Abstract
Since fulminant type 1 diabetes was reported as a distinct subtype of type 1 diabetes in 2000, the Committee on Type 1 diabetes, Japan Diabetes Society has continuously recruited patients and conducted genomic research to elucidate the genetic basis of fulminant type 1 diabetes. The contribution of the human leukocyte antigen complex (HLA) to genetic susceptibility to fulminant type 1 diabetes was compared with that of other subtypes in 2009. The alleles and haplotypes associated with fulminant type 1 diabetes were found to be different from acute-onset and slowly progressive type 1 diabetes. DRB1*15:01-DQB1*06:02, a protective haplotype against acute-onset type 1 diabetes, does not provide protection against fulminant type 1 diabetes and DRB1*08:02-DQB1*03:02, a susceptible haplotype to acute-onset type 1 diabetes, does not confer susceptibility to fulminant type 1 diabetes. Recently, the first genome-wide association study (GWAS) of fulminant type 1 diabetes was performed in Japanese individuals. A strong association was observed with multiple single nucleotide polymorphisms (SNPs) in the HLA region, and the strongest association was observed with rs9268853 in the class II DR region. In addition, 11 SNPs outside the HLA region showed some evidence of association with the disease. In particular, rs11170445 in CSAD/lnc-ITGB7-1 on chromosome 12q13.13 showed an association at a genome-wide significance level. Fine mapping revealed that rs3782151 in CSAD/lnc-ITGB7-1 showed the lowest P value. CSAD/lnc-ITGB7-1 was found to be strongly associated with susceptibility to fulminant, but not classical, autoimmune type 1 diabetes, implicating this locus in the distinct phenotype of fulminant type 1 diabetes.
Keywords: Genetics, Fulminant type 1 diabetes, Genome-wide association study (GWAS), Human leukocyte antigen (HLA), Cysteine sulfinic acid decarboxylase (CSAD), Integrin subunit beta 7 (ITGB7)
Introduction
Since the first report on fulminant type 1 diabetes in 2000 [1], continuous effort led by the Committee on Type 1 diabetes, Japan Diabetes Society, revealed genetics, etiology and pathogenesis of fulminant type 1 diabetes. This review discusses recent progress in the genetics of fulminant type 1 diabetes in comparison with that of classical autoimmune type 1 diabetes.
Type 1 diabetes is a multifactorial disease caused by the destruction of insulin-producing β cells of the pancreas. The resulting absolute insulin deficiency requires exogenous insulin for survival, and long-term complications can disturb quality of life and shorten life span. Most cases of type 1 diabetes are thought to be of autoimmune etiology. Fulminant type 1 diabetes is also immune-mediated, but studies on genetics and pathogenesis suggest a difference between fulminant and classical autoimmune type 1 diabetes [2]. Type 1 diabetes develops as a consequence of a combination of genetic predisposition, environmental factors, and precipitating events [3]. Identification of genes associated with predisposition to common multifactorial diseases, such as diabetes, is important in establishing effective methods for prediction, prevention, and intervention in diseases.
Studies on type 1 diabetes genetics started in the 1970s and revealed the key contribution of the human leukocyte antigen (HLA) region to type 1 diabetes susceptibility. Family studies and candidate gene approaches discovered additional non-HLA loci associated with type 1 diabetes. Starting in 2007, genome-wide association study (GWAS) dramatically increased the number of loci associated with type 1 diabetes to over 60 [4] (Table 1). However, few of these loci have yet to be fine-scale mapped to causative variants or specific genes.
Table 1.
Loci associated with onset of Type 1 diabetes identified in Caucasian populations
| Reference | Identified loci |
|---|---|
| WTCCC a) | HLA-DRB1, INS, CTLA4, PTPN22, IL2RA, IFIH1, PPARG, KCNJ11, TCF7L2 |
| Todd et al.b) | PHTF1-PTPN22, ERBB3, CLEC16A, C12orf30 |
| Hakonarson et al.c) | HLA-DRB1, HLA-DQA2, CLEC16A, INS, PTPN22 |
| Hakonarson et al.d) | SUOX–IKZF4 |
| Concannon et al.e) | INS, IFIH1, CLEC16A, UBASH3A |
| Cooper et al.f) | PTPN22, CTLA4, HLA, IL2RA, ERRB3, C12orf30, CLEC16A, PTPN2 |
| Grant et al.g) | EDG7, BACH2, GLIS3, UBASH3A, RASGRP1 |
| Barrett et al.h) | MHC, PTPN22, INS, C10orf59, SH2B3, ERBB3, CLEC16A, CTLA4, PTPN2, IL2RA, IL27, C6orf173, IL2, ORMDL3, GLIS3, CD69, IL10, IFIH1, UBASH3A, COBL, BACH2, CTSH, PRKCQ, C1QTNF6, PGM1 |
| Wallace et al.i) | DLK1, TYK2 |
| Bradfield et al.j) | LMO7, EFR3B, 6q27, TNFRSF11B, LOC100128081, FOSL2 |
a: the Wellcome Trust Case Control Consortium. Nature. 2007; 447:661–78
b: Todd JA et al. Nat Genet. 2007; 39: 857–64
c: Hakonarson H et al. Nature. 2007; 448: 591–4
d: Hakonarson H et al. Diabetes. 2008; 57: 1143–6
e: Concannon P et al. Diabetes. 2008; 57: 2858–61
f: Cooper JD et al. Nat Genet. 2008; 40: 1399–401
g: Grant SF et al. Diabetes. 2009; 58: 290–5
h: Barrett JC et al. Nat Genet. 2009; 41: 703–7
i: Wallace C et al. Nat Genet. 2010; 42: 68–71
j: Bradfield JP et al. PLoS Genet. 2011; 7: e1002293
Almost all large-scale studies on type 1 diabetes to date have been performed in Caucasian populations because a large number of samples from cases and controls are required to identify disease-causing variants with a modest effect, as in the case of non-HLA genes for type 1 diabetes. In the Japanese population, the number of samples was limited because of the very low incidence (less than 1/10 of that in Caucasians) of type 1 diabetes [5]. To overcome this limitation, seven leading groups in the field of genetics of type 1 diabetes in Japan established the Study Group on Type 1 Diabetes Genetics in 2003 [6], leading to the identification of susceptibility genes for type 1 diabetes with special reference to similarities and differences between Japanese and Caucasian populations [7] (Table 2). More recently, the Committee on Type 1 diabetes, Japan Diabetes Society has been conducting genome research on type 1 diabetes, and has contributed significantly to studies on the genetics of fulminant type 1 diabetes in comparison with classical autoimmune type 1 diabetes.
Table 2.
Association of candidate genes in non-HLA region for type 1 diabetes in Japanese and Caucasian populations
| Gene | Variant | Association | ||
|---|---|---|---|---|
| Caucasian | Japanese | Remarks | ||
| INS | Class I VNTR | Yes | Yesa) | Frequency of disease-susceptible class I haplotype is very high in Japanese population |
| CTLA4 | rs3087243 | Yes | Restrictedb) | Association is concentrated in a subtype of type 1 diabetes with autoimmune thyroid disease (AITD) |
| PTPN22 | R620W (rs2476601) | Yes | Unknownc) |
Disease-susceptible Trp620 allele is absent in Asians rs2488457 showed association with type 1 diabetes in Japanese population |
| IL2RA | rs11594656 | Yes | Nod) |
The minor frequencies of the SNPs are very low in Japanese population rs706778 and rs3118470 showed association with type 1 diabetes in Japanese population |
| rs41295061 | Yes | Nod) | ||
| ERBB3 | rs2292399 | Yes | Yese) | |
| CLEC16A | rs2903692 | Yes | Yese) | |
| IFIH1 | rs1990760 | Yes | Nof) | |
| IL7R | rs6897932 | Yes | Yesf) | |
a: Awata T et al. J Clin Endocrinol Metab 2007; 92: 1791–5
b: Ikegami H al. J Clin Endocrinol Metab 2006; 91: 1087–92
c: Kawasaki E et al. Am J Med Genet 2006; 140: 586–93
d: Kawasaki E et al. J Clin Endocrinol Metab 2009; 94: 947–52
e: Awata T et al. J Clin Endocrinol Metab 2009; 94: 231–5
f: Yamashita H et al. Diabetes Metab Res Rev 2011; 27: 844–8
Fulminant type 1 diabetes has been reported mostly in East Asian populations, which represents up to 20% of adult-onset type 1 diabetes in Japan [2] and 7% of Korean type 1 diabetes [8], but only a limited number of cases have been reported in Caucasian populations [9]. Therefore, the genetics of fulminant type 1 diabetes can only be investigated in East Asian populations, particularly in the Japanese population.
Familial clustering of type 1 diabetes
The degree of familial clustering of a disease can be estimated from λs, which is calculated by dividing the lifetime risk in siblings of type 1 diabetic probands by that in the general population. In Caucasian populations, λs is estimated at 15 (6%/0.4%) [10]. In the Japanese population, the frequencies of type 1 diabetes in siblings of diabetic probands (1.3–3.8%) are much higher than those in the general population (0.01–0.02%) [11]. λs is estimated to be more than 65—much higher than that in Caucasian populations—indicating that type 1 diabetes clusters in families, even in countries with low incidence, such as Japan. Familial clustering of fulminant type 1 diabetes has not been clarified due to the limited number of patients.
HLA region
In addition to linkage studies and candidate gene approaches, GWASs have also demonstrated the absence of other loci comparable to the HLA region in effect size. HLA is reported to account for approximately 40–50% of familial aggregation of type 1 diabetes [12, 13]. The highly polymorphic nature of each HLA locus as well as the strong linkage disequilibrium (LD) between different HLA loci makes association analyses complicated and assessment of risk labor intensive and challenging.
The susceptibility to type 1 diabetes is conferred mainly by HLA class II DR and DQ genes [14]. However, substantial differences in alleles and haplotypes conferring susceptibility to type 1 diabetes have been reported among different ethnic groups. The DR3 (DRB1*03:01-DQB1*02:01) and DR4 (DRB1*04:01-DQB1*03:02) haplotypes were positively associated with type 1 diabetes in Caucasian populations, whereas the DR4 (DRB1*04:05-DQB1*04:01) and DR9 (DRB1*09:01-DQB1*03:03) haplotypes are associated with the disease in Japanese and most East Asian populations [15–17]. The difference in HLA haplotypes associated with type 1 diabetes between Japanese and Caucasian populations can be explained by the presence or absence of haplotypes in each population [7, 18].
Very low frequency of fulminant type 1 diabetes in Caucasian populations makes such a trans-racial comparison in fulminant type 1 diabetes impossible. Characteristics of HLA in fulminant type 1 diabetes, however, can be clarified by comparing different subtypes of type 1 diabetes in Japanese populations where all three subtypes of type 1 diabetes, acute-onset, fulminant, and slowly progressive, are present. A nationwide effort conducted by the Committee on Type 1 diabetes clarified similarities and differences in the contribution of HLA to genetic susceptibility to three subtypes [19]. Class II HLA is associated with all three subtypes, but the alleles, haplotypes, and genotypes associated with the disease are markedly different among the three subtypes. The alleles and haplotypes associated with acute-onset and slowly progressive type 1 diabetes were similar, whereas those associated with fulminant type 1 diabetes were mostly different from those in the other two subtypes of type 1 diabetes (Figs. 1, 2) [19]. Subsequent studies by the Committee with a larger number of samples with fulminant type 1 diabetes confirmed the association between fulminant type 1 diabetes and class II HLA and suggested a difference in HLA between fulminant type 1 diabetes with and without GAD antibodies [20].
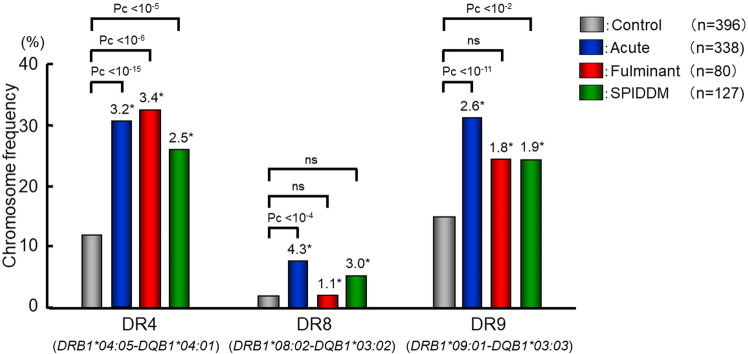
Fig. 1.
Frequencies of DRB1-DQB1 haplotypes conferring susceptibility to autoimmune type 1 diabetes in patients with acute-onset, fulminant, and slowly progressive type 1 diabetes, and control subjects. *: odds ratio. Data based on Ref. 19
Fig. 2.
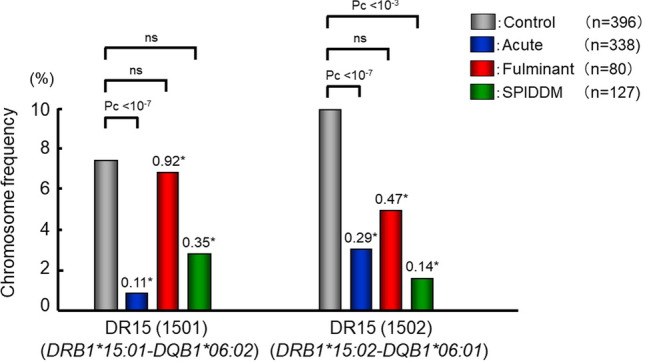
Frequencies of DRB1-DQB1 haplotypes providing protection against autoimmune type 1 diabetes in patients with acute-onset, fulminant and slowly progressive type 1 diabetes, and control. *: odds ratio. Data based on Ref. 19
The similarities and differences between fulminant and other subtypes can be summarized as follows: (1) DRB1*15:01-DQB1*06:02, a protective haplotype against acute-onset type 1 diabetes does not provide protection against fulminant type 1 diabetes. (2) DRB1*08:02-DQB1*03:02, a susceptible haplotype to acute-onset type 1 diabetes, does not confer susceptibility to fulminant type 1 diabetes. (3) DRB1*04:05-DQB1*04:01 and DRB1*09:01-DQB1*03:03 were associated with fulminant as well as acute-onset and slowly progressive type 1 diabetes. The fact that neither DRB1*1501-DQB1*0602 nor DRB1*08:02-DQB1*03:02 affected susceptibility and only Asian-specific DRB1*0405-DQB1*0401 and DRB1*09:01-DQB1*03:03 conferred susceptibility to fulminant type 1 diabetes may explain the reason why most people with fulminant type 1 diabetes are from East Asian countries [21], and only a limited number of cases were reported in the Caucasian populations.
Non-HLA genes
Except for HLA, the genetic susceptibility to fulminant type 1 diabetes is largely unknown. To identify susceptibility genes for fulminant type 1 diabetes, the Committee on Type 1 diabetes performed the first GWAS in the Japanese population, and identified variants in CSAD/lnc-ITGB7-1 on chromosome 12q13.13 associated with susceptibility to fulminant type 1 diabetes [22].
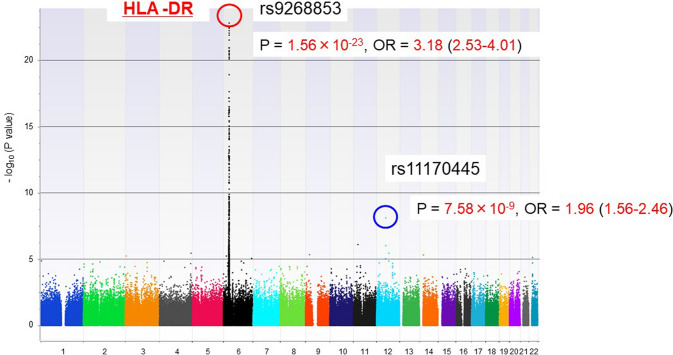
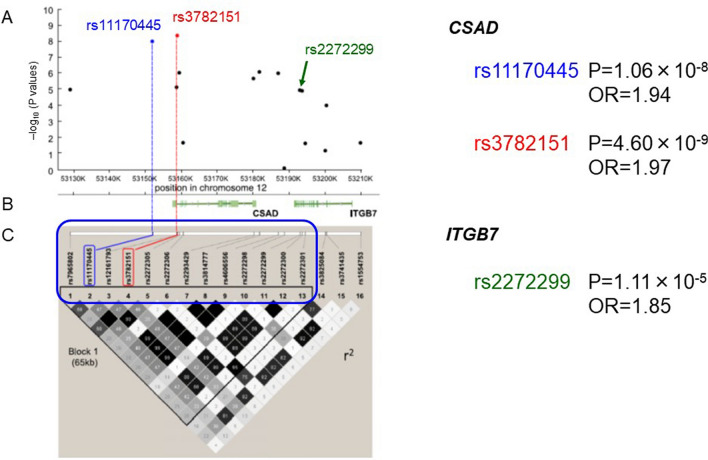
Genotyping for 600,307 SNPs was performed with unrelated Japanese patients with fulminant type 1 diabetes (n = 257) and healthy controls (n = 419) using the Axiom Genome-Wide ASI 1 Array, which is the first array designed to maximize genomic coverage of rare alleles of a consensus East Asian genome. A highly significant association was observed for multiple SNPs in the HLA region on chromosome 6, and the strongest association was found for rs9268853 in the HLA class II DR region (Fig. 3). In addition, 11 SNPs outside the HLA region showed some evidence of association with the disease. In particular, rs11170445 on chromosome 12q13.13 was associated with genome-wide significance (P = 7.58 × 10−9, OR 1.96). Fine mapping of the region surrounding rs11170445 identified multiple SNPs with low P values, and the strongest association was observed for rs3782151 (P = 4.60 × 10−9, OR 1.97) (Fig. 4). LD analysis identified a 65-kb LD block containing three protein-coding genes (CSAD, ZNF740, and ITGB7). A haplotype association test using all 13 SNPs in the LD block suggested that the minor A allele of rs3782151 in CSAD is primarily associated with susceptibility to fulminant type 1 diabetes.
Fig. 3.
Manhattan plot presenting the P values across the genome. The -log10 P values from 426,851 SNPs in 257 fulminant type 1 diabetes case subjects and 419 control subjects plotted according to their physical positions on successive chromosomes. A top-hit SNP in the HLA region is circled in red. An SNP associated with fulminant type 1 diabetes with genome-wide significance outside the HLA region is circled in blue. Data based on Ref. [22]
Fig. 4.
a The –log10 (P values) for 16 tagged SNPs from the CSAD-ITGB7 region compared between 257 patients with fulminant type 1 diabetes and 419 healthy individuals. K, thousands. b Genomic location of RefSeq genes and their intron and exon structures (NCBI). c Haploview plot of LD between markers. OR odds ratio. Data based on Ref. [22]
To examine the interaction of SNPs in CSAD with HLA, we performed a regression analysis using a top-hit SNP in the HLA region, rs9268853, as a covariate. The results showed that rs11170445 exhibited a strong association with fulminant type 1 diabetes (P = 6.23 × 10−7). In addition, no heterogeneity in the strength of the association was observed depending on the presence or absence of susceptible HLA haplotypes (Fig. 5).
Fig. 5.
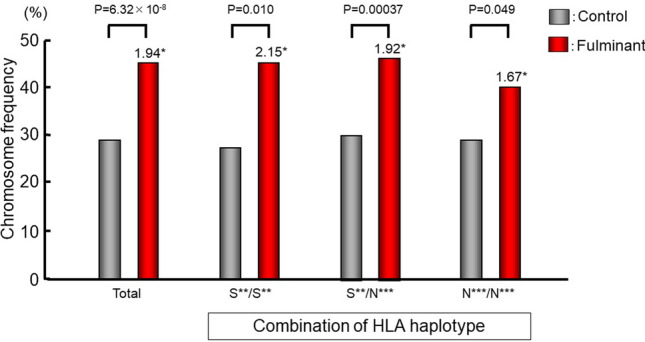
Effect of HLA DR-DQ haplotypes on the association of rs3782151 in CSAD with fulminant type 1 diabetes. *: odds ratio. S**: susceptible haplotypes, DRB1*04:05-DQB1*04:01 or DRB1*09:01-DQB1*03:03. N**: non-susceptible haplotypes, haplotypes other than DRB1*04:05-DQB1*04:01 and DRB1*09:01-DQB1*03:03. Data based on Ref. [22]
The association of rs3782151 with autoimmune type 1 diabetes was weak compared with its very strong association with fulminant type 1 diabetes (Fig. 6). The frequency of the minor allele at rs3782151 was significantly higher in fulminant type 1 diabetes than in autoimmune type 1 diabetes, suggesting that the association of rs3782151 in CSAD with type 1 diabetes is unique to the fulminant subtype. In Caucasian populations, autoimmune type 1 diabetes was reported to be associated with a variant in ITGB7 [23]. To investigate the contribution of the CSAD-ITGB7 region to type 1 diabetes in different ethnic groups, rs3782151 in CSAD and rs2272299 in ITGB7 was studied in control subjects (n = 225) and patients with autoimmune type 1 diabetes (n = 229) of Caucasian descent. Autoimmune type 1 diabetes was associated with rs2272299 in ITGB7, but not with rs3782151 in CSAD in Caucasian populations (Fig. 6). This tendency is similar to that found in the Japanese population (Fig. 6). A meta-analysis of the two populations showed the association of rs2272299 in ITGB7 [summary OR 1.78 (95% CI 1.41–2.26), P = 1.52 × 10−6), but not rs3782151 in CSAD [summary OR 1.17 (0.99–1.38), NS], with autoimmune type 1 diabetes. These findings suggest that two distinct loci for type 1 diabetes exist in the CSAD-ITGB7 region—one in CSAD for the fulminant subtype and the second in ITGB7 for the autoimmune subtype.
Fig. 6.
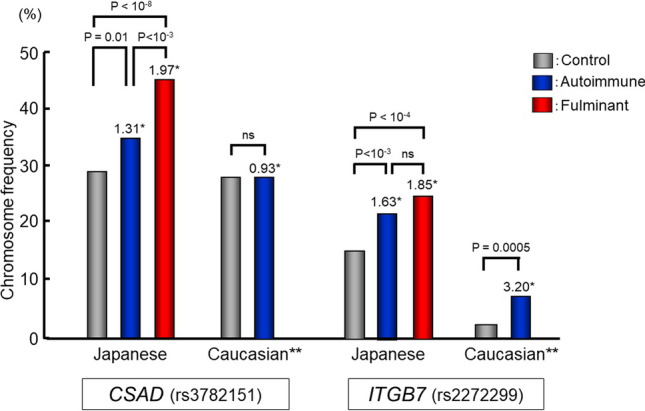
Association of CSAD (rs3782151) and ITGB7 (rs2272299) with autoimmune and fulminant type 1 diabetes in Japanese and Caucasian populations. Due to the absence of fulminant type 1 diabetes in Caucasian populations, only autoimmune type 1 diabetes is described in Caucasian populations. *: odds ratio. **: samples of Caucasian populations(n = 454, 225 controls and 229 patients with autoimmune type 1 diabetes) were kindly provided by George Eisenbarth, Barbara Davis Center for Childhood Diabetes, Aurora, Co. USA (Noso S et al. J Genet Syndr Gene Ther 2013;4:204). Data in Japanese are based on Ref. [22]
CSAD encodes cysteine sulfinic acid decarboxylase, which is a key enzyme in taurine synthesis. Taurine has been reported to exert anti-inflammatory and cytoprotective effects by attenuating apoptosis and stimulating antioxidant activity [24–28]. Taurine is also important for mitochondrial function. Recent studies have indicated that taurine modification of tRNALeu(UUR) is defective in mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) [29]. Taurine supplementation prevented the stroke-like episodes in MELAS [30]. The contribution of taurine to the protection of pancreatic islets from destruction has been reported in both type 1 diabetes and streptozotocin-induced apoptosis [31, 32], suggesting that CSAD variants might contribute to fulminant type 1 diabetes by impairing the protection of pancreatic islets. To identify causal variants in CSAD, the CSAD region was re-sequenced in 32 participants who were homozygous for the risk allele at rs3782151 and 31 single nucleotides were identified. One nonsynonymous variant (His288Arg) was identified in the coding region, but the extremely low frequency of this variant suggests that the variants in the protein-coding region of CSAD are unlikely to be a common cause of fulminant type 1 diabetes.
The CSAD region encodes not only CSAD but also a long noncoding (lnc) RNA known as lnc-ITGB7-1 [33]. The top-hit SNP rs3782151 has been reported to be a cis eQTL of ITGB7 in Caucasian populations [34]. In addition, several SNPs flanking rs3782151 in the CSAD/lnc-ITGB7-1 region have been reported to be cis eQTLs of ITGB7 in the Japanese population [35, 36]. ITGB7 is involved in the migration, entry, and adhesion of lymphocytes in inflamed organs, including the pancreas [37–41]. Disease-associated minor alleles increase ITGB7 expression [35, 36], which suggests that the CSAD/lnc-ITGB7-1 region contributes to fulminant type 1 diabetes through an increase in ITGB7 expression and the acceleration of tissue destruction.
Conclusion
Genetic susceptibility to fulminant type 1 diabetes is distinct from classical autoimmune diabetes in both HLA and non-HLA genes. HLA associations with fulminant type 1 diabetes are qualitatively different from those with other subtypes of type 1 diabetes in that DRB1*15:01- DQB1*06:02, a well-known protective haplotype against acute-onset type 1 diabetes, does not provide protection and DRB1*08:02-DQB1*03:02, a susceptible haplotype to acute-onset type 1 diabetes, does not confer susceptibility to fulminant type 1 diabetes. The CSAD/lnc-ITGB7-1 region on chromosome 12q13.13 was identified by GWAS as the first non-HLA susceptibility locus for fulminant type 1 diabetes that was unique to fulminant, but not for classical autoimmune type 1 diabetes. Further studies are needed to clarify the underlying mechanisms of the contribution of identified SNPs to the development of fulminant as well as classical type 1 diabetes.
Acknowledgements
The authors thank all of the participants in the project, the members of the Study Group on Type 1 Diabetes Genetics and the Committee on Type 1 diabetes, Japan Diabetes Society.
Compliance with ethical standards
Conflict of interest
The authors have no financial conflicts of interest to disclose for this manuscript.
Ethics policy
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Imagawa A, Hanafusa T, Miyagawa J, Matsuzawa Y. A novel subtype of type 1 diabetes mellitus characterized by a rapid onset and an absence of diabetes related antibodies. Osaka IDDM study group. N Engl J Med. 2000;342(5):301–307. doi: 10.1056/NEJM200002033420501. [DOI] [PubMed] [Google Scholar]
- 2.Imagawa A, Hanafusa T. Fulminant type 1 diabetes: a novel clinical entity requiring special attention by all medical practitioners. Nat Clin Prac Endocrinol Metab. 2007;3:36–45. doi: 10.1038/ncpendmet0351. [DOI] [PubMed] [Google Scholar]
- 3.Gianani R, Eisenbarth GS. The stages of type 1A diabetes: 2005. Immunol Rev. 2005;204:232–249. doi: 10.1111/j.0105-2896.2005.00248.x. [DOI] [PubMed] [Google Scholar]
- 4.Bakay M, Pandey R, Grant SFA, Hakonarson H. The genetic contribution to type 1 diabetes. Curr Diab Rep. 2019;19:116. doi: 10.1007/s11892-019-1235-1. [DOI] [PubMed] [Google Scholar]
- 5.Karvonen M, Viik-Kajander M, Moltchanova E, Libman I, LaPorte R, Tuomilehto J. Incidence of childhood type 1 diabetes worldwide. Diabetes mondiale (DiaMond) project group. Diabetes Care. 2000;23:1516–1526. doi: 10.2337/diacare.23.10.1516. [DOI] [PubMed] [Google Scholar]
- 6.Ikegami H, Awata T, Kawasaki E, Kobayashi T, Maruyama T, Nakanishi K, Shimada A, Amemiya S, Kawabata Y, Kurihara S, Tanaka S, Kanazawa Y, Mochizuki M, Ogihara T. Japanese study group on type 1 diabetes genetics: the association of CTLA4 polymorphism with type 1 diabetes is concentrated in patients complicated with autoimmune thyroid disease: a multi-center collaborative study in Japan. J Clin Endocrinol Metab. 2006;91:1087–1092. doi: 10.1210/jc.2005-1407. [DOI] [PubMed] [Google Scholar]
- 7.Ikegami H, Kawabata Y, Noso S, Fujisawa T, Ogihara T. Genetics of type 1 diabetes in Asian and Caucasian populations. Diabetes Res Clin Pract. 2007;77(Suppl. 1):S116–S121. doi: 10.1016/j.diabres.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 8.Cho YM, Kim JT, Ko KS, Koo BK, Yang SW, Park MH, Lee HK, Park KS. Fulminant type 1 diabetes in Korea: high prevalence among patients with adult-onset type 1 diabetes. Diabetologia. 2007;50:2276–2279. doi: 10.1007/s00125-007-0812-z. [DOI] [PubMed] [Google Scholar]
- 9.Moreau C, Drui D, Arnault-Ouary G, Charbonnel B, Chaillous L, Cariou B. Fulminant type 1 diabetes in Caucasians: a report of three cases. Diabetes Metab. 2008;34:529–532. doi: 10.1016/j.diabet.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Rish N. Assessing the role of HLA-linked and unlinked determinants of disease. Am J Hum Genet. 1987;40:1–14. [PMC free article] [PubMed] [Google Scholar]
- 11.Ikegami H, Oihara T. Genetics of insulin-dependent diabetes mellitus. Endocrine J. 1996;43:605–613. doi: 10.1507/endocrj.43.605. [DOI] [PubMed] [Google Scholar]
- 12.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EA, Bingley PJ. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the United Kingdom. J Clin Endocrinol Metab. 2004;89:4037–4043. doi: 10.1210/jc.2003-032084. [DOI] [PubMed] [Google Scholar]
- 14.Todd J, Bell JI, MacDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 15.Ikegami H, Kawaguchi Y, Yamato E, Kuwata S, Tokunaga K, Noma Y, Shima K, Ogiharaet T. Analysis by the polymerase chain reaction of histocompatibility leukocyte antigen-DR9-linked susceptibility to insulin-dependent diabetes mellitus. J Clin Endocrinol Metab. 1992;75:1381–1385. doi: 10.1210/jcem.75.5.1358911. [DOI] [PubMed] [Google Scholar]
- 16.Kawabata Y, Ikegami H, Kawaguchi Y, Fujisawa T, Shintani M, Ono M, Nishino M, Uchigata Y, Lee I, Ogihara T. Asian-specific HLA haplotypes reveal heterogeneity of the contribution of HLA-DR and -DQ haplotypes to susceptibility to type 1 diabetes. Diabetes. 2002;51:545–551. doi: 10.2337/diabetes.51.2.545. [DOI] [PubMed] [Google Scholar]
- 17.Thomson G, Valdes AM, Noble JA, et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens. 2007;70:110–127. doi: 10.1111/j.1399-0039.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- 18.Ikegami H, Noso S, Babaya N, Hiromine Y, Kawabata Y. Genetic basis of type 1 diabetes: similarities and differences between East and West. Rev Diabet Stud. 2008;5:64–72. doi: 10.1900/RDS.2008.5.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawabata Y, Ikegami H, Awata T, Imagawa A, Maruyama T, Kawasaki E, Tanaka S, Shimada A, Osawa H, Kobayashi T, Hanafusa T, Tokunaga K, Makino H. Committee on type 1 diabetes, Japan diabetes society. Diabetologia. 2009;52:2513–2521. doi: 10.1007/s00125-009-1539-9. [DOI] [PubMed] [Google Scholar]
- 20.Tsutsumi C, Imagawa A, Ikegami H, Makino H, Kobayashi T, Hanafusa T. Japan diabetes society committee on type 1 diabetes mellitus research. Class II HLA genotype in fulminant type 1 diabetes: a nationwide survey with reference to glutamic acid decarboxylase antibodies. J Diabetes Investig. 2012;3:62–69. doi: 10.1111/j.2040-1124.2011.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imagawa A, Hanafusa T. Fulminant type 1 diabetes–an important subtype in East Asia. Diabetes Metab Res Rev. 2011;27:959–964. doi: 10.1002/dmrr.1236. [DOI] [PubMed] [Google Scholar]
- 22.Kawabata Y, Nishida N, Awata T, Kawasaki E, Imagawa A, Shimada A, Osawa H, Tanaka S, Takahashi K, Nagata M, Yasuda H, Uchigata Y, Kajio H, Makino H, Yasuda K, Kobayashi T, Hanafusa T, Tokunaga K, Ikegami H. Genome-wide association study confirming a strong effect of HLA and identifying variants in CSAD/lnc-ITGB7–1 on chromosome 12q13.13 associated with susceptibility to fulminant type 1 diabetes. Diabetes. 2019;68:665–675. doi: 10.2337/db18-0314. [DOI] [PubMed] [Google Scholar]
- 23.Evangelou M, Smyth DJ, Fortune MD, Burren OS, Walker NM, Guo H, Onengut-Gumuscu S, Chen W-M, Concannon P, Rich SR, Todd JA, Wallace C. A method for gene-based pathway analysis using genome wide association study summary statistics reveals nine new type 1 diabetes associations. Genet Epidemiol. 2014;38:661–670. doi: 10.1002/gepi.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirdah MM. Protective and therapeutic effectiveness of taurine in diabetes mellitus: a rationale for antioxidant supplementation. Diabetes Metab Syndr. 2015;9:55–64. doi: 10.1016/j.dsx.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Lambert IH, Kristensen DM, Holm JB, Mortensen OH. Physiological role of taurine–from organism to organelle. Acta Physiol (Oxf) 2015;213:191–212. doi: 10.1111/apha.12365. [DOI] [PubMed] [Google Scholar]
- 26.Huxtable RJ. Physiological actions of taurine. Physiol Rev. 1992;72:101–163. doi: 10.1152/physrev.1992.72.1.101. [DOI] [PubMed] [Google Scholar]
- 27.Marcinkiewicz J, Kontny E. Taurine and inflammatory diseases. Amino Acids. 2014;46:7–20. doi: 10.1007/s00726-012-1361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C, Cha Y-N. Taurine chloramine produced from taurine under inflammation provides anti-inflammatory and cytoprotective effects. Amino Acids. 2014;46:89–100. doi: 10.1007/s00726-013-1545-6. [DOI] [PubMed] [Google Scholar]
- 29.Fakruddin M, Wei F, Suzuki T, et al. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 2018;22:482–496. doi: 10.1016/j.celrep.2017.12.051. [DOI] [PubMed] [Google Scholar]
- 30.Ohsawa Y, Hagiwara H, Nishimatsu S, et al. Taurine supplementation for prevention of stroke-like episodes in MELAS: a multicenter, open-label, 52-week phase III trial. J Neurol Neurosurg Psychiatry. 2019;90:529–536. doi: 10.1136/jnnp-2018-317964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arany E, Strutt B, Romanus P, Remacle C, Reusens B, Hill DJ. Taurine supplement in early life altered islet morphology, decreased insulitis and delayed the onset of diabetes in non-obese diabetic mice. Diabetologia. 2004;47:1831–1837. doi: 10.1007/s00125-004-1535-z. [DOI] [PubMed] [Google Scholar]
- 32.Lin S, Yang J, Wu G, Liu M, Lv Q, Yang Q, Hu J. Inhibitory effects of taurine on STZ-induced apoptosis of pancreatic islet cells. Adv Exp Med Biol. 2013;775:287–297. doi: 10.1007/978-1-4614-6130-2_24. [DOI] [PubMed] [Google Scholar]
- 33.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narahara M, Higasa K, Nakamura S, Tabara Y, Kawaguchi T, Ishii M, Matsubara K, Matsuda F, Yamada R. Large-scale East-Asian eQTL mapping reveals novel candidate genes for LD mapping and the genomic landscape of transcriptional effects of sequence variants. PLoS ONE. 2014;9:e100924. doi: 10.1371/journal.pone.0100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higasa K, Miyake N, Yoshimura J, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet. 2016;61:547–553. doi: 10.1038/jhg.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, Holzmann B, Weissman IL, Hamann A, Butcher EC. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 38.Streeter PR, Berg EL, Rouse BT, Bargatze RF, Butcher EC. A tissue-specific endothelial cell molecule involved in lymphocyte homing. Nature. 1988;331:41–46. doi: 10.1038/331041a0. [DOI] [PubMed] [Google Scholar]
- 39.Hänninen A, Taylor C, Streeter PR, Stark LS, Sarte JM, Shizuru JA, Simell O, Michie SA. Vascular addressins are induced on islet vessels during insulitis in nonobese diabetic mice and are involved in lymphoid cell binding to islet endothelium. J Clin Invest. 1993;92:2509–2515. doi: 10.1172/JCI116859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faveeuw C, Gagnerault MC, Lepault F. Expression of homing and adhesion molecules in infiltrated islets of Langerhans and salivary glands of nonobese diabetic mice. J Immunol. 1994;152:5969–5978. [PubMed] [Google Scholar]
- 41.Yang XD, Sytwu HK, McDevitt HO, Michie SA. Involvement of beta 7 integrin and mucosal addressin cell adhesion molecule-1 (MAdCAM-1) in the development of diabetes in obese diabetic mice. Diabetes. 1997;46:1542–1547. doi: 10.2337/diacare.46.10.1542. [DOI] [PubMed] [Google Scholar]