Abstract
Sex-specific synaptic connectivity is beginning to emerge as a remarkable, but little explored feature of animal brains. We describe here a novel mechanism that promotes sexually dimorphic neuronal function and synaptic connectivity in the nervous system of the nematode Caenorhabditis elegans. We demonstrate that a phylogenetically conserved, but previously uncharacterized Doublesex/Mab-3 related transcription factor (DMRT), dmd-4, is expressed in two classes of sex-shared phasmid neurons specifically in hermaphrodites but not in males. We find dmd-4 to promote hermaphrodite-specific synaptic connectivity and neuronal function of phasmid sensory neurons. Sex-specificity of DMD-4 function is conferred by a novel mode of posttranslational regulation that involves sex-specific protein stabilization through ubiquitin binding to a phylogenetically conserved but previously unstudied protein domain, the DMA domain. A human DMRT homolog of DMD-4 is controlled in a similar manner, indicating that our findings may have implications for the control of sexual differentiation in other animals as well.
Research organism: C. elegans
Introduction
Across both invertebrate and vertebrate phyla, adult animals usually display striking sexually dimorphic features (Fairbairn, 2013). These features range from morphological secondary sex characteristics such as pigmentation patterns to complex behavioral programs executed by one sex but not the other. Many sexually dimorphic morphologies and behaviors arise during specific phases of juvenile development, indicating the existence of developmental timing mechanisms that must intersect with the genetic programs that control sexual identity to produce sexually dimorphic features and structures of the adult animal (Faunes and Larraín, 2016; Tena-Sempere, 2013).
The existence of sexually dimorphic behaviors suggests the existence of sexually dimorphic features in the underlying neural circuits that govern such behaviors. The recently reconstructed synaptic wiring diagram of an adult C. elegans male provides novel vistas on the sex-specificity of nervous system anatomy, revealing the existence of sex-specific synaptic connections between neurons present in both sexes (‘sex-shared neurons’)(Cook et al., 2019; Jarrell et al., 2012; White et al., 1986). Moreover, specific sexually dimorphic circuits comprised of sex-shared neurons have been shown to generate behavioral dimorphisms in C. elegans (Barr et al., 2018; Emmons, 2018; Oren-Suissa et al., 2016; Ryan et al., 2014; White et al., 2007). How are sex-shared neurons genetically instructed to undergo sexually dimorphic maturation that results in changes in sex-specific synaptic connectivity and behavior?
While sexual differentiation is a vital part of development across a breadth of species, the upstream components of genetic pathways known to trigger sex determination are strikingly divergent. Multiple distinct primary sex determination mechanisms have arisen at various points in evolutionary history (autosomal sex determination, male or female heterogamy, fertilization status, or environmental cues), all with the ultimate downstream goal of producing stable mating types (Fairbairn, 2013). Nevertheless, in many metazoans, these divergent modes of sex determination employ, at some step of the sexual differentiation process, member(s) of the DMRT (Doublesex and MAB-3 related transcription factor) family of transcription factors (Kopp, 2012; Matson and Zarkower, 2012). As their name implies, DMRT family members were first identified in D. melanogaster (Doublesex) and C. elegans (MAB-3) based on their roles in directing sexually dimorphic development (Baker and Ridge, 1980; Shen and Hodgkin, 1988). DMRT family members were subsequently found to also control sexual differentiation of gonadal structures in vertebrates (Kopp, 2012; Matson and Zarkower, 2012; Raymond et al., 1998).
DMRT-encoding genes evolved from three ancestral clusters of genes that were present in the ancestor of bilateria and radiated independently in multiple distinct lineages unique to each phylum (Mawaribuchi et al., 2019; Volff et al., 2003; Wexler et al., 2014). Those DMRT proteins most extensively characterized in controlling sex-specific differentiation in the brain – mab-3, mab-23, and dmd-3 in C. elegans and doublesex in Drosophila (Lints and Emmons, 2002; Mason et al., 2008; Rideout et al., 2010; Siehr et al., 2011; Taylor et al., 1994; Verhulst and van de Zande, 2015; Yi et al., 2000) – are examples of such radiated DMRTs. Since those DMRTs have no clear orthologs outside their respective phyla, the question arises to what extent the phylogenetically more deeply conserved DMRT proteins are involved in sexual differentiation in the brain.
We describe here the function of a previously uncharacterized DMRT gene, dmd-4, that is deeply conserved across phylogeny. We show that dmd-4 is expressed in a specific set of neurons and in a subset of sensory neurons in a sex-specific manner. We show that DMD-4 is required to generate sexually dimorphic connectivity and behavioral output of sex-shared neurons. Unexpectedly, we found that sexually dimorphic expression of DMD-4 is established post-translationally via a previously uncharacterized, phylogenetically conserved domain, the DMA domain, which we identify as a ubiquitin-binding domain both in C. elegans and H. sapiens. Ubiquitin binding appears to be critical to stabilize DMD-4 protein in all cell types, but this ubiquitin-mediated stabilization is dynamically controlled in a cell-, time- and sex-specific manner to sculpt sexually dimorphic expression of DMD-4 in phasmid neurons during sexual maturation.
Results
Expression pattern of dmd-4
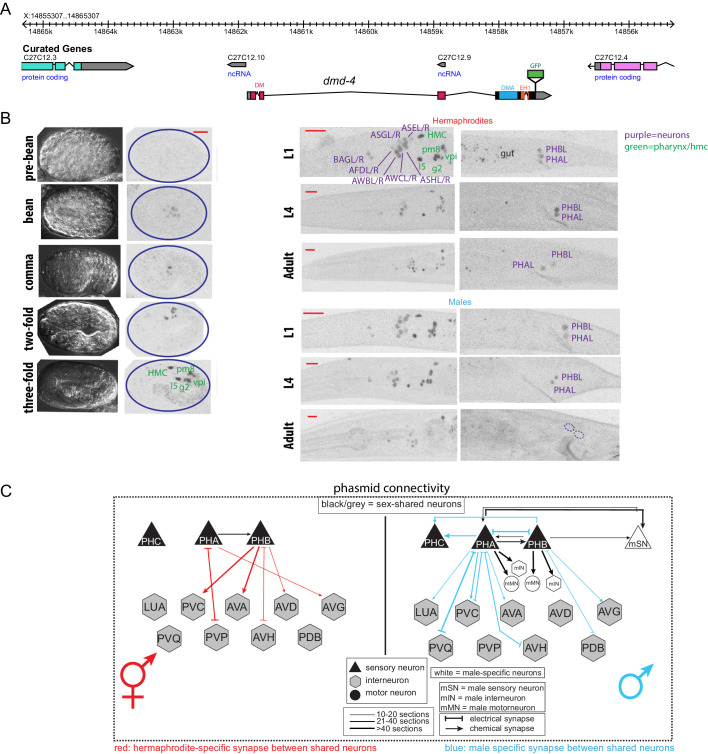
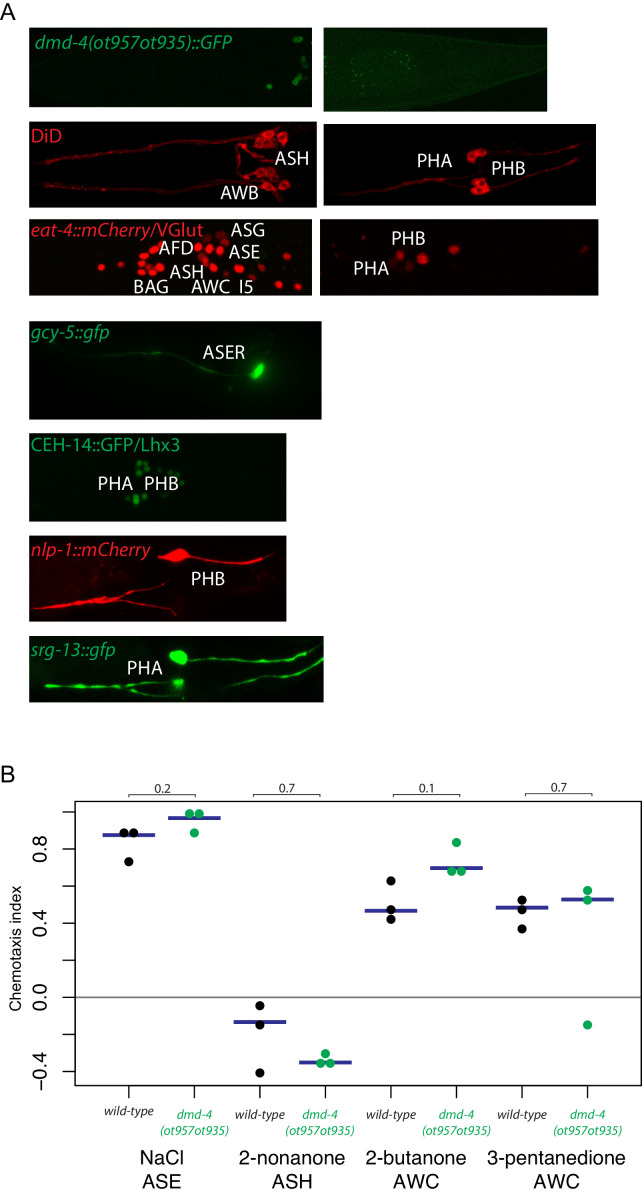
DMD-4 is one of ten DMRT proteins encoded by the C. elegans genome, only two of which, DMD-4 and DMD-5, are phylogenetically conserved in vertebrates (Volff et al., 2003; Wexler et al., 2014). Neither the expression nor the function of DMD-4 have previously been described. To determine the expression pattern of dmd-4, we used CRISPR/Cas9 genome engineering to tag the endogenous dmd-4 locus at the 3’ end of its coding region with gfp. We examined expression in sexually mature, adult animals and identified two sites of sexually dimorphic expression in the tail of the animal. DMD-4::GFP is expressed in PHA and PHB phasmid sensory neuron pairs of adult hermaphrodites, but not adult males (Figure 1A,B; Figure 1—figure supplement 1). Electron micrographic reconstruction of the synaptic connectivity of males and hermaphrodites revealed that these neurons display highly dimorphic synaptic connectivity (Cook et al., 2019; Jarrell et al., 2012; Figure 1C).
Figure 1. DMD-4 shows both sexually dimorphic and non-dimorphic expression.
(A) Schematic of the GFP-tagged dmd-4 locus in the dmd-4 (ot935) allele. The DM, DMA, and EH1 domains of DMD-4 are indicated in the colored boxes and labeled above the locus. (B) DMD-4::GFP is expressed in head sensory neurons and pharyngeal cells/the head mesodermal cell (hmc) in both sexes, and the phasmid sensory neurons PHA and PHB in both sexes until adulthood, when it is degraded in the male phasmids. Expression in the pharyngeal cells onsets during the ‘bean’ stage of embryogenesis. GFP is shown as color-inverted black and white. All scale bars (red horizontal bars) indicate 10 microns. Head images are maximum intensity projections of the entire worm; tail images are maximum intensity images of one half of the tail to clearly display phasmid nuclei. (C) Sexually dimorphic connectivity of the PHA and PHB neurons based on electron micrograph reconstruction (Cook et al., 2019; Jarrell et al., 2012).
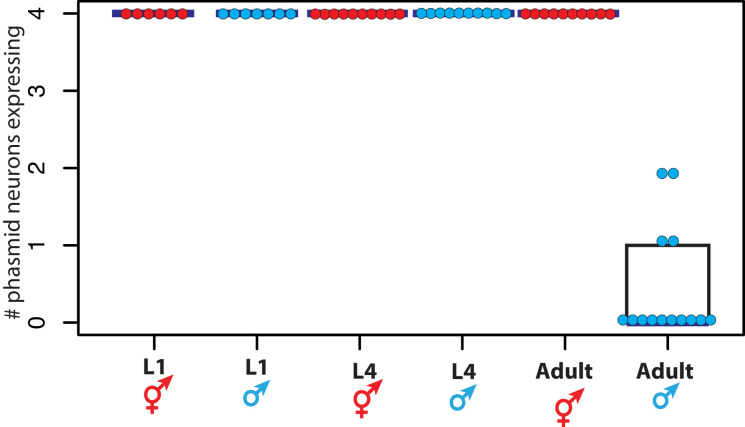
Figure 1—figure supplement 1. DMD-4::GFP is dimorphically expressed in the phasmid neurons.
In addition to this sexually dimorphic expression, we observed sexually non-dimorphic expression of DMD-4 in a subset of head sensory neurons (AFD, AWB, AWC, ASE, ASG, ASH, BAG), a single pharyngeal neuron (I5) and a number of MS blastomere derived cells - the head mesodermal cell (hmc), a single pharyngeal muscle (pm8), a pharyngeal gland cell pair (g2L/R) and four of the six pharyngeal intestinal valve cells (Figure 1B).
The onset of sexual differentiation in the C. elegans nervous system occurs at different stages of embryonic and postembryonic development (Barr et al., 2018; Emmons, 2005; Sulston and Horvitz, 1977; Sulston et al., 1983). To address the timing of sexually dimorphic DMD-4 expression, we examined expression of the gfp-tagged dmd-4 locus throughout all developmental stages. DMD-4::GFP fluorescence is first detectable in mid-embryonic development, with a pattern that is restricted to all the pharyngeal and the hmc cells described above. Onset of expression in the non-pharyngeal nervous system is first observed at around hatching in the same set of head and tail sensory neurons as observed in the adult stage, including the phasmid neurons PHA and PHB. During larval development, dmd-4::gfp expression is the same in both sexes until the L4 to adult molt when DMD-4::GFP protein starts to disappear from PHA and PHB in the male, but not the hermaphrodite (Figure 1—figure supplement 1). We conclude that sexually dimorphic dmd-4::gfp expression in the adult PHA and PHB neurons is the result of male-specific downregulation of dmd-4::gfp from these neurons. These dynamics contrast with the ontogeny of other sexually dimorphically expressed genes in the C. elegans nervous system, which often are not expressed at all until sexual maturation and then turn on in a sex-specific manner (e.g. daf-7 in ASJ, flp-13, eat-4 in PHC, srj-54 in AIM)(Hilbert and Kim, 2017; Lawson et al., 2019; Serrano-Saiz et al., 2017). Moreover, the hermaphrodite-specificity of DMD-4 expression also contrasts with the strict male-specificity of other DMRT proteins known to be dimorphically expressed in C. elegans (Lints and Emmons, 2002; Mason et al., 2008; Oren-Suissa et al., 2016; Yi et al., 2000).
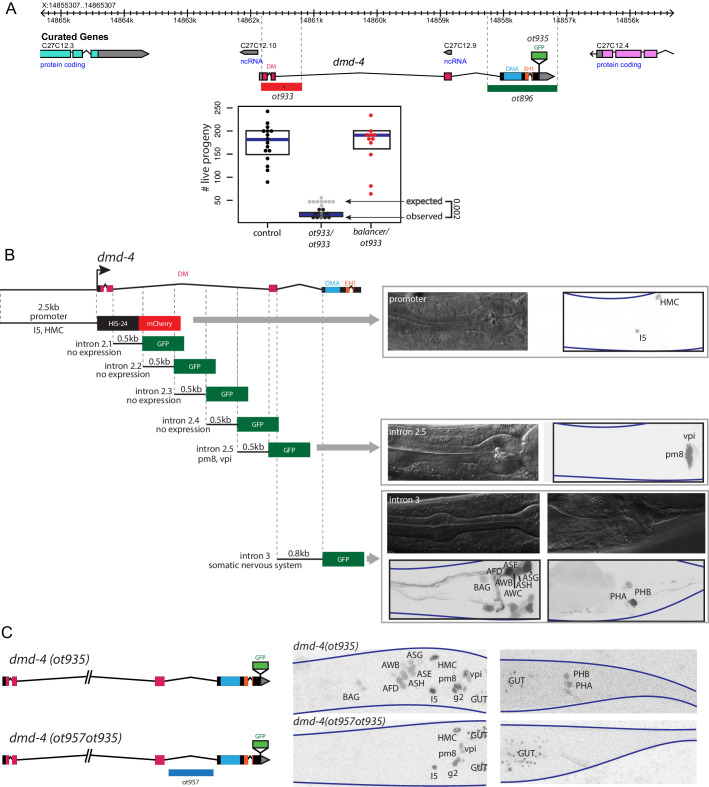
Generation of a nervous system-specific knock-out allele of dmd-4
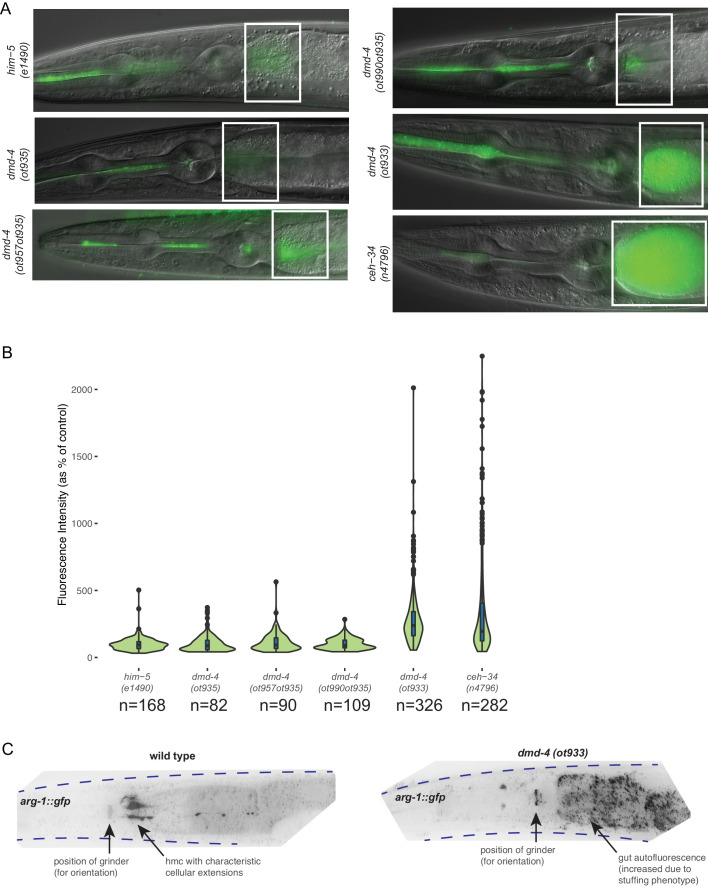
To assess dmd-4 function in the nervous system and, specifically, in the phasmid neurons, we used CRISPR/Cas9 genome engineering to delete the first two exons of the gene which code for most of the DNA binding domain (Figure 2A). This putative null allele (ot933) results in highly penetrant embryonic lethality (Figure 2A). A deletion allele generated by the C. elegans knockout consortium (tm1951), that eliminates parts of the DM domain, also results in lethality. Null mutant animals that escape embryonic lethality are scrawny and display a stuffed anterior intestine (Figure 2—figure supplement 1A,B). The stuffed anterior intestine may relate to developmental defects of the hmc, which fail to express a cell fate marker (arg-1::gfp) in dmd-4 mutants (Figure 2—figure supplement 1C). Another deletion allele that we generated using CRISPR/Cas9 leaves the DNA binding domain unaffected, but deletes other domains of the protein (DMA domain and EH1 domain)(Figure 2A), but this allele (ot896) results in no embryonic lethality.
Figure 2. Generation of a viable cis-regulatory allele of dmd-4.
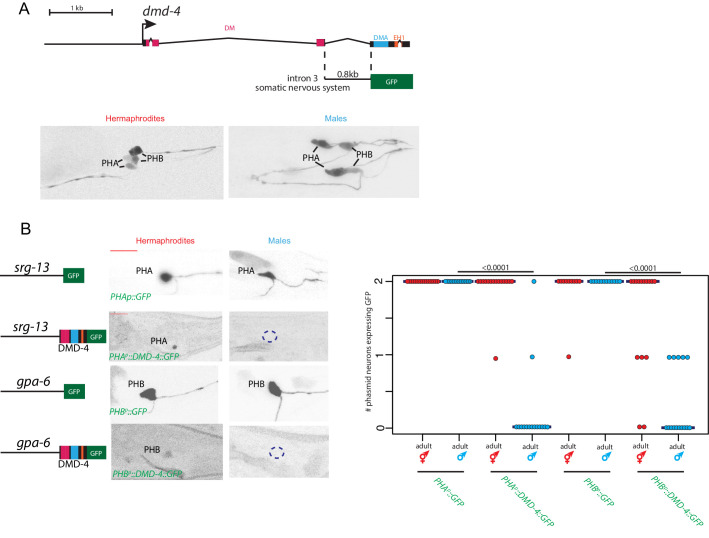
(A) The ot933 probable null allele of dmd-4 results in embryonic lethality and severe feeding defects in mutants that survive to adulthood. Deletion alleles generated by CRISPR/Cas9 are shown in the context of the dmd-4 locus above, the penetrance of ot933 embryonic lethality was quantified below by comparing the number of expected ot933 homozygous progeny from a balanced heterozygote mother (25% of total brood) with observed ot933 homozygous progeny. The difference between the observed number of ot933 homozygous progeny and expected homozygous progeny gives a measure of the penetrance of embryonic lethality. We observe a mean of 52% embryonic lethality, and comparing observed to expected ot933 progeny counts is significant by Wilcoxon rank-sum test. Each dot represents the brood size of one hermaphrodite, with blue bars indicating median and black boxes indicating quartiles. We show both observed and expected numbers of ot933 progeny in the figure panel. (B) The enhancers for I5/hmc expression, pharyngeal muscle expression, and somatic nervous system expression are located in separable regions of the dmd-4 promoter and introns. All constructs we examined are schematized, those that showed expression (promoter, intron 2.5, and intron 3) are shown in adult hermaphrodites to the right. GFP and mCherry are shown as color-inverted black and white, boundaries of the heads and tails of animals are marked with navy lines in all panels. Expressing cells are labeled on the GFP/mCherry images. (C) Generation of the dmd-4 (ot957ot935) nervous system null allele. Deletion of the third intron is schematized in the context of the dmd-4 locus above, color-inverted black and white GFP images of dmd-4 (ot935[DMD-4::GFP]) and dmd-4 (ot957ot935) hermaphrodites are shown below.
Figure 2—figure supplement 1. DMD-4 expression in the pharynx is required for proper anterior intestinal function.
Figure 2—figure supplement 2. General neuron identity is unaffected in dmd-4 (ot957ot935) mutants.
We sought to generate a nervous system-specific allele of the dmd-4 locus that we would expect to be viable and thus allow us to study the role of dmd-4 in the sexually dimorphic phasmid sensory neurons. To this end, we aimed to identify (and then delete) the cis-regulatory element required for dmd-4 expression in the nervous system. Through the fusion of individual segments of the dmd-4 locus to gfp, we identified the third intron of the dmd-4 locus as containing the cis-regulatory information for all nervous system expression (Figure 2B). Using CRISPR/Cas9 genome engineering, we deleted this enhancer in the context of the dmd-4::gfp allele (dmd-4(ot957ot935)), and indeed found that in these animals dmd-4::gfp expression was lost in all head and tail neurons at all developmental stages, but unaffected in the pharynx and hmc (Figure 2C). Most importantly, dmd-4(ot957ot935) mutants showed none of the lethality or intestinal stuffing phenotypes associated with removal of the gene from all tissues (Figure 2—figure supplement 1A,B), allowing us to characterize the function of dmd-4 in the nervous system.
Using this nervous system-specific allele, we evaluated several markers of neuronal identity to assess whether neurons that express dmd-4 were properly specified. We found no evidence of defects in general neuronal specification or morphology; all head sensory neurons as well as the phasmid neurons were generated in dmd-4(ot957ot935) mutant hermaphrodites and had a normal appearance of axon and dendrite morphologies (Figure 2—figure supplement 2). Furthermore, we examined transgenic reporters for neurotransmitter identity, neuropeptides, and receptors, and found no loss or misexpression of these genes in the amphid and phasmid neurons of dmd-4(ot957ot935) mutant animals (Figure 2—figure supplement 2). Lastly, behaviors associated with the function of head amphid sensory neurons also appear unaffected (Figure 2—figure supplement 2).
dmd-4 mutants display defects in sexually dimorphic synaptic connectivity
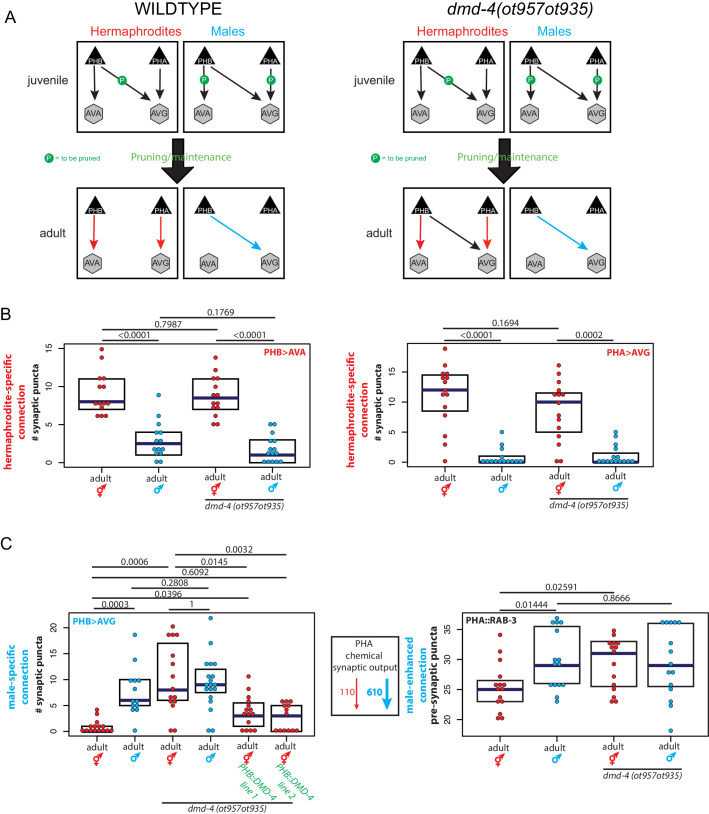
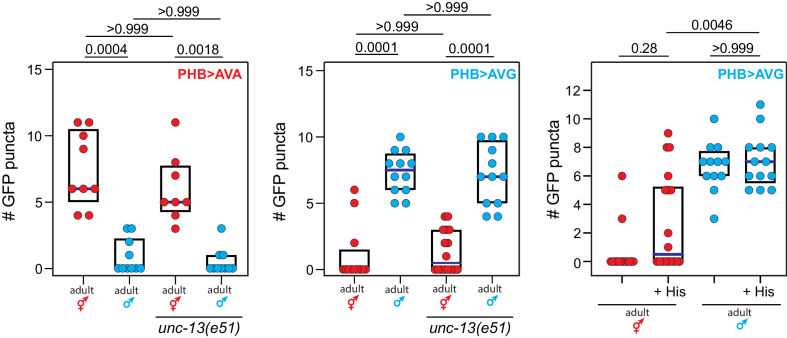
The sex-shared phasmid sensory neurons are notable for their strikingly dimorphic synaptic connectivity patterns (Cook et al., 2019; Jarrell et al., 2012; Figure 1C). Not only do both the PHA and PHB neurons generate synaptic connections to male-specific neurons, but they also display dimorphic synaptic connections to neurons that are shared between the two sexes (Figure 1C). We have previously used in vivo synapse labeling techniques, GRASP (GFP-Reconstitution Across Synaptic Partners)(Feinberg et al., 2008) and iBLINC (Desbois et al., 2015), to show that juvenile stages exhibit non-dimorphic ‘sex-hybrid’ connectivity and that during sexual maturation, some synapses are then pruned, while others are maintained (Figure 3A; Oren-Suissa et al., 2016). Unlike many pruning/maintenance events during nervous system development in other animal species (Katz and Callaway, 1992; Katz and Shatz, 1996; Morgan et al., 2011; Vonhoff and Keshishian, 2017), this synaptic maintenance is not dependent on sex-specific patterns of neuronal activity. We arrived at this conclusion by genetic silencing of the phasmid neurons or reducing synaptic signaling in unc-13 mutant animals, neither of which affects sexually dimorphic synapse pruning/maintenance (Figure 3—figure supplement 1).
Figure 3. dmd-4 is required for sexually dimorphic synaptic connectivity.
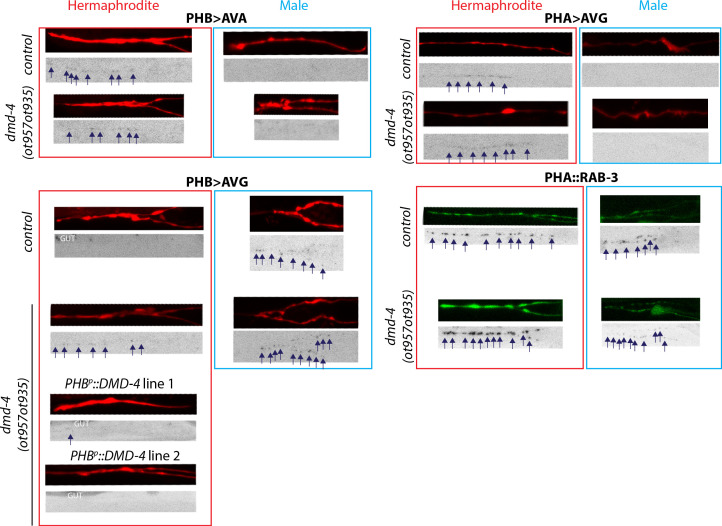
(A) Sexually dimorphic connectivity of the phasmid neurons in wild-type animals and dmd-4 (ot957ot935) animals (quantified in B, representative images shown in Figure 3—figure supplement 2) is schematized. Adult sexual dimorphism is generated by synaptic pruning of juvenile ‘sex-hybrid’ connectivity (Oren-Suissa et al., 2016). (B) The hermaphrodite-specific connections between PHB>AVA and PHA>AVG are unaffected in dmd-4 (ot957ot935) mutants. Each dot represents one animal (red = hermaphrodite, cyan = male, in all figures), blue bars show median, black boxes represent quartiles. P values are shown above horizontal black bars by Wilcoxon rank-sum test with Bonferroni corrections for multiple testing where applicable (see Materials and methods). Representative images are shown in Figure 3—figure supplement 2. (C) The male-specific PHB>AVG connection is not pruned in dmd-4 (ot957ot935) mutant hermaphrodites, and this phenotype can be rescued by expressing dmd-4 cDNA specifically in the PHB neurons (‘PHB::DMD-4’). We quantified two independent transgenic lines for mutant rescue. dmd-4 (ot957ot935) mutant hermaphrodites also show an increased number of BFP puncta in the BFP::RAB-3 construct labeling pre-synaptic vesicles. The sexual dimorphism of PHA chemical synaptic output is shown on the left, based on the number of EM sections where PHA is presynaptic in both sexes, 110 sections in hermaphrodites and 610 sections in males, (Cook et al., 2019). We term this connectivity ‘male-enhanced’ as it is present in both sexes but stronger in the male. Representative images are shown in Figure 3—figure supplement 2.
Figure 3—figure supplement 1. Phasmid synaptic pruning is not activity-dependent.
Figure 3—figure supplement 2. dmd-4 is required for dimorphic synaptic connectivity.
Using these GRASP reagents we assessed a subset of the hermaphrodite-specific and male-specific connections in the adult phasmid neurons of dmd-4(ot957ot935) mutant animals. We observed selective defects in sexually dimorphic synaptic connectivity. While hermaphrodite-specific synaptic connections (between PHA>AVG and PHB>AVA) are unaffected in dmd-4 mutants (Figure 3A,B, Figure 3—figure supplement 2), male-specific synapses (PHB>AVG) failed to be pruned in hermaphrodite dmd-4 mutants (Figure 3A,C, Figure 3—figure supplement 2; Oren-Suissa et al., 2016). We also developed a PHA-specific transgene using a synaptic vesicle-associated protein, RAB-3, to visualize all presynaptic zones of PHA as discrete BFP puncta (Figure 3C). Transgenic animals show the predicted sexual dimorphism based on electron micrographic reconstruction of PHA synaptic connectivity, with wild-type males displaying more synaptic outputs than wild-type hermaphrodites (Figure 1C, Figure 3C, Figure 3—figure supplement 2; Cook et al., 2019; Jarrell et al., 2012). In dmd-4(ot957ot935) mutant animals this sexual dimorphism is also lost, corroborating the notion that synapses fail to be pruned (Figure 3C, Figure 3—figure supplement 2).
We rescued the PHB>AVG synaptic pruning defect by expressing dmd-4 cDNA under the control of a PHB-specific promoter, confirming the cell-autonomous role of dmd-4 in the presynaptic neuron to control sex-specific synaptic pruning (Figure 3C, Figure 3—figure supplement 2). Taken together our results indicate that dmd-4 functions in the phasmid neurons to promote pruning of phasmid synaptic connections in hermaphrodite animals during sexual maturation.
Behavioral defects of dmd-4 mutants
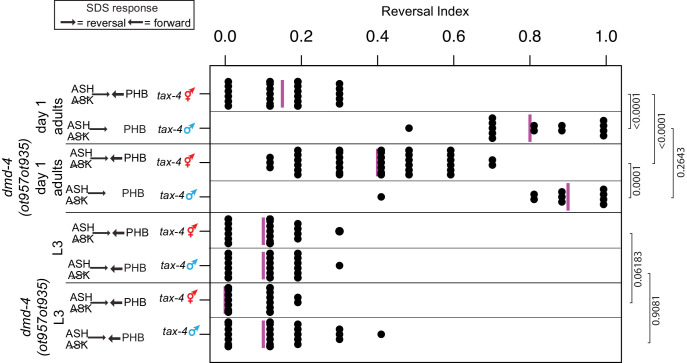
Sexually dimorphic connectivity of the phasmid neurons gives rise to sexually dimorphic behavioral outputs in adult animals. In larval animals, the phasmid neurons function to mediate chemosensory avoidance behavior (Hilliard et al., 2002; Zou et al., 2017), a functional output which is maintained in adult hermaphrodites, but lost in adult males (Oren-Suissa et al., 2016). dmd-4 mutants exhibit no defects in SDS avoidance in juvenile stages, before sexual maturation (Figure 4A). However, as adults, these animals display a partial masculinization of SDS avoidance behavior, such that mutant hermaphrodites were less responsive to the noxious stimulus, although still intermediate relative to control adult males (Figure 4A). It is possible that this defect is the result of lack of proper pruning of synapses, but it could also be the result of other functions of dmd-4 in the phasmid neurons.
Figure 4. dmd-4 affects sexually dimorphic behavior.
Noxious chemosensory avoidance is mediated by the PHB neuron (Hilliard et al., 2002), and sexually dimorphic in adults (Oren-Suissa et al., 2016). Left panel shows predicted synaptic input into avoidance behavior by relevant amphid and phasmid neurons. All experiments were done using tax-4 (p678) mutants to disable amphid input and uncover phasmid function, as previously described (Hilliard et al., 2002). dmd-4 (ot957ot935) adult hermaphrodites show reduced chemosensory avoidance behavior, but this is not evident in larval animals. Each dot represents the average reversal index of one animal over 10 experimental trials, median shown with vertical magenta bar. P values shown to the right by Wilcoxon rank-sum test with Bonferroni corrections for multiple testing (where applicable; see Materials and methods).
Control of spatiotemporal and sexual specificity of DMD-4 expression
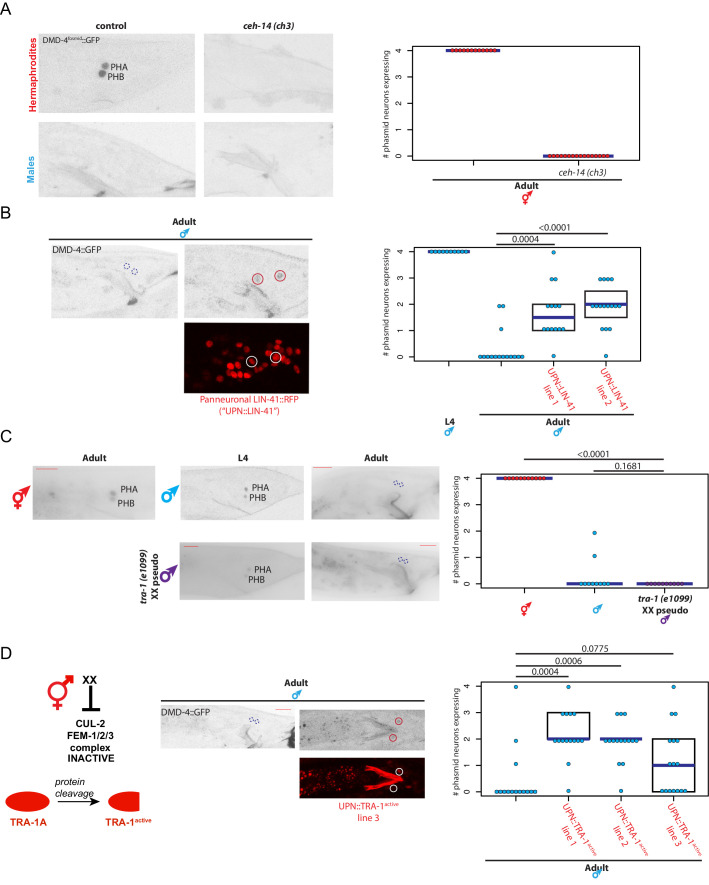
We sought to place the regulation of DMD-4 expression in the context of other regulatory pathways that control spatial, sexual and temporal aspects of gene expression in the nervous system. First, to ask how dmd-4 expression is established specifically in the phasmid neurons in an initially non-sex-specific manner at hatching, we turned to terminal selector type transcription factors, master regulatory factors that control multiple aspect of a neuron’s identity (Hobert, 2016). Based on previous mutant analysis, the ceh-14 LIM homeobox transcription factor is a terminal selector for the phasmid neurons (Kagoshima et al., 2013; Serrano-Saiz et al., 2013). We find that in ceh-14 null mutants, dmd-4::gfp expression is selectively lost in the phasmid neurons in both sexes during larval stages and in the adult stage in hermaphrodites (Figure 5A). This is also consistent with the observation that dmd-4(ot957ot935) mutant hermaphrodites display masculinization of SDS avoidance behavior (Figure 4A), a phenotype observed in ceh-14 mutant hermaphrodites (Oren-Suissa et al., 2016).
Figure 5. dmd-4 is regulated by spatial, temporal, and sex-specific cues in the phasmid neurons.
(A) DMD-4 expression is lost in hermaphrodite animals when the ceh-14 terminal selector of phasmid neuron fate is mutated. Male expression is unaffected. Color-inverted black and white GFP images are shown to the left in all panels, quantification of phasmid neurons expressing DMD-4 in wild-type and mutant animals is shown to the right in all panels. Horizontal blue line indicates median. (B) Constitutive, panneuronal expression of the LIN-41 heterochronic gene maintains DMD-4 in the adult male phasmids. Color-inverted black and white GFP images of adult males are shown in addition to the panneuronal, RFP-tagged LIN-41 overexpression (‘UPN::LIN-41’). L4 male quantification of DMD-4 expression in the phasmids (from Figure 1B) is shown again for reference comparison to control adults (siblings of transgenic animals) and LIN-41 overexpressing males. We quantified two independent transgenic lines. P values are shown above horizontal black bars by Wilcoxon rank-sum test with Bonferroni corrections for multiple testing where applicable (see Materials and methods) in all panels. (C) DMD-4 is degraded in the phasmid neurons of tra-1 (e1099) XX pseudomales in adulthood, but not in the L4 stage. (D) Constitutive expression of proteolytically cleaved TRA-1active feminizes adult male phasmid neurons and maintains DMD-4 expression. The hermaphrodite sex determination pathway represses the CUL-2/FEM-1/2/3 ubiquitin ligase pathway, resulting in TRA-1 stabilization and proteolytic cleavage, which is schematized to the left. Color-inverted black and white GFP images of adult males are shown in addition to the panneuronal, RFP-tagged TRA-1active in the center (‘UPN::TRA-1active::RFP’). We examined three independent transgenic lines; two had a significant effect on DMD-4 expression in the male phasmid neurons and one did not.
We found that the well-characterized heterochronic gene regulatory pathway (Ambros and Horvitz, 1984; Rougvie and Moss, 2013) affects the timing of the downregulation of DMD-4::GFP in male phasmid neurons at the adult stage. During the third larval stage, the heterochronic regulator LIN-41, a RNA-binding protein, is down-regulated to allow progression into the final larval stage and then adulthood (Slack et al., 2000), which is one of several developmental time windows during which sexual maturation events occur in several distinct parts of the nervous system (Lawson et al., 2019; Pereira et al., 2019). We generated transgenic animals that constitutively express LIN-41 in the nervous system and asked whether ectopically maintaining LIN-41 throughout all larval stages to adulthood is sufficient to maintain DMD-4 expression in the adult male phasmids. Indeed, we found that DMD-4 was ectopically maintained in these transgenic male animals (Figure 5B), demonstrating that the heterochronic pathway functions to regulate the timing of DMD-4 degradation.
To address how the sexual specificity of dmd-4::gfp downregulation is controlled, we turned to the TRA-1 master regulatory factor of sex determination, a transcription factor expressed in somatic cells of the hermaphrodite but degraded in males (Schvarzstein and Spence, 2006; Zarkower and Hodgkin, 1992). Manipulation of TRA-1 activity has been previously found to be required for the establishment of a number of sexually dimorphic gene expression programs in the nervous system, as well as sexually dimorphic synaptic connectivity (Lee and Portman, 2007; Mowrey et al., 2014; Oren-Suissa et al., 2016; White et al., 2007). We found that genetic removal of tra-1, in tra-1(e1099) XX pseudomales, leads to a downregulation of DMD-4 expression in hermaphrodite phasmid neurons at the adult stage (Figure 5C). Conversely, force-expression of an engineered, activated version of the TRA-1 protein which escapes protein degradation (Schvarzstein and Spence, 2006) in the phasmid neurons of both sexes results in ectopic DMD-4 expression in male phasmid neurons (Figure 5D). We conclude that the transcriptional regulator TRA-1 normally acts to promote DMD-4::GFP expression in hermaphrodite phasmid neurons upon sexual maturation, while its absence in adult male phasmids results in DMD-4 loss.
DMD-4 downregulation is controlled post-transcriptionally
Our implication of TRA-1, a transcription factor, in maintaining DMD-4 expression in hermaphrodite, but not male phasmid neurons (where TRA-1 is not expressed) suggested a transcriptional control mechanism for DMD-4 downregulation. However, in the context of our analysis of the cis-regulatory control regions of dmd-4 expression, we noted that the transcriptional reporter containing the nervous system enhancer for dmd-4 expression (Figure 2B), was expressed in the phasmid neurons of both adult hermaphrodite and adult male animals (Figure 6A). This suggested that the regulatory information generating DMD-4 sexual dimorphism is not encoded on the transcriptional level. To define the potential mechanism of posttranscriptional regulation, we expressed gfp tagged dmd-4 cDNA with a heterologous 3’ UTR under control of a non-dimorphic PHA-specific and PHB-specific promoter. In transgenic animals, these constructs show dimorphic GFP fluorescence in the phasmid neurons, identical to the endogenous DMD-4::GFP expression pattern (Figure 6B). Since the dmd-4 cDNA alone confers male-specific downregulation, sexually dimorphic DMD-4 presence may be controlled on the level of stability/degradation of the protein. In the next section, we describe experiments that demonstrate that a specific domain and amino acids within the DMD-4 protein can indeed be made responsible for DMD-4::GFP downregulation and reveal an unanticipated regulatory mechanism.
Figure 6. Sexually dimorphic degradation of DMD-4.
(A) Dimorphic expression is not regulated on the level of transcription. The enhancer driving DMD-4 expression in the somatic nervous system is schematized above (same as shown in Figure 2B), color-inverted black and white GFP images in adult hermaphrodite and male are shown below. (B) Driving dmd-4 cDNA under heterologous promoters in the phasmid neurons recapitulates the endogenous expression pattern. Heterologous promoters driving gfp with and without the dmd-4 cDNA are shown to the left in adult hermaphrodites and males, quantification is shown to the right. Scale bar (horizontal red bar) denotes 10 microns. For quantification, each dot represents the number of phasmid neurons expressing GFP in one animal (maximum is two neurons for PHAL/R or PHBL/R), red = hermaphrodite and cyan = male. Horizontal blue bars indicate median. P values are shown above horizontal black bars by Wilcoxon rank-sum test.
The ubiquitin-binding DMA domain controls DMD-4 stability
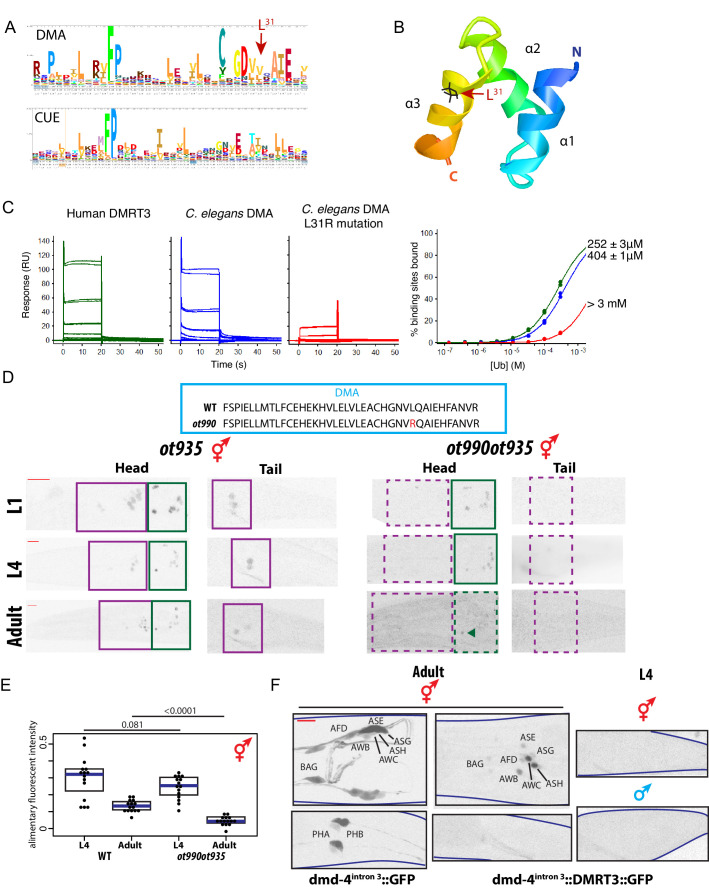
To assess which part of the protein may be responsible for sexually dimorphic DMD-4 protein expression, we turned to the phylogenetically conserved DMA domain (for DM associated). The DMA domain is found associated with the DM domain in most of the phylogenetically conserved DMRT proteins and is also present in the sole homolog of the most primitive DMRT protein from corals (Miller et al., 2003). However, the function of the DMA domain has not previously been examined. Pairwise alignments of profile hidden Markov models using HHsearch (Soding, 2005) reveals that the DMA domain shows sequence similarity to CUE domains (for coupling of ubiquitin to ER degradation), a subtype of the ubiquitin-associated (UBA) family of domains (Kang et al., 2003; Figure 7A). We produced a homology model of the DMD-4 DMA domain based on known CUE-ubiquitin domain structures (Kang et al., 2003) using the program I-TASSER (Zheng et al., 2019). The homology model showed an appropriate domain structure, with all inwardly oriented residues of hydrophobic character arranged as a well-packed core, with charged and polar residues at the surface (Figure 7B). The DMA domain contains three alpha-helices; by analogy with CUE domains, helices 1 and 3 would be expected to form a binding surface for direct non-covalent contact with ubiquitin.
Figure 7. The conserved DMA domain is a novel, ubiquitin binding degron.
(A) Weighted sequence alignment of DMA domains and CUE domains across phyla. The similarities behind these two sequence profiles is demonstrated by pairwise alignments of profile hidden Markov models using HHsearch (Soding, 2005) and is noted in the PF03474 Pfam record of the DMA domain. (B) Homology model of the DMD-4 DMA domain based on the known structure of a ubiquitin-binding CUE domain (Kang et al., 2003). The three alpha-helices are labeled; helices 1 and 3 form a binding surface for contact with ubiquitin. The leucine 31 side chain mutated in panel D is highlighted in black; note the position on the ubiquitin-binding surface. (C) SPR binding analysis of the C. elegans dmd-4 and human Dmrt3 DMA domains shows that both bind to monoubiquitin, and this binding is lost in the L31R mutation (see panel A). (D) CRISPR/Cas9 mutation of leucine 31 (see panel B) to arginine, which we predicted to impair DMA binding to ubiquitin, promotes DMD-4::GFP degradation. ot935 is the gfp-tagged dmd-4 locus and the ot990 allele carries the leucine 31 mutation (to Arg) as indicated. Color-inverted black and white GFP images of L1/L4/adult hermaphrodite heads and tails are shown below, purple boxes indicate nervous system expression, green boxes indicate pharyngeal expression. Dashed boxes indicate missing expression. I5 sometimes maintains DMD-4::GFP expression in adults (green arrowhead). Scale bars (red horizontal bars) denote 10 microns. (E) DMD-4::GFP fluorescence intensity in gfp tagged animals (ot935) and ot935 animals in which the ot990 mutation is incorporated (shown in panel D) is quantified in a defined region of the posterior pharynx (alimentary tissues excluding I5) in L4 and adult wildtype and mutant hermaphrodites. Each dot represents fluorescence intensity in one animal (normalized to background fluorescence), blue bars indicate medians, black boxes indicate quartiles. P values are shown above horizontal black bars by Wilcoxon rank-sum test. (F) Expressing human DMRT3 under the dmd-4intron3 enhancer ("dmd-4intron3::DMRT3::GFP") recapitulates DMD-4 expression in the head neurons, but in the phasmid neurons DMRT3 is degraded constitutively and is absent in both sexes in both larval and adult stages (compared to expression of the enhancer alone, 'dmd-4intron3::GFP'). Color-inverted black and white GFP images of adult hermaphrodite heads and tails and L4 hermaphrodite and male tails are shown, red scale bar indicates 10 microns.
Figure 7—figure supplement 1. Analysis of DMA domain dimerization.
Figure 7—figure supplement 2. Disrupting the DMA domain promotes DMD-4::GFP degradation.
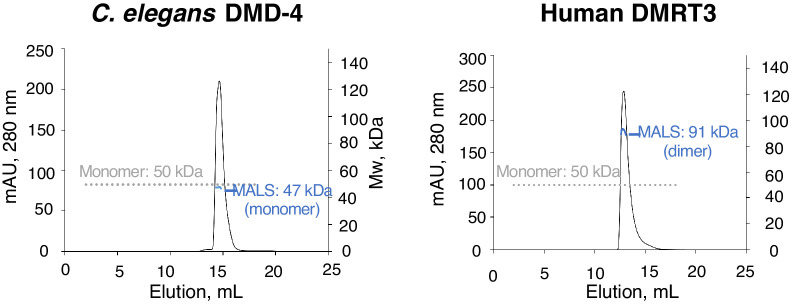
To assess the ability of the DMD-4 DMA domain to bind ubiquitin, we expressed the DMA domain as a maltose-binding protein (MBP) fusion in bacteria, and assessed its binding to human ubiquitin (which differs in only one of the 76 amino acids from C. elegans ubiquitin) by Surface Plasmon resonance (SPR) analysis. The DMD-4 DMA domain bound monoubiquitin with an affinity of ~400 µM, which is similar to known affinities for CUE-ubiquitin binding (Hurley et al., 2006; Figure 7C). CUE domains have also been found to dimerize (Azurmendi et al., 2010; Prag et al., 2003). Testing whether the DMA domain of DMD-4 may display the same feature, we used size exclusion chromatography with multi-angle static light scattering (SEC-MALS). We found that the DMD-4 DMA domain behaves as a monomer (Figure 7—figure supplement 1).
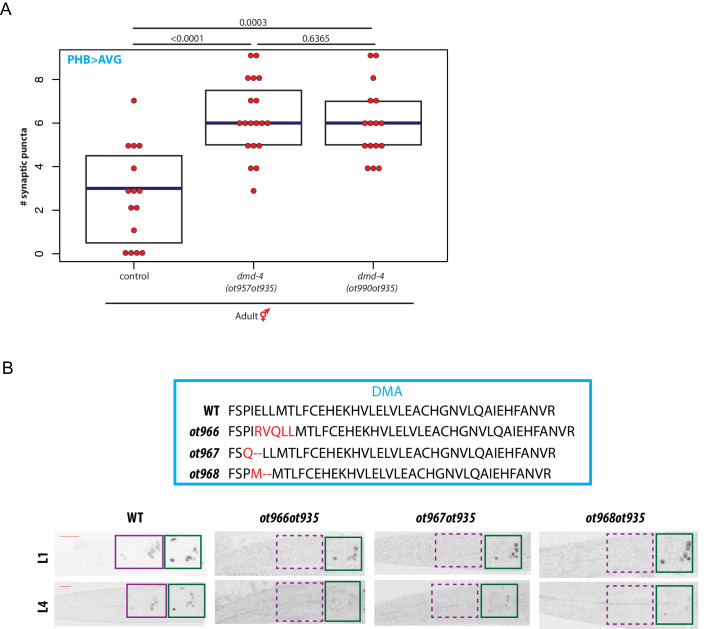
To test the functional significance of ubiquitin binding, we used our homology model to identify Leu31 (numbering relative to the beginning of the DMA domain) as a residue expected to make direct contact with ubiquitin (Figure 7B) and mutated this residue to Arg in order to disrupt ubiquitin binding. Based on the outward-facing position of this residue in our homology model, we do not anticipate that mutating it results in structural changes to the domain. As expected, the in vitro binding of the mutated DMA domain to ubiquitin was disrupted, with the affinity of mutant DMD-4 DMA for ubiquitin reduced from 404 µM to >3 mM (Figure 7C). We engineered this mutation into the endogenous gfp-tagged dmd-4 locus (ot935) using CRISPR/Cas9 to generate the dmd-4(ot990ot935) allele. Our guiding hypothesis was that through an abrogation of ubiquitin binding, we may perhaps prevent the degradation of DMD-4 in male PHA/PHB neurons. Much to our surprise, we observed the exact opposite result; we find that in dmd-4(ot990ot935) animals, DMD-4 expression is lost in the phasmid neurons in both sexes and at all stages of development (Figure 7D). Expression in the head sensory neurons was also lost, again in both sexes of all stages. Consistent with the phenotype of the ot957 null allele, the ot990 allele also results in defective pruning on the PHB>AVG synaptic connection in hermaphrodites (Figure 7—figure supplement 1A). This suggests that the binding of the DMD-4 DMA domain to ubiquitin is required to promote the stability of DMD-4 in the nervous system, and that degradation of DMD-4 in the male phasmids at the larval to adult transition is possibly generated by loss of the ubiquitin-based stabilization mechanism.
Intriguingly, the DMA domain mutation that disrupts ubiquitin binding does not affect expression or DMD-4::GFP stability in the alimentary system (pharynx and hmc) throughout embryonic and larval development; however, expression is reduced at the larval to adult transition (Figure 7D). Hence, these animals do not display the embryonic lethality associated with dmd-4 null alleles and in spite of the reduction of protein in the adult, they show no stuffed anterior intestine (Figure 2—figure supplement 2A, B).
We sought to recapitulate these results independently with other mutations in the DMA domain. Rather than introducing a mutation that is predicted to leave the overall structure of the DMA domain intact, we used CRISPR/Cas9 to mutate the first alpha helix of the DMA domain, which is predicted to affect the overall structural integrity of the DMA domain fold. We isolated three different small insertions/deletions in this helix, ot966, ot967 and ot968, each in the context of the gfp-tagged dmd-4 allele (Figure 7—figure supplement 1B). We find that these alleles behave similarly to the Leu>Arg missense mutation that is predicted to disrupt ubiquitin binding. DMD-4::GFP is destabilized in the somatic nervous system of both sexes at all stages, but expression in alimentary tissue is unaffected during embryonic and larval development (Figure 7—figure supplement 1B).
Taken together, the stability of DMD-4 in both neurons and non-neuronal tissues is promoted by ubiquitin binding to the DMA domain, but the need to promote DMD-4 stability by ubiquitin binding is different in different tissues and at different times (permanently in the nervous system, but not until adulthood in the alimentary system).
Phylogenetic conservation of DMA domain properties
To test whether the DMA domain of vertebrate DMRT proteins may carry properties similar to the DMD-4 DMA domain, we examined DMRT3, the closest vertebrate homolog of DMD-4 (Wexler et al., 2014). We find that the purified DMA domain of human DMRT3 also binds ubiquitin in vitro through SPR analysis, with an affinity similar to that of the DMD-4 DMA domain (Figure 7E). However, unlike DMD-4, but like other CUE domains, the DMRT3 domain can homodimerize (Figure 7—figure supplement 1).
To test whether the full-length human DMRT3 (hDMRT3) protein is subjected to a similar posttranscriptional regulation as C. elegans DMD-4, we used the neuronal enhancer of dmd-4 to express GFP-tagged hDMRT3 in the dmd-4(+) neurons. We found that like DMD-4, hDMRT3::GFP is not degraded in the amphid head neurons, but that in phasmid neurons hDMRT3::GFP is degraded (Figure 7F). Unexpectedly, this degradation appears to be not subjected to sexual or temporal control, as we fail to observe hDMRT-3::GFP expression in the phasmid neurons of both sexes at all times (Figure 7F).
Discussion
Sexual differentiation is controlled by an exceptionally diverse range of genetic mechanisms within the animal kingdom. Members of the DMRT transcription factor family are presently the only known factors commonly involved in sexual differentiation across the animal kingdom (Kopp, 2012; Matson and Zarkower, 2012). However, the DMRT family has radiated in distinct animal phyla, generating clade-specific family members. For example, the well-studied doublesex gene is an arthropod-specific DMRT gene (Mawaribuchi et al., 2019). We have described here the expression and function of a DMRT gene, C. elegans dmd-4, that in contrast to doublesex is conserved throughout the animal kingdom and find that this gene plays a critical role in sculpting sexually dimorphic synaptic connectivity patterns. DMD-4 protein is expressed and functions sex-specifically in the phasmid sensory neurons where it is required to promote hermaphrodite-specific synapse pruning.
We have previously reported that other DMRT proteins (dmd-5 and dmd-11) also control sexually dimorphic synaptic pruning in the phasmid circuit (Oren-Suissa et al., 2016), but the function and expression of DMD-4 differs from the previously described cases in three ways. First, loss of DMD-4 results in aberrant maintenance of a normally sexually dimorphic synaptic connection, whereas loss of DMD-5 and DMD-11 resulted in ectopic pruning of a synaptic connection. Second, DMD-5 and DMD-11 function in a post-synaptic neuron (the AVG interneuron, a sex-specific target of the phasmid neurons) to modulate synaptic pruning, whereas we find that DMD-4 functions pre-synaptically. Finally, DMD-4 is present specifically in hermaphrodites, which stands in contrast not only to the male-specificity of DMD-5 and DMD-11 and all other dimorphically expressed DMRT proteins in C. elegans (mab-3, mab-23, dmd-3)(Lints and Emmons, 2002; Mason et al., 2008; Oren-Suissa et al., 2016; Yi et al., 2000).
In addition to the sexually dimorphic functions in the hermaphrodite phasmid neurons, we found that dmd-4 is required for animal viability, apparently due to a non-sexually dimorphic requirement in the feeding organ of the worm. The dmd-4 orthologs DMRT93B (Arthropods) and Dmrt3 (vertebrates) have also been implicated in development of feeding organs based on both expression and function, with mouse Dmrt3 knockout mutants unable to survive past weaning due to feeding defects (Ahituv et al., 2007; Inui et al., 2017; Panara et al., 2019). Apart from non-dimorphic expression in the alimentary system, DMD-4 is also expressed in head sensory neurons of both sexes. While we did not observe a requirement of DMD-4 for general features of neuron specification, we have not examined all possible functions of DMD-4, for example, the synaptic wiring of these cells.
Perhaps the most unanticipated results of our studies relate to the control of sexual specificity of DMD-4 function in phasmid neurons via sex-specific protein degradation of DMD-4. Sex-specific transcription factor degradation is not unprecedented. Not only has it been observed for the TRA-1 master regulator of sexual identity in C. elegans (Schvarzstein and Spence, 2006), but a recent study in Drosophila also revealed sex-specific degradation of the Lola transcription factor (Sato et al., 2019). We reveal here a different mechanism that involves an evolutionarily ancient domain, the DMA domain. Phylogenetic analysis of the DMRT gene family suggests that the presence of a DMA domain was characteristic of the three ancestral gene clusters, and arose before the evolution of eumetazoans, but this domain has not been maintained in all extant DMRT clusters (Mawaribuchi et al., 2019; Wexler et al., 2014). In C. elegans, dmd-4 represents the only gene that encodes a DMA domain, which we identify here as required for protein stability and able to bind monoubiquitin, consistent with its sequence similarity to the CUE domains of the UBA family. We find that the H. sapiens Dmrt3 DMA domain is also able to non-covalently bind monoubiquitin and that H. sapiens Dmrt3 appears to also be subjected to posttranslational regulation in worms, suggesting a conserved molecular function of the DMA domain. CUE domains have been associated with a breadth of regulatory functions as a result of their association with ubiquitin: dimerization, subcellular trafficking, covalent ubiquitylation, and protein complex formation (Bagola et al., 2013; Chen et al., 2006; Davies et al., 2003; Prag et al., 2003). Intriguingly, certain types of non-covalent ubiquitin-binding domains have been shown to stabilize proteins (Flick et al., 2006; Heessen et al., 2005; Heinen et al., 2011; Heinen et al., 2010). For example, the non-covalent ubiquitin-binding domain of a yeast transcription factor, Met4, interacts with ubiquitin molecules that are covalently attached elsewhere on the protein to prevent polyubiquitin chain extension and proteasomal degradation of Met4 (Flick et al., 2006). Consequently, disruption of this non-covalent ubiquitin binding domain of Met4 results in degradation of Met4. Based on the Met4 precedent, we hypothesize that ubiquitin binding to DMD-4 may also prevent protein degradation.
The apparently ubiquitin-dependent stabilization of DMD-4 is susceptible to three parameters: sexual identity, cell-type and time. In wild-type animals, DMD-4 is only degraded in the phasmid neurons, not in other neuronal or non-neuronal cell types. In addition, this degradation only occurs in males and it only occurs at a specific time of development. Based on the degradation of DMD-4 in phasmid neurons in both sexes at all times upon mutation of the presumptive ubiquitin binding site , we hypothesize that the DMD-4/ubiquitin interaction may be subject to dynamic control in the phasmid neurons of wildtype animals. For example, this stabilizing effect may be disrupted sex-specifically in males, thereby permitting degradation of the protein.
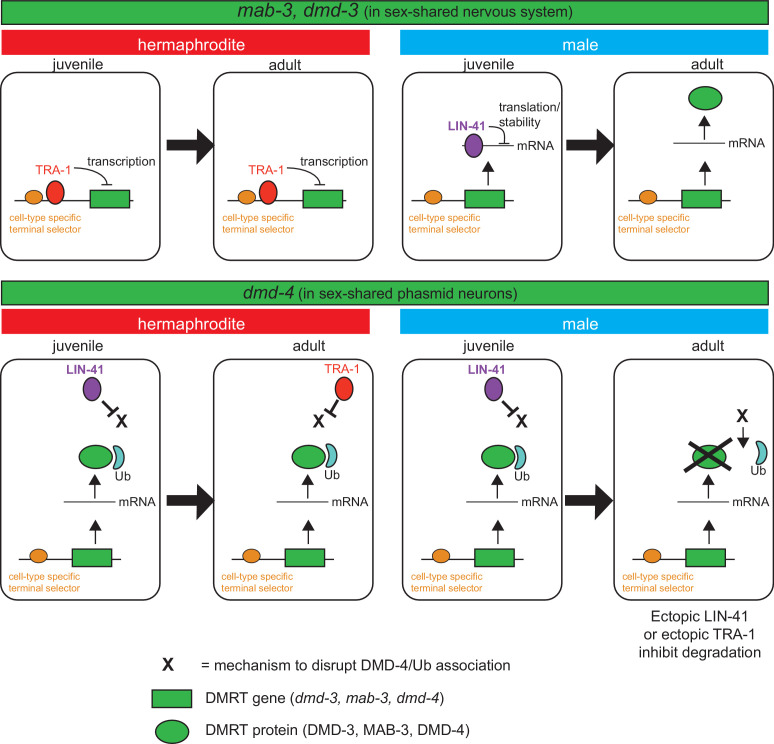
Both the heterochronic regulatory pathway as well as the sex determination pathway control the dynamic nature of this process. Since the forced expression of LIN-41 (and hence the forced retention of a juvenile state) suppresses DMD-4 degradation, it appears that an adult-specific process, controlled by the heterochronic pathway (e.g. a protein phosphorylation event) allows the disruption of the DMD-4/ubiquitin interaction. This destabilization may be the ‘default state’, but may be antagonized by LIN-41 in juveniles in both sexes and then by TRA-1 in adult hermaphrodites (schematized in Figure 8). Consistent with this notion, either forced expression of LIN-41 or TRA-1 prevents destabilization in male adult phasmid neuron.
Figure 8. Spatiotemporal specificity and regulation of DMD-4 expression differs from the regulation of other C. elegans DMRT genes.
Similar pathways are used to control the spatial, temporal and sexual specificity of C. elegans DMRT genes, but these pathways are utilized in a fundamentally distinct manner for different DMRT proteins, ranging from transcriptional to posttranslation regulation. See text for more discussion.
Strikingly, in the head sensory neurons, there appears to be no dynamic control of stability. In that cellular context, DMD-4 may always require ubiquitin binding for its stabilization. However, in the pharynx and hmc, the situation is again different. While ubiquitin binding is also required to stabilize the protein, this binding is only required in the adult stage to stabilize DMD-4 in both sexes. We infer this from the observation that the ubiquitin binding mutation only destabilizes DMD-4 in adult, but not in juvenile stages. While we clearly do not yet fully understand the mechanisms that lead to this spatiotemporal regulation of DMD-4 protein level, our studies have identified novel means by which a DMRT protein in specific, and sexual dimorphisms more broadly, are controlled in the nervous system in a manner that depends on cell type, time of development and sexual identity. The deeply conserved nature of the DMA domain suggest that these regulatory mechanisms are conserved in other species as well.
It is fascinating to note the diversity of mechanisms by which the spatial, temporal and sexually specificity of DMRT proteins is controlled, particularly in the context of one and the same species. While similar pathways are used to control the spatial, temporal and sexual specificity, some of these regulatory factors are utilized in a fundamentally distinct manner, as schematized in Figure 8 for different DMRTs in C. elegans. As demonstrated before for mab-3, dmd-3 and here for dmd-4, the spatial specificity of expression (i.e. the cell types which display dimorphic DMRT expression) is controlled by terminal selector-type transcription factors that generally control neuronal identity (Pereira et al., 2019). The heterochronic pathway and the sex determination pathway then sculpt the sexual and temporal specificity of these genes, but in a mechanistically different manner. The sexual specificity of mab-3 and dmd-3 is controlled by direct transcriptional repression by the TRA-1 protein, which prevents transcription of these genes in hermaphrodites (Mason et al., 2008; Pereira et al., 2019; Yi et al., 2000). Temporal specificity is controlled by the posttranscriptional repression of mab-3 and dmd-3 mRNA via a member of the heterochronic pathway (specifically, the mRNA binding protein LIN-41), therefore resulting in male- and adult-specific expression of MAB-3 and DMD-3 protein (Figure 8; Pereira et al., 2019). In striking contrast, dmd-4 is transcribed and the protein accumulates in both sexes in juvenile states, but its sexual specificity is regulated by protein stabilization and it appears to be this protein stabilization phenomenon that is controlled by presently unknown means via the LIN-41 and TRA-1 protein (Figure 8). The largely unexplored patterns of expression of vertebrate DMRT proteins - particularly those that contain a DMA domain (DMRT3, DMRT4, DMRT5) - may reveal similarly complex pattern of regulatory control.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Caenorhabditis elegans) | him-5 (e1490) V; dmd-4 (ot935) X | This work | OH15814 ‘DMD-4::GFP’ |
Figure 1 |
| Genetic reagent (C. elegans) | dmd-4 (ot896) X | This work | OH15302 | Figure 2 |
| Genetic reagent (C. elegans) | dmd-4(ot933)/tmC24 X | This work | OH16260 | Figure 2 |
| Genetic reagent (C. elegans) | ccIs4251; stIs10539 | CGC | SD1633 |
Figure 2
ccIs4251 [(pSAK2) myo-3p::GFP::LacZ::NLS + (pSAK4) myo-3p::mitochondrial GFP + dpy-20(+)] I; stIs10539 [dmd-4p::HIS-24::mCherry + unc-119(+)] |
| Genetic reagent (C. elegans) | otEx6774; him-5 (e1490) | This work | OH14506 |
Figure 2
dmd-4int3::GFP, pRF4 |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH15908 | Figure 2 |
| Genetic reagent (C. elegans) | otIs612; him-5 (e1490) | Oren-Suissa et al., 2016 | OH13575 ‘PHB > AVA’ |
Figure 3
MVC12 (flp-18p::nlg-1::gfp11)15 ng/ul, MVC6 (gpa-6p::nlg-1::gfp1- 10)15 ng/ul, MVC11 (flp- 18 p::mcherry)10 ng/ul, MVC15 (nlp- 1::mcherry)10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otIs612; him-5 (e1490); dmd-4 (ot957ot935) X | This work | OH16014 ‘PHB > AVA’ |
Figure 3
MVC12 (flp-18p::nlg-1::gfp11) 15 ng/ul, MVC6 (gpa-6p::nlg-1::gfp1- 10)15 ng/ul, MVC11 (flp- 18 p::mcherry)10 ng/ul, MVC15 (nlp- 1::mcherry)10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otEx6347; him-5 (e1490) V | Oren-Suissa et al., 2016 | OH13696 ‘PHA > AVG’ |
Figure 3
srg-13::BirA::nrx-1 25 ng/ul, inx-18p::AP::nlg-1 25 ng/ul, inx-18p::wcherry 10 ng/ul, unc-122::streptavidin::2xsfGFP 25 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otEx6347; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16067 ‘PHA > AVG’ |
Figure 3
srg-13::BirA::nrx-1 25 ng/ul, inx-18p::AP::nlg-1 25 ng/ul, inx-18p::wcherry 10 ng/ul, unc-122::streptavidin::2xsfGFP 25 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otIs614; him-5 (e1490) | Oren-Suissa et al., 2016 | OH13577 ‘PHB > AVG’ |
inx-18p::NLG-1::GFP1130 ng/ul, MVC6 gpa-6::NLG-1:::GFP1-10 30 ng/ul, MVC15 nlp-1::mcherry 5 ng/ul, inx-18::wcherry 10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otIs614; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16017 ‘PHB > AVG’ |
Figure 3
inx-18p::NLG-1::GFP11 30 ng/ul, MVC6 gpa-6::NLG-1:::GFP1-10 30 ng/ul, MVC15 nlp-1::mcherry 5 ng/ul, inx-18::wcherry 10 ng/ul, pRF4 50 ng/ul |
| Genetic reagent (C. elegans) | otEx7431; otIs614; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16217 ‘PHBp::DMD-4 line 1’ |
Figure 3
gpa-6p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | otEx7433; otIs614; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16208 ‘PHBp::DMD-4 line 2’ |
Figure 3
gpa-6p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | otIs702; him-5 (e1490) V | This work | OH15565 ‘PHA::RAB-3’ |
Figure 3
srg-13p::RAB-3::BFP, srg-13p::gfp, ttx-3::mCherry |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot935ot957) X | This work | OH16030 ‘PHA::RAB-3’ |
Figure 3
srg-13p::RAB-3::BFP, srg-13p::gfp, ttx-3::mCherry |
| Genetic reagent (C. elegans) | tax-4 (p678); him-5 (e1490) | Oren-Suissa et al., 2016 | OH15339 | Figure 4 |
| Genetic reagent (C. elegans) | tax-4 (p678); him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16115 | Figure 4 |
| Genetic reagent (C. elegans) | wgIs418; him-5 (e1490) V | This work | OH16268 |
Figure 5
DMD-4fosmid::gfp |
| Genetic reagent (C. elegans) | wgIs418; ceh-14 (ch3) X | This work | OH14503 |
Figure 5
DMD-4fosmid::gfp |
| Genetic reagent (C. elegans) | otEx7429; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH16159 ‘UPN::LIN-41 line 1’ |
Figure 5
UPN::LIN-41A, rab-3p::tagRFP, pRF4 |
| Genetic reagent (C. elegans) | otEx7451; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH16235 ‘UPN::LIN-41 line 2’ |
Figure 5
UPN::LIN-41A, rab-3p::tagRFP, pRF4 |
| Genetic reagent (C. elegans) | tra-1 (e1099) III/hT2 [bli-4(e937) umnIs36] (I;III); dmd-4 (ot935) X | This work | OH16194 | Figure 5 |
| Genetic reagent (C. elegans) | otEx7352; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH15952 ‘UPN::TRA-1active line 1’ |
Figure 5
UPN::tagRFP::TRA-1active, pRF4 |
| Genetic reagent (C. elegans) | otEx7353; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH16158 ‘UPN::TRA-1active line 2’ |
Figure 5
UPN::tagRFP::TRA-1active, pRF4 |
| Genetic reagent (C. elegans) | otEx7354; him-5 (e1490) V; dmd-4 (ot935) X | This work | OH15993 ‘UPN::TRA-1active line 3’ |
Figure 5
UPN::tagRFP::TRA-1active, pRF4 |
| Genetic reagent (C. elegans) | otIs627; him-8 (e1489) | This work | OH14022 ‘PHAp::GFP’ |
Figure 6
srg-13p::GFP |
| Genetic reagent (C. elegans) | otEx7435; him-5 (e1490) V ; dmd-4 (ot957ot935) X | This work | OH16177 ‘PHAp::DMD-4::GFP’ |
Figure 6
srg-13p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | otEx6456; him-5 (e1490) V | This work | OH13892 ‘PHBp::GFP’ |
Figure 6
gpa-6p::GFP |
| Genetic reagent (C. elegans) | otEx7433; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16175 ‘PHBp::DMD-4::GFP’ |
Figure 6
gpa-6p::DMD-4::GFP, ttx-3p::GFP |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot990ot935) X | This work | OH16125 | Figure 7 |
| Genetic reagent (C. elegans) | nIs175 [ceh-28p::4xNLS::GFP + lin-15AB(+)] IV; ceh-34(n4796) V | CGC | MT15695 | Figure 2—figure supplement 1 |
| Genetic reagent (C. elegans) | otIs518; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16054 | Figure 2—figure supplement 2 |
| Genetic reagent (C. elegans) | ntIs1; dmd-4 (ot957ot935) X | This work | OH16225 | Figure 2—figure supplement 2 |
| Genetic reagent (C. elegans) | wgIs73; him-5 (e1490) V; dmd-4 (ot957ot935) X | This work | OH16018 | Figure 2—figure supplement 2 |
| Genetic reagent (C. elegans) | otIs612; unc-13(e51) I;him-5 (e1490) V | This work | MOS14 | Figure 3—figure supplement 1 |
| Genetic reagent (C. elegans) | otEx6913; unc-13(e51) I; him-5 (e1490) V | This work | MOS234 |
Figure 3—figure supplement 1
inx-18p::NLG-1::GFP11, MVC6 gpa-6::NLG-1:::GFP1-10, MVC15 inx-18::wcherry, pRF4 |
| Genetic reagent (C. elegans) | etyEx2; otIs614; him-5 (e1490) V | This work | MOS243 |
Figure 3—figure supplement 1
gpa-6::HisCl1, myo-2::GFP |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot966ot0935) X | This work | OH16013 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot967ot935) X | This work | OH16015 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) | him-5 (e1490) V; dmd-4 (ot968ot935) X | This work | OH16016 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) |
otIs614; him-5 (e1490) V; dmd-4 (ot990ot935) X | This work | OH16771 | Figure 7—figure supplement 2 |
| Genetic reagent (C. elegans) |
ccIs4443 IV; him-5 (e1490) V | This work | OH16545 |
Figure 2—figure supplement 1
arg-1::GFP + dpy-20(+) |
| Genetic reagent (C. elegans) |
ccIs4443 IV; him-5 (e1490) V; dmd-4 (ot933) X | This work | OH16770 | Figure 2—figure supplement 1 |
Strains
Wild-type strains were C. elegans variety Bristol, strain N2. Worms were maintained by standard methods (Brenner, 1974). Worms were grown at 20C on nematode growth media (NGM) plates seeded with bacteria (E. coli OP50) as a food source, with the exception of temperature sensitive alleles, which were maintained at 15C. Mutant and transgenic strains used are listed in the Key Resources Table.
CRISPR/Cas9-mediated genome engineering
The ot935, ot933, and ot990 alleles were generated using Cas9 protein, tracrRNA, and crRNA from IDT, as previously described (Dokshin et al., 2018). The GFP repair template for ot935 was amplified from pPD95.75, and it was fused in-frame immediately before the endogenous dmd-4 stop codon using gRNA GTTTTCAACCGTATTACTTG.
ot933 is a 556 bp insertion/deletion beginning at +four relative to the dmd-4 start codon and was generated using gRNA: TTGTTGTGTATATATCACAT. With 100 bp flanks (underlined), the wild-type sequence is (caps-lock indicates exons):
atctacagaccgagcaagcgaaagaggcggaatcacaaattgttgtgtatatatcacattggctattcatttttgttattagctgatctatagataATGATGATCGGTAATCTACATGTATTCCCAAACGGACGAATCGAAAGAGAACGGAAACCTAAATGTGCCAGgtaagagaagctttgtaaactaaatactctaacaactgcaaccattttgtaaatgttccagATGCAGAAATCACGGACTGGTCAGTTGGCTGAAAGGACACAAGCGGCATTGCAAGTACAAgtaagttgaagcggaacacttctgcaaaaaatattgtttagtttaagcatttataactagaatattgtgttacaatggctcagtatcaatcaacattaattcattttgcacacgttctctatgaagaatataggttttggaatggggttaaaatacatgcttctttttcattgtacataatttatgtttacgattgtgagttataaaaatgtagttttttttctaacaactaatcgacttgataacggttttggtgaaaattgaaaattgaaaaaattcataatttcagtagaatatctaggcacttctaattagtaaaaattgattctgaatgctgaagtagaaaataatttaatttttattaaaataaacgtatacagtatatttttttaaacgattattgataatagtctagtctcccacattatatatatatatattaatcgttgataaatttgaagtaggataattttataacacaac
And the mutant sequence is (not underlined indicates insertion):
atctacagaccgagcaagcgaaagaggcggaatcacaaattgttgtgtatatatcacattggctattcatttttgttattagctgatctatagataATGAaataatcgtttaaaatatttttttaaacgattattgataatagtctagtctcccacattatatatatatatattaatcgttgataaatttgaagtaggataattttataacacaac
ot990 is a substitution in the dmd-4 DMA domain generated using gRNA TGGCTTGAAGTACATTTCCA and a ssODN repair template (Dokshin et al., 2018). The mutated DMA nucleotide sequence is (bold indicates arginine codon):
TTCTCACCAATTGAACTCCTGATGACTTTATTTTGTGAACACGAAAAGCACGTGCTCGAACTTGTGCTCGAAGCCTGCCATGGAAATGTACGTCAAGCCATTGAACATTTTGCAAATGTGAGA.
The ot896 and ot957 deletion alleles were generated using dpy-5 coCRISPR and Cas9 plasmid as previously described (Arribere et al., 2014). ot896 is a 916 bp deletion/insertion from +3293 to +4208 relative to the dmd-4 start codon. gRNAs used were AATCTGCTCGTGGGATTGAT and TATACACCTACACAGAAAAA.
With 100 bp flanks (underlined), the wild-type sequence is (caps-lock indicates exons):
GgactatccaatgcccaaaaaatgataaagccagtaacaaaatctttatcattagtgccttgagttcacatgtttcatatattatttttttattacaagctgaatttgcaaaacgtactctccttctcctttcatttcatcaacttataaattgaataactcgaaaattgattaagtttatcgatatgtttgtgccacttactcgctagttagtgcaacttcgcagttttcatttccggtcagttcatatgcagaaatttacgctcttggtgttttttttttcaatttgaataatttttgttcaaaaaaaagcaaggattcatctatatctctttatacggagggtggagcaagtaacaggattatcaactttttttttgcatttatctcgtgagagcttatgtgataagctagcgaaaaaatttcccaagttttttctctttttttttgaatgaaaatgagtgtttagtgattccctttttctgtgtaggtgtataaatctgctcgtgggattgatcggctaatagaaaaaaatttattttaaactatcgtcctaaaataatatattcagAGTTGTTGCTGGGCAAGCAATTGATAGGCTCCCTCAAGGTCCAGTTTGGAACACAGCTGGTGGTGAAGACGAAGATATGGACTACTTGGATGAAGAAATCGAAGAAAAACCGCCAACTCCAGTGTCAGAGGAACTCGTCCCTTTGAAACGGAAGAAAGTAGAGGAGACATACGAATTGTCGAGCTTCTCACCAATTGAACTCCTGATGACTTTATTTTGTGAACACGAAAAGCACGTGCTCGAACTTGTGCTCGAAGCCTGCCATGGAAATGTACTTCAAGCCATTGAACATTTTGCAAATGTGAGACGAGTGAAAAATATTAATCAAATGAAGCTATTCGCTGCAGCAACAAGGATGGATCACGTGTTTCCAAATATGACAAACTTTATACTTCCAAAGCAATCTTTTCTCATTGATTCGTTGTTAGAGgtaagataaagaaactttaaactttttataatttatattacatactatctgaaaccggcaattttcagCAACCAACATTCCCAGTCTCCAGTCAAGCATCTACAAGTACTAAAAC
And the mutant sequence is (no underline indicates insertion):
GgactatccaatgcccaaaaaatgataaagccagtaacaaaatctttatcattagtgccttgagttcacatgtttcatatattatttttttattacaagcgagcagatttatatttaaactttttataatttatattacatactatctgaaaccggcaattttcagCAACCAACATTCCCAGTCTCCAGTCAAGCATCTACAAGTACTAAAAC
ot957 is a 510 bp deletion removing +3201 to +3710 relative to the dmd-4 start codon, made in the context of ot935 using gRNAs TATACACCTACACAGAAAAA and AATTGCTCTTGGACTACGGT.
With 100 bp flanks (underlined), the wild-type sequence is (caps-lock indicates exons):
atgctagttattgtattacaaattgcacctgtaattctcttgaatcatttttttttcacttgcgatatcaaagtctttatccttcggatgccggactatccaatgcccaaaaaatgataaagccagtaacaaaatctttatcattagtgccttgagttcacatgtttcatatattatttttttattacaagctgaatttgcaaaacgtactctccttctcctttcatttcatcaacttataaattgaataactcgaaaattgattaagtttatcgatatgtttgtgccacttactcgctagttagtgcaacttcgcagttttcatttccggtcagttcatatgcagaaatttacgctcttggtgttttttttttcaatttgaataatttttgttcaaaaaaaagcaaggattcatctatatctctttatacggagggtggagcaagtaacaggattatcaactttttttttgcatttatctcgtgagagcttatgtgataagctagcgaaaaaatttcccaagttttttctctttttttttgaatgaaaatgagtgtttagtgattccctttttctgtgtaggtgtataaatctgctcgtgggattgatcggctaatagaaaaaaatttattttaaactatcgtcctaaaataatatattcagAGTTGTTGCTGGGCAAGCAATTGATAGGCTCCCTCAAGGTCCAGTTT
And the mutant sequence is (ot957 is a clean deletion with no inserted sequences):
AtgctagttattgtattacaaattgcacctgtaattctcttgaatcatttttttttcacttgcgatatcaaagtctttatccttcggatgccggactatcggctaatagaaaaaaatttattttaaactatcgtcctaaaataatatattcagAGTTGTTGCTGGGCAAGCAATTGATAGGCTCCCTCAAGGTCCAGTTT
ot966, ot967, and ot968 are small indels in the DMA domain generated using gRNA TGGCTTGAAGTACATTTCCA
The wildtype DMA domain sequence is:
TTCTCACCAATTGAACTCCTGATGACTTTATTTTGTGAACACGAAAAGCACGTGCTCGAACTTGTGCTCGAAGCCTGCCATGGAAATGTACTTCAAGCCATTGAACATTTTGCAAATGTGAGA
The ot966 sequence is:
TTCTCACCAATTCGTGTTCAACTCCTGATGACTTTATTTTGTGAACACGAAAAGCACGTGCTCGAACTTGTGCTCGAAGCCTGCCATGGAAATGTACTTCAAGCCATTGAACATTTTGCAAATGTGAGA
The ot967 sequence is:
TTCTCACAACTCCTGATGACTTTATTTTGTGAACACGAAAAGCACGTGCTCGAACTTGTGCTCGAAGCCTGCCATGGAAATGTACTTCAAGCCATTGAACATTTTGCAAATGTGAGA
The ot968 sequence is:
TTCTCACCAATGATGACTTTATTTTGTGAACACGAAAAGCACGTGCTCGAACTTGTGCTCGAAGCCTGCCATGGAAATGTACTTCAAGCCATTGAACATTTTGCAAATGTGAGA
Resultant amino acid sequences of ot966, ot967, and ot968 are given in Figure 7—figure supplement 2B.
Cloning and constructs
The LIN-41A and TRA-1active cDNAs used in this study were reverse transcribed from C. elegans total RNA using the SuperScript III First-Strand Synthesis System (Thermo-Fisher). TRA-1active is a shortened version of the TRA-1A cDNA (860aa) predicted to encode the proteolytically activated version of TRA-1A generated by C-terminal cleavage in hermaphrodites (Schvarzstein and Spence, 2006). This truncated protein was previously shown to have feminizing activity when expressed under control of its own regulatory elements (Schvarzstein and Spence, 2006).
Restriction-Free Cloning was used to insert TRA-1active into UPN::RFP (to generate pEAB70) and LIN-41A into UPN driver vector (to generate pHS41). UPN is a concatenated panneuronal promoter (containing promoter fragments from unc-11, rgef-1, ehs-1, and ric-19), and was generated and cloned by Ev Yemini (Yemini et al., submitted). pHS41 [UPN::LIN-41A] was cloned by HaoSheng Sun.
The DMD-4 cDNA was obtained from the Dharmacon C. elegans ORF collection and cloned using Restriction-Free Cloning into pPD95.75. Gibson assembly was used to clone the srg-13 and gpa-6 promoters (Oren-Suissa et al., 2016) into the SphI/XmaI sites.
The human DMRT3 cDNA was codon-optimized (https://worm.mpi-cbg.de/codons/cgi-bin/optimize.py), synthesized by GeneWiz, and cloned using Restriction-Free Cloning into dmd-4intron 3::gfp to generate pEAB71.
Microscopy
Worms were anesthetized using 100 mM of sodium azide (NaN3) and mounted on 5% agar on glass slides. Worms were analyzed by Nomarski optics and fluorescence microscopy, using a Zeiss 880 confocal laser-scanning microscope. Multidimensional data was reconstructed as maximum intensity projections using Zeiss Zen software. For GRASP and RAB-3 experiments, animals were imaged using 63x objective and puncta were quantified by scanning the original full z-stack for distinct dots in the area where the processes of the two neurons overlap. GRASP and RAB-3 experiments were scored blinded to genotype for mutant analysis and rescue array analysis. For fluorescence quantification experiments, animals were imaged using 40x objective with fixed imaging settings, and quantification was performed on maximum intensity projections using the Zeiss Zen software by quantifying the mean fluorescence intensity in rectangular area of the posterior pharynx (containing all posterior muscle and intestinal valve cells that express DMD-4), and normalizing this value to the image background (an identical rectangular area directly anterior). Sex of L1 animals was determined using rectal epithelial cell morphology.
Behavioral analysis
SDS-avoidance behavior
SDS avoidance assay was based on procedures described (Hilliard et al., 2002). A small drop of solution containing either the repellent (0.1% SDS in M13 buffer) or buffer (M13 buffer: 30 mM Tris-HCl pH 7.0, 100 mM NaCl, 10 mM KCl) is delivered near the tail of an animal while it moves forward. Once in contact with the tail, the drop surrounds the entire animal by capillary action and reaches the anterior amphid sensory organs. Drop was delivered using 10 uL glass calibrated pipets (VWR international) pulled by hand on a flame to reduce the diameter of the tip. The capillary pipette was mounted in a holder with rubber tubing and operated by mouth. Assayed worms were transferred individually to fresh non-wet unseeded NGM plates. Each assay started with testing the animals with drops of M13 buffer alone. The response to each drop was scored as reversing or not reversing. The avoidance index is the number of reversal responses divided by the total number of trials. An Inter Stimuli Interval of at least two minutes was used between successive drops to the same animal. Each animal was tested 10 times. Two experimental replicates were performed for each experiment.
Chemotaxis assays
The response to NaCl gradients was assayed as previously described (Bargmann and Horvitz, 1991). Briefly, 10 ml of buffered agar (20 g/l agar, 1 mM CaCl2, 1 mM MgSO4 and 5 mM KPO4) was poured into 10 cm diameter petri dishes. To establish the chemical gradient, we applied 10 μl of odorant to the attractant/repulsion spot and 10 μl of double-distilled H2O to the control spot. The odorant was allowed to diffuse for 14–16 hr at room temperature before the assay. To increase the steepness of the gradient, another 4 μl of odorant or water was added to the same spots 4 hr before the assay. We applied a 1 μl drop of 1 M sodium azide to both attractant and control spots 10 min before the assay to immobilize worms that reached these areas. Synchronized animals were washed three times with CTX solution (1 mM CaCl2, 1 mM MgSO4 and 5 mM KPO4) and 100–200 animals were placed in the center of the assay plate in a minimal volume of buffer. Animals were allowed to move about the agar surface for 1 hr, after which assay plates were placed at 4°C overnight. The distribution of animals across the plate was then determined and a chemotaxis index was calculated as the number of animals at the odorant minus the number of animals at the control spot, divided by the total number of animals. Animals that did not leave the initial inner circle were not included in the count of total number of animals, as these animals were dead or had movement defects. N2 animals were used as controls.
Neuronal silencing using the histamine chloride channel system
We silenced the PHB neurons by expressing HisCl1 channel (Pokala et al., 2014) in the PHB neurons, using a gpa-6 driver, as previously described (Oren-Suissa et al., 2016). Transgenic animals were picked at the L4 stage and placed on NGM plates containing 10 mM histamine with OP50 bacteria as food source. As a control, animals were placed on NGM plates containing OP50 bacteria but no histamine. GRASP connectivity was assessed ~24 hr later when the animals had reached adulthood.
Biochemical analysis
Protein expression and purification
C. elegans and H. sapiens DMA domain cDNAs were synthesized by GeneWiz, codon optimized for E. coli. These cDNAs were cloned into the KpnI site of pMal-c5x (NEB) to produce maltose binding protein fusions, and transformed into BL21 cells. E. Coli BL21/DE3 cells were grown in Luria Broth with 100 ug/mL ampicillin. When OD at 600 nm of 1.0 was attained, the cells were induced with 300 uM IPTG and grown for an additional 2 hr. Cells were harvested by centrifugation and sonicated in 20 mM HEPES, pH 7.4, 200 mM NaCl, 1 mM EDTA and protease inhibitors. After clarification by centrifugation, 5 mL of amylose resin (New England BioLabs) equilibrated in HEPES 20 mM pH 7.4, NaCl 200 mM was added to ~60 mL of clear lysate and subjected to gentle rocking at 4°C for 2 hr. The bound resin was packed onto a column and washed with at least 15 column volumes of 20 mM HEPES, pH 7.4, 200 mM NaCl. Proteins were eluted with 30 mL of the HEPES 20 buffer supplemented with 10 mM maltose. This eluent was concentrated and loaded onto a Superose 200 10/300 GL Increase (GE Healthcare) gel filtration column. Proteins were eluted at 0.5 mL/min with 10 mM HEPES, pH 7.4, 150 mM NaCl, and appropriate fractions were pooled and concentrated.
SPR binding experiments
SPR binding experiments were performed using a Biacore T100 biosensor equipped with a Series S CM4 sensor chip. Human DMRT, DMD-4 and its L31R mutant were immobilized over individual flow cells using amine-coupling chemistry in 10 mM HEPES-OH, pH 7.4, 150 mM NaCl, 0.005% Tween-20 at 32°C using a flow rate of 20 μL/min. Dextran surfaces were activated for 7 min using equal volumes of 0.1 M NHS(N-Hydroxysuccinimide) and 0.4 M EDC(1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide) for 10 min. Each protein of interest was immobilized at 50 μg/mL in 10 mM sodium acetate, pH 4.25 for 7 min each. The immobilized surface was blocked using a 3 min injection of 1.0 M ethanolamine, pH 8.5 resulting in immobilizations at ~3,000 RU. An unmodified surface was used as a reference flow cell to subtract bulk shift refractive index changes.
Binding experiments were performed at 25°C in a running buffer containing 10 mM Tris-Cl pH 7.4, 150 mM NaCl, and 0.01% (v/v) Tween-20. Ubiquitin was prepared in the same buffer at a stock concentration of 300 μM and serially diluted using a three-fold dilution series from 300 to 0.137 μM. Ubiquitin analyte samples were tested in duplicate in order of increasing concentration. In each binding cycle, analytes were injected over the three immobilized surfaces at 50 μL/min for 20 s, followed by 180 s of dissociation phase, a running buffer wash step and a buffer injection at 100 μL/min for 60 s. Buffer was used instead of an analyte sample every two binding cycles to double reference the binding responses by removing systematic noise and instrument drift. The binding signal between 15 and 19 s was fit against the ubiquitin concentration using a 1:1 interaction model to calculate the KD. The data was processed and analyzed using Scrubber 2.0 (BioLogic Software).
SEC-MALS
Size exclusion chromatography with multi-angle static light scattering (SEC-MALS) was performed using an AKTA FPLC system with a Superdex 200 Increase 10/300 GL column (GE Healthcare), equilibrated with 10 mM HEPES, pH 7.4, 150 mM NaCl, with a flow rate of 0.5 mL/min. Each protein was injected at a concentration of 3 mg/mL, using an injection volume of 100 μL. UV absorbance was monitored at 280 nm using the AKTA UV detector and light-scattering and refractive index data for molecular weight determination was collected with an Optilab T-rEX and a DAWN Heleos-II detector (Wyatt Technology). Molecular weights were calculated using the Astra 6.1 software (Wyatt Technologies) using a Zimm-plot.
Quantification of intestinal stuffing
To assay the degree of anterior intestinal stuffing in the dmd-4 mutants dmd-4(ot935), dmd-4(ot957ot935), dmd-4(ot990ot935), and dmd-4(ot933) relative to the him-5(e1490) control and the positive control ceh-34(n4796) (which is required for pharyngeal development and was used as a positive control [Hirose et al., 2010]), embryos synchronized by hypochlorite treatment were plated onto NGM plates seeded with an E. coli OP50 strain that contains the GFP-expressing plasmid pFPV25.1 (OP50-GFP, CGC). Worms were grown to early adulthood (72 hr after egg prep, at 20°C). In order to reduce background signal, worms were washed twice in 15 mL of M9 and centrifuged at 300 x g for 1 min to separate and discard any fluorescent bacteria in suspension. Worms were then run through the Union Biometrica COPAS FP-250 Biosorter system to measure GFP signal as a function of worm length. Fluorescence data was acquired and processed with the Union Biometrica FlowPilot software (version 1.5.9.2). Events detected by the COPAS system were filtered attending to the time of flight (i.e. worm length) and extinction (i.e. worm opacity) to include only adult males and young adult hermaphrodites in the data set and exclude gravid hermaphrodites, as the condition of gravidity produces its own genotype-independent stuffed phenotype. Any data point without an associated parabolic time-of-flight trace was also excluded. A non-parabolic trace indicates a bent worm in the flow cell, which produces skewed fluorescence data. The green fluorescence trace measures green signal as a function of length, and the area under this curve corresponds to the total amount of fluorescence from the OP50-GFP in the anterior intestine, as confirmed by 63x-objective epifluorescent imaging (Zeiss Axio Imager.Z2). To normalize data, all data points were divided by the mean of the control group (him-5(e1490)).
Statistics
Plots for expression data were generated in R using the beeswarm and ggplot2 packages. Statistical tests were performed in R as detailed in figure legends, in addition to post-hoc Bonferroni corrections to adjust p-values for number of pairwise tests in all cases where more than two pairwise statistical tests were performed.
Acknowledgements
We thank Chi Chen for generating transgenic strains, the CGC (supported by the NIH P40 OD010440) for strains, and members of the Hobert laboratory for comments on the manuscript. This research was supported by NIH R37 NS039996, an NRSA Kirschstein predoctoral fellowship to Emily Bayer (F31NS096863) and the Howard Hughes Medical Institute.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Oliver Hobert, Email: or38@columbia.edu.
Claude Desplan, New York University, United States.
Piali Sengupta, Brandeis University, United States.
Funding Information
This paper was supported by the following grants:
Howard Hughes Medical Institute to Oliver Hobert.
National Institutes of Health F31NS096863 to Emily A Bayer.
National Institutes of Health R37 NS039996 to Oliver Hobert.
Additional information
Competing interests
Reviewing Editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Writing - original draft, Writing - review and editing.
Formal analysis, Investigation.
Formal analysis, Investigation.
Formal analysis, Investigation.
Formal analysis, Investigation.
Formal analysis, Investigation.
Conceptualization, Supervision, Writing - review and editing.
Conceptualization, Supervision, Funding acquisition, Writing - review and editing.
Conceptualization, Formal analysis, Investigation.
Conceptualization, Supervision, Funding acquisition, Writing - original draft, Project administration, Writing - review and editing.
Additional files
Data availability
All strains are listed in the Key Resources Table and are either deposited at the CGC or available upon request.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLOS Biology. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azurmendi HF, Mitra S, Ayala I, Li L, Finkielstein CV, Capelluto DG. Backbone (1)H, (15)N, and (13)C resonance assignments and secondary structure of the tollip CUE domain. Molecules and Cells. 2010;30:581–585. doi: 10.1007/s10059-010-0145-5. [DOI] [PubMed] [Google Scholar]
- Bagola K, von Delbrück M, Dittmar G, Scheffner M, Ziv I, Glickman MH, Ciechanover A, Sommer T. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Molecular Cell. 2013;50:528–539. doi: 10.1016/j.molcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Baker BS, Ridge KA. Sex and the single cell on the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-H. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Barr MM, García LR, Portman DS. Sexual dimorphism and sex differences in Caenorhabditis elegans Neuronal Development and Behavior. Genetics. 2018;208:909–935. doi: 10.1534/genetics.117.300294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its cue domain, RING finger, and an E2-binding site. PNAS. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LT, Bayer EA, Duerr JS, Bülow HE, Hobert O, Hall DH, Emmons SW. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019;571:63–71. doi: 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BA, Topp JD, Sfeir AJ, Katzmann DJ, Carney DS, Tall GG, Friedberg AS, Deng L, Chen Z, Horazdovsky BF. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. Journal of Biological Chemistry. 2003;278:19826–19833. doi: 10.1074/jbc.M301059200. [DOI] [PubMed] [Google Scholar]
- Desbois M, Cook SJ, Emmons SW, Bülow HE. Directional Trans-Synaptic labeling of specific neuronal connections in live animals. Genetics. 2015;200:697–705. doi: 10.1534/genetics.115.177006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC. Robust genome editing with short Single-Stranded and long, partially Single-Stranded DNA donors in Caenorhabditis elegans. Genetics. 2018;210:781–787. doi: 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons SW. Male development. WormBook. 2005;1:22. doi: 10.1895/wormbook.1.33.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons SW. Neural circuits of sexual behavior in Caenorhabditis elegans. Annual Review of Neuroscience. 2018;41:349–369. doi: 10.1146/annurev-neuro-070815-014056. [DOI] [PubMed] [Google Scholar]
- Fairbairn DJ. Odd Couples: Extraordinary Differences Between the Sexes in the Animal Kingdom. Princeton University Press; 2013. [Google Scholar]
- Faunes F, Larraín J. Conservation in the involvement of heterochronic genes and hormones during developmental transitions. Developmental Biology. 2016;416:3–17. doi: 10.1016/j.ydbio.2016.06.013. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Flick K, Raasi S, Zhang H, Yen JL, Kaiser P. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nature Cell Biology. 2006;8:509–515. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]
- Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Molecular Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Heinen C, Garner TP, Long J, Böttcher C, Ralston SH, Cavey JR, Searle MS, Layfield R, Dantuma NP. Mutant p62/SQSTM1 UBA domains linked to Paget's disease of bone differ in their abilities to function as stabilization signals. FEBS Letters. 2010;584:1585–1590. doi: 10.1016/j.febslet.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Heinen C, Acs K, Hoogstraten D, Dantuma NP. C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nature Communications. 2011;2:191. doi: 10.1038/ncomms1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert ZA, Kim DH. Sexually dimorphic control of gene expression in sensory neurons regulates decision-making behavior in C. elegans. eLife. 2017;6:e21166. doi: 10.7554/eLife.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI, Bazzicalupo P. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Current Biology. 2002;12:730–734. doi: 10.1016/S0960-9822(02)00813-8. [DOI] [PubMed] [Google Scholar]
- Hirose T, Galvin BD, Horvitz HR. Six and eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. PNAS. 2010;107:15479–15484. doi: 10.1073/pnas.1010023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Terminal selectors of neuronal identity. Current Topics in Developmental Biology. 2016;116:455–475. doi: 10.1016/bs.ctdb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochemical Journal. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Tamano M, Kato T, Takada S. CRISPR/Cas9-mediated simultaneous knockout of Dmrt1 and Dmrt3 does not recapitulate the 46,XY gonadal dysgenesis observed in 9p24.3 deletion patients. Biochemistry and Biophysics Reports. 2017;9:238–244. doi: 10.1016/j.bbrep.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. The connectome of a decision-making neural network. Science. 2012;337:437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Cassata G, Tong YG, Pujol N, Niklaus G, Bürglin TR. The LIM homeobox gene ceh-14 is required for phasmid function and neurite outgrowth. Developmental Biology. 2013;380:314–323. doi: 10.1016/j.ydbio.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/S0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- Katz LC, Callaway EM. Development of local circuits in mammalian visual cortex. Annual Review of Neuroscience. 1992;15:31–56. doi: 10.1146/annurev.ne.15.030192.000335. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kopp A. Dmrt genes in the development and evolution of sexual dimorphism. Trends in Genetics. 2012;28:175–184. doi: 10.1016/j.tig.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson H, Vuong E, Miller RM, Kiontke K, Fitch DH, Portman DS. The makorin lep-2 and the lncRNA lep-5 regulate lin-28 to schedule sexual maturation of the C. elegans nervous system. eLife. 2019;8:e43660. doi: 10.7554/eLife.43660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Portman DS. Neural sex modifies the function of a C. elegans sensory circuit. Current Biology. 2007;17:1858–1863. doi: 10.1016/j.cub.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Lints R, Emmons SW. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes & Development. 2002;16:2390–2402. doi: 10.1101/gad.1012602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Rabinowitz JS, Portman DS. dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development. 2008;135:2373–2382. doi: 10.1242/dev.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nature Reviews Genetics. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawaribuchi S, Ito Y, Ito M. Independent evolution for sex determination and differentiation in the DMRT family in animals. Biology Open. 2019;8:bio041962. doi: 10.1242/bio.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SW, Hayward DC, Bunch TA, Miller DJ, Ball EE, Bardwell VJ, Zarkower D, Brower DL. A DM domain protein from a coral, Acropora millepora, homologous to proteins important for sex determination. Evolution and Development. 2003;5:251–258. doi: 10.1046/j.1525-142X.2003.03032.x. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Soto F, Wong RO, Kerschensteiner D. Development of cell type-specific connectivity patterns of converging excitatory axons in the retina. Neuron. 2011;71:1014–1021. doi: 10.1016/j.neuron.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowrey WR, Bennett JR, Portman DS. Distributed effects of biological sex define sex-typical motor behavior in Caenorhabditis elegans. Journal of Neuroscience. 2014;34:1579–1591. doi: 10.1523/JNEUROSCI.4352-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Suissa M, Bayer EA, Hobert O. Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature. 2016;533:206–211. doi: 10.1038/nature17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panara V, Budd GE, Janssen R. Phylogenetic analysis and embryonic expression of panarthropod dmrt genes. Frontiers in Zoology. 2019;16:23. doi: 10.1186/s12983-019-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Aeschimann F, Wang C, Lawson H, Serrano-Saiz E, Portman DS, Großhans H, Hobert O. Timing mechanism of sexually dimorphic nervous system differentiation. eLife. 2019;8:e42078. doi: 10.7554/eLife.42078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. PNAS. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, Horazdovsky BF, Hurley JH. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/S0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nature Neuroscience. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Moss EG. Developmental transitions in C. elegans larval stages. Current Topics in Developmental Biology. 2013;105:153–180. doi: 10.1016/B978-0-12-396968-2.00006-3. [DOI] [PubMed] [Google Scholar]
- Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, Sengupta P, Portman DS. Sex, age, and hunger regulate behavioral prioritization through dynamic modulation of chemoreceptor expression. Current Biology. 2014;24:2509–2517. doi: 10.1016/j.cub.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ito H, Yokoyama A, Toba G, Yamamoto D. Partial proteasomal degradation of lola triggers the male-to-female switch of a dimorphic courtship circuit. Nature Communications. 2019;10:166. doi: 10.1038/s41467-018-08146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Developmental Cell. 2006;11:733–740. doi: 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell. 2013;155:659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Oren-Suissa M, Bayer EA, Hobert O. Sexually dimorphic differentiation of a C. elegans Hub Neuron Is Cell Autonomously Controlled by a Conserved Transcription Factor. Current Biology. 2017;27:199–209. doi: 10.1016/j.cub.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. 1988;54:1019–1031. doi: 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- Siehr MS, Koo PK, Sherlekar AL, Bian X, Bunkers MR, Miller RM, Portman DS, Lints R. Multiple doublesex-related genes specify critical cell fates in a C. elegans male neural circuit. PLOS ONE. 2011;6:e26811. doi: 10.1371/journal.pone.0026811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Molecular Cell. 2000;5:659–669. doi: 10.1016/S1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Taylor BJ, Villella A, Ryner LC, Baker BS, Hall JC. Behavioral and neurobiological implications of sex-determining factors in Drosophila. Developmental Genetics. 1994;15:275–296. doi: 10.1002/dvg.1020150309. [DOI] [PubMed] [Google Scholar]
- Tena-Sempere M. Keeping puberty on time: novel signals and mechanisms involved. Current Topics in Developmental Biology. 2013;105:299–329. doi: 10.1016/B978-0-12-396968-2.00011-7. [DOI] [PubMed] [Google Scholar]
- Verhulst EC, van de Zande L. Double nexus--doublesex is the connecting element in sex determination. Briefings in Functional Genomics. 2015;14:396–406. doi: 10.1093/bfgp/elv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Zarkower D, Bardwell VJ, Schartl M. Evolutionary dynamics of the DM domain gene family in metazoans. Journal of Molecular Evolution. 2003;57:S241–S249. doi: 10.1007/s00239-003-0033-0. [DOI] [PubMed] [Google Scholar]
- Vonhoff F, Keshishian H. Activity-Dependent synaptic refinement: new insights from Drosophila. Frontiers in Systems Neuroscience. 2017;11:23. doi: 10.3389/fnsys.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- Wexler JR, Plachetzki DC, Kopp A. Pan-metazoan phylogeny of the DMRT gene family: a framework for functional studies. Development Genes and Evolution. 2014;224:175–181. doi: 10.1007/s00427-014-0473-0. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the Nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society B: Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, Jorgensen EM. The sensory circuitry for sexual attraction in C. elegans males. Current Biology. 2007;17:1847–1857. doi: 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-X. [DOI] [PubMed] [Google Scholar]
- Zheng W, Zhang C, Bell EW, Zhang Y. I-TASSER gateway: a protein structure and function prediction server powered by XSEDE. Future Generation Computer Systems. 2019;99:73–85. doi: 10.1016/j.future.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Cheng H, Li S, Yue X, Xue Y, Chen S, Kang L. Polymodal responses in C. elegans Phasmid Neurons Rely on Multiple Intracellular and Intercellular Signaling Pathways. Scientific Reports. 2017;7:42295. doi: 10.1038/srep42295. [DOI] [PMC free article] [PubMed] [Google Scholar]