Abstract
Background and Aims
Vitamin E (tocochromanol) is a lipid-soluble antioxidant and an essential nutrient for human health. Among cereal crops, barley (Hordeum vulgare) contains a high level of vitamin E, which includes both tocopherols and tocotrienols. Although the vitamin E biosynthetic pathway has been characterized in dicots, such as Arabidopsis, which only accumulate tocopherols, knowledge regarding vitamin E biosynthesis in monocots is limited because of the lack of functional mutants. This study aimed to obtain gene knockout mutants to elucidate the genetic control of vitamin E composition in barley.
Methods
Targeted knockout mutations of HvHPT and HvHGGT in barley were created with CRISPR/Cas9-enabled genome editing. High-performance liquid chromatography (HPLC) was performed to analyse the content of tocochromanol isomers in transgene-free homozygous Hvhpt and Hvhggt mutants.
Key Results
Mutagenesis efficiency among T0 regenerated plantlets was 50–65 % as a result of two simultaneously expressed guide RNAs targeting each gene; most of the mutations were stably inherited by the next generation. The transgene-free homozygous mutants of Hvhpt and Hvhggt exhibited decreased grain size and weight, and the HvHGGT mutation led to a shrunken phenotype and significantly lower total starch content in grains. HPLC analysis revealed that targeted mutation of HvHPT significantly reduced the content of both tocopherols and tocotrienols, whereas mutations in HvHGGT completely blocked tocotrienol biosynthesis in barley grains. Transient overexpression of an HvHPT homologue in tobacco leaves significantly increased the production of γ- and δ-tocopherols, which may partly explain why targeted mutation of HvHPT in barley grains did not eliminate tocopherol production.
Conclusions
Our results functionally validated that HvHGGT is the only committed gene for the production of tocotrienols, whereas HvHPT is partly responsible for tocopherol biosynthesis in barley.
Keywords: Barley (Hordeum vulgare), CRISPR/Cas9, HvHGGT, HvHPT, genome editing, tocopherol, tocotrienol, vitamin E
INTRODUCTION
Vitamin E (tocochromanol) is a potent lipid-soluble antioxidant and an essential component of the human diet. Many human diseases, such as cardiovascular disease and certain cancers, are associated with insufficient vitamin E intake (Bramley et al., 2000). Because of its beneficial effect, the daily requirement of vitamin E for humans has been raised to 15–30 mg (DellaPenna, 2005). To meet the demand for human consumption, it is necessary to modify the level and composition of vitamin E in food crops via genome engineering and precision plant breeding.
Vitamin E is classified into two categories, tocopherols (T) and tocotrienols (T3), both of which contain a polar chromanol head group linked to an isoprenoid (phytyl) side chain. There are four T forms (α-T, β-T, γ-T and δ-T) and four T3 forms (α-T3, β-T3, γ-T3 and δ-T3), which are based on the number and position of methyl groups on the chromanol head group (Schneider, 2005; Falk and Munne-Bosch, 2010). Tocopherols and tocotrienols differ in their degree of unsaturation: tocopherols have a fully saturated phytyl tail arising from the C20 isoprenoid intermediate phytyl diphosphate (PDP); tocotrienols have an unsaturated side chain with three trans double bonds that is derived from the C20 isoprenoid intermediate geranylgeranyl diphosphate (GGDP). Although tocopherol and tocotrienol both function as potent antioxidants, they exhibit distinctive biological properties. Tocopherols, typically in the α form, are widely distributed in photosynthetic plants, whereas tocotrienols occur primarily in the seed endosperm of most monocots and a limited number of dicots (Horvath et al., 2006; Falk and Munne-Bosch, 2010; Yang et al., 2011). In vitro studies indicate that α-tocopherol is the most active form of vitamin E, interacting with the polyunsaturated acyl groups of lipids, stabilizing membranes, and scavenging and quenching various reactive oxygen species. Therefore, tocopherols play an important role in protecting oxygenic phototrophs from lipid peroxidation (Maeda et al., 2005). However, in some cases, tocotrienols possess superior biological properties to tocopherols, including hypocholesterolaemic, anticancer and neuroprotective activities, which are not often observed in tocopherols (Sen et al., 2007; Aggarwal et al., 2010; Chin et al., 2016).
Tocopherols and tocotrienols are synthesized in plastids of plant cells (Yang et al., 2011; Lushchak and Semchuk, 2012) and their biosynthetic pathways have been elucidated (Schneider, 2005; Chen et al., 2006). Following the synthesis of tyrosine and its derivative 4-hydroxyphenylpyruvate (HPP) via the shikimate pathway, p-hydroxyphenylpyruvate dioxygenase catalyses the conversion of HPP to homogentisate (HGA). The committed step in tocopherol biosynthesis has traditionally been considered the condensation of HGA and PDP to form 2-methyl-6-phytyl-1,4-benzoquinol (MPBQ), as catalysed by homogentisate phytyltransferase (HPT). Tocotrienol biosynthesis involves GGDP rather than PDP, which is different from that of tocopherol synthesis. GGDP is directly condensed with HGA by homogentisate geranylgeranyl transferase (HGGT), producing 2-methyl-6-geranylgeranyl-1,4-benzoquinol (MGGBQ). In tocopherol and chlorophyll biosynthesis, GGDP reductase carries out the conversion of GGDP to PDP (Keller et al., 1998). Subsequent reactions in tocotrienol and tocopherol syntheses are similar, including ring methylations and ring cyclization, which are catalysed by the common enzymes 2-methyl-6-phytylbenzoquinone methyltransferase, tocopherol cyclase and γ-tocopherol methyltransferase (γ-TMT).
HGGT and HPT, two committed enzymes in the vitamin E biosynthesis pathway, are the main targets for metabolic engineering of both tocopherols and tocotrienols in plants. In dicots, such as Arabidopsis, the vitamin E biosynthesis pathway has been well established (Collakova and DellaPenna, 2003; Sattler et al., 2004; Gilliland et al., 2006; Valentin et al., 2006). Deletion in the HPT (VTE2) gene results in total tocopherol deficiency in all tissues (Sattler et al., 2004), whereas overexpression of Arabidopsis HPT (HPT1) causes a 4.4-fold increase in total tocopherol levels in leaves compared to that in the wild-type (Collakova and DellaPenna, 2003). Furthermore, the content of α-tocopherol has been increased by co-overexpression of HPT1 and the gene encoding γ-TMT, resulting in a 12-fold increase in vitamin E activity. However, only modest increases in vitamin E contents in the seeds of soybeans (Glycine max) and Arabidopsis occurred by enhancing the expression of HPT (Savidge et al., 2002; Collakova and DellaPenna, 2003; Karunanandaa et al., 2005). HGGT, which probably evolved in monocots, is predicted to be functionally divergent from HPT, which is conserved in both monocot and dicot species (Cahoon et al., 2003). Overexpression of barley HGGT in Arabidopsis, soybeans and tobacco results in considerable accumulation of total vitamin E, primarily in the form of tocotrienols (Cahoon et al., 2003; Konda et al., 2020), although expression of rice HGGT in soybeans only led to a slight accumulation of tocotrienols (Kim et al., 2011). Our recent work has demonstrated that overexpression of HvHGGT can significantly enhance tocotrienol levels and antioxidant activities while altering the composition of vitamin E in transgenic barley grains (Chen et al., 2017). Moreover, a recent joint linkage and genome-wide association study in 5000 lines of US maize (Zea mays) showed that allelic variation in 14 genes is responsible for 56–93 % of tocochromanol production in grains (Diepenbrock et al., 2017). In hexaploid oat (Avena sativa), two of five HPT (VTE2) homologues were identified as highly correlated with tocochromanol accumulation through deep sequencing and orthology-guided assembly (Gutierrez-Gonzalez and Garvin, 2016). Surprisingly, among the three HGGT homologues, one did not correlate with tocochromanol content, whereas the others had negative correlation coefficients (Gutierrez-Gonzalez and Garvin, 2016). Although transgenic overexpression of HPT or HGGT has been conducted to increase vitamin E content or alter the composition of vitamin E in monocot plants, the lack of functional mutants in the main step of the vitamin E pathway has impeded our understanding of tocochromanol biosynthesis in agronomically important cereal crops.
Barley (Hordeum vulgare), the fourth most important cereal crop in the world, is an excellent source of biologically active nutrients, including dietary fibre, β-glucans, phenolic acids, sterols, tocopherols and tocotrienols (Andersson et al., 2008; Cai et al., 2016). Barley and oats contain higher levels of tocochromanols compared with other cereals (Gutierrez-Gonzalez and Garvin, 2016). In particular, barley contains eight naturally occurring isomers of tocochromanol, and thus serves as a valuable source of vitamin E and an appropriate crop plant for genetic manipulation of vitamin E levels and composition. Despite the aforementioned progress in studying vitamin E biosynthesis in plants, there are few reports on metabolic engineering of tocopherol and tocotrienol biosynthesis in barley, which is partly because of the low efficiency of barley transformation and the lack of effective genome engineering tools. Recently, the CRISPR/CRISPR-associated protein 9 nuclease (Cas9) system has emerged as an efficient and versatile tool for genome editing and precision plant breeding (Xie et al., 2015; Shen et al., 2018; Cui et al., 2020).
In the present study, barley HvHGGT and HvHPT mutants were successfully created with CRISPR/Cas9-enabled genome editing for genetic and developmental analyses. Phenotypic analysis of grains showed that the transgene-free homozygous Hvhpt and Hvhggt mutants had decreased grain size and total starch content. Analysis of vitamin E components revealed that HvHGGT was the only committed gene that produced tocotrienols, whereas HvHPT was not the only gene that controls the synthesis of tocopherols in barley.
MATERIALS AND METHODS
gRNA design and vector construction
Gene sequences of barley HvHPT (HORVU7Hr1G110990) and HvHGGT (HORVU7Hr1G114330) were downloaded from Ensembl Plants (http://plants.ensembl.org/Hordeum_vulgare/Info/Index). Based on these sequences, four single guide RNAs (gRNAs) were designed and synthesized for targeting four distinct protospacer regions using the web tool CRISPR-P (http://cbi.hzau.edu.cn/crispr/) (Lei et al., 2014). Editing constructs containing the tRNA–gRNA fragment were generated according to the method of Xie et al. (2015). The pGTR plasmid was used as a template to amplify the gRNA–tRNA fusion. pRGEB32, a binary vector, carries an hpt selection marker and a cloning site for insertion of the gRNA–tRNA fragment. Two tRNA–gRNA fragments were individually cloned into pRGEB32, resulting in two editing constructs: PTG-HPT/Cas9 (gRNA1 + gRNA2) and PTG-HGGT/Cas9 (gRNA3 + gRNA4) (the primers used for the DNA constructs are listed in Supplementary Data Table S1 and detailed sequences of the two PTG constructs are provided in Table S2). These constructs were sequenced to ensure the accuracy of each tRNA–gRNA insert before they were electroporated into Agrobacterium tumefaciens strain EHA105 for barley transformation.
Plant material and genetic transformation
Wild-type spring barley plants (H. vulgare L. ‘Golden Promise’), obtained from the Australian Centre for Plant Functional Genomics at the University of Adelaide, were grown from October to April in experimental fields under natural conditions at the Agricultural Experiment Station of Zhejiang University, Hangzhou, Zhejiang Province, China. Caryopses were harvested 2–3 weeks after pollination. The immature scutella, 1.5–2 mm in size, were obtained from barley embryos after removal of the embryo axis and used as explants for Agrobacterium-mediated transformation following the procedure of Harwood (2014). Transgenic calli were induced from infected immature scutella on hygromycin (50 mg L–1) containing medium, and plantlets resistant to hygromycin were regenerated. Regenerated plants at the seedling stage were grown for 12–16 weeks in a growth chamber with a 16-h light/8-h night cycle, a temperature of 23 °C and 70 % humidity. Subsequently, transgenic plants were grown until maturity under natural light in 6-inch pots in a glasshouse.
Molecular characterization of transgenic plants
Genomic DNA was extracted from leaves sampled from putative transgenic regenerants ~4 months after genetic transformation; at this stage, plantlets are 8–15 cm high. To identify transgenic lines, T0 regenerated plantlets were first screened for the presence of the gRNA/Cas9 transgene using the primer pair pU3-Forward (5′-TGGGTACGTTGGAAACCACG-3′) and pUBI10-Reverse (5′-GTTTGTTGGTCGCCGTTAGG-3′). PCR was carried out using Taq DNA polymerase (Vazyme Biotech, Jiangsu, China) with an XP thermal cycler (Bioer Technology, Hangzhou, China). The cycling conditions comprised an initial denaturation step at 95 °C for 5 min, followed by 32 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min, and a final extension step at 72 °C for 8 min. The product was analysed by 1.5 % (w/v) agarose gel electrophoresis. To detect mutagenesis at gRNA target sites, DNA samples positive for gRNA/Cas9 fragments were again assessed by mutation genotyping using specific primers (Supplementary Data Table S3). The PCR product was examined by 2 % agarose gel electrophoresis and stained with ethidium bromide to detect chromosomal-fragment deletions. To detect indels at desired sites, the PCR product was further separated by 14 % SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis). Selected PCR products were sequenced directly by Shanghai Boshang Biology Company (Shanghai, China) or purified using a DNA Purification Kit (Sangon Biotech, Shanghai, China) and then transferred into the pClone007 simple vector (TsingKe Biotech, Beijing, China) for TA cloning (TA is short for thymine and adenine). Ligation products were transformed into the Escherichia coli strain DH5α, and single colonies were selected for plasmid isolation and DNA sequencing.
Morphological properties of barley grains
The phenotypic observation of grain development in wild-type barley and mutants (hvhpt15 and hvhggt13) was performed at 1, 2, 3 and 4 weeks after pollination. Whole grains or dehusked grains and their longitudinal section were imaged with a trinocular stereo microscope (Nikon, SMZ745T, Japan). Mature grains from gRNA/Cas9 transgene-free, homozygous T2 mutant lines were used for the analysis of grain width (with ImageJ software) and thousand-grain weight.
Mature barley grains were fractured transversely using a razor blade and tweezers. The resulting cross-sections of wild-type barley grains (outer endosperm regions) and their mutants were fixed on an aluminium specimen holder with double graphite tape. Samples were sputter-coated (Hitachi E-1010, Japan) with gold and examined with a scanning electron microscope (Hitachi S-3000N) at 20 kV at room temperature of 25 °C.
Measurement of starch content
Total starch content was determined enzymatically using the Total Starch Analysis Kit (Megazyme, Bray, Ireland). The amylose content was assayed with the Amylose/Amylopectin Assay Kit (Megazyme) based on the glucose oxidase–peroxidase method, as recommended by the manufacturer.
Gene expression analysis
Tissues from wild-type and transgenic lines were homogenized in liquid nitrogen, and then total RNA was prepared with an EZNA Plant RNA kit following the manufacturer’s instructions (Omega, Norcross, GA, USA). One microgram of total RNA was digested with a gDNA Remover Kit and reversely transcribed into cDNA in ReverTra Ace qPCR RT Master Mix (Toyobo, Kyoto, Japan) in a 20-µL reaction volume. Transcript analysis was conducted using semi-quantitative reverse transcriptase PCR (RT-PCR), and HvACTIN was used as the control gene for all cDNA samples. The primer sequences for all barley genes determined are listed in Supplementary Data Table S4.
Vitamin E measurement by high-performance liquid chromatography (HPLC)
Vitamin E was extracted according to previously described methods, with minor modifications (Chen et al., 2017). Leaves or mature grains of barley were ground to a fine powder in liquid nitrogen, and 50 mg of powder was extracted in 1.5 mL methanol by vortexing vigorously for 10 min at room temperature. The samples were centrifuged at 10 000 g for 5 min, and the supernatant was transferred to a new tube. The pellet was re-extracted twice. All three supernatants were pooled, filtered through a 0.22-μm membrane, and dried under a vacuum using a centrifugal evaporator (Labconco). The residues were dissolved in 100 µL methanol/isopropanol (1 : 1, v/v) and centrifuged at 14 000 g for 5 min. Vitamin E was determined using an Agilent 1200 HPLC (Agilent Technologies) equipped with a Phenomenex Kinetex F5 100A column (2.6 μm, 150 × 4.6 mm; Phenomenex) and a G1321A fluorescence detector. The mobile phase used was methanol/H2O (85 : 15, v/v) at a flow rate of 1.0 mL min−1. Sample components were detected by fluorescence with excitation at 298 nm and emission at 328 nm (Grebenstein and Frank, 2012). Tocopherols and tocotrienols were quantified against external standard curves using authentic compounds (ChromaDex).
Phylogenetic analysis
The amino acid sequences of HPTs and HGGTs from eight plant species (Hordeum vulgare, Triticum aestivum, Zea mays, Oryza sativa, Arabidopsis thaliana, Sorghum bicolor, Brachypodium distachyon and Avena sativa) were used to construct a phylogenetic tree. The accession numbers for these sequences were retrieved from the Phytozome (v12.1.6), UniProtKB or NCBI database. Multiple sequence alignments were performed using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/), and subsequent phylogenetic analysis was conducted using the maximum-likelihood method in the MEGA7 program.
Transient gene expression in tobacco leaves
A full-length coding sequence of an HPT homologue (HORVU2Hr1G117600) was amplified (primers are listed in Supporting Data Table S3) using RT-PCR from mRNAs extracted from grains (14 d after pollination) of barley (‘Morex’) and cloned into the pH7FWG2.0 vector under control of the 35S promoter. The resulting DNA construct was electroporated into Agrobacterium tumefaciens strain EHA105. Agrobacterium cultures, carrying the empty vector (pH7FWG2.0) and 35S: HORVU2Hr1G117600 were infiltrated into 1-month-old tobacco (Nicotiana benthamiana) leaves using a needleless syringe. The plants were incubated at 24 °C under a 16-h/8-h light–dark cycle. Five days after infiltration, the control and infiltrated leaves were collected for tocochromanol extraction and HPLC analyses.
Statistical analysis
All experiments were repeated at least three times. The data shown represent the mean ± s.e. Asterisks indicate a significant difference between the wild-type and mutant plants at *P < 0.05 or **P < 0.01, as determined by Student’s t-tests.
RESULTS
Design and delivery of PTG editing constructs into barley plants
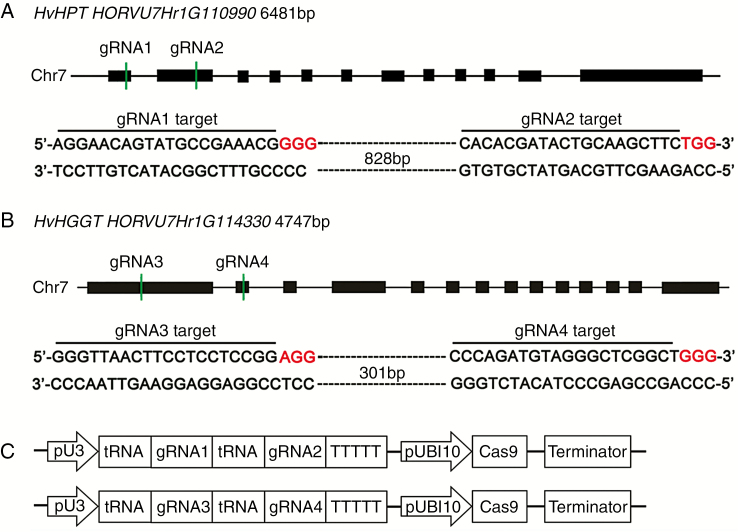
Genomic sequences of barley HPT (HvHPT, HORVU7Hr1G110990) and HGGT (HvHGGT, HORVU7Hr1G114330) genes from the Ensembl Plants database showed that HvHPT (6481 bp) has 12 exons and 11 introns and that HvHGGT (4747 bp) has 14 exons and 13 introns (Fig. 1A, B). To effectively carry out targeted mutations of these two genes, four single gRNAs were designed to target specific sites in the protein-coding region with low sequence homology (Fig. 1A, B; Supplementary Data Fig. S1A). gRNA1 and gRNA2 were expected to target the first and second exons of HvHPT, respectively, whereas gRNA3 and gRNA4 were designed to target the first two exons of HvHGGT. As shown in Fig. 1C, two gRNAs were assembled into a single vector using the polycistron tRNA–gRNA (PTG) strategy (Xie et al., 2015), generating two constructs, PTG-HPT/Cas9 and PTG-HGGT/Cas9 (Table S2), for barley transformation and genome editing. The two constructs were independently transformed into immature barley embryos via Agrobacterium-mediated transformation. We obtained ~700 and 550 embryo-derived calli exhibiting hygromycin resistance for the PTG-HPT/Cas9 and PTG-HGGT/Cas9 constructs, respectively. A total of 26 primary plantlets for PTG-HPT/Cas9 and 17 for PTG-HGGT/Cas9 were regenerated from the transformed calli (Table S5).
Fig. 1.
Schematic diagram of HvHPT and HvHGGT genes targeted by four specific gRNAs. (A) Gene structure of HvHPT with gRNA1 and gRNA2 targeting sites. The distance between the two target sites is 831 bp. (B) Gene structure of HvHGGT with gRNA3 and gRNA4 targeting sites. The distance between the two target sites is 304 bp. (C) Schematic of the tRNA–gRNA fragments inserted into the pRGEB32 vector. Black rectangles indicate protein-coding regions. The PAM, protospacer-adjacent motif, is shown in red. The green vertical lines indicate the relative location of gRNA targeting sites; the overlined nucleotides indicate the target sequence for each gRNA. pUBI10, rice ubiquitin promoter; pU3, rice U3 snoRNA promoter.
Targeted mutagenesis of HvHPT and HvHGGT induced by CRISPR/Cas9 in the T0 generation
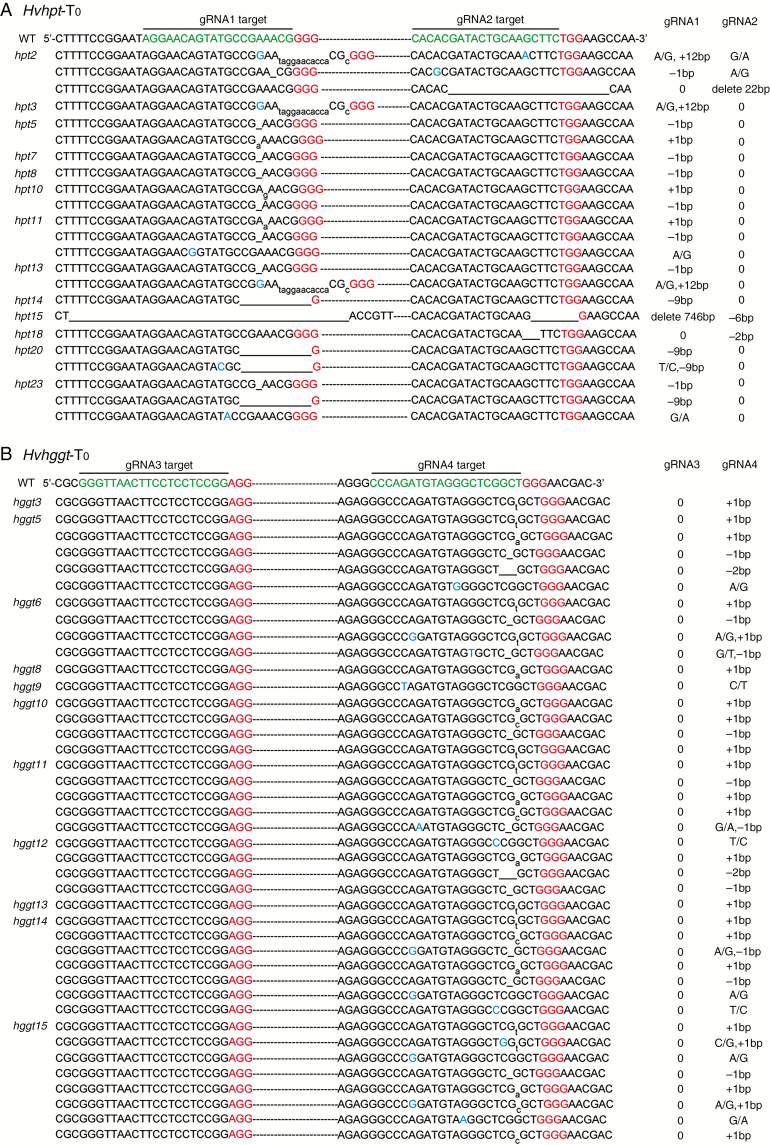
To examine targeted mutagenesis in T0 plants, the targeted protospacer regions of HvHPT and HvHGGT were amplified by PCR for DNA sequencing (10–15 amplicons per primary mutant plant; see Supplementary Data Table S3 for primer sequences), and the resulting sequences were aligned against the wild-type DNA sequence. The sequencing data revealed 13 mutant lines for Hvhpt and 11 for Hvhggt (Table S5). Mutagenesis efficiency among T0 regenerated plantlets was 50–65 % as a result of two simultaneously expressed gRNAs targeting each gene. In the PTG-HPT/Cas9 transgenic lines (Fig. 2A), site-specific indels (insertions and deletions) occurred at 0–7 bp upstream of the protospacer-adjacent motif (PAM) or at imprecise sites, thus eliminating PAMs or a genomic sequence far from PAMs. A fragment deletion of up to 746 bp at a single target site was also observed in line hpt15. Among the 13 Hvhpt mutant lines, targeted mutagenesis at gRNA1 target sites was much higher than that at gRNA2 sites (Fig. 2A), indicating wide variation in gRNA1 and gRNA2 mutagenic efficiency. In the PTG-HGGT/Cas9 primary transgenic lines (Fig. 2B), single-base substitutions or small indels (+1, −1 or −2 bp) were only observed at the gRNA4 target site. Notably, all of these small indels occurred specifically 3 bp upstream of the PAM. Among all T0 Hvhggt mutant plants, no mutation was detected at the gRNA3 target site (Fig. 2). In general, the mutations induced in the barley HvHPT and HvHGGT genes consisted predominantly of 1-bp indels.
Fig. 2.
Mutation types in primary transgenic plants revealed by DNA sequencing. (A) Mutant genotypes at gRNA1 and gRNA2 target sites (targeting HvHPT). (B) Mutant genotypes at gRNA3 and gRNA4 target sites (targeting HvHGGT). WT, wild-type; target sites are shown in green type; single-base substitutions are shown in blue type; the PAM motif is shown in red type; insertions are shown with lowercase letters (subscript); and deletions are indicated by underscoring.
Quantitative analysis of the T0 mutant plants showed that the mutation rate induced by gRNA4 (64.7 %) was much higher than those induced by gRNA1 (46.2 %) and gRNA2 (11.5 %) and that gRNA3 did not induce indels at the target site (Table 1). No homozygous mutations were identified in T0 mutant plants (Table 1). As heterozygous plants may also be chimeric, heterozygosity and chimerism cannot readily be distinguished in a given plant. Therefore, all of the primary mutant lines for Hvhpt and Hvhggt were categorized as heterozygous/chimeric. These results indicated a high percentage of heterozygous/chimeric mutants in the T0 generation and that the four gRNAs varied in genome-editing performance, suggesting that gRNA design is important for mutagenesis efficiency.
Table 1.
The mutagenesis frequencies of HvHPT and HvHGGT gene mutations in the T0 generation.
| Gene | gRNA ID | WT | Homozygous | Heterozygous/ chimeric |
|---|---|---|---|---|
| HvHPT | gRNA1 | 53.8 % | ND | 12 (46.2 %) |
| gRNA2 | 88.5 % | ND | 3 (11.5 %) | |
| HvHGGT | gRNA3 | 0 | ND | ND |
| gRNA4 | 35.3 % | ND | 11 (64.7 %) |
ND, not detected.
Characterization of chimeric lines in T0 transgenic plants and selection of ‘transgene-free’ homozygous barley mutants
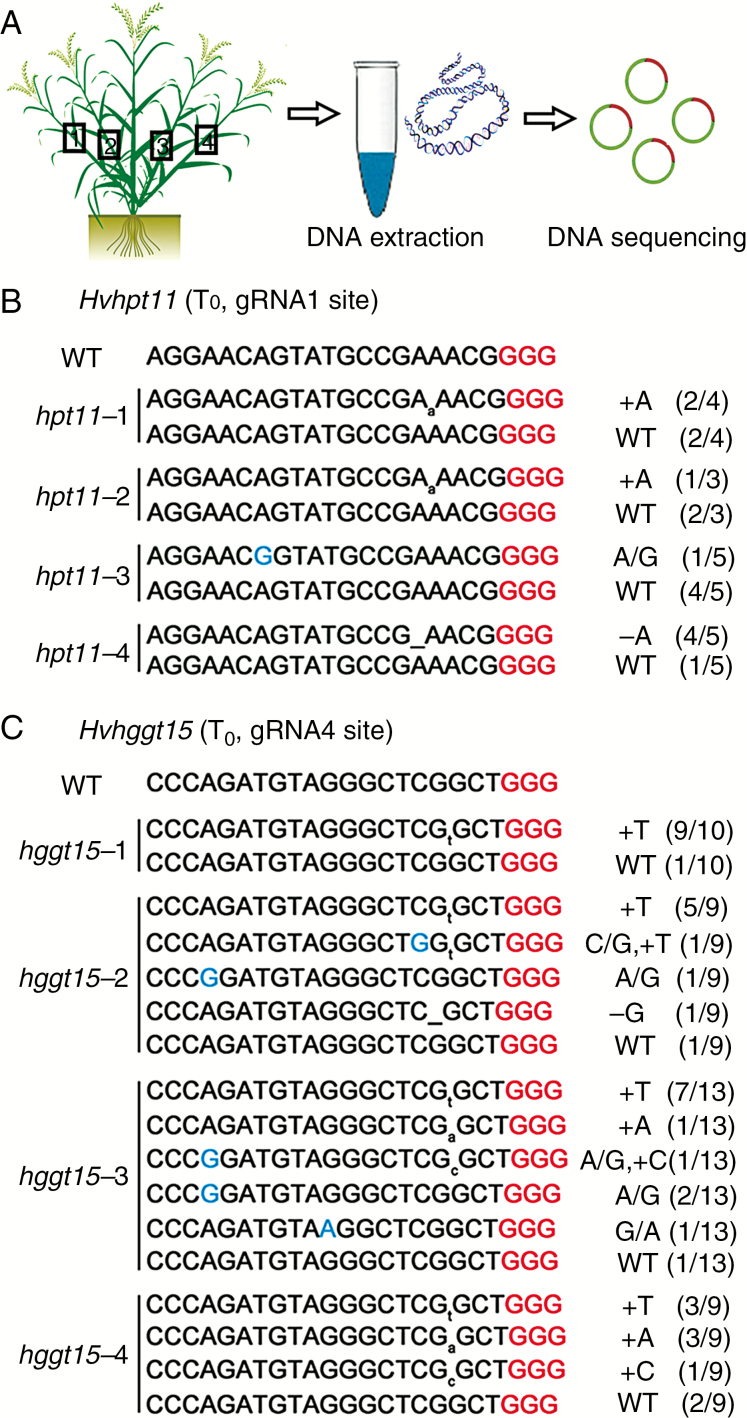
To investigate whether chimeras were present in a tiller or multiple tillers of a mutant line, genomic DNA from four different tillers of each T0 mutant plant was extracted, and the target sites were individually sequenced (Fig. 3A). The wild-type sequence was found in all tillers (Fig. 3B, C). In line hpt11, mutation types varied among the four tillers, but each tiller had only one mutation type (Fig. 3B). This result suggested the existence of chimeras among the different tillers. However, one to five different mutation types were detected in individual tillers of the line hggt15 (Fig. 3C), suggesting that this phenomenon could be present not only in different tillers but also in leaves from the same tiller. Among 13 hpt mutants and 11 hggt mutants, at least seven lines were chimeric (Fig. 2). Overall, more than half of the mutant plants were chimeric, and chimeras were found in the same tiller or different tillers.
Fig. 3.
Primary transgenic plants are mostly chimeric at the target sites of HvHPT or HvHGGT. (A) Extraction of genomic DNA from four different tillers of each mutant plant for sequencing. (B) Mutation patterns of four individual tillers detected in the T0 hpt11 mutant. (C) Mutation patterns of four individual tillers detected in the T0 hggt15 mutant. WT, wild-type; single-base substitutions are shown in blue type; the PAM motif is shown in red type; insertions are shown with lowercase letters (subscript); and deletions are indicated by underscoring.
Because chimeras were observed in T0 transgenic lines, the inheritance and consistency of targeted mutations were further analysed in the T1 generation. To assess whether the gRNA/Cas9 transgene was able to induce new mutations in progeny plants, we chose seedlings of the T1 generation that were derived from one tiller of a chimeric primary transgenic line (hggt15-3) (Fig. 3C). Most mutation types of the T0 plants were faithfully inherited in the T1 generation, such as a 1-bp (+A, +T or +C) insertion (Supplementary Data Fig. S2). However, not all mutation types were transmitted from the T0 to T1 generation, including base substitutions (A/G and G/A), which were not detected in the latter generation (Fig. S2). Notably, trans-generational editing occurred in T1 plants, and the presence of the gRNA/Cas9 transgene induced new mutation types at target sites, including 1- to 5-bp deletions (−G, −CG, −GCCG, −GGCTC), 2-bp insertions (+TT) and T/C substitutions, although no new mutations occurred in transgene-free plants (Fig. S2). Therefore, plants with the gRNA/Cas9 transgene displayed more mutation types compared with the gRNA/Cas9-free plants, suggesting that it is necessary to obtain transgene-free plants for fixed mutations.
To avoid possible new mutation types produced in gRNA/Cas9 transgenic plants, homozygous mutations for HvHPT or HvHGGT without any transgenic elements were screened in the T1 plants via PCR and DNA sequencing. Among at least 44 plants of each T1 line tested, 12–27 plants without the Cas9 transgene were obtained for each mutant line (Table 2). There were 14 Cas9-free homozygotes for hpt mutants, including seven hpt15 plants [large-fragment deletion (−746 bp)], five hpt14 plants [9-bp deletion (−CGAAACGGG)] and two hpt11 plants [1-bp indel (+G or −A)]. The number of Cas9-free homozygotes for hggt5, hggt10, hggt13 and hggt15 lines were 15, 14, 15 and seven, respectively (Table 2). Notably, all of the transgene-free hggt homozygotes displayed only a 1-bp indel (+T, +A, or −G). Therefore, ‘transgene-free’ homozygous Hvhpt and Hvhggt mutants with different mutation types were obtained in the T1 generation.
Table 2.
Selection and characterization of ‘transgene-free’ homozygous barley mutants
| T-DNA constructs | T1 mutant lines | No. of plants tested | No. of Cas9-free plants | No. of heterozygotes (Cas9-) | No. of homozygotes (Cas9-) | Mutation pattern of homozygotes (Cas9-) |
|---|---|---|---|---|---|---|
| PTG-HPT/Cas9 | hpt11 | 146 | 27 | 4 | 2 | +1 bp (+G); −1 bp (−A) |
| hpt14 | 54 | 12 | 7 | 5 | −9 bp (−CGAAACGGG) | |
| hpt15 | 48 | 21 | 13 | 7 | −746 bp (large-fragment deletion) | |
| PTG-HGGT/Cas9 | hggt5 | 52 | 15 | 0 | 15 | +1 bp (+A); +1 bp (+T); −1 bp (−G) |
| hggt10 | 50 | 16 | 2 | 14 | +1 bp (+A); −1 bp (−G); +1 bp (+A)/ −1 bp (−G) | |
| hggt13 | 44 | 15 | 0 | 15 | +1 bp (+T) | |
| hggt15 | 54 | 17 | 4 | 7 | +1 bp (+A) |
Amino acid sequence analysis and phenotypic observation of Hvhpt and Hvhggt homozygous mutants
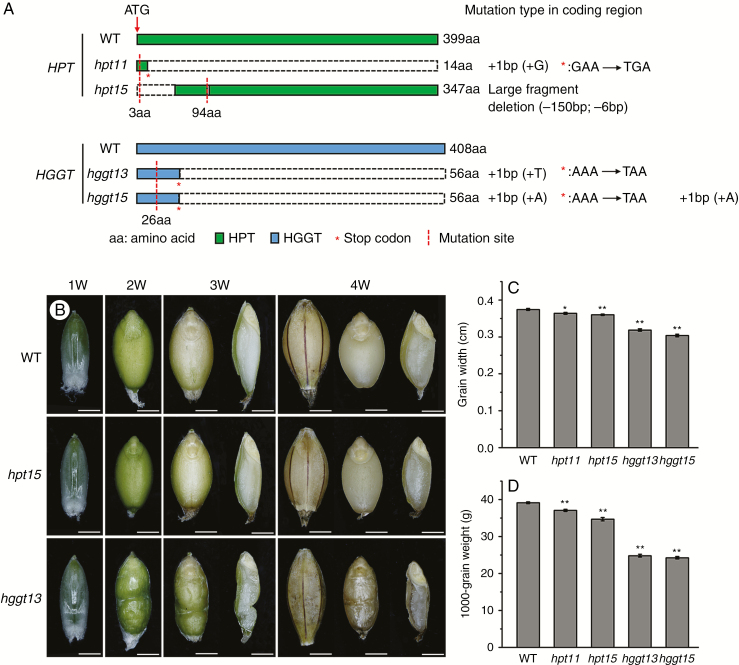
DNA sequencing analysis showed that hpt11 carried a single nucleotide insertion, leading to a nonsense mutation at amino acid (aa) 14 (GAA to TGA; Glu to stop) of the 399-residue polypeptide (Fig. 4A). For hpt15, a 50-aa deletion occurred at amino acid position 3 and a 2-aa deletion occurred at amino acid 94 (Fig. 4A). Regarding hggt13 and hggt15, a single nucleotide insertion of A or T resulted in a nonsense mutation at amino acid 56 (AAA to TAA, Lys to stop) of the 408-residue polypeptide (Fig. 4A). These deletion, frameshift or in-frame mutations caused truncation near the N-terminus or led to missense mutations in the respective peptides. These two transgene-free homozygous hpt lines (hpt11 and hpt15) and two hggt mutant lines (hggt13 and hggt15) were used for further study.
Fig. 4.
Amino acid sequence analysis and grain phenotypic observation of barley hpt and hggt mutants. (A) Amino acid sequence analysis of barley hpt and hggt mutants. (B) Developing grains of the wild-type (WT) and mutants (hvhpt15 and hvhggt13). (C) Grain width (cm). (D) Thousand-grain weight (g). Phenotypic observation of grain development was performed at 1, 2, 3 and 4 weeks (W) after pollination. Whole grains or dehusked grains and their longitudinal section were imaged with a trinocular stereo microscope. Mature grains of transgene-free, homozygous T2 mutant lines were used for the analysis of grain width (with ImageJ software) and thousand-grain weight. Means and s.e. of at least three replicates are presented, with n > 45 for each repeat. Asterisks indicate a significant difference between the wild-type and mutant lines at *P < 0.05 or **P < 0.01, as determined by Student’s t-tests. Scale bars = 2.6 mm.
Phenotypic observation of T2 mutant plants revealed that the developing grains of the hggt13 homozygous mutant possessed a severely shrunken phenotype, whereas the grain size of the hpt15 mutant was slightly reduced in comparison to that of the wild-type (Fig. 4B). Further analysis of the mature grains showed that the average grain width decreased by 3–4 % in hpt mutants and by 15–19 % in hggt mutants compared to that of the wild-type (Fig. 4C). Moreover, 1000-grain weights were decreased by 5–11 % in hpt mutants and by 37–38 % in hggt mutants (Fig. 4D). Therefore, both homozygous mutations, particularly the Hvhggt mutation, significantly decreased grain size and weight, indicating a significant role of HvHPT and HvHGGT in barley grain development.
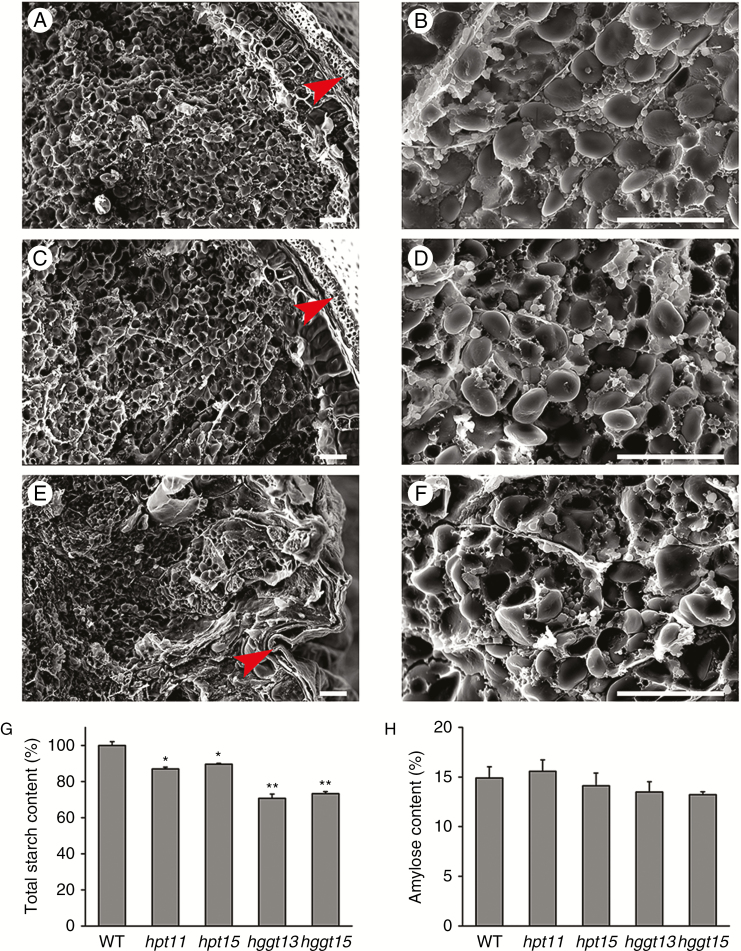
Additionally, scanning electron microscopy of cross-sections of the grains showed that the grain husk of hggt mutants was shrunken in comparison with that of the wild-type and hpt (Fig. 5A, C, E). Furthermore, the starch granules of hggt mutants were irregularly arranged compared with that of the wild-type and hpt (Fig. 5B, D, F). Because grains consist mainly of starch, we presumed that the shrunken kernel phenotype of the Hvhggt mutant might be related to starch accumulation in barley. To investigate the effects of HvHPT and HvHGGT mutation on starch levels, the contents of total starch and amylose were determined for the mature barley grains. The results showed that the total starch content decreased significantly by 10–13 % in hpt mutants and by 27–30 % in hggt mutants compared with that of the wild-type (Fig. 5G). However, the ratio of amylose to total starch content was not significantly different between that of the wild-type and the mutants (Fig 5H). The above results suggested that HvHGGT is involved in starch accumulation in barley grains.
Fig. 5.
Morphology of cross-section of grains and the content of total starch and amylose in barley. Scanning electron micrographs of barley grains in the wild-type (WT) (A and B), hpt (C and D), and hggt (E and F) mutants. (G) Total starch content (%). (H) Amylose content (%). Mature grains of the WT, and hpt and hggt mutants were used for microscopy and starch content measurement. Means and s.e. of at least three replicates are presented. Asterisks indicate a significant difference between the WT and mutants at *P < 0.05 or **P < 0.01, as determined by Student’s t-tests. Red arrowheads, grain husk. Scale bars = 50 µm.
CRISPR/Cas9-induced mutations significantly altered vitamin E contents in mature grains of T2 homozygous mutants
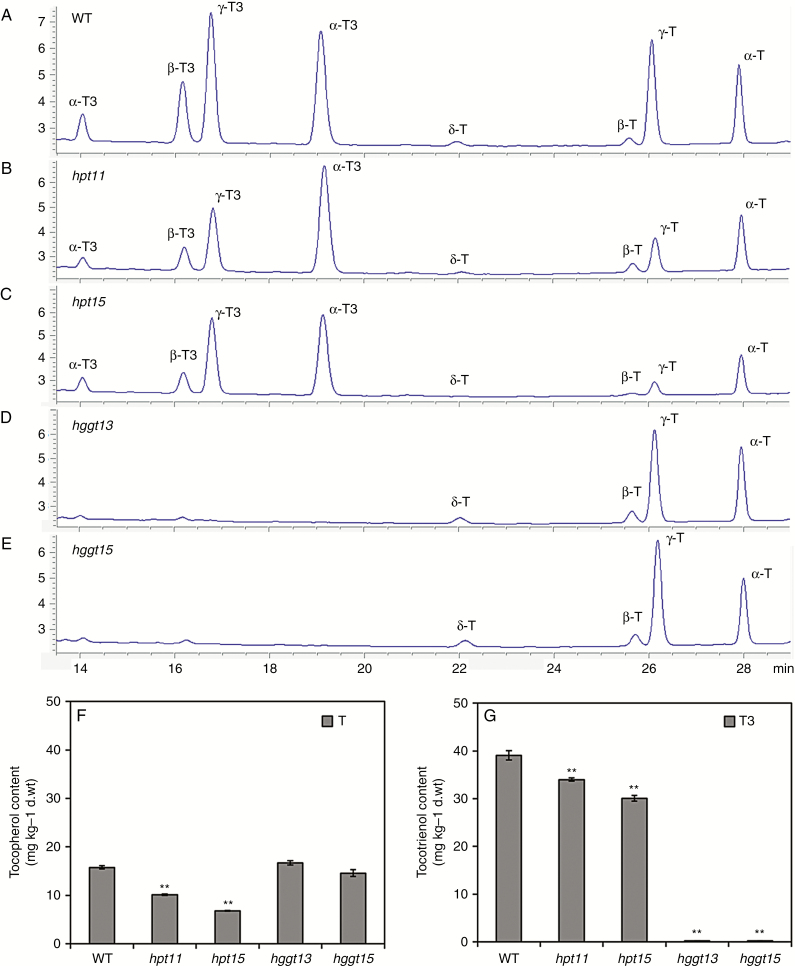
To investigate whether CRISPR/Cas9-mediated targeted mutagenesis of HvHPT and HvHGGT affected the vitamin E metabolic pathway, tocochromanols were extracted from the T2 mature grains of homozygous mutants and analysed using HPLC for the contents of eight vitamin E isomers (Fig. 6A-E). As shown in Fig. 6A, the peaks of the eight isomers varied greatly in wild-type barley (the peaks for γ and α isomers were predominant, whereas the δ and β peaks were much smaller). Peaks in the hpt11 and hpt15 mutant lines were similar (Fig. 6B, C). The peaks for γ-T and α-T were smaller than those in the wild-type, and a modest decline was observed for δ-T3, β-T3, γ-T3 and α-T3 peaks. Notably, peaks for the four isomers of tocotrienol were almost undetectable in both the hggt13 and the hggt15 mutant lines, whereas peaks for the four isomers of tocopherol were similar to those in the wild-type (Fig. 6A, D, E).
Fig. 6.
Tocopherol and tocotrienol levels in mature grains of Hvhpt and Hvhggt T2 mutant lines. (A–E) HPLC profiles of tocopherol and tocotrienol isomers in the wild-type, and hpt and hggt mutants. (F) The content of tocopherol (T). (G) The content of tocotrienol (T3). Mature grains of transgene-free homozygous T2 mutant lines were used for measurements and 50 mg of grain powder was used in each replicate. Means and s.e. of at least three replicates are presented. Asterisks indicate a significant difference between the wild-type and mutant lines at *P < 0.05 or **P < 0.01, as determined by Student’s t-tests.
Quantitative analysis confirmed that α-T3 was the predominant form of vitamin E in wild-type barley, followed by γ-T3, α-T, γ-T and β-T3 and that the contents of the β-T and δ isoforms were very low (Supplementary Data Table S6). Mutations in HvHPT significantly decreased the tocopherol content by 36–57 % and tocotrienol content by 13–23 % (Fig. 6F, G). Isoforms α and γ were still the major isoforms of vitamin E in lines hpt11 and hpt15, and the contents of α-T, γ-T and γ-T3 were reduced by 25–43, 68–88 and 32–50 %, respectively (Table S6). In contrast, no significant difference was observed for α-T and γ-T contents in HvHGGT mutant lines compared with that of the wild-type (Fig. 6F; Table S6). Because α-T and γ-T account for 95 % of the total tocopherols, we found no significant difference in total tocopherol content between wild-type and hggt mutants. Nonetheless, the tocotrienol content did decrease to almost zero in the hggt mutant lines (Fig. 6G), indicating that HvHGGT is the gene responsible for the synthesis of tocotrienols in barley grains. Indeed, the mutation in HvHGGT almost completely blocked tocotrienol biosynthesis but did not affect tocopherol biosynthesis. In contrast, the mutation in HvHPT did not completely block tocopherol biosynthesis in barley grains. One possible explanation is the existence of an HPT homologue or the existence of other pathway genes in the barley genome that contribute to tocopherol biosynthesis.
Phylogenetic and functional analyses of HPT and HGGT homologous genes in barley
To determine whether an unknown homologue of HvHPT for tocopherol biosynthesis exists in the barley genome, we searched the barley genome database and identified a coding sequence (HORVU2Hr1G117600) that shared 79.8 % nucleotide identity (73.3 % amino acid identity) with HvHPT (Supplementary Data Fig. S1B). Furthermore, the translated amino acid sequence contained a UbiA prenyltransferase domain that is conserved in both HPT and HGGT proteins (Fig. S1). Phylogenetic analysis of the amino acid sequences indicated that HPT and HGGT belonged to two specific subgroups with distinct clustering (Fig. S3). Among the eight species examined, HPTs were found in both monocots and dicots, whereas HGGTs were present only in the monocots assessed. The sequence similarity of barley HPT to that of wheat and oat HPT was higher than that of the Brachypodium, Sorghum, maize and rice genes, although the deduced amino acid sequence of HORVU2Hr1G117600 was closest to that of the rice HPT among monocots. We isolated the full-length cDNA of HORVU2Hr1G117600 via reverse transcription from mRNAs extracted from the barley grains (14 d after pollination). To test whether the possible HPT homologue (HORVU2Hr1G117600) has a potential role in tocopherol biosynthesis, we performed transient overexpression of HORVU2Hr1G117600 in tobacco leaves by agro-infiltration. HPLC analysis showed that α-T and γ-T were the major tocopherol isoforms of vitamin E in tobacco leaves, and overexpression of HORVU2Hr1G117600 significantly increased the production of γ- and δ-tocopherol isoforms (Fig. S4). This result suggested that the HPT homologue (HORVU2Hr1G117600) may function in tocopherol biosynthesis in barley grains.
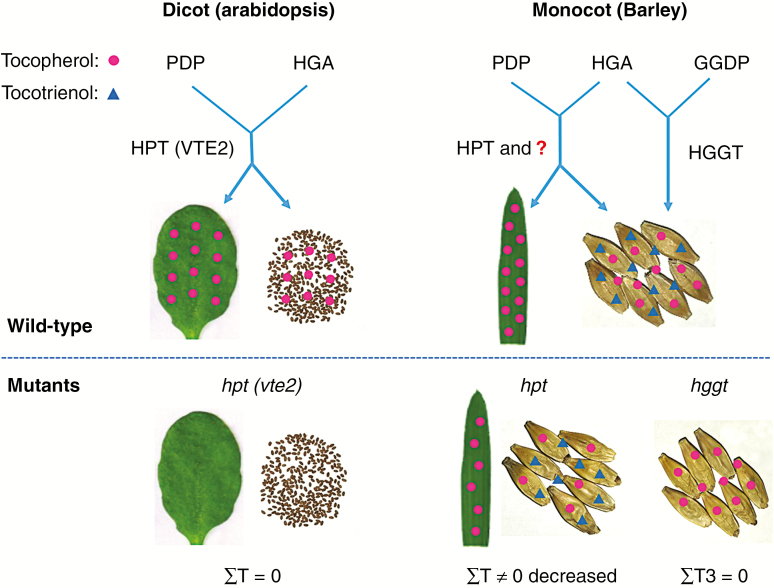
To further determine whether HORVU2Hr1G117600 or HGGT contributes to tocopherol biosynthesis in barley, we examined their expression in the leaves and grains of hpt mutants. The results showed that the expression of HORVU2Hr1G117600 and HGGT was only found in grains, but not in leaves of the wild-type and mutant lines (Supplementary Data Fig. S5A). Furthermore, analysis of leaf tocochromanols showed a significant decrease in the total tocopherol content in line hpt15 of 41 % of that in the wild-type, and no tocotrienols were detected in hpt15 or the wild-type (Fig. S5B). Regardless, tocopherols still accumulated in hvhpt mutants when HvHGGT and HORVU2Hr1G117600 were not expressed in leaves, suggesting that they may not contribute to vitamin E biosynthesis in barley leaves. Taken this evidence together, HORVU2Hr1G117600 may share HvHPT function in barley grains, but not in leaves, implying that other biosynthesis pathway-related genes might be involved in the accumulation of tocopherols in barley (Fig. 7).
Fig. 7.
Vitamin E biosynthesis in barley and Arabidopsis. HPT (homogentisate phytyltransferase) and HGGT (homogentisate geranylgeranyl transferase) are the committed-step enzymes for the biosynthesis of tocopherols and tocotrienols, respectively. The question mark indicates an HPT homologue or other vitamin E biosynthesis pathway-related genes that may participate in tocopherol production in barley and other monocots. HGA, homogentisate; PDP, phytyl diphosphate; GGDP, geranylgeranyl diphosphate; T, tocopherol; T3, tocotrienol; hpt, HPT mutant; vte2, HPT mutant in Arabidopsis; hggt, HGGT mutant. Red circles represent tocopherols; blue triangles represent tocotrienols.
DISCUSSION
Targeted mutagenesis of HPT and HGGT genes in barley via CRISPR/Cas9-mediated genome editing
Barley is a transformation-recalcitrant species, for which genome editing is often an inefficient, laborious and time-consuming process. To improve editing efficiency, we used the PTG strategy (Xie et al., 2015) and employed two gRNAs to target HvHPT or HvHGGT to increase the success rate. Mutagenesis efficiency among T0 regenerated plantlets was 50–65 % (Supplementary Data Table S5), which is comparable to that of a previous study on barley using the Agrobacterium-mediated transfer of the Cas9 construct, with a frequency of at least 78 % of Cas9-induced mutations in the primary generation (Kapusi et al., 2017), but it is higher than the other three studies in barley, which yielded mutation rates of only 10–23, 44 and 14–25 %, respectively (Lawrenson et al., 2015; Holme et al., 2017; Yang et al., 2020). Another important utility of two gRNAs for targeting one gene is to achieve chromosomal-fragment deletion between two gRNA target sites, which has been shown in barley (Kapusi et al., 2017). In our study, the targeted mutation efficiency of the four gRNAs we designed varied greatly (Table 1). For HPT, because the mutation efficiency of gRNA2 was much lower than that of gRNA1, more transgenic lines were needed to increase the odds of achieving fragment deletion. For HGGT, gRNA3 caused no indels at the target site. Recently, a transient assay system using protoplasts was used to identify the most effective gRNA candidate or gRNA combinations before transformation (Liang et al., 2017), which would further contribute to improving editing efficiency for crop species, such as maize, barley and wheat.
Stable inheritance of target gene mutations is a major advantage of utilizing the CRISPR/Cas9 system for genome editing and precision breeding. In agreement with the previous studies, we noticed that small indels were the most frequent type of mutation induced by CRISPR/Cas9, and most of the mutations found in T0 barley plants were inherited by the T1 generation (Fig. 2; Supplementary Data Fig. S2) (Feng et al., 2014; Zhang et al., 2014; Lawrenson et al., 2015). However, a few mutation types, such as the base substitution (A/G) that occurred in the T0 generation, were absent in the T1 generation (Fig. S2). It is possible that this type of mutation existed in somatic cells, which do not contribute to germ-line development. Because more than half of the T0 mutant plants were chimeric (Figs 2, 3), the T1 segregation patterns derived from the T0 chimeras will be diverse and unpredictable. In the T0 generation, the CRISPR/Cas9-induced targeted gene modification events may occur relatively late in callus development and/or present multiple cells with different genetic transformation events incorporated during the production of a single transformant. In contrast, transgenic plants with only one type of mutation probably resulting from a single genetic transformation and mutation event occurred in a single embryogenic barley cell.
Additionally, plants with an active gRNA/Cas9 transgene might generate more mutation types than transgene-free plants in subsequent generations (Supplementary Data Fig. S2). Such a situation confirms that the CRISPR/Cas9 system has the sustaining ability to continue gene editing as long as the target site is the wild-type (Feng et al., 2014; Zhang et al., 2014). Usually, gRNA/Cas9 DNA is randomly integrated into the plant genome after entering the nucleus, which may increase the chance of off-target mutation (Lawrenson et al., 2015), induce gene inactivation or instability, and trigger regulatory concerns regarding genetically modified organisms (Jones, 2015). Therefore, it is necessary to remove the gRNA/Cas9 transgene to fix the targeted mutation and to generate transgene-free plants for breeding purposes. Fortunately, exogenous elements, such as single gRNA, Cas9 and the associated selectable marker can be readily removed in later generations via genetic segregation through selfing or backcrossing. Thus far, the CRISPR/Cas9 system has been applied to a variety of plant species to generate transgene-free plants, such as rice (Li et al., 2016; Shen et al., 2018), wheat (Liang et al., 2017) and barley (Lawrenson et al., 2015; Holme et al., 2017; Kapusi et al., 2017; Yang et al., 2020) among others. In the present study, we successfully performed targeted mutagenesis of two committed genes in vitamin E biosynthesis in barley via CRISPR/Cas9-mediated genome editing. Transgene-free homozygous Hvhpt and Hvhggt mutants could be utilized to understand the genetic control of vitamin E composition in monocots.
Distinctive functions of HvHPT and HvHGGT in barley vitamin E biosynthesis
In the vitamin E biosynthetic pathway, HPT catalyses the committed step of tocopherol biosynthesis to produce MPBQ, and HGGT catalyses tocotrienol biosynthesis to produce MGGBQ (Schneider, 2005; Chen et al., 2006). Tocopherol biosynthesis has been characterized in the dicot Arabidopsis thaliana. Because of the absence of a tocotrienol pathway in Arabidopsis, mutations in HPT (VTE2) result in total tocopherol deficiency in all tissues, as well as reduced seed longevity (Sattler et al., 2004; Yang et al., 2011). In contrast, studies regarding the vitamin E biosynthetic pathway in monocots are extremely limited because of the lack of functional mutants. Our results showed that the HvHGGT knockout resulted in undetectable levels of tocotrienols in barley grains without affecting the tocopherol content (Fig. 6), indicating that HvHGGT encodes the only committed enzyme that controls the step for GGDP condensation with HGA to produce tocotrienols in barley. Furthermore, the developing grains of hggt mutants exhibited a shrunken phenotype and low total starch content (Figs 4, 5), implying that the HvHGGT mutation may also influence starch biosynthesis during grain development. In comparison, the targeted mutation of HvHPT significantly reduced the content of both tocopherol and tocotrienol (Fig. 6). In Arabidopsis, a mutation to the HPT (VTE2) gene results in total tocopherol deficiency in seeds and seedlings, and the vte2 mutants display severe seedling growth defects during germination (Sattler et al., 2004). As major lipid-soluble antioxidants in chloroplasts, tocopherols play specific functions in the plant redox-active pathway (Sattler et al., 2004). Additionally, tocopherol is required for the plant response to most abiotic stresses, such as high-intensity light stress (Havaux et al., 2005; Maeda and DellaPenna, 2007). In barley, the reduced tocopherol level in the hpt mutants may lead to a partially disrupted redox-active pathway and to tolerance to abiotic stresses, which might indirectly influence the accumulation of tocotrienol during grain development.
Because the knockout of HvHPT reduced the tocopherol content ~50 % compared to that of the wild-type grains (Fig. 6), HvHPT is partly responsible for tocopherol biosynthesis in barley, implying that potential HvHPT homologues or other genes might be involved in tocopherol production in barley. In oats, five different homologues/paralogues of the HPT gene were expressed and two of them were identified as highly correlated with tocopherol accumulation by deep sequencing and orthology-guided assembly (Gutierrez-Gonzalez et al., 2013; Gutierrez-Gonzalez and Garvin, 2016). Based on the barley genome database, a possible HvHPT homologue, HORVU2Hr1G117600, was found and it was specifically expressed in the barley grains (Supplementary Data Fig. S5). Transient overexpression of HORVU2Hr1G117600 in tobacco leaves significantly increased the production of γ- and δ-tocopherol isoforms (Fig. S4), suggesting that HORVU2Hr1G117600 may share the HvHPT function. This result may partly explain why the targeted mutation in HvHPT did not eliminate tocopherol production in barley grains.
Although HORVU2Hr1G117600 was not expressed in leaves, tocopherol still accumulated in considerable amounts in the leaves of hpt mutants, suggesting that HORVU2Hr1G117600 may not contribute to tocopherol biosynthesis in barley leaves. A recent study showed that many novel genes involved in fatty acid metabolism, chlorophyll metabolism and chloroplast function were potentially involved in the variation of tocopherol content in maize kernels (Wang et al., 2018). Interestingly, in a joint linkage and genome-wide association study of 5000 US maize lines revealed a lack of association between the HPT locus and tocopherol traits (Diepenbrock et al., 2017). In contrast, two novel large-effect loci (chlorophyll biosynthetic genes por1 and por2) were identified as strongly correlated to tocopherol content in maize grains. These two genes encode homologues of protochlorophyllide reductase, which is involved in a highly regulated step in chlorophyll biosynthesis. Other studies have also noted that tocopherol biosynthesis is related to the chlorophyll biosynthesis pathway (Valentin et al., 2006; Zhang et al., 2015; Wang et al., 2018). Regarding the potential role of HORVU2Hr1G117600 on tocopherol biosynthesis in grains but not in leaves, we propose that other non-tocopherol pathway-related genes might be involved in the accumulation of tocopherols in barley.
HGGT is predicted to be functionally divergent from HPT, which is conserved in both monocot and dicot species (Cahoon et al., 2003). Overexpression of monocot HGGT in Arabidopsis can functionally replace HPT in the biosynthesis of vitamin E (Yang et al., 2011). However, we detected no HvHGGT expression in barley leaves, even though tocopherol was still synthesized in considerable amounts in hpt mutant lines (Supplementary Data Fig. S5). The results suggested that HvHGGT cannot replace the tocopherol biosynthesis function of HvHPT in barley. Our study functionally validated that HvHGGT is the only gene committed to produce tocotrienols, whereas HvHPT is partly responsible for tocopherol biosynthesis in barley. Hence, the vitamin E biosynthetic pathway has diverged between dicots and monocots, and the HPT homologue (HORVU2Hr1G117600) or other biosynthesis pathway-related genes may participate in the metabolic fluxes of tocopherol accumulation in barley and other monocots (Fig. 7). Therefore, this study expands our understanding of the vitamin E biosynthesis pathway in agronomically important cereal grains.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: Comparison of amino acid sequences for barley genes. Figure S2: The trans-generational influence of Cas9 on the mutation type in T1 transgenic barley plants. Figure S3: Phylogenetic relationship among the amino acid sequences of HPT and HGGT of barley and seven other plant species. Figure S4: Transient overexpression of HORVU2Hr1G117600 in tobacco leaves by agro-infiltration. Figure S5: Semi-quantitative RT-PCR analysis of genes and the content of tocochromanols in leaves and grains. Table S1: Primers used for making PTG editing constructs. Table S2: PTG sequences in the two editing constructs. Table S3: Primers for gene amplification. Table S4: Primers for semi-quantitative RT-PCR of genes. Table S5: Summary of results after barley genetic transformation. Table S6: The content of tocochromanol isomers in HvHPT and HvHGGT T2 grains of transgenic lines.
ACKNOWLEDGEMENTS
We thank Xinhang Jiang (College of Life Sciences, Zhejiang University, China) for his technical assistance with the HPLC analyses of tocopherols and tocotrienols of barley grains.
FUNDING
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31971932 and 31771776), China Agriculture Research System (CARS-05-05A) and the Science Foundation of Zhejiang Province (Grant No. LGN18C130001).
LITERATURE CITED
- Aggarwal BB, Sundaram C, Prasad S, Kannappan R. 2010. Tocotrienols, the vitamin E of the 21st century: its potential against cancer and other chronic diseases. Biochemical Pharmacology 80: 1613–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AAM, Lampi AM, Nystrom L, et al. 2008. Phytochemical and dietary fiber components in barley varieties in the HEALTHGRAIN diversity screen. Journal of Agricultural and Food Chemistry 56: 9767–9776. [DOI] [PubMed] [Google Scholar]
- Bramley PM, Elmadfa I, Kafatos A, et al. 2000. Vitamin E. Journal of the Science of Food and Agriculture 80: 913–938. [Google Scholar]
- Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ. 2003. Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nature Biotechnology 21: 1082–1087. [DOI] [PubMed] [Google Scholar]
- Cai S, Han Z, Huang Y, Hu H, Dai F, Zhang G. 2016. Identification of quantitative trait loci for the phenolic acid contents and their association with agronomic traits in Tibetan wild barley. Journal of Agricultural and Food Chemistry 64: 980–987. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu C, Shi B, et al. 2017. Overexpression of HvHGGT enhances tocotrienol levels and antioxidant activity in barley. Journal of Agricultural and Food Chemistry 65: 5181–5187. [DOI] [PubMed] [Google Scholar]
- Chen S, Li H, Liu G. 2006. Progress of vitamin E metabolic engineering in plants. Transgenic Research 15: 655–665. [DOI] [PubMed] [Google Scholar]
- Chin KY, Pang KL, Soelaiman IN. 2016. Tocotrienol and its role in chronic diseases. Advances in Experimental Medicine and Biology 928: 97–130. [DOI] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D. 2003. Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiology 131: 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hu X, Liang G, et al. 2020. Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9. Plant Biotechnology Journal 10.1111/pbi.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D. 2005. A decade of progress in understanding vitamin E synthesis in plants. Journal of Plant Physiology 162: 729–737. [DOI] [PubMed] [Google Scholar]
- Diepenbrock CH, Kandianis CB, Lipka AE, et al. 2017. Novel loci underlie natural variation in vitamin E levels in maize grain. The Plant Cell 29: 2374–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Munne-Bosch S. 2010. Tocochromanol functions in plants: antioxidation and beyond. Journal of Experimental Botany 61: 1549–1566. [DOI] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, et al. 2014. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 111: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland LU, Magallanes-Lundback M, Hemming C, et al. 2006. Genetic basis for natural variation in seed vitamin E levels in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 103: 18834–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenstein N, Frank J. 2012. Rapid baseline-separation of all eight tocopherols and tocotrienols by reversed-phase liquid-chromatography with a solid-core pentafluorophenyl column and their sensitive quantification in plasma and liver. Journal of Chromatography A 1243: 39–46. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Gonzalez JJ, Garvin DF. 2016. Subgenome-specific assembly of vitamin E biosynthesis genes and expression patterns during seed development provide insight into the evolution of oat genome. Plant Biotechnology Journal 14: 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Gonzalez JJ, Tu ZJ, Garvin DF. 2013. Analysis and annotation of the hexaploid oat seed transcriptome. BMC Genomics 14: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood WA. 2014. A protocol for high-throughput Agrobacterium-mediated barley transformation. Methods in Molecular Biology 1099: 251–260. [DOI] [PubMed] [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dörmann P. 2005. Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. The Plant Cell 17: 3451–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme IB, Wendt T, Gil-Humanes J, et al. 2017. Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Molecular Biology 95: 111–121. [DOI] [PubMed] [Google Scholar]
- Horvath G, Wessjohann L, Bigirimana J, et al. 2006. Accumulation of tocopherols and tocotrienols during seed development of grape (Vitis vinifera L. cv. Albert Lavallee). Plant Physiology and Biochemistry 44: 724–731. [DOI] [PubMed] [Google Scholar]
- Jones HD. 2015. Regulatory uncertainty over genome editing. Nature Plants 1: 14011. [DOI] [PubMed] [Google Scholar]
- Kapusi E, Corcuera-Gómez M, Melnik S, Stoger E. 2017. Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Frontiers in Plant Science 8: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanandaa B, Qi Q, Hao M, et al. 2005. Metabolically engineered oilseed crops with enhanced seed tocopherol. Metabolic Engineering 7: 384–400. [DOI] [PubMed] [Google Scholar]
- Keller Y, Bouvier F, d’Harlingue A, Camara B. 1998. Metabolic compartmentation of plastid prenyllipid biosynthesis–evidence for the involvement of a multifunctional geranylgeranyl reductase. European Journal of Biochemistry 251: 413–417. [DOI] [PubMed] [Google Scholar]
- Kim YH, Lee YY, Kim YH, et al. 2011. Antioxidant activity and inhibition of lipid peroxidation in germinating seeds of transgenic soybean expressing OsHGGT. Journal of Agricultural and Food Chemistry 59: 584–591. [DOI] [PubMed] [Google Scholar]
- Konda AR, Nazarenus TJ, Nguyen H, et al. 2020. Metabolic engineering of soybean seeds for enhanced vitamin E tocochromanol content and effects on oil antioxidant properties in polyunsaturated fatty acid-rich germplasm. Metabolic Engineering 57: 63–73. [DOI] [PubMed] [Google Scholar]
- Lawrenson T, Shorinola O, Stacey N, et al. 2015. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biology 16: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. 2014. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Molecular Plant 7: 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li J, Meng X, Zong Y, et al. 2016. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nature Plants 2: 16139. [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, et al. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nature Communications 8: 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak VI, Semchuk NM. 2012. Tocopherol biosynthesis: chemistry, regulation and effects of environmental factors. Acta Physiologiae Plantarum 34: 1607–1628. [Google Scholar]
- Maeda H, DellaPenna D. 2007. Tocopherol functions in photosynthetic organisms. Current Opinion in Plant Biology 10: 260–265. [DOI] [PubMed] [Google Scholar]
- Maeda H, Sakuragi Y, Bryant DA, Dellapenna D. 2005. Tocopherols protect Synechocystis sp. strain PCC 6803 from lipid peroxidation. Plant Physiology 138: 1422–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. 2004. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. The Plant Cell 16: 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B, Weiss JD, Wong YH, et al. 2002. Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiology 129: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. 2005. Chemistry and biology of vitamin E. Molecular Nutrition & Food Research 49: 7–30. [DOI] [PubMed] [Google Scholar]
- Sen CK, Khanna S, Roy S. 2007. Tocotrienols in health and disease: the other half of the natural vitamin E family. Molecular Aspects of Medicine 28: 692–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wang C, Fu Y, et al. 2018. QTL editing confers opposing yield performance in different rice varieties. Journal of Integrative Plant Biology 60: 89–93. [DOI] [PubMed] [Google Scholar]
- Valentin HE, Lincoln K, Moshiri F, et al. 2006. The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. The Plant Cell 18: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xu S, Fan Y, et al. 2018. Beyond pathways: genetic dissection of tocopherol content in maize kernels by combining linkage and association analyses. Plant Biotechnology Journal 16: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences of the United States of America 112: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zhong X, Li Q, et al. 2020. Mutation of the d-hordein gene by RNA-guided Cas9 targeted editing reducing the grain size and changing grain compositions in barley. Food Chemistry 311: 125892. [DOI] [PubMed] [Google Scholar]
- Yang W, Cahoon RE, Hunter SC, et al. 2011. Vitamin E biosynthesis: functional characterization of the monocot homogentisate geranylgeranyl transferase. The Plant Journal: for Cell and Molecular Biology 65: 206–217. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang W, Ren G, et al. 2015. Chlorophyll synthase under epigenetic surveillance is critical for vitamin E synthesis, and altered expression affects tocopherol levels in Arabidopsis. Plant Physiology 168: 1503–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, et al. 2014. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnology Journal 12: 797–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.