Abstract
Reliably producing a competent oocyte entails a deeper comprehension of ovarian follicle maturation, a very complex process that includes meiotic maturation of the female gamete, the oocyte, together with the mitotic divisions of the hormone-producing somatic cells. In this report, we investigate murine ovarian folliculogenesis in vivo using publicly available time-series microarrays from primordial to antral stage follicles. Manually curated protein interaction networks were employed to identify autocrine and paracrine signaling between the oocyte and the somatic cells (granulosa and theca cells) at multiple stages of follicle development. We established plausible protein-binding interactions between expressed genes that encode secreted factors and expressed genes that encode cellular receptors. Some computationally identified signaling interactions are well established, such as the paracrine signaling from the oocyte to the somatic cells through the oocyte-secreted growth factor Gdf9, while others are novel connections in term of ovarian folliculogenesis, such as the possible paracrine connection from somatic-secreted factor Ntn3 to the oocyte receptor Neo1. Additionally, we identified several of the likely transcription factors that might control the dynamic transcriptome during ovarian follicle development, noting that the YAP/TAZ signaling pathway is very active in vivo. This novel dynamic model of signaling and regulation can be employed to generate testable hypotheses regarding follicle development that could be validated experimentally, guiding the improvement of culture media to enhance in vitro ovarian follicle maturation and possibly novel therapeutic targets for reproductive diseases.
Electronic supplementary material
The online version of this article (10.1007/s43032-019-00075-8) contains supplementary material, which is available to authorized users.
Keyword: Dynamic signaling, Dynamic regulation ovarian follicle development, Inter-cellular communication
Introduction
The production of a competent female germ cell, the oocyte, that can undergo fertilization requires highly orchestrated paracrine, autocrine, endocrine, and juxtacrine signaling that occurs between the oocyte and the supporting somatic cells, granulosa, and theca cells. This complex biological structure formed by the oocyte and surrounding somatic cells is called the ovarian follicle. During ovarian follicle maturation, a primordial follicle (50 μm diameter in the mouse) that is composed from a handful of cells grows into an antral follicle (500 μm), in order to attain a competent oocyte. Attempts to separate the different cellular components of the follicle to study follicle maturation lead to different behavior of the individual cell types. Unfortunately, little can be controlled in vivo to learn the effects of different biological variables (e.g., effects of hormones, extracellular matrix stiffness) on the follicle maturation. Thus, several in vitro systems that mimic in vivo ovarian follicle development [1–3] have led to some of the most significant advances in reproductive biology [4]. Up to date, only organ-on-a-chip technology that combines ovarian tissue, fallopian tubes, and uterus [5]—EVATAR™—has been able to mimic the menstrual cycle including ovulation. While with EVATAR™, the effects of hormones or the extracellular matrix stiffness could be systematically studied, and it suffers from limitations similar to in vivo models: lacking capabilities to understand intercellular and intracellular communications between the different ovarian follicle cell types. One of the main hindrances is difficulty determining how paracrine (e.g., between the oocyte and granulosa cells) and autocrine (e.g., oocyte ligands that affect the oocyte) communication between the different follicle cell types occurs. While some of the intercellular ligands such as GDF-9 and BMP-15 are well-established [6], many have yet to be characterized. Similarly, once a given ligand binds to its corresponding receptors, a complex signaling cascade is initiated through several biochemical mechanisms (e.g., phosphorylation, protein binding, calcium release) that ends in the activation or deactivation of transcriptional programs (i.e., intracellular communication). Transcription factors (TFs) are the mediators of cytoplasmic-to-nucleus signaling via translocation between the two cellular compartments. Once a TF translocates to the nucleus, directly or in the form of a protein complex, it binds DNA and initiates or inhibits transcription. Therefore, TFs are potent regulators of the cellular phenotype. For instance, in the ovarian follicle, the transcription factor FOXL2 is a marker of granulosa cells and essential for proper ovarian follicle development [7].
In the recent years, advances in high-throughput techniques have allowed for collection of large amounts of information about the ovarian follicle transcriptome [8–12]. Analysis of these large biological datasets requires statistical and computational methods to identify the processes that are associated with the manifest phenotypes. Yet these transcriptional data have not been explored to their maximum potential. Currently, there are methods to computationally identify the more plausible TFs that are regulating a given phenotype [13, 14]. Similarly, given a set of genes that are expressed in a given cell, the most likely genes that encode for ligands and receptors that present in a cell type could be identified, using well-curated biological databases (e.g., DIP [15], MetaCore).
The main aim of this manuscript is the creation of datasets that can drive generation of new hypotheses regarding the autocrine and paracrine ligand requirements for in vitro grown ovarian follicles and that can be tested and validated experimentally. To derive these hypotheses, we devised a systems biology approach to computationally identify the key intra- and intercellular dynamic processes during murine ovarian folliculogenesis in vivo between and among the different follicular cell types (e.g., oocyte, granulosa cells, cumulus, mural cells) involved in each developmental stage (e.g., primordial to primary ovarian follicles). Specifically, we initially identified ligands, receptors, and transcriptional regulators involved in ovarian follicle development, and subsequently characterized the dynamic associations between these proteins, which were classified based on the cell type within the follicle and also the timing throughout ovarian follicle development.
Methods
Inter-Cellular Networks
Murine genes that encode for secreted proteins and receptors were identified using the GeneGO database (Advance Search 2.0). Protein-protein interactions between secreted proteins and receptors were obtained from manually curated databases, GENEGO and DIP [15]. Autocrine and paracrine connections were deemed possible if either one of the members of the interaction, either the ligand or the receptor, had a statistically significant change for the corresponding transition under consideration—see Table S3 from Peñalver Bernabé and colleagues for more details [16]—and that the corresponding receptor or ligand was at least present in the microarray for the very same transition. The specificity of each gene that encodes for a ligand or a receptor was previously identified—i.e., a gene is only transcribed by the oocyte, by the mural cells [16]. All the intercellular graphs were plotted with Cytoscape [17].
Most Likely Transcription Factors
Computationally predicted targets of TFs were detected by exploring whether the TF position weighted matrices (PWMs) could bind to the consensus mammalian promoter regions of a given gene [18] between −2000 to 2000 base pairs with respect to the transcription starting site (TSS) of the given gene. We used two different TF-binding site search programs, FIMO [13] and BEEML [14], to establish the targets of a TF. Agreement between the results of FIMO and BEEML, using a cutoff of p value ≤ 10–4 and E-score ≥ 0.3, respectively, was deemed as an indication that a given TF could bind to the promoter region of a gene. The list of explored TFs using FIMO and BEEML was obtained from a combination of different sources: (i) TFs that have experimentally identified position weighted matrices (PWMs) in vertebrates that were reported in TRANSFAC[19, 20]; (ii) the nonoverlapping PWMs that were in the CIS-BP database [21] but not in the TRANSFAC, including the non-inferred PWMs in the CIS-BP dataset—a total 3216 PWMs from 1164 different TFs. We additionally added connections between TFs and target genes that were in the Ovarian Kaleidoscope database [22], and from conserved motifs in mammals [23]. We established whether a TF was active at a given stage by determining the significance of the ratio between genes that were significantly changing for a given cell type and stage compared with nonsignificant genes using a hypergeometric distribution [24]. A total of 500 bootstrapping samples with the same number of the nonsignificant genes for a given cell type and stage than the number of significant genes were selected. Medians for p values were reported after been corrected for multiple comparisons using false discovery rate method [25].
Results
Identification of the Ligands and Receptors that Drive Inter-Cellular Signaling During Ovarian Follicle Development
Understanding intercellular communications during ovarian follicle maturation in vivo entails the identification of the secreted proteins (i.e., ligands) and available receptors in the oocyte and in the somatic cells (e.g., granulosa and theca cells) that support the oocyte’s growth and maturation. We established the most likely ligands and receptors during ovarian follicle development by characterizing the set of statistically significant genes that encode for ligands and receptors during ovarian follicle maturation. We mined several publicly available time-series transcriptomics—i.e., oocytes [12], somatic cells including granulosa and theca cells [16], cumulus and mural cells collected during antrum formation [11], and cumulus cells during the oocyte competence acquisition [26]. Combination of all these data sets led to a list of the significant transcribed ligands and receptors in each individual cell type (e.g., oocyte and each somatic cell type).
We identified 100 genes that encode for ligands and 95 genes that encode for receptors that could potentially regulate intercellular communication during ovarian follicle development (File S1). [Note for easier reading, we will substitute “genes that encode for ligands or receptors” to just ligands or receptors]. Some of the ligands and receptors were transcribed in multiple cell types, e.g., Dnc in the oocyte and somatic cells and Efna2 in mural and cumulus granulosa cells; yet others were very specific (e.g., Wnt10a in cumulus cells). More than half of the ligands, a total of 59, were cell-specific: 12 to the oocyte, 19 to somatic cells, 8 to cumulus granulosa cells, 10 to mural granulosa cells, and 10 to cumulus granulosa cells during the oocyte transition from a chromatin non-surrounded nucleolus (NSN) to a surrounded chromatic nucleolus (SN). More than 60% of the receptors were specific (i.e., 12, 24, 3, 12, and 7 in the oocyte and somatic, cumulus and mural granulosa cells and in cumulus cells during the oocyte transition from NSN to SN, respectively). In terms of the number of stages that a given ligand or a receptor was actively transcribed during the transition, some intercellular signaling genes were more ubiquitous, as they were transcribed in multiple ovarian follicle stages (e.g., Apoa4 from secondary to large antral follicles, Igf1 from primary to large antral follicles), while others were very specific, such as Bmp15 and Wnt6 that were only actively transcribed during the primordial to primary transition or in the primary to secondary transition, respectively. Taken together, a combination of transcriptomics data and manually curated database analytics revealed that more than 100 ligands and receptors are likely involved in the intercellular communications. More than half of these factors were cell-specific and only a few of them were stage-specific.
Constructions of Inter-Cellular Signaling Networks During Ovarian Follicle Maturation
Combination of multiple datasets and manually curated databases led to the identification for a total of 1663 connections between the 100 ligands and 95 receptors (File S1). Interactions between present ligands and receptors, i.e., those whose transcriptional levels were above the array background, were deemed plausible if an experimentally validated interaction between them was previously reported and recorded in any of the selected manually curated databases, i.e., Metacore and DIP [15]. Out of those interactions, 46% of the connections were autocrine signaling, mostly between somatic cells (45%)—note that these somatic autocrine connections could be within the same somatic cell or between two different somatic cells within the same follicle or even between two different follicles. In terms of possible paracrine signaling relationships, more than 22% were initiated from a ligand produced by the oocyte. Interestingly, 290 of the 1663 connections that we identified were between specific genes that encode for ligands and receptors (i.e., only significant in one cell type) and 40% of them were autocrine signaling, mostly within somatic cells (65 somatic ligand to somatic receptor). Independent of the cellular origin of the ligand-receptor pair, we observed that several ligands could have a widespread effect and target multiple receptors that were present in multiple cell types and during distinct stages of ovarian follicle development. For instance, Vegfa has multiple possible receptors (e.g., App, Ftl1, integrins) that like Vegfa are ubiquitous, as they are transcribed by multiple cell types (e.g., granulosa, cumulus and mural cells) and at different stages during ovarian follicle development (e.g., primordial and secondary follicles). Based on this myriad of possibilities, Vegfa could bind to its receptors in 37 different manners. More prominently, Thbs1 presented 55 possible distinct connections, most of them during the small to large antral transition. Yet, some ligands were specific, as they only bind 1 or 2 receptors, e.g., Shh or Rspo2 (Fig. 1, File S1). These results underscore the complex and intricate intercellular communication during ovarian follicle development and provide a roadmap for testing multifactorial outcomes that in the past could only be addressed one-by-one.
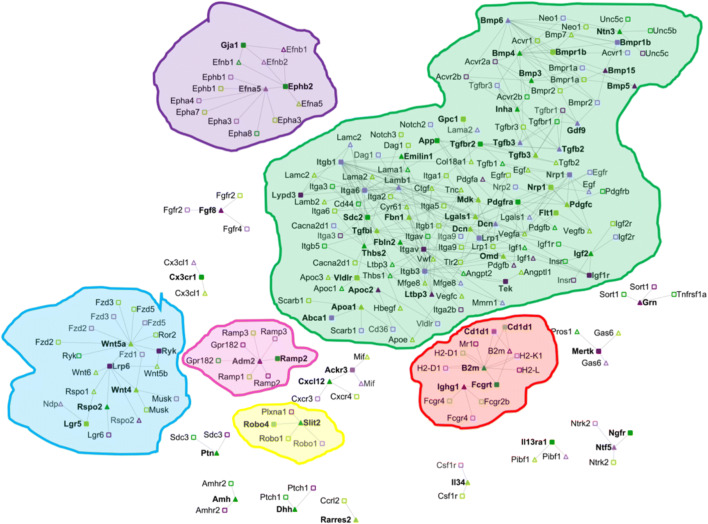
Fig. 1.
Intercellular networks between oocytes and somatic cells primordial to primary transition. Receptors only identified as significant (FC ≥ 2 and p value ≤ 0.01) in the somatic cells are represented as darker green squares (e.g., App, Tgfbr2); ligands only identified as significant (FC ≥ 2 and p value ≤ 0.01) in the somatic cells are represented as darker green triangles (e.g., Amh, Thbs2); Receptors only identified as significant (FC ≥ 2.5 and p value ≤ 0.01) in the oocyte are represented as darker purple squares (e.g., Itga9, Bmpr1b); ligands only identified as significant (FC ≥ 2.5 and p value ≤ 0.01) in the oocyte are represented as darker purple triangles (e.g., Tac1, Bmp5). Lighter colors indicate that the receptor or ligand are also significant in other cell types during ovarian follicle development (e.g., Nrp1, Tgfb3, Bmpr1a, Gdf9). Bold text corresponds to genes whose abundance change during the specific transition (e.g., primordial to primary), such as Gdf9, Neo1; otherwise, those genes are presented in the follicles during the given transition yet their abundance is not changing (e.g., Egfr, Sdc3) are not bolded. Connections between the different ligand receptors are only present if at least one of member of the pair is changing its expression during a given developmental stage. The connections and the references for each of the edges in the network can be found in File S1
Inter-Cellular Signaling Networks In Vivo During the Primordial to Primary Transition
The primordial to primary transition was the most complex of all the stages during ovarian follicle development. The majority of inter-cellular communications identified, a total of 1663, occurred during the transition from primordial to primary ovarian follicles (22% of the connections). Multiple hallmarks during this transition were present in the transcriptomics data (e.g., zona pellucida formation and gap connections, Table S1), and multiple genes known to be involved in the development of primary ovarian follicles from primordial ovarian follicles were identified in the transcriptional data (e.g., Zp1, Gja4, Amhr, Supplemental Note1).
The transcriptional activity and the number of paracrine and autocrine communications between somatic cells surpassed those of the oocyte (Table S1). Out of all the autocrine and paracrine communications, only a few of them were cell-specific, i.e., between ligands and receptors predicted as uniquely present in one cell type (File S1). Only 11 oocyte autocrine, 15 somatic autocrine, 12 oocyte-somatic, and 28 somatic-oocyte paracrine communications were specific. Our results recovered well-known ligands in this stage, such as Gdf9, Bmp15, and Amh, as well as ligands that have not been previously reported in the literature and have not been characterized during the primordial to primary ovarian follicle transition, such as Adam2 and Ntn3 (Fig. 1).
The intricacy and complexity of the primordial to primary transition are clearly depicted in the inter-cellular networks, which were divided into several subnetworks. The largest subnetwork (shown in green in Fig. 1) included well-known ligands and receptors from the Tgf family (e.g., Gdf9, Bmp4, Inha) and from the Bmp family and also contained a substantial core of diverse extracellular-binding protein families, such as integrins (e.g., Itga6, Itgb1), laminin (e.g., Lama1, Lamab1), and collagen (e.g., Col18a1). Only the integrins and laminins transcribed in the oocyte (i.e., Itga5, Itgb1, Lamb1) significantly changed in this transition (p value < 0.01, File S1). Some ligands, such as Dcn—a proteoglycan identified in the later stages of follicle development [27]—and Nrp1 were produced by both the oocyte and the somatic cells. Additionally, this green subnetwork contains connections not previously studied in relation to ovarian follicle development, such as Ntn3 and its corresponding receptors, e.g., Unc5b, Unc5c, and Neo1. According to our computational model, Ntn3 is a somatically produced ligand that interacted with receptors in the oocyte and the somatic cells. Ntn3 functions have been described in other developmental processes, such as axonal growth [28, 29]. Similarly, its receptors Unc5b and Unc5 are known to participate in angiogenesis [30, 31] and are anti-apoptotic [32], and Neo1 is related to cellular growth [33].
Other medium size networks (e.g., purple subnetwork) encoded the Ephrin and Wnt families. Oocyte-secreted Efna5 ligand interacted with Epha1 and Gji somatic receptors, which were among the top genes whose transcriptional abundance changed the most (File S1). Mice who lack Efna5 ligand are subfertile [34]. This purple subnetwork also encompassed paracrine communication from somatic ligands Wnt4 and Wnt5 to oocyte-specific receptors Lrp6 and Ryk. Wnt4 signaling regulates the expression of Amh, and mice that lack Wnt4 suffer from premature ovarian failure [35]. Rspo2, a somatic-specific ligand also binds to the oocyte-specific receptor Lrp6. The red subnetwork contained a somatic autocrine communication between B2m and Fcgrt, the latter also being involved in a specific oocyte-somatic communication via the oocyte ligand Ighg1. B2m and Igh1 have been previously studied in relation to ovarian cancer biology [36, 37], but this research has not been extended to ovarian follicle development.
Finally, two other smaller and disconnected networks were related with the Ramp and the Robo families of receptors. Ramp2 was identified as a somatic-specific receptor that could bind to the oocyte-specific ligand Adm2 from the pink subnetwork. Adm2 prevents oocyte atresia by regulating cell-cell interactions in the cumulus-oocyte complex [38]. The somatic-specific receptor Silt2 could bind to the nonspecific somatic ligand Robo4 from the yellow subnetwork. This interaction between Silt2 and Robo is known to occur at the time of the formation of the ovarian follicle and diminished the rate of the number of proliferating oocytes [39]. Finally, neurotrophic soluble growth factors were also identified as important during the primordial to primary transition, corroborating previous studies [40] on their involvement in the transition of squamous somatic cells to cuboidal [41] through the oocyte-somatic specific paracrine communication between Ntf5 and Ngfr.
In conclusion, the most difficult transition to reproduce in vitro, from primordial to primary follicle, was indeed the most intricate and complex transition compared with the subsequent transitions (e.g., primary to secondary). The number of somatically driven intercellular communications during ovarian follicle development surpassed those that were oocyte driven. Interestingly, intercellular communications during primordial to primary transition were highly dependent on ovarian follicle extracellular matrix composition (e.g., integrins, collagens), which suggests a role for mechano-transduction and cell-matrix based signaling.
Inter-Cellular Signaling Networks In Vivo during the Primary to Secondary Transition
During the transition from primary to secondary stages, the follicle acquires up to 10 layers of granulosa cells [42], the formation of the theca layer commences and the follicle acquires hormone-producing capabilities. Several of the transcripts involved in those developmental pathways could be recapitulated from the recompiled ensemble transcriptional data, such as Cyp17a1 (Supplemental Note2). Numerous transcripts were changing during the primary to secondary transition, most likely due to the addition of the theca cells, yet the complexity of the inter-cellular signaling network was reduced compared with the primordial to primary transition (Table S1, Fig. S1). The majority of the intercellular communications during the primary to secondary transition were autocrine communications between somatic cells, followed by paracrine signaling between ligands secreted by the somatic cells to receptors in the oocyte. Only 21 of the intercellular communications were specific, i.e., there were between ligands or receptors only expressed in the oocyte or somatic cells. For instance, Angpt2 was produced by the somatic cells and interacted with integrins Itga5 and Itgb5 that were present only in somatic cells and with an oocyte specific receptor, Tek. Angpt2, Igf2, Pros1, and Thbs2 were the only cell-specific ligands, and H2-D1, H2-L, Tgfbr2, and Sdc3 is the only transcripts encoding for cell-specific receptors that were significantly altered during the primary to secondary transition (File S1).
In terms of subnetworks, communities highlighted in green, pink, and blue during the primordial to primary transition (Fig. 1) were diminished in terms of the number of connections between the transcripts and the yellow and purple communities were not present at all (Fig. S1). Similarly, the somatic-oocyte paracrine and somatic autocrine communications of Amh were not identified either. Interestingly, a somatic-specific subnetwork of the Edn family appeared during the primary to the secondary transition, in agreement with prior studies on the role of endothelin in ovarian follicle development [43]. Thus, based on our computational predictions, intercellular communications between the oocyte and the somatic cells during the primary to secondary transition were still actively transcribed and several of them aligned with prior research. Importantly, the number of connections during this transition was reduced by more than 60% compared with the primordial to primary transition and were driven by somatic cells.
Inter-Cellular Signaling Networks In Vivo during the Secondary to Small Antral Transition
Only a few secondary follicles sensitive to hormones FSH and LH transition into small antral follicles, avoiding atresia, the default pathway [44]. Follicles start producing androgens in the theca cells and estrogens in the granulosa cells [45], the antral cavity emerges, which is filled with hyaluronic acid and proteoglycans [46, 47] such as versican and perlican [48], and the theca layer becomes vascularized [49]. Multiple genes that are known to play a role during this transition were significantly changing at the transcriptional level, e.g., Fshr, Vcan (File S2, Supplementary Note3), although the number of downregulated genes exceeded the number of upregulated transcripts (Table S1). The complexity of the intercellular signaling network during the secondary to small antral transition was similar to the primary to secondary transitions, and therefore had fewer intercellular connections than the primordial to primary transition (Fig. S2, Table S1, File S1). The majority of intercellular communications were somatic autocrine interactions, and only 12 of them were cell specific. Out of the significantly changing ligands, Inha was the only one whose transcriptional abundance increased in this stage; Rspo2 and Wnt9 were significantly downregulated (File S2).
There were distinct changes during the secondary to small antral transition compared with the two prior transitions. For instance, the major blue subnetwork during the primordial to primary transition (Fig. 1) was divided into two smaller subnetworks, one of them highly enriched in members of the Tgfb family (Fig. S2). While the Ephrin family networks appeared again at this stage, the Edn subnetwork, important in the primary to secondary transition, disappeared. Taken together, the large phenotypical changes during the secondary to small antral transition were associated with a downregulated transcriptional transition and similar number of autocrine and paracrine inter-cellular communications. Most of the important subnetworks at this stage, such as Ephrin and Wnt pathways, promote cellular growth and align well with the expansion in the number of granulosa cells as the secondary follicle transition into the antral stage.
Inter-Cellular Signaling Networks In Vivo During the Small to Large Antral Transition
At the end of this transition, the oocyte is competent to resume meiosis [50], and the large antral cavity that provides enough oxygen and nutrients to the oocyte is fully formed. Transcripts from genes that are known to participate during the antrum formation were present in the transcriptomic data that we collected (e.g., Hspg2, Star, and Hsd3b1, Supplemental Note4, File S2). The oocyte was mostly transcriptionally silent, yet the somatic cells were transcriptionally active—even more so than in any other prior stage during ovarian follicle development, and the number of downregulated genes excessed the number of upregulated transcripts (Table S1). Opposite to the two prior transitions—from primary to small antral follicles—the complexity of the signaling network increased substantially, and communications were led by somatic cells (Fig. 2). Indeed, all the autocrine communications emerged between somatic cells and the majority of the paracrine signaling was through somatically produced ligands. Only 5 intercellular communications were between actively changed and cell-specific transcripts (i.e., oocyte or somatic cells).
Fig. 2.
Intercellular networks between oocytes and somatic cells from small antral to large antral transition. Legend as in Fig. 2. Yellow nodes indicate ligands secreted by the endocrine systems and present in the blood accessible to the growing antral follicle (e.g., Lhb, Ins2, Fsh). The connections and the references for each of them can be found in File S1
The large and complex green subnetwork that involved members of the Tgfb family, integrins, and vascular signals during the primordial to primary transition (Fig. 1) appeared again during the formation of the antral cavity (Fig.2). The Eph family subnetwork (highlighted in purple in Fig.1) contained more nodes and more connections at this stage compared to the primordial to the primary transition. On the other hand, the blue subnetwork—mostly enriched in Wnt ligands—and the red subnetwork—associated with Ramp receptors—decreased their importance during the antral cavity formation and the subnetwork associated with the Robo family (yellow subnetwork) had completely disappeared. Interestingly, it is plausible that the deactivation of the Slit-Robo signaling pathway might promote the essential apoptosis of granulosa cells for the formation of the antral cavity to avoid oocyte hypoxia [51]. The connections pertaining to the Edn families were significant again at this stage, as they were during the primary to secondary transition (Fig. S1), and the transcriptional levels of the gene that encode for the Lhcgr receptor were significantly increasing for the first time during ovarian follicle development. Also, other endocrine communications, such as from Fsh to Fshr or from Ins2 to its somatic receptors, were present at this stage as well. To summarize mechano-transduction and apoptotic signaling play an important role during antral cavity formation, which was mostly driven by somatic cells’ autocrine and paracrine signaling.
Inter-Cellular Signaling Networks In Vivo During the Small Antral to Large Antral Transition Between Oocyte and Mural and Cumulus Granulosa Cells
At the phenotypic level, one of the most important biological processes during antrum formation is the differentiation of the granulosa cells into mural and cumulus granulosa cells [50]. Several genes involved in this stage were present in the publicly available transcriptomic data of mural and cumulus cells [11], such as Cd34 and Has2 (Supplemental Note5, File S2). The number of downregulated and upregulated genes was very comparable in cumulus granulosa cells and in mural granulosa cells (Table S1). Interestingly, the number of total transcripts that were significantly changed in the cumulus cells far exceeded those of the oocyte, somatic cells (granulosa and theca cells), or mural granulosa cells (Table S1). More than a third of the significantly altered genes in the cumulus cell transcriptomic data were specifically produced by only cumulus granulosa cells. The transcripts from mural granulosa cells exhibited a similar ratio of specificity, a clear indication of different functions that cumulus and mural granulosa cells have during antrum formation.
The number of paracrine and autocrine signals was substantial, with almost all the autocrine signaling equally divided between mural or cumulus granulosa cells (Table S1, Fig. 3). Several paracrine communication pathways were initiated by non-significantly changing oocyte ligands to receptors in both mural and cumulus cells and the order of magnitude of paracrine communications for cumulus and mural granulosa cells were comparable, with a limited number towards non-significantly changing receptors in the oocyte.
Fig. 3.
Intercellular networks between oocytes and cumulus and mural cells from small antral to large antral transition. Legend as in Fig. 2. Red, cumulus cells; blue, mural cells; yellow ligands from the endocrine system. The connections and the references for each of them can be found in File S1. Oocyte: FC ≥ 2.5, p value ≤ 0.01; cumulus and mural granulosa cells: FC ≥ 2 and p value ≤ 0.01. The connections and the references for each of them can be found in File S1
Cumulus and mural intercellular signaling pathways were distinct and specific during ovarian follicle development. For instance, while the Gdf9 transcript was detected in cumulus cells, mural cells, and oocyte, it was only actively changing in cumulus cells. Several ligands (e.g., Wnt11, Tgfa, Inhba) and receptors (e.g., Pdgfrb, Acvr1c and Acvr2a) were specifically produced by the cumulus cells, while ligands such Epha5, Graba1, Sort1, and Pfgfd and receptors such as Fzd4, Il10rb, Cd44, or Gabrb2 were specific to mural cells. Some of these ligands and receptors have been previously reported (e.g., Pdgfrb) [11], yet Graba1 and its receptor Gabrb2, involved in gamma-aminobutyric acid (GABA), have not been reported in the ovarian follicle literature. The autocrine signaling between the ligand Pdgf and the receptor Pdgfrb only occurred in cumulus cells, and the paracrine communication between cumulus ligand Wnt11 and mural receptor Fzd4 was specific as well. At the transcriptional level, the mural autocrine communication between the ligand Lamc1 and the receptor Cd44 was important, with abrupt transcriptional increases for both genes (FC = 265, p value (fdr) = 5*10− 17 and FC = 135, p value (fdr) = 9*10− 18, respectively). In genome-wide association studies, Lamc1 has been associated with premature ovarian failure [52], and Cd44 is well-known for its anti-apoptotic properties during the antral stage [53].
At the subnetwork level, the green subnetwork during the primordial to primary transition (Fig. 1) gained new members of the Tgf, Pdgf, and integrin families. More members of the Apoa family were detected, although fragmented from the large green subnetwork (Fig. 3). Interestingly, more members of the Fgf family emerged as well as multiple small subnetworks related to Kitl, Il10, Il13ra1, and Gdnf. The Wnt family became mostly focused on Fdz receptors and also gained more connections. Amh signaling completely vanished at this mural-cumulus-oocyte network stage (Fig. 3). Thus, the transition from small to large antral stage presented distinct transcription profiles and intercellular communication in mural and cumulus granulosa cells, underscoring their distinct functionalities during this transition, some of which may be underexplored (e.g., GABA signaling in mural cells).
Inter-Cellular Signaling Networks In Vivo During the NSN to SN Transition in the Oocyte
Oocyte maturation is required for successful egg fertilization. One of the hallmarks required for achieving oocyte maturation is chromatin condensation [54], i.e., chromatin condensation in the oocyte nucleolus, from a non-surrounded nucleolus (NSN) to a surrounded nucleolus (SN) [55]. This transition cannot be achieved by denuded oocytes and requires the presence of cumulus granulosa cells in the form of a cumulus-oocyte complex (COC) [56]. Analysis of the available transcriptomics data during the NSN to SN transition agreed with the current understating of the changes associated with oocyte maturation (Supplemental Note6).
Cumulus cells were orchestrating the oocyte NSN to SN transition with a total of 237 intercellular signaling communications, out of which 162 were autocrine communications between cumulus cells, 55 were paracrine communications led by cumulus cell ligands (Table S1). There was no autocrine communication between the oocyte and no paracrine communication led by ligands secreted by the oocyte. While multiple genes that encode for cumulus ligands increased their transcription rates during this stage (e.g., Wnt10a, Ltb, Il6, Ereg, Camp, and Apln), their associated receptors did not significantly change their transcription levels (i.e., Aplnr, Cd14, Erbb2, Erbb4, Il6ra, Il6st, Ltbr, Robo1, Robo2, Fzd1, and Lrp5). Additionally, Gpr182 and Epha8, cumulus cell specific genes according to our models, were significantly upregulated in cumulus cells. As expected, the oocyte was completely silent at the transcriptional level (File S2).
As expected, the intercellular signaling network structure during the oocyte NSN to SN transition had little overlap with the primordial to primary intercellular network transition. The large subnetwork during the primordial to primary transition (marked in blue in Fig. 1) contained less nodes and less interactions during the oocyte NSN to SN transition (Fig. S3). For instance, the cumulus-specific ligand Fd6 and its associated connections were non-present. Similarly, Serpine1, Ins2, Erg, and Tgf families were separated from the majority of the components of the core blue subnetwork (Fig.1), which still contained a large number of intercellular communications through integrins (e.g., Itga7). Other parts of the primordial to primary blue subnetwork completely vanished, such as the Gdf9, Bmp, and Inhibin families. Additionally, multiple small subnetworks only appeared during this stage (i.e., Ifnr1, Cd47 and Gpr182; Il6; Camp), while others disappeared (e.g., Ramp1, Akrc receptor, and Fgf ligand families). Interestingly, Robo, which was only present during the primordial to primary transition (Fig. 1), became significant again in the cumulus cells during the oocyte NSN to SN transition. Thus, the intimate relationship between the oocyte and cumulus cells during the oocyte NSN to SN transition was characterized by cumulus-driven inter-cellular signaling with several subnetworks that promote cumulus cell expansion, e.g., Ifnr1 [57], and avoid apoptosis, e.g., Robo [51].
Identification of the most Likely TFs that Control the Significant Genes in each Follicular Cell Type During In Vivo Follicle Maturation
Finally, we identified the most likely TFs that could regulate the significantly changing genes during ovarian follicle development in vivo for each cell type and each follicular stage (Fig. 4, File S3). Employing experimental, manually curated databases, e.g., Metacore, Ovarian kaleidoscope [22], or computationally, e.g., FIMO [13] and BEEML [14], we determined genes that a given TF could transcriptionally regulate. We identified a total of 891 position-weighted matrices (PWM) that could potentially bind to the area around the transcription starting site (TSS) of each gene that was significantly changing their transcriptional abundance at any given stage and cell type during ovarian follicle maturation. This was out of the 3216 PWMs available in the database ensembled. Only 1.7% and 3.5% of the PWMs were on average predicted to be active (fdr-corrected p value < 0.05) in the oocyte and in the somatic cells (granulosa and theca cells), respectively. Yet the higher percentages of predicted active transcription factors occurred during the primordial to the primary transition (3.3% and 7.5% in the oocyte and somatic cells, respectively). Thus, the complexity during the primordial to primary transition in terms of the number of autocrine and paracrine communications between the oocyte and the somatic cells (intercellular communication) was mirrored by the number of TFs that were active during this same stage (intracellular communications).
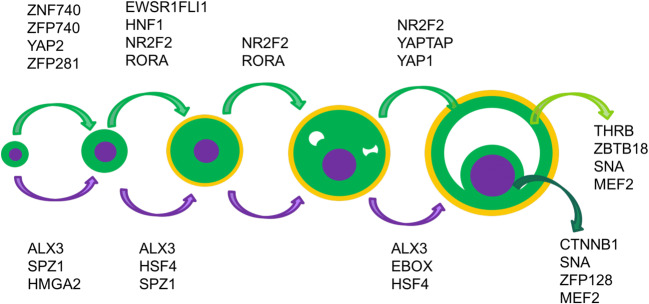
Fig. 4.
Representative active TFs during ovarian follicle development in the oocyte, somatic cells, cumulus and mural cells. The rest of the transcription factors are summarized in File S3
Our computational approach revealed TFs known to play a role during ovarian follicle development [58]. For instance, well-known oocyte-specific TFs, such as TCF7, ARX, POU3F1 [22], and the HOX family [59] were recovered as transcriptional regulators using our computational approach (File S3). Yet, our computational approached also identified new transcriptional regulators. For instance, we predicted that the TF ALX3 regulates oocyte transcriptional program from primordial to antral follicles, with the exception of the secondary to small antral transition—most likely due to the lack of power from the close similarity between secondary follicles and small antral follicles. ALX3 is involved in cell differentiation and female ALX3-deficient mice present with reduced fertility [60]. Other examples of non-previously reported TFs that regulated oocyte development included SPZ1, from the primordial to secondary follicle transition and HSF4 from the primary to antral transitions. Some TFs were specific to distinct ovarian follicle transitions. For instance, the TF MHGA2 was significant during the early transitions, i.e., the primordial to primary transition, while the TF EBOX was identified as a very likely regulator during the last ovarian follicle stage, during the small antral to large antral transition (File S3).
Similarly, we also identified TFs that could regulate the differentiation and development of somatic (granulosa and theca cells) cells during ovarian follicle development. The model identified FOXL2, NFKB, and the HEY family, known to be active in the transcriptional regulation of granulosa cells [61–63], RORA activity, which is a TF specific to theca cells [49], and the active transcriptional regulation control of SP1 in cumulus cells [64] during the oocyte NSN to SN transition. Importantly, novel TFs that have not been previously reported to be active in somatic cells (granulosa and theca cells) during ovarian follicle development were predicted by our computational model, including several zinc finger TFs (e.g., ZFP281 and ZFP740) and YAP signaling TFs that regulated the initial activation of somatic cells in primordial follicles. Additionally, YAP signaling was activated in somatic cells (granulosa and theca cells) during the small to large antral transition. NR2F2 and RORA were among the most significant TFs from the primordial to the small antral transition, with NR2F2 regulating the transcription of a large group of genes during antrum formation. While some TFs were common in the mural and cumulus cells (e.g., SNA and MEF2), others were uniquely active either in cumulus cells (e.g., CTNNB1 and ZFP128) or in mural cells (e.g., THRB and ZBTB18), as expected due to the different functions that these two cells types have during ovarian follicle development.
Discussion
Signals that control ovarian follicle development and enable the formation of a competent oocyte are not fully understood yet due to difficulty characterizing the intracellular and intercellular communications among and between the oocyte and its surrounding somatic cells (granulosa and theca cells). In this report, we have presented the most plausible intercellular communication networks during in vivo follicle maturation, as well as the most likely TFs that control and regulate ovarian follicle development at each follicular stage and in each follicular cell type using available dynamic transcriptomic data. These intercellular networks and intracellular regulation, which should be confirmed experimentally, can help to generate testable hypotheses in the laboratory that can enable to better understand this complex system.
Not only has our computational approach been able to recapitulate the presence of well-known ligands and receptors, including the cell type that produce them (e.g., oocyte or somatic cells) and the exact ovarian follicle developmental stage (e.g., from primordial to antral stage), but we also identified novel ligands and receptors that were not known to play a role during ovarian follicle development. For instance, several families of ligands and receptors that are well-known to intervene during follicle development such as the Bmp, Inh, and Tgfb families and their corresponding receptors, the Bmpr, Acvr, and Tgfbr [6, 65], as well as mechano-transduction receptors such as integrins [66] were included in our inter-cellular signaling networks [67]. Yet, our transition-specific networks also portray other families that have not been explored in the context of ovarian follicle development. For instance, the role of the Efn family and its receptors Eph [68], or the functions associated with the binding of the Thbs family [69] to integrins present in the oocyte plasma membrane, especially during the primordial to the primary transition, should be further studied. Additionally, there are several unique genes such as Gnr, Ighg1, Ndp, Ntn3, Pibf1, Pros1, Sct, App, Neo1, Tyro3, Ptprz1, or Phtr1, which have not been fully examined in their connection to ovarian follicle development. Moreover, the combinatorial possibilities are enormous: there are ligands capable to bind to the same receptor in the oocyte and somatic cells such as Amh, while others bind to multiple receptors, such as Vegfa. There are ligands whose genes are expressed in both cell types and have receptors in both cell types as well, such as Dnc. All this complex intercellular communication supports previous challenges to grow individual primordial follicles in 3D alginate hydrogels, while it is possible to grow them in ovarian tissue [1, 2] or groups of primordial follicles [70], where the somatic and ovarian cells provide all those ligands.
Supporting the experimental observations of the difficulties growing primordial follicles in vitro by themselves, the most entangled communication between the different cell types occurs during the transition from the primordial to primary stages through ligands that were mostly secreted by somatic cells. While the oocyte autocrine and oocyte-somatic paracrine communication proportions decreased during follicle maturation, from 64 to 0, somatic autocrine and somatic-oocyte paracrine communications were maintained or slightly decreased (Table S1, Fig. S4). These results highlight the growing importance of somatic cells in controlling the intercellular communications between the oocyte and the somatic cells as the follicle progresses from the primordial to antral stage. Additionally, our intercellular networks from the primordial to primary transition indicated that this early stage may entail several very convoluted communications between the oocyte and the surrounding somatic cells or very likely the stromal cells in the ovary, as primordial follicles can grow in ex vivo ovaries, and clearly underscored the complex interaction between the TGF-beta family and mechano-transduction. Importantly, from the intercellular networks, there are several possible candidates that maybe further explored experimentally by adding them to the follicle culture media to grow in vitro primordial follicles, such as Ntn3 or Omd.
Finally, using available experimental data and computational methods, we were able to also recover TFs that have been studied previously during ovarian follicle maturation, as well as others not that well understood, such as zinc finger proteins. Our results indicate that the YAP/TAZ signaling pathway is indeed active in somatic cells (i.e., granulosa and theca cells) in the primordial to primary transition and during granulosa cell expansion (Fig. 4). YAP/TAZ activity is an indication of cell proliferation and growth [71], which correlates with the significant cellular expansion that the granulosa cells undergo during ovarian follicle development. Yet, primordial follicles are under arrested growth due to the activation of the Hippo pathway, which in turn represses YAP/TAZ activation [72]. In fact, the YAP/TAZ pathway is activated spontaneously when arrested primordial follicles are removed from their native ovarian microenvironment [72]. While it is not clear how YAP/TAZ regulation could overcome Hippo repression, one plausible mechanism is through Akt dephosphorylation of YAP in activation in primordial follicles [73] and through a FSH-mediated PKA activation in secondary follicles [74]. The final size of the antral follicle might be regulated by the activation of the Hippo pathway. Expansion of the granulosa cells increases the number of granulosa cell-cell interactions [75], and mechanical stress is capable of activating the Hippo pathway [76], thus repressing the YAP/TAZ activation and subsequently avoiding the continuation of the granulosa cell proliferation.
While our computational approach is very powerful to disentangle the complex intercellular communications during ovarian follicle maturation, several pitfalls should be noted. For instance, though the intercellular connections apparently only affect a given follicle, secreted molecules, either proteins or metabolites by the oocyte and somatic cells, are capable of altering the processes in other surrounding follicles as well, directly (i.e., by binding in the receptors active in other somatic cells) or indirectly (i.e., by altering the endocrine system). These differences cannot be unraveled with the currently available experimental data. Moreover, as the transcriptomics data comes from surviving follicles, all the competition effects that cause some follicles to undergo atresia and not enter the recruiting pool are not represented in the intercellular networks depicted in this article. Finally, the current networks are based on transcriptional abundance—we have employed mRNA levels as a proxy of protein activation—and they can be highly improved by obtaining proteomic measurements at each follicular transition for each individual cell type (e.g., oocyte, granulosa, theca, mural, and cumulus cells). Finally, using transcription factor activation assays [77], transcriptional factors that are active can be followed, as shown previously by Zhou and colleagues who demonstrated that NFKB is indeed active in ovarian follicles [78].
In summary, systems reproductive biology approaches reveal the potentially key ligands and receptors associated with each cell type at each transition during ovarian follicle development, yet also predict the complex autocrine and paracrine communications between the oocyte and surrounding supporting cells that would lead to the production of a competent oocyte. We expect that our computational predictions will allow for the development of novel data-driven hypotheses that should be experimentally validated to increase our comprehension of ovarian follicle development, which can potentially be applied to guide the development of novel treatments for fertility disorders, such as polycystic ovarian syndrome, or for fertility preservation.
Electronic supplementary material
(DOCX 749 kb)
Acknowledgments
We thank Dr. Lei Lei, Sarah Kiesewetter for their invaluable technical support and Dr. Nadereh Jafari, Director of the Genomics Core Facility (Center for Genetic Medicine, Northwestern University) and Dr. Simon Lin and Dr. Gang Feng. Additionally, we would like to thank Dr. Ariella Shikanov and Andrea Jones for their insightful comments and throughout editing this manuscript.
Author Contributions
BPB designed the computational approached and performed all the computational study. BPB, TKW, LJB and LDS interpreted the results and wrote the manuscript.
Funding
This work has been mainly supported by NIH/NICHD 2 U54 HD041857–07 for study design and collection, analysis and interpretation of the data. BPB was supported NIH/NIGMS 2 T32 GM008449–16.
Compliance with Ethical Standards
Conflict of Interests
The authors declare that they have no conflict of interest.
Ethics Approval
Animals were treated in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the established IACUC protocol at Northwestern University.
Consent to Publish
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Teresa Woodruff, Email: tkw@northwestern.edu.
Linda J. Broadbelt, Email: broadbelt@northwestern.edu
Lonnie D. Shea, Email: ldshea@umich.edu
References
- 1.O'Brien MJ, Pendola JK, Eppig JJ. A revised protocol for in vitro development of mouse oocytes from primordial follicles dramatically improves their developmental competence. Biol Reprod. 2003;68(5):1682–1686. doi: 10.1095/biolreprod.102.013029. [DOI] [PubMed] [Google Scholar]
- 2.Eppig JJ, OBrien MJ. Development in vitro of mouse oocytes from primordial follicles. Biol Reprod. 1996;54(1):197–207. doi: 10.1095/biolreprod54.1.197. [DOI] [PubMed] [Google Scholar]
- 3.Xu M, West E, Shea LD, Woodruff TK. Identification of a stage-specific permissive in vitro culture environment for follicle growth and oocyte development. Biol Reprod. 2006;75(6):916–923. doi: 10.1095/biolreprod.106.054833. [DOI] [PubMed] [Google Scholar]
- 4.Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocrine reviews. 2009;30(6):624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao S, Coppeta JR, Rogers HB, et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nature Communications. 2017;8. 10.1038/ncomms14584. [DOI] [PMC free article] [PubMed]
- 6.Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132(2):191–206. doi: 10.1530/rep.1.01074. [DOI] [PubMed] [Google Scholar]
- 7.Uhlenhaut NH, Treier M. Foxl2 - function in ovarian development. Mol Genet Metab. 2006;88(3):225–234. doi: 10.1016/j.ymgme.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Chronowska E. High-Throughput Analysis of Ovarian Granulosa Cell Transcriptome. Biomed Res Int. 2014. 10.1155/2014/213570. [DOI] [PMC free article] [PubMed]
- 9.Yoon SJ, Kim KH, Chung HM, et al. Gene expression profiling of early follicular development in primordial, primary, and secondary follicles. Fertil Steril. 2006;85(1):193–203. doi: 10.1016/j.fertnstert.2005.07.1296. [DOI] [PubMed] [Google Scholar]
- 10.Skory RM, Bernabe BP, Galdones E, Broadbelt LJ, Shea LD, Woodruff TK. Microarray analysis identifies COMP as the most differentially regulated transcript throughout in vitro follicle growth. Mol Reprod Dev. 2013;80(2):132–144. doi: 10.1002/mrd.22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wigglesworth K, Lee KB, Emori C, Sugiura K, Eppig JJ. Transcriptomic diversification of developing cumulus and mural granulosa cells in mouse ovarian follicles. Biol Reprod. 2015;92(1):23. doi: 10.1095/biolreprod.114.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan H, O'Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286(2):493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Stormo GD. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat Biotechnol. 2011;29(6):480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic acids research. 2000;28(1):289–291. doi: 10.1093/Nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penalver Bernabe B, Thiele I, Galdones E, et al. Dynamic genome-scale cell-specific metabolic models reveal novel inter-cellular and intra-cellular metabolic communications during ovarian follicle development. BMC Bioinformatics. 2019;20(1):307. doi: 10.1186/s12859-019-2825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/Gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie XH, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3 ' UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24(1):238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weirauch MT, Yang A, Albu M, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leo CP, Vitt UA, Hsueh AJ. The Ovarian Kaleidoscope database: an online resource for the ovarian research community. Endocrinology. 2000;141(9):3052–3054. doi: 10.1210/endo.141.9.7679. [DOI] [PubMed] [Google Scholar]
- 23.Schulz MH, Devanny WE, Gitter A, Zhong S, Ernst J, Bar-Joseph Z. DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. Bmc Syst Biol. 2012;6. 10.1186/1752-0509-6-104. [DOI] [PMC free article] [PubMed]
- 24.Sui SJH, Mortimer JR, Arenillas DJ, et al. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33(10):3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995:289–300.
- 26.Charlier C, Montfort J, Chabrol O, et al. Oocyte-somatic cells interactions, lessons from evolution. Bmc Genomics. 2012;13:560. doi: 10.1186/1471-2164-13-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adam M, Saller S, Strobl S, et al. Decorin is a part of the ovarian extracellular matrix in primates and may act as a signaling molecule. Hum Reprod. 2012;27(11):3249–3258. doi: 10.1093/humrep/des297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang JS, Yi MJ, Zhang W, Feinleib JL, Cole F, Krauss RS. Netrins and neogenin promote myotube formation. J Cell Biol. 2004;167(3):493–504. doi: 10.1083/jcb.200405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Copeland NG, Gilbert DJ, Jenkins NA, Tessier-Lavigne M. Netrin-3, a mouse homolog of human NTN2L, is highly expressed in sensory ganglia and shows differential binding to netrin receptors. J Neurosci. 1999;19(12):4938–4947. doi: 10.1523/JNEUROSCI.19-12-04938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Le Noble F, Yuan L, et al. The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature. 2004;432(7014):179–186. doi: 10.1038/nature03080. [DOI] [PubMed] [Google Scholar]
- 31.Larrivee B, Freitas C, Trombe M, et al. Activation of the UNC5B receptor by Netrin-1 inhibits sprouting angiogenesis. Genes Dev. 2007;21(19):2433–2447. doi: 10.1101/gad.437807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozmadenci D, Feraud O, Markossian S, et al. Netrin-1 regulates somatic cell reprogramming and pluripotency maintenance. Nat Commun. 2015;6:7398. doi: 10.1038/ncomms8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson NH, Key B. Neogenin: one receptor, many functions. Int J Biochem Cell Biol. 2007;39(5):874–878. doi: 10.1016/j.biocel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 34.Buensuceso AV, Son AI, Zhou R, Paquet M, Withers BM, Deroo BJ. Ephrin-A5 Is required for optimal fertility and a complete ovulatory response to gonadotropins in the female mouse. Endocrinology. 2016;157(2):942–955. doi: 10.1210/en.2015-1216. [DOI] [PubMed] [Google Scholar]
- 35.Prunskaite-Hyyrylainen R, Shan J, Railo A, et al. Wnt4, a pleiotropic signal for controlling cell polarity, basement membrane integrity, and antimullerian hormone expression during oocyte maturation in the female follicle. FASEB J. 2014;28(4):1568–1581. doi: 10.1096/fj.13-233247. [DOI] [PubMed] [Google Scholar]
- 36.Yang HS, Li Y, Deng HX, Peng F. Identification of beta2-microglobulin as a potential target for ovarian cancer. Cancer Biol Ther. 2009;8(24):2323–2328. doi: 10.4161/cbt.8.24.9982. [DOI] [PubMed] [Google Scholar]
- 37.Qian J, Ji F, Ye X, et al. IGHG1 promotes motility likely through epithelial-mesenchymal transition in ovarian cancer. Chin J Cancer Res. 2018;30(2):282–290. doi: 10.21147/j.issn.1000-9604.2018.02.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang CL, Wang HS, Soong YK, Huang SY, Pai SY, Hsu SY. Regulation of oocyte and cumulus cell interactions by intermedin/adrenomedullin 2. J Biol Chem. 2011;286(50):43193–43203. doi: 10.1074/jbc.M111.297358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickinson RE, Hryhorskyj L, Tremewan H, et al. Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction. 2010;139(2):395–407. doi: 10.1530/REP-09-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dissen GA, Hirshfield AN, Malamed S, Ojeda SR. Expression of neurotrophins and their receptors in the mammalian ovary is developmentally-regulated - changes at the time of folliculogenesis. Endocrinology. 1995;136(10):4681–4692. doi: 10.1210/en.136.10.4681. [DOI] [PubMed] [Google Scholar]
- 41.Dissen GA, Romero C, Hirshfield AN, Ojeda SR. Nerve growth factor is required for early follicular development in the mammalian ovary. Endocrinology. 2001;142(5):2078–2086. doi: 10.1210/en.142.5.2078. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen T, Peters H. Proposal for a classification of oocytes and follicles in the mouse ovary. J Reprod Fertil. 1968;17(3):555–557. doi: 10.1530/jrf.0.0170555. [DOI] [PubMed] [Google Scholar]
- 43.Bridges PJ, Cho J, Ko C. Endothelins in regulating ovarian and oviductal function. Front Biosci (Schol Ed) 2011;3:145–155. doi: 10.2741/s140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21(2):200–214. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 45.Wood JR, Strauss JF., 3rd Multiple signal transduction pathways regulate ovarian steroidogenesis. Rev Endocr Metab Disord. 2002;3(1):33–46. doi: 10.1023/A:1012748718150. [DOI] [PubMed] [Google Scholar]
- 46.Jensen CE, Zachariae F. Studies on the mechanism of ovulation: isolation and analysis of acid mucopolysaccharides in bovine follicular fluid. Acta Endocrinol. 1958;27(3):356–368. doi: 10.1530/acta.0.0270356. [DOI] [PubMed] [Google Scholar]
- 47.Gebauer H, Lindner HR, Amsterdam A. Synthesis of heparin-like glycosaminoglycans in rat ovarian slices. Biol Reprod. 1978;18(3):350–358. doi: 10.1095/biolreprod18.3.350. [DOI] [PubMed] [Google Scholar]
- 48.Eriksen GV, Carlstedt I, Morgelin M, Uldbjerg N, Malmstrom A. Isolation and characterization of proteoglycans from human follicular fluid. Biochem J. 1999;340:613–620. doi: 10.1042/0264-6021:3400613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young JM, McNeilly AS. Theca: the forgotten cell of the ovarian follicle. Reproduction. 2010;140(4):489–504. doi: 10.1530/REP-10-0094. [DOI] [PubMed] [Google Scholar]
- 50.Mehlmann LM. Stops and starts in mammalian oocytes: recent advances in understanding the regulation of meiotic arrest and oocyte maturation. Reproduction. 2005;130(6):791–799. doi: 10.1530/rep.1.00793. [DOI] [PubMed] [Google Scholar]
- 51.Dickinson RE, Myers M, Duncan WC. Novel regulated expression of the SLIT/ROBO pathway in the ovary: possible role during luteolysis in women. Endocrinology. 2008;149(10):5024–5034. doi: 10.1210/en.2008-0204. [DOI] [PubMed] [Google Scholar]
- 52.Pyun JA, Cha DH, Kwack K. LAMC1 gene is associated with premature ovarian failure. Maturitas. 2012;71(4):402–406. doi: 10.1016/j.maturitas.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 53.Kaneko T, Saito H, Toya M, Satio T, Nakahara K, Hiroi M. Hyaluronic acid inhibits apoptosis in granulosa cells via CD44. J Assist Reprod Gen. 2000;17(3):162–167. doi: 10.1023/A:1009470206468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albertini DF, Sanfins A, Combelles CM. Origins and manifestations of oocyte maturation competencies. Reprod BioMed Online. 2003;6(4):410–415. doi: 10.1016/S1472-6483(10)62159-1. [DOI] [PubMed] [Google Scholar]
- 55.Zuccotti M, Piccinelli A, Rossi PG, Garagna S, Redi CA. Chromatin organization during mouse oocyte growth. Mol Reprod Dev. 1995;41(4):479–485. doi: 10.1002/mrd.1080410410. [DOI] [PubMed] [Google Scholar]
- 56.De la Fuente R, Eppig JJ. Transcriptional activity of the mouse oocyte genome: companion granulosa cells modulate transcription and chromatin remodeling. Dev Biol. 2001;229(1):224–236. doi: 10.1006/dbio.2000.9947. [DOI] [PubMed] [Google Scholar]
- 57.Jang YJ, Park JI, Moon WJ, Dam PTM, Cho MK, Chun SY. Cumulus cell-expressed type I interferons induce cumulus expansion in mice. Biol Reprod. 2015;92(1). 10.1095/biolreprod.114.122770. [DOI] [PubMed]
- 58.Sirotkin AV. Transcription factors and ovarian functions. J Cell Physiol. 2010;225(1):20–26. doi: 10.1002/jcp.22248. [DOI] [PubMed] [Google Scholar]
- 59.Du H, Taylor HS. The role of hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb Perspect Med. 2015;6(1):a023002. doi: 10.1101/cshperspect.a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lakhwani S, Garcia-Sanz P, Vallejo M. Alx3-deficient mice exhibit folic acid-resistant craniofacial midline and neural tube closure defects. Dev Biol. 2010;344(2):869–880. doi: 10.1016/j.ydbio.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Zhang CP, Yang JL, Zhang J, et al. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology. 2011;152(6):2437–2447. doi: 10.1210/en.2010-1182. [DOI] [PubMed] [Google Scholar]
- 62.Pisarska MD, Barlow G, Kuo FT. Minireview: roles of the forkhead transcription factor FOXL2 in granulosa cell biology and pathology. Endocrinology. 2011;152(4):1199–1208. doi: 10.1210/en.2010-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu S, Nishi Y, Yanase T, Nawata H, Fuller PJ. Transrepression of estrogen receptor beta signaling by nuclear factor-kappab in ovarian granulosa cells. Mol Endocrinol. 2004;18(8):1919–1928. doi: 10.1210/me.2004-0021. [DOI] [PubMed] [Google Scholar]
- 64.Worrad DM, Ram PT, Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein. TBP Development. 1994;120(8):2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- 65.Knight PG, Glister C. Local roles of TGF-beta superfamily members in the control of ovarian follicle development. Anim Reprod Sci. 2003;78(3–4):165–183. doi: 10.1016/S0378-4320(03)00089-7. [DOI] [PubMed] [Google Scholar]
- 66.Woodruff TK, Shea LD. The role of the extracellular matrix in ovarian follicle development. Reprod Sci. 2007;14(8 Suppl):6–10. doi: 10.1177/1933719107309818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallardo TD, John GB, Shirley L, et al. Genomewide discovery and classification of candidate ovarian fertility genes in the mouse. Genetics. 2007;177(1):179–194. doi: 10.1534/genetics.107.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buensuceso AV, Deroo BJ. The ephrin signaling pathway regulates morphology and adhesion of mouse granulosa cells in vitro. Biology of reproduction. 2013;88(1):25. doi: 10.1095/biolreprod.112.100123. [DOI] [PubMed] [Google Scholar]
- 69.Hatzirodos N, Hummitzsch K, Irving-Rodgers HF, Harland ML, Morris SE, Rodgers RJ. Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. Bmc Genomics. 2014;15(40). 10.1186/1471-2164-15-40. [DOI] [PMC free article] [PubMed]
- 70.Hornick JE, Duncan FE, Shea LD, Woodruff TK. Multiple follicle culture supports primary follicle growth through paracrine-acting signals. Reproduction. 2013;145(1):19–32. doi: 10.1530/REP-12-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lei QY, Zhang H, Zhao B, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28(7):2426–2436. doi: 10.1128/Mcb.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawamura K, Cheng Y, Suzuki N, et al. Hippo signaling disruption and Akt stimulation of ovarian follicles for infertility treatment. Proc Natl Acad Sci U S A. 2013;110(43):17474–17479. doi: 10.1073/pnas.1312830110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J, Kawamura K, Cheng Y, et al. Activation of dormant ovarian follicles to generate mature eggs. Proc Natl Acad Sci U S A. 2010;107(22):10280–10284. doi: 10.1073/pnas.1001198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu FX, Zhang YF, Park HW, et al. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes & development. 2013;27(11):1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aplin AE, Howe AK, Juliano RL. Cell adhesion molecules, signal transduction and cell growth. Curr Opin Cell Biol. 1999;11(6):737–744. doi: 10.1016/S0955-0674(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 76.Gerard C, Goldbeter A. The balance between cell cycle arrest and cell proliferation: control by the extracellular matrix and by contact inhibition. Interface Focus. 2014;4(3). 10.1098/Rsfs.2013.0075. [DOI] [PMC free article] [PubMed]
- 77.Weiss MS, Penalver Bernabe B, Shin S, et al. Dynamic transcription factor activity and networks during ErbB2 breast oncogenesis and targeted therapy. Integr Biol (Camb) 2014;6(12):1170–1182. doi: 10.1039/c4ib00086b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou H, Decker JT, Lemke MM, et al. Synergy of paracrine signaling during early-stage mouse ovarian follicle development in vitro. Cell Mol Bioeng. 2018;11(5):435–450. doi: 10.1007/s12195-018-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 749 kb)