Abstract
The hemagglutinin-esterase (HE) protein of betacoronavirus lineage A is a secondary receptor in the infection process and is involved in the emergence of new betacoronavirus genotypes with altered host specificity and tissue tropism. We previously reported a novel recombinant bovine coronavirus (BCoV) strain that was circulating in dairy cattle in China, but this virus was not successfully isolated, and the genetic characteristics of BCoV are still largely unknown. In this study, 20 diarrheic faecal samples were collected from a farm in Liaoning province that had an outbreak of calf diarrhea (≤ 3 months of age) in November 2018, and all of the samples tested positive for BCoV by RT-PCR. In addition, a BCoV strain with a recombinant HE (designated as SWUN/A1/2018) and another BCoV strain with a recombinant HE containing an insertion (designated as SWUN/A10/2018) were successfully isolated in cell culture (TCID50: 104.25/mL and 104.73/mL, respectively). Unexpectedly, we identified the emergence of a novel BCoV variant characterized by a 12-nt bovine gene insertion in the receptor-binding domain in a natural recombinant HE gene, suggesting a novel evolutionary pattern in BCoV.
Electronic supplementary material
The online version of this article (10.1007/s00705-020-04840-y) contains supplementary material, which is available to authorized users.
Betacoronavirus lineage A includes human coronavirus HKU1, human coronavirus OC43 (HCoV-OC43), equine coronavirus, mouse hepatitis virus (MHV), canine respiratory coronavirus (CRCoV), porcine hemagglutinating encephalomyelitis virus, and bovine coronavirus (BCoV). CRCoV and HCoV-OC43 are closely genetically related to BCoV [1, 2]. Members of this lineage have a unique gene encoding a surface hemagglutinin-esterase (HE) protein that is not found in other coronaviruses The HE protein serves as a secondary receptor in the infection process [3] and is involved in the emergence of novel genotypes and in shifts in host specificity and tissue tropism [4]. In addition, HE protein can induce the production of neutralizing antibodies against the BCoV [5].
BCoV causes gastrointestinal and respiratory diseases in cattle, leading to serious economic losses all over the word [6–8]. No consistent genetic or antigenic markers have been identified for discriminating BCoV in different clinical syndromes [9, 10], and the virus is thought to only has one genotype [10, 11]. BCoV can infect wild ruminant animals, including alpacas, sambar deer, and giraffes [8, 12, 13] and probably can be transmitted to dogs [1, 14, 15]. Interestingly, a coronavirus isolated from human diarrheic samples has been shown to be related to BCoV [16], indicating its public health significance.
Recently, we reported the wide circulation of a novel BCoV strain with a recombinant HE in dairy cattle in China [17]. In this study, in order to monitor the epidemic of the recombinant strain, a total of 20 diarrheic faecal samples were collected from a farm that had an outbreak of diarrhea in calves three months of age or younger in Liaoning province in November, 2018, and all of the samples tested positive for BCoV by RT-PCR as described previously [17]. Full-length HE genes (1275 nt) were successfully cloned from 18 of the 20 BCoV-positive samples as described previously [18–20], and PCR products were purified and cloned into the pMD19-T simple vector prior to sequencing. A phylogenetic tree based on the complete HE gene sequences of BCoVs using the neighbor-joining method showed that all 18 HE gene sequences (GenBank accession numbers: MN982182-MN982197, MN982198, and MN982199) together with 12 Chinese BCoV HE genes [17] clustered in a large independent branch (Fig. 1), and three of the 18 HE gene sequences (GenBank accession numbers: MN982186, MN982190, and MN982199) clustered in an independent small branch. Further analysis showed that all 18 HE genes identified in this study had undergone the same recombination event identified in our previous report [17]. This suggests an increase in the prevalence of this recombinant, since it was found in only two out of five strains detected in the earlier study (samples collected in January 2018 [17]) but was present in 18 of the 18 isolates from the same farm characterized in this study (samples collected in November 2018). This indicates that the HE recombinant BCoV strain is still present in Liaoning, China, and may be increasing in prevalence.
Fig. 1.
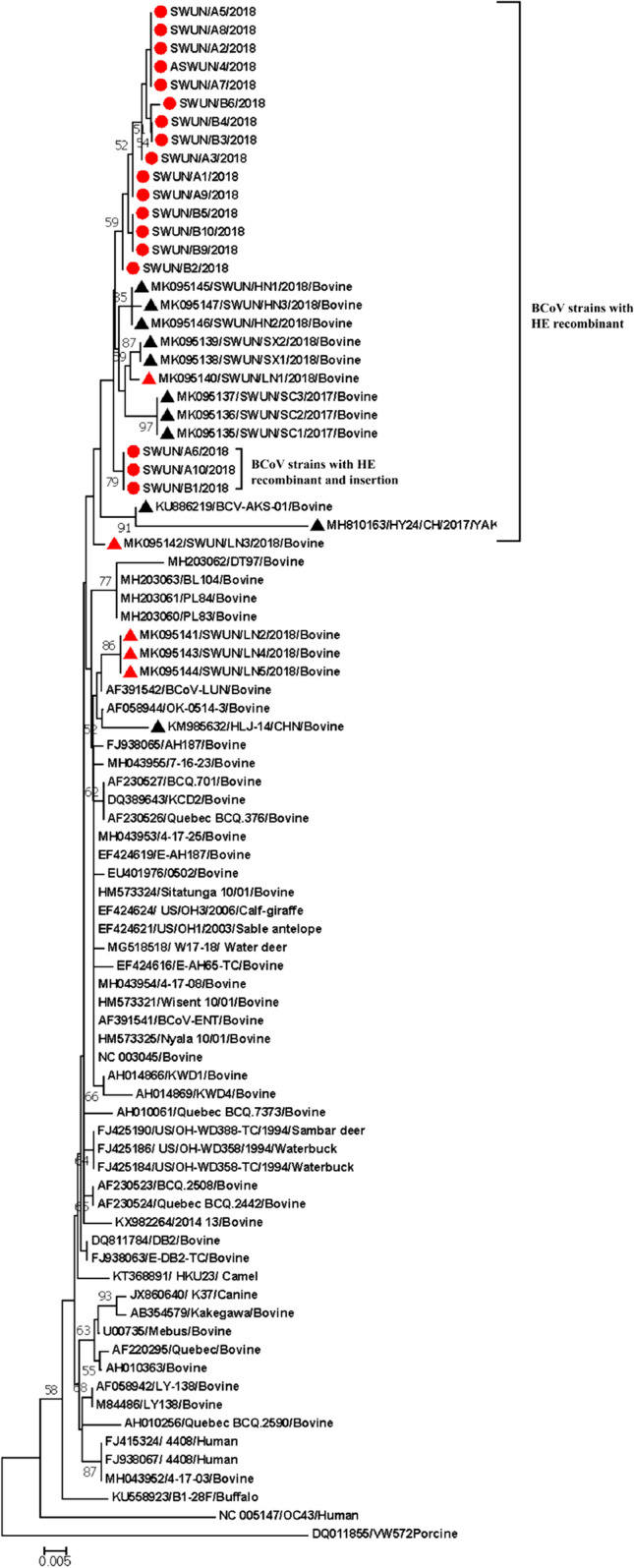
Phylogenetic tree based on the deduced 424-aa sequence of the complete HE protein of BCoV. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method with bootstrap values calculated for 1,000 replicates. The strains in this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles. The Liaoning strains are indicated in red (red circles and red triangles). The BCoV strain with recombinant HE and the insertion strain with recombinant HE with an insertion are indicated in the figure
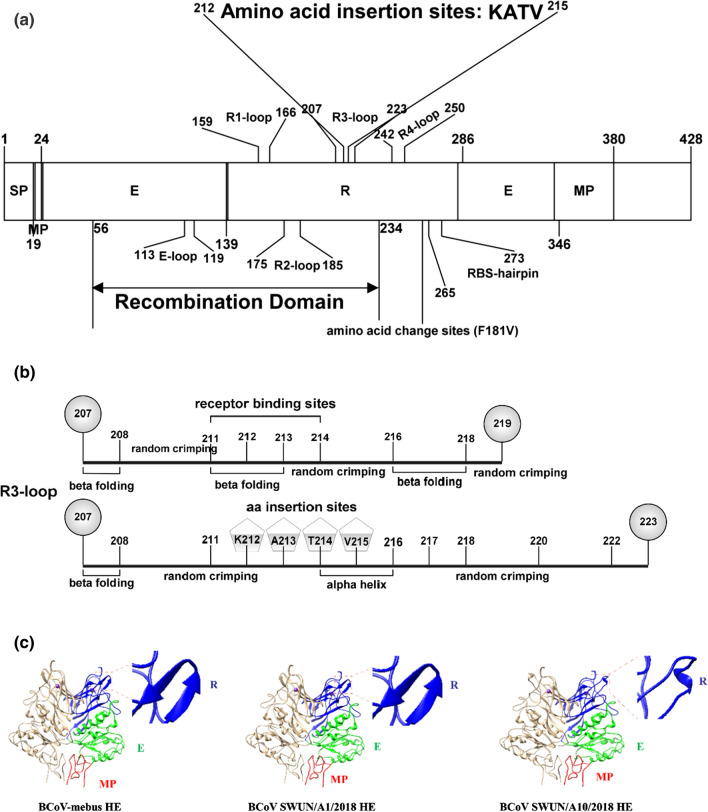
The BCoV HE receptor-binding domain is composed of six surface loops, five of which are grafted onto the beta-sandwich core of the lectin domain, the R1-loop, R2-loop, R3-loop, R4-loop, and the RBS-hairpin, and the other is present on the esterase domain, the E-loop (Fig. 2A). The 18 isolates with a recombinant HE were sequenced and found to contain the same amino acid (aa) variation (F181V) in the R2-loop that was described in our previous report [17]. The R2-loop in the lectin domain acts in ligand binding, and aa substitutions in this domain may affect receptor binding. For example, HCoV-OC43 may have evolved during adaptation of BCoV to a human host by gradually losing its lectin activity due to progressive accumulation of aa mutations in the R2-loop of the lectin-binding domain in the HE protein [2]. Unexpectedly, three isolates that clustered in an independent small branch all contained an identical insertion of 12 nucleotides (AAGGCTACTGTT), resulting in four additional amino acids in the R3-loop (Fig. 2B). This sequence was not present in any of the available BCoV HE sequences in the GenBank database. Interestingly, the inserted nucleotides were identified as originating from the bovine host, matching the sequence of a transcript variant of ORF159 on Bos taurus chromosome 16 C1 (GenBank accession no. XM027565276). It is worth noting that the R3-loop is composed of 13 aa (aa 207 -219) in BCoV HE, and residues F 211, L 212, S 213, and N 214 are essential for receptor-ligand interaction [3]. A 3D model constructed based on the crystal structure of bovine coronavirus hemagglutinin-esterase (SMTL ID: 3cl4.1) using the online software SWISSMODEL (https://www.swissmodel.expasy.org/interactive) showed that the position of the 4-aa insertion in the R3-loop between F 211 and F 212 in HE in the recombinant BCoV strains would alter the spatial conformation of the receptor-binding site relative to that in the HE recombinant strains and prototype strains lacking this insertion (Fig. 2B and C). MHV is also a member of betacoronavirus lineage A, and a variant has been reported with an aa insertion in the R3-loop that causes a change in its receptor specificity to 4-O-Ac-Sia from 9-O-Ac-Sia [4]. It is reasonable to speculate that the 4-aa insertion in the R3-loop in HE may affect HE receptor binding in the BCoV strain. The betacoronavirus HE protein is involved in receptor binding, host spectrum and tissue tropism [2, 4, 21], and it would be valuable to further investigate the biological significance of this HE variant.
Fig. 2.
A. Predicted structure of the recombinant HE protein containing the insertion. The insertion is located in the R3-loop of HE. E, esterase domain; MP, membrane-proximal domain; R, lectin domain; SP, signal peptide. B. Predicted structure of the R3-loop of the recombinant HE containing the insertion. The receptor binding site is at aa 211–214, and the 4-aa (KATV) insertion between F 211 and F 212, would cause a change in the structure of he R3-loop from FLS (beta folding)-NT (random crimping)-KYY (beta folding) for aa positions 211–218 to FKA (random crimping)-TVL (alpha helix)-SNTKYY (random crimping) for positions aa 211–222. C. Predicted crystal structures of the BCoV HE proteins (aa residues 19–380) from three strains: BCoV mebus prototype strain (GenBank accession number: U00735.2), SWUN/A1/2019 with recombinant HE, and SWUN/A10/2019 with recombinant HE with the insertion. The 3D models were constructed based on the crystal structure of bovine coronavirus hemagglutinin-esterase (SMTL ID: 3cl4.1) using the biological online software SWISSMODEL (https://www.swissmodel.expasy.org/interactive). These proteins have identical structures in the esterase domain and the membrane-proximal domain. SWUN/A10/2019 differs from the other two strains in the lectin domain. The boxed portions of the structures indicate the different conformations of the same R domain of BCoV strains. The domains are color-coded: lectin domain (R, blue); esterase domain (E, green); membrane-proximal domain (MP, red)
Betacoronavirus HE may have arisen from an influenza-C-like HE fusion protein (HEF), with transformation of the betacoronavirus HE from a trimer into a dimer for increased plasticity of the receptor-binding site (RBS) compared to that of HEF [3]. The integration of a bovine gene into other viruses has been reported previously. For example, insertion of the bovine SMT3B gene into the genome of bovine viral diarrhea virus correlates with the cytopathogenicity of the virus [22]. Interestingly, the inserted sequence in HE in this study may have originated from bovine chromosomal DNA, which may provide an evolutionary advantage for BCoV, and its identification should enhance our current understanding of the genetic evolution of BCoV.
The BCoV strain with a recombinant HE (designated as SWUN/A1/2018) and BCoV strain with a recombinant HE containing an insertion (designated as SWUN/A10/2018) successfully isolated in Vero cell culture. A cytopathic effect (CPE) was observed after three blind passages, and typical CPE characterized by cell rounding, cell lysis, and detachment from the culture flask was observed after 48 h at passage 6. After plaque purification, the TCID50 values of strains SWUN/A1/2018 and SWUN/A10/2018 were calculated as 104.25/mL and 104.73/mL, respectively, by the Reed-Muench method [23]. To better understand the genetic evolution of SWUN/A10/2018, genomic RNA was successfully obtained from strains SWUN/A1/2018 and SWUN/A10/2018 (GenBank accession number: MN982198 and MN982199). The two complete genomes shared 99.4% nt sequence identity with each other, and 97.0%–98.7% nt sequence identity with all 47 other BCoV genomes with sequences in the GenBank database. Phylogenetic trees constructed by the neighbor-joining method based on the full-length genome sequence and the ORF1a, ORF1b and S genes showed that the two strains have a close genetic relationship (see appendix). Thus, the BCoV SWUN/A10/2018 strain might have evolved from the SWUN/A1/2018 strain.
In summary, we report a novel bovine coronavirus variant with an insertion of 12 nucleotides in the HE gene that results in the addition of four amino acids in the receptor-binding portion of the HE protein. This disruption could alter the receptor-binding site by changing its spatial conformation, and it may reflect an emerging pattern in the evolution of BCoV. The betacoronavirus HE gene is involved in the emergence of new betacoronavirus genotypes, host specificity, and tissue tropism [2, 4, 21]. Thus, further investigation of the biological and epidemiological characteristics of this variant BCoV strain is warranted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix Figure 1 Phylogenetic tree based on complete genome sequences. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the previously identified Chinese BCoV strains are indicated by triangles (TIF 4592 kb)
Appendix Figure 2 Phylogenetic tree based on the complete sequence of the ORF1a gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 4557 kb)
Appendix Figure 3 Phylogenetic tree based on the complete sequence of the ORF1b gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 4455 kb)
Appendix Figure 4 Phylogenetic tree based on the complete sequence of the S gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3461 kb)
Appendix Figure 5 Phylogenetic tree based on the complete sequence of the E gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3790 kb)
Appendix Figure 6 Phylogenetic tree based on the complete sequence of the M gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3545 kb)
Appendix Figure 7 Phylogenetic tree based on the complete sequence of the N gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3547 kb)
Acknowledgements
This work was funded by the 13th Five-Year Plan National Science and Technology Support Program (Grant number 2016YFD0500907), the Innovation Team for Emerging Animal Diseases on the Qinghai-Tibet Plateau, Southwest Minzu University, and Fundamental Research Funds for the Central Universities, Southwest Minzu University (Grant number 3300220239).
Data availability statement
The data set supporting the conclusions of this article is available in the GenBank database.
Compliance with ethical standards
Ethical approval
This study did not involve animal experiments other than collecting faecal samples from diarrhoeic dairy calves during visits to farms for clinical treatment.
Conflict of interest
All authors report no conflict of interest related to the submitted work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hua Yue, Email: yhua900@163.com.
Cheng Tang, Email: tangcheng101@163.com.
References
- 1.Erles K, Shiu K-B, Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 2007;124(1–2):78–87. doi: 10.1016/j.virusres.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakkers MJ, Lang Y, Feitsma LJ, Hulswit RJ, de Poot SA, van Vliet AL, et al. Betacoronavirus adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe. 2017;21(3):356–366. doi: 10.1016/j.chom.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng Q, Langereis MA, van Vliet AL, Huizinga EG, de Groot RJ. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci. 2008;105(26):9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langereis MA, Zeng Q, Heesters BA, Huizinga EG, Groot RJD. The murine coronavirus hemagglutinin-esterase receptor-binding site: a major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012;8(1):e1002492. doi: 10.1371/annotation/a1e2a2e4-df03-40db-b10b-fd0cfcf78d3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deregt D, Babiuk LA. Monoclonal antibodies to bovine coronavirus: characteristics and topographical mapping of neutralizing epitopes on the E2 and E3 glycoproteins. Virology. 1987;161(2):0–420. doi: 10.1016/0042-6822(87)90134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azizzadeh M, Shooroki HF, Kamalabadi AS, Stevenson MA. Factors affecting calf mortality in Iranian Holstein dairy herds. Prev Vet Med. 2012;104(3–4):335–340. doi: 10.1016/j.prevetmed.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Bok M, Miño S, Rodriguez D, Badaracco A, Nuñes I, Souza SP, et al. Molecular and antigenic characterization of bovine Coronavirus circulating in Argentinean cattle during 1994–2010. Vet Microbiol. 2015;181(3–4):221–229. doi: 10.1016/j.vetmic.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson KK, Pendell DL. Market impacts of reducing the prevalence of bovine respiratory disease in United States beef cattle feedlots. Front Vet Sci. 2017;4:189. doi: 10.3389/fvets.2017.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saif LJ. Bovine respiratory coronavirus. Vet Clin Food Anim Pract. 2010;26(2):349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodnik JJ, Ježek J, Starič J. Coronaviruses in cattle. Trop Anim Health Prod. 2020 doi: 10.1007/s11250-020-02354-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ukena A. Serological characterization of genotypically distinct enteric and respiratory bovine coronaviruses. New York: Kansas State University; 2015. [Google Scholar]
- 12.Alekseev KP, Vlasova AN, Jung K, Hasoksuz M, Zhang X, Halpin R, et al. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J Virol. 2008;82(24):12422–12431. doi: 10.1128/JVI.01586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung J-Y, Kim H-R, Bae Y-C, Lee O-S, Oem J-K. Detection and characterization of bovine-like coronaviruses from four species of zoo ruminants. Vet Microbiol. 2011;148(2–4):396–401. doi: 10.1016/j.vetmic.2010.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneshima T, Hohdatsu T, Hagino R, Hosoya S, Nojiri Y, Murata M, et al. The infectivity and pathogenicity of a group 2 bovine coronavirus in pups. J Vet Med Sci. 2007;69(3):301–303. doi: 10.1292/jvms.69.301. [DOI] [PubMed] [Google Scholar]
- 15.Erles K, Toomey C, Brooks HW, Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310(2):216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Herbst W, Kousoulas K, Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol. 1994;44(2):152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keha A, Xue L, Yan S, Yue H, Tang C. Prevalence of a novel bovine coronavirus strain with a recombinant hemagglutinin/esterase gene in dairy calves in China. Transbound Emerg Dis. 2019;00:1–11. doi: 10.1111/tbed.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gélinas A-M, Boutin M, Sasseville AM-J, Dea S. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns49 protein. Virus Res. 2001;76(1):43–57. doi: 10.1016/S0168-1702(01)00243-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau SK, Lee P, Tsang AK, Yip CC, Tse H, Lee RA, et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. 2011;85(21):11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez N, Brandão PE, de Souza SP, Barrera M, Santana N, de Arce HD, et al. Molecular and phylogenetic analysis of bovine coronavirus based on the spike glycoprotein gene. Infect Genet Evol. 2012;12(8):1870–1878. doi: 10.1016/j.meegid.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo PC, Lau SK, Yip CC, Huang Y, Tsoi H-W, Chan K-H, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80(14):7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi F, Ridpath JF, Berry ES. Insertion of a bovine SMT3B gene in NS4B and duplication of NS3 in a bovine viral diarrhea virus genome correlate with the cytopathogenicity of the virus. Virus Res. 1998;57(1):1–9. doi: 10.1016/S0168-1702(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 23.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27(3):493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix Figure 1 Phylogenetic tree based on complete genome sequences. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the previously identified Chinese BCoV strains are indicated by triangles (TIF 4592 kb)
Appendix Figure 2 Phylogenetic tree based on the complete sequence of the ORF1a gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 4557 kb)
Appendix Figure 3 Phylogenetic tree based on the complete sequence of the ORF1b gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 4455 kb)
Appendix Figure 4 Phylogenetic tree based on the complete sequence of the S gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3461 kb)
Appendix Figure 5 Phylogenetic tree based on the complete sequence of the E gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3790 kb)
Appendix Figure 6 Phylogenetic tree based on the complete sequence of the M gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3545 kb)
Appendix Figure 7 Phylogenetic tree based on the complete sequence of the N gene. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed by the neighbor-joining method, with bootstrap values calculated for 1,000 replicates. The strains from this study are indicated by circles, and the other Chinese BCoV strains are indicated by triangles (TIF 3547 kb)
Data Availability Statement
The data set supporting the conclusions of this article is available in the GenBank database.