Abstract
The North Pacific Marine Salmon Diet Database is an open-access relational database built to centralize and make accessible salmon diet data through a standardized database structure. The initial data contribution contains 21,862 observations of salmon diet, and associated salmon biological parameters, prey biological parameters, and environmental data from the North Pacific Ocean. The data come from 907 unique spatial areas and mostly fall within two time periods, 1959–1969 and 1987–1997, during which there are more data available compared to other time periods. Data were extracted from 62 sources identified through a systematic literature review, targeting peer-reviewed and gray literature. The purpose of this database is to consolidate data into a common format to address gaps in our ecological understanding of the North Pacific Ocean, particularly with respect to salmon. This database can be used to address a variety of questions regarding salmon foraging, productivity, and marine survival. The North Pacific Marine Salmon Diet Database will continue to grow in the future as more data are digitized and become available.
Subject terms: Marine biology, Food webs, Ichthyology
| Measurement(s) | Gastric Content • diet |
| Technology Type(s) | digital curation • microscopy |
| Factor Type(s) | predator capture method • year of data collection • longitude • latitude • predator life stage • predator lowest taxonomic level • predator characteristics • predator replicates • type of diet data • prey life stage • prey lowest taxonomic level • prey characteristics |
| Sample Characteristic - Organism | Oncorhynchus mykiss • Oncorhynchus kisutch • Oncorhynchus tshawytscha • Oncorhynchus nerka • Oncorhynchus keta • Oncorhynchus gorbuscha |
| Sample Characteristic - Environment | ocean |
| Sample Characteristic - Location | North Pacific Ocean |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12981989
Background & Summary
Even though salmon spend 1–6 years of their life in the marine environment, this phase of their life cycle is poorly understood compared to their freshwater phase1. There are limited data on how salmon are distributed, what they feed on, and what threats to survival they may face during this phase, which includes nearshore and offshore components. The marine phase is hypothesized to contain salmon population bottlenecks2, and research has shown that salmon smolt to adult survival rates can be less than 1% for some stocks in the North Pacific3. There is also evidence that Pacific salmon marine survival has been declining over the past several decades in certain areas, especially more southern stocks4,5. Therefore, it is becoming urgent that researchers understand what is happening to salmon during the marine phase of their life cycle, especially after they have moved offshore, because this phase tends to be data-poor compared to the early marine coastal phase.
One of the most important factors affecting salmon survival is the presence and abundance of suitable prey. Although it is difficult to assess prey distribution across ocean basins, information on prey presence and abundance can be derived from salmon diets. Marine diet data has the potential to give insight into food webs, niche overlap among species/stocks, competition, health, and changing ocean conditions6–8. Since the early 1900s, researchers have been examining the diets of Pacific salmon to provide information on species biology and the conditions that salmon face in the ocean9,10. Although there have been some reviews of salmon diet data in the North Pacific, these have been limited in time and space and the data are not normally made publicly available7,8,11–15.
Salmon diet data have been collected using a variety of methodologies employed by researchers from countries across the North Pacific. These data are scattered across the peer-reviewed and gray literature, making collation challenging. However, there is high value in collating these data, considering the costliness and difficulty of conducting fieldwork in the open ocean. Synthesizing these data can reveal important information about salmon open ocean life history experience and further understanding of the potential impacts of changing oceans on salmon productivity. Additionally, more comprehensive, accurate and robust diet data will be an asset to ecosystem models, which are increasingly being applied in ecosystem-based management16. As salmon face an uncertain future with climate change17,18, this is a critical time to consolidate available knowledge in order to advance research on salmon marine ecology.
The goal of this project was to develop an open-access database framework for collating marine salmon diet data, alongside available salmon biological data, prey biological data, and environmental data. We compiled an initial contribution of salmon stomach content diet data from offshore areas using a systematic literature review, followed by quality control and standardization procedures for two time periods: 1959–1969 and 1987–1997. These decades were selected partially because they are time periods in which there are a larger quantity of data available on salmon diets. This database will continue to grow as more sources are identified and added and can be used as a tool by researchers to study salmon marine survival and North Pacific ecosystem dynamics.
Methods
Systematic literature review
In order to identify sources that contained salmon diet data, in the form of stomach contents, a systematic literature review was performed using database keyword searches of ProQuest: Aquatic Sciences and Fisheries Abstracts (https://search.proquest.com/asfa), Web of Science: Core Collection and Web of Science: Zoological Record (https://webofknowledge.com). Not all salmon diet studies are part of the peer-reviewed literature and many North Pacific researchers have published data through the North Pacific Anadromous Fish Commission (NPAFC) and the defunct International North Pacific Fisheries Commission (INPFC). Therefore, database search results were supplemented with relevant INPFC documents and bulletins (https://npafc.org/inpfc/), NPAFC documents (https://npafc.org/npafc-documents/) and bulletins (https://npafc.org/bulletin/), and relevant bibliographic references found within these documents and bulletins (Table 1).
Table 1.
The number of salmon diet data sources identified from a systematic literature review.
| Search terms | Source | Results before filtering | Results after filtering |
|---|---|---|---|
|
(Chinook OR “Oncorhynchus tshawytscha” OR coho OR “Oncorhynchus kisutch” OR sockeye OR “Oncorhynchus nerka” OR pink OR “Oncorhynchus gorbuscha” OR chum OR “Oncorhynchus keta” OR steelhead OR “Oncorhynchus mykiss”) AND (marine OR ocean* OR coast* OR “Gulf of Alaska” OR “Bering Sea”) AND (stomach OR gut* OR “prey composition” OR “diet* composition” OR “composition of diet” OR “composition of prey”) AND (diet* OR prey OR food) |
Proquest: Aquatic Sciences and Fisheries Abstracts | 410 | 23 |
| Web of Science: Core Collection | 182 | 6 | |
| Web of Science: Zoological Record | 142 | 12 | |
| North Pacific Anadromous Fish Commission Documents | 23 | ||
| North Pacific Anadromous Fish Commission Bulletins | 16 | ||
| International North Pacific Fish Commission Documents | 5 | ||
| International North Pacific Fish Commission Bulletins | 6 | ||
| Total = 62 |
A keyword search was used to identify sources in three online databases (Proquest: Aquatic Sciences and Fisheries Abstracts, Web of Science: Core Collection, Web of Science: Zoological Record), which contained most of the peer-reviewed literature. The former steelhead species name “Salmo gairdneri”, when included as a search term, did not provide any more relevant sources that met our criteria. A manual search through the NPAFC and INPFC documents and bulletins provided most of the gray literature. A total of 62 unique sources met the qualifications for database entry.
The database keyword searches identified 591 unique sources. Sources were filtered for relevance and excluded based on the following criteria:
-
(i)
the source did not have salmon stomach content diet data from between 1959–1969 or 1987–1997 for the marine environment, as defined by the area beyond the Riverine Coastal Domain (~15 km)19 (556 sources);
-
(ii)
the source did not have extractable diet data and authors did not respond to inquiries (1 source);
-
(iii)
the source was a review, in which case the original sources were used, if possible and relevant, to extract data (2 sources);
-
(iv)
the source overlapped completely with another source, i.e., the data were reported using the exact same metrics for the same samples as another source (2 sources).
The database search was supplemented with sources that met the same criteria from the INPFC documents and bulletins and the NPAFC documents and bulletins, bringing the total number of unique sources to 62 (Online-only Table 1).
Online-Only Table 1.
The sources included in the database.
| Source_id | First Author | Year | Reference Number |
|---|---|---|---|
| 1 | Andrievskaya, L. D. | 1966 | 24 |
| 2 | Carlson, H. R. | 1976 | 25 |
| 3 | Chuchukalo, V. L. | 1995 | 26 |
| 4 | Davis, N. D. | 1996 | 27 |
| 5 | Davis, N. D. | 1998 | 28 |
| 6 | Davis, N. D. | 2000 | 29 |
| 7 | Dulepova, E. P. | 2003 | 30 |
| 8 | Fukataki, H. | 1967 | 31 |
| 9 | Glebov, I. I. | 1998 | 32 |
| 10 | Ito, J. | 1964 | 33 |
| 11 | Kaeriyama, M. | 2000 | 34 |
| 12 | Kaeriyama, M. | 2004 | 15 |
| 13 | Kanno, Y. | 1971 | 35 |
| 14 | Karpenko, V. I. | 2007 | 13 |
| 15 | Manzer, J. I. | 1968 | 36 |
| 16 | Perry, R. I. | 1996 | 37 |
| 17 | Tadokoro, K. | 1996 | 38 |
| 18 | Takeuchi, I. | 1972 | 39 |
| 19 | Ueno, M. | 1969 | 40 |
| 20 | Volkov, A. F. | 1995 | 41 |
| 21 | Auburn, M. E. | 2000 | 42 |
| 22 | Aydin, K. Y. | 1998 | 43 |
| 23 | Brodeur, R. D. | 2007 | 11 |
| 24 | Daly, E. A. | 2015 | 44 |
| 25 | Davis, N. D. | 2003 | 45 |
| 26 | Kawamura, H. | 1998 | 46 |
| 27 | Qin, Y. | 2016 | 8 |
| 28 | Starovoytov, A. N. | 2007 | 14 |
| 29 | Ueno, Y. | 1992 | 47 |
| 30 | Ueno, Y. | 1992 | 48 |
| 31 | Waddell, B. J. | 1992 | 49 |
| 32 | Andrievskaya, L. D. | 1970 | 50 |
| 33 | Atcheson, M. E. | 2012 | 51 |
| 34 | Carlson, H. R. | 1996 | 52 |
| 35 | Davis, N. D. | 2005 | 53 |
| 36 | Myers, K. W. | 1996 | 54 |
| 37 | Myers, K. W. | 1995 | 55 |
| 38 | Sturdevant, M. V. | 1997 | 56 |
| 39 | Walker, R. V. | 1993 | 57 |
| 40 | Suzuki, T. | 1994 | 58 |
| 41 | Shimazaki, K. | 1969 | 59 |
| 42 | Weitkamp, L. | 2004 | 60 |
| 43 | Starovoytov, A. N. | 2003 | 61 |
| 44 | LeBrasseur, R. J. | 1966 | 62 |
| 45 | LeBrasseur, R. J. | 1966 | 63 |
| 46 | LeBrasseur, R. J. | 1966 | 64 |
| 47 | Ishida, Y. | 1999 | 65 |
| 48 | Tamura, R. | 1999 | 66 |
| 49 | Seki, J. | 1998 | 67 |
| 50 | Suzuki, T. | 1995 | 68 |
| 51 | Andrievskaya, L. D. | 1974 | 69 |
| 52 | Andrievskaya, L. D. | 1970 | 70 |
| 53 | Chuchukalo, V. I. | 1994 | 71 |
| 54 | Gorbatenko, K. M. | 1996 | 72 |
| 55 | Kayev, A. M. | 1993 | 73 |
| 56 | Klovatch, N. V. | 2002 | 74 |
| 57 | Shershnev, A. P. | 1982 | 75 |
| 58 | Tutubalin, B. G. | 1992 | 76 |
| 59 | Volkov, A. F. | 1996 | 77 |
| 60 | Volkov, A. F. | 1994 | 78 |
| 61 | Fisheries Agency of Japan | 1966 | 79 |
| 62 | Davis, N. D. | 1990 | 80 |
Sources are listed in order of their source_id number, which corresponds to their source_id in the database. For each source, the first author, year of publication and the reference number, which corresponds to the final ‘References’ section, are listed.
Data extraction
For each salmon diet sample, we extracted the following data (if available): source metadata (e.g., publication year, title, authors) (Online-only Table 2), salmon capture method (Online-only Table 3), site information (time, location), salmon information (e.g., taxonomy, life stage, sex), salmon replicates, type of diet data (e.g., percent weight of prey, total number of prey), and prey information (e.g., taxonomy, life stage, quantity) (Online-only Table 4). A diet sample is defined as a distinct sample in time and space from a specific source and is entered into the database exactly as it is reported in the source. A sample can contain diet data from one salmon or more than one salmon when individuals were grouped together for diet analysis (up to 2,215 in this data compilation). If different diet metrics were reported for the same diet sample (e.g., number of prey and volume of prey), then all metrics were entered into the database. If the data were not available in table format, but figure format only, then the data were extracted using WebPlotDigitizer (https://apps.automeris.io/wpd/).
Online-Only Table 2.
The metadata extracted for each source.
| Column | Explanation |
|---|---|
| source_id | A unique number that is generated and assigned to each source (e.g., 1, 2, 3…etc.) |
| publication_year | Publication year in the format: YYYY |
| title | Title of the source |
| author1 | First author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author2 | Second author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author3 | Third author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author4 | Fourth author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author5 | Fifth author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author6 | Sixth author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author7 | Seventh author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author8 | Eighth author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author9 | Ninth author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| author10 | Tenth author of the source in the following format: M. C. Graham or M. Graham (first and middle names are abbreviated by initials and a period while last names are completely spelled out) |
| url | URL associated with source, if applicable |
| citation | The full citation for the source |
| source_notes | Any additional notes about the source; for example, if the data might be overlapping with another source, this is indicated in this attribute |
| data_processing_notes | Notes about how the data were processed – in the lab or field, were quantities measured using scales or visually estimated, etc. |
| date_entered | The date the source was added to the database |
| entered_by | The full name of the person who entered the data (e.g., Caroline Graham) |
This table corresponds to the ‘Sources’ file in the figshare repository.
Online-Only Table 3.
The data extracted for each salmon gear type.
| Column | Explanation |
|---|---|
| gear_type_predator_id | A unique number that is generated and assigned to each unique predator gear type for each source (e.g., 1, 2, 3…etc.) |
| gear_type | The most basic description of the type of gear given in the source (e.g., trawl, gillnet, longline) |
| gear_length_value | The gear length |
| gear_length_min | If there are a range of gear lengths, then this attribute represents the minimum length |
| gear_length_max | If there are a range of gear lengths, then this attribute represents the maximum length |
| gear_length_units | The units associated with the gear length; units are fully spelled and plural (e.g., meters instead of meter) |
| gear_width_value | The gear width |
| gear_width_min | If there are a range of gear widths, then this attribute represents the minimum width |
| gear_width_max | If there are a range of gear widths, then this attribute represents the maximum width |
| gear_width_units | The units associated with the gear width; units are fully spelled and plural (e.g., meters instead of meter) |
| gear_depth_value | The gear depth; if the gear is reported to be deployed at the surface then the depth value is assigned to 0 |
| gear_depth_min | If there are a range of gear depths, then this attribute represents the minimum depth; if the gear is reported to be deployed at the surface then the depth value is assigned to 0 |
| gear_depth_max | If there are a range of gear depths, then this attribute represents the maximum depth |
| gear_depth_units | The units associated with the gear depth; units are fully spelled and plural (e.g., meters instead of meter) |
| mesh_size_value | The gear mesh size |
| mesh_size_min | If there are a range of gear mesh sizes, then this attribute represents the minimum mesh size |
| mesh_size_max | If there are a range of gear mesh sizes, then this attribute represents the maximum mesh size |
| mesh_size_units | The units associated with the mesh size; units are fully spelled and plural (e.g., millimeters instead of millimeter) |
| fishing_depth_value | The fishing depth; if fishing is reported to be at the surface then the depth value is assigned to 0 |
| fishing_depth_min | If there are a range of fishing depths, then this attribute represents the minimum fishing depth; if fishing is reported to be at the surface then the depth value is assigned to 0 |
| fishing_depth_max | If there are a range of fishing depths, then this attribute represents the maximum fishing depth |
| fishing_depth_units | The units associated with the fishing depth; units are fully spelled and plural (e.g., meters instead of meter) |
| tow_speed_value | The gear tow speed |
| tow_speed_min | If there are a range of gear tow speeds, then this attribute represents the minimum tow speed |
| tow_speed_max | If there are a range of gear tow speeds, then this attribute represents the maximum tow speed |
| tow_speed_units | The units associated with the tow speed; units are fully spelled and plural (e.g., knots instead of knot) |
| duration_deployment_value | The gear duration of deployment |
| duration_deployment_min | If there are a range of gear durations of deployment, then this attribute represents the minimum duration of deployment |
| duration_deployment_max | If there are a range of gear durations of deployment, then this attribute represents the maximum duration of deployment |
| duration_deployment_units | The units associated with the duration of deployment; units are fully spelled and plural (e.g., minutes instead of minute) |
| gear_notes | Any additional comments on the gear |
This table corresponds to the ‘Gear_type_predator’ file in the figshare repository.
Online-Only Table 4.
The diet data extracted for each salmon predator.
| Column | Explanation |
|---|---|
| predator_id | A unique number that is generated and assigned to each predator sample |
| source_id | This number corresponds with the source_id from Online-only Table 2 |
| year_min | YYYY; if there is just one value for the year then it is found in this attribute; if there are a range of values for the year then the minimum value is found in this attribute |
| year_max | YYYY; if there are a range of values for the year then the maximum value is entered into this attribute |
| warm_cool_years | Either ‘warm’, ‘cool’ or NA; this attribute will only have a value if the samples are explicitly reported as being from a warm versus cool year(s) and can only be uniquely identified this way |
| odd_even_years | Either ‘odd, ‘even’ or NA; this attribute will only have a value if the samples are explicitly reported as being from an odd versus even year(s) and can only be uniquely identified this way |
| season_min | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there is just one value for the season then it is entered into this attribute; if there are a range of values for the season then the minimum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| season_max | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there are a range of values for the season then the maximum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| month_min | Month names are completely spelled out with the first letter capitalized; if there is just one value for the month then it is entered into this attribute; if there are a range of values for the month then the minimum value is found in this attribute |
| month_max | Month names are completely spelled out with the first letter capitalized; if there are a range of values for the month then the maximum value is found in this attribute |
| date_min | Expressed using year, month, and day; if there is just one value for the date then it is entered into this attribute; if there are a range of values for the date then the minimum value is found in this attribute |
| date_max | Expressed using year, month, and day; if there are a range of values for the date then the maximum value is found in this attribute |
| time_min | HH:MM:SS; if there is just one value for the time then it is found in this attribute; if there are a range of values for the time then the minimum value is found in this attribute |
| time_max | HH:MM:SS; if there are a range of values for the time then the maximum value is found in this attribute |
| lat_min | If there is just one value for the latitude then it is found in this attribute;if there are a range of values for the latitude then the minimum value is found in this attribute; values are in decimal degrees format |
| lat_max | If there are a range of values for the latitude then the maximum value is found in this attribute; values are in decimal degrees format |
| lon_min | If there is just one value for the longitude then it is entered into this attribute; if there are a range of values for the longitude then the minimum value is entered into this attribute; values are in decimal degrees format |
| lon_max | If there are a range of values for the longitude then the maximum value is found in this attribute; values are in decimal degrees format |
| predator_lowest_taxonomic_level | The lowest taxonomic level reported in the source; if a source reports to the species level then this attribute includes both the genus and species names (e.g., Oncorhynchus nerka); only scientific names are reported, not common names |
| predator_life_stage | Either ‘juvenile’, ‘adult’ or NA |
| predator_life_stage_min | If there are a mixture of juveniles and adults, then ‘juvenile’ is entered here |
| predator_life_stage_max | If there are a mixture of juveniles and adults, then ‘adult’ is entered here |
| predator_freshwater_age | An integer to indicate the number of years spent living in freshwater |
| predator_freshwater_age_min | If there are a mixture of freshwater ages, then this attribute represents the minimum age; an integer to indicate the number of years spent living in freshwater |
| predator_freshwater_age_max | If there are a mixture of freshwater ages, then this attribute represents the maximum age; an integer to indicate the number of years spent living in freshwater |
| predator_ocean_age | An integer to indicate the number of years spent living in the ocean |
| predator_ocean_age_min | If there are a mixture of ocean ages, then this attribute represents the minimum age; an integer to indicate the number of years spent living in the ocean |
| predator_ocean_age_max | If there are a mixture of ocean ages, then this attribute represents the maximum age; an integer to indicate the number of years spent living in the ocean |
| predator_maturity | Either ‘juvenile’, ‘immature’, ‘maturing’ ‘mature’, ‘kelt’ (for steelhead), or NA |
| predator_maturity_min | If there are a mixture of maturity levels, then the minimum maturity level is found here |
| predator_maturity_max | If there are a mixture of maturity levels, then the maximum maturity level is found here |
| predator_length_value_cm | The length of a predator in centimeters (could be either fork length or total length); only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_length_min_cm | The minimum length of a predator in centimeters (could be either fork length or total length) if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_length_max_cm | The maximum length of a predator in centimeters (could be either fork length or total length) if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_value_g | The weight of a predator in grams; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_min_g | The minimum weight of a predator in grams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_max_g | The maximum weight of a predator in grams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_subsample_id | A unique number that is generated and assigned to predator samples if a source reports the diets of individual predators with no unique identifiers; values are assigned for each source starting from 1 and increasing by a value of 1 each time (e.g., 1,2,3…); if unique subsample_ids are not required then the default value is 0 |
| predator_sex | Either ‘male’, ‘female’ or ‘unspecified’ |
| hatchery_wild | Either ‘hatchery’, ‘wild’ or ‘unspecified’ |
| predator_replicates | The total number of predator replicates per sample |
| predator_notes | Any additional comments on the predator |
| gear_type_predator_id1 | This id number corresponds with the gear_type_predator_id from Online-only Table 3 |
| gear_type_predator_id2 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 2 types of gear used to sample predators |
| gear_type_predator_id3 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 3 types of gear used to sample predators |
| gear_type_predator_id4 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 4 types of gear used to sample predators |
| gear_type_predator_id5 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 5 types of gear used to sample predators |
| type_diet_data | The diet metric reported in the source (e.g., percent weight of prey, index of relative importance) |
| diet_data_units | The diet data units (e.g., percent); if the diet data are reported as a number then the units are left as blank |
| formula | The formula for the diet metric, if applicable; this is for metrics such as the index of relative importance or the stomach content index because they may be calculated differently in different sources |
| prey_lowest_taxonomic_level | The lowest taxonomic level reported in the source; if a source reports to the species level then this attribute should include both the genus and species names (e.g., Calanus pacificus); in some cases the lowest taxonomic level is not a scientific name – like ‘gelatinous’ or ‘zooplankton_collective’ or ‘miscellaneous’ |
| prey_kingdom | The kingdom based on the lowest taxonomic level |
| prey_phylum | The phylum based on the lowest taxonomic level |
| prey_class | The class based on the lowest taxonomic level |
| prey_order | The order based on the lowest taxonomic level |
| prey_family | The family based on the lowest taxonomic level |
| prey_genus | The genus based on the lowest taxonomic level |
| prey_species | The species based on the lowest taxonomic level |
| value_diet_data | Diet data values for the specific prey item and the sample |
| prey_sex | Either ‘male’, ‘female’ or ‘unspecified’ |
| prey_life_stage | The prey life stage |
| prey_life_stage_min | If there are a mixture of life stages, then this attribute represents the minimum life stage |
| prey_life_stage_max | If there are a mixture of life stages, then this attribute represents the maximum life stage |
| prey_length_value_mm | The length of prey in millimeters; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_length_min_mm | The minimum length of prey in millimeters if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_length_max_mm | The maximum length of prey in millimeters if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_weight_value_mg | The weight of prey in milligrams; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_weight_min_mg | The minimum weight of prey in milligrams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_weight_max_mg | The maximum weight of prey in milligrams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_notes | Any additional comments on prey |
This table corresponds to the ‘Diet_data’ file in the figshare repository.
For each source, we extracted data as it was presented in the sources in almost all cases, and therefore it was extracted according to the data resolution of the source. For example, if the sample location was presented as a station, it was extracted as a station with specific geographical coordinates, but if it was presented as a transect or area, then it was extracted as a transect or area with latitude and/or longitude minimums and maximums. If geographical coordinates were not specified, then they were estimated based on survey maps or descriptions present in the source. Prey taxonomy was reported to different resolutions across sources (e.g., Copepoda versus Neocalanus cristatus). In order to keep the lowest data resolution reported in the source while also being able to compare across sources, each prey item was reported at all possible taxonomic levels (kingdom, phylum, class, order, family, genus, species). In addition to the salmon diet data, if the source presented additional related data for salmon biological parameters (i.e., variables) (Online-only Table 5), prey biological parameters (Online-only Tables 6 and 7), or environmental parameters (Online-only Table 8), these data were extracted as well. For detailed information about the different types of data extracted and the extraction methodology see Online-only Tables 2–8.
Online-Only Table 5.
The associated salmon biological data for each salmon sample.
| Column | Explanation |
|---|---|
| predator_id | A unique number that is generated and assigned to each predator sample |
| source_id | This number corresponds with the source_id from Online-only Table 2 |
| year_min | YYYY; if there is just one value for the year then it is found in this attribute; if there are a range of values for the year then the minimum value is found in this attribute |
| year_max | YYYY; if there are a range of values for the year then the maximum value is entered into this attribute |
| warm_cool_years | Either ‘warm’, ‘cool’ or NA; this attribute will only have a value if the samples are explicitly reported as being from a warm versus cool year(s) and can only be uniquely identified this way |
| odd_even_years | Either ‘odd, ‘even’ or NA; this attribute will only have a value if the samples are explicitly reported as being from an odd versus even year(s) and can only be uniquely identified this way |
| season_min | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there is just one value for the season then it is entered into this attribute; If there are a range of values for the season then the minimum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| season_max | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there are a range of values for the season then the maximum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| month_min | Month names are completely spelled out with the first letter capitalized; if there is just one value for the month then it is entered into this attribute; If there are a range of values for the month then the minimum value is found in this attribute |
| month_max | Month names are completely spelled out with the first letter capitalized; if there are a range of values for the month then the maximum value is found in this attribute |
| date_min | Expressed using year, month, and day; if there is just one value for the date then it is entered into this attribute; if there are a range of values for the date then the minimum value is found in this attribute |
| date_max | Expressed using year, month, and day; if there are a range of values for the date then the maximum value is found in this attribute |
| time_min | HH:MM:SS; if there is just one value for the time then it is found in this attribute; if there are a range of values for the time then the minimum value is found in this attribute |
| time_max | HH:MM:SS; if there are a range of values for the time then the maximum value is found in this attribute |
| lat_min | If there is just one value for the latitude then it is found in this attribute; if there are a range of values for the latitude then the minimum value is found in this attribute; values are in decimal degrees format |
| lat_max | If there are a range of values for the latitude then the maximum value is found in this attribute; values are in decimal degrees format |
| lon_min | If there is just one value for the longitude then it is entered into this attribute; if there are a range of values for the longitude then the minimum value is entered into this attribute; values are in decimal degrees format |
| lon_max | If there are a range of values for the longitude then the maximum value is found in this attribute; values are in decimal degrees format |
| predator_lowest_taxonomic_level | The lowest taxonomic level reported in the source; if a source reports to the species level then this attribute includes both the genus and species names (e.g., Oncorhynchus nerka); only scientific names are reported, not common names |
| predator_life_stage | Either ‘juvenile’, ‘adult’ or NA |
| predator_life_stage_min | If there are a mixture of juveniles and adults, then ‘juvenile’ is entered here |
| predator_life_stage_max | If there are a mixture of juveniles and adults, then ‘adult’ is entered here |
| predator_freshwater_age | An integer to indicate the number of years spent living in freshwater |
| predator_freshwater_age_min | If there are a mixture of freshwater ages, then this attribute represents the minimum age; an integer to indicate the number of years spent living in freshwater |
| predator_freshwater_age_max | If there are a mixture of freshwater ages, then this attribute represents the maximum age; an integer to indicate the number of years spent living in freshwater |
| predator_ocean_age | An integer to indicate the number of years spent living in the ocean |
| predator_ocean_age_min | If there are a mixture of ocean ages, then this attribute represents the minimum age; an integer to indicate the number of years spent living in the ocean |
| predator_ocean_age_max | If there are a mixture of ocean ages, then this attribute represents the maximum age; an integer to indicate the number of years spent living in the ocean |
| predator_maturity | Either ‘juvenile’, ‘immature’, ‘maturing’ ‘mature’, ‘kelt’ (for steelhead), or NA |
| predator_maturity_min | If there are a mixture of maturity levels, then the minimum maturity level is found here |
| predator_maturity_max | If there are a mixture of maturity levels, then the maximum maturity level is found here |
| predator_length_value_cm | The length of a predator in centimeters (could be either fork length or total length); only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_length_min_cm | The minimum length of a predator in centimeters (could be either fork length or total length) if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_length_max_cm | The maximum length of a predator in centimeters (could be either fork length or total length) if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_value_g | The weight of a predator in grams; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_min_g | The minimum weight of a predator in grams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_max_g | The maximum weight of a predator in grams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_subsample_id | A unique number that is generated and assigned to predator samples if a source reports the diets of individual predators with no unique identifiers; values are assigned for each source starting from 1 and increasing by a value of 1 each time (e.g., 1,2,3…); if unique subsample_ids are not required then the default value is 0 |
| predator_sex | Either ‘male’, ‘female’ or ‘unspecified’ |
| hatchery_wild | Either ‘hatchery’, ‘wild’ or ‘unspecified’ |
| predator_replicates | The total number of predator replicates per sample |
| predator_notes | Any additional comments on the predator |
| gear_type_predator_id1 | This id number corresponds with the gear_type_predator_id from Online-only Table 3 |
| gear_type_predator_id2 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 2 types of gear used to sample predators |
| gear_type_predator_id3 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 3 types of gear used to sample predators |
| gear_type_predator_id4 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 4 types of gear used to sample predators |
| gear_type_predator_id5 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 5 types of gear used to sample predators |
| biological_parameter | The predator biological parameter reported in the source (e.g., total length, fork length, body weight) |
| predator_bio_notes | Any additional comments on the predator biological parameters |
| value | The biological parameter value |
| mean | The biological parameter mean |
| error | The error associated with the biological parameter mean |
| min | If there are a range of values for the biological parameter this attribute represents the minimum value |
| max | If there are a range of values for the biological parameter this attribute represents the maximum value |
| units | The units associated with the biological parameter; units are fully spelled and plural (e.g., centimeters instead of centimeter) |
This table corresponds to the ‘Predator_biological_data’ file in the figshare repository.
Online-Only Table 6.
The data extracted for each prey gear type.
| Column | Explanation |
|---|---|
| gear_type_prey_id | A unique number that is generated and assigned to each unique prey gear type for each source (e.g., 1, 2, 3…etc.) |
| gear_type | The most basic description of the type of gear given in the source (e.g., bongo net); if the prey is a diet item, then the gear_type is ‘predator’ to indicate that it was not collected from the environment but instead in a salmon stomach |
| gear_length_value | The gear length |
| gear_length_min | If there are a range of gear lengths, then this attribute represents the minimum length |
| gear_length_max | If there are a range of gear lengths, then this attribute represents the maximum length |
| gear_length_units | The units associated with the gear length; units are fully spelled and plural (e.g., meters instead of meter) |
| gear_width_value | The gear width |
| gear_width_min | If there are a range of gear widths, then this attribute represents the minimum width |
| gear_width_max | If there are a range of gear widths, then this attribute represents the maximum width |
| gear_width_units | The units associated with the gear width; units are fully spelled and plural (e.g., meters instead of meter) |
| gear_depth_value | The gear depth; if gear is reported to be deployed at the surface then the depth value is assigned to 0 |
| gear_depth_min | If there are a range of gear depths, then this attribute represents the minimum depth; if gear is reported to be deployed at the surface then the depth value is assigned to 0 |
| gear_depth_max | If there are a range of gear depths, then this attribute represents the maximum depth |
| gear_depth_units | The units associated with the gear depth; units are fully spelled and plural (e.g., meters instead of meter) |
| mesh_size_value | The gear mesh size |
| mesh_size_min | If there are a range of gear mesh sizes, then this attribute represents the minimum mesh size |
| mesh_size_max | If there are a range of gear mesh sizes, then this attribute represents the maximum mesh size |
| mesh_size_units | The units associated with the mesh size; units are fully spelled and plural (e.g., millimeters instead of millimeter) |
| fishing_depth_value | The fishing depth; if fishing is reported to be at the surface then the depth value is assigned to 0 |
| fishing_depth_min | If there are a range of fishing depths, then this attribute represents the minimum fishing depth; if fishing is reported to be at the surface then the depth value is assigned to 0 |
| fishing_depth_max | If there are a range of fishing depths, then this attribute represents the maximum fishing depth |
| fishing_depth_units | The units associated with the fishing depth; units are fully spelled and plural (e.g., meters instead of meter) |
| tow_speed_value | The gear tow speed |
| tow_speed_min | If there are a range of gear tow speeds, then this attribute represents the minimum tow speed |
| tow_speed_max | If there are a range of gear tow speeds, then this attribute represents the maximum tow speed |
| tow_speed_units | The units associated with the tow speed; units are fully spelled and plural (e.g., knots instead of knot) |
| duration_deployment_value | The gear duration of deployment |
| duration_deployment_min | If there are a range of gear durations of deployment, then this attribute represents the minimum duration of deployment |
| duration_deployment_max | If there are a range of gear durations of deployment, then this attribute represents the maximum duration of deployment |
| duration_deployment_units | The units associated with the duration of deployment; units are fully spelled and plural (e.g., minutes instead of minute) |
| gear_notes | Any additional comments on the gear |
This table corresponds to the ‘Gear_type_prey’ file in the figshare repository.
Online-Only Table 7.
The associated prey biological data for each prey sample.
| Column | Explanation |
|---|---|
| prey_id | A unique number that is generated and assigned to each prey sample |
| predator_id | A unique number that is generated and assigned to each predator sample |
| source_id | This number corresponds with the source_id from Online-only Table 2 |
| year_min | YYYY; if there is just one value for the year then it is found in this attribute; if there are a range of values for the year then the minimum value is found in this attribute |
| year_max | YYYY; if there are a range of values for the year then the maximum value is entered into this attribute |
| warm_cool_years | Either ‘warm’, ‘cool’ or NA; this attribute will only have a value if the samples are explicitly reported as being from a warm versus cool year(s) and can only be uniquely identified this way |
| odd_even_years | Either ‘odd, ‘even’ or NA; this attribute will only have a value if the samples are explicitly reported as being from an odd versus even year(s) and can only be uniquely identified this way |
| season_min | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there is just one value for the season then it is entered into this attribute; If there are a range of values for the season then the minimum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| season_max | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there are a range of values for the season then the maximum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| month_min | Month names are completely spelled out with the first letter capitalized; if there is just one value for the month then it is entered into this attribute; If there are a range of values for the month then the minimum value is found in this attribute |
| month_max | Month names are completely spelled out with the first letter capitalized; if there are a range of values for the month then the maximum value is found in this attribute |
| date_min | Expressed using year, month, and day; if there is just one value for the date then it is entered into this attribute; if there are a range of values for the date then the minimum value is found in this attribute |
| date_max | Expressed using year, month, and day; if there are a range of values for the date then the maximum value is found in this attribute |
| time_min | HH:MM:SS; if there is just one value for the time then it is found in this attribute; if there are a range of values for the time then the minimum value is found in this attribute |
| time_max | HH:MM:SS; if there are a range of values for the time then the maximum value is found in this attribute |
| lat_min | If there is just one value for the latitude then it is found in this attribute; if there are a range of values for the latitude then the minimum value is found in this attribute; values are in decimal degrees format |
| lat_max | If there are a range of values for the latitude then the maximum value is found in this attribute; values are in decimal degrees format |
| lon_min | If there is just one value for the longitude then it is entered into this attribute; if there are a range of values for the longitude then the minimum value is entered into this attribute; values are in decimal degrees format |
| lon_max | If there are a range of values for the longitude then the maximum value is found in this attribute; values are in decimal degrees format |
| predator_lowest_taxonomic_level | The lowest taxonomic level reported in the source; if a source reports to the species level then this attribute includes both the genus and species names (e.g., Oncorhynchus nerka); only scientific names are reported, not common names |
| predator_life_stage | Either ‘juvenile’, ‘adult’ or NA |
| predator_life_stage_min | If there are a mixture of juveniles and adults, then ‘juvenile’ is entered here |
| predator_life_stage_max | If there are a mixture of juveniles and adults, then ‘adult’ is entered here |
| predator_freshwater_age | An integer to indicate the number of years spent living in freshwater |
| predator_freshwater_age_min | If there are a mixture of freshwater ages, then this attribute represents the minimum age; an integer to indicate the number of years spent living in freshwater |
| predator_freshwater_age_max | If there are a mixture of freshwater ages, then this attribute represents the maximum age; an integer to indicate the number of years spent living in freshwater |
| predator_ocean_age | An integer to indicate the number of years spent living in the ocean |
| predator_ocean_age_min | If there are a mixture of ocean ages, then this attribute represents the minimum age; an integer to indicate the number of years spent living in the ocean |
| predator_ocean_age_max | If there are a mixture of ocean ages, then this attribute represents the maximum age; an integer to indicate the number of years spent living in the ocean |
| predator_maturity | Either ‘juvenile’, ‘immature’, ‘maturing’ ‘mature’, ‘kelt’ (for steelhead), or NA |
| predator_maturity_min | If there are a mixture of maturity levels, then the minimum maturity level is found here |
| predator_maturity_max | If there are a mixture of maturity levels, then the maximum maturity level is found here |
| predator_length_value_cm | The length of a predator in centimeters (could be either fork length or total length); only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_length_min_cm | The minimum length of a predator in centimeters (could be either fork length or total length) if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_length_max_cm | The maximum length of a predator in centimeters (could be either fork length or total length) if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_value_g | The weight of a predator in grams; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_min_g | The minimum weight of a predator in grams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_weight_max_g | The maximum weight of a predator in grams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of diet data from a source |
| predator_subsample_id | A unique number that is generated and assigned to predator samples if a source reports the diets of individual predators with no unique identifiers; values are assigned for each source starting from 1 and increasing by a value of 1 each time (e.g., 1,2,3…); if unique subsample_ids are not required then the default value is 0 |
| predator_sex | Either ‘male’, ‘female’ or ‘unspecified’ |
| hatchery_wild | Either ‘hatchery’, ‘wild’ or ‘unspecified’ |
| predator_replicates | The total number of predator replicates per sample |
| predator_notes | Any additional comments on the predator |
| gear_type_predator_id1 | This id number corresponds with the gear_type_predator_id from Online-only Table 3 |
| gear_type_predator_id2 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 2 types of gear used to sample predators |
| gear_type_predator_id3 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 3 types of gear used to sample predators |
| gear_type_predator_id4 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 4 types of gear used to sample predators |
| gear_type_predator_id5 | This id number corresponds with the gear_type_predator_id from Online-only Table 3; this attribute is required if there are at least 5 types of gear used to sample predators |
| prey_lowest_taxonomic_level | The lowest taxonomic level reported in the source; if a source reports to the species level then this attribute includes both the genus and species names (e.g., Calanus pacificus); in some cases the lowest taxonomic level is not a scientific name – like ‘gelatinous’ or ‘zooplankton_collective’ or ‘miscellaneous’ |
| prey_kingdom | The kingdom based on the lowest taxonomic level |
| prey_phylum | The phylum based on the lowest taxonomic level |
| prey_class | The class based on the lowest taxonomic level |
| prey_order | The order based on the lowest taxonomic level |
| prey_family | The family based on the lowest taxonomic level |
| prey_genus | The genus based on the lowest taxonomic level |
| prey_species | The species based on the lowest taxonomic level |
| prey_life_stage | The prey life stage |
| prey_life_stage_min | If there are a mixture of life stages, then this attribute represents the minimum life stage |
| prey_life_stage_max | If there are a mixture of life stages, then this attribute represents the maximum life stage |
| prey_length_value_mm | The length of prey in millimeters; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_length_min_mm | The minimum length of prey in millimeters if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_length_max_mm | The maximum length of prey in millimeters if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_weight_value_mg | The weight of prey in milligrams; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_weight_min_mg | The minimum weight of prey in milligrams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_weight_max_mg | The maximum weight of prey in milligrams if there are a range of sizes; only reported if there is no other way to determine life stage, or if length or weight categories are the only way to uniquely identify samples of prey data from a source |
| prey_sex | Either ‘male’, ‘female’ or ‘unspecified’ |
| prey_subsample_id | A unique number that is generated and assigned to prey samples if a source reports the biological parameters of individual prey with no unique identifiers; values are assigned for each source starting from 1 and increasing by a value of 1 each time (e.g., 1,2,3…) |
| prey_replicates | The total number of prey replicates per sample |
| prey_notes | Any additional comments on the prey |
| gear_type_prey_id1 | This id number corresponds with the gear_type_prey_id from Online-only Table 6; if the prey is part of a diet data sample then the id should be 1 which corresponds to the ‘predator’ gear type (i.e. sample came from a predator stomach) |
| gear_type_prey_id2 | This id number corresponds with the gear_type_prey_id from Online-only Table 6; this attribute is required if there are at least 2 types of gear used to sample predators |
| gear_type_prey_id3 | This id number corresponds with the gear_type_prey_id from Online-only Table 6; this attribute is required if there are at least 3 types of gear used to sample predators |
| gear_type_prey_id4 | This id number corresponds with the gear_type_prey_id from Online-only Table 6; this attribute is required if there are at least 4 types of gear used to sample predators |
| gear_type_prey_id5 | This id number corresponds with the gear_type_prey_id from Online-only Table 6; this attribute is required if there are at least 5 types of gear used to sample predators |
| biological_parameter | The prey biological parameter reported in the source (e.g., body length, body weight) |
| prey_bio_notes | Any additional comments on the prey biological parameters |
| value | The biological parameter value |
| mean | The biological parameter mean |
| error | The error associated with the biological parameter mean |
| min | If there are a range of values for the biological parameter this attribute represents the minimum value |
| max | If there are a range of values for the biological parameter this attribute represents the maximum value |
| units | The units associated with the biological parameter; units are fully spelled and plural (e.g., centimeters instead of centimeter) |
This data could come from diet samples or from environmental samples of potential prey items. This table corresponds to the ‘Prey_biological_data’ file in the figshare repository.
Online-Only Table 8.
The associated environmental data for each salmon or prey sample.
| Column | Explanation |
|---|---|
| environmental_data_id | A unique number that is generated and assigned to each environmental data point |
| source_id | This number corresponds with the source_id from Online-only Table 2 |
| year_min | YYYY; if there is just one value for the year then it is found in this attribute; if there are a range of values for the year then the minimum value is found in this attribute |
| year_max | YYYY; if there are a range of values for the year then the maximum value is entered into this attribute |
| warm_cool_years | Either ‘warm’, ‘cool’ or NA; this attribute will only have a value if the samples are explicitly reported as being from a warm versus cool year(s) and can only be uniquely identified this way |
| odd_even_years | Either ‘odd, ‘even’ or NA; this attribute will only have a value if the samples are explicitly reported as being from an odd versus even year(s) and can only be uniquely identified this way |
| season_min | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there is just one value for the season then it is entered into this attribute; If there are a range of values for the season then the minimum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| season_max | Either ‘spring’, ‘summer’, ‘autumn’, or ‘winter’; if there are a range of values for the season then the maximum value is entered into this attribute; this attribute will only have a value if there is no value for the month/date and the source explicitly defines the temporal sampling period by season |
| month_min | Month names are completely spelled out with the first letter capitalized; if there is just one value for the month then it is entered into this attribute; If there are a range of values for the month then the minimum value is found in this attribute |
| month_max | Month names are completely spelled out with the first letter capitalized; if there are a range of values for the month then the maximum value is found in this attribute |
| date_min | Expressed using year, month, and day; if there is just one value for the date then it is entered into this attribute; if there are a range of values for the date then the minimum value is found in this attribute |
| date_max | Expressed using year, month, and day; if there are a range of values for the date then the maximum value is found in this attribute |
| time_min | HH:MM:SS; if there is just one value for the time then it is found in this attribute; if there are a range of values for the time then the minimum value is found in this attribute |
| time_max | HH:MM:SS; if there are a range of values for the time then the maximum value is found in this attribute |
| lat_min | If there is just one value for the latitude then it is found in this attribute; if there are a range of values for the latitude then the minimum value is found in this attribute; values are in decimal degrees format |
| lat_max | If there are a range of values for the latitude then the maximum value is found in this attribute; values are in decimal degrees format |
| lon_min | If there is just one value for the longitude then it is entered into this attribute; if there are a range of values for the longitude then the minimum value is entered into this attribute; values are in decimal degrees format |
| lon_max | If there are a range of values for the longitude then the maximum value is found in this attribute; values are in decimal degrees format |
| environmental_data_type | The environmental data type reported in the source (e.g., temperature, salinity) |
| measurement_depth | The depth associated with the environmental parameter measurement; if the measurement in reported to be at the surface (e.g., sea surface temperature) then the measurement depth value is assigned to 0 |
| depth_units | The units associated with the measurement depth; units are fully spelled and plural (e.g., meters instead of meter) |
| environmental_subsample_id | A unique number that is generated and assigned to environmental samples if there are no other unique identifiers; values are assigned for each source starting from 1 and increasing by a value of 1 each time (e.g., 1,2,3…) |
| environmental_notes | Any additional comments on the environmental parameter measurement |
| value | The value of the environmental parameter |
| mean | The mean of the environmental parameters |
| error | The error associated with the environmental parameter mean |
| min | If there are a range of values for the environmental parameter this attribute represents the minimum value |
| max | If there are a range of values for the environmental parameter this attribute represents the maximum value |
| environmental_units | The units associated with the environmental parameter; units are fully spelled and plural (e.g., micrograms per liter instead of microgram per liter) |
This table corresponds to the ‘Environmental_data’ file in the figshare repository.
Database framework
We built an open-access relational database in MySQL v8.0.18 called the “North Pacific Marine Salmon Diet Database”20,21. The North Pacific Marine Salmon Diet Database contains all of the extracted data noted above: diet data, salmon biological data, prey biological data, and environmental data. This database also allows for inclusion of prey biological data that are not associated with a salmon sample. For example, if a researcher conducted a zooplankton tow for potential prey and they have biological data for these potential prey (e.g., length, weight), these data can be added to the database. In this database, all data are linked by site, which has both a temporal and spatial component. All data are also related to a source in order to distinguish related data and trace its origins. While the database was built specifically to house North Pacific salmon diet data from the marine environment, the database structure was designed to easily be applied to other predator and prey interactions with only slight modifications.
Data Records
The North Pacific Marine Salmon Diet Database currently contains 6,869 diet observations from 6,305 unique diet samples of over 69,942 salmon. Types of diet data included percent weight of prey, absolute weight of prey, average weight of prey, percent volume of prey, percent number of prey, absolute number of prey, average number of prey, frequency of occurrence (numerical and percent), stomach content index, index of fullness, and index of relative importance. The database also houses 11,965 observations of salmon biological parameters for 6,172 unique salmon samples. One observation means one biological parameter measured for one sample of salmon, which can contain one or many fish. Salmon biological parameters include length, weight, daily ration, empty stomachs, male/female ratio and many others. Additionally, the database includes 238 observations of prey biological parameters from 112 unique prey taxonomic categories. Prey biological parameters include body length, body weight, body width and size index. Finally, the database contains 2,790 observations of environmental parameters. Environmental parameters include temperature, salinity and others. These data are available in a static figshare repository22. The most current version of the relational database, associated documentation, and data are available in a dynamic GitHub repository, which will be updated as more sources are identified and added to the database21. Data were visualized using R statistical software v3.6.123.
Spatial and temporal coverage
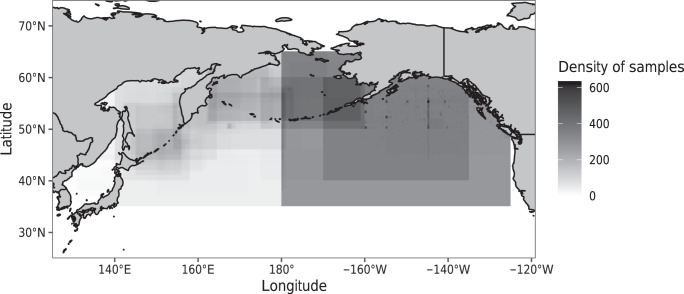
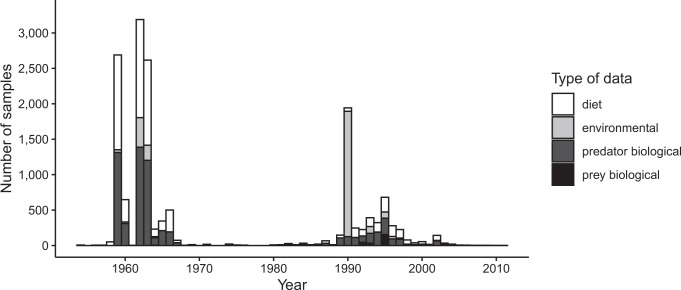
Diet samples were collected at 751 unique spatial locations, which included areas (polygons), transects, and point locations across the North Pacific from the California Current to the Sea of Japan (Fig. 1). Salmon biological data were collected from 709 locations, prey biological data from 4 locations, and environmental data from 446 locations. Salmon biological data were reported across the entire North Pacific, while prey biological data were sparsely reported from a few locations in the Gulf of Alaska and the Sea of Okhotsk/Kuril Islands. Environmental data were mainly available from the eastern and central North Pacific Ocean. Since our search was focused on specific time periods, most of the data we collected fell within our specified decadal periods: 1959–1969 and 1987–1997 (Fig. 2). However, some sources reported data from other time periods and these data were also included in the database. While diet data and salmon biological data were consistently reported across the temporal range, environmental data and prey biological data were inconsistently reported.
Fig. 1.
The spatial distribution of diet samples across the North Pacific Ocean. The density of diet samples, in the form of points, lines, and polygons (rectangles) based on the latitude and longitude minimum and maximum values.
Fig. 2.
The number of samples for each type of data reported across the temporal range 1950–2011. If a single sample consisted of data from multiple years, then the median year was selected to represent the sample and half years were rounded down.
Salmon and prey species coverage
Sockeye (Oncorhynchus nerka), pink (Oncorhynchus gorbuscha) and chum (Oncorhynchus keta) were reported most frequently in our database, while coho (Oncorhynchus kisutch), Chinook (Oncorhynchus tshawytscha) and steelhead (Oncorhynchus mykiss) were reported less frequently (Table 2). The most commonly reported prey groups were amphipods, fish (Class Actinopterygii), euphausiids, cephalopods (Subclass Coleoidea), and copepods (Table 3). The category ‘miscellaneous’ was also commonly reported, although it usually made up just a small percentage of the diets. Within the diet data reported in the database, there are 186 unique prey taxa, meaning the lowest taxonomic classifications of prey items. Only 18.5% of salmon diet data were reported to the species level, while the majority were reported to higher taxonomic levels (e.g., Amphipoda, Decapoda, Euphausiacea).
Table 2.
The number of diet samples in the database for each species of salmon.
| Salmon species | Number of samples |
|---|---|
| Sockeye (Oncorhynchus nerka) | 2287 |
| Pink (Oncorhynchus gorbuscha) | 1768 |
| Chum (Oncorhynchus keta) | 1526 |
| Coho (Oncorhynchus kisutch) | 293 |
| Chinook (Oncorhynchus tshawytscha) | 271 |
| Steelhead (Oncorhynchus mykiss) | 160 |
Table 3.
The number of diet samples containing each prey taxonomy.
| Prey taxonomy | Number of samples |
|---|---|
| Amphipoda | 2446 |
| Actinopterygii | 2436 |
| Euphausiacea | 2153 |
| Miscellaneous | 2026 |
| Coleoidea | 2009 |
| Copepoda | 1460 |
| Pteropoda | 697 |
| Limacina | 595 |
| Themisto japonica | 584 |
| Thysanoessa longipes | 525 |
| Limacina helicina | 438 |
| Brachyura | 389 |
| Neocalanus cristatus | 353 |
| Clione limacina | 319 |
| Hyperiidae | 299 |
| Hemilepidotus | 290 |
| Decapoda | 241 |
| Anomura | 200 |
| Gelatinous collective | 195 |
| Euphausia pacifica | 180 |
| Myctophidae | 179 |
| Hyperia medusarum | 148 |
| Polychaeta | 140 |
| Calanoida | 131 |
| Chaetognatha | 122 |
| Cephalopoda | 118 |
| Parasagitta elegans | 115 |
| Primno macropa | 110 |
| Sagitta | 96 |
| Euphausiidae | 90 |
| Rhynchonereella angelini | 79 |
| Eucalanus bungii | 74 |
| Neocalanus plumchrus | 73 |
| Insecta | 68 |
| Thysanoessa raschii | 68 |
| Oikopleura | 67 |
| Gadus chalcogrammus | 60 |
| Clupea pallasii | 56 |
| Gammaridae | 55 |
| Themisto pacifica | 55 |
| Gonatopsis borealis | 49 |
| Crustacea | 47 |
| Hyperia galba | 44 |
| Mallotus villosus | 43 |
| Gonatus kamtschaticus | 40 |
| Mysida | 40 |
| Ammodytes hexapterus | 39 |
| Ammodytidae | 35 |
| Thysanoessa inspinata | 35 |
| Gonatidae | 33 |
| Hexagrammidae | 32 |
| Cnidaria | 29 |
| Pleurogrammus | 28 |
| Harpacticoida | 27 |
| Themisto | 27 |
| Berryteuthis magister | 26 |
| Engraulis japonicus | 26 |
| Nekton collective | 26 |
| Thysanoessa inermis | 26 |
| Tunicata | 25 |
| Appendicularia | 23 |
| Leuroglossus schmidti | 22 |
Prey taxonomy refers to the lowest taxonomic level identified by the source. Prey taxonomies that were reported in less than 20 samples were excluded.
Technical Validation
Standardization procedures were used to verify and collate the data. Since some of the taxonomic records were outdated, the taxonomies were verified and updated using the World Register of Marine Species (http://www.marinespecies.org/). For types of diet data that should add to a cumulative percentage of 100 (e.g., percent weight, percent volume), diet samples were excluded if the cumulative percent of prey was above 105% or below 95%. If the cumulative percentage did not add to 100 but still fell within this range, diet data values for that sample were rescaled to add to 100. For other metrics, including absolute and average weight and number of prey, as well as numerical frequency of occurrence, we consulted a salmon diet expert to determine if our highest values were reasonable to find in adult salmon stomachs (V. Zahner, pers. comm.).
Usage Notes
The North Pacific Marine Salmon Diet Database contains observations from many different sources, which report data with varying levels of detail. The method for data digitalization was to extract data as it was presented in the sources, or as close to it as possible. In this way, we have provided a more complete set of data that can be used for a variety of studies on salmon ecology and North Pacific ecosystems and left the manipulation and interpretation of data up to the discretion of the user.
The North Pacific Marine Salmon Diet Database is publicly available and can be used under the license of CC BY, meaning that the work can be distributed, remixed, adapted and built upon with acknowledgement of authors. There are a number of ways to access the information contained in the database. A static copy of the initial data contribution to the North Pacific Marine Salmon Diet Database described in this paper is available in a figshare repository in the form of seven csv files (‘Diet_data’, ‘Prey_biological_data’, ‘Sources’, ‘Predator_biological_data’, ‘Environmental_data’, ‘Gear_type_prey’, ‘Gear_type_predator’)22. In this case, ‘predator’ refers to salmon, but the database was designed so that it could easily be applied to other predator and prey interactions and thus we used the predator-prey terminology. Detailed information about the variables in the database can be found in Online-only Tables 2–8, which corresponds to the csv files in the figshare repository. To avoid making assumptions about how the data will be used, we have included all available data from the sources. This means that in some cases the same diet samples are reported multiple times using different diet metrics. In the figshare repository, there is R v3.6.1 code (‘Preprocessing_data’) to exclude overlapping data based on diet metrics of interest: presence/absence, weight/volume, frequency of occurrence, and numerical.
A dynamic version of the North Pacific Marine Salmon Diet Database can be accessed through a GitHub repository and will be updated as more historic data are digitized and made available by the Pelagic Ecosystems Laboratory at the University of British Columbia21. The GitHub repository contains updated versions of the files found in the figshare repository, in addition to documentation for the North Pacific Marine Salmon Diet Database ('npmsdd_documentation’), a data entry template (‘npmsdd_data_entry_template’), and a dumped version of the MySQL relational database (‘npmsdd_MySQL_database_dump’) for users that are interested in working with the database in the relational database format.
Acknowledgements
This project was made possible by funding from the Ambrose Monell and G. Unger Vetlesen Foundations, and the University of British Columbia. We would like to acknowledge Raymond Ng for his guidance on the database design, Sally Taylor for her help in the literature review process, Tim Cashion for his assistance with figures, and Marina Espinasse and Carmen Manuel for their assistance with data entry. We would also like to acknowledge Svetlana Naydenko, Alexey Khoruzhiy and Alexei Somov for helping to locate sources of data and Tim Spesivy and Kyoko Adachi for translating Russian and Japanese sources.
Online-only Tables
Author contributions
Caroline Graham: systematic literature review, data extraction and standardization, database structure and creation, drafting and revision of the article. Evgeny A. Pakhomov: project concept, project funding, revision of the article. Brian P. V. Hunt: project concept, project funding, database structure, revision of the article.
Code availability
The figshare repository contains R v3.6.1 code for downstream data processing (‘Preprocessing_data’), and code to create the figures that appear in this publication (‘Publication_figures’)22. The GitHub repository contains the most up-to-date code for downstream data processing21.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pacific Salmon Life Histories. (eds. Groot, C & Margolis, L.) (University of British Columbia Press, 1991).
- 2.Beamish RJ, Mahnken C. A critical size and period hypothesis to explain natural regulation of salmon abundance and the linkage to climate and climate change. Prog. Oceanogr. 2001;49:423–437. doi: 10.1016/S0079-6611(01)00034-9. [DOI] [Google Scholar]
- 3.Bradford MJ. Comparative review of Pacific salmon survival rates. Can. J. Fish. Aquat. Sci. 1995;52:1327–1338. doi: 10.1139/f95-129. [DOI] [Google Scholar]
- 4.Mueter FJ, Peterman RM, Pyper BJ. Corrigendum: Opposite effects of ocean temperature on survival rates of 120 stocks of Pacific salmon (Oncorhynchus spp.) in northern and southern areas. Can. J. Fish. Aquat. Sci. 2003;60:757–757. doi: 10.1139/f03-063. [DOI] [Google Scholar]
- 5.Zimmerman MS, et al. Spatial and temporal patterns in smolt survival of wild and hatchery coho salmon in the Salish Sea. Mar. Coast. Fish. 2015;7:116–134. doi: 10.1080/19425120.2015.1012246. [DOI] [Google Scholar]
- 6.Dale KE, Daly EA, Brodeur RD. Interannual variability in the feeding and condition of subyearling Chinook salmon off Oregon and Washington in relation to fluctuating ocean conditions. Fish. Oceanogr. 2017;26:1–16. doi: 10.1111/fog.12180. [DOI] [Google Scholar]
- 7.Davis ND, et al. Review of BASIS salmon food habits studies. North Pacific Anadromous Fish Comm. Bull. 2009;5:197–208. [Google Scholar]
- 8.Qin Y, Kaeriyama M. Feeding habits and trophic levels of Pacific salmon (Oncorhynchus spp.) in the North Pacific Ocean. North Pacific Anadromous Fish Comm. Bull. 2016;6:469–481. doi: 10.23849/npafcb6/469.481. [DOI] [Google Scholar]
- 9.Chapman, W. M. The Pilchard Fishery of the State of Washington in 1936 with Notes on the Food of the Silver and Chinook Salmon off the Washington Coast. Biological Report No. 36C (State of Washington, Division of Scientific Research, Department of Fisheries, 1936).
- 10.Silliman RP. Fluctuations in the diet of the Chinook and silver salmons (Oncorhynchus tschawytscha and O. kisutch) off Washington, as related to the troll catch of salmon. Copeia. 1941;1941:80–87. doi: 10.2307/1437436. [DOI] [Google Scholar]
- 11.Brodeur RD, Daly EA, Schabetsberger RA, Mier KL. Interannual and interdecadal variability in juvenile coho salmon (Oncorhynchus kisutch) diets in relation to environmental changes in the northern California Current. Fish. Oceanogr. 2007;16:395–408. doi: 10.1111/j.1365-2419.2007.00438.x. [DOI] [Google Scholar]
- 12.Brodeur, R. D. A Synthesis of the Food Habits and Feeding Ecology of Salmonids in Marine Waters of the North Pacific. INPFC Doc; FRI-UW-9016. (Fisheries Research Institute, University of Washington, 1990).
- 13.Karpenko VI, Volkov F, Koval MV. Diets of Pacific salmon in the Sea of Okhotsk. Bering Sea, and Northwest Pacific Ocean. North Pacific Anadromous Fish Comm. Bull. 2007;4:105–116. [Google Scholar]
- 14.Starovoytov AN. Trends in abundance and feeding of chum salmon in the Western Bering Sea. North Pacific Anadromous Fish Comm. Bull. 2007;4:45–51. [Google Scholar]
- 15.Kaeriyama M, et al. Change in feeding ecology and trophic dynamics of Pacific salmon (Oncorhynchus spp.) in the central Gulf of Alaska in relation to climate events. Fish. Oceanogr. 2004;13:197–207. doi: 10.1111/j.1365-2419.2004.00286.x. [DOI] [Google Scholar]
- 16.Jamieson, G., Livingston, P. & Zhang, C.-I. Report of Working Group 19 on Ecosystem-based Management Science and its Application to the North Pacific. PICES Scientific Report 37 (North Pacific Marine Science Organization, 2010).
- 17.Schoen ER, et al. Future of Pacific salmon in the face of environmental change: Lessons from one of the world’s remaining productive salmon regions. Fisheries. 2017;42:538–553. doi: 10.1080/03632415.2017.1374251. [DOI] [Google Scholar]
- 18.Healey M. The cumulative impacts of climate change on Fraser River sockeye salmon (Oncorhynchus nerka) and implications for management. Can. J. Fish. Aquat. Sci. 2011;68:718–737. doi: 10.1139/f2011-010. [DOI] [Google Scholar]
- 19.Carmack E, Winsor P, Williams W. The contiguous panarctic Riverine Coastal Domain: A unifying concept. Prog. Oceanogr. 2015;139:13–23. doi: 10.1016/j.pocean.2015.07.014. [DOI] [Google Scholar]
- 20.MySQL version 8.0.18. MySQL, https://www.mysql.com/ (2019).
- 21.Graham, C, Pakhomov, E. A., & Hunt, B. P. V. North Pacific Marine Salmon Diet Database. GitHub, https://github.com/mcarolinegraham/North_Pacific_Marine_Salmon_Diet_Database (2020). [DOI] [PMC free article] [PubMed]
- 22.Graham C, Pakhomov EA, Hunt, BPV. 2020. A salmon diet database for the North Pacific Ocean. figshare. [DOI] [PMC free article] [PubMed]
- 23.R Core Development Team. R: A language and environment for statistical computing, version 3.6.1. The R Project for Statistical Computinghttps://www.r-project.org/ (2019).
- 24.Andrievskaya LD. Food relationships of the Pacific salmon in the sea. Vopr. Ikhtiologii. 1966;6:84–90. [Google Scholar]
- 25.Carlson HR. Foods of juvenile sockeye salmon, Oncorhynchus nerka, in the inshore coastal waters of Bristol Bay, Alaska, 1966–67. Fish. Bull. 1976;74:458–462. [Google Scholar]
- 26.Chuchukalo, V. L., Volkov, A. F., Efimkin, A. Y. & Kuznetsova, N. A. Feeding and Daily Rations of Sockeye Salmon (Oncorhynchus nerka) During the Summer Period. NPAFC Doc. 125 (Pacific Research Institute of Fisheries Oceanography (TINRO), 1995).
- 27.Davis, N. D., Takahashi, M. & Ishida, Y. The 1996 Japan-U.S. Cooperative High-seas Salmon Research Cruise of the Wakatake maru and a Summary of 1991-1996 Results. NPAFC Doc. 194; FRI-UW-9617 (Fisheries Research Institute, University of Washington; National Research Institute of Far Seas Fisheries, 1996).
- 28.Davis, N. D., Aydin, K. Y. & Ishida, Y. Diel Feeding Habits and Estimates of Prey Consumption of Sockeye, Chum, and Pink Salmon in the Bering Sea in 1997. NPAFC Doc. 363; FRI-UW-9816 (Fisheries Research Institute, University of Washington; National Research Institute of Far Seas Fisheries, 1998).
- 29.Davis ND, Aydin KY, Ishida Y. Diel catches and food habits of sockeye, pink, and chum salmon in the Central Bering Sea in summer. North Pacific Anadromous Fish Comm. Bull. 2000;2:99–109. [Google Scholar]
- 30.Dulepova, E. P. & Dulepov, V. I. Interannual and Interregional Analysis of Chum Salmon Feeding Features in the Bering Sea and Adjacent Pacific Waters of Eastern Kamchatka. NPAFC Doc. 728 (Pacific Research Fisheries Centre, TINRO-Centre, 2003).
- 31.Fukataki H. Stomach contents of the pink salmon, Oncorhynchus gorbuscha (Walbaum), in the Japan Sea during the spring season of 1965. Bull. Jap. Sea Reg. Fish. Res. Lab. 1967;17:49–66. [Google Scholar]
- 32.Glebov, I. I. Chinook and Coho Salmon Feeding Habits in the Far Eastern Seas in the Course of Yearly Migration Cycle. NPAFC Doc. 378 (Pacific Research Fisheries Centre TINRO-Centre, 1998).
- 33.Ito J. Food and feeding habits of Pacific salmon (genus Oncorhynchus) in their oceanic life. Bull. Hokkaido Reg. Fish. Res. Lab. 1964;29:85–97. [Google Scholar]
- 34.Kaeriyama M, et al. Feeding ecology of sockeye and pink salmon in the Gulf of Alaska. North Pacific Anadromous Fish Comm. Bull. 2000;2:55–63. [Google Scholar]
- 35.Kanno Y, Hamai I. Food of salmonid fish in the Bering Sea in summer of 1966. Bull. Fac. Fish. Hokkaido Univ. 1971;22:107–128. [Google Scholar]
- 36.Manzer JI. Food of Pacific salmon and steelhead trout in the Northeast Pacific Ocean. J. Fish. Res. Board Canada. 1968;25:1085–1089. doi: 10.1139/f68-094. [DOI] [Google Scholar]
- 37.Perry RI, Hargreaves NB, Waddell BJ, Mackas DL. Spatial variations in feeding and condition of juvenile pink and chum salmon off Vancouver Island, British Columbia. Fish. Oceanogr. 1996;5:73–88. doi: 10.1111/j.1365-2419.1996.tb00107.x. [DOI] [Google Scholar]
- 38.Tadokoro K, Ishida Y, Davis ND, Ueyanagi S, Sugimoto T. Change in chum salmon (Oncorhynchus keta) stomach contents associated with fluctuation of pink salmon (O. gorbuscha) abundance in the central subarctic Pacific and Bering Sea. Fish. Oceanogr. 1996;5:89–99. doi: 10.1111/j.1365-2419.1996.tb00108.x. [DOI] [Google Scholar]
- 39.Takeuchi I. Food animals collected from the stomachs of three salmonid fishes (Oncorhynchus) and their distribution in the natural environments in the northern North Pacific. Bull. Hokkaido Reg. Fish. Res. Lab. 1972;38:1–119. [Google Scholar]
- 40.Ueno M, Kosaka S, Ushiyama H. Food and feeding behavior of Pacific salmon—II. Sequential change of stomach contents. Bull. Japanese Soc. Sci. Fish. 1969;35:1060–1066. doi: 10.2331/suisan.35.1060. [DOI] [Google Scholar]
- 41.Volkov, A. F., Chuchukalo, V. I., Efimkin, A. Y. Feeding of Chinook and Coho Salmon in the Northwestern Pacific Ocean. NPAFC Doc. 124 (Pacific Research Institute of Fisheries Oceanography, 1995).
- 42.Auburn ME, Ignell SE. Food habits of juvenile salmon in the Gulf of Alaska July–August 1996. North Pacific Anadromous Fish Comm. Bull. 2000;2:89–97. [Google Scholar]
- 43.Aydin, K. Y. Abiotic and Biotic Factors Influencing Food Habits of Pacific Salmon in the Gulf of Alaska. In Technical report: Workshop of Climate Change and Salmon Production (ed. Myers, K. W.) 39–40 (North Pacific Anadromous Fish Commission, 1998).
- 44.Daly EA, Brodeur RD. Warming ocean conditions relate to increased trophic requirements of threatened and endangered salmon. PLoS One. 2015;10:e0144066. doi: 10.1371/journal.pone.0144066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis, N. D., Armstrong, J. L. & Myers, K. W. Bering Sea Salmon Food Habits: Diet Overlap in Fall and Potential for Interactions Among Salmon. SAFS-UW-0311 (Fisheries Research Institute, School of Aquatic and Fisheries Sciences, University of Washington, 2003).
- 46.Kawamura H, Miyamoto M, Nagata M, Hirano K. Interaction between chum salmon and fat greenling juveniles in the coastal Sea of Japan off northern Hokkaido. North Pacific Anadromous Fish Comm. Bull. 1998;1:412–418. [Google Scholar]
- 47.Ueno Y. Deepwater migrations of chum salmon (Oncorhynchus keta) along the Pacific coast of northern Japan. Can. J. Fish. Aquat. Sci. 1992;49:2307–2312. doi: 10.1139/f92-253. [DOI] [Google Scholar]
- 48.Ueno Y, Seki J, Shimizu IP, Shershnev A. Large juvenile chum salmon Oncorhynchus keta collected in coastal waters of Iturup Island. Nippon Suisan Gakkaishi. 1992;58:1393–1397. doi: 10.2331/suisan.58.1393. [DOI] [Google Scholar]
- 49.Waddell BJ, Morris JFT, Healey MC. The abundance, distribution, and biological characteristics of Chinook and coho salmon on the fishing banks off southwest Vancouver Island, May 18-30, 1989 and April 23-May 5, 1990. Can. Tech. Rep. Fish. Aquat. Sci. 1992;1891:1–113. [Google Scholar]
- 50.Andrievskaya LD. The feeding of Pacific salmon fry in the sea. Proceedings of the Pacific Research Institute of Fisheries and Oceanography. 1970;64:73–80. [Google Scholar]
- 51.Atcheson ME, Myers KW, Beauchamp DA, Mantua NJ. Bioenergetic response by steelhead to variation in diet, thermal habitat, and climate in the North Pacific Ocean. Trans. Am. Fish. Soc. 2012;141:1081–1096. doi: 10.1080/00028487.2012.675914. [DOI] [Google Scholar]
- 52.Carlson, H. R. et al. Cruise Report of the F/V Great Pacific Survey of Young Salmon in the North Pacific–Dixon Entrance to Western Aleutians—July–August 1996. NPAFC Doc. 222 (Auke Bay Laboratory, Alaska Fisheries Science Center, National Marine Fisheries Service, National Oceanic and Atmospheric Administration, 1996).
- 53.Davis ND, Fukuwaka M, Armstrong JL, Myers KW. Salmon food habits studies in the Bering Sea. 1960 to present. North Pacific Anadromous Fish Comm. Tech. Rep. 2005;6:24–28. [Google Scholar]
- 54.Myers, K. W. & Aydin, K. Y. The 1996 International Cooperative Salmon Research Cruise of the Oshoro maru and a Summary of 1994-1996 Results. NPAFC Doc. 195; FRI-UW-9613 (University of Washington, Fisheries Research Institute, 1996).
- 55.Myers, K. W. et al. Migrations, Abundance, and Origins of Salmonids in Offshore Waters of the North Pacific – 1995. NPAFC Doc. 152; FRI-UW-9613 (University of Washington, Fisheries Research Institute, 1995).
- 56.Sturdevant, M. V, Ignell, S. E. & Morris, J. Diet of Juvenile Salmon off Southeastern Alaska, October-November 1995. NPAFC Doc. 275 (Auke Bay Laboratory, Alaska Fisheries Science Center, National Marine Fisheries Service, National Oceanic and Atmospheric Administration, 1997).
- 57.Walker, R. V. Summary of Cooperative U.S.-Japan High Seas Salmonid Research Aboard the Japanese Research Vessel Oshoro Maru, 1993. NPAFC Doc. 21 (Fisheries Research Institute, University of Washington, 1993).
- 58.Suzuki T, et al. Feeding selectivity of juvenile chum salmon in the Japan Sea Coast of Northern Honshu. Sci. Reports Hokkaido Salmon Hatch. 1994;48:11–16. [Google Scholar]
- 59.Shimazaki K, Mishima S. On the diurnal change of the feeding activity of salmon in the Okhotsk Sea. Bull. Fac. Fish. Hokkaido University. 1969;20:82–93. [Google Scholar]
- 60.Weitkamp, L. A. Ocean Conditions, Marine Survival, and Performance of Juvenile Chinook (Oncorhynchus tshawytscha) and Coho (O. kisutch) Salmon in Southeast Alaska. PhD thesis, University of Washington (2004).
- 61.Starovoytov AN. Chum salmon (Oncorhynchus keta (Walbaum)) in the Far East Seas - biological description of the species 2. Diet composition and trophic linkages of chum salmon in the Far East Seas and adjacent waters of the Northwest Pacific Ocean. Izv. TINRO. 2003;133:3–34. [Google Scholar]
- 62.LeBrasseur, R. J. & Doidge, D. A. Stomach Contents of Salmonids Caught in the Northeastern Pacific Ocean - 1959 & 1960. In Circular, Statistical Series. vol. 3 (Fisheries Research Board of Canada, 1966).
- 63.Lebrasseur, R. J. & Doidge, D. A. Stomach Contents of Salmonids Caught in the Northeastern Pacific Ocean – 1962. In Circular, Statistical Series. vol. 4 (Fisheries Research Board of Canada, 1966).
- 64.Lebrasseur, R. J. & Doidge, D. A. Stomach Contents of Salmonids Caught in the Northeastern Pacific Ocean - 1963 & 1964. In Circular, Statistical Series. vol. 5 (Fisheries Research Board of Canada, 1966).
- 65.Ishida Y, Davis ND. Chum salmon feeding habits in relation to growth reduction. Salmon Rep. Ser. 1999;47:104–110. [Google Scholar]
- 66.Tamura R, Shimazaki K, Ueno Y. Trophic relations of juvenile salmon (genus Oncorhynchus) in the Okhotsk Sea and Pacific waters off the Kuril Islands. Salmon Rep. Ser. 1999;47:138–168. [Google Scholar]
- 67.Seki J, Shimizu I. Diel migration of zooplankton and feeding behavior of juvenile chum salmon in the central Pacific coast of Hokkaido. Bull. Nat. Salmon Resour. Cent. 1998;1:13–27. [Google Scholar]
- 68.Suzuki T, Fukuwaka M, Kawana M, Ohkuma K, Seki J. Investigation on survival mechanism of juvenile chum salmon during the early sea life in 1994. Salmon Database. 1995;3:59–68. [Google Scholar]
- 69.Andrievskaya LD. The feeding of pink salmon in the wintering areas in the Sea of Japan. Izv. TINRO. 1974;90:97–110. [Google Scholar]
- 70.Andrievskaya LD. Feeding of Pacific salmon juveniles in the Sea of Okhotsk. Izv. TINRO. 1970;78:105–115. [Google Scholar]
- 71.Chuchukalo, V. I., Volkov, A. F., Efimkin, Ay. & Blagoderov, A. I. Distribution and feeding of the Chinook salmon (Oncorhynchus tschawytscha) in the northwest Pacific. Izv. TINRO, 137–141 (1994).
- 72.Gorbatenko KM. Food and feeding habits of juvenile pink and chum salmons in the epipelagic zone of the Okhotsk Sea in winter. Izv. TINRO. 1996;199:234–243. [Google Scholar]
- 73.Kayev AM, Chupakhin VM, Fedotova NA. Feeding peculiarities and interrelationships between juvenile salmons in coastal waters of the Etorofu Island. Vopr. Ikhtiologii. 1993;33:215–224. [Google Scholar]
- 74.Klovatch, N. V. Ecological Consequences of Large-scale Breeding Operations of Chum Salmon (Oncorhynchus keta). PhD extended summary (VNIRO, 2002).
- 75.Shershnev AP, Chupakhin VM, Rudnev VA. Some features of the ecology of young Sakhalin and Iturup pink salmon Oncorhynchus gorbuscha (Walbaum) (Salmonidae) during marine period of life. Vopr. Ikhtiologii. 1982;22:441–448. [Google Scholar]
- 76.Tutubalin, B. G. & Chuchukalo, V. I. The Feeding of Genus Oncorhynchus Pacific Salmons in the North Pacific During the Winter-Spring Period. In Living Resources of the Pacific Ocean: Collected Papers (eds. Gristenko, O. F., Churikov, A. A. & Klovach, N. V.) 77–85 (VNIRO, 1992).
- 77.Volkov AF. Food and feeding habits of young Pacific salmon in the Okhotsk Sea during the autumn-winter period. Okeanologiya. 1996;36:80–85. [Google Scholar]
- 78.Volkov AF. Food and feeding habits of pink, chum and sockeye salmon during their anadromous migrations. Izv. TINRO. 1994;116:128–137. [Google Scholar]
- 79.Fisheries Agency of Japan. Report on research by Japan for the International North Pacific Fisheries Commission during the year 1965. International North Pacific Fisheries Commission Ann. Rep., 42–55 (1965).
- 80.Davis, N. D. U.S.-Japan Cooperative High Seas Salmonid Research in 1990: Summary of Research Aboard the Japanese Research Vessel Hokuho Maru, 4 June to 19 July. INPFC Doc.; FRI-UW-9010. (Fisheries Research Institute, University of Washington, 1990).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Graham C, Pakhomov EA, Hunt, BPV. 2020. A salmon diet database for the North Pacific Ocean. figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The figshare repository contains R v3.6.1 code for downstream data processing (‘Preprocessing_data’), and code to create the figures that appear in this publication (‘Publication_figures’)22. The GitHub repository contains the most up-to-date code for downstream data processing21.