Abstract
Background: Sepiapterin reductase (SPR) is an important regulator of the biosynthesis of tetrahydrobiopterin (BH4), which has been shown to be a promoter of different kinds of tumors. This study aims to investigate the role of SPR in breast cancer and to explore its molecular mechanism. Methods: SPR expressions in breast cancer tissues with different pathological stages were compared with the corresponding pericarcinomatous tissues and were analyzed using immunohistochemical staining and western blot. SPR knockdown was performed in MDA-MB-231 and MDA-MB-468 triple-negative breast cancer cells using specific siRNAs. Quantitative real-time PCR and western blot were used to determine the efficiency of the SPR knockdown. The intracellular BH4 levels were measured using HPLC, and the intracellular ROS levels were measured using an ROS detection kit. Clone formation and cell proliferation assays were used to study the effect of the SPR knockdown on cell proliferation. Annexin V/PI double staining, cell mitochondria isolation, and western blot were performed to study the effect of the SPR knockdown on cell apoptosis. ROS scavenger NAC was used to inhibit increased ROS caused by the SPR knockdown. Results: SPR is highly expressed in breast cancer tissues compared with the pericarcinomatous tissues and positively correlated with the pathological stages. The knockdown of SPR causes decreased intracellular BH4 and increased intracellular ROS and inhibits the proliferation of MDA-MB-231 and MDA-MB-468 cells. The knockdown of SPR also induces mitochondrial pathway-mediated apoptosis. NAC suppresses the SPR knockdown-caused cell apoptosis and cell death. Conclusions: SPR promotes the proliferation of breast cancer cells. The knockdown of SPR suppresses the proliferation of breast cancer cells by inducing ROS-mediated apoptosis.
Keywords: Sepiapterin reductase, breast cancer, ROS, apoptosis
Introduction
Metabolic reprogramming is a hallmark of cancer cells [1]. Dissecting the complex regulatory network is critical for target discovery and the development of anti-cancer medicines. Tetrahydrobiopterin (BH4; 6R-L-erythro-5,6,7,8-tetrahydrobiopterin) is involved in the process of phenylalanine catabolism [2], lipid metabolism [3], and immunometabolism [4], which is a regulator of cell metabolism. In addition, several studies has previously demonstrated the role of BH4 in the growth, angiogenesis, and migration of cancer cells [5-7]. Sepiapterin reductase (SPR) is an important regulator of the biosynthesis of BH4, which has been demonstrated to be significantly correlated with the proliferation of human neuroblastoma (NB) [8] and hepatocellular carcinoma [9]. However, SPR’s biological functions and its underlying mechanisms in most cancer types are still unclear. Among these undetermined cancer types, breast cancer is still the leading causes of cancer-related death in women [10]. Whether SPR and BH4 are involved in the development of breast cancer is of interest.
In addition, it has been reported that BH4 is highly redox-sensitive and easily oxidized [11,12], and it is also an effective ROS scavenger [13-15]. ROS is a common inducer of mitochondrial pathway-mediated apoptosis. When cells are stimulated by ROS, cytochrome C is released through the mitochondrial membrane into the cytoplasms, and this is one of the most critical mitochondrial pathway steps. In this step, the Bcl-2 family proteins such as Bax are activated to bind to the mitochondrial outer membrane, forming a channel facilitating substance exchange between the mitochondria and the cytoplasms, which causes the release of cytochrome C. At the same time, the Bcl-2 itself can inhibit the function of Bax and hinder the process that plays an anti-apoptotic role [16].
In this study, we report the role of SPR in facilitating the proliferation of breast cancer. The knockdown of SPR suppresses the production of BH4 and induces apoptosis through the ROS-mediated mitochondrial pathway. Thus, our study provides evidence for SPR as a therapeutic target in the treatment of breast cancer.
Materials and methods
Cell culture
MDA-MB-231 and MDA-MB-468 cell lines were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 50 U/mL penicillin, and 50 U/mL streptomycin. All the cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Human breast cancer specimens and immunohistochemical staining
29 human breast cancer specimens and the corresponding pericarcinomatous tissues were collected from the Department of Surgical Oncology of the First Affiliated Hospital of Bengbu Medical College. The SPR expressions were determined using immunochemistry.
Western blot
Cell lysates were prepared in a RIPA buffer (20 mM Tris-HCl, pH 7.5, 0.1% (w/v) sodium lauryl sulfate, 0.5% (w/v) sodium deoxycholate, 135 mM NaCl, 1% (v/v) Triton X-100, 10% (v/v) glycerol, 2 mM EDTA), supplemented with a protease inhibitor and a phosphatase inhibitor (Beyotime, Jiangsu, China). The total protein concentration was determined using the Bradford Bio-Rad assay (Hercules, CA, USA) and quantified using a BioTek SynergyMx (Winooski, VT, USA). The cell lysates in the SDS-sample buffer were incubated for 15 min at 95°C, and equal amounts of total protein were analyzed using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot. The antibodies used in this study were: rabbit anti-SPR antibody, purchased from Abcam (Cambridge science park, England), rabbit anti-p-mTOR (Ser2448), rabbit anti-p-p70S6k (Ser371), rabbit anti-cytochrome C, and anti-rabbit IgG, HRP-linked antibody, purchased from Cell Signaling Technology, Danvers, USA, rabbit anti-mTOR, rabbit anti-p70S6k, rabbit anti-caspase 3, rabbit anti-cleaved caspase 3, rabbit anti-caspase-9, rabbit anti-cleaved caspase-9, rabbit anti-PARP, rabbit anti-cleaved PARP, rabbit anti-Bcl-2, rabbit anti-Bax, and rabbit anti-β-actin, obtained from Proteintech (Wuhan, China). All the primary antibodies were used at 1:1000, and all the secondary antibodies were used at 1:10,000.
siRNA and transfection
SPR siRNA (siSPR#1, siSPR#2 and siSPR#3) and non-targeting siRNA (NC) were purchased from Shanghai Genomeditech Co., Ltd. (Shanghai, China). The of siSPR#1, siSPR#2, and siSPR#3 sequences were 5’-GACUGCUGCUUAUCAACAATT-3’, 5’-GCUGCUCGUGAUAUGCUGUTT-3’ and 5’-CCCAACTGAAGTGAACAACTA-3’ respectively. The transfection of the SPR siRNA was performed using the lipofectamine 3000 transfection reagent (Invitrogen, USA), according to the manufacturer’s instructions.
Quantitative real-time PCR (qRT-PCR)
The total cellular RNA was isolated with the TRIzol® Reagent (Vazyme Biotech, Jiangsu, China) and reverse transcribed with the HiScript QRT SuperMix for quantitative PCR (qPCR; Vazyme Biotech, Jiangsu, China). The mRNA level was measured with the SYBR Green master mix (Vazyme Biotech, Jiangsu, China). The amount of mRNA for each gene was standardized with the internal control (GAPDH mRNA). Each treatment group was compared with the control group to show the relative mRNA level. The primer sequences for quantitative qRT-PCR are provided in Table 1.
Table 1.
Primer sequences for qRT-PCR
| Gene | Sequences |
|---|---|
| GAPDH | Forward Sequence: GTCTCCTCTGACTTCAACAGCG |
| Reverse Sequence: ACCACCCTGTTGCTGTAGCCAA | |
| SPR | Forward Sequence: GACCTGAAAGTGGTGCTGGCAG |
| Reverse Sequence: GAGGAAGCCTTTGGAAACATCCC |
Clone formation assay
After transfection of the SPR siRNA for 48 h, 1000 cells were plated in 6-well culture plates. After 14 days’ culture, the cells were stained with crystal violet solution (Beyotime, Jiangsu, China). The number of colonies was then counted macroscopically.
Cell proliferation assay
After transfection of the SPR siRNA for 48 h, 5×104 cells were seeded into 12-well culture plates. The cells were counted using an automatic counter (Countstar) every 24 h. The proliferation curves were produced using GraphPad Prism 8 software.
Annexin V/PI double staining
After transfection of the SPR siRNA for 48 h, the apoptotic cells were identified using an Annexin V-FITC Apoptosis Detection Kit (Vazyme Biotech, Jiangsu, China) in accordance with the manufacturer’s instructions. The flow cytometry analysis was performed immediately after the supravital staining. The data acquisition and analysis were performed in a Becton Dickinson FACS-Calibur flow cytometer using Cell Quest software (Franklin Lakes). Tp block the ROS, the cells were treated with 5 mmol/L N-acetylcysteine (NAC, Beyotime).
BH4 detection
The cells were lysed using trichloroacetic acid containing 10 mmol/l dithiothreitol. The lysates were subjected to differential oxidation in acidic (0.2 mol/l trichloroacetic acid) or alkalytic (0.1 mol/l NaOH) solutions containing 2.5% I2/10% KI or 0.9% I2/1.8% KI, as shown previously. After centrifugation, 20 μl supernatant was injected into an HPLC system with a fluorescent detector. Excitation and emission wavelengths of 350 and 450 nm, respectively, were used to detect the fluorescent BH4 and its oxidized species, as previously shown [17,18].
ROS detection
After transfection of the SPR siRNA for 48 h, the cells were collected and alkalytic intracellular ROS level was determined using a Reactive Oxygen Species Assay Kit (Beyotime, Jiangsu, China) according to the manufacturer’s instructions.
Cell mitochondria isolation
After the transfection of the SPR siRNA for 48 h, cell mitochondria isolation was performed using a Cell Mitochondria Isolation Kit (Beyotime, Jiangsu, China) according to the manufacturer’s instructions. Briefly, the cells were washed with PBS and digested with trypsin, then the cells were centrifuged at a speed of 100-200 g for 5 min to collect alkalytic cells. We then transferred the cells to clean tubes and homogenized the cells 10-30 times. The cell homogenate was centrifuged at a speed of 600 g at 4°C for 10 min. We transferred the supernate to another clean tube and centrifuged it at a speed of 11000 g at 4°C for 10 min. The supernate was cytosol, and the deposit was mitochondria.
Statistical analysis
All of the results were presented as the mean ± SD from triplicate experiments performed in a parallel manner unless otherwise indicated. Statistically significant differences (One-way ANOVAs followed by Bonferroni’s Multiple Comparison Test) were determined using GraphPad Prism 8 software. A value of P < 0.05 was considered significant, and values of P < 0.01, P < 0.001 and P < 0.0001 were considered highly significant.
Results
SPR expression is up-regulated in tumor tissues and positively correlates with pathological stages
To investigate the role SPR plays in the progression of breast cancer, 29 breast cancer patients with different pathological stages were recruited as the study cohort. These patients’ detailed information is shown in Table 2, and the SPR expressions in the different pathological stages were analyzed using immunohistochemical staining and western blot. Compared with the pericarcinomatous tissues, the SPR expressions in the tumor tissues were significantly higher (Figure 1A). In addition, the SPR expressions were also positively correlated with the pathological stages (Figure 1B). Thus, these results suggest that SPR might facilitate the progression of breast cancer.
Table 2.
The characteristics of the patients
| Characteristics | Sub-groups | Number |
|---|---|---|
| Age (years) | < 50 | 12 |
| ≥ 50 | 17 | |
| Tumor size (cm) | < 2 | 13 |
| ≥ 2 | 16 | |
| TNM stage | I-II | 19 |
| III-IV | 10 | |
| ER status | Negative | 14 |
| Positive | 15 | |
| PR status | Negative | 17 |
| Positive | 12 | |
| HER-2 status | Negative | 14 |
| Positive | 15 | |
| Lymph nodal status | Negative | 18 |
| Positive | 11 |
Figure 1.
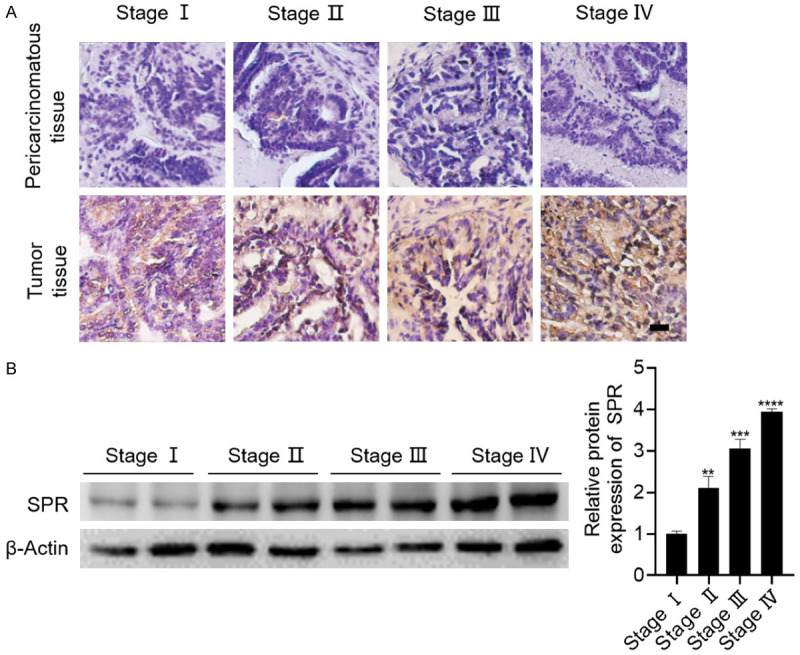
SPR is highly expressed in breast cancer tissues and positively correlated with pathological stage. A. Representative pictures of the immunohistochemical staining of SPR in breast cancer tissue with different pathological stages compared with the corresponding pericarcinomatous tissues. B. The SPR expressions in the breast cancer tissues with different pathological stages were analyzed using western blot. The data are represented as the mean ± SD in triple. **P < 0.01; ***P < 0.001; ****P < 0.0001.
The knockdown of SPR inhibits the proliferation of breast cancer cells
To elucidate the role SPR plays in breast cancer, two triple-negative breast cancer cell lines, MDA-MB-231 and MDA-MB-468, were adopted. We first analyzed the SPR expressions in the MDA-MB-231 and MDA-MB-468 cells compared with the non-tumorigenic mammary gland epithelial cell line MCF10A. As shown in Figure 2A, the SPR expressions in the MDA-MB-231 and MDA-MB-468 cells were significantly higher. To further study the role of SPR in these two cell lines, a knockdown of SPR using siRNA was performed. Among the three siSPR sequences, the knockdown efficiency of siSPR#3 was consistent and achieved 80% in both breast cancer cell lines at both the mRNA and protein levels (Figure 2B and 2C). Thus, we chose siSPR#3 to use in our following experiments. We first tested the effect of the SPR knockdown on the clone formation and proliferation. The SPR knockdown inhibited the formation of cell clones and suppressed the proliferation of both breast cancer lines (Figure 2D and 2E). The mTOR signaling pathway, which is closely related to cell proliferation, was also suppressed by the SPR knockdown, as shown by the decreased expression of phosphorylated mTOR and phosphorylated p70S6k (Figure 2F). Taken together, these results indicate that the knockdown of SPR inhibits the proliferation of breast cancer cells.
Figure 2.
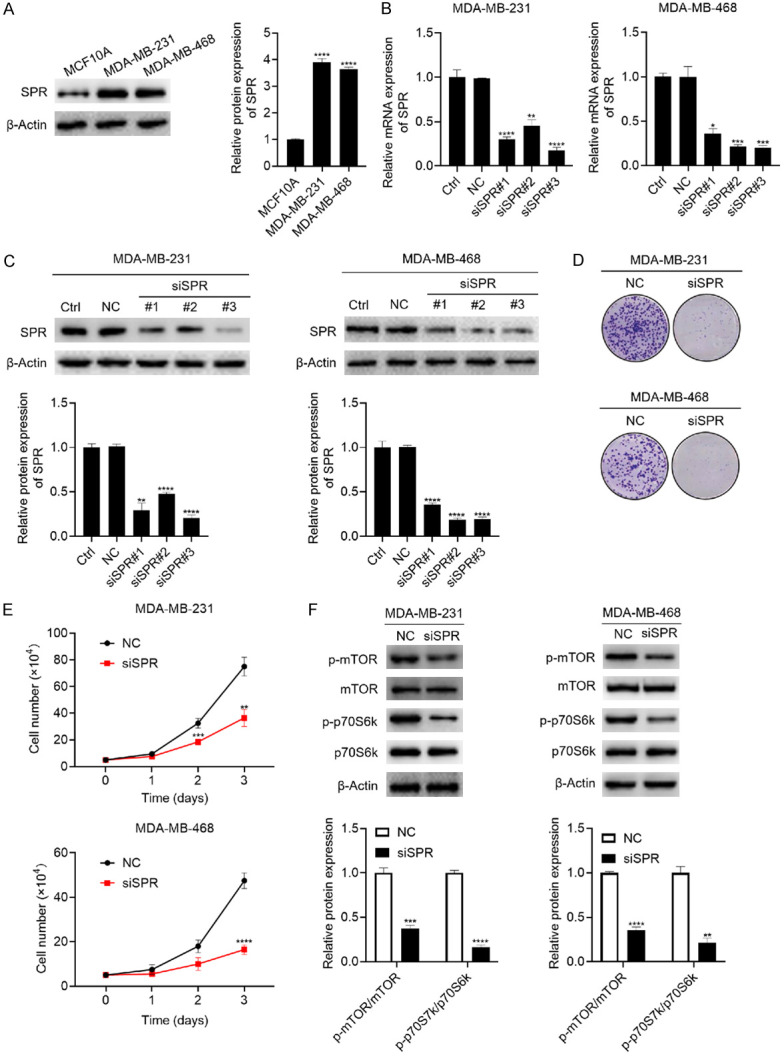
The knockdown of SPR inhibited the proliferation of MDA-MB-231 and MDA-MB-468 breast cancer cells. (A) The SPR expressions in the non-tumorigenic mammary gland epithelial cell lines MCF10A and MDA-MB-231 and MDA-MB-468 triple-negative breast cancer cells were analyzed using western blot. (B, C) The MDA-MB-231 and MDA-MB-468 breast cancer cells were transfected with NC, siSPR#1, siSPR#2, and siSPR#3 for 4 h, then replaced with fresh DMEM containing 10% FBS for another 48 h culture. mRNA and protein samples were harvested, the mRNA expression of SPR was analyzed using qRT-PCR (B), and the SPR protein expression was analyzed using western blot (C). (D) After the transfection of siSPR#3, 1000 cells were seeded into 6-well plates and cultured for 14 days, then the cells were stained with a crystal violet solution. (E) After the transfection of siSPR#3, 5×104 cells were seeded into 12-well-plates and cultured for 3 days, and the cells were counted every 24 h. (F) After the transfection of siSPR#3 and culturing the cells to 90% confluence, the cells were harvested, and the mTOR, p-mTOR, p70S6k, and p-p70S6k expressions were analyzed using western blot. The data are represented as the mean ± SD in triple. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
The knockdown of SPR induces apoptosis in breast cancer cells
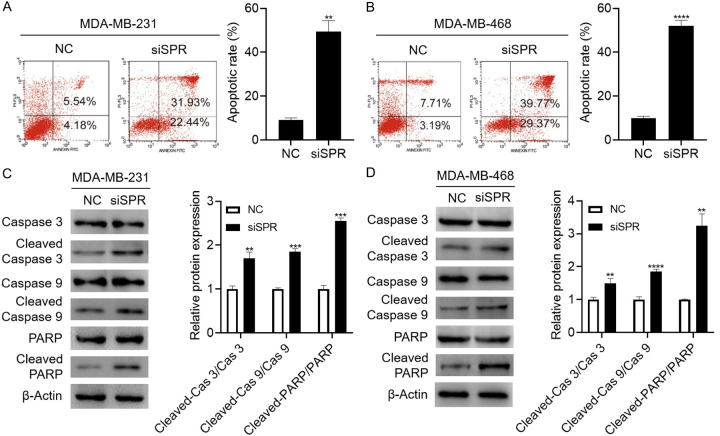
To determine whether the marked reduction in cell proliferation caused by the SPR knockdown could be attributed to the activation of apoptotic cell death, flow cytometry was performed to determine the effect of the SPR knockdown on the cell apoptosis. As shown in Figure 3A and 3B, the SPR knockdown indeed increased the apoptotic cells in both breast cancer cell lines. The cleavages of caspase 3, caspase-9, and PARP, three markers of cell apoptosis, were also increased by the SPR knockdown, which confirmed the cell apoptosis at the molecular level (Figure 3C and 3D). Thus, these results suggest that SPR knockdown induces apoptosis in MDA-MB-231 and MDA-MB-468 breast cancer cells.
Figure 3.
SPR knockdown induces apoptosis in MDA-MB-231 and MDA-MB-468 breast cancer cells. A, B. After the transfection of siSPR#3, the cells were stained with PI and Annexin V, and the apoptotic cells were analyzed using flow cytometry. C, D. After the transfection of siSPR#3 and culturing the cells to 90% confluence, the cells were harvested and the caspase-3, cleaved-caspase 3, caspase-9, cleaved-caspase-9, PARP, and cleaved-PARP expressions were analyzed using western blot. The data are represented as the mean ± SD in triple. **P < 0.01; ***P < 0.001; ****P < 0.0001.
The knockdown of SPR activates the mitochondrial apoptotic pathway in breast cancer cells
Next, we investigated the underlying mechanism of the SPR knockdown that caused the suppressed proliferation of breast cancer cells. It was reported that BH4 is an effective scavenger of ROS. Thus, we hypothesized that the SPR knockdown induced oxidative stress in the breast cancer cells. We first tested the effect of the SPR knockdown on the intracellular BH4 levels in both the MDA-MB-231 and MDA-MB-468 breast cancer cells. As expected, the intracellular BH4 levels were significantly reduced in these two breast cancer cell lines (Figure 4A). In contrast, the intracellular ROS level was increased by the SPR knockdown (Figure 4B). As ROS is a common inducer of the mitochondrial pathway of apoptosis, we hypothesized that SPR induced apoptosis via the mitochondrial pathway. The results showed that the SPR knockdown suppressed the expression of Bcl-2, an anti-apoptotic protein, and increased the expression of Bax, a pro-apoptotic protein (Figure 4C). Furthermore, the SPR knockdown significantly promoted the release of cytochrome C from the mitochondria to the cytosol (Figure 4D). These results strongly demonstrate that SPR knockdown activates the mitochondrial apoptotic pathway.
Figure 4.
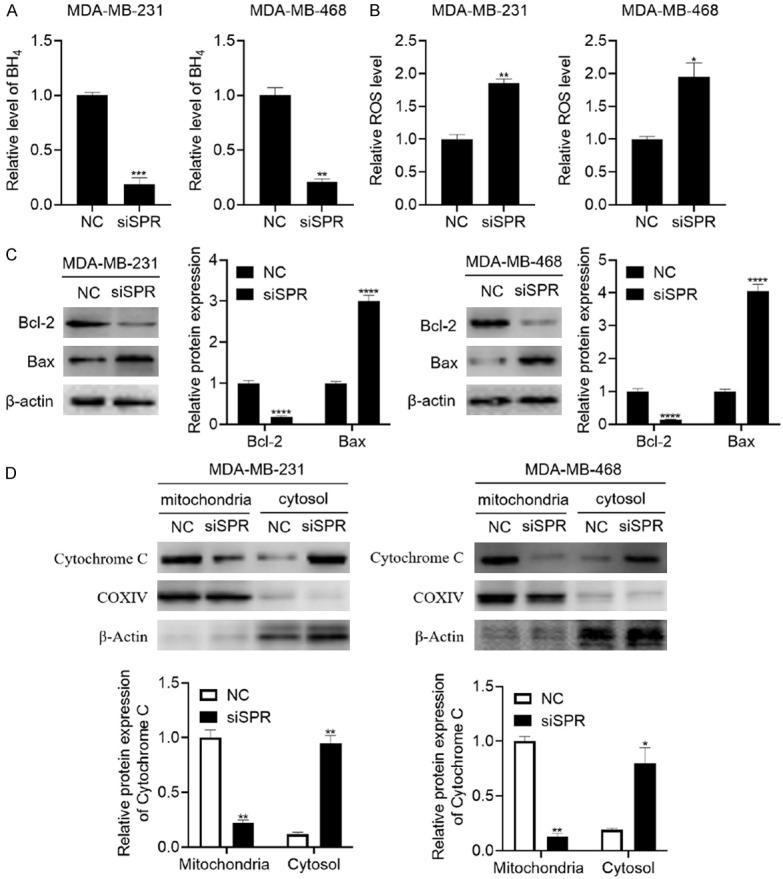
SPR knockdown activates the mitochondrial apoptotic pathway. A. After the transfection of siSPR#3, the cells were lysed and BH4 was measured using HPLC. B. After the transfection of siSPR#3 and culturing the cells to a 90% confluence, the cells were harvested, and the intracellular ROS level was measured using an ROS assay kit. C. After the transfection of siSPR#3 and culturing the cells to a 90% confluence, the cells were harvested and the Bcl-2 and Bax expressions were analyzed using western blot. D. After the transfection of siSPR#3 and culturing the cells to 90% confluence, the mitochondria and cytosol were isolated and the expression of cytochrome C was measured using western blot. The data are represented as the mean ± SD in triplicate. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
The ROS scavenger NAC partially reverses SPR knockdown-induced apoptosis and proliferation inhibition
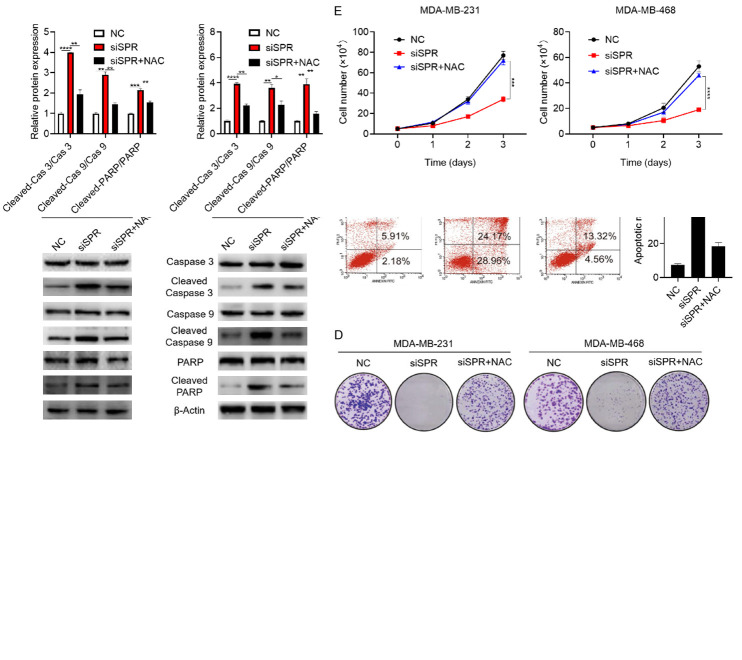
The above results showed that SPR knockdown increases the intracellular ROS levels. In order to study the role of ROS in SPR knockdown-induced apoptosis and proliferation inhibition, the ROS scavenger NAC was used to clear the intracellular ROS. As shown in Figure 5A, NAC indeed inhibited the increase of the intracellular ROS levels caused by the SPR knockdown. Then, Annexin V/PI staining was performed. The results showed that NAC decreased the apoptotic cells caused by the SPR knockdown (Figure 5B). At the molecular level, the increased cleavages of caspase 3, caspase-9, and PARP induced by the SPR knockdown were also suppressed by NAC (Figure 5C). Furthermore, the clone formation and cell proliferation inhibited by the SPR knockdown were both attenuated by NAC (Figure 5D and 5E). Taken together, these results indicate that the ROS scavenger NAC partially reverses SPR knockdown-induced apoptosis and proliferation inhibition.
Figure 5.
NAC partially reverses the SPR knockdown induced apoptosis and proliferation inhibition. (A) The cells were transfected with siSPR#3 for 4 h and replaced fresh DMEM containing 10% FBS for 24 h, then 5 mM NAC was added and cultured for another 24 h. The cells were harvested, and the intracellular ROS level was measured using a ROS assay kit. (B) The cells were treated as mentioned above in (A), then the cells were stained with PI/Annexin V, and the apoptotic cells were analyzed using flow cytometry. (C) The cells were treated as mentioned above in (A), then the cells were harvested and the caspase-3, cleaved-caspase-3, caspase-9, cleaved-caspase-9, PARP, and cleaved-PARP expressions were analyzed using western blot. (D) The cells were transfected with siSPR#3 for 4 h and replaced with fresh DMEM containing 10% FBS for 48 h to 80% confluence, then 1000 cells were seeded into 6-well plates, and the medium was replaced with fresh medium with or without 5 mM NAC every 2 days for 14 days. Then the cells were stained with crystal violet solution. (E) The cells were treated as mentioned above in (D) and the number of cells was counted every day for 3 days. The data are represented as the mean ± SD in triplicate. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
Although the biological functions of SPR in BH4 biosynthesis have been well studied, SPR’s potential roles and underlying mechanisms in the progression of cancers are still not widely recognized, including in breast cancer. In our present study, we compared the expressions between human breast cancer tissues and their corresponding pericarcinomatous tissues and investigated the role of SPR in the development of breast cancer cells. According to our results, the SPR expression in breast cancer tissues was significantly higher than in their corresponding pericarcinomatous tissues. The knockdown of SPR inhibited the production of BH4 and induced ROS-mediated apoptosis through the mitochondrial pathway. Importantly, the knockdown of SPR obviously suppressed the proliferation of breast cancer cells. These anti-breast cancer effects of SPR knockdown were attenuated by ROS scavenger NAC, which highlighted the role of ROS in the SPR knockdown-mediated anti-breast cancer effect. Thus, our study provides evidence for SPR as a therapeutic target for breast cancer.
Previous studies have clarified the role of SPR in NB [8] and hepatocellular carcinoma [9]; however, the underlying mechanisms still were not fully illustrated. BH4 can also function as an anti-oxidant [13-15], but the role of SPR in regulating oxidative stress in cancer cells, including breast cancer cells, is still unclear. Our results showed that the knockdown of SPR causes reduced BH4 production and increased ROS levels inside breast cancer cells. ROS is a common inducer of mitochondrial pathway-mediated apoptosis [19]. Bcl-2 is an anti-apoptotic molecule, and Bax, caspase-3, caspase-9 are pro-apoptotic molecules. When mitochondrial apoptotic pathway is activated, MMP will decrease and Cytochrome C will be released to cytosol [20]. In present study, knockdown of SPR promoted the expression of activated caspase 3, caspase-9, and Bax, inhibited the expression of Bcl-2. In addition, the cytochrome C released from the mitochondria to cytosol was also increased by the SPR knockdown. These results strongly indicate that the SPR knockdown caused the mitochondrial pathway-mediated apoptosis. Thus, our study demonstrates the role SPR plays in the proliferation of breast cancer cells and provides a possible therapeutic strategy for breast cancer.
Acknowledgements
This research project was financially supported by the Key Project of Education, Department of Anhui Province (no. KJ2017A245 and no. KJ2019A0342), the Science and Technology Innovation Guidance Project of Bengbu (no. 20190309), and the Science and Technology Development Fund of Bengbu Medical College (no. BYKF1879).
Disclosure of conflict of interest
None.
References
- 1.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang XL, Liu HZ, Shao YX, Peng MZ, Zhang W, Li D, Li XZ, Cai YN, Tan T, Lu XS, Xu JN, Su XY, Lin YT, Liu ZC, Huang YG, Zeng CH, Tang YP, Liu L. A novel GTPCH deficiency mouse model exhibiting tetrahydrobiopterin-related metabolic disturbance and infancy-onset motor impairments. Metabolism. 2019;94:96–104. doi: 10.1016/j.metabol.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Watschinger K, Werner ER. Alkylglycerol monooxygenase. IUBMB Life. 2013;65:366–72. doi: 10.1002/iub.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey JD, Diotallevi M, Nicol T, McNeill E, Shaw A, Chuaiphichai S, Hale A, Starr A, Nandi M, Stylianou E, McShane H, Davis S, Fischer R, Kessler BM, McCullagh J, Channon KM, Crabtree MJ. Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep. 2019;28:218–230. e7. doi: 10.1016/j.celrep.2019.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho YR, Kim SH, Ko HY, Kim MD, Choi SW, Seo DW. Sepiapterin inhibits cell proliferation and migration of ovarian cancer cells via down-regulation of p70S6K-dependent VEGFR-2 expression. Oncol Rep. 2011;26:861–7. doi: 10.3892/or.2011.1335. [DOI] [PubMed] [Google Scholar]
- 6.Chen LY, Zeng X, Wang JH, Briggs SS, O’Neill E, Li JL, Leek R, Kerr DJ, Harris AL, Cai SJ. Roles of tetrahydrobiopterin in promoting tumor angiogenesis. Am J Pathol. 2010;177:2671–80. doi: 10.2353/ajpath.2010.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronin SJF, Seehus C, Weidinger A, Talbot S, Reissig S, Seifert M, Pierson Y, McNeill E, Longhi MS, Turnes BL, Kreslavsky T, Kogler M, Hoffmann D, Ticevic M, da Luz Scheffer D, Tortola L, Cikes D, Jais A, Rangachari M, Rao S, Paolino M, Novatchkova M, Aichinger M, Barrett L, Latremoliere A, Wirnsberger G, Lametschwandtner G, Busslinger M, Zicha S, Latini A, Robson SC, Waisman A, Andrews N, Costigan M, Channon KM, Weiss G, Kozlov AV, Tebbe M, Johnsson K, Woolf CJ, Penninger JM. The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature. 2018;563:564–568. doi: 10.1038/s41586-018-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange I, Geerts D, Feith DJ, Mocz G, Koster J, Bachmann AS. Novel interaction of ornithine decarboxylase with sepiapterin reductase regulates neuroblastoma cell proliferation. J Mol Biol. 2014;426:332–46. doi: 10.1016/j.jmb.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Du HZ, Zhan MX, Wang HX, Chen P, Du DY, Liu XY, Huang XX, Ma PC, Peng DZ, Sun L, Yuan ST, Ding J, Lu LG, Jiang JW. Sepiapterin reductase promotes hepatocellular carcinoma progression via FoxO3a/Bim signaling in a nonenzymatic manner. Cell Death Dis. 2020;11:248. doi: 10.1038/s41419-020-2471-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding SA, Jemal A, Siegel RL. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 11.Moncada S, Palmer RM, Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991;43:109–42. [PubMed] [Google Scholar]
- 12.Furchgott RF, Vanhoutte PM. Endothelium-derived relaxing and contracting factors. Wien Klin Wochenschr. 1991;103:405–11. [PubMed] [Google Scholar]
- 13.Kojima S, Ona S, Iizuka I, Arai T, Mori H, Kubota K. Antioxidative activity of 5,6,7,8-tetrahydrobiopterin and its inhibitory effect on paraquat-induced cell toxicity in cultured rat hepatocytes. Free Radic Res. 1995;23:419–30. doi: 10.3109/10715769509065263. [DOI] [PubMed] [Google Scholar]
- 14.Vásquez-Vivar J, Hogg N, Martásek P, Karoui H, Pritchard KA Jr, Kalyanaraman B. Tetrahydrobiopterin-dependent inhibition of superoxide generation from neuronal nitric oxide synthase. J Biol Chem. 1999;274:26736–42. doi: 10.1074/jbc.274.38.26736. [DOI] [PubMed] [Google Scholar]
- 15.Ihlemann N, Rask-Madsen C, Perner A, Dominguez H, Hermann T, Køber L, Torp-Pedersen C. Tetrahydrobiopterin restores endothelial dysfunction induced by an oral glucose challenge in healthy subjects. Am J Physiol Heart Circ Physiol. 2003;285:H875–82. doi: 10.1152/ajpheart.00008.2003. [DOI] [PubMed] [Google Scholar]
- 16.Bleicken S, Wagner C, García-Sáez AJ. Mechanistic differences in the membrane activity of Bax and Bcl-xL correlate with their opposing roles in apoptosis. Biophys J. 2013;104:421–31. doi: 10.1016/j.bpj.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalupsky K, Cai H. Endothelial dihydrofolate reductase: critical for nitric oxide bioavailability and role in angiotensin II uncoupling of endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2005;102:9056–61. doi: 10.1073/pnas.0409594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oak JH, Cai H. Attenuation of angiotensin II signaling recouples eNOS and inhibits nonendothelial NOX activity in diabetic mice. Diabetes. 2007;56:118–26. doi: 10.2337/db06-0288. [DOI] [PubMed] [Google Scholar]
- 19.Badrinath N, Yoo SY. Mitochondria in cancer: in the aspects of tumorigenesis and targeted therapy. Carcinogenesis. 2018;39:1419–1430. doi: 10.1093/carcin/bgy148. [DOI] [PubMed] [Google Scholar]
- 20.Hoye AT, Davoren JE, Wipf P, Fink MP, Kagan VE. Targeting mitochondria. Acc Chem Res. 2008;41:87–97. doi: 10.1021/ar700135m. [DOI] [PubMed] [Google Scholar]