Abstract
Plants integrate a variety of biotic and abiotic factors for optimal growth in their given environment. While some of these responses are local, others occur distally. Hence, communication of signals perceived in one organ to a second, distal part of the plant and the coordinated developmental response require an intricate signaling system. To do so, plants developed a bipartite vascular system that mediates the uptake of water, minerals, and nutrients from the soil; transports high-energy compounds and building blocks; and traffics essential developmental and stress signals. One component of the plant vasculature is the phloem. The development of highly sensitive mass spectrometry and molecular methods in the last decades has enabled us to explore the full complexity of the phloem content. As a result, our view of the phloem has evolved from a simple transport path of photoassimilates to a major highway for pathogens, hormones and developmental signals. Understanding phloem transport is essential to comprehend the coordination of environmental inputs with plant development and, thus, ensure food security. This review discusses recent developments in its role in long-distance signaling and highlights the role of some of the signaling molecules. What emerges is an image of signaling paths that do not just involve single molecules but rather, quite frequently an interplay of several distinct molecular classes, many of which appear to be transported and acting in concert.
Keywords: lipids, Long-distance signaling, nucleic acids, phloem, plant development and stress response, proteins
Introduction
The vascular system is fundamental for systemic transport of energy-rich molecules, building blocks, and nutrients in the plant. It comprises the xylem for unidirectional translocation of water and minerals from roots to shoots and the phloem, whose predominant role is the transport of photoassimilates from source to sink tissues. The xylem pulls water and minerals up through xylem vessels, driven by the water potential gradient between the soil and the atmosphere surrounding the plant. The phloem consists of three main components: the companion cells (CCs), the sieve elements (SEs), and the parenchyma cells. The main conduit for phloem transport, the SEs, are enucleated cells separated by porous sieve plates, through which long-distance transport occurs by bulk flow according to Muench’s Pressure-Flow Hypothesis [1]. The Pressure-Flow Hypothesis attributes phloem transport to the difference in osmotic pressure between source tissues with high concentrations of sugars and sink tissues with low sugar concentration. While the SEs have few organelles and are thought not to perform transcription or translation, the neighboring CCs are fully functional cells that load and unload photoassimilates and other macromolecules into and out of the SEs. The loading of photoassimilates can follow several paths, namely, active apoplastic transport facilitated by Sugar Will Eventually be Exported Transporters (SWEETs) and Sucrose Transporters (SUTs); passive symplastic loading through plasmodesmata (PD) between mesophyll, CCs, and the SE; or polymer trapping dependent on the generation of a concentration gradient via the synthesis of raffinose and stachyose from sucrose [2–5]. Unloading occurs through and is controlled by specific PD [6,7]. Beyond sugars, the phloem is also a conduit for remobilization of carbon and nitrogen from source tissues to sink tissues, often in the form of amino acids [8,9]. These processes occur, for example, during leaf senescence but also as a consequence of de novo amino acid synthesis [9]. Amino acids are thought to be loaded into the phloem via the apoplastic mechanism, regulated by transporters like Usually Multiple Acids Move In and Out Transporters (UMAMITs) for export out of leaf cells and Amino Acid Permeases (AAPs) for import into the phloem. Whereas symplastic mechanisms seem to predominate during unloading, with amino acids passing through PD from the SEs into sink tissues [8]. As a result, the phloem is critical for the reallocation of carbon and nitrogen assimilates for the systemic distribution of energy and nutrients in the plant.
Another critical function of the phloem is long-distance signaling, which is necessary for the systemic coordination of plant development under normal conditions as well as during abiotic stress. It is also essential for the response to biotic factors, such as viruses, mutualistic and pathogenic microbiota, fungi, herbivores, and other pathogens such as nematodes and sucking/piercing insects. Our knowledge of phloem anatomy, development, and function was thoroughly reviewed in 2013 [10]. Here we will focus specifically on the review and update of various phloem mobile macromolecules and their roles as systemic signals.
Mobile macromolecules and the regulation of their transport
In addition to photoassimilates, other macromolecules, including nucleic acids, peptides, proteins, hormones, and lipids have been identified in the phloem [11,12]. Their presence elevates the function of the phloem from simple energy transport to an integral signaling conduit.
Nucleic acids
Nucleic acids, more specifically RNA species, are among some of the most well-studied phloem-mobile signaling molecules. For a more detailed review of RNA trafficking, see recent publications by Ham and Lucas [13], Lin and Chen [14], and Kehr and Kragler [15]. Representatives of all types of RNA, such as mRNA, microRNA (miRNA), small interfering RNA (siRNA), and other non-coding RNA (ncRNA) have been identified in the phloem of many plant species, including Arabidopsis, rice, barley, pumpkin, watermelon, cucumber, and several others [16–24]. While the existence of mRNA and ncRNA in the vasculature system hints at their mobility and possible role in plant development, identification alone does not conclusively prove movement or physiological function. Using predominantly grafting approaches, numerous studies have provided evidence for phloem-mobile mRNA [25,26] and ncRNA [27–30]. Furthermore, data suggest environmental conditions may affect transcript mobility independent of changes in gene expression, implicating mobile transcripts as possible stress response signals [26,31].
While some evidence suggest that mRNA mobility can be ascribed to transcript abundance and stability [32], other studies have identified selective mechanisms, including specific sequence motifs, that determine systemic transport of nucleic acids [33–36]. The untranslated regions of StBEL5 appear to influence transcript stability for transport and impact translation and delivery to distal tissues [33]. Similarly, a cis-acting element within the 102 nucleotides at the 5′ end of FLOWERING LOCUS T (FT) is critical for the systemic transport of Arabidopsis FT RNA [37]. RNA-binding proteins play an important role in the translocation of some RNA species in the phloem; they contain motifs, like CU-rich polypyrimidine-binding regions, which can facilitate RNA binding and transport [34,35,38]. Additionally, other sequence motifs and secondary structures, such as tRNA-like structures, may also promote transcript mobility in the phloem [39]. Degradation mechanisms at the destination organs have also been suggested as the driving force for transcript signal selection, a hypothesis that further expands on the role of transcript stability for long-distance signaling [40].
Phloem-mobile RNA species show directionality, target specificity, and stress responsiveness. Further characterizations of selected phloem-localized RNA species have linked RNA to particular stresses and developmental signals.
Hormones and lipids
The phloem has been implicated in the systemic translocation of phytohormones, including auxins, gibberellins (GAs), cytokinins (CKs), jasmonic acid (JA), and abscisic acid (ABA) [41–44]. As a conduit for photoassimilates, the phloem is an aqueous, hydrophilic environment, and yet hydrophobic compounds including fatty acids, hormones, and lipids have been identified in the phloem [12,45,46]. While some of these hormones and other molecules may act as independent mobile signals or translocate as conjugates, lipophilic compounds may interact with proteins to facilitate their transport and signaling activity [45,47,48]. Protein-associated long-distance lipid transport is a widely accepted mechanism for signaling in animal systems [49–52]. Lipid interactions are necessary for the activation and function of transcription factors (TFs), interaction with receptors, and other signaling components, but have been understudied in plants [53–58]. However, several examples of lipid interaction with TFs have recently been published in plants as well: phospholipid–TF complexes play roles in flowering [59,60], the circadian clock [61], nuclear localization [62], and lipid metabolism [63]. Hence, some roles for protein–lipid complexes in systemic signaling in plants echo those in animal systems.
Proteins and peptides
Several proteomics studies have identified hundreds up to thousands of proteins in the phloem, despite little evidence for translation in the sieve tube, which suggests that peptides and proteins are loaded for transport as possible systemic signals [23,64–68].
It has been debated whether these non-sugar macromolecules diffuse into the phloem by accident [69–71] or enter through targeted transport [72,73]. At least in some cases these macromolecules have been shown to be carefully regulated, intentionally transported, and essential for successful plant development. For example, when proteins from pumpkin phloem exudate were introduced into the rice sieve tube, some proteins were transported shootward in the direction of bulk flow, whereas some moved rootward. This suggests that movement was destination-driven with a possible regulatory role for interacting proteins [74]. The PD likely play a crucial regulatory role in the loading and unloading of macromolecules into and out of the phloem stream, as they are known to mediate transport between cells [75]. Comparative proteomics has identified fluctuations in phloem protein composition under various environmental conditions, providing further evidence for controlled trafficking [66,76].
One of the specifically regulated and important regulatory proteins, FT (20 kDa), for example, is phloem-mobile and exhibits essential, tissue-specific function [77–79]. FT export into SEs for long-distance translocation is partly dependent on FT INTERACTING PROTEIN 1 (FTIP1), suggesting a mechanism for selective protein transport into the phloem [80]. Additionally, an MCTP-SNARE (multiple C2 domain and transmembrane protein-soluble N-ethylmaleimide-sensitive factor protein attachment protein receptor) complex composed of SYNTAXIN OF PLANTS 121 (SYP121) and QUIRKY (QKY) acts in a mechanism parallel to the FTIP1-mediated loading to transport FT into the SE [81]. Further regulation during phloem unloading at distal tissues or binding to spatially specific receptors, may contribute to systemic transport and specificity for delivery of physiologically relevant signals [82]. Taken together, these findings suggest tightly regulated and highly selective processes for the systemic trafficking of mobile signals like FT and other signaling proteins outlined in Table 1. While non-specific diffusion may account for some proteins in the phloem, it is evident that multiple mobile proteins, critical for development and stress response, are deliberately transported. These findings, along with convincing case studies of mobile proteins with physiological function, to be discussed throughout this review, demonstrate the controlled, purposeful translocation of systemic protein signals in the phloem
Table 1. Phloem-localized systemic signals.
| Mobile signal | Abbreviation | Species class | Developmental process | Plant species | References |
|---|---|---|---|---|---|
| Transition from vegetative to generative growth | |||||
| FLOWERING LOCUS T | FT | Protein | Flowering | Arabidopsis thaliana, Cucurbita moschata, Curcurbita maxima, Oryza sativa, Solanum lycopersicum, Zea mays, Glycine max, Nicotiana tabacum, Gossypium hirsutum, Solanum tuberosum | [77,78,82,83–94,95] |
| Nucleic acid (mRNA) | [37,96,97] | ||||
| CENTRORADIALIS | ATC | Protein | Flowering | Arabidopsis thaliana | [98] |
| Gibberellins | GA | Hormone | Flowering | Arabidopsis thaliana, Lolium temulentum | [41,42,99,100,99–110] |
| Cytokinins | CK | Hormone | Flowering | Arabidopsis thaliana, Elaeis guineensis | [41–44,100] |
| SINGLE FLOWER TRUSS | SFT | Protein | Flowering, leaf maturation, termination, and abscission | Solanum lycopersicum | [87,88,111] |
| Tuber development | |||||
| BEL1-RELATED HOMEOTIC PROTEIN 5 | BEL5 | Nucleic acid (mRNA) | Tuber development (promotion) | Solanum tuberosum | [112] |
| BEL1-RELATED HOMEOTIC PROTEIN 11 | BEL11 | Nucleic acid (mRNA) | Tuber development (inhibition) | Solanum tuberosum | [113] |
| BEL1-RELATED HOMEOTIC PROTEIN 29 | BEL29 | Nucleic acid (mRNA) | Tuber development (inhibition) | Solanum tuberosum | [113] |
| MicroRNA 156 | miR156 | Nucleic acid (miRNA) | Tuber development (promotion) | Solanum tuberosum | [114] |
| MicroRNA 172 | miR172 | Nucleic acid (miRNA) | Tuber development (promotion) | Solanum tuberosum | [115] |
| POTATO HOMEOBOX 1 | POTH1 | Nucleic acid (mRNA) | Tuber development (promotion) | Solanum tuberosum | [116] |
| SELF-PRUNING 6A | SP6A | Protein | Tuber development/photoperiod (promotion) | Solanum tuberosum | [95] |
| Nutrient status | |||||
| MicroRNA 399 | miR399 | Nucleic acid (miRNA) | Phosphate starvation | Arabidopsis thaliana, Brassica napus, Cucurbita maxima | [117,118] |
| C-TERMINALLY ENCODED PEPTIDE (CEP) DOWNSTREAM 1 | CEPD1 | Protein | Nitrogen acquisition | Arabidopsis thaliana | [119] |
| CEP DOWNSTREAM 2 | CEPD2 | Protein | Nitrogen acquisition | Arabidopsis thaliana | [119] |
| CEPD-LIKE 2 | CEPDL2 | Protein | Nitrogen acquisition | Arabidopsis thaliana | [120] |
| ELONGATED HYPOCOTYL 5 | HY5 | Protein | Nitrogen acquisition and carbon fixation | Arabidopsis thaliana | [121] |
| Root development | |||||
| CYCLOPHILIN 1 | CYP1 | protein | Photosynthetic status/light intensity, root morphology/nutrient acquisition | Solanum lycopersicum | [122,123] |
| INDOLE-3-ACETIC ACID 18 | IAA18 | Ncleic acid (mRNA) | Auxin signaling, lateral root formation | Arabidopsis thaliana | [124] |
| INDOLE-3-ACETIC ACID 28 | IAA28 | Nucleic acid (mRNA) | Auxin signaling, lateral root formation | Arabidopsis thaliana | [124] |
| Auxin | - | Hormone | Gravitropism/halotropism | Arabidopsis thaliana | [41,125,126] |
| MicroRNA 2111 | miR2111 | nucleic acid (miRNA) | Nodulation | Lotus japonicus | [127] |
| Cytokinins | CK | Hormone | Nodulation | Arabidopsis thaliana, Lotus japonicus | [41–44,128] |
| Biotic stress | |||||
| DEFECTIVE IN INDUCED RESISTANCE 1 | DIR1 | Protein | Biotic stress, pathogen defense | Arabidopsis thaliana, Cucumis sativus | [79,129] |
| Dehydroabietinal | DHA | Lipid | Biotic stress, pathogen defense | Arabidopsis thaliana | [12,130,131,132] |
| Azelaic acid | AZA | Lipid | Biotic stress, pathogen defense | Arabidopsis thaliana | [12,133,129,131,134,132] |
| glycerol-3-phosphate | G3P | Lipid | Biotic stress, pathogen defense | Arabidopsis thaliana | [12,135,133,131,132] |
| ACYL-COA BINDING PROTEIN 3 | ACBP3 | protein | Biotic stress, pathogen defense | Arabidopsis thaliana | [136,137] |
| ACYL-COA BINDING PROTEIN 6 | ACBP6 | protein | Biotic stress, pathogen defense | Arabidopsis thaliana, Cucurbita maxima, Oryza sativa | [138,136,137] |
| 12-oxo-phytodienoic acid | OPDA | Lipid | Wounding, pathogen defense | Arabidopsis thaliana | [12,139,140,132] |
| Jasmonic acid | JA | Hormone | Wounding, pathogen defense | Arabidopsis thaliana, Nicotiana attenuata, Solanum lycopersicum | [139–144] |
| Calcium | Ca2+ | Ion/electrical signal | Wounding, pathogen defense | Arabidopsis thaliana | [145] |
| Abiotic stress | |||||
| Abscisic acid | ABA | Hormone | Water status | Glycine max, Xanthium strumarium, Ricinus communis, Citrus sinensis x Poncirus trifoliata, Solanum lycopersicum, Arabidopsis thaliana | [146–152] |
| CLAVATA3/EMBRYO-SURROUNDING REGION-RELATED 25 | CLE25 | Protein | Water status | Arabidopsis thaliana | [153] |
| PHLOEM LIPID ASSOCIATED FAMILY PROTEIN | PLAFP | Protein | Water status | Arabidopsis thaliana, Brassica oleracea | [12,132,154,155] |
| Phosphatidic acid | PtdOH | Lipid | Water status | Arabidopsis thaliana | [12,132,154,155] |
Species and references highlight studies in which the relevant signal is phloem-mobile and functions in the associated developmental process.
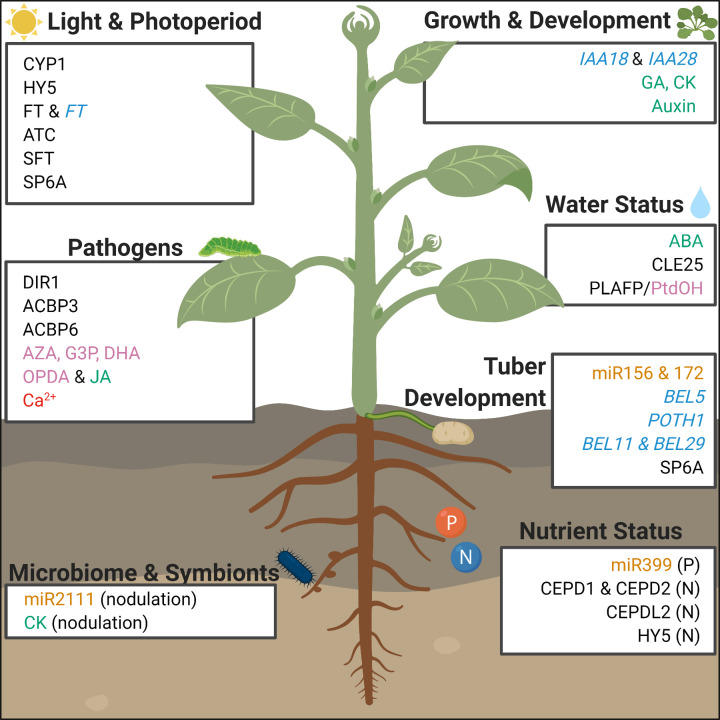
Table 1 lists all phloem-associated systemic signals by substance class with references and the plant species they were observed in. An overview of these phloem-associated systemic signals and their roles in plant development is provided in Figure 1.
Figure 1. Overview of long-distance signaling in the phloem.
Mobile signals involved in plant development and stress response are listed: proteins (all-caps; black), lipophilic compounds (pink), ions (red), miRNA (orange), mRNA (italicized; light blue), hormones (green). Abbreviations, plants species, and references for all compounds are detailed in Table 1.
The variation among phloem-localized macromolecules begs the question: how does this diversity of compounds—nucleic acids, proteins, hormones, lipids, ions, and their complexes—function in plant systems? What physiological demands dictate which molecular species is required?
Phloem-mobile macromolecules act in a variety of processes to coordinate plant development under both normal and challenging conditions. Long-distance signals influence aspects of plant morphology and physiology by participating in diverse genetic, biochemical, and even epigenetic mechanisms [101]. In the following sections, systemic signals in the phloem and their impact on the genetic and biochemical regulation of plant development are discussed. What will become obvious is that, often, not a single macromolecule but a combination of different signal classes can trigger the developmental response.
Transition to flowering: vegetative to generative growth
Major developmental shifts require the careful integration of external and internal cues. Long-distance coordination and systemic signaling have been linked to changes in leaf phenotypes including pinnation and lobe patterns, trichome development, GA-related leaf development, and palisade tissue shape to optimize light absorption [99, 100, 102–105]. Control of leaf morphology is among the first examples of phloem-mobile mRNAs influencing plant physiology. These foundational studies pointed to the ability of long-distance RNA transport to play a functional role in plant development.
One of the most important and dramatic developmental changes in plants is the transition from vegetative to generative growth. This transition is controlled by the photoperiod, which is sensed in the leaves. A mobile signal, often referred to as florigen, is required to transmit information about photoperiod from the leaves to the shoot apical meristem (SAM) where flower development can begin. Depending on the plant species, phytohormones like GA, CKs, and ethylene participate in the regulation of flowering, either independently, as phloem-mobile signals themselves, or in FT/CONSTANS (CO)-dependent mechanisms [106–110, 156]. In many plant species, the key florigenic signal is the protein FT. In Arabidopsis, long day conditions and circadian clock components promote the transcription of CONSTANS in the leaf vasculature, which then interacts with the FT promoter to initiate its expression [157]. Several studies have conclusively shown that FT protein or its homolog in other plants is mobile and is central to the transition to reproductive growth [77,78,83–94]. Upon arrival at the SAM, it interacts with the bZIP transcription factor FD to initiate expression of genes necessary for floral development [158]. Interestingly, specific Phosphatidylcholine (PtdCho) molecular species exhibit diurnal oscillations in the SAM; FT preferentially binds the species that predominates during the day. [59]. A recent structural characterization of Arabidopsis FT suggests PtdCho as a mediating ligand to facilitate FT/FD/14-3-3 protein interaction to form the Florigen Activation Complex (FAC) and its binding to DNA to promote flowering [60]. Conversely, Arabidopsis CENTRORADIALIS (ATC) may act to counterbalance FT activity for floral initiation. Like FT, ATC is expressed in vascular tissue and ATC is graft-transmissible, indicating long-distance mobility. ATC may compete with FT to interact with the FD, which then inhibits flowering [98].
Despite extensive evidence for FT protein, more recent research also suggested movement of FT mRNA. Non-translatable FT RNA is independently phloem-mobile, facilitates the movement of other RNAs, and can overcome SAM viral exclusion mechanisms [37,96,97]. The regulatory mechanisms for the transition from vegetative to reproductive growth are summarized in Figure 2.
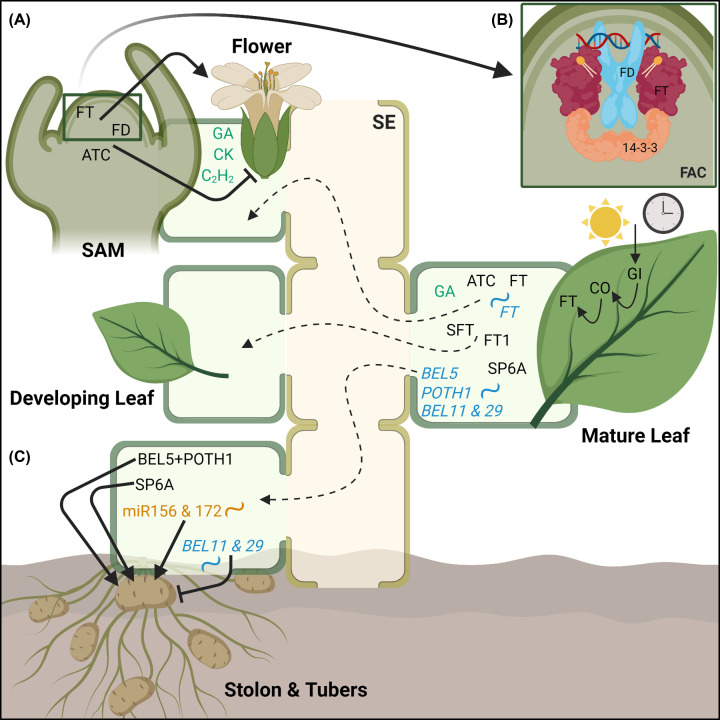
Figure 2. Phloem-mobile signals in shoot development.
(A) Transition to flowering: FT and its homologs are expressed in the leaves in a photoperiod-controlled mechanism through GIGANTEA (GI), other circadian clock components and CO. FT and FT-like proteins, as well as FT mRNA transcripts, are transported in the phloem to the SAM to promote the transition from vegetative to reproductive growth. FT interacts with FD to promote flowering, whereas ATC, a second mobile flowering regulator, competes with FT to inhibit flowering. (B) The FAC comprises FT, 14-3-3 protein, and the TF FD. Complex formation and binding to DNA is mediated by interaction with PtdCho, as shown by the cartoon of the proposed structure from Nakamura et al. [60]. Phytohormones GAs, CKs, and ethylene (C2H2) also play a role in flowering in some plant species. Other FT-like proteins, such as SFT (tomato) and FT1 (cotton), not only act in flowering but also in leaf maturation, termination, and abscission signaling. (C) Tuber formation: BEL5, 11, and 29 and POTH1 mRNA, as well as SP6A protein and miR156 and 172, move in the SE of the phloem to stolon tissue, where they regulate tuber development. A BEL5–POTH1 complex, SP6A, miR156 and miR172 promote tuber formation, whereas BEL11 and BEL29 suppress it. Abbreviations, plants species and references for all compounds are detailed in Table 1.
FT is pivotal for the transition from vegetative to generative growth. However, its role is not simply confined to the induction of flowering. FT and its homologs control additional seasonal and developmental responses (Figure 2). The mobile florigen hormone in tomato, SINGLE FLOWER TRUSS (SFT), affects leaf maturation, termination, and abscission [87,88,111]. Overexpression of the cotton FT-like gene GhFT1 promotes lateral shoot outgrowth and early flower abscission [93]. These studies, along with other examples, highlight FT and FT-like proteins as crucial not only in flowering but also in the overall balance between vegetative and reproductive growth and development [159].
Development of underground organs
Tuber development
Plants develop several bulbous underground storage organs. Some are derived from roots; others however, despite their location, are derived from specialized shoots. Tuber formation in potato is coordinated, in part, by day length-associated signals in above ground tissue. Several phloem-mobile transcripts, including multiple StBELs and POTATO HOMEOBOX 1 (POTH1) mRNAs as well as miR156 and 172, work in concert to regulate tuber formation in potato [112–116]. Facilitated by short-day induced RNA-binding proteins at its 3′ untranslated region, StBEL5 transcripts are transported from leaves to stolon, where the StBEL5–POTH1 complex induces gibberellic acid-, CK-, and auxin-related genes for tuber formation [33,112,160–163]. Phylogenetically related transcripts StBEL11 and StBEL29 mirror StBEL5 in phloem mobility and accumulation during short days; however StBEL11 and StBEL29 seem to act antagonistically to StBEL5 and reduce tuber yield [113]. These three transcripts may work in conjunction to balance cell growth during the transition from stolon to tuber in potatoes and coordinate tuber formation according to photoperiod by translocating in the phloem from leaves to stolon. An FT ortholog in potato, SELF-PRUNING 6A (StSP6A), is another promising phloem-mobile candidate for photoperiod-regulated systemic control of tuberization and may be a target of StBEL5 [95,164]. Mechanisms for tuber initiation and development, including these phloem-mobile players, are further evaluated and reviewed in [165–167]. Again, the interplay between multiple mobile species, mRNA, miRNA and proteins, defines converging paths and efficiency of long-distance signaling.
Root development and nutrient deficiency
Root development is regulated by the availability of nutrients from the soil as well as the abundance of energy carriers and building blocks generated above ground. Similarly, nutrient availability affects shoot development as well. Hence, plants tightly regulate nutrient sensing, acquisition, and allocation to maintain their health and productivity. Phloem transport plays an important role in the systemic signaling of nutrient levels. For example, iron content in phloem is determined, in part, by the loading activity of OLIGOPEPTIDE TRANSPORTER 3 (OPT3) and iron itself influences the expression of Fe uptake genes like IRON-REGULATED TRANSPORTER 1 (IRT1) and FERRIC REDUCTION OXIDASE 2 (FRO2) in the roots, which then modulates Fe distribution between source and sink tissues along with other, as of yet, unidentified shoot-borne signals [168–170]. Furthermore, limited sulfur, copper, iron, and phosphorus availability affects proteins, miRNA and other small RNA species in the phloem [171–173]. As such, mobile RNAs and peptides are involved in communicating nutrition status and coordinating mineral allocation systemically.
Under phosphate starvation, plants employ several mechanisms to maintain homeostasis, including lipid remodeling as well as reallocation of excess phosphate from the roots to developing tissues [174,175]. Under Pi-deplete conditions in shoots expression of miRNA 399 (miR399) increases. The miR399 then translocates in the phloem from shoot to root where it suppresses PHOSPHATE 2 (PHO2) expression, via the gene silencing mechanism. The absence of PHO2, an E2 ubiquitin-conjugating enzyme that represses phosphate uptake, allows for an increase in Pi loading into the xylem for transport to shoots (Figure 3, orange) [117, 118, 176–179]. In addition, under phosphate limited conditions, many mRNA transcripts, including hormone receptors, TFs, and Pi signaling genes like PHO2, as well as lncRNA transcripts like the target mimic of miRNA399 INDUCED BY PHOSPHATE STARVATION 1 (IPS1), are differentially expressed in source phloem and subsequently transported to sinks—developing leaves, the shoot apex, and root tips—in a tissue-specific manner [31,34,40]. While the exact mechanism and function of these mobile transcripts is still unknown, it is clear that a variety of RNA species are involved in the systemic early response to phosphate deficiency by coordinating nutrient perception between roots and shoots.
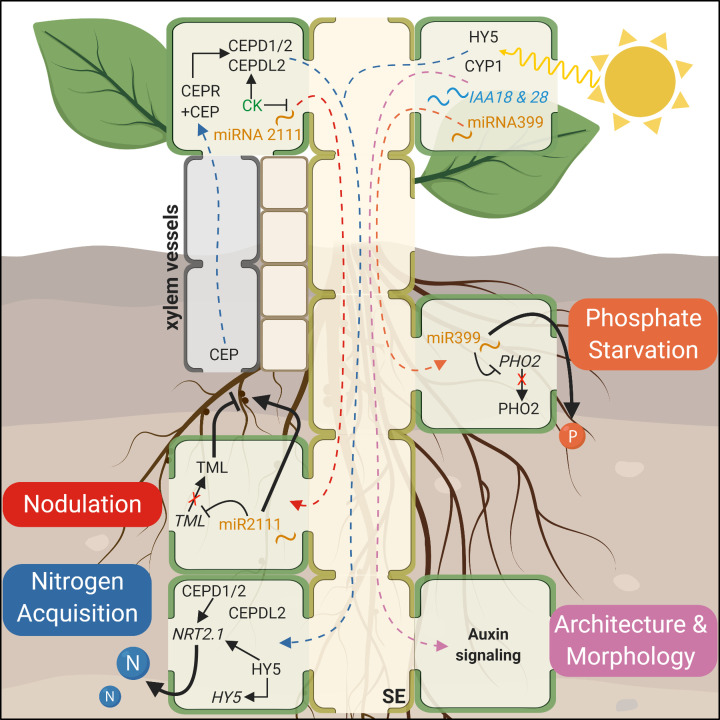
Figure 3. Phloem-mobile signals in root development.
Nodulation (red arrows): In unmodulated roots, shoot-derived miR2111 translocates to the roots, where it inhibits TML, thereby promoting susceptibility to nodulation. Following sufficient nodulation, a root-derived CK-dependent signal represses miR2111 production, which restricts further nodule formation. Nitrogen acquisition (blue arrows): Under nitrogen-starved conditions, CEP translocates in the xylem from roots to shoots, where it is sensed by its receptor, CEPR. The CEP–CEPR module promotes the production of CEPD1 and 2 proteins, and, together with CK, the expression of CEPDL2. These proteins are transported in the phloem to the roots to regulate the expression of the nitrogen transporters NRT2.1, thus influencing nitrogen uptake and transport. HY5 integrates carbon fixation and photosynthetic status with nutrient acquisition by translocating from leaves to roots, where it auto-activates its expression and promotes NRT2.1-mediated nitrogen uptake. During phosphate starvation (orange arrows), miR399 is expressed in the leaves and translocates in the SE to the roots, where it suppresses expression of PHO2, allowing an increase in phosphate uptake and loading. Architecture and root morphology (pink arrows): CYP1 and IAA 18 and 28 transcripts translocate from shoot to root, where they coordinate light and photosynthetic status in the leaves with root development and nutrient acquisition in the roots. Abbreviations, plants species and references for all compounds are detailed in Table 1.
Roots sense nitrogen availability in the soil and adjust their architecture to improve nutrient uptake. Systemic N-demand signaling involves a xylem-mobile peptide called C-TERMINALLY ENCODED PEPTIDE (CEP), which signals nitrogen starvation from the roots to the shoots [180,181]. CEP originates in the roots, accumulates in the vascular tissue in the leaf, and likely diffuses from the xylem into the phloem where it is sensed by CEP RECEPTOR (CEPR) protein. Downstream of the CEP-CEPR module, CEP DOWNSTREAM 1 and 2 (CEPD1 and CEPD2) are loaded into the phloem in the shoot and translocate to the roots. Once there, they increase the expression of NITRATE TRANSPORTER 2.1 (NRT2.1), but only if nitrate is available in the surrounding soil [119]. A third peptide, CEPD-LIKE 2 (CEPDL2) is expressed in the leaf vasculature in response to low nitrogen. In this case, both CKs and CEP appear to regulate gene expression. CEPDL2 then translocates to the root, where it affects both nitrate-uptake and transport through the regulation of the expression of several high-affinity nitrate transporters [120]. CEPD1/2 and CEPDL2 appear to be part of a dual N-sensing and response system, with CEPD1/2 responding to the root N status and CEPDL2 responding to the shoot status (Figure 3, blue). This root-to-shoot-to-root circuit utilizes both xylem and phloem to systemically coordinate nitrogen status across the whole plant.
Nitrogen acquisition in the roots is also informed by light cues and carbon fixation in the shoots. Multiple phloem mobile signals contribute to the integration of shoot-localized photosynthetic processes and nutrient uptake in the roots. The bZIP transcription factor ELONGATED HYPOCOTYL 5 (HY5), which regulates carbon assimilation in the shoots, translocates to the roots where it auto-activates its own expression [121]. The subsequent increase in HY5 TFs in the roots promotes NRT2.1 expression and potentiates NRT2.1-facilitated transport of nitrate from the soil [121]. Shoot-derived CYCLOPHILIN 1 (CYP1) in tomatoes integrates information about light intensity and photosynthetic status in the shoots with morphology and nutrient acquisition in the roots by influencing auxin response (Figure 3, pink) [122,123]. In addition, auxin directly controls root architecture as well. The transcripts of INDOLE-3-ACETIC ACID 18 (IAA18) and IAA28, which are expressed in leaf vasculature, were identified in Arabidopsis phloem and affect lateral root formation [124]. As has become clear in previous paragraphs, many developmental processes are not regulated by a single long-distance signal but rather an interplay of complementary signals, consisting of proteins, hormones, and nucleic acids.
Nodulation
Plants are unable to fix atmospheric nitrogen on their own, so some species, namely legumes, form specialized root organs called nodules that function to facilitate mutualistic interactions with nitrogen-fixing soil bacteria. Root nodulation and symbiosis with rhizobia is controlled, in part, by systemic signaling, including a phloem-mobile miRNA species, microRNA 2111 (miR2111), that regulates susceptibility (Figure 3, red). TOO MUCH LOVE (TML), the target of miR2111, restricts nodule formation [127]. However, in the absence of rhizobia, miR2111 is expressed in leaf CCs and translocates to the roots, where mature miR2111 post-transcriptionally regulates TML, which down-regulates genes that affect nodulation factor perception and thus inhibits repression of nodulation [182]. Following exposure to rhizobia, a CK-dependent signal from roots inhibits miR2111 production in the shoot, which allows TML to limit nodule formation [127,128]. This long-distance circuit balances infection and nodulation for successful symbiosis.
Biotics stress and systemic acquired resistance
Pathogen infection triggers both a local and a systemic response. The latter often involves Systemic Acquired Resistance (SAR), a mechanism triggered by immune responses to local pathogen infection, which then allows distal tissues in plants to become resistant. While researchers have yet to unambiguously determine the mobile signal(s) for SAR, several components have been identified that contribute to the coordination of pathogen and biotic stress response (Figure 4, right).
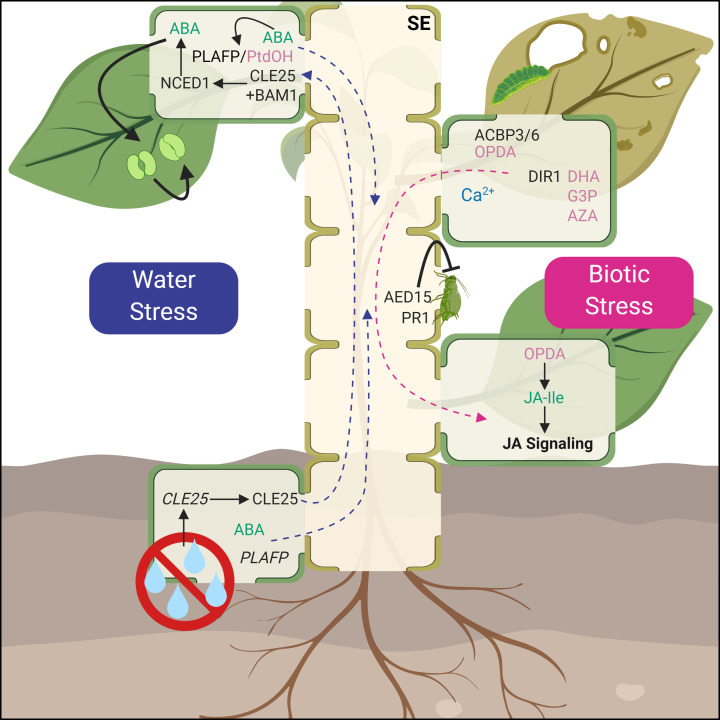
Figure 4. Phloem-mobile signals during water stress and biotic stress.
Water stress (blue arrows) signals, derived in both the roots and the shoots, are shown on the left. When water limitation is sensed in the roots, CLE25 translocates in the SE to leaf tissues and is perceived by BARELY ANY MERISTEM 1 (BAM1). The CLE25-BAM1 module promotes the expression NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3), which promotes synthesis of the phytohormone, ABA. ABA signaling regulates the closing of stomata (light green) in response to water stress. PLAFP and phosphatidic acid (PtdOH) are produced in response to ABA and may play a role in systemic drought response. Both ABA and PLAFP could be root- and/or shoot-derived. Biotic stress (magenta arrows; right half): AED15 and PR1 act directly as anti-herbivory or antimicrobial agents in phloem sap. Several proteins and lipophilic compounds, as well as electrical signals like Ca2+, are produced in wounded tissues and act in systemic signaling for resistance in distal tissues. ACBP 3 and 6 affect the fatty acid composition in the phloem, including oxylipins. The phloem-mobile, defense-response protein DIR1 may facilitate the transport of SAR signals like DHA into and through the phloem. Mobile signals such as DHA, AZA, or G3P derivatives move to distal tissues, where they can trigger defense response signals like JA signaling, involving the bioactive JA conjugate JA–Isoleucine (JA–Ile), and induce resistance systemically. Abbreviations, plants species, and references for all compounds are detailed in Table 1.
Comparative proteomics uncovered 16 proteins induced in phloem exudates after SAR induction. Conversely, the same study also found that 46 phloem proteins are suppressed under SAR [76]. Some of the identified SAR-responsive phloem proteins, like PATHOGENESIS RELATED 1 (PR1) and APOPLASTIC ENHANCED DISEASE SUSCEPTIBILITY 1-DEPENDENT 15 (AED15) are hypothesized to act as antimicrobial or anti-herbivory agents in phloem sap directly. However, other proteins are thought to be important for SAR long-distance signaling, either as signal carriers or as signals themselves [76]. Among these are several predicted lipid-interacting proteins, including DEFECTIVE IN INDUCED RESISTANCE 1 (DIR1), Acyl-CoA Binding Proteins (ACBPs), and a major latex-like protein (MLP). In addition, several lipophilic compounds such as dehydroabietinal (DHA), azelaic acid (AZA), and glycerol-3-phosphate (G3P) derivatives have been found to play a role in SAR [12,129, 130, 133, 135]. The accumulation of these proteins in the phloem during SAR suggests a role for lipid-binding proteins in conveying a lipophilic compound as part of the SAR signal.
DIR1 is a lipid transfer protein that has been shown to be phloem mobile during SAR and is required for systemic resistance [79,183]. While DIR1 alone does not induce resistance in distant tissues, it may be required for the transport of other SAR signals like DHA [131]. Additionally, AZELAIC ACID INDUCED 1 (AZI1) and EARLY ARABIDOPSIS ALUMINUM INDUCED 1 (EARLI1) may play a role in the loading and transport of AZA as a mobile SAR signal [134].
Thioredoxin H (TRXH) proteins have been identified in phloem sap during SAR in Arabidopsis as well as other plants species like rice [76,138]. While the function of TRXH in the phloem is unknown, it has been linked to pathogen immunity by regulating the oligomerization of NONEXPRESSER OF PR GENES 1 (NPR1) [184]. It is possible that the phloem-localized TRXH protein could act to regulate other mobile SAR signals, like DIR1.
Among the phloem-localized, wound-response genes are acyl-CoA binding proteins ACBP3 and ACBP6. ACBP6 is expressed in CCs in response to wounding, and the protein is found in phloem exudates [136]. The same is true for ACBP3. Grafting studies indicate that ACBP3 is phloem-mobile and moves from shoots to roots. Furthermore, the absence of ACBP3 impairs defense response in both locally wounded and distal tissue [137]. ACBPs play a role in maintaining acyl-CoA pools and lipid metabolism. Along these same lines, both ACBP3 and ACBP6 affect the fatty acid composition of the phloem, more specifically a group of defense-related fatty acids, the oxylipins such as 12-oxo-phytodienoic acid (OPDA) and methyl jasmonate [137]. These oxylipins have long been suggested as mobile signals for wounding, pathogenesis, and SAR [139, 141–143]. Jasmonate is involved both locally and distally for the transmission and perception of systemic wounding signals [144]. The JA precursor OPDA and its derivatives have been shown to translocate via the phloem from wounded shoots to undamaged root tissue, where these precursors are converted into bioactive JA-Ile for systemic defense response [12,140].
As was seen in the previously discussed long-distance signaling paths, a combination of compounds, in this case proteins, small molecules and lipids, are essential to mediate a response. Their exact interplay remains to be determined. In addition, beyond chemical signals, electrical signaling appears to contribute to systemic wound signaling as well. A glutamate/glutamate receptor/calcium ion module acts to propagate signals from local to distal leaves. In this mechanism, Ca2+ ions participate as a phloem mobile signal to coordinate wound response systemically [145].
Water stress and ABA signaling
Systemic signaling for abiotic stress is understudied compared with biotic stress responses like SAR. Nonetheless, hormones, proteins, and peptides in the phloem seem to play an important role in the coordination of systemic abiotic stress response as well (Figure 4, left). One crucial environmental factor/stress is the availability of water. We can distinguish between different water stresses: too much, too little, or inaccessibility due to high salt/osmotic properties of the soil or freezing temperatures. Whether water stress perception predominates in the roots or the leaves is an active area of research [185].
ABA is a phytohormone produced in response to drought-, salt-, osmotic-, and freezing-based water limitation as well as for seed and root development, acting both locally and systemically [186]. The long-distance transport of ABA has been attributed to both the xylem [187–190] and the phloem [146–149], with reports of both root- and shoot-derived ABA pools during water stress [150–152]. One of the water-stress responses, the ABA-mediated stomatal closure is regulated, in part, by a root-derived, vascular-mobile CLAVATA3/EMBRYO-SURROUNDING REGION-RELATED (CLE) protein CLE25. When dehydration is sensed in the roots, CLE25 is expressed in root vascular tissue, and the protein is transported to leaves where it is sensed by BARELY ANY MERISTEM 1 (BAM1) and 3 receptor-like kinases and affects the expression of an ABA synthesis enzyme NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3) [153]. Whether the root-to-shoot translocation of CLE25 occurs in xylem or phloem remains to be determined [191] The increase in ABA is sensed by its receptor PYRABACTIN RESISTANCE (PYR)/PYR-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTOR (RCAR), and the protein phosphatase 2C enzyme ABSCISIC ACID INSENSITIVE 1 (ABI1) is sequestered, in part, by PHOSPHOLIPASE Dα1-derived Phosphatidic acid (PtdOH) [192]. With ABI1 no longer available to dephosphorylate and repress SNF1-RELATED KINASE 2 (SnRK2) activity, SnRK2s can then phosphorylate ABA-Response Element (ABRE) Binding/ABRE Binding Factors (AREB/ABF) TFs, which initiate the expression of drought-responsive genes by interacting with ABA response elements in promoter regions. These findings indicate that both, the hormone ABA as well as the protein CLE25 are long-distance signals mediating the water-stress response. Alternatively, hydraulic signals have been proposed instead of root-derived ABA as an indicator of water limitation [193].
Recent data suggest that protein–lipid complexes may be involved in drought signaling as well. Several small lipid-binding proteins have been identified in the phloem and were proposed to play a role in long-distance signaling [12,132,154]. PHLOEM LIPID ASSOCIATED FAMILY PROTEIN (PLAFP) may be involved in drought response and ABA signaling. PLAFP is expressed in the vasculature. Its expression increases in response to treatment with ABA as well as the drought mimic polyethylene glycol [154]. PLAFP interacts specifically with PtdOH, a known stress signal involved in ABA signaling and detected in phloem exudate [12,192,154, 194–196]. These data suggest the PLAFP–PtdOH complex as a possible phloem-mobile signal for the systemic coordination of drought response [132,155].
These findings further substantiate previous results that long distance signaling, rather than relying on single molecules, often encompasses multiple converging pathways and the interaction between proteins, RNA, small molecules, and even lipids.
Conclusion
The last few decades have shown that the plant phloem is far more complex than originally anticipated: in addition to sugars, it contains a vast array of small molecules, hormones, proteins, lipids, and nucleic acids that facilitate tightly coordinated developmental and stress responses. The heterogeneity of systemic-driven processes requires a diversity of phloem-associated molecular species. The examples discussed in this review aim to highlight this multiplicity: it is not a single molecule or class of molecules that predominates long-distance signaling, but rather a diverse arsenal of signal classes interact to systemically coordinate plant development. The role of these, often multipronged, signaling processes remains to be resolved—perhaps they confer redundancy to ensure some minimum response or modularity to fine-tune the intensity and specificity of the response. While much has been learned, phloem signaling continues to be an important field of active research essential not only to understand how plants respond to changes in the environment but also to ensure food security.
Acknowledgements
Figures were generated with the professional version of BioRender.com. Colors were adjusted to accommodate readers with color-perception issues using the following simulator: https://www.color-blindness.com/coblis-color-blindness-simulator/
Abbreviations
- AAP
amino acid permease
- ABA
abscisic acid
- ABF
ABA-response element binding factor
- ABI1
abscisic acid insensitive 1
- ABRE
ABA-response element
- ACBP
Acyl-CoA binding protein
- AED15
apoplastic enhanced disease susceptibility 1-dependent 15
- AREB
ABA-Response Element Binding
- ATC
arabidopsis centroradialis
- AZA
azelaic acid
- AZI1
aza induced 1
- BAM1
barely any meristem 1
- BEL
bel 1-related homeotic proteins
- CC
companion cell
- CEP
c-terminally encoded peptide
- CEPD
cep downstream
- CEPDL2
cepd-like 2
- CEPR
cep receptor
- CK
cytokinin
- CLE25
clavata3/emryo-surrounding region-related
- CO
constans
- CYP1
cyclophilin 1
- DHA
dehydroabietinal
- DIR1
defective in induced resistance 1
- FAC
florigen activation complex
- FRO2
ferric reduction oxidase 2
- FT
flowering locus T
- FTIP1
FT interacting protein 1
- G3P
glycerol-3-phosphate
- GA
gibberellin
- HY5
elongated hypocotyl 5
- IAA
indole-3-acetic acid
- IPS1
induced by phosphate starvation 1
- IRT1
iron-regulated transporter 1
- JA
jasmonic acid
- MCTP-SNARE
multiple C2 domain and transmembrane protein-soluble N-ethylmaleimide-sensitive factor protein attachment protein receptor
- miR2111
microRNA 2111
- miR399
microRNA 399
- miRNA
microRNA
- MLP
major latex-like protein
- NCED3
nine-cis-epoxycarotenoid dioxygenase 3
- ncRNA
non-coding RNA
- NPR1
nonexpressor of pr genes 1
- NRT2.1
nitrate transporter 2.1
- OPDA
12-oxo-phytodienoic acid
- OPT3
oligopeptide transporter 3
- PD
plasmodesmata
- PHO2
phosphate 2
- PLAFP
phloem lipid-associated family protein
- POTH1
potato homeobox 1
- PR1
pathogenesis related 1
- PtdCho
phosphatidylcholine
- PtdOH
phosphatidic acid
- PYR/PYL/RCAR
pyrabactin resistance/pyr-like/regulatory components of aba receptor
- QKY
quirky
- SAM
shoot apical meristem
- SAR
systemic acquired resistance
- SE
sieve element
- SFT
single flower truss
- siRNA
small interfering RNA
- SnRK2
snif1-related kinase 2
- StSP6A
solanum tuberosum self-pruning 6A
- SUT
sucrose transporter
- SWEET
sugar will eventually be exported transporter
- SYP121
syntaxin of plants 121
- TF
transcription factor
- TML
too much love
- TRXH
thioredoxin H
- UMAMIT
usually multiple acids move in and out transporter
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported, in part, by the USDA-NIMMS [grant number NC1203]; the NSF [grant number #1841251 (to S.H.B.)]; and the USDA-NIFA [grant number NNF 2015-38420-23697 (to A.M.K.)].
References
- 1.Muench E. (1930) Die Stoffbewegungen in der Pflanze, Fischer, Jena, Germany [Google Scholar]
- 2.Zhang C. and Turgeon R. (2018) Mechanisms of phloem loading. Curr. Opin. Plant Biol. 43, 71–75 10.1016/j.pbi.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 3.Turgeon R. (2010) The role of phloem loading reconsidered. Plant Physiol. 152, 1817–1823 10.1104/pp.110.153023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turgeon R. and Wolf S. (2009) Phloem transport: cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 60, 207–221 [DOI] [PubMed] [Google Scholar]
- 5.Zhang C., Han L., Slewinski T.L., Sun J., Zhang J., Wang Z.Y. et al. (2014) Symplastic phloem loading in poplar. Plant Physiol. 166, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross-Elliott T.J., Jensen K.H., Haaning K.S., Wager B.M., Knoblauch J., Howell A.H. et al. (2017) Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. Elife 6, e24125 10.7554/eLife.24125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truernit E. (2017) Plant physiology: unveiling the dark side of phloem translocation. Curr. Biol. 27, R348–R350 [DOI] [PubMed] [Google Scholar]
- 8.Tegeder M. and Hammes U.Z. (2018) The way out and in: phloem loading and unloading of amino acids. Curr. Opin. Plant Biol. 43, 16–21 [DOI] [PubMed] [Google Scholar]
- 9.Troncoso-Ponce M.A., Cao X., Yang Z. and Ohlrogge J.B. (2013) Lipid turnover during senescence. Plant Sci 205–206, 13–19 [DOI] [PubMed] [Google Scholar]
- 10.Lucas W.J., Groover A., Lichtenberger R., Furuta K., Yadav S.R., Helariutta Y. et al. (2013) The plant vascular system: evolution, development and functions. J. Integr. Plant Biol. 55, 294–388 10.1111/jipb.12041 [DOI] [PubMed] [Google Scholar]
- 11.Atkins C.A., Smith P.M. and Rodriguez-Medina C. (2011) Macromolecules in phloem exudates–a review. Protoplasma 248, 165–172 10.1007/s00709-010-0236-3 [DOI] [PubMed] [Google Scholar]
- 12.Guelette B.S., Benning U.F. and Hoffmann-Benning S. (2012) Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 63, 3603–3616 10.1093/jxb/ers028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ham B.K. and Lucas W.J. (2017) Phloem-mobile RNAs as systemic signaling agents. Annu. Rev. Plant Biol. 68, 173–195 10.1146/annurev-arplant-042916-041139 [DOI] [PubMed] [Google Scholar]
- 14.Liu L. and Chen X. (2018) Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4, 869–878 10.1038/s41477-018-0288-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehr J. and Kragler F. (2018) Long distance RNA movement. New Phytol. 218, 29–40 10.1111/nph.15025 [DOI] [PubMed] [Google Scholar]
- 16.Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G. and Hedrich R. (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55, 746–759 10.1111/j.1365-313X.2008.03555.x [DOI] [PubMed] [Google Scholar]
- 17.Sasaki T., Chino M., Hayashi H. and Fujiwara T. (1998) Detection of several mRNA species in rice phloem sap. Plant Cell Physiol. 39, 895–897 10.1093/oxfordjournals.pcp.a029451 [DOI] [PubMed] [Google Scholar]
- 18.Doering-Saad C., Newbury H.J., Bale J.S. and Pritchard J. (2002) Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J. Exp. Bot. 53, 631–637 10.1093/jexbot/53.369.631 [DOI] [PubMed] [Google Scholar]
- 19.Gaupels F., Buhtz A., Knauer T., Deshmukh S., Waller F., van Bel A.J. et al. (2008) Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J. Exp. Bot. 59, 3297–3306 10.1093/jxb/ern181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz-Medrano R., Xoconostle-Cazares B. and Lucas W.J. (1999) Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126, 4405–4419 [DOI] [PubMed] [Google Scholar]
- 21.Omid A., Keilin T., Glass A., Leshkowitz D. and Wolf S. (2007) Characterization of phloem-sap transcription profile in melon plants. J. Exp. Bot. 58, 3645–3656 10.1093/jxb/erm214 [DOI] [PubMed] [Google Scholar]
- 22.Doering-Saad C., Newbury H.J., Couldridge C.E., Bale J.S. and Pritchard J. (2006) A phloem-enriched cDNA library from Ricinus: insights into phloem function. J. Exp. Bot. 57, 3183–3193 10.1093/jxb/erl082 [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Medina C., Atkins C.A., Mann A.J., Jordan M.E. and Smith P.M. (2011) Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.). BMC Plant Biol. 11, 36 10.1186/1471-2229-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo S., Zhang J., Sun H., Salse J., Lucas W.J., Zhang H. et al. (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45, 51–58 10.1038/ng.2470 [DOI] [PubMed] [Google Scholar]
- 25.Notaguchi M., Higashiyama T. and Suzuki T. (2015) Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol. 56, 311–321 10.1093/pcp/pcu210 [DOI] [PubMed] [Google Scholar]
- 26.Thieme C.J., Rojas-Triana M., Stecyk E., Schudoma C., Zhang W., Yang L. et al. (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1, 15025 10.1038/nplants.2015.25 [DOI] [PubMed] [Google Scholar]
- 27.Yoo B.C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y.M. et al. (2004) A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000 10.1105/tpc.104.023614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolstyko E., Lezzhov A. and Solovyev A. (2019) Identification of miRNA precursors in the phloem of Cucurbita maxima. PeerJ 7, e8269 10.7717/peerj.8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S., Sun L. and Kragler F. (2009) The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150, 378–387 10.1104/pp.108.134767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gai Y.P., Zhao H.N., Zhao Y.N., Zhu B.S., Yuan S.S., Li S. et al. (2018) MiRNA-seq-based profiles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 8, 812 10.1038/s41598-018-19210-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Zheng Y., Ham B.K., Chen J., Yoshida A., Kochian L.V. et al. (2016) Vascular-mediated signalling involved in early phosphate stress response in plants. Nat. Plants 2, 16033 10.1038/nplants.2016.33 [DOI] [PubMed] [Google Scholar]
- 32.Calderwood A., Kopriva S. and Morris R.J. (2016) Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell 28, 610–615 10.1105/tpc.15.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A.K., Lin T. and Hannapel D.J. (2009) Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol. 151, 1831–1843 10.1104/pp.109.144428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z., Zheng Y., Ham B.K., Zhang S., Fei Z. and Lucas W.J. (2019) Plant lncRNAs are enriched in and move systemically through the phloem in response to phosphate deficiency. J. Integr. Plant Biol. 61, 492–508 10.1111/jipb.12715 [DOI] [PubMed] [Google Scholar]
- 35.Ham B.K., Brandom J.L., Xoconostle-Cazares B., Ringgold V., Lough T.J. and Lucas W.J. (2009) A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21, 197–215 10.1105/tpc.108.061317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris R.J. (2018) On the selectivity, specificity and signalling potential of the long-distance movement of messenger RNA. Curr. Opin. Plant Biol. 43, 1–7 10.1016/j.pbi.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 37.Li C., Zhang K., Zeng X., Jackson S., Zhou Y. and Hong Y. (2009) A cis element within flowering locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. J. Virol. 83, 3540–3548 10.1128/JVI.02346-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y., Ham B.K., Chong Y.H., Yeh S.D. and Lucas W.J. (2020) A plant SMALL RNA-BINDING PROTEIN 1 family mediates cell-to-cell trafficking of RNAi signals. Mol. Plant 13, 321–335 10.1016/j.molp.2019.12.001 [DOI] [PubMed] [Google Scholar]
- 39.Zhang W., Thieme C.J., Kollwig G., Apelt F., Yang L., Winter N. et al. (2016) tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28, 1237–1249 10.1105/tpc.15.01056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walther D. and Kragler F. (2016) Limited phosphate: mobile RNAs convey the message. Nat. Plants 2, 16040 10.1038/nplants.2016.40 [DOI] [PubMed] [Google Scholar]
- 41.Hoad G.V. (1995) Transport of hormones in the phloem of higher plants. Plant Growth Regul. 16, 173–182 10.1007/BF00029538 [DOI] [Google Scholar]
- 42.Hoffmann-Benning S. (2015) Transport and function of lipids in the plant phloem. AOCS Lipid Library http://lipidlibrary.aocs.org/Biochemistry/content.cfm?ItemNumber=41357 [Google Scholar]
- 43.Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H. and Sakakibara H. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83 10.1093/jxb/erm157 [DOI] [PubMed] [Google Scholar]
- 44.Kudo T., Kiba T. and Sakakibara H. (2010) Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52, 53–60 10.1111/j.1744-7909.2010.00898.x [DOI] [PubMed] [Google Scholar]
- 45.Benning U., Tamot B., Guelette B. and Hoffmann-Benning S. (2012) New aspects of phloem-mediated long-distance lipid signaling in plants. Front. Plant Sci. 3, 53 10.3389/fpls.2012.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valim M.F. and Killiny N. (2017) Occurrence of free fatty acids in the phloem sap of different citrus varieties. Plant Signal. Behav. 12, e1327497 10.1080/15592324.2017.1327497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koo A.J., Gao X., Jones A.D. and Howe G.A. (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986 10.1111/j.1365-313X.2009.03924.x [DOI] [PubMed] [Google Scholar]
- 48.Ruan J., Zhou Y., Zhou M., Yan J., Khurshid M., Weng W. et al. (2019) Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20, 2479–2494 10.3390/ijms20102479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glatz J.F., Borchers T., Spener F. and van der Vusse G.J. (1995) Fatty acids in cell signalling: modulation by lipid binding proteins. Prostaglandins Leukot. Essent. Fatty Acids 52, 121–127 10.1016/0952-3278(95)90010-1 [DOI] [PubMed] [Google Scholar]
- 50.Charbonneau D., Beauregard M. and Tajmir-Riahi H.A. (2009) Structural analysis of human serum albumin complexes with cationic lipids. J. Phys. Chem. B 113, 1777–1784 10.1021/jp8092012 [DOI] [PubMed] [Google Scholar]
- 51.Blaner W.S. (1989) Retinol-binding protein: the serum transport protein for vitamin A. Endocr. Rev. 10, 308–316 10.1210/edrv-10-3-308 [DOI] [PubMed] [Google Scholar]
- 52.Natanson A.O. and Kon I. (1979) Vitamin A transport in the blood: retinol-binding serum protein and its biological role. Vopr. Pitan. 3–12 [PubMed] [Google Scholar]
- 53.Tontonoz P., Hu E. and Spiegelman B.M. (1994) Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79, 1147–1156 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- 54.Wahli W. and Michalik L. (2012) PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 23, 351–363 10.1016/j.tem.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 55.Nagy L. and Szanto A. (2005) Roles for lipid-activated transcription factors in atherosclerosis. Mol. Nutr. Food Res. 49, 1072–1074 10.1002/mnfr.200500097 [DOI] [PubMed] [Google Scholar]
- 56.Musille P.M., Kohn J.A. and Ortlund E.A. (2013) Phospholipid–driven gene regulation. FEBS Lett. 587, 1238–1246 10.1016/j.febslet.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musille P.M., Pathak M., Lauer J.L., Hudson W.H., Griffin P.R. and Ortlund E.A. (2012) Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nat. Struct. Mol. Biol. 19, 532–S2 10.1038/nsmb.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janda C.Y., Waghray D., Levin A.M., Thomas C. and Garcia K.C. (2012) Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 10.1126/science.1222879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura Y., Andres F., Kanehara K., Liu Y.C., Dormann P. and Coupland G. (2014) Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 5, 3553 10.1038/ncomms4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura Y., Lin Y.C., Watanabe S., Liu Y.C., Katsuyama K., Kanehara K. et al. (2019) High-resolution crystal structure of Arabidopsis FLOWERING LOCUS T illuminates its phospholipid-binding site in flowering. iScience 21, 577–586 10.1016/j.isci.2019.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S.C., Nusinow D.A., Sorkin M.L., Pruneda-Paz J. and Wang X. (2019) Interaction and regulation between lipid mediator phosphatidic acid and circadian clock regulators. Plant Cell 31, 399–416 10.1105/tpc.18.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao H., Wang G., Guo L. and Wang X. (2013) Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell 25, 5030–5042 10.1105/tpc.113.120162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai G., Kim S.C., Li J., Zhou Y. and Wang X. (2020) Transcriptional regulation of lipid catabolism during seedling establishment. Mol. Plant 13, 984–1000 10.1016/j.molp.2020.04.007 [DOI] [PubMed] [Google Scholar]
- 64.Giavalisco P., Kapitza K., Kolasa A., Buhtz A. and Kehr J. (2006) Towards the proteome of Brassica napus phloem sap. Proteomics 6, 896–909 10.1002/pmic.200500155 [DOI] [PubMed] [Google Scholar]
- 65.Lin M.K., Lee Y.J., Lough T.J., Phinney B.S. and Lucas W.J. (2009) Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol. Cell. Proteomics 8, 343–356 10.1074/mcp.M800420-MCP200 [DOI] [PubMed] [Google Scholar]
- 66.Carella P., Wilson D.C., Kempthorne C.J. and Cameron R.K. (2016) Vascular sap proteomics: providing insight into long-distance signaling during stress. Front. Plant Sci. 7, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anstead J.A., Hartson S.D. and Thompson G.A. (2013) The broccoli (Brassica oleracea) phloem tissue proteome. BMC Genomics 14, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez-Cobollo R.M., Filippis I., Bennett M.H. and Turnbull C.G. (2016) Comparative proteomics of cucurbit phloem indicates both unique and shared sets of proteins. Plant J. 88, 633–647 [DOI] [PubMed] [Google Scholar]
- 69.Oparka K.J. and Cruz S.S. (2000) THE GREAT ESCAPE: phloem transport and unloading of macromolecules1. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 323–347 [DOI] [PubMed] [Google Scholar]
- 70.Paultre D.S.G., Gustin M.P., Molnar A. and Oparka K.J. (2016) Lost in transit: long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell 28, 2016–2025 10.1105/tpc.16.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulz A. (2017) Long-distance trafficking: lost in transit or stopped at the gate? Plant Cell 29, 426–430 10.1105/tpc.16.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spiegelman Z., Golan G. and Wolf S. (2013) Don’t kill the messenger: long-distance trafficking of mRNA molecules. Plant Sci. 213, 1–8 10.1016/j.plantsci.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 73.Lough T.J. and Lucas W.J. (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57, 203–232 10.1146/annurev.arplant.56.032604.144145 [DOI] [PubMed] [Google Scholar]
- 74.Aoki K., Suzui N., Fujimaki S., Dohmae N., Yonekura-Sakakibara K., Fujiwara T. et al. (2005) Destination-selective long-distance movement of phloem proteins. Plant Cell 17, 1801–1814 10.1105/tpc.105.031419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stahl Y. and Faulkner C. (2016) Receptor complex mediated regulation of symplastic traffic. Trends Plant Sci. 21, 450–459 10.1016/j.tplants.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 76.Carella P., Merl-Pham J., Wilson D.C., Dey S., Hauck S.M., Vlot A.C. et al. (2016) Comparative proteomics analysis of phloem exudates collected during the induction of systemic acquired resistance. Plant Physiol. 171, 1495–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I. et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- 78.Turck F., Fornara F. and Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 10.1146/annurev.arplant.59.032607.092755 [DOI] [PubMed] [Google Scholar]
- 79.Champigny M.J., Isaacs M., Carella P., Faubert J., Fobert P.R. and Cameron R.K. (2013) Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 4, 230 10.3389/fpls.2013.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu L., Liu C., Hou X., Xi W., Shen L., Tao Z. et al. (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 10, e1001313 10.1371/journal.pbio.1001313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu L., Li C., Teo Z.W.N., Zhang B. and Yu H. (2019) The MCTP-SNARE complex regulates florigen transport in Arabidopsis. Plant Cell 31, 2475–2490 10.1105/tpc.18.00960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoo S.C., Chen C., Rojas M., Daimon Y., Ham B.K., Araki T. et al. (2013) Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J. 75, 456–468 10.1111/tpj.12213 [DOI] [PubMed] [Google Scholar]
- 83.Mathieu J., Warthmann N., Kuttner F. and Schmid M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060 10.1016/j.cub.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 84.Jaeger K.E. and Wigge P.A. (2007) FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17, 1050–1054 10.1016/j.cub.2007.05.008 [DOI] [PubMed] [Google Scholar]
- 85.Lin M.K., Belanger H., Lee Y.J., Varkonyi-Gasic E., Taoka K., Miura E. et al. (2007) FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19, 1488–1506 10.1105/tpc.107.051920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tamaki S., Matsuo S., Wong H.L., Yokoi S. and Shimamoto K. (2007) Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036 [DOI] [PubMed] [Google Scholar]
- 87.Lifschitz E. and Eshed Y. (2006) Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57, 3405–3414 [DOI] [PubMed] [Google Scholar]
- 88.Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z. et al. (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U.S.A. 103, 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meng X., Muszynski M.G. and Danilevskaya O.N. (2011) The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23, 942–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kong F., Liu B., Xia Z., Sato S., Kim B.M., Watanabe S. et al. (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun H., Jia Z., Cao D., Jiang B., Wu C., Hou W. et al. (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 6, e29238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harig L., Beinecke F.A., Oltmanns J., Muth J., Muller O., Ruping B. et al. (2012) Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 72, 908–921 10.1111/j.1365-313X.2012.05125.x [DOI] [PubMed] [Google Scholar]
- 93.Li C., Zhang Y., Zhang K., Guo D., Cui B., Wang X. et al. (2015) Promoting flowering, lateral shoot outgrowth, leaf development, and flower abscission in tobacco plants overexpressing cotton FLOWERING LOCUS T (FT)-like gene GhFT1. Front. Plant Sci. 6, 454 10.3389/fpls.2015.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A. et al. (2008) Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49, 1645–1658 10.1093/pcp/pcn154 [DOI] [PubMed] [Google Scholar]
- 95.Navarro C., Abelenda J.A., Cruz-Oro E., Cuellar C.A., Tamaki S., Silva J. et al. (2011) Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122 10.1038/nature10431 [DOI] [PubMed] [Google Scholar]
- 96.Li C., Gu M., Shi N., Zhang H., Yang X., Osman T. et al. (2011) Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Sci. Rep. 1, 73 10.1038/srep00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jackson S.D. and Hong Y. (2012) Systemic movement of FT mRNA and a possible role in floral induction. Front. Plant Sci. 3, 127 10.3389/fpls.2012.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang N.C., Jane W.N., Chen J. and Yu T.S. (2012) Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J. 72, 175–184 10.1111/j.1365-313X.2012.05076.x [DOI] [PubMed] [Google Scholar]
- 99.Gomi K. and Matsuoka M. (2003) Gibberellin signalling pathway. Curr. Opin. Plant Biol. 6, 489–493 10.1016/S1369-5266(03)00079-7 [DOI] [PubMed] [Google Scholar]
- 100.Huntley R.P., Jones L.H. and Hanke D.E. (2002) Cytokinins and gibberellins in sap exudate of the oil palm. Phytochemistry 60, 117–127 10.1016/S0031-9422(02)00099-7 [DOI] [PubMed] [Google Scholar]
- 101.Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R. and Baulcombe D.C. (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875 10.1126/science.1187959 [DOI] [PubMed] [Google Scholar]
- 102.Kim M., Canio W., Kessler S. and Sinha N. (2001) Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289 10.1126/science.1059805 [DOI] [PubMed] [Google Scholar]
- 103.Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P. et al. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 10.1101/gad.11.23.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haywood V., Yu T.S., Huang N.C. and Lucas W.J. (2005) Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68 10.1111/j.1365-313X.2005.02351.x [DOI] [PubMed] [Google Scholar]
- 105.Munekage Y.N., Inoue S., Yoneda Y. and Yokota A. (2015) Distinct palisade tissue development processes promoted by leaf autonomous signalling and long-distance signalling in Arabidopsis thaliana. Plant Cell Environ. 38, 1116–1126 10.1111/pce.12466 [DOI] [PubMed] [Google Scholar]
- 106.Turnbull C. (2011) Long-distance regulation of flowering time. J. Exp. Bot. 62, 4399–4413 10.1093/jxb/err191 [DOI] [PubMed] [Google Scholar]
- 107.Eriksson S., Bohlenius H., Moritz T. and Nilsson O. (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18, 2172–2181 10.1105/tpc.106.042317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jang S., Torti S. and Coupland G. (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60, 614–625 10.1111/j.1365-313X.2009.03986.x [DOI] [PubMed] [Google Scholar]
- 109.King R.W., Moritz T., Evans L.T., Junttila O. and Herlt A.J. (2001) Long-day induction of flowering in Lolium temulentum involves sequential increases in specific gibberellins at the shoot apex. Plant Physiol. 127, 624–632 10.1104/pp.010378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hisamatsu T. and King R.W. (2008) The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot. 59, 3821–3829 10.1093/jxb/ern232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y. et al. (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. U.S.A. 106, 8392–8397 10.1073/pnas.0810810106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Banerjee A.K., Chatterjee M., Yu Y., Suh S.G., Miller W.A. and Hannapel D.J. (2006) Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457 10.1105/tpc.106.042473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ghate T.H., Sharma P., Kondhare K.R., Hannapel D.J. and Banerjee A.K. (2017) The mobile RNAs, StBEL11 and StBEL29, suppress growth of tubers in potato. Plant Mol. Biol. 93, 563–578 10.1007/s11103-016-0582-4 [DOI] [PubMed] [Google Scholar]
- 114.Bhogale S., Mahajan A.S., Natarajan B., Rajabhoj M., Thulasiram H.V. and Banerjee A.K. (2014) MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 164, 1011–1027 10.1104/pp.113.230714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin A., Adam H., Diaz-Mendoza M., Zurczak M., Gonzalez-Schain N.D. and Suarez-Lopez P. (2009) Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136, 2873–2881 10.1242/dev.031658 [DOI] [PubMed] [Google Scholar]
- 116.Mahajan A., Bhogale S., Kang I.H., Hannapel D.J. and Banerjee A.K. (2012) The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Mol. Biol. 79, 595–608 10.1007/s11103-012-9931-0 [DOI] [PubMed] [Google Scholar]
- 117.Lin S.I., Chiang S.F., Lin W.Y., Chen J.W., Tseng C.Y., Wu P.C. et al. (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling1[W][OA]. Plant Physiol. 147, 732–746 10.1104/pp.108.116269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pant B.D., Buhtz A., Kehr J. and Scheible W.R. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738 10.1111/j.1365-313X.2007.03363.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M. and Matsubayashi Y. (2017) Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3, 17029 10.1038/nplants.2017.29 [DOI] [PubMed] [Google Scholar]
- 120.Ota R., Ohkubo Y., Yamashita Y., Ogawa-Ohnishi M. and Matsubayashi Y. (2020) Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 11, 641 10.1038/s41467-020-14440-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen X., Yao Q., Gao X., Jiang C., Harberd N.P. and Fu X. (2016) Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646 10.1016/j.cub.2015.12.066 [DOI] [PubMed] [Google Scholar]
- 122.Spiegelman Z., Ham B.K., Zhang Z., Toal T.W., Brady S.M., Zheng Y. et al. (2015) A tomato phloem-mobile protein regulates the shoot-to-root ratio by mediating the auxin response in distant organs. Plant J. 83, 853–863 10.1111/tpj.12932 [DOI] [PubMed] [Google Scholar]
- 123.Spiegelman Z., Shahar A. and Wolf S. (2017) Down-regulation of SlCyp1 in the phloem reduces auxin response and photosynthetic rate in tomato (Solanum lycopersicum) plants. Plant Signal. Behav. 12, e1338224 10.1080/15592324.2017.1338224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Notaguchi M., Wolf S. and Lucas W.J. (2012) Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 54, 760–772 10.1111/j.1744-7909.2012.01155.x [DOI] [PubMed] [Google Scholar]
- 125.Woodward A.W. and Bartel B. (2005) Auxin: regulation, action, and interactions. Ann. Bot. 95, 707–735 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Adamowski M. and Friml J. (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsikou D., Yan Z., Holt D.B., Abel N.B., Reid D.E., Madsen L.H. et al. (2018) Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236 10.1126/science.aat6907 [DOI] [PubMed] [Google Scholar]
- 128.Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H. and Kawaguchi M. (2014) Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5, 4983 10.1038/ncomms5983 [DOI] [PubMed] [Google Scholar]
- 129.Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J. and Greenberg J.T. (2009) Priming in systemic plant immunity. Science 324, 89–91 10.1126/science.1170025 [DOI] [PubMed] [Google Scholar]
- 130.Chaturvedi R., Venables B., Petros R.A., Nalam V., Li M., Wang X. et al. (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 71, 161–172 10.1111/j.1365-313X.2012.04981.x [DOI] [PubMed] [Google Scholar]
- 131.Shah J., Chaturvedi R., Chowdhury Z., Venables B. and Petros R.A. (2014) Signaling by small metabolites in systemic acquired resistance. Plant J. 79, 645–658 10.1111/tpj.12464 [DOI] [PubMed] [Google Scholar]
- 132.Barbaglia A.M. and Hoffmann-Benning S. (2016) Long-distance lipid signaling and its role in plant development and stress response. Subcell. Biochem. 86, 339–361 10.1007/978-3-319-25979-6_14 [DOI] [PubMed] [Google Scholar]
- 133.Yu K., Soares J.M., Mandal M.K., Wang C., Chanda B., Gifford A.N. et al. (2013) A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3, 1266–1278 10.1016/j.celrep.2013.03.030 [DOI] [PubMed] [Google Scholar]
- 134.Cecchini N.M., Steffes K., Schlappi M.R., Gifford A.N. and Greenberg J.T. (2015) Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nat. Commun. 6, 7658 10.1038/ncomms8658 [DOI] [PubMed] [Google Scholar]
- 135.Chanda B., Xia Y., Mandal M.K., Yu K., Sekine K.T., Gao Q.M. et al. (2011) Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427 10.1038/ng.798 [DOI] [PubMed] [Google Scholar]
- 136.Ye Z.W., Lung S.C., Hu T.H., Chen Q.F., Suen Y.L., Wang M. et al. (2016) Arabidopsis acyl-CoA-binding protein ACBP6 localizes in the phloem and affects jasmonate composition. Plant Mol. Biol. 92, 717–730 10.1007/s11103-016-0541-0 [DOI] [PubMed] [Google Scholar]
- 137.Hu T.H., Lung S.C., Ye Z.W. and Chye M.L. (2018) Depletion of Arabidopsis ACYL-COA-BINDING PROTEIN3 affects fatty acid composition in the phloem. Front. Plant Sci. 9, 2 10.3389/fpls.2018.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Du B., Wei Z., Wang Z., Wang X., Peng X., Chen R. et al. (2015) Phloem-exudate proteome analysis of response to insect brown plant-hopper in rice. J. Plant Physiol. 183, 13–22 10.1016/j.jplph.2015.03.020 [DOI] [PubMed] [Google Scholar]
- 139.Gasperini D., Chauvin A., Acosta I.F., Kurenda A., Stolz S., Chetelat A. et al. (2015) Axial and radial oxylipin transport. Plant Physiol. 169, 2244–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schulze A., Zimmer M., Mielke S., Stellmach H., Melnyk C.W., Hause B. et al. (2019) Wound-induced shoot-to-root relocation of JA-Ile precursors coordinates Arabidopsis growth. Mol. Plant 12, 1383–1394 10.1016/j.molp.2019.05.013 [DOI] [PubMed] [Google Scholar]
- 141.Thorpe M.R., Ferrieri A.P., Herth M.M. and Ferrieri R.A. (2007) 11C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226, 541–551 10.1007/s00425-007-0503-5 [DOI] [PubMed] [Google Scholar]
- 142.Sato C., Aikawa K., Sugiyama S., Nabeta K., Masuta C. and Matsuura H. (2011) Distal transport of exogenously applied jasmonoyl-isoleucine with wounding stress. Plant Cell Physiol. 52, 509–517 10.1093/pcp/pcr011 [DOI] [PubMed] [Google Scholar]
- 143.Jimenez-Aleman G.H., Scholz S.S., Heyer M., Reichelt M., Mithofer A. and Boland W. (2015) Synthesis, metabolism and systemic transport of a fluorinated mimic of the endogenous jasmonate precursor OPC-8:0. Biochim. Biophys. Acta 1851, 1545–1553 10.1016/j.bbalip.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 144.Koo A.J. and Howe G.A. (2009) The wound hormone jasmonate. Phytochemistry 70, 1571–1580 10.1016/j.phytochem.2009.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Toyota M., Spencer D., Sawai-Toyota S., Jiaqi W., Zhang T., Koo A.J. et al. (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–1115 10.1126/science.aat7744 [DOI] [PubMed] [Google Scholar]
- 146.Setter T.L., Brun W.A. and Brenner M.L. (1980) Effect of obstructed translocation on leaf abscisic acid, and associated stomatal closure and photosynthesis decline. Plant Physiol. 65, 1111–1115 10.1104/pp.65.6.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zeevaart J.A. and Boyer G.L. (1984) Accumulation and transport of abscisic acid and its metabolites in ricinus and xanthium. Plant Physiol. 74, 934–939 10.1104/pp.74.4.934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Setter T.L. and Brun W.A. (1981) Abscisic acid translocation and metabolism in soybeans following depodding and petiole girdling treatments. Plant Physiol. 67, 774–779 10.1104/pp.67.4.774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhong W., Hartung W., Komor E. and Schobert C. (1996) Phloem transport of abscisic acid in Ricinus communis L seedlings. Plant Cell Environ. 19, 471–477 10.1111/j.1365-3040.1996.tb00339.x [DOI] [Google Scholar]
- 150.Manzi M., Lado J., Rodrigo M.J., Zacarias L., Arbona V. and Gomez-Cadenas A. (2015) Root ABA accumulation in long-term water-stressed plants is sustained by hormone transport from aerial organs. Plant Cell Physiol. 56, 2457–2466 10.1093/pcp/pcv161 [DOI] [PubMed] [Google Scholar]
- 151.McAdam S.A.M., Manzi M., Ross J.J., Brodribb T.J. and Gómez-Cadenas A. (2016) Uprooting an abscisic acid paradigm: shoots are the primary source. Plant Signal. Behav. 11, e1169359 10.1080/15592324.2016.1169359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hartung W., Sauter A. and Hose E. (2002) Abscisic acid in the xylem: where does it come from, where does it go to? J. Exp. Bot. 53, 27–32 10.1093/jexbot/53.366.27 [DOI] [PubMed] [Google Scholar]
- 153.Takahashi F., Suzuki T., Osakabe Y., Betsuyaku S., Kondo Y., Dohmae N. et al. (2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238 10.1038/s41586-018-0009-2 [DOI] [PubMed] [Google Scholar]
- 154.Barbaglia A.M., Tamot B., Greve V. and Hoffmann-Benning S. (2016) Phloem proteomics reveals new lipid-binding proteins with a putative role in lipid-mediated signaling. Front. Plant Sci. 7, 563 10.3389/fpls.2016.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Koenig A.M., Benning C. and Hoffmann-Benning S. (2020) Lipid trafficking and signaling in plants. In Lipid Signaling and Metabolism(Ntambi J.M., ed.), Elsevier [Google Scholar]
- 156.Iqbal N., Khan N.A., Ferrante A., Trivellini A., Francini A. and Khan M.I.R. (2017) Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8, 475 10.3389/fpls.2017.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Andres F. and Coupland G. (2012) The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- 158.Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y. et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- 159.Pin P.A. and Nilsson O. (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755 10.1111/j.1365-3040.2012.02558.x [DOI] [PubMed] [Google Scholar]
- 160.Chen H., Rosin F.M., Prat S. and Hannapel D.J. (2003) Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 132, 1391–1404 10.1104/pp.103.022434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hannapel D.J., Sharma P. and Lin T. (2013) Phloem-mobile messenger RNAs and root development. Front. Plant Sci. 4, 257 10.3389/fpls.2013.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin T., Sharma P., Gonzalez D.H., Viola I.L. and Hannapel D.J. (2013) The impact of the long-distance transport of a BEL1-like messenger RNA on development. Plant Physiol. 161, 760–772 10.1104/pp.112.209429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cho S.K., Sharma P., Butler N.M., Kang I.H., Shah S., Rao A.G. et al. (2015) Polypyrimidine tract-binding proteins of potato mediate tuberization through an interaction with StBEL5 RNA. J. Exp. Bot. 66, 6835–6847 10.1093/jxb/erv389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sharma P., Lin T. and Hannapel D.J. (2016) Targets of the StBEL5 transcription factor include the FT ortholog StSP6A. Plant Physiol. 170, 310–324 10.1104/pp.15.01314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Suarez-Lopez P. (2013) A critical appraisal of phloem-mobile signals involved in tuber induction. Front. Plant Sci. 4, 253 10.3389/fpls.2013.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hannapel D.J., Sharma P., Lin T. and Banerjee A.K. (2017) The multiple signals that control tuber formation. Plant Physiol. 174, 845–856 10.1104/pp.17.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Natarajan B., Kondhare K.R., Hannapel D.J. and Banerjee A.K. (2019) Mobile RNAs and proteins: prospects in storage organ development of tuber and root crops. Plant Sci. 284, 73–81 10.1016/j.plantsci.2019.03.019 [DOI] [PubMed] [Google Scholar]
- 168.Zhai Z., Gayomba S.R., Jung H.I., Vimalakumari N.K., Pineros M., Craft E. et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26, 2249–2264 10.1105/tpc.114.123737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vert G.A., Briat J.F. and Curie C. (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 132, 796–804 10.1104/pp.102.016089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kobayashi T. and Nishizawa N.K. (2012) Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152 10.1146/annurev-arplant-042811-105522 [DOI] [PubMed] [Google Scholar]
- 171.Gutierrez-Carbonell E., Lattanzio G., Albacete A., Rios J.J., Kehr J., Abadia A. et al. (2015) Effects of Fe deficiency on the protein profile of Brassica napus phloem sap. Proteomics 15, 3835–3853 10.1002/pmic.201400464 [DOI] [PubMed] [Google Scholar]
- 172.Buhtz A., Pieritz J., Springer F. and Kehr J. (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10, 64 10.1186/1471-2229-10-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huen A.K., Rodriguez-Medina C., Ho A.Y.Y., Atkins C.A. and Smith P.M.C. (2017) Long-distance movement of phosphate starvation-responsive microRNAs in Arabidopsis. Plant Biol. (Stuttg) 19, 643–649 10.1111/plb.12568 [DOI] [PubMed] [Google Scholar]
- 174.Chien P.S., Chiang C.P., Leong S.J. and Chiou T.J. (2018) Sensing and signaling of phosphate starvation: from local to long distance. Plant Cell Physiol. 59, 1714–1722 10.1093/pcp/pcy148 [DOI] [PubMed] [Google Scholar]
- 175.Moellering E.R. and Benning C. (2011) Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci. 16, 98–107 10.1016/j.tplants.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 176.Delhaize E. and Randall P.J. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107, 207–213 10.1104/pp.107.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L. and Chiou T.J. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141, 1000–1011 10.1104/pp.106.078063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F. and Su C. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis[W]. Plant Cell 412–421 10.1105/tpc.105.038943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bari R., Datt Pant B., Stitt M. and Scheible W.R. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999 10.1104/pp.106.079707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ohyama K., Ogawa M. and Matsubayashi Y. (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55, 152–160 10.1111/j.1365-313X.2008.03464.x [DOI] [PubMed] [Google Scholar]
- 181.Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H. and Matsubayashi Y. (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346 10.1126/science.1257800 [DOI] [PubMed] [Google Scholar]
- 182.Gautrat P., Mortier V., Laffont C., De Keyser A., Fromentin J., Frugier F. et al. (2019) Unraveling new molecular players involved in the autoregulation of nodulation in Medicago truncatula. J. Exp. Bot. 70, 1407–1417 10.1093/jxb/ery465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Isaacs M., Carella P., Faubert J., Rose J.K. and Cameron R.K. (2016) Orthology analysis and in vivo complementation studies to elucidate the role of DIR1 during systemic acquired resistance in Arabidopsis thaliana and Cucumis sativus. Front. Plant Sci. 7, 566 10.3389/fpls.2016.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C. et al. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Scharwies J.D. and Dinneny J.R. (2019) Water transport, perception, and response in plants. J. Plant Res. 132, 311–324 10.1007/s10265-019-01089-8 [DOI] [PubMed] [Google Scholar]
- 186.Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J., Mishra R.K. et al. (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8, 161 10.3389/fpls.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schachtman D.P. and Goodger J.Q. (2008) Chemical root to shoot signaling under drought. Trends Plant Sci. 13, 281–287 10.1016/j.tplants.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 188.Davies W.J. and Zhang J. (1991) Root signals and the regulation of growth and development of plants in drying soil. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 55–76 10.1146/annurev.pp.42.060191.000415 [DOI] [Google Scholar]
- 189.Zhang J. (1987) Control of stomatal behaviour by abscisic acid which apparently originates in the roots. J. Exp. Bot. 38, 1174–1181 10.1093/jxb/38.7.1174 [DOI] [Google Scholar]
- 190.Jiang F. and Hartung W. (2008) Long-distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. J. Exp. Bot. 59, 37–43 10.1093/jxb/erm127 [DOI] [PubMed] [Google Scholar]
- 191.McLachlan D.H., Pridgeon A.J. and Hetherington A.M. (2018) How Arabidopsis talks to itself about its water supply. Mol. Cell 70, 991–992 10.1016/j.molcel.2018.06.011 [DOI] [PubMed] [Google Scholar]
- 192.Zhang W., Qin C., Zhao J. and Wang X. (2004) Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 9508–9513 10.1073/pnas.0402112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Christmann A., Weiler E.W., Steudle E. and Grill E. (2007) A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 52, 167–174 10.1111/j.1365-313X.2007.03234.x [DOI] [PubMed] [Google Scholar]
- 194.Mishra G., Zhang W., Deng F., Zhao J. and Wang X. (2006) A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266 10.1126/science.1123769 [DOI] [PubMed] [Google Scholar]
- 195.Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R. et al. (2009) Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377 10.1105/tpc.108.062992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Testerink C. and Munnik T. (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375 10.1016/j.tplants.2005.06.002 [DOI] [PubMed] [Google Scholar]