Highlights
-
•
The study shows the effect of feature extraction on classification results after using the image contrast enhancement technique in X-ray images.
-
•
Assessment of classification performances with a small number of features selected over X-ray images with the help of meta-heuristic algorithms.
-
•
It offers an approach that helps the diagnosis of covid-19 on X-ray images.
Keywords: COVID-19, BPSO, BGWO, Pneumonia, Deep learning models
Abstract
COVID-19 is a disease that causes symptoms in the lungs and causes deaths around the world. Studies are ongoing for the diagnosis and treatment of this disease, which is defined as a pandemic. Early diagnosis of this disease is important for human life. This process is progressing rapidly with diagnostic studies based on deep learning. Therefore, to contribute to this field, a deep learning-based approach that can be used for early diagnosis of the disease is proposed in our study. In this approach, a data set consisting of 3 classes of COVID19, normal and pneumonia lung X-ray images was created, with each class containing 364 images. Pre-processing was performed using the image contrast enhancement algorithm on the prepared data set and a new data set was obtained. Feature extraction was completed from this data set with deep learning models such as AlexNet, VGG19, GoogleNet, and ResNet. For the selection of the best potential features, two metaheuristic algorithms of binary particle swarm optimization and binary gray wolf optimization were used. After combining the features obtained in the feature selection of the enhancement data set, they were classified using SVM. The overall accuracy of the proposed approach was obtained as 99.38%. The results obtained by verification with two different metaheuristic algorithms proved that the approach we propose can help experts during COVID-19 diagnostic studies.
1. Introduction
COVID-19 is a pandemic disease that has affected about 6.2 million people as of early June. This disease has caused many deaths around the world. COVID-19 is highly contagious and continues to spread rapidly with common symptoms such as fever, cough, muscle pain, and weakness. In addition to the tests performed for the diagnosis of this disease, infected individuals are detected using radiology images. Currently, real-time transcriptase-polymerase chain reaction (RT-PCR) is the accepted standard diagnostic method. Since it is a new type of virus, vaccination studies are continuing and deep learning-based approaches that can help experts diagnose this disease will enable the process to progress faster. When the studies about COVID-19 diagnosis with deep learning are examined, both X-ray and computed tomography (CT) images are used with the data sets created for the disease. It was observed that the images with COVID-19 diagnosis are limited in number in these studies.
In the studies we examined, studies using X-ray images for the diagnosis of COVID-19 are as follows:
-
•
Hemdan et al. [1] classified the status of being positive or negative on X-ray images with 7 deep learning models in their study called Covidx-Net. In the study, it was stated that the VGG19 model gave better results with 90% success compared to other models.
-
•
Toğaçar et al. [2] trained MobileNet and SqueezeNet with X-ray images for COVID-19 diagnosis. A stacked data set was obtained before training. The features were extracted from the models trained with these data and these features were selected with the help of the SMO algorithm. Selected features were classified by SVM. As a result of the classification, it was stated that an overall accuracy of 99.27% was achieved.
-
•
Zhang et al. [3] detected COVID-19 by performing anomaly detection on X-ray images. They used 18-layer ResNet in their work. Images were separated into COVID and non−COVID using the binary cross entropy loss function in the network structure established with a 2-class structure. The success of their work was given as 95.18%.
-
•
Afshar et al. [4] also defined COVID-19 on X-ray images by using a capsule networks-based framework. The approach they recommend includes many capsules and convolutional layers. It was stated that this approach achieved success of 98.3%.
-
•
Apostolopoulos et al. [5] carried out diagnosis of COVID-19 disease on X-ray images with the transfer learning approach on convolutional neural networks. Success of 98.75% was achieved in the transfer learning process with VGG19.
-
•
Ozturk et al. [6] achieved success of 98.08% on X-ray images by using DarkNet with 17 convolutional layers.
-
•
Pereira et al. [7] defined COVID-19 with both its deep neural network and its texture properties obtained by using various feature extraction methods. It was reported to have achieved an 0.89 score for F-score.
-
•
Uçar et al. [8] achieved 98.3% success in their study using SqueezeNet and Bayesian optimization.
The studies which used CT images for the detection of COVID-19 are as follows:
-
•
Ardakani et al. [9] classified COVID and non−COVID classes using 10 well-known deep learning models. In their study, ResNet and Xception models provided the best results.
-
•
Barstugan et al. [10,11] performed the classification of the features they obtained by using feature extraction algorithms during image processing operations from CT images with machine learning methods. In another study conducted by the same team, the feature fusion and ranking method was applied to the features obtained from deep learning models and then classified with SVM.
-
•
Yan et al. [12] performed segmentation to determine COVID-19 on CT images.
-
•
Hasan et al. [13] used LSTM Neural Network Classifier as a classifier in their study using Q-Deformed Entropy and Deep Learning Features.
-
•
Singh et al. [14] used multi-objective differential evolution in determining the parameters of convolutional neural networks, and thus classified CT images with COVID-19.
The motivation for the study is to propose an approach that effectively classifies COVID-19, pneumonia and healthy lung X-ray images by combining deep learning and meta-heuristic algorithms for early diagnosis of this disease that is important for human life. It is known that X-ray images contain a high amount of noise and are low-density grayscale images. For this reason, the contrast on X-rays obtained from some machines and boundary representations may be weak. It is quite challenging to extract features from these X-ray images. The quality of these images can be improved by applying some contrast enhancement techniques. Thus, feature extraction from these images can be performed more efficiently and comfortably. In this study, we focused on an image processing method that provides the best contrast. After trying many techniques, we decided on the image contrast enhancement algorithm (ICEA). We obtained the enhancement data set using this technique. We ran this data set with the best known deep learning models and extracted the feature vectors of the data set. Another source of motivation for our study is that there is no study about contrast among the other studies and there are a limited number of studies that perform feature extraction with deep learning models.
We can summarize our contribution to this field as follows:
-
1.
The study presents a contrast-adjusted data set containing 3 classes of COVID-19, pneumonia, and normal images for the use of researchers.
-
2.
The study shows the effect of feature extraction on classification results after using the image contrast enhancement technique in X-ray images.
-
3.
Assessment of classification performances with a small number of features selected on X-ray images with the help of meta-heuristic algorithms.
Our study is organized as follows for this purpose. In Section 2, datasets, models, methods and MH-CovidNet approach are explained. Experimental studies are shown in Section 3. At the end of the study, there is a discussion and conclusion.
2. Datasets, models, methods, proposed approach
2.1. Datasets
2.1.1. Original dataset
The data set we created for the study includes COVID-19, pneumonia and normal X-ray images. As known, it is very difficult to find an open source data set since COVID-19 is a new disease type. For this reason, the images in openly shared data sets were combined when creating the data set. The first data set we received is the data set presented by Joseph Paul Cohen [15] on Github. This data set contains 145 images labeled COVID-19. The second dataset we used for COVID-19 is the dataset that was made publicly available in Kaggle by Rahman et al. [16] with 219 images. By combining these data sets, a data set with 364 images was obtained for COVID-19. For pneumonia and normal chest X-ray images, the data set prepared by Kermany et al. [17] was used. In order to increase the performance of the models, an equal number of images were selected for each class. Since cCOVID-19 images are limited in number, 364 images were selected for other classes, and the original data set was created.
2.1.2. Enhancement dataset
In the process of creating the enhancement data set, contrast enhancement was performed on each image in the original dataset separately by using the image contrast enhancement algorithm (ICEA). In this way, the noise in the original data set was removed and the best contrast was achieved. The ICEA is one of the image processing techniques developed as a solution to the contrast enhancement problem. In this study, it was used for the first time on X-ray images. The algorithm is explained in Section 2.3.1.
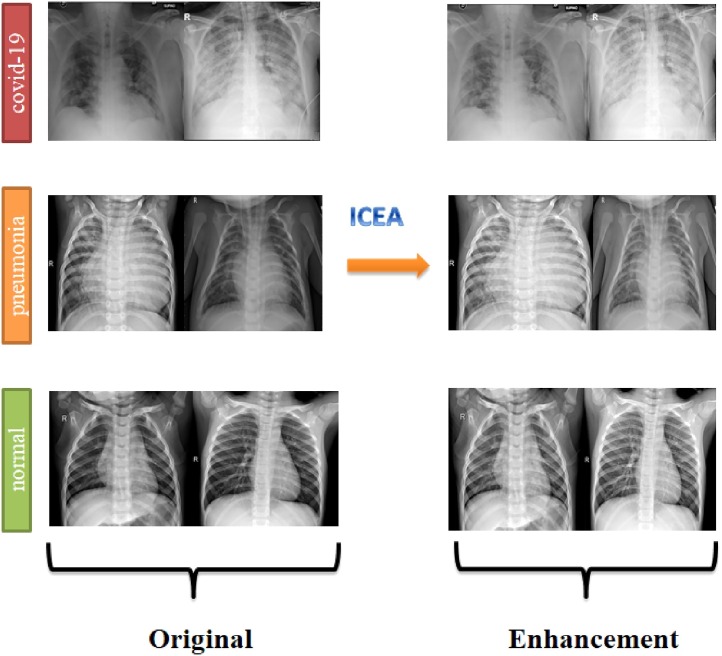
In the approach proposed for the study, the results will be examined in both data sets. In experimental studies, 70% of the data set was used as training data and 30% as test data. In the final steps of the study, the consistency of the study results was tested by using k-fold cross validation for the Enhancement data set. COVID-19 chest images in the original dataset and enhancement data set are shown in Fig. 1 .
Fig. 1.
Dataset samples from original and enhancement data set.
2.2. Deep learning models
2.2.1. AlexNet
AlexNet [18], an 8-Layer CNN network, was first announced in 2012 with an award in the ImageNet competition. After this competition, it was proven that the image properties obtained from CNN architectures can exceed the properties obtained by classical methods. In the AlexNet structure, there is a 11 × 11 convolution window on the first layer. The input size before this layer was determined as 227 × 227. In the second layer, this convolution window is first reduced to 5 × 5 and then to 3 × 3. Also, 2-step stride and max pooling layers are added. There is an output layer of 4096 after the convolution layer in the last layer. The layer named “FC8” that comes after this is the layer where we obtained the 1000 feature vectors used in our application. In this model, RELU is used as activation function instead of Sigmoid.
2.2.2. VGG19
VGG is a convolutional neural network model proposed by K. Simonyan and A. Zisserman from the University of Oxford [19]. The model achieved 92.7% top 5 test accuracy on more than 14 million image datasets of 1000 classes in ImageNet. This model improved on AlexNet by using 3 × 3 core size filters one after the other, instead of large core size filters. The input size in the first layer is 224 × 224. After 3 × 3 convolution layers and max pooling layers, two 4096 fully connected layers are found in the structure of this model. As with the AlexNet model, this model also has an “FC8” layer, which we used for feature extraction.
2.2.3. GoogleNet
The GoogleNet model was introduced in 2015 as a deep learning model that emerged with the idea that existing neural networks should go deeper [20]. This network model consists of modules. Each module consists of different-sized convolution and max-pooling layers. Each module is called 'inception'. Although the model consisting of a total of 9 inception blocks has computational complexity, the speed and performance of the model were increased with the improvements. In this model, 1000 features were extracted using the "loss3-classifier" layer in our study.
2.2.4. ResNet
The ResNet [21] model is a deep learning model developed by Microsoft Research Team that won the 2015 “ImageNet Large Scale Visual Recognition Challenge (ILSVRC)” competition with an error rate of 3.57%. Each layer of a ResNet consists of several blocks. With this model, when the residual layer structure is determined, the number of parameters calculated is reduced compared to other models. In this model, 1000 features were extracted by using the “fc1000” layer for feature extraction in our study.
In our study, these deep learning models were used for feature extraction. From each of them, 1000 features were obtained and selected with feature selection of meta-heuristic algorithms and classified with SVM. The parameter values used in the models are given in Table 1 . Model structures are summarized in Table 2 .
Table 1.
Model parameters.
| Model | Image Size | Optimization | Mini Batch | Momentum | Learning Rate |
|---|---|---|---|---|---|
| AlexNet | 227 × 227 | Stochastic Gradient Descent (SGD) | 64 | 0.9 | 1e-5 |
| VGG19 | 224 × 224 | Stochastic Gradient Descent (SGD) | 64 | 0.9 | 1e-5 |
| GoogleNet | 224 × 224 | Stochastic Gradient Descent (SGD) | 64 | 0.9 | 1e-5 |
| ResNet | 224 × 224 | Stochastic Gradient Descent (SGD) | 64 | 0.9 | 1e-5 |
Table 2.
Structure of Models.
| Models | Size (M) | #layers | Model description |
|---|---|---|---|
| AlexNet | 238 | 8 | 5 conv+3 fc layers |
| VGG19 | 560 | 19 | 16 conv+3 fc layers |
| GoogleNet | 40 | 22 | 21 conv+1 fc layers |
| ResNet | 235 | 50 | 49 conv+1 fc layers |
Table 1 contains the parameters for each model that we used in our study. For example, while the input image size of the AlexNet model is 227 × 227, the input size of the others is 224 × 224. The momentum of the stochastic gradient descent (SGD) optimization algorithm used for each model was determined as 0.9. The minibatch value is determined as 64 for each model. This value can be 128 or 256 depending on the performance of the hardware that the applications are running on. It is a significant parameter that needs to be adjusted since it requires a lot of memory. The learning rate used for all models is 1e-5. All these parameters were obtained through experimental experience. Table 2 shows the dimensions of all models, the number of layers in the models, and the names of these layers, respectively.
2.3. Methods
2.3.1. Image contrast enhancement algorithm
This algorithm used to create the enhancement dataset is a new algorithm proposed by Ying et al. [22] to provide accurate contrast enhancement. This algorithm works as follows; first, the weight matrix is designed for image fusion using lighting prediction techniques. The camera response model is then presented to synthesize multiple exposure images. Then, in regions where the original image is underexposed, the best exposure ratio is found for good exposure of the synthetic image. Finally, the input image and the synthetic image are combined according to the weight matrix to obtain an enhanced image. The main formulas used in the algorithm are given in Eqs. (1) and (2). The publication [22] can be reviewed for detailed information about the algorithm. The images are combined as in Eq. (4) to obtain a well-exposed image of all pixels.
| (1) |
Where, N represents the number of images, Pi represents the i-th image in the exposure set, Wi represents the i-th image's weight map, c is the index of the three-color channels, and R is the result of enhancement. Pi is obtained from Eq. (2).
| (2) |
Where g is called Brightness Transform Function (BTF) and ki is the exposure ratio.
The Beta-Gamma Connection Model in Eq. (3) was used as BTF in our study.
| (3) |
Where β and γ are parameters that can be calculated from camera parameters a, b, and exposure ratio k. We used a constant parameter as in the original study (a = −0.3293, b = 1.1258).
At the end of the algorithm, the enhancement image is obtained by using Eq. (4).
| (4) |
2.4. Feature selection
Feature selection is a critical component in data science. High dimensional data causes some undesirable situations in applied models. These include 1) training time increases with increased features, and 2) causes overfitting in models
Selecting effective features with feature selection helps prevent such undesirable situations Although there are many feature selection algorithms, feature selection with meta-heuristic algorithms has been widely used recently. Therefore, in our approach, we chose to use two swarm-based meta-heuristic algorithms for feature selection. The parameter values used for these algorithms were obtained by examining the studies using these algorithms. Among the features obtained using deep neural networks, the most effective ones are selected using these meta-heuristic algorithms. Binary versions of the algorithms are preferred. Algorithms choose features according to their flows during the study. A fitness value is obtained by sending these features to the fitness function. In each iteration, features that will provide a better value than this fit value are sought by the algorithm. At the end of the algorithm's work, the features with the best value are selected. In our study, the number of features obtained with the help of each model at the beginning is 1000. The features selected from these 1000 features were classified with the SVM classifier in the next step.
2.4.1. Binary particle swarm optimization (BPSO)
Particle swarm optimization (PSO) [23] is a meta-heuristic algorithm that results from modeling swarm movements of animals such as birds and fish. In this algorithm, there are two important arguments of pbest and gbest values used to update the speed and position information of candidate solutions in the swam. Eqs. for the algorithm are [24];
| (5) |
Where rand is a random number uniformly distributed between 0 and 1.
| (6) |
Where x is the solution, pbest is personal best and gbest is global best solution, F(.) is fitness function and t is the number of iterations.
BPSO is the binary version of this algorithm. For the fitness function specified in the equations, K-nearest neighbor classifier [25] error rate was used in our application. The BPSO algorithm used for the application can be accessed at [26]. Parameter values are; N = 20; T = 100; c1 = 2; c2 = 2; Vmax = 6; Wmax = 0.9; and Wmin = 0.4.
2.4.2. Binary Gray Wolf Optimization (BGWO)
This is an optimization algorithm that mimics the hunting strategy and social leadership of gray wolves proposed by Mirjalili in 2014 [27]. The group size is between 5 and 12 individuals. The hierarchy of gray wolves comprises four groups: alpha, beta, delta and omega wolves. Leader wolves are called alpha. Alpha wolves are the best wolves to manage the other wolves in the group and are usually responsible for making decisions about hunting, sleeping place, waking time and so on. The second in the social group's social hierarchy is the beta wolf. Beta is the leading wolf’s (alpha) assistant in many events. Delta wolf is the compulsory third wolf to comply with alpha and beta wolves, and can only rule omega wolves. In other words, the omega wolf is the lowest level of gray wolf [27]. Mathematical equations for the models developed for the hunting strategies of wolves are given in Eq. (8).
| (7) |
Where Xp is the position of prey, A is the coefficient vector, and D is defined as
| (8) |
Where C is the coefficent vector, and X is the position of the gray wolf.
The position updates of the gray wolves occur as in Eq. (9).
| (9) |
Feature selection was made with BGWO [28], which is a binary version of this algorithm. Again, as in BPSO, K-nearest neighbor [25] error rate was used as the fitness function for this algorithm. The algorithm can be accessed at [29]. Parameter values used for this algorithm; population: 20, and iteration: 100.
2.5. Support Vector Machine (SVM)
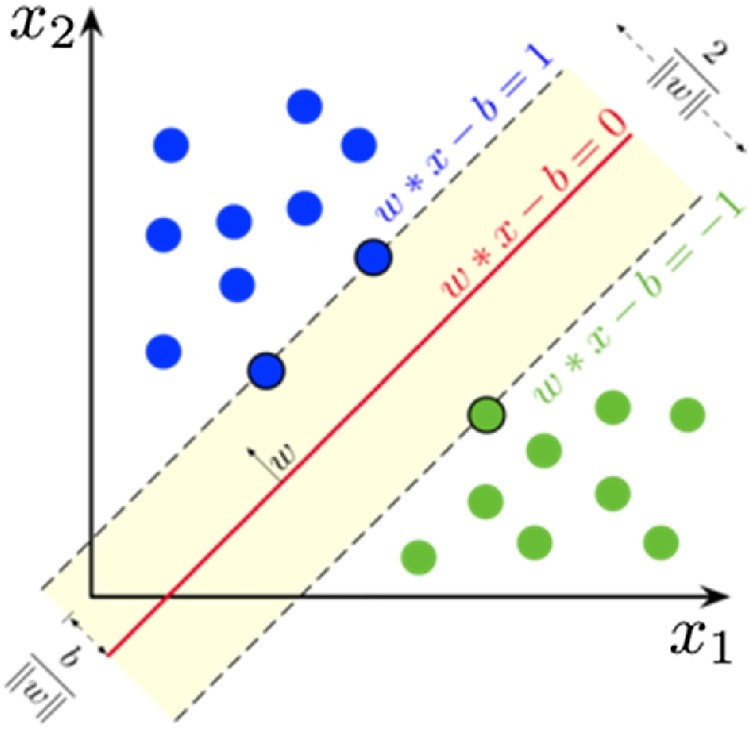
SVM [30], a supervised learning model, is particularly effective for classification, numerical prediction and pattern recognition tasks. SVMs find a line between different classes of data to maximize the distance of a line or hyper plane to the next closest data points. In other words, the support vector machines calculate a maximum margin limit, which leads to a homogeneous division of all data points. Eqs. (10) and (11) represent formulas for a line or hyper plane, respectively. SVM [31,32] should find weights so that the data points are separated according to a decision rule. SVM is demonstrated in Fig. 2 .
| (10) |
| (11) |
where w is the normal vector to the hyperplane, x is the input vector, and y is the correct output of the SVM for ith training example.
Fig. 2.
Support Vector Machine.
* Default parameters of the Matlab program were used for SVM.
2.6. Proposed approach
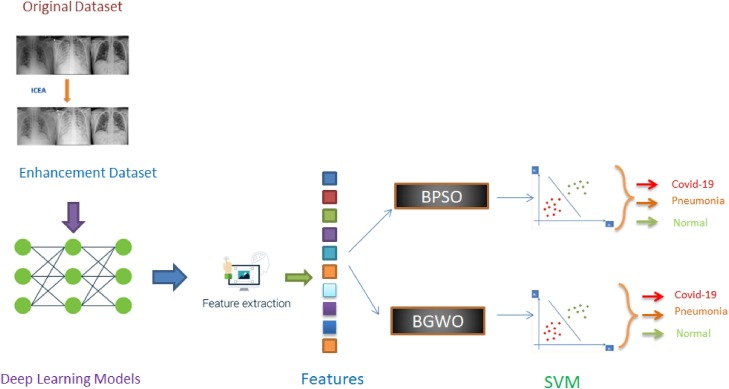
Since we use deep learning algorithms and meta-heuristic algorithms together in our proposed approach, we thought it would be appropriate to name it MH-CovidNet. Our approach will be referred to by this name from now on. The MH-CovidNet approach aims to distinguish COVID-19, pneumonia, and normal X-ray images using features. The MH-CovidNet proposed in our study to achieve this consists of 4 stages. After the data set is created, a new data set was obtained with the image enhancement method ICEA proposed by Ying [22]. This is the first stage of this approach. The purpose of this phase is to provide better quality pictures. In the second step of the approach, deep neural networks are trained with both the original and enhancement datasets. The models obtained as a result of training of deep neural networks with the data set in 3 classes were used to extract the features in the third step. At this stage, 1000 features are extracted for each image in each model. First of all, these features were subjected to classification with SVM. In the last stage of our approach, we tried to select the most effective features by using BPSO and BGWO meta-heuristic algorithms among the 1000 features obtained from deep neural networks with the enhancement data set. These 1000 features were obtained from the "FC8" layer in the Alexnet and VGG19 network, while "loss3-classifier" was used in the GoogleNet network and the "FC1000" layer was used in the ResNet network. These features are recorded as ".mat" extensions and will be published in the repo specified in the open source code section. Feature selection is made with BPSO and BGWO meta-heuristic algorithms, and classification is done with SVM. In addition, the effective features obtained from each algorithm are combined among themselves, and the success of multiple classifications is increased. Fig. 3 shows the proposed approach.
Fig. 3.
Graphical abstract of MH-CovidNet.
To briefly summarize Fig. 3 and MH-CovidNet:
In our study, a 3-class data set of COVID-19, normal, and pneumonia was created from the X-ray images we obtained from open sources. Pre-processing was performed on this data set using the ımage contrast enhancement algorithm (ICEA) [15]. The newly obtained data set was trained with deep learning models such as AlexNet, VGG19, GoogleNet, and ResNet. Feature extraction was completed using the trained models. With the help of two different meta-heuristic algorithms of binary particle swarm optimization (BPSO) and binary gray wolf optimization (BGWO), the most effective features were selected. If we explain the process here a little more; features obtained from each model are subjected to feature selection through the BPSO algorithm. Selected features are classified by SVM. Then, the features of the two models that provide the highest accuracy in SVM from the features obtained with the help of BPSO are combined and the feature selection is made with BPSO. The same process is done for the features of the models providing the lowest accuracy. We decided it would be appropriate to use a second meta-heuristic algorithm to verify the reliability of these results obtained with BPSO. Therefore, we used the BGWO algorithm for the same operations. In other words, these two algorithms work independently of each other and perform operations such as feature selection and combination on their own.
3. Experimental analysis and results
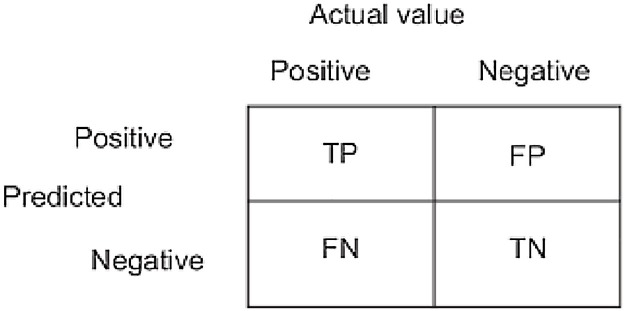
The application developed for the study was developed in the Matlab environment. The computer running the application has features such as 16 GB RAM, I7 processor and GeForce 1070 graphics card. Performance metrics [33,34] are calculated from the confusion matrix obtained in the experimental results. These metrics are Sensitivity (Se), Specificity (Sp), F-score (F-Scr), Precision (Pre), and Accuracy (Acc). True Positive (TP), False Positive (FP), True Negative (TN), and False Negative (FN) values are used to calculate the metrics. Equations for metric values calculated with these values are given in 12−16. For a 2-class structure, these values are shown in Fig. 4 for the confusion matrix.
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
Fig. 4.
Confusion matrix for 2-class.
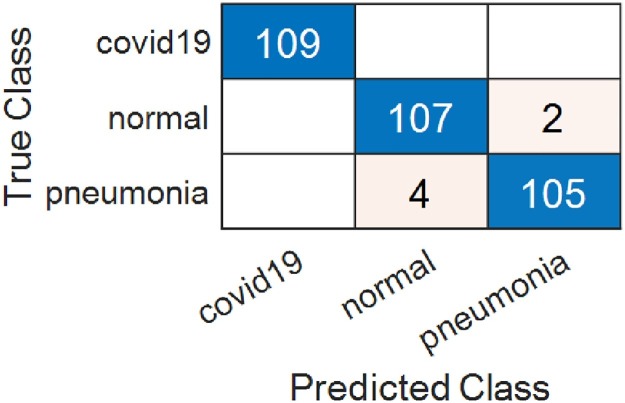
In experimental studies, 30% of the data was used for testing and 70% for training at every stage of the approach. In the final steps of the study, the consistency of the study result was tested using k-fold cross-validation with a k value of 5 for the feature dataset obtained from the enhancement data set. The first stage of the application is the stage of creating the enhancement data set by applying the ICEA method to the original data set. In the second stage, each deep neural network was trained with original and enhancement data and the models were recorded as “* .mat” files separately. In the first step of this stage, results from trained models were obtained using the original data set and SVM. In experimental studies using the original dataset, AlexNet achieved 97.55%, VGG19 98.16%, GoogleNet 95.10% and ResNet achieved overall accuracy rate of 95.71%. In Fig. 5 , the confusion matrices resulting from the SVM classification of the feature dataset that we obtained after training the original dataset on VGG19 are shown.
Fig. 5.
Confusion matrix obtained from VGG19 for the original dataset.
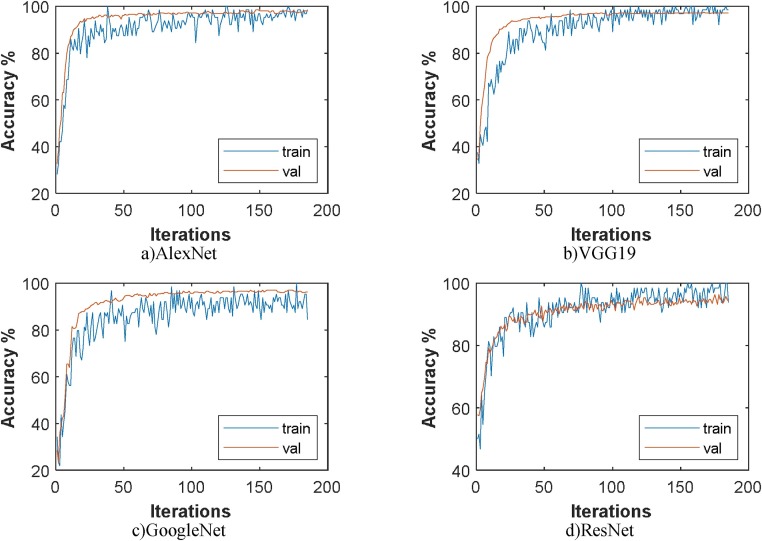
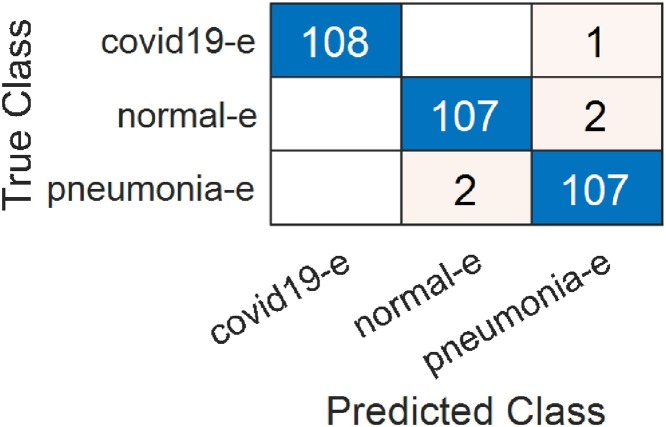
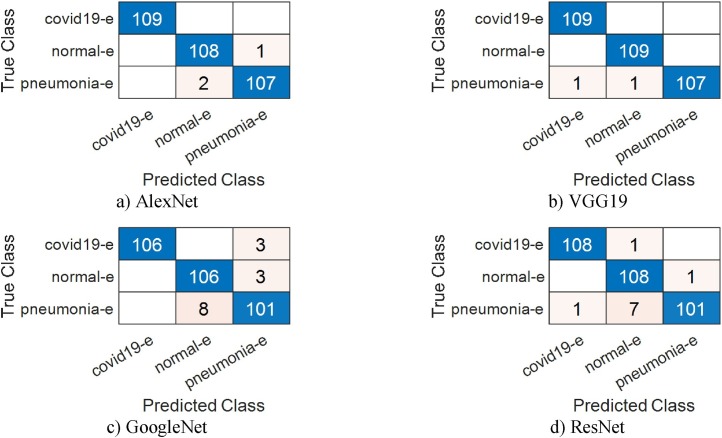
In the second step, classification was made using enhancement data set and SVM. In this step, AlexNet achieved 97.55%, VGG19 98.47%, GoogleNet 96.94% and ResNet achieved 96.94% overall accuracy rate. It is possible to say that the enhancement technique provides successful enhancement for GoogleNet and ResNet. Fig. 6 shows the training and validation graphics of deep learning models obtained as a result of separating the enhancement dataset as training and validation data while running the models. In Fig. 7 , the confusion matrices resulting from the SVM classification of the feature dataset that we obtained after training the enhancement dataset on VGG19 are shown.
Fig. 6.
Training and validation accuracy of the models on the enhancement dataset.
Fig. 7.
Confusion matrix obtained from VGG19 for the enhancement dataset.
The experimental results obtained for both data sets are shown in Table 3 .
Table 3.
Metric values of the confusion matrix of models.
| Original Data |
Enhancement Data |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Classes | F-Scr (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) | F-Scr (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) |
| AlexNet | COVID-19 | 99.54 | 99.09 | 99.69 | 97.55 | 98.16 | 98.16 | 98.77 | 97.55 |
| Pneumonia | 96.83 | 95.53 | 97.85 | 97.71 | 97.27 | 98.47 | |||
| Normal | 96.26 | 98.09 | 97.55 | 96.77 | 97.22 | 97.85 | |||
| VGG19 | COVID-19 | 100 | 100 | 100 | 98.16 | 99.53 | 100 | 99.69 | 98.47 |
| Pneumonia | 97.27 | 96.39 | 98.16 | 98.16 | 98.16 | 98.77 | |||
| Normal | 97.22 | 98.13 | 98.16 | 97.71 | 97.27 | 98.47 | |||
| GoogleNet | COVID-19 | 98.16 | 98.16 | 98.77 | 95.1 | 99.09 | 98.19 | 99.38 | 96.94 |
| Pneumonia | 94.59 | 92.92 | 96.33 | 96.33 | 96.33 | 97.55 | |||
| Normal | 92.52 | 94.28 | 95.10 | 95.37 | 96.26 | 96.94 | |||
| ResNet | COVID-19 | 98.60 | 100 | 99.08 | 95.71 | 98.63 | 98.18 | 99.08 | 96.94 |
| Pneumonia | 94.59 | 92.92 | 96.33 | 96.83 | 95.53 | 97.85 | |||
| Normal | 94.00 | 94.44 | 96.02 | 95.32 | 97.14 | 96.94 | |||
In the last step of the second stage, the enhancement dataset was split using the k-fold cross validation method to prove the accuracy of the previous step. In the previous step, 30% of the data was used as test data. The number of k-folds used for each model is 5. The overall accuracy achieved by AlexNet at this step was 97.98. This value was 97.55 in the previous step. The results obtained by this network are stable in both steps. Another network, VGG19, achieved an overall accuracy of 98.71 in this step and the value in the previous step was 98.47. When we look at the results for the remaining two models, the overall accuracy values for GoogleNet and ResNet are 95.6 and 96.61, respectively. The values for these networks in the previous step was 96.94. The results for these networks are stable in this step. Results obtained with k-fold cross validation confirmed the results obtained with 30% test data in the previous step. This shows that the proposed approach is reliable. Fig. 8 shows the some of confusion matrices obtained in this step. The analysis results in this step are given in Table 4 .
Fig. 8.
Confusion matrices with the method of 5-fold cross-validation for enhancement data.
Table 4.
Metric values of the confusion matrix of models (cross validation).
| Model & Data Type | Classes | F-Scr (%) | Se. (%) | Sp. (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) |
|---|---|---|---|---|---|---|---|
| AlexNet & Enhancement Data | COVID-19 | 99.44 | 99.17 | 99.86 | 99.72 | 99.63 | 97.98 |
| Pneumonia | 97.40 | 98.07 | 98.35 | 96.74 | 98.26 | ||
| Normal | 97.10 | 96.70 | 98.76 | 97.50 | 98.07 | ||
| VGG19 & Enhancement Data | COVID-19 | 99.58 | 99.45 | 99.86 | 99.72 | 99.72 | 98.71 |
| Pneumonia | 98.36 | 99.17 | 98.76 | 97.56 | 98.90 | ||
| Normal | 98.20 | 97.52 | 99.45 | 98.88 | 98.80 | ||
| GoogleNet & Enhancement Data | COVID-19 | 98.48 | 98.07 | 99.45 | 98.89 | 98.99 | 95.6 |
| Pneumonia | 94.76 | 96.97 | 96.15 | 92.65 | 96.42 | ||
| Normal | 93.55 | 91.75 | 97.80 | 95.42 | 95.78 | ||
| ResNet & Enhancement Data | COVID-19 | 98.06 | 97.52 | 99.31 | 98.61 | 98.71 | 96.61 |
| Pneumonia | 96.62 | 98.35 | 97.39 | 94.96 | 97.71 | ||
| Normal | 95.13 | 93.95 | 98.21 | 96.33 | 96.79 |
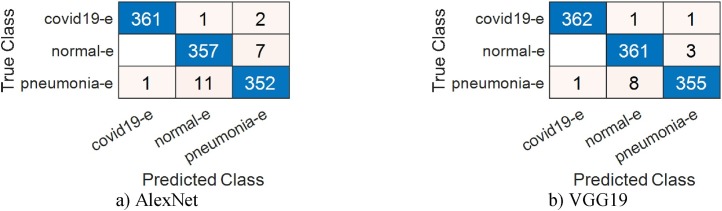
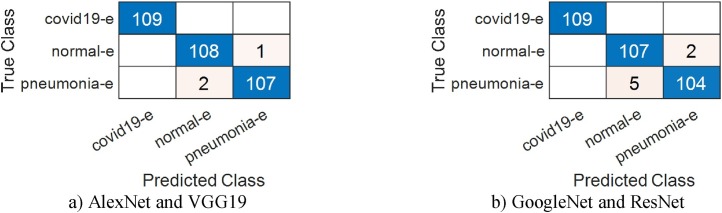
In the third stage of our approach, the performance of each algorithm is evaluated separately. The purpose of this stage is to show the effects of the properties obtained from these algorithms on the classification success. For this reason, this stage consists of 2 sub-stages. In the first step of the first sub-stage, effective properties were selected with BPSO. In this step, 499 features were selected by BPSO among the 1000 AlexNet features and were first classified and then classified with SVM. The classification obtained for this network had overall accuracy of 99.08%. Also, the classification success for the detection of COVID-19 data with the AlexNet model was 100%. Among the features of the VGG19 model, 488 features were selected and an overall accuracy of 99.38% was obtained. The percentage of success achieved for COVID-19 data is 99.69%. The number of features chosen by BPSO for the other GoogleNet and ResNet models was 488 and 477 respectively, while overall accuracy was 95.71% and 96.94%. In the second step of this first sub-stage, the features selected by BPSO from AlexNet and VGG19 models with high accuracy were classified with SVM after being combined. The newly obtained features value is 987. By using 30% of these features as test data, an overall accuracy of 99.08% was achieved. Also, the classification success for the detection of COVID-19 data with the combined features was 100%. While the features obtained through BPSO from GoogleNet and ResNet are combined and the 965-feature value is obtained, an overall accuracy of 97.85% was obtained as a result of classification of these features with SVM. This value is higher than the previous performance results obtained by these models. This confirmed the positive effect of the features selected by BPSO on the classification success percentage. Again, in order to verify the reliability of this phase in the third step, SVM classification was done with the combined features using the cross-validation method. In this step, which used 5 as a kfold value, a success rate of 99.08% was achieved for Combined AlexNet and VGG19, while a success rate of 97.06% was achieved for Combined GoogleNet and ResNet. Data analysis tables for this sub-stage are given Table 5, Table 6 . Confusion matrices are shown in Fig. 9, Fig. 10 .
Table 5.
Metric values obtained using the BPSO method.
| Model & Data Type | Classes | Total of Features | Test Data % | F-Scr (%) | Se. (%) | Sp. (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) |
|---|---|---|---|---|---|---|---|---|---|
| AlexNet & Enhancement Data | COVID-19 | 499 | 30 | 100 | 100 | 100 | 100 | 100 | 99.08 |
| Pneumonia | 98.63 | 99.08 | 99.08 | 98.18 | 99.08 | ||||
| Normal | 98.61 | 98.16 | 99.54 | 99.07 | 99.08 | ||||
| VGG19 & Enhancement Data | COVID-19 | 488 | 30 | 99.54 | 100 | 99.54 | 99.09 | 99.69 | 99.38 |
| Pneumonia | 99.54 | 100 | 99.54 | 99.09 | 99.69 | ||||
| Normal | 99.07 | 98.16 | 100 | 100 | 99.38 | ||||
| GoogleNet & Enhancement Data | COVID-19 | 488 | 30 | 98.60 | 97.24 | 100 | 100 | 99.08 | 95.71 |
| Pneumonia | 95.06 | 97.24 | 96.33 | 92.98 | 96.63 | ||||
| Normal | 93.51 | 92.66 | 97.24 | 94.39 | 95.71 | ||||
| ResNet & Enhancement Data | COVID-19 | 477 | 30 | 99.08 | 99.08 | 99.54 | 99.08 | 99.38 | 96.94 |
| Pneumonia | 96 | 99.08 | 96.33 | 93.10 | 97.24 | ||||
| Normal | 95.73 | 92.66 | 99.54 | 99.01 | 97.24 |
Table 6.
Metric values obtained using the BPSO method on combined features.
| Model | Classes | Total of Features | Test Data % | F-Scr (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) | k-fold | F-Scr (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AlexNet & VGG19 | COVID-19 | 987 | 30 | 100 | 100 | 100 | 99.08 | k=5 | 99.58 | 99.72 | 99.72 | 99.08 |
| Pneumonia | 98.63 | 98.18 | 99.08 | 98.90 | 98.36 | 99.26 | ||||||
| Normal | 98.61 | 99.07 | 99.08 | 98.75 | 99.16 | 99.17 | ||||||
| GoogleNet & ResNet | COVID-19 | 965 | 30 | 100 | 100 | 100 | 97.85 | k=5 | 98.89 | 99.44 | 99.26 | 97.06 |
| Pneumonia | 96.83 | 95.53 | 97.85 | 96.35 | 94.69 | 97.52 | ||||||
| Normal | 96.74 | 98.11 | 97.85 | 95.96 | 97.18 | 97.34 |
Fig. 9.
Confusion matrices obtained using the BPSO method.
Fig. 10.
Confusion matrices obtained using the BPSO method; (a) by combining the features of the AlexNet model with the VGG19 model (30% test data) (b) by combining the features of the GoogleNet model with the ResNet model (30% test data).
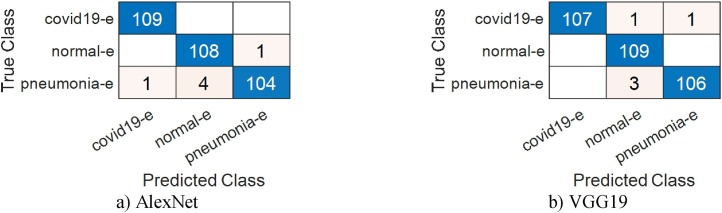
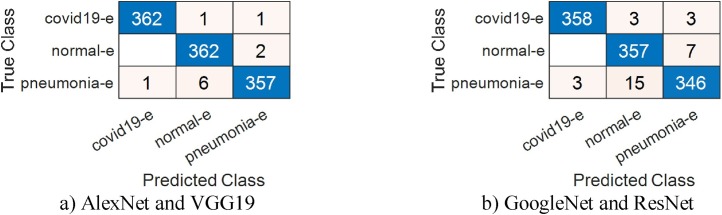
In the last stage of this phase, all feature selection and classification procedures performed in the previous stage were done by BGWO and SVM. The percentage of test data used is 30%. A total of 575 features were selected from AlexNet features by BGWO algorithm and an overall accuracy of 98.16% was obtained. The percentage of classification success for COVID-19 data is 99.69%. The number of effective features selected from VGG19 is 627 and overall accuracy is 98.47%. The percentage of success for classifying COVID-19 data for this network is 99.38%. Finally, 662 and 572 features were selected by BGWO, respectively, for GoogleNet and ResNet, with 96.33% and 96.94% overall accuracy values obtained. Similar to BPSO, the features selected by this algorithm were combined among themselves for models. SVM was used again for the classification of combined features. The number of features obtained by combining AlexNet and VGG19 features is 1202 and overall accuracy is 99.08%. On the other hand, the number of features obtained by combining GoogleNet and ResNet features is 1234, overall accuracy is 97.24%. Attempts were made to verify the reliability of these results once again with the cross-validation method. In this method, where the k-fold number was determined as 5, the classification accuracy percentage obtained with the combined properties of AlexNet and VGG19 was 98.99%, while this ratio was 97.16% for GoogleNet and ResNet. The results are consistent with the results obtained using 30% test data. The analysis results in this stage are given in Table 7, Table 8 . Confusion matrices are given in Fig. 11, Fig. 12 .
Table 7.
Metric values obtained using the BGWO method.
| Model &Data Type | Classes | Total of Features | Test Data % | F-Scr (%) | Se. (%) | Sp. (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) |
|---|---|---|---|---|---|---|---|---|---|
| AlexNet & Enhancement Data | COVID-19 | 575 | 30 | 99.54 | 100 | 99.54 | 99.09 | 99.69 | 98.16 |
| Pneumonia | 97.73 | 99.08 | 98.16 | 96.42 | 98.47 | ||||
| Normal | 97.19 | 95.41 | 99.54 | 99.04 | 98.16 | ||||
| VGG19 & Enhancement Data | COVID-19 | 627 | 30 | 99.074 | 98.16 | 100 | 100 | 99.38 | 98.47 |
| Pneumonia | 98.19 | 100 | 98.16 | 96.46 | 98.77 | ||||
| Normal | 98.14 | 97.24 | 99.54 | 99.06 | 98.77 | ||||
| GoogleNet & Enhancement Data | COVID-19 | 662 | 30 | 99.09 | 100 | 99.08 | 98.19 | 99.38 | 96.33 |
| Pneumonia | 95.45 | 96.33 | 97.24 | 94.59 | 96.94 | ||||
| Normal | 94.39 | 92.66 | 98.16 | 96.19 | 96.33 | ||||
| ResNet & Enhancement Data | COVID-19 | 572 | 30 | 98.16 | 98.16 | 99.08 | 98.16 | 98.77 | 96.94 |
| Pneumonia | 96.88 | 100 | 96.78 | 93.96 | 97.85 | ||||
| Normal | 95.73 | 92.66 | 99.54 | 99.01 | 97.24 |
Table 8.
Metric values obtained using the BGWO method on combined features.
| Model | Classes | Total of Features | Test Data % | F-Scr (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) | k-fold | F-Scr (%) | Pre. (%) | Acc. (%) | Overall Acc. (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AlexNet & VGG19 | COVID-19 | 1202 | 30 | 99.07 | 100 | 99.38 | 99.08 | k=5 | 99.58 | 99.72 | 99.72 | 98.99 |
| Pneumonia | 99.54 | 99.09 | 99.69 | 98.77 | 98.1 | 99.17 | ||||||
| Normal | 98.63 | 98.18 | 99.08 | 98.61 | 99.16 | 99.08 | ||||||
| GoogleNet & ResNet | COVID-19 | 1234 | 30 | 99.54 | 99.09 | 99.69 | 97.24 | k=5 | 98.75 | 99.16 | 99.17 | 97.16 |
| Pneumonia | 96.46 | 93.16 | 97.55 | 96.61 | 95.2 | 97.71 | ||||||
| Normal | 95.69 | 100 | 97.24 | 96.11 | 97.19 | 97.43 |
Fig. 11.
Confusion matrices obtained using the BGWO method.
Fig. 12.
Confusion matrices obtained using the BGWO method; (a) by combining the features of the AlexNet model with the VGG19 model (k fold value = 5). (b) by combining the features of the GoogleNet model with the ResNet model (k fold value = 5).
The studies performed on X-ray images so far and the comparative table of our study is given in Table 9 .
Table 9.
Comparison of the proposed MH-CovidNet with other existing deep learning methods.
| Study | Method used | Image Pre-Processing | Constrat Enhancement | Number of Cases | Accuracy % | Feature Size | Computation Time | |
|---|---|---|---|---|---|---|---|---|
| Hemdan et al [1] | VGG19 | No | No | 25 COVID, 25 non-COVID, type of image:jpg and png | 90 | Not Available | Max:2645 s | |
| Toğaçar et al. [2] | MobileNet and SqueezeNet | Yes | No | 295 COVID, 98 pneumonia, 65 normal, type of image:jpg and png | 99.27 | Min:663 Max:1357 | Not Available | |
| Zhang et al. [3] | ResNet | No | No | 100 COVID, 1431 pneumonia, type of image:jpg and png | 95.18 | Not Available | Not Available | |
| Afshar et al. [4] | CapsulNet | No | No | 94,323 x-ray images, type of image: png | 98.3 | Not Available | Not Available | |
| Apostolopoulos et al. [5] | VGG19 | No | No | 224 COVID, 700 pneumonia,504 normal, type of image:jpg and png | 98.75 | Not Available | Not Available | |
| Ozturk et al. [6] | DarkNet | No | No | 127 COVID, 500 pneumonia, 500 normal, type of image:jpg and png | 98.08 | Not Available | Not Available | |
| Uçar et al. [7] | SqueezeNet and Bayesian optimization | Yes | No | 76 COVID, 1591 pneumonia, 1203 normal, type of image:jpg and png | 98.3 | Not Available | Max:2395 s | |
| Proposed MH-CovidNet Approach | AlexNet | Yes | Yes | 364 COVID, 364 pneumonia, 364 normal, type of image:jpg | 97.55 | 1000 | Max:2500 s | |
| VGG19 | 98.47 | 1000 | ||||||
| GoogleNet | 96.94 | 1000 | ||||||
| ResNet | 96.94 | 1000 | ||||||
| BPSO | AlexNet | 99.08 | 499 | |||||
| VGG19 | 99.38 | 488 | ||||||
| GoogleNet | 95.71 | 488 | ||||||
| ResNet | 96.94 | 477 | ||||||
| BGWO | AlexNet | 98.16 | 575 | |||||
| VGG19 | 98.47 | 627 | ||||||
| GoogleNet | 96.33 | 662 | ||||||
| ResNet | 96.94 | 572 | ||||||
When we look at the studies done so far, the approach we propose achieved the best value. It should be emphasized that this success was achieved by using fewer numbers of features than other models. In addition, the non-balanced class problem is avoided by keeping the number of cases in the class equal. For detailed information about the studies, see the introduction section.
4. Discussion
In this study, deep learning models and meta-heuristic algorithms were used together for classification of 3 classes using COVID-19, pneumonia and normal lung X-ray images. AlexNet and VGG19, both deep learning models, provided better results in experimental studies using both the original and enhancement datasets compared to other models. When we look at the initial values of the results obtained using the features obtained from GoogleNet and ResNet models and the results obtained with the approach we propose, there was not much change. However, when the features obtained from these two models were combined after feature selection with the help of meta-heuristic algorithms, an increase was observed in the success value. In the study, the most effective features were selected with the help of two meta-heuristic algorithms that verify each other. In order to confirm the accuracy of the results obtained, we tried to prove the reliability of the approach we proposed using holdout validation and k-fold cross validation methods.
The advantages of the study include showing the effect of image preprocessing on classification success, reducing the computation time by selecting the most effective features with the help of meta-heuristic algorithms, and showing the performance of different deep learning models for COVID-19 disease diagnosis. Another advantage is that the MH-CovidNet approach developed a different solution in this area since there is no previous study about contrast enhancement in diagnosing COVID-19 disease in X-ray images. The disadvantages are that not every deep learning model is able to achieve sufficient success with the proposed approach, and it is necessary to investigate new deep learning models that will ensure success. Studies are ongoing about the diagnosis of COVID-19. For this reason, all studies will contribute to the process. It is obvious that different models and methods should be attempted to diagnose the disease.
5. Conclusion
COVID-19, which is a rapidly spreading disease in the world, will continue to affect our lives for a long time if vaccine studies do not succeed in the near future. Researchers continue to investigate methods for diagnosis and treatment in this regard. The primary purpose of our study is to contribute to this research. For this purpose, we created a 3-class dataset, which included COVID-19, pneumonia and normal X-ray lung images we obtained from open sources. The created data set was pre-processed and a new data set was obtained. Deep learning models of AlexNet, VGG19, GoogleNet, and ResNet, trained with this data set, were used for feature extraction. Then, the most effective features were selected from the extracted features with the help of meta-heuristic algorithms. Selected features were classified with the SVM classifier. The features of the models that provided the highest performance were combined among themselves, and the features of the models that provided the lowest performance were combined. Again, classification was done with SVM. When we look at the results obtained, 99.38% overall accuracy was obtained as a result of selecting and classifying the features obtained from the VGG19 model with the help of the BPSO algorithm. Another successful model was found to be AlexNet. Since the approach was proven to be reliable by considering different criteria, it is predicted that it can be used to provide another idea for experts during the diagnosis of COVID-19 disease. In order to contribute to this field in future studies, the plan is to continue studies using image processing and different deep learning models.
Open source code
Information about source codes, datasets, and related analysis results used in this study will be given at this web link. https://github.com/mcanayaz
Ethical approval
This article does not contain any data, or other information from studies or experimentation, with the involvement of human or animal subjects.
CRediT authorship contribution statement
Murat Canayaz: Methodology, Software, Validation, Investigation, Data curation, Writing - original draft, Visualization, Project administration, Conceptualization, Validation, Formal analysis, Resources, Writing - review & editing, Supervision, Visualization, Funding acquisition.
Acknowledgment
This study was supported by Scientific Research Projects Department Project No. FBA-2018-6915 from Van Yuzuncu Yıl University.
Acknowledgments
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.E.E.D. Hemdan, M.A. Shouman, M.E. Karar, COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images. (2020) arXiv preprint arXiv:2003.11055.
- 2.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Z. Jianpeng, X. Yuton, L. Yi, S. Chunhua, X. Yong, COVID-19 Screening on Chest X-ray Images Using Deep Learning based Anomaly Detection. (2020) arXiv preprint arXiv:2003.12338.
- 4.A. Parnian, H. Shahin, N. Farnoosh, O. Anastasia, P. Konstantinos, M. Arash, COVID-CAPS: A Capsule Network-based Framework for Identification of COVID-19 cases from X-ray Images. (2020). arXiv preprint arXiv:2004.02696. [DOI] [PMC free article] [PubMed]
- 5.Apostolopoulos I., Mpesiana T. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020 doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira R., Bertolini D., Teixeira L., Silla C., Costa Y. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020;194:105532. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ucar F., Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ardakani A., Kanafi A., Acharya U., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. Barstugan, U. Ozkaya, S. Ozturk, Coronavirus (COVID-19) Classification Using CT Images by Machine Learning Methods, (2020), arXiv preprint arXiv:2003.09424.
- 11.U. Ozkaya, S. Ozturk, M. Barstugan, Coronavirus (COVID-19) Classification using Deep Features Fusion and Ranking Technique, (2020), arXiv preprint arXiv:2004.03698.
- 12.Q. Yan, B. Wang, D. Gong, C. Luo,W.. Zhao, J. Shen,Q.. Shi, S. Jin, L. Zhang, Z. You, COVID-19 Chest CT Image Segmentation — A Deep Convolutional Neural Network Solution. (2020) arXiv preprint arXiv:2004.10987.
- 13.Hasan A., AL-Jawad M., Jalab H., Shaiba H., Ibrahim R., AL-Shamasneh A. Classification of Covid-19 coronavirus, pneumonia and healthy lungs in CT scans using Q-Deformed entropy and deep learning features. Entropy. 2020;22:517. doi: 10.3390/e22050517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh D., Kumar V., Kaur M.V. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020:1–11. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J.P. Cohen, P. Morrison, L. Dao, COVID-19 image data collection, (2020) arXiv:2003.11597, https://github.com/ieee8023/covid-chestxray-dataset.
- 16.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N., Reaz M.B.I. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 17.Kermany D., Zhang K., Goldbaum M. 2018. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification, Mendeley Data; p. v2.https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia? [DOI] [Google Scholar]
- 18.Krizhevsky A., Sutskever I., Hinton G. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017;60:84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 19.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition. (2014) arXiv 1409.1556.
- 20.Szegedy C. Going deeper with convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Boston, MA; 2015. pp. 1–9. [DOI] [Google Scholar]
- 21.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, NV; 2016. pp. 770–778. [DOI] [Google Scholar]
- 22.Ying Z., Li G., Ren Y., Wang R., Wang W. A new image contrast enhancement algorithm using exposure fusion framework. In: Felsberg M., Heyden A., Krüger N., editors. vol. 10425. Springer; Cham: 2017. (Computer Analysis of Images and Patterns. CAIP 2017. Lecture Notes in Computer Science). [Google Scholar]
- 23.Kennedy J., Eberhart R. Particle swarm optimization. Proceedings of IEEE International Conference on Neural Networks. IV. 1995:1942–1948. doi: 10.1109/ICNN.1995.488968. [DOI] [Google Scholar]
- 24.Too J., Abdullah A., Saad N.M. A new co-evolution binary particle swarm optimization with multiple inertia weight strategy for feature selection. Informatics. 2019;6(2):21. doi: 10.3390/informatics6020021. [DOI] [Google Scholar]
- 25.Tahir M.A., Bouridane A., Kurugollu F. Simultaneous feature selection and feature weighting using Hybrid Tabu Search/K-nearest neighbor classifier. Pattern Recognit. Lett. 2007;28(4):438–446. doi: 10.1016/j.patrec.2006.08.016. [DOI] [Google Scholar]
- 26.Too J., Abdullah A., Saad N.M., Tee W. EMG feature selection and classification using a Pbest-guide binary particle swarm optimization. Computation. 2019;7(1):12. doi: 10.3390/computation7010012. [DOI] [Google Scholar]
- 27.Mirjalili S., Mirjalili S.M., Lewis A. Grey wolf optimizer. Adv. Eng. Softw. 2014;69:46–61. [Google Scholar]
- 28.Emary E., Zawbaa H.M., Hassanien A.E. Binary grey wolf optimization approaches for feature selection. Neurocomputing. 2016;172:371–381. doi: 10.1016/j.neucom.2015.06.083. [DOI] [Google Scholar]
- 29.Too J., Abdullah A., Saad N.M., Ali N.M., Tee W. A new competitive binary grey wolf optimizer to solve the feature selection problem in EMG signals classification. Computers. 2018;7(4):58. doi: 10.3390/computers7040058. [DOI] [Google Scholar]
- 30.Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 31.Awad M., Khanna R. In: Support Vector Machines for Classification BT - Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers. Awad M., Khanna R., editors. Apress; Berkeley, CA: 2015. pp. 39–66. [DOI] [Google Scholar]
- 32.Doğan Ü., Glasmachers T., Igel C. A unified view on multi-class support vector classification. J. Mach. Learn. Res. 2016;17(45):1–32. [Google Scholar]
- 33.Cengil E., Çınar A. A new approach for image classification: convolutional neural network. Eur. J. Teach. Educ. 2016;6:96–103. [Google Scholar]
- 34.Comert Z. Fusing fine-tuned deep features for recognizing different tympanic membranes. Biocybern. Biomed. Eng. 2020;40:40–51. doi: 10.1016/j.bbe.2019.11.001. [DOI] [Google Scholar]