Summary
Hand-foot-and-mouth disease is a common childhood illness primarily caused by coxsackievirus A16 (CVA16), for which there are no current vaccines or treatments. We identify three CVA16-specific neutralizing monoclonal antibodies (nAbs) with therapeutic potential, 18A7, 14B10, and NA9D7. We present atomic structures of these nAbs bound to all three viral particle forms --the mature virion, A-particle and empty particle-- and show that each Fab can simultaneously occupy the mature virion. Additionally, 14B10 or NA9D7 provide 100% protection against lethal CVA16 infection in a neonatal mouse model. 18A7 binds to a non-conserved epitope present in all three particles, whereas 14B10 and NA9D7 recognize broad protective epitopes but only bind the mature virion. NA9D7 targets an immunodominant site, which may overlap the receptor binding site. These findings indicate that CVA16 vaccines should be based on mature virions and that these antibodies could be used to discriminate optimal virion-based immunogens.
Keywords: coxsackievirus A16, cryo-EM structure, therapeutic antibody, vaccine design
In Brief
He et al. describe a variety of atomic structures for three forms of coxsachievirus A16 complexed with three distinct neutralizing mAbs, and map the neutralization sites. They suggest that the mature virion is the optimal immunogen for vaccine development and design an immune assay to specifically quantify such virion state.
Graphical Abstract
Introduction
The coxsackievirus A16 (CVA16), a non-enveloped, single-stranded RNA picornavirus belonging to the Picornaviridae family, is the dominant pathogen of hand, foot, and mouth disease (HFMD) inflicting infants and younger children worldwide(Fields et al., 2007). Since the first outbreak of CVA16-associated HFMD in Canada in 1957(Robinson et al., 1958), CVA16 infections have spread to Asia, Africa, Europe, and South America, where severe complications and fatal cases have been reported(CHANG et al., 1999; Wang et al., 2004). Furthermore, CVA16 can co-circulate and co-infect with other enteroviruses such as enterovirus 71 (EV71), another major pathogen of HFMD, leading to serious clinical outcomes and viral genetic recombination(Liu et al., 2014; Mao et al., 2014). Although effective inactivated EV71 vaccine has been recently marketed, it cannot provide cross-protection against CVA16 infection(Li et al., 2016; Li et al., 2014; Zhu et al., 2014; Zhu et al., 2013), and no antiviral therapy or vaccine is available for CVA16. This underscores the necessity and urgency to develop an effective treatment for CVA16 infection.
Picornaviruses generally undergo numerous structural transitions during their life cycle; several different particle types—procapsid, mature virion, A-particles and empty particles—have been isolated and well-characterized. These particles are different in component, structure, and immunogenicity, and therefore, vaccine development needs to consider the particulate form that would act best as an effective vaccine immunogen(Baggen et al., 2018; Ren et al., 2013; Ren et al., 2015; Wang et al., 2012; Xu et al., 2017; Zheng et al., 2019; Zhu et al., 2018a; Zhu et al., 2018c). For poliovirus, two distinct antigenic forms exist, designated C-antigens and D-antigens. The C-antigen is found on A-particles and empty particles, whereas the D-antigen is expressed on the mature virion and is regarded as a more potent immunogen for vaccine development(Ferguson et al., 1993). Indeed, previous immunological characterizations have shown that the CVA16 mature virion can induce a higher neutralizing antibody titer than the empty particle(Chong et al., 2012), suggesting that it may contain the major immunodominant epitope(s) and that its abundant content will influence the effectiveness of a CVA16 vaccine.
Vaccines usually prevent virus infection by inducing high-titers of potent neutralizing antibodies (nAbs); antibody epitopes on the virus, in turn, serve as structure templates for the rational design or selection of appropriate vaccine immunogens. The enterovirus must transition from a mature virion to an A-particle to penetrate the cell membrane and release its genome into the cytosol of the host cell (Baggen et al., 2018; Shingler et al., 2013). Such transitions are accompanied by structural rearrangement and changes in particle antigenicity(Zheng et al., 2019; Zhu et al., 2018c). For the picornaviruses EV71, CVA10, EVD68 and HRV, some nAbs show cross-reactivity to the different conformational states of the viral particles(Dong et al., 2017; Plevka et al., 2014; Zheng et al., 2019; Zhu et al., 2018b; Zhu et al., 2018c). However, there is no structural information available in the literature regarding the immune-complex or viral neutralization epitopes for CVA16. Given that different CVA16 particle states display antigenic diversity, it remains unclear whether nAbs recognize a specific CVA16 viral conformation or define a common site that must remain intact during capsid transformation.
Here, we systematically analyzed the functional properties of three CVA16 nAbs—18A7, 14B10 and NA9D7—and identified the molecular basis of nAb-mediated neutralization for CVA16. We show that 18A7 neutralizes only restricted strains of CVA16 and binds to the viral 5-fold vertex of all three types of the CVA16 particle. In contrast, 14B10 and NA9D7 both exhibit broad neutralization activities and therapeutic potential against CVA16 infection but bind exclusively to mature virions surrounding viral 3-fold and 2-fold axes, respectively. These results support the use of CVA16 mature virions for vaccine development, and an assay based on antibodies 18A7 and NA9D7 allows the quantification of immunogen constituents.
Results
Characterization of anti-CVA16 nAbs 18A7, 14B10, and NA9D7
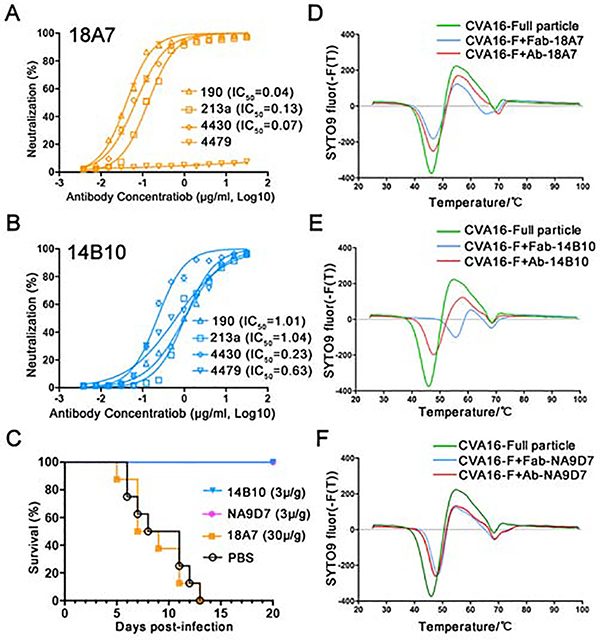
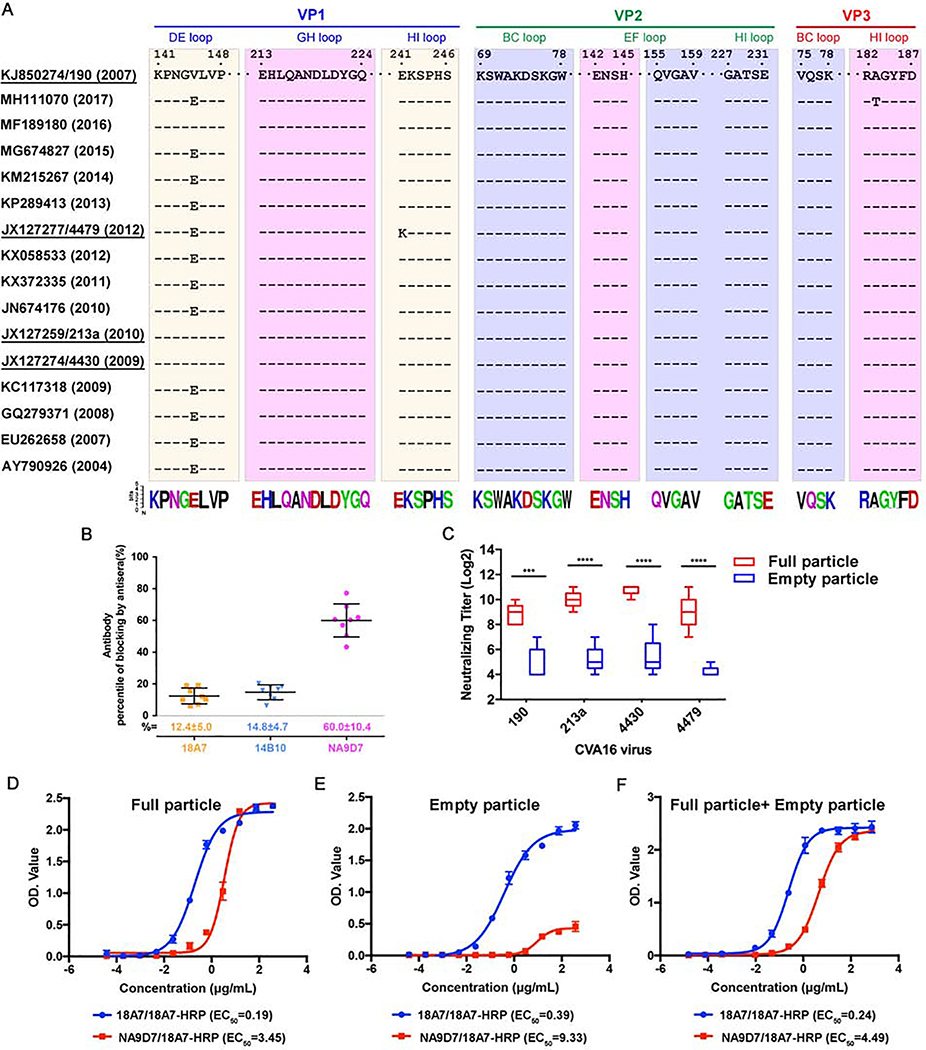
To investigate the antibody-mediated neutralization of CVA16 and the antigenic profiles across different particle forms, we selected three murine nAbs for functional and structural analysis: two nAbs, 18A7 and 14B10, were developed in this study; one nAb, NA9D7, from a panel of 48 murine mAbs in our previous work(Ye et al., 2016). Prior to structure determination at high-resolution, these three nAbs were identified to bind distinct regions of CVA16 capsid by a preliminary cryo-EM analysis. First, using a binding assay with various viral particle forms, we found that 18A7 exhibits comparable binding efficiency to full particles (including genomic RNA) and empty particles, whereas 14B10 and NA9D7 bind preferentially to full particles (Figures S1A–S1C). Through cell-based neutralization assays using a local epidemic strain, CVA16/190 (GenBank accession no. KJ850274, also used for the subsequent structural analysis), we found that 18A7 possessed higher neutralizing activity than 14B10 and NA9D7, with half-maximum inhibitory concentrations (IC50) of 0.04, 1.01, and 1.96 μg/mL, respectively (Figures 1A, 1B and S1D). Further neutralization assays showed that 18A7 could neutralize 213a and 4430 strains but not the 4479 strain, while14B10 and NA9D7 could neutralize all three strains but with a lower efficacy than 18A7 (Figures 1A, 1B and S1D).
Figure 1. Characterization of anti-CVA16 NAbs 18A7, 14B10, and NA9D7.
(A and B) Cross-neutralizing efficacy of 18A7 (A) and 14B10 (B) against CVA16 strains 190, 213a, 4430, and 4479 were evaluated using an in vitro micro-neutralization assay in human rhabdomyosarcoma (RD) cells. The neutralization efficacy was evaluated using the IC50 values calculated from the inhibition ratio plotted against antibody concentration.
(C) In vivo protective efficacies of antibodies against CVA16. One-day-old mice were challenged with CVA16 and then treated with 18A7, 14B10, or NA9D7 after virus infection. The control group were treated with PBS. Mouse survival rates were monitored and recorded daily for 20 days.
(D-F) Stabilities of CVA16 full particles and their immune complexes with 18A7 (D), 14B10 (E), or NA9D7 (F) determined by thermal stability assays using the SYTO9 dye to detect RNA exposure. The first derivatives are shown and the experiments were independently repeated in triplicates.
See also Figure S1.
We next sought to assess the correlation between in vitro neutralization and in vivo protection. Inoculation with either 14B10 or NA9D7 provided 100% protection against lethal infection with CVA16 in a neonatal mouse model at a dosage of 3 μg/g, whereas 18A7 generated no protection, even at a 10-fold higher dosage (30 μg/g) (Figure 1C). The inconsistent outcome that nAb 18A7 neutralized CVA16 in human RD cell model whereas provided no protection in mouse might result from some different host factors, of which human SCARB2 and PSGL-1 were identified as CVA16 receptors, but in contrast murine ones exhibited no such function in the mouse model(Nishimura et al., 2009; Yamayoshi et al., 2009). Fluorescence-based thermal stability assays revealed a ~1°C higher melting temperature when the full particle was bound with the 18A7 Fab fragment or full-length antibody as compared with the unbound full particle (Figure 1D). In contrast, 14B10 Fab and the full-length antibody stabilized the full particle by 10°C and 1.8°C, respectively, and NA9D7 Fab and the full -length antibody stabilized the full particle by 2.5°C or 1.8°C, respectively (Figure 1E and 1F). These results indicate that virions exhibited enhanced thermal stability through engagement with either the Fab or the full-length antibody for all three nAbs, with a particular efficacy observed in the 14B10 Fab. Overall, these findings show the differences in interaction characteristics of the three nAbs and suggest distinct mechanisms of virus neutralization.
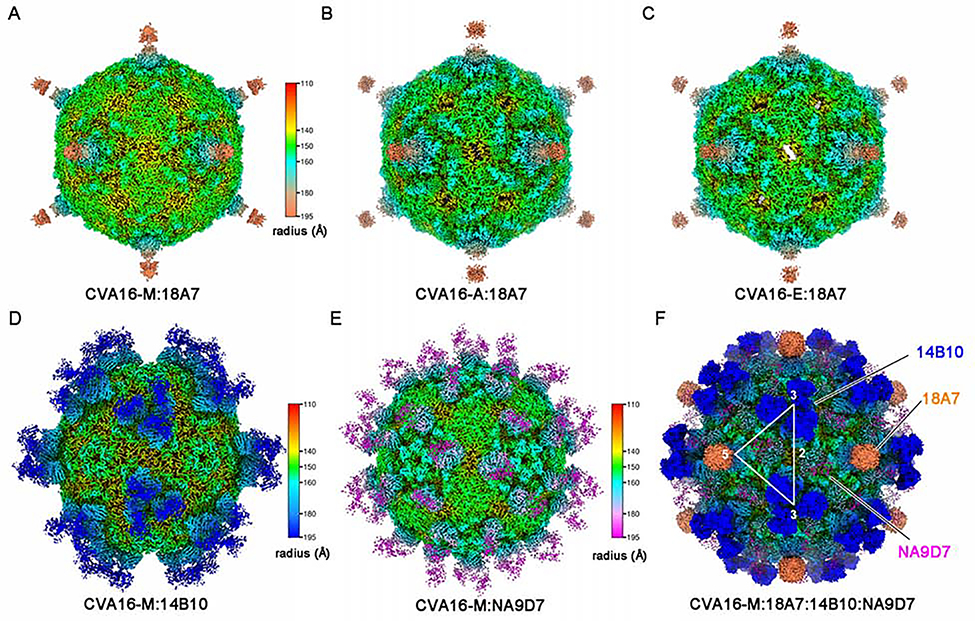
Purified full and empty particles of CVA16 were separately subjected to cryo-EM inspection and 2D classification for structural analysis. The full particle fraction was primarily composed of mature virions (CVA16-M, 73% occupancy in total particles) with some A-particles (CVA16-A, 14%) and empty particles (CVA16-E, 13%). Comparatively, the CVA16 empty particle fraction nearly exclusively consisted of empty particles (Figures S2A and S2B). Other enterovirus studies(Fan et al., 2017; Zhu et al., 2018a) also confirmed the coexistence of three particle forms in purified fractions. The CVA16 full particle fraction was complexed separately with Fab 18A7, 14B10, or NA9D7 for cryo-EM structure reconstruction. In line with the binding profiles of the three nAbs (Figures S1A–S1C), 18A7 bound to all three types of CVA16 particles (Figures S2C–S2E) with structures resolved at 2.65 Å, 3.07 Å, and 3.13 Å for CVA16-M:18A7, CVA16:A-18A7, and CVA16-E:18A7, respectively (Figures 2A–2C, S3, S4D–S4F, Table 1 and Video S1). Contrastingly, 14B10 and NA9D7 exclusively bound to mature virions, with structures of CVA16-M:14B10 and CVA16-M:NA9D7 solved at 3.30 Å and 3.23 Å, respectively (Figures 2D, 2E, S2F–S2K, S4G, S4H, Table 1, Videos S2 and S3).
Figure 2. cryo-EM structures of immune complexes of CVA16-M:18A7, CVA16-A:18A7, CVA16-E:18A7, CVA16-M:14B10, CVA16-M:NA9D7 and CVA16-M: 18A7:14B10:NA9D7.
(A-F) Radially colored surface views down a two-fold axis of the cryo-EM density maps of the immune complexes CVA16-M:18A7 (A), CVA16-A:18A7 (B), CVA16-E:18A7 (C), CVA16-M:14B10 (D), CVA16-M:NA9D7 (E) and CVA16-M:18A7:14B10:NA9D7 (F). Fabs 18A7, 14B10, and NA9D7 bind the capsid around its 5-, 3- and 2-fold axes, respectively. These axes are denoted by the corresponding numbers with the triangle demarking an asymmetric unit.
See also Figures S2–S4 and Table 1 and Videos S1–S4.
Table 1. Cryo-EM data collection, refinement and validation statistics.
The Table shows cryo-EM data collection, reconstruction, and model validation results and values. See also Figures S2–S4.
| Mature virion (CA16-M) | A-particle (CA16-A) | Empty particle (CA16-E) | CA16-M:18A7 | CA16-M:18A7-local | CA16-A:18A7 | CA16-E:18A7 | CA16-M:14B10 | CA16-M:NA9D7 | CA16-M:18A7:14B10:NA9D7 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Data collection and processing | ||||||||||
| Magnification | x93,000 | x93,000 | x93,000 | x130,000 | x130,000 | x130,000 | x130,000 | x93,000 | x93,000 | x93,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 40 | 40 | 40 | 56 | 56 | 56 | 56 | 40 | 40 | 40 |
| Defocus range (μm) | −0.5 to −4.2 | −0.5 to −4.2 | −0.4 to −5.5 | −0.3 to −2.9 | −0.3 to −2.9 | −0.3 to −2.9 | −0.3 to −2.9 | −0.4 to −4.2 | −0.5 to −4.7 | −0.5 to −4.6 |
| Pixel size (Å) | 1.128 | 1.128 | 1.128 | 1.307 | 1.307 | 1.307 | 1.307 | 1.128 | 1.120 | 1.120 |
| Symmetry imposed | I2 | I2 | I2 | I2 | C1 | I2 | I2 | I2 | I2 | I2 |
| Initial particle images (no.) | 29,497 | 29,497 | 14,507 | 101,235 | 726,696 | 101,235 | 101,235 | 38,833 | 52,713 | 38,967 |
| Final particle images (no.) | 17,781 | 3,409 | 11,574 | 60,558 | 57,409 | 10,246 | 10,406 | 10,580 | 45,235 | 27,819 |
| Map resolution (Å) | 3.56 | 3.33 | 3.43 | 2.65 | 3.67 | 3.07 | 3.13 | 3.30 | 3.06 | 3.78 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 3.0–5.0 | 3.0–5.0 | 3.0–5.0 | 2.5–4.5 | 3.0–5.0 | 2.5–4.5 | 2.5–4.5 | 2.5–4.5 | 3–5.0 | 3.0–5.0 |
| Refinement | ||||||||||
| Initial model used (PDB code) | 5C4W | 4JGY | 5C9A | 5C4W | 5C4W | 4JGY | 5C9A | 5C4W | 5C4W | n/a |
| Model resolution (Å) | 3.72 | 3.51 | 3.53 | 2.75 | 3.71 | 3.17 | 3.23 | 3.40 | 3.32 | n/a |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | |
| Model resolution range (Å) | 3.0–5.0 | 3.0–5.0 | 3.0–5.0 | 2.5–4.5 | 3.0–5.0 | 2.5–4.5 | 2.5–4.5 | 2.5–4.5 | 3–5.0 | |
| Map sharpening B factor (Å2) | −148.4 | −170.8 | −204.7 | −124.7 | −90.0 | −116.4 | −126.7 | −173.5 | −90.0 | −220.7 |
| Model composition | n/a | |||||||||
| Non-hydrogen atoms | 6,444 | 5,087 | 4,972 | 6,433 | 10,754 | 5,072 | 49,62 | 8,125 | 8,181 | |
| Protein residues | 827 | 648 | 636 | 826 | 1,329 | 646 | 635 | 1,045 | 1,055 | |
| Ligands | 1 | 0 | 0 | 1 | 5 | 0 | 0 | 1 | 1 | |
| B factors (Å2) | ||||||||||
| Protein | 98.01 | 101.06 | 93.84 | 35.93 | 75.63 | 54.81 | 60.51 | 71.06 | 83.62 | |
| Ligand | 105.89 | 36.93 | 65.80 | 65.93 | 57.87 | |||||
| R.m.s. deviations | ||||||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.006 | 0.006 | 0.007 | 0.006 | 0.006 | 0.006 | 0.009 | |
| Bond angles (°) | 0.780 | 0.709 | 0.804 | 0.798 | 1.124 | 0.765 | 0.735 | 0.868 | 0.869 | |
| Validation | ||||||||||
| MolProbity score | 1.16 | 1.38 | 1.51 | 1.28 | 2.19 | 1.16 | 1.59 | 1.60 | 1.41 | |
| Clashscore | 2.98 | 3.27 | 3.24 | 3.15 | 9.36 | 2.29 | 3.86 | 4.18 | 3.28 | |
| Poor rotamers (%) | 0.71 | 0.18 | 0.36 | 1.14 | 2.05 | 0.72 | 0.18 | 0.34 | 0.67 | |
| Ramachandran plot | ||||||||||
| Favored (%) | 97.67 | 96.20 | 94.19 | 97.30 | 92.72 | 97.14 | 93.70 | 93.96 | 95.86 | |
| Allowed (%) | 2.33 | 3.80 | 5.81 | 2.58 | 6.74 | 2.7 | 6.30 | 5.94 | 4.14 | |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.12 | 0.54 | 0.16 | 0.00 | 0.10 | 0.00 |
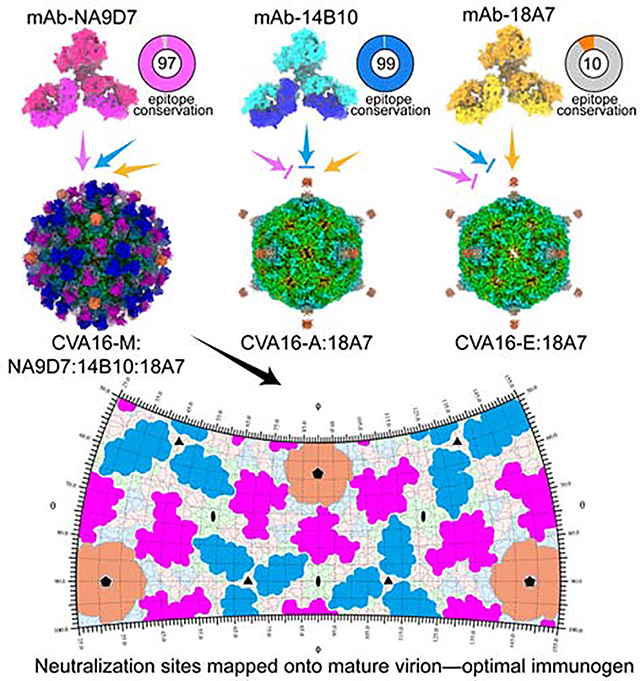
The density maps of the three 18A7-bound immune complexes show that a single 18A7 Fab engages one 5-fold vertex of the icosahedral capsid and therefore only 12 Fabs can bind to each viral particle (Figures 2A–2C). In contrast, three 14B10 Fabs and two NA9D7 Fabs target the virus capsid around its 3-fold and 2-fold axes, respectively, and thus up to 60 copies of 14B10 or NA9D7 Fabs can bind to each mature virion (Figures 2D and 2E). Because 18A7, 14B10 and NA9D7 bind to disparate loci, we further successfully obtained a quadruple immune complex (CVA16-M:18A7:14B10:NA9D7) of the CVA16 virion fully bound with each of the three Fabs (Figures S3J–S3I) and determined the cryo-EM structure at 3.67 Å. The density map confirms that the three Fabs can simultaneously bind to one mature virion without any steric hindrance (Figures 2F, S2L–S2N, S4I and Video S4).
Interaction analysis of CVA16 immune complexes
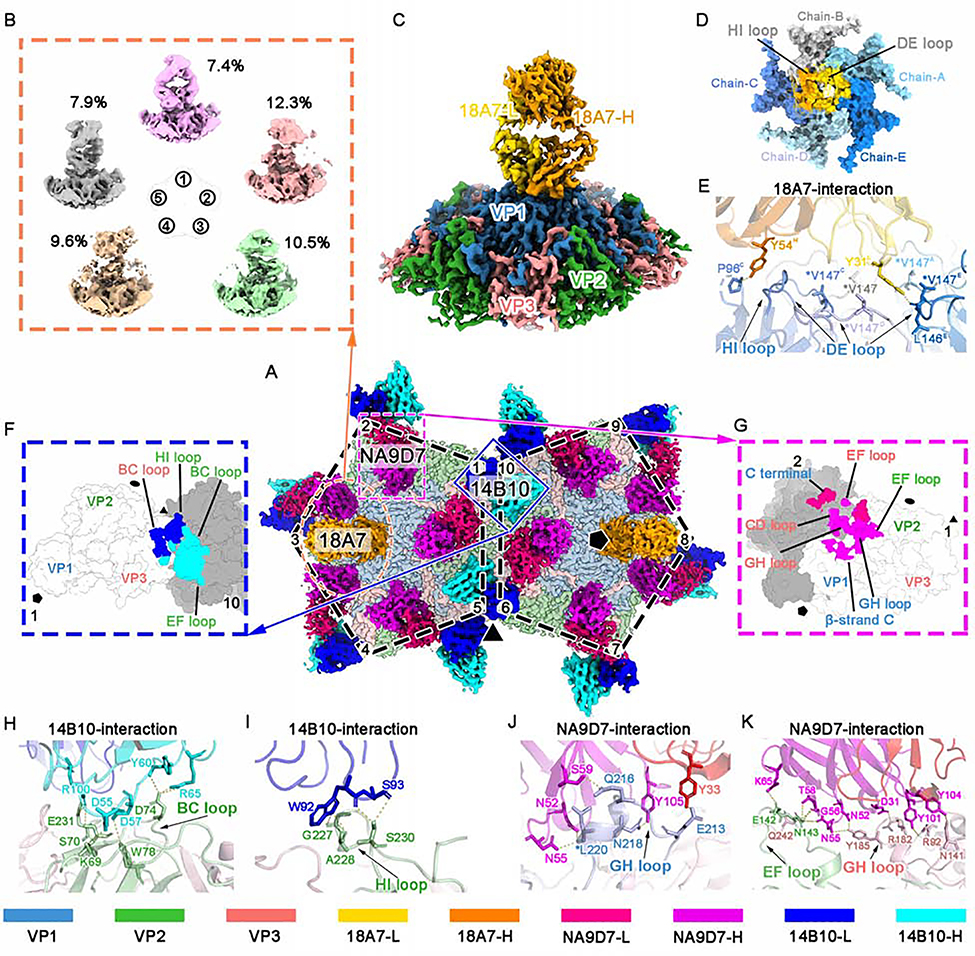
To elucidate the binding footprints of the three nAbs, we segmented out 10 asymmetric units of the quadruple immune-complex that reflected all the virion-Fab interacting scenarios (Figure 3A). Note that binding of one 18A7 Fab against 5 VP1 capsid proteins causes a symmetry mismatch in the arrangement of the 18A7 Fab with the capsid, such that regular icosahedral reconstruction results in an incomplete density of the 18A7 Fab at lower resolution (Figures 2A–2C). This is in contrast to the complete Fab densities of 14B10 and NA9D7 (Figures 2D and 2E), which comply with the capsid symmetry. Similar symmetry mismatch has been observed in other Fab-capsid structure determinations, including EV71(Lee et al., 2013), human rhinovirus (HRV)(Hewat and Blaas, 2006), cucumber mosaic virus (CMV)(Bowman et al., 2002), and human papillomavirus (HPV)(Li et al., 2017); only partial density with these Fabs at a lower resolution can be resolved by the regular strategy. To overcome symmetry mismatch and reveal high-resolution details of antibody-capsid interactions, we extracted the 12 5-fold vertex regions of each CVA16-M:18A7 particle as sub-particles and subjected all sub-particles to 3D classification and refinement(Ilca et al., 2015). Five stable 3D classes emerged, each corresponding to binding of the 18A7 Fab at the 5-fold vertex in five orientations with 72°interval in one round. These five sub-particles share a similar occupancy ratio (7% to 12%) among the total sub-particle population (Figures 3B and S3), suggesting that the 18A7 Fab engages with each 5-fold vertex of the virion in a random manner, with a total of five possible orientations. Our highest resolution structure of the sub-particle is at 3.67 Å, which was obtained by refining sub-particles in class #5 (CVA16-M:18A7-local) (Figures 3C, S3, S4J, Table 1 and Video S1). Based on this sub-particle reconstruction and the above-described high-resolution structures of CVA16 with and without bound Fabs, we built atomic models for the Fab variable domains and the capsid although we excluded the constant domains for atomic modeling due to their structural flexibility (Figures S4G, S4H and S4J).
Figure 3. Sub-particle reconstruction of the CVA16-M:18A7 immune complex and interaction analysis of CVA16-M:18A7, CVA16-M:14B10, and CVA16-M:NA9D7.
(A) Surface view of the cryo-EM densities of two adjacent five-fold vertexes of CVA16-M:18A7:14B10:NA9D7, containing 10 asymmetric units. Fabs bound to these ten asymmetric units include two 18A7 Fabs (orange), 10 14B10 (heavy chain: cyan, light chain: cyan) and 10 NA9D7 (heavy chain: magenta, light chain: red). Icosahedral 2-, 3-, and 5-fold axes are indicated by black ellipse, triangle, and pentagon symbols, respectively.
(B) Sub-particle reconstruction of the CVA16-M:18A7 five-fold vertex yielded five well-defined 3D classes from different number of sub-particles (percentages indicated) showing different Fab binding directions.
(C and D) Surface views of the final sub-particle reconstruction of the five-fold vertex showing CVA16-M:18A7 binding (resolution 3.67 Å) viewed from side (C) and top (D). In d, the Fab density is removed to reveal the footprint of Fab 18A7 on a surface area buried by the Fab heavy (gold) and light (orange) chains.
(E) Close-up of the interface between the capsid and Fab 18A7 with residues participating in the interactions labeled and their side chains shown as sticks on the ribbon models.
(F and G) Footprints of Fabs 14B10 (F) and NA9D7 (G) on the CVA16 viral capsid rendered as surface representation.
(H and I) Close-up views of the interface between the capsid and either the heavy (H) or the light chain (I) of Fab 14B10.
(J and K) Fab NA9D7 interacts with the GH loop of VP1 (J), the EF loop of VP2, and the GH loop of VP3 (K). Hydrogen bonds and salt bridges are marked by yellow dashed lines. Side chains involved in the interaction between the antigen and the antibody are labeled and shown as sticks. Asterisks mark the position of an escape mutation.
Proteins are colored according to the codes at the bottom.
See also Figures S3 and S5 and Table S1.
Our asymmetric sub-particle reconstruction of the 18A7-CVA16 immune complex shows that 18A7 interacts, intriguingly, with all five VP1s at the 5-fold vertex region in the viral capsid, which buries a large surface area of ~1,184 Å2. 18A7 also interacts with all six of the complementarity-determining regions (CDRs) of the nAb (Table S1). The light chain accounts for ~69% of the buried surface area, and dominates the interaction through its involvement with all five VP1s, whereas the heavy chain accounts for ~31%, and interacts only with two VP1s (termed chain-B and chain-C) (Figures 3C, 3D and Table S1). The 18A7 neutralization sites cover the DE loop (K141-P148), the HI loop (E241-S246), and the BC loop (P96) of VP1, where residues V147, P148, P244, and H245 from three VP1s elaborately interact with the loops of the CDRs via numerous van der Waals contacts. In addition, P96 from the BC loop of chain-C targets Y54 of the CDR heavy chain loop 2 (H2) of 18A7 through a strong hydrogen bond (Figures 3D, 3E and Table S1). Finally, we note that virus-escape mutants, generated under immune pressure from 18A7, bear a V147A mutation at the virus-antibody binding interface. Surface electrostatic potential analysis suggests that the interaction sites mostly comprise positively charged residues (Figure S5A) that associate with CVA16 binding to cellular receptors, such as heparan sulfate and human P-selectin glycoprotein ligand-1, putative receptors for enteroviruses (Figure S5B) (Nishimura et al., 2009; Zhang et al., 2017).
Our structure of the 14B10-CVA16-M immune complex reveals that one 14B10 Fab binds across every two adjacent protomer around the 3-fold region (Figures 3A and 3F). The 14B10 Fab covers a buried surface area of ~761 Å2, of which the heavy chain accounts for ~56% (~425 Å2). The interaction sites include VP3 from one protomer (referred to as protomer 1, Figure 3F) and VP2 from the adjacent protomer (protomer 10). Twelve hydrogen bonds, 7 salt bridges, and dozens of van der Waals contacts contribute to the interaction between 14B10 and the capsid. Of note, the donors of the available hydrogen bonds and salt bridges contributed by the VP2 protein of protomer 10 are mediated mostly by the BC loop and the HI loop (Figures 3F, 3H, 3I and Table S1). Meanwhile, D74 of the BC loop of VP2, and S230 and E231 of the HI loop of VP2 make a number of interactions with 14B10, indicating that these are critical sites for 14B10 neutralization (Figures 3H, 3I and Table S1).
Our structure of the NA9D7-CVA16 immune complex reveals that 60 copies of the NA9D7 Fab binds across two adjacent protomers at the 2-fold region (Figures 3A and 3G). The NA9D7 Fab has an interaction area of ~1200 Å2 across two protomers, of which ~69% (~824 Å2) is attributed to protomer 1 and ~31% (~376 Å2) to protomer 2. NA9D7 Fab extensively interacts with VP1, VP2, and VP3, specifically with the GH loop of VP1, the EF loop of VP2, and the GH loop of VP3 totaling 16 hydrogen bonds and 2 salt bridges (Figures 3G, 3J, 3K and Table S1). Among these interactions, 12 bonds are formed by each NA9D7 Fab binding to VP3 from its own protomer, whereas the other six bonds are donated from the adjacent protomer. Notably, L220 from VP1 and N143 from VP2 interact with multiple residues of Fab NA9D7 (Figures 3J, 3K and Table S1). CVA16 escape mutants of NA9D7 harbor mutations in residue 220 of VP1 (L220V, L220S, or L220F), suggesting a critical role of L220 in the virus-antibody interaction.
Structural analysis of neutralization sites for all three particle forms of CVA16
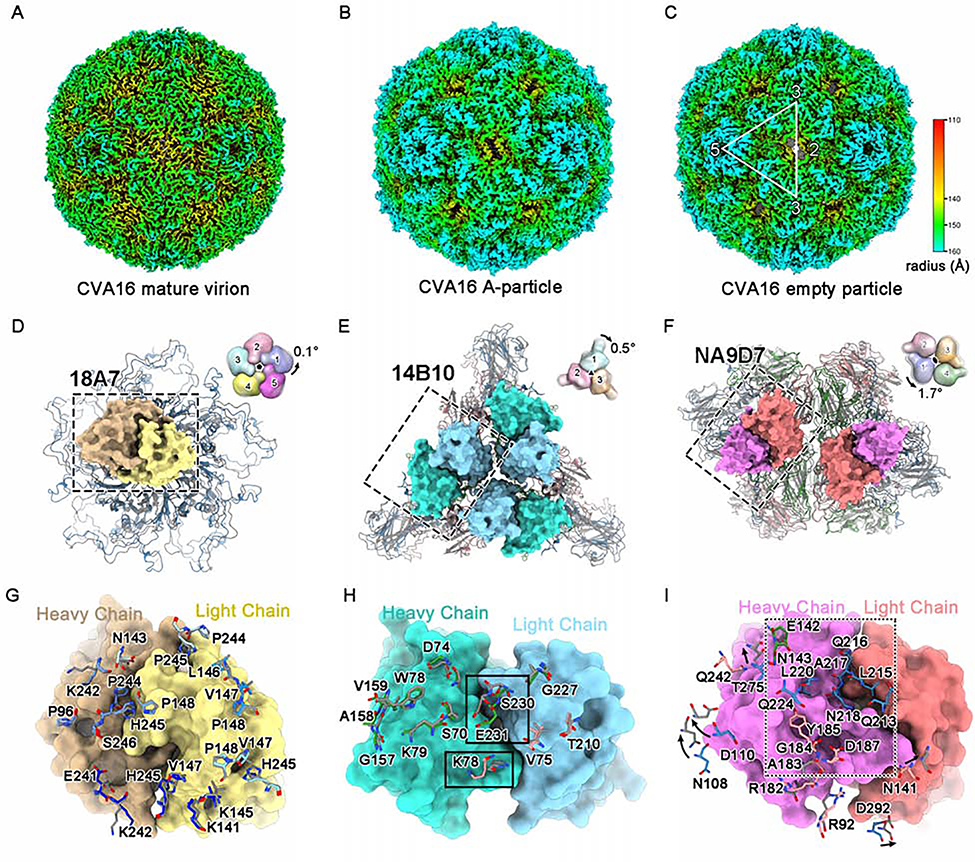
To assess the three neutralization sites altered upon viral capsid transformation, we first determined the structures of the CVA16 mature virion, the A-particle and the empty particle at a resolution of 3.56 Å, 3.33 Å, and 3.43 Å, respectively (Figures 4A–4C, S4A–S4C, and Table 1). We then compared the binding sites of the nAbs across the three particle forms. Of note, 14B10 and NA9D7 recognize neither the A-particle nor the empty particle. Our structures of the CVA16 mature virion and the A-particle are nearly identical to the corresponding virion and A-particle structures reported in other studies(Ren et al., 2013; Ren et al., 2015) with RMSD values of 0.81 Å and 0.73 Å, respectively, for all of the Cα atoms of the VP proteins (Figures S6A and S6B). In contrast, the cryo-EM structure of the empty particle showed some conspicuous differences to that of the reported crystal structure (pdb code: 5C9A) (RMSD = 2.11 Å), which was derived from a sample that had been treated before crystallization with formaldehyde (Figure S6C) (Ren et al., 2015). Our cryo-EM structure of the A-particle is nearly identical to that of the empty particle (RMSD = 0.47 Å), with both demonstrating differences to the mature virion (RMSD = 2.38 Å and 1.87Å); these differences resulted from capsid transformation, including the collapsed pocket factor-binding regions, the expanded 2-fold channels, and the lost VP4s (Figures S6D–S6F). Finally, superimpositions of the protomers of the viral particles with and without bound nAbs show neither global nor local conformational changes upon nAb binding (Figures S6G–S6K).
Figure 4. Structural and antigenic comparisons among different types of CVA16 particles.
(A-C) Radially colored surface views of the cryo-EM density maps of the CVA16 mature virion (A), A-particle (B) and empty particle (C).
(D-F) Structural comparisons of the mature virion and A-particle (which is highly similar to the empty particle) on their viral 5-fold (D), 3-fold (E) and 2-fold (F) vertexes, respectively. The variable regions of one 18A7 Fab (heavy chain: orange, light chain: yellow), three 14B10 Fabs (heavy chain: cyan, light chain: light blue) and two NA9D7 Fabs (heavy chain: magenta, light chain: pink) bound to the viral 5-fold (D), 3-fold (E) and 2-fold (F) vertexes are shown as surfaces. Diagrams in the upper-right corner depict the structural discrepancy for different protomers located in 5-fold, 3-fold, or 2-fold vertexes, with global rotation against the corresponding icosahedral axes between the mature virion (colored) and the A-particle (gray, 50% transparency). The axes are located at the center of the diagrams and are perpendicular to the page.
(G-I) Close-up views and structural comparisons of the binding sites of 18A7 (G), 14B10 (H), and NA9D7 (I) between the mature virion and the A-particle. The 18A7 binding sites cover five VP1s localized to the 5-fold vertex of the viral capsid (blue) (G). (H) Important differences in the residues between the mature virion and A-particle (rectangle). (I) NA9D7 epitope residues of the mature virion that were not interpreted in the A-particle model are boxed in the dashed rectangle. VP1, VP2, VP3, and VP4 of the CVA16 mature virion are colored blue, green, red, and yellow, respectively. The A-particle model is in gray. The amino acid residues of CVA16 involved in nAb-capsid interactions in (G-I) are shown as sticks.
See also Figure S6.
We next interrogated the structural variance of the three types of CVA16 particles that pivot on the 5-, 3- and 2-fold vertexes, respectively (Figures 4D–4F). Intriguingly, structural alignment of the viral vertex pivots of the mature virion and the A-particle (whose structure is nearly identical to that of the empty particle) showed almost identical 5-fold and 3-fold channels, with only a 0.1°anti-clockwise rotation and a 0.5°clockwise rotation, respectively. Yet, there is significant variations for the 2-fold channel, with an anti-clockwise rotation of 1.7°(Figures 4D–4F). Most of the residues involved in the binding with 18A7 are related to the viral 5-fold channel and are located at the DE and HI loops of two VP1s (chain-B and chain-C). In addition, these residues show a highly similar conformation between the mature virion and the A-particle (RMSD = 0.36 Å) (Figure 4G). In contrast, the binding sites for 14B10 and NA9D7 nAbs in the mature virion are significantly different for the A-particle and the empty particle, with RMSD values of ~2.1 Å and ~4.8 Å, respectively (Figures 4H and 4I). Remarkably, the most deviated Cα atoms in the 14B10 epitope between the mature virion and the A-particle have a space shift of ~2.5 Å for K78 of VP3 and ~2.3 Å for S230-E231 of VP2 (Figure 4H). However, in the NA9D7 binding region, the surface loops (VP1: GH loop, VP2: EF loop and VP3: GH loop) are flexible, with incontinuous densities for both the A-particle and the empty particle. Additionally, residues N108, D110, T275, and D292 of VP1, and N141 of VP3 in the mature virion move outward when transforming into the A-particle, reaching a maximal Ca shift ~4.3 Å (Figure 4I). Together, these results indicate that, besides the existence of shared antigenic sites (18A7 binding site) of all three types of particle forms, the mature virion structure possesses some unique epitopes and may serve as a more efficacious immunogenic vaccine candidate.
Antibodies for identifying constituents of vaccine immunogens
To explore sequence conservation and the underlying immune-dominance of the epitopes reciprocals to nAbs 18A7, 14B10 and NA9D7, we performed multiple sequence alignments of VP genes from all the 157 virus strains of CVA16 in GenBank, and measured the blocking ratios of nAbs binding to full particle using native virus-infected human sera. As shown in the loop regions covering the three nAbs’ epitopes from the 16 representative VP sequences, nAb 18A7 recognizes a discontinuous epitope spanning the DE loop and the HI loop of two adjacent VP1 capsid proteins (Figures 4G and 5A). Of note, 18A7 could not neutralize the 4479 strain when residues at positions 145 and 241 of VP1 are glutamine and lysine, respectively; but are reactive with 190 strain with valine and glutamic acid at these positions. These results suggest that the 18A7 epitope is less sequence conserved among native virus isolates, despite that it holds integrity across the three capsid forms of the 18A7-reactive viruses. In contrast, 14B10 and NA9D7 can neutralize all four of the tested CVA16 strains and have highly conserved (~99% and ~97%, respectively) strategic epitope residues across all of the aligned 157 CVA16 strains (Figure 5A).
Figure 5. Conservation analysis of neutralization epitopes and ELISA of purified CVA16 particles.
(A) Sequence alignment showing the epitopes of the three Fabs covering 16 strains of CVA16. The neutralization epitopes are boxed as cream (18A7), purple (14B10) and pink (NA9D7), respectively. The amino acid residue conservation is depicted beneath the boxes using the Weblogo representation of the alignment of 157 CVA16 strains.
(B) Competitive ELISA of 18A7, 14B10, and NA9D7. Antibodies were conjugated to HRP and then used to block the binding of CVA16-positive human sera to CVA16 particles that were pre-coated on ELISA plates. The percentage of blockage is expressed as the mean ± standard deviation (SD).
(C) Cross-strain in vitro neutralizing efficacies of antisera elicited by different types of CVA16 particles. BALB/c mice were vaccinated intraperitoneally with CVA16 strain 190 full particles or empty particles. The neutralizing efficacies of the antisera collected from mice against CVA16 strains 190, 213a, 4430, and 4479 were evaluated by neutralization assay, with neutralizing titers expressed as mean ± SD. Statistical significance was analyzed by an unpaired Student’s t-test (***P < 0.001, n=5).
(D-F) Reactivities of CVA16 particles by two modes of double-antibody sandwich ELISAs. The ELISA systems used 18A7 paired with 18A7-HRP or 18A7 paired with NA9D7-HRP as the capture and detection antibody for the detection of plenary particles and mature virions, respectively. CVA16 full particle (D), empty particle (E) and 1:1 mixture (F) were tested in the two ELISA systems. EC50 values were calculated with curves generated by nonlinear regression fitted.
Naturally acquired anti-CVA16 human sera could block nAbs 18A7, 14B10, and NA9D7 from binding to the CVA16 full particles at blocking rates of 12.4%, 14.8%, and 60%, respectively. This shows that NA9D7, but not 18A7 or 14B10, likely binds a dominant, immunogenic site of CVA16. The NA9D7 site thus may serve as a major target for an effective vaccine constituent that solely exists at the mature virion and may disrupt transformation to A-particles and empty particles (Figure 5B). It is worth mentioning that the presence of murine NA9D7 could not directly reflect on the antibody generation in human albeit it partially competes the binding of the CVA16 reactive human sera in competition ELISA. It would be necessary to explore whether the category of NA9D7-like antibody could be dominantly elicited in human upon the native infection of CVA16 virus.
We next verified the unique epitopes in the mature virions by evaluating antibody responses after vaccination with purified CVA16 (190 strain) full particles and empty particles in mice. As expected, full particles elicited 1.83- to 2.14-times higher neutralizing antibody titers than those of the empty particles for all four virus strains (P < 0.001). The neutralization titers from the full particles of the CVA16 190 strain was 28.8, and cross-neutralization against the 213a, 4430, and 4479 strains was achieved at 210.0,210.8, and 29.0, respectively; all of these values surpass the putative protection level of a neutralization antibody titer suggested in previous studies (Figure 5C)(Cai et al., 2013; Chong et al., 2012). Taken together, we conclude that the CVA16 mature virion harbors the predominant immunogenic site, at least in the region around the 2-fold axis of the intact virion, where it accommodates a 2-fold channel that is open during capsid transformation and associated with genome release(Ren et al., 2013; Wang et al., 2012). Thus, virus strain(s) bearing the conserved neutralization epitope(s) and possessing a maximum proportion of the mature virion during cell culture may be selected as a candidate(s) for CVA16 vaccine development.
Given that these three nAbs provide distinctive indicators when neutralization is achieved, we next designed two double-antibody sandwich ELISA assays to monitor particle proportion and mature virion stability during CVA16 immunogen production. In the first ELISA assay, 18A7, which was expected to bind/capture all virus particle forms, hereafter referred to as the “capture antibody,” was immobilized to the microplate. For the second, we used NA9D7 as the capture antibody to exclusively harvest the mature virions in the sample. Then, for both assays, 18A7 was labeled with secondary antibody to horseradish peroxidase (HRP), termed 18A7-HRP, and used as the detection antibody. The EC50 values, which are calculated from curves of OD vs. the input particle concentration, showed comparable reactivities for the full particles, the empty particles, and their mixture (v:v=1:1) in the 18A7/18A7-HRP ELISA, but read 3.45, 9.33 and 4.49 μg/mL for these three samples respectively, indicative of differences in the abundance of mature virions in these samples (Figures 5D–5F). A correlation of the mature virions and the plenary particles using these two ELISAs can thus be used to quantify the effective constituents of vaccine immunogens and their stability during vaccine development.
Discussion
High-resolution structures of viruses and their immune complexes provide valuable information for vaccine discovery and design. For pathogens such as HIV-1, RSV, and influenza, the neutralizing antibodies elicited by vaccines act almost exclusively through one or two enveloped glycoproteins or specific sites with conformational integrity(Laursen et al., 2018; McLellan et al., 2013; Xu et al., 2018). Thus, immunogen screening using conformation as a basis is essential for effective vaccine development. However, the enterovirus life cycle—encompassing the transformation of procapsids, mature virions, A-particles and empty particles—means that different components, structures, and immunogenicity can exist and thereby influence the utility of a vaccine, depending on the stage of the life cycle during treatment(Baggen et al., 2018; Shingler et al., 2013). The poliovirus D-antigen expressed on the mature virion is an important determinant of antigenicity and immunogenicity, owing to the regnant and integrated neutralizing epitopes; this contrasts with the C-antigen on A-particles and empty particles, which are known to generate poor immunogenic responses to viral infection(Ferguson et al., 1993). Similar to other studies(Chong et al., 2012), we show in the current study that the mature virions of the full particles of CVA16 induce higher neutralizing antibody titers than the other particle types, possibly due to their expression of protected antigenic sites. Our neutralizing antibodies, together with structural information of their interaction, further clarified the mature virion-specific neutralizing epitopes of 14B10 and NA9D7 nAbs. We show that both epitopes are highly conserved in most CVA16 strains, with the NA9D7 epitope being an immunodominant antigenic site (Figures 5A–5C), however, the antibody was produced in mice by the immunization of CVA16 cell culture, whether it reflects a similar situation in the antibody reservoir of a virus-infected human should be further interrogated. Collectively, these findings suggest that the mature virion of CVA16 should be targeted for vaccine design, and at least the structural integrity of NA9D7-like epitope that exclusively remains in mature virion should be well-characterized and rationally introduced to the vaccine immunogen. Our findings of three distinctive nAbs call for an approach of sandwiching ELISA of 18A7:18A7-HRP and 18A7:NA9D7-HRP) to quantify the proportion of mature virions present in vaccine preparation (Figures 5D–5F).
Previous structural studies have depicted how picornaviruses attach and gain entry into the cell(Hogle et al., 1985; Ren et al., 2013; Wang et al., 2012; Zheng et al., 2019). Entry of the mature virion is initiated through receptor-mediated binding, which initiates endocytosis and triggers the formation of a cell-entry intermediate or an A-particle(Baggen et al., 2018; Bergelson and Coyne, 2013; Johnson, 2010). Neutralization can occur at multiple steps of the infection process. For picornavirus EV71, EV-D68, and HRV, nAbs can cross-react with different conformational states near the icosahedral 3-fold axis, and thereby promote the untimely uncoating of the virus and its genome release(Dong et al., 2017; Plevka et al., 2014; Zheng et al., 2019). Here, using biochemical and structural analyses, we show that 14B10 and NA9D7 nAbs cannot bind to the A-particle or empty particle only the mature virion (Figures S1B, S1C, S2G and S2J). Furthermore, the nAbs’ binding either in Fab or full-length antibody form can stabilize the full particles as manifested by 1.0–2.5 °C higher melting temperature than unbound full particles in fluorescence-based thermal stability assays (Figures 1D–1F), exceptionally, 14B10 Fab can even stabilize by 10°C, much higher than 14B10 full-length antibody (1.8 °C), the discrepancy might ari se from only ⅓ 14B10 binding sites coordinated around 3-fold axis being occupied with full-length antibodies due to the steric hindrance (Figures S5C and S5E). Taken together with the structural information of all three Fabs binding to the virus across two or more protomers (Figures 3D, 3F and 3G), we demonstrate the three nAbs may neutralize CVA16 by restricting the conformational changes of the mature virion, such changes that are vital for viral entry and RNA release. As for the potential for full-length antibody bivalently interacting with the capsid, we fitted multiple full-length antibody models (PDB no. 1IGY) (Harris et al., 1998) to representative Fab densities within the cryo-EM structures of the immune complexes CVA16-M:14B10 and CVA16-M:NA9D7. The fitted models showed that two full-length antibodies of either 14B10 or NA9D7 are impossible to bivalently interact with two adjacent regions coordinated with 3-fold or 2-fold axis (Figures. S5C, S5D, S5F and S5G), even the hinge linking two Fabs in the full-length antibody is highly flexible.
NAbs of picornaviruses commonly target the exposed loop regions of the viral capsid, some of which overlap with the viral receptor binding sites. Although several receptors have been identified for many picornaviruses(Nishimura et al., 2009; Yamayoshi et al., 2009), the binding structural details of the CVA16 receptor is still poorly understood. However, the binding sites of other virus types have been outlined and lend important structural insight. For instance, the poliovirus and rhinovirus receptors on the viral surface cover the BC and HI loops on VP1 around the 5-fold icosahedral vertices of the capsid(Belnap et al., 2000; Hewat et al., 2000; Xing et al., 2000). In addition, studies have shown that EV71 and CVA16 use SCARB2 and PSGL-1 as functional receptors for cell entry, with the binding site of the EV71 SCARB2 receptor on the southern rim of the canyon of the VP1 GH and VP2 EF loops (Figure S5B)(Zhou et al., 2019). Here, we show that the neutralization epitope of 14B10 is located on the VP3 BC loop and the VP2 BC, EF, and HI loops at the 3-fold vertex. In contrast, we show that NA9D7 mainly targets the VP1 GH and VP2 EF loops surrounding the 2-fold axis (Figures 3F, 3G and S5B). These findings suggest that these loops may be involved in SCARB2 receptor binding, and function in CVA16 attachment to the cell surface. In addition, the footprint of 18A7 covers the DE and HI loops of VP1 at the 5-fold vertex, where it associates with CVA16 binding to other identified receptors, PSGL-1 and heparan sulfate (HS) (Figure 3D and S5B)(Nishimura et al., 2009; Zhang et al., 2017). Thus, the structural atlas of the three nAbs may help to identify the receptor binding site for CVA16.
STAR⋆METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ningshao Xia (nsxia@xmu.edu.cn).
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
BALB/c mice were obtained from the Slac Laboratory Animal Co., Shanghai, China. All mice were maintained in a specific-pathogen-free facility of Xiamen University. For infection studies, mice were maintained in BSL2 facility under the following housing conditions. The mice were housed in a temperature-controlled (20–25 °C), air-conditioned room on a 12-hr light-dark cycle, and supplied with corn cob bedding (1/4-inch depth), irradiated diet and sterilized water. All animal experimental protocols were approved by the Xiamen University Laboratory Animal Center (XMULAC) and were conducted in strict compliance with the guidelines of the Xiamen University Institutional Committee for Care and Use of Laboratory Animals (approval code: XMULAC20160049). Details of mouse information can be found in the “Preparation of antibodies and antibody fragments”, “In vivo protection assay” and “Vaccine preparation and immunization of mice” subsections.
Cell lines and viruses
Human rhabdomyosarcoma (RD) cells were obtained from the American Type Culture Collection (ATCC, CCL-136) and maintained in minimal essential medium (MEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco). The CVA16/190 strain (GenBank accession no. KJ850274) was isolated from an HFMD clinical specimen in Taiwan. The CVA16/213a strain (GenBank accession no. JX127259), the CVA16/4430 strain (GenBank accession no. JX127274) and the CVA16/4479 strain (GenBank accession no. JX127277) were isolated from HFMD clinical specimens in Fujian. The CVA16/190 strain was used for all biochemical and structural analyses. The CVA16/213a, 4430, and 4479 strains were used only in the in vitro neutralization assay. Viruses were stored in aliquots at −80°C in our lab oratory.
METHODS DETAILS
Virus production and purification
CVA16 was grown in RD cells at a multiplicity of infection (MOI) of 0.2 at 37C for 3 days. After infection, the virus was centrifuged to remove cell debris at 6,000 ×g for 30 min and precipitated using 7% (w/v) polyethylene glycol (PEG) 6,000 and 0.3 M NaCl in PBS at 4°C overnight. After centrifugation, the virus was resuspended in PBS and then loaded onto a linear 15% to 45% (w/v) sucrose density gradient in a Beckman SW41 Ti rotor at 135,000 ×g for 4 h. Virus fractions were independently collected and dialyzed against PBS, and further concentrated by an Ultra-4 centrifugal concentrator. The quantity and quality of CVA16 particles were assessed by negative-staining electron microscopy.
Preparation of antibodies and antibody fragments
To obtain mAbs 18A7, 14B10 (named NA14B10 in our original mAb panel), and NA9D7, 8-week-old female BALB/c mice were immunized subcutaneously with the supernatant of CVA16 cultures emulsified in Freund’s adjuvant and boosted twice at 2-week intervals. After the final boost, splenic cells from immunized mice were fused with myeloma cells (Sp2/0) and the hybridoma supernatants were screened using a neutralization assay against CVA16. Antibodies were purified from mouse ascites fluid using protein A affinity columns. The sequences of the variable regions of the heavy and light chains of the antibodies were determined by cloning and sequencing. To obtain antibody Fab fragments, mAbs were digested with 10/00 (w/w) papain in 20 mM PB (pH 7.0) containing 30 mM L-Cys and 50 mM EDTA at 37°C for 12 h. The re sulting Fab fragments were separated from the Fc fragments using a DEAE column.
In vitro neutralization assay
RD cells were pre-seeded into 96-well plates at 2 × 104 cells per well. Antibodies or antisera collected from mice were serially diluted 2-fold and incubated with an equal volume of CVA16 (104 TCID50 per well) at 37°C for 1 h. Each mixture was then a dded to cells and incubated at 37°C for 24 h. After incubat ion, an enzyme-linked immunosorbent spot (Elispot) assay was performed(Hou et al., 2015). The inhibition rate of the antibody was calculated using the following formula: Neutralization % = (1-[Ntest - Ncell control]) / (Nvirus control -Ncell control) × 100%. In this formula, Ntest, Ncell control and Nvirus control are the average number of spots in the test wells, cell control wells, and virus control wells, respectively. The IC50 was calculated from nonlinear regression fitting curves using GraphPad Prism version 7.0. The neutralization titers were defined as the highest dilution giving >50% neutralization. The experiments were performed in triplicate.
In vivo protection assay
Groups of 1-day-old mice (n = 8) were challenged intracranially (i.c.) with 105 TCID50 CVA16 at 24 h before intraperitoneal (i.p.) infection with antibodies. The mice in the control group were treated with PBS buffer. All mice were monitored and recorded daily until 20 days’ post-infection.
Particle stability thermal release assay
A ThermoFluor assay(Walter et al., 2012) was performed with an MX3005P RT-PCR instrument (Agilent). The presence of RNA was detected with an environment-sensitive fluorescent dye, SYTO9 (Invitrogen). Each 50-μl reaction mixture was prepared in thin-walled PCR plates (Agilent), containing 1 μg of CVA16 full particles and 5 μM SYTO9 in PBS buffer. The fluorescence level was recorded in triplicate at 0.5°C intervals from 25°C to 99°C.
Generation and sequencing of the escape mutant viruses
For the first round of infection, antibodies (0.2 mg/ml) were incubated with 106 TCID50 of CVA16 at room temperature for 1 h and then at 37°C for 2 h. The mixtures were added to RD cells and incubated at 37°C until cytopathic effect (CPE) developed. The cultures were harvested and subjected to three freeze-thaw cycles, and then clarified by centrifugation. For the second and third rounds of infection, the antibody concentration was increased to 0.8 mg/ml. After selection, the resultant resistant viruses were purified from plaques and cloned into RD cells. The P1 region of the viral genome was amplified by RT-PCR using primers (forward 5’ -TTAAAACAGCCTGTGGGT- 3’ and reverse 5’ - CGTTGTTATCTTGTCTCTACTAGT-3’) and verified by DNA sequencing.
Vaccine preparation and immunization of mice
The immunogenicity of the CVA16 virus was evaluated in 6-week-old female BALB/c mice. Purified CVA16 full particles and empty particles were diluted in PBS with an equal volume of aluminum adjuvant. Groups of mice (n = 5 per group) were respectively vaccinated with CVA16 full particles, CVA16 empty particles, or aluminum adjuvant (control) at days 0 and 21. Each mouse was intraperitoneally immunized with 1.5 μg per dose in 0.5-ml samples. Antisera were inactivated by incubation at 56C for 30 min and stored at −20°C for in vitro neutralization assays.
Binding ELISA
ELISA plates were pre-coated with the purified CVA16 full particles or empty particles at 50 ng per well at 4°C overnight and blocked with 5% skim milk in PBS at 37°C for 2 h. Antibodies were added into the wells at various concentrations and incubated at 37°C for 1 h. Each well was then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse (GAM-HRP; 1:5000 dilution) IgG antibodies at 37°C for 30 min. For color development, the plates were incubated with a TMB chromogen substrate solution at 37C for 15 min, and the reaction was terminated with 2 M H2SO4. The absorbance was measured at A450/620.
Competitive ELISA
CVA16-positive human sera (1:100 dilution) were added into wells pre-coated with the purified CVA16 full particles at 37°C for 30 min. T he HRP-conjugated mAbs 18A7, 14B10, or NA9D7 were then added respectively. After 30 min of incubation at 37°C, the plates were subjected to color development as described for the Binding ELISA (Methods). The absorbance was measured at A450/620. Percentage inhibition (PI) was calculated as: PI (%) = 100 – [(ODsample/ ODcontrol) × 100].
Sandwich ELISA
Samples of CVA16 (1) full particles + empty particles (1:1 mass), (2) full particles, or (3) empty particles were diluted in PBS in 5-fold serial dilutions, and then added to wells pre-coated with mAb NA9D7 or 18A7 (200 ng per well) and incubated at 37°C for 1 h. The HRP-conjugated mAb 18A7 was used as the detecting antibody (1:2,000 dilution). After 30 min at 37°C, the plates were subjected to color development, as described for the Binding ELISA (Methods). Absorbance was measured at A450/620.
Cryo-EM sample preparation and data collection
Purified 18A7, 14B10, or NA9D7 Fab fragments were incubated with CVA16 full particle at room temperature for 30 min at a molecular ratio of 72 Fab per viral particle. For the quadruple immune complex (CVA16-M:18A7:14B10:NA9D7), three Fabs were simultaneously mixed with the CVA16 full particle using the same ratio as above. A 3-μl aliquot of purified full particle, empty particle or immune complex was applied to freshly glow-discharged holey carbon Quantifoil Cu grids (R2/2, 200 mesh, Quantifoil Micro Tools) and then blotted for 6 s before plunge-freezing the grids into liquid ethane cooled by liquid nitrogen inside a Vitrobot Mark IV (Thermo Fisher Scientific) at nearly 100% humidity and 8°C. The cryo-grids were initially che cked at 300 kV with the FEI Tecnai F30, with only good quality grids selected for data collection. Cryo-EM images acquired with the FEI Titan Kiros (Thermo Fisher Scientific) EM were recorded on a Gatan K2 Summit direct electron detector at a nominal 130,000× magnification, corresponding to a calibrated physical pixel size of 1.307 Å (0.6535 Å for super-resolution pixel size). Movies comprise 40 frames, with a total dose of 56 e−/Å2 at an exposure time of 8 s. Data were semi-automatically collected using SerialEM(Mastronarde, 2003). Cryo-EM images acquired with the FEI Tecnai F30 were recorded on the Falcon II or III direct electron detector at a nominal 93,000× magnification, corresponding to a pixel size of 1.128 Å or 1.120 Å, respectively. A total electron dose of about 40 e−/Å2 was fractionated into 17 or 39 frames in every movie at an exposure time of 1 s. Data were automatically collected using FEI EPU.
Three-dimensional reconstruction
Drift and beam-induced motion correction were performed with MotionCor2(Zheng et al., 2017). Contrast transfer function (CTF) fitting and phase-shift estimation were carried out with Gctf(Zhang, 2016). Micrographs with astigmatism, significant drift, or contamination were discarded before reconstruction. Particles were automatically picked and screened using cisTEM(Grant et al., 2018). The initial 3D models for each dataset were generated with a random model method using AUTO3DEM(Yan et al., 2007). Several rounds of reference-free 2D classifications and unsupervised 3D classifications were executed using Relion 2.1(Kimanius et al., 2016). Sorted particles were then subjected to final homogenous refinement using Relion 2.1, cisTEM, or cryoSPARC v2.4.2(Punjani et al., 2017). The resolution of all density maps was determined by the gold-standard Fourier shell correlation curve with a cutoff at 0.143. Local map resolution was estimated with ResMap(Kucukelbir et al., 2014).
Sub-particle classification and refinement for CVA16-M:18A7
To resolve the structures of 18A7 Fab at high resolution, we used Relion symmetry expansion and Scipion(De la Rosa-Trevin et al., 2016) to exact and perform sub-particle reconstruction as previously described(Ilca et al., 2015). Briefly, after 3D refinement with imposition of icosahedral symmetry, we extracted sub-particles from the 5-fold region in a box-size of 140×140 pixels and expanded the sub-particles with I2 symmetry. The extracted sub-particles were used to generate the initial model using relion_reconstruct. 3D classification was then performed without alignment. Five classes (#1, #5, #6, #7, #8) revealed distinguished features of the Fab fragment, of which class #5 reconstruction had the highest quality. Further refinement of class #5 led to a structure at 3.67 Å resolution. The resolution was assessed by Fourier shell correlation curve with a cutoff at 0.143 from two independent half-sets of the sub-particles. The work flow is depicted in Figure S3.
Model building and refinement
As the constant domain of the Fab fragment exhibited very weak densities, we only built the variable domain model for structural analysis (Figure S8). The initial atomic models for the variable domain of the Fab fragments (18A7, 14B10 and NA9D7) were generated from homology modeling by Accelrys Discovery Studio software(Studio, 2009). Combining with the crystal structures of the CVA16 mature virus (pdb code: 5C4W), A particle (pdb code: 4JGY), and empty particle (pdb code: 5C9A), we initially fitted the templates into the corresponding segmented volume (enclosing a protomer) of the final cryoEM maps (viral particles or immune complexes) using Chimera(Pettersen et al., 2004) and further corrected and adjusted manually by real-space refinement in Coot(Emsley and Cowtan, 2004). The built models were then refined using phenix.real_space_refine in PHENIX(Adams et al., 2010). These operations were executed iteratively until the problematic regions, Ramachandran outliers, and poor rotamers were in favor. After several cycles of refinement, the resulting models were fitted into the map of six neighboring protomers. These total of seven protomers were taken together for further refinement to optimize the clashes. The final atomic models were validated using Molprobity(Chen et al., 2010). Model statistics are summarized in the Table 1. Sequence alignment was performed with Clustal Omega on the EBI server (https://www.ebi.ac.uk/Tools/msa/clustalo/). The Fab-capsid buried surface area and interaction interfaces were calculated using the CCP4(Collaborative, 1994) program suite and the PISA server (www.ebi.ac.uk/pdbe/pisa). For the intermolecular interactions, maximal cut-off distances of 4.0 Å were used for hydrogen bonding, salt-bridge formation and other contacts. The roadmap in graphical abstract was generated using Rivem(Xiao and Rossmann, 2007). All figures were generated with ChimeraX(Goddard et al., 2018), Chimera, or Pymol(Schrodinger, 2010).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments are indicated in the figure legend for Figure 5. Statistical significance was determined as p < 0.05 using GraphPad Prism 7.0 and 95% confidence level was applied. Data were analyzed by unpaired Student’s t-test analysis, and each group was composed of 5 values (represent 5 mice per group). The neutralizing titers were expressed as mean ± SD.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The cryo-EM density maps and corresponding atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB), respectively. The accession codes are: CVA16 mature virion (EMD-0887, PDB: 6LHA); CVA16 A-particle (EMD-0888, PDB: 6LHB); CVA16 empty particle (EMD-0889, PDB: 6LHC); CVA16-M:18A7(EMD-0890, PDB: 6LHK); CVA16-M:18A7-local (EMD-0897, PDB: 6LHT); CVA16-A:18A7 (EMD-0891, PDB: 6LHL); CVA16-E:18A7 (EMD-0892, PDB: 6LHO); CVA16-M:14B10 (EMD-0894, PDB: 6LHP); CVA16-M:NA9D7 (EMD-0895, PDB: 6LHQ); CVA16-M:18A7:14B10:NA9D7 (EMD-0898).
Supplementary Material
Table S1. Interactions between different Fabs and CVA16 mature virion, Related to Figure 3.
Video S1. Overall structure and high-resolution features of CVA16-M:18A7 immune complex, Related to Figures 2 and 3.
Depicts the global reconstructed features of CVA16-M:18A7 at the level of 1.5σ and 3.5σ. The 3.67 Å high-resolution features of CVA16-M:18A7-local was shown.
Video S2. Overall structure of CVA16-M:14B10 immune complex, Related to Figure 2.
Depicts the global reconstructed features of CVA16-M:14B10, and close-up view of the 14B10 binding region.
Video S3. Overall structure of CVA16-M:NA9D7 immune complex, Related to Figure 2.
Depicts the global reconstructed features of CVA16-M:NA9D7, and close-up view of the 9D7 binding region.
Video S4. Overall structure of CVA16-M:18A7:14B10:NA9D7 immune complex, Related to Figure 2.
Depicts the global reconstructed features of CVA16-M: 18A7:14B10:NA9D7
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal Antibody-18A7 | This study | N/A |
| Mouse Monoclonal Antibody-14B10 | This study | N/A |
| Mouse Monoclonal Antibody-NA9D7 | This study | N/A |
| Bacterial and Virus Strains | ||
| CVA16/190 strain | This study (Infectious clone) | GenBank no. KJ850274 |
| CVA16/213a strain | This study (Infectious clone) | GenBank no. JX127259 |
| CVA16/4430 strain | This study (Infectious clone) | GenBank no. JX127274 |
| CVA16/4479 strain | This study (Infectious clone) | GenBank no. JX127277 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SYTO9 | Invitrogen | S34854 |
| Critical Commercial Assays | ||
| QIAamp Mini viral RNA Extraction Kit | Qiagen | 52904 |
| Deposited Data | ||
| Cryo-EM structure of CVA16 mature virion | This paper | EMD-0887, PDB: 6LHA |
| Cryo-EM structure of CVA16 A-particle | This paper | EMD-0888, PDB: 6LHB |
| Cryo-EM structure of CVA16 empty particle | This paper | EMD-0889, PDB: 6LHC |
| Cryo-EM structure of CVA16-M:18A7 | This paper | EMD-0890, PDB: 6LHK |
| Cryo-EM structure of CVA16-A:18A7 | This paper | EMD-0891, PDB: 6LHL |
| Cryo-EM structure of CVA16-E:18A7 | This paper | EMD-0892, PDB: 6LHO |
| Cryo-EM structure of CVA16-M:18A7-local | This paper | EMD-0897, PDB: 6LHT |
| Cryo-EM structure of CVA16-M:14B10 | This paper | EMD-0894, PDB: 6LHP |
| Cryo-EM structure of CVA16-M:NA9D7 | This paper | EMD-0895, PDB: 6LHQ |
| Cryo-EM structure of CVA16-M:18A7:14B10:NA9D7 | This paper | EMD-0898 |
| Experimental Models: Cell Lines | ||
| Myeloma cells (Sp2/0-Ag-14) | This study | N/A |
| RD cells | ATCC | CCL-136 |
| Experimental Models: Organisms/Strains | ||
| BALB/c mice | Slac Laboratory Animal Co., shanghai, China | N/A |
| Software and Algorithms | ||
| Prism 7.0 | GraphPad Software | N/A |
| MotionCor2 | (Zheng et al., 2017) | http://msg.ucsf.edu/em/software/motioncor2.html |
| Gctf | (Zhang, 2016) | https://en.wikibooks.org/w/index.php?title=Software_Tools_For_Molecular_Microscopy&stable=0#Gctf |
| RELION2.1 | (Kimanius et al., 2016) | http://www2.mrc-lmb.cam.ac.uk/relion |
| ResMap | (Kucukelbir et al., 2014) | http://resmap.sourceforge.net |
| AUTO3DEM | (Yan et al., 2007) | http://cryoem.ucsd.edu/wikis/software/start.php?id=auto3dem:home |
| Chimera | (Pettersen et al., 2004) | http://www.cgl.ucsf.edu/chimera |
| PyMOL | (Schrodinger, 2010) | https://pymol.org/2/ |
| cryoSPARC | (Punjani et al., 2017) | https://cryosparc.com |
| ChimeraX | (Goddard et al., 2018) | https://www.cgl.ucsf.edu/chimerax/ |
| COOT | (Emsley and Cowtan, 2004) | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| PHENIX | (Adams et al., 2010) | http://phenix-online.org |
| Molprobity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu |
| CCP4 | (Collaborative, 1994) | http://www.ccp4.ac.uk |
| cisTEM | (Grant et al., 2018) | https://cistem.org |
| Scipion | (De la Rosa-Trevín et al., 2016) | https://scipion-em.github.io/docs/ |
| Localized reconstruction | (Ilca et al., 2015) | https://github.com/OPIC-Oxford/localrec |
| Rivem | (Xiao and Rossmann, 2007) | http://bilbo.bio.purdue.edu/~viruswww/Rossmann_home/softwares/river_programs/rivem.php |
| Other | ||
| Quantifoil R 2/2 Cu grids | Quantifoil | Q2100CR2 |
Highlights.
Atomic models show CVA16 can simultaneously bind three distinct potent nAbs
The neutralization sites vary across three forms of CVA16
CVA16 mature virion bearing conserved epitopes is the optimal vaccine immunogen
nAb-based assay allows quantification of mature virions for vaccine development
Acknowledgements
We thank Drs. Xin Zhang and Shenghai Chang for cryo-EM data acquisition in Center of Cryo-Electron Microscopy, Zhejiang University. This work was supported by a grant from the Major Program of National Natural Science Foundation of China (no. 81991490), the National Science and Technology Major Projects for Major New Drugs Innovation and Development (no. 2018ZX09711003–005), the National Science and Technology Major Project of Infectious Diseases (2017ZX10304402–002-003), the National Natural Science Foundation of China (no. 31670933 and 81801646), the China Postdoctoral Science Foundation (no. 2018M640599 and 2019T120557) and the Principal Foundation of Xiamen University (no. 20720190117), grants from by the National Institutes of Health (grants R37-GM33050, GM071940, DE025567 and AI094386). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. We also thank Nguyen Titania for proofreading the manuscript.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB., Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, and Grosse-Kunstleve RW (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D: Biological Crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen J, Thibaut H, Strating JR, and van Kuppeveld FJ. (2018). The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol, 1. [DOI] [PubMed] [Google Scholar]
- Belnap DM, McDermott BM, Filman DJ, Cheng N, Trus BL, Zuccola HJ, Racaniello VR., Hogle JM, and Steven AC (2000). Three-dimensional structure of poliovirus receptor bound to poliovirus. Proceedings of the National Academy of Sciences 97, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson JM, and Coyne CB (2013). Picornavirus entry. In Viral Entry into Host Cells (Springer; ), pp. 24–41. [DOI] [PubMed] [Google Scholar]
- Bowman VD, Chase ES, Franz AW, Chipman PR., Zhang X, Perry KL, Baker TS, and Smith TJ (2002). An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. Journal of virology 76, 12250–12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Liu Q, Huang X, Li D, Ku Z, Zhang Y, and Huang Z (2013). Active immunization with a Coxsackievirus A16 experimental inactivated vaccine induces neutralizing antibodies and protects mice against lethal infection. Vaccine 31, 2215–2221. [DOI] [PubMed] [Google Scholar]
- CHANG L-Y, LIN T-Y, HUANG Y-C, TSAO K-C, SHIH S-R, KUO M-L, NING H-C, CHUNG P-W, and KANG C-M (1999). Comparison of enterovirus 71 and coxsackievirus A16 clinical illnesses during the Taiwan enterovirus epidemic, 1998. The Pediatric infectious disease journal 18, 1092–1096. [DOI] [PubMed] [Google Scholar]
- Chen VB., Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P, Guo M-S, Lin FH-Y, Hsiao K-N, Weng S-Y, Chou A-H, Wang J-R, Hsieh S-Y, Su IJ, and Liu C-C (2012). Immunological and biochemical characterization of coxsackie virus A16 viral particles. PLoS One 7, e49973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative CP (1994). The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography 50, 760. [DOI] [PubMed] [Google Scholar]
- De la Rosa-Trevín J, Quintana A, Del Cano L, Zaldivar A, Foche I, Gutiérrez J, Gómez-Blanco J, Burguet-Castell J, Cuenca-Alba J, and Abrishami V (2016). Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. Journal of structural biology 195, 93–99. [DOI] [PubMed] [Google Scholar]
- Dong Y, Liu Y, Jiang W, Smith TJ., Xu, Z., and Rossmann, M.G. (2017). Antibody-induced uncoating of human rhinovirus B14. Proceedings of the National Academy of Sciences 114, 8017–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fan C, Ye X, Ku Z, Kong L, Liu Q, Xu C, Cong Y, and Huang Z (2017). Beta-propiolactone inactivation of coxsackievirus A16 induces structural alteration and surface modification of viral capsids. Journal of virology 91, e00038–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Wood DJ, and Minor PD. (1993). Antigenic structure of poliovirus in inactivated vaccines. Journal of general virology 74, 685–690. [DOI] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, and Howley PM. (2007). Fields virology, 5th edn (Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; ). [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). cisTEM, user-friendly software for single-particle image processing. Elife 7, e35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Skaletsky E, and McPherson A (1998). Crystallographic structure of an intact IgG1 monoclonal antibody. Journal of molecular biology 275, 861–872. [DOI] [PubMed] [Google Scholar]
- Hewat EA, and Blaas D (2006). Nonneutralizing human rhinovirus serotype 2-specific monoclonal antibody 2G2 attaches to the region that undergoes the most dramatic changes upon release of the viral RNA. Journal of virology 80, 12398–12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewat EA, Neumann E, Conway JF, Moser R, Ronacher B, Marlovits TC, and Blaas D (2000). The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. The EMBO journal 19, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle J, Chow M, and Filman D (1985). Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229, 1358–1365. [DOI] [PubMed] [Google Scholar]
- Hou W, Yang L, He D, Zheng J, Xu L, Liu J, Liu Y, Zhao H, Ye X, and Cheng T (2015). Development of a coxsackievirus A16 neutralization test based on the enzyme-linked immunospot assay. Journal of virological methods 215, 56–60. [DOI] [PubMed] [Google Scholar]
- Ilca SL, Kotecha A, Sun X, Poranen MM, Stuart DI, and Huiskonen JT (2015). Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat Commun 6, 8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE (2010). Cell entry by non-enveloped viruses, Vol 343 (Springer Science & Business Media; ). [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nature methods 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen NS, Friesen RH, Zhu X, Jongeneelen M, Blokland S, Vermond J, van Eijgen A, Tang C, van Diepen H, and Obmolova G (2018). Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 362, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Cifuente JO, Ashley RE, Conway JF, Makhov AM, Tano Y, Shimizu H, Nishimura Y, and Hafenstein S (2013). A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody. Journal of virology 87, 11363–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-X, Song Y-F, Wang L, Zhang X-F, Hu Y-S, Hu Y-M, Xia J-L, Li J, and Zhu F-C (2016). Two-year efficacy and immunogenicity of Sinovac Enterovirus 71 vaccine against hand, foot and mouth disease in children. Expert review of vaccines 15, 129–137. [DOI] [PubMed] [Google Scholar]
- Li R, Liu L, Mo Z, Wang X, Xia J, Liang Z, Zhang Y, Li Y, Mao Q, and Wang J (2014). An inactivated enterovirus 71 vaccine in healthy children. New England Journal of Medicine 370, 829–837. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Gu Y, Song S, He M, Shi J, Liu X, Wei S, Li J, and Yu H (2017). Crystal structures of two immune complexes identify determinants for viral infectivity and type-specific neutralization of human papillomavirus. MBio 8, e00787–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wu S, Xiong Y, Li T, Wen Z, Yan M, Qin K, Liu Y, and Wu J (2014). Co-circulation and genomic recombination of coxsackievirus A16 and enterovirus 71 during a large outbreak of hand, foot, and mouth disease in Central China. PloS one 9, e96051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q, Wang Y, Yao X, Bian L, Wu X, Xu M, and Liang Z (2014). Coxsackievirus A16: epidemiology, diagnosis, and vaccine. Human vaccines & immunotherapeutics 10, 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2003). SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microscopy and Microanalysis 9, 1182–1183. [Google Scholar]
- McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, and Zheng A (2013). Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, and Shimizu H (2009). Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nature medicine 15, 794. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Plevka P, Lim P-Y, Perera R, Cardosa J, Suksatu A, Kuhn RJ, and Rossmann MG (2014). Neutralizing antibodies can initiate genome release from human enterovirus 71. Proceedings of the National Academy of Sciences 111, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods 14, 290. [DOI] [PubMed] [Google Scholar]
- Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, and Zhao Y (2013). Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4, 1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wang X, Zhu L, Hu Z, Gao Q, Yang P, Li X, Wang J, Shen X, and Fry EE (2015). Structures of coxsackievirus A16 capsids with native antigenicity: implications for particle expansion, receptor binding, and immunogenicity. Journal of virology 89, 10500–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C, Doane FW, and Rhodes A (1958). Report of an outbreak of febrile illness with pharyngeal lesions and exanthem: Toronto, summer 1957—isolation of group A coxsackie virus. Canadian Medical Association Journal 79, 615. [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L (2010). The PyMOL molecular graphics system. Version 1, 0. [Google Scholar]
- Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, and Hafenstein S (2013). The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS pathogens 9, e1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studio D (2009). version 2.5. Accelrys Inc: San Diego, CA, USA. [Google Scholar]
- Walter TS, Ren J, Tuthill TJ, Rowlands DJ, Stuart DI, and Fry EE (2012). A plate-based high-throughput assay for virus stability and vaccine formulation. Journal of virological methods 185, 166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Y, Lu FL, Wu M-H, Lee C-Y, and Huang L-M (2004). Fatal coxsackievirus A16 infection. The Pediatric infectious disease journal 23, 275–276. [DOI] [PubMed] [Google Scholar]
- Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, and Porta C (2012). A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nature structural & molecular biology 19, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, and Rossmann MG (2007). Interpretation of electron density with stereographic roadmap projections. Journal of structural biology 158, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Tjarnlund K, Lindqvist B, Kaplan GG, Feigelstock D, Cheng RH, and Casasnovas JM (2000). Distinct cellular receptor interactions in poliovirus and rhinoviruses. The EMBO Journal 19, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Acharya P, Kong R, Cheng C, Chuang G-Y, Liu K, Louder MK, O’Dell S, Rawi R, and Sastry M (2018). Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nature medicine 24, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zheng Q, Li S, He M, Wu Y, Li Y, Zhu R, Yu H, Hong Q, and Jiang J (2017). Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody. Nat Commun 8, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, and Koike S (2009). Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nature medicine 15, 798. [DOI] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, and Baker TS (2007). AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. Journal of structural biology 157, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Yang L, Jia J, Han J, Li S, Liu Y, Xu L, Zhao H, Chen Y, and Li Y (2016). Development of sandwich ELISAs that can distinguish different types of coxsackievirus A16 viral particles. Applied microbiology and biotechnology 100, 2809–2815. [DOI] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. Journal of structural biology 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi J, Ye X, Ku Z, Zhang C, Liu Q, and Huang Z (2017). Coxsackievirus A16 utilizes cell surface heparan sulfate glycosaminoglycans as its attachment receptor. Emerging microbes & infections 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Zhu R, Xu L, He M, Yan X, Liu D, Yin Z, Wu Y, Li Y, and Yang L (2019). Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization. Nature microbiology 4, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhao Y, Kotecha A, Fry EE, Kelly JT, Wang X, Rao Z, Rowlands DJ, Ren J, and Stuart DI (2019). Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. Nature microbiology 4, 414. [DOI] [PubMed] [Google Scholar]
- Zhu F, Xu W, Xia J, Liang Z, Liu Y, Zhang X, Tan X, Wang L, Mao Q, and Wu J (2014). Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. New England Journal of Medicine 370, 818–828. [DOI] [PubMed] [Google Scholar]
- Zhu F-C, Meng F-Y, Li J-X, Li X-L, Mao Q-Y, Tao H, Zhang Y-T, Yao X, Chu K, and Chen Q-H (2013). Efficacy, safety, and immunology of an inactivated alum-adjuvant enterovirus 71 vaccine in children in China: a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet 381, 2024–2032. [DOI] [PubMed] [Google Scholar]
- Zhu L, Sun Y, Fan J, Zhu B, Cao L, Gao Q, Zhang Y, Liu H, Rao Z, and Wang X (2018a). Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating. Nat Commun 9, 4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Xu K, Wang N, Cao L, Wu J, Gao Q, Fry EE, Stuart DI, Rao Z, and Wang J (2018b). Neutralization mechanisms of two highly potent antibodies against human enterovirus 71. mBio 9, e01013–01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, and Li Z (2018c). Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10. Sci Adv 4, eaat7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Interactions between different Fabs and CVA16 mature virion, Related to Figure 3.
Video S1. Overall structure and high-resolution features of CVA16-M:18A7 immune complex, Related to Figures 2 and 3.
Depicts the global reconstructed features of CVA16-M:18A7 at the level of 1.5σ and 3.5σ. The 3.67 Å high-resolution features of CVA16-M:18A7-local was shown.
Video S2. Overall structure of CVA16-M:14B10 immune complex, Related to Figure 2.
Depicts the global reconstructed features of CVA16-M:14B10, and close-up view of the 14B10 binding region.
Video S3. Overall structure of CVA16-M:NA9D7 immune complex, Related to Figure 2.
Depicts the global reconstructed features of CVA16-M:NA9D7, and close-up view of the 9D7 binding region.
Video S4. Overall structure of CVA16-M:18A7:14B10:NA9D7 immune complex, Related to Figure 2.
Depicts the global reconstructed features of CVA16-M: 18A7:14B10:NA9D7
Data Availability Statement
Data Resources
The cryo-EM density maps and corresponding atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB), respectively. The accession codes are: CVA16 mature virion (EMD-0887, PDB: 6LHA); CVA16 A-particle (EMD-0888, PDB: 6LHB); CVA16 empty particle (EMD-0889, PDB: 6LHC); CVA16-M:18A7(EMD-0890, PDB: 6LHK); CVA16-M:18A7-local (EMD-0897, PDB: 6LHT); CVA16-A:18A7 (EMD-0891, PDB: 6LHL); CVA16-E:18A7 (EMD-0892, PDB: 6LHO); CVA16-M:14B10 (EMD-0894, PDB: 6LHP); CVA16-M:NA9D7 (EMD-0895, PDB: 6LHQ); CVA16-M:18A7:14B10:NA9D7 (EMD-0898).