Abstract
Cervical cancer is one of the foremost common cancers in women. Human papillomavirus (HPV) infection remains a major risk factor of cervical cancer. In addition, numerous other genetic and epigenetic factors also are involved in the underlying pathogenesis of cervical cancer. Recently, it has been reported that apolipoprotein B mRNA editing enzyme catalytic polypeptide like (APOBEC), DNA-editing protein plays an important role in the molecular pathogenesis of cancer. Particularly, the APOBEC3 family was shown to induce tumor mutations by aberrant DNA editing mechanism. In general, APOBEC3 enzymes play a pivotal role in the deamination of cytidine to uridine in DNA and RNA to control diverse biological processes such as regulation of protein expression, innate immunity, and embryonic development. Innate antiviral activity of the APOBEC3 family members restrict retroviruses, endogenous retro-element, and DNA viruses including the HPV that is the leading risk factor for cervical cancer. This review briefly describes the pathogenesis of cervical cancer and discusses in detail the recent findings on the role of APOBEC in the molecular pathogenesis of cervical cancer.
Keywords: Cytidine deaminase, DNA/RNA editing, HPV, Viral restriction, Mutation
Highlights
-
•
APOBEC enzymes deaminate cytidine to uridine and control diverse biological processes including viral restriction.
-
•
APOBEC3, DNA/RNA-editing enzyme plays an important role in the molecular pathogenesis of cervical cancer.
-
•
APOBEC3-mediated DNA editing leads to the accumulation of somatic mutations in tumors and HPV genome.
-
•
Deregulation of APOBEC3 family genes cause genomic instability and result in drug resistance, and immune-evasion in tumors.
1. Introduction
Cervical cancer is the fourth most common cancer in women and accounts for ~5,70,000 new cases and ~3,11,000 deaths worldwide in the year 2018 [1]. Persistent infection with high-risk HPVs remains a major risk factor of cervical cancer [2]. Most HPV infections are cleared within months by the immune system but a few high-risk subtypes like HPV16 and HPV18 persist and express viral oncogenes E6 and E7 which lead to increased genomic instability, accumulation of somatic mutations, and integration of HPV into the host genome resulting in cervical cancer [3]. The uterine cervix displays various pathophysiological conditions which include, cervical incompetence, pelvic inflammatory disease (PID), cervicitis, cervical polyps, cervical warts, cervical dysplasia, and cervical cancer. Cervical warts and cervical cancer are caused by HPV infection. Persistent HPV infection progresses from cervical warts to benign cervical dysplasia and cervical intraepithelial neoplasia (CIN) into invasive cervical carcinoma [4]. In this review, we discussed the role of APOBECs in the restriction of HPV and the accumulation of somatic mutation and the associated genetic factors involved in cervical cancer pathogenesis.
2. Cervical cancer and HPV
Cervical cancer is dived into two types: cervical squamous cell carcinomas (CSCCs), which is derived from squamous cells, and cervical adenoma, arising from the glandular cells of the cervix [4]. The CSCCs account for more than 80% of cervical cancers with higher morbidity and mortality. It is developed through a defined series of preneoplastic lesions to increasing cellular dysplasia, CIN, and to squamous cell carcinoma [5]. Human papillomaviruses belonging to the family Papillomaviridae were shown to be strongly associated with cervical cancer. More than 200 types of papillomaviruses have been detected in humans and among them, ~40 HPV types can infect the epithelial and mucosa lining of the anogenital tract. The genital and mucosal types belong to the genus alpha-papillomavirus that also contains some cutaneous types, which cause genital warts (condyloma). The CSCC-associated HPVs are divided into low-and high-risk types which are mainly causing genital warts and associated with invasive cervical cancer types, respectively [2,3]. Of the high-risk HPV types, HPV 16 and 18 are responsible for around 90% of all cervical cancer cases. Women infected with high-risk types 16 and 18 have a greater chance of progressing to invasive cancer than those infected with low-risk HPV types. Despite the low or high risk, all the HPVs infect the epithelial cells particularly keratinocytes and immortalize them [5].
The genome of all HPVs contains the following three functional parts: the early (E) region that encodes proteins (E1–E7) necessary for viral replication; the late (L) region that encodes the structural proteins (L1–L2) that are required for virion assembly; and a largely non-coding part that is referred to as the long control region, which contains cis-elements that are necessary for the replication and transcription of viral DNA. E1 and E2 proteins initiate viral DNA replication and also act as transcriptional activators [6]. The virus expresses early genes (E5, E6, and E7). The E5 protein induces evasion of immune response and relieves the cellular dependence on growth factors and hence leading to enhanced cell proliferation [7]. The E6 protein binds with cellular tumor suppressor protein p53 and, induces degradation of the p53 protein [8,9]. The E7 protein forms complex with retinoblastoma protein (pRb) and degrade the pRb through the ubiquitin-proteasome pathway [[10], [11], [12]]. The next step is to infect the supra-basal layer where, the virus expresses the late genes, initiating replication of the circular viral genome, and formation of structural proteins. As the virus reaches the upper layers of the epidermis or mucosa, complete viral particles are assembled and released. Viral capsid proteins (L1 and L2) are expressed to assemble the virus progeny in cells upon their differentiation and E4 is also expressed to complete its life cycle when the infected cell enters the upper epithelial layers [13]. HPVs also alter the expression of multiple genes and activate various pathways including growth factor-mediated receptor, Notch, RAS, and PI3K/Akt/mTOR pathway to stimulate host cell survival and proliferation leading to carcinogenesis [7].
3. APOBEC: a mutator
Over time, various somatic mutations accumulate in a cell and most of them are “benign mutations” which are biologically neutral and not implicated in cancer development [14,15]. Along with many other mutations there exist “driver mutations” which lead to the progression of a transformed clone. These mutations fuel tumor adaptation, escape from immunosurveillance, and render treatment resistance [16]. These multiple mutational processes can be due to endogenous and exogenous mutagen exposures, aberrant DNA editing, replication errors, and defective DNA repair. The propensity of increased tumor mutations by aberrant DNA editing has been attributed to the activity of APOBEC DNA-editing proteins [14,15,17,18].
The APOBEC-induced mutagenesis promotes divergence in the genome which often results in evolving many variants with drug resistance, and immune-escape capacity [16]. A recent integrated bioinformatic and biochemical analyses of mesoscale genomic features in the mutational landscape of different human cancers affirmed that various recurrent cancer-associated hotspot mutations earlier claimed as “driver mutations” are possible “passenger mutations”. These “passenger hotspot mutations” are likely to be induced by the APOBEC3A in the DNA stem-loops. On the other hand, the APOBEC-signature recurrent mutations found outside of stem-loops were reported to be accumulated in many validated driver genes and may anticipate new driver genes in cancer [19].
APOBEC genes are a superfamily of cytidine deaminases that deaminate cytidine to uridine in DNA and RNA controlling diverse biological processes including viral restriction. APOBEC family genes are often deregulated in cancers resulting in undesirable mutagenesis and their overexpression causes genomic instability in cancer cells making them the most significant proto-oncogenes. Studying the expression and functions of APOBECs in cancer cells and their ability to induce somatic mutation is being explored more in recent times. Moreover, germline variations present in these genes augment their mutagenic ability making them more prolific mutators, and these mutations likely to fuel the tumorigenesis [20].
3.1. APOBEC family genes and cytosine deamination
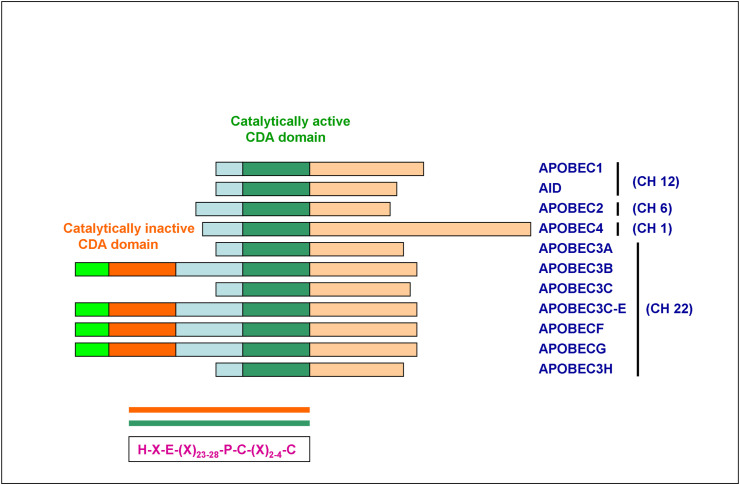
A total of 11 APOBEC family genes are present in the human genome. Activation-induced cytidine deaminases (AID s) including APOBEC1 and APOBEC4 are on chromosome 12 and 1, respectively, and APOBEC2 on chromosome 6. APOBEC3 gene cluster comprising of 7 members (APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D E, APOBEC3F, APOBEC3G, and APOBEC3H) reside at the chromosomal loci 22q13. Each member of the APOBEC family exerts a differential function at the cellular level [21] (Fig. 1 & Table 1 ).
Fig. 1.
Structural organization of APOBEC family genes
The schematic diagram shows the protein structures of the APOBEC family members. The domains and position of the evolutionarily conserved residues are shown. The color-coded in orange is the catalytically inactive CDA domain and green is the catalytically active CDA domain covalently linked on the same peptide in APOBEC3 family members 3B, 3C-E, 3F, and 3G. Their chromosomal localization is mentioned on the right side. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Genomic structure, function and sequence specificity of APOBEC superfamily.
| Gene | Chromosomal location | No of exons | Function | Tissue specific expression | Cellular localization | Target sequence | Strand bias | Deaminase substrate |
|---|---|---|---|---|---|---|---|---|
| AID | 12p13 | 5 | Immunoglobin diversification | Activated B cells | Nuclear but predominantly cytoplasmic | 5′ WRCY | Non-transcribed strand during transcription | ssDNA |
| APOBEC1 | 12p13.1 | 5 | Lipid metabolism and transport | Gastrointestinal tract | Nuclear but predominantly cytoplasmic | 5′ AC | – | ssDNA, RNA |
| APOBEC2 | 3 | Mitochondrial function | Differentiated skeletal and cardiac muscle | Unknown | ||||
| APOBEC3A | 22q13.1 | 5 | Viral restriction | Monocytes/macrophages, non-progenitor cells | Cell wide | 5′ TCW | Lagging strand | ssDNA, RNA |
| APOBEC3B | 22q13.1 | 8 | Viral restriction | IFN/-activated liver cells | Nuclear | 5′ TCW | Lagging strand | ssDNA |
| APOBEC3C | 22q13.1 | 4 | Viral restriction | Immune centers, peripheral blood cells | Cell wide | 5′ TCW | Deamination of C > U occurs at minus strand viral DNA and the resulting G > A mutation is fixed in the plus strand | ssDNA |
| APOBEC3D | 22q13.1 | 7 | Viral restriction | Immune centers, peripheral blood cells | Cytoplasmic | 5′ TCW | ssDNA | |
| APOBEC3F | 22q13.1 | 8 | Viral restriction | Immune centers, peripheral blood cells, IFN/-activated liver cells | Cytoplasmic | 5′ TCW | ssDNA | |
| APOBEC3G | 22q13.1 | 8 | Viral restriction | Immune centers, peripheral blood cells, IFN/-activated liver cells | Cytoplasmic | 5′ CC | ssDNA | |
| APOBEC3H | 22q13.1 | 5 | Viral restriction | Immune centers, peripheral blood cells | Cell wide | 5′ TCW | ssDNA | |
| APOBEC4 | 1q25.3 | 2 | Unknown | |||||
3.2. Structural elements of APOBEC proteins
The APOBEC family members display difference importantly in the catalytic domains (one/two) which contain a conserved zinc-binding deaminase motif, characterized by the conserved amino acid sequences H-X-E-X (23–28)-P-C-X (2–4)-C (X is any amino acid) in which the histidine (H) and cysteine (C) residues bind zinc at the active site and the glutamic acid residue (E) has an important function in proton shuttling during deamination [[21], [22]]. The cytosine is deaminated to uracil by APOBECs through the zinc-mediated hydrolytic mechanism, in which the conserved glutamic acid in the zinc-binding deaminase motif deprotonates water, resulting in zinc-stabilized hydroxide ion which attacks the 4-position of the cytosine nucleobase replacing amine group (NH2) with a carbonyl group (C O). The hallmark property of APOBEC enzymes is their sequence preference at the 5′ of the target cytosine site during deamination which is unique to each APOBEC enzyme – 5′ WRCY for AID1 (W – A or T; R – Purine), 5′ AC for APOBEC1, 5′ CC for APOBEC3G and 5′ TC for all the other enzymes (APOBEC3A, B, C, D, F, and H) [23,24] (Fig. 2 & Table 1).
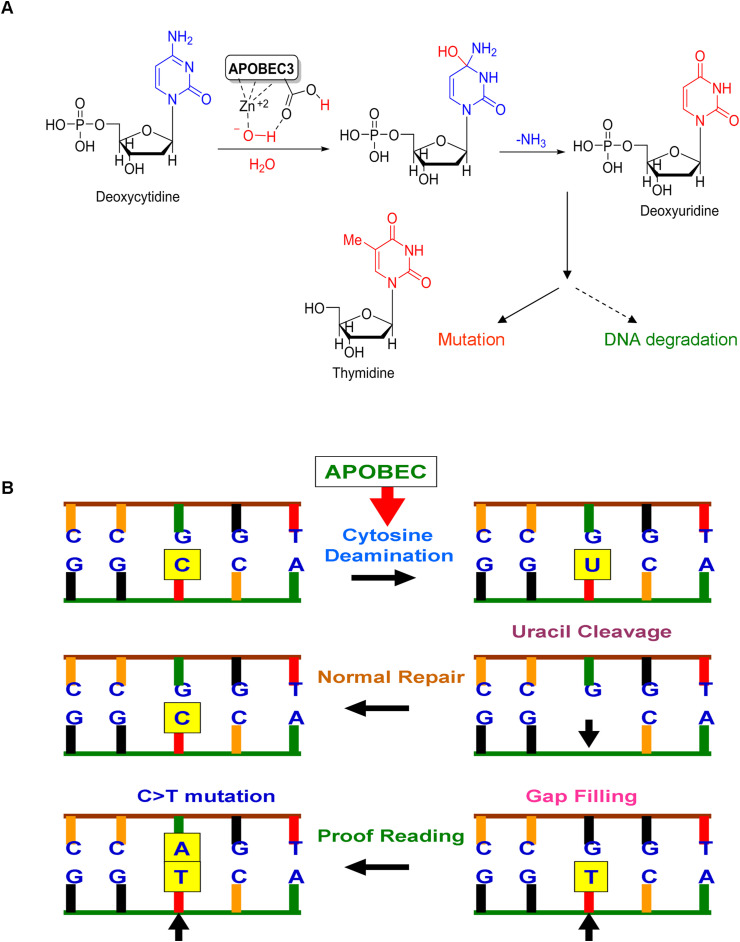
Fig. 2.
Mechanism of APOBEC-mediated cytosine deamination and APOBEC-induced mutations in the host genome.
A) APOBEC-mediated catalytic activity. APOBEC family enzymes catalyze the hydrolytic reaction of cytosine to uracil (C-to-U) and induce DNA degradation or mutations if the APOBEC3-mediated conversion of cytosine to uracil is not repaired. B) Induction of C > T mutation by APOBEC. The illustration shows the C > T mutation resulting from a series of biochemical changes including cytosine deamination and defect in proofreading during replication.
The structural and conformational features of APOBEC proteins dictate their cytosine deamination activity. The core catalytic site of cytidine deaminase (CDA) domain of all 11 APOBECs shares a conserved zinc-dependent deaminase sequence motif (ZDD) within an α-β-α a super-secondary structural element that comprises a five-stranded mixed β sheet that is stabilized by α helices (Fig. 1 & Table 1). Variations in the length, composition, and spatial location of these conserved secondary structural features regulate the function, substrate selection, and catalytic deamination of APOBEC enzymes. Particularly, the loops adjacent to the catalytic site determine the functional differences in substrate recognition and catalytic site interaction with the substrate. Loop 7 has a conserved sequence motif which is the principal determinant of sequence specificity in AID/APOBEC enzymes with additional interactions with loop 1, 3, and 5 [25].
The interaction of single-strand DNA/RNA with APOBECs is mediated through shallow grooves on the protein surface that leads to the catalytic site which is lined with patches of positively charged (basic) and aromatic (hydrophobic) amino acid residues that stabilize interactions with the negatively charged nucleic acid backbone and stack with nucleic acid bases, respectively [24]. In addition, many of APOBECs form functionally relevant high molecular weight homo-and hetero complexes which are required for their catalytic activity. For example, APOBEC1 forms complex with APOBEC1 complementation factor (A1CF) which is an RNA binding protein co-factor, AID is functionally active in both homodimer and monomer forms. APOBEC2, APOBEC3A, and APOBEC3C are monomeric in cells while APOBEC3B, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H are multimeric. Such oligomerization influences the orientation of loops involved in nucleic acid binding and catalytic site access regulating the kinetics of deamination [26,27].
3.3. Functions of APOBEC
The APOBECs are not essential for the development and growth but they control diverse vital biological processes including immunity, viral restriction, metabolism, and regulation of gene expression. The first member of this family is APOBEC1, which deaminates apolipoprotein B messenger RNA to generate a premature stop codon resulting in two forms of proteins ApoB-100 and ApoB-48 which affects lipid metabolism and transport [28]. The AID plays a key role in adaptive immunity causing multiple mutations required for immunoglobin diversification and recombination processes known as somatic hypermutation (SHM), class switch recombination (CSR), and gene conversion (GC) [29]. APOBEC3 subfamily proteins contribute to innate immunity by restricting viral infection (both DNA and RNA viruses) and retrotransposition. Among APOBEC3 proteins, APOBEC3G shows higher-level expression than other APOBEC3s contributing anti-HIV activity while APOBEC3D, 3F, and 3H also contribute to HIV restriction activity [30]. Particularly, APOBEC3A, 3B, 3C, 3F, and 3G restrict DNA viruses including hepatitis B virus (HBV), HPV, adeno-associated virus (AAV), herpes simplex-1 (HSV-1), and Epstein−Barr virus (EBV) [31]. In addition, APOBEC3 proteins inhibit the transposition of two major classes of retroelements including elements with long terminal repeats (LTRs) and non-LTR retroelements such as long interspersed nuclear elements (LINE) and short interspersed nuclear elements (SINE). The AID may play a role in DNA methylation changes at certain loci during cellular differentiation and establishment of pluripotency [32]. The APOBEC mutational signatures have been shown to be enriched in tumor subclones, suggesting a vital role in fueling subclonal expansions that result in tumor divergence particularly intratumor heterogeneity and hence targeting the APOBEC family members may pave way for inhibiting the evolution, adaptation, and drug resistance of tumor [33]. This phenomenon was mainly observed in HIV. The APOBEC-induced mutagenesis promotes divergence in the genome which often results in evolving many variants with drug resistance, and immune-escape capacity [16]. A recent integrated bioinformatic and biochemical analyses of mesoscale genomic features in the mutational landscape of different human cancers affirmed that various recurrent cancer-associated hotspot mutations earlier claimed as “driver mutations” possibly “passenger mutations”. These “passenger hotspot mutations” are likely to be induced by the APOBEC3A in the DNA stem-loops. These results suggest that studies planning to identify disease/cancer-associated mutations need to integrate mesoscale genomic features into computational models [19].
The biological function of APOBECs is greatly regulated by cellular localization and nucleocytoplasmic trafficking. The enzymes, APOBEC1 and AID have nuclear localization signals (NLS) and chromosomal maintenance 1 (CRM1)-dependent nuclear export signals (NES) to execute the editing of mRNA and immunoglobin genes, respectively. When not needed, APOBEC1 is retained in the cytoplasm by interacting with A1CF and AID with eukaryotic elongation factor 1α (eEF1α) and heat shock protein 90 (Hsp90). The APOBECs which involve viral restriction are retained in the cytoplasm through cytoplasmic retention signal (CRS) whereas APOBEC3B is predominantly localized in the nucleus due to the presence of NLS [24]. APOBEC3G and 3F notably localize in cytoplasmic microdomains (non-membrane structures) called mRNA-processing bodies or P-bodies [34]. Therefore, regulation of APOBECs expression and their subcellular localization affects the host genomic integrity with the accumulation of unnecessary mutations due to nuclear localization of APOBECs leading to various diseases including cancer. The APOBEC3A has been implicated in inducing RNA-editing in monocytes and macrophages [35]. Recently, in a murine model of melanoma, it has been demonstrated that APOBEC3B overexpression in tumors could enhance resistance to chemotherapy however, concurrently increases sensitivity to immune checkpoint blockade. Nevertheless, APOBEC3B-mediated mutations could generate heteroclitic neoepitopes in vaccine cells that induced T cell responses de novo manner and this resulted in significant cures in subcutaneous and intracranial tumor models. These results suggested that an increased mutational load in tumor cell vaccines could enhance their immunogenicity to propel therapy for cancer which can be carried out in combination with immune checkpoint blockade [36]. Functional details of APOBEC2 and APOBEC4 are yet to be defined.
4. APOBEC3 in viral restriction
The APOBEC3 genes are implicated in the first-line defense against many exogenous and endogenous retroviruses. Generally, they are interferon-inducible genes, involved in the mechanisms of innate defense against exogenous viruses and endogenous retroelements [37]. The deaminase activity of APOBEC3s involves the removal of the exocyclic amine group from deoxycytidine to form deoxyuridine followed either by the degradation of uracilated viral DNA via UNG2 (uracil DNA glycosylase)-dependent pathway or by the induction of deleterious mutations in the viral genome [34]. Human retroviruses including human immunodeficiency virus (HIV), human T-lymphotropic virus (HTLV), and simian foamy virus (SFV) are often predominantly restricted by APOBEC3G and also by APOBEC3F and APOBEC3H which are highly expressed in immune cells. Upon HIV infection, these APOBEC3s are expressed by CD4+T cells and packed into virions leading to proviral DNA mutations [34]. Deaminase activity of APOBEC3s pertains not only to retroviruses and retroelements but also to DNA viruses. APOBEC3A, 3B, 3C, 3F, and 3G act as restriction factors for DNA viruses, including AAV, HBV, HPV, HSV-1, and EBV [23]. On the other hand, a recent study revealed that retroviruses could drive the rapid evolution of the mammalian APOBEC3 gene. Upon examination of the genomes of 160 mammalian species, Ito et al. identified 1420 AID/APOBEC-related genes and showed that APOBEC3 genes have been amplified in mammals with the history of germline colonization by endogenous retroviruses (ERVs). Furthermore, they also showed that the ERVs genome carried signature mutations in the entire genome. In most of the mammalian species with expanded APOBEC3 repertoires and the ERVs were enriched for G-to-A mutations [38] (Table 2 ). Recently APOBEC was shown to be involved in host-dependent RNA editing of the SARS-CoV-2 transcriptome that it is likely to alter the fate of both coronavirus and the infected human host [39]. It would be interesting to understand whether APOBEC expression is induced by the SARS-CoV-2 virus and its high-level expression has any role in treatment outcome in cancer patients infected with SARS-CoV-2.
Table 2.
Viral restriction property of the seven APOBEC3 enzymes.
| APOBEC3 Genes |
Retro viruses |
Retro elements |
Hepatitis DNA viruses |
Herpes viruses |
Papillomavirus |
|||
|---|---|---|---|---|---|---|---|---|
| HIV-1 | HTLV-1 | HERVs | SFV | |||||
| APOBEC3A | + | + | + | + | ||||
| APOBEC3B | + | |||||||
| APOBEC3C | + | + | + | + | ||||
| APOBEC3D | + | |||||||
| APOBEC3F | + | + | + | |||||
| APOBEC3G | + | + | + | + | + | |||
| APOBEC3H | + | + | + | + | ||||
5. The functional role of APOBEC3 in human cancers
Apart from the restriction of the viral genome, there is an emerging consensus that these APOBEC enzymes could also cause mutations in the cellular genome at replication forks or within transcription bubbles depending on the physiological state of the cell and the cell cycle phase at which they are expressed [23]. As illustrated in Fig. 2, in the normal repair process of APOBEC deamination, cytosine residue is restored again in the DNA sequences by removal of uracil by DNA glycosylase and replacing the cytosine in the abasic site whereas occasionally, the abasic site is replaced with thymine resulting in a GT pair, which will be changed to AT during replication. This will result in a mutation in which a GC pair is replaced by AT [40]. Uncontrolled activities of APOBEC at the cellular level resulting in DNA hypermutation and promiscuous RNA editing associated with the development of cancer [24]. Generally, DNA mismatch repair is driven towards early-replication and gene-rich chromosomal domains. Recently, the DNA mismatch repair was shown to promote APOBEC3-mediated diffuse hypermutation in malignant tumors which is many-fold higher than the common carcinogenic agents including tobacco smoke and UV exposure [41]. Moreover, chromothripsis and kataegis are frequently detected in human cancer. This is likely to arise from the telomere crisis (a period of genome instability during carcinogenesis while exhaustion of the telomere reserve makes unstable dicentric chromosomes). Kataegis observed at chromothriptic breakpoints was demonstrated to be APOBEC3B-dependant while the chromothripsis was shown to be TREX1-driven [42]. Overexpressed APOBECs enhance the propensity for mutation recurrence at specific positions in a distinctive sequence motif in many types of cancer [18]. Conversely, overexpression of APOBEC3B was shown to be associated with improved progression-free survival (PFS) and moderately with overall survival (OS) in clear cell ovarian cancer (CCOC). This has been confirmed with APOBEC3B activity, uracil processing to sensitivity to cisplatin and carboplatin, and the PDX model. It is conceivable that DNA damage caused by APOBEC3B sensitizes the CCOC cells to some additional cisplatin-induced genotoxic stress. It suggests that APOBEC3B could play as a molecular determinant and it can also be a predictive biomarker for the treatment response to platinum-based therapy [43]. A recent study found that endogenous APOBEC3B overexpression could clearly distinguish the HPV-negative low-grade oral epithelial dysplasias and oral cancers as the former showed intermediate APOBEC3B expression levels, while the latter showed higher expression of APOBEC3B levels. Importantly, APOBEC3B levels were highest in grade II and III oral cancers suggesting that APOBEC3B overexpression may provide a marker for advanced grade oral dysplasia and cancer [44].
6. APOBEC signature mutations in human cancer
The somatic mutations occur in cancer mainly due to the multiple mutational processes including the infidelity of DNA replication machinery, exogenous or endogenous mutagen exposures, enzymatic modification of DNA. and defective DNA repair. Such kinds of different mutational processes generate unique combinations of mutation types, termed “mutational signatures”. Mutational signature characterization could generally reveal the episodic of APOBEC mutagenesis in human cancer and recently it has been well-studied in human cancer cell lines [45,46]. Although these recent studies could not bring out initiating factors for the bursts, it concluded that mobilization of retrotransposons may contribute. Catalog of somatic mutations in cancer (COSMIC) provided a comprehensive mutational catalog by analyzing characteristics of mutational patterns and different substitutions (C > A, C > G, C > T, T > A, T > C, and T > G). It classified nearly 30 signature mutations based on the types of substitution, bases immediately 5′ and 3’ to each mutated base, etiology for the mutational processes underlying the signature, and other mutational features.
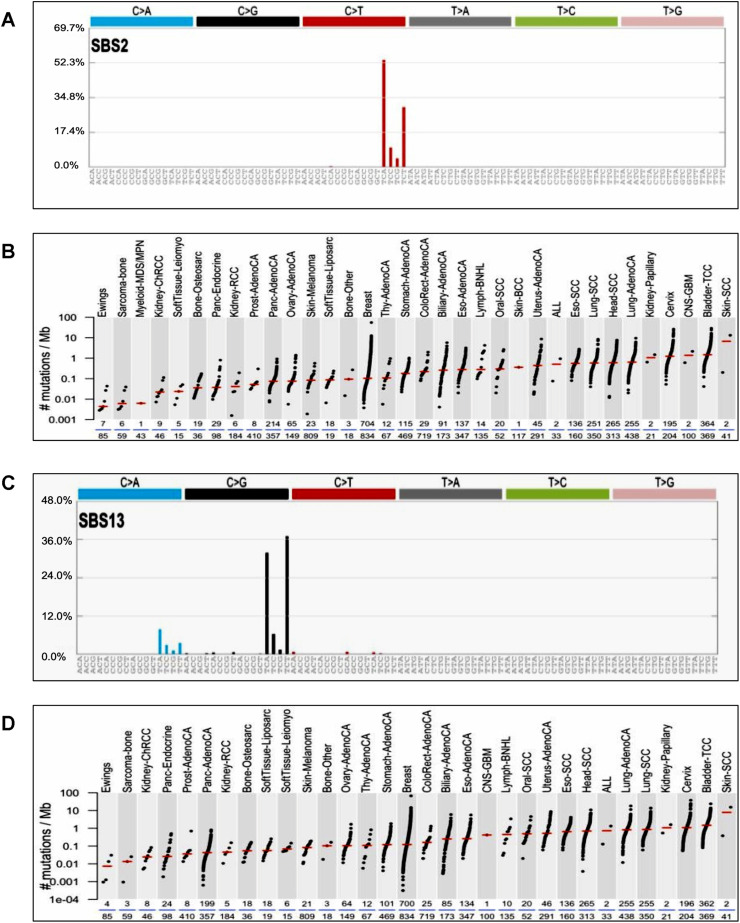
The COSMIC database classified the tumor mutations across the spectrum of human cancer types by analyzing 10,952 exomes and 1048 whole-genomes across 40 distinct types of human cancer using The Cancer Genome Atlas (TCGA), and International Cancer Genome Consortium (ICGC) databases. Among the 30 signature mutations, signature 2 and 13 have been found to be implicated in mutations with the 5′-TCW contexts (W = Adenine or Thymine), and have been attributed to the activity of APOBEC family genes specifically APOBEC1, APOBEC3A, and APOBEC3B in human cancers. Both SBS2 and SBS13 signatures are common in cervical cancers among the various other cancer types (Fig. 3 ). Based on the similarities in the sequence context of cytosine mutations, caused by APOBEC enzymes, APOBEC1, APOBEC3A, and/or APOBEC3B appear more likely to be predominant in human cancers (COSMIC). The TCGA data showed a significant presence of the APOBEC mutation pattern in cervical, bladder, head and neck, and lung cancers, reaching 68% of all mutations in some samples [47]. Among the 7 APOBEC3 family members, both APOBEC3A and APOBEC3B can generate mutations in cancer cells. There is growing evidence establishing the mutagenic ability of these two APOBEC enzymes in various cancer types.
Fig. 3.
APOBEC SBS 2 and 13 signature mutations in cervical cancer.
A) APOBEC SBS2 signature mutations. SBS2 mutations are generated directly by DNA replication across uracil or by error-prone polymerases replicating across abasic sites generated by base excision repair removal of uracil. TCA context mutations are more prevalent >50% than TCC, TCG, and TCT. B) Distribution of APOBEC SBS2 Signature mutations found in human cancers. APOBEC3 SBS2 signature mutations are more frequently observed in cervical and bladder cancers. C) APOBEC SBS13 signature mutations.SBS13 mutations are generated by error-prone polymerases (such as REV1) replicating across abasic sites generated by base excision repair removal of uracil. TCA and TCT context mutations are more common in SBS13 signature mutation. D) Distribution of APOBEC SBS13 signature mutations in human cancers.SBS13 is usually found in the same samples as SBS2. APOBEC3 SBS13 signature mutations are commonly observed in cervical and bladder cancers. (The figure was generated using COSMIC data base).
APOBEC3A is a single-domain enzyme (only one zinc-dependent cytidine deaminase domains (ZD-CDAs)) with the highest catalytic activity among human APOBEC3 proteins and a known restriction factor for the retroelement LINE-1 and HPV [[48], [49], [50], [51]]. Upon overexpression in heterologous systems, APOBEC3A hypermutate nuclear DNA (nuDNA) resulting in the formation of double-stranded DNA breaks (DSBs) and apoptosis. The group also reported that the repeated APOBEC3A-induced DNA damage will be a major driving force behind iterative somatic mutations across many cancer types [52]. On the other hand, endogenous APOBEC3A expression is reported to be largely specific to myeloid lineage cell types and, upon natural induction by interferon-α, it localizes to the cytoplasmic compartment [53].
APOBEC3B has two ZD-CDAs in which only the carboxyl-terminal CDA has cytosine deamination activity and N-terminal CDA is inactive. Like APOBEC3A, APOBEC3B also plays a crucial role in retrovirus and endogenous retrotransposon restriction by hyperediting complementary DNA intermediates and has been reported to be involved with tumor evolution and metastasis. APOBEC3B is expressed almost exclusively in the nucleus. Elevated APOBEC3B expression was reported in diverse forms of cancer tissues and cell lines and has been associated with poor clinical outcomes for estrogen receptor-positive breast cancer, lung cancer, multiple myeloma, and renal cell carcinoma [53]. Endogenous APOBEC3B has been correlated with elevated levels of genomic uracil, increased mutation frequencies, and C-to-T transitions in breast cancer [54]. APOBEC3B is upregulated and its preferred target sequence is reported to be frequently mutated and clustered in various cancers [55]. In addition, Caval et al. reported that repeated APOBEC3A-induced DNA damage was the major driving force behind iterative somatic mutation across many cancer types revealing the significance of both APOBEC3A and APOBEC3B in cancer mutagenesis [52]. In addition to the antiviral effect, recent pan-cancer transcriptomic analysis established the immune and proliferative function of APOBEC3 [56]. Integrative genomic analyses of APOBEC-mutational signature in multiple human cancers showed that the expression level of the uc011aoc (A3AB deletion isoform, APOBEC3A_B) transcribed from the APOBEC3A/B chimera was associated with a greater burden of APOBEC-mutational signature mainly in breast cancer, while its germline deletion was linked with higher expression of uc011aoc in other cancer types. The deletion was shown to be correlated with increased APOBEC-mutational signature, neo-antigen loads, and T cells (CD8+) composition mostly in breast cancer [57].
7. APOBEC3 and cervical cancer
The APOBEC3 genes play a potential oncogenic role in cancers with viral etiology particularly in cervical cancer [58]. It has been indicated that APOBEC activity is induced in virally infected cells and APOBEC signature mutations occur high frequently in cervical cancer [59]. This implies that the APOBEC3 signature in CSCC may indeed be related to human papilloma viral infections. Cells infected with HPV, launch an innate immune response by releasing cytokines such as interferons and interleukins which trigger the expression of AID/APOBEC proteins [60]. Infection of the keratinocyte cell line with HPV E6 results in increased expression of APOBEC3A, APOBEC3B, and APOBEC3H as a result of the cytokine-mediated inflammatory response which in turn hyperedit HPV DNA and induce off-target damage to cellular DNA [61]. Periyasamy and colleagues reported that loss of p53 activity through its mutation or HPV-16 E6/E7-mediated downregulation increased the expression of APOBEC3B and promote the increased mutagenic capacity of normal and cancer cells. Generally, p53 represses A3B expression and cytosine deaminase activity in cancer cells, through a p21-dependent mechanism and that loss of p53 activity through its mutation or HPV-16 E6/E7-mediated downregulation causes APOBEC3B upregulation [62].
Furthermore, transient expressions of HPV E6 and E7 proteins have each been demonstrated to enhance the expression of APOBEC3 family members. The APOBEC3A was shown to restrict infection with artificially-generated HPV16 pseudovirions while the APOBEC3A activity was unblocked by HPVs. Moreover, the HPV genomes contain lesser than the predicted APOBEC3 recognition-sites [61,[63], [64], [65], [66]]. A recent study showed that thousands of HPV16 variants that exhibit various nucleotide changes are consistent with APOBEC3-mediated action. With no exception, APOBEC3-mediated mutagenesis extensively causes various mutational drift. Besides, it has been shown that frequent nucleotide changes in the host cellular genome are caused by the expression of APOBEC3. Therefore, HPV sequence integration during malignant transformation is accompanied by enhanced APOBEC3A levels [67]. Consistently, not only cervical squamous cell carcinomas but also other HPV-associated malignancies exhibit APOBEC3 mutational signatures [47,54,55,68]. A study comparing the HPV+ and HPV- head and neck squamous cell carcinoma found significantly increased expression of APOBEC3B and increased level of TCW mutations in HPV positive tumors than HPV negative tumors suggesting a strong link between HPV and APOBEC-mediated mutagenesis of cellular genes [59]. On the other hand, keratinocyte differentiation has been shown to induce APOBEC3A, B, and mitochondrial hypermutation establishing the hypermutations in the carcinogenesis of cervical cancer [69]. A recent study identified four de novo mutational signatures, among them one matched with the APOBEC-associated single base substitution 2 (SBS2). Furthermore, HPV-associated cervical infections pose a bigger burden of somatic HPV16 APOBEC3-induced mutations however, they are more likely to be non-malignant or consecutively they are cleared suggesting that APOBEC3-induced somatic mutations in the HPV16 genome were significantly correlated with viral clearance [70,71]. It has been demonstrated that APOBEC3A and HPV16 E7 protein could interact with CUL2, suggesting that the E7-CUL2 complex formed during HPV infection may regulate cellular APOBEC3A levels and E7-stabilized APOBEC3A had intact enzymatic activity. This finding suggests that the HPV E7 deregulates APOBEC3A levels and hence provides the molecular mechanism behind the cellular triggering of APOBEC3 mutations in HPV-positive malignancies [72].
7.1. Host genetic variations in HPV-induced cervical cancer
HPV exhibits a high degree of cellular tropism for squamous epithelial cells of the cervix and is associated with clinical manifestations ranging from benign hyperplastic lesions to invasive cancer [5]. Prolonged HPV infection in squamous cells makes CSCC more commonly occurred (80–85%) than adenocarcinoma (10–15%) [73]. Genetic variations of the host could potentially contribute to the inter-individual and inter-ethnic differences in HPV persistence and cervical cancer risk. The polymorphisms in the host genetic factors and their corresponding protein products, either alone or in cooperation with HPV, confer a proliferative advantage to the cancer cell. These genetic variations also form complex interactions with various endogenous and environmental factors, significantly influencing cervical carcinogenesis [74]. In addition to inherited genetic variations, somatically acquired genetic aberrations induce malignant transformation of a neoplastic clone.
The development of cervical cancer and the persistence of HPV infection depends on inherited genetic factors to a significant degree. Storey et al. reported that the p53 codon 72 arginine variant was more susceptible to HPV E6-mediated degradation and individuals carrying homozygous arginine alleles had a higher chance to have an HPV-associated squamous cell carcinoma of the cervix [75]. Cervical carcinoma displays familial aggregation and a pattern of decreasing familial relative risk correlating with the degree of biological relatedness, supporting the idea that genetic factors are a major cause of familial aggregation. Many genetic risk factors have been identified but with weak effects on carcinogenesis. The most prominent among the known risk factors are the human leukocyte antigen (HLA) class II haplotypes associated with increased risk [76]. Several other candidate genes that are part of different pathways have also been suggested to influence cervical cancer heritability. Wang et al. studied the effect of 7146 single-nucleotide polymorphisms (SNPs) in 305 genes with a presumed function in DNA repair, viral infection and, cell entry, on cervical cancer and found that polymorphisms in 4 DNA repair genes (FANCA, EXO1, CYBA, and XRCC1) and 2 immune response genes (IRF3 and TLR2) were significantly associated with CIN3 or cervical cancer when compared to disease-free controls. Polymorphisms, S427T in IRF3, and Q399R in XRCC1 were significantly associated with HPV persistence when women with persistent HPV infection were compared with random control subjects [77].
Many studies focused more on the association of inherited variations in immune response genes with cervical cancer risk as host innate and adaptive immune responses are responsible for the regression, persistence, or progression of HPV infection [78]. In relevance to persistent HPV infection and cervical carcinogenesis, a family of APOBEC enzymes with DNA/RNA editing capability has been analyzed as they are involved in various immune functions including restriction of viral replication, antigen presentation, and maturation of host immune receptors which are highly polymorphic in nature [79]. DNA editing ability of APOBECs increases undesirable mutations in cancer cells and this could be augmented its polymorphic nature. These results suggest that inherited genetic variations in APOBEC family genes contribute to HPV persistence and the accumulation of somatic mutations aiding cervical carcinogenesis.
7.2. Genome-wide association studies (GWAS): single nucleotide polymorphisms (SNPs) implicated in cervical cancer risk
Despite heritable genetic factors in cervical cancer, hypothesis-driven candidate gene studies in cervical cancer were mostly inconsistent or deficient in replicating independently. However, genome-wide association studies (GWAS) revealed several novel susceptibility loci that were previously not identified in cervical cancer reflecting the strength of GWAS analysis. Subsequent functional analysis and pathway-based analyses of associated SNPs could faithfully provide new insights into the cervical cancer pathogenesis [80]. A meta-analysis of previous research reports with case-control studies on cervical cancer-associated SNPs revealed that there were 42 SNPs associated with the risk of cervical cancer. In addition, CYP1A1 SNP (A4889G, rs1048943) was strongly associated with cervical cancer both in the allele effect model and dominant and recessive effect model. As some SNPs in cervical cancer were tested only in a few studies and they are likely subjected to publication bias thus more studies are required to confirm these loci [81]. Analysis of the association of cervical cancer and HLA haplotype revealed a strong association of cervical cancer risk with protective HLA haplotypes which were decided by the HLA-DRB1 at amino-acid positions 13 and 71 and HLA-B at position 156. Moreover, common genetic variants-based determination of 36% of liability of HPV-associated cervical pre-cancer and cancer showed that women within the cut-off of the top 10% genetic risk scores tend to be >7.1% risk. Those cases in the top 5% get roughly >21.6% risk for cervical cancer developing in the future. However, future studies with a valid screening method may be required to test genetic risk prediction to confirm these results [82]. A recent GWAS study on East Asian cervical cancer populations with 2609 cases and 4712 control samples in the discovery stage and 1461 cases and 3295 control cases in the experimental stage found two novel-significant associations at 5q14 and 7p11 which correspondingly lead two SNPs, rs59661306 and rs7457728, respectively. Further analysis revealed that the former SNP in 5q14 was found to be connected with ARRDC3 (arrestin domain-containing 3) gene promoter and ARRDC3 knockdown in HeLa cells resulted in significantly reduced cell growth which also impaired susceptibility to pseudovirion infection (HPV16). These results suggested that the ARRDC3 may be required for the entry of HPV into the cell [83].
7.3. APOBEC3A/3B deletion polymorphism and cervical cancer
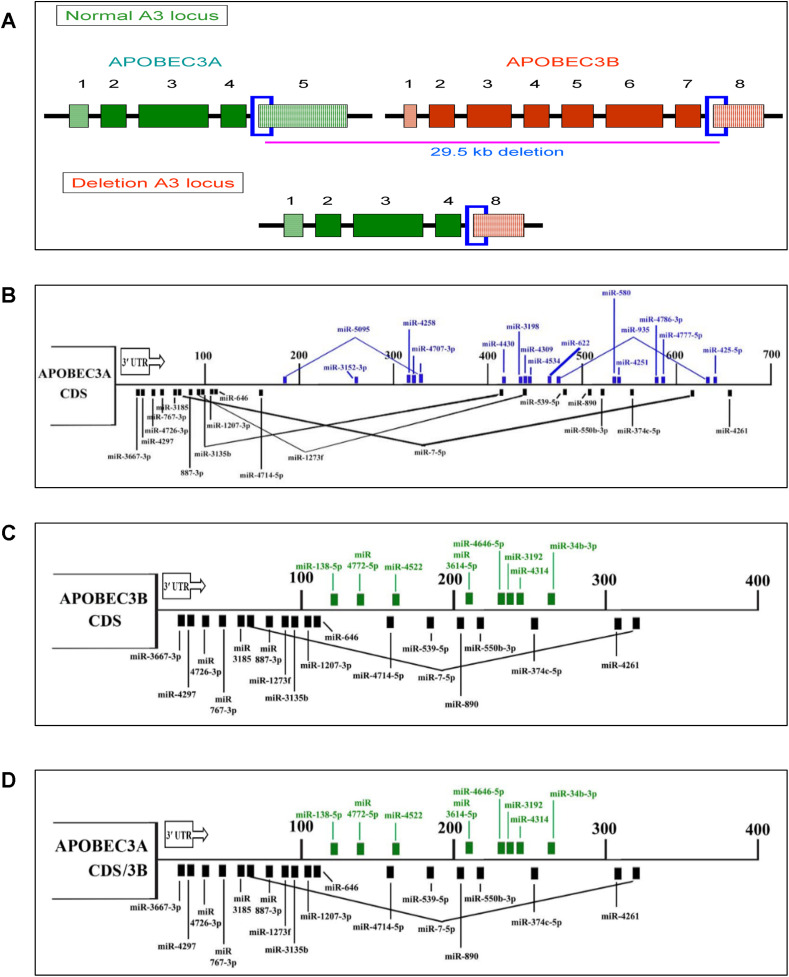
Recently, a common germline deletion polymorphism in the APOBEC3 gene cluster was frequently reported to be involved in increasing cancer susceptibility (Fig. 4 ). This polymorphism is the deletion of a 29.5 kb fragment spanning the fifth exon of APOBEC3A and the eighth exon of APOBEC3B. APOBEC3A and APOBEC3B genes are adjacent to each other and in the same orientation on chromosome 22. The 29.5 kb genomic deletion occurs between identical 370-bp segments spanning intron 4/exon5 of APOBEC3A and intron 7/exon8 of APOBEC3B resulting in a hybrid APOBEC3A/3B gene. This hybrid gene is predicted to transcribe a fusion transcript splicing APOBEC3A coding sequence with the 3′ UTR of APOBEC3B resulting in a complete loss of the APOBEC3B coding sequence. The fusion transcript encodes a protein that has an amino acid sequence identical to APOBEC3A and is more stable, resulting in higher intracellular APOBEC3A levels [52,84] and this could be due to altered post-transcriptional regulation by microRNAs as described later.
Fig. 4.
APOBEC3A/3B deletion polymorphism and miRNA-mediated regulation of APOBEC3A, B and APOBEC3A/B fusion transcripts
A) Schematic diagram of APOBEC3A/3B deletion polymorphism. Illustration on top shows both APOBEC3A (Green color) and APOBEC3B (Orange color) and 29.6 kb deletion border region marked a blue color box. The diagram in the bottom shows the APOBEC3A/3B fusion transcript. B), C) & D) Diagram shows the 3′-UTR map of miRNAs that modulates the post-transcriptional regulation of APOBEC3A, APOBEC3B, and APOBEC3A/B fusion transcript, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The APOBEC3A/3B germline deletion allele confers cancer susceptibility through increased intracellular APOBEC3A levels and this increased activity of APOBEC-dependent mutational processes result in higher mutation burden per cancer genome. The carriers of the deletion allele show more APOBEC signature mutations than non-carriers in a wide range of cancers [52,84]. As listed in Table 3 , a differential frequency of this deletion polymorphism has been reported in various populations; it is rare in African and European populations but common in the East Asian population [85]. The prevalence of APOBEC3A/3B deletion polymorphism has been reported to be 40.1% in Indonesia, 6% in Europe, 37% in East Asians, 34.5% in India, 57.1% in America, and 93% in Oceania [86].
Table 3.
Association of APOBEC3A/3B deletion polymorphism in human cancers.
| S.No | Cancer | Population | Association | Reference |
|---|---|---|---|---|
| 1 | Breast cancer | South East Iranian | Associated | [94] |
| 2 | European | Associated | [84] | |
| 3 | Chinese | Associated | [95] [96] |
|
| 4 | Malaysian | Associated | [96] | |
| 5 | Indian | Associated | ||
| 6 | No | [86] | ||
| 7 | Swedish | No | [88] | |
| 8 | Moroccan | No | [89] | |
| 9 | Dutch | No | [90] | |
| 10 | Norwegian | No | [97] | |
| 11 | Ovarian cancer | Chinese | Associated | [98] |
| 12 | Pancreatic cancer | Chinese | Associated | [99] |
| 13 | Bladder cancer | European | No | [100] |
| 14 | Japanese | No | ||
| 15 | Cervical cancer | Indian | No | [86] |
| 16 | Oral cancer | Indian | No | [86] |
7.4. Significance of APOBEC3A/3B deletion polymorphism in cervical cancer
The APOBEC 3A/3B polymorphism plays a potential oncogenic role in cancers with viral etiology as APOBEC3 enzymes participate in the innate antiviral activity and mediate the restriction of retroviruses, endogenous retro-element, and DNA viruses like HPV. The viral restriction ability of APOBEC3A and APOBEC3B is hindered by the deletion polymorphism which is evidenced by its association with increased viral infection in various populations [87]. Indeed, APOBEC3A/3B deletion polymorphism was reported to reduce the restriction of HPV in cervical cancer [50] in an increased HPV infection in cells with catalytically inactive APOBEC3A [63], suggesting its involvement in promoting carcinogenesis due to decreased HPV restriction and increased somatic mutations.
The APOBEC3A/3B fusion transcript is reported to be more stable and expressed in 10- 20-fold higher than the steady-state levels [52]. The major difference between wild-type APOBEC3A and the fusion APOBEC3A/3B is that the 3′ UTR of the former is replaced with APOBEC3B 3′ UTR. The transfection of the APOBEC3A gene with APOBEC3B 3′ UTR yielded 20-fold higher expression demonstrating that APOBEC3A expression levels of the wild-type and fusion transcripts are primarily controlled by the UTRs [52]. Modulation of post-transcriptional regulation by microRNAs (miRNAs) could be the reason for such overexpression of APOBEC3A/3B fusion transcript.
7.5. Deregulation of miRNA-mediated post-transcriptional regulation of APOBEC3A/3B fusion transcript
MicroRNAs (miRNAs) are a class of small, single-stranded non-coding RNAs of 18–25 nucleotides in length, involved in post-transcriptional regulation of gene expression through partial complementary binding with the 3′ UTR of target mRNAs, leading either to their translational repression or sometimes mRNA degradation. The predicted miRNAs binding to the original transcripts of APOBEC3A, APOBEC3B, and the APOBEC3A/3B fusion transcript were shown in Fig. 4. Analyzing the differential expression of miRNAs potentially binding to both APOBEC3A and APOBEC3B 3′ UTR may be helpful to decipher the reason for highly stable APOBEC3A/3B fusion transcript. However, the role of APOBEC3A/3B deletion polymorphism in carcinogenesis is still uncertain as there are so many reports stating that this particular polymorphism is not associated with cancer risk and other viral infections [[88], [89], [90], [91], [92], [93]] (Table 4 ). Our in-silico analysis of miRNAs regulating APOBEC3A and APOBEC3B showed that 8 miRNAs specific to APOBEC3B might regulate the APOBEC3A/3B fusion transcript instead of the original 17 APOBEC3A-specific miRNAs, that could enhance the stability to the fusion transcript [86]. We also observed that the miRNAs predicted to have stronger repressing ability were shown to be downregulated in many cancers leading to enhanced stability of APOBEC3A/3B mRNA. In our miRNA expression profiling, we further observed that miR-34b-3p was downregulated in cervical tumors which may result in loss of repression of APOBEC3A/3B fusion transcript by miRNA resulting in the increased level of the APOBEC3A/3B fusion protein and somatic mutations [86]. A complete expression profiling of all these predicted miRNAs and analyzing the stability of APOBEC3A/3B fusion transcript may help in elucidating the role of this deletion polymorphism in tumorigenesis.
Table 4.
Association of APOBEC3A/3B deletion polymorphism with viral restriction.
| S.No | Virus | Disease | Population | Association | Reference |
|---|---|---|---|---|---|
| 1 | Hepatitis B Virus (HBV) | Hepatocellular carcinoma | Chinese | Associated | [12] |
| Chronic HBV infection | Japanese | No | [101] | ||
| Moroccan | No | [92] | |||
| 2 | Human Immunodeficiency Virus (HIV) | AIDS | American | Associated | [102] |
| Indonesia | Associated | [87] | |||
| Japanese | No No |
[91] [93] |
|||
| Indian | No | [93] | |||
| 3 | Human papillomavirus | Cervical cancer | Indian | No | [86] |
8. Summary and future prospective
In summary, the APOBEC is the underlying protein molecule in the pathogenesis of cervical cancer through the HPV restriction and accumulation of mutations in cancer driver genes. Moreover, it has been observed that cancer incidence is modulated by the distribution of the APOBEC3A/3B deletion polymorphism in the South East Asian population [52]. But it is counter-intuitive because of less possibility of a higher mutation rate in the complete deletion of APOBEC3B which is a strong nuclear DNA mutator enzyme. Moreover, the association of APOBEC3A/3B deletion polymorphism with cervical squamous cell carcinoma is still not investigated in detail. Therefore, in the future, this deletion polymorphism needs to be investigated in different populations to understand its true association with cancer risk, viral restriction, and mutagenesis.
Author contribution statement
SR and AKM wrote the manuscript. AKM, SR and AK Munirajan participated in its design and coordination. HN and II critically reviewed and edited the manuscript. AK Munirajan conceived the study, read, and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Declaration of competing interest
The authors have no conflict of interest to declare.
Acknowledgments
We apologize to the colleagues whose work could not be cited owing to the limitation of space. The authors wish to thank Dr. G. Arunkumar for critical reading of this manuscript and Dr. A.K. Mohanakrishnan for his help in drawing the chemical structures. This work was supported by NIG-JOINT (1A2019) to Dr. A.K. Munirajan. Dr. S. Revathidevi was supported by a researh fellowship from University Grant Commission, Government of India. We gratefully acknowledge the department/institute infrastructural facilities supported through UGC-SAP, DST-FIST and DHR-MRU grants and individual research grants to Dr. A.K. Munirajan from DBT, ICMR and DAE-BRNS, Government of India.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cruz-Gregorio A., Aranda-Rivera A.K., Pedraza-Chaverri J. Human papillomavirus-related cancers and mitochondria. Virus Res. 2020;286:198016. doi: 10.1016/j.virusres.2020.198016. [DOI] [PubMed] [Google Scholar]
- 3.Pal A., Kundu R. Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front. Microbiol. 2020;10:3116. doi: 10.3389/fmicb.2019.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X., Wang W., Hu K., Zhang F., Hou X., Yan J., et al. Wang W. A risk stratification for patients with cervical cancer in stage IIIC1 of the 2018 FIGO staging system. Sci. Rep. 2020;10:362. doi: 10.1038/s41598-019-57202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willemsen A., Bravo I.G. Origin and evolution of papillomavirus (onco)genes and genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374:20180303. doi: 10.1098/rstb.2018.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham S.V. Keratinocyte differentiation-dependent human papillomavirus gene regulation. Viruses. 2017;9:245. doi: 10.3390/v9090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Wu J., Ling M.T., Zhao L., Zhao K.-N. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol. Canc. 2015;14:87. doi: 10.1186/s12943-015-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werness B.A., Levine A.J., Howley P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 9.Scheffner M., Werness B.A., Huibregtse J.M., Levine A.J., Howley P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 10.Dyson N., Howley P.M., Munger K., Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 11.Munger K., Werness B.A., Dyson N., Phelps W.C., Harlow E., Howley P.M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer S.N., Wazer D.E., Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Canc. Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 13.Maglennon G.A., Doorbar J. The biology of papillomavirus latency. Open Virol. J. 2012;6:190–197. doi: 10.2174/1874357901206010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suda K., Nakaoka H., Yoshihara K., Ishiguro T., Tamura R., Mori Y., Tanaka K. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24:1777–1789. doi: 10.1016/j.celrep.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 15.Moore L., Leongamornlert D., Coorens T.H., Sanders M.A., Ellis P., Dentro S.C., Maura F. The mutational landscape of normal human endometrial epithelium. Nature. 2020;580:640–646. doi: 10.1038/s41586-020-2214-z. [DOI] [PubMed] [Google Scholar]
- 16.Venkatesan S., Rosenthal R., Kanu N., McGranahan N., Bartek J., Quezada S.A., Hare J., Harris R.S., Swanton C. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 2018;29:563–572. doi: 10.1093/annonc/mdy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Stratton M.R. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Stratton M.R. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buisson R., Langenbucher A., Bowen D., Kwan E.E., Benes C.H., Zou L., Lawrence M.S. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. 2019;364 doi: 10.1126/science.aaw2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox E.J., Prindle M.J., Loeb L.A. Do mutator mutations fuel tumorigenesis? Canc. Metastasis Rev. 2013;32:353–361. doi: 10.1007/s10555-013-9426-9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navaratnam N., Sarwar R. An overview of cytidine deaminases. Int. J. Hematol. 2006;83:195–200. doi: 10.1532/IJH97.06032. [DOI] [PubMed] [Google Scholar]
- 22.Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 23.Siriwardena S.U., Chen K., Bhagwat A.S. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 2016;116:12688–12710. doi: 10.1021/acs.chemrev.6b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salter J.D., Bennett R.P., Smith H.C. The APOBEC protein family: united by structure, divergent in function. TIBS (Trends Biochem. Sci.) 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betts L., Xiang S., Short S.A., Wolfenden R., Carter C.W. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Chen Y., Li M., Carpenter M.A., McDougle R.M., Luengas E.M., Macdonald P.J., Harris R.S., Mueller J.D. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J. Mol. Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brar S.S., Sacho E.J., Tessmer I., Croteau D.L., Erie D.A., Diaz M. Activation-induced deaminase, AID, is catalytically active as a monomer on single-stranded DNA. DNA Repair. 2008;7:77–87. doi: 10.1016/j.dnarep.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Contois J.H., McConnell J.P., Sethi A.A., Csako G., Devaraj S., Hoefner D.M., Warnick G.R. Apolipoprotein B and cardiovascular disease risk: position statement from the AACC lipoproteins and vascular diseases division working group on best practices. Clin. Chem. 2009;55:407–419. doi: 10.1373/clinchem.2008.118356. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Woo C.J., Iglesias-Ussel M.D., Ronai D., Scharff M.D. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 30.Chaipan C., Smith J.L., Hu W.-S., Pathak V.K. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J. Virol. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stavrou S., Ross S.R. APOBEC3 proteins in viral immunity. J. Immunol. 2015;195:4565–4570. doi: 10.4049/jimmunol.1501504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez P.M., Teater M., Chambwe N., Kormaksson M., Redmond D., Ishii J., et al. Shaknovich R. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep. 2015;12:2086–2098. doi: 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanton, C., McGranahan, N., Starrett, G.J., & Harris, R.S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Canc. Discov., 5, 704-712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed]
- 34.Willems L., Gillet N.A. APOBEC3 interference during replication of viral genomes. Viruses. 2015;7:2999–3018. doi: 10.3390/v7062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., Baysal B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015;6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Driscoll C.B., Schuelke M.R., Kottke T., Thompson J.M., Wongthida P., Tonne J.M., et al. Vile R.G. APOBEC3B-mediated corruption of the tumor cell immunopeptidome induces heteroclitic neoepitopes for cancer immunotherapy. Nat. Commun. 2020;11:790. doi: 10.1038/s41467-020-14568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira V.C., Soares M.A. The role of cytidine deaminases on innate immune responses against human viral infections. BioMed Res. Int. 2013;2013:683095. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito J., Gifford R.J., Sato K. Retroviruses drive the rapid evolution of mammalian APOBEC3 genes. Proc. Natl. Acad. Sci. U.S.A. 2020;117:610–618. doi: 10.1073/pnas.1914183116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Giorgio S., Martignano F., Torcia M.G., Mattiuz G., Conticello S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2 in humans. Science Advances. 2020 doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhter J., Ali Aziz M.A., Al Ajlan A., Tulbah A., Akhtar M. Breast cancer: is there a viral connection? Adv. Anat. Pathol. 2014;21:373–381. doi: 10.1097/PAP.0000000000000037. [DOI] [PubMed] [Google Scholar]
- 41.Mas-Ponte D., Supek F. DNA mismatch repair promotes APOBEC3-mediated diffuse hypermutation in human cancers. Nat. Genet. 2020;52:958–968. doi: 10.1038/s41588-020-0674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maciejowski J., Chatzipli A., Dananberg A., Chu K., Toufektchan E., Klimczak L.J.…de Lange T. APOBEC3-dependent kataegis and TREX1-driven chromothripsis during telomere crisis. Nat. Genet. 2020;52:884–890. doi: 10.1038/s41588-020-0667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serebrenik A.A., Argyris P.P., Jarvis M.C., Brown W.L., Bazzaro M., Vogel R.I., et al. Harris R.S. The DNA cytosine deaminase APOBEC3B is a molecular determinant of platinum responsiveness in clear cell ovarian cancer. Clin. Canc. Res. 2020;26:3397–3407. doi: 10.1158/1078-0432.CCR-19-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Argyris P.P., Wilkinson P.E., Jarvis M.C., Magliocca K.R., Patel M.R., Vogel R.I., et al. Harris R.S. Endogenous APOBEC3B overexpression characterizes HPV-positive and HPV-negative oral epithelial dysplasias and head and neck cancers. Mod. Pathol. 2020 doi: 10.1038/s41379-020-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarvis M.C., Ebrahimi D., Temiz N.A., Harris R.S. Mutation signatures including APOBEC in cancer cell lines. Journal of National Cancer Institute Cancer Spectr. 2018;2:pky002. doi: 10.1093/jncics/pky002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petljak M., Alexandrov L.B., Brammeld J.S., Price S., Wedge D.C., Grossmann S., Stratton M.R. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell. 2019;176:1282–1294. doi: 10.1016/j.cell.2019.02.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Gordenin D.A. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carpenter M.A., Li M., Rathore A., Lackey L., Law E.K., Land A.M., Leonard B., Shandilya S.M., Bohn M.F., Schiffer C.A., Brown W.L., Harris R.S. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogerd H.P., Wiegand H.L., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vartanian J.P., Guétard D., Henry M., Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 51.Silvas T.V., Hou S., Myint W., Nalivaika E., Somasundaran M., Kelch B.A., Matsuo H., Kurt Yilmaz N., Schiffer C.A. Substrate sequence selectivity of APOBEC3A implicates intra-DNA interactions. Sci. Rep. 2018;8:7511. doi: 10.1038/s41598-018-25881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caval V., Suspène R., Shapira M., Vartanian J.-P., Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat. Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 53.Starrett G.J., Luengas E.M., McCann J.L., Ebrahimi D., Temiz N.A., Love R.P., et al. Harris R.S. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., et al. Harris R.S. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burns M.B., Temiz N.A., Harris R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng J.C.F., Quist J., Grigoriadis A., Malim M.H., Fraternali F. Pan-cancer transcriptomic analysis dissects immune and proliferative functions of APOBEC3 cytidine deaminases. Nucleic Acids Res. 2019;47:1178–1194. doi: 10.1093/nar/gky1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z., Wen W., Bao J., Kuhs K.L., Cai Q., Long J., et al. Guo X. Integrative genomic analyses of APOBEC-mutational signature, expression and germline deletion of APOBEC3 genes, and immunogenicity in multiple cancer types. BMC Med. Genom. 2019;12:131. doi: 10.1186/s12920-019-0579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith N.J., Fenton T.R. The APOBEC3 genes and their role in cancer: insights from human papillomavirus. J. Mol. Endocrinol. 2019;62:R269–R287. doi: 10.1530/JME-19-0011. [DOI] [PubMed] [Google Scholar]
- 59.Henderson S., Chakravarthy A., Su X., Boshoff C., Fenton T.R. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Read S.A., Douglas M.W. Virus induced inflammation and cancer development. Canc. Lett. 2014;345:174–181. doi: 10.1016/j.canlet.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 61.Vieira V.C., Leonard B., White E.A., Starrett G.J., Temiz N.A., Lorenz L.D., et al. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. mBio. 2014;5 doi: 10.1128/mBio.02234-14. e02234-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Periyasamy M., Singh A.K., Gemma C., Kranjec C., Farzan R., Leach D.A., Ali S. p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res. 2017;45:11056–11069. doi: 10.1093/nar/gkx721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warren C.J., Xu T., Guo K., Griffin L.M., Westrich J.A., Lee D., Lambert P.F., Santiago M.L., Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 2015;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z., Wakae K., Kitamura K., Aoyama S., Liu G., Koura M., et al. Muramatsu M. APOBEC3 deaminases induce hypermutation in human papillomavirus 16 DNA upon beta interferon stimulation. J. Virol. 2014;88:1308–1317. doi: 10.1128/JVI.03091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warren C.J., Van Doorslaer K., Pandey A., Espinosa J.M., Pyeon D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evolution. 2015;1:vev015. doi: 10.1093/ve/vev015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warren C.J., Westrich J.A., Van Doorslaer K., Pyeon D. Roles of APOBEC3A and APOBEC3B in human papillomavirus infection and disease progression. Viruses. 2017;9:233. doi: 10.3390/v9080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondo S., Wakae K., Wakisaka N., Nakanishi Y., Ishikawa K., Komori T., Yoshizaki T. APOBEC3A associates with human papillomavirus genome integration in oropharyngeal cancers. Oncogene. 2017;36:1687–1697. doi: 10.1038/onc.2016.335. [DOI] [PubMed] [Google Scholar]
- 68.Wallace N.A., Münger K. The curious case of APOBEC3 activation by cancer-associated human papillomaviruses. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakae, K., Nishiyama, T., Kondo, S., Izuka, T., Que, L., Chen, C.,… Muramatsu M. Keratinocyte differentiation induces APOBEC3A, 3B, and mitochondrial DNA hypermutation. Sci. Rep., 8, 9745. doi: 10.1038/s41598-018-27930-z. [DOI] [PMC free article] [PubMed]
- 70.Mirabello L., Yeager M., Yu K., Clifford G.M., Xiao Y., Zhu B., et al. Raine-Bennett T. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu, B., Xiao, Y., Yeager, M., Clifford, G., Wentzensen, N., Cullen, M.,...Mirabello, L. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun., 11, 886. doi: 10.1038/s41467-020-14730-1. [DOI] [PMC free article] [PubMed]
- 72.Westrich J.A., Warren C.J., Klausner M.J., Guo K., Liu C.W., Santiago M.L., Pyeon D. Human papillomavirus 16 E7 stabilizes APOBEC3A protein by inhibiting cullin 2-dependent protein degradation. J. Virol. 2018;92 doi: 10.1128/JVI.01318-17. e01318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Snijders P.J.F., Steenbergen R.D.M., Heideman D.A.M., Meijer C.J.L.M. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J. Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- 74.Tan S.C., Ankathil R. Genetic susceptibility to cervical cancer: role of common polymorphisms in apoptosis-related genes. Tumour Biology. 2015;36:6633–6644. doi: 10.1007/s13277-015-3868-2. [DOI] [PubMed] [Google Scholar]
- 75.Storey A., Thomas M., Kalita A., Harwood C., Gardiol D., Mantovani F., Banks L. Role of a p53 polymorphism in the development of human papilloma-virus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 76.Ivansson E.L., Juko-Pecirep I., Erlich H.A., Gyllensten U.B. Pathway-based analysis of genetic susceptibility to cervical cancer in situ: HLA-DPB1 affects risk in Swedish women. Gene Immun. 2011;12:605–614. doi: 10.1038/gene.2011.40. [DOI] [PubMed] [Google Scholar]
- 77.Wang S.S., Bratti M.C., Rodríguez A.C., Herrero R., Burk R.D., Porras C., et al. Hildesheim A. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. JID (J. Infect. Dis.) 2009;199:20–30. doi: 10.1086/595563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X., Jiang J., Shen H., Hu Z. Genetic susceptibility of cervical cancer. Journal of Biomedical Research. 2011;25:155–164. doi: 10.1016/S1674-8301(11)60020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moris A., Murray S., Cardinaud S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 2014;5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen D., Gyllensten U. Lessons and implications from association studies and post-GWAS analyses of cervical cancer. Trends Genet. 2015;31:41–54. doi: 10.1016/j.tig.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Wang S., Sun H., Jia Y., Tang F., Zhou H., Li X., et al. Li S. Association of 42 SNPs with genetic risk for cervical cancer: an extensive meta-analysis. BMC Med. Genet. 2015;16:25. doi: 10.1186/s12881-015-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leo P.J., Madeleine M.M., Wang S., Schwartz S.M., Newell F., Pettersson-Kymmer U., et al. Brown M.A. Defining the genetic susceptibility to cervical neoplasia-A genome-wide association study. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takeuchi F., Kukimoto I., Li Z., Li S., Li N., Hu Z., Shi Y. Genome-wide association study of cervical cancer suggests a role for ARRDC3 gene in human papillomavirus infection. Hum. Mol. Genet. 2019;28:341–348. doi: 10.1093/hmg/ddy390. [DOI] [PubMed] [Google Scholar]
- 84.Nik-Zainal S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli ., Stratton M.R. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kidd J.M., Newman T.L., Tuzun E., Kaul R., Eichler E.E. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:584–592. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Revathidevi S., Manikandan M., Rao A.K.D.M., Vinothkumar V., Arunkumar G., Rajkumar K.S., Munirajan A.K. Analysis of APOBEC3A/3B germline deletion polymorphism in breast, cervical and oral cancers from South India and its impact on miRNA regulation. Tumor Biol. 2016;37:11983–11990. doi: 10.1007/s13277-016-5064-4. [DOI] [PubMed] [Google Scholar]
- 87.Prasetyo A.A., Sariyatun R., Reviono, Sari Y., Hudiyono, Haryati S., Kageyama S. The APOBEC3B deletion polymorphism is associated with prevalence of hepatitis B virus, hepatitis C virus, Torque Teno virus, and Toxoplasma gondii co-infection among HIV-infected individuals. J. Clin. Virol. 2015;70:67–71. doi: 10.1016/j.cell.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 88.Göhler S., Da Silva Filho M.I., Johansson R., Enquist-Olsson K., Henriksson R., Hemminki K., Lenner P., Försti A. Impact of functional germline variants and a deletion polymorphism in APOBEC3A and APOBEC3B on breast cancer risk and survival in a Swedish study population. J. Canc. Res. Clin. Oncol. 2016;142:273–276. doi: 10.1007/s00432-015-2038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marouf C., Göhler S., Filho Miguel Inacio D.S., Hajji O., Hemminki K., Nadifi S., Försti A. Analysis of functional germline variants in APOBEC3 and driver genes on breast cancer risk in Moroccan study population. BMC Canc. 2016;16:165. doi: 10.1186/s12885-016-2210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J., Sieuwerts A.M., Look M.P., Van Der Vlugt-Daane M., Meijer-Van Gelder M.E., Foekens J.A., Hollestelle A., Martens J.W.M. The 29.5 kb APOBEC3B deletion polymorphism is not associated with clinical outcome of breast cancer. PloS One. 2016;11:1–15. doi: 10.1371/journal.pone.0161731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Imahashi M., Izumi T., Watanabe D., Imamura J., Matsuoka K., Ode H., Naoe T. Lack of association between intact/deletion polymorphisms of the APOBEC3B gene and HIV-1 risk. PloS One. 2014;9 doi: 10.1371/journal.pone.0092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ezzikouri S., Kitab B., Rebbani K., Marchio A., Wain-Hobson S., Dejean A., Vartanian J.P., Pineau P., Benjelloun S. Polymorphic APOBEC3 modulates chronic hepatitis B in Moroccan population. J. Viral Hepat. 2013;20:678–686. doi: 10.1111/jvh.12042. [DOI] [PubMed] [Google Scholar]
- 93.Itaya S., Nakajima T., Kaur G., Terunuma H., Ohtani H., Mehra N., Kimura A. No evidence of an association between the APOBEC3B deletion polymorphism and susceptibility to HIV infection and AIDS in Japanese and Indian populations. JID (J. Infect. Dis.) 2010;202:815–816. doi: 10.1086/655227. [DOI] [PubMed] [Google Scholar]
- 94.Rezaei M., Hashemi M., Hashemi S.M., Mashhadi M.A., Taheri M. APOBEC3 deletion is associated with breast cancer risk in a sample of southeast Iranian population. International Journal of Molecular and Cellular Medicine. 2015;4:103–108. [PMC free article] [PubMed] [Google Scholar]
- 95.Long J., Delahanty R.J., Li G., Gao Y.T., Lu W., Cai Q., et al. Zheng W. A common deletion in the APOBEC3 genes and breast cancer risk. J. Natl. Cancer Inst. 2013;105:573–579. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wen W.X., Soo J.S.-S., Kwan P.Y., Hong E., Khang T.F., Mariapun S., Teo S.H. Germline APOBEC3B deletion is associated with breast cancer risk in an Asian multi-ethnic cohort and with immune cell presentation. Breast Canc. Res. 2016;18:56. doi: 10.1186/s13058-016-0717-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gansmo L.B., Romundstad P., Hveem K., Vatten L., Nik-Zainal S., Lønning P.E., Knappskog S. APOBEC3A/B deletion polymorphism and cancer risk. Carcinogenesis. 2018;39:118–124. doi: 10.1093/carcin/bgx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qi G., Xiong H., Zhou C. APOBEC3 deletion polymorphism is associated with epithelial ovarian cancer risk among Chinese women. Tumour Biology. 2014;35:5723–5726. doi: 10.1007/s13277-014-1758-7. [DOI] [PubMed] [Google Scholar]
- 99.Liang J.-W., Shi Z.-Z., Shen T.-Y., Che X., Wang Z., Shi S.-S., Wang M.R. Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization. PloS One. 2014;9 doi: 10.1371/journal.pone.0114616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Middlebrooks C.D., Banday A.R., Matsuda K., Udquim K.-I., Onabajo O.O., Paquin A., Prokunina-Olsson L. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet. 2016;48:1330–1338. doi: 10.1038/ng.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abe H., Ochi H., Maekawa T., Hatakeyama T., Tsuge M., Kitamura S., et al. Chayama K. Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection. Hepatol. Res. 2009;39:1159–1168. doi: 10.1111/j.1872-034X.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- 102.An P., Johnson R., Phair J., Kirk G.D., Yu X.-F., Donfield S., Winkler C.A. APOBEC3B deletion and risk of HIV-1 acquisition. JID (J. Infect. Dis.) 2009;200:1054–1058. doi: 10.1086/605644. [DOI] [PMC free article] [PubMed] [Google Scholar]