Abstract
Background
Phytohormones are small molecules that regulate virtually every aspect of plant growth and development, from basic cellular processes, such as cell expansion and division, to whole plant environmental responses. While the phytohormone levels and distribution thus tell the plant how to adjust itself, the corresponding growth alterations are actuated by cell wall modification/synthesis and internal turgor. Plant cell walls are complex polysaccharide-rich extracellular matrixes that surround all plant cells. Among the cell wall components, cellulose is typically the major polysaccharide, and is the load-bearing structure of the walls. Hence, the cell wall distribution of cellulose, which is synthesized by large Cellulose Synthase protein complexes at the cell surface, directs plant growth.
Scope
Here, we review the relationships between key phytohormone classes and cellulose deposition in plant systems. We present the core signalling pathways associated with each phytohormone and discuss the current understanding of how these signalling pathways impact cellulose biosynthesis with a particular focus on transcriptional and post-translational regulation. Because cortical microtubules underlying the plasma membrane significantly impact the trajectories of Cellulose Synthase Complexes, we also discuss the current understanding of how phytohormone signalling impacts the cortical microtubule array.
Conclusion
Given the importance of cellulose deposition and phytohormone signalling in plant growth and development, one would expect that there is substantial cross-talk between these processes; however, mechanisms for many of these relationships remain unclear and should be considered as the target of future studies.
Keywords: Cellulose, auxin, brassinosteroid, ABA, ethylene, jasmonate, cytoskinin, microtubules
INTRODUCTION
Phytohormones are small molecules produced by plants to signal environmental alterations and support developmental progression. The major phytohormone classes include abscisic acid (ABA), auxins, brassinosteroids (BRs), cytokinin (CK), ethylene (ET), gibberellins, jasmonic acid (JA), salicylic acid (SA) and, more recently, strigolactone (SL) (Davière and Achard, 2016). The biosynthetic and signalling pathways for these phytohormone classes have been outlined through genetic, chemical and structural studies. While these pathways have been explored independently, the last decade has provided ample examples of extensive crosstalk between phytohormone signalling and biosynthetic pathways (Kuppusamy et al., 2009; Bai et al., 2012), suggesting that a highly interconnected hormone signalling framework is of critical importance to regulate plant growth.
While phytohormones control assorted aspects of plant growth, development and responses to the environment, the primary actuators of plant growth comprise two intimately linked processes: turgor-mediated cell expansion, and the synthesis, deposition and remodelling of plant cell walls. Cell walls consist largely of polysaccharides, proteins, water-soluble material and, in some cases, polyphenolics (Somerville et al., 2004; Cosgrove, 2005; Burton et al., 2010). Of these components, the polysaccharides are typically the main constituents and are divided into three broad classes: cellulose, hemicelluloses and pectins. Additionally, plant cell walls can be subclassified based on thickness, composition and tissue distribution into primary cell walls, which surround nearly all plant cells, and secondary cell walls, which are substantially thickened cellulose and lignin-rich structures found primarily in the plant vasculature. Cellulose is often the principal polysaccharide of plant cell walls and, with a tensile strength similar to that of steel, contributes the load-bearing strength necessary to counteract the substantial turgor pressure that arises in growing plant cells (Somerville et al., 2004; Cosgrove, 2005). Directed cell expansion is therefore governed primarily by cellulose orientation, which largely renders mechanical anisotropy (Eng and Sampathkumar, 2018).
Cellulose synthesis
Cellulose consists of β-(1→4)-linked glucans that are typically arranged into para-crystalline microfibrils via intermolecular hydrogen bonds. This polysaccharide is produced by Cellulose Synthase A (CESA) protein complexes (CSCs) at the plasma membrane of all land plants (Lampugnani et al., 2019). The CSCs are thought to consist of three unique, but structurally related, CESA subunits (Schneider et al., 2016), which serve as the catalytic subunits of CSCs (Purushotham et al., 2016; Cho et al., 2017). In Arabidopsis thaliana, the three subunits CESA1, 3 and CESA6-like CESAs produce cellulose in primary walls (Desprez et al., 2007; Persson et al., 2007). By contrast, another set of CESAs, CESA4, 7 and 8, form a CSC that is active during secondary wall cellulose synthesis (Taylor et al., 2003). The CESAs are synthesized at the endoplasmic reticulum (ER) and assembled into CSCs either at the ER or at the Golgi apparatus (Polko and Kieber, 2019) (Fig. 1) with the aid of the STELLO1 and 2 proteins (STL) (Zhang et al., 2016). After assembly, the CSCs are secreted via the trans-Golgi network (TGN) to the plasma membrane, a process which requires TGN acidic pH (Luo et al., 2015), a dynamic actin cytoskeleton and actin-mediated trafficking (Sampathkumar et al., 2013; Zhang et al., 2019), the FRAGILE FIBER1 (FRA1) kinesin (Zhu et al., 2015), the exocyst complex via PATROL1 (Zhu et al., 2018), and a pair of proteins referred to as SHOU4 and SHOU4-like (Polko et al., 2018). CSCs are preferentially delivered to plasma membrane sites that coincide with cortical microtubules (CMTs) of growing cells (Gutierrez et al., 2009).
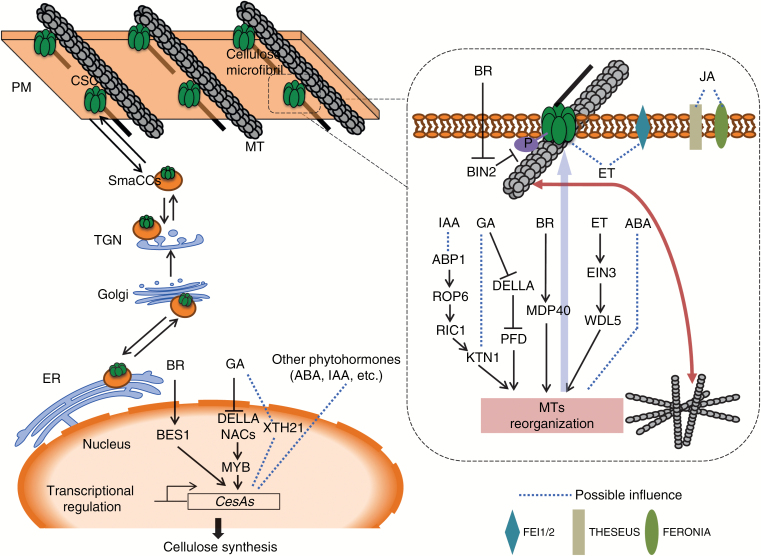
Fig. 1.
The impacts of phytohormones on cellulose synthesis and microtubule (MT) organization. Cellulose is synthesized by plasma membrane (PM)-localized cellulose synthase (CESA) complexes (CSC) that move along underlying cortical MT. CESAs are synthesized at the endoplasmic reticulum (ER) and then transported via the Golgi and the trans-Golgi Network (TGN) to the PM, possibly via small CESA compartments (SmaCCs). Phytohormones regulate CESA expression. For example, the brassinosteroid (BR)-regulated transcription factor BES1 can directly bind to the promoter of CESAs, thus activating expression. Also, the gibberellin (GA)-mediated DELLA-NAC signalling pathway may regulate CESA expression and secondary wall biosynthesis. Besides transcriptional regulation, phytohormones can control post-translational modifications of CESAs. Here, BIN2, a negative regulator of BR signalling, negatively regulates CSC activity and cellulose synthesis by phosphorylating CESA1 in Arabidopsis. Some PM-localized receptor kinases provide links between hormone signalling and cellulose and cell wall synthesis. FEI1 and 2, two leucine-rich repeat receptor kinases, might interact with ACS, an important component for ethylene (ET) synthesis, which may regulate cellulose synthesis and cell expansion. The protein kinases THESEUS (THE1) and FERONIA (FER) might connect jasmonic acid (JA) signalling, cellulose synthesis and cell wall integrity. Notably, most of the links between phytohormone levels, signalling and cellulose synthesis are indirect and much work is needed to consolidate the connections and underlying mechanisms. MT organization and dynamics are regulated by phytohormones. For instance, auxin (IAA) impacts MT organization via the IAA-ABP1-ROP6-RIC1-KTN1 pathway. Moreover, prefoldin (PFD) proteins can interact with DELLAs and then directly influence MT dimerization and behaviour. The data presented here derive from current studies on Arabidopsis.
After delivery, the CSCs are probably activated, possibly via post-translational modifications (Speicher et al., 2018). Their activation would lead to cellulose extrusion into the apoplast and incorporation of the microfibrils into the cell wall structure. This incorporation would effectively immobilize the microfibrils and therefore force the active CSCs to move forward in the plasma membrane. Indeed, newly delivered CSCs are typically immobile for about 1 min (Gutierrez et al., 2009; Zhang et al., 2019) after which they become motile (Paredez et al., 2006; Diotallevi and Mulder, 2007; Watanabe et al., 2015). The direction of movement is templated by CMTs (Paredez et al., 2006; Watanabe et al., 2015, 2018) (Fig. 1). The CSCs functionally associate with CMTs through several proteins (Schneider et al., 2016). Here, CELLULOSE SYNTHASE INTERACTIVE (CSI1) and COMPANION OF CELLULOSE SYNTHASE1 (CC1) and 2 are important factors that maintain CSCs on CMT tracks (Bringmann et al., 2012; Li et al., 2012; Endler et al., 2015; Schneider et al., 2017; Kesten et al., 2019). In addition, the CELLULOSE-MICROTUBULE UNCOUPLING1 (CMU1) and 2 contribute to the stability of the microtubules to withstand forces generated by the moving CSCs (Liu et al., 2016). Hence, as the templating devices for cellulose synthesis, CMTs may drive mechanical wall anisotropy to steer cell expansion. However, cellulose microfibril orientation does not always align with CMTs (Himmelspach et al., 2003), and coordinated CSC behaviour occurs in the absence of CMTs (Paredez et al., 2006). It is therefore plausible that certain wall-related features or membrane restrictions may also contribute to maintenance of CSC coordination (Baskin, 2001; Schneider et al., 2017; Chan and Coen, 2020).
The speed of CSC movement is likely to represent the catalytic rate of the complexes (Schneider et al., 2016). Primary wall CSCs typically move bi-directionally at a rate of 150–500 nm/min, which corresponds to ~300–1000 glucose residues per glucan chain per minute (Paredez et al., 2006). Mutations in several CSC-associated proteins impact these rates. For example, mutations of the endo-glucanase KORRIGAN (KOR1) and CHITINASE-LIKE1 (CTL1) lead to reduced CSC speeds with corresponding reductions in cellulose content (Nicol et al., 1998; Paredez et al., 2008; Sánchez-Rodríguez et al., 2012). Several other components are closely linked to cellulose production and impact cellulose levels, such as members of the glycosylphosphatidylinositol (GPI) anchored proteins COBRA (COB) (Brown et al., 2005; Roudier et al., 2005) and a protein of unknown function called KOBITO (KOB1) (Pagant et al., 2002). Once the CSC produces a cellulose microfibril of sufficient length, typically around 3 µm if one assumes a CSC speed of 300 nm/min and a lifetime of just below 10 min (Sampathkumar et al., 2013), the CSC is stalled through an unknown mechanism and awaits internalization. This internalization process is driven by the TPLATE complex (Gadeyne et al., 2014; Sánchez-Rodríguez et al., 2018) with aid of the Adapter-Protein complex (AP2) (Bashline et al., 2013, 2015). The internalization may lead to a recycling route via the TGN, or perhaps the intermediate membrane compartments referred to as small CESA compartments (smaCCs) (Gutierrez et al., 2009)/microtubule-associated cellulose synthase compartments (MASCs) (Crowell et al., 2009), or to degradation via the lytic vacuole.
Based on the critical importance of phytohormone signalling and cellulose biosynthesis in coordinating plant growth and development, it is anticipated that numerous functional relationships exist between these two processes (Sánchez-Rodríguez et al., 2010). In the following sections, we outline how cellulose synthesis interconnects with hormone production and signalling and how these relationships impact plant growth. We also provide concrete steps to improve our understanding of cross-talks between cellulose biosynthesis and phytohormone signal transduction.
BR signalling and cellulose biosynthesis
Brassinosteroids (BRs) are a key class of growth-promoting phytohormones that impact physiological processes across the entire plant life cycle, including seedling germination, root meristem maintenance, cell expansion, stomatal development and responses to environmental perturbations (Kim et al., 2012; Hu and Yu, 2014; Espinosa-Ruiz et al., 2017; Krishna et al., 2017). The BR synthesis and signalling pathways have been extensively reviewed elsewhere (Kim and Wang, 2010; Wang et al., 2014; Planas-Riverola et al., 2019). In brief, BRs are perceived by the BRASSINOSTEROID INSENSITIVE 1 (BRI1) receptor kinase, which interacts with and reciprocally transphosphorylates BRI1 Associated receptor Kinase 1 (BAK1) (Wang et al., 2008). This interaction promotes phosphorylation of the cytosolic Brassinosteroid Signaling Kinases (BSKs) and CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) (Tang et al., 2008). These kinases activate BRI1 SUPPRESOR 1 (BSU1) phosphatase (Kim et al., 2009), which in turn inactivates BRASSINOSTEROID INSENSITIVE 2 (BIN2). Inactivation of BIN2 and subsequent dephosphorylation of BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1) transcription factors by Protein Phosphatase 2A (PP2A) family members regulate the expression of numerous target genes in the plant genome (Sun et al., 2010). In the absence of BRs, BIN2 is autophosphorylated and phosphorylates BZR1 and BES1 to promote their interaction with 14-3-3 proteins, and to sequestration in the cytosol (Gampala et al., 2007; Ryu et al., 2007). This leads to the degradation of BES1 and BZR1 by the 26S proteasome (Zhu et al., 2017) and thus prevents transcriptional regulation of BR responsive genes (He et al., 2002).
Relationships between BR signalling and cellulose production are evident in elongating cotton fibres, where cellulose production is intimately linked to fibre elongation (Haigler et al., 2012). Exogenous Epi-Brassinolide (BL, an active form of BR) application caused fibre elongation in Gossypium herbaceum (cotton), and exogenous Brassinazole (BRZ, a BR biosynthesis inhibitor) inhibited cotton fibre elongation (Sun et al., 2005), indicating that BRs control fibre elongation. PAGODA1 (PAG1), a cytochrome p450 homologue in cotton, hydroxylates and deactivates endogenous BR metabolites (Zuoren et al., 2014), and pag1 activation-tag mutants displayed reduced fibre elongation with concomitant defects in cell wall biosynthesis that could be rescued by exogeneous BL application. Additionally, cotton fibre secondary cell walls were thicker upon exogenous BL treatment, and overexpression of the BRI1 receptor in cotton led to transcriptional upregulation of GhCESA1, while exogenous application of BRZ and an antisense insertion of BRI1 had the opposite effect on cell wall thickness (Sun et al., 2015). These results indicate a role for BRs in the regulation of secondary cell wall deposition (Sun et al., 2015).
BR signalling directly influences the transcriptional regulation of CESA genes involved in primary and secondary cell wall cellulose deposition in Arabidopsis (Xie et al., 2011). For example, biomass accumulation and cellulose content decreased in the BR-signalling mutant bri1-301 and in the BR synthesis mutant det2-1 (a point mutation in a steroid 5-alpha reductase, rendering plants incapable of BR biosynthesis) (Xie et al., 2011). Conversely, cellulose content increased in plants overexpressing BRI1 or BES1. Upon BR treatment, genes implicated in primary wall (CESA1, 2, 5, 6 and 9) and secondary wall (CESA4, 7 and 8) cellulose deposition were transcriptionally upregulated (Xie et al., 2011). Chromatin immunoprecipitation using a BES1 antibody followed by quantitative PCR (qPCR) showed that BES1 could bind to promoter regions of these CESAs (Xie et al., 2011). These observations suggest that the BR transcriptional regulation programme participates in directly controlling CESA gene expression in response to BR signalling.
The CSC is also post-translationally regulated in a BR-dependent manner by BIN2 in Arabidopsis (Sánchez-Rodríguez et al., 2017) (Fig. 1). Crystalline cellulose content was significantly reduced in the hypermorphic bin2-1 mutant, and this effect was alleviated by the BIN2-specific kinase inhibitor bikinin (BK) (Sánchez-Rodríguez et al., 2017), indicating that BIN2 activity negatively regulates CSC activity. CSC motility was also reduced in both the det2-1 and the bin2-1 mutant backgrounds, and these motility defects could be rapidly rescued by addition of BL and BK, respectively. Subsequent in vitro protein kinase assays with synthetic peptides demonstrated that recombinantly expressed BIN2 phosphorylates CESA1 at position T157 in a priming phosphorylation-dependent manner. BIN2-mediated phosphorylation of CESA1 also directly modulated CSC activity in vivo. Phosphonull mutants of CESA1 at position T157 (T157A) exhibited CSC velocities comparable to that of the wild type in the bin2-1 mutant backgrounds, indicating that this site is responsible for altered CSC motility at the plasma membrane when BRs are not present or when BRs are not perceived by the cell (Sánchez-Rodríguez et al., 2017).
Overall, these studies indicate that BRs stimulate cellulose synthesis. When BRs are perceived by the cell, CESA transcripts accumulate, CSC activity is increased and cellulose content increases. However, in the absence of BRs, BIN2 protein kinase is active and phosphorylates at least one component of the CSC, thus negatively modulating CSC activity. It is plausible that BIN2 here reduces cellulose production as cells do not require sustained directed cell growth after they have fully expanded. Hence, this would be an effective way to coordinate BR signalling with wall polysaccharide deposition. It is also possible that other protein kinases in the BR signalling cascade, such as BRI1, BAK1 and/or CDG1, directly regulate CSC components, but this hypothesis remains to be tested. For example, the CESA1T157 phosphorylation event that is mediated by BIN2 requires a priming phosphorylated residue at CESA1S162 (Sánchez-Rodríguez et al., 2017). These BR-regulated kinases may represent reasonable enzymes to mediate such a phosphorylation event.
BR signalling also impacts the dynamics and orientation of CMTs that guide CSCs. In rapidly expanding epidermal cells of etiolated hypocotyls, CMTs are typically transversely oriented, leading to transverse cellulose microfibril orientation (Paredez et al., 2006). However, mutants that are compromised in BR synthesis and signalling typically display longitudinally oriented CMT arrays (Wang et al., 2012). Moreover, MT orientation could be changed from longitudinal to transverse in the det2-1 mutant when seedlings were treated with BL; however, such MT reorientation could not be accomplished in the bri1-116 mutant that is insensitive to BL (Wang et al., 2012). Further treatments of these mutants with the MT-destabilizing drug oryzalin indicated that BR synthesis or signalling mutants exhibit more stable and hence less dynamic CMTs (Wang et al., 2012; Liu et al., 2018). This increase in stability of CMTs prevents their reorientation from longitudinal to transverse in expanding root and hypocotyl cells.
The molecular mechanisms behind the BR-controlled MT dynamics are not clear. However, recent studies have identified some components associated with this process. For example, MICROTUBULE DESTABLILIZING PROTEIN 40 (MDP40) is a direct transcriptional target of BZR1, and MDP40 is induced upon BL treatment (Wang et al., 2012). MDP40 co-localizes with CMTs, and importantly, MDP40 RNAi lines displayed enhanced stability and reduced reorientation rates of the MT array, similar to what was observed in BR signalling and synthesis mutants (Wang et al., 2012). Hence, MDP40 plays a critical role in this process. BIN2 may also directly interact with MTs and increase MT stability. This suggestion is based on the observation that CMTs were less sensitive to oryzalin in the bin2-1 mutant (Liu et al., 2018). Overall, these observations indicate that BR synthesis and signalling promote a dynamic and transversely oriented CMT array to support transverse deposition of cellulose microfibrils as well as anisotropic growth.
ABA signalling and cellulose biosynthesis
Abscisic acid (ABA) is an important phytohormone that regulates assorted processes in the plant life cycle, including seed development, stomatal conductance, seedling growth, and responses to abiotic and biotic stresses (Cutler et al., 2010). ABA synthesis and signalling pathways have been extensively reviewed elsewhere (Cutler et al., 2010; Munemasa et al., 2015; Yang et al., 2017; Yan and Chen, 2017). In brief, the first steps of ABA synthesis occur in plastids with subsequent steps in the cytoplasm, with C40 carotenoid serving as a synthetic precursor (Seo and Koshiba, 2002). Zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NCED) and ABA-aldehyde oxidase (AAO) are key ABA biosynthetic enzymes, with NCED functioning as a rate-limiting enzyme for ABA biosynthesis (Seo and Koshiba, 2002; Hauser et al., 2017). Exposure of plants to various types of abiotic stresses, such as salt or drought stress, causes rapid accumulation of endogenous ABA levels and increased expression of ABA biosynthesis-related genes (Xiong et al., 2002). ABA is perceived by members of the PYRABACTIN RESISTANT (PYR)/PYR-LIKE (PYL)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) protein family (Park et al., 2009), which directly bind to ABA and undergo a conformational change to inhibit the activity of Protein Phosphatase 2C (PP2C) isoforms (Melcher et al., 2009; Santiago et al., 2009). This triggers autophosphorylation and activation of SNF1-RELATED PROTEIN KINASE 2 (SnRK2) isoforms, resulting in phosphorylation of numerous downstream targets, including the ABA-RESPONSIVE ELEMENT BINDING FACTOR (ABF) transcription factors that mediate ABA-regulated transcriptional reprogramming (Fujii et al., 2009). Due to the central role of ABA in coordinating growth responses to abiotic stress and the fact that the CSC responds to abiotic stress conditions (Gutierrez et al., 2009; Endler et al., 2015), it is anticipated that ABA may play a role in regulating cellulose biosynthesis, particularly under stress conditions. However, clear mechanistic links between cellulose biosynthesis and ABA signalling are lacking.
Defects in the secondary wall CESAs affect ABA biosynthesis and signalling in Arabidopsis. The lew2-1 mutant of CESA8/IRX1 was more drought and salt stress tolerant than control plants and exhibited an increased ABA content, possibly due to the enhanced expression of the ABA synthesis-related gene SDR1/ABA2. Similarly, the ABA-responsive gene RD29A was up-regulated in lew2-1. This result indicated that ABA biosynthesis and signalling are constitutive in lew2-1 mutants, perhaps because the defects in water transport or vascular function in the mutant resulted in constant plant stress that is signalled via ABA (Chen Chen et al., 2005). In line with this hypothesis, mutations in the three secondary wall CESAs, CESA4, 7 or 8, enhanced resistance against the soil-borne bacterium Ralstonia solanacearum and the necrotrophic fungus Plectosphaerella cucumerina in an ABA-dependent manner (Hernández-Blanco et al., 2007). Furthermore, perturbation in ABA synthesis and signalling in cesa7 or cesa8 mutants resulted in enhanced developmental defects (Hernández-Blanco et al., 2007), corroborating that ABA signalling is important for plant development when secondary wall cellulose content is impaired. Nevertheless, it is unclear how the secondary wall cellulose defects are translated into ABA changes and if other secondary wall defects would lead to similar ABA changes.
A potential connection between ABA and cellulose synthesis was suggested in cotton. The basis for this association was that several ABA-responsive motifs were present in the promoter of Gossypium hirsutum CESA4 (GhCESA4). However, ABA treatment resulted only in modest expression changes of the CESA4 gene (Kim et al., 2011). In addition, endogenous ABA levels did not correlate with cellulose accumulation during the development of cotton fibres. Instead, the ratio of ABA/IAA levels and secondary cell wall thickening in the fibres correlated well (You-Ming et al., 2001), perhaps indicative of how cell wall production may be controlled via hormone cross-talks.
Abiotic stress impacts cellulose synthesis (Gutierrez et al., 2009; Endler et al., 2015; Wang et al., 2016). Abiotic stresses, such as salt and drought stress, induce rapid increases in ABA content (Xiong et al., 2002), suggesting that ABA may play a role in regulating cellulose biosynthesis under such stress conditions. The plant-specific CC1 and CC2 proteins sustain cellulose synthesis and plant growth under salt stress by promoting the re-establishment of CMT arrays and the re-localization of CESAs to the plasma membrane (Endler et al., 2015). However, whether ABA signalling is necessary for CC1/2-mediated cellulose synthesis and salt tolerance remains unclear, and might represent an interesting avenue to explore cellulose synthesis mechanisms under abiotic stress. This line of investigation may also help to infer whether and how ABA signalling could influence cellulose synthesis.
Another potential link between salt stress, ABA and cellulose synthesis is KOB1, which is a putative glycosyltransferase-like protein (Pagant et al., 2002). The kob1 mutant displayed severe growth defects, reduced cellulose synthesis and disordered cellulose microfibril orientations (Pagant et al., 2002). Interestingly, the KOB1 mutant allele abi8 was isolated in a screen for ABA-resistant seed germination mutants. The abi8 mutant displayed abnormal stomatal activities and ABA-responsive gene expression, indicating that KOB1 might be associated with ABA-related processes (Brocard-Gifford et al., 2004). Moreover, KOB1 gene expression was suppressed in the root epidermis and cortex during salt stress (Dinneny et al., 2008), indicating a link between salt stress, ABA and cellulose synthesis. However, these observations are only correlative and it is unclear how KOB1 impacts stomata function, ABA synthesis and cellulose production (Pagant et al., 2002; Lertpiriyapong and Sung, 2003). Thus, it will be interesting to study how KOB1 functions in response to salt stress, and it would also be useful to cross kob1 with CESA and CMT marker lines to determine whether CSC activity and MT behaviour is altered in the kob1 mutant under abiotic stress or exogenous ABA treatment.
While salt stress impacts CMT organization and dynamics (Endler et al., 2015), there are some indications that ABA can also directly affect MT behaviour. Studies in leek leaf epidermal cells revealed that 20 µm ABA treatment for 14–18 h altered CMT array orientation and promoted longitudinal organization in leaf epidermal cells (Seung et al., 2013). In Arabidopsis hypocotyl cells, 1 µm ABA treatment for 2 weeks resulted in CMT depolymerization (Takatani et al., 2015), which also led to ectopic epidermal cell outgrowths (Takatani et al., 2015). However, it is perhaps unlikely that these changes are due to a direct effect of ABA as treatment times are extensive.
Stomata close in response to abiotic stress to avoid water loss, and ABA serves as an essential phytohormone to regulate this process (Tallman, 2004). Qu et al. (2018) found that ABA treatment led to MT depolymerization in guard cells and decreased stomatal aperture, while the additional application of paclitaxel, an MT-stabilizing agent, partially suppressed the ABA-induced MT depolymerization and stomatal closure. This indicates that depolymerization of a guard cell MT array might be required for ABA-induced stomatal closure (Qu et al., 2018), and is in line with the results of Takatani et al. (2015), to indicate that ABA may be able to depolymerize CMTs. ABA-triggered CMT depolymerization could in this way change cell wall organization to alter growth. For example, such changes may alter cell wall polysaccharide deposition or properties to facilitate rapid stomatal opening/closing in response to abiotic stresses. A click-based approach (Anderson et al., 2012) to monitor certain cell wall polymer alterations could perhaps be used to monitor such ABA-induced structural changes in real-time. Notably, similar effects on CMT behaviour have been observed under salt and osmotic stress, i.e. CMT depolymerization and CMT re-orientation (Wang et al., 2007; Endler et al., 2015). It may thus be interesting to determine whether salt- and/or osmotic-stress-induced CMT depolymerization is dependent on ABA signalling. This hypothesis could be tested in a nced3 mutant background to abolish ABA induction or ABA-insensitive mutants, such as abi1-1, to interrupt ABA signalling.
Gibberellin signalling and cellulose biosynthesis
Gibberellins (GAs) are essential phytohormones that play important regulatory roles in plant growth and development (Yamaguchi, 2008). GAs promote plant growth by stimulating directed cell expansion, which should influence cellulose deposition and CMT organization (Bringmann et al., 2012). GA synthesis and signalling pathways have been extensively reviewed elsewhere (Davière and Achard, 2013; Binenbaum et al., 2018). GA biosynthesis is well understood, with most of the enzymes identified, i.e. GA 20-oxidase (GA20ox) and GA 2-oxidase (GA2ox) (Hedden and Thomas, 2012).
GA signal transduction is largely mediated via the DELLA proteins, a group of proteins containing the conserved DELLA amino acid sequence in their N-terminal domain. The Arabidopsis genome encodes five DELLA proteins [GAI, RGA, RGA-LIKE 1 (RGL1), RGL2 and RGL3], whereas only a single DELLA protein, SLENDER RICE 1 (SLR1), has been identified in rice (Locascio et al., 2013a). DELLA proteins serve as repressors of GA signal transduction and inhibit GA-responsive gene transcription in the absence of GA, leading to the suppression of GA-mediated plant growth and development. Once GA is produced, it is perceived by the GA receptor GID1 and forms a GA–GID1–DELLA ternary protein complex. The DELLA proteins are then polyubiquitinated by the E3 ubiquitin ligase SLEEPY1 (SLY1) and subsequently degraded in a 26S proteasome-dependent manner, which relieves the suppression of GA responsive genes (Wang and Deng, 2011). GA signalling through DELLA proteins transmits a range of environmental and developmental signals by interacting with multiple transcription factors to control gene expression (Davière and Achard, 2013).
The effect of GA on cellulose synthesis has been studied in several plant species, including cotton and rice. For instance, enhanced GA production led to increased secondary wall thickness and cellulose content in cotton fibres. This increase was probably mediated through elevated sucrose synthase activity, which presumably produced more UDP-glucose to fuel cellulose synthesis (Bai et al., 2014). While that study provided indirect evidence for how GA could impact cellulose synthesis, other studies point in similar directions. A recent study in Sorghum bicolor showed that the Sorghum sp. dwf1-1 mutant, containing a genetic lesion in GA20-oxidase, exhibited a dwarfed phenotype and reduced cellulose content, indicating that suppression of GA biosynthesis-related genes impacts cellulose synthesis (Petti et al., 2015). Additionally, exogenous treatment with the GA biosynthesis inhibitors daminozide or chlorochlorine chloride resulted in decreased cellulose synthesis in Sorghum. They also demonstrated that gene expression levels of the primary wall CESAs, SbCESA1/3/6, increased after GA application (Petti et al., 2015). However, the mechanistic basis of GA-regulated transcription of the CESA genes in Sorghum is unclear.
In rice, the GA-deficient mutant sdl1, which contains a mutation in GA20-oxidase, exhibited thinner sclerenchyma cell walls, reduced secondary wall CESA expression levels and reduced cellulose content. Notably, genetic disruption of SLR1 (the DELLA protein in rice) or exogenous GA application restored the secondary wall CESA expression levels and cellulose content, indicating that GA also promotes cellulose production in rice (Huang et al., 2015). Interestingly, SLR1 directly interacted with NAC29/NAC31, two members of the NAC transcription factor (TF) family, which are transcriptional regulators required for secondary wall formation. This interaction occurred in the absence of GA and, given the nature of DELLAs, thus effectively prevented the activation of downstream TFs, such as MYB61, and consequently repressed CESA expression and inhibited cellulose biosynthesis (Huang et al., 2015). In the presence of GA, SLR1 was degraded, which activated secondary wall-regulating TFs and secondary wall cellulose synthesis (Huang et al., 2015). Therefore, it was inferred that the GA-mediated DELLA-NAC signalling pathway is essential for regulation of the secondary wall biosynthesis (Huang et al., 2015).
Another link between GA and cellulose synthesis is mediated by Xyloglucan Endotransglucosylase/Hydrolase 21 (XTH21). XTH21 expression was induced by gibberellic acid (GA3) treatment, but not by other hormones, in Arabidopsis (Liu et al., 2007). Mutations in XTH21 led to several alterations of cell wall properties, including reduced mass-average molecular weight of xyloglucan, depressed expression levels of CESA2/4, decreased cellulose content and reduced cell expansion (Liu et al., 2007). These observations suggested that XTH21 might play a role in the cell elongation and plant growth processes by affecting cellulose synthesis and cell wall modelling. However, if XTH21 has a direct effect on cellulose synthesis, or whether these phenotypes are a side-effect from its effect on xyloglucans, and whether GA could function as a coordinator in this process remain unclear. Similarly, the impact of GA on XTH21 expression might be mediated by DELLA-related TFs but could also be mediated by other indirect effects, which provides research directions to further study the regulatory mechanism of GA-related cellulose synthesis and cell wall formation.
GA also affects CMT organization and dynamics. Indeed, early studies indicated that the CMT array orientation is influenced by GA content (Baluška et al., 1993). GA deficiency caused by naturally occurring mutations in the d5 mutant, or application of the GA biosynthesis inhibitor paclobutrazol, caused disordered MT arrays in the root cortex of maize. These changes could be restored by exogenous GA treatment, indicating that GA plays a role in CMT organization (Baluška et al., 1993).
More recent studies showed that DELLA proteins may connect GA and CMT organization. For example, prefoldin proteins can interact with DELLA proteins and this interaction can directly influence CMT behaviour (Locascio et al., 2013b) (Fig. 1). Here, the prefoldin complex, which typically functions in the cytoplasm, played an important role in tubulin dimer assembly. In the absence of GA, prefoldin was largely localized to the nucleus and directly interacted with DELLA proteins, which in turn impaired the tubulin folding chaperone function of prefoldin and caused CMT array disorganization. When GA was present, DELLA proteins were degraded and the prefoldin complex localized to the cytoplasm, resulting in increased tubulin dimer production and organized CMT alignment (Locascio et al., 2013b).
MT severing, which is facilitated by MT-severing proteins, is critical for CMT array organization and dynamics and is important for various physiological processes, such as mitosis, meiosis and morphogenesis in plants (Sharp and Ross, 2012; Lindeboom et al., 2013). Mutations in KTN1 (ktn1, fra2, lue1), a katanin-like MT-severing protein, led to decreased cellulose and hemicellulose content, as well as reduced fibre cell length and cell wall thickness (Burk et al., 2001). Additionally, ktn1 mutants exhibited abnormal CMT array structure and altered cellulose microfibril orientation (Burk and Ye, 2002). Notably, ktn1 mutants also displayed enhanced expression levels of GA synthesis-related genes, including GA4 and GA5, which are two key genes in GA synthesis (Meier et al., 2001). These findings indicate a cross-talk between CMT organization, cellulose synthesis and GA signalling and/or synthesis. Another link between KTN1 and GA was revealed in the ga1-1 mutant, a GA-deficient mutant that had reduced levels of KTN1 transcripts. This decrease could be restored by exogenous GA3 application, suggesting that KTN1 expression is modulated by GA level in plants (Bouquin et al., 2003).
Together, these results indicate that GA signalling and CMT behaviour are coordinated to regulate cellulose deposition and cell elongation, with proteins such as DELLA and KTN1 potentially serving as coordinators of these processes. It will be interesting to use relevant mutants or inhibitors for GA synthesis and/or signalling to monitor cellulose production, the properties of cell wall components and MT dynamics. Such studies would at least begin to unravel the relationship between GA signalling and cellulose production.
Auxin signalling and cellulose biosynthesis
Auxins impact basic cellular growth processes, such as cell division and cell expansion, but also control a variety of tissue- and whole-plant-level responses, such as responses to gravitropism, apical dominance, light-related tropisms and lateral organ development. These aspects of auxin synthesis and signalling have been extensively reviewed elsewhere (Lavy and Estelle, 2016; Matthes et al., 2019; Gallei et al., 2020).
In light of the diverse processes that are controlled by auxin, it is not surprising that the perception and cellular distributions of auxin are quite complex. At least two key signalling modules are suggested to control a number of classical auxin responses. The first of these modules consists of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) proteins, the AUXIN RESPONSE FACTOR (ARF) transcriptional activators and the Aux/IAA transcriptional repressors (Lavy and Estelle, 2016). In this signalling module, Aux/IAA proteins heterodimerize with ARF transcription factors to repress auxin-regulated genes under conditions in which auxin is absent. Auxin forms a ternary complex between the TIR1/AFB proteins, and the degron domain of Aux/IAA proteins, thereby serving as ‘molecular glue’ to stabilize this interaction (Dharmasiri et al., 2005; Tan et al., 2007). This ternary complex results in ubiquitination of the Aux/IAA proteins, their subsequent degradation by the 26S proteasome and ARF transcriptional activation. This core pathway mediates complex transcriptional reprogramming to facilitate auxin responses (Lavy and Estelle, 2016), but it is important to note that a large number of ARF, Aux/IAA and TIR1/AFB protein combinations exist in plant genomes, so the resulting transcriptional response is probably tuned in various cell types (Calderón Villalobos et al., 2012).
Auxin treatment leads rapidly to apoplast acidification (Ren and Gray, 2015), resulting in cell wall loosening through the activation of expansins (Lehman et al., 2017) and providing a direct relationship between auxin signalling, the cell wall and growth. The molecular basis for the well-known ‘acid growth hypothesis’ and its relationship to the core auxin signalling pathway has until recently remained relatively ill defined. SMALL AUXIN UP-REGULATED RNA (SAUR) genes are among the earliest transcriptionally induced genes that are responsive to auxin (Hagen and Guilfoyle, 2002). The resulting SAUR proteins localize to the plasma membrane, where they specifically inhibit Protein Phosphatase 2C D-subfamily isoforms from dephosphorylating T947 within the autoinhibitory domain of plasma membrane-localized H+-ATPases (Spartz et al., 2014). Due to this activity of SAUR proteins, the H+-ATPase is activated and pumps protons into the extracellular matrix, thus stimulating acidification of the apoplast and cell wall loosening.
A final important point regarding auxin signalling is that auxin concentrations are not uniform throughout the plant but are tightly spatially controlled by a sophisticated network of auxin transport proteins. Members of the PIN-FORMED (PIN) transporter family largely control this process. The PIN proteins are polarly localized to the apical or basal domains of plant cells and serve as auxin efflux carriers that transport auxin to concentrate this hormone in a rootward direction in root cells or a shootward direction in shoots. Members of the AUXIN1/LIKE AUXIN1 (AUX/LAX) and ATP Binding Cassette subfamily B (ABCB) transporters also contribute to polar auxin transport and the establishment of localized auxin gradients in various tissues (Geisler et al., 2017).
Although there is a clear relationship between auxin action and cell wall metabolism (Arsuffi and Braybrook, 2018), few direct links between auxin signalling and cellulose biosynthesis have been established. One important link concerns the relationship between auxin transport and cellulose deposition. Although there is clear evidence that auxin transporter polar localization requires constant recycling of vesicles to the target membrane (Grunewald and Friml, 2010), computational simulations have suggested that limiting lateral membrane diffusion of PIN transporters is critical for maintenance of PIN polar localization (Heisler et al., 2010). In a screen for mutants that resulted in mis-localized PIN transporters, mutations in Arabidopsis CESA3 were identified (Feraru et al., 2011). Similar PIN mis-localization was also observed in other primary wall cesa mutants and after treatment with the cellulose synthesis inhibitors isoxaben and DCB (Feraru et al., 2011). Indeed, the temperature-sensitive CESA1 mutant rsw1-1 also displayed altered PIN localization in arabidopsis roots, which leads to substantial changes in CESA and auxin reporter gene expression (Lehman and Sanguinet, 2019). In contrast, the pattern of PIN1 localization remained unaltered in the shoot meristems of the cesa3 point mutant (cev1), although the overall intensity of PIN1 signal was significantly reduced (Sampathkumar et al., 2013). These results suggest that cellulose deposition is required to limit the lateral diffusion of polarly localized PIN auxin transporters, and therefore that cellulose deposition impacts auxin distributions in the plants.
Based on the critical functions of auxin in regulating cell division, cell expansion, and directed growth in response to gravistimulation or light-mediated responses, this phytohormone would be expected to impact the orientation and dynamics of CMT arrays that guide CSCs at the plasma membrane. However, such impact has been substantially debated, in terms of both how auxin affects the CMT array dynamics as well as the associated molecular mechanisms. For example, auxin can cause transverse to longitudinal reorientation of CMT arrays in both Arabidopsis root and hypocotyl epidermal cells (Chen et al., 2014). Further work in their study indicated that CMT reorientation was dependent upon AUXIN BINDING PROTEIN1 (ABP1) and its putative downstream signalling components ROP6-GTPase, ROP-INTERACTIVE1 (RIC1) and KTN1 (Chen et al., 2014). However, subsequent studies have called into question the validity of ABP1 as an auxin receptor (Gao et al., 2015), so the conclusions should be interpreted with caution. In contrast to the dark-grown hypocotyl CMT responses described above, auxin reproducibly induces transverse CMT arrays in light-grown hypocotyls. Several recent studies have indicated that the formation of such transverse CMT arrays in light-grown hypocotyls is dependent upon the TIR1/ABF auxin co-receptors (True and Shaw, 2020) and that the establishment of transverse arrays could be blocked by dominant mutations in AUXIN RESPONSIVE2 (AXR2), a member of the IAA transcriptional repressor family. These observations indicate that at least in light-grown hypocotyls, canonical auxin transcriptional responses are necessary for longitudinal to transverse CMT array re-orientation, although it should be noted that growth stimulation through the application of the proton-ATPase stimulating toxin fuciscoccin could produce similar results (Adamowski et al., 2019).
Based on the critical functions of auxin in regulating cell division, cell expansion, and directed growth in response to gravistimulation or light-mediated responses, this phytohormone would be expected to impact the orientation and dynamics of CMT arrays that guide CSCs at the plasma membrane. However, such impact has been substantially debated, in terms of both how auxin affects the CMT array dynamics as well as the associated molecular mechanisms. For example, auxin can cause transverse to longitudinal reorientation of CMT arrays in both Arabidopsis root and hypocotyl epidermal cells (Chen et al., 2014). Further work in their study indicated that CMT reorientation was dependent upon AUXIN BINDING PROTEIN1 (ABP1) and its putative downstream signalling components ROP6-GTPase, ROP-INTERACTIVE1 (RIC1) and KTN1 (Chen et al., 2014). However, subsequent studies have called into question the validity of ABP1 as an auxin receptor (Gao et al., 2015), so the conclusions should be interpreted with caution. In contrast to the dark-grown hypocotyl CMT responses described above, auxin reproducibly induces transverse CMT arrays in light-grown hypocotyls. Several recent studies have indicated that the formation of such transverse CMT arrays in light-grown hypocotyls is dependent upon the TIR1/ABF auxin co-receptors (True and Shaw, 2020) and that the establishment of transverse arrays could be blocked by dominant mutations in AUXIN RESPONSIVE2 (AXR2), a member of the IAA transcriptional repressor family. These observations indicate that at least in light-grown hypocotyls, canonical auxin transcriptional responses are necessary for longitudinal to transverse CMT array re-orientation, although it should be noted that growth stimulation through the application of the proton-ATPase stimulating toxin fuciscoccin could produce similar results (Adamowski et al., 2019).
Overall, there is indirect evidence relating auxin signalling to cellulose biosynthesis, the patterning of cellulose deposition and the role of cellulose in establishing polar auxin distributions. Further work will, however, require careful simultaneous analysis of CSC and CMT dynamics in the presence of auxin or the plethora of chemical and genetic tools that are available to perturb auxin synthesis and signalling. Additionally, it will be important to understand how the inhibition of protein phosphatase signalling mediated by SAUR proteins impacts the post-translational phosphorylation of CSC components upon auxin treatment. Based on these connections and the important role of auxin in controlling plant cell growth, we anticipate the uncovering of many mechanistic links between auxin synthesis, signalling and cellulose biosynthesis.
Cytokinin signalling and cellulose biosynthesis
Cytokinins are a family of prenylated adenosine derivatives that regulate various plant growth and development processes, often in concert with other phytohormones, such as cell division, cellular differentiation, leaf phyllotaxis and vascular development (Kieber and Schaller, 2018). Cytokinins are perceived through a complex signalling relay system that is reminiscent of bacterial two-component response regulator systems (Kieber and Schaller, 2018). Cytokinins bind to endoplasmic reticulum-localized histidine kinase (HK) receptors, and cytokinin binding leads to activation of the HK receptor kinase domain and autophosphorylation on a key conserved histidine residue. The phosphate is subsequently transferred to an aspartate residue in the HK receiver domain, and then to Authentic Histidine Phosphotransferase (AHP) proteins, which serve as an intermediate between HK receptors and Response Regulator (RR) proteins. Two types of RR subgroups exist, type A and B. Type B RRs contain a receiver domain as well as an Myb-like DNA binding domain. These proteins are phosphorylated by AHP proteins, become activated and mediate cytokinin-induced transcriptional responses. Type A RRs do not contain a DNA-binding domain, and probably serve as negative regulators of cytokinin signal transduction. Type A RR transcripts are induced by cytokinin through the action of type B RRs and probably compete with type B RRs for the phosphorylation signal that is transduced by AHPs.
Direct links between cytokinin signalling and cellulose metabolism are sparse, but it is likely that cytokinins partake in responses to cell wall biosynthesis inhibition. Isoxaben is a potent inhibitor of cellulose biosynthesis that has been extensively used to examine the signalling events that are elicited upon cell wall damage. Recent work has demonstrated that isoxaben treatment causes reduced expression of cell cycle markers, such as Cyclin B1;1 and Cyclin D3;1 in the root apical meristem of Arabidopsis seedlings (Gigli-Bisceglia et al., 2018). Due to the well-known role of cytokinin in regulating cell division, the authors of that study used metabolomics to investigate how isoxaben treatment impacted cytokinin biosynthesis and found that the treatment caused a substantial decrease in total cytokinin content as well as trans-zeatin. They additionally demonstrated that isoxaben treatment caused transcriptional up-regulation of the cytokinin-degrading enzyme genes CYTOKININ OXIDASE 2 and 3 (CKX2/3). These observations suggest a model in which cellulose biosynthesis inhibition triggers the degradation of cytokinins, thus preventing cellular proliferation in the root tissues (Gigli-Bisceglia et al., 2018).
Overall, while there may be an involvement of cytokinin signalling in the response to cell wall damage, the direct effects of cytokinins on cellulose biosynthesis are largely unknown and merit further investigation. In this light, it is important to note that CESA proteins are trafficked to the cell plate during the process of cell division, which presents a potentially interesting relationship with cytokinin signalling. Here, CSCs are initially localized to the plasma membrane, but after the initiation of phragmoplast formation, CESA proteins are re-localized to the developing cell plate (Miart et al., 2014). However, the signal that initiates this process remains unknown. It may therefore be useful to investigate whether cytokinin signalling plays a role in this re-localization process, either directly or indirectly.
Ethylene signalling and cellulose biosynthesis
Ethylene (ET) is a gaseous hormone that can function during different growth and developmental phases, such as germination, senescence, abscission and fruit ripening, as well as responses against external stresses (Chen et al., 2005). The biosynthetic pathway of ET has been well studied in plants, consisting of the conversion of methionine to S-AdoMet by S-AdoMet synthetase (SAMS), and then synthesis of ACC from S-AdoMet by ACC synthase (ACS). The resulting ACC molecule is oxidized to ET by ACC oxidase (ACO) (Wang et al., 2002).
After synthesis, ET is perceived by membrane-localized ET receptors, including ETR1, ETR2, ERS1, ERS2 and EIN4 in Arabidopsis. Upon ET binding, these receptors and the downstream Raf-like kinase CTR1 are inactivated, resulting in the activation of an ET response. Members of the ETHYLENE INSENSITIVE3/ETHYLENE-INSENSITIVE3-LIKE (EIN3/EIL) transcription factor family serve as important regulators of ET-responsive gene expression (Alonso and Stepanova, 2004; Y. F. Chen et al., 2005; Kendrick and Chang, 2008). ET treatment classically results in shorter, thicker hypocotyls and roots as well as exaggerated apical hooks in the etiolated hypocotyls, which is termed the ‘triple response’ (Guzmán and Ecker, 1990). However, ET can also promote cell elongation in light-grown plants (Smalle et al., 1997), indicating that ET treatment has complex effects on cell expansion and plant growth. This process requires the rearrangement of plant cell wall components, suggesting a relationship between ET and cellulose synthesis.
Early studies suggested a relationship between ET signalling and cellulose biosynthesis through the observation that ET treatment led to alterations in cellulose microfibril and CMT orientations from transverse to longitudinal in the third internode of pea (Lang et al., 1982). A more direct link between ET synthesis/signalling and cellulose synthesis occurs in cev1, a mutant corresponding to CESA3 in Arabidopsis (Ellis and Turner, 2001; Ellis et al., 2002). cev1 exhibited increased ET levels and constitutive expression of the ET-responsive genes PDF1.2 and CHI-B, indicating that the disruption of cellulose synthesis stimulated ET production and ET signal transduction (Ellis and Turner, 2001; Ellis et al., 2002). The shorter hypocotyl and root lengths observed in the cev1 mutant could be partially rescued by mutating ETR1, suggesting that ET signalling was required for these phenotypic defects. Moreover, growth defects and ectopic lignification caused by isoxaben, an inhibitor of cellulose biosynthesis, were related to ET signalling, as the expression of the ET-responsive gene PDF1.2 was induced by isoxaben treatment (Caño-Delgado et al., 2003). Overall, these observations indicate that ET signalling is at least partially responsible for the growth inhibition caused by the disruption of cellulose biosynthesis (Ellis and Turner, 2001; Ellis et al., 2002; Caño-Delgado et al., 2003).
Mutations in the chitinase-like gene CTL1 exhibited reduced hypocotyl and root lengths as well as an increased number of root hairs (Zhong et al., 2002). Interestingly, the ctl1 mutant also caused enhanced ET production, and exogenous application of the ET biosynthesis inhibitor aminoethoxyvinyl glycine (AVG) or the ET perception inhibitor Ag+ could partially restore the altered growth phenotype (Zhong et al., 2002). CTL1 and its homologue CTL2 are transcriptionally co-expressed with primary and secondary wall CESAs, respectively, indicating that these genes may participate in cellulose synthesis (Persson et al., 2005). Indeed, the ctl1 mutant (also known as pom1) exhibited reduced CSC velocity, disorganized CSC movement and reduced cellulose content. CTL1/2 potentially play a role in cellulose synthesis by binding to the newly formed cellulose microfibrils to promote their proper assembly, which can subsequently facilitate interactions between cellulose and hemicellulose (Sánchez-Rodríguez et al., 2012). Overall, these observations suggest that the phenotypic defects elicited by cellulose biosynthesis inhibition are related to ET, but how the mutation of CTL1 affects ET production and how ET could modulate the CTL1-mediated plant growth remains unclear.
Another important link between ET, cellulose synthesis and cell expansion occurs through two leucine-rich repeat receptor kinases, FEI1 and FEI2. The fei1 fei2 double mutant exhibited defects in anisotropic cell expansion, cellulose synthesis and shorter, swollen roots under high sucrose conditions or isoxaben treatment (Xu et al., 2008). Interestingly, this suite of phenotypic defects was partially rescued by the application of ET synthesis inhibitors, such as aminoisobutytic acid (AIB) or aminooxyacetic acid (AOA), but not by perturbation of the ET signalling pathway, indicating that FEI1/2 may play a role in regulating cellulose synthesis and cell expansion via an ET synthesis-related pathway. Xu et al., (2008) proposed that FEI1/2 might directly interact with ACS protein isoforms, which would then localize in the plasma membrane and generate a signal to regulate cell wall biosynthesis (Xu et al., 2008). Consistently, Tsang et al., (2011) found that responses to cellulose biosynthesis inhibition are dependent on ACC biosynthesis, but do not require ET perception (Tsang et al., 2011).
As mentioned above, ET treatment elicits transverse to longitudinal reorientation of CMT arrays, which can consequently affect the orientation of cellulose microfibril deposition in pea (Lang et al., 1982). Similar ET-triggered CMT reorientation effects were observed in other species, including Arabidopsis (Le et al., 2004). With the advent of better chemical and genetic tools, the molecular basis for ET-mediated CMT reorientation is slowly being elucidated. For example, ACC treatment caused reduced hypocotyl length and CMT reorientation in Arabidopsis. However, genetic disruption of WAVE-DAMPENED2-LIKE5 (WDL5), a CMT-stabilizing protein, inhibited ACC-mediated CMT reorientation and restored hypocotyl elongation, indicating that WDL5 is a mediator of ET-induced CMT reorientation. Additionally, EIN3, a key ET signalling TF, directly regulates WDL5 transcription to affect CMT behaviour and cell elongation in etiolated Arabidopsis hypocotyls (Sun et al., 2015). Their study sheds light on the regulatory mechanism for ET-mediated CMT organization and suggested that CMT orientation represents a link between ET signalling and ET-mediated inhibition of cell elongation in etiolated hypocotyls. It has also been proposed that ET plays an essential role in promoting the re-establishment of organized CMT arrays after salt stress by regulating the expression of WDL5 (Dou et al., 2018). Together, these studies suggest that ET regulates CMT organization through the transcriptional regulation of WDL5.
In summary, there are clear links between ET signalling and cellulose biosynthesis; this is especially well defined in the context of CMT re-orientation and during the response to cellulose biosynthesis inhibition. To further understand these potential ET-mediated regulatory mechanisms, it will be meaningful to use the wealth of ET signalling and synthesis chemical inhibitors or mutants to examine whether and how cellulose production, CSC activity and CMT dynamics are affected to elucidate the regulatory mechanisms of ET-mediated cellulose synthesis and cell elongation. It will be particularly useful to investigate the localization and dynamics of CSCs under these conditions and simultaneously visualize their functional association with CMTs under the same conditions.
JA signalling and cellulose biosynthesis
Jasmonic acid (JA) and its active conjugate jasmonate-isoleucine (JA-Ile) are key regulators of abiotic/biotic stress responses as well as aspects of plant growth and development (Wasternack and Hause, 2013; Khan et al., 2016). JA biosynthesis is mediated by a series of enzymes located in multiple organelles (Wasternack and Hause, 2013). JA and its conjugate JA-Ile accumulate after mechanical wounding, herbivory damage or necrotrophic pathogen infection (Acosta and Farmer, 2010). JA-Ile is perceived by the F-box protein CORONATINE INSENSITIVE 1 (COI1) and serves as a bridging ligand between COI1 and the transcriptional repressor JASMONATE ZIM DOMAIN (JAZ) proteins to form the JA co-receptor complex. Binding of JA-Ile to COI1 and JAZ proteins leads to the ubiquitination and proteasome-mediated degradation of JAZ proteins, resulting in an alleviation of transcriptional repression and transcription of JA responsive genes (Chini et al., 2007; Sheard et al., 2010) in a process that is similar to the TIR1/auxin/IAA protein system.
Many of the triggers of JA synthesis and signalling involve cell wall perturbation or damage. Accordingly, several cellulose-deficient mutants or treatment with cellulose inhibitory drugs, such as isoxaben and DCB, exhibit elevated JA levels and constitutive expression of JA responsive genes (Ellis et al., 2002; Gigli-Bisceglia et al., 2018). JA accumulation typically initiates a transcriptional cascade that activates defence responses at the expense of plant growth (Guo et al., 2018). Interestingly, JA accumulation in cellulose-deficient mutants and/or plants treated with isoxaben is partially responsible for the growth inhibition observed in these plants. For example, reduced root growth in the cev1 mutant was partially rescued in the cev1 coil double mutant, confirming that JA signalling is responsible for some of the reduced growth in cellulose-deficient mutants (Ellis et al., 2002).
JA accumulation in response to cell wall damage is also dependent on cell wall integrity sensing mechanisms, which are mediated by members of the Cathanathus roseus receptor-like kinase 1-like protein (CrRLK1L) family, including THESEUS (THE1) and FERONIA (FER) (Hématy et al., 2007; Westermann et al., 2019) (Fig. 1). FER acts as a negative regulator of JA signalling by phosphorylating and destabilizing MYC2, which is a major JA signalling transcriptional regulator (Guo et al., 2018). THE1 is required for cell wall damage-induced JA accumulation, and the1-1 mutants demonstrate a complete loss of JA accumulation, both in cellulose-deficient mutants and in response to isoxaben. In line with this observation, the gain-of-function THE1 allele the1-4 shows hyperaccumulation of JA, both in cellulose-deficient mutants and in response to isoxaben (Engelsdorf et al., 2018).
How THE1 perceives the cell wall damage, and how signalling components downstream of THE1 are involved in the initiation of JA biosynthesis, however, are unknown. One possibility is that osmotic and mechanical stress at the plasma membrane may activate responses after cell wall damage. Cell walls counteract turgor pressure inside the cells and without the support of intact cell walls, the cells are likely to burst or deform. Isoxaben treatment and cell wall damage result in weaker cell walls, leading to changes in turgor pressure, osmotic stress and mechanical displacement of the plasma membrane. Therefore, mechanical stress at the plasma membrane is a potential candidate stimulus for the activation of THE1. Indeed, the addition of osmoticum to balance the osmotic pressure across the plasma membrane attenuated JA accumulation in response to isoxaben treatment (Gigli-Bisceglia et al., 2018). Additionally, JA induction after cell wall damage is partially dependent on the plasma membrane-localized mechanosensitive Ca2+ channel MID1-COMPLEMENTING ACTIVITY1 (MCA1), which functions downstream of THE1 in cell wall damage perception. It is mechanistically unclear how MCA1 activates JA synthesis. However, intracellular Ca2+/cation levels are strongly linked to activation of JA biosynthesis (Lenglet et al., 2017). Finally, it should be noted that RAPID ALKALINIZATION FACTOR34 (RALF34) was recently demonstrated to bind directly to the THE1 extracellular domain, and this binding event is necessary to mediate THE1 signalling (Gonneau et al., 2018). Although it is unclear how RALF34–THE1 signalling is activated upon the perception of cell wall damage, this receptor–ligand pair represents a key step toward understanding the relationships between THE1 signalling and perception of cell wall damage. Elucidating the relationship between osmoregulation after cell wall damage and JA biosynthesis initiation will be critical for our understanding of the mechanisms that maintain cell wall integrity.
JA accumulation in response to cell wall damage also regulates cell wall composition and remodelling, probably through a feedback mechanism. For example, cellulose deficiency-induced ectopic lignification is dependent on JA signalling (Denness et al., 2011). Furthermore, JA accumulation results in increased expression of cell wall-modifying enzymes, such as pectin methylesterases, upon pathogen infection (Bethke et al., 2015), and exogeneous JA treatment or overexpression of COI1 results in increased expression of pectin-modifying genes, strengthening evidence that JA regulates cell wall composition and remodelling (Bömer et al., 2018). The impact of JA accumulation on cellulose biosynthesis is currently unclear. However, JA treatment leads to depolymerization of CMTs (Z. B. Yang et al., 2017), suggesting that JA probably regulates cellulose biosynthesis.
Based on these observations, it is likely that JA signalling serves as either an inhibitory signal for cellulose biosynthesis and/or arises as a consequence of cellulose biosynthesis inhibition. Further experiments requiring careful imaging of CSC dynamics and quantification of cellulose content after cell wall damage, JA treatment, and in mutants lacking JA biosynthesis and signalling components will be necessary to elucidate the impact of JA on cellulose biosynthesis. It will also be important to resolve the signalling connections between THE1, MCA1 and other unidentified downstream components to understand how JA signalling impacts cell wall integrity sensing in the context of cellulose biosynthesis.
SA and SL signalling and cellulose biosynthesis
In addition to the ‘classic’ phytohormones introduced above, salicylic acid (SA) and strigolactone (SL) are important plant hormones in defence and growth, although their impacts on cellulose biosynthesis are poorly understood.
SA is a small phenolic signalling molecule that is important in plant defence against biotic stresses across many plant species. Moreover, SA also regulates other physiological processes, such as seed germination, growth, flowering and abiotic stress responses (Hayat et al., 2010; Rivas-San Vicente and Plasencia, 2011). Two main SA synthetic pathways have been identified in plants: the isochorismate synthase (ICS) pathway in the chloroplast and the phenylalanine ammonia lyase (PAL) pathway in the cytoplasm (Chen et al., 2009). Much of the current understanding regarding SA signal transduction has focused on its role in biotic stress. A classic SA-mediated disease resistance pathway is exemplified in the activation of NONEXPRESSOR OF PR-1 (NPR1), which subsequently interacts with downstream TFs, such as TGAs and WRKYs, by SA. This, in turn, regulates the expression of defence-related genes, including PATHOGENESIS RELATED 1 (PR1). An NPR1-independent SA signal transduction route has also been identified; however, the regulatory mechanism remains unclear (Lu, 2009; Kachroo and Robin, 2013). Moreover, SA signalling is antagonistic to ET/JA signalling during pathogen exposure (Kunkel and Brooks, 2002). These aspects of SA signalling, the underlying molecular mechanisms and the role of SA signalling in different physiological responses have been reviewed extensively elsewhere (Hayat et al., 2010; Rivas-San Vicente and Plasencia, 2011).
There is only sparse information about how SA and cellulose synthesis intersect. A microarray study of Arabidopsis suspension culture cells indicated that expression of SA-responsive genes did not change upon treatment with the cellulose biosynthesis inhibitors isoxaben and thaxtomin A (Duval and Beaudoin, 2009). As mentioned above, mutants of CESA4/7/8 in Arabidopsis exhibited enhanced resistance to several pathogens in an ABA-dependent manner. The same study concluded that the enhanced resistance was independent of SA signalling (Hernández-Blanco et al., 2007). Nevertheless, there are clear relationships between SA and other cell wall components, including callose and lignin. Callose is often synthesized to reinforce the plant cell wall to reduce cell damage under stress conditions. Unexpectedly, the powdery mildew resistant 4 (pmr4) mutant with defects in a callose synthase exhibited enhanced resistance to pathogens, while the powdery mildew susceptibility of the pmr4 mutant was increased by introducing mutations in the SA-signalling pathway (Nishimura et al., 2003). In Brachypodium distachyon seedlings, 2 weeks of 100 µm SA treatment led to reduced growth rates and lignin content in leaves, while cellulose content and CESA transcript abundance were not significantly altered (Napoleão et al., 2017). Hence, SA may change cell wall components, but very little data link the hormone to cellulose production.
SL comprises a relatively new class of plant hormones, initially found in root exudates, that functions in regulating plant growth and development, specifically in rhizosphere communication and in shoot branching (Xie et al., 2010; Brewer et al., 2013). SL is synthesized via a complex pathway that uses carolactone as a precursor scaffold that is structurally elaborated on by a variety of enzymes, such as MORE AXILLARY GROWTH (MAX) enzymes, at least in Arabidopsis, to produce a suite of biologically active SL molecules. SL is synthesized in both roots and stems, and is transported through the xylem. SL signalling, biosynthesis and function have been extensively reviewed elsewhere (Brewer et al., 2013; Seto and Yamaguchi, 2014; Lopez-Obando et al., 2015; Waters et al., 2017).
SL biosynthesis and signalling mutants exhibit dwarf and shoot branching phenotypes, as shown in the max mutants of Arabidopsis, the htd1 mutant of rice and the rms mutant in pea (Beveridge et al., 1996; Zou et al., 2006; Brewer et al., 2013). However, whether synthesis of cellulose or other cell wall-related components are affected in those mutants has not been thoroughly examined. A genetic study by Ramírez and Pauly (2019) indicates that the dwarfism, collapsed xylem and low cellulose content of the Arabidopsis irx1 and irx3 mutants, which contain mutations in secondary CESA genes CESA8 and CESA7, respectively, could be partially rescued by introducing the max4 mutation, a key SL biosynthetic gene. However, growth and developmental defects in procuste1 (prc1) caused by mutations in CESA6, a primary cell wall CESA gene, were not relevant to MAX4 (Ramírez and Pauly, 2019). These observations suggest that secondary wall defects, caused by cellulose biosynthesis defects, could lead to irregular plant growth possibly via the activation of the SL biosynthetic pathway. UDP-glucose, which can be produced by sucrose synthase (SuSy) or via cytosolic invertase plus UDP-glucose pyrophosphorylase (UGP), serves as a substrate for CESAs during cellulose synthesis (Verbančič et al., 2018). Upon GR24 treatment, a synthetic analogue of SL, UGP was de-phosphorylated in rice, indicating that UGP might be a potential target for SL signalling that in turn may regulate cellulose synthesis (Chen et al., 2014).
Future work towards understanding SA/SL–cellulose synthesis relationships could begin by examining plant growth, cellulose content analyses, cellulose synthesis-related gene expression analysis and CSC behaviour in SA/SL-treated plants or in SA/SL-related mutants. These experiments might provide more direct information about the relationship between cellulose synthesis and SA/SL signalling. It may also be interesting to explore whether SA/SL could function together with other hormones in regulating cellulose biosynthetic processes.
A framework to better understand the role of hormones in cellulose synthesis
The immediate importance of hormone action and cellulose synthesis on plant growth and morphogenesis indicate that there should be direct links between these processes. While some mechanistic details behind such connections are emerging, there are still surprisingly little data directly linking phytohormone signalling and cellulose biosynthesis. Transcriptional regulation of cellulose-related genes by TFs that are connected to hormone signalling has been explored in some detail, and transcriptional regulation of cellulose biosynthesis is particularly well established for biosynthesis of secondary wall cellulose. Here, combinatorial hormone cocktails induce master-regulators, i.e. TFs that can either directly or indirectly activate a battery of biosynthetic genes, including secondary wall CESA genes, that produce different types of secondary walls (Taylor-Teeples et al., 2015). Such knowledge is grossly under-represented for the primary wall-producing cell wall genes, including that of the CESAs. For example, while BES1 directly binds and activates several of the CESA genes in Arabidopsis (Xie et al., 2011), over-expression of BES1 does not result in ectopic primary wall production, contrasting with what is observed for the secondary wall-inducing master regulators, such as the NAC TFs VND6 and 7 (Yamaguchi et al., 2010). Nevertheless, members of the APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family TF subfamily could replace secondary walls with primary wall-like structures and did activate primary wall CESA genes in Arabidopsis (Sakamoto et al., 2018). While it is unclear if these AP2/ERF TFs are induced by ET, they represent a key step in understanding how primary cell wall cellulose deposition is regulated.
A major path to identify cellulose synthesis-associated components has been to utilize transcriptional co-expression analysis, i.e. the notion that two genes that are involved in the same process may be transcriptionally coordinated (Persson et al., 2005; Usadel et al., 2009). Intuitively, this approach should be applicable to identify TFs that control primary CESA expression as it is likely that once a CESA-activating TF is expressed, the CESA genes should be coordinately expressed. Indeed, several key TFs controlling secondary wall synthesis, including those acting on the secondary wall CESA genes, are transcriptionally co-expressed with the secondary wall CESA genes (Ruprecht and Persson, 2012). Therefore, it may be expected that mutations in at least some of the TFs that are co-expressed with the primary wall CESAs would show phenotypes related to defects in primary wall cellulose synthesis. Notably, we ordered T-DNA lines for many of the TFs (>50) that showed close co-expression relationships with the primary wall CESAs but failed to detect any abnormal growth phenotypes as compared to wild-type plants. This is perhaps a reasonable outcome, as mutations in the VND-related TFs did not show any major defects in growth despite being master-regulators for secondary wall synthesis (Kubo et al., 2005). Instead, the VNDs were fused to dominant activator (VP16) or repressor (SRDX) tags and were over-produced in plants, which provided the basis to demonstrate that these TFs control secondary wall production (Yamaguchi et al., 2010). Similarly, the AP2/ERF TFs that induce ectopic primary wall-like structures performed this function more efficiently when fused to VP16 fragments (Sakamoto et al., 2018). Hence, by over-producing such versions of the co-expressed TFs, including those linked to hormone signalling, it may be possible to identify new transcriptional activators of primary wall cellulose synthesis genes and perhaps alter or control primary wall production.
Post-transcriptional regulation may also play a role in controlling cellulose biosynthesis. Previous work has shown that primary wall CESA genes could be regulated via anti-sense transcripts in barley, and that these anti-sense transcripts could be processed into small RNAs (Held et al., 2008). The anti-sense transcripts increased during the later stages of leaf elongation, indicating that CSC-associated genes and presumably other cellulose-related genes may be post-transcriptionally down-regulated during this stage in leaf development. While there is currently no direct link to hormone signalling or synthesis for such anti-sense transcripts, hormones influence the production of small RNAs (D’Ario et al., 2017; Liu et al., 2018) and could thus be prominent players for this type of cellulose regulation, particularly due to changing hormone concentrations along the leaf developmental axis. While the small RNA area and cellulose synthesis remain underexplored, this area will be an interesting path to investigate in the coming years.
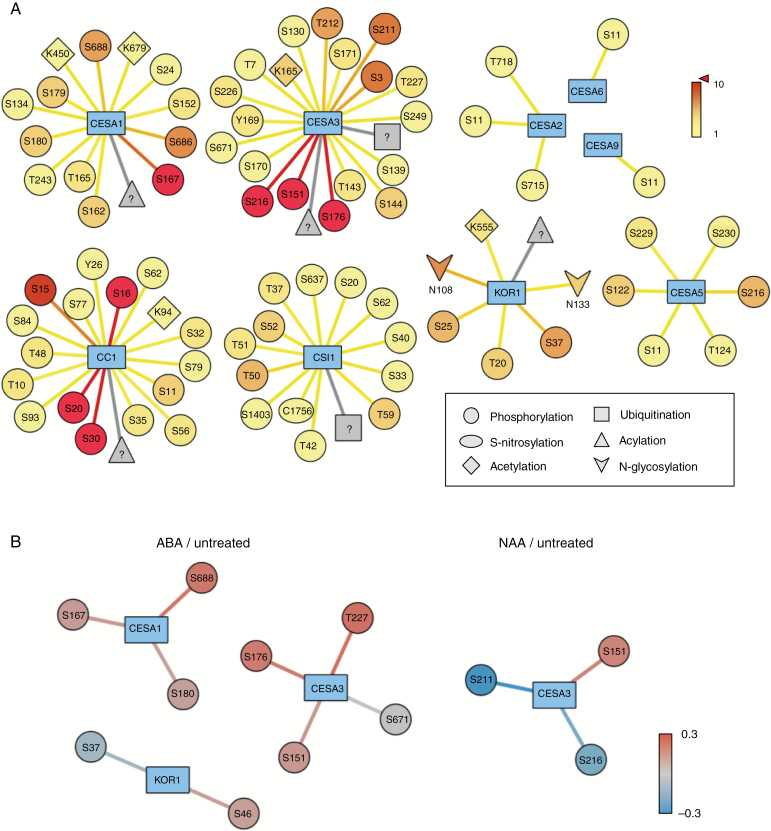
One likely avenue for hormonal control of cellulose synthesis is via post-translational modifications (PTMs). Indeed, many of the proteins associated with the CSC are subject to numerous PTMs, including phosphorylation, N-glycosylation, lysine acetylation and S-acylation (Kumar et al., 2016; Cruz et al., 2019) (Fig. 2A). Defects in those modifications can cause either mis-localization, for example when KORRIGAN is under-glycosylated (Rips et al., 2014), or changes in CSC activity, i.e. when CESA phosphorylation sites are mutated (Chen et al., 2010, 2016). Currently, only one study directly links CSC PTMs with hormone signalling (Sánchez-Rodríguez et al., 2017). As outlined above, this study revealed that BIN2 phosphorylates CESA1 and that such phosphorylation inhibits CSC motility. Despite the number of reported sites of phosphorylation in the CESAs and associated proteins, BIN2 is the only kinase that has been shown to directly phosphorylate a CSC-associated protein. Given the number of kinases and phosphatases associated with hormone signalling, it may be anticipated that some of these proteins directly change the catalytic rate by which cellulose is made and thus may regulate the phosphorylation status of CSC-associated proteins. One way to assess the potential role of hormone signalling on PTMs would be to investigate whether the abundance of modified peptides is changed in the cellulose-related proteins after hormone treatment or inhibition. Using the Functional Analysis Tools for Post-Translational Modifications database (Cruz et al., 2019), we undertook such a survey and found that multiple phosphorylation sites within CESA1, CESA3 and KOR1 are responsive to either ABA or auxin treatment, which are the only hormone-related quantitative analyses available in this database (Fig. 2B). These data indicate that at least certain types of phytohormone treatments may impact phosphorylation status of proteins that drive cellulose synthesis. Notably, BIN2 was also found to be co-expressed with the CESAs (Sánchez-Rodríguez et al., 2017). While kinases or phosphatases are not necessarily transcriptionally co-regulated with their substrates, this type of approach may also be of interest to research in this area.
Fig. 2.
Post-translational modifications and phytohormone-regulated phosphorylation events associated with CSC subunits in Arabidopsis: The FAT-PTM database was queried for each of the CSC protein components using the post-translational modification and quantitative phosphorylation search functions (Cruz et al., 2019) to identify post-translational modifications that occur in CSC components and phosphorylation events that are altered upon treatment with phytohormones. (A) In each diagram, the central blue node represents a protein of interest with its Arabidopsis gene name indicated. Peripheral nodes represent post-translational modification events according to the symbol key in the lower right corner with their respective residue indices indicated. Edges and peripheral nodes are coloured based on the number of experimentally supported spectral observations, where warmer colours represent increased spectral support. Grey symbols represent a post-translational modification that was experimentally associated with a protein but has an unresolved modification site. In these cases, the residue index is indicated by ‘?’ . (B) Phosphorylation events in CSC components impacted by ABA treatment or NAA treatment. In each diagram, the central blue node represents the protein of interest, with its Arabidopsis gene name indicated. Peripheral nodes represent phosphorylation events with their respective residue indices indicated. Edges and peripheral nodes are coloured based on quantitative log2 fold-abundance changes, where warmer colours represent increased phosphopeptide abundance and cooler colours represent decreased abundance.
While phosphorylation and glycosylation are two major PTMs, there are many other types of additions. One could envisage that the turnover rate and/or recycling of the CESAs, and associated proteins, may be an important factor to control cellulose production. A major regulator of protein turnover and internalization of proteins is ubiquitination (Vierstra, 2009), and large-scale proteomic studies have identified CSC-associated proteins as ubiquitination targets (Kim et al., 2013; Johnson and Vert, 2016). There are, however, no studies that have indicated that ubiquitination of the CESAs impact their function. Nevertheless, this could be one mechanism that could regulate internalization and recycling, or degradation, of the CSC. Such regulation has for example been shown for PIN2 (Leitner et al., 2012), where mono-ubiquitination causes PIN2 removal from the plasma membrane and further ubiquitination at the endomembrane determines the fate of the protein. Perhaps a similar mechanism could also drive the trafficking of the CESA complex and its associated proteins.
Overall, the observations presented in this review suggest that there are numerous connections between phytohormone signalling and cellulose biosynthesis or responses to cellulose biosynthesis inhibition. However, what is now urgently needed are mechanisms that directly connect signalling components of phytohormone signalling pathways to the transcriptional, post-transcriptional and post-translational control of the CSC or its direct regulators. Fortunately, ample well-characterized chemical and genetic tools are available to facilitate this goal, and these resources could probably be combined with robust cell biological tools, such as CSC and MT marker lines, to understand how phytohormone signalling impacts the in vivo behaviour of the CSC. Additionally, these chemical and genetic tools could be utilized to biochemically ascertain how cellulose content or cell wall composition is impacted by phytohormone synthesis and signalling. Finally, these tools could be coupled with PTM enrichment methods and quantitative mass spectrometry to generate a systems-level view of how phytohormone signalling impacts PTM networks controlling cellulose biosynthesis. Based on the substantial importance of both phytohormone signalling and plant cell wall biogenesis in controlling plant growth, development and responses to the environment, we anticipate that these lines of research will reveal substantial mechanistic interconnections between cellulose deposition and phytohormone signal transduction.
FUNDING
L.W. is supported by the China Scholarship Council. G.A.K. is a recipient of a Swiss National Science Foundation grant (P400PB_180834 / 1). S.P. is funded through Australian Research Council FT and DP grants (DP190101941; FT160100218). I.S.W. is supported by a National Science Foundation CAREER Award (MCB 1750359).
Literature cited
- Acosta IF, Farmer EE. 2010. Jasmonates. The Arabidopsis Book 8: e0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M, Li L, Friml J. 2019. Reorientation of cortical microtubule arrays in the hypocotyl of Arabidopsis thaliana is induced by the cell growth process and independent of auxin signaling. International Journal of Molecular Sciences 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN. 2004. The ethylene signaling pathway. Science 306: 1513–1515. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Wallace IS, Somerville CR. 2012. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proceedings of the National Academy of Sciences of the United States of America 109: 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsuffi G, Braybrook SA. 2018. Acid growth: an ongoing trip. Journal of Experimental Botany 69: 137–146. [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, et al. 2012. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nature Cell Biology 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai WQ, Xiao YH, Zhao J, et al. 2014. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall eeposition in cotton fibers. PLoS ONE 9: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F, Brailsford RW, Hauskrecht M, Jackson MB, Barlow PW. 1993. Cellular dimorphism in the maize root cortex: involvement of microtubules, ethylene and gibberellin in the differentiation of cellular behaviour in postmitotic growth zones. Botanica Acta 106: 394–403. [Google Scholar]
- Bashline L, Li S, Anderson CT, Lei L, Gu Y. 2013. The endocytosis of cellulose synthase in arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiology 163: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Li S, Zhu X, Gu Y. 2015. The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proceedings of the National Academy of Sciences of the United States of America 112: 12870–12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. 2001. On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215: 150–171. [DOI] [PubMed] [Google Scholar]
- Bethke G, Thao A, Xiong G, et al. 2015. Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 28: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. 1996. Branching in pea: action of genes Rms3 and Rms4. Plant Physiology 110: 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binenbaum J, Weinstain R, Shani E. 2018. Gibberellin localization and transport in plants. Trends in Plant Science 23: 410–421. [DOI] [PubMed] [Google Scholar]
- Bömer M, O’Brien JA, Pérez-Salamó I, et al. 2018. COI1-dependent jasmonate signalling affects growth, metabolite production and cell wall protein composition in Arabidopsis. Annals of Botany 122: 1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T, Mattsson O, Næsted H, Foster R, Mundy J. 2003. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. Journal of Cell Science 116: 791–801. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA. 2013. Diverse roles of strigolactones in plant development. Molecular Plant 6: 18–28. [DOI] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Perssona S. 2012. POM-POM2/CELLULOSE SYNTHASE INTERACTING1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR. 2004. The Arabidopsis thaliana abscisic acid-insensitive8 locus encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16: 406–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. 2005. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. 2001. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Ye ZH. 2002. Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14: 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Gidley MJ, Fincher GB. 2010. Heterogeneity in the chemistry, structure and function of plant cell walls. Nature Chemical Biology 6: 724–732. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LIA, Lee S, De Oliveira C, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nature Chemical Biology 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M. 2003. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant Journal 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Chan J, Coen E. 2020. Interaction between autonomous and microtubule guidance systems controls cellulose synthase trajectories. Current Biology 30: 941–947.e2. [DOI] [PubMed] [Google Scholar]
- Chen F, Jiang L, Zheng J, et al. 2014. Identification of differentially expressed proteins and phosphorylated proteins in rice seedlings in response to strigolactone treatment. PLoS ONE 9: e93947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR. 2010. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proceedings of the National Academy of Sciences of the United States of America 107: 17188–17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jia H, Zhao H, et al. 2016. Anisotropic cell expansion is affected through the bidirectional mobility of cellulose synthase complexes and phosphorylation at two critical residues on CESA3. Plant Physiology 171: 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H, et al. 2014. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE. 2005. Ethylene signal transduction. Annals of Botany 95: 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hong X, Zhang H, et al. 2005. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant Journal 43: 273–283. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B. 2009. Biosynthesis of salicylic acid in plants. Plant Signaling and Behavior 4: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Cho SH, Purushotham P, Fang C, et al. 2017. Synthesis and self-assembly of cellulose microfibrils from reconstituted cellulose synthase. Plant Physiology 175: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, et al. 2009. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. The Plant Cell Online 21: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz ER, Nguyen H, Nguyen T, Wallace IS. 2019. Functional analysis tools for post-translational modification: a post-translational modification database for analysis of proteins and metabolic pathways. Plant Journal 99: 1003–1013. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61: 651–6 79. [DOI] [PubMed] [Google Scholar]
- D’Ario M, Griffiths-Jones S, Kim M. 2017. Small RNAs: big impact on plant development. Trends in Plant Science 22: 1056–1068. [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P. 2013. Gibberellin signaling in plants. Development (Cambridge) 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P. 2016. A pivotal role of DELLAs in regulating multiple hormone signals. Molecular Plant 9: 10–20. [DOI] [PubMed] [Google Scholar]
- Denness L, McKenna JF, Segonzac C, et al. 2011. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in arabidopsis. Plant Physiology 156: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, et al. 2007. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, et al. 2008. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945. [DOI] [PubMed] [Google Scholar]
- Diotallevi F, Mulder B. 2007. The cellulose synthase complex: a polymerization driven supramolecular motor. Biophysical Journal 92: 2666–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou L, He K, Higaki T, Wang X, Mao T. 2018. Ethylene signaling modulates cortical microtubule reassembly in response to salt stress. Plant Physiology 176: 2071–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval I, Beaudoin N. 2009. Transcriptional profiling in response to inhibition of cellulose synthesis by thaxtomin A and isoxaben in Arabidopsis thaliana suspension cells. Plant Cell Reports 28: 811–830. [DOI] [PubMed] [Google Scholar]
- Ellis C, Ellis C, Karafyllidis I, et al. 2002. The Arabidopsis mutant. Society 14: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. 2001. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, et al. 2015. A mechanism for sustained cellulose synthesis during salt stress. Cell 162: 1353–1364. [DOI] [PubMed] [Google Scholar]
- Eng RC, Sampathkumar A. 2018. Getting into shape: the mechanics behind plant morphogenesis. Current Opinion in Plant Biology 46: 25–31. [DOI] [PubMed] [Google Scholar]
- Engelsdorf T, Gigli-Bisceglia N, Veerabagu M, et al. 2018. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Science Signaling 11. [DOI] [PubMed] [Google Scholar]
- Espinosa-Ruiz A, Martínez C, De Lucas M, et al. 2017. TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development (Cambridge) 144: 1619–1628. [DOI] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Kleine-Vehn J, et al. 2011. PIN polarity maintenance by the cell wall in Arabidopsis. Current Biology 21: 338–343. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, et al. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, et al. 2014. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704. [DOI] [PubMed] [Google Scholar]
- Gallei M, Luschnig C, Friml J. 2020. Auxin signalling in growth: Schrödinger’s cat out of the bag. Current Opinion in Plant Biology 53: 43–49. [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, et al. 2007. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Developmental Cell 13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. 2015. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proceedings of the National Academy of Sciences of the United States of America 112: 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Aryal B, Di Donato M, Hao P. 2017. A critical view on ABC transporters and their interacting partners in auxin transport. Plant and Cell Physiology 58: 1601–1604. [DOI] [PubMed] [Google Scholar]
- Gigli-Bisceglia N, Engelsdorf T, Strnad M, et al. 2018. Cell wall integrity modulates arabidopsis thaliana cell cycle gene expression in a cytokinin-and nitrate reductase-dependent manner. Development (Cambridge) 145: 1–14. [DOI] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, et al. 2018. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Current Biology 28: 2452–2458.e4. [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J. 2010. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO Journal 29: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Nolan TM, Song G, et al. 2018. FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Current Biology 28: 3316–3324.e6. [DOI] [PubMed] [Google Scholar]
- Guo Q, Yoshida Y, Major IT, et al. 2018. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 115: E10768–E10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. 2009. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nature Cell Biology 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. 2002. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Molecular Biology 49: 373–385. [PubMed] [Google Scholar]
- Haigler CH, Betancur L, Stiff MR, Tuttle JR. 2012. Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Frontiers in Plant Science 3: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Li Z, Waadt R, Schroeder JI. 2017. SnapShot: abscisic acid signaling. Cell 171: 1708.e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat Q, Hayat S, Irfan M, Ahmad A. 2010. Effect of exogenous salicylic acid under changing environment: a review. Environmental and Experimental Botany 68: 14–25. [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. 2002. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. 2012. Gibberellin biosynthesis and its regulation. Biochemical Journal 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, et al. 2010. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biology 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Penning B, Brandt AS, et al. 2008. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proceedings of the National Academy of Sciences of the United States of America 105: 20534–20539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Sado PE, Van Tuinen A, et al. 2007. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Current Biology 17: 922–931. [DOI] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, et al. 2007. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelspach R, Williamson RE, Wasteneys GO. 2003. Cellulose microfibril alignment recovers from DCB-induced disruption despite microtubule disorganization. Plant Journal 36: 565–575. [DOI] [PubMed] [Google Scholar]
- Hu Y, Yu D. 2014. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26: 4394–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wang S, Zhang B, et al. 2015. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Vert G. 2016. Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiology 171: 1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Robin GP. 2013. Systemic signaling during plant defense. Current Opinion in Plant Biology 16: 527–533. [DOI] [PubMed] [Google Scholar]
- Kendrick MD, Chang C. 2008. Ethylene signaling: new levels of complexity and regulation. Current Opinion in Plant Biology 11: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesten C, Wallmann A, Schneider R, et al. 2019. The companion of cellulose synthase 1 confers salt tolerance through a Tau-like mechanism in plants. Nature Communications 10: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan GA, Vogiatzaki E, Glauser G, Poirier Y. 2016. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiology 171: 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. 2018. Cytokinin signaling in plant development. Development (Cambridge, England) 145: 1–7. [DOI] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD. 2013. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Murai N, Fang DD, Triplett BA. 2011. Functional analysis of Gossypium hirsutum cellulose synthase catalytic subunit 4 promoter in transgenic Arabidopsis and cotton tissues. Plant Science 180: 323–332. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Yu S, et al. 2009. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nature Cell Biology 11: 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY. 2012. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Wang Z-Y. 2010. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annual Review of Plant Biology 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Krishna P, Prasad BD, Rahman T. 2017. Brassinosteroid action in plant abiotic stress tolerance. In: Russinova E, Cano-Delgado A, eds Methods in molecular biology. Totowa, NJ: Humana Press Inc., 193–202. [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, et al. 2005. Transcription switches for protoxylem and metaxylem vessel formation. Genes and Development 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Wightman R, Atanassov I, et al. 2016. S-acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353: 166–169. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. 2002. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology 5: 325–331. [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Walcher CL, Nemhauser JL. 2009. Cross-regulatory mechanisms in hormone signaling. Plant Molecular Biology 69: 375–381. [DOI] [PubMed] [Google Scholar]
- Lampugnani ER, Flores-Sandoval E, Tan QW, Mutwil M, Bowman JL, Persson S. 2019. Cellulose synthesis – central components and their evolutionary relationships. Trends in Plant Science 24: 402–412. [DOI] [PubMed] [Google Scholar]
- Lang JM, Eisinger WR, Green PB. 1982. Effects of ethylene on the orientation of microtubules and cellulose microfibrils of pea epicotyl cells with polylamellate cell walls. Protoplasma 110: 5–14. [Google Scholar]
- Lavy M, Estelle M. 2016. Mechanisms of auxin signaling. Development (Cambridge) 143: 3226–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, Van Der Straeten D, Verbelen JP. 2004. Position and cell type-dependent microtubule reorientation characterizes the early response of the Arabidopsis root epidermis to ethylene. Physiologia Plantarum 121: 513–519. [Google Scholar]
- Lehman TA, Sanguinet KA. 2019. Auxin and cell wall crosstalk as revealed by the Arabidopsis thaliana cellulose synthase mutant radially swollen 1. Plant and Cell Physiology 60: 1487–1503. [DOI] [PubMed] [Google Scholar]
- Lehman TA, Smertenko A, Sanguinet KA. 2017. Auxin, microtubules, and vesicle traffcking: conspirators behind the cell wall. Journal of Experimental Botany 68: 3321–3329. [DOI] [PubMed] [Google Scholar]
- Leitner J, Petrášek J, Tomanov K, et al. 2012. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proceedings of the National Academy of Sciences of the United States of America 109: 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenglet A, Jaślan D, Toyota M, et al. 2017. Control of basal jasmonate signalling and defence through modulation of intracellular cation flux capacity. New Phytologist 216: 1161–1169. [DOI] [PubMed] [Google Scholar]
- Lertpiriyapong K, Sung ZR. 2003. The elongation defective1 mutant of Arabidopsis is impaired in the gene encoding a serine-rich secreted protein. Plant Molecular Biology 53: 581–595. [DOI] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y. 2012. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proceedings of the National Academy of Sciences USA 109: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom JJ, Nakamura M, Hibbel A, et al. 2013. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 342: 1245533. [DOI] [PubMed] [Google Scholar]
- Liu H, Yu H, Tang G, Huang T. 2018. Small but powerful: function of microRNAs in plant development. Plant Cell Reports 37: 515–528. [DOI] [PubMed] [Google Scholar]
- Liu X, Yang Q, Wang Y, Wang L, Fu Y, Wang X. 2018. Brassinosteroids regulate pavement cell growth by mediating BIN2-induced microtubule stabilization. Journal of Experimental Botany 69: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YB, Lu SM, Zhang JF, Liu S, Lu YT. 2007. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226: 1547–1560. [DOI] [PubMed] [Google Scholar]
- Liu Z, Schneider R, Kesten C, et al. 2016. Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Developmental Cell 38: 305–315. [DOI] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D. 2013a Genomic analysis of della protein activity. Plant and Cell Physiology 54: 1229–1237. [DOI] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D. 2013b Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Current Biology 23: 804–809. [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, Ligerot Y, Bonhomme S, Boyer FD, Rameau C. 2015. Strigolactone biosynthesis and signaling in plant development. Development (Cambridge) 142: 3615–3619. [DOI] [PubMed] [Google Scholar]
- Lu H. 2009. Dissection of salicylic acid-mediated defense signaling networks. Plant Signaling and Behavior 4: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Scholl S, Doering A, et al. 2015. V-ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis. Nature Plants 1: 15094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P. 2019. Auxin EvoDevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Molecular Plant 12: 298–320. [DOI] [PubMed] [Google Scholar]
- Meier C, Bouquin T, Nielsen ME, et al. 2001. Gibberellin response mutants identified by luciferase imaging. Plant Journal 25: 509–519. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miart F, Desprez T, Biot E, et al. 2014. Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant Journal 77: 71–84. [DOI] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI. 2015. Mechanisms of abscisic acid-mediated control of stomatal aperture. Current Opinion in Plant Biology 28: 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoleão TA, Soares G, Vital CE, et al. 2017. Methyl jasmonate and salicylic acid are able to modify cell wall but only salicylic acid alters biomass digestibility in the model grass Brachypodium distachyon. Plant Science 263: 46–54. [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. 1998. A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO Journal 17: 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. 2003. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972. [DOI] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, et al. 2002. Kobito1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in arabidopsis. Plant Cell 14: 2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Persson S, Ehrhardt DW, Somerville CR. 2008. Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiology 147: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville C, Ehrhardt D. 2006. Visualization of cellulose synthase with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Park SS-Y, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C. Science 324: 1068–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, et al. 2007. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. 2005. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National Academy of Sciences of the United States of America 102: 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C, Hirano K, Stork J, DeBolt S. 2015. Mapping of a cellulose-deficient mutant named dwarf1-1 in Sorghum bicolor to the green revolution gene gibberellin20-oxidase reveals a positive regulatory association between gibberellin and cellulose biosynthesis. Plant Physiology 169: 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegoń-Putze I, Bosch N, Ibanḛs M, Cano-Delgado AI. 2019. Brassinosteroid signaling in plant development and adaptation to stress. Development (Cambridge) 146: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko JK, Barnes WJ, Voiniciuc C, et al. 2018. SHOU4 proteins regulate trafficking of cellulose synthase complexes to the plasma membrane. Current Biology 28: 3174–3182.e6. [DOI] [PubMed] [Google Scholar]
- Polko JK, Kieber JJ. 2019. The regulation of cellulose biosynthesis in plants. Plant Cell 31: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham P, Hyun S, Díaz-Moreno SM, Kumar M, Nixon BT, Bulone V. 2016. A single heterologously expressed plant cellulose synthase isoform is sufficient for cellulose microfibril formation in vitro. Proceedings of the National Academy of Sciences of the United States of America 113: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Song P, Hu Y, et al. 2018. Regulation of stomatal movement by cortical microtubule organization in response to darkness and ABA signaling in Arabidopsis. Plant Growth Regulation 84: 467–479. [Google Scholar]
- Ramírez V, Pauly M. 2019. Genetic dissection of cell wall defects and the strigolactone pathway in Arabidopsis. Plant Direct 3: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM. 2015. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Molecular Plant 8: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rips S, Bentley N, Jeong IS, Welch JL, von Schaewen A, Koiwa H. 2014. Multiple n-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26: 3792–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J. 2011. Salicylic acid beyond defence: its role in plant growth and development. Journal of Experimental Botany 62: 3321–3338. [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, et al. 2005. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C, Persson S. 2012. Co-expression of cell wall-related genes: new tools and insights. Frontiers in Plant Science 3: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. 2007. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Somssich M, Nakata MT, et al. 2018. Complete substitution of a secondary cell wall with a primary cell wall in Arabidopsis. Nature Plants 4: 777–783. [DOI] [PubMed] [Google Scholar]
- Sampathkumar A, Gutierrez R, McFarlane HE, et al. 2013. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiology 162: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Bauer S, Hématy K, et al. 2012. CHITINASE-LIKE1/POM-POM1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24: 589–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Ketelaar KD, Schneider R, et al. 2017. BRASSINOSTEROID INSENSITIVE2 negatively regulates cellulose synthesis in Arabidopsis by phosphorylating cellulose synthase 1. Proceedings of the National Academy of Sciences of the United States of America 114: 3533–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Rubio-Somoza I, Sibout R, Persson S. 2010. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends in Plant Science 15: 291–301. [DOI] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Shi Y, Kesten C, et al. 2018. The cellulose synthases are cargo of the TPLATE adaptor complex. Molecular Plant 11: 346–349. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, et al. 2009. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668. [DOI] [PubMed] [Google Scholar]
- Schneider R, Hanak T, Persson S, Voigt CA. 2016. Cellulose and callose synthesis and organization in focus, what’s new? Current Opinion in Plant Biology 34: 9–16. [DOI] [PubMed] [Google Scholar]
- Schneider R, Tang L, Lampugnani ER, et al. 2017. Two complementary mechanisms underpin cell wall patterning during xylem vessel development. Plant Cell 29: 2433–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Koshiba T. 2002. Complex regulation of ABA biosynthesis in plants. Trends in Plant Science 7: 41–48. [DOI] [PubMed] [Google Scholar]
- Seto Y, Yamaguchi S. 2014. Strigolactone biosynthesis and perception. Current Opinion in Plant Biology 21: 1–6. [DOI] [PubMed] [Google Scholar]
- Seung D, Webster MW, Wang R, Andreeva Z, Marc J. 2013. Dissecting the mechanism of abscisic acid-induced dynamic microtubule reorientation using live cell imaging. Functional Plant Biology 40: 224–236. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Ross JL. 2012. Microtubule-severing enzymes at the cutting edge. Journal of Cell Science 125: 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. 2010. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Van Der Straeten D. 1997. Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proceedings of the National Academy of Sciences of the United States of America 94: 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, et al. 2004. Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, et al. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher TL, Li PZ, Wallace IS. 2018. Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants 7: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ma Q, Mao T. 2015. Ethylene regulates the arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiology 169: 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, et al. 2005. Brassinosteroid regulates fiber development on cultured cotton ovules. Plant and Cell Physiology 46: 1384–1391. [DOI] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Fokar M, et al. 2015. Brassinosteroid signaling affects secondary cell wall deposition in cotton fibers. Industrial Crops and Products 65: 334–342. [Google Scholar]
- Takatani S, Hirayama T, Hashimoto T, Takahashi T, Motose H. 2015. Abscisic acid induces ectopic outgrowth in epidermal cells through cortical microtubule reorganization in Arabidopsis thaliana. Scientific Reports 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallman G. 2004. Are diurnal patterns of stomatal movement the result of alternating metabolism of endogenous guard cell ABA and accumulation of ABA delivered to the apoplast around guard cells by transpiration? Journal of Experimental Botany 55: 1963–1976. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, et al. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, et al. 2008. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. 2003. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences of the United States of America 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, De Lucas M, et al. 2015. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JH, Shaw SL. 2020. Exogenous auxin induces transverse microtubule arrays through TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX receptors. Plant physiology 182: 892–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang DL, Edmond C, Harrington JL, Nühse TS. 2011. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiology 156: 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, et al. 2009. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant, Cell and Environment 32: 1633–1651. [DOI] [PubMed] [Google Scholar]
- Verbančič J, Lunn JE, Stitt M, Persson S. 2018. Carbon supply and the regulation of cell wall synthesis. Molecular Plant 11: 75–94. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2009. The ubiquitin-26S proteasome system at the nexus of plant biology. Nature Reviews Molecular Cell Biology 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wang C, Li J, Yuan M. 2007. Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant and Cell Physiology 48: 1534–1547. [DOI] [PubMed] [Google Scholar]
- Wang F, Deng XW. 2011. Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Research 21: 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. 2002. Ethylene biosynthesis and signaling networks. Plant Cell 14: 131–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, McFarlane HE, Persson S. 2016. The impact of abiotic factors on cellulose synthesis. Journal of Experimental Botany 67: 543–552. [DOI] [PubMed] [Google Scholar]
- Wang W, Bai MY, Wang ZY. 2014. The brassinosteroid signaling network – a paradigm of signal integration. Current Opinion in Plant Biology 21: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, et al. 2008. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Developmental Cell 15: 220–235. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T. 2012. Arabidopsis MICROTUBULE DESTABILIZING PROTEIN40 Is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24: 4012–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell LM, et al. 2015. Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Schneider R, Barkwill S, et al. 2018. Cellulose synthase complexes display distinct dynamic behaviors during xylem transdifferentiation. Proceedings of the National Academy of Sciences of the United States of America 115: E6366–E6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC. 2017. Strigolactone signaling and evolution. Annual Review of Plant Biology 68: 291–322. [DOI] [PubMed] [Google Scholar]
- Westermann J, Streubel S, Franck CM, Lentz R, Dolan L, Boisson-Dernier A. 2019. An evolutionarily conserved receptor-like kinases signaling module controls cell wall integrity during tip growth. Current Biology 29: 3899–3908.e3. [DOI] [PubMed] [Google Scholar]
- Xie L, Yang C, Wang X. 2011. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. Journal of Experimental Botany 62: 4495–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kaori Y, Koichi Y. 2010. The strigolactone story. Annual Review of Phytopathology 48: 93–117. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. 2002. Cell signaling during cold, drought, and salt stress. Plant Cell 14: S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. 2008. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in arabidopsis. Plant Cell 20: 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Goué N, Igarashi H, et al. 2010. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiology 153: 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation. Annual Review of Plant Biology 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yan A, Chen Z. 2017. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Reports 36: 689–703. [DOI] [PubMed] [Google Scholar]
- Yang W, Zhang W, Wang X. 2017. Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnology Journal 15: 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, He C, Ma Y, Herde M, Ding Z. 2017. Jasmonic acid enhances al-induced root growth inhibition. Plant Physiology 173: 1420–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You-Ming Y, Chu-Nian X, Bao-Min W, Jun-Zhen J. 2001. Effects of plant growth regulators on secondary wall thickening of cotton fibres. Plant Growth Regulation 35: 233–237. [Google Scholar]
- Zhang W, Cai C, Staiger CJ. 2019. Myosins XI are involved in exocytosis of cellulose synthase complexes. Plant Physiology 179: 1537–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nikolovski N, Sorieul M, et al. 2016. Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nature Communications 7: 2041-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Kays SJ, Schroeder BP, Ye ZH. 2002. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ganguly A, Baskin TI, et al. 2015. The fragile fiber1 kinesin contributes to cortical microtubule-mediated trafficking of cell wall components. Plant Physiology 167: 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Li Y, Cao DM, et al. 2017. The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Molecular Cell 66: 648–657.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Li S, Pan S, Xin X, Gu Y. 2018. CSI1, PATROL1, and exocyst complex cooperate in delivery of cellulose synthase complexes to the plasma membrane. Proceedings of the National Academy of Sciences of the United States of America 115: E3578–E3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, et al. 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant Journal 48: 687–698. [DOI] [PubMed] [Google Scholar]
- Zuoren Y, Zhang C, Yang X, et al. 2014. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytologist 203: 437–448. [DOI] [PubMed] [Google Scholar]