Abstract
Radiopharmaceutical dosimetry depends on the localization in space and time of radioactive sources and requires the estimation of the amount of energy emitted by the sources deposited within targets. In particular, when computing resources are not accessible, this task can be performed using precomputed tables of specific absorbed fractions (SAFs) or S values based on dosimetric models. The aim of the OpenDose collaboration is to generate and make freely available a range of dosimetric data and tools. Methods: OpenDose brings together resources and expertise from 18 international teams to produce and compare traceable dosimetric data using 6 of the most popular Monte Carlo codes in radiation transport (EGSnrc/EGS++, FLUKA, GATE, Geant4, MCNP/MCNPX, and PENELOPE). SAFs are uploaded, together with their associated statistical uncertainties, in a relational database. S values are then calculated from monoenergetic SAFs on the basis of the radioisotope decay data presented in International Commission on Radiological Protection Publication 107. Results: The OpenDose collaboration produced SAFs for all source region and target combinations of the 2 International Commission on Radiological Protection Publication 110 adult reference models. SAFs computed from the different Monte Carlo codes were in good agreement at all energies, with SDs below individual statistical uncertainties. Calculated S values were in good agreement with OLINDA/EXM 2.0 (commercial) and IDAC-Dose 2.1 (free) software. A dedicated website (www.opendose.org) has been developed to provide easy and open access to all data. Conclusion: The OpenDose website allows the display and downloading of SAFs and the corresponding S values for 1,252 radionuclides. The OpenDose collaboration, open to new research teams, will extend data production to other dosimetric models and implement new free features, such as online dosimetric tools and patient-specific absorbed dose calculation software, together with educational resources.
Keywords: nuclear medicine, dosimetry, open-access database, Monte Carlo methods
The necessity of dosimetry in nuclear medicine is increasing with the remarkable development of targeted radionuclide therapy (1).
Absorbed doses in nuclear medicine are usually calculated using the MIRD formalism (2), with the following time-independent formulation:
| Eq. 1 |
where is the mean absorbed dose (Gy) delivered to target tissue, is the total number of nuclear transformations in the source (Bqs), is the mean absorbed dose to a given target per nuclear disintegration in the source (S value; GyBq−1s−1), and is the dose integration period after administration of the radioactive material. S values are obtained from reference human models and used for model-based dosimetry or adjusted to the geometry of a specific patient (3). The calculation of S values (Eq. 2) requires a clear definition of the geometry of the model and isotope decay characteristics:
| Eq. 2 |
where is the specific absorbed fraction (SAF; kg−1) for radiation type i and and are the energy (J) and yield (Bq−1s−1), respectively, of radiation type i.
Historically, SAFs and S values were computed from mathematic models and were the basis for International Commission on Radiological Protection (ICRP) reports for radiopharmaceutical dosimetry (4). Since the publication of ICRP Publication 103 (5), the ICRP recommends using voxel-based models, such as those presented in ICRP Report 110 (ICRP 110) (6). This is a logical evolution that accounts for the increasing availability of a refined description of human geometry. In addition, recent developments proposed polygonal mesh models (7), which can overcome voxel-based model limitations in the description of thin structures such as organ walls.
For decades, the reference S values for mathematic models were included in MIRDOSE and then in OLINDA/EXM (http://www.doseinfo-radar.com). They were easily accessible, and most dosimetric results presented to the Food and Drug Administration or European Medicines Agency for approval of a new radiopharmaceutical were obtained using these codes. OLINDA/EXM 1.0 is no longer available, and OLINDA/EXM 2.0 is part of Hermes Medical Solutions (http://vueinnovations.com/olinda). Moreover, SAFs and S values are no longer available on the Radiation Dose Assessment Resource website (http://www.doseinfo-radar.com/).
Recently, ICRP Report 133 (ICRP 133) (8) presented SAFs for 79 sources and 43 targets of the ICRP 110 computational models. On the basis of this dataset, Andersson et al. (9) proposed IDAC-Dose 2.1 desktop software for performing absorbed dose calculations.
SAFs are generated through intensive Monte Carlo simulations requiring substantial computational power. As a consequence, there is a delay between the publication of a model and its corresponding SAFs on the order of years. This delay might be amplified in coming years as the number of models and their level of complexity increase (10).
The OpenDose project is based on the idea that a collaborative approach enables this challenge to be addressed by offering a responsive network of shared dosimetric resources and expertise from several teams involved in radiopharmaceutical dosimetry. This approach allows the production and comparison of data with different codes, increasing the robustness of the results. The OpenDose collaboration strives to produce and disseminate dosimetric data that are traceable and reproducible and include associated uncertainties (11).
MATERIALS AND METHODS
Collaboration
The first meeting of the collaboration was held at the European Association of Nuclear Medicine Congress in Barcelona, Spain, in 2016. At this meeting, the structure of the OpenDose collaboration and its scientific priorities were defined and a collaboration agreement in which the teams committed to generate freely accessible dosimetric data that meet the FAIR Guiding Principles was drafted (12).
OpenDose currently includes 18 research teams at 30 institutes and has a steering committee composed of 1 representative per team. The collaboration organizes annual meetings to present results and discuss future actions.
Framework
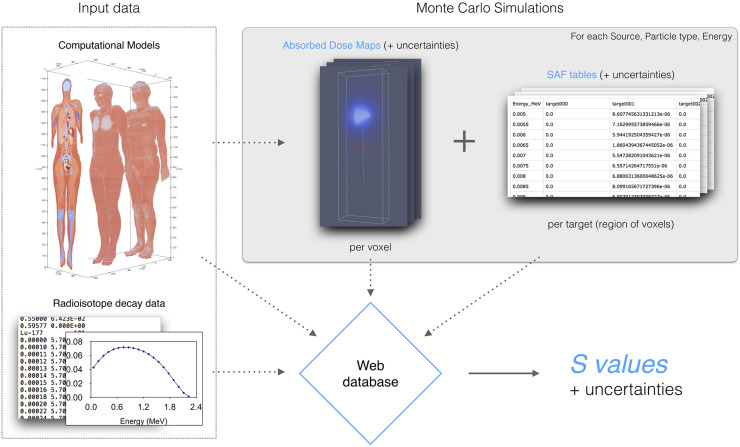
A common framework (Fig. 1) has been defined to harmonize the different Monte Carlo codes and different units and formats used by the teams. Within this framework, each team is responsible for its own Monte Carlo simulations but uses the same input models and delivers the data in a common output format. Therefore, each collaboration member can independently contribute to data generation. The data produced are then collected and centralized in a database along with the models and radioisotope decay data. A dedicated website allows access to the database, displays SAFs, allows the calculation of S values, and allows downloading of the results.
FIGURE 1.
OpenDose framework. SAFs produced by Monte Carlo simulations are stored in database along with input data. Web application allows calculation, display, and downloading of SAFs and S values.
Here we describe the details of the OpenDose framework.
Models and Radioisotope Decay Data
The first models considered within the collaboration were those defined in ICRP 110 (6). These voxel-based models represent the average anatomy of male and female subjects. The adult female is described by 299 × 137 × 348 voxels for a size of 1.775 × 1.775 × 4.84 mm, corresponding to a height of 1.63 m and a mass of 60 kg. The adult male is described by 254 × 127 × 222 voxels for a size of 2.137 × 2.137 × 8.0 mm, corresponding to a height of 1.76 m and a mass of 73 kg.
These models comprise 141 segmented regions (20,000 target source combinations) and 53 different media. Because uncertainties from different regions cannot be summed after simulation, as deposited energies in adjacent regions are likely to be correlated, 31 compound regions were added to regroup parts of an organ (e.g., the kidneys are composed of regions 89–94). These additional 31 compound regions are the same as the target compound regions and “Total body except organ contents” defined in Tables D.1 and C.1 of ICRP 110, respectively (6).
The radioisotope decay data of ICRP Publication 107 (13) were considered for S value calculation. The ICRP Publication 107 provides electronic data for 1,252 radioisotopes.
Monte Carlo Simulations and Specific Absorbed Fractions
The variety of research teams offers a broad experience over 6 of the most widely used Monte Carlo codes in radiopharmaceutical dosimetry: EGSnrc/EGS++ (14), FLUKA (15), GATE (16), Geant4 (17), MCNP/MCNPX (18), and PENELOPE (19). These codes come from different research fields and implement different approaches to simulate radiation transport in matter, effectively offering the independent production of data. This approach allows verification of the production pipeline and increases the robustness of data.
Monte Carlo simulations estimate energy deposition in the models as a function of source location, particle type, and energy. Source location can be any region of the selected model from which particles are emitted isotropically. Monte Carlo simulations were performed for monoenergetic photons and electrons. A list of 91 energies ranging from 5 keV to 10 MeV was defined on the basis of the logarithmic energy distribution of β-emitters in ICRP Publication 107 (13). Physics models, the use of variance reduction techniques, and any other parameters are the choice of each team. However, to ensure the traceability and reproducibility of data, each team must keep track of all simulation parameters and files. At least primary particles are recommended as a trade-off between good statistics and computation time. Statistical uncertainties are estimated for all quantities (energy deposited, absorbed dose) during the run time of the simulations using the history-by-history method (20). Each simulation provides absorbed doses and uncertainties at the individual voxel level (as binary matrices) as well as for target regions (as ASCII files).
From the simulated absorbed dose to a target, the SAF is calculated by dividing the absorbed dose by the initial energy (J) of the particles and by the number of simulated particles. Each team records the SAFs (and corresponding uncertainties) in a specific comma-separated values format with a specific naming convention to ease integration into the database.
Database
SAFs are stored in a relational Structured Query Language database as a function of parameters such as model (name, version), source and target (name, volume, mass), particle type, energy, and number of simulated particles. In addition to these parameters, data provenance (contact name, date) is stored to ensure traceability. The radioisotope decay data are also stored in the database to enable the calculation of S values.
S Value Calculation
A Python script was developed to calculate S values from SAFs, for any radioisotope, using Equation 2. First, SAFs are extracted from the database and averaged over all Monte Carlo codes, increasing the robustness of the calculation. Then, SAFs are linearly interpolated to the energy of each radiation type of the radioisotope considered and multiplied by the corresponding yield (Bq−1s−1) and energy (J). For β-emission spectra, SAFs are integrated over energy bins. α-particles and α-recoil nuclei are considered to deposit their energy locally. Finally, all emission contributions are summed to obtain the S value for a selected model, source, target, and radioisotope. The S value is also calculated per particle type (photon, electron, alpha-particle) to provide more insight into the S value composition.
At each step of the calculation, the uncertainty of the S value is computed as
| Eq. 3 |
where and are the energy (J) and yield (Bq−1s−1), respectively, of radiation type i (for a given isotope) and is the SAF statistical uncertainty (kg−1) for radiation type i.
The computation is performed on demand to use the most up-to-date data (SAFs and nuclear data). No S values are stored in the database.
Website
A dedicated website was developed to provide access to all OpenDose data. A website has the advantages of being easily accessible on every device (without any installation) and being easy to keep up to date.
Verification of Produced Data
To assess the variability of results among the different Monte Carlo codes, comparisons were performed using 1 model for a restricted number of sources and energies. SAFs were computed by all teams for the ICRP 110 adult female model using the Liver and Blood Vessels Trunk as sources and monoenergetic photons and electrons of 0.05, 0.1, 0.2, 0.5, 1, 2, and 5 MeV. The Liver was chosen for its relevance in radiopharmaceutical dosimetry and its compact shape, whereas the Blood Vessels Trunk was chosen to test a more spatially complex source.
The SAFs for the source Liver were compared with the data published in ICRP 133 (8).
For the calculation of S values from the SAFs (Eq. 2), the database must contain at least 1 value for each model, source, target, and energy combination. To meet this requirement, simulations were run with a reduced number of particles with Geant4 10.5 to reach a complete dataset. S values for a source located in the Liver of the female model, 6 targets, and 3 radioisotopes commonly used in nuclear medicine (131I, 177Lu, and 90Y) were compared with IDAC-Dose 2.1 (9) and OLINDA/EXM 2.0 (21).
RESULTS
Current Production Status
Producing a complete dataset for 1 model (e.g., the ICRP 110 adult female) requires approximately 30,000 simulations, representing approximately 750,000 CPU hours of computation. By sharing the workload and the computing resources of the teams (https://www.ziemowit.hpc.polsl.pl/en, https://cc.in2p3.fr/en, https://www.calmip.univ-toulouse.fr, and https://nci.org.au), the collaboration can produce data for a model in a few months. Some developments to take advantage of large grid infrastructures have also been made (22).
As of October 2019, the database held 11,177,347 entries, representing data spread over 2 models, 141 sources, 172 targets, 2 particle types (photons and electrons), and 91 energies produced with 9 different Monte Carlo code versions.
Verification of Produced Data
For photons, SAFs from all of the teams are in good agreement at all energies for both sources. For example, SAFs have relative SDs ranging from 0.67% at 5 keV to 0.23% at 5 MeV for Liver Blood Vessels Trunk and from 0.69% at 5 keV to 0.16% at 5 MeV for Spleen Liver. For electrons, SAFs have higher statistical fluctuations, especially at low energy and for targets at large distances from the source. In particular, statistical uncertainties of SAFs for Brain Liver are close to 100% at less than 200 keV. As a consequence, the SAF SDs between the different Monte Carlo codes can reach 65.34% for Brain Liver and electrons of 100 keV and as low as 1.56% at 5 MeV. These high relative differences correspond to negligible absolute differences. For the target Spleen, which is closer to the Liver, the relative SDs range from 22.86% at 5 keV to 2.0% at 5 MeV.
SAFs from this preliminary assessment were also compared with data published in ICRP 133 (8). The overall agreement between OpenDose and ICRP 133 values was good, with mean relative differences of 1.65% for photons and 8.2% for electrons of greater than 200 keV. However, some large differences were noted for low-energy electrons, with a maximum relative difference of +120% for Thyroid Liver at 100 keV. This large difference likely was due to high statistical fluctuations in the data. Unfortunately, no information on the SAF uncertainties is given in ICRP 133. ICRP 133 values for Brain Liver at energies of less than 200 keV are equal to 0.0, which may indicate a lower statistical power of the ICRP 133 simulations than of the OpenDose data. These mean relative differences did not include Liver Liver SAF values, for which a systematic difference of 22% was found. This finding is explained by the modification of some target masses in ICRP 133 (Tables A.1 and A.2 from (8)) to add blood content; the original 1.400 kg for the Liver in the ICRP 110 adult female model was changed to 1.810 kg.
SAF values from a selection of different Monte Carlo codes are available in Supplemental Tables 1–8 (supplemental materials are available at http://jnm.snmjournals.org), along with SAFs from ICRP 133 for comparison. The “OpenDose mean” gives the mean and SD of all available Monte Carlo code values.
S values are available for the 2 models, 141 sources, 172 targets, and 1,252 radioisotopes considered so far within the OpenDose collaboration. Table 1 shows S values obtained for the adult female model and the comparison with IDAC-Dose 2.1 (9) and OLINDA/EXM 2.0 (21). In general, good agreement was found. In particular, OpenDose and IDAC-Dose 2.1 values were very close, as expected, as they use the same input model (ICRP 110), whereas OLINDA/EXM 2.0 values are calculated for a nonuniform rational basis spline adult female model based on ICRP Publication 89 (23). A systematic difference from IDAC-Dose 2.1 for Liver Liver can be explained by the mass adjustment made in the ICRP 133 SAF calculation.
TABLE 1.
OpenDose S Values Compared with IDAC-Dose 2.1 and OLINDA/EXM 2.0 for 131I, 177Lu, and 90Y in Liver of ICRP 110 Female Model
|
131I |
177Lu |
90Y |
|||||||
| Target | OpenDose | IDAC-Dose 2.1 | OLINDA/EXM 2.0 | OpenDose | IDAC-Dose 2.1 | OLINDA/EXM 2.0 | OpenDose | IDAC-Dose 2.1 | OLINDA/EXM 2.0 |
| Brain | 1.492e−8 ± 9.5e−11 | 1.46e−8 | 1.47e−8 | 8.423e−10 ± 4.6e−12 | 8.44e−10 | 8.10e−10 | 1.084e−10 ± 2.6e−12 | 1.08e−10 | 4.27e−11 |
| Gallbladder wall | 6.355e−6 ± 1.2e−8 | 6.33e−6 | 2.43e−6 | 1.100e−6 ± 2.8e−9 | 1.09e−6 | 2.52e−7 | 2.228e−5 ± 2.5e−8 | 2.20e−5 | 5.16e−8 |
| Liver | 2.842e−5 ± 4.7e−9 | 2.20e−5 | 2.89e−5 | 1.753e−5 ± 1.3e−9 | 1.36e−5 | 1.76e−5 | 1.015e−4 ± 7.7e−9 | 7.83e−5 | 1.03e−4 |
| Pancreas | 2.165e−6 ± 2.9e−9 | 2.18e−6 | 2.15e−6 | 2.255e−7 ± 5.0e−10 | 2.27e−7 | 2.34e−7 | 4.518e−7 ± 1.3e−9 | 4.61e−7 | 1.54e−6 |
| Spleen | 7.654e−7 ± 1.7e−9 | 7.69e−7 | 4.12e−7 | 7.655e−8 ± 2.3e−10 | 7.69e−8 | 3.81e−8 | 7.824e−9 ± 5.3e−11 | 7.94e−9 | 2.35e−9 |
| Thyroid | 1.443e−7 ± 4.5e−10 | 1.44e−7 | 1.74e−7 | 1.282e−8 ± 9.9e−11 | 1.29e−8 | 1.61e−8 | 1.287e−9 ± 2.8e−11 | 1.43e−9 | 4.86e−10 |
Values are mean ± SD.
Website
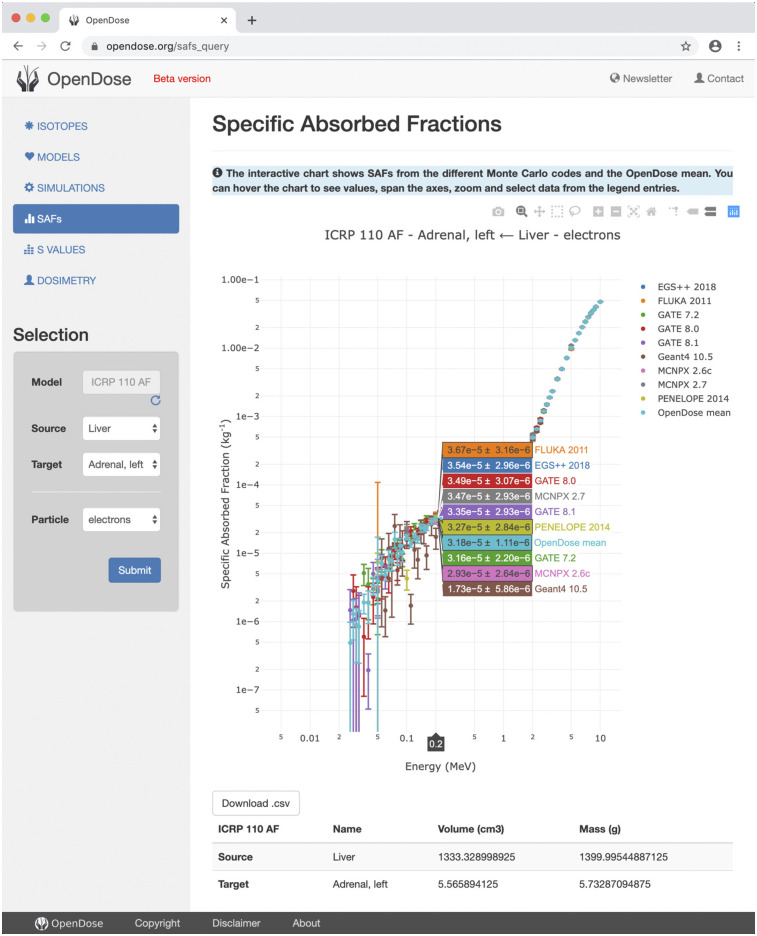
The OpenDose website is online at www.opendose.org. It provides information on the project, allows access to the SAF database, and allows calculation of the S values for 1,252 radionuclides. Results are displayed with interactive plots and can be downloaded in a comma-separated values format. Figure 2 shows the SAF section of the website, with results from all of the Monte Carlo codes. The available data should be used for research purposes only, and the website content is licensed under a Creative Commons Attribution-ShareAlike 4.0 International license.
FIGURE 2.
SAF section of OpenDose website allows display and downloading of data from all Monte Carlo codes.
DISCUSSION
The generation of dosimetric data represents a computing challenge because of the resources and time required to perform simulations. The idea of a collaborative framework designed for this purpose required a preliminary verification of the precision of the dosimetric estimations of the different Monte Carlo codes used within the community.
The comparison exercise proposed to members of the collaboration demonstrated that SAFs produced with different Monte Carlo codes exhibit low dispersion and are in good agreement with published ICRP 133 data. The uncertainties of the SAFs or the S values estimated by OpenDose are a good indicator of the limitations of the values themselves. For example, data for electrons of low energy show statistical limitations for targets far from the source. This information is not present in any other published SAF data. Calculated S values also show good agreement with IDAC-Dose 2.1, which uses the same model and radioisotope decay scheme. OpenDose and IDAC-Dose 2.1 S values differ only for some sources because ICRP integrated organ mass adjustments. OpenDose provides data corresponding to the models used in the Monte Carlo simulations, without any mass adjustments, to avoid any calculation approximation or radiation transport simplifications and to maintain a strict framework for data traceability and reproducibility.
The decay data presented in ICRP Publication 107 do not include corresponding standard uncertainties, so the reported standard uncertainty of the S value (Eq. 3) is purely statistical. For many common radionuclides used in nuclear medicine, the uncertainties in the decay data can be the dominant source of the uncertainty of the S value and should be included in any uncertainty estimation (24). In the future, nuclear data with reported standard uncertainties will be used in OpenDose (25).
During the last meeting (October 2019), the steering committee decided to include, as part of the OpenDose collaboration products, 2 open-source software products currently under development. The first implements model-based dosimetry and will be integrated in a future section of the website, whereas the second implements patient-specific dosimetry in a 3DSlicer (https://www.slicer.org) extension and will be available for downloading.
The future vision of OpenDose is to make the website a global resource for internal dosimetry, including dosimetric data, software, and education.
CONCLUSION
The aim of the OpenDose project is to establish a long-term international collaboration to generate an open-access database for nuclear medicine dosimetry. The collaboration includes 18 teams with expertise in radiopharmaceutical dosimetry, Monte Carlo methods, and high-performance computing. The initial task set for all partners to produce a set of simulations for a specific model allowed the definition of a common computational framework and the establishment of a unified format for results. OpenDose has now produced SAFs and uncertainties for the 2 ICRP 110 adult models, covering all source and target combinations, 2 particle types (photons and electrons), and 91 energies. Data production will continue and extend to other computational models.
The dedicated website is online at www.opendose.org. It allows easy access to the SAF database and the calculation of S values for 1,252 radionuclides. S values generated for the models considered so far are close to those already available in the literature. A new website section for performing model-based dosimetry calculations is under development.
The collaboration is open to new research teams willing to participate in any OpenDose-related activity, from the production of dosimetric data to the development of the website and dosimetry tools.
DISCLOSURE
This work was supported by the National Measurement System of the U.K. Department for Business, Energy and Industrial Strategy. This work was performed within the framework of SIRIC LYriCAN Grant INCa-INSERM-DGOS-12563 and LABEX PRIMES (ANR-11-LABX-0063) of Université de Lyon, within the program Investissements d’Avenir (ANR-11-IDEX-0007) operated by the ANR. Part of this work was supported by the Euratom Research and Training Program, 2014–2018, under grant agreement 755523 (MEDIRAD). Calculations were partially performed on the Ziemowit computer cluster in the Laboratory of Bioinformatics and Computational Biology, Silesian University of Technology, created in EU Innovative Economy Program POIG.02.01.00-00-166/08 and expanded in the POIG.02.03.01-00-040/13 project. This project was undertaken with the assistance of resources and services from the National Computational Infrastructure (NCI), which is supported by the Australian Government. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can the practice of dosimetry in nuclear medicine be facilitated?
PERTINENT FINDINGS: The OpenDose collaboration brings together the resources and expertise of many research teams involved in radiopharmaceutical dosimetry. The www.opendose.org website provides a global repository for internal dosimetry, including data, software, and education material.
IMPLICATIONS FOR PATIENT CARE: The OpenDose project will help the practice of internal dosimetry by providing open-access resources.
Supplementary Material
Acknowledgments
The authors acknowledge the support of France Grilles, the EGI Infrastructure, the Biomed Virtual Organization, and the Virtual Imaging Platform for providing computing resources and services. The authors also acknowledge Yves-Marie Gouesnard for his contribution to the website.
The OpenDose collaborators include Ernesto Amato, Department of Biomedical and Dental Sciences, University of Messina, Messina, Italy, and Department of Morphologic and Functional Imaging, University of Messina, Messina, Italy; Lucrezia Auditore, Department of Biomedical and Dental Sciences, University of Messina, Messina, Italy, and Department of Morphologic and Functional Imaging, University of Messina, Messina, Italy; Sorina Camarasu-Pop, Université de Lyon-CREATIS-CNRS UMR 5220-Inserm U1044-INSA Lyon-Université Lyon 1-Centre Léon Bérard, Lyon, France; Frederic Cervenansky, Université de Lyon-CREATIS-CNRS UMR 5220-Inserm U1044-INSA Lyon-Université Lyon 1-Centre Léon Bérard, Lyon, France; José M. Clavijo, Centro de Isótopos, La Habana, Cuba; Marco A. Coca Pérez, MEDSCAN, Nuclear Medicine and PET/CT Centre, Concepción, Chile; Marta Cremonesi, Medical Physics Unit, IEO, European Institute of Oncology IRCCS, Milan, Italy; José M. Fernández-Varea, Facultat de Física, Universitat de Barcelona, Barcelona, Spain; Ludovic Ferrer, Institut de Cancérologie de l’Ouest, St. Herblain, France; Telma C. F. Fonseca, Post-Graduate Program in Nuclear Sciences and Techniques, Department of Nuclear Engineering, UFMG, Belo Horizonte, Brazil; Didier Franck, Institut de Radioprotection et de Sûreté Nucléaire (IRSN), Fontenay-aux-Roses, France; Silvano Gnesin, Institute of Radiation Physics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland; Antonio Italiano, Istituto Nazionale di Fisica Nucleare, Sezione di Catania, Italy; Nico Lanconelli, University of Bologna, Bologna, Italy; Kamil Matusik, Department of Systems Biology and Engineering, Silesian University of Technology, Gliwice, Poland; Massimiliano Pacilio, Azienda Ospedaliera Universitaria Policlinico Umberto I, Roma, Italy; Isabelle Perseil, Département du Système d’Information, Inserm, Paris, France; Daniele Pistone, Department of Mathematics, Computer Science, Physics and Earth Science, University of Messina, Messina, Italy; Janick Rüegger, Radiology and Nuclear Medicine Department, Luzerner Kantonsspital, Switzerland; Leonel A. Torres Aroches, Centro de Isótopos, La Habana, Cuba; and Thiago V.M. Lima, Institute of Radiation Physics, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland and Radiology and Nuclear Medicine Department, Luzerner Kantonsspital, Switzerland.
REFERENCES
- 1.Stokke C, Gabiña PM, Solný P, et al. Dosimetry-based treatment planning for molecular radiotherapy: a summary of the 2017 report from the Internal Dosimetry Task Force. EJNMMI Phys. 2017;4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolch WE, Eckerman KF, Sgouros G, et al. A generalized schema for radiopharmaceutical dosimetry: standardization of nomenclature. MIRD Pamphlet No. 21. J Nucl Med. 2009;50:477–484. [DOI] [PubMed] [Google Scholar]
- 3.Divoli A, Chiavassa S, Ferrer L, Barbet J, Flux GD, Bardiès M. Effect of patient morphology on dosimetric calculations for internal irradiation as assessed by comparisons of Monte Carlo versus conventional methodologies. J Nucl Med. 2009;50:316–323. [DOI] [PubMed] [Google Scholar]
- 4.Mattsson S, Johansson L, Leide Svegborn S, et al. Radiation dose to patients from radiopharmaceuticals: a compendium of current information related to frequently used substances. ICRP Publication 128. Ann ICRP. 2015;44:7–321. [DOI] [PubMed] [Google Scholar]
- 5.International Commission on Radiological Protection. The 2007 recommendations of the International Commission on Radiological Protection. ICRP Publication 103. Ann ICRP. 2007;37:1–332. [DOI] [PubMed] [Google Scholar]
- 6.Menzel HG, Clement C, DeLuca P. Realistic reference phantoms: an ICRP/ICRU joint effort—a report of adult reference computational phantoms. ICRP Publication 110. Ann ICRP. 2009;39:1–164. [DOI] [PubMed] [Google Scholar]
- 7.Kim CH, Yeom YS, Nguyen TT, et al. New mesh-type phantoms and their dosimetric applications, including emergencies. Ann ICRP. 2018;47:45–62. [DOI] [PubMed] [Google Scholar]
- 8.Bolch WE, Jokisch D, Zankl M, et al. The ICRP computational framework for internal dose assessment for reference adults: specific absorbed fractions. ICRP Publication 133. Ann ICRP. 2016;45:5–73. [DOI] [PubMed] [Google Scholar]
- 9.Andersson M, Johansson L, Eckerman K, Mattsson S. IDAC-Dose 2.1, an internal dosimetry program for diagnostic nuclear medicine based on the ICRP adult reference voxel phantoms. EJNMMI Res. 2017;7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu XG. An exponential growth of computational phantom research in radiation protection, imaging, and radiotherapy: a review of the fifty-year history. Phys Med Biol. 2014;59:R233–R302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gear JI, Cox MG, Gustafsson J, et al. EANM practical guidance on uncertainty analysis for molecular radiotherapy absorbed dose calculations. Eur J Nucl Med Mol Imaging. 2018;45:2456–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerman K, Endo A. Nuclear decay data for dosimetric calculations. ICRP Publication 107. Ann ICRP. 2008;38:7–96. [DOI] [PubMed] [Google Scholar]
- 14.Kawrakow I, Mainegra-Hing E, Rogers DWO, Tessier F, Walters BRB. The EGSnrc Code System: Monte Carlo Simulation of Electron and Photon Transport. NRCC Report PIRS-701. Ottawa, Ontario, Canada: NRC Canada; 2017.
- 15.Böhlen TT, Cerutti F, Chin MPW, et al. The FLUKA code: developments and challenges for high energy and medical applications. Nuclear Data Sheets. 2014;120:211–214. [Google Scholar]
- 16.Sarrut D, Bardiès M, Boussion N, et al. A review of the use and potential of the GATE Monte Carlo simulation code for radiation therapy and dosimetry applications. Med Phys. 2014;41:064301. [DOI] [PubMed] [Google Scholar]
- 17.Agostinelli S, Allison J, Amako K, et al. Geant4: a simulation toolkit. Nucl Instrum Methods Phys Res A. 2003;506:250–303. [Google Scholar]
- 18.Waters LS, McKinney GW, Durkee JW, et al. The MCNPX Monte Carlo radiation transport code. AIP Conf Proc. 2007;896:81–90. [Google Scholar]
- 19.Salvat F, Fernández-Varea JM, Sempau J. PENELOPE-2011: A Code System for Monte Carlo Simulation of Electron and Photon Transport. Paris, France: OECD Nuclear Energy Agency; 2011. [Google Scholar]
- 20.Walters BRB, Kawrakow I, Rogers DWO. History by history statistical estimators in the BEAM code system. Med Phys. 2002;29:2745–2752. [DOI] [PubMed] [Google Scholar]
- 21.Stabin MG, Siegel JA. RADAR dose estimate report: a compendium of radiopharmaceutical dose estimates based on OLINDA/EXM Version 2.0. J Nucl Med. 2018;59:154–160. [DOI] [PubMed] [Google Scholar]
- 22.Chauvin M, Mathieu G, Camarasu-Pop S, Bonnet A, Bardiès M, Perseil I. Enabling large scale data production for OpenDose with GATE on the EGI infrastructure. In: 2019 19th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGRID). Piscataway, NJ: IEEE; 2019:658–665. [Google Scholar]
- 23.International Commission on Radiological Protection. Basic anatomical and physiological data for use in radiological protection reference values. ICRP Publication 89. Ann ICRP. 2002;32:5–265. [PubMed] [Google Scholar]
- 24. Working Group 1 of the Joint Committee for Guides in Metrology. Evaluation of Measurement Data: Guide to the Expression of Uncertainty in Measurement. Sèvres, France: JCGM; 2008.
- 25.Tuli JK. Evaluated Nuclear Structure Data File: A Manual for the Preparation of Data Sets. Upton, NY: National Nuclear Data Center; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.