Abstract
Human embryogenesis frequently coinciding with cell division mistakes contributing to pervasive embryonic aneuploidy/mosaicism. While embryo self-correction was elegantly demonstrated in mouse models, human studies are lacking. Here we are witness to human embryos ability to eliminate/expel abnormal blastomeres as cell debris/fragments. Each blastocyst and its corresponding debris were separated and underwent whole genome amplification. Seven of the 11 pairs of blastocysts and their corresponding cell debris/fragments revealed discordant results. Of the 9 euploid blastocysts, four showed euploid debris, while in the others, the debris were aneuploid. In the remaining pairs, the debris showed additional aneuploidy to those presented by their corresponding blastocyst. The observed ability of human embryos to self-correction doubts many invasive and non-invasive preimplantation testing for aneuploidy at the blastocyst stage, rendering high rate of false positive (discarding “good” embryos) by identifying the cell-free DNA originated from the expelled cell debris, as aneuploidy/mosaic blastocyst.
Keywords: Self-correction, PGT, Human embryo, Blastocyst, Mosaicism
Introduction
The first stages of human embryogenesis are characterized by rapid cell proliferation, which frequently coinciding with cell division mistakes, generating changes in chromosome content, e.g. aneuploidy [1–3]. Oocyte meiotic aneuploidies may result from non-disjunction or premature separation of a chromosome into sister chromatids [4], and appears in the whole embryo’s cells [4, 5]. Along with errors in meiosis, mitotic errors during post-zygotic cell division contribute to pervasive aneuploidy in human embryos [6]. Mitotic mistakes are common, with the highest occurrence through the first three cleavages after fertilization [7]. Consequently, the majority of the human preimplantation embryos show aneuploidies that appears mainly as a diploid–aneuploid mosaicism. As with meiotic errors, mitotic mistakes also decrease with embryonic development. This was elegantly observed while re-analyzing blastocyst-stage embryos, which were detected as aneuploid at the cleavage stage (Day-3) [8–10].
Mosaicism has been reported in, as high as 50% of cleavage- and blastocyst-stage embryos derived from IVF [11]. Preliminary studies suggest that “mosaic” embryos display low rates of concordance between multiple trophectoderm (TE) biopsies. Moreover, “mosaic” embryos demonstrate increased cell proliferation and cell death in comparison to euploid embryos, both observations suggestive of significant self-correction abilities of embryos [12–14].
Santos et al. have suggested that embryo self-correction may result from increased aneuploidy cells death, or decreased cell division rate [15]. These suggested mechanisms might be supported by observations of aneuploid blastomeres leaving the blastocyst following the activation of apoptotic pathways [16]. Daughtry et al. [17] have demonstrated that Rhesus embryos overcome chromosome instability during preimplantation development by encapsulation of chromosome-containing cellular fragments into micronuclei and their elimination via cellular fragmentation. In their elegant mice study, Bolton et al. shed additional light onto the embryo’s ability to self-correct [14]. Treating mouse embryos with a spindle assembly checkpoint inhibitor during the 4- to 8-cell division, the authors generated chimeric embryos with euploid and aneuploid cells, which they followed via live-embryo imaging and single cell tracking. They found that aneuploid cells in the fetal lineage (i.e., inner cell mass producing the fetus) were eliminated by apoptosis, while those in the placental lineage (i.e., the TE) did show proliferative defects though survived. In a recently published study from the same group, Singla et al. [18] have demonstrated that aneuploid cells are preferentially eliminated from the embryonic lineage in a p53-dependent process involving both autophagy and apoptosis before, during and after implantation. It might be therefore argued, that early embryos development could be controlled by the establishment of a cell death program to ensure the elimination of damaged cells [19], while maintaining an optimal balance between survival and apoptotic signals.
Studies of cell death and survival in preimplantation embryos use mainly, mice models, aiming to overcome the ethical concerns limiting human embryos research. However, species specific differences might limit the extrapolation of results from mouse to human embryos. For example, Haouzi et al. [20] showed that while mouse and human embryos express components of the apoptotic and survival pathways during early embryonic development, their expression profile differs.
Recently, we presented a case of a cleaving human embryo from one to- two cells, with one cell hatching out from the zone pellucida, and thereafter their development into two blastocysts [21]. The two blastocysts were separated and sent to a comprehensive molecular analysis (polymorphic markers and array-CGH), demonstrating identical twins. The embryo in the zona pellucida showed normal balanced chromosomal profile, while the embryo that was expelled from the zona pellucida showed unbalanced chromosomal profile. In the present case, we were evident of ‘normalization’ or ‘self-correction’ of the chromosomally abnormal human preimplantation embryo by splitting in to two embryos, already in the first cell-division.
Even more intriguing, is the ability of human “mosaic” blastocysts to implant and develop to a viable pregnancy, though, with higher rates of miscarriage than euploid blastocyst [22]. One explanation for this ‘self-correction’ ability of an abnormal mosaic embryo to actually develop into a viable pregnancy, has been the natural apoptosis of abnormal cells. Of notice, finding direct evidence for any corrective mechanisms during early human development is extremely challenging. To overcome these limitations, we used affected / discarded human embryos undergoing preimplantation genetic testing for monogenic aberrations (PGT-M) and gender related disorders. In our PGT-M program, DNA extraction is obtained during the cleavage stage, where one blastomere from Day 3 embryo is extracted and undergo genetic diagnosis. In our routine clinical practice, following Day-3 biopsy, healthy embryos are transferred on Day-4 or 5, and the affected embryos are discarded. When these embryos were cultured until Day-5-6, we noticed that some of the blastocysts expel cell debris/ fragments within the zona pellucida (Fig. 1).
Fig. 1.
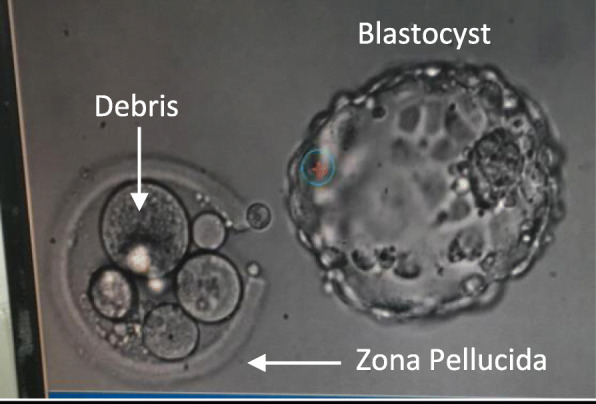
Day 5 hatched blastocyst and its original zona pellucida containing leftovers of cell debris
Prompted by this observation, we sought to examine the DNA content of these debris and their corresponding blastocyst (affected embryos that their day-3 blastomere biopsy revealed single-gene defect, and were donated for research by the couples), aiming to prove the hypothesis, that human embryos have the ability to self- correction and can eliminate/expel abnormal blastomere (the debris).
Material and methods
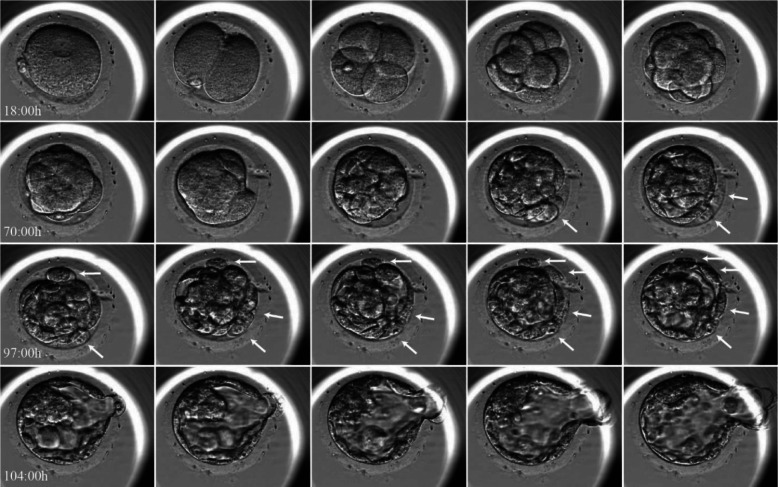
High-quality blastocysts [23], in which their Day-3 blastomere biopsy revealed an affected embryo with single-gene defect, were donated by couples undergoing PGT-M treatment at the Sheba Medical Center. Only blastocysts cultured in closed system using the time-lapse EmbryoScope™ incubator, that following hatching leaved cell debris/fragments within the zona pellucida were analyzed (Fig. 2 and Additional file 1). Each blastocyst and its corresponding debris were separated and underwent whole genome amplification (WGA).
Fig. 2.
Time-lapse EmbryoScope™ photography of embryo expelling cell debris/cell fragments within the zona pellucida. Arrows pointing the cell debris
WGA
Full-genomic amplification of the DNA was carried out by WGA-PCR PicoPlex SingleCell WGA Kit (Rubicon Genomics) [24]. The quality and quantity of DNA received during amplification were controlled by electrophoresis using 1% agarose gel.
Array-CGH
WGA products were processed referring to the protocol of Agilent oligonucleotide array-based CGH for single cell G4410–90003 Revision B0, October 2018. These products were fluorescently labelled with controls (Human Reference DNA Female/Male) according to the instructions of SureTag Complete DNA Labeling Kit (Agilent technologies, CA, USA), and then competitively hybridized to G9500A GenetiSure Pre-Screen Complete kit (8 × 60) Agilent technologies, CA, USA) [25].
Interpretation of array-CGH results
Subsequent data analysis was performed according to the manufacturer recommended single cell analysis method. We only reported whole chromosome trisomies or monosomies.
The study required no modification of patient’s routine follow-up or treatment. Informed consent was obtained from all patients before participation in the study, and the study was approved by our Institutional Clinical Research Committee (IRB SMC-19-6140).
Results
Twenty women (age 33.7 + 4.8 yrs) achieved 175 blastocysts that were cultured in closed system using the time-lapse photography (EmbryoScope™ incubator) in our PGT-M program, from August 2017 to March 2020. The recorded films were analysed and revealed that 112 (64%) embryos expel cell debris/ fragments outside the intact embryo (Fig. 2 and Additional file 1).
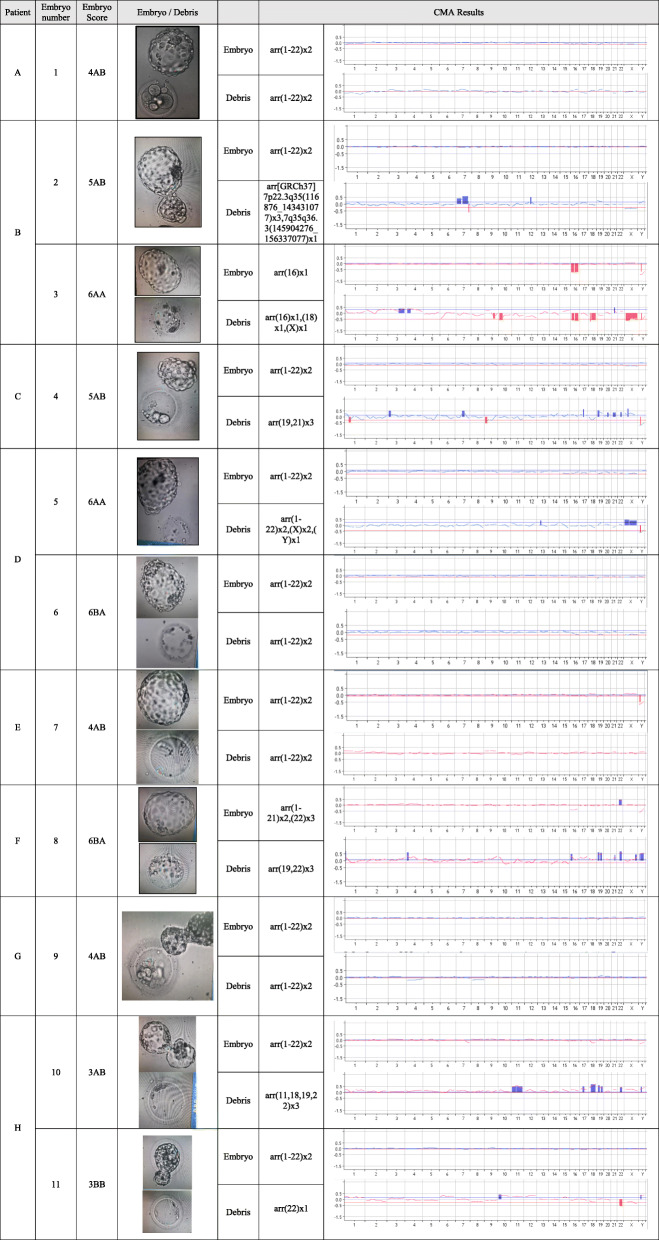
Eight patients (mean women age 32.4 + 3.2 years, range 27–37 years) undergoing IVF/ICSI cycle for PGT-M, donated 11 high-quality blastocysts, which were found to be affected following Day-3 blastomere biopsy. The blastocyst morphological grading [23], their appearances, and the CGH results of the blastocysts and their corresponding cell debris/fragments are presented in Table 1.
Table 1.
The appearance and CGH results of the blastocysts and their corresponding cell debris/fragments
Seven out of the 11 pairs of blastocysts/cell debris revealed discordant results. Of the 9 euploid blastocysts, the cell debris of four were euploid (embryos #1,6,7,9), four others demonstrated trisomies 19,21 (embryo #4); trisomies 11,18,19,22(embryo #10); trisomy 7(embryo #2); and monosomy 22(embryo #11), and one was XXY(embryo #5). In the remaining two pairs, one blastocyst was monosomic 16 (embryo #3) and the debris monosomic 16, 18 and X, while the other showed trisomy 22 (embryo #8) and its debris revealed trisomy 22, in addition to trisomy 19.
Discussion
In the present study of extended embryo culture, we are witness of human embryos self-correction mechanism, discovered by their ability to eliminate/expel abnormal blastomeres as cell debris/fragments (Fig. 2 and Additional file 1). Seven (63.6%) out of the 11 pairs of blastocysts expelled cell debris with additional chromosomal rearrangements. Moreover, of the 9 euploid blastocysts, 5 (55.5%) expelled aneuploidy debris.
Women’s fecundity decreases gradually with increasing age, coincident with increased embryonic aneuploidy and spontaneous miscarriages rates [26]. These observations have led to the attractively logical, but still equivocal hypothesis of PGT-A, that the transfer of only euploid embryos should improve IVF outcomes [27, 28]. Even more perturbing, is the reported efficacy of noninvasive PGT-A (niPGT-A) in the spent culture media of human blastocysts by analyzing the cell-free DNA. Huang et al. [29] have reported a zero false-negative rate for niPGT-A, with both the positive predictive value and specificity found to be much higher than TE biopsy PGT-A. Since the concordance rates for both embryo ploidy and chromosome copy numbers were higher for niPGT-A, they suggested that niPGT-A is less prone to errors associated with embryo mosaicism and is more reliable than TE-biopsy PGT-A.
The observed ability of human embryos to self-correction doubts many invasive and ni-PGT-A at the blastocyst stage, rendering high rate of false positive (discarding “good” embryos), by identifying the cell-free DNA originated from the expelled cell debris as aneuploidy/mosaic blastocyst. In their editorial, Gleicher and Barad [30] have provided further explanation to the pitfalls of niPGT-A, e.g. “TE and ICM are assumed to leak into spent media, but TE is in direct contact with medium and ICM is not”. Our observation of human embryo ability to self-correction and expulsion of aneuploidy cells to the culture media, and the reported incongruity between TE and ICM biopsies [12], further highlights the unreliability of ni-PGT-A.
Huang et al. [29] have further speculated that “at least for the euploid embryos, the leakage of DNA from the euploid cells outweighs that of the apoptotic aneuploid cells; otherwise, niPGT-A would not be able to report the euploid embryos successfully”. This assumption was not confirmed in our study, but the opposite. Five out of the 9 euploid whole blastocysts examined revealed different CGH results in their expelled cell-debris: trisomies 19,21; trisomies 11,18,19,22; trisomy 7; monosomy 22, and XXY.
Low-level mosaicism is a common feature of early human development. A recently published study on single-cell genomic data revealed widespread mosaic aneuploidies, with 80% embryos harboring at least one putative aneuploid cell, with no significant enrichment of aneuploid cells in the TE compared to the ICM [31]. These observations conclusively demonstrate that mosaicism in blastocyst-stage embryos is basically a normal physiological phenomenon that can be found in almost all embryos [14, 31].
In conclusion, the present study sheds more light on human embryogenesis and its capability for self-correction. Before adopting any future diagnostic procedure aiming to improve embryo selection during an IVF cycle, the aforementioned discussed physiological/embryological observations should be considered in an attempt to improve the test validity.
Supplementary information
Additional file 1. Time-lapse EmbryoScope™ photography of embryo expelling cell debris/cell fragments within the zona pellucida (https://youtu.be/3RNUJ4iW0IE).
Acknowledgements
The authors would like to thank Ms. Moran Madari for her contributions to Data collection and secretarial assistance.
Authors’ contributions
R.O. Designed the study, wrote the first paper draft, edited it, proof read the paper and took part in discussions regarding the results. C.S. Conducted and retrieved the embryological data, proof read the paper and took part in discussions regarding the results. S.R. and A.J.G. conducted the WGA experiments, proof read the paper and took part in discussions regarding the WGA interpretations. H.S. Proof read the paper and took part in discussions regarding the results. A.A. Participated in designing the study, conducted the embryological work and retrieved the data, proof read the paper and took part in discussions regarding the results. The author(s) read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by our institutional review board.
Consent for publication
Not applicable.
Competing interests
RO is the journal EIC. CS, SR, AJG, HS, AA have nothing to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Raoul Orvieto, Email: Raoul.orvieto@sheba.health.gov.il.
Chen Shimon, Email: Chen.Shimon@sheba.health.gov.il.
Shlomit Rienstein, Email: Shlomit.Rienstein@sheba.health.gov.il.
Anat Jonish-Grossman, Email: Anat.Grossman@sheba.health.gov.il.
Hagit Shani, Email: Shani.Hagit@sheba.health.gov.il.
Adva Aizer, Email: Adva.aizer@sheba.health.gov.il.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12958-020-00650-8.
References
- 1.Delhanty JD, Griffin DK, Handyside AH, Harper J, Atkinson GH, Pieters MH, Winston RM. Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination by fluorescent in situ hybridisation (FISH) Hum Mol Genet. 1993;2:1183–1185. doi: 10.1093/hmg/2.8.1183. [DOI] [PubMed] [Google Scholar]
- 2.Angell RR, Aitken RJ, van Look PF, Lumsden MA, Templeton AA. Chromosome abnormalities in human embryos after in vitro fertilization. Nature. 1983;303:336–338. doi: 10.1038/303336a0. [DOI] [PubMed] [Google Scholar]
- 3.Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2:366. doi: 10.1016/S0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- 4.Fragouli E, Wells D, Delhanty JD. Chromosome abnormalities in the human oocyte. Cytogenet Genome Res. 2011;133:107–118. doi: 10.1159/000323801. [DOI] [PubMed] [Google Scholar]
- 5.Mantzouratou A, Delhanty JD. Aneuploidy in the human cleavage stage embryo. Cytogenet Genome Res. 2011;133:141–148. doi: 10.1159/000323794. [DOI] [PubMed] [Google Scholar]
- 6.Bielanska M, Tan SL, Ao A. Chromosomal mosaicism throughout human preimplantation development in vitro: incidence, type, and relevance to embryo outcome. Hum Reprod. 2002;17:413–419. doi: 10.1093/humrep/17.2.413. [DOI] [PubMed] [Google Scholar]
- 7.Vanneste T, Voet C, LeCaignec M, Ampe P, Konings C, Melotte S, Debrock M, Amyere M, Vikkula F, Schuit JP, Fryns G, Verbeke T, D'Hooghe Y, Moreau JR. Vermeesch. Chromosome instability is common in human cleavage-stage embryos Nat. Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 8.Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- 9.Li M, DeUgarte CM, Surrey M, Danzer H, DeCherney A, Hill DL. Fluorescence in situ hybridization reanalysis of day-6 human blastocysts diagnosed with aneuploidy on day 3. Fertil Steril. 2005;84:1395–1140. doi: 10.1016/j.fertnstert.2005.04.068. [DOI] [PubMed] [Google Scholar]
- 10.Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, Trounson AO. Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod. 2000;15:1781–1786. doi: 10.1093/humrep/15.8.1781. [DOI] [PubMed] [Google Scholar]
- 11.Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update. 2014;20:571–581. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 12.Orvieto R, Shuly Y, Brengauz M, Feldman B. Should pre-implantation genetic screening be implemented to routine clinical practice? Gynecol Endocrinol. 2016;32(6):506–508. doi: 10.3109/09513590.2016.1142962. [DOI] [PubMed] [Google Scholar]
- 13.Victor AR, Griffin DK, Brake AJ, Tyndall JC, Murphy AE, Lepkowsky LT, Lal A, Zouves CG, Barnes FL, McCoy RC, Viotti M. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum Reprod. 2019;34(1):181–192. doi: 10.1093/humrep/dey327. [DOI] [PubMed] [Google Scholar]
- 14.Bolton H, Graham SJ, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, Voet T, Zernicka-Goetz M. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;29(7):11165. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos MA, Teklenburg G, Macklon NS, Van Opstal D, Schuring-Blom GH, Krijtenburg PJ, de Vreeden-Elbertse J, Fauser BC, Baart EB. The fate of the mosaic embryo: chromosomal constitution and development of day 4, 5 and 8 human embryos. Hum Reprod. 2010;25:1916–1926. doi: 10.1093/humrep/deq139. [DOI] [PubMed] [Google Scholar]
- 16.Mantikou E, Wong KM, Repping S, Mastenbroek S. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta. 1822;2012:1921–1930. doi: 10.1016/j.bbadis.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, Torkenczy KA, Adey A, Yan M, Gao L, Park B, Nevonen KA, Carbone L, Chavez SL. Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 2019;29:367–382. doi: 10.1101/gr.239830.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singla S, Iwamoto-Stohl LK, Zhu M, Zernicka-Goetz M. Autophagy-mediated apoptosis eliminates Aneuploid cells in a mouse model of chromosome Mosaicism. Nat Commun. 2020;11:2958. doi: 10.1038/s41467-020-16796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boumela I, Assou S, Aouacheria A, Haouzi D, Dechaud H, De Vos J, Handyside A, Hamamah S. Involvement of BCL2 family members in the regulation of human oocyte and early embryo survival and death: gene expression and beyond. Reproduction. 2011;141:549–561. doi: 10.1530/REP-10-0504. [DOI] [PubMed] [Google Scholar]
- 20.Haouzi D, Boumela I, Chebli K, Hamamah S. Global, Survival, and apoptotic Transcriptome during Mouse and Human Early Embryonic Development. Biomed Res Int. 2018;5895628. 10.1155/2018/5895628 eCollection 2018. [DOI] [PMC free article] [PubMed]
- 21.Orvieto R, Yonish M, Shuly Y, Shavit T. Do mosaic embryos have the ability of self-correction? J Clin Med Images. 2020;V3(5):1–2. [Google Scholar]
- 22.Popovic M, Dhaenens L, Boel A, et al. Chromosomal mosaicism in human blastocysts: the ultimate diagnostic dilemma. Hum Reprod Update. 2020;26(3):313–334. doi: 10.1093/humupd/dmz050. [DOI] [PubMed] [Google Scholar]
- 23.Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270–1283. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 24.Takarabio. GenetiSure Pre-Screen Complete Protocol. Available from: https://www.takarabio.com/assets/documents/User%20Manual/PicoPLEX%20WGA%20Kit%20Protocol-At-A-Glance-070717.pdf.
- 25.Agilent. Oligo aCGH/ChIP-Chip Hybridization Kit Protocol. Available from: https://www.agilent.com/en/product/cgh-cgh-snp-microarray-platform/cgh-cgh-snp-microarray-kits-reagents/oligo-acgh-chip-on-chip-hybridization-kit-228433.
- 26.ACOG Committee on Gynecologic Practice; Practice Committee of the ASRM Female age-related fertility decline. Committee Opinion No. 589. Obstet Gynecol. 2014;123:719–721. doi: 10.1097/01.AOG.0000444440.96486.61. [DOI] [PubMed] [Google Scholar]
- 27.Orvieto R, Gleicher N. Preimplantation genetic testing for aneuploidy (PGT-A)-finally revealed. J Assist Reprod Genet. 2020;37(3):669–672. doi: 10.1007/s10815-020-01705-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gleicher N, Orvieto R. Is the hypothesis of preimplantation genetic screening (PGS) still supportable? A review. J Ovarian Res. 2017;10(1):21. doi: 10.1186/s13048-017-0318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Bogale B, Tang Y, Lu S, Xie XS, Racowsky C. Noninvasive preimplantation genetic testing for aneuploidy in spent medium may be more reliable than trophectoderm biopsy. Proc Natl Acad Sci U S A. 2019;116:14105–14112. doi: 10.1073/pnas.1907472116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gleicher N, Barad DH. Not even noninvasive cell-free DNA can rescue preimplantation genetic testing. Proc Natl Acad Sci U S A. 2019;116(44):21976–21977. doi: 10.1073/pnas.1911710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starostik MR, Sosina OA, McCoy RC. Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism. Genome Res. 2020;30(6):814–825. doi: 10.1101/gr.262774.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Time-lapse EmbryoScope™ photography of embryo expelling cell debris/cell fragments within the zona pellucida (https://youtu.be/3RNUJ4iW0IE).
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.