Abstract
Targeted therapeutic agents such as poly (ADP-ribose) polymerases (PARP) inhibitors have emerged in treating cancers associated with germline BRCA mutations. Recently studies demonstrated the effectiveness of PARP inhibitors in treating patients with somatic BRCA mutations. Somatic mutations in 122 Chinese breast or ovarian cancer patients without BRCA, PTEN and TP53 mutations were screened using multigene sequencing panel. The five most frequent pathogenic or likely pathogenic mutated genes identified in breast cancer patients were PIK3CA (28.6%), TP53 (16.9%), MAP3K1 (14.3%), GATA3 (14.3%) and PTEN (5.2%). The five most frequently mutated genes identified in ovarian patients were TP53 (52.9%), KRAS (23.5%) and PIK3CA (11.8%), BRCA1 (5.9%) and RB1 (5.9%). Somatic PIK3CA and TP53 mutations were common events in both germline BRCA-negative breast and ovarian cancer patients. In contrast, somatic screening of BRCA mutations in BRCA-negative breast cancer patients has limited value. The results highlight the benefit of somatic testing to guide future research directions on other targeted therapies for breast and ovarian malignancies.
Keywords: Breast cancer, ovarian cancer, BRCA-negative, somatic mutations, multigene panel
Introduction
The link between genetic mutations and cancer pathogenesis has long been extensively studied, and genetic testing is taking on an increasingly important role to reduce the disease burden of breast cancer. The discoveries of BRCA germline mutations, which are associated with an estimated 20% of hereditary breast cancers, fueled the excitement for potential breast cancer treatment target [1-4]. Targeted therapeutic agents such as poly(ADP-ribose) polymerases (PARP) inhibitors have emerged with better outcomes in treating cancers with BRCA mutations and those with other homologous recombination (HR) deficiencies [5,6].
The first PARP inhibitor was FDA-approved initially for ovarian cancer patients with germline BRCA mutations. Subsequent studies demonstrated their effective in those with somatic BRCA mutations and other HR deficiencies, which are present in up to 20% of serous ovarian cancers [7]. Olaparib and talazoparib are FDA-approved PARP inhibitors for metastatic breast cancer patients with germline BRCA mutations [8,9] and metastatic triple negative breast cancer patient with somatic BRCA1 mutation [10]. Therefore, it is postulated that somatic BRCA-mutated tumors might also respond to this new class of therapeutics similar to that in ovarian cancer setting. A phase II clinical trial is currently investigating the effectiveness of olaparib in both germline and somatic BRCA-positive breast cancer patients [11]. Other than PARP inhibitors, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) inhibitor, alpelisib, has been used to treat patients with PIK3CA-mutated, hormonal receptor-positive, HER2-negative advanced breast cancer [12]. Several phase I and II clinical trials are conducted to investigate the efficacy of PIK3CA and AKT inhibitors on breast cancer patients with somatic mutations in the PI3K/AKT/mTOR pathway.
Somatic mutation analysis of 173 genes using in 2,433 breast tumors identified 13,084 somatic mutations which were predicted to affect protein sequence [13]. Another study identified mutations in 93 protein-coding cancer genes in 560 paired breast tumors and normal tissues by whole genome sequencing [14]. PIK3CA and TP53 were the most frequently mutated genes found in breast tumors among these studies [13,14]. Notably, the rate of somatic BRCA mutations in breast malignancy is rather low, implying that other mechanisms, such as epigenetics, might be a more important event in somatic breast cancers with BRCAness [14-18]. On the other hand, large-scale genetic mutation analysis from The Cancer Genome Atlas (TCGA) revealed TP53 mutation was also found in 96% of high-grade serous ovarian cancer tissue samples [19] and commonly seen in different populations [20,21]. Therefore, the presence of somatic BRCA mutations or other mutated genes must be ascertained so as to offer appropriate targeted therapies to improve treatment outcome in breast or ovarian cancers.
The objective of our study is to uncover the landscape of somatic mutations in germline (BRCA, PTEN and TP53) mutation-negative breast and ovarian cancer patients and identity the common somatic mutations which can potentially be targeted for therapeutic purposes.
Methods
Ethics statement
All human tissue samples in this study were used according to the Declaration of Helsinki. Written informed consent was obtained from all participants recruited in this study. This study was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority West Cluster and other contributing hospitals in Hong Kong.
Samples and selection criteria
122 tumor tissues of breast or ovarian cancer tested negative for germline BRCA1, BRCA2, TP53, and PTEN mutations were retrieved for this study. They were recruited by the Hong Kong Hereditary and High Risk Breast Cancer Program from March 2007 to October 2017. The selection criteria were described previously with modification [4]. High-risk patients who were germline BRCA/TP53/PTEN negative and had not received any neoadjuvant chemotherapy were included.
DNA extraction & multigene panel sequencing
Genomic DNA was extracted from tissues with DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Extracted tumor DNA was subjected to Human Breast Cancer Panel from QIAseq Targeted DNA Panel (DHS-001Z, Qiagen) with 93 breast cancer predisposition genes. Sequencing libraries were prepared according the manufacturer’s instructions. The quality and quantity of the re-purified DNA were assessed by Qubit Fluorometer (Thermo Fisher Scientific, Massachusetts, US). DNA was enzymatically fragmented and ligated with molecular barcodes and sample indexing adaptor. Ligated DNA was further enriched by PCR with a universal forward primer and the specific targeting primers from above panel which allows the targets and barcodes to enrich sufficiently. Another round of universal PCR amplification was carried out with platform specific adaptor sequences added for final completion of the library construct. The amplicon products of each sample were subjected to quality check with the use of Agilent DNA 1000 Kit on a 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA). The libraries were sequenced on MiSeq or NextSeq (Illumina, San Diego, CA) with QIAseq A Read 1 Primer I and the minimum sequencing depth was 50-fold and average depth of 500.
Data analysis
The bioinformatics analysis was performed on a Cray XC30 supercomputer (Cray, Seattle, WA). Paired sequencing reads were mapped to human reference genome sequence GRCh37/hg19 using BWA-MEM v0.7.7 [22] and default parameters. Post-alignment primer clipping and unique molecular identifier (UMI) extraction were performed using BAMClipper [23] adapted for single primer extension dataset. Samples having at least 75% of gene-specific primers with at least 100 detected UMI per primer were considered to pass quality control and subjected to variant calling by FreeBayes v1.0.2-15 [24]. Called variants with variant allelic fraction (VAF) at least 10% and sequencing depth at least 500× were annotated by Ensembl Variant Effect Predictor v75 [25]. A high stringency is adopted in order to eliminate the heterogeneity within tumors and also noise parameters.
Variant interpretation
With reference to the public databases including gnomAD (https://gnomad.broadinstitute.org/), dbSNP (http://www.ncbi.nlm.nih.gov/SNP) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), variants were interpreted and assigned classifications according to the four-tier terminology system of the joint guidelines from Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists [26]: Tier I (variants with strong clinical significance); Tier II (variants with potential clinical significance); Tier III (variants of unknown significance) and Tier IV (benign or likely benign). Mutation variants that present in 1,000 Genomes resource or causing in-frame consequences were not analyzed. In addition, SNPs were excluded if the allele frequency ≥ 0.01% in East Asian population in the gnomAD database. Passenger mutations or mutations with high mutation background (e.g. MUC16 and KMT2C) were omitted as they were seen in almost 99% of our samples.
Statistical analysis
Clinicopathological variables from pathogenic/likely pathogenic mutation carriers and non-carriers were tabulated in contingency tables. Statistical tests suitable for categorical data were then considered. Since some variables had expected values less than 5 in some cells, and most of the variable did not have natural ordering, Fisher’s exact test was finally adopted. Significance level was set at 5%. A p-value less than 0.05 would then indicate the rejection of null hypothesis of independence of variables. The computation was performed using R (version 3.4.2, Foundation for Statistical Computing, Vienna, Austria).
Results
Study population
Of 108 breast cancer patients, the mean age of diagnosis was 54.1 years (range, 25-86 years). Most of the patients were diagnosed at stage I (31.5%) and II (47.2%), and 14.8% of the patients had stage III. Majority of the tumors were invasive ductal carcinoma (82/108, 75.9%) and were grade 2 tumors (49/108, 50.5%). In particular, 43 (39.8%) had at least one first- or second-degree relative with BRCA-related cancer (other than breast or ovarian cancer) and 37 (34.3%) had family history of breast cancer. Clinicopathological characteristics of the breast cancer patients were listed in Table 1A.
Table 1A.
Clinicopathologic data of the breast cohort patients (N=108)
| Sex | |
| Female | 108 (100%) |
| Age (Mean/Range) | 54.05 (25-86) |
| Diagnosis Age | |
| <35 | 5 (4.63%) |
| 35-44 | 35 (32.41%) |
| 45-54 | 25 (23.15%) |
| 55-64 | 14 (12.96%) |
| ≥65 | 29 (26.85%) |
| Bilateral Breast | 18 (16.67%) |
| Personal cancers | |
| Breast Cancer only | 90 (83.33%) |
| Both Breast & Ovarian or GYN cancers | 3 (2.78%) |
| Breast cancer with multiple other cancers | 15 (13.89%) |
| Menopause | |
| Yes | 54 (50.00%) |
| Family History (1st or 2nd degree) | |
| Breast Cancer | 37 (34.26%) |
| Ovarian Cancer | 4 (3.70%) |
| BRCA Related Cancer (other than Breast & Ovarian) | 43 (39.81%) |
| Histology of Breast tumors | |
| Ductal | 82 (75.93%) |
| Ductal + Lobular/Medullary | 3 (2.78%) |
| In situ carcinoma | 7 (6.48%) |
| Lobular | 6 (5.56%) |
| Medullary | 1 (0.93%) |
| Others/Unclassified | 9 (8.33%) |
| Molecular Subtypes of Breast tumors | |
| Hormonal + | 75 (69.44%) |
| TNBC | 19 (17.59%) |
| Her2 | 14 (12.96%) |
| Stage | |
| Stage 0 | 7 (6.48%) |
| Stage I | 34 (31.48%) |
| Stage II | 51 (47.22%) |
| Stage III | 16 (14.81%) |
| Invasive Grade | |
| Grade 1 | 15 (15.46%) |
| Grade 2 | 49 (50.52%) |
| Grade 3 | 33 (34.02%) |
| Not stated | 4 |
| DCIS only | 7 |
Among 14 patients with ovarian tumors, the mean age of diagnosis was 45.3 years (range, 16-78 years). Most of the tumors are at high grade (8/14, 61.5%), 6 (42.9%) had endometrioid and 5 (35.7%) had serous cancer. Majority of the patients were diagnosed with stage I (4/14, 30.8%) and III (7/14, 53.9%). The patients’ characteristics are summarized in Table 1B.
Table 1B.
Clinicopathologic data of the ovarian cohort patients (N=14)
| Sex | |
| Female | 14 (100%) |
| Age (Mean/Range) | 45.29 (16-78) |
| Diagnosis Age | |
| <35 | 3 (21.43%) |
| 35-44 | 2 (14.29%) |
| 45-54 | 6 (42.86%) |
| 55-64 | 2 (14.29%) |
| ≥65 | 1 (7.14%) |
| Personal cancers | |
| Ovarian Cancer | 14 (100%) |
| Menopause | |
| Yes | 9 (64.29%) |
| Family History (1st or 2nd degree) | |
| Breast Cancer | 2 (14.29%) |
| Ovarian Cancer | 0 (0%) |
| BRCA Related Cancer (other than Breast & Ovarian) | 2 (14.29%) |
| Histology of Ovarian tumors | |
| Serous | 5 (35.71%) |
| Mucinous | 2 (14.29%) |
| Endometrioid | 6 (42.86%) |
| Metastatic adenocarcinoma | 1 (7.14%) |
| FIGO Stage | |
| I | 4 (30.77%) |
| II | 1 (7.69%) |
| III | 7 (53.85%) |
| IV | 1 (7.69%) |
| Not stated | 1 |
| Grade | |
| 1 | 1 (7.69%) |
| 2 | 4 (30.77%) |
| 3 | 8 (61.54%) |
| Not stated | 1 |
Mutation landscapes in breast tumors
With our customized bioinformatics pipeline for variant calling, 369 somatic mutations in 67 cancer predisposition genes were predicted to affect protein sequences in Tier I-IV. Among these mutations, there were 9 small insertions or deletions, 38 caused frameshift termination or early termination, and 19 were splice site variants. Each tumor had an average of 3.4 coding mutations and 25 of them harbored at least 5 coding mutations. Six of them were devoid of any mutation. PIK3CA (29.6%) and TP53 (25.9%) mutations dominated the somatic mutation landscape of breast tumors. Five other common genes were SYNE1 (20.4%), BRCA2 (16.7%), MSH2 (13.0%), BRCA1 (11.1%) and MLH1 (11.1%) (Figure 1).
Figure 1.
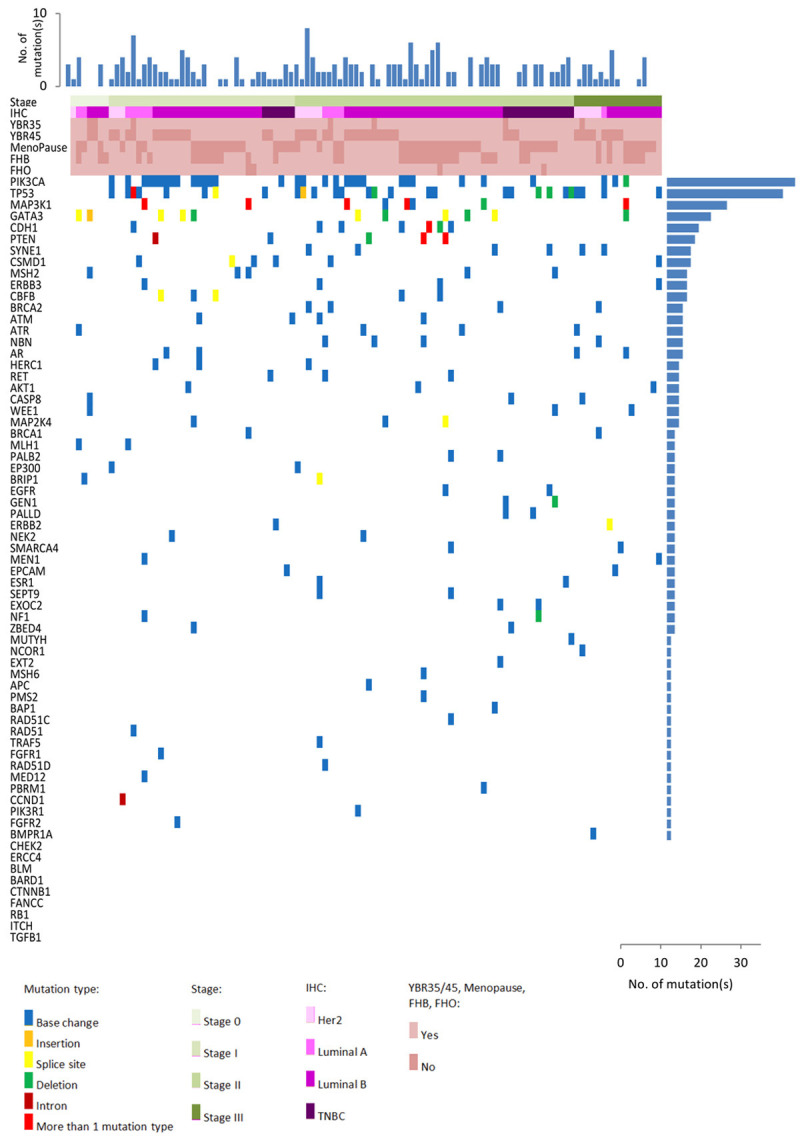
Heatmap of mutation identified in breast tumor tissues. Heatmap of most frequent somatic mutations identified in this study according to subtype. Mutation types are color-coded according to the legend. Top panel: the number of mutations found in each tumor is shown. Right Panel: The number of mutations identified in each gene is shown. Left Panel: Different types of gene mutations carried by each tumors indicated. Abbreviation: IHC: Immunohistochemistry; YBR: Young Breast Cancer; FHB: Family history of breast cancer; FHO: Family history of ovarian cancer.
Altogether 77 Tier I/II variants corresponding to 15 cancer predisposition genes were identified (Table 2), in which 49.1% of the breast tumors had at least one Tier I or Tier II variants. The most frequent mutated genes were PIK3CA (28.6%), TP53 (16.9%), MAP3K1 (14.3%), GATA3 (14.3%) and PTEN (5.2%). PIK3CA c.3140A>G (11.1%), GATA3 c.925-3_925-2delCA (5.6%) and PIK3CA c.1633G>A (4.6%) were common in breast tumors. Also, the major mutations were the cancer driver genes involved in AKT signaling (37.7%), genome integrity (20.8%), transcription regulation (18.2%) and MAPK signaling pathways (18.2%) (Figure 2). Apart from Tier I/II mutations, a total of 144 variants of Tier III in 55 cancer susceptibility genes from 66 breast tumors were identified (Table S1).
Table 2.
Tier I and Tier II mutation variants called in Breast cohort
| Gene | HVGS | Frequency |
|---|---|---|
| AKT1 | c.49G>A; p.Glu17Lys | 3 |
| BRIP1 | c.1936-1G>C | 1 |
| CBFB | c.133C>T; p.Gln45Ter | 1 |
| CBFB | c.165+2_165+3insT | 1 |
| CBFB | c.79-1G>T | 1 |
| CDH1 | c.1345C>T; p.Gln449Ter | 1 |
| CDH1 | c.2173delC; p.Leu725CysfsTer45 | 1 |
| CSMD1 | c.10039+1G>C | 1 |
| GATA3 | c.1085delT; p.Ile362ThrfsTer43 | 1 |
| GATA3 | c.1103_1104delAA; p.Lys368AsnfsTer3 | 1 |
| GATA3 | c.1223_1224insT; p.Pro409AlafsTer99 | 1 |
| GATA3 | c.1278delA; p.Ser427ProfsTer49 | 1 |
| GATA3 | c.1299_1318delACACCACCCCTCCAGCATGG; p.His435ArgfsTer66 | 1 |
| GATA3 | c.925-3_925-2delCA | 6 |
| GEN1 | c.562_563delAA; p.Lys188GlufsTer2 | 1 |
| MAP2K4 | c.219-2A>G | 1 |
| MAP2K4 | c.274G>T; p.Glu92Ter | 1 |
| MAP3K1 | c.1080_1081insC; p.Val361ArgfsTer24 | 1 |
| MAP3K1 | c.1152+1G>A | 1 |
| MAP3K1 | c.1594C>T; p.Arg532Ter | 1 |
| MAP3K1 | c.1624_1625insA; p.Thr542AsnfsTer17 | 1 |
| MAP3K1 | c.2262delT; p.Gly756AlafsTer6 | 1 |
| MAP3K1 | c.2479_2488delGTTACTACAG; p.Val827TyrfsTer9 | 1 |
| MAP3K1 | c.2867_2870delTTCA; p.Val956GlufsTer11 | 1 |
| MAP3K1 | c.3315_3316insCA; p.Ile1106GlnfsTer12 | 1 |
| MAP3K1 | c.3387_3396delCTCCAGTATT; p.Asn1129LysfsTer16 | 1 |
| MAP3K1 | c.3814_3817delAAAC; p.Lys1272ArgfsTer2 | 1 |
| MAP3K1 | c.3982+1_3982+2insG | 1 |
| MSH6 | c.2194C>T; p.Arg732Ter | 1 |
| NEK2 | c.952C>T; p.Arg318Ter | 1 |
| PALB2 | c.3113G>A; p.Trp1038Ter | 1 |
| PIK3CA | c.1624G>A; p.Glu542Lys | 1 |
| PIK3CA | c.1633G>A; p.Glu545Lys | 5 |
| PIK3CA | c.3140A>G; p.His1047Arg | 12 |
| PIK3CA | c.3140A>T; p.His1047Leu | 3 |
| PIK3CA | c.353G>A; p.Gly118Asp | 1 |
| PTEN | c.127_149delGAAGGCGTATACAGGAACAATAT; p.Glu43Ter | 1 |
| PTEN | c.238A>T; p.Lys80Ter | 1 |
| PTEN | c.396delT; p.Val133Ter | 1 |
| PTEN | c.405_406insA; p.Cys136MetfsTer44 | 1 |
| TP53 | c.216_217insC; p.Val73ArgfsTer76 | 1 |
| TP53 | c.321C>G; p.Tyr107Ter | 1 |
| TP53 | c.488A>G; p.Tyr163Cys | 1 |
| TP53 | c.493delC; p.Gln165SerfsTer5 | 1 |
| TP53 | c.559+2T>C | 1 |
| TP53 | c.635_636delTT; p.Phe212SerfsTer3 | 1 |
| TP53 | c.723delC; p.Cys242AlafsTer5 | 2 |
| TP53 | c.734G>A; p.Gly245Asp | 2 |
| TP53 | c.817C>T; p.Arg273Cys | 1 |
| TP53 | c.916C>T; p.Arg306Ter | 2 |
Figure 2.
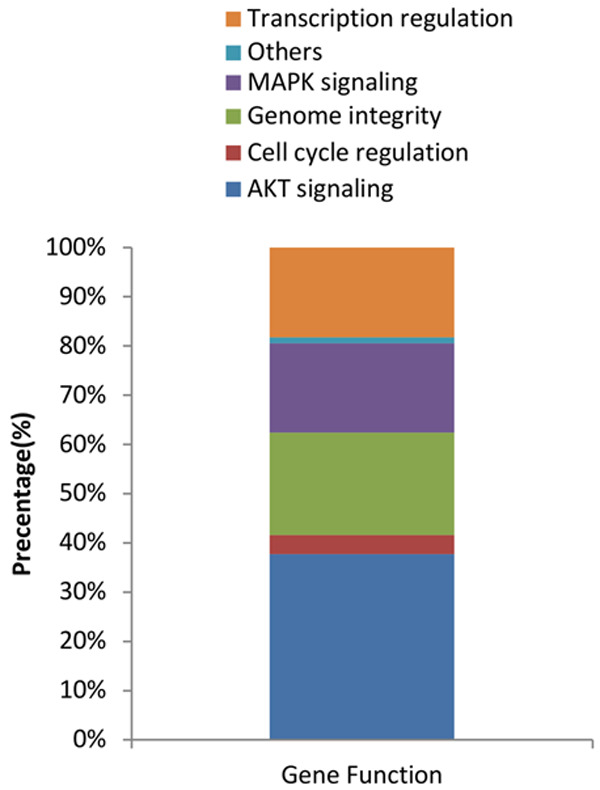
Identification of 15 mutated gene function. Genes carried mutations were classified according to functions: transcription regulation; MAPK signaling; genome instability; cell cycle regulation and AKT signaling.
A total of 221 mutation variants (Tier I/II: 77; Tier III: 144) from 108 tumors were identified (Table 3). Surprisingly, 6 of the tumors had no somatic mutation found and the rest carried an average of 2.2 mutations. Among these 221 mutation variants, 13 dominant recurrence mutation variants were seen in 46.2% of the tumors (50/108) and none of them have been reported in gnomAD within the East Asian population (Table S2). Of note, there were 6 BRCA mutations in Tier III (BRCA1:2; BRCA2:4) but none was in Tier I/II, in which 4 of the BRCA mutations were confirmed to be germline. Interestingly, there was only one case with both Tier III somatic BRCA1 and BRCA2 mutations.
Table 3.
Mutation frequency (N=221) of different genes in breast cohort
| Gene (s) | Tier I/II (N=77) | Tier III (N=144) | Total (N=221) |
|---|---|---|---|
| AKT1 | 3 (3.9%) | - | 3 (1.36%) |
| APC | - | 1 (0.69%) | 1 (0.45%) |
| AR | - | 4 (2.78%) | 4 (1.81%) |
| ATM | - | 4 (2.78%) | 4 (1.81%) |
| ATR | - | 4 (2.78%) | 4 (1.81%) |
| BAP1 | - | 1 (0.69%) | 1 (0.45%) |
| BMPR1A | - | 1 (0.69%) | 1 (0.45%) |
| BRCA1 | - | 2 (1.39%) | 2 (0.9%) |
| BRCA2 | - | 4 (2.78%) | 4 (1.81%) |
| BRIP1 | 1 (1.3%) | 1 (0.69%) | 2 (0.9%) |
| CASP8 | - | 3 (2.08%) | 3 (1.36%) |
| CBFB | 3 (3.9%) | 2 (1.39%) | 5 (2.26%) |
| CCND1 | - | 1 (0.69%) | 1 (0.45%) |
| CDH1 | 2 (2.6%) | 6 (4.17%) | 8 (3.62%) |
| CSMD1 | 1 (1.3%) | 5 (3.47%) | 6 (2.71%) |
| EGFR | - | 2 (1.39%) | 2 (0.9%) |
| EP300 | - | 2 (1.39%) | 2 (0.9%) |
| EPCAM | - | 2 (1.39%) | 2 (0.9%) |
| ERBB2 | - | 2 (1.39%) | 2 (0.9%) |
| ERBB3 | - | 5 (3.47%) | 5 (2.26%) |
| ESR1 | - | 2 (1.39%) | 2 (0.9%) |
| EXOC2 | - | 2 (1.39%) | 2 (0.9%) |
| EXT2 | - | 1 (0.69%) | 1 (0.45%) |
| FGFR1 | - | 1 (0.69%) | 1 (0.45%) |
| FGFR2 | - | 1 (0.69%) | 1 (0.45%) |
| GATA3 | 11 (14.29%) | - | 11 (4.98%) |
| GEN1 | 1 (1.3%) | 1 (0.69%) | 2 (0.9%) |
| HERC1 | - | 3 (2.08%) | 3 (1.36%) |
| MAP2K4 | 2 (2.6%) | 1 (0.69%) | 3 (1.36%) |
| MAP3K1 | 11 (14.29%) | 4 (2.78%) | 15 (6.79%) |
| MED12 | - | 1 (0.69%) | 1 (0.45%) |
| MEN1 | - | 2 (1.39%) | 2 (0.9%) |
| MLH1 | - | 2 (1.39%) | 2 (0.9%) |
| MSH2 | - | 5 (3.47%) | 5 (2.26%) |
| MSH6 | 1 (1.3%) | - | 1 (0.45%) |
| MUTYH | - | 1 (0.69%) | 1 (0.45%) |
| NBN | - | 4 (2.78%) | 4 (1.81%) |
| NCOR1 | - | 1 (0.69%) | 1 (0.45%) |
| NEK2 | 1 (1.3%) | 1 (0.69%) | 2 (0.9%) |
| NF1 | - | 2 (1.39%) | 2 (0.9%) |
| PALB2 | 1 (1.3%) | 1 (0.69%) | 2 (0.9%) |
| PALLD | - | 2 (1.39%) | 2 (0.9%) |
| PBRM1 | - | 1 (0.69%) | 1 (0.45%) |
| PIK3CA | 22 (28.57%) | 10 (6.94%) | 32 (14.48%) |
| PIK3R1 | - | 1 (0.69%) | 1 (0.45%) |
| PMS2 | - | 1 (0.69%) | 1 (0.45%) |
| PTEN | 4 (5.19%) | 3 (2.08%) | 7 (3.17%) |
| RAD51 | - | 1 (0.69%) | 1 (0.45%) |
| RAD51C | - | 1 (0.69%) | 1 (0.45%) |
| RAD51D | - | 1 (0.69%) | 1 (0.45%) |
| RET | - | 3 (2.08%) | 3 (1.36%) |
| SEPT9 | - | 2 (1.39%) | 2 (0.9%) |
| SMARCA4 | - | 2 (1.39%) | 2 (0.9%) |
| SYNE1 | - | 6 (4.17%) | 6 (2.71%) |
| TP53 | 13 (16.88%) | 16 (11.11%) | 29 (13.12%) |
| TRAF5 | - | 1 (0.69%) | 1 (0.45%) |
| WEE1 | - | 3 (2.08%) | 3 (1.36%) |
| ZBED4 | - | 2 (1.39%) | 2 (0.9%) |
Clinical and pathological association
Association of clinical and pathological parameters and dominant gene mutations from 77 Tier I/II mutation variants were analyzed by Fisher Exact test. PIK3CA mutations were common in Luminal A tumors (P=0.0294). Mutations in GATA3 (P=0.0172) were significantly associated with ER positivity. For other mutations, there were no significant association between the mutation and young onset age, patient status, staging and menopausal status.
Somatic single nucleotide variants (SNVs)
566 SNVs were detected, of which 340 of them were unique variants. The median SNV was 5.29 and ranged from 1-18 SNVs per tumor, where five patients got >10 SNVs and three patients got one SNV only. Early-onset and HER2+ tumors often acquired a G to T transversion while tumors from menopause patients yielded mostly a T to G transversion (Figure 3 and Table 4). The identified SNVs include 21 nonsense mutation variants in 15 genes. Recurrent mutations were seen in BMPR1A, MAP3K1, PALB2 and TP53 genes. For missense mutations, 322 mutations were found in 69 different genes, in which SYNE1 (n=24) and TP53 (n=24) were common. Among them, 72 of these SNVs were listed in the “Candidate Cancer Gene Database” category A potential cancer drivers [27] and 471 of the SNVs from 49 genes which were actively relevant to cancer according to “Cancer Gene Census” database [28].
Figure 3.
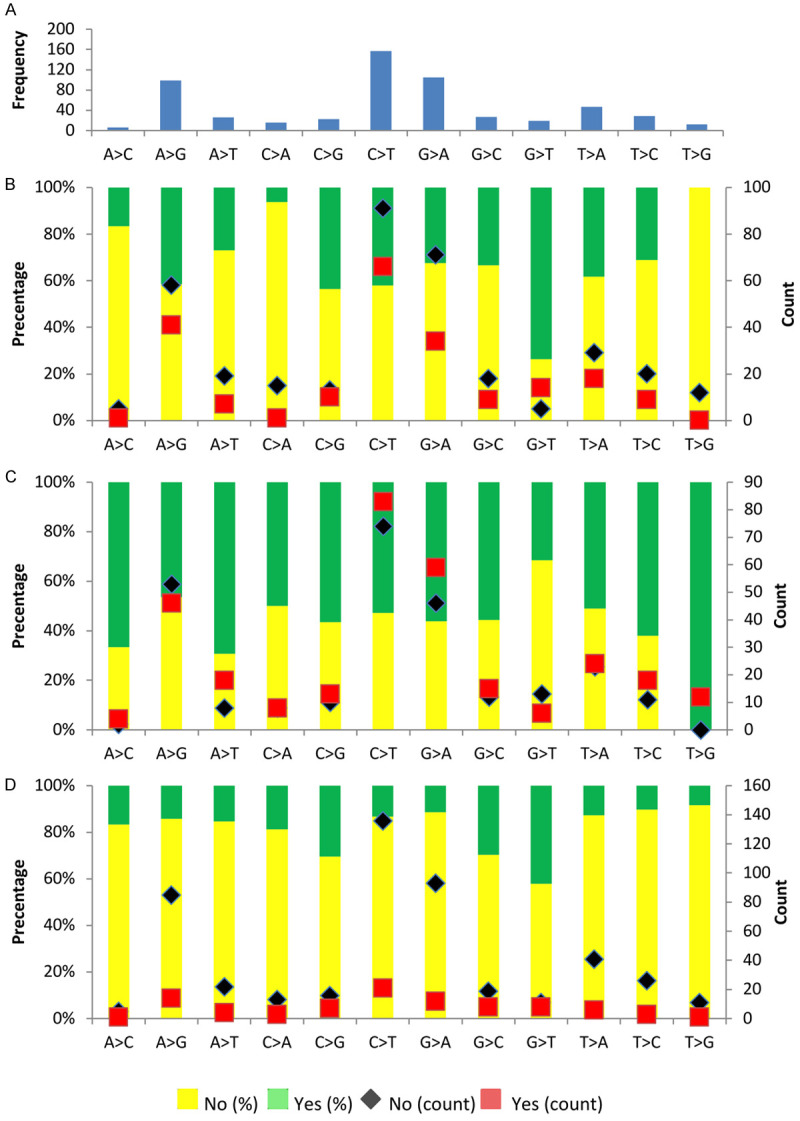
Nucleotide mutational profile. A. Frequency of nucleotide change. B. Distribution of YBR45. C. Distribution of Menopause. D. Distribution of Her2.
Table 4.
Frequency of transition and transversion mutations
| To | |||||
|---|---|---|---|---|---|
|
| |||||
| A | C | G | T | ||
| From | A | 6 (4.58%) | 99 (75.57%) | 26 (19.85%) | |
| C | 16 (8.16%) | 23 (11.73%) | 157 (80.1%) | ||
| G | 105 (83.33%) | 2 (1.59%) | 19 (15.08%) | ||
| T | 47 (53.41%) | 29 (32.95%) | 12 (13.64%) | ||
Mutation spectrum of breast tumors versus ovarian tumors
Analysis revealed 17 Tier I/II mutation variants in all 14 tumors, and each tumor carried an average of 1.21 mutations. The mutation signature of ovarian tumors was different from breast tumors (Table 5), the most frequently mutated genes were TP53 (52.9%), KRAS (23.5%), PIK3CA (11.8%), BRCA1 (5.9%) and RB1 (5.9%). Of note, Tier I/II BRCA1 somatic mutation was seen in ovarian tumor (1/14, 7.1%) but not in breast tumor. The complete list of Tier I-III variants of ovarian cancers was shown in Table S3.
Table 5.
Mutation spectrum of breast and ovarian tumors
| Breast (N=108) | Ovarian (N=14) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Tier I/II | Tier III | Total | Tier I/II | Tier III | Total | |
| APC | 1 | 1 | ||||
| AR | 4 | 4 | ||||
| ATM | 4 | 4 | 1 | 1 | ||
| ATR | 4 | 4 | 1 | 1 | ||
| BRCA1 | 2 | 2 | 1 | 1 | 2 | |
| BRCA2 | 4 | 4 | ||||
| BRIP1 | 1 | 1 | 2 | |||
| CBFB | 3 | 2 | 5 | |||
| CDH1 | 2 | 6 | 8 | |||
| CSMD1 | 1 | 5 | 6 | |||
| ERBB3 | 5 | 5 | 1 | 1 | ||
| EXOC2 | 2 | 2 | 1 | 1 | ||
| EXT2 | 1 | 1 | ||||
| GATA3 | 11 | 11 | ||||
| HERC1 | 3 | 3 | 1 | 1 | ||
| KRAS | 4 | 4 | ||||
| MAP2K4 | 2 | 1 | 3 | 1 | 1 | |
| MAP3K1 | 11 | 4 | 15 | 2 | 2 | |
| MSH2 | 5 | 5 | ||||
| NBN | 4 | 4 | ||||
| NF1 | 2 | 2 | 1 | 1 | ||
| PALB2 | 1 | 1 | 2 | 2 | 2 | |
| PIK3CA | 22 | 10 | 32 | 2 | 2 | |
| PTEN | 4 | 3 | 7 | 1 | 1 | |
| RAD51C | 1 | 1 | ||||
| RB1 | 1 | 1 | ||||
| SYNE1 | 6 | 6 | ||||
| TP53 | 13 | 16 | 29 | 9 | 2 | 11 |
Discussion
In Hong Kong, approximately 10% of the breast cancer cases are inherited with BRCA mutations [4] and these patients are offered risk reduction surgery or targeted therapy such as PARP inhibitors. However, limited study has investigated the prevalence of somatic mutation in germline BRCA mutation-negative breast and ovarian cancer patients, who may potentially benefit from other targeted therapies. Interpretation of somatic variants are likely to impact clinical managements including estimation of the sensitivity and resistance of specific drug treatments. With the increase demand of genetic information, screening of germline and somatic mutations have been incorporated into the routine clinical practice for treatment decision making in different cancer types including breast cancer [29].
Somatic PIK3CA and TP53 mutations were found in 27-40% and 23-34% of breast cancer respectively [30-32]. In a large cohort study (n=1,794), somatic TP53 mutation rate was lowered in older age groups (>59 years) [33], similar as in our cohort, TP53 mutations were seen in 40% young breast patient’s (≤ 45 years) tumor, while 18% were age >45 years. On the contrary, there was one study reported that 20% of the tumors had no association with early-onset breast cancer [31]. Patients with somatic TP53 mutations had a poor overall survival in ER-positive than in ER-negative patients [33]. Docetaxel has been suggested as a better therapeutic option than anthracyclines in treating TP53-mutated breast cancer patients than the wild-type patients.
Somatic PIK3CA mutations, another common mutated gene, was identified in 20.4% of our breast tumors. Studies showed that 25-46.5% of breast cancer patients had PI3K mutations were significantly associated with ER-positive tumors [34-39]. Similarly, mutation hotspots E542K, E545K and H1047R mutations were also observed in our cohort [38]. Furthermore, several retrospective and prospective studies had contradictory conclusions on prognostic and predictive values of PIK3CA mutations in breast cancer tumors [40-43]. PIK3CA mutations can co-exist with other PI3K-enhancing mechanisms, such as HER2 amplification. HER2-targeted therapy is suggested in PIK3CA-mutated patients [44]. However, some studies indicated that PIK3CA mutations may predict resistance to trastuzumab [45,46]. The antitumor activity of combining BKM120 and trastuzumab were promising in patients with HER2-positive advanced or metastatic breast cancer developed resistant to trastuzumab [47]. To develop PI3K inhibitors as novel therapeutics for HER2-positive advanced or metastatic breast cancer, we still need to overcome challenge of maximizing efficacy of these agents with minimum side effects.
TP53 and KRAS are the most dominated somatic mutations in our ovarian cancer cohort. TP53 mutations were seen in 90-96% of high-grade ovarian tumors [19,48]. Several on-going phase I/II clinical trials are conducted to assess the efficacy of TP53 activators (APR-246 and MK-1775-004) in treating platinum-sensitive ovarian cancer and platinum-resistant high-grade serous ovarian cancer [49,50]. Also, the use of AMG 510 in treating KRAS c.34G>T mutation, a common mutation in solid tumors, showed promising results in patients with non-small-cell lung carcinoma [51,52]. Also, a mRNA-derived KRAS-targeted vaccine, mRNA-5671, targeting both KRAS c.35G>A and c.34G>T mutations is undergoing Phase I trial [51]. Herein, somatic mutation screening is likely beneficial for ovarian patients without germline mutations to provide better treatment strategies in the future.
Several clinical trials of olaparib revealed promising results on high-grade serous ovarian cancer patients with germline or somatic BRCA mutation, the median PFS was improved in olaparib group than placebo (11.2 months vs 4.3 months) [53]. Besides, clinical trials of other PARP inhibitors (rucaparib and niraparib) have extended the study to platinum-based therapy in high-grade ovarian cancer patient regardless of BRCA status and yield positive results. On the other hand, the prevalence of somatic BRCA mutations in sporadic breast cancer was around 3.5% [54], yet very little information in hereditary breast cancer is available. In this testing cohort, no somatic BRCA mutation was identified, hence, we believed that BRCA-negative breast cancer patients are unlikely to carry somatic BRCA mutation. In light of a relatively low reported frequency in hereditary breast cancer, somatic BRCA mutation testing for non-BRCA breast cancer patients are of limited value, unlike as in ovarian cancer.
In conclusion, characterization of somatic mutations in breast and ovarian tumors could provide insights into tumorigenesis and reveal candidates for targeted therapeutics. We found that somatic PIK3CA and TP53 mutations were common events in germline mutation-negative breast cancer patients and had distinct spectrum than in ovarian cancer. Results from this study exemplify the necessity of somatic testing in breast and ovarian cancer patients, besides germline mutation screening, and to guide future research directions on other targeted therapies.
Acknowledgements
This study was supported by Asian Fund for Cancer Research, Health and Medical Research Fund (03143406), Seed Fund for Basic Research (201611159186), Dr Ellen Li Charitable Foundation, Kerry Kuok Foundation and Hong Kong Hereditary Breast Cancer Family Registry. We thank Wing Pan Luk and Ling Hiu Fung for assisting in statistical analysis.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Sharma P, Klemp JR, Kimler BF, Mahnken JD, Geier LJ, Khan QJ, Elia M, Connor CS, McGinness MK, Mammen JM, Wagner JL, Ward C, Ranallo L, Knight CJ, Stecklein SR, Jensen RA, Fabian CJ, Godwin AK. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat. 2014;145:707–714. doi: 10.1007/s10549-014-2980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 4.Kwong A, Shin VY, Au CH, Law FB, Ho DN, Ip BK, Wong AT, Lau SS, To RM, Choy G, Ford JM, Ma ES, Chan TL. Detection of germline mutation in hereditary breast and/or ovarian cancers by next-generation sequencing on a four-gene panel. J Mol Diagn. 2016;18:580–594. doi: 10.1016/j.jmoldx.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faraoni I, Graziani G. Role of BRCA mutations in cancer treatment with Poly(ADP-ribose) Polymerase (PARP) inhibitors. Cancers (Basel) 2018;10:487. doi: 10.3390/cancers10120487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moschetta M, George A, Kaye SB, Banerjee S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol. 2016;27:1449–1455. doi: 10.1093/annonc/mdw142. [DOI] [PubMed] [Google Scholar]
- 8.Exman P, Barroso-Sousa R, Tolaney SM. Evidence to date: talazoparib in the treatment of breast cancer. Onco Targets Ther. 2019;12:5177–5187. doi: 10.2147/OTT.S184971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 10.Pascual T, Gonzalez-Farre B, Teixidó C, Oleaga L, Oses G, Ganau S, Chic N, Riu G, Adamo B, Galván P, Vidal M, Soy D, Urbano Á, Muñoz M, Prat A. Significant clinical activity of olaparib in a somatic BRCA1-mutated triple-negative breast cancer with brain metastasis. JCO Precis. 2019:1–6. doi: 10.1200/PO.19.00012. [DOI] [PubMed] [Google Scholar]
- 11.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M, Carmichael J. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 12.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B, Yamashita T, Lu YS, Inoue K, Takahashi M, Papai Z, Longin AS, Mills D, Wilke C, Hirawat S, Juric D SOLAR-1 Study Group. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 13.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, Tsui DW, Liu B, Dawson SJ, Abraham J, Northen H, Peden JF, Mukherjee A, Turashvili G, Green AR, McKinney S, Oloumi A, Shah S, Rosenfeld N, Murphy L, Bentley DR, Ellis IO, Purushotham A, Pinder SE, Borresen-Dale AL, Earl HM, Pharoah PD, Ross MT, Aparicio S, Caldas C. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, Ju YS, Smid M, Brinkman AB, Morganella S, Aure MR, Lingjaerde OC, Langerod A, Ringner M, Ahn SM, Boyault S, Brock JE, Broeks A, Butler A, Desmedt C, Dirix L, Dronov S, Fatima A, Foekens JA, Gerstung M, Hooijer GK, Jang SJ, Jones DR, Kim HY, King TA, Krishnamurthy S, Lee HJ, Lee JY, Li Y, McLaren S, Menzies A, Mustonen V, O’Meara S, Pauporte I, Pivot X, Purdie CA, Raine K, Ramakrishnan K, Rodriguez-Gonzalez FG, Romieu G, Sieuwerts AM, Simpson PT, Shepherd R, Stebbings L, Stefansson OA, Teague J, Tommasi S, Treilleux I, Van den Eynden GG, Vermeulen P, Vincent-Salomon A, Yates L, Caldas C, van’t Veer L, Tutt A, Knappskog S, Tan BK, Jonkers J, Borg A, Ueno NT, Sotiriou C, Viari A, Futreal PA, Campbell PJ, Span PN, Van Laere S, Lakhani SR, Eyfjord JE, Thompson AM, Birney E, Stunnenberg HG, van de Vijver MJ, Martens JW, Borresen-Dale AL, Richardson AL, Kong G, Thomas G, Stratton MR. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, Lanchbury JS, Stemke-Hale K, Hennessy BT, Arun BK, Hortobagyi GN, Do KA, Mills GB, Meric-Bernstam F. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–1089. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lancaster JM, Wooster R, Mangion J, Phelan CM, Cochran C, Gumbs C, Seal S, Barfoot R, Collins N, Bignell G, Patel S, Hamoudi R, Larsson C, Wiseman RW, Berchuck A, Iglehart JD, Marks JR, Ashworth A, Stratton MR, Futreal PA. BRCA2 mutations in primary breast and ovarian cancers. Nat Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]
- 18.Rhiem K, Todt U, Wappenschmidt B, Klein A, Wardelmann E, Schmutzler RK. Sporadic breast carcinomas with somatic BRCA1 gene deletions share genotype/phenotype features with familial breast carcinomas. Anticancer Res. 2010;30:3445–3449. [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Bonazzoli E, Bellone S, Choi J, Dong W, Menderes G, Altwerger G, Han C, Manzano A, Bianchi A, Pettinella F, Manara P, Lopez S, Yadav G, Riccio F, Zammataro L, Zeybek B, Yang-Hartwich Y, Buza N, Hui P, Wong S, Ravaggi A, Bignotti E, Romani C, Todeschini P, Zanotti L, Zizioli V, Odicino F, Pecorelli S, Ardighieri L, Silasi DA, Litkouhi B, Ratner E, Azodi M, Huang GS, Schwartz PE, Lifton RP, Schlessinger J, Santin AD. Mutational landscape of primary, metastatic, and recurrent ovarian cancer reveals c-MYC gains as potential target for BET inhibitors. Proc Natl Acad Sci U S A. 2019;116:619–624. doi: 10.1073/pnas.1814027116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Shao D, Li L, Wu M, Ma S, Tan X, Zhong S, Guo F, Wang Z, Ye M. Germline and somatic mutations of multi-gene panel in Chinese patients with epithelial ovarian cancer: a prospective cohort study. J Ovarian Res. 2019;12:80. doi: 10.1186/s13048-019-0560-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. 2013;1303:1–3. [Google Scholar]
- 23.Au CH, Ho DN, Kwong A, Chan TL, Ma ESK. BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing. Sci Rep. 2017;7:1567. doi: 10.1038/s41598-017-01703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv e-prints. 2012 [Google Scholar]
- 25.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, Nikiforova MN. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, american society of clinical oncology, and college of american pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbott KL, Nyre ET, Abrahante J, Ho YY, Isaksson Vogel R, Starr TK. The candidate cancer gene database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015;43:D844–848. doi: 10.1093/nar/gku770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, Lipkin SM, Syngal S, Wollins DS, Lindor NM. American society of clinical oncology policy statement update: genetic and genomic testing for cancer susceptibility. J. Clin. Oncol. 2015;33:3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 30.Behring M, Vazquez AI, Cui X, Irvin MR, Ojesina AI, Agarwal S, Manne U, Shrestha S. Gain of function in somatic TP53 mutations is associated with immune-rich breast tumors and changes in tumor-associated macrophages. Mol Genet Genomic Med. 2019;7:e1001. doi: 10.1002/mgg3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Encinas G, Maistro S, Pasini FS, Katayama ML, Brentani MM, Bock GH, Folgueira MA. Somatic mutations in breast and serous ovarian cancer young patients: a systematic review and meta-analysis. Rev Assoc Med Bras. 2015;61:474–483. doi: 10.1590/1806-9282.61.05.474. [DOI] [PubMed] [Google Scholar]
- 32.Li V, Li K, Li JT. TP53 mutations as potential prognostic markers for specific cancers: analysis of data from the cancer genome atlas and the international agency for research on cancer TP53 database. J Cancer Res Clin Oncol. 2019;145:625–636. doi: 10.1007/s00432-018-2817-z. [DOI] [PubMed] [Google Scholar]
- 33.Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Borresen-Dale AL. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 34.Li G, Guo X, Chen M, Tang L, Jiang H, Day JX, Xie Y, Peng L, Xu X, Li J, Wang S, Xiao Z, Dai L, Wang J. Prevalence and spectrum of AKT1, PIK3CA, PTEN and TP53 somatic mutations in Chinese breast cancer patients. PLoS One. 2018;13:e0203495. doi: 10.1371/journal.pone.0203495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng L, Zhu X, Sun Y, Wang J, Zhong X, Li J, Hu M, Zheng H. Prevalence and prognostic role of PIK3CA/AKT1 mutations in chinese breast cancer patients. Cancer Res Treat. 2019;51:128–140. doi: 10.4143/crt.2017.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, Trink B, Sidransky D. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7:R609. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 40.Lai YL, Mau BL, Cheng WH, Chen HM, Chiu HH, Tzen CY. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15:1064–1069. doi: 10.1245/s10434-007-9751-7. [DOI] [PubMed] [Google Scholar]
- 41.Lerma E, Catasus L, Gallardo A, Peiro G, Alonso C, Aranda I, Barnadas A, Prat J. Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch. 2008;453:133–139. doi: 10.1007/s00428-008-0643-4. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjold B, Rutqvist LE, Skoog L, Stal O. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13:3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 43.Maruyama N, Miyoshi Y, Taguchi T, Tamaki Y, Monden M, Noguchi S. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13:408–414. doi: 10.1158/1078-0432.CCR-06-0267. [DOI] [PubMed] [Google Scholar]
- 44.Loibl S, Minckwitz Gv, Schneeweiss A, Paepke S, Lehmann A, Rezai M, Zahm DM, Sinn P, Khandan F, Eidtmann H, Dohnal K, Heinrichs C, Huober J, Pfitzner B, Fasching PA, Andre F, Lindner JL, Sotiriou C, Dykgers A, Guo S, Gade S, Nekljudova V, Loi S, Untch M, Denkert C. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. Int J Clin Oncol. 2014;32:3212–3220. doi: 10.1200/JCO.2014.55.7876. [DOI] [PubMed] [Google Scholar]
- 45.Rimawi MF, De Angelis C, Contreras A, Pareja F, Geyer FC, Burke KA, Herrera S, Wang T, Mayer IA, Forero A, Nanda R, Goetz MP, Chang JC, Krop IE, Wolff AC, Pavlick AC, Fuqua SAW, Gutierrez C, Hilsenbeck SG, Li MM, Weigelt B, Reis-Filho JS, Kent Osborne C, Schiff R. Low PTEN levels and PIK3CA mutations predict resistance to neoadjuvant lapatinib and trastuzumab without chemotherapy in patients with HER2 over-expressing breast cancer. Breast Cancer Res Treat. 2018;167:731–740. doi: 10.1007/s10549-017-4533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Black JD, Lopez S, Cocco E, Bellone S, Altwerger G, Schwab CL, English DP, Bonazzoli E, Predolini F, Ferrari F, Ratner E, Silasi DA, Azodi M, Schwartz PE, Santin AD. PIK3CA oncogenic mutations represent a major mechanism of resistance to trastuzumab in HER2/neu overexpressing uterine serous carcinomas. Br J Cancer. 2015;113:1020–1026. doi: 10.1038/bjc.2015.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saura C, Bendell J, Jerusalem G, Su S, Ru Q, De Buck S, Mills D, Ruquet S, Bosch A, Urruticoechea A, Beck JT, Di Tomaso E, Sternberg DW, Massacesi C, Hirawat S, Dirix L, Baselga J. Phase Ib study of Buparlisib plus Trastuzumab in patients with HER2-positive advanced or metastatic breast cancer that has progressed on trastuzumab-based therapy. Clin Cancer Res. 2014;20:1935–1945. doi: 10.1158/1078-0432.CCR-13-1070. [DOI] [PubMed] [Google Scholar]
- 48.Sugino K, Tamura R, Nakaoka H, Yachida N, Yamaguchi M, Mori Y, Yamawaki K, Suda K, Ishiguro T, Adachi S, Isobe M, Yamaguchi M, Kashima K, Motoyama T, Inoue I, Yoshihara K, Enomoto T. Germline and somatic mutations of homologous recombination-associated genes in Japanese ovarian cancer patients. Sci Rep. 2019;9:17808. doi: 10.1038/s41598-019-54116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.p53 Activation in Platinum-Resistant High Grade Serous Ovarian Cancer, a Study of PLD With APR-246. https://clinicaltrials.gov/ct2/show/ NCT03268382.
- 50.A study of MK-1775 in combination with paclitaxel and carboplatin versus paclitaxel and carboplatin alone for participants with platinum-sensitive ovarian tumors with the P53 gene mutation (MK-1775-004) https://clinicaltrials.gov/ct2/show/ NCT01357161.
- 51.Mullard A. Cracking KRAS. Nat Rev Drug Discov. 2019;18:887–891. doi: 10.1038/d41573-019-00195-5. [DOI] [PubMed] [Google Scholar]
- 52.Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B. 2019;9:871–879. doi: 10.1016/j.apsb.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W, Shapira-Frommer R, Safra T, Matei D, Fielding A, Spencer S, Dougherty B, Orr M, Hodgson D, Barrett JC, Matulonis U. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 54.Lang GT, Shi JX, Hu X, Zhang CH, Shan L, Song CG, Zhuang ZG, Cao AY, Ling H, Yu KD, Li S, Sun MH, Zhou XY, Huang W, Shao ZM. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int J Cancer. 2017;141:129–142. doi: 10.1002/ijc.30692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


