Abstract
Microvascular endothelial cells possess versatile functions and their roles in a variety of viral infections have been documented. Porcine reproductive and respiratory syndrome virus (PRRSV) infection induces severe lung inflammatory lesions in piglets, which is manifested as pulmonary endothelial dysfunction. However, the underlying mechanism of PRRSV affecting porcine pulmonary microvascular endothelial cells (PMECs) remains unknown. This study aimed to evaluate the susceptibility of PMECs to PRRSV. Primary PMECs were isolated and purified from piglet lungs, and the expression of three PRRSV receptors was characterized using immunofluorescence. Overt cytopathic effects of the PRRSV strain HN in PMECs were observed at day five post-infection, and PRRSV antigens in PMECs were determined at both RNA and protein levels using immunofluorescence and quantitative RT-PCR assays. The viral antigen significantly increased at 96 hr post-infection, and infectious virus was recovered from the supernatant of the infected PMECs. The results show that PMECs can be infected with the PRRSV strain HN, and that their receptor expression pattern is different from that of alveolar macrophages. The results of this study shed light on the potential roles of PMECs in PRRSV infection and provide a comprehensive understanding of the pathogenesis underlying its severe manifestation.
Keywords: cytopathic effects, infection, microvascular endothelial cells, porcine reproductive and respiratory syndrome virus, receptor
Microvascular endothelial cells are known for their unique anatomical position, various biological functions, as well as important roles in numerous viral diseases [2, 12, 15]. Porcine reproductive and respiratory syndrome virus (PRRSV) infection causes huge economic losses to the pig industry worldwide. As evidenced by classical symptoms such as skin cyanosis [13], interstitial pneumonia [19], and viral antigen distribution in the endothelium [9] of PRRSV-infected piglets, endothelial injury and dysfunction should be involved in its pathogenesis, which has been highlighted in some literature. For example, porcine endothelial cells in the endometrium and carotid artery had been demonstrated to have a high susceptibility to PRRSV [4, 18]. PRRSV infection could lead to severe endothelial dysfunction due to unbalanced expression of regulatory proteins, HSP90 and caveolin-1 [23], over-expression of inflammatory cytokines in vascular endothelial cells [8], and impairment of edema resolution by down-regulating the expression of aquaporin 1 and 5 in endothelial cells and secretory cells of terminal bronchiole [24].
It is well known that endothelial cells from different tissues or organs display extensive structural and functional heterogeneity [1], and they may be differently susceptible to PRRSV. Considering that serious lung injuries are the main damages mediated by PRRSV in piglets, research on the interaction between PRRSV and porcine pulmonary microvascular endothelial cells (PMECs) is crucial to elucidating the underlying mechanism of pathogenesis. However, there is so far, no conclusive evidence indicating that PMECs are susceptible to PRRSV. Therefore, in this present study, primary PMECs were isolated and purified from piglet lungs, and their susceptibility to the highly pathogenic PRRSV strain HN was investigated.
MATERIALS AND METHODS
Virus
The PRRSV strain HN used in the study was obtained from Dr. Zhanzhong Zhao from the Chinese Academy of Agricultural Sciences, and grown on African green monkey kidney MARC-145 cells, which were cultivated in Dulbecco’s Modified Eagle’s Medium (Cat #H2387, DMEM, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Cat #10099-141, FBS, Gibco), 2 mM L-glutamine, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in an incubator humidified with 5% CO2. The PRRSV strain was propagated and titrated on the MARC-145 cells before inoculation into porcine PMECs. At 72 hr post-infection (hpi), a viral titer of 4.4 logs TCID50 was detected in the infected MARC-145 cells.
Cell culture
Piglets (approximately 10-day-old) were purchased from Beijing Center for Specific Pathogen-free Swine Breeding and Management, and tested negative for PRRSV. Porcine PMECs were isolated from the piglet lungs and prepared as described previously with some modifications [10]. All procedures were approved by the Animal Care and Protection Committee of Beijing University of Agriculture (BUA_ZT201901). In brief, lungs were sterilely removed from anesthetized and heparinized piglets and placed into ice-cold PBS. The visceral pleura was stripped from each lung lobe, and the peripheral lung tissue without visible tubes and connective tissue was dissected and pooled. The samples were finely minced and washed, and the resulted fragments were digested in 0.2% collagenase type II (Cat # LS004174 Worthington Biochemical, Lakewood, NJ, USA) solution for 60 min, and the digested solution was then filtered using a 70-µm nylon mesh. The filtrate was centrifuged at 1,000 rpm and room temperature, and the cell pellet was resuspended in the complete medium (DMEM with 20% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin). Cells were inoculated onto 6-well plates, and after a 2-hr incubation, the non-adherent and dead cells were selectively washed away. The complete medium was replaced every two to three days, and 90% confluent cells were passaged using trypsinization. Cell culture was identified by using immunofluorescence staining for platelet endothelial cell adhesion molecule (PECAM-1, the anti-PECAM-1 antibody, Cat #ab186720, Abcam, Cambridge, MA, USA).
Cytopathic effects
Cytopathic effects (CPE) of the PRRSV strain HN in porcine PMECs were examined using the 3rd–4th passage cell culture. Cells were sub-cultured on 24-well plates and divided into two groups of five replicates. After removing the medium, infected group was inoculated with 0.1 ml of PRRSV solution per well for 1 hr, and the negative control (mock-infected) group was incubated with 0.1 ml maintenance medium (DMEM with 2% FBS) per well. Cells were washed five times with PBS and incubated in the maintenance medium, and replaced every 48 hr. Morphology of cell was observed under a microscope and recorded every day.
Immunofluorescence
Porcine PMECs (4th–5th passage) were seeded into 24-well plates for immunofluorescence. To detect viral antigens, cells were incubated for 12 hr and then infected with the PRRSV strain HN. Immunofluorescence staining for PRRSV GP5 and N proteins was carried out at 48 hpi using polyclonal antibodies (Cat #bs-4504R and Cat #23941R, Bioss, Beijing, China). To detect the receptors of PRRSV, the PMECs were fixed with 4% paraformaldehyde after a 24-hr incubation, and a routine indirect immunofluorescence method was performed. Anti-CD151 (Cat #sc-18753-R, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-CD163 (Cat #MCA2311GA, AbD Serotec, Kidlington, Oxford, UK), and anti-CD169 (Cat #MCA2316GA, AbD Serotec) antibodies and goat anti-mouse (Cat #bs-0296G-FITC, Bioss) and goat anti-rabbit (Cat #bs-0296G-FITC, Bioss) FITC-conjugated secondary antibodies were used.
Quantitative reverse transcription PCR (qRT-PCR)
To determine the kinetic viral loads of the PRRSV strain HN in porcine PMECs, PRRSV N gene expression was quantitatively analyzed using real-time reverse transcription PCR (RT-PCR). PMECs were seeded into 6-well plates and the cellular RNA was extracted using TRIzol (Invitrogen, Cat #15596026) at 24, 48, 72 and 96 hpi. Following digestion with DNase I to eliminate genomic DNA contamination, reverse transcription was performed using 2 µg of RNA and M-MLV reverse transcriptase (Cat #2641A, Takara, Kusatsu, Japan). Real-time RT-PCR assay was performed in triplicate using 1 µl of the RT product and according to the following method: an initial denaturation at 95°C for 5 min; 45 cycles of denaturation at 95°C for 15 sec and annealing at 65°C for 30 sec, melting curve analysis was performed in the range of 65–95°C. Primer pairs for reverse transcription are listed in Table 1. PCR products were separated on 2% agarose gel for further analysis.
Table 1. Primer pairs used for quantitative RT-PCR in this study.
| Target gene | Primer pair | Sequence (5′ to 3′) | Base number | Amplicon size |
|---|---|---|---|---|
| β-Actin | Forward | TGCGGGACATCAAGGAGAAGC | 21 | 273 bp |
| Reverse | ACAGCACCGTGTTGGCGTAGAG | 22 | ||
| PRRSV N | Forward | ATCGCCCAACAAAACCAGTC | 20 | 239 bp |
| Reverse | TGCGTCGGCAAACTAAACTC | 20 |
PRRSV, porcine reproductive and respiratory syndrome virus.
Virus titration
To further characterize the infectivity of the PRRSV isolated from the porcine PMECs, titration for the PRRSV strain HN was performed as previously described [14]. Briefly, the PMECs (6th passage) and MARC-145 cells were seeded into 6-well plates and infected with the PRRSV strain HN. At various time points post-infection, the cell supernatant was collected in duplicate and titrated on MARC-145 cells in eight replicates. The titers were calculated using the Muench–Reed method [11].
Statistical analysis
Statistical comparisons were performed using variance analysis. Differences were considered to be significant or extremely significant when P values were less than 0.05 or 0.01, respectively.
RESULTS
Biological characterization of porcine PMECs
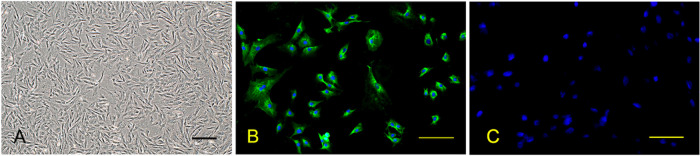
Cultured porcine PMECs quickly adhered to the plates after seeding. By differential adherence at 2 hr post-seeding, predominant cells with spindle-shaped appearance were purified, further grown in monolayers, and the morphology was maintained to achieve a growth rate of at least 7 passages (Fig. 1A). PMECs were identified by using positive immunofluorescence staining for PECAM-1 (Fig. 1B).
Fig. 1.
Morphological observation and identification using PECAM-1 immunofluorescence of porcine pulmonary microvascular endothelial cells (PMECs). (A) PMECs from piglet lungs were observed under a light microscope; (B) Porcine PMECs of passage 3 were stained with the anti-PECAM-1 antibody at 24 hr after cultivation; (C) Negative control of PECAM-1 immunofluorescence; Bar=100 µm.
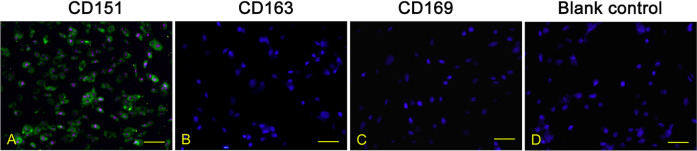
PRRSV infection of the host cells is a receptor-mediated endocytosis and replication process. To characterize receptor expression of PRRSV in porcine PMECs, an indirect immunofluorescence assay was used to measure CD151, CD163, and CD169. As shown in Fig. 2, expression of CD151 was detected in porcine PMECs, whereas both CD163 and CD169 were not detected in the cells.
Fig. 2.
Identification of porcine reproductive and respiratory syndrome virus receptors in porcine pulmonary microvascular endothelial cells using immunofluorescence staining. Cells were seeded into 24-well plates and fixed after 24 hr of incubation. Indirect immunofluorescence was performed using anti-CD151 (A), anti-CD163 (B), and anti-CD169 antibodies (C). Bar=100 µm.
CPE of the PRRSV strain HN in porcine PMECs
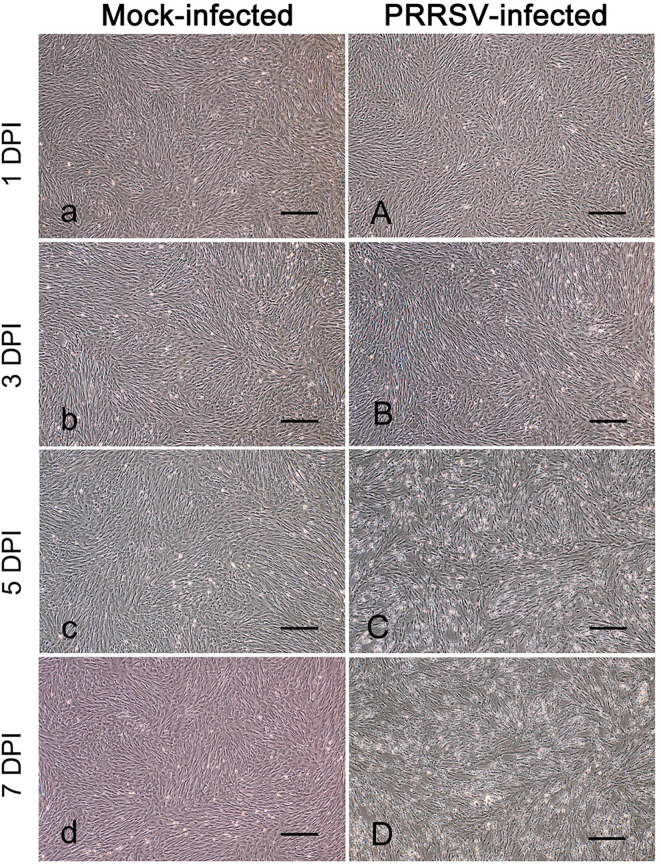
To test the susceptibility of porcine PMECs to PRRSV, cells were infected or mock-infected with the PRRSV strain HN and observed daily for CPE. From five days post-infection (dpi) onwards, increased cell degeneration was observed and some cells shriveled and detached from the wall of the wells. At 7 dpi, a large number of cells were dead and detached, and the monolayers were disrupted (Fig. 3A–D). However, the mock-infected control cells maintained their normal morphology with intact monolayers throughout the incubation period (Fig. 3A–D).
Fig. 3.
Observation of cytopathic effects (CPE) in porcine reproductive and respiratory syndrome virus strain HN-infected porcine pulmonary microvascular endothelial cells. Cells were sub-cultured on 24-well plates and infected or mock-infected with PRRSV strain HN. Cellular morphology was observed and photographs were taken every day. Bar=100 µm. DPI: days post-infection.
Viral antigens of the PRRSV strain HN in porcine PMECs
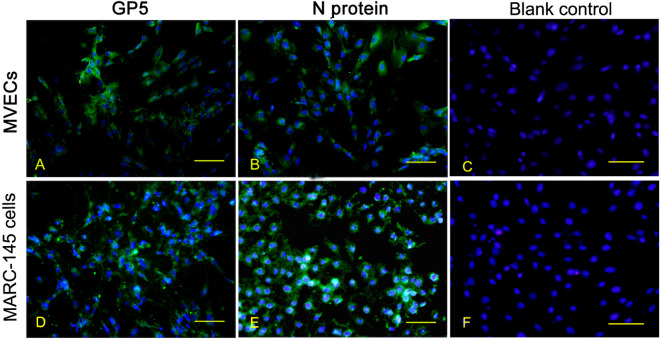
To confirm the PRRSV strain HN infection, porcine PMECs were further analyzed using immunofluorescence for the viral antigen expression of GP5 and N proteins. At 24 hpi, nearly all of the PRRSV-infected PMECs, like MARC-145 cells, were stained positively with the antibodies against PRRSV GP5 and N proteins (Fig. 4).
Fig. 4.
Immunofluorescence detection of viral antigens in porcine reproductive and respiratory syndrome virus (PRRSV)-infected porcine pulmonary microvascular endothelial cells. Cells were seeded into 24-well plates for 12 hr and infected with the PRRSV strain HN. Staining for PRRSV GP5 protein and N protein was carried out at 48 hpi. Bar=100 µm. hpi: hours post-infection. MARC-145 cells: African green monkey embryonic renal epithelial cells.
Replication kinetics of the PRRSV strain HN in porcine PMECs
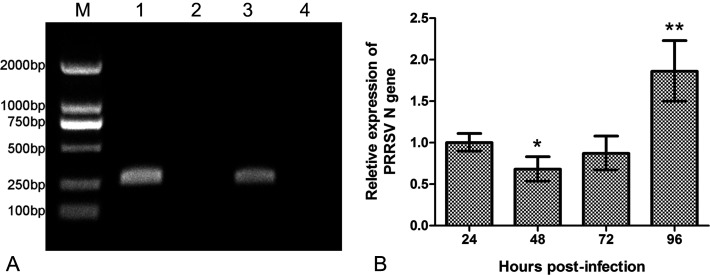
To evaluate the replication efficiency of the PRRSV strain HN in porcine PMECs, PRRSV N gene expression in virus-infected cells was detected using qRT-PCR. Results indicated that the relative expression level at 48 hpi was significantly lower than that at 24 hpi, and there was no significant difference in the expression levels between 72 and 24 hpi. However, at 96 hpi, the expression level was extremely higher compared to that at 24 hpi (P<0.01) (Fig. 5B).
Fig. 5.
Determination of porcine reproductive and respiratory syndrome virus (PRRSV) replication kinetics in porcine pulmonary microvascular endothelial cells by using qRT-PCR assay. (A) Agarose gel electrophoresis analysis of the PCR products. Lane M: DL2000 marker, lane 1: β-Actin, lane 2: negative control of β-Actin, lane 3: PRRSV N protein, lane 4: negative control of PRRSV N protein. (B) Relative ratios of PRRSV N protein gene expression to β-Actin. * denotes a significant difference (P<0.05) between 48 and 24 hpi; ** denotes an extremely significant difference (P<0.01) between 96 and 24 hpi.
Infectivity of PRRSV from porcine PMECs
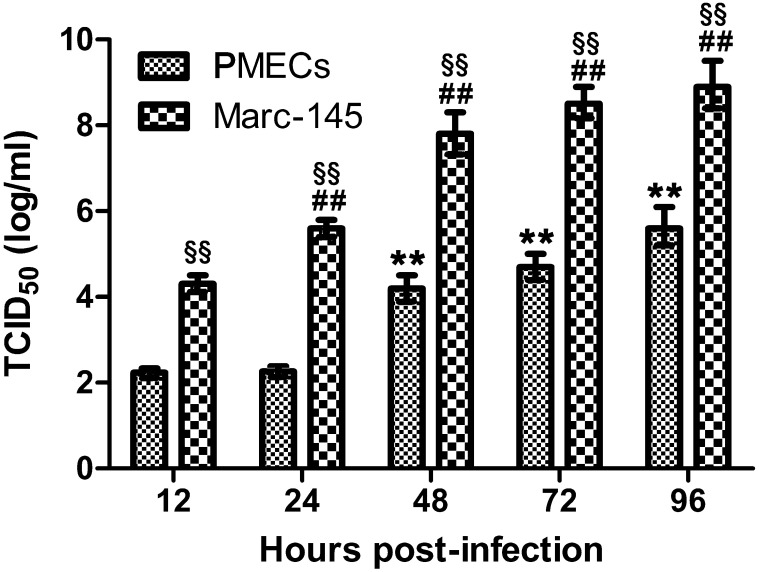
Infectivity of PRRSV from infected porcine PMECs was evaluated by using the titration assay. Result indicates that infectious viruses were recovered from the cell supernatant. Although both displayed a time dependent manner, viral titers from MARC-145 cells were significantly higher compared to porcine PMECs at all time points post-infection (all P<0.01) (Fig. 6).
Fig. 6.
Replication efficiencies of porcine reproductive and respiratory syndrome virus (PRRSV) in porcine pulmonary microvascular endothelial cells (PMECs) MARC-145 cells at different time points post-infection. Porcine PMECs were seeded on 6-well plates and infected with PRRSV strain HN. At 12, 24, 48, 72 and 96 hpi, cell supernatant was collected and titrated on MARC-145 cells. ** and ## denote extremely significant differences (P<0.01) compared to PMECs and MARC-145 cells at 12 hpi, respectively, and §§ denotes extremely significant differences (P<0.01) between PMECs and MARC-145 cells at the same time points.
DISCUSSION
PRRSV is one of the most important pathogens that severely affecting the global pig industry. Traditionally, monocytes and macrophages are considered as the main target cells in PRRSV infection. However, PRRSV-induced complicated pathological changes cannot be simply attributed to the monocyte-macrophage lineage. Understanding on the target specificity of PRRSV to PMECs is critical to clarify its mechanism of pathogenesis. Results obtained in the present study indicate that although CD163 and CD169 were not expressed in PMECs, overt CPE were observed, viral antigens were detected, and infectious viruses were recovered.
Both the CPE result and the viral antigen detection evidence as obtained through immunofluorescence and by qRT-PCR assays respectively, indicate the infection of PRRSV in porcine PMECs. However, overt CPE appeared about three days later in porcine PMECs compared to MARC-145 cells. A remarkable increase of viral replication until 4 dpi at least partially accounted for overt CPE appearing at 5 dpi indicating its lower replication efficiency in porcine PMECs compared to in macrophages or MARC-145 cells [5], and that PRRSV may need more time to adapt to PMECs. Nevertheless, PRRSV infection could cause dysfunction of PMECs and the PRRSV pathogenicity roles in pulmonary injuries warrant further study.
Although both overt CPE and remarkable replication in porcine PMECs did not occur over three days after infection, infectious PRRSV was detected in the supernatants as early as 12 hpi. This result indicates that porcine PMECs were not only susceptible to PRRSV infection but also became the hosts of infectious PRRSV. To compare the viral replication efficiency in PMECs with that in MARC-145 cells, the supernatants from infected PMECs and control cells were titrated on MARC-145 cells. The lower viral titers from PMECs showed that the replication rate of the PRRSV strain HN was slower compared to that in MARC-145 cells. This could be attributed to the fact that the PRRSV strain HN which was chronically grown on MARC-145 cells, had achieved better adaptability.
Heparin sulfate, CD169 (sialoadhesin), CD163 (scavenger receptor), CD151, and vimentin have been identified as important receptors of PRRSV that perform diverse functions in different stages of viral infection [16] and are the main targets of investigation in alveolar macrophages or MARC-145 cells. Heparin sulfate and vimentin have been reported to be expressed on various endothelial cells [3, 4, 7], and the distributions of the remaining three receptor molecules in different cells are highly discrepant. Therefore, CD151, CD163, and CD169 were detected in this study. Out of the three, CD151 was the only one that was positively expressed in porcine PMECs, which is in agreement with other reports [17, 20]. Feng et al. [4] also found that CD151 and CD169, rather than CD163 were expressed in porcine endometrial endothelial cells, which displayed a high susceptibility to PRRSV. We speculate that the mechanism of infection of PRRSV in endothelial cells is different from that in alveolar macrophages or MARC-145 cells, and that the roles of CD151, heparin sulfate, and vimentin on PRRSV infection in PMECs warrant further investigation. Moreover, CD169 and CD163 are responsible for PRRSV endocytosis [21] and the uncoating of virus particles [22], respectively, and CD151 is mainly involved in cellular functions, such as cell signaling and activation, which suggests that PRRSV infection-induced functional changes of porcine PMECs should be the focus of further research.
Although the endothelial cells from several tissues were demonstrated to be susceptible to PRRSV [4, 18], some contrary results have been published by other researchers. For example, five strains of PRRSV were experimentally proven not capable of replication in cultures using large-vessel endothelial cells isolated from the aorta and pulmonary artery [6], from which the authors addressed the importance of undertaking further investigation with fetal and microvasculature-derived endothelium and other PRRSV strains. In conclusion, endothelial cells from different organs or tissues display significant heterogeneity and diverse tropisms toward PRRSV infection.
In summary, our results show that the PRRSV strain HN can infect porcine PMECs. To the best of our knowledge, this is the first study demonstrating the susceptibility of porcine PMECs to PRRSV. Our findings would serve as a crucial evidence to better understand PRRSV infection while providing new insights for the investigation of its pathogenesis.
Acknowledgments
We sincerely thank Dr. Zhanzhong Zhao from the Chinese Academy of Agricultural Sciences for providing the PRRSV strain HN. We gratefully acknowledge the financial support from the National Natural Science Foundation (No. 31672597) and the Beijing Natural Science Foundation (No. 6202003).
REFERENCES
- 1.Aird W. C.2012. Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2: a006429. doi: 10.1101/cshperspect.a006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalrymple N. A., Mackow E. R.2014. Virus interactions with endothelial cell receptors: implications for viral pathogenesis. Curr. Opin. Virol. 7: 134–140. doi: 10.1016/j.coviro.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dave J. M., Bayless K. J.2014. Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation 21: 333–344. doi: 10.1111/micc.12111 [DOI] [PubMed] [Google Scholar]
- 4.Feng L., Zhang X., Xia X., Li Y., He S., Sun H.2013. Generation and characterization of a porcine endometrial endothelial cell line susceptible to porcine reproductive and respiratory syndrome virus. Virus Res. 171: 209–215. doi: 10.1016/j.virusres.2012.11.015 [DOI] [PubMed] [Google Scholar]
- 5.Gray D. K., Dvorak C. M. T., Robinson S. R., Murtaugh M. P.2019. Characterization of age-related susceptibility of macrophages to porcine reproductive and respiratory syndrome virus. Virus Res. 263: 139–144. doi: 10.1016/j.virusres.2019.01.015 [DOI] [PubMed] [Google Scholar]
- 6.Howerth E. W., Murphy M. D., Roberts A. W.2002. Failure of porcine reproductive and respiratory syndrome virus to replicate in porcine endothelial cell cultures. J. Vet. Diagn. Invest. 14: 73–76. doi: 10.1177/104063870201400117 [DOI] [PubMed] [Google Scholar]
- 7.Kreuger J., Matsumoto T., Vanwildemeersch M., Sasaki T., Timpl R., Claesson-Welsh L., Spillmann D., Lindahl U.2002. Role of heparan sulfate domain organization in endostatin inhibition of endothelial cell function. EMBO J. 21: 6303–6311. doi: 10.1093/emboj/cdf638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Wu Z., Liu K., Qi P., Xu J., Wei J., Li B., Shao D., Shi Y., Qiu Y., Ma Z.2017. Proteomic analysis of the secretome of porcine alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. Proteomics 17: 1700080. doi: 10.1002/pmic.201700080 [DOI] [PubMed] [Google Scholar]
- 9.Li Z., He Y., Xu X., Leng X., Li S., Wen Y., Wang F., Xia M., Cheng S., Wu H.2016. Pathological and immunological characteristics of piglets infected experimentally with a HP-PRRSV TJ strain. BMC Vet. Res. 12: 230. doi: 10.1186/s12917-016-0854-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magee J. C., Stone A. E., Oldham K. T., Guice K. S.1994. Isolation, culture, and characterization of rat lung microvascular endothelial cells. Am. J. Physiol. 267: L433–L441. [DOI] [PubMed] [Google Scholar]
- 11.Ramakrishnan M. A.2016. Determination of 50% endpoint titer using a simple formula. World J. Virol. 5: 85–86. doi: 10.5501/wjv.v5.i2.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahni S. K.2007. Endothelial cell infection and hemostasis. Thromb. Res. 119: 531–549. doi: 10.1016/j.thromres.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 13.Scruggs D. W., Sorden S. D.2001. Proliferative vasculopathy and cutaneous hemorrhages in porcine neonates infected with the porcine reproductive and respiratory syndrome virus. Vet. Pathol. 38: 339–342. doi: 10.1354/vp.38-3-339 [DOI] [PubMed] [Google Scholar]
- 14.Shang Y., Wang G., Yin S., Tian H., Du P., Wu J., Chen Y., Yang S., Jin Y., Zhang K., Lu Z., Liu X.2013. Pathogenic characteristics of three genotype II porcine reproductive and respiratory syndrome viruses isolated from China. Virol. J. 10: 7. doi: 10.1186/1743-422X-10-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shcheglovitova O. N., Skliankina N. N., Babaiants A. A., Frolova I. S., Beliaev D. L., Ershov F. I.2011. [Adhesion molecules expressed in vascular endothelial cells in natural immunity against viral infections]. Vestn. Akad. Med. Nauk SSSR 10: 54–60 (in Russian). [PubMed] [Google Scholar]
- 16.Shi C., Liu Y., Ding Y., Zhang Y., Zhang J.2015. PRRSV receptors and their roles in virus infection. Arch. Microbiol. 197: 503–512. doi: 10.1007/s00203-015-1088-1 [DOI] [PubMed] [Google Scholar]
- 17.Sincock P. M., Fitter S., Parton R. G., Berndt M. C., Gamble J. R., Ashman L. K.1999. PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J. Cell Sci. 112: 833–844. [DOI] [PubMed] [Google Scholar]
- 18.Sun G. L., Cai X. H., Zheng S. M., Sun G., Rong F. L.2010. Experimental studies on the culture of porcine reproductive and respiratory syndrome virus in porcine arterial endothelial cells. Chin. Vet. Sci. 40: 997–1001. [Google Scholar]
- 19.Szeredi L., Szentirmai C.2008. Proliferative and necrotising pneumonia and severe vascular lesions in pigs naturally infected with porcine circovirus type 2. Acta Vet. Hung. 56: 101–109. doi: 10.1556/avet.56.2008.1.10 [DOI] [PubMed] [Google Scholar]
- 20.Van Gorp H., Van Breedam W., Delputte P. L., Nauwynck H. J.2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89: 2943–2953. doi: 10.1099/vir.0.2008/005009-0 [DOI] [PubMed] [Google Scholar]
- 21.Vanderheijden N., Delputte P. L., Favoreel H. W., Vandekerckhove J., Van Damme J., van Woensel P. A., Nauwynck H. J.2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J. Virol. 77: 8207–8215. doi: 10.1128/JVI.77.15.8207-8215.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch S. K. W., Calvert J. G.2010. A brief review of CD163 and its role in PRRSV infection. Virus Res. 154: 98–103. doi: 10.1016/j.virusres.2010.07.018 [DOI] [PubMed] [Google Scholar]
- 23.Yan M., Hou M., Liu J., Zhang S., Liu B., Wu X., Liu G.2017. Regulation of iNOS-derived ROS generation by HSP90 and Cav-1 in porcine reproductive and respiratory syndrome virus-infected swine lung injury. Inflammation 40: 1236–1244. doi: 10.1007/s10753-017-0566-9 [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Yan M., Gu W., Chen A., Liu J., Li L., Zhang S., Liu G.2018. Downregulation of aquaporins (AQP1 and AQP5) and Na, K-ATPase in porcine reproductive and respiratory syndrome virus-infected pig lungs. Inflammation 41: 1104–1114. doi: 10.1007/s10753-018-0762-2 [DOI] [PubMed] [Google Scholar]