Abstract
Endometriosis remains a challenge to understand and to diagnose. This is an observational cross-sectional pilot study to characterize the gut and vaginal microbiome profiles among endometriosis patients and control subjects without the disease and to explore their potential use as a less-invasive diagnostic tool for endometriosis. Overall, 59 women were included, n = 35 with endometriosis and n = 24 controls. Rectal and vaginal samples were collected in two different periods of the menstrual cycle from all subjects. Gut and vaginal microbiomes from patients with different rASRM (revised American Society for Reproductive Medicine) endometriosis stages and controls were analyzed. Illumina sequencing libraries were constructed using a two-step 16S rRNA gene PCR amplicon approach. Correlations of 16S rRNA gene amplicon data with clinical metadata were conducted using a random forest-based machine-learning classification analysis. Distribution of vaginal CSTs (community state types) significantly differed between follicular and menstrual phases of the menstrual cycle (p = 0.021, Fisher’s exact test). Vaginal and rectal microbiome profiles and their association to severity of endometriosis (according to rASRM stages) were evaluated. Classification models built with machine-learning methods on the microbiota composition during follicular and menstrual phases of the cycle were built, and it was possible to accurately predict rASRM stages 1–2 verses rASRM stages 3–4 endometriosis. The feature contributing the most to this prediction was an OTU (operational taxonomic unit) from the genus Anaerococcus. Gut and vaginal microbiomes of women with endometriosis have been investigated. Our findings suggest for the first time that vaginal microbiome may predict stage of disease when endometriosis is present.
Electronic supplementary material
The online version of this article (10.1007/s43032-019-00113-5) contains supplementary material, which is available to authorized users.
Keywords: Endometriosis, Pathogenesis, Microbiome, Vaginal microbiome, Diagnosis
Introduction
Endometriosis is a chronic gynecological condition defined as the presence of endometrial glands and/or stroma in ectopic sites that leads to a severe inflammatory state [1]. Pathogenesis of endometriosis is not yet fully understood, and different genetic, hormonal, environmental, and immunological factors have been implicated [2]. Studies have shown a significant association of aberrant immune response and maintenance of disease activity in women with endometriosis [3]. Although distinct immunological abnormalities have been reported in patients with disease, the specific mechanism in the pathogenesis of endometriosis is not yet fully understood [4, 5].
The non-surgical diagnosis of endometriosis remains an immense challenge. Although ovarian endometrioma and deep endometriosis are easily identified by means of transvaginal ultrasound (TVUS) and MRI (magnetic resonance imaging), superficial disease is currently not detected by any of these tools [6, 7]. Surgery is the only way to diagnose all cases, and it is considered the gold standard with definitive confirmation obtained with histology [7]. A noninvasive test for diagnosing all types of endometriosis does not yet exist, despite several studies evaluating biomarkers from blood, peritoneal fluid, and endometrial samples [8–10].
The concept that the human microbiome influences homeostasis and alters states of disease and health has been explored. Microbiota contribute to the equilibration of the immune system and help to maintain host homeostasis [11]. Recently, advances in technology and the use of next generation sequencing (NGS) facilitated the study of the human microbiome and its impact or response to different disease conditions [12]. The role of the gut microbiome in inflammatory and autoimmune diseases is well established but not yet fully understood [13]. Intestinal microbiota dysbioses are correlated not only with bowel inflammatory disorders such as Crohn’s disease but also with non-intestinal-related autoimmune diseases, such as multiple sclerosis, type-1 diabetes, Grave’s diseases, systemic lupus erythematosus, autism spectrum disorders, and psoriasis [14, 15].
Evaluation of the gut microbiome has been utilized as a potential and putative noninvasive diagnostic tool in colorectal cancer; furthermore, there is an optimistic perspective that in the near future, the gut microbiome might be a diagnostic tool for other chronic inflammatory gastrointestinal disorders such ulcerative colitis, Crohn’s disease, irritable bowel syndrome (IBS), and inflammatory bowel disease (IBD) [16, 17].
More recently, evaluation of the vaginal microbiota has been suggested to be useful for identification of common diseases in the upper genital tract [18]. Mycoplasma genitalium has been noted to play a role in the immune tolerance process and in modulating local immune response in women with endometriosis. Cells from the peritoneal fluid from women with endometriosis manifested a downregulation of genes associated with the inflammatory response. This downregulation profile was higher when Mycoplasma genitalium was present in the uterine cervix [19].
Given the inflammatory condition of patients with endometriosis and its similarities with autoimmune disorders [20, 21], the main goals of this pilot study are to characterize the microbiome from the gut and vagina of women with diagnosed endometriosis compared with women who do not have the disease and to explore the potential of using the gut and/or vaginal microbiome profiles as a diagnostic tool for endometriosis.
Methods
Study Approval, Subjects, and Sample Collection
The Internal Review Boards of the University of Sao Paulo (CAPPesq #814.743) and the Massachusetts Institute of Technology (COUHES protocol # 1107004572) approved this protocol. Written informed consent was obtained from all participants. This is an observational cross-sectional study and it was based on the STROBE (Strengthening the Reporting of Observational studies in Epidemiology) statement from the EQUATOR (Enhancing the Quality and Transparency of Health Research) guideline. All consecutive patients between the ages of 21–49 years old who underwent laparoscopy between 2012 and 2014 due to suspected or confirmed endometriosis by imaging (TVUS or MRI) or previous surgery were included in the study. Patients were excluded if they were on hormones (GnRH analogues, progestogens, combined oral contraceptives) and antibiotics in the past 3 months prior to sample collections; if they had active systemic infection, a history of autoimmune diseases, active vaginosis, or history of sexually transmitted diseases; and if they had acquired or primary immunodeficient diseases (including HIV), pregnancy, and malignant neoplasia. Control subjects (group B) included women who do not have endometriosis and any inflammatory conditions and who underwent laparoscopic surgery for other gynecologic indications.
Rectal and vaginal samples were collected from all patients (study and control) 2 months prior to surgery and during two different phases of the menstrual cycle—menstrual period (days 1–3 of the cycle) and follicular phase (days 8–12 of the cycle). The menstrual and follicular phases were selected as they represented known higher and lower inflammation states during the menstrual cycle [22]. All samples were obtained using Sterile Catch-All sample collection swabs (Fisher Scientific, Pittsburgh, PA, USA). Rectal samples were collected by swabbing the rectal tissue at a depth of 3 cm. Vaginal samples were collected in the mid-vagina at the site of the vaginal introitus. No speculums or lubricants were used prior to collection. Immediately after collection, all swabs were immersed in the lysis buffer of Mobio PowerBead tubes (MoBio Laboratories Inc., Carlsbad, CA, USA) and held against the sides of the tube three times for 20 s each to ensure transfer of biological material. All tubes were then transferred on wet ice to a clinical lab in less than 2 h and then stored at − 80 °C until extraction. Vaginal exams were performed after all vaginal collections to ensure that patients did not have any active vaginal infections.
DNA Extraction, 16S rRNA Amplicon Library, Sequencing Data Processing and OTU Analysis, and Vaginal Community State Type Assignment
All samples were extracted using the MoBio Powersoil extraction kit (MoBio Laboratories Inc., Carlsbad, CA, USA). Paired-end Illumina sequencing libraries were constructed using a two-step 16S rRNA gene PCR amplicon approach described previously elsewhere [23]. The v4 variability region of the 16S rRNA gene was amplified using the 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT) primers. All paired-end libraries were multiplexed into one lane and sequenced on the Illumina MiSeq platform at the Biomicro Center (MIT, Cambridge, MA). Libraries were sequenced with 2 × 250 bases. From the sequencing data, forward and reverse reads were merged and filtered by estimated error per read using USEARCH (version 8). Further processing and clustering of 97% sequence-based operational taxonomic units (OTUs) was performed using an in-house pipeline (S1 Text). OTUs represent clusters of organisms, grouped by DNA sequence similarity of a specific taxonomic marker gene (16S rRNA) [23]. OTUs were taxonomically classified using the Ribosomal Database Project with a confidence cutoff of 0.5 [24].
Microbiomes were categorized according to community state types (CSTs). Studies about profile vaginal and gut microbiomes have showed that there is a large variation among individuals as well as within individuals over a period of time. In order to gain greater understanding of their clinical utility, efforts were made to categorize the microbiome profiles into smaller clusters of microbiomes, known as CSTs.
Previous studies have already used CSTs to summarize the microbial communities observed in the human vagina and the human gut [25].
As previously published, vaginal CSTs are in this way divided: CST I is dominated by Lactobacillus crispatus; CST II by Lactobacillus gasseri; CST III by Lactobacillus inners; CST IV is the only one not dominated by Lactobacillus spp, with higher diversity of predominantly anaerobic bacteria; and CST V is dominated by Lactobacillus jensenii [26].
To determine if the five CSTs and eight sub-CSTs observed in other vaginal microbiome studies were present in this study, CST assignment was determined using a model trained on data derived from previous vaginal microbiome studies (S3 Text). CST assignment was not determined using hierarchical clustering of samples within our study alone. Species level classification is required for discriminating between many of the Lactobacillus-dominated CST groups. In order to perform the CST assignment, an environment-specific species level classification table was generated using speciateIT and MCclassifier. After the species table was generated, a support vector machine-learning model, previously trained on data derived from multiple vaginal microbiome studies, was utilized to predict the CST of each sample (S3 Text).
Statistical Analysis
16S rRNA amplicon data was used to generate a random forest-based machine-learning classification to identify correlations between OTU relative abundances and clinical metadata (disease state, self-reported pain) [27]. All OTU abundance data were analyzed with log transformation, and all metadata categories were analyzed without transformation.
All statistical tests were performed using custom scripts and publically available statistical packages (S4 Text). A power calculation was not performed to estimate the sample size given that this is a pilot study and no previous studies have yet been published.
Results
A total of 64 women with endometriosis (n = 40) and without endometriosis (n = 24) were included in this study. Five patients from the study group (group A) were excluded due to missing data. Patient demographics and disease characteristics are summarized in Table 1. There was no statistical difference in the age and BMI between the study and control groups. However, race did show statistically significant differences between the two groups (p = 0.008, chi square test). It is important to note that our subject cohort included white, mixed race, and black subjects, 61%, 22%, and 16.9% of the cohort, respectively. It is possible that the CST distributions observed in this study are due to more than half of the subjects being white. Endometriosis is more common in Caucasian and Asian populations, and CST distribution has been suggested to vary by race [26, 33]. As this is the first time that we know of, that the distribution of CSTs has been investigated in a South American population, we highlight these findings with caution, as the sample size was limited.
Table 1.
Clinical features of included subjects
| Group | ||||
|---|---|---|---|---|
| Variable | Endometriosis (n = 35) | Control (n = 24) | Total | p |
| (n = 59) | ||||
| Age (years) | ||||
| Mean (SD) | 34.9 (6.8) | 35.25 (6.9) | 35.0 (6.8) | 0.866* |
| Median (min.; max.) | 36 (21; 49) | 35 (21; 47) | 35 (21; 49) | |
| Race, n (%) | ||||
| White | 26 (74.3%) | 10 (41.7%) | 36 (61.0%) | 0.008** |
| Black | 6 (17.1%) | 4 (16.6%) | 10 (17.0%) | |
| Mulato | 3 (8.6%) | 10 (41.7%) | 13 (22.0%) | |
| BMI | ||||
| Mean (SD) | 24.8 (4.5) | 24.3 (2.7) | 24.6 (3.8) | 0.728✢ |
| Median (min.; max.) | 23.3 (19.1; 35.6) | 23.7 (20.6; 30.9) | 23.7 (19.1; 35.6) | |
| ASRM stage, n (%) | ||||
| I or II | 21 (60%) | 21 (60%) | ||
| III or IV | 14 (40%) | 14 (40%) | ||
| Dysmenorrhea | ||||
| Mean (SD) | 6.9 (3.2) | 1.8 (3.3) | 4.9 (4.1) | < 0.001✢ |
| Median (min.; max.) | 8 (0; 10) | 0 (0; 9) | 6 (0; 10) | |
| Dyspareunia | ||||
| Mean (SD) | 4.4 (3.4) | 1.8 (2.9) | 3.3 (3.4) | 0.011✢ |
| Median (min.; max.) | 6 (0; 10) | 0 (0; 10) | 3 (0; 10) | |
| Infertility, n (%) | ||||
| Yes | 12 (34.3%) | 0 (0%) | 12 (20.3%) | 0.001*** |
| No | 23 (65.7%) | 24 (100%) | 47 (79.7%) | |
*Student test; **Chi square test; ***Fisher’s exact test; ✢ Mann-Whitney test
Also, as expected, there were statistically significant differences among groups regarding clinical symptoms such as dyspareunia (p = 0.011, Mann-Whitney test), dysmenorrhea (p < 0.001, Mann-Whitney test), and infertility (p = 0.001, Fisher’s exact test). Control subjects (n = 24) included 18 women submitted to tubal ligation (75%), two cystectomies for ovarian benign teratoma–dermoid cyst (8%), one diagnostic laparoscopy for pelvic pain (4%), one hysterectomy for uterine bleeding (4%), one myomectomy (4%), and one cystectomy for ovarian serous cystadenoma (4%).
Among the 35 study patients, 13 patients were found to have deep bowel endometriosis (37.1%), 14 with deep retrocervical endometriosis (40%), four with deep bladder endometriosis (11%), two with ovarian endometriomas alone (5.7%), one with peritoneal superficial endometriosis (2.8%), and one with abdominal wall endometriotic nodule (2.8%). When staged using the rASRM endometriosis classification system (rASRM, 1996), nine patients were found to have rASRM stage 1 (25.7%), 12 stage 2 (34.2%), four stage 3 (11.4%), and ten stage 4 (28.5%). For comparative analysis of patients and controls with this relatively small patient cohort, patients with rASRM (revised American Society for Reproductive Medicine, 1996) stages 1 and 2 were grouped together as rASRM stage 1–2, as were patients with stages 3 and 4 for rASRM stage 3–4, following common conventions [28].
Given the correlation between the gut microbiome and other chronic inflammatory disorders observed in other studies [16, 17], we investigated if the rectal microbiome profile was predictive of patients having deep bowel endometriosis. A random forest-based classification model was built using the rectal microbiome during the menstrual phase of 10 patients with deep bowel endometriosis and 12 patients with other presentations of the disease. The accuracy of the classification models was determined using area under the curve (AUC) analysis of receiver operating characteristic curves. This model while predictive of site of disease (p value = 0.03) was determined to not be sufficiently accurate to purse in the context of this study (AUC = 0.29).
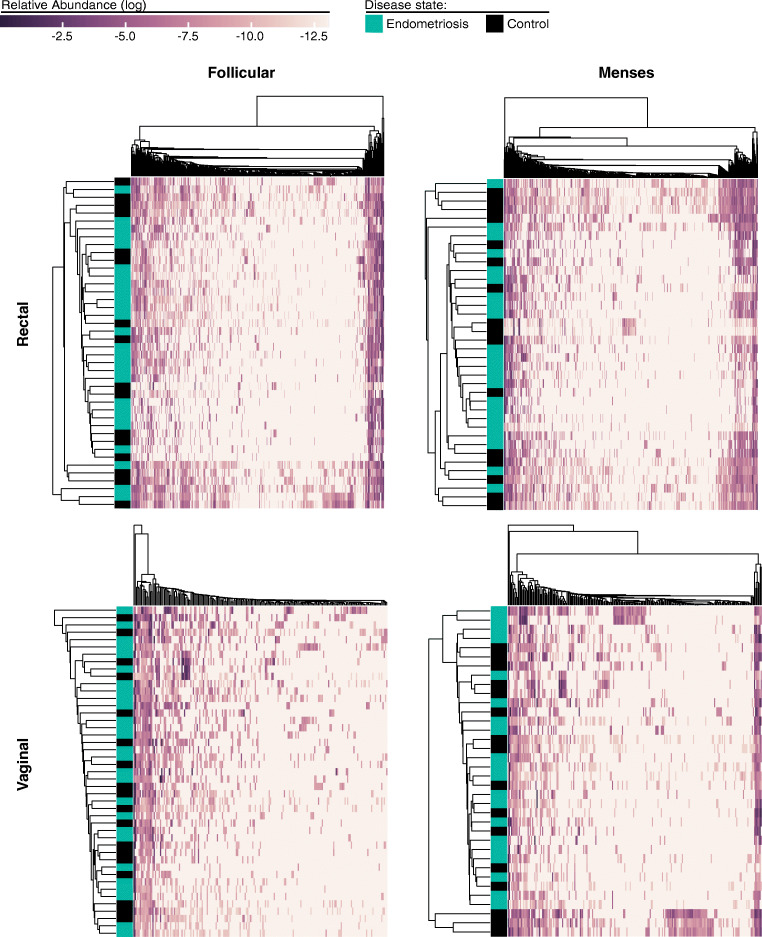
Rectal and vaginal samples collected during the menstrual and follicular phases of the menstrual cycle from the study and control patients underwent 16S rRNA analysis. In this cohort of women, there were no differences in the gut and vaginal microbiomes between study and control groups for both periods of the menstrual cycle—follicular and menstrual phases (Fig. 1). In order to better understand the correlation between the microbiome and status of disease—endometriosis patients and control subjects—a hierarchical linkage clustering was performed based on the CSTs composition and OTU abundance of samples for each sample type in the two periods of the cycle they were sampled.
Fig. 1.
Rectal and vaginal microbial communities do not cluster by disease state. We illustrate the hierarchical linkage clustering of samples based upon the community composition and OTU abundance for patient rectal and vaginal samples collected during the follicular and menses time points. Patient disease status is indicated as endometriosis or control group, blue and black respectively. OTU abundances are displayed as log relative abundance
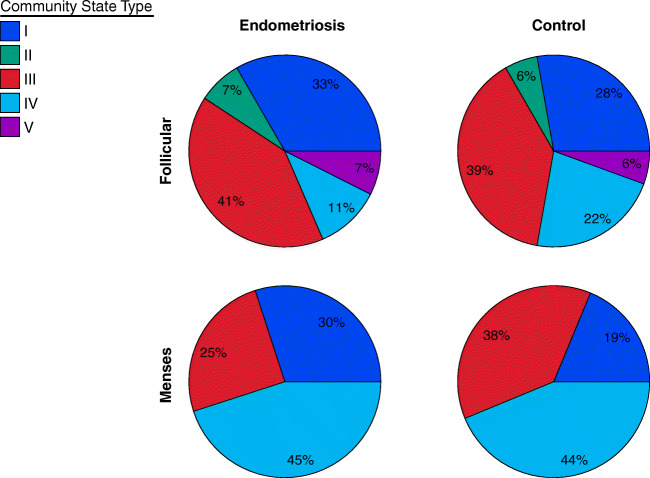
The distribution of vaginal CSTs differed significantly between follicular and menstrual phases of the menstrual cycle within the same individuals. There was an increase in the number of patients with CST IV microbiomes during menstrual phase for both endometriosis (by 30%) and control subjects (by 25%), while CSTs II and V were lost during the menstrual phase (p = 0.021, Fisher’s exact test) (Fig. 2). A two-sided Fisher exact test for count data was used in R (version 3.3.1) to test the significance of CST distributions between groups of patients.
Fig. 2.
The distribution of vaginal community state types differs between follicular and menses phases of the menstrual cycle. Distribution of vaginal community state types observed in endometriosis and control subjects during follicular and menses phases. The number of subjects included is 27, 18, 21, and 16; top left, right, bottom left, and right, respectively
Although not statistically significant, the ratio of CST I microbiome, which is dominated by Lactobacillus crispatus, was found to be in lower ratio in the control subjects compared with endometriosis patients, especially in the menstrual period (19% vs 30%, p = 0.57, Fisher’s exact test) as illustrated in Fig. 2.
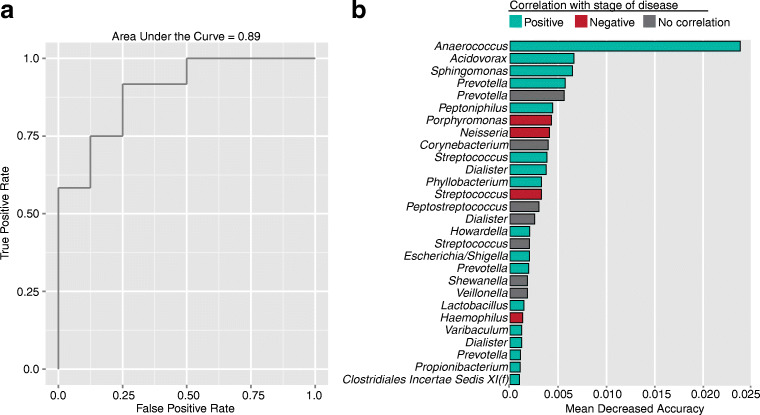
The vaginal microbiome profile was predictive of severity of endometriosis (according to rASRM stages) A random forest-based classification model was built using the vaginal microbiome during the menstrual phase of 12 patients with rASRM stages 1–2, and 8 patients with rASRM stages 3–4 of endometriosis demonstrated that the vaginal microbiome was surprisingly predictive of advanced disease (p value = 0.019) (Fig. 3), despite the fact that there were no significant differences when comparing patients with either stage of disease to control patients. The accuracy of the classification models was determined using AUC analysis. The highest contributing feature of the classification was an OTU of genus Anaerococcus (phylum Firmicutes) (Fig. 3). Contributions of specific OTUs to the classification analysis were ranked using mean decrease accuracy (S2 Text). Associations between log-transformed OTU abundance and disease state were calculated using a two-tailed t test for independent samples.
Fig. 3.
Microbiota composition predicts stage of disease. Here we illustrate a accuracy, as determined by AUC analysis of ROC curves of the model built to predict stages 1–2 versus stages 3–4 endometriosis from the composition of the vaginal microbial community during the menses time point. b The top contributing OTU features for this model, ranked by Mean Decreased Accuracy. Taxonomic level for each OTU displayed genus except in the case of a single OTU where f indicates the family level. Correlations of OTU abundance with stage of disease, calculated using a two-sided t test, are indicated as positive (blue, t statistic > 0 and p value < 0.05), negative (red, t statistic < 0 and p value < 0.05), and no correlation (gray, p value > 0.05)
The relative abundance of the top predictive OTU significantly differed between stages 1–2 and stages 3–4 disease subjects using a univariate analysis (Fig. 4). There was no significant difference in abundance for this OTU between the stages 1–2 or stages 3–4 disease and control subject groups. The significance of OTU abundance between discrete categories was calculated using the Mann-Whitney U test and a Benjamini-Hochberg false discovery rate correction. Only OTUs with a relative abundance of at least 10−6 were considered.
Fig. 4.
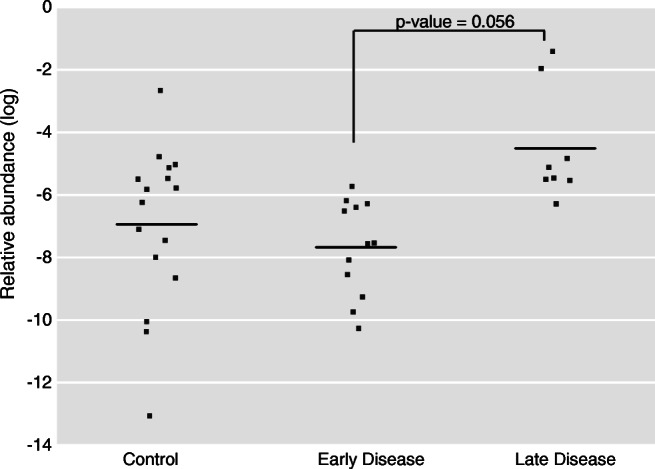
The relative abundance of the top predictive feature differs between stages 1–2 and stages 3–4 endometriosis patients. We illustrate the abundance in vaginal samples during the menses time point of the most predictive feature from our machine learning model, an OTU classified as belonging to the genera Anaerococcus. Log relative abundances for this OTU are shown for control, stages 1–2 stage endometriosis and stages 3–4 endometriosis subjects. Corrected p values shown were calculated using the Mann-Whitney U test with a Benjamini-Hochberg false discovery rate correction as described in the materials and methods
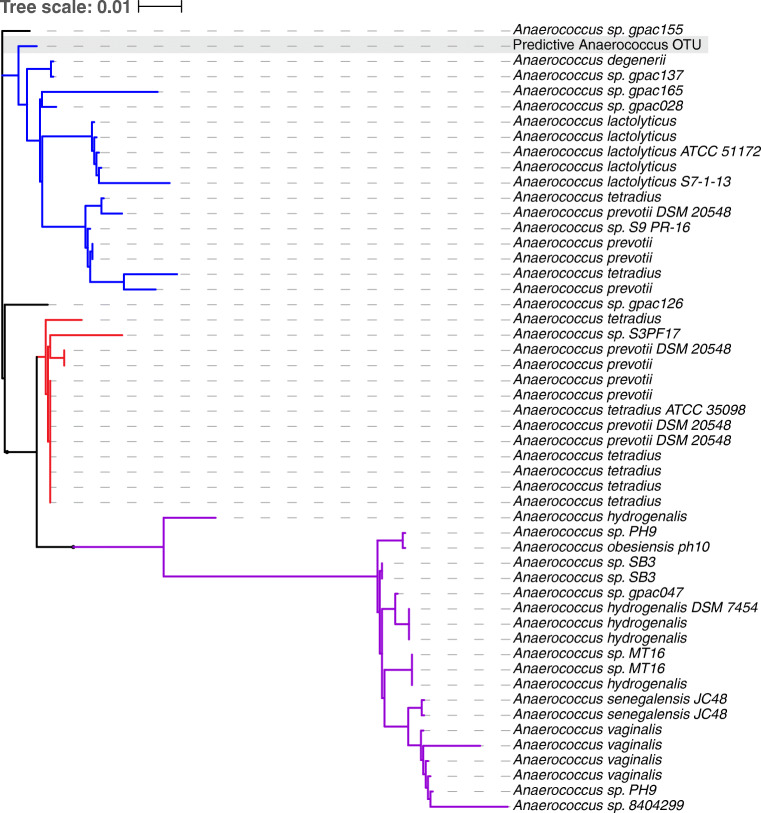
The species most closely related to this highly predictive Anaerococcus OTU were Anaerococcus lactolyticus and Anaerococcus degenerii species when compared with cultured representatives of these genera in the SILVA database (Fig. 5) [29]. 16S rRNA gene sequence similarity was utilized to investigate the phylogenetic relationship between the top predictive OTU identified in the classification models and other closely related representatives of the same genus (S5 Text).
Fig. 5.
The phylogenetic relationship between the predictive Anaerococcus OTU and other cultured representatives of this genus. We illustrate the phylogenetic relationship between the Anaerococcus OTU identified in this study as predictive of stage of disease (highlighted with a gray box) and Anaerococcus representatives in the SILVA database. There are three major clades: a clade including the predictive OTU (blue), A. lactoylticus, A. tetradius, and A. prevotii strains; a clade of A. tetradius and A. prevotii strains (red); and a clade of A. hydrogenalis, A. senegalensis, and A
vaginalis strains (purple).
Discussion
The gut microbiota has been indicated in the pathogenesis of endometriosis [30]. Similarly, vaginal microbiota has been attributed to be useful in identifying genital tract disorders [18]. However, there is a lack of studies investigating the connection between the vaginal and the gut microbiomes with endometriosis in humans. Recently, in an animal study, some data was published providing new insights on the association of endometriosis and gut microbiota using high-throughput sequencing technology. Authors revealed changes in the gut microbiota during endometriosis development in a murine model [31]. Their findings showed an elevation in the ratio Firmicutes/Bacteroidetes in mice with endometriosis, indicating that endometriosis may induce dysbiosis in the host [31]. Women with endometriosis have not been assessed until now.
The preliminary results from this pilot study revealed that the relative abundance of a specific OTU classified as Anaerococcus was predictive of endometriosis rASRM stage of disease when assessed during menstruation. Interestingly, it is the same period of the menstrual cycle that other biomarkers have been shown, with reliability and higher sensitivity, to identify endometriosis patients [8]. Patients with endometriosis rASRM stages 1–2 were differentiated from patients with endometriosis rASRM stages 3–4 using the relative abundance of an Anaerococcus OTU. This information may prove useful for future research regarding the role of the vaginal microbiome and Anaerococcus species specifically, in the pathogenesis and diagnosis of advanced stages of endometriosis.
The vaginal microbiome sampled from different locations within the same individual has been shown to vary [18]. Chen et al. explored the vaginal microbiome in three different locations—lower third of vagina, posterior fornix, and cervical mucus—in a healthy reproductive-age Chinese population. The microbiome profile from the samples collected in the lower third of the vagina, which is very similar to the collection protocol used in the present study, comprised only of CSTs I, III, and IV [18]. The same two CSTs II and V that were lost in our cohort during the menstrual period were both absent in Chen et al.’s cohort. Curiously, none of those patients were in the menstrual phase of the cycle in the occasion of sample collection [18]. This may suggest differences in the vaginal microbiome profile between the two populations, considering that a cohort of reproductive-age women without endometriosis was also included in the present pilot study as controls. Conversely, the microbiome profile of the disease and control cohorts in the present study revealed the presence of all five CSTs in the samples collected during follicular phase, revealing the loss of CSTs II and V in the samples collected during menstruation.
The distribution of CSTs obtained from Brazilian patients in this study was similar to those of other studies conducted with women of European descent. It was found that the CSTs distributions of bacteria for both endometriosis and control subjects during the follicular phase were the same of those observed in non-pregnant reproductive age women of European descent [32–35]. Moreover, the definition of each CST in the present study is in agreement with that described by Ravel et al. and Gajer et al. [26, 33], as well as with the one described by Chen et al., which included only Chinese women [18].
Interestingly, in addition to differences in the distribution of CSTs, a flux directed mainly into CST IV during the menstrual period was observed (Fig. 2). CST IV is defined by a vaginal community that is not dominated by Lactobacillus but is instead a heterogeneous community of anaerobic bacteria [26, 35]. Two Lactobacillus-dominated CSTs, II and V, dominated by high abundances of Lactobacillus gasseri and L. jensenii, respectively, were lost during the menstrual phase in both groups of women (Fig. 2). The loss of CST II and V during menstrual phase observed in this study could be related to a lack of power due to a small sample size used to identify these CSTs in both periods. However, an alternative hypothesis could be the fall in estrogen levels that occur during menstruation [35]. It has been previously demonstrated that during follicular phase, which is associated with higher estrogen levels and increased vaginal secretions, there is an accumulation of glycogen that potentially culminates in the growth of Lactobacillus species [36, 37]. Interestingly, CST I (Lactobacillus crispatus-dominated) appears to be in lower ratio among control subjects compared with endometriosis patients, especially in the menstrual period. Although this result is not statistically significant, probably due to the small sample size included in this pilot study, it is possible that it is reflective of the population if one takes into consideration known biological properties from Lactobacillus crispatus. This lower ratio of CST I might be related to the inflammatory profile and response observed in endometriosis. It has been demonstrated that a specific strain of Lactobacillus crispatus grants an anti-inflammatory phenotype to dendritic cells (DCs) by upregulating anti-inflammatory/regulatory IL-10 cytokine production as well as inducing CD4(+) CD25(+) FOXP3(+) T cells at great dosage [38]. It is already known that endometriosis itself may upregulate IL-10 yield [20] and induce CD4(+) CD25(+) FOXP3(+) T cells [39]. It is plausible that Lactobacillus crispatus is somehow involved within the local immune response of patients with endometriosis. This potential immunological role of Lactobacillus crispatus could explain why CST I (Lactobacillus crispatus-dominated) might be in higher concentration among patients with endometriosis compared with patients without the disease. Future studies should consider investigating the possible role of Lactobacillus crispatus in patients with endometriosis. The use of bigger cohorts would make it possible to identify statistical differences in the concentration of CST I between women with and without endometriosis. Additionally, immunological aspects should be measured to understand the role of the vaginal microbiome within immune response regulation in endometriosis.
Given that there was no difference in the vaginal and gut microbiome profiles between endometriosis and control patients assessed during the follicular and menstrual phases within this cohort of patients, two hypotheses were raised. First, this lack of difference might reflect the high inter-subject and intra-subject variability of the gut and vaginal microbiomes as previously reported [26, 35, 40]. Second, this is a pilot study with a small sample size and current results might be related to a lack of power. This might also be the case for the results we had comparing the gut microbiome of patients with bowel endometriosis with patients presenting with other sites of endometriosis lesions. However, we hope that including this information will encourage future studies with larger cohorts to investigate this hypothesis.
Ideally, a prospective, collaborative work, including women from different populations will fulfill the gaps left by this pilot study.
Our findings suggest for the first time that the vaginal microbiome may predict stage of disease when endometriosis is present.
Conclusion
An OTU within the genus Anaerococcus differed significantly in abundance between women with endometriosis rASRM stages 1–2 and stages 3–4 during menstrual phase, predicting these different stages of disease. Moreover, L. crispatus might be involved in immune response regulation within endometriosis patients. This is the first study investigating both gut and vaginal microbiome in patients with endometriosis and control women without the disease. Current findings bring new insights on the pathogenesis of endometriosis and the further potential use of the human vaginal microbiome as a diagnostic tool. As this is a pilot study, it has limitations, specially the small sample size, and therefore these current finding should be considered with caution until more data from larger cohorts are published.
Electronic Supplementary Material
(DOCX 19 kb)
Acknowledgments
The authors would like to acknowledge the support of Marta Privato for providing invaluable organizational and communication support over the course of this research.
The authors would like to acknowledge the support of Pawel Gajer and Jacque Ravel for providing access to speciateIT and McClassifier, the trained support vector machine learning model utilized for CST assignment in this study.
Funding Information
Library preparation and sequencing efforts through the MIT BioMicro Center were funded by the National Institute of Environmental Health Sciences of the NIH under award P30-ES002109.
This study was funded by grants from the Brazilian Council for Scientific and Technological Development (CNPq). Specifically, the Massachusetts Institute of Technology (MIT)/Brazil Seed Fund (Process number: CNPq-MIT 457125/2012-8).
Data Availability
The raw sequencing data of the vaginal and rectal samples used in this study as well as the associated metadata have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject PRJNA424567.
Compliance with Ethical Standards
The Internal Review Boards of the University of Sao Paulo (CAPPesq #814.743) and the Massachusetts Institute of Technology (COUHES protocol # 1107004572) approved this protocol. Written informed consent was obtained from all participants.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98(3):511–519. doi: 10.1016/j.fertnstert.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laganà AS, Vitale SG, Salmeri FM, Triolo O, Ban Frangež H, Vrtačnik-Bokal E, Stojanovska L, Apostolopoulos V, Granese R, Sofo V. Unus pro omnibus, omnes pro uno: a novel, evidence-based, unifying theory for the pathogenesis of endometriosis. Med Hypotheses. 2017;103:10–20. doi: 10.1016/j.mehy.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 3.Riccio LDGC, Santulli P, Marcellin L, Abrão MS, Batteux F, Chapron C. Immunology of endometriosis. Best Pract Res Clin Obstet Gynaecol. 2018 Feb 8. pii: S1521–6934(18)30028–2. [DOI] [PubMed]
- 4.Senturk LM, Arici A. Immunology of endometriosis. J Reprod Immunol. 1999;43(1):67e83. doi: 10.1016/S0165-0378(98)00079-5. [DOI] [PubMed] [Google Scholar]
- 5.Christodoulakos G, Augoulea A, Lambrinoudaki I, Sioulas V, Creatsas G. Pathogenesis of endometriosis: the role of defective 'immunosurveillance'. Eur J Contracept Reprod Health Care. 2007;12(3):194e202. doi: 10.1080/13625180701387266. [DOI] [PubMed] [Google Scholar]
- 6.Abrao MS, Gonçalves MO, Dias JA, Jr, Podgaec S, Chamie LP, Blasbalg R. Comparison between clinical examination, transvaginal sonography and magnetic resonance imaging for the diagnosis of deep endometriosis. Hum Reprod. 2007;22(12):3092–3097. doi: 10.1093/humrep/dem187. [DOI] [PubMed] [Google Scholar]
- 7.Dunselman GA, Vermeulen N, Becker C, Calhaz-Jorge C, D'Hooghe T, De Bie B, Heikinheimo O, Horne AW, Kiesel L, Nap A, Prentice A, Saridogan E, Soriano D, Nelen W; European society of human reproduction and embryology. ESHRE guideline: management of women with endometriosis. Hum Reprod 2014 Mar;29(3):400–412. [DOI] [PubMed]
- 8.O DF, Flores I, Waelkens E, D'Hooghe T. Noninvasive diagnosis of endometriosis: review of current peripheral blood and endometrial biomarkers. Best Pract Res Clin Obstet Gynaecol 2018 Jul;50:72–83. doi: 10.1016/j.bpobgyn.2018.04.001. Epub 2018 Apr 13. [DOI] [PubMed]
- 9.Nisenblat V, Prentice L, Bossuyt PM, Farquhar C, Hull ML, Johnson N. Combination of the non-invasive tests for the diagnosis of endometriosis. Cochrane Database Syst Rev. 2016 Jul 13;7:CD012281. 10.1002/14651858.CD012281 Review. [DOI] [PMC free article] [PubMed]
- 10.Borrelli GM, Abrão MS, Mechsner S. Can chemokines be used as biomarkers for endometriosis? A systematic review. Hum Rep. 2014;29(2):253–266. doi: 10.1093/humrep/det401. [DOI] [PubMed] [Google Scholar]
- 11.Peterson CT, Sharma V, Elmen L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015;179:363–377. doi: 10.1111/cei.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers GB, Bruce KD. Next-generation sequencing in the analysis of human microbiota: essential considerations for clinical application. Mol Diagn Ther. 2010 Dec 1;14(6):343-. [DOI] [PubMed]
- 13.Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360:j5145. doi: 10.1136/bmj.j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiely CJ, Pavli P, O'Brien CL. The role of inflammation in temporal shifts in the inflammatory bowel disease mucosal microbiome. Gut Microbes. 2018;15:1–25. doi: 10.1080/19490976.2018.1448742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Opazo MC, Ortega-Rocha EM, Coronado-Arrázola I, Bonifaz LC, Boudin H, Neunlist M, Bueno SM, Kalergis AM, Riedel CA. Intestinal microbiota influences non-intestinal related autoimmune diseases. Front Microbiol. 2018;9:432. doi: 10.3389/fmicb.2018.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan V, Peppelenbosch MP, Konstantinov SR. Human fecal microbiome-based biomarkers for colorectal cancer. Cancer Prev Res (Phila) 2014;7(11):1108–1111. doi: 10.1158/1940-6207.CAPR-14-0273. [DOI] [PubMed] [Google Scholar]
- 17.Quigley EMM. Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet? Nat Rev Gastroenterol Hepatol. 2017;14(5):315–320. doi: 10.1038/nrgastro.2017.29. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, Li F, Yu X, Feng Q, Wang Z, Xie H, Chen X, Zeng C, Wen B, Zeng L, du H, Tang H, Xu C, Xia Y, Xia H, Yang H, Wang J, Wang J, Madsen L, Brix S, Kristiansen K, Xu X, Li J, Wu R, Jia H. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017;8(1):875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos GB, Marques LM, Rezende IS, Barbosa MS, Abrão MS, Timenetsky J. Mycoplasma genitalium can modulate the local immune response in patients with endometriosis. Fertil Steril. 2018;109(3):549–560. doi: 10.1016/j.fertnstert.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Podgaec S, Abrao MS, Dias JA, Rizzo LV, De Oliveira RM, Baracat EC. Endometriosis: an inflammatory disease with a Th2 immune response component. Hum Reprod. 2007;22(5):1373–1379. doi: 10.1093/humrep/del516. [DOI] [PubMed] [Google Scholar]
- 21.Kvaskoff M, Mu F, Terry KL, Harris HR, Poole EM, Farland L, Missmer SA. Endometriosis: a high-risk population for major chronic diseases? Hum Reprod Update. 2015;21(4):500–516. doi: 10.1093/humupd/dmv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maybin JA, Critchley HOD. Menstrual physiology: implications for endometrial pathology and beyond. Hum Reprod Update. 2015;21(6):748–761. doi: 10.1093/humupd/dmv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preheim SP, Perrotta AR, Martin-Platero AM, Gupta A, Alm EJ. Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl Environ Microbiol. 2013;79(21):6593–6603. doi: 10.1128/AEM.00342-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks JP, Buck GA, Chen G, Diao L, Edwards DJ, Fettweis JM, Huzurbazar S, Rakitin A, Satten GA, Smirnova E, Waks Z, Wright ML, Yanover C, Zhou YH. Changes in vaginal community state types reflect major shifts in the microbiome. Microb Ecol Health Dis. 2017;28(1):1303265. doi: 10.1080/16512235.2017.1303265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, Mcculle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci. 2010;108 (Supplement 1):4680–7. [DOI] [PMC free article] [PubMed]
- 27.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, Schauer DB, Ward DV, Korzenik JR, Xavier RJ, Bousvaros A, Alm EJ. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7(6):e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beste MT, Pfäffle-Doyle N, Prentice EA, Morris SN, Lauffenburger DA, Isaacson KB, et al. Molecular network analysis of endometriosis reveals a role for c-Jun—regulated macrophage activation. Sci Transl Med. 2014;6(222):222ra16--222ra16. [DOI] [PMC free article] [PubMed]
- 29.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;gks1219. [DOI] [PMC free article] [PubMed]
- 30.Laschke MW, Menger MD. The gut microbiota: a puppet master in the pathogenesis of endometriosis? Am J Obstet Gynecol. 2016 Jul;215(1):68.e1–4. [DOI] [PubMed]
- 31.Yuan M, Li D, Zhang Z, Sun H, An M, Wang G. Endometriosis induces gut microbiota alterations in mice. Hum Reprod. 2018;33(4):607–616. doi: 10.1093/humrep/dex372. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Hansmann MA, Davis CC, Suzuki H, Brown CJ, Pierson JD, et al. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol Med Microbiol. 2010;58(2):169–181. doi: 10.1111/j.1574-695X.2009.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anahtar MN, Byrne EH, Fichorova RN, Kwon DS, Anahtar MN, Byrne EH, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital article cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42(5):965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2(1):4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UME, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52-132ra52. [DOI] [PMC free article] [PubMed]
- 36.Farage M, Maibach H. Lifetime changes in the vulva and vagina. Arch Gynecol Obstet. 2006;273(4):195–202. doi: 10.1007/s00404-005-0079-x. [DOI] [PubMed] [Google Scholar]
- 37.Nunn KL, Forney LJ. Unraveling the dynamics of the human vaginal microbiome. Yale J Biol Med. 2016;89:331–337. [PMC free article] [PubMed] [Google Scholar]
- 38.Eslami S, Hadjati J, Motevaseli E, Mirzaei R, Farashi Bonab S, Ansaripour B, Khoramizadeh MR. Lactobacillus crispatus strain SJ-3C-US induces human dendritic cells (DCs) maturation and confers an anti-inflammatory phenotype to DCs. APMIS. 2016;124(8):697–710. doi: 10.1111/apm.12556. [DOI] [PubMed] [Google Scholar]
- 39.Podgaec S, Rizzo LV, Fernandes LF, Baracat EC, Abrao MS. CD4(+) CD25(high) Foxp3(+) cells increased in the peritoneal fluid of patients with endometriosis. Am J Reprod Immunol. 2012;68(4):301–308. doi: 10.1111/j.1600-0897.2012.01173.x. [DOI] [PubMed] [Google Scholar]
- 40.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2015;15(7). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 19 kb)
Data Availability Statement
The raw sequencing data of the vaginal and rectal samples used in this study as well as the associated metadata have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject PRJNA424567.