Abstract
Immunoglobulin G (IgG) glycosylation is a key post-translational modification in regulating IgG function. It is therefore a prominent target for biomarker discovery and a critical quality attribute of antibody-based biopharmaceuticals. A common approach for IgG glycosylation analysis is the measurement of tryptic glycopeptides. Glycosylation stability during sample processing is a key prerequisite for an accurate and robust analysis yet has hitherto hardly been studied. Especially, acid hydrolysis of sialic acids may be a source for instability. Therefore, we investigated acid denaturation, centrifugal vacuum concentration, and glycopeptide storage regarding changes in the IgG glycosylation profile. Intravenous IgG was analyzed employing imaginable deviations from a reference method and stress conditions. All glycosylation features —sialylation, galactosylation, bisection, and fucosylation—remained unchanged for most conditions. Only with prolonged exposure to acidic conditions at 37 °C, sialylation decreased significantly and subtle changes occurred for galactosylation. Consequently, provided that long or intense heating in acidic solutions is avoided, sample preparation for bottom-up glycoproteomics does not introduce conceivable biases.
Keywords: glycoproteomics, glycosylation stability, immunoglobulin glycosylation, acid hydrolysis, glycopeptide storage
Introduction
Glycosylation is a post-translational modification critical to the stability, metabolism, pharmacokinetics, and function of antibodies.1−3 The large market share of antibody-based biotherapeutics and the key role of antibodies in immunity result in an imminent importance of antibody glycosylation.4 For example, antibody glycosylation is a critical quality attribute of biotherapeutics because it affects their safety and efficacy.2,5 Due to its importance in (patho)physiology, antibody glycosylation has been explored for biomarker identification in autoimmune diseases and neurology and as a therapeutic target in autoimmune skin diseases.6−8
A robust analysis of antibody glycosylation is essential in the biopharmaceutical industry and for biological and clinical studies.9,10 For immunoglobulin G (IgG), conserved N-glycosylation is found in the Fc region of the heavy chain. IgG Fc glycosylation crucially impacts the key interaction with Fc gamma receptors and modulates complement activation.11 However, evidence is also growing for a functional role of glycosylation in the variable domain of the Fab region.6
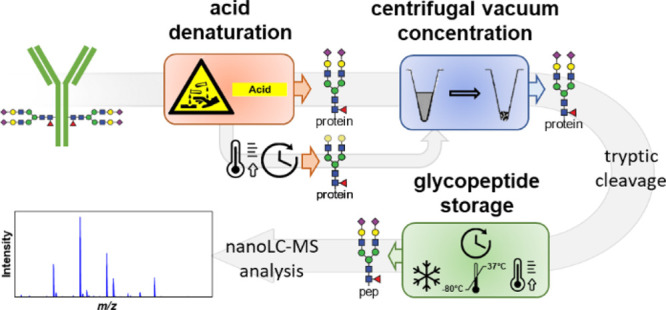
Antibody N-glycans can be robustly analyzed on different levels, each approach featuring its specific advantages and limitations.10,12 Bottom-up workflows provide site- and protein-specific relative quantitation with a high sensitivity. Proteolytic cleavage of IgG, typically using trypsin, results in medium-sized glycopeptides, which are analyzed by liquid chromatography-mass spectrometry (LC-MS). Prior denaturation is essential for obtaining correct glycosylation profiles.13 Many classical protein denaturation approaches are applicable, but applying 100 mM formic acid (FA) at room temperature (RT) followed by centrifugal evaporation allows subsequent efficient IgG cleavage and glycopeptide generation.14 Reduction and alkylation of cysteines can further improve digestion efficiency but is not required for IgG glycoprofiling. High-throughput approaches are important to enable large clinical studies. For IgG Fc N-glycopeptide analysis, a reference workflow is available.14
Even though glycans are considered rather stable, loss of sialic acids during sample preparation and MS analysis has been reported.15−17 Especially under acidic conditions at elevated temperatures, N-acetylneuraminic acid (NANA) is released from glycans by hydrolysis.18 Regarding hydrolysis or degradation of other monosaccharides, much less information is available. Stability issues may negatively impact accuracy and precision, potentially forming an obstacle to biopharmaceutical, biological, or clinical studies. Especially for a loss of fucose, small biases may already lead to misinterpretation of biological or pharmaceutical studies. Therefore, monosaccharide hydrolysis under common sample processing conditions, especially during acid exposure, needs to be excluded.
Few papers have studied N-glycan stability of processed samples under different storage conditions. For example, dried blood spot storage for up to 6 weeks at RT or −80 °C did not influence the glycosylation profile.19 The impact of sample processing on glycopeptide stability in bottom-up workflows has not been investigated, even though they can include acidic conditions. We applied a reference workflow to intravenous immunoglobulin (IVIg) with minor to extreme variation of the experimental conditions, in order to investigate method robustness and the stability of N-glycosylation profiles.
Experimental Section
Materials
Nanogam (IVIg) of 50 mg/mL was kindly shared by Sanquin Research (Amsterdam, Netherlands). Sequencing-grade trypsin was purchased from Promega (Madison, USA). V-bottom 96-well microplates were purchased from Greiner Bio-One (Frickenhausen, Germany). Acetic acid (glacial), ammonium bicarbonate (ABC, >99.5%), and acetonitrile (ACN, for LC-MS) were purchased from Sigma Aldrich (Steinheim, Germany). FA (for MS, ∼98%) was purchased from Thermo Fischer Scientific (Landsmeer, Netherlands). Trifluoroacetic acid (TFA, >99.7%) was purchased from Merck (Darmstadt, Germany). Deionized water was generated at 18.2 MΩ by using a Q-Gard 2 system (Millipore, Amsterdam, Netherlands).
Sample Preparation
IVIg glycoprofiles were analyzed according to a previously published method, albeit without affinity purification.14 In short, IVIg was denatured in 100 mM FA for 6 min at RT and then dried using a centrifugal vacuum concentrator (SpeedVac, RVC 2-33 CDplus; Christ) at 60 °C to complete dryness for 2 h. Dried glycoprotein samples were resuspended in freshly prepared 25 mM ABC and digested with 10 ng/μL sequencing-grade trypsin dissolved in ice-cold 25 mM ABC at 37 °C for 18 h with an enzyme/protein ratio of 1/25. Afterward, plates were sealed, and the tryptic digest was stored at −20 °C until measured by LC-MS.
Acidic Conditions
Six aliquots of IVIg each were incubated in sealed plates with 100 mM FA under the following conditions: for 2 weeks at −80,–20, and 37 °C and for 2 days at RT and 37 °C. Additionally, a control for the impact of three freeze–thaw cycles was included. All samples were additionally incubated for 6 min at RT to achieve the necessary denaturation.
Centrifugal Vacuum Concentration
Glycan stability was studied under diverse centrifugal vacuum concentrator settings: 60 °C for 1, 2, and 3 h; 60 °C with infrared for 1, 2, and 3 h; and 20 °C (RT) for 2 and 3 h. Infrared radiation increases the heat transfer to the samples, and therefore, it is used to accelerate the process. Each condition was performed in triplicate.
Postprocessing Storage Conditions
Stability of the tryptic glycopeptides was evaluated for different storage temperatures, time of storage, and exposure to light. Postprocessing, samples were stored under the following conditions: for 2 weeks at −80,–20, and 4 °C, RT, 37, and 50 °C in the dark, and RT with exposure to light; and for 2 days at 4 °C, RT in the dark, and RT with exposure to light. Storage experiments were performed in quadruplicate. After the last timepoint for all storage conditions, all samples were measured.
NanoRP-LC-ESI-MS
The LC-MS method was applied as previously reported with slight modifications.14,15 200 nL of the tryptic digests were separated by nano reverse phase (RP)-LC using a Dionex UltiMate 3000 nanoLC system (Thermo Fisher Scientific, Breda, Netherlands). After trapping on an Acclaim PepMap 100 C18 5 mm × 300 μm trap column (Thermo Fisher Scientific), a nanoEase MZ Peptide BEH C18 column of 75 μm × 100 mm, featuring 130 Å pores and 1.7 μm particles, was used for separation at 600 nL/min (Waters, Milford, USA). The binary gradient was from 3.0 to 21.7% B from 0.0 to 4.5 min, linear from 21.7 to 50% B from 4.5 to 5.5 min, isocratic at 50.0% B for 2.5 min, linear from 50 to 3% from 8 to 9 min, and re-equilibration at 3% for 2.5 min. Solvents A and B were an aqueous 0.1% TFA solution and 95% ACN, respectively. Also, electrospray ionization mass spectrometry (ESI-MS) parameters were as reported before.14
LC-MS Data Processing
The obtained spectra were transformed into mzXML files using MSConvertGUI (ProteoWizard). The data were aligned and calibrated, and analytes were quantified using LaCyTools (version 1.1.0 alpha) as previously described14,20 (see the Supporting Information). The list of alignment and calibration masses is shown in Tables S-1 and S-2, respectively. A total of 136 glycopeptide compositions were targeted, of which 39 were quantified after curation (Tables S-3 and S-4). Subsequently, the derived traits were calculated independently for each subclass: galactosylation, sialylation, fucosylation, and bisection were calculated similarly to previously described21 (Supporting Information).
Statistical Analysis
Derived traits for each studied condition were compared to the reference condition—drying down at 60 °C for 2 h14 or storage at −80 °C for 2 weeks—with a parametric, unpaired t-test. Bonferroni’s multiple comparison correction was applied. Statistical analysis was performed using GraphPad Prism, version 8.1.1, for Windows (GraphPad Software, San Diego, California, USA).
Results and Discussion
Glycopeptide stability may be compromised by hydrolysis of glycosidic bonds under acidic conditions. Prominently, sialic acids may be lost from glycans. In addition, the activity of contaminating proteases or (residual) side activity of trypsin may degrade the peptide backbone. These phenomena may lead to biases in glycoprofiling or decreased sensitivity. Therefore, sample processing for bottom-up glycosylation analysis encompasses three potentially critical steps—one, the exposure to mild acids during denaturation and/or immunoprecipitation; two, the evaporation for buffer exchange that may concentrate the acid and expose the sample to additional heating; and three, the necessary storage of the final (glyco)peptide mixture, for example, in autosamplers, especially in large high-throughput studies with prolonged processing and storage times. Using IVIg, we tested the impact of (accidental) changes to the protocol using foreseeable deviations and extreme stress conditions. Glycosylation traits for the reference conditions were comparable to those previously reported for Dutch market IVIg.21
Acidic Conditions
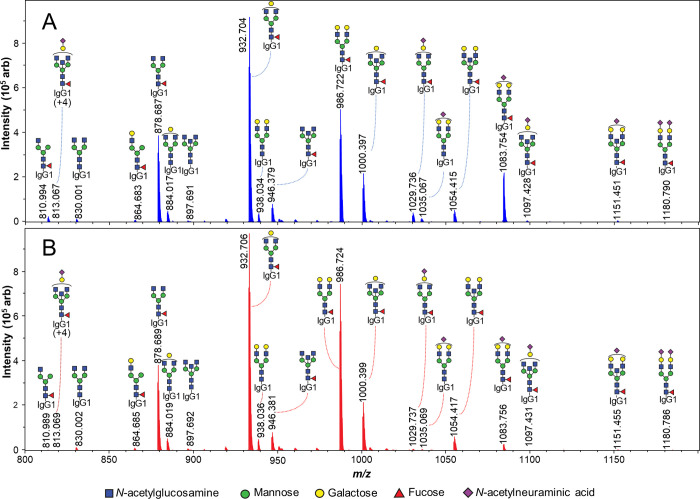
Mild acid exposure (100 mM FA, pH 2.3) did not change the glycosylation features when storing the samples frozen or even at RT for a limited period (Figure 2). Moreover, exposing the samples to multiple freeze–thaw cycles did not cause any changes in glycosylation either. When acidic conditions were applied at an elevated temperature (37 °C) and for a rather long period, expectedly, we detected a prominent decrease in sialylation. The difference in means for IgG1 sialylation was −1.33 ± 0.06% after 2 days and −5.91 ± 0.02% after 2 weeks in 100 mM FA (Figures 1 and 2A). This could be observed as an increased signal of asialylated glycopeptides compared to corresponding sialylated structures. Similar trends were obtained for IgG2 and IgG4 (Figure S1).
Figure 2.
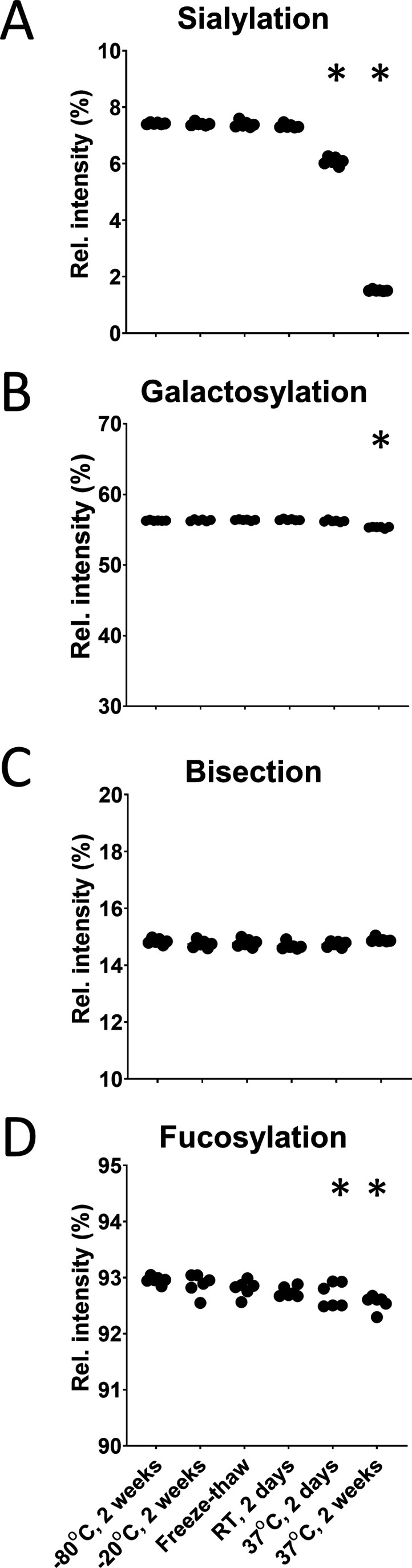
Stability of glycosylation features under acidic conditions (100 mM FA). All of the features are highly stable under conditions that may accidentally occur during sample processing. (A) Sialylation decreases only if IVIg is stressed at a high temperature. (B) Galactosylation decreases only very slightly even under the most extreme conditions (37 °C, 2 weeks). (C) Bisection does not change even under severe acidic stress. (D) Fucosylation apparently decreases very slightly due to minor interdependencies with sialic acids. Actual fucosylation is unaffected (Figure S6). * Changes in the respective glycosylation feature compared to the reference condition (−80 °C).
Figure 1.
MS spectra of IgG1 tryptic N-glycopeptides from IVIg after acid exposure (elution time 81 to 93 s). (A) Reference protocol (stored at −80 °C) and (B) sample stressed at 37 °C for 2 weeks. Sialylated glycoforms, prominently G2FS at m/z 1083.754, decrease in relative intensity after extended acid stress, while expected asialylation products, such as G2F at m/z 986.722, increase.
No major differences in galactosylation were found. Again, only under stress conditions of 37 °C, a slight decrease was observed after 2 days, −0.76 ± 0.16% for IgG4, and after 2 weeks, −0.96 ± 0.05% for IgG1 (Figure 2B), −0.54 ± 0.05% for IgG2, and −1.91 ± 0.10% for IgG4 (Figure S1). However, neither fucosylation nor bisection was found to be affected by acid exposure (Figure 2C,D, Figure S2). The minor changes in apparent fucosylation result from the low abundance of afucosylated, sialylated species. While for neutral species, four of 12 afucosylated species can be quantified, for sialylated species, this ratio is 1 in 6. This small bias toward neutral species in afucosylated glycans leads to a minor positive correlation between fucosylation and sialylation—only 0.4% at an almost complete loss of sialic acids. Actual fucosylation is stable as G0/G0F and G1/G1F ratios remain unchanged (Figure S6).
In summary, glycopeptides are highly stable under the conditions applied for acid denaturation. No changes occur under expectable deviations from standard practice. Even at RT, glycosylation is stable under mild acidic conditions for an extended period. Only deliberate acid exposure at increased temperature for a longer period will result in noteworthy desialylation.
Centrifugal Vacuum Concentration
Centrifugal vacuum concentration and other freeze-drying techniques are also common in glycoproteomics. They serve for buffer exchange and to increase the concentration of analytes. In our reference protocol, acidity increases during vacuum concentration, as water evaporates faster than FA, potentially leading to glycan instability. Settings that may affect IVIg glycosylation profiles are heat and/or the use of infrared radiation to speed up this process and the time of exposure of the samples to these conditions. Results for each condition were compared to vacuum centrifugation at 60 °C for 2 h, which is part of the reference method.14
A wide range of vacuum centrifugation conditions did not cause any noteworthy differences (Figure S3). An incidental −0.17% difference in IgG2 bisection is far below any consequence in a biological context and might well be an undersampling artifact. Surprisingly, glycosylation was not affected under vacuum by exposure to heat (up to 60 °C) and infrared irradiation for up to 3 h. The samples usually freeze during the drying down process. Therefore, analytes may not be exposed to enough energy to induce the degradation processes observed at elevated temperatures under atmospheric pressure.
Postprocessing Storage Conditions
After samples have been processed, they may be directly measured by nanoLC-MS or briefly stored. Sample storage must prevent modification of features of interest in order not to bias the measurement. Samples were stored at −80 °C for 2 weeks as the reference condition. The storage conditions were on the one hand chosen to simulate real-life situations and on the other hand to stress samples. Temporary storage in −20 °C freezers, cooled autosamplers, or fridges and (accidental) extended thawing at RT are examples of expectable conditions or deviations. Stressing samples at elevated temperatures was applied to mimic long-term storage effects.
All traits, except fucosylation, showed some minor fluctuations in this experiment (Figure S4). However, all differences remained well under 1% relative abundance and thus far below any functional relevance. Additionally, effects were mostly inconsistent between IgG subclasses and did not show trends related to increased temperature stress, indicating complex multifactorial causes. Considering that sample storage at 37 °C for 2 weeks did not induce any relevant changes in the glycosylation features, the processed samples could likely be stored frozen or at 4 °C for months.
Total intensity of all glycopeptides was stable among the different storage conditions. This indicates that glycopeptides are not measurably degraded, for example, by unspecific proteolysis. A slight increase in intensity was observed as temperatures increased. An increase in the concentration of target glycopeptides by continued specific tryptic cleavage could explain this observation (Figure S5).
Conclusions
IgG glycosylation was very stable under critical sample processing conditions: acid denaturation, centrifugal vacuum concentrator settings, and intermediate-term storage of tryptic digests. This indicates a high robustness of bottom-up glycoproteomics workflows. Within reasonable limits, and sometimes even under stress, IgG glycoprofiles remained stable. As expected, sialic acids were labile under a combination of elevated temperatures, acidic conditions, and prolonged exposure. To a far lesser extent, galactosylation decreased under such stress conditions. Bisection and fucosylation were unaffected by any tested condition. Thus, unintentional variation in or deviation from the reference protocol, representative of common IgG glycoprofiling practice, will not result in any degradation-related biases or imprecision.
Acknowledgments
The authors thank Jan Nouta and Carolien Koeleman for expert technical assistance. This research is supported by the EU Marie Curie Innovative Training Network (Analytics for Biologics project, grant agreement No. 765502).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00656.
Experimental details and supporting figures: (Figure S-1) extracted ion chromatogram (EIC) of sialylated and asialylated IgG1 glycoforms after acid exposure and heat; (Figure S-2) graphs comparing acidic conditions; (Figure S-3) graphs comparing centrifugal vacuum concentrator conditions; (Figure S-4) graphs comparing postprocessing storage conditions; (Figure S-5) total intensity comparing postprocessing storage conditions; and (Figure S-6) graphs comparing fucosylated and nonfucosylated asialylated species under different acidic conditions (PDF).
Supporting Information Tables: (Table S-1) alignment list; (Table S-2) calibration list; (Table S-3) analyte list; and (Table S-4) relative abundances (xlsx).
Author Contributions
All authors designed the experiments and wrote the manuscript. M.A.-M. performed and evaluated the experiments, supervised by D.F. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): Manuela Amez Martn is employed by Ludger Ltd.
Supplementary Material
References
- Zheng K.; Bantog C.; Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs 2011, 3, 568–576. 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch D.; Tejada M. L. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology 2015, 25, 1325–1334. 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur R.; Einarsdottir H. K.; Vidarsson G. IgG-effector functions: ″the good, the bad and the ugly″. Immunol. Lett. 2014, 160, 139–144. 10.1016/j.imlet.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Chung J. Special issue on therapeutic antibodies and biopharmaceuticals. Exp. Mol. Med. 2017, 49, e304. 10.1038/emm.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakae Y.; Satoh T.; Yagi H.; Yanaka S.; Yamaguchi T.; Isoda Y.; Iida S.; Okamoto Y.; Kato K. Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fcgamma receptor IIIa. Sci. Rep. 2017, 7, 13780. 10.1038/s41598-017-13845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergroesen R. D.; Slot L. M.; van Schaik B. D. C.; Koning M. T.; Rispens T.; van Kampen A. H. C.; Toes R. E. M.; Scherer H. U. N-Glycosylation Site Analysis of Citrullinated Antigen-Specific B-Cell Receptors Indicates Alternative Selection Pathways During Autoreactive B-Cell Development. Front Immunol. 2019, 10, 2092. 10.3389/fimmu.2019.02092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J.; Streich L.; Pinto S.; Pronto-Laborinho A.; Nimtz M.; Conradt H. S.; de Carvalho M. Exploring Cerebrospinal Fluid IgG N-Glycosylation as Potential Biomarker for Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2019, 56, 5729–5739. 10.1007/s12035-019-1482-9. [DOI] [PubMed] [Google Scholar]
- Mihai S.; Albert H.; Ludwig R. J.; Iwata H.; Björck L.; Collin M.; Nimmerjahn F. In vivo enzymatic modulation of IgG antibodies prevents immune complex-dependent skin injury. Exp. Dermatol. 2017, 26, 691–696. 10.1111/exd.13163. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Woen S.; Wang T.; Liau B.; Zhao S.; Chen C.; Yang Y.; Song Z.; Wormald M. R.; Yu C.; Rudd P. M. Challenges of glycosylation analysis and control: an integrated approach to producing optimal and consistent therapeutic drugs. Drug Discovery Today 2016, 21, 740–765. 10.1016/j.drudis.2016.01.006. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Falck D.; Wuhrer M. Monitoring of immunoglobulin N- and O-glycosylation in health and disease. Glycobiology 2020, 30, 226–240. 10.1093/glycob/cwz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y.; Barb A. W. A synopsis of recent developments defining how N-glycosylation impacts immunoglobulin G structure and function. Glycobiology 2020, 30, 214–225. 10.1093/glycob/cwz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo S.; Pérez-Robles R.; Jakes C.; Ribeiro da Silva M.; Millán Martín S.; Farrell A.; Navas N.; Bones J. Comparing different domains of analysis for the characterisation of N-glycans on monoclonal antibodies. J. Pharm. Anal. 2020, 10, 23–34. 10.1016/j.jpha.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck D.; Jansen B. C.; Plomp R.; Reusch D.; Haberger M.; Wuhrer M. Glycoforms of Immunoglobulin G Based Biopharmaceuticals Are Differentially Cleaved by Trypsin Due to the Glycoform Influence on Higher-Order Structure. J. Proteome Res. 2015, 14, 4019–4028. 10.1021/acs.jproteome.5b00573. [DOI] [PubMed] [Google Scholar]
- Falck D.; Jansen B. C.; de Haan N.; Wuhrer M. High-Throughput Analysis of IgG Fc Glycopeptides by LC-MS. Methods Mol. Biol. 2017, 1503, 31–47. 10.1007/978-1-4939-6493-2_4. [DOI] [PubMed] [Google Scholar]
- Reusch D.; Haberger M.; Falck D.; Peter B.; Maier B.; Gassner J.; Hook M.; Wagner K.; Bonnington L.; Bulau P.; Wuhrer M. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 2: Mass spectrometric methods. mAbs 2015, 7, 732–742. 10.1080/19420862.2015.1045173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S.; Wada Y.; Tanaka K. Derivatization for stabilizing sialic acids in MALDI-MS. Anal. Chem. 2005, 77, 4962–4968. 10.1021/ac050287o. [DOI] [PubMed] [Google Scholar]
- Toyoda M.; Ito H.; Matsuno Y. K.; Narimatsu H.; Kameyama A. Quantitative derivatization of sialic acids for the detection of sialoglycans by MALDI MS. Anal. Chem. 2008, 80, 5211–5218. 10.1021/ac800457a. [DOI] [PubMed] [Google Scholar]
- Chuanxiang W.; Lian X.; Lijie L.; Fengli Q.; Zhiwei S.; Xianen Z.; Jinmao Y. A sensitive and efficient method for determination of N-acetylhexosamines and N-acetylneuraminic acid in breast milk and milk-based products by high-performance liquid chromatography via UV detection and mass spectrometry identification. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1011, 14–23. 10.1016/j.jchromb.2015.12.032. [DOI] [PubMed] [Google Scholar]
- Vreeker G. C. M.; Bladergroen M. R.; Nicolardi S.; Mesker W. E.; Tollenaar R. A. E. M.; van der Burgt Y. E. M.; Wuhrer M. Dried blood spot N-glycome analysis by MALDI mass spectrometry. Talanta 2019, 205, 120104. 10.1016/j.talanta.2019.06.104. [DOI] [PubMed] [Google Scholar]
- Jansen B. C.; Falck D.; de Haan N.; Hipgrave Ederveen A. L.; Razdorov G.; Lauc G.; Wuhrer M. LaCyTools: A Targeted Liquid Chromatography–Mass Spectrometry Data Processing Package for Relative Quantitation of Glycopeptides. J. Proteome Res. 2016, 15, 2198–2210. 10.1021/acs.jproteome.6b00171. [DOI] [PubMed] [Google Scholar]
- Fokkink W. J. R.; Falck D.; Santbergen T. C. M.; Huizinga R.; Wuhrer M.; Jacobs B. C. Comparison of Fc N-Glycosylation of Pharmaceutical Products of Intravenous Immunoglobulin G. PLoS One 2015, 10, e0139828. 10.1371/journal.pone.0139828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.