Abstract
Despite the use of immune checkpoint blockade (ICB) therapy for hepatocellular carcinoma (HCC), developing more effective immunotherapy and predicting HCC’s response to ICB therapy remain top priorities. Ribosomal protein S3A (RPS3A) is a multifunctional molecule, but its association with tumor immune cell infiltration and prognosis in HCC patients is unknown. Thus, we aimed to investigate the correlation of RPS3A with HCC immune cell infiltration and prognosis to explore novel therapeutic strategies and prognostic biomarkers for this disease. Here, we showed that RPS3A expression levels were higher in HCC cell lines and samples than in normal hepatocytes and adjacent tumor-free tissues, respectively, and patients with high RPS3A expression had worse overall and recurrence-free survival durations than did patients with low expression. Moreover, single-sample gene set enrichment analysis (ssGSEA) and immunohistochemistry demonstrated a strongly negative correlation between RPS3A expression and tumor immune cell infiltration. Meanwhile, RPS3A was revealed to be positively correlated with that of most examined immune checkpoint molecules. GSEA also suggested that high RPS3A expression promoted the biological processes related to tumorigenesis, metastasis, and immunosuppression. Finally, RPS3A-based nomograms were constructed and exhibited better predictive accuracy for HCC prognosis and more net clinical benefits when compared with traditional prognosis-prediction staging systems. In short, these findings suggest that high RPS3A expression correlates with low tumor immune cell infiltration and poor prognosis in HCC patients. Furthermore, RPS3A-based nomograms are robust HCC prognostic predictors. RPS3A therefore may serve as a therapeutic target in and predict the efficacy of ICB therapy for HCC.
Keywords: Ribosomal protein S3A, hepatocellular carcinoma, tumor immune cell infiltration, prognosis, nomogram
Introduction
The incidence of hepatocellular carcinoma (HCC) is increasing the most rapidly among all cancers in 2020, and its 5-year relative survival rate is the second lowest [1]. The dismal prognosis for this disease is the consequence of a high frequency of metastasis or recurrence after curative treatment [2]. Currently, the programmed cell death protein 1 inhibitors (nivolumab and pembrolizumab) and the emerging immune checkpoint blockade (ICB) therapy are approved by the U.S. Food and Drug Administration as second-line therapeutic strategies for advanced HCC [3]. Despite the promising applications of immunotherapy, the objective response rate for it is still unsatisfactory, and precise methods of evaluating the response of HCC to immunotherapy are lacking [4]. Therefore, exploring novel immune-related candidate biomarkers and potential drug targets for prognosis prediction and effective treatment is warranted.
Parsing the tumor microenvironment, especially the characteristics of tumor immune cell infiltration, can increase insight into the likelihood of immunotherapy response or survival [5]. Scholars have discussed the possibility of optimizing cancer immunotherapy based on the presence of tumor-infiltrating lymphocytes and programmed death-ligand 1 expression in the tumor microenvironment [6]. Because the liver has a tightly controlled immunological network that eliminates gastrointestinal pathogens while maintaining immune tolerance, the tumor immune microenvironment in HCC patients are more complicated and heterogeneous [7]. Researchers have increasingly studied the roles of various types of tumor-infiltrating immune effector cells in the evolution of HCC, and enhancing ICB therapy for HCC by improving the infiltration of antitumor immune cells in tumors seems plausible [8,9]. Therefore, molecules that can reflect or regulate tumor immune cell infiltration would be useful to potentiate or predict the effectiveness of immunotherapy for HCC.
Accumulating evidence suggests that members of the ribosomal protein family have roles in tissue infiltration of immune-related cells in some diseases [10-14]. Reportedly, phosphorylated ribosomal protein S6 can increase the accumulation of lymphocytes and CD138+ plasma cells in salivary glands [12]. Also, ribosomal protein S19 can be released from apoptotic tumor cells and facilitate the recruitment of myeloid-derived suppressor cells to tumors [13]. Another member of this protein family, ribosomal protein S3A (RPS3A), plays a crucial role in regulating translational initiation and protein synthesis and can be recruited for multiple extraribosomal functions, the so-called ‘one gene-dual function’ [15]. Besides its potential role in regulation of cell differentiation and apoptosis [16-19], RPS3A is also reported to be involved in oncogenic transformation of cells like Rat-1 fibroblasts and B cells [20,21]. However, the immune-related function of RPS3A, especially in regulation of tumor immune cell infiltration, is not fully understood.
In this context, we attempted to reveal the correlation of RPS3A with HCC immune cell infiltration and prognosis to develop novel therapeutic strategies and prognostic biomarkers for this disease. In the present study, we investigated the association of RPS3A expression with clinicopathological features of HCC patients. We hypothesized that the positive relationship between high RPS3A expression and poor clinicopathological characteristics may be attributed to its effect on tumor immune cell infiltration. We first used quantitative reverse transcription polymerase chain reaction (qRT-PCR), Western blotting, and immunohistochemistry (IHC) to determine RPS3A expression profile in HCC samples and cell lines. Subsequently, we performed single-sample gene set enrichment analysis (ssGSEA) and IHC to explore the correlations of RPS3A expression with infiltration patterns for various tumor-infiltrating immune cells. In addition, the association of RPS3A expression level with immune checkpoint molecules expression was analyzed based on the public database and confirmed using PCR array. We also conducted GSEA to identify biological pathways in which RPS3A may be involved. Additionally, given the high accuracy of nomograms in predicting survival and distant metastases in cancer patients [22], we constructed prognostic nomograms integrating the RPS3A expression level and other important prognostic factors to achieve more reliable prognosis prediction for HCC patients compared to traditional staging systems.
Materials and methods
Patient enrollment and follow-up
154 patients who underwent surgery for HCC from January 2009 to January 2010 in Zhongshan Hospital, Fudan University (Shanghai, China) were enrolled in this study for IHC staining. Another 70 tumor tissues from HCC patients who received hepatectomy at our hospital in 2012 were selected to detect the mRNA expression of RPS3A and various immune checkpoint molecules. Patients were selected according to the following inclusion and exclusion criteria: 1) no history of other malignancies, 2) no prior cancer treatments before operations, 3) no extrahepatic metastases found before surgery, 4) no evidence of infection or other inflammatory conditions other than viral hepatitis, 5) primary curative resection, and 6) a definite postoperative pathological diagnosis of HCC.
Conventional clinicopathological variables such as age, sex, ascites, liver cirrhosis, preoperative laboratory indexes, and postoperative pathology reports were collected from electronic medical records. Laboratory indexes mainly included alanine aminotransferase (ALT), hepatitis B virus surface antigen (HBsAg), carcinoembryonic antigen (CEA), aspartate aminotransferase (AST), α-fetoprotein (AFP), and carbohydrate antigen 19-9 (CA19-9) level. Tumor number, size, encapsulation, differentiation, and microvascular invasion were obtained from pathology reports. Tumors were staged according to the seventh edition of the TNM classification by the American Joint Committee on Cancer and the Barcelona Clinic Liver Cancer (BCLC) staging system [23,24].
The follow-up protocol, generally consisting of routine blood examination, liver function testing, serological tumor biomarker measurement, abdominal ultrasound, and a chest X-ray, was conducted every 3 months for the first year, every 3-6 months for the following 2 years, and then once a year. Enhanced computed tomography or magnetic resonance imaging was implemented for patients suspected of having recurrence. Overall survival (OS) and recurrence-free survival (RFS) were defined as the interval from the date of operation to the date of death and that of recurrence (or last follow-up visit), respectively. The study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University (Y2018-155), and written informed consent was obtained from each subject.
Cell lines
The human HCC cell lines MHCC97H and MHCCLM3 were obtained from Zhongshan Hospital, Fudan University. The HCC cell lines SMMC-7721, SK-Hep1, Huh7, Hep3B, and PLC/PRF/5 and the normal hepatocyte line L-02 were purchased from the cell bank of the Chinese Academy of Sciences.
qRT-PCR
Total RNA was extracted from eight cell lines using TRIzol reagent (Invitrogen). Subsequently, cDNA was synthesized using a PrimeScript RT reagent kit (Takara Bio). Real-time PCR was then performed using an ABI Prism 7500 Sequence Detection system (Applied Biosystems) using SYBR Premix ExTaq (Takara Bio). The primers used for quantitative reverse transcription PCR analyses were RPS3A-forward: 5’-CGAGAGGTGCAGACAAATGA-3’; RPS3A-reverse: 5’-CGAAGACATCATGGAGAGGATAAA-3’; β-actin-forward: 5’-GGACCTGACTGACTACCTCAT-3’; and β-actin-reverse: 5’-CGTAGCACAGCTTCTCCTTAAT-3’. Relative quantification of gene expression was performed using the 2-ΔΔCT method.
Western blotting
Cells were lysed in RIPA extraction reagent (Pierce Biotechnology) supplemented with protease inhibitors (Roche) and phosphatase inhibitors (Sigma-Aldrich). Twenty micrograms of protein per well was loaded, electrophoresed on 10% gels, and electrotransferred to membranes (Millipore). After blocking with 5% nonfat dry milk, membranes were blotted with primary antibodies against RPS3A (1:1000; Abcam) and GAPDH (1:1000; Sigma-Aldrich) and then incubated with secondary antibodies (1:5000; Jackson ImmunoResearch Laboratories). The final gel images were obtained using a Tanon-5200 Chemiluminescent Imaging System.
Immunohistochemistry
Construction of tissue microarrays was described previously [25]. In brief, deparaffinized sections of tissue samples were immersed in 3% H2O2 and then incubated with an antibody against RPS3A (1:100; Abcam) or CD8 (1:250; Abcam) overnight. The next day, slides were incubated with a secondary antibody and diaminobenzidine solution (Dako Denmark A/S). Nuclear staining was performed using Harris hematoxylin. Afterward, a semiquantitative histological score was assigned independently by two pathologists blinded to the data to evaluate the slides. The final RPS3A expression level was classified based on the product of the staining intensity score (0, -; 1, +; 2, ++; and 3, +++) and staining extent (0-100%). The H-scores were ranked using a four-grade scale: strong (150~300%), moderate (100~150%), weak (0~100%), and negative (0).
PCR array
70 HCC tissues were immersed in TRIzol (Invitrogen) to extract tumor RNA. The cDNA was synthesized in a reverse transcription system using the purified RNA. For PCR array, a RT2 Profiler PCR Array (BioTNT) was used to achieve simultaneous mRNA expression examination of RPS3A and immune checkpoint molecules. Briefly, cDNA was added to a 384-well plate of the PCR array according to the manufacturer’s instructions. Subsequently, RT-PCR was performed on an ABI Prism 7500 platform using the RT2 SYBR Green qPCR Master mix. The cycle threshold (CT) values of the target genes were normalized by the housekeeping genes, and the relative mRNA expression levels of the target genes were calculated using the ΔCT method. The patients were divided into low and high RPS3A groups according to the median value of RPS3A mRNA expression. The primer sequences used for PCR array were listed in Table S1.
Acquisition of gene expression data from The Cancer Genome Atlas
RNA sequencing data obtained using a HiSeq system (Illumina) and the corresponding clinical data for 371 patients with liver cancer were obtained from The Cancer Genome Atlas (TCGA) website (https://portal.gdc.cancer.gov/). Ten patients whose pathological diagnoses were not HCC and five patients whose survival times were showed as zero were excluded. The scale method in the limma package in the R computing language (version 3.6.0) was applied to normalization of the mRNA expression data for these patients [26].
ssGSEA
Immune-cell infiltration levels in tumors were estimated using ssGSEA with the gsva package in the R language. ssGSEA can determine the immune cell population in a tumor sample according to gene expression data [27,28]. In total, 24 types of immune cells could be discriminated in our study using the deconvolution method: T helper 17 (Th17) cells, neutrophils, dendritic cells (DCs), eosinophils, gamma delta T cells, regulatory T cells, immature DCs, macrophages, plasmacytoid DCs, T cells, Th1 cells, Th2 cells, T follicular helper cells, natural killer (NK) cells, NK CD56bright cells, NK CD56dim cells, activated DCs, B cells, mast cells, CD8+ T cells, cytotoxic cells, T helper cells, effector memory T cells, and central memory T cells. Spearman rank correlation analysis was then performed to examine relationships between the specific gene expression levels and corresponding immune-cell infiltration levels. The ggplot2 package in the R language was used to show plots of immune cell types in which correlation coefficients were greater than 0.2.
GSEA
To identify the biological pathways that differed significantly in high and low RPS3A-expressing HCC patients, GSEA (version 4.0.3) was conducted according to the MSigDB molecular signatures database (version 7). C2 (curated) and c7 (immunologic) gene sets were imported from MsigDB into GSEA software for further analyses of the biological processes involving RPS3A. The significance threshold was set at a P value less than 0.05.
Statistical analysis
Data were presented as means (± standard deviation) or counts (percentages) and analyzed statistically using SPSS software (version 23.0; IBM). The Student t-test or Mann-Whitney U test was used based on the data distribution to compare continuous variables in two groups. Differences in categorical variables were analyzed using the Pearson chi-square test or Fisher exact test. A survival curve was plotted using the Kaplan-Meier method, and prognostic differences were examined using a log-rank test. Variables associated with OS or RFS were identified using a univariate Cox regression model, and significant factors were subjected to further multivariate Cox regression analysis in a stepwise manner. The pheatmap and corrplot package in the R computing language were applied to analyzing the association between RPS3A expression levels and immune checkpoint molecule expression levels. Two-sided P values less than 0.05 were considered significant.
Establishment and assessment of nomograms
Nomograms were established using the rms package in the R language based on the results of multivariate analyses. The predictive accuracy of different prognostic models was compared using a concordance index (C-index), a calibration curve, and decision curve analysis. The Hanley-McNeil test was applied to examining the differences in C-indexes [29].
Results
The RPS3A expression pattern and its correlation with HCC prognosis
We first explored the RPS3A expression pattern in HCC cell lines and samples. qRT-PCR and Western blotting demonstrated that both mRNA and protein expression for RPS3A were markedly higher in HCC cell lines than in normal hepatocytes (L-02) (Figure 1A and 1B). Furthermore, in IHC staining for RPS3A of 154 paired HCC and adjacent tumor-free tissue samples obtained from the study patients, we ranked RPS3A as negative, weak, moderate, or strong (Figure 1C and 1D). Compared with the tumor-free tissue samples, the HCC samples had significantly higher percentages of moderate (28.57%) and strong (22.73%) staining of RPS3A (P = 0.021) (Figure 1E).
Figure 1.
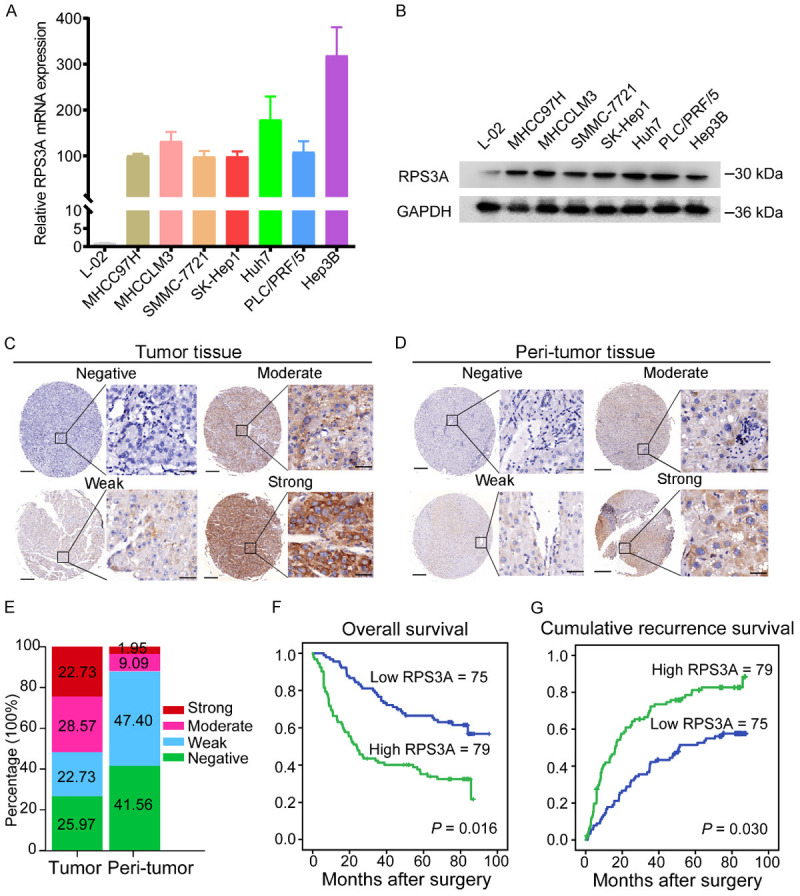
Patterns of RPS3A expression in HCC cell lines and samples and its correlation with prognosis for HCC. A, B. Relative RPS3A mRNA and protein expression levels in seven HCC cell lines and a normal hepatocyte line (L-02). C, D. Representative IHC images of negative, weak, moderate, and strong RPS3A staining in HCC and adjacent tumor-free tissue samples. Scale bars, 200 µm (left lane) and 20 µm (right lane). E. Percentages of different IHC staining grades for RPS3A in HCC and adjacent tumor-free tissue samples. F, G. Kaplan-Meier OS and RFS curves for HCC patients with low (n = 75) and high (n = 79) RPS3A expression. Prognostic differences are analyzed using a log-rank test.
Next, based on the RPS3A expression grades in tumor samples, we defined 79 HCC patients with strong or moderate staining as the high RPS3A group and the remaining 75 patients as the low RPS3A group. Survival analyses demonstrated that patients with high RPS3A expression experienced obviously worse OS and RFS durations than did those with low RPS3A expression (P = 0.016 and P = 0.030, respectively) (Figure 1F and 1G). Thus, we hypothesized that high RPS3A expression in HCC patients may indicate a poor prognosis.
Correlation between RPS3A expression level and HCC immune cell infiltration
To determine whether the effect of RPS3A expression on HCC prognosis is attributable to tumor immune cell infiltration, we performed ssGSEA to assess the association between the immune cell infiltration pattern and RPS3A level based on transcriptome profiling data for 356 HCC patients in TCGA database. Spearman correlation analyses revealed that high RPS3A expression was mainly associated with low infiltration of several immune cell types (Figure 2A), especially Th17 cells (r = -0.42, P < 0.001; Figure 2B). Neutrophils (r = -0.3, P < 0.001) and DCs (r = -0.24, P < 0.001) were the other two of the top three immune cell types that were negatively correlated with RPS3A expression level (Figure 2C and 2D). We observed only weakly positive correlations of RPS3A expression level with infiltration of other types of immune cells, such as NK CD56bright (r = 0.28), NK CD56dim (r = 0.27), T follicular helper (r = 0.26), and Th2 (r = 0.22) cells (Figure S1).
Figure 2.
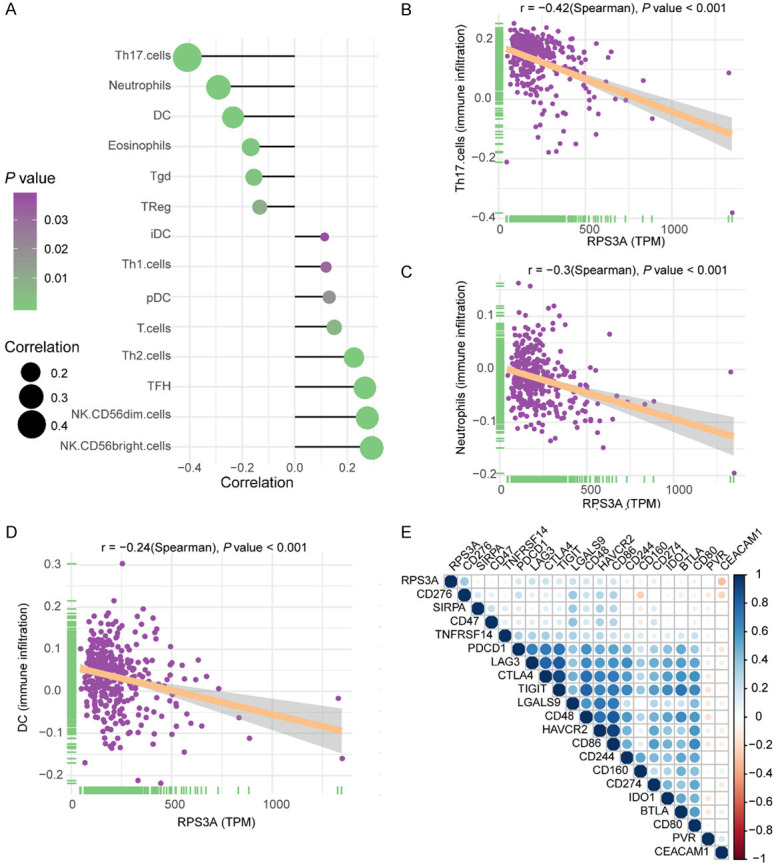
Correlation of RPS3A expression with specific tumor-infiltrating immune cell types and immune checkpoint molecules in HCC patients. A. Correlation of RPS3A expression with the infiltration levels for various immune cell types. Left, immune cells negatively correlated with RPS3A expression; right, immune cells positively associated with RPS3A expression. B-D. Correlation of RPS3A expression with infiltration of Th17 cells, neutrophils, and DCs. E. Correlation of RPS3A expression with immune checkpoint molecules expression. The color and size of each circle in the grids represent the corresponding correlation coefficient value (blue, positive correlation; red, negative correlation).
Additionally, we investigated the relationship between RPS3A and various immune checkpoint molecules. In correlation analyses, expression of most of the immune checkpoints examined was positively correlated with RPS3A expression, such as CD276 (r = 0.36, P = 1.53E-12), galectin 9 (LGALS9, r = 0.22, P = 3.77E-05), cytotoxic T lymphocyte-associated protein 4 (CTLA4, r = 0.21, P = 4.81E-05), lymphocyte-activation gene 3 (LAG3, r = 0.21, P = 5.19E-05), and CD86 (r = 0.20, P = 0.000203) (Table S2 and Figure 2E). Only one immune checkpoint molecule in our study, carcinoembryonic antigen cell adhesion molecule 1, was significantly negatively correlated with RPS3A expression (r = -0.30, P = 6.13E-09).
The association of high RPS3A expression level with low tumor immune cell infiltration and high immune checkpoint molecules expression were further confirmed in our own tumor samples. Among the various tumor-infiltrating immune cells, the CD8+ cytotoxic T cell (CTL) is the most important immune cell to suppress tumors [30]. Hence, we performed IHC staining to investigate the association between RPS3A expression and tumor infiltration degree of CD8+ CTL. The representative IHC images shown in Figure 3A confirmed the negative correlation between RPS3A expression and CD8+ CTL infiltration in HCC. As expected, 65% (49 of 75) of tumor with low RPS3A expression tended to have high CD8+ CTL infiltration, while negative or weak CD8 staining was observed in about 63% (50 of 79) of tumor with high RPS3A expression (Table 1). In addition, the results of PCR array demonstrated that there was a distinct transcriptional profile of immune checkpoint molecules between HCC patients with high and low RPS3A mRNA expression (Figure 3B). Correlation analyses in our patients indicated that the mRNA expressions of the CD276 (r = 0.85, P < 0.001), LGALS9 (r = 0.81, P < 0.001), CTLA4 (r = 0.95, P < 0.001), LAG3 (r = 0.81, P < 0.001), CD86 (r = 0.96, P < 0.001), CD48 (r = 0.93, P < 0.001), HAVCR2 (r = 0.95, P < 0.001), PDCD1 (r = 0.93, P < 0.001), TIGIT (r = 0.95, P < 0.001) were significantly positively correlated with RPS3A expression (Figure 3C-K). Hence, on the basis of the showed results, there is a direct pathogenic link between RPS3A expression and induced tumor immune evasion.
Figure 3.
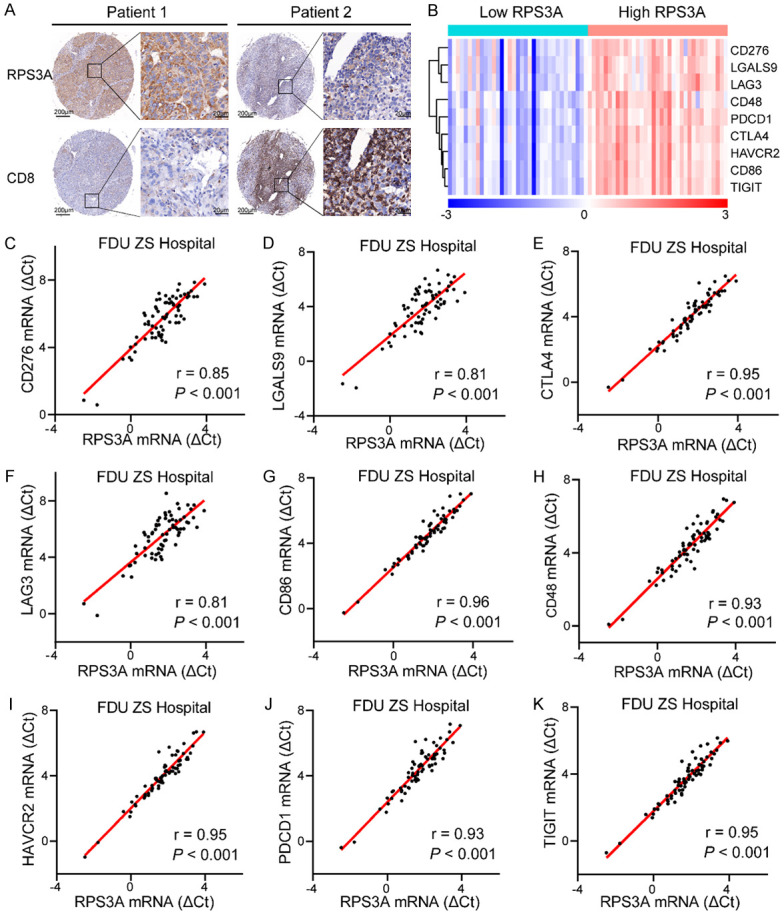
Association of high RPS3A expression with low tumor immune cell infiltration and high immune checkpoint molecules expression confirmed in HCC patients from Zhongshan Hospital. A. Representative IHC images of RPS3A and CD8 staining in the tumor tissues of two HCC patients. B. The heatmap of nine immune checkpoint molecules mRNA expression levels between HCC patients with low and high RPS3A expression. The median value of RPS3A mRNA expression was served as the cut-off point to assign 70 HCC patients to low and high RPS3A groups. C-K. Correlation between RPS3A mRNA expression and immune checkpoint molecules CD276, LGALS9, CTLA4, LAG3, CD86, CD48, HAVCR2, PDCD1, and TIGIT expression.
Table 1.
Correlations between RPS3A and CD8 expression levels in 154 HCC patients
| RPS3A expression | P value | ||||
|---|---|---|---|---|---|
|
| |||||
| -/+ | ++/+++ | Total | |||
| CD8 expression | -/+ | 26 (16.9%) | 50 (32.5%) | 76 (49.4%) | |
| ++/+++ | 49 (31.8%) | 29 (18.8%) | 78 (50.6%) | ||
| Total | 75 (48.7%) | 79 (51.3%) | 154 (100%) | < 0.001 | |
Involvement of biological pathways and processes in high and low RPS3A-expressing tumors
To identify the potential involvement of biological pathways and processes in HCC patients with high RPS3A expression, we analyzed RPS3A expression based on the RNA sequencing profiles of TCGA and c2 gene sets in the MSigDB database using GSEA. Of the 5501 gene sets in c2, the gene signatures “Anderson LIVER CANCER KRT19 UP”, “YAMASHITA LIVER CANCER WITH EPCAM UP”, “HEDENFALK BREAST CANCER BRACX UP”, “LI AMPLIFIED IN LUNG CANCER”, and “LUI THYROID CANCER CLUSTER 3” were enriched in patients with high RPS3A expression (Figure 4A). In addition, several signatures related to tumorigenesis and metastasis were positively correlated with RPS3A expression, such as “BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2”, “DANG MYC TARGETS UP”, and “REACTOME PERK REGULATES GENE EXPRESSION” (Figure 4B). Furthermore, to examine the association of RPS3A expression with other immune-related molecules in the HCC immune microenvironment, we looked at c7 gene sets using GSEA. Of the 4872 gene sets in c7, the gene signatures “GSE11924 TH1 VS TH2 CD4 T CELL DN”, “GSE26030 TH1 VS TH17 RESTIMULATED DAY5 POST POLARIZATION DN”, “GSE27786 NKTCELL VS NEUTROPHIL DN”, “GSE32423 IL7 VS IL7 IL4 NAIVE CD8 TCELL DN”, and “GSE4748 CTRL VS LPS STIM DC 3H DN” were significantly downregulated in high RPS3A-expressing tumors (Figure 4C). Collectively, these results further verified that high RPS3A expression in HCCs may promote the biological processes associated with tumorigenesis and metastasis and create an immunosuppressive tumor microenvironment in HCC patients.
Figure 4.
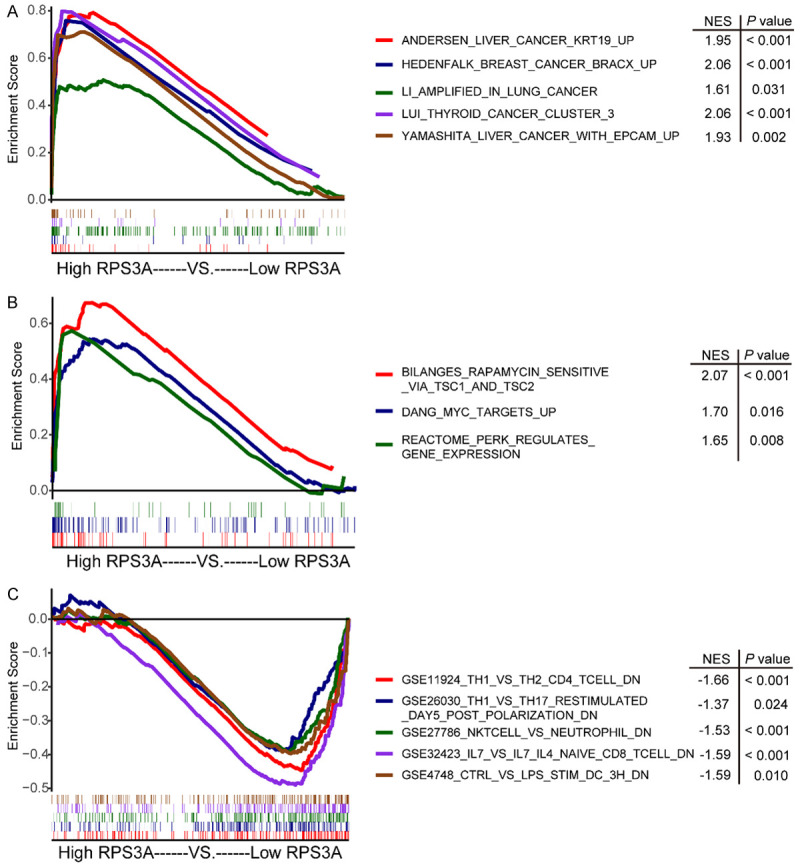
Biological pathways and processes that RPS3A may be involved in. A, B. GSEA of c2 (curated) gene sets from the MSigDB molecular signatures database in high versus low RPS3A-expressing tumors. C. GSEA of c7 (immunologic) gene sets from MSigDB in high and low RPS3A-expressing tumors. The different curve colors represent different gene signatures. The lower horizontal bars mark the genes in corresponding gene sets.
Association of RPS3A expression with clinicopathological characteristics of HCC patients
Considering the significance of RPS3A in the tumor immune microenvironment, we subsequently analyzed the association of RPS3A expression in tumor samples with clinicopathological features of the HCC patients. As shown in Table S3, the RPS3A level was positively correlated with the AFP level (P = 0.008), presence of liver cirrhosis (P = 0.025), and AST level (P = 0.021). Its correlation with other factors was not statistically significant.
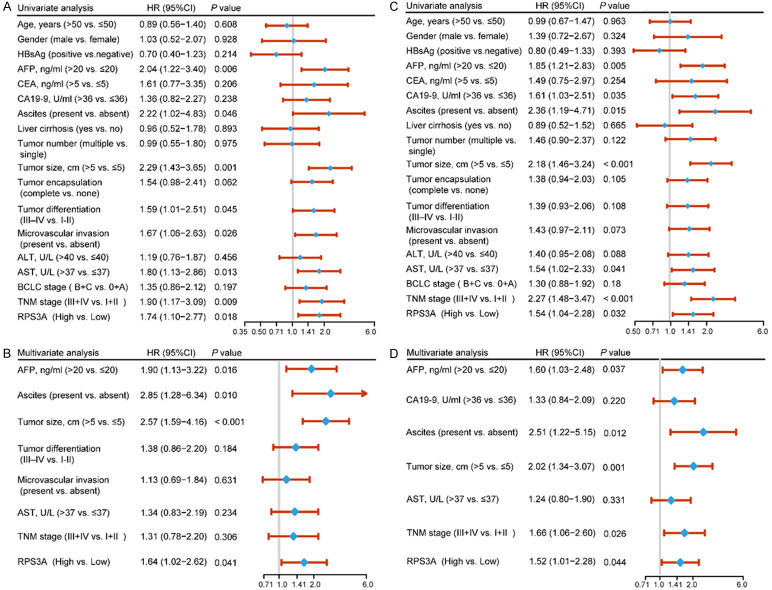
In order to clarify the prognostic value of RPS3A, we carried out univariate and multivariate analyses of prognosis in the HCC patients in our cohort (Figure 5). The results of univariate analyses demonstrated that RPS3A expression level in tumor samples as well as TNM stage, the AST level, tumor size, ascites, AFP level, and were highly associated with OS and RFS. In addition, microvascular invasion and tumor differentiation were considerably associated with OS of HCC, whereas the CA19-9 level was considered to be a high risk factor for RFS (Figure 5A and 5C). Furthermore, multivariate Cox regression analyses demonstrated that RPS3A expression was an independent prognostic predictor and exhibited robust predictive capacity for OS (hazard ratio (HR) = 1.64, 95% confidence interval (CI): 1.02-2.62; P = 0.041) and RFS (HR = 1.52, 95% CI: 1.01-2.28; P = 0.044). Meanwhile, AFP level, ascites, and tumor size were regarded as independent prognostic predictors for both OS and RFS. However, TNM stage only exhibited independent prognostic ability for RFS in HCC patients (Figure 5B and 5D).
Figure 5.
Univariate and multivariate Cox regression analysis of clinicopathological factors associated with survival and recurrence in HCC patients. A, B. Univariate and multivariate analyses of the clinicopathological features and RPS3A expression level for OS in HCC patients. The forest plot visualizes the hazard ratio (blue dot) and 95% CI (red horizontal line) for each variable. C, D. Univariate and multivariate analyses of the clinicopathological features and RPS3A expression level for RFS in HCC patients.
Establishment of RPS3A-based prognostic nomograms for HCC
The results described above demonstrated that high RPS3A expression correlated with low tumor immune cell infiltration and unfavorable prognosis in HCC patients. To provide more precise prediction of OS and RFS in HCC patients, which may contribute to clinical decision-making by physicians, we integrated tumor RPS3A expression level with other independent prognostic indicators mentioned above to establish nomograms for OS and RFS. The nomogram for OS combined RPS3A expression level, AFP level, ascites, and tumor size, whereas that for RFS combined these four variables and TNM stage (Figure 6A and 6B). Afterward, we constructed calibration curves to evaluate the accuracy of our nomograms. In the prediction of 3- and 5-year OS and RFS, the curves showed good consistency of predicted and actual observed outcomes (Figure 6C-F).
Figure 6.
Establishment of RPS3A-based prognostic nomograms for HCC. A. Nomogram for OS integrating RPS3A expression level, AFP level, ascites, and tumor size. The value of each indicator is obtained according to the upper point scale, and 3- and 5-year OS can be predicted by drawing a vertical line from the total point axis to the corresponding probability scale. B. Nomogram for RFS integrating RPS3A expression level, AFP level, ascites, tumor size, and TNM stage. C, D. Calibration curves for the nomogram for 3- and 5-year OS. The y-axis and x-axis represent actual and nomogram-predicted OS, respectively. The dashed line represents the optimal consistency of actual and predictive values. E, F. Calibration curves for the nomogram for 3- and 5-year RFS.
Predictive performance of RPS3A-based prognostic nomograms
To corroborate the good performance of RPS3A-based prognostic nomograms, we used the C-index to compare our constructed nomograms with conventional prognosis-prediction staging systems. As shown in Table 2, the nomograms had robust predictive ability for both OS (C-index = 0.701, 95% CI: 0.640-0.762) and RFS (C-index = 0.702, 95% CI: 0.647-0.757). More importantly, the nomograms for OS and RFS performed better at C-index than did the TNM (P < 0.001) and BCLC (P < 0.001) staging systems.
Table 2.
C-indices of RPS3A-based nomograms and independent prognostic factors in OS and RFS prediction for HCC patients
| Variable | Overall survival | Recurrence-free survival | ||
|---|---|---|---|---|
|
|
|
|||
| C-index (95% CI) | P value | C-index (95% CI) | P value | |
| TNM | 0.572 (0.520-0.624) | 0.579 (0.536-0.622) | ||
| BCLC | 0.541 (0.483-0.599) | 0.542 (0.491-0.593) | ||
| Nomogram | 0.701 (0.640-0.762) | 0.702 (0.647-0.757) | ||
| Nomogram vs. TNM | < 0.001 | < 0.001 | ||
| Nomogram vs. BCLC | < 0.001 | < 0.001 | ||
BCLC: Barcelona Clinic Liver Cancer; CI: confidence interval; C-index: concordance index; TNM: Tumor-Nodes-Metastases.
Furthermore, decision curve analysis showed that when compared with the TNM and BCLC staging systems, RPS3A-based nomograms exhibited a wider range of predictive thresholds and more net clinical benefits for OS and RFS at 3 and 5 years (Figure 7A-D). Taken together, these results revealed that RPS3A-based prognostic nomograms may be better for prognosis for HCC and have more net clinical benefits when compared with traditional staging systems.
Figure 7.
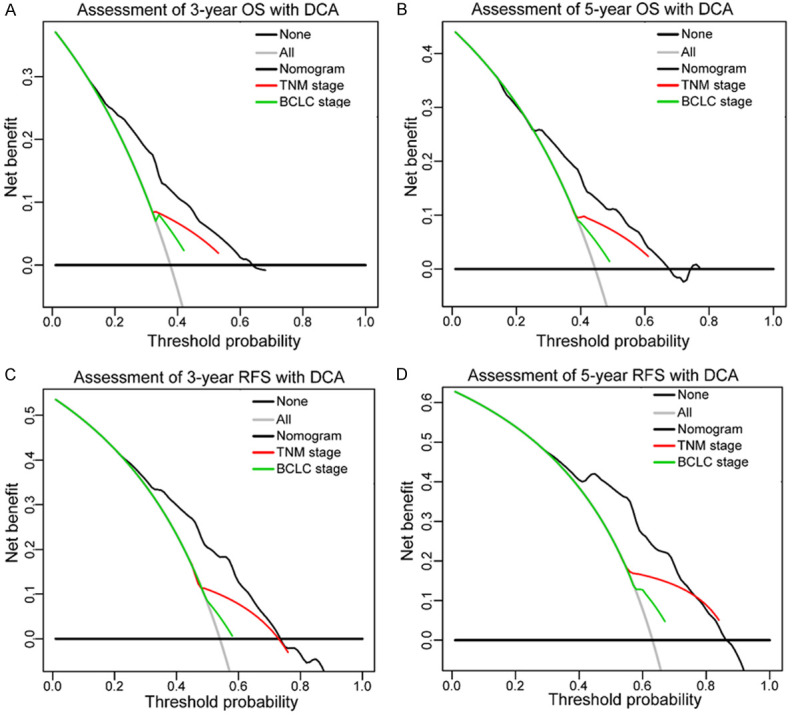
Decision curve analysis of the RPS3A-based nomograms, TNM stage, and BCLC stage for 3- and 5-year OS (A, B) and RFS (C, D). Lines for the different models (black, nomogram; red, TNM stage; green, BCLC stage) represent the corresponding net benefit (y-axis) and range of predictive thresholds of the probability of OS or RFS (x-axis). The horizontal black line at the bottom and the gray line indicate the assumptions that no and all patients would experience the event, respectively.
Discussion
RPS3A is an important component of the eukaryotic ribosomal small subunit (40S) and has multiple biological functions [15]. As described herein, we identified RPS3A as a potential modulator of tumor immune cell infiltration in HCC patients and highlighted the notable prognostic relevance of RPS3A expression to this disease. To the best of our knowledge, this is the first demonstration that HCC patients with high RPS3A expression are predisposed to a low immune infiltration status and to worse prognosis than are patients with low RPS3A expression.
The aberrantly high expression of RPS3A in HCC samples and cell lines we observed, consistent with that in other studies [31,32], suggested a role for this molecule in HCC carcinogenesis and progression. This suspicion was supported by our findings that higher RPS3A expression was closely related to worse OS and RFS of HCC. Previous studies demonstrated the participation of RPS3A in initiation of translation-related domains of hepatitis C virus and RPS3A-dependent enhancement of the hepatitis B virus X protein-induced oncogenic signaling pathway [33,34]. Because hepatitis B and C viruses are the main viral etiologies in HCC patients [35], RPS3A’s possible involvement in aberrant hepatocyte transformation and even tumorigenesis is plausible. Indeed, our results of GSEA showed significant enrichment of several cancer-related gene sets in high RPS3A-expressing tumors. Among these upregulated signatures, phosphorylated extracellular signal-regulated kinase (p-ERK) is reported to play a crucial role in promoting the proliferation, invasion, and metastasis of HCC [36]. Also, the proto-oncogene MYC is extensively involved in various oncogenic pathways, including the ERK/c-Myc pathway [37]. Meanwhile, we found that high RPS3A expression seemed to activate the oncogenic mammalian target of rapamycin (mTOR) signing pathway and affect the sensitivity of tumors to rapamycin by interacting with tuberous sclerosis complexes 1 (TSC1) and 2 (TSC2). Thus, our results set the groundwork for a deeper understanding of HCC-promoting mechanisms of RPS3A, suggesting that RPS3A may exert its tumor-promoting effect through activation of the ERK and mTOR pathways.
Tumor immune cell infiltration is associated with tumor progression and immunotherapy responses [5-7]. In our study, we observed a strongly negative relationship between RPS3A expression and the infiltration of several immune cell types, suggesting a relatively low degree of tumor immune cell infiltration in HCC patients with high RPS3A expression. Of the immune cell types that were significantly negatively correlated with RPS3A expression, the top three were Th17 cells, neutrophils, and DCs. Th17 cells can potentiate antitumor immune response by facilitating the recruitment of CD8+ T cells, NK cells, and DCs to the tumor microenvironment [38-40]. Neutrophils are the main immune cells involved in the inflammatory process and can participate in tumor-suppressing activities, such as activation of antitumor T cells and inhibition of cancer metastasis [41,42]. DCs are antigen-presenting cells that can induce potent antitumor immune response by priming naïve T cells [43]. The IHC staining in our own patients also showed a negative association between RPS3A expression and tumor infiltration of CD8+ CTL, which is the main tumor-killing immune cell [30]. The mechanism of RPS3A in regulating immune cell infiltration is not described in the literature, but authors reported that it can inhibit the activity of poly (ADP-ribose) polymerase [19]. The deletion of poly (ADP-ribose) polymerase is reported to reduce Th17 cells infiltration in the murine central nervous system [44]. Hence, it holds promise for increasing tumor immune cell infiltration via targeting of RPS3A. In addition, the positive correlation between expression of RPS3A and that of most immune checkpoint molecules that we observed in both public database and our own HCC samples suggested that this multifunctional ribosomal protein may promote the synthesis or expression of immunosuppressive molecules through unknown mechanisms. Thus, RPS3A may be a biomarker for predicting the responsiveness of HCC to ICB therapy. Another important issue is whether tumor immune cell infiltration-inducing effect of RPS3A-targeting therapy is synergistic with ICB therapy. Furthermore, the downregulated gene signatures related to immune systems in high RPS3A-expressing tumors suggested that RPS3A has immunosuppressive functions besides regulation of immune cell infiltration. Given the complex information in gene sets and lack of definite descriptions of signatures, studies designed to determine the exact mechanism of RPS3A in regulating recruitment of specific immune cell populations to tumors or other immune-related processes should be conducted in the future.
Analysis of the association between RPS3A and clinicopathological features in HCC patients demonstrated that the RPS3A expression level was related to the AFP level, a recognized prognostic factor for HCC. Also, the positive correlation of RPS3A with liver cirrhosis, which is accompanied by perturbed immunity [7], reflected a possible effect of RPS3A on immune regulation of the liver from another aspect.
The immunosuppressive and tumor-promoting roles of RPS3A may partially explain that HCC patients with high RPS3A expression have the worst prognoses. Therefore, we tried to find more prognostic value of RPS3A and its possible clinical translation in HCC prognosis prediction. Multivariate Cox regression analysis demonstrated that RPS3A expression was indeed an independent predictor of OS and RFS in HCC patients. Moreover, the improved predictive accuracy and increased net clinical benefits of RPS3A-based nomograms for OS and RFS make the nomograms promising prognostic prediction systems for HCC. The clinical application of these nomograms should be seriously considered and prospectively validated in the future.
Some limitations of our study should be acknowledged. This was a retrospective study and the number of patients enrolled was relatively small, although we exploited the data of HCC patients from TCGA database to support our study. Also, cohort-specific biases should be considered as all HCC patients were from a single medical center. Thus, a prospective, multicenter study is required to externally validate our results.
Taken together, our findings reveal that high RPS3A expression is associated with low tumor immune cell infiltration and poor OS and RFS in HCC patients. Furthermore, the positive correlation of RPS3A expression with immune checkpoint molecules expression and its potential tumor-promoting role suggest that RPS3A is a therapeutic target and can predict the effectiveness of ICB therapy for HCC. Finally, an RPS3A-based nomogram can serve as a robust prognostic predictor for HCC patients after hepatectomy.
Acknowledgements
We thank the China Scholarship Council for supporting Dr. Chenhao Zhou as a visiting PhD candidate at The University of Texas MD Anderson Cancer Center. We also thank Tamara Locke at the Department of Scientific Publications at the University of Texas MD Anderson Cancer Center for editing this manuscript.
This work was supported by the Shanghai International Science and Technology Collaboration Program (18410721900), the National Natural Science Foundation of China (81472672), the National Key Research and Development Program of China (2017YFC1308604 and 2017YFC0908402).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Han X, Yu X, Xu Z, Yang G, Liu B, Xiu P. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37:213. doi: 10.1186/s13046-018-0893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawkins J, Webster RM. The hepatocellular carcinoma market. Nat Rev Drug Discov. 2019;18:13–14. doi: 10.1038/nrd.2018.146. [DOI] [PubMed] [Google Scholar]
- 4.Hou J, Zhang H, Sun B, Karin M. The immunobiology of hepatocellular carcinoma in humans and mice: basic concepts and therapeutic implications. J Hepatol. 2020;72:167–182. doi: 10.1016/j.jhep.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smyth MJ, Ngiow SF, Ribas A, Teng MW. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 7.Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222–232. doi: 10.1038/s41590-018-0044-z. [DOI] [PubMed] [Google Scholar]
- 8.Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:681–700. doi: 10.1038/nrgastro.2015.173. [DOI] [PubMed] [Google Scholar]
- 9.Cariani E, Missale G. Immune landscape of hepatocellular carcinoma microenvironment: implications for prognosis and therapeutic applications. Liver Int. 2019;9:1608–1621. doi: 10.1111/liv.14192. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Tang Y, Tang PMK, Lv J, Huang XR, Carlsson-Skwirut C, Da Costa L, Aspesi A, Frohlich S, Szczesniak P, Lacher P, Klug J, Meinhardt A, Fingerle-Rowson G, Gong R, Zheng Z, Xu A, Lan HY. Blocking macrophage migration inhibitory factor protects against cisplatin-induced acute kidney injury in mice. Mol Ther. 2018;26:2523–2532. doi: 10.1016/j.ymthe.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv J, Huang XR, Klug J, Frohlich S, Lacher P, Xu A, Meinhardt A, Lan HY. Ribosomal protein S19 is a novel therapeutic agent in inflammatory kidney disease. Clin Sci (Lond) 2013;124:627–637. doi: 10.1042/CS20120526. [DOI] [PubMed] [Google Scholar]
- 12.Nayar S, Campos J, Smith CG, Iannizzotto V, Gardner DH, Colafrancesco S, Pipi E, Kollert F, Hunter KJ, Brewer C, Buckley CD, Bowman SJ, Priori R, Valesini G, Juarez M, Fahy WA, Fisher BA, Payne A, Allen RA, Barone F. Phosphatidylinositol 3-kinase delta pathway: a novel therapeutic target for Sjogren’s syndrome. Ann Rheum Dis. 2019;78:249–260. doi: 10.1136/annrheumdis-2017-212619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markiewski MM, Vadrevu SK, Sharma SK, Chintala NK, Ghouse S, Cho JH, Fairlie DP, Paterson Y, Astrinidis A, Karbowniczek M. The ribosomal protein S19 suppresses antitumor immune responses via the complement C5a receptor 1. J Immunol. 2017;198:2989–2999. doi: 10.4049/jimmunol.1602057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T. Molecular mechanism of monocyte predominant infiltration in chronic inflammation: mediation by a novel monocyte chemotactic factor, S19 ribosomal protein dimer. Pathol Int. 2000;50:863–871. doi: 10.1046/j.1440-1827.2000.01132.x. [DOI] [PubMed] [Google Scholar]
- 15.Naora H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol. 1999;77:197–205. doi: 10.1046/j.1440-1711.1999.00816.x. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y, He Y, Li C, Mu W, Zou Y, Liu C, Qian S, Zhang F, Pan J, Wang Y, Huang H, Pan D, Yang P, Mei J, Zeng R, Tang QQ. RPS3A positively regulates the mitochondrial function of human periaortic adipose tissue and is associated with coronary artery diseases. Cell Discov. 2018;4:52. doi: 10.1038/s41421-018-0041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell L, Naora H, Naora H. Down-regulated RPS3a/nbl expression during retinoid-induced differentiation of HL-60 cells: a close association with diminished susceptibility to actinomycin D-stimulated apoptosis. Cell Struct Funct. 2000;25:103–113. doi: 10.1247/csf.25.103. [DOI] [PubMed] [Google Scholar]
- 18.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song D, Sakamoto S, Taniguchi T. Inhibition of poly(ADP-ribose) polymerase activity by Bcl-2 in association with the ribosomal protein S3a. Biochemistry. 2002;41:929–934. doi: 10.1021/bi015669c. [DOI] [PubMed] [Google Scholar]
- 20.Kashuba E, Yurchenko M, Szirak K, Stahl J, Klein G, Szekely L. Epstein-Barr virus-encoded EBNA-5 binds to Epstein-Barr virus-induced Fte1/S3a protein. Exp Cell Res. 2005;303:47–55. doi: 10.1016/j.yexcr.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 21.Kho CJ, Zarbl H. Fte-1, a v-fos transformation effector gene, encodes the mammalian homologue of a yeast gene involved in protein import into mitochondria. Proc Natl Acad Sci U S A. 1992;89:2200–2204. doi: 10.1073/pnas.89.6.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callegaro D, Miceli R, Bonvalot S, Ferguson P, Strauss DC, Levy A, Griffin A, Hayes AJ, Stacchiotti S, Pechoux CL, Smith MJ, Fiore M, Dei Tos AP, Smith HG, Mariani L, Wunder JS, Pollock RE, Casali PG, Gronchi A. Development and external validation of two nomograms to predict overall survival and occurrence of distant metastases in adults after surgical resection of localised soft-tissue sarcomas of the extremities: a retrospective analysis. Lancet Oncol. 2016;17:671–680. doi: 10.1016/S1470-2045(16)00010-3. [DOI] [PubMed] [Google Scholar]
- 23.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 24.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 25.Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, Xu Y, Li YW, Tang ZY. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J. Clin. Oncol. 2007;25:2586–2593. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Zhao Y, Dai Y, Cheng JN, Gong Z, Feng Y, Sun C, Jia Q, Zhu B. Immune landscape of colorectal cancer tumor microenvironment from different primary tumor location. Front Immunol. 2018;9:1578. doi: 10.3389/fimmu.2018.01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flecken T, Schmidt N, Hild S, Gostick E, Drognitz O, Zeiser R, Schemmer P, Bruns H, Eiermann T, Price DA, Blum HE, Neumann-Haefelin C, Thimme R. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology. 2014;59:1415–1426. doi: 10.1002/hep.26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuda M, Kondoh N, Tanaka K, Ryo A, Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, Horiuchi S, Shichita M, Yamamoto N, Yamamoto M. Enhanced expression of translation factor mRNAs in hepatocellular carcinoma. Anticancer Res. 2000;20:2489–2494. [PubMed] [Google Scholar]
- 32.Kim MY, Park E, Park JH, Park DH, Moon WS, Cho BH, Shin HS, Kim DG. Expression profile of nine novel genes differentially expressed in hepatitis B virus-associated hepatocellular carcinomas. Oncogene. 2001;20:4568–4575. doi: 10.1038/sj.onc.1204626. [DOI] [PubMed] [Google Scholar]
- 33.Lim KH, Kim KH, Choi SI, Park ES, Park SH, Ryu K, Park YK, Kwon SY, Yang SI, Lee HC, Sung IK, Seong BL. RPS3a over-expressed in HBV-associated hepatocellular carcinoma enhances the HBx-induced NF-kappaB signaling via its novel chaperoning function. PLoS One. 2011;6:e22258. doi: 10.1371/journal.pone.0022258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malygin AA, Shatsky IN, Karpova GG. Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosc) 2013;78:53–59. doi: 10.1134/S0006297913010069. [DOI] [PubMed] [Google Scholar]
- 35.Ng CKY, Di Costanzo GG, Terracciano LM, Piscuoglio S. Circulating cell-free DNA in hepatocellular carcinoma: current insights and outlook. Front Med (Lausanne) 2018;5:78. doi: 10.3389/fmed.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Yang C, Wu J, Nan Y. ADAMTS8 targets ERK to suppress cell proliferation, invasion, and metastasis of hepatocellular carcinoma. Onco Targets Ther. 2018;11:7569–7578. doi: 10.2147/OTT.S173360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Lin C, Liu Y, Jiang Y. HMGB1 promotes HCC progression partly by downregulating p21 via ERK/c-Myc pathway and upregulating MMP-2. Tumour Biol. 2016;37:4399–4408. doi: 10.1007/s13277-015-4049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryczek I, Zhao E, Liu Y, Wang Y, Vatan L, Szeliga W, Moyer J, Klimczak A, Lange A, Zou W. Human TH17 cells are long-lived effector memory cells. Sci Transl Med. 2011;3:104ra100. doi: 10.1126/scitranslmed.3002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Orozco N, Muranski P, Chung Y, Yang XO, Yamazaki T, Lu S, Hwu P, Restifo NP, Overwijk WW, Dong C. T helper 17 cells promote cytotoxic T cell activation in tumor immunity. Immunity. 2009;31:787–798. doi: 10.1016/j.immuni.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kryczek I, Wei S, Szeliga W, Vatan L, Zou W. Endogenous IL-17 contributes to reduced tumor growth and metastasis. Blood. 2009;114:357–359. doi: 10.1182/blood-2008-09-177360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponzetta A, Carriero R, Carnevale S, Barbagallo M, Molgora M, Perucchini C, Magrini E, Gianni F, Kunderfranco P, Polentarutti N, Pasqualini F, Di Marco S, Supino D, Peano C, Cananzi F, Colombo P, Pilotti S, Alomar SY, Bonavita E, Galdiero MR, Garlanda C, Mantovani A, Jaillon S. Neutrophils driving unconventional T cells mediate resistance against murine sarcomas and selected human tumors. Cell. 2019;178:346–360. e324. doi: 10.1016/j.cell.2019.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castano Z, Juan BP, Spiegel A, Pant A, DeCristo MJ, Laszewski T, Ubellacker JM, Janssen SR, Dongre A, Reinhardt F, Henderson A, Del Rio AG, Gifford AM, Herbert ZT, Hutchinson JN, Weinberg RA, Chaffer CL, McAllister SS. IL-1beta inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat Cell Biol. 2018;20:1084–1097. doi: 10.1038/s41556-018-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banchereau J, Palucka AK. Dendritic cells as therapeutic vaccines against cancer. Nat Rev Immunol. 2005;5:296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- 44.Kamboj A, Lu P, Cossoy MB, Stobart JL, Dolhun BA, Kauppinen TM, de Murcia G, Anderson CM. Poly(ADP-ribose) polymerase 2 contributes to neuroinflammation and neurological dysfunction in mouse experimental autoimmune encephalomyelitis. J Neuroinflammation. 2013;10:49. doi: 10.1186/1742-2094-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.