Abstract
234 diagnostic formalin-fixed paraffin-embedded (FFPE) blocks from homogeneously treated patients with locally advanced head and neck squamous cell carcinoma (HNSCC) within a multicentre phase III clinical trial were characterised. The mutational spectrum was examined by next generation sequencing in the 26 most frequent oncogenic drivers in cancer and correlated with treatment response and survival. Human papillomavirus (HPV) status was measured by p16INK4a immunohistochemistry in oropharyngeal tumours. Clinicopathological features and response to treatment were measured and compared with the sequencing results. The results indicated TP53 as the most mutated gene in locally advanced HNSCC. HPV-positive oropharyngeal tumours were less mutated than HPV-negative tumours in TP53 (p < 0.01). Mutational and HPV status influences patient survival, being mutated or HPV-negative tumours associated with poor overall survival (p < 0.05). No association was found between mutations and clinicopathological features. This study confirmed and expanded previously published genomic characterization data in HNSCC. Survival analysis showed that non-mutated HNSCC tumours associated with better prognosis and lack of mutations can be identified as an important biomarker in HNSCC. Frequent alterations in PI3K pathway in HPV-positive HNSCC could define a promising pathway for pharmacological intervention in this group of tumours.
Subject terms: Cancer genetics, Genetics, Genomics, Personalized medicine, Cancer, Head and neck cancer
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common neoplasia in the developed world1. It constitutes a heterogeneous disease of tumours of the upper aerodigestive tract with different pathogenic origins and clinical prognosis. Tobacco smoking and alcohol consumption are still the most classical risk factors2 followed by viral infection3,4.
Most HNSCC are diagnosed as locally advanced disease (stage III or IV) and therefore multidisciplinary treatment strategies include surgery, radiotherapy (RT), chemotherapy (CT) and targeted therapy. However, treatment with chemoradiotherapy (CTRT) has become the standard of care after the publication of a large pool analysis5. With the aim of improving the clinical benefit, the addition of cetuximab, an IgG1 chimeric monoclonal antibody against epidermal growth factor receptor (EGFR), concomitant with RT was explored, resulting in longer progression-free survival (PFS) and overall survival (OS) compared to RT alone, although a direct comparison with CTRT has not been evaluated yet6.
The role of induction chemotherapy has remained a subject of controversy. The combination of docetaxel-cisplatin and 5-fluorouracil (TPF) has emerged as the most active regimen in locally advanced disease, showing better results than PF, although it did not show a convincing survival benefit in induction regimens compared with historical data of treatment with concomitant chemoradiotherapy alone7–9. Induction chemotherapy to improve organ preservation and survival may be an alternative to CTRT. The addition of cetuximab to radiation therapy in patients with laryngeal cancer stage III and IVA that respond to TPF could improve functional laryngeal preservation10, although randomized phase III trials did not find that induction chemotherapy provided benefit in time-to-treatment failure or OS11–14. On the other hand, a randomized phase II–III study suggested that adding TPF induction chemotherapy to CTRT resulted in a higher rate of radiological complete response compared with concurrent CTRT alone, improving PFS and OS by induction TPF15. The fact that patient populations in these trials were very heterogeneous, questions induction chemotherapy’s benefit thus, subgroups that will have a benefit from it need to be identified.
Next-generation sequencing (NGS) has helped to identify genetic alterations that could be used as a molecular vulnerability for therapeutic discovery and target optimization. In addition, they could have a prognosis utility as biomarkers of response in different tumour types including head and neck squamous cell carcinomas16,17. For instance, the analysis of The Cancer Genome Atlas (TCGA) described the molecular landscape of HPV-positive and HPV-negative HNSCC as having molecular alterations not reported before18. Since the first description of the recurrently mutated genes in HNSCC19, additional studies have included other genes such as TP53, NOTCH1, PIK3CA, CDKN2A, CCDN1, HRAS, FAT1, FBXW7 and FGFR3, among others20,21. For this reason, targeted sequencing has become a flexible tool to study those genes previously reported as mutated in HNSCC21.
To contribute to the understanding of how somatic mutations influence the outcome of HNSCC treatment, we have studied a panel of 26 genes (Table S1) by next-generation sequencing in a homogenously treated locally advanced HNSCC Spanish cohort. In this study we report some mutations linked with detrimental outcome and their presence in relation to HPV presence.
Results
Cohort characteristics
234 FFPE blocks with diagnostic biopsies from HNSCC patients within a multicentre phase III clinical trial were incorporated in this study (Fig. S1). Clinical demographic factors such as age, gender, disease site and tumour stage are consistent between the whole cohort within the clinical trial and the subsequent random selection due to FFPE block availability in this study. Overall, most were from men (89.7%), with pharyngeal carcinoma (65.4%) and diagnosed in tumour stage IV-A (71.4%) with an average of 57 years old (Table 1). Clinicopathologic features by locations are shown in Table 1.
Table 1.
Clinicopathologic characteristic of the 234 HNSCC patients included in the study: overall and by subtypes.
| Overall | Subtypes | ||||
|---|---|---|---|---|---|
| Variable | N (%) | Oropharynx | Hypopharynx | Larynx | Oral cavity |
| N (%) | N (%) | N (%) | N (%) | ||
| 234 (100) | 90 (38.5) | 63 (26.9) | 41 (17.5) | 40(17.1) | |
| Age (years ± SD) | 57.42 ± 6.9 | 59.46 ± 6.7 | 56.61 ± 8.1 | 58.34 ± 5.7 | 57.55 ± 6.9 |
| Sex | |||||
| Man | 209 (89.7) | 76 (36.4) | 59 (28.2) | 40 (19.1) | 34 (16.3) |
| Woman | 24 (10.3) | 14 (58.3) | 3 (12.5) | 1 (4.2) | 6 (25.0) |
| Unk | 1 | 0 | 1 | 0 | 0 |
| TNM | |||||
| III | 20 (8.7) | 11 (55.0) | 1 (5.0) | 4 (20.0) | 4 (20.0) |
| IVA | 165 (71.4) | 64 (38.8) | 43 (26.1) | 29 (17.6) | 29 (17.6) |
| IVB | 46 (19.9) | 15 (32.6) | 17 (37.0) | 8 (17.4) | 6 (13.0) |
| Unk | 3 | 0 | 2 | 0 | 1 |
| Grade | |||||
| Well differentiated | 32 (15.1) | 13 (40.6) | 8 (25.0) | 3 (9.4) | 8 (25.0) |
| Moderately differentiated | 102 (48.1) | 41 (40.2) | 15 (14.7) | 27 (26.5) | 19 (18.6) |
| Poorly differentiated | 78 (36.8) | 28 (35.9) | 32 (41.0) | 9 (11.5) | 9 (11.5) |
| Unk | 22 | 8 | 8 | 2 | 4 |
| Histology | |||||
| Keratinizing | 94 (48.0) | 40 (42.6) | 19 (20.2) | 13 (13.8) | 22 (23.4) |
| Non-keratinizing | 100 (51.0) | 36 (36.0) | 31 (31.0) | 25 (25.0) | 8 (8.0) |
| Undifferentiated | 2 (1.0) | 1 (50.0) | 0 (0) | 0 (0) | 1 (50.0) |
| Unk | 38 | 13 | 13 | 3 | 9 |
| HPV | |||||
| Positive | 13 (6.7) | 13 (100.0) | 0 (0) | 0 (0) | 0 (0) |
| Negative | 182 (93.3) | 63 (34.6) | 50 (27.5) | 38 (20.9) | 31 (17.0) |
| Unk | 39 | 14 | 13 | 3 | 9 |
TNM classification system stands for tumour, node and metastasis.
SD standard deviation, Unk unknown.
Considering only oropharyngeal tumours (see “Methods” section), 13 samples (17.1%) were HPV-positive based on p16 immunohistochemistry (IHC). According to its grade, HPV-positive samples were statistically associated with poorly differentiated (p = 0.016) and TP53 wild-type (p = 0.009) tumours (Table 2).
Table 2.
HPV association in oropharyngeal tumours with clinicopathological characteristics.
| Variable | HPV-negative | HPV-positive | p-value |
|---|---|---|---|
| N (%) | N (%) | ||
| 63 (82.9) | 13 (17.1) | ||
| Age (years ± SD) | 57.08 (7.11) | 58.46 (6.40) | 0.562*1 |
| Sex | |||
| Man | 55 (84.6) | 10 (15.4) | 0.388*2 |
| Woman | 8 (72.7) | 3 (27.3) | |
| TNM | |||
| III | 5 (71.4) | 2 (28.6) | 0.166*3 |
| IVA | 45 (80.4) | 11 (19.6) | |
| IVB | 13 (100.0) | 0 (0.0) | |
| Grade | |||
| Well differentiated | 10 (83.3) | 2 (16.7) | 0.016*3 |
| Moderately differentiated | 36 (94.7) | 2 (5.3) | |
| Poorly differentiated | 16 (64.0) | 9 (36.0) | |
| Histology | |||
| Keratinizing | 36 (90.0) | 4 (10.0) | 0.177*3 |
| Non-keratinizing | 26 (74.3) | 9 (25.7) | |
| Undifferentiated | 1 (100.0) | 0 (0.0) | |
| Mutational status | |||
| Non-mutated | 15 (78.9) | 4 (21.1) | 0.726*2 |
| Mutated | 48 (84.2) | 9 (15.8) | |
| TP53 mutated | |||
| Non-mutated | 18 (66.7) | 9 (33.3) | 0.009*2 |
| Mutated | 45 (91.8) | 4 (8.2) | |
| PIK3CA mutated | |||
| Non-mutated | 57 (85.1) | 10 (14.9) | 0.178*2 |
| Mutated | 6 (66.7) | 3 (33.3) | |
TNM classification system stands for tumour, node and metastasis.
HPV status based on p16 + IHC could only be measured in 76 (84.4%) out of the 90 oropharyngeal tumour samples.
SD standard deviation, Unk unknown.
*1Mann-Whitney U test.
*2Fisher’s exact test.
*3Chi-square test.
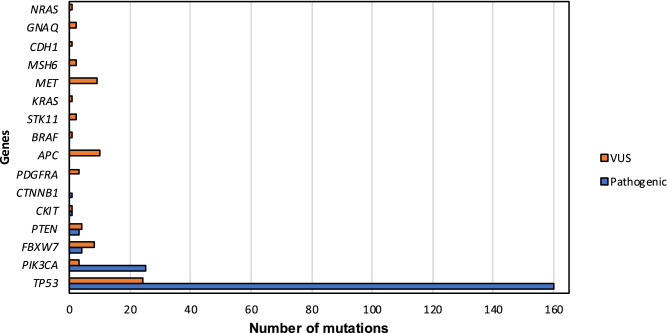
Targeted panel sequencing in HNSCC FFPE blocks identified 162 samples (69.23%) with previously described pathogenic mutations whereas 46 (19.66%) did not carry any mutation and 26 (11.11%) showed variants of uncertain clinical significance (VUS). 194 pathogenic mutations and 72 VUS were found in the sequencing of the 234 FFPE blocks. All samples were sequenced > 5000 × (7074 ± 10,516). Globally, the most mutated gene was TP53 (61.1%) followed by PIK3CA (10.3%), FBXW7 (1.7%), PTEN (1.3%) and CKIT and CTNNB1 (both with 0.43%) (Fig. 1). 144 out of 162 (88.89%) mutated tumours had TP53 mutations either alone or with others. Most of the pathogenic variants were missense (55.67%), followed by stop-gained (18.04%), frameshift (14.95%), splice-donor (8.76%) and in-frame deletions (2.58%) (Table S2).
Figure 1.
Number of mutations found in the sequencing of 234 HNSCC by TruSight Tumor 26 panel. Blue bars represent pathogenic mutations while orange bars show variants of uncertain significance (VUS).
Association of mutations with clinical variables
General comparison of the mutational status and tumour characteristics such as location, grade and histology, did not show any significant difference (p > 0.05) (Table 3). However, considering variants of uncertain significance, women were associated with a lower percentage of mutation than men in our cohort (p = 0.002) (Table 3).
Table 3.
Association between mutational status and clinicopathological characteristics.
| Variable | Normal | Pathogenic | VUS* | p-valuea | p-valueb |
|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | All | Normal vs mutant | |
| 46 (19.66) | 162 (69.23) | 26 (11.11) | |||
| Age (years ± SD) | 57.02 (7.65) | 57.68 (6.95) | 56.44 (6.50) | 0.683*1 | 0.941*1 |
| Sex | |||||
| Man | 39 (18.7) | 152 (72.7) | 18 (8.6) | 0.002*2 | 0.290*2 |
| Woman | 7 (29.2) | 10 (41.7) | 7 (29.2) | ||
| Unk | 0 | 0 | 1 | ||
| TNM | |||||
| III | 5 (25.0) | 13 (65.0) | 2 (10.0) | 0.777*2 | 0.781*2 |
| IVA | 32 (19.4) | 113 (68.5) | 20 (12.1) | ||
| IVB | 8 (17.4) | 35 (76.1) | 3 (6.5) | ||
| Unk | 1 | 1 | 1 | ||
| Location | |||||
| Oropharynx | 16 (17.8) | 66 (73.3) | 8 (8.9) | 0.644*2 | 0.582*2 |
| Hypopharynx | 14 (22.2) | 43 (68.3) | 6 (9.5) | ||
| Larynx | 8 (19.5) | 29 (70.7) | 4 (9.8) | ||
| Oral cavity | 8 (20.0) | 24 (60.0) | 8 (20.0) | ||
| Grade | |||||
| Well differentiated | 7 (21.9) | 24 (75.0) | 1 (3.1) | 0.578*2 | 0.588*2 |
| Moderately differentiated | 17 (16.7) | 73 (71.6) | 12 (11.7) | ||
| Poorly differentiated | 17 (21.8) | 52 (66.7) | 9 (11.5) | ||
| Unk | 5 | 13 | 4 | ||
| Histology | |||||
| Keratinizing | 14 (14.9) | 72 (76.6) | 8 (8.5) | 0.346*2 | 0.652*2 |
| Non-keratinizing | 21 (21.0) | 67 (67.0) | 12 (12.0) | ||
| Undifferentiated | 1(50.0) | 1(50.0) | 0 (0.0) | ||
| Unk | 10 | 22 | 6 | ||
TNM classification system stands for tumour, node and metastasis.
SD standard deviation, Unk unknown.
aInitially, statistical analysis was done comparing normal, pathogenic mutation and variants of uncertain clinical significance (VUS), p-value.
bTo avoid bias with VUS, a direct comparison only between normal and pathogenic mutation was done, p-value.
*Variant of uncertain clinical significance.
*1Kruskal–Wallis H test.
*2Fisher’s exact test.
*3Chi-square test.
Mutational profile and HPV presence in oropharyngeal tumours
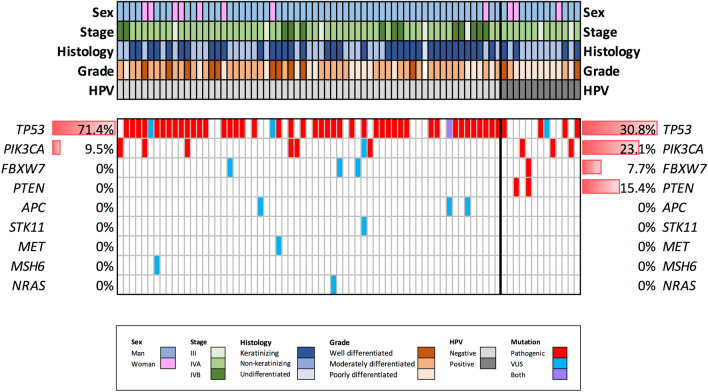
HPV mutational profile in oropharyngeal tumours is shown in Fig. 2. HPV-positive samples presented slightly more pathogenic mutations than HPV-negative (76.2% versus 69.2%, p = 0.762) (Table 2). Despite the fact that TP53 was the most frequently mutated gene in both groups, these mutations were more recurrent in HPV-negative tumours (71.4% in HPV-negative and 30.8% in HPV-positive), difference statistically significant (p = 0.009). Conversely, the second most mutated gene, PIK3CA, although more represented in HPV-positive tumours (9.5% in HPV-negative versus 23.1% in HPV-positive), did not show any statistically significant difference (p = 0.178). While HPV-negative tumours did not present pathogenic mutations in other genes, PTEN was the third most commonly mutated in HPV-positive tumours (15.4%), followed by FBXW7 (7.7%).
Figure 2.
Mutational landscape plot divided into HPV-negative (left) and HPV-positive (right) oropharyngeal tumours. Legend represents different colours according to its clinical features. Percentage of pathogenic mutations in each gene is indicated at the edges by their HPV profile.
Mutational status and response to treatment
After induction chemotherapy, 188 (80.34%) patients were similarly randomized: 95 (50.53%) to conventional treatment and 93 (49.47%) to the experimental arm. Preliminary data indicated that the two regimens showed similar survival, response rates, toxicity and locoregional control22. For that reason, both arms were evaluated within the same group as final response (or response after randomization). Evaluation of the two time-point responses according to the mutational profile did not show any statistical difference (Table 4). There was, however, a tendency between mutated tumours and complete response at the end of the treatment taking VUS into consideration, p = 0.096 (Table S3).
Table 4.
Association between mutational status and response after induction and randomization (final response), without VUS.
| Variable | Normal | Pathogenic | OR | 95% CI | p-value |
|---|---|---|---|---|---|
| N (%) | N (%) | ||||
| Response after induction | 37 (21.5) | 135 (78.5) | |||
| Complete | 5 (13.5) | 19 (14.1) | 0.954 | 0.330–2.753 | 0.931 |
| Partial/stabilization | 32 (86.5) | 116 (85.9) | |||
| Final response | 21 (17.9) | 96 (82.1) | |||
| Complete | 13 (61.9) | 61 (63.5) | 0.932 | 0.352–2.469 | 0.888 |
| Partial/stabilization | 8 (38.1) | 35 (36.5) |
p-value significant if p < 0.05 and size effect indicated by the odd ratio (OR) with 95% confidence interval (CI). 172 (73.5%) and 117 (50.0%) patients were evaluable after induction chemotherapy or randomization respectively.
Considering only HPV profile in oropharyngeal tumours, there were no differences between HPV-positive and HPV-negative individuals either after induction chemotherapy (p = 0.396) or randomization (p = 0.914) (Table 5).
Table 5.
Association between HPV status and response after induction and randomization (final response).
| Variable | HPV-negative | HPV-positive | OR | 95% CI | p-value |
|---|---|---|---|---|---|
| N (%) | N (%) | ||||
| Response after induction | 52 (80.0) | 13 (20.0) | |||
| Complete | 7 (70.0) | 3 (30.0) | 0.519 | 0.114–2.362 | 0.396 |
| Partial/stabilization | 45 (81.8) | 10 (18.2) | |||
| Final response | 33 (78.6) | 9 (21.4) | |||
| Complete | 19 (79.2) | 5 (20.8) | 1.086 | 0.246–4.793 | 0.914 |
| Partial/stabilization | 14 (77.8) | 4 (22.2) |
p-value significant if p < 0.05 and size effect indicated by the odd ratio (OR) with 95% confidence interval (CI). From initial 76 oropharyngeal tumours with HPV determination, 65 (85.5%) of them were evaluable for response after induction and 42 (55.3%) for final response.
Finally, an exploratory analysis was performed using the two most mutated genes in the study: TP53 and PIK3CA (Table S4). Analysing patients with mutations in those genes alone or within other genes and the clinical response, indicated that none of the TP53 subgroups were associated in any of the clinical trial treatment timepoints (p > 0.05). By contrast, considering only PIK3CA mutations, a statistically positive association was found in the complete response group after induction chemotherapy (p = 0.024). However, this finding was not corroborated in final response group (p = 0.235) (Table S4) what could suggest that this could be a false positive result taken into consideration multiple testing and sample size bias.
Poeta’s23 and Neskey’s24 classification in patients harbouring TP53 mutations in relation with clinical response before and after randomization did not show any statistically significant association (p > 0.05, Table S5).
HPV, mutational status and clinical outcome
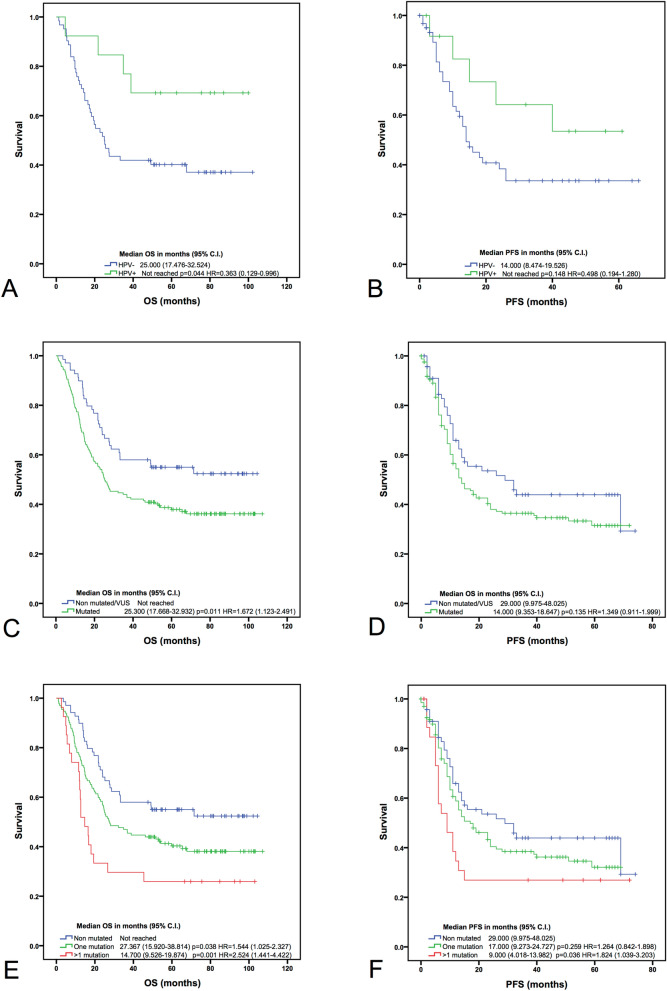
HPV-positive oropharyngeal tumours showed higher OS compared with HPV-negative (p = 0.044). This tendency was also shown in PFS, however, without statistically significant results (p = 0.148, HR = 0.498 (0.194–1.280)) (Fig. 3A,B).
Figure 3.
Kaplan–Meier survival curves. HPV status in oropharyngeal tumours and overall survival (OS) (A) and progression free survival (PFS) (B), mutational status in all the samples and OS (C) and PFS (D), number of mutations in all the samples and their OS (E) and PFS (F). Median with 95% confidence interval (CI), log rank test p-values and hazard ratios (HR) with 95% CI, are shown in each plot.
Moreover, OS was correlated with the mutational status. Patients without mutations in the selected genes had a better OS than patients with mutated tumours (p = 0.011, HR = 1.672 (1.123–2.491)) (Fig. 3C). This difference was also observed in PFS without statistically significant results (p = 0.135, HR = 1.349 (0.911–1.999)) (Fig. 3D).
A correlation with the number of mutations also showed that tumours with one mutation had lower OS (p = 0.038, HR = 1.544 (1.025–2.327)) than non-mutated patients, with the exception of PFS (p = 0.259, HR = 1.264 (0.842–1.898)) (Fig. 3E,F). Equally, tumours with more than one mutation showed lower OS (p = 0.001, HR = 2.524 (1.441–4.422)) and PFS (p = 0.036, HR = 1.824 (1.039–3.203)) than non-mutated samples. Conversely, the differences between tumours with one or more mutations were not statistically significant (p > 0.05).
Finally, we compared TP53 mutations based on Poeta’s23 and Neskey’s24 stratification models with OS and PFS (Fig. S2). No association was observed between low-risk/high risk mutations or non-disruptive/disruptive mutations and survival in these patients (Fig. S2).
Discussion
As most of the head and neck cancers are diagnosed at a locally advanced stage the identification of biomarkers of response is a main goal to optimize treatment and reduce side effects. In recent years, induction chemotherapy has been shown to produce a benefit in organ preservation without a clear improvement in survival. In addition, this approach led to a high toxicity, particularly when concurrent radiotherapy was given with high doses of cisplatin. At present, very few predictive biomarkers of response have been described. For this reason, we proposed a study of the mutational status in 26 of the most common altered genes in cancer with next-generation sequencing in a homogeneously treated representative Spanish cohort of HNSCC from the phase III clinical trial TTCC-2007-0122.
The epidemiology characteristics of the HNSCC patients included in our study were similar to other series reported from the same region: the ratio between sexes is 9:1 in detriment of men, and most of the patients were diagnosed at stage IV25. p16 IHC, a surrogate of HPV infection in oropharyngeal tumours, showed that HPV was present in 17.1% of samples, a lower percentage than previously reported in Europe26 but with similar location to other Southern European countries in oropharynx27.
Globally, the most mutated gene in our series was TP53 (61.1%). We observed a statistically significant lower percentage of mutated TP53 in HPV-positive oropharyngeal tumours (71.4%) than in HPV-negative (30.8%) as has been previously reported in HNSCC28,29. These results could be explained if TP53 sequestration by the viral oncoprotein E6 prevents gaining mutations in this gene under selective pressure of30,31. Comparing to other series, there was a higher percentage of TP53 mutations in HPV-positive tumours29. This fact could be explained by the coexistence of viral infection and other aetiological factors such as tobacco smoking and alcohol consumption during tumourigenesis32; these data were not collected in this study. TCGA data described 85% of TP53 mutation in HPV-negative tumours and only 3% in HPV-positive ones18. However, the sample population was very different with a high predominance of oral cavity tumours (62%) and mainly heavy smokers.
PI3K/AKT/mTOR has been reported as the most mutated pathway in HNSCC (13% to 56%), regardless of the HPV status18. PIK3CA gene, that encodes the catalytic subunit of the family, has been reported with an average mutational rate of 10.53% in HNSCC33, similar to the 10.25% found in this cohort, and with a higher frequency in laryngeal tumours34. Mutations in this gene have also been related to HPV-positive tumours4. Our results corroborate this fact, being PIK3CA more frequently mutated in HPV-positive tumours (23.1% versus 9.5% in HPV-negative oropharyngeal tumours), similar to previously described data35. We did not, however, see an increased percentage in laryngeal carcinoma. 73% of the mutations in PIK3CA are commonly located in 3 hotspots (E542K, E545K and H1047R/L)36, result also found in 76% of PIK3CA mutated samples in our study, emphasising the accuracy of using the targeted panel in HNSCC.
Mutations in FBXW7: an E3 ubiquitin ligase member of the F-box protein family, have been previously observed in HNSCC19. This tumour suppressor gene targets NOTCH1, being an important protein in cell proliferation control. Previous studies found FBXW7 mutated in 5% of HNSCC37,38 and a higher percentage of mutations was previously considered as a prevalent event in HPV-positive tumours39. Our cohort confirmed these results in FBXW7 with a similar percentage only found in HPV-positive tumours (7.7%).
PTEN was the third most mutated gene in 15.4% of the HPV-positive oropharyngeal tumours while not mutations were found in HPV-negative ones. Contrary to our results, TCGA study showed PTEN mutated in 12% of HPV-negative tumours and 6% of HPV-positive18. Apart from PTEN, there were other genes which mutated at a lower percentage in our series, such as CKIT or CTNNB1 (both mutated at less than 1% and only in non-oropharyngeal HPV-negative tumours), have been reported in HNSCC in varied percentages30,38,40. Together with PIK3CA, our result enhances the hypothesis of higher prevalence of PI3K pathway activated mutations in HPV-positive tumours41.
Overall, excluding TP53 mutations, recurrent alterations in PIK3CA, PTEN and FBXW7 genes, all belonging to the PI3K/AKT/mTOR pathway, could define a potential new target for pharmacological intervention in HNSCC, as it has been suggested in other publications42.
In terms of survival, HPV-positive oropharyngeal tumours were associated with better prognosis, showing an increased OS and PFS compared to HPV-negative tumours as it was previously defined26,43–47. Secondly, the presence of mutation in the targeted genes was associated with inferior outcome demonstrated by the presence of detrimental OS. These results could be an indirect measure of tumour aggressiveness, as has been reported in other series43,47. Moreover, the fact that carriers of tumours with more than one mutation have lower OS than those with non-mutated tumours reinforces this concept.
Lastly, there was a lack of association between mutational status and response after treatment. This can indicate that, excluding genetic-driven druggable targets, HNSCC mutational profile is not related to any clinical response but is a matter of mutational burden as is shown in the survival analyses. Similarly, there was no association between TP53 mutations stratified by Poeta’s23 and Neskey’s models24 and response to treatment or survival. These classification systems can serve as an important tool in individualizing and improving treatment for high TP53 mutated tumours, as it was previously identified in a subset of high-risk patients with a decreased response to platinum-based therapies48. Nevertheless, these classification models did not have any implication on outcome in our cohort.
Overall, our data strongly support and expand previously published studies exploring the presence and prognosis of mutations in this population. We have characterized the mutational profile of HPV-positive/HPV-negative oropharyngeal HNSCC in a representative cohort of patients. In this context apart from TP53 mutations, frequent alterations in PIK3CA, PTEN and FBXW7 genes, define possible pathways for pharmacological intervention. Finally, survival analysis showed that mutational status in the tumour could define patient prognosis, and may potentially be used as biomarkers to stratify patients for more intensive treatment. However, larger studies should be performed to confirm these results aiming at stratifying patients to different therapeutic interventions.
Methods
Samples
234 FFPE blocks with diagnostic biopsies from HNSCC patients were included in this study. A consort diagram reporting the dropout is shown in Fig. S1. All samples belong to the clinical trial TTCC-2007-01 entitled: “Open label randomized, multi-centre phase III trial of TPF plus concomitant treatment with cisplatin and radiotherapy versus concomitant cetuximab and radiotherapy in locally advanced, unresectable head and neck cancer”, ClinicalTrials.gov identifier: NCT0071639122.
TTCC-2007-01 trial design and data collection
It was a non-inferiority, randomized and controlled study with a parallel assignment intervention model and an endpoint of safety/efficacy, carried out between 2008 and 2013. The follow-up of the clinical trial finished in November 2016. According to protocol, written informed consent was obtained from living subjects and the protocol was approved by the University Hospital of Salamanca and the ethical committees of each hospital in accordance with the 1964 Helsinki declaration and its later amendments.
Eligible patients: histologically or cytologically confirmed, previously untreated unresectable locally advanced (Stage III–IV) tumours (from oral cavity, oropharynx, larynx, hypopharynx), ECOG performance status 0–1. Unresectable disease was determined by Northern California Oncology Group in measurable disease. Treatment: docetaxel, cisplatin, 5-fluorouracil (TPF)-based induction chemotherapy (T 75 mg/m2 d1, P 75 mg/m2 d1, F 750 mg/m2 CI d 1–5 q 21 d + G-CSF & ciprofloxacin, by 3 cycles; then, if objective response achieved, they were randomized to: conventional radiotherapy (RT) up to 70 Gy + P 100 mg/m2 d 1–22–43 vs conventional RT up to 70 Gy + cetuximab 400/250 mg/m2 weekly until the completion of RT, and they were stratified by primary tumour site. Surgery after RT (neck dissection) was allowed. The primary endpoint was non-inferiority of cetuximab-radiotherapy versus cisplatin-radiotherapy in terms of overall survival. Response rate, loco-regional control and toxicity in both arms were considered secondary objectives. Preliminary data of this trial did not show any difference in terms of survival or response rates, toxicity and loco-regional control as secondary end points in the two regimens22.
Clinical data were compiled in a case report form by medical oncologists involved in the clinical trial. All data were treated with the security measures established in compliance with the Protection of Personal Data Organic Law 15/1999, 13th December, and safe-keeping at the University Hospital of Salamanca in its specific server.
DNA extraction
Percentage of tumour cells was measured in haematoxylin–eosin tissue sections by central pathologist. Between four and ten 10 µm FFPE sections from diagnosis blocks were treated with deparaffinization solution (Qiagen, Hilden, Germany) and DNA extraction was done using QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany).
DNA quality evaluation and targeted NGS
Following TruSight Tumor 26 Reference Guide (Illumina, San Diego, USA), DNA quality was measured by qPCR. Comparing FFPE-gDNA amplification potential with a reference non-FFPE gDNA (QCT), delta Cq value was used to predict the dilution required for each sample.
TruSight Tumor 26 panel includes a set of 174 amplicons in complete exons of 26 cancer-associated genes (Table S1). This panel was selected due to its exceptional success rate using minimal DNA input even from FFPE samples where genetic material is often degraded. Following steps of hybridization with the oligo pool, removing unbound oligos and extension and ligation with bound oligos, an amplification of the libraries were performed. PCR products were checked on a 4% TBE agarose gel and finally the libraries were cleaned up by AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA). PCR products were quantified using Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA) and libraries were normalized at 4 nM in a final pool. Sequencing was performed in a NextSeq 500 System (Illumina, San Diego, USA).
Data were transformed in BaseSpace platform and the VCF file format were read in the Variant Studio Software (Illumina, San Diego, USA). Following Illumina recommendations, somatic variants over 5% of frequency, with yields at least 1000 × cumulative coverage between the 2 strands and considered from the software of PASS filter were reported. Those variants of uncertain significance were considered pathogenic if at least two in silico prediction tools (SIFT and PolyPhen) classified them as deleterious/probably damaging49, and they were defined as likely pathogenic in the Catalogue Of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic) or the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/clinvar) databases.
Assessment of HPV status
In the original study protocol, the assessment of HPV status was carried out by p16 immunohistochemistry (IHC), a surrogate marker for HPV infection50 as the gold-standard technique. FFPE sections were deparaffinized and exposed to 10 mM citrate buffer antigen retrieval at 92 °C for 30 min and then they were stained using a p16INK4a mouse monoclonal antibody (Cell Marque, Rocklin, CA, USA). Percentage of p16 staining was measured and only those tumours > 70% nuclear and cytoplasmic p16+ were considered positive. 33 samples were considered HPV-positive following this methodology: 13 oropharyngeal, 4 hypopharyngeal, 2 laryngeal and 9 oral cavity tumours. However, after the publication of the guidelines from the college of American pathologists, p16 IHC is only recommended in oropharyngeal tumours but other locations, where DNA/RNA viral determination should be performed as a confirmatory test51. Since there was not more DNA from all the samples after the library preparation, only oropharyngeal tumours with > 70% p16 positive staining were considered HPV-positive.
Statistical analyses
Statistical analysis compared categorical parameters and mutational status by the Chi-square or Fisher’s exact tests; while in continuous nonparametric variables, the Mann–Whitney U or Kruskal–Wallis H tests were used. p-values were calculated excluding missing values and they were considered statistically significant when p < 0.05. Significant variables were included in the logistic regression analysis and size effects were indicated by odds ratio (OR) with their 95% confidence interval (95% CI). Mutational status was classified as presence or absence of mutations, number of mutations (none, one or more than one) and the status of TP53 and PIK3CA (mutant or wild-type). Response was divided in two groups of treatment: after induction chemotherapy and after chemo/cetuximab plus radiotherapy (final response) due to the similar outcome in both arms22. Response was classified in both groups as complete response versus partial response/stabilization. No progressions were shown in the cohort.
Survival analysis was done according to the overall survival (OS) and progression-free survival (PFS) by Kaplan–Meier plots and log-rank test p-values were calculated in all the curves. Median was indicated in those plots in which it was achieved. Hazard-ratio was calculated to measure the risk of the event with its 95% confidence interval (95% CI) by Cox regression. Median follow-up in OS was 32.23 months while in PFS it was 15.31 months.
Due to high prevalence in TP53 mutations, we applied Poeta’s and Neskey’s classifications stratifying the mutations according to its change and functional effect, allowing a better comprehensive understanding on their relevance in clinical outcome. Following Poeta’s classification23, TP53 mutations were divided in two categories: disruptive and non-disruptive according to their functional effects on the p53 protein. Additionally, according to Neskey’s model24, also named as Evolutionary Action score of TP53-coding variants (EAp53), missense mutations were stratified into high-risk and low-risk through an in-silico scoring (https://mammoth.bcm.tmc.edu/EAp53/). Then, comparative analysis was performed in response to treatment, OS and PFS.
All these tests were conducted using SPSS software version 21.0 (SPSS Inc., Chicago) and GraphPad Prism software version 6.0 (GraphPad Software Inc., California).
Supplementary information
Acknowledgements
Authors would like to thank the individuals who consented to participate in this study and their relatives, and all their colleagues from the Spanish Group of Treatment of Head and Neck Cancer (TTCC) who have participated in this study and are not included in the list of authors. We would also like to thank the pathologist technician María del Carmen Rodríguez for its implication in the study, Dr Eva Maria Sánchez Tapia and Dr Elena Bueno-Martínez for technical support; and Roger Townsend for English editing.
Author contributions
All authors have read and agree to the published version of the manuscript. Conceptualization, R.S.T., R.G.S. and J.J.C.H.; methodology, J.F.M. and J.P.G.; validation, J.F.M. and J.P.G.; formal analysis, J.F.M. and A.O.; investigation, J.F.M., J.P.G., R.S.T, R.M., J.R.C., C.G.G., L.I., A.C.M., J.C.A.K., M.T., S.V., M.A.G. and E.D.B. ;resources, J.F.M., J.P.G, R.S.T, R.M., J.R.C., C.G.G., L.I., A.C.M., J.C.A.K., M.T., S.V., M.A.G., E.D.B., J.J.C.H. and R.G.S.; data curation, J.F.M., J.P.G. and R.S.T.; writing—original draft preparation, J.F.M., A.O., R.G.S. and J.J.C.H.; writing—review and editing, all authors; visualization, J.F.M., A.O., R.G.S. and J.J.C.H.; supervision, R.G.S. and J.J.C.H.; project administration, .G.S. and J.J.C.H.; funding acquisition, .G.S. and J.J.C.H.
Funding
This research was funded by the health research program of the “Instituto de Salud Carlos III” (PI14/00071) co financed with FEDER founds and for the Health Regional Management of the Junta de Castilla y León (GRS1385/A/16). J. Fernández-Mateos was partially supported by a predoctoral research grant from the Consejería de Educación—Junta de Castilla y León and the European Social Fund to CC-B (EDU/1084/2012).
Competing interests
J.J.C.H. declares conflict of interest in advisory role: Merck, MSD, BMS, Novartis and conferences with fee: Merck, BMS, MSD, Roche, Astra Zeneca, Novartis. However, the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The rest of the authors declare no competing interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rogelio González-Sarmiento, Email: gonzalez@usal.es.
Juan Jesús Cruz-Hernández, Email: jjcruz@usal.es.
Supplementary information
is available for this paper at 10.1038/s41598-020-72927-2.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J. Clin. 2019 doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Sturgis EM, Wei Q, Spitz MR. Descriptive epidemiology and risk factors for head and neck cancer. Semin. Oncol. 2004;31:726–733. doi: 10.1053/j.seminoncol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Lechner M, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV-tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pignon JP, le Maître A, Maillard E, Bourhis J, MACH-NC Collaborative Group Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): An update on 93 randomised trials and 17,346 patients. Radiother. Oncol. 2009;92(1):4–14. doi: 10.1016/j.radonc.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Bonner JA, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard P, et al. Taxane-cisplatin-fluorouracil as induction chemotherapy in locally advanced head and neck cancers: An individual patient data meta-analysis of the meta-analysis of chemotherapy in head and neck cancer group. J. Clin. Oncol. 2013;31:2854–2860. doi: 10.1200/JCO.2012.47.7802. [DOI] [PubMed] [Google Scholar]
- 8.Posner MRMD, et al. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. N. Engl. J. Med. 2007;357:1705–1715. doi: 10.1056/NEJMoa070956. [DOI] [PubMed] [Google Scholar]
- 9.Lorch JH, et al. Induction chemotherapy with cisplatin and fluorouracil alone or in combination with docetaxel in locally advanced squamous-cell cancer of the head and neck: Long-term results of the TAX 324 randomised phase 3 trial. Lancet Oncol. 2011;12:153–159. doi: 10.1016/S1470-2045(10)70279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesía R, et al. Could the addition of cetuximab to conventional radiation therapy improve organ preservation in those patients with locally advanced larynx cancer who respond to induction chemotherapy? An organ preservation Spanish head and neck cancer cooperative group. Int. J. Radiat. Oncol. 2016;97:473–480. doi: 10.1016/j.ijrobp.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Cohen EEW, et al. Phase III randomized trial of induction chemotherapy in patients with N2 or N3 locally advanced head and neck cancer. J. Clin. Oncol. 2014;32:2735–2743. doi: 10.1200/JCO.2013.54.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitt R, et al. A randomized phase III trial comparing induction chemotherapy followed by chemoradiotherapy versus chemoradiotherapy alone as treatment of unresectable head and neck cancer. Ann. Oncol. 2014;25:216–225. doi: 10.1093/annonc/mdt461. [DOI] [PubMed] [Google Scholar]
- 13.Haddad R, et al. Induction chemotherapy followed by concurrent chemoradiotherapy (sequential chemoradiotherapy) versus concurrent chemoradiotherapy alone in locally advanced head and neck cancer (PARADIGM): A randomised phase 3 trial. Lancet Oncol. 2013;14:257–264. doi: 10.1016/S1470-2045(13)70011-1. [DOI] [PubMed] [Google Scholar]
- 14.Geoffrois L, Martin L, Garaud P, Raucourt DD, Miny J, Maingon P, et al. Induction docetaxel platinum 5-FU (TPF) followed by cetuximab-radiotherapy (cetux-RT) versus concurrent chemo-radiotherapy (CT/RT) in patients with N2b/c-N3 non operated stage III-IV squamous cell cancer of the head and neck (SCCHN): Results of the GORTEC 2007–02 phase III randomized trial. J. Clin. Oncol. 2016;34(Suppl):6000. doi: 10.1200/JCO.2016.34.15_suppl.6000. [DOI] [Google Scholar]
- 15.Ghi MG, et al. Concomitant chemoradiation (CRT) or cetuximab/RT (CET/RT) versus induction Docetaxel/ Cisplatin/5-Fluorouracil (TPF) followed by CRT or CET/RT in patients with locally advanced squamous cell carcinoma of head and neck (LASCCHN). A randomized phase III fac. J. Clin. Oncol. 2014;32:6004–6004. doi: 10.1200/jco.2014.32.15_suppl.6004. [DOI] [Google Scholar]
- 16.Rizzo G, Black M, Mymryk JS, Barrett JW, Nichols AC. Defining the genomic landscape of head and neck cancers through next-generation sequencing. Oral Dis. 2015;21:e11–e24. doi: 10.1111/odi.12246. [DOI] [PubMed] [Google Scholar]
- 17.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Network TCGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W, Califano JA. Sequencing the head and neck cancer genome: Implications for therapy. Ann. N. Y. Acad. Sci. 2014;1333:33–42. doi: 10.1111/nyas.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabatabaeifar S, Kruse TA, Thomassen M, Larsen MJ, Sørensen JA. Use of next generation sequencing in head and neck squamous cell carcinomas: A review. Oral Oncol. 2014;11:1035–1040. doi: 10.1016/j.oraloncology.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Hitt R, Mesia R, Grau J. Randomized phase III trial of induction chemotherapy (ICT) with docetaxel-cisplatin-5fluorouracil (DCF) followed by cisplatin-radiotherapy (CRT) or cetuximab-radiotherapy (CetRT) in patients (pts) with locally advanced unresectable head and neck cancer. J. Clin. Oncol. 2016;34:6001. doi: 10.1200/JCO.2016.34.15_suppl.6001. [DOI] [Google Scholar]
- 23.Luana Poeta M, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2007;25:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neskey DM, et al. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck cancer. Cancer Res. 2015;7:1527–1536. doi: 10.1158/0008-5472.CAN-14-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seijas-Tamayo R, et al. Epidemiological characteristics of a Spanish cohort of patients diagnosed with squamous cell carcinoma of head and neck: Distribution of risk factors by tumor location. Clin. Transl. Oncol. 2016;18:1114–1122. doi: 10.1007/s12094-016-1493-1. [DOI] [PubMed] [Google Scholar]
- 26.D’Souza G, et al. Effect of HPV on head and neck cancer patient survival, by region and tumor site: A comparison of 1362 cases across three continents. Oral Oncol. 2016;62:20–27. doi: 10.1016/j.oraloncology.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baboci L, et al. Low prevalence of HPV-driven head and neck squamous cell carcinoma in North-East Italy. Papillomavirus Res. 2016;2:133–140. doi: 10.1016/j.pvr.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaykalova DA, et al. Novel insight into mutational landscape of head and neck squamous cell carcinoma. PLoS ONE. 2014;9:1–9. doi: 10.1371/journal.pone.0093102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westra WH, Taube JM, Poeta ML, Begum S, Sidransky D, Koch WM. Inverse relationship between human papillomavirus-16 infection and disruptive p53 gene mutations in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2008;14:366–369. doi: 10.1158/1078-0432.CCR-07-1402. [DOI] [PubMed] [Google Scholar]
- 30.Chung CH, et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 2015;26:1216–1223. doi: 10.1093/annonc/mdv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: Its role in pathogenesis and clinical implications. Clin. Cancer Res. 2009;15:6758–6762. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- 32.Zevallos JP, Yim E, Brennan P, Liu AY, Taylor JM, Weissler M, et al. Molecular profile of human papillomavirus positive oropharyngeal squamous cell carcinoma stratified by smoking status. Int. J. Radiat. Oncol. Biol. Phys. 2016;94:864. doi: 10.1016/j.ijrobp.2015.12.022. [DOI] [Google Scholar]
- 33.Mountzios G, Rampias T, Psyrri A. The mutational spectrum of squamous-cell carcinoma of the head and neck: Targetable genetic events and clinical impact. Ann. Oncol. 2014;25:1889–1900. doi: 10.1093/annonc/mdu143. [DOI] [PubMed] [Google Scholar]
- 34.Tabatabaeifar S, Kruse TA, Thomassen M, Larsen MJ, Sørensen JA. Use of next generation sequencing in head and neck squamous cell carcinomas: A review. Oral Oncol. 2014;50:1035–1040. doi: 10.1016/j.oraloncology.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin. Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Ginkel JH, de Leng WWJ, de Bree R, van Es RJJ, Willems SM. Targeted sequencing reveals TP53 as a potential diagnostic biomarker in the post-treatment surveillance of head and neck cancer. Oncotarget. 2016;7:61575. doi: 10.18632/oncotarget.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Er T-K, et al. Molecular characterization of oral squamous cell carcinoma using targeted next generation sequencing. Oral Dis. 2015;7:872–878. doi: 10.1111/odi.12357. [DOI] [PubMed] [Google Scholar]
- 39.Rusan M, Li YY, Hammerman PS. Genomic landscape of human papillomavirus-associated cancers. Clin. Cancer Res. 2015;21:2009–2019. doi: 10.1158/1078-0432.CCR-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saba NF, et al. Mutation and transcriptional profiling of formalin-fixed paraffin embedded specimens as companion methods to immunohistochemistry for determining therapeutic targets in oropharyngeal squamous cell carcinoma (OPSCC): A pilot of proof of principle. Head Neck Pathol. 2015;9:223–235. doi: 10.1007/s12105-014-0566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon C, et al. PIK3CA, HRAS and PTEN in human papillomavirus positive oropharyngeal squamous cell carcinoma. BMC Cancer. 2013;13:602. doi: 10.1186/1471-2407-13-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saada-Bouzid E, Le Tourneau C. Beyond EGFR targeting in SCCHN: Angiogenesis, PI3K, and other molecular targets. Front. Oncol. 2019;9:74. doi: 10.3389/fonc.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verri C, et al. Mutational profile from targeted NGS predicts survival in LDCT screening-detected lung cancers. J. Thorac. Oncol. 2017;6:922–931. doi: 10.1016/j.jtho.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coordes A, et al. Meta-analysis of survival in patients with HNSCC discriminates risk depending on combined HPV and p16 status. Eur. Arch. Oto-Rhino-Laryngol. 2016;273:2157–2169. doi: 10.1007/s00405-015-3728-0. [DOI] [PubMed] [Google Scholar]
- 45.Dayyani F, et al. Meta-analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC) Head Neck Oncol. 2010;2:15. doi: 10.1186/1758-3284-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ragin CCR, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: Review and meta-analysis. Int. J. Cancer. 2007;121:1813–1820. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- 47.Dubot C, et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur. J. Cancer. 2018;91:47–55. doi: 10.1016/j.ejca.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 48.Osman A, et al. Evolutionary action score of TP53 coding variants is predictive of platinum response in head and neck cancer patients. Cancer Res. 2015;7:1205–1215. doi: 10.1158/0008-5472.CAN-14-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB. In silico analysis of missense substitutions using sequence-alignment based methods. Hum. Mutat. 2008;29:1327–1336. doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas J, Primeaux T. Is p16 immunohistochemistry a more cost-effective method for identification of human papilloma virus-associated head and neck squamous cell carcinoma? Ann. Diagn. Pathol. 2012;16:91–99. doi: 10.1016/j.anndiagpath.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Lewis JS, et al. Human papillomavirus testing in head and neck carcinomas: Guideline from the College of American Pathologists. Arch. Pathol. Lab. Med. 2018;142:559–597. doi: 10.5858/arpa.2017-0286-CP. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.