via TPD52 in CC cells.
Figure 5.
MiR-634 exerted its role through targeting TPD52 in CC cells. (A-N) Hela and SiHa cells were transfected with NC mimics, miR-634 mimics, miR-634 mimics+pcDNA, or miR-634 mimics+TPD52. (A-D) QRT-PCR and Western blotting were conducted to analyze the levels of TPD52 mRNA and protein in Hela and SiHa cells. (E-L) CCK-8, plate clone, flow cytometry, and transwell assays were executed to evaluate the proliferation, colony formation, cell cycle progression, apoptosis, migration, or invasion of Hela and SiHa cells. (M and N) Western blotting was performed to assess the levels of cyclin D1, c-caspase 3, MMP-2, and MMP-9 in Hela and SiHa cells. *P < 0.05.
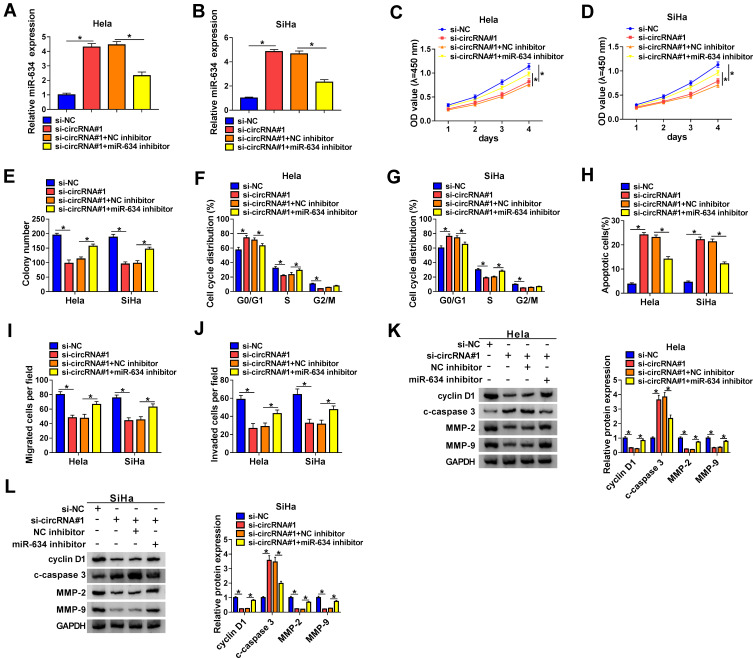
MiR-634 Inhibitor Abolished hsa_circ_0084927 Silencing-Mediated Effect on the Malignancy of CC Cells
In consideration of the targeting relationship between hsa_circ_0084927 and miR-634 in CC cells, we further verified whether hsa_circ_0084927 regulated the malignancy of CC cells via miR-634. We found that the upregulation of miR-634 in hsa_circ_0084927-inhibited Hela and SiHa cells was partly overturned by miR-634 inhibitor (Figure 6A and B). Moreover, the downregulation of miR-634 reversed the inhibitory influence of hsa_circ_0084927 knockdown on proliferation and colony formation of Hela and SiHa cells (Figure 6C-E). Furthermore, miR-634 silencing overturned the accelerative influence of hsa_circ_0084927 inhibition on cell cycle arrest and apoptosis of Hela and SiHa cells (Figure 6F-H). Also, silenced miR-634 expression abolished the inhibition of migration and invasion of Hela and SiHa cells caused by hsa_circ_0084927 knockdown (Figure 6I and J). Likewise, miR-634 inhibition reversed hsa_circ_0084927 knockdown-mediated effects on the levels of cyclin D1, c-caspase 3, MMP-2, and MMP-9 in Hela and SiHa cells (Figure 6K and L). These resulted indicated that hsa_circ_0084927 regulated the malignancy of CC cells via sponging miR-634.
Figure 6.
Hsa_circ_0084927 regulated the malignancy of CC cells through miR-634. (A-L) Hela and SiHa cells were transfected with si-NC, si-circRNA#1, si-circRNA#1+NC inhibitor, or si-circRNA#1+miR-634 inhibitor. (A and B) Expression of miR-634 in Hela and SiHa cells were examined with qRT-PCR. (C-J) CCK-8, plate clone, flow cytometry, and transwell assays were performed to determine the proliferation, colony formation, cell cycle progression, apoptosis, migration, or invasion of Hela and SiHa cells. (K and L) The levels of cyclin D1, c-caspase 3, MMP-2, and MMP-9 in Hela and SiHa cells were measured with Western blotting. *P < 0.05.
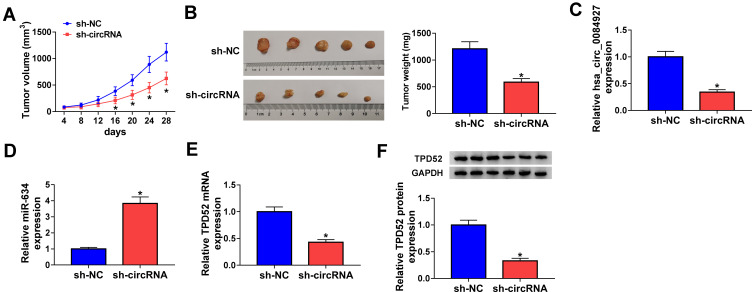
Hsa_circ_0084927 Inhibition Decreased Tumor Growth in vivo
To further explore the impact of hsa_circ_0084927 on tumor growth in vivo, we established xenograft nude mouse models by injecting SiHa cells carrying sh-NC or sh-circRNA into nude mice. We observed that tumor volume and weight were reduced in the sh-circRNA group relative to the sh-NC group (Figure 7A and B). Moreover, qRT-PCR exhibited that hsa_circ_0084927 was downregulated while miR-634 was upregulated in mice tumor tissues of the sh-circRNA group relative to the sh-NC group (Figure 7C and D). Also, we observed that TPD52 mRNA and protein levels were reduced in mice tumor tissues of the sh-circRNA group in comparison to the sh-NC group (Figure 7E and F). These results indicated that hsa_circ_0084927 knockdown could reduce tumor growth in vivo.
Figure 7.
Hsa_circ_0084927 knockdown could reduce tumor growth in vivo. (A and B) In comparison to the sh-NC group, tumor volume and weight were reduced in hsa_circ_0084927-inhibited nude mice. (C-E) Hsa_circ_0084927, miR-634, and TPD52 mRNA levels were examined with qRT-PCR in mice tumor tissues. (F) TPD52 protein level was detected by Western blotting in mice tumor tissues. *P < 0.05.
Discussion
Recently, the role of circRNAs in tumorigenesis and advancement has attracted widespread attention. CircRNAs can be used as tumor diagnostic markers or therapeutic targets due to their unique molecular structure and cell/tissue-specific and stage-specific expression.29 Nevertheless, the role of a large number of circRNAs in CC advancement is unclear.
Hsa_circ_0084927 is a circRNA, which has been pointed out to be freakishly expressed in certain tumors. Studies have revealed that hsa_circ_0084927 is upregulated in malignant pleural effusion,30 laryngeal cancer,31 and CC.9 Herein, hsa_circ_0084927 was upregulated in CC tissues and cells. High hsa_circ_0084927 expression was associated with the FIGO stage, lymph node metastasis, and depth of cervical invasion, implying that hsa_circ_0084927 might be a novel biomarker for CC diagnosis. Downregulation of hsa_circ_0084927 decreased tumor growth in vivo and induced cell cycle arrest, apoptosis, suppressed colony formation, proliferation, invasion, and migration of CC cells in vitro. Nevertheless, hsa_circ_0084927 was revealed to be downregulated in resistant-small cell lung cancer cells, and hsa_circ_0084927 overexpression elevated the chemosensitivity of resistant-small cell lung cancer cells,8 which might be associated with cell specificity.
Accumulating researches have proved that circRNAs modulate cellular function as miRs sponges.32 For instance, hsa_circ_0084927 elevated the chemosensitivity of resistant-small cell lung cancer cells through suppressing TGF-β signaling via sponging miR-93-5p.8 Herein, we discovered that hsa_circ_0084927 regulated TPD52 expression via sponging miR-634 in CC cells. Tan et al pointed out that miR-634 could increase cell sensitivity to temozolomide through the Raf-ERK pathway by targeting CYR61 in glioma.33 Another study demonstrated that miR-634 elevated the radiosensitivity of breast cancer cells through downregulating STAT3.34 MiR-634 has been revealed to exert a suppressive role in gastric cancer,16 pancreatic cancer,17 hepatocellular cancer,35 and CC.18 In the current study, hsa_circ_0084927 and miR-634 had a negative correlation in CC tissues. MiR-634 inhibitor overturned hsa_circ_0084927 silencing-mediated influence on the malignancy of CC cells. Therefore, we concluded that hsa_circ_0084927 regulated the malignancy of CC cells via sponging miR-634.
TPD52 was reported as an oncogene in a range of tumors.21,36,37 In CC, lncRNA HULC silencing reduced TPD52 expression, which inhibited cancer cell invasion and migration.22 Also, miR-15a-3p reduced the radioresistance of CC cells by targeting TPD52.38 However, TPD52 was revealed as a tumor repressor in renal cell cancer, which might be related to tissue specificity. In this study, we discovered that TPD52 served as a target for miR-634 and was positively correlated with hsa_circ_0084927 in CC. Moreover, TPD52 elevation could abolish the inhibition of the malignancy of CC cells caused by miR-634 mimics. Therefore, we concluded that hsa_circ_0084927 modulated the malignancy of CC cells via regulating the miR-634/TPD52 axis. In the future, the downstream molecular mechanism of the hsa_circ_0084927/miR-634/TPD52 axis can be further explored.
In summary, we discovered a new mechanism by which hsa_circ_0084927 elevated TPD52 expression via sponging miR-634, which facilitated the advancement of CC, manifesting that hsa_circ_0084927 acted as a promising target for CC treatment.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Dong J, Su M, Chang W, Zhang K, Wu S, Xu T. Long non-coding RNAs on the stage of cervical cancer (Review). Oncol Rep. 2017;38(4):1923–1931. doi: 10.3892/or.2017.5905 [DOI] [PubMed] [Google Scholar]
- 3.Wright JD, Chen L, Tergas AI, et al. Population-level trends in relative survival for cervical cancer. Am J Obstet Gynecol. 2015;213(5):670. doi: 10.1016/j.ajog.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi: 10.1016/S0140-6736(18)32470-X [DOI] [PubMed] [Google Scholar]
- 5.Chen L-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. [DOI] [PubMed] [Google Scholar]
- 6.Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022 [DOI] [PubMed] [Google Scholar]
- 7.Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111. doi: 10.1186/s12943-019-1040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Yang Y, Wu J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 2020;27(5):1709–1727. doi: 10.1038/s41418-019-0455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong J, Jiang H, Shu C, et al. Integrated analysis of circular RNA-associated ceRNA network in cervical cancer: observational study. Medicine. 2019;98(34):e16922. doi: 10.1097/MD.0000000000016922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 12.Du W, Feng Z, Sun Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochem Biophys Res Commun. 2018;507(1–4):198–202. doi: 10.1016/j.bbrc.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 13.Lian Y, Xiong F, Yang L, et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253. doi: 10.1186/s13046-018-0918-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X, Huang T, Zhang C, et al. Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle. 2019;18(16):1908–1924. doi: 10.1080/15384101.2019.1632641 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Li S, Zeng A, Hu Q, Yan W, Liu Y, You Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro-Oncology. 2017;19(1):55–65. doi: 10.1093/neuonc/now129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo J, Zhang CD, An JX, et al. Expression of miR-634 in gastric carcinoma and its effects on proliferation, migration, and invasion of gastric cancer cells. Cancer Med. 2018;7(3):776–787. doi: 10.1002/cam4.1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D, Wu X, Zhao J, Zhao X. MicroRNA-634 functions as a tumor suppressor in pancreatic cancer via directly targeting heat shock-related 70-kDa protein 2. Cancer Med. 2019;17(5):3949–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong J, Liu R, Wang X, Jiang H, Zhang Y. MiR-634 decreases cell proliferation and induces apoptosis by targeting mTOR signaling pathway in cervical cancer cells. Artif Cells Nanomed Biotechnol. 2016;44(7):1694–1701. doi: 10.3109/21691401.2015.1080171 [DOI] [PubMed] [Google Scholar]
- 19.van Duin M, van Marion R, Vissers K, et al. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer. 2005;44(4):438–449. doi: 10.1002/gcc.20259 [DOI] [PubMed] [Google Scholar]
- 20.Kumamoto T, Seki N, Mataki H, et al. Regulation of TPD52 by antitumor microRNA-218 suppresses cancer cell migration and invasion in lung squamous cell carcinoma. Int J Oncol. 2016;49(5):1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin W, Shi L, Mao Y. MicroRNA-449b-5p suppresses cell proliferation, migration and invasion by targeting TPD52 in nasopharyngeal carcinoma. J Biochem. 2019;166(5):433–440. doi: 10.1093/jb/mvz057 [DOI] [PubMed] [Google Scholar]
- 22.Lu W, Wan X, Tao L, Wan J. Long non-coding RNA HULC promotes cervical cancer cell proliferation, migration and invasion via miR-218/TPD52 axis. OncoTargets Ther. 2020;13:1109–1118. doi: 10.2147/OTT.S232914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han G, Fan M, Zhang X. microRNA-218 inhibits prostate cancer cell growth and promotes apoptosis by repressing TPD52 expression. Biochem Biophys Res Commun. 2015;456(3):804–809. doi: 10.1016/j.bbrc.2014.12.026 [DOI] [PubMed] [Google Scholar]
- 24.Sun YS, Thakur K, Hu F, Zhang JG, Wei ZJ. Icariside II inhibits tumorigenesis via inhibiting AKT/Cyclin E/CDK 2 pathway and activating mitochondria-dependent pathway. Pharmacol Res. 2020;152:104616. doi: 10.1016/j.phrs.2019.104616 [DOI] [PubMed] [Google Scholar]
- 25.Sun YS, Thakur K, Hu F, Cespedes-Acuña CL, Zhang JG, Wei ZJ. Icariside II suppresses cervical cancer cell migration through JNK modulated matrix metalloproteinase-2/9 inhibition in vitro and in vivo. Biomed Pharmacother. 2020;125:110013. doi: 10.1016/j.biopha.2020.110013 [DOI] [PubMed] [Google Scholar]
- 26.Zhang YY, Zhang F, Zhang YS, et al. Mechanism of Juglone-induced cell cycle arrest and apoptosis in Ishikawa human endometrial cancer cells. J Agric Food Chem. 2019;67(26):7378–7389. doi: 10.1021/acs.jafc.9b02759 [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, Zhang YY, Sun YS, et al. Asparanin A from L. induces G0/G1 cell cycle arrest and apoptosis in human endometrial carcinoma ishikawa cells via mitochondrial and PI3K/AKT signaling pathways. J Agric Food Chem. 2020;68(1):213–224. [DOI] [PubMed] [Google Scholar]
- 28.Lv M, Ou R, Zhang Q, et al. MicroRNA-664 suppresses the growth of cervical cancer cells via targeting c-Kit. Drug Des Devel Ther. 2019;13:2371–2379. doi: 10.2147/DDDT.S203399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristensen LS, Hansen TB, Venø MT, Circular KJ. RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen Y, Wang Y, Xing Z, Liu Z, Hou Z. Microarray expression profile and analysis of circular RNA regulatory network in malignant pleural effusion. Cell Cycle. 2018;17(24):2819–2832. doi: 10.1080/15384101.2018.1558860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8(2):932–939. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan Z, Zhao J, Jiang Y. MiR-634 sensitizes glioma cells to temozolomide by targeting CYR61 through Raf-ERK signaling pathway. Cancer Med. 2018;7(3):913–921. doi: 10.1002/cam4.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B, Kuai F, Chen Z, et al. miR-634 decreases the radioresistance of human breast cancer cells by targeting. Cancer Biother Radiopharm. 2020;35(3):241–248. doi: 10.1089/cbr.2019.3220 [DOI] [PubMed] [Google Scholar]
- 35.Zhang CZ, Cao Y, Fu J, Yun JP, Zhang MF. miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol. 2016;10(10):1532–1541. doi: 10.1016/j.molonc.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goto Y, Nishikawa R, Kojima S, et al. Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett. 2014;588(10):1973–1982. doi: 10.1016/j.febslet.2014.04.020 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Wang J, Gao R, et al. Downregulation of microRNA-449 promotes migration and invasion of breast cancer cells by targeting tumor protein D52 (TPD52). Oncol Res. 2017;25(5):753–761. doi: 10.3727/096504016X14772342320617 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Wu Y, Huang J, Xu H, Gong Z. Over-expression of miR-15a-3p enhances the radiosensitivity of cervical cancer by targeting tumor protein D52. Biomed Pharmacother. 2018;105:1325–1334. doi: 10.1016/j.biopha.2018.06.033 [DOI] [PubMed] [Google Scholar]