Abstract
Background
To evaluate the diagnostic performance of apparent diffusion coefficient (ADC) histogram parameters for differentiating the genetic subtypes in lower-grade diffuse gliomas and explore which segmentation method (ROI-1, the entire tumor ROI; ROI2, the tumor ROI excluding cystic and necrotic portions) performs better.
Materials and Methods
We retrospectively evaluated 56 lower-grade diffuse gliomas and divided them into three categories: IDH-wild group (IDHwt, 16cases); IDH mutant with the intact 1p or 19q group (IDHmut/1p19q+, 18cases); and IDH mutant with the 1p/19q codeleted group (IDHmut/1p19q−, 22cases). Histogram parameters of ADC maps calculated with the two different ROI methods: ADCmean, min, max, mode, P5, P10, P25, P75, P90, P95, kurtosis, skewness, entropy, StDev, and inhomogenity were compared between these categories using the independent t test or Mann–Whitney U test. For statistically significant results, a receiver operating characteristic (ROC) curves were constructed, and the optimal cutoff value was determined by maximizing Youden's index. Area under the curve (AUC) results were compared using the method of Delong et al.
Results
The inhomogenity from the two different ROI methods for distinguishing IDHwt gliomas from IDHmut gliomas both showed the biggest AUC (0.788, 0.930), the optimal cutoff value was 0.229 (sensitivity, 81.3%; specificity, 75.0%) for the ROI-1 and 0.186 (sensitivity, 93.8%; specificity, 82.5%) for the ROI-2, and the AUC of the inhomogenity from the ROI-2 was significantly larger than that from another segmentation, but no significant differences were identified between the AUCs of other same parameters from the two different ROI methods. For the differentiaiton of IDHmut/1p19q− tumors and IDHmut/1p19q+ tumors, with the ROI-1, the ADCmode showed the biggest AUC (AUC: 0.784; sensitivity, 61.1%; specificity, 90.9%), with the ROI-2, and the skewness performed best (AUC, 0.821; sensitivity, 81.8%; specificity, 77.8%), but no significant differences were identified between the AUCs of the same parameters from the two different ROI methods.
Conclusion
ADC values analyzed by the histogram method could help to classify the genetic subtypes in lower-grade diffuse gliomas, no matter which ROI method was used. Extracting cystic and necrotic portions from the entire tumor lesions is preferable for evaluating the difference of the intratumoral heterogeneity and classifying IDH-wild tumors, but not significantly beneficial to predicting the 1p19q genotype in the lower-grade gliomas.
1. Introduction
Glioma is the most common neuroepithelial tumor in the brain which accounts for 80% of the malignant brain tumors. The severity of gliomas is further distinguished by malignant grades (I to IV) on the basis of the histopathological and clinical criteria [1]. The grade II and III gliomas are sometimes described as lower-grade gliomas, which present approximately one-third of all gliomas. Lower-grade gliomas form a biologically heterogeneous group of tumors. They are usually less aggressive tumors with a longer, indolent clinical course, but a subset of these gliomas will progress to glioblastoma (WHO grade IV gliomas) within months [2, 3]. Histology alone is often insufficient to make accurate prognostic estimates, and tumors belonging to the same WHO grade may display different malignant behavior, depending on their molecular profile. The Cancer Genome Atlas (TCGA) Analysis Working Group grouped LGGs into three robust molecular classes on the basis of mutations in isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2, hereafter collectively referred to as IDH) and codeletion of chromosomes 1p and 19q. LGGs without IDH mutation are associated with the most aggressive clinical behavior and worst outcome, similar to that of glioblastomas (WHO grade IV). LGGs with IDH mutation and 1p/19q codeletion are associated with the most favorable clinical outcome and possibly improved sensitivity to procarbazine, lomustine, and vincristine chemotherapy compared with noncodeleted neoplasms. LGGs with IDH mutation and no 1p/19q codeletion are associated with an intermediate outcome, worse than those with 1p/19q codeletion, but far more favorable than IDHwt neoplasms [3]. This molecular classification has been integrated into the 2016 WHO classification of brain tumors [4].
Diffusion-weighted imaging (DWI) is a physiologic imaging modality that exploits the diffusion of water molecules to create contrast between tissues. Apparent diffusion coefficient (ADC) calculated from DWI is used as a quantitative parameter to assess the grade of restrictive diffusion and to provide information about tissue structure and cellularity [5, 6]. Previous studies have demonstrated the ability of ADC to differentiate the IDH-wild gliomas from the mutant ones, the 1p19q codeleted gliomas from those noncodeleted ones [7–9]. However, previous studies were limited to using the mean value of ADCs based on regions of interest (ROIs) from a single representative slice of a lesion or region of interest from tumor volume, which may dilute or even mask the small but important differences between different disease entities. Additionally, they may not precisely depict the tumor status due to the intrinsic heterogeneous environment of tumors. Histogram analysis of the whole lesion may offer multiple parameters containing not only the quantitative accumulated ADC parameters, such as percentiles, minimal and maximal values, and mode but also the distribution parameters, such as the kurtosis, skewness, range, StDev, inhomogenity, and entrophy, thus providing more information about the tumor heterogeneity than the mean values [10, 11]. In previous studies, two main ROI placement methods of the ADC histogram were used, including the tumor ROI excluding cystic and necrotic portions [12–14] and the entire tumor ROI containing cystic and necrotic areas [11, 15–17]. The theoretical basis of the former mentioned method is that necrotic and cystic-appearing areas may increase ADC values, which may be a confusing factor for differentiating the subtypes of gliomas based on ADC maps, but the latter method contained all compositions of the tumor, theoretically, it can better assess the heterogeneity of the tumor in its entirety. So, the purposes of this retrospective study were to evaluate whether the ADC values analyzed by the histogram method could help to classify IDH-wild tumors from IDH-mutated ones as well as IDHmut-NonCodel tumors from IDHmut-Codel ones in lower-grade diffuse gliomas and determine which segmentation method performs better.
2. Materials and Methods
2.1. Patients
We searched the electronic hospital information system and picture archiving and communication system to identify patients from January 2016 to August 2019 who met the following inclusion criteria: (1) final histopathologic results were WHO grade II–III diffuse gliomas on the basis of the WHO classification for tumors of the central nervous system; (2) diffusion-weighted MRI, T2-weighted, and postcontrast T1-weighted anatomical scan performed at initial diagnosis and prior to any surgery; (3) and known IDH1 mutation and 1p/19q codeletion status. On the other hand, patients were excluded for the poor DWI images quality, which influence the consequent image analysis. The institutional review board at the First Affiliated Hospital of Chongqing Medical University approved this study. Thus, 56 consecutive patients were included in the final study cohort. The patients were divided into the following categories: IDH wild-group (IDHwt), IDH mutant with the intact 1p or 19q group (IDHmut/1p19q+), and IDH mutant with the 1p/19q codeleted group (IDHmut/1p19q−).
There were 40 cases in the IDH-mutated group (18 men, 22 women; age range, 23-66 years; mean age, 41.5 ± 10.5 years; WHO grade II gliomas, n = 32; grade III gliomas, n = 8). Among the IDH-mutated group, there were 18 cases of IDHmut/1p19q+gliomas (8 men, 10 women; age range, 24-59 years; mean age, 40.8 ± 9.8 years; WHO grade II gliomas, n = 15; grade III gliomas, n = 3) and 22 cases of the IDHmut/1p19q− group (10 men, 12 women; age range, 23-66 years; mean age, 42.0 ± 11.2 years; WHO grade II glomas, n = 17; grade III gliomas, n = 5). There were 16 cases in the IDHwt group (9 men, 7women; age range, 21-73 years; mean age, 51.9 ± 16.0 years; WHO grade II gliomas, n = 5; grade III gliomas, n = 11).
2.2. ADC Histogram Measurement
Each case was investigated using DWI (multishot echo-planar-imaging sequence with b values of 0 and 1000 s/mm2) obtained with a 3.0 T MRI scanner (Signa HDxt, GE Medical System, WI). The ADC maps were digitally transferred from the picture archiving and communication system workstation to a personal computer and processed with an in-house software (Firevoxel, available at https://wp.nyu.edu/firevoxel/). For each case, the ROI was manually drawn by two independent radiologists with no knowledge of the final pathologic results. 2 different types of segmentation were completed: ROI-1, the entire tumor ROI containing all compositions; ROI-2, the entire tumor ROI excluding cystic and necrotic areas (cystic or necrotic portions met conditions: first, no enhancement with the contrast agent in the T1-weighted images and second, high intensity, like cerebrospinal fluid (CSF), in the T2-weighted images). According to Kang and Lue's methods, the tumor boundaries were defined with reference to the high-signal-intensity areas thought to represent the tumor tissue on the T2-weighted images [9, 11]. The ROIs were placed carefully inside the mass to avoid regions influenced by the partial volume effect, and the position of the ROIs was verified using postcontrast T1-weighted images, T2-weighted images, and T2-FLAIR imaging. Then, the following accumulated ADC parameters: mean ADC (ADCmean), maximum ADC (ADCmax), minimum ADC (ADCmin), mode ADC (ADCmode), 5th (P5 ADC), 10th (P10 ADC), 25th (P25 ADC), 75th (P75 ADC), 90th (P90 ADC), and 95th (P95 ADC) were calculated, as well as distribution parameters—the kurtosis, skewness, entropy, StDev, and inhomogenity—were also estimated. Skewness reflects the asymmetry of the distribution, being positive if more values lie to the left of the mean, and negative if the opposite. Kurtosis is a measure of the peakedness of the distribution, and a higher kurtosis indicates a sharper peak of the histogram. In case of a normal distribution, skewness equals 0 and kurtosis equals 3. Entropy, StDev, and inhomogenity represent the statistical measure of variation that can be used to characterize the image texture [13]. For further analyses of ADC histogram parameters, the results of two readers were averaged. The two different ROI segmentations and the corresponding ADC histograms are shown in Figures 1–3.
Figure 1.
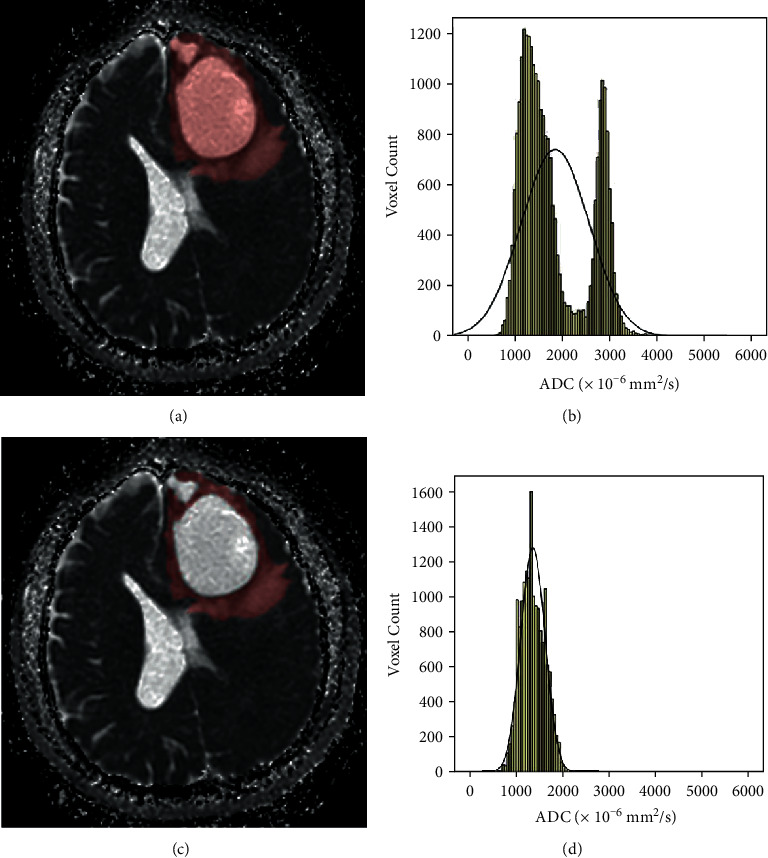
Freehand segmentation of IDH-wild LGG. (a) The ROI-1 (entire tumor ROI) delineated on an ADC map, (b) the corresponding ADC histogram of the ROI-1, (c) the ROI-2 (tumor ROI excluding cystic and necrotic portions) delineated on an ADC map, and (d) the corresponding ADC histogram of the second segmentation (color should be used for this figure).
Figure 2.
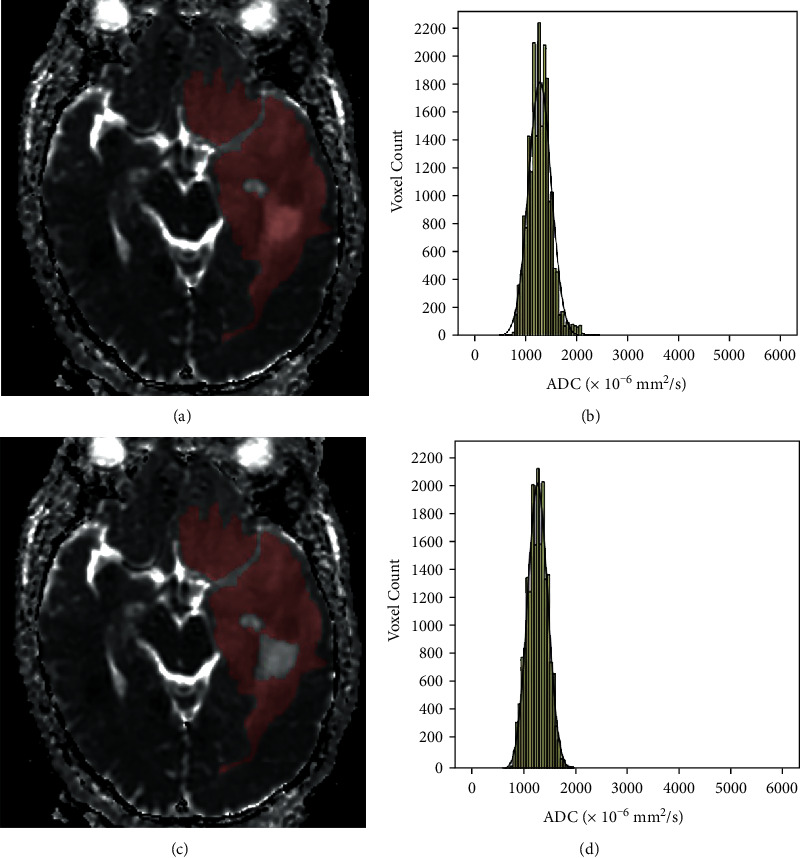
Freehand segmentation of IDH mutant with intact 1p or 19q LGG. (a) The ROI-1 delineated on an ADC map, (b) the corresponding ADC histogram of the ROI, (c) the ROI-2 delineated on an ADC map, and (d) the corresponding ADC histogram of the ROI (color should be used for this figure).
Figure 3.
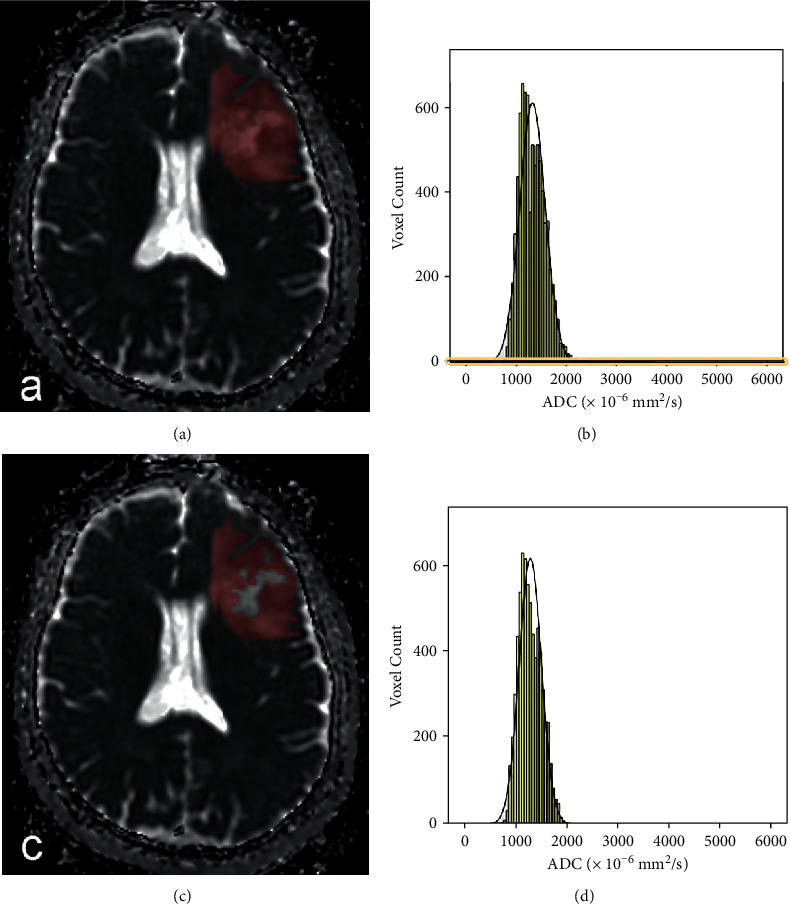
Freehand segmentation of IDH mutant with 1p/19q co- deleted LGG. (a) the ROI-1 delineated on an ADC map, (b) the corresponding ADC histogram of the ROI, (c) theROI-2 delineated on an ADC map, and (d) the corresponding ADC histogram of the ROI (color should be used for this figure).
2.3. Histopathology and Molecular Analysis
All tissue samples underwent analysis at The First Affiliated Hospital of Chongqing Medical University's neuropathology department and center for molecular medicine testing according to the World Health Organization (WHO) 2016 guidance on immunohistochemistry testing for glioma. IDH R132H immunonegative tumors underwent multiple gene Sanger sequencing. The 1p/19q codeletion status was determined by fluorescence in situ hybridization-specific probes for the 1p36 and 19q13 loci.
2.4. Statistical Analysis
All statistical testing was performed with SPSS 22 (IBM) and MedClac Version15.6.1. The intraclass correlation coefficient (ICC) was calculated to evaluate interobserver agreement of all histogram parameters (κ = 0.00–0.20, poor correlation; κ = 0.21–0.40, fair correlation; κ = 0.41–0.60, moderate correlation; κ = 0.61–0.80, good correlation; κ = 0.81–1.00, excellent correlation) [15, 16]. The Shapiro-Wilk test was used to check whether the measurement data followed a normal distribution. Normally distributed continuous variables were compared using the independent t test, and nonnormally distributed continuous variables were compared using the Mann–Whitney U test between different molecular groups. For statistically significant results, receiver operating characteristic (ROC) curves were constructed to determine the optimal threshold for each histogram parameter to differentiate the molecular subtypes, and the optimal cutoff value was determined by maximizing Youden's index. Area under the curve (AUC) results were compared using the method of Delong et al. The results with p values of less than 0.05 were considered to be statistically significant.
3. Results
3.1. Comparison of Clinical Characteristics of the 56 Patients
The mean age was greater in the IDHwt group than in the IDHmut group (t = 2.422, p = 0.025). The proportion of grade III gliomas in the IDHwt group was statistically significantly larger than in the IDHmut group (Fisher's exact test p < 0.001). Compared with the IDHmut/1p19q+ group, neither the greater mean age or larger proportion of grade III gliomas in the IDHmut/1p19q− group was statistically significant (t = −0.362, p = 0.716 for age, Fisher's exact test p = 0.709 for the proportion of grade III gliomas). The histological and molecular characteristics of the patient population are listed in Table 1.
Table 1.
Comparison of clinical characteristics of the 56 patients.
| Patient group | ||||
|---|---|---|---|---|
| IDHwt | IDHmut | |||
| Total | IDHmut/1p19q+ | IDHmut/1p19q− | ||
| WHO grade | ||||
| II | 5 | 32 | 15 | 17 |
| III | 11 | 8 | 3 | 5 |
| Total | 16# | 40# | 18## | 22## |
| Age (years, X ± SD) | 51.9 ± 16.0∗ | 41.5 ± 10.5∗ | 40.8 ± 9.8∗∗ | 42.0 ± 11.2∗∗ |
# Pearson's chi-square test p < 0.001, ##Fisher's exact test p = 0.709; ∗t = 2.422, p = 0.025; ∗∗t = −0.362, p = 0.716.
3.2. Comparison of ADC Histogram Parameters for Each Method
When applying the ROI-1 segmentation, the interobserver agreements were good to excellent for all parameters in the three groups, with ICCs of 0.980-0.999, 0.868-0.999, and 0.809-0.999. With the second segmentation,the interobserver agreements were moderate to excellent for all parameters in the three groups, with ICCs of 0.880-0.987, 0.792-0.957, and 0.808-0.965.
For the IDHwt tumors, the ADCmean (p = 0.027), P75ADC (p = 0.016), P90ADC (p = 0.009), P95ADC (p = 0.005), ADCmax (p = 0.001), StDev (p = 0.003), inhomogenity (p = 0.006), and range (p = 0.001) obtained from the ROI-1 were proved to be significantly greater than those from the second segmentation. Other histogram parameters did not show significant differences between the two segmentation methods (Table 2).
Table 2.
ADC histogram parameters of the IDHwt tumors using different ROI methods.
| Values (×10−6 mm2/s) | The tumor ROI | The tumor ROI excluding cystic and necrotic portions | Test values | p values |
|---|---|---|---|---|
| ADCmean | 1646.10 ± 402.18 | 1368.54 ± 254.39 | -2.333a | 0.027 |
| ADCmin | 518.50 ± 226.12 | 485.38 ± 195.34 | -0.443a | 0.661 |
| P5 ADC | 972.53 ± 207.95 | 880.19 ± 168.79 | -1.379a | 0.178 |
| P10 ADC | 1066.38 ± 239.79 | 966.75 ± 184.50 | -1.317a | 0.198 |
| P25 ADC | 1281.41 ± 306.36 | 1142.50 ± 220.39 | -1.472a | 0.151 |
| P50 ADC | 1606.94 ± 442.95 | 1368.63 ± 267.23 | -1.843a | 0.075 |
| P75 ADC | 2022.41 ± 580.95 | 1593.25 ± 304.42 | -2.617a | 0.016 |
| P90 ADC | 2298.56 ± 620.70 | 1767.56 ± 331.19 | -2.848a | 0.009 |
| P95 ADC | 2403.84 ± 609.61 | 1864.00 ± 342.97 | -3.085a | 0.005 |
| ADCmax | 3606.75 ± 1159.31 | 2450.25 ± 417.17 | -3.755a | 0.001 |
| ADCmode | 1616.19 ± 583.11 | 1517.06 ± 533.69 | -0.502a | 0.620 |
| Kurtosis | -0.527 (-1.027, 0.480) | −0.401 ± 0.429 | -0.075b | 0.956 |
| Skewness | 0.434 ± 0.689 | 0.065 ± 0.310 | -1.957a | 0.060 |
| StDev | 388.630 (343.670, 689.601) | 303.055 ± 72.511 | -2.902b | 0.003 |
| Inhomogenity | 0.278 ± 0.069 | 0.220 ± 0.033 | -3.047a | 0.006 |
| Range | 3087.31 ± 1130.76 | 1966.56 ± 389.39 | -3.799a | 0.001 |
| Entropy | 3.968 ± 0.224 | 4.082 ± 0.199 | -1.527a | 0.137 |
a t value; bZ value.
For the IDHmut/1p19q+ tumors, only the ADCmax (p = 0.044) differed significantly between the two segmentation methods, and other histogram parameters did not show significant differences between the two segmentation methods (Table 3).
Table 3.
ADC histogram parameters for IDHmut/1p19q+ tumors using different ROI methods.
| Values (×10−6 mm2/s) | The tumor ROI | The tumor ROI excluding cystic or necrotic portions | Test values | p values |
|---|---|---|---|---|
| ADCmean | 1575.71 ± 286.14 | 1511.41 ± 253.75 | 0.713a | 0.481 |
| ADCmin | 740.69 ± 123.11 | 751.83 ± 131.62 | -0.26a | 0.795 |
| P5 ADC | 1085.02 ± 149.95 | 1077.67 ± 151.22 | 0.147a | 0.884 |
| P10 ADC | 1176.92 ± 179.65 | 1163.22 ± 177.72 | 0.230a | 0.820 |
| P25 ADC | 1355.22 ± 235.42 | 1332.89 ± 232.28 | 0.287a | 0.776 |
| P50 ADC | 1560.53 ± 286.86 | 1526.11 ± 280.79 | 0.364a | 0.718 |
| P75 ADC | 1805.81 ± 430.58 | 1687.00 ± 301.50 | 0.959a | 0.344 |
| P90 ADC | 1981.53 ± 471.58 | 1811.67 ± 307.63 | 1.280a | 0.209 |
| P95 ADC | 2096.78 ± 485.56 | 1879.68 ± 302.98 | 1.609a | 0.117 |
| ADCmax | 2957.83 ± 1009.01 | 2415.72 ± 362.81 | 2.145a | 0.044 |
| ADCmode | 1571.61 ± 349.80 | 1575.94 ± 355.41 | -0.037a | 0.971 |
| Kurtosis | -0.342 (-0.518, 1.988) | -0.354 (-0.482, 0.080) | -0.253b | 0.800 |
| Skewness | -0.200 (-0.446, 0.912) | -0.227 (-0.526, 0.183) | -0.886b | 0.376 |
| StDev | 288.203 (215.733, 366.08) | 248.611 ± 58.938 | -1.487b | 0.137 |
| Inhomogenity | 0.172 (0.155, 0.244) | 0.163 ± 0.025 | -1.361b | 0.173 |
| Range | 2232.56 ± 1076.62 | 1688.11 ± 406.17 | 2.007a | 0.057 |
| Entropy | 3.970 ± 0.281 | 4.054 ± 0.246 | -0.944a | 0.352 |
a, t value; b, Z value.
For the IDHmut/1p19q− tumors, the greater P90ADC (p = 0.023), P95ADC (p = 0.005), ADCmax (p = 0.004), kurtosis (p = 0.002), skewness (p = 0.001), StDev (p = 0.002), inhomogenity (p = 0.004), and range (p = 0.011) calculated from the ROI-1 were all statistically significant than those from the second segmentation, and other histogram parameters did not show significant differences between the two segmentation methods (Table 4).
Table 4.
ADC histogram parameters for IDHmut/1p19q− tumors using different ROI methods.
| Values(×10−6 mm2/s) | The tumor ROI | The tumor ROI excluding cystic or necrotic portions | Test values | p values |
|---|---|---|---|---|
| ADCmean | 1353.09 ± 131.59 | 1296.48 ± 128.26 | -1.445a | 0.156 |
| ADCmin | 670.09 ± 116.37 | 672.82 ± 125.13 | -0.075a | 0.941 |
| P5 ADC | 974.11 ± 70.45 | 966.09 ± 69.62 | -0.380a | 0.706 |
| P10 ADC | 1033.61 ± 77.99 | 1024.23 ± 81.31 | -0.391a | 0.698 |
| P25 ADC | 1152.87 ± 101.56 | 1137.09 ± 100.86 | -0.518a | 0.607 |
| P50 ADC | 1306.20 ± 131.69 | 1276.00 ± 129.73 | -0.766a | 0.448 |
| P75 ADC | 1499.43 ± 159.83 | 1432.64 ± 153.36 | -1.414a | 0.165 |
| P90 ADC | 1728.45 ± 226.92 | 1585.68 ± 170.46 | -2.360a | 0.023 |
| P95 ADC | 1925.86 ± 344.50 | 1677.45 ± 180.02 | -2.998a | 0.005 |
| ADCmax | 2776.43 ± 743.58 | 2220.59 ± 367.02 | -3.144a | 0.004 |
| ADCmode | 1158.50 (1093.50, 1329.88) | 1170.50 (1093.50, 1328.25) | -0.094b | 0.925 |
| Kurtosis | 1.191 (0.242, 2.290) | 0.102(-0.309, 0.328) | -3.157b | 0.002 |
| Skewness | 0.984 ± 0.646 | 0.378 (0.231, 0.642) | -3.898b | 0.001 |
| StDev | 293.603 ± 97.224 | 217.231 ± 43.743 | -3.360a | 0.002 |
| Inhomogenity | 0.197 (0.165,0.251) | 0.166 ± 0.025 | -2.900b | 0.004 |
| Range | 1939.25 (1391.00, 2733.13) | 1547.77 ± 405.23 | -2.547b | 0.011 |
| Entropy | 3.933 ± 0.225 | 3.950 ± 0.249 | 1.450a | 0.155 |
a t value; bZ value.
3.3. Ability to Differentiate the Genetic Subtypes of Diffuse Lower-Grades Gliomas
The P75ADC (p = 0.017), P90ADC (p = 0.015), P95ADC (p = 0.025), ADCmax (p = 0.028), StDev (p = 0.002), inhomogenity (p = 0.001), and range (p = 0.007) from the ROI-1 were all statistically significant greater in the IDHwt group than in the IDHmut group, and the ADCmin (p = 0.006) and kurtosis (p = 0.007) in the IDHwt group were smaller (Table 5).
Table 5.
Comparison of ADC histogram parameters of the two different ROIs between IDHwt gliomas and IDHmut gliomas.
| Values (×10−6 mm2/s) | IDHwt group, n = 16 | IDHmut group, n = 40 | Test values | p values |
|---|---|---|---|---|
| ADCmean1 | 1646.10 ± 402.18 | 1419.95 (1279.30, 1565.89) | -1.796b | 0.073 |
| ADCmean2 | 1368.54 ± 254.39 | 1341.13 (1242.53, 1547.90) | -0.109b | 0.913 |
| ADCmin1 | 518.50 ± 226.12 | 701.86 ± 123.14 | -3.067a | 0.006 |
| ADCmin2 | 485.38 ± 195.34 | 708.38 ± 132.54 | -4196a | <0.001 |
| P5 ADC1 | 972.53 ± 207.95 | 1010.00 (929.86, 1094.88) | -0.852b | 0.394 |
| P5 ADC2 | 880.19 ± 168.79 | 1009.00 (913.75, 1076.75) | -2.648b | 0.008 |
| P10 ADC1 | 1066.38 ± 239.79 | 1073.25 (987.88, 1161.63) | -0.381b | 0.703 |
| P10 ADC2 | 966.75 ± 184.50 | 1069.50 (970.75, 1173.75) | -1.904b | 0.057 |
| P25 ADC1 | 1281.41 ± 306.36 | 1205.75 (1104.88, 1336.38) | -0.689b | 0.491 |
| P25 ADC2 | 1142.50 ± 220.39 | 1178.50 (1081.50, 1331.75) | -1.904b | 0.301 |
| P50 ADC1 | 1606.94 ± 442.95 | 1352.75 (1255.63, 1586.38) | -1.560b | 0.119 |
| P50 ADC2 | 1368.63 ± 267.23 | 1320.50 (1225.00, 1547.25) | -01.109b | 0.913 |
| P75 ADC1 | 2022.41 ± 580.95 | 1586.50 (1411.63, 1764.75) | -2.385b | 0.017 |
| P75 ADC2 | 1593.25 ± 304.42 | 1481.50 (1356.50, 1749.00) | -0.580b | 0.562 |
| P90 ADC1 | 2298.56 ± 620.70 | 1815.25 (1615.50, 2003.50) | -2.430b | 0.015 |
| P90 ADC2 | 1767.56 ± 331.19 | 1687.38 ± 264.32 | 0.953a | 0.345 |
| P95 ADC1 | 2403.84 ± 609.61 | 2002.78 ± 417.24 | 2.415a | 0.025 |
| P95 ADC2 | 1864.00 ± 342.97 | 1768.46 ± 260.47 | 1.135a | 0.261 |
| ADCmax1 | 3606.75 ± 1159.31 | 2716.50 (2161.63, 3409.75) | -2.195b | 0.028 |
| ADCmax2 | 2450.25 ± 417.17 | 2308.40 ± 373.60 | 1.242a | 0.220 |
| ADCmode1 | 1616.19 ± 583.11 | 1285.00 (1139.50, 1619.25) | -1.324b | 0.185 |
| ADCmode2 | 1517.06 ± 533.69 | 1273.50 (1142.25, 1610.25) | -0.517b | 0.605 |
| Kurtosis1 | -0.527 (-1.027, 0.480) | 0.661 (-0.397, 2.273) | -2.721b | 0.007 |
| Kurtosis2 | −0.401 ± 0.429 | -0.126 (-0.374, 0.304) | -2.512b | 0.012 |
| Skewness1 | 0.434 ± 0.689 | 0.664 ± 0.831 | -0.979a | 0.332 |
| Skewness2 | 0.065 ± 0.310 | 0.198 ± 0.522 | -0.955a | 0.344 |
| StDev1 | 388.630 (343.670, 689.60) | 278.864 (217.080, 357.755) | -3.156b | 0.002 |
| StDev2 | 303.055 ± 72.511 | 231.352 ± 51.862 | 4.110a | <0.001 |
| Inhomogenity1 | 0.278 ± 0.069 | 0.190 (0.160, 0.231) | -3.347b | 0.001 |
| Inhomogenity2 | 0.220 ± 0.033 | 0.165 ± 0.025 | 6.048a | <0.001 |
| Range1 | 3087.31 ± 1130.76 | 1941.25 (1406.00, 2788.38) | -2.721b | 0.007 |
| Range2 | 1966.56 ± 389.39 | 1610.93 ± 406.61 | 2.991a | 0.004 |
| Entropy1 | 3.968 ± 0.224 | 3.950 ± 0.249 | 0.218a | 0.825 |
| Entropy2 | 4.082 ± 0.199 | 4.034 ± 0.203 | 0.798a | 0.428 |
1ROI-1; 2ROI-2; at value; bZ value.
The StDev (p < 0.001), inhomogenity (p < 0.001), and range (p = 0.004) from the ROI-2 differed significantly between IDHwt gliomas and IDHmut gliomas and decreased with the IDH mutation, but the ADCmin (p < 0.001), P5 ADC (p = 0.008), and kurtosis (p = 0.012) in the IDHwt gliomas were lower than the other group (Table 5).
The ADCmean (p = 0.006), P5ADC (p = 0.008), P10ADC (p = 0.005), P25 ADC (p = 0.003), P50ADC (p = 0.002), P75ADC (p = 0.009), P90ADC (p = 0.048), and ADCmode (p = 0.002) from the ROI-1 were significantly lower in IDHmut/1p19q−gliomas, and the skewness (p = 0.007) was greater in IDHmut/1p19q−gliomas; the similar results were obtained from the ROI-2, but the P95 ADC (p = 0.019) from the ROI-2 was also significantly lower in IDHmut/1p19q−gliomas, and the kurtosis (p = 0.037) from the ROI-2 was lager in IDHmut/1p19q−gliomas (Table 6).
Table 6.
Comparison of ADC histogram parameters of the two different ROIs between the IDHmut/1p19q− group and IDHmut/1p19q+ group.
| Values (×10−6 mm2/s) | IDHmut/1p19q+, n = 18 | IDHmut/1p19q−, n = 22 | Test values | p value |
|---|---|---|---|---|
| ADCmean1 | 1575.71 ± 286.14 | 1353.09 ± 131.59 | 3.048a | 0.006 |
| ADCmean2 | 1511.411 ± 253.75 | 1296.48 ± 128.26 | 3.268a | 0.003 |
| ADCmin1 | 740.69 ± 123.11 | 670.09 ± 116.37 | 1.860a | 0.071 |
| ADCmin2 | 751.83 ± 131.62 | 672.82 ± 125.13 | 1.941a | 0.060 |
| P5 ADC1 | 1085.02 ± 149.95 | 974.11 ± 70.45 | 2.888a | 0.008 |
| P5 ADC2 | 1077.67 ± 151.22 | 966.09 ± 69.62 | 2.890a | 0.008 |
| P10 ADC1 | 1176.92 ± 179.65 | 1033.61 ± 77.99 | 3.150a | 0.005 |
| P10 ADC2 | 1163.22 ± 177.72 | 1024.23 ± 81.31 | 3.066a | 0.006 |
| P25 ADC1 | 1355.22 ± 235.42 | 1152.87 ± 101.56 | 3.397a | 0.003 |
| P25 ADC2 | 1332.89 ± 232.28 | 1137.09 ± 100.86 | 3.329a | 0.003 |
| P50 ADC1 | 1560.53 ± 286.86 | 1306.20 ± 131.69 | 3.474a | 0.002 |
| P50 ADC2 | 1526.11 ± 280.79 | 1276.00 ± 129.73 | 3.487a | 0.002 |
| P75 ADC1 | 1805.81 ± 430.58 | 1499.43 ± 159.83 | 2.862a | 0.009 |
| P75 ADC2 | 1687.00 ± 301.50 | 1432.64 ± 153.36 | 3.252a | 0.003 |
| P90 ADC1 | 1981.53 ± 471.58 | 1728.45 ± 226.92 | 2.088a | 0.048 |
| P90 ADC2 | 1811.67 ± 307.63 | 1585.68 ± 170.46 | 2.786a | 0.010 |
| P95 ADC1 | 2096.78 ± 485.56 | 1925.86 ± 344.50 | 1.300a | 0.201 |
| P95 ADC2 | 1879.68 ± 302.98 | 1677.45 ± 180.02 | 2.494a | 0.019 |
| ADCmax1 | 2957.83 ± 1009.01 | 2776.43 ± 743.58 | 0.654a | 0.517 |
| ADCmax2 | 2415.72 ± 362.81 | 2220.59 ± 367.02 | 1.681a | 0.101 |
| ADCmode1 | 1571.61 ± 349.80 | 1158.50 (1093.50, 1329.88) | -3.059b | 0.002 |
| ADCmode2 | 1575.94 ± 355.41 | 1170.50 (1093.50, 1328.25) | -3.086b | 0.002 |
| Skewness1 | -0.200 (-0.446, 0.912) | 0.984 ± 0.646 | -2.644b | 0.007 |
| Skewness2 | -0.227 (-0.526, 0.183) | 0.378 (0.231, 0.642) | -2.093b | <0.001 |
| Kurtosis1 | -0.342 (-0.518, 1.988) | 1.191 (0.242, 2.290) | -1.930b | 0.054 |
| Kurtosis2 | -0.354 (-0.482, 0.080) | 0.102 (-0.309, 0.328) | -3.453b | 0.037 |
| StDev1 | 288.203 (215.736, 366.088) | 293.603 ± 97.224 | -0.214b | 0.828 |
| StDev2 | 248.611 ± 58.938 | 217.231 ± 43.743 | 1.932a | 0.061 |
| Inhomogenity1 | 0.172 (0.155, 0.244) | 0.197 (0.165, 0.251) | -1.142b | 0.253 |
| Inhomogenity2 | 0.163 ± 0.025 | 0.166 ± 0.025 | -0.346a | 0.731 |
| Range1 | 2232.56 ± 1076.52 | 1939.25 (1391.00, 2733.13) | -0.381b | 0.703 |
| Range2 | 1688.11 ± 406.17 | 1547.77 ± 405.23 | 1.089a | 0.283 |
| Entropy1 | 3.970 ± 0.281 | 3.933 ± 0.225 | 0.467a | 0.643 |
| Entropy2 | 4.054 ± 0.246 | 3.950 ± 0.249 | 0.527a | 0.601 |
1ROI-1; 2ROI-2; at value; bZ value.
3.4. ROC Analysis and Comparation
ADC histogram parameters from the two different ROI methods were evaluated for their ability to discriminate the genetic subtypes using ROC analysis (Tables 7 and 8). The inhomogenity from the two different ROI methods for distinguishing IDHwt gliomas from IDHmut gliomas both showed the biggest AUC (0.788, 0.930), and the optimal cutoff value was 0.229 (sensitivity, 81.3%; specificity, 75.0%) for the ROI-1 and 0.186 (sensitivity, 93.8%; specificity, 82.5%) for the ROI-2, respectively. In the pairwise comparison of ROC curves with the AUC, a major finding was that the AUC of the inhomogenity from the ROI-2 was significantly larger than that from the ROI-1, but no significant differences were identified between the AUCs of other same parameters from the two different ROI methods (Table 7).
Table 7.
ROC Curve analysis of different parameters for distinguishing IDHwt gliomas from IDHmut gliomas.
| Parameter | ROI | AUC | Specificity (%) | Sensitivity (%) | Cutoff value | p value |
|---|---|---|---|---|---|---|
| ADCmin | ROI-1 | 0.749 | 87.5 | 62.5 | 560.00 | 0.180 |
| ROI-2 | 0.831 | 90.0 | 62.5 | 543.00 | ||
|
| ||||||
| P5 ADC | ROI-1 | — | — | — | — | — |
| ROI-2 | 0.728 | 57.5 | 81.2 | 980.00 | ||
|
| ||||||
| P75 ADC | ROI-1 | 0.705 | 68.8 | 70.0 | 1688.25 | — |
| ROI-2 | — | — | — | — | ||
|
| ||||||
| P90 ADC | ROI-1 | 0.709 | 68.8 | 77.5 | 1962.00 | — |
| ROI-2 | — | — | — | — | ||
|
| ||||||
| P95 ADC | ROI-1 | 0.709 | 68.8 | 72.5 | 2164.75 | — |
| ROI-2 | — | — | — | — | ||
|
| ||||||
| ADCmax | ROI-1 | 0.689 | 81.3 | 57.5 | 2849.00 | — |
| ROI-2 | — | — | — | — | ||
|
| ||||||
| Kurtosis | ROI-1 | 0.734 | 87.5 | 56.2 | -0.487 | 0.826 |
| ROI-2 | 0.716 | 47.5 | 87.5 | -0.042 | ||
|
| ||||||
| StDev | ROI-1 | 0.772 | 87.5 | 65.0 | 321.702 | 0.646 |
| ROI-2 | 0.803 | 68.8 | 80.0 | 272.351 | ||
|
| ||||||
| Inhomogenity | ROI-1 | 0.788 | 81.3 | 75.0 | 0.229 | 0.013 |
| ROI-2 | 0.930 | 93.8 | 82.5 | 0.186 | ||
|
| ||||||
| Range | ROI-1 | 0.734 | 81.3 | 60.0 | 2144.5 | 0.838 |
| ROI-2 | 0.747 | 68.8 | 82.5 | 1977.5 | ||
Table 8.
ROC curve analysis of different parameters for distinguishing IDHmut/1p19q− tumors and IDHmut/1p19q+ tumors.
| Parameter | ROI | AUC | Specificity (%) | Sensitivity (%) | Cutoff value | p value |
|---|---|---|---|---|---|---|
| ADCmean | ROI-1 | 0.715 | 55.6 | 95.5 | 1546.32 | 0.151 |
| ROI-2 | 0.758 | 72.2 | 81.8 | 1387.97 | ||
|
| ||||||
| P5 ADC | ROI-1 | 0.735 | 50.0 | 100.0 | 1108.00 | 1.00 |
| ROI-2 | 0.735 | 44.4 | 100.0 | 1110.00 | ||
|
| ||||||
| P10 ADC | ROI-1 | 0.749 | 50.0 | 100.0 | 1177.50 | 1.00 |
| ROI-2 | 0.749 | 50.0 | 100.0 | 1191.00 | ||
|
| ||||||
| P25ADC | ROI-1 | 0.766 | 66.7 | 86.4 | 1246.75 | 1.00 |
| ROI-2 | 0.766 | 50.0 | 100.0 | 1345.00 | ||
|
| ||||||
| P50 ADC | ROI-1 | 0.763 | 55.6 | 100.0 | 1581.25 | 1.00 |
| ROI-2 | 0.763 | 67.7 | 86.4 | 1384.00 | ||
|
| ||||||
| P75 ADC | ROI-1 | 0.710 | 50.0 | 100.0 | 1788.50 | 1.00 |
| ROI-2 | 0.710 | 66.7 | 81.8 | 1540.00 | ||
|
| ||||||
| P90 ADC | ROI-1 | 0.669 | 44.4 | 90.9 | 1984.00 | 1.00 |
| ROI-2 | 0.669 | 55.6 | 90.9 | 1812.50 | ||
|
| ||||||
| P95ADC | ROI-1 | — | — | — | — | — |
| ROI-2 | 0.689 | 61.1 | 86.4 | 1979.00 | ||
|
| ||||||
| ADCmode | ROI-1 | 0.784 | 61.1 | 90.9 | 1448.75 | 0.729 |
| ROI-2 | 0.787 | 61.1 | 90.9 | 1450.00 | ||
|
| ||||||
| Skewness | ROI-1 | 0.747 | 100.0 | 44.4 | -0.164 | 0.178 |
| ROI-2 | 0.821 | 81.8 | 77.8 | 0.186 | ||
|
| ||||||
| Kurtosis | ROI-1 | — | — | — | — | — |
| ROI-2 | 0.694 | 86.4 | 55.6 | -0.349 | ||
For the differentiation of IDHmut/1p19q− tumors and IDHmut/1p19q+ tumors, the ADCmode from the ROI-1 showed the biggest AUC (AUC, 0.784; sensitivity, 61.1%; specificity, 90.9%), and the skewness from the ROI-2 showed the biggest AUC (AUC, 0.821; sensitivity, 81.8%; specificity, 77.8%), but no significant differences were identified between the AUCs of the same parameters from the two different ROI methods (Table 8).
4. Discussion
The results obtained from this study suggest that the ADC values analyzed by the histogram method can help to classify IDH-wild tumors from IDH-mutated tumors as well as IDHmut-Codel tumor from IDHmut-NonCodel tumors in lower-grade diffuse gliomas, no matter which ROI method is used. The StDev, inhomogenity, and range from the two different ROI methods were both larger in IDH-wild tumors compared with the IDH-mutated tumors, and the ADCmin and kurtosis in the IDH-wild group were smaller. Some different results appeared in the two different ROI methods, and one of these was that the P5ADC from the ROI-2 in the IDH-wild tumors was lower than the mutated ones, while P5ADC from ROI-1 showed no significant difference in these two types of tumors. Another difference was that the P75ADC, P90ADC, P95ADC, and ADCmax from ROI-1 were larger in IDH-wild tumors, while the same parameters from the ROI-2 showed no significant differences in the two sorts of tumor. For identifying the 1p19q-Codel ones in the IDH-mutated tumors, we found that the ADCmean, P5ADC, P10ADC, P25ADC, P50ADC, P75ADC, P90ADC, and ADCmode from the two different ROI methods were both significantly lower in the 1p19q -Codel tumors; meanwhile, the skewness was greater in this group. Compared with ROI-1, the second segmentation gave us more parameters valuable for the differentiation, such as the P95ADC and kurtosis, the P95ADC was significantly lower in the 1p19q–Codel tumors, and the kurtosis was lager in the 1p19q–Codel ones.
It is widely recognized that the pathological heterogeneity may manifest as radiologic heterogeneity on ADC maps [5], and differences in ADCs are mainly attributed to the tumor cellularity but also to the presence of necrosis or cysts [6, 11, 18]. Consequently, the ADCs within a given tumor can vary widely between different regions of that tumor [10]. The voxels with low ADC value are reportedly well correlated with highly cellular components within the tumor, which reflects tumor proliferative rate and aggressiveness [5], whereas the higher frequency of voxels with high ADC values reflect cystic, necrotic, or myxoid components. In other words, the larger intratumoral heterogeneity is the wider ADC values distribute [15]. Our results demonstrated that the IDH wild-type gliomas showed a lower ADCmin, likely representing higher cellularity and aggressiveness, consistent with some previous studies [7, 9, 19], as well as appeared larger higher end values, inhomogenity, and StDev, likely representing more common cystic and necrotic portions, which might be associated with higher grade features and larger intratumoral heterogenitiy [11, 20]; meanwhile, in the present study, we found that no matter which segmentation was used, the parameters reflecting the intratumoral heterogenitiy, such as the inhomogenity and StDev, performed better than the conventional cumulative ADC values, so it is valuable and important to evaluate the intratumoral heterogenitiy using quantitative parameters. It was interesting that the entire tumor ROI provided some additional statistically significant parameters than the other segmentation, such as the higher end ADC values. This result suggests that, to a certain degree, the necrotic and cystic components may facilitate the discrimination between IDH-mutated tumors and IDH-wild gliomas, but their performance is weaker than the inhomogenity. The inhomogenity calculated from the ROI-2 performed better than that from the ROI-1, although the diagnostic performance of the other parameters obtained from the two methods showed no statistically significant differences, which reveals that extracting cystic and necrotic portions from the entire tumor lesions is better for evaluating the difference of the intratumoral heterogeneity and more helpful to classify IDH-wild tumors in the lower-grade gliomas.
For the discrimination between the IDHmut-NonCodel tumors and the IDHmut-Codel tumors, an important finding in our study was the increase of restrictive diffusion in the IDHmut-Codel tumors. This result parallels those of previous studies reporting lower minimum ADC and a lower mean histogram ADC in tumors with 1p/19q loss compared to those without [14, 21]. There are several possible explanations for this result. One of them is the presence of calcification, which is common in IDHmut-Codel tumors [22], that may limit water content as well as hinder water movement [20], and another is that the IDHmut-Codel tumors are often highly cellular lesions with closely packed, relatively small cells in central regions and prominent secondary structure formation, which may also delay the passage of small molecules [23]. When applying the ROI-1, the ADCmode performed best at predicting the 1p19q genotype in this study, which was lower in the IDHmut-Codel tumors. ADCmode means the value that appears most frequently in a set of ADC values, and this result may also be due to the larger diffusion restriction in the IDHmut-Codel tumors. With the second segmentation, the skewness showed the biggest AUC, which was larger in IDHmut-Codel tumors. The skewness reflects the asymmetry of the ADC distribution, more pixels have lower ADC values and lie to the left of the mean of the histogram, and the skewness is more positive. Therefore, this result indicates that the IDHmut-Codel tumors contain more voxels with ADC values below the mean of the histogram, which may also be associated with the increase of restrictive diffusion, but the performance of the same parameters from the two different method showed no statistically significant differences.
As for the comparison of histogram parameters calculated from the two different segmentations, we found that almost all ADC histogram parameters calculated from the whole tumor ROIs tended to be larger than those from the other segmentation. Statistically, significant differences were found in the higher end values of cumulative ADC histograms and some histogram distribution characteristics from these two different methods in IDH-wild gliomas and IDHmut-Codel tumors, such as P75ADC, P90ADC, P95ADC, ADCmax, StDev, and inhomogenity. This result may be associated with the following reasons: the first one is that the IDH wild-type gliomas show larger intratumoral heterogeneity due to its increased cell proliferation and necrosis [7, 20], so the entire tumor ROI contained areas of necrosis and systs will lead to more higher ADC values, larger StDev, and inhomogenity. As for the codeleted tumors, the signal intensity on MR images is more heterogeneous than the noncodeleted ones [20, 24]; as a result, the StDev and inhomogenity obtained from the entire tumor ROI were lager. Conversely, the similar results of the two ROI segmentations in noncodeleted tumors can be explained by a relative homogeneity in these gliomas [20], so extracting cystic and necrotic portions from the ROIs does not cause any obvious differences.
Some limitations of this study should be considered. First, this was a retrospective study with a small study population. Therefore, the usefulness of the ADC histogram analysis should be prospectively examined in a larger and more balanced study population. Second, the ROIs were manually determined. Automatic segmentation algorithms may facilitate the procedure. Third, how well the ADC histogram analysis works in data independent from the current data was not evaluated, and our findings must be validated in further studies. Finally, it is known that the conventional ADC value derived from the monoexponential model is influenced by a combination of both diffusion and perfusion effects. Intravoxel incoherent motion (IVIM) imaging may tease out each component from the total DWI signal, so it may be more valuable to compare the ADC values derived from intravoxel incoherent motion (IVIM), but this idea needs the more widespread application of this technology.
In conclusion, our study has shown the utility of ADC histogram analysis in the characterization of the IDH-wild tumors and the IDH-mutated ones as well as the IDHmut-Codel tumors and the IDHmut-NonCodel ones in the lower-grade diffuse gliomas. Although the entire tumor ROI provided some additional statistically significant parameters than the other segmentation in the characterization of the IDH-wild tumors and the IDH-mutated ones, such as the higher end ADC values, the inhomogenity calculated from the tumor ROIs excluding cystic and necrotic portions performed best in the differentiation between these two kinds of tumors, and this suggests that extracting cystic and necrotic portions from the entire tumor lesions is needed for preferably evaluating the difference of the intratumoral heterogeneity and more helpful to classify IDH-wild tumors in the lower-grade gliomas. For predicting the 1p19q genotype in the lower-grade gliomas, when applying the entire tumor ROI, the ADCmode performed best, while applying the second method, the skewness showed the best performance, but the performance of the same parameters from the two different methods showed no statistically significant differences. Further prospective study with a more general patient population is warranted.
Acknowledgments
This research was supported by the National Natural Sciences Foundation of China (81171366).
Data Availability
The clinical data used to support the findings of this study were provided by Department of Radiology, the First Affiliated Hospital of Chongqing Medical University, and cannot be made freely available. Access to these data will be considered by the author upon request, with permission from the Director of the Department of Radiology of this hospital.
Additional Points
Highlights. (i) Lower-grade gliomas form a biologically heterogeneous group of tumors, whose clinical behavior, outcome, and therapeutic sensitivity are, to some extent, depending on their molecular profile. (ii) ADC values analyzed by the histogram method could help to classify IDH-wild tumors from IDH-mutated tumors as well as IDHmut-Codel tumors from IDHmut-NonCodel tumors in lower-grade diffuse gliomas. (iii) Extracting cystic and necrotic portions from the entire tumor lesions can preferably evaluate the difference of the intratumoral heterogeneity and classifying IDH-wild ones in the lower-grade gliomas.
Conflicts of Interest
None declared.
Authors' Contributions
Dan Liu and Shuai-Xiang Gao contributed equally to this work.
References
- 1.Ceccarelli M., Barthel F. P., Malta T. M., et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki H., Aoki K., Chiba K., et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nature Genetics. 2015;47(5):458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Networ. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopes M. B. S. The 2017 World Health Organization classification of tumors of the pituitary gland: a summary. Acta Neuropathologica. 2017;134(4):521–535. doi: 10.1007/s00401-017-1769-8. [DOI] [PubMed] [Google Scholar]
- 5.Padhani A. R., Liu G., Mu-Koh D., et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11(2):102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iima M., le Bihan D. Clinical intravoxel incoherent motion and diffusion MR imaging: past, present, and future. Radiology. 2016;278(1):13–32. doi: 10.1148/radiol.2015150244. [DOI] [PubMed] [Google Scholar]
- 7.Wu C.-C., Jain R., Radmanesh A., et al. Predicting genotype and survival in glioma using standard clinical MR imaging apparent diffusion coefficient images: a pilot study from the cancer genome atlas. AJNR American Journal of Neuroradiology. 2018;39(10):1814–1820. doi: 10.3174/ajnr.A5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thust S. C., Hassanein S., Bisdas S., et al. Apparent diffusion coefficient for molecular subtyping of non-gadolinium-enhancing WHO grade II/III glioma: volumetric segmentation versus two-dimensional region of interest analysis. European Radiology. 2018;28(9):3779–3788. doi: 10.1007/s00330-018-5351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leu K., Ott G. A., Lai A., et al. Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II-III diffuse gliomas. Journal of Neuro-Oncology. 2017;134(1):177–188. doi: 10.1007/s11060-017-2506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surov A., Hamerla G., Meyer H. J., Winter K., Schob S., Fiedler E. Whole lesion histogram analysis of meningiomas derived from ADC values. Correlation with several cellularity parameters, proliferation index KI 67, nucleic content, and membrane permeability. Magnetic Resonance Imaging. 2018;51:158–162. doi: 10.1016/j.mri.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Kang Y., Choi S. H., Kim Y. J., et al. Gliomas: histogram analysis of apparent diffusion coefficient maps with standard- or high-b-value diffusion-weighted MR imaging--correlation with tumor grade. Radiology. 2011;261(3):882–890. doi: 10.1148/radiol.11110686. [DOI] [PubMed] [Google Scholar]
- 12.Surov A., Ginat D. T., Lim T., et al. Histogram analysis parameters apparent diffusion coefficient for distinguishing high and low-grade meningiomas: a multicenter study. Translational Oncology. 2018;11(5):1074–1079. doi: 10.1016/j.tranon.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suo S., Zhang K., Cao M., et al. Characterization of breast masses as benign or malignant at 3.0T MRI with whole-lesion histogram analysis of the apparent diffusion coefficient. Journal of Magnetic Resonance Imaging. 2016;43(4):894–902. doi: 10.1002/jmri.25043. [DOI] [PubMed] [Google Scholar]
- 14.Latysheva A., Emblem K. E., Brandal P., et al. Dynamic susceptibility contrast and diffusion MR imaging identify oligodendroglioma as defined by the 2016 WHO classification for brain tumors: histogram analysis approach. Neuroradiology. 2019;61(5):545–555. doi: 10.1007/s00234-019-02173-5. [DOI] [PubMed] [Google Scholar]
- 15.Nakajo M., Fukukura Y., Hakamada H., et al. Whole-tumor apparent diffusion coefficient (ADC) histogram analysis to differentiate benign peripheral neurogenic tumors from soft tissue sarcomas. Journal of Magnetic Resonance Imaging. 2018;48(3):680–686. doi: 10.1002/jmri.25987. [DOI] [PubMed] [Google Scholar]
- 16.Umanodan T., Fukukura Y., Kumagae Y., et al. ADC histogram analysis for adrenal tumor histogram analysis of apparent diffusion coefficient in differentiating adrenal adenoma from pheochromocytoma. Journal of Magnetic Resonance Imaging. 2017;45(4):1195–1203. doi: 10.1002/jmri.25452. [DOI] [PubMed] [Google Scholar]
- 17.Lewis M. A., Ganeshan B., Barnes A., et al. Filtration-histogram based magnetic resonance texture analysis (MRTA) for glioma IDH and 1p19q genotyping. European Journal of Radiology. 2019;113:116–123. doi: 10.1016/j.ejrad.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 18.le Bihan D., Iima M. Diffusion magnetic resonance imaging: what water tells us about biological tissues. PLoS Biology. 2015;13(7, article e1002203) doi: 10.1371/journal.pbio.1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villanueva-Meyer J. E., Wood M. D., Choi B. S., et al. MRI features and IDH mutational status of grade II diffuse gliomas: impact on diagnosis and prognosis. AJR. American Journal of Roentgenology. 2018;210(3):621–628. doi: 10.2214/AJR.17.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smits M., van den Bent M. J. Imaging correlates of adult glioma genotypes. Radiology. 2017;284(2):316–331. doi: 10.1148/radiol.2017151930. [DOI] [PubMed] [Google Scholar]
- 21.Johnson D. R., Diehn F. E., Giannini C., et al. Genetically defined oligodendroglioma is characterized by indistinct tumor borders at MRI. AJNR American Journal of Neuroradiology. 2017;38(4):678–684. doi: 10.3174/ajnr.A5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamauchi T., Ohno M., Matsushita Y., et al. Radiological characteristics based on isocitrate dehydrogenase mutations and 1p/19q codeletion in grade II and III gliomas. Brain Tumor Pathology. 2018;35(3):148–158. doi: 10.1007/s10014-018-0321-4. [DOI] [PubMed] [Google Scholar]
- 23.Wesseling P., van den Bent M., Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta Neuropathologica. 2015;129(6):809–827. doi: 10.1007/s00401-015-1424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J. W., Park C. K., Park S. H., et al. Relationship between radiological characteristics and combined 1p and 19q deletion in World Health Organization grade III oligodendroglial tumours. Journal of Neurology, Neurosurgery & Psychiatry. 2011;82(2):224–227. doi: 10.1136/jnnp.2009.178806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The clinical data used to support the findings of this study were provided by Department of Radiology, the First Affiliated Hospital of Chongqing Medical University, and cannot be made freely available. Access to these data will be considered by the author upon request, with permission from the Director of the Department of Radiology of this hospital.


