ABSTRACT
The question of how phenotypic and genomic complexity are inter‐related and how they are shaped through evolution is a central question in biology that historically has been approached from the perspective of animals and plants. In recent years, however, fungi have emerged as a promising alternative system to address such questions. Key to their ecological success, fungi present a broad and diverse range of phenotypic traits. Fungal cells can adopt many different shapes, often within a single species, providing them with great adaptive potential. Fungal cellular organizations span from unicellular forms to complex, macroscopic multicellularity, with multiple transitions to higher or lower levels of cellular complexity occurring throughout the evolutionary history of fungi. Similarly, fungal genomes are very diverse in their architecture. Deep changes in genome organization can occur very quickly, and these phenomena are known to mediate rapid adaptations to environmental changes. Finally, the biochemical complexity of fungi is huge, particularly with regard to their secondary metabolites, chemical products that mediate many aspects of fungal biology, including ecological interactions. Herein, we explore how the interplay of these cellular, genomic and metabolic traits mediates the emergence of complex phenotypes, and how this complexity is shaped throughout the evolutionary history of Fungi.
Keywords: fungi, complexity, multicellularity, secondary metabolism, genome evolution
I. INTRODUCTION
Although less often considered than animals or plants, the ecological success of Fungi is equally impressive, and comprises a broad diversity of lineages and life styles that populate every corner of our planet (Naranjo‐Ortiz & Gabaldón, 2019a, b ), with most estimates predicting millions of extant fungal species (Hawksworth, 2001; Blackwell, 2011; Aime & Brearley, 2012; Hawksworth & Lücking, 2017). At the heart of this success lies a series of interwoven cellular and biochemical traits, which are ultimately determined by the genome. The question of how phenotypic complexity is related to genomic complexity, and how these change during evolution, historically has been approached from the perspective of animals and plants. However, in recent years fungi have increasingly been the focus of research aiming to understand the origin and evolution of genomic and morphological complexity. Fungi originated from a flagellated ancestor, but most current diversity encompasses non‐flagellated cells that often grow in a form of simple multicellularity called the mycelium, a true cellular network that sometimes extends over large areas. From this mycelial growth, many fungal organisms can switch to a unicellular growth form (e.g. yeast), often depending on the environmental conditions or on the stage of their life cycle. In several lineages throughout the fungal tree of life, mycelial growth has been abandoned, often completely but sometimes only to be recovered later. Some other lineages have taken multicellularity one step further, originating complex fruiting bodies whose macroscopic morphologies compete in intricacy and beauty with those of plants and animals. Complex multicellularity implies coordination of different cell types to form tissues and the existence of a tightly regulated developmental program. To achieve such levels of complexity, fungi have developed specific structural and regulatory systems that are still not fully understood. From a biochemical perspective, fungi present a truly vast diversity, with levels of complexity that are comparable or surpass those of other eukaryotic clades. Fungal organisms are osmotrophs and their cells are generally in contact with the surrounding environment. The relationship of a fungus with its immediate environment is defined by an array of secreted proteins and metabolites. The origin and diversification of these secreted metabolites is of great practical interest, given the often powerful effects they have on other organisms. Furthermore, production of these metabolites is tightly regulated and localized within the mycelial network, which saves resources, protects the fungus from damage from highly toxic intermediate metabolites and opens up a wide array of phenotypes in their interactions with other organisms.
This cellular and biochemical diversity is ultimately reflected in the highly dynamic nature of fungal genomes. Fungi are ideal subjects for genomic studies, since they tend to have highly compact genomes [theyrarely reach genome sizes of giga base pairs (Gbp)], are usually haploid, and can often be grown in axenic conditions. Consequently, the number of fully sequenced fungal genomes is now in the order of thousands. Hence, in combination with decades of studies in genetics, biochemistry and cell biology, fungal genomics is revolutionizing our understanding of this group (Scazzocchio, 2014). The study of the genetic repertoire of a growing diversity of fungi is unveiling a metabolic landscape far wider and intriguing than we could have imagined two decades ago. In addition, evolutionary genomic analyses are not only helping us to identify the components in the mycelial and multicellular fungal machinery, but also are uncovering what processes may drive the evolution of fungal genomes. In this respect, fungi have recently challenged evolutionary paradigms adopted from the study of animals and plants. For instance, non‐vertical evolutionary processes such as horizontal gene transfer or hybridization seem to be far more common in fungi than previously anticipated, and prokaryotic paradigms such as that of pangenomes seem also to be applicable to fungi. In this review, we discuss recent advances in our understanding of the evolution of phenotypic and genomic complexity within the fungal kingdom, focusing on the cellular and biochemical traits that have driven the success of fungi in the biosphere. In so doing we emphasize, when known, the genomic features that underlie those traits as well as their evolution.
II. CELLULAR COMPLEXITY
The last fungal common ancestor was likely a flagellated, unicellular organism with a saprotrophic or parasitoid lifestyle (James, Porter & Martin, 2014; Karpov et al., 2014a; Powell & Letcher, 2014; Naranjo‐Ortiz & Gabaldón, 2019b). However, very early in the evolution of fungi, a shift towards more complex forms of cellular organization occurred. This shift made possible the conquest of non‐aquatic environments, leading to loss of the flagellum in several lineages and explosive radiation (Liu, Hodson & Hall, 2006; Berbee, James & Strullu‐Derrien, 2017; Naranjo‐Ortiz & Gabaldón, 2019b). Even though many fungal lineages have independently returned to a (non‐flagellated) unicellular lifestyle, most extant fungi exist (at least for a sizeable fraction of their life cycle) as networks of filamentous cells able to grow indefinitely and in intricate patterns (Fig. 1). These networks are populated by an array of nuclei that may or may not be genetically identical. Non‐identical nuclei within a population are subjected to ecological pressures that have the potential to affect the phenotype of the whole network (Pontecorvo, 1956; James et al., 2008). The sheer physical size of fungal networks allows the simultaneous exploitation of several nutrient sources, a huge advantage for an otherwise sessile microbe. These properties allow filamentous fungi effectively to influence a far greater volume than would be suggested by simple measures of their biomass; they can thus be seen as true territorial microbes (Simonin et al., 2012; Fricker et al., 2017). In addition, many fungi are able to form tissues out of their filamentous cells, which generally act as support structures for the dissemination of propagative spores (Kües, Khonsuntia & Subba, 2018). The origin of this complex multicellularity presents many similarities with that of plants and animals, and many unique characteristics that we are only now starting to unravel (Niklas, 2014; Nagy, Kovács & Krizsán, 2018). In this section we discuss the evolutionary transition from a unicellular to mycelial form, the implications that a hyphal lifestyle has for fungal biology, and current knowledge regarding the development of complex multicellularity in the different lineages where this trait is found.
Figure 1.
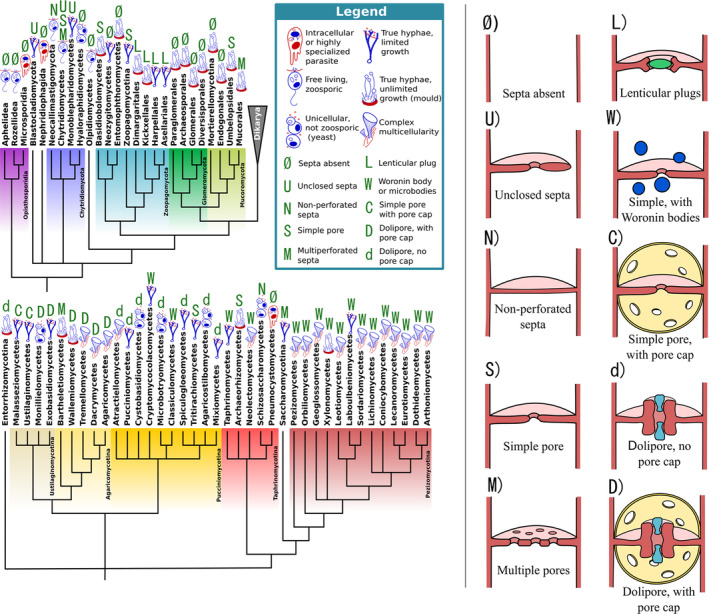
Cellular complexity and septal pore structure across the kingdom Fungi. Phylogenetic tree showing the main fungal lineages and their evolutionary relationships. Each group is associated with an icon and one or more letters. Icons show the highest level of cellular complexity described within the group. Letters show the type of pore structures described within the group; these are illustrated on the right of the figure. Ø, several groups are not known to produce septa, including various zoosporic and parasitic groups, as well as certain zygomycetous lineages. N, non‐perforated septa are rare in fungi, restricted mostly to the Neocallimastigomycotina, where they appear at the base of rhizoids. U, unclosed septa appear in certain groups of zoosporic fungi, either within true hyphae or as pores separating the main cellular body and the rhizoid. They typically present a central pore in addition to the unclosed septal division. S, simple septa with a main central pore. M, septation with multiple pores of variable size. L, lenticular plugs with variable morphology are a synapomorphic trait of the Kicxellomycotina. W, simple pores associated with small organelles that are able to block the pore. The most common form of this type of pore architecture is Woronin bodies of the Pezizomycotina, although other lineages have developed morphologically similar structures independently. C, d, D, most members of the Basidiomycota possess either dolipores and/or parenthesomes (also known as pore caps). Dolipores are barrel‐like swellings of the septal wall that delimit a pore. The pore is often blocked by occlusive bodies. Parenthesomes are membranous structures derived from the endoplasmic reticulum that surround the septal pore. Their morphology and ultrastructure is highly variable and has been used as a taxonomic trait.
(1). Unicellular fungi
Members of several fungal lineages have a mostly unicellular organization, a trait that can be both ancestral and derived within a group. Zoosporic lineages tend to exist as swimming individual cells with a saprotrophic or predatory–parasitic lifestyle. The swimming cell typically attaches to a substrate or host and produces feeding structures that range from amoeboid phagocytic protrusions in the Aphelidea (Gromov, 2000; Karpov et al., 2014b; Letcher et al., 2017) to polycentric rhizoids in many members of the Chytridiomycota (Powell & Letcher, 2014). Information regarding these groups is scarce, at least at the cellular, biochemical, ecological and phylogenetic levels. Environmental studies indicate that the described diversity in these lineages represents only a small fraction of the real diversity of the group (Lara, Moreira & López‐García, 2010; Jones et al., 2011; Rojas‐Jimenez et al., 2017; Tedersoo et al., 2017; Karpov et al., 2018). Several genomic studies have been conducted in these groups, but the lack of a solid phylogeny makes it difficult to obtain an accurate global picture. Despite their morphological simplicity, zoosporic lineages seem to have genome sizes and gene numbers comparable to or larger than filamentous fungi (Fig. 2). This seems to be valid for the Blastocladiomycota and Monoblepharidomycetes (Chytridiomycota), and for several species of Chytridiomycetes. The genomes of highly specialized anaerobes in the Neocallimastigomycotina are even larger, having genomes ranging from approximately 40 Mbp and 11000 genes (in Piromyces sp. E2) to almost 200 Mbp and 20200 genes (in Neocallimastix californiae). This genome size is rather high for fungi, and comparable to mushroom‐forming Agaricomycotina (Fig. 2). Many other members of the zoosporic lineages, however, have small genomes and gene content. Several members of the Chytridiomycetes fall into this category. The Opisthosporidia (Aphelidea, Rozellidea and Microsporidia) (Karpov et al., 2014a) are highly specialized parasites and show a high degree of genomic compaction. It is difficult to specify whether these differences in genome sizes within the zoosporic lineages result from reductions or expansions relative to their last common ancestor, as the only sequenced non‐fungal member of Holomycota, Fonticula alba, has a genome size similar to that of zoosporic fungi with small genomes. These observations suggest that there is little correlation in Fungi between cellular complexity and genomic characteristics such as haploid genome size or the number of protein‐coding genes. Some small‐genome chytrids appear to be parasites, which might suggest that adaptation to a parasitic lifestyle has driven genome reduction relative to a saprotrophic ancestor with a larger genome.
Figure 2.
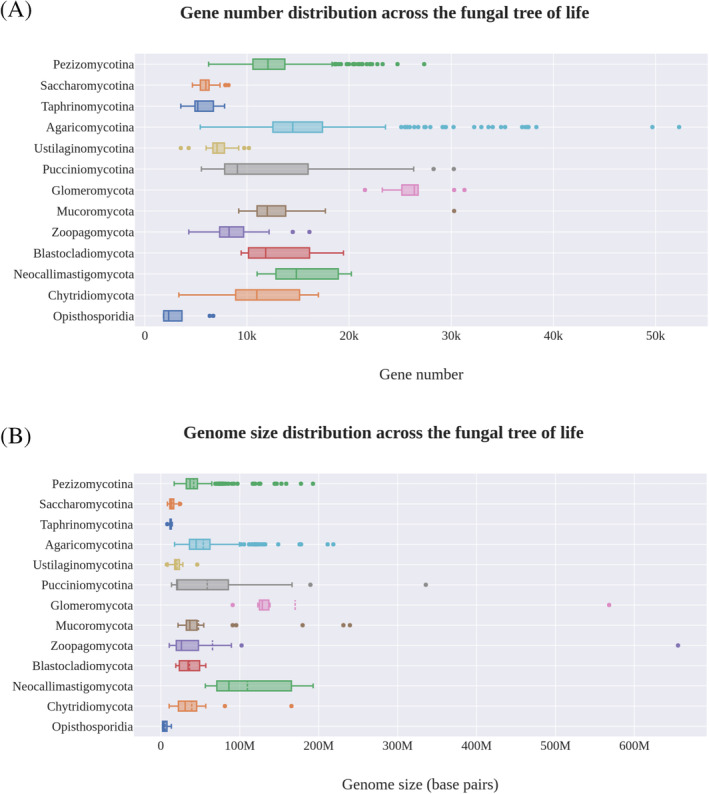
Gene number and genome size across the fungal kingdom. (A) Distribution of the number of genes per genome across different lineages. (B) Distribution of genome size across different lineages. All numbers were obtained from Mycocosm, accessed in June 2019.
In many fungal lineages, a unicellular lifestyle is a secondarily acquired trait, resulting from reduction or complete loss of the ability to form multicellular structures. Within Ascomycota, this reduction in complexity can be observed in several lineages of the Taphrinomycotina (Schizosaccharomycetes, Taphrinomycetes, Pneumocystidomycetes), and Pezizomycotina (black yeasts and related groups in the Eurotiomycetes and Dothideomycetes; symbiotic Ophiocordyceps in the Sordariomycetes and Symbiotaphrina within Xylonomycetes), and it represents a major evolutionary transition in Saccharomycotina. However, it must be noted that filamentous lineages within the Taphrinomycotina do not diverge much in terms of genome size or gene numbers from their yeast‐forming relatives. Taphrinomycotina is the sister group to a clade comprising both the filamentous Pezizomycotina and the yeast‐forming Saccharomycotina. Inferring the ancestral genomic characteristics of early Ascomycota is difficult, with the only trait that we can infer with certainty being their filamentous nature. Unicellular or mostly unicellular forms are very common across the Basidiomycota. In this group, it is generally acknowledged that thallus reduction has occurred in groups that are primarily biotrophic parasites (Begerow et al., 2014; Wang et al., 2015a, b ; McLaughlin et al., 2017; Oberwinkler, 2017). This lifestyle generally implies a high degree of genome compaction, reduction of many signalling and structural components, and loss of secondary metabolism pathways.
(2). The hyphal cell as a living pipe
The basic cellular unit for most described fungi is the hypha, a walled cylindrical multinucleated cell that is highly polarized. Cell polarization is necessary for hyphal growth, although not all fungal polarized cells are hyphae (e.g. the yeast Saccharomyces cerevisiae). Polarized growth has been widely studied in S. cerevisiae, and a core of proteins have been identified (Arkowitz & Bassilana, 2011; Riquelme, 2013; Martin & Arkowitz, 2014; Diepeveen et al., 2018). This core toolkit seems to be relatively well conserved across all fungal lineages, although no individual component appears to be completely essential (Diepeveen et al., 2018). Within the kingdom, true hyphal growth is an evolutionary novelty of the ‘terrestrial fungi’, a monophyletic clade that includes the phyla Zoopagomycota, Mucoromycota, Glomeromycota, Ascomycota and Basidiomycota. True mycelial growth is also observed in some members of the Blastocladiomycota (e.g. Allomyces) (James et al., 2014) and the Monoblepharidomycetes (e.g. Gonapodya) (Dee et al., 2015). Due to controversies regarding the phylogenetic placement of Blastocladiomycota (Tanabe, Watanabe & Sugiyama, 2005; Sekimoto et al., 2011; Ebersberger et al., 2012; Tretter et al., 2013; Ren et al., 2016; Spatafora et al., 2016), it is currently unknown whether the ability to form hyphae in this group has a shared evolutionary origin with the terrestrial fungi. By contrast, hyphal growth in Monoblepharidomycetes clearly has an independent origin (Dee et al., 2015).
The hypha of most filamentous fungi is organized around an organelle called the Spitzenkörper (SPK) (Steinberg, 2007; Arkowitz & Bassilana, 2011; Riquelme & Sánchez‐León, 2014; Lin et al., 2014b; Takeshita, 2016; Steinberg et al., 2017b; Riquelme et al., 2018). The SPK is composed of a collection of vesicles originating in the Golgi apparatus that contain the enzymes, lipids and polysaccharides required for the synthesis of membranes and cell wall. Comparative studies have revealed the conservation of this molecular machinery across all Dikarya, regardless of whether they present hyphal growth or not, but information outside this group is very limited. For instance, most zygomycetous fungi present a less‐organized aggregation of vesicles named the apical vesicle crescent (AVC) (Fisher & Roberson, 2016). This structure has been studied mostly using electron microscopy, and thus equivalence between SPK and AVC components at the molecular level is poorly known (Roberson et al., 2011; Henk & Fisher, 2012; Fisher et al., 2018). The SPK seems to be present in Basidiobolus (Roberson et al., 2011) and Conidiobolus (Fisher et al., 2018), which are early‐diverging lineages within the Entomophthoromycotina. Members of the Blastocladiomycota (e.g. Allomyces, Blastocladiella) present a morphologically recognizable SPK (Vargas, Aronson & Roberson, 1993; Srinivasan, Vargas & Roberson, 1996; McDaniel & Roberson, 1998; James et al., 2014). The presence of the organelle in these lineages suggests that the last common ancestor of all terrestrial fungi could have had an SPK that was subsequently lost or modified into an AVC in the zygomycetous fungi, although it is currently impossible to rule out whether the SPK in these lineages arose independently.
Another widespread trait in filamentous fungi is the presence of septa, which are transversal modifications of the cell wall that allow the selective passage of cytoplasmic components between adjacent cells. At least in Pezizomycotina, septa‐divided cells show a certain degree of biochemical and regulatory autonomy (Bleichrodt et al., 2012; Steinberg et al., 2017b; Tegelaar & Wösten, 2017), which is a prerequisite for complex multicellularity. Septal structure varies widely among groups (Fig. 1), ranging from incomplete pseudosepta in some Blastocladiomycota (Meyer & Fuller, 1985) to highly sophisticated membranous barriers in Agaricomycetes (Orlovich & Ashford, 1994; Muller et al., 1998; van Driel et al., 2009; McLaughlin et al., 2015). The distribution and function of these structures within the mycelium is also highly variable, and many lineages within the zygomycetous fungi lack them or restrict their presence to specific structures or senescent hyphae (Benny, Humber & Voigt, 2014; Redecker & Schüβler, 2014). The formation of septa requires the activity of actin rings and chitin synthases (Harris, 2001; Rittenour, Si & Harris, 2009; Lin et al., 2014b; Riquelme et al., 2018). Most filamentous members of Dikarya possess seven genes encoding chitin synthases (Pacheco‐Arjona & Ramirez‐Prado, 2014). However, some plant‐pathogenic fungi have expanded this repertoire, while yeasts tend to reduce it (Pacheco‐Arjona & Ramirez‐Prado, 2014). Outside Dikarya, however, the number and distribution of these enzymes is highly variable, with no recognizable pattern (Pacheco‐Arjona & Ramirez‐Prado, 2014). Septal barriers in their different forms have emerged independently in several lineages, deriving from different cellular components (peroxisome‐derived Woronin bodies in Pezizomycotina, endoplasmic reticulum‐derived dolipores in most Basidiomycota), although the ontology of these structures is not fully understood for some groups (Benny et al., 2014; Redecker & Schüβler, 2014; Nguyen et al., 2017). Neolecta is a genus of filamentous Taphrinomycotina that has septal pores that are morphologically similar and perhaps homologous to Woronin bodies (Landvik et al., 2003; Healy et al., 2013; Nguyen et al., 2017). The Kickxellomycotina have sophisticated septal plugs of unclear ontogeny whose morphology varies among the different clades (Tretter et al., 2014).
Fungi must grow in an expansive way to explore and exploit their territory, a complex task at which they are extremely proficient (Bebber et al., 2007; Asenova et al., 2016). To achieve this, simple cylindrical hyphae are not sufficient, both in terms of exploiting the available space and in growth speed (Simonin et al., 2012; Fricker et al., 2017). Instead, fungi grow by forming branching patterns by generating novel cylindrical hyphae from already established ones. Very little is known about the molecular intricacies of this process (Harris, 2008, 2011; Lin et al., 2014b; Riquelme et al., 2018). Establishment of a new branching hyphal tip seems to share many components with yeast budding (Harris, 2008, 2011), but there are considerable differences between the main tip and the branches that stem from it (Momany, 2002; Riquelme & Bartnicki‐Garcia, 2004; Harris, 2011; Lin et al., 2014b). Additionally, the establishment of a true network requires the ability to grow in a convergent pattern through hyphal tip fusion, or anastomosis. This process requires complex cell recognition mechanisms, preventing fusion between genetically dissimilar hyphae during vegetative growth (Saupe, 2000; Glass & Kaneko, 2003; Hall et al., 2010; Ishikawa et al., 2012; Zhang et al., 2014; Fleißner & Herzog, 2016; Daskalov et al., 2017). After fusion, the genetic compatibility of the newly formed heterokaryon is evaluated, resulting in programmed cell death if certain requirements are not met. In Pezizomycotina, this check point is based on the presence of highly polymorphic het loci, which have homologs in organisms as distant as Basidiomycota (Van der Nest et al., 2014). A minimum of 24 different proteins are known to be involved in this process in Neurospora, including proteins involved in vesicular transport, membrane components, cell wall integrity, mitogen‐activated protein kinase (MAPK) cascades and numerous transcription factors (Aldabbous et al., 2010; Fu et al., 2011; Jonkers et al., 2014). However, while some of these components such as het appear to be conserved over long evolutionary distances, others are taxonomically restricted to Pezizomycotina (Riquelme et al., 2011; Herzog, Schumann & Fleißner, 2015). Hyphal fusion is essential for fungal sex (Ni et al., 2011; Teixeira et al., 2017). In zygomycetous fungi, this fusion generates meiotically reduced zygospores, with the two original hyphae acting as de facto gametes (Benny et al., 2014; Lee & Heitman, 2014; Lee & Idnurm, 2017). In Dikarya, fusion establishes dikaryotic hyphae that later will initiate meiotic recombination (Lee et al., 2010; Ni et al., 2011; Heitman, Sun & James, 2013). Homologs of mating (MAT) systems and meiotic machinery can also be found in the genomes of Microsporidia, Chytridiomycota, Blastocladiomycota and even Glomeromycota, for which sexual structures have never been observed (Lee et al., 2010; Heitman et al., 2013; Tang et al., 2016).
(3). The mycelium as a living network
Hyphae acquire a network organization once they reach a critical size, through generation of lateral branches and anastomoses. The structure of this network is typically highly dynamic, and responds to fluctuations in the environment or biotic interactions (Simonin et al., 2012). The whole mycelium should be considered an independent organism that presents global and local responses to stimuli. Fungi, like many other eukaryotes, undergo apoptosis and senescence (Hamann, Brust & Osiewacz, 2008; Sharon et al., 2009; Shlezinger, Doron & Sharon, 2011; Shlezinger, Goldfinger & Sharon, 2012). Apoptosis helps the mycelium to dismantle and reuse components of network regions that are not useful, such as those that have already depleted the nutrients in their immediate surroundings (Hamann et al., 2008; Sharon et al., 2009; Shlezinger et al., 2012). The fungal apoptotic machinery shows deep homology with that of metazoans, and thus it is safe to assume that this process also exists in non‐Dikarya fungi (Sharon et al., 2009; Shlezinger et al., 2012).
Unlike plants, nutrient transport in filamentous fungi is performed entirely through cytoplasmic currents, which can also transport cellular components, including fresh nuclei for growing hyphal tips (Tlalka et al., 2007; Fricker et al., 2008, 2017; Lew, 2011; Simonin et al., 2012). While transport in vascular plants has clear directionality (water and inorganic nutrients are absorbed in the roots and travel to the rest of the plant, while photosynthetic products flow in the opposite direction), fungi are able to create a more flexible flux. This allows fungi to allocate different limiting nutrients from distant sources across heterogeneous environments (Fricker et al., 2008, 2017; Simonin et al., 2012; Boberg et al., 2014), akin to the movement of goods and people along roads (Bebber et al., 2007). Movement of cytoplasmic components throughout the mycelial network is an active process, which uses cytoplasmic waves to transport over large distances (Tlalka et al., 2007; Fricker et al., 2008, 2017; Lew, 2011) and cytoskeleton‐based movement for short‐range movements or against the main cytoplasmic current (Fricker et al., 2008; Lichius, Berepiki & Read, 2011; Takeshita, 2016). Septate fungi can exert an additional layer of control over the flux, as they can selectively block nodes of the network to limit the harm caused by injuries or to regulate the movement of cellular components (Palma‐Guerrero et al., 2008; Jedd & Pieuchot, 2012; Fricker et al., 2017; Steinberg et al., 2017a, c ).
Like any multicellular organism, the mycelium must be able to sense a wide array of physical and chemical signals. Fungal sensory systems are functionally very similar to those found in plants. For instance, fungi are able to detect the ratio of different wavelengths of light to adjust to their surroundings, similar to phototropin‐mediated signalling in plants (Fuller, Loros & Dunlap, 2015; Fischer et al., 2016; Schumacher, 2017). In many fungi, light can affect expression of genes, including those involved in important processes such as reproduction, morphogenesis, virulence and metabolism (Corrochano & Garre, 2010; Kamada et al., 2010; Idnurm, 2013; Fuller et al., 2015; Fischer et al., 2016; Schumacher, 2017; Adam et al., 2018; Wang et al., 2018). Fungi possess well‐studied circadian clocks, too (Dunlap & Loros, 2004, 2006; Liu & Bell‐Pedersen, 2006; Salichos & Rokas, 2010; Fuller et al., 2015). Whether seasonal cycles exist in fungi remains unclear, but a combination of wavelength‐ratio sensing and circadian clocks, both of which exist in fungi, are involved in such processes in plants (Searle & Coupland, 2004; Andrés & Coupland, 2012; Johansson & Staiger, 2015). Transcriptomic changes in response to light, including circadian cycles, are mediated in all studied fungi by white collar complex proteins (He et al., 2002; Dunlap & Loros, 2004; Olmedo et al., 2013; Fuller et al., 2015; Fischer et al., 2016). These are zinc‐finger transcription factors that include chromophore‐binding domains, similar to the non‐homologous phototropins in plants. White collar proteins are found in all main lineages of the kingdom (Corrochano & Garre, 2010; Fuller et al., 2015), while some yeast and specialized parasitic lineages have lost them secondarily. These are not the only light‐sensitive proteins in fungi. Opsin‐like proteins in fungi are involved in regulation of the sexual cycle and in pathogenesis in some species, including Fusarium fujikuroi (Hypocreales) (García‐Martínez et al., 2015; Adam et al., 2018) and Blastocladiella emersonii (Blastocladiomycota) (Scheib et al., 2015). Despite the ancient nature of this protein family, information about the biological roles of its members is still very fragmented.
Fungi are able to sense gravity and show, in some cases (e.g. aerial sporangia), strong gravitropism or gravity‐based morphogenic patterns. Gravity perception in fungi has been studied extensively in Phycomyces blakeensianus (Mucorales), where it is mediated by a combination of statoliths made of oxalate crystals and buoyant lipid structures (Schimek et al., 1999; Eibel et al., 2000; Göttig & Galland, 2014). Gravitropism in Phycomyces is apparently mediated by a differential flux of H+ and Ca2+ in the mycelium (Živanović, 2005, 2012, 2013; Göttig & Galland, 2014). However, statoliths in this fungus seem to have originated from a recent bacterial gene transfer (Nguyen et al., 2018). In the absence of crystalline structures in other lineages, it has been proposed that nuclei themselves might act as statolith‐like structures in Agaricomycotina (Monzer, 1995, 1996; Moore et al., 1996; Kern, Mendgen & Hock, 1997). However, biophysical data suggest that the density of the nuclei in Ascomycota might not be sufficient for them to function as statoliths (Grolig, Döring & Galland, 2006). Buoyancy systems have been identified in Ascomycota, Basidiomycota, Mucoromycotina, Mortierellomycotina and Glomeromycota (Grolig et al., 2006) and the identified components seem to be well conserved across long evolutionary distances. Fungi are also able to detect electric fields and respond to them in a Ca2+‐dependent manner (Gow, 1984; Lever et al., 1994). These responses are well described in both filamentous and zoosporic fungi, suggesting a considerable degree of evolutionary conservation, but unfortunately little is known about their molecular mechanisms.
Structural damage is another important factor to which fungal organisms must respond appropriately. At a local level, fungi respond to mycelial breakage by closing their septa to constrain cytoplasmic loss, a response which is often followed by the promotion of branching and sporulation (Maruyama, Escaño & Kitamoto, 2010; Hernández‐Oñate et al., 2012; Medina‐Castellanos et al., 2014, 2018). Injury is able to induce coordinated responses across the mycelium, such as increasing the production of toxic metabolic compounds, as shown by arthropod grazing experiments (Rohlfs et al., 2007; Yin et al., 2012; Caballero Ortiz, Trienens & Rohlfs, 2013; Döll et al., 2013; Rohlfs, 2014; Atriztán‐Hernández et al., 2018; Künzler, 2018). There are several injury‐signalling pathways described in fungi. Reactive oxygen species (ROS) are one of the most important responses to any kind of damage in fungi (Hernández‐Oñate et al., 2012; Medina‐Castellanos et al., 2014, 2018; Hernández‐Oñate & Herrera‐Estrella, 2015). Not only do ROS induce responses in the fungus, but they are also used as a chemical weapon against invasive biological agents. ROS also play important roles in cell differentiation signalling, as discussed below (Section II.4). Extracellular ATP is another important injury‐response molecule that acts through a Ca2+‐mediated cascade (Hernández‐Oñate et al., 2012; Medina‐Castellanos et al., 2014, 2018). ATP in the surrounding medium is rare, and thus the presence of this molecule can be used by the fungus as a signal of cytoplasm leakage. It is well established that the presence of cell wall components induces defence responses in plants, but such reactions in response to the enzymatic degradation of their own cell walls are not described in fungi. However, we consider that chitosans, partially deacetylated forms of chitin, are likely recognized by fungi as a signal of cell wall damage. Chitosan itself is known to have antifungal properties (Palma‐Guerrero et al., 2008, 2009; Lopez‐Moya & Lopez‐Llorca, 2016). However, fungi that are specialized mycoparasites, arthropod pathogens or nematophagous are virtually impervious to this compound. Due to their trophic lifestyle, these fungi possess high chitinolytic activities, and thus are unlikely to respond to the presence of chitin‐derived products in the surrounding medium as if they were a signal of damage to their own cells. The fungus Rhizopus also has a high tolerance to chitosan (Lopez‐Moya & Lopez‐Llorca, 2016) and, as in other members of the Mucorales, possesses high concentrations of chitosan in its own cell walls (Battaglia et al., 2011). Chitosan is not toxic to animals, indeed, in humans chitosan is marketed as weight‐loss supplement (Saper, Eisenberg & Phillips, 2004; Mhurchu et al., 2005), but it can elicit innate immune responses in both animals (Zaharoff et al., 2007; Li et al., 2013) and plants (Benhamou & Thériault, 1992; Sathiyabama, Akila & Charles, 2014; Malerba & Cerana, 2016). Thus, we propose that the antifungal activity of chitosan could result from over‐activation of fungal defence mechanisms rather than inherent toxicity. Oxylipin signalling also plays an important role in damage responses in fungi (Brodhun & Feussner, 2011). Importantly, all of these signalling pathways are well conserved among fungi, plants and animals (Hernández‐Oñate et al., 2012; Hernández‐Oñate & Herrera‐Estrella, 2015). Finally, complex multicellular fungi have been shown to possess action potential‐like electric signals that travel across large distances in response to direct damage (Adamatzky, 2018); a similar phenomenon is well studied in plants (Fromm & Lautner, 2007; Katicheva et al., 2014; Vodeneev, Akinchits & Sukhov, 2015).
Besides responding to external stimuli, fungi can generate a wide array of extracellular chemicals that are used to coordinate mycelial behaviour. Oxylipins are a diverse class of molecules derived from poly‐unsaturated fatty acids that play important signalling roles in virtually all eukaryotes, including animals (Andreou, Brodhun & Feussner, 2009), plants (Mosblech, Feussner & Heilmann, 2009; Wasternack & Feussner, 2018) and fungi (Brodhun & Feussner, 2011). However, we still have a very incomplete knowledge about the role of oxylipins in fungal biology. It is clear that these compounds regulate important processes, including morphological switches, secondary metabolism, pathogenesis, the sex cycle, and defence against grazing (Brodhun & Feussner, 2011; Kretschmer, Wang & Kronstad, 2012; Künzler, 2018). Additionally, parasitic fungi can synthesise oxylipins that exert biological activity in their hosts (Noverr, Erb‐Downward & Huffnagle, 2003; Wilson et al., 2004; Tsitsigiannis & Keller, 2007; Brodhun & Feussner, 2011; Christensen & Kolomiets, 2011; Fischer & Keller, 2016). Expansions in the genes encoding oxylipins have been proposed to be important developments in the ability of some fungal species to invade plant tissues (Tsitsigiannis & Keller, 2006, 2007; Gao et al., 2011). Screening for conidiation‐defective mutants in Aspergillus led to the discovery of regulatory polyketide synthetases (PKSs) and non‐ribosomal peptide synthetases (NRPSs) (Lee & Adams, 1994, 1995; Lo et al., 2012; Soid‐Raggi et al., 2016; Riquelme et al., 2018). These regulatory secondary metabolites were subsequently described in other filamentous Ascomycota, such as Fusarium fujikuroi (Wiemann et al., 2012; Riquelme et al., 2018). Since these classes of compounds are known to possess a wide range of biological activities, the possibility that some fungi might have adapted their own regulatory extracellular metabolites to disrupt the biology of other fungi is intriguing, particularly in terms of the search for novel antimycotic agents. However, PKS and NRPS biosynthetic clusters are reduced or absent in some lineages, such as yeasts (Dujon, 2010, 2015) and certain biotrophic plant pathogens (Kämper et al., 2006; Perlin et al., 2015).
(4). Complex multicellularity
The ability to form multicellular structures emerged independently in several lineages of eukaryotes. Complex multicellularity, which implies the coordination of different cell types to form tissues, has emerged in Metazoa, Streptophyta, Chlorophyta, Rhodophyta, Ochrophyta and Fungi (Niklas, 2014). Fungi are peculiar in this regard, as their complex multicellularity is almost always restricted to fruiting bodies, i.e. reproductive structures that are usually intermittent. In all cases, fruiting bodies are formed from dikaryotic mycelium, generating a vegetative hyphal tissue that protects fertile hyphae, where meioisis takes place. These structures are named ascomata in the Ascomycota (Pöggeler, Nowrousian & Kück, 2006; Engh & Nowrousian, 2010; Lord & Read, 2011; Kües et al., 2018) and basidiomata in the Basidiomycota (Kües, 2000; Kües & Liu, 2000; Hibbett et al., 2014; Kües & Navarro‐González, 2015; Kües et al., 2018). Even in fungi that produce fruiting bodies, these structures are not indispensable for reproduction and dispersal, given the possibility to propagate asexually. Multicellular structures have evolved independently at least twice in fungi (Nguyen et al., 2017; Kües et al., 2018; Nagy et al., 2018) and is only present in three lineages: Neolectomycetes, Pezizomycotina and Agaricomycetes.
The first and least well‐known group to have developed complex multicellularity is the class Neolectomycetes (Taphrinomycotina) (Landvik et al., 2003; Healy et al., 2013; Kurtzman & Sugiyama, 2015). The Taphrinomycotina is an early‐branching lineage of Ascomycota that is sister to the group formed by the Pezizomycotina (which tend to be filamentous and often possess complex fruiting bodies) plus the Saccharomycotina (which possess a highly reduced thallus). Genomic analysis of Neolecta irregularis shows a very reduced genome, comparable to other members of the Taphrinomycotina (Nguyen et al., 2017). Despite having a yeast‐like genome size and number of protein‐coding genes, Neolecta form true, albeit simple, fruiting bodies (Landvik et al., 2003; Healy et al., 2013). Neolecta shares approximately 1000 genes with filamentous Pezizomycotina that are absent in yeast‐like members of the Taphrinomycotina, with this set mostly enriched in genes relating to endomembrane systems (Nguyen et al., 2017). Some studies have suggested that the fossil Prototaxites [420–370 million years ago (Mya)] (Hueber, 2001; Selosse, 2002) is affiliated to Neolectomycetes based on structural characters (Honegger et al., 2018). Under that interpretation, Prototaxites represents fruiting bodies or vegetative thalli of an unspecified lineage within the Ascomycota, and is probably a member of the Taphrinomycotina. Thus, either modern members of Neolectomycetes are secondarily simplified, or the lineage leading to Prototaxites evolved considerably increased complexity.
Multicellular fruiting bodies are known in most classes within the Pezizomycotina (Liu & Hall, 2004; Schmitt, 2011), with the lack of correlation between morphological complexity and phylogeny suggesting they are an ancestral trait. Some of the most complex fruiting bodies within this subphylum belong to the Pezizomycetes, which are recovered as sister to the rest of the group or to the rest of the group minus Orbiliomycetes by most phylogenies (Liu & Hall, 2004; Spatafora et al., 2006; Prieto et al., 2013). It is unclear whether the molecular basis of multicellular fruiting bodies in Pezizomycotina is homologous to that of the complex structures in Neolectomycetes, but if so, this would imply a multicellular common ancestor for all Ascomycota. The basic fruiting body morphology in Pezizomycotina is a cup‐like structure with asci oriented towards the concavity (apothecia). This basic body plan has become elaborated in many groups to form a bottle‐like (perithecia) or completely closed (cleistothecia) architecture (Liu & Hall, 2004; Schoch et al., 2009; Kües et al., 2018). Cleistothecia often act as both protective and dissemination structures. It is important to note that sex or sexual structures have never been described for many Pezizomycotina, with the literature referring to these fungi as fungi imperfecti or Deuteromycetes. Genomic evidence of meiotic recombination suggests that sex does occur in these fungi, albeit under unknown circumstances (Lee et al., 2010; Ni et al., 2011; Heitman et al., 2013). This was confirmed with the discovery of sexual cycles in Penicillium (Houbraken, Frisvad & Samson, 2010) and Aspergillus (O'Gorman, Fuller & Dyer, 2009; Swilaiman et al., 2013), widely studied fungi that were thought to be asexual for more than a century. Ascomata size is highly variable, ranging from less than a millimeter to several centimeters (Schmitt, 2011; Kües et al., 2018). The relationships between phylogeny and fruiting body morphology are poorly understood in Pezizomycotina, although it is generally acknowledged that closed ascomatas (Pezizomycetes and lichen‐forming Lecanoromycetes) are derived forms (Liu & Hall, 2004; Schoch et al., 2009; Schmitt, 2011). The genetics of fruiting body development have been well studied in several model species, allowing the identification of developmental mutants (Nowrousian et al., 2007; Dirschnabel et al., 2014; Teichert et al., 2017; Trail et al., 2017). Comparative transcriptomic analyses in the Sordariomycetes (Trail et al., 2017) and Pezizomycetes (Murat et al., 2018) suggest that the regulatory machinery of the fruiting body is well conserved, at least within these groups. Many questions remain regarding ascomata development in Pezizomycotina. Similarly to Neolecta, some ascomata‐forming Pezizomycotina have gene numbers similar to those of yeast species. For instance, the Périgord truffle Tuber melanosporum possesses approximately 7500 protein‐coding annotated genes (Martin et al., 2010), not dissimilar to the approximately 6000 genes of S. cerevisiae. Lichen‐forming fungi tend to produce a macroscopic thallus that includes their symbionts, often in an organized layered structure. This organization, in many cases resulting in a well‐defined morphology, has been interpreted as complex multicellularity by some authors (Grube & Hawksworth, 2007; Sanders & de los Rios, 2012, 2017). Lichen genomics is still in its infancy, and to date there is no comprehensive study of developmental programs in lichen species. It is likely that lichen thalli will share regulatory pathways with the ascomata developmental program, a hypothesis that will undoubtedly be addressed in the near future.
Agaricomycotina is the other main group with complex multicellularity, and the one whose fruiting bodies (basidiomata or basidiocarp) are most familiar to humans (de Mattos‐Shipley et al., 2016). We can differentiate between two main fruiting body architectures (Hibbett, 2006; Millanes et al., 2011; Shirouzu et al., 2013; Hibbett et al., 2014; Oberwinkler, 2014; Weiss et al., 2014; Kües & Navarro‐González, 2015). Gelatinous fruiting bodies are morphologically more simple and are found in several lineages (Tremellomycetes, Dacrymycetes, Cantharellales, Auriculariales), probably with independent origins. These fruiting bodies lack a true tissue organization and tend to form very simple amorphous structures. By contrast, the Holobasidiomycetes are a monophyletic group that includes most members of the Agaricomycetes and whose fruiting bodies have true tissues and often distinct morphologies. The tissue of the basidiocarp is formed by several different types of hyphae embedded in an extracellular matrix. These cell types differ from vegetative assimilative hyphae in the thickness of their cell walls, the frequency of lateral branching, and the distribution of clamp connections, etc. (Manocha, 1965; Volz & Niederpruem, 1969; Kennedy & Larcade, 1971; States, 1975; Mol, Vermeulen & Wessels, 1990; Nakagiri & Ito, 1991). The proportion of these different hyphae varies between the stipe and the cap, and determines the mechanical properties of the basidioma. Different forms of organization are known in certain species, typically corresponding with morphologically distinct regions of the fruiting body. Virtually nothing is known regarding the molecular basis of this cell differentiation or if there are other cell types identifiable (e.g. by biochemical or immunological markers). Most descriptions of tissue organization in mushroom‐forming fungi come from historical works that make use of often outdated and inconsistent terminology, greatly hindering comparisons among species. Comparative analyses show that fruiting body‐forming Agaricomycetes have expanded sets of genes encoding kinases and several families of ubiquitin‐signalling pathways, and have an increased frequency of alternative splicing (Krizsan et al., 2019). Similar traits have evolved independently in multicellular metazoans and plants. There is evidence for a conserved developmental program in Agaricomycotina (Stajich et al., 2010; Plaza et al., 2014; Cheng et al., 2015; Nowrousian, 2018), which emerged independently from that found in Ascomycota (Nguyen et al., 2017; Nagy et al., 2018; Kües et al., 2018). Transcriptomic analyses in Coprinopsis cinerea suggest that gene expression in fruiting bodies follows a highly conservative pattern early in fruiting body development, compared with production of the vegetative mycelium or late basidioma (Cheng et al., 2015). The development of the fruiting body shows wide differences in gene expression compared with the development of the vegetative mycelium, including overexpresion of many genes involved directly or indirectly in cell wall remodelling, DNA synthesis, ribosomes, lipid metabolism and hydrophobins (Ohm et al., 2010b; Krizsan et al., 2019). Environmental factors influence fruiting body development through cyclic AMP (cAMP), rat sarcoma (Ras) and MAPK cascades (Palmer & Horton, 2006; Nowrousian, 2018; Sakamoto, 2018). Additionally, some groups (e.g. the genus Armillaria) are able to form truly multicellular vegetative structures called rhizomorphs, which are thread‐like aggregations of vegetative hyphae that allow the relocation of nutrients over very large distances (Motta, 1969; Agerer & Iosifidou, 2004; Morrison, 2004). Transcriptomic analyses of Armillaria rhizomorphs suggest an origin via redeployment of the fruiting body developmental program (Sipos et al., 2017).
III. GENOME COMPLEXITY
The main driver for the acquisition of morphological complexity in fungi is protection of the sexual structures and dissemination of spores. For Ascomycota and Basidiomycota, the dikaryotic stage involves several phenotypic traits that sets them apart from the monokaryon, although in a different way for each group. Sexual recombination in fungi is typically sporadic, and sexual stages have not been identified for many fungal lineages. The existence of sexual and parasexual cycles opens the possibility for recombinant lineages, including the formation of inter‐species hybrids (Peter et al., 2018). Hybridization is starting to be recognized as an important source of genetic diversity in fungal species. This has deep implications, particularly in the fields of fungal epidemiology and fungus–plant interactions (Stukenbrock, 2016; Möller & Stukenbrock, 2017; Feurtey & Stukenbrock, 2018; Giordano et al., 2018). The typically clonal nature of many fungal populations imposes another type of challenge: if sex is uncommon in these fungi, how do they adapt to an ever‐changing environment? While spontaneous mutations and horizontal gene transfer might provide a certain level of variability, none of these phenomena seem to be as prevalent in fungi as they are in prokaryotes. Mutations in particular need to become fixed in a population, which in many fungi would mean a whole mycelium. However, fungi seem to be able to tolerate high levels of chromosomal mutations such as polyploidies and aneuploidies, particularly under stressful conditions (Cogliati et al., 2012; Li et al., 2012; Bennett, Forche & Berman, 2014; Kravets et al., 2014; Gerstein & Berman, 2015; Berman, Wertheimer & Stone, 2016; Todd, Forche & Selmecki, 2017). From a phenotypic point of view, impairment in regulatory networks caused by chromosomic aberrations has the potential to affect signalling pathways controlling morphological complexity. This gene dosage alteration also has the potential to induce changes in metabolism, which might in turn generate new phenotypes. Chromosomal aberrations emerge spontaneously and are reversible, and thus could help fungi to adapt to new conditions in a rapid and transitory way. It is important to note that sex and chromosomal aberrations are interconnected. Aberrations to chromosomes can potentially impair meiotic recombination, or by contrast might be responsible for the stabilization of highly divergent hybrid genomes (Aminnejad et al., 2012; Forche, 2012; Morrow & Fraser, 2013). Genomics is starting to explore these processes, revealing new challenges and opportunities in this field. Figure 3 illustrates the different sources of genomic variability that can be identified within a single mycelium. In this section we discuss all these chromosome alterations, as well as their physiological and evolutionary relevance.
Figure 3.
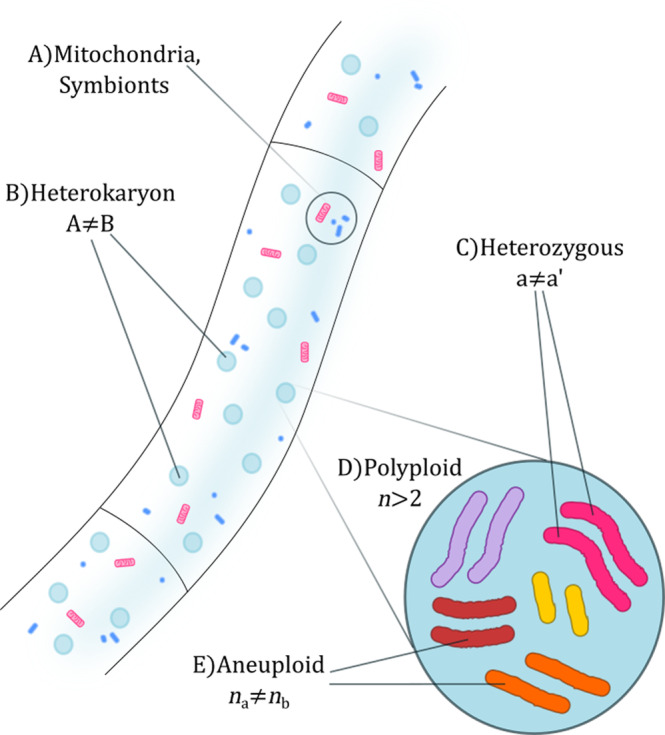
Deviations from standard genome structure in fungi. A, Many fungi harbour symbiotic associations with bacteria or other eukaryotes whose DNA is sequenced alongside the fungal DNA. B, Filamentous fungi can possess populations of nuclei with sequence differences, a phenomenon known as heterokaryosis. C, Sequence differences between recombinant chromosomes are possible, known as heterozygosis. D, Most eukaryotes possess a number of chromosome copies equal to either one (haploid) or two (diploid). Copy numbers above two are known as polyploidies. E, Chromosome number might vary among chromosomes (aneuploidies). All these phenomena carry important biological consequences and might occur at the same time, making genome assembly and analyses more difficult.
(1). Hybridization
The use of S. cerevisiae as a research model in genetics and biochemistry led to the identification of their sexual cycle and of metabolic traits that could be used as makers for different lineages. This enabled the discovery that several Saccharomyces strains, classified as independent species, were in fact hybrids (Dujon, 2010; Borneman et al., 2011; Morales & Dujon, 2012; Hittinger, 2013; Kumaran, Yang & Leu, 2013; Walther, Hesselbart & Wendland, 2014; Leducq et al., 2016). Due to the difficulty of defining species boundaries in fungi, the definition of hybrids is similarly unclear. Here, we refer to hybrids as any fungal lineage that has emerged from mating between two lineages whose divergence exceeds that typically found across the most distant strains of well‐recognized species (e.g. ~1% in S. cerevisiae; Peter et al., 2018). The advent of genome sequencing led to a revolution in yeast research, and showed that the ability of Saccharomyces to hybridize was not exceptional within Saccharomycotina. Hybrid yeasts seem to be common in industrial environments and some of them are particularly relevant as fermenters in the food and beverage industry (James et al., 2005; Hellborg & Piskur, 2009; Morales & Dujon, 2012; Walther et al., 2014; Borneman et al., 2014). Hybrid yeasts can also be found among clinical isolates; the Candida parapsilosis species complex is a particularly interesting example, with numerous described hybrid isolates that apparently possess higher pathogenic capabilities than their non‐hybrid relatives (Pryszcz et al., 2014, 2015; Gabaldón, Naranjo‐Ortíz & Marcet‐Houben, 2016; Mixão & Gabaldón, 2018). The inability to identify the parental strains for some hybrids might reflect the increased success of the hybrid in taking over the ecological niche of the parental strains (Pryszcz et al., 2015; Depotter et al., 2016). Hybridization is thus likely to be a powerful driver for adaptation to novel environments, including new hosts. However, it is important to note that we still know very little about the physiology of yeasts in natural environments, and while this hypothesis is indeed attractive, it is currently very difficult to demonstrate.
Hybridization outside Saccharomycotina has been described, although sampling in other groups is certainly not as extensive. The Cryptococcus neoformans species complex contains several hybrids between distantly related strains (Aminnejad et al., 2012; Cogliati et al., 2012; Li et al., 2012). For the AD serotypes of this species complex, one of the parental species appears to be geographically restricted to certain areas in Africa, while the hybrid has spread all over the world, suggesting that hybridization provided the pathogen with a selective advantage (Cogliati et al., 2012; Li et al., 2012). Mating between Cr. neoformans and Cr. gatti is possible in laboratory conditions, producing a viable offspring that possesses a highly unstable genome (Aminnejad et al., 2012). The hybrid rapidly loses and rearranges chromosomes in a manner similar to the parasexual cycle of Candida albicans, suggesting that hybridization could promote rapid adaptation by generating highly volatile genomic configurations (Aminnejad et al., 2012; Forche, 2012; Morrow & Fraser, 2013). Population analysis in Coccidioides (Eurotiomycetes), another genus including human pathogens, detected recent hybridization (Neafsey et al., 2010). Hybridization is common in the grass endophyte Epichlöe/Neotyphodium (Sordariomycetes) (Hamilton, Faeth & Dowling, 2009; Saari & Faeth, 2012; Shoji et al., 2015), and it has been shown that some of these events enhance its ability to colonize grass under stressful conditions. Numerous examples of hybridizations, both in vitro and in natural populations, have been described for plant pathogens in the Basidiomycota and Ascomycota (Park & Wellings, 2012; Stukenbrock et al., 2012; Sriswasdi et al., 2016; Stukenbrock, 2016). In this regard, introduction of crops to new territories, global trade, and movement of people around the globe provide routes for new contacts between otherwise geographically isolated populations that may potentially favour the formation of novel hybrid strains that could become emergent pathogens (Gonthier et al., 2004; Stukenbrock et al., 2011; Stukenbrock, 2016; Möller & Stukenbrock, 2017; Mixão & Gabaldón, 2018).
(2). Heterokaryosis
The number of nuclei in a fungal colony can easily be in the order of thousands (Roper et al., 2012, 2013). Hence, the assumption that all nuclei are genotypically identical is likely to be an oversimplification. The coexistence of two or more genetically distinct nuclear populations within a syncytium is referred to as heterokaryosis. If the phenotypic characteristics of these different populations of nuclei are different, variations in their proportions could translate into phenotypic variation in the whole colony. This was proposed and demonstrated on the basis of experiments using wild heterokaryotic Penicillium (Jinks, 1952; Strom & Bushley, 2016). As mentioned above, fungal mycellia can cover large areas while maintaining cytoplasm continuity (Sipos, Anderson & Nagy, 2018). Heterokaryosis would then affect local responses to stimuli within different areas of the same mycelium (Jany & Pawlowska, 2010; Roper et al., 2012).
At least theoretically, heterokaryons are expected to be unstable (Hallatschek & Nelson, 2008; Roper et al., 2012, 2013). If nuclear populations spread differentially based on their relative fitness and simple diffusion, then one of the populations eventually should be out‐competed by the other or disappear due to stochastic effects, in a similar manner to alleles within a population. ‘Nuclear death’ in filamentous fungi has been described; nuclei from senescent mycelia enter apoptosis and their nutrients are recycled (Maheshwari, 2005). This implies that nuclei with low fitness will not simply become diluted within a population. Phenotypic heterogeneity in nuclei sharing the same cytoplasm can be better understood in terms of population dynamics. For example, under the right conditions, such as growth in supplemented media, mutant nuclei can out‐compete the wild phenotype (Ryan & Lederberg, 1946; Maheshwari, 2005). On the other hand, nuclei carrying different mutations may complement each other, as shown for the carotenoid biosynthetic pathway in Phycomyces (De la Guardia et al., 1971; Sanz et al., 2002; Strom & Bushley, 2016). Mixed nuclear populations with distinct genetic backgrounds might, in theory, become stable under so‐called Black Queen scenarios (Morris, 2015) (Fig. 4). In this scenario, in a simple community of two members (A and B), if A loses the ability to perform a certain essential task that can be fulfilled sufficiently by B, then B will be ‘trapped’ and unable to lose that function, as this would mean the collapse of the community. If the same happens for another essential function, but this time in B, A and B would be mutually dependent of each other for survival. This situation was artificially generated in a classic experiment with Neurospora (Beadle & Coonradt, 1944; Strom & Bushley, 2016). The population nature of the heterokaryon adds a new layer of phenotypic complexity without involving the development of complex regulatory mechanisms (Maheshwari, 2005; Roper et al., 2012, 2013, 2015; Anderson et al., 2013; Dundon et al., 2016; Strom & Bushley, 2016) or increasing effective genome size, since each nucleus contains roughly the same genetic information. Experimental evidence in Neurospora tetrasperma suggests that, at least in this species, nuclear populations are kept at controlled proportions that vary during the life cycle of the fungus (Roper et al., 2012, 2013, 2015; Johannesson & Samils, 2014). Despite the astounding growth speed of this mould, reaching >5 mm/h in optimal conditions (Ryan, Beadle & Tatum, 1943), and its asynchronous nuclear division, the heterokaryon is stable over long periods of time. It is important to note that nuclear division in Neurospora is not restricted to hyphal tips, and active cytoplasmic currents provide the growing tip with fresh nuclei generated throughout the colony (Maheshwari, 2005; Roper et al., 2012, 2015). Cytoplasmic currents prevent stochastic extinction by actively mixing the cytoplasm of the whole colony (Roper et al., 2013, 2015; Johannesson & Samils, 2014). In Eremothecium, however, nuclear migration is mediated by the cytoskeleton (Gladfelter, 2006; Anderson et al., 2013; Dundon et al., 2016; Gibeaux et al., 2017). The ability to transport nuclei from distant regions in the network would provide a steady supply of nuclei, maintaining directed growth even if comparatively less‐fit nuclei are present, as is likely to be the case to some degree in any nuclear population (Nobre et al., 2014; Anderson et al., 2015). It is important to mention here that, despite cytoplasm continuity, nuclei seem to control cytoplasmic territories with considerable autonomy, a situation also known as ‘cells within cells’ (Nair et al., 2010; Roper et al., 2012, 2013; Anderson et al., 2013; Roberts & Gladfelter, 2015). Classic protoplasm fusion experiments force this autonomy to the point of allowing nuclei from even different fungal phyla to share a cytoplasm (Peberdy, 1979, 1989; Kavanagh & Whittaker, 1996; Strom & Bushley, 2016).
Figure 4.
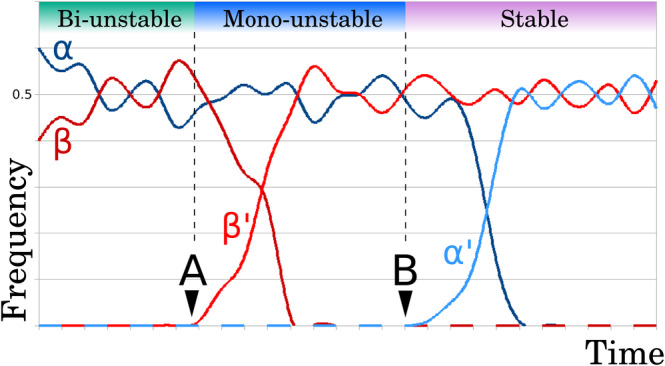
Black Queen population dynamics. Assume a hypothetical population of two types of nuclei, α and β within a single cytoplasm. If the frequency at which each type of nucleus divides is the same, the population would become bi‐unstable and would tend to collapse into a single type due to stochastic variation alone (drift). At point A in time a β mutant nucleus (β') appears that has a deletion in some key cellular function. Since both α and β can encode the mutated function, β' can survive. However, since β' presents the same characteristics as β, but makes savings in terms of the resources that were originally required for the function, β' would eventually out‐compete β. Once β has disappeared from the population, α and β' reach a new unstable equilibrium. β' should divide faster than α, but β' it still needs α to maintain a minimum level of the mutated cellular function. The population at this stage is mono‐unstable, as the only possible course is that drift drives β' extinct. At point B, a second cellular function deletion occurs within the α population (α'). Again, α' tends to out‐compete α. However, since α' and β' present complementary mutations, the new population is stable. Once this stage has been achieved, the system can only evolve towards further complementary reduction of its pair of components or towards the generation of a new nuclear population that restores the mutated functions by recombination of α' and β'.
Filamentous fungi can fuse their hyphae during their normal sexual cycle, and formation of dikaryotic hyphae is a defining trait of Dikarya. If the fused hyphae are too divergent these mixtures might become trapped, unable to undergo meiosis. Fungi have genetic mechanisms to prevent such unions (Saupe, 2000; Glass & Kaneko, 2003; Micali & Smith, 2006; Ishikawa et al., 2012; Van der Nest et al., 2014; Daskalov et al., 2017), although these barriers are not insurmountable. As mentioned above, compatibility is evaluated by check‐point mechanisms that can induce apoptosis. Sporadically, however, this system could be overcome and inter‐species heterokaryons may be formed. Furthermore, environmental factors, such as certain mycoviruses (Wu et al., 2017), are able to lower the thresholds of these recognition systems. The presence of short haploid regions in an apparently diploid genome is suggestive of the existence of two non‐recombining sub‐genomes and is compatible with heterokaryosis. Additionaly, significant deviations in the expected 1:1 proportions for reference versus alternative alleles might be used to identify heterokaryotic genomes in some cases. Successive formation of heterokaryons might allow the fungus to ‘update’ its genome dynamically, fusing its cytoplasm with new individuals as conditions change (Beadle & Coonradt, 1944; Strom & Bushley, 2016). However this carries the risk of being invaded by a faster dividing nuclear population, as well as being exposed to viruses and other infectious elements (Saupe, 2000; Aanen et al., 2008; Johannesson & Samils, 2014; Strom & Bushley, 2016). Even temporary, exotic unions might have long‐lasting effects by promoting genome rearrangements and transfer of genetic material (Kinsey, 1990; James et al., 2008; Van Der Does & Rep, 2012; Soanes & Richards, 2014). Heterokaryosis might emerge spontaneously without the need for mating or high heterozygosity, for instance through ploidy changes (Anderson et al., 2015). This is the case for the filamentous saccharomycotine Eremothecium gossypii (syn. Ashbya gossypii), which possesses hyphae whose nuclei divide independently (Gladfelter, 2006; Nair et al., 2010; Anderson et al., 2013; Dundon et al., 2016) and form populations with varying karyotypes. The proportion of nuclei carrying abnormal chromosome numbers varies under stressful conditions and with mycelial age (Fisher et al., 2012). Under environmental conditions that positively select for chromosome aberrations, the presence of normal nuclei within the cytoplasm might help buffer any deleterious effects (Toledo‐Hernández et al., 2013). While most experimental work has been carried out in filamentous Ascomycota, the dikaryon phase in these organisms is usually short lived. In Basidiomycota, on the other hand, the dikaryon forms most of the vegetative thallus. In most cases, the fungus controls nuclear division tightly by forcing synchronization through clamp connections (Shepherd, Orlovich & Ashford, 1993; Iwasa, Tanabe & Kamada, 1998; Maheshwari, 2005; Raudaskoski & Kothe, 2010). Even so, nuclear competition and population dynamics have been described and studied in Heterobasidion (Garbelotto et al., 2004; James et al., 2008; James, Johansson & Johannesson, 2009; Garbelotto & Gonthier, 2013; Giordano et al., 2018) and Termitomyces (Nobre et al., 2014). Arbuscular mycorrhizal fungi also seem to show high levels of heterokaryosis in nature (Bever & Wang, 2005; Boon et al., 2015; Wyss et al., 2016; Mathieu et al., 2018), forming nuclear populations that are mantained over time through the production of highly multinucleated spores (Bever & Wang, 2005; Jany & Pawlowska, 2010; Chagnon, 2014; Boon et al., 2015). However, there are considerable discrepancies among studies and methods and in the amount of estimated divergence (Kuo et al., 2014; Lin et al., 2014a; Ropars & Corradi, 2015). In these fungi, nuclear populations vary depending on the nature of the fungal host, which might explain the apparent low specificity of mycorrhizae–plant interactions (Angelard et al., 2014; Chagnon, 2014).
(3). Aneuploidy
Aneuploidy is the presence of different ploidy levels within the same genome, usually affecting entire chromosomes or large chromosomal regions. Aneuploidies can emerge spontaneously within populations (Torres, Williams & Amon, 2008) and tend to have dramatic fitness effects due to imbalances in gene dosage and in the formation of multipolar meiotic and mitotic divisions (Torres et al., 2008; Oromendia, Dodgson & Amon, 2012; Kumaran et al., 2013; Bonney, Moriya & Amon, 2015; Dodgson et al., 2016). However, under certain conditions aneuploidies might provide a selective advantage (Kravets et al., 2014; Bennett et al., 2014; Gerstein & Berman, 2015; Berman et al., 2016; Todd et al., 2017). For instance, in a medium containing a toxic compound, aneuploid cells that increase the dosage of genes related to detoxification could tolerate higher concentrations, thereby being fitter than their euploid counterparts. This has been observed in fungal pathogens acquiring resistance to antifungal drugs (Sionov et al., 2010; Farrer et al., 2013; Morrow & Fraser, 2013; Sun et al., 2014; Harrison et al., 2014; Anderson et al., 2017; Ksiezopolska & Gabaldón, 2018). While the same phenotypic effect would be possible with tandem gene duplications, the frequency of these mutations is much lower. Aneuploidies can easily revert to a euploid state if the stressful condition is transitory. If not, mutations that reduce the deleterious effects of the altered ploidy state while keeping the advantageous phenotype will be selected. Thus, aneuploidies serve as transitory, intermediate states during the process of adaptation to novel conditions (Farrer et al., 2013; Morrow & Fraser, 2013; Harrison et al., 2014; Hirakawa et al., 2015; Berman et al., 2016; Anderson et al., 2017). Finally, aneuploidies can result from unstable polyploidies. The best‐studied example of this is the parasexual cycle of C. albicans (Saccharomycotina) (Whelan et al., 1985; Forche et al., 2008; Forche, 2012; Brown et al., 2014; Hickman et al., 2015). This process is not related to mating in filamentous fungi that leads to the formation of a heterokaryon, also known as the parasexual cycle (Pontecorvo, 1956; Daskalov et al., 2017) (Fig. 5). In C. albicans, fusion of two diploid cells by non‐meiotic mating results in an effective tetraploid state. The tetraploid is genomically unstable, and suffers concerted chromosome loss that recovers stability. Thus, the parasexual cycle promotes aneuploidies and helps this pathogenic yeast to adapt to the host immune system, as well as to pharmacological treatments (Bennett et al., 2014; Harrison et al., 2014; Gerstein & Berman, 2015). A parasexual cycle in C. albicans has apparently evolved through loss of part of the meiotic recombinatory machinery, but aneuploidy might still be an important genome stabilizer in cases in which meiotic recombination is impaired, such as hybridization (see Section III.1). Aneuploid populations are also well known for other members of Saccharomycotina isolated from industrial environments, such as Saccharomyces (Borneman et al., 2011; Walther et al., 2014; Zhu, Sherlock & Petrov, 2016) and Brettanomyces (Hellborg & Piskur, 2009; Borneman et al., 2014; Avramova et al., 2018), as well as in the frog‐killing chytrid Batrachochytrium dendrobatidis (Joneson et al., 2011; Rosenblum et al., 2013).
Figure 5.
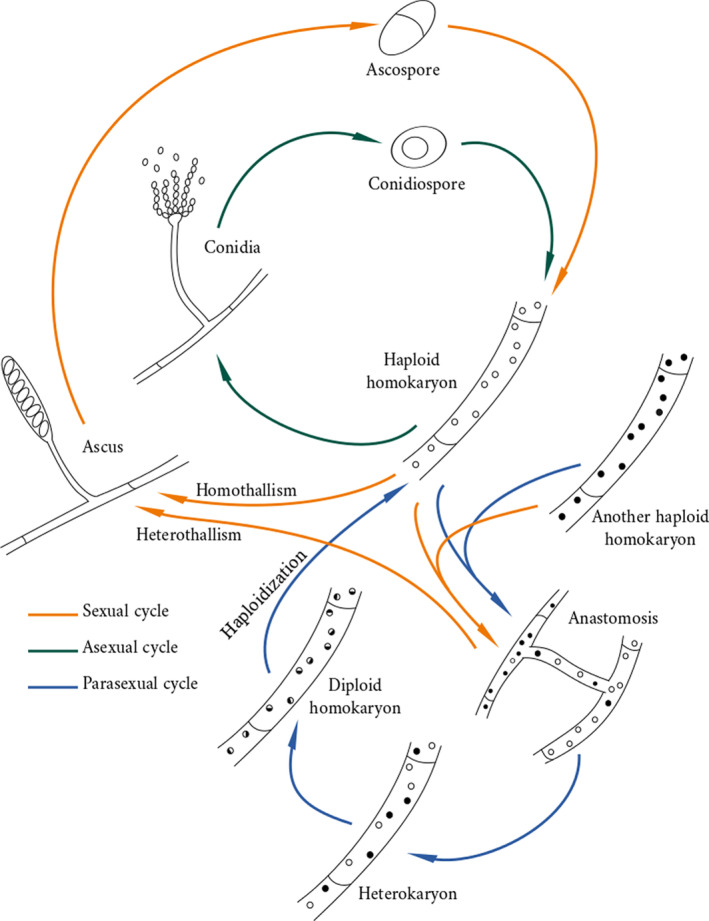
Life cycle of a generic Pezizomycotina. In most cases the vegetative thallus is formed by a homokaryotic mycelium. Sexual cycle: a homokaryotic mycelium might transform into an ascoid mycelium, where nuclei undergo meiosis and form ascospores in asci. This change might occur after fusion with another compatible mycelium, producing recombination between two strains (heterothallism), or occur in the nuclear population within the mycelium without genetic exchange (homothallism). Asexual cycle: a vegetative mycelium can form conidia that produce conidiospores by simple mitosis. Parasexual cycle: two compatible vegetative mycelia might fuse through anastomosis, interchanging nuclei and forming an haploid heterokaryon. At some point, the different nuclei might fuse to form a diploid homokaryon that undergoes concerted chromosome loss to regain a haploid state in the absence of meiosis. This process returns the mycelium to a haploid homokaryon state.
Over short evolutionary timescales, chromosome aberrations possess some other emergent ecological properties. Even in highly homogeneous environments, such as liquid laboratory cultures, small microniches that impose differential selective pressures might emerge (Rosenzweig et al., 1994; Ibarra, Edwards & Palsson, 2002; Wortel et al., 2016). If that is the case, two or more reversible chromosome states might coexist. Polyploidies might produce meiotic and mitotic impairments and involve higher nitrogen and phosphorus costs per cell division, which should slow growth under optimal conditions (Otto, 2007; Schoenfelder & Fox, 2015; Scott et al., 2017). It is important to note that such conditions are met almost exclusively in laboratory settings. It is then reasonable to assume that putative polyploids or aneuploids that have been grown in axenic cultures for long periods could have streamlined their genome towards a haploid or diploid state that maximizes growth rate. Non‐canonical chromosome conformations in environmental fungi might even prevent growth in experimental conditions, thus contributing to plate count anomalies (Staley & Konopka, 1985; Zak & Visser, 1996; Bridge & Spooner, 2001; Anderson & Cairney, 2004). This could explain why culture‐based diagnostic methods of fungal infections are prone to negative results (Ostrosky‐zeichner, 2012). Genome sequencing projects tend to focus on reference strains or on strains that a particular laboratory uses as a model. In both cases, it is very likely that these strains have been grown for years in non‐limiting conditions. Even in cases in which an isolate with chromosome aberrations can be sequenced, this requires specialized experimental and computational approaches that are far from standard, involving expertise and increased costs. Polyploid strains, particularly allopolyploids, produce highly fragmented assemblies that often present an inflated assembly size, due to their high heterozygosity (Kajitani et al., 2014; Safonova, Bankevich & Pevzner, 2015; Pryszcz & Gabaldón, 2016; Huang, Kang & Xu, 2017). The same holds for supernumerary chromosomes, which might pass unnoticed as a collection of highly fragmented scaffolds within an otherwise typical assembly. Because of this, many hybrids, polyploids and aneuploids might have been already sequenced but still not described (M. A. Naranjo‐Ortiz, M. Molina‐Marín, V. Mixào & T. Galbadón, in preparation).
(4). Polyploidy
As in plants, fungi are able to undergo autopolyploidization, in which all chromosomes have the same genotype at the moment of duplication; or allopolyploidization, in which the chromosomes are genetically distinct (Otto, 2007; Albertin & Marullo, 2012; Todd et al., 2017). Over longer timescales, these events provide plenty of opportunities for innovation through extensive gene duplication followed by subfunctionalization and neofunctionalization (Conant & Wolfe, 2008; Albertin & Marullo, 2012; Magadum et al., 2013). However, autopolyploidization does not provide genotypic innovation in the short term, a necessity for fixing this mutation within a population. Despite the lack of genotypic novelty, autopolyploidy might still provide advantages. This process usually produces larger cells with a reduced surface area to volume ratio (Otto, 2007; Schoenfelder & Fox, 2015). This has implications for membrane transport, which in turn affects general metabolism. An increased size might be selectively advantageous against certain selective pressures, such as phagocytic predation. This seems to be the case for titan cells in Cryptococcus, which are polyploid vegetative cells resistant to attack by the vertebrate immune system, from which infectious diploid and aneuploid cells emerge (Okagaki & Nielsen, 2012; Gerstein et al., 2015). Finally, a putative tetraploid state has been described for a widespread strain of the microsporidian Nosema ceranae, although the putative selective advantages in this case remain unclear (Pelin et al., 2015).
Unlike in plants, for which many ancient polyploidization events have been identified, very few have been identified in fungi (Campbell et al., 2016). There is evidence of an ancient allopolyploidization event that occurred 100 Mya in the Saccharomycetaceae (Saccharomycotina), affecting the common ancestor of the genera Saccharomyces, Nakaseomyces, Kazachstania and Naumovozyma (Wolfe & Shields, 1997; Hittinger, 2013; Marcet‐Houben & Gabaldón, 2015). The order Mucorales (Mucoromycotina) seems to have experienced at least two well‐characterized events of this kind, an ancient event affecting the ancestor of Mucor and Phycomyces (Corrochano et al., 2016), and a more recent one within the genus Rhizopus (Ma et al., 2009). The apparent scarcity of ancient polyploidization in fungi, compared with animals or plants, is at odds with their expected higher plasticity and is likely the result of greater difficulty in detecting them (Campbell et al., 2016). Finally, an additional whole‐genome duplication event has been described for the hyperhalotolerant black yeast Hortaea werneckii (Lenassi et al., 2013; Sinha et al., 2017). This event has contributed to the expansion of cationic transporters, important in surviving high salinity. The origin of this whole‐genome duplication has been ascribed to an inter‐species hybridization event (Gostinčar et al., 2018). We are confident that many ancient whole‐genome duplication events resulting from either auto‐ or allopolyploidization events, are yet to be identified and will be discovered once sufficient genome sampling is available for enough fungal clades. Very likely, some of these events will correlate with the emergence of important traits and ecological transitions within the group, just as has been observed for plants and metazoans. However, cytogenetic studies in fungi are much less prevalent than in plants and animals, and require more costly approaches such as pulse‐field electrophoresis. In comparison with these groups, fungi tend to have shorter generation times and larger population sizes, higher ecological dependency on the speed their nuclei can complete meiosis, more active haploid estates and an absence of embryonic programs. All these traits translate into much faster evolutionary rates, higher chromosome plasticity and higher selective pressures for a reduced genome, while having less freedom in this regard due to higher genome compaction. These methodological and biological factors make the detection of footprints of ancient polyploidies such as synteny, chromosome numbers or widespread and phylogenetically restricted gene duplication more difficult than in plants (Jiao et al., 2011; Leitch & Leitch, 2012; Carretero‐Paulet et al., 2015; Del Pozo & Ramirez‐Parra, 2015; Soltis et al., 2015) and animals (Van de Peer, Taylor & Meyer, 2003; Van de Peer, 2004; Dehal & Boore, 2005; Kenny et al., 2016; Schwager et al., 2017; Li et al., 2018).
IV. METABOLIC COMPLEXITY
The last cornerstone defining the complexity of fungal life lies in the diversity of their metabolic pathways. This enzymatic diversity, coupled with their cellular versatility, gives fungi access to a vast array of substrates. Additionally, fungi are often able to synthesize bioactive compounds that play key roles in their interactions with the rest of the biosphere. One of the main physiological achievements of fungi is their ability finely to localize these biochemical pathways within their mycelial networks. An illustrative example concerns the appresoria of many parasitic fungi. The appresorium itself is a form of hypha, with a particular structure that enables it to exert pressure at a highly localized point. Many fungi complement this physical assault with hydrolytic enzymes that weaken the host cell walls to facilitate invasion. From an evolutionary perspective, new metabolic capabilities typically emerge by some form of gene duplication, followed by functional differentiation between the duplicates. In this regard, chromosomal aberrations and other genome architecture changes have a great impact on gene dosage and can lead to the stabilization of gene duplicates. Gene loss and duplication is a powerful way to increase metabolic diversity, but it is limited by the requirement for pre‐existing genes. Acquisition of novel genes through horizontal gene transfer (HGT), often from distantly related genomes, could provide a source of immediate and radical novelty. In addition, novelty might emerge through the establishment of new regulatory networks, often through changes in the physical organization of the genome. Finally, recent studies focusing on intraspecific genomic variation offer a new perspective on the dynamic nature of fungal genomes, as genes are gained, lost and exchanged across partially isolated populations. In this section we will discuss the evolutionary processes that shape the biochemical dimensions of fungal biology with special emphasis on the origin, diversification and functional implications of their vast secondary metabolism.
(1). Secondary metabolites in fungal biology
The filamentous nature of most fungi allows these organisms to explore and control a certain territory. This control requires some form of dissuasion, that in the case of fungi is achieved through limiting nutrient access to competitors and, more importantly, through the production of toxic secondary metabolites (Keller, 2015, 2018; Bills & Gloer, 2016). These toxins protect the fungus from invertebrate grazers (Caballero Ortiz et al., 2013; Rohlfs, 2014), delimit exclusion areas for other competing mycelial networks (Boddy & Hiscox, 2016), and regulate the so‐called ‘mycosphere’ (Boer et al., 2005; Warmink & van Elsas, 2008; Boersma et al., 2010; Nazir et al., 2010; Haq et al., 2014), i.e. the microbial populations associated with the hyphal surface and its immediate proximity. In parasitic fungi, secondary metabolites can be used to inflict direct damage to the host (necrotrophic strategies) (Arunachalam & Doohan, 2013), mitigate host immune responses (Williams et al., 2011), induce changes in host tissues (Bömke & Tudzynski, 2009) or, in the case of animal parasites, induce behavioural changes (Boyce et al., 2019). Not all secondary metabolites are toxins, however. Other known functions include cell signalling, pigmentation, osmotic protection or metal chelation (Bills & Gloer, 2016; Rokas, Wisecaver & Lind, 2018). Although the diversity of bioactive compounds synthesized by fungi is enormous, they ultimately fall into a few broad categories of enzymatic pathways (Zhong & Xiao, 2009; Keller, 2015; Bills & Gloer, 2016). The main groups of secondary metabolites are small nuclear‐encoded peptides, non‐ribosomal peptides (NRPs), polyketides (PKs), terpenoids and derivatives from the shikimate pathway (Bills & Gloer, 2016; Rokas et al., 2018). Innovations in secondary metabolism might emerge by duplication and modification of existing pathways or by acquisition of novel enzymes. Secondary metabolites might confer important selective advantages to Fungi even when produced at very low concentrations, albeit only under certain conditions. On the other hand, production of these compounds usually implies an important investment in terms of energy and nutrient cost, not to mention the potential effects of the toxin itself on the producer fungus (Chanda et al., 2009; Brakhage, 2013; Keller, 2018). Notably, most of these functions are dependent on responses to these compounds by other organisms, implying that their selective advantages might disappear if the target organisms respond differently. This has important consequences from the perspective of population genetics. Whenever the ability to synthesize a successful secondary metabolite appears in a genome, the associated selective advantage would drive the fixation of these genetic components in the population. Such evolutionary dynamics are ideal for the proliferation and establishment of rare evolutionary events, including HGT (Richards et al., 2011; Wisecaver, Slot & Rokas, 2014; Wisecaver & Rokas, 2015). Analogously, if the conferred benefit is restricted to uncommon circumstances, it is very likely that the absence of selection will result in loss of the ability to produce the compound in some subpopulations (Spatafora & Bushley, 2015; Bills & Gloer, 2016; Rokas et al., 2018). This volatility further complicates evolutionary studies, as it will lead to patchy distributions in signals that are commonly used to discern putative gene duplications and transfers (Wisecaver et al., 2014).
(2). Metabolic gene clusters
A peculiarity of fungal genomes is the tendency to group together genes that are functionally related, particularly those encoding enzymes from the same metabolic pathway. Clustering in non‐metabolic genes has also been described and seems to be an important evolutionary process in many fungal lineages (Marcet‐Houben & Gabaldón, 2019). There are clear differences regarding the evolutionary dynamics between metabolic and non‐metabolic gene clusters (Marcet‐Houben & Gabaldón, 2019), but most studies have focused on the former. Metabolic gene clusters exist in other eukaryotes, particularly in plants (Boycheva et al., 2014; Nützmann, Huang & Osbourn, 2016), but not in the diversity and abundance found in fungal genomes. Some important pathways can be found clustered in most Fungi (Slot & Hibbett, 2007; Slot, 2017), while others appear clustered only in some species or strains (Inglis et al., 2013; Slot, 2017; Rokas et al., 2018). Secondary metabolism pathways require the coordinated action of a wide set of enzymes, which has been invoked to explain why these pathways appear clustered in fungal genomes (Bills & Gloer, 2016; Rokas et al., 2018). However, some pathways that are clustered in fungi appear unclustered in other eukaryotes, suggesting that clustering itself carries an associated selective advantage in fungi (Wisecaver et al., 2014; Slot, 2017; Rokas et al., 2018; Marcet‐Houben & Gabaldón, 2019). Metabolic gene clusters often include more than enzyme‐encoding genes, and may involve other genes such as transporters or transcription factors. The presence of genes in the same cluster should facilitate coordinated expression of the different components, similarly to the situation for bacterial operons (Fischbach, Walsh & Clardy, 2008; Brakhage, 2013; Keller, 2018). Intermediate compounds in these pathways and many final products that will be exported extracellularly are often highly toxic, and coordinated expression might be necessary to avoid hazardous accumulation in the cytoplasm (Wisecaver et al., 2014; Keller, 2015; Bills & Gloer, 2016; Slot, 2017; Rokas et al., 2018). This same logic applies when considering events that might break the cluster apart. For instance, accumulation of toxic intermediates has been proposed to ensure cluster completeness during HGT events (Lawrence, 1999; Fischbach et al., 2008; Wisecaver et al., 2014; McGary, Slot & Rokas, 2013; Slot, 2017; Krause et al., 2018). In addition, if the pathway genes are scattered across the genome, aneuploidies or chromosome loses could affect its components differentially, rendering the pathway unbalanced or incomplete. Horizontally transferred bacterial operons or polycistrons would necessarily appear tightly clustered (Lawrence, 1999; Fischbach et al., 2008), although relatively few clusters seem to have this origin (Wisecaver et al., 2014; Marcet‐Houben & Gabaldón 2019). Importantly, many gene clusters contain genes that protect the fungus from the toxicity of the cluster products (Bills & Gloer, 2016; Slot, 2017; Rokas et al., 2018). There are some cases in which more than one cluster is required for the synthesis of a particular compound, representing an exception to the otherwise general rule of ‘one cluster, one secondary metabolite’ (Lo et al., 2012; Bills & Gloer, 2016).
Due to the aforementioned factors, tight regulation of cluster expression appears essential (Brakhage, 2013; Slot, 2017). Epigenetic mechanisms have emerged as key regulators of fungal secondary metabolism, a situation probably facilitated by the physical proximity of their components (Gacek & Strauss, 2012; Brakhage, 2013; Slot, 2017; Keller, 2018). Based on this, mutation or inhibition of chromatin signalling components are beginning to be used to induce the expression of cryptic metabolic clusters (Cichewicz, 2010; Brakhage & Schroeckh, 2011; Yin & Keller, 2011; Brakhage, 2013; Keller, 2018). Global regulators affect the expression of a wide array of clusters in response to external or internal stimuli in Pezizomycotina. The best‐studied global regulators are transcription factors from the Velvet complex (Sarikaya Bayram et al., 2010; Yin & Keller, 2011; Bayram & Braus, 2012; López‐Berges et al., 2013; Lan et al., 2014; Schumacher et al., 2015; Estiarte et al., 2016; Akhberdi et al., 2018). Velvet complex proteins are present across all groups of fungi, but their role in regulating secondary metabolism has barely been studied outside Pezizomycotina (Bayram & Braus, 2012; Todd et al., 2014). Velvet complex is also important for sexual development in both Ascomycota and Basidiomycota. External factors include nutrient availability, light, pH or injury, while internal factors refer to specific cell types, senescence or the sexual cycle (Bok & Keller, 2004; Yin & Keller, 2011; Brakhage, 2013; Estiarte et al., 2016; Keller, 2018). These regulators allow the fungus to coordinate the cost‐effective production of secondary metabolites under specific conditions, and in a coordinated manner. Some of these regulators seem to be evolutionarily conserved across large phylogenetic distances, although functional differences might exist (Bok & Keller, 2004; Brakhage, 2013; Jain & Keller, 2013; Estiarte et al., 2016). Similar regulators might exist outside Pezizomycotina that remain to be discovered. Additionally, to prevent the organism producing its whole arsenal at the same time, each cluster must respond differentially to many other regulatory elements (Brakhage, 2013; Bills & Gloer, 2016; Keller, 2018). Many clusters contain transcription factors that regulate their own expression (Brakhage, 2013; Bills & Gloer, 2016; Slot, 2017; Rokas et al., 2018; Marcet‐Houben & Gabaldón, 2019). This provides greater independence in terms of the complexity hypothesis (Jain, Rivera & Lake, 1999), and enhances the chances of successful HGT (Wisecaver & Rokas, 2015; Rokas et al., 2018). It has been found that transcriptional factors included in a cluster can sometimes affect the expression of other clusters (Martín, 2017; Keller, 2018). Such cluster cross talk has important evolutionary implications, as the phenotypic effects of a recently acquired cluster are potentially far greater than the mere production of its metabolite. Fungal genomes reveal a much broader diversity of gene clusters than described secondary metabolites, indicating that many fungi encode clusters that are only expressed under unknown conditions (Chiang et al., 2009; Brakhage, 2013; Gerhards et al., 2015; Bills & Gloer, 2016; Keller, 2018). Some of these clusters might even be inhibited by others (Brakhage, 2013; Keller, 2018), and thus deletion of known clusters could be an overlooked strategy for mining novel compounds (Gerke & Braus, 2014).
Quite often, clusters show conservation across different fungi. Such clusters show a set of genes that appear physically linked within a given chromosome region, but whose order is often highly variable. This phenomenon is known as mesosynteny and seems to be a peculiarity of genomes in the Pezizomycotina (Hane et al., 2011). Many evolutionarily widespread gene clusters present patchy taxonomic distributions, which has sometimes been explained as being the result of HGT (Khaldi et al., 2008; Marcet‐Houben & Gabaldón, 2010; Slot & Rokas, 2011; Wisecaver et al., 2014; Wisecaver & Rokas, 2015; Slot, 2017; Rokas et al., 2018; Marcet‐Houben & Gabaldón, 2019) or convergent evolution (Marcet‐Houben & Gabaldón, 2019). Gene loss is likely common in gene clusters, which can explain patchy distribution patterns. Even different species of the same genus might contain radically different repertoires of metabolic clusters, as observed in Penicillium species (Malmstrøm, Christophersen & Frisvad, 2000; Marcet‐Houben et al., 2012; Julca et al., 2016; de Vries et al., 2017; Koul & Singh, 2017). A particular gene cluster might exist in a given species, but be absent in the analysed strain (Kelly & Ward, 2018; Plissonneau, Hartmann & Croll, 2018; Syme et al., 2018; McCarthy & Fitzpatrick, 2019). Finally, the ability to produce particular metabolites might arise independently through convergent evolution. For instance, a recent study has found that cicada pathogens in the genus Massospora (Entomophthorales) are able to produce the psychotropic psilocybin, previously known only from ‘magic mushrooms’ in the Agaricomycetes (Boyce et al., 2019). Inability to detect certain expected intermediate compounds suggests that Massospora have probably acquired the ability to synthesize psychotropic compounds through an independent pathway. The composite nature of gene clusters opens many evolutionary possibilities. Clusters might mutate, duplicate, divide into two, combine, or lose or gain new genes, occasionally through HGT (Fischbach et al., 2008; Martín & Liras, 2016; Slot, 2017; Rokas et al., 2018; Olarte et al., 2019). Each of these possibilities is rife with methodological complications for evolutionary studies (Bull et al., 1993; Castresana, 2007; Fischbach et al., 2008). However, it is important to note that most clusters are in silico predictions, detected based on co‐linearity. This clearly biases our knowledge against unclustered biosynthetic pathways, as those must be detected using more laborious techniques. Even worse, it is difficult to estimate how many of the predicted clusters are genuine. While this has not been evaluated in Fungi, analyses on plant metabolic clusters using patterns of transcriptional coexpression suggest that most of the predicted clusters are false positives (Wisecaver et al., 2017).
Pezizomycotina are the best known group of fungi that produce secondary metabolites, and most of the information regarding the evolutionary dynamics of clusters, their regulatory expression and even their derived pharmacology comes from members of this clade. Free‐living Pezizomycotina tend to possess a higher diversity of PKS and NRPS gene clusters (Rokas et al., 2018). HGT seems to play a much more important role in the evolution of metabolic diversity in Pezizomycotina than in other fungi (Schmitt & Lumbsch, 2009; Fitzpatrick, 2012; Wisecaver et al., 2014; Rokas et al., 2018), and there is convincing evidence for a high frequency of cluster gene transfer among members of this subphylum (Wisecaver et al., 2014; Bills & Gloer, 2016). Lichen‐forming fungi produce an even greater diversity of compounds, with several classes not found in other filamentous Ascomycota (Boustie & Grube, 2005; Stocker‐Wörgötter, 2008; Molnár & Farkas, 2010). One peculiarity of these organisms is that their secondary metabolites often accumulate in crystalline forms in extracellular spaces. However, due to the paucity of available lichen genomes (McDonald et al., 2013; Meiser et al., 2017) and the recent discovery of several groups of novel endolichenic organisms (Spribille et al., 2016), assigning metabolic pathways to a particular mycobiont is not straightforward, and thus the full scope of cryptic secondary metabolism is much less understood than for non‐lichenic fungi (Stocker‐Wörgötter, 2008; Molnár & Farkas, 2010; Bills & Gloer, 2016).
Members of the Agaricomycotina are the other main group of secondary metabolite‐producing fungi (Schüffler & Anke, 2009; Zhong & Xiao, 2009; Quin, Flynn & Schmidt‐Dannert, 2014; Stadler & Hoffmeister, 2015; Lin et al., 2019). HGT seems to be relatively rare among this group, and cluster gene duplication appears to be the main evolutionary force behind metabolic diversification (Wisecaver et al., 2014). Agaricomycetes produce a relative abundance of alkaloid and terpenoid compounds and a reduced amount of NRPs, compared to Pezizomycotina (Brakhage, 2013; Bills & Gloer, 2016; Rokas et al., 2018). The diversity of secondary metabolites in Agaricomycetes is not fully explored, mostly due to limitations on de novo gene prediction strategies that are optimized for the Pezizomycotina (Quin et al., 2014), although thousands of compounds have been chemically identified from this group (Schüffler & Anke, 2009; Zhong & Xiao, 2009; Stadler & Hoffmeister, 2015). Information regarding genetic regulation of secondary metabolism in Agaricomycetes is very limited. Genomic analyses for Ganoderma sinense suggest that global regulatory proteins control secondary metabolism through epigenomic mechanisms, including Velvet complex proteins similar to those found in Pezizomycotina (Zhu et al., 2015).
Gene cluster composition and evolutionary dynamics is markedly different in other fungal lineages. Within Ascomycota, the Saccharomycotina are notable for having lost virtually all their secondary metabolism arsenal. This reduction even affects the Velvet complex, whose components are completely absent in S. cerevisiae and C. albicans, but present in Yarrowia lipolytica (Bayram & Braus, 2012). This metabolic reduction is shared by other yeast‐forming groups in the Taphrinomycotina and Basidiomycota mostly due to genome changes associated with their biotrophic lifestyles, although the complete loss of Velvet complex seems rare (Bayram & Braus, 2012).
Secondary metabolite production is poorly studied in zygomycetous and zoosporic lineages. Arbuscular mycorrhizae fungi in the Glomeromycotina are known to produce secondary metabolites (e.g. isoprenoids) that mediate communication with plant roots (Strack et al., 2003). Little is known regarding the diversity, structure or regulation of these metabolic clusters, but genome sequencing suggests that these fungi have a reduced repertoire of secondary metabolic pathways compared to other zygomycetous lineages (Morin et al., 2019), similar to biotrophic plant parasites in the Dikarya. Most members of the Mucoromycotina and Mortierellomycotina have a typical mould lifestyle (filamentous, saprotrophic, free‐living) and as such they are capable of secreting secondary metabolites. However, the predicted metabolic diversity in these groups lags behind that of Pezizomycotina and Agaricomycotina (Lebreton et al., 2018; Rokas et al., 2018). For example, comparative analyses of five Mucor species revealed only one NRPS and one PKS gene cluster. Surprisingly, though, the genome of Mortierella alpina (Mortierellomycotina) contains an expansion of small secreted nuclear encoded peptides (defensines) (Wu, Gao & Zhu, 2014). Small peptides are very difficult to predict accurately by de novo gene prediction models and it is entirely possible that their diversity, particularly in zygomycetous fungi, has been largely overlooked. Many zygomycetous fungi exhibit associations with cytoplasmic bacteria with the ability to produce secondary metabolites (Partida‐Martinez et al., 2007). Little is known regarding the metabolic production of members of the Zoopagomycota or the zoosporic lineages. Alkaloid compounds have been described in several members of the Entomophthoromycotina, with different activities in their insect hosts (Claydon, 1978; Wrońska et al., 2018; Boyce et al., 2019). However, very little is known regarding the genomic organization of these pathways. Most analysed species (Zoopagomycotina, Kickxellomycotina, Blastocladiomycota and Chytridiomycota) present no more than one or two NRPS genes, although the mycoparasite Dimargaris cristalligena (Kickxellomycotina) is remarkable for possessing 27 of these genes (Ahrendt et al., 2018). Most analysed species in this study show highly reduced thalli and a biotrophic lifestyle, which seems to be common in this group and could explain their reduced secondary metabolism. It is well known that secondary metabolism is highly compartmentalized in Pezizomycotina (Chanda et al., 2009; Martín, Ullán & García‐Estrada, 2010; Roze, Chanda & Linz, 2011; Kistler & Broz, 2015), which should help the fungus to limit self‐toxicity. Thus, the absence of a septal apparatus in most zygomycetes might limit the development of their metabolic repertoire. It is noteworthy that Dimargaris belongs to the only zygomycetous group that shows well‐developed septa. Zoosporic lineages seem to contain a secondary metabolism pool similar to that of zygomycetous fungi (Spatafora & Bushley, 2015) and Velvet complex proteins (Bayram & Braus, 2012), but very few details are known. Interestingly, methylation patterns in zygomycetous lineages and the Neocallimastigomycotina are unusual, as they show widespread gene activation mediated by adenine N6 methylation (Mondo et al., 2017). This would suggest that global epigenetic regulators others than Velvet complex might be capable of regulating secondary metabolism in early‐diverging fungi.
(3). Genes on the move: horizontal gene transfer in Fungi
After the publication of the first drafts of the human genome, the field of HGT in eukaryotes entered its ‘Dark Age’. The human genome paper included the claim that more than 200 genes were putatively acquired from bacteria via HGT (Lander et al., 2001). However, this finding was later rejected as an artefact resulting from insufficient sampling in the genomic comparisons (Stanhope et al., 2001; Salzberg, 2017). This led to a decade in which HGT detection in eukaryotic genomes was faced with severe skepticism, with most genome studies not even exploring that possibility. Yet, evidence began to accumulate supporting a not‐uncommon occurrence of HGT in some eukaryotic lineages, including fungi. The detection of HGT events usually requires finding incongruences between a gene phylogeny and the known species tree (Galtier & Daubin, 2008; Leigh et al., 2011; Grant & Katz, 2014; Wisecaver & Hackett, 2014; Katz, 2015; Nguyen et al., 2015; Soucy, Huang & Gogarten, 2015; Szöllősi et al., 2015; Naranjo‐Ortíz et al., 2016; Wisecaver et al., 2016; Dupont & Cox, 2017). This requirement has been traditionally difficult to meet for most eukaryotic lineages, since genomic studies have long been focused on just a few economically important species, limiting taxonomic coverage. However, fungi were an early exception to this. With a taxonomically robust backbone of sequenced genomes and DNA generally coming from axenic cultures, HGT claims in Fungi are relatively robust. Once it became possible to search for HGT in a wide taxonomic range of Fungi, it was revealed that these evolutionary phenomena affect Pezizomycotina preferentially (Marcet‐Houben & Gabaldón, 2010; Wisecaver et al., 2014; Gluck‐Thaler et al., 2015), at least compared with Saccharomycotina and Basidiomycota. Furthermore, in some cases, HGT has been shown to have been key in the appearance of important adaptations, as shown in Neocallimastigomycotina and its outstanding carbohydrate‐degrading enzymatic pool (Garcia‐Vallvé, Romeu & Palau, 2000; Rosewich & Kistler, 2000; Murphy et al., 2019).
Despite the increasing evidence for HGT in Fungi, a key question remains unsolved. The exact mechanism that allows transference and integration of external genetic material into the genome of a fungus is not understood (Andersson, 2005, 2009; Richards et al., 2006, 2011; Soanes & Richards, 2014; Husnik & McCutcheon, 2017). Fungal cells are surrounded by a thick cell wall and do not have phagotrophic capabilities, thus ingested microbes cannot be a source of HGT, as posited by the ‘you are what you eat’ hypothesis (Doolittle, 1998). However, reports of in vitro recombination between bacteria and yeasts have been published (Heinemann & Sprague, 1989, 1991; Inomata, Nishikawa & Yoshida, 1994; Sawasaki, Inomata & Yoshida, 1996; Moriguchi et al., 2013; Suzuki, Moriguchi & Yamamoto, 2015). Agrobacterium‐like bacteria can be used to transform filamentous fungi in the laboratory, suggesting that a similar process could mediate HGT in nature (Lacroix et al., 2006; Jiang et al., 2013; Lacroix & Citovsky, 2016). HGT can span long portions of a chromosome, or even whole chromosomes, which allows the transfer of groups of genes that are clustered together. Some of these genetic elements might have evolved for mobility, acting as transferable cassettes of functionally linked genes. Such is the case for mobile chromosomes in certain plant pathogens, such as Fusarium and Zymoseptoria (Akagi et al., 2009; Coleman et al., 2009; Mehrabi et al., 2011; Van Der Does & Rep, 2012; Vlaardingerbroek et al., 2016; Mehrabi, Mirzadi Gohari & Kema, 2017). These regions contain many genes related to host‐specific pathogenicity, and their acquisition allows non‐pathogenic strains to be infective in a new host. While the mechanisms of these genetic exchanges are not fully understood, we are starting to have a better picture of what type of genes are transferred. The ‘complexity hypothesis’ (Jain et al., 1999) describes the gene content of an organism in terms of network connectivity and posits that new members can be more easily added to the boundaries of existing networks. This implies that simple transporters, simple enzymatic pathways or secondary metabolism genes are more likely to be transferred than highly interconnected proteins in the network. Indeed, some types of enzymes, such as amino acid racemases seem particularly prone to HGT in fungi and other microbial eukaryotes, although their physiological role in the receiving organisms remains to be clarified (Marcet‐Houben & Gabaldón, 2010; Naranjo‐Ortíz et al., 2016). At the other extreme, genes related to information processing, such as translation or transcription, are among the most recalcitrant genes in terms of HGT. Comparative genomic studies support the paradigm of the complexity hypothesis (Marcet‐Houben & Gabaldón, 2010; Wisecaver et al., 2014; Wisecaver & Rokas, 2015).
Given the difficulties in detecting HGT, it is likely that a fair portion of HGT events are simply not detected. In general, it is far easier to detect an event if the resulting phylogenetic incongruence is large, such as that caused by interdomain transfers (Galtier & Daubin, 2008; Marcet‐Houben & Gabaldón, 2010; Leigh et al., 2011; Haegeman et al., 2014; Naranjo‐Ortíz et al., 2016). In the hypothetical scenario of HGT between relatively closely related species (for instance, members of the same family), alternative hypotheses such as differential gene loss or incomplete lineage sorting cannot be ruled out easily. In addition, ancient HGT events might have an untraceable phylogenetic signal, perhaps to be expected given that transferred genes may undergo periods of rapid accumulation of changes to accommodate to their new environments, or that databases might lack adequate representation of the donor group. HGT might occur several times independently for the same gene, either through sequential transfers or by independent transfers from a phylogenetically close donor. These routes would create complex phylogenetic patterns that could prove difficult to interpret. In addition, some protein families evolve in ways that result in intricate molecular phylogenies that are difficult to resolve. For example, NRPSs are modular multidomain enzymes that can evolve by losing, duplicating or shuffling functional domains, rather than simply by point mutations (Weber & Marahiel, 2001; Marahiel, 2009; Hur, Vickery & Burkart, 2012). Many protein families display extremely low sequence conservation, which makes the reconstruction of accurate molecular phylogenies difficult, even over small evolutionary distances (Ponting, 2017). This is true for secreted peptidases in Fungi (Poppe et al., 2015; Krishnan et al., 2018). Both NRPSs and peptidases are well‐known virulence factors in many fungi or have roles relevant to certain niches, are compatible with the complexity hypothesis, and have a huge diversity in filamentous fungi, making them prime candidates for HGT events.
HGT can play important roles in the adaptation of species to novel niches or lifestyles, and has been shown to be involved in short‐term evolutionary transitions. For example, the acquisition of a toxin‐encoding gene by Pyrenophora tritici‐repentis from Stagonospora nodorum turned a fungus causing occasional spots in wheat leaves into a devastating pest in a matter of decades (Friesen et al., 2006; Oliver & Solomon, 2008). HGT between plant pathogens seems to be fairly widespread and, in certain cases, patterns of repeated HGT between the same groups have been reported (Khaldi et al., 2008; Armijos‐Jaramillo, Sukno & Thon, 2014; Bettini et al., 2014; Gluck‐Thaler et al., 2015; Qiu et al., 2016; Yin et al., 2016). HGT has been involved in drastic shifts in lifestyle, such as the acquisition of an entomopathogenic habit from a grass endophyte lifestyle in the genus Metarrhizium (Zhang et al., 2019). This mounting evidence suggests that HGT is an important factor promoting the rise of novel plant pathogens. Another example of recent acquisition of novel genes with functional implications concerns fungi involved in beverage and food‐production environments. Wine strains of S. cerevisiae contain genes that are not present in beer strains and are probably related to adaptation to their particular industrial environment (Novo et al., 2009; Galeote et al., 2010; Borneman et al., 2011). Similarly, several Penicillium species growing on cheese have been shown to contain recently acquired genes that are adaptive in this particular environment (Ropars et al., 2015). These examples highlight the power of HGT in enabling microbial adaptation to novel niches, including those related to domestication.
While ancient events are difficult to detect, there is clear evidence that some fungal groups have been affected by HGT over long evolutionary timescales. Many yeasts in the Saccharomycotina possess a horizontally transferred cytoplasmic dihydroorotate dehydrogenase that allows for the biosynthesis of pyrimidines in anoxic conditions (Gojkovic et al., 2004). HGT of high‐affinity nitrate transporters from Oomycota has been proposed to be an important feature for the land hegemony of Dikarya (Slot & Hibbett, 2007). A large fraction of the polyketide synthase genes in Lecanoromycetes seem to arise from Actinobacteria, followed by gene expansions (Schmitt & Lumbsch, 2009). Some of these genes are involved in the synthesis of ecologically relevant secondary metabolites such as mycotoxins or antibiotics. Terpenoids and alkaloids are also widespread families of secondary metabolites whose biosynthetic pathways have dispered across the fungal kingdom through HGT (Marcet‐Houben & Gabaldón, 2016; Reynolds et al., 2018; Jia et al., 2019). However, some studies suggest that these may be exceptional findings, and that, overall, HGT events in eukaryotes are evolutionarily short lived (Katz, 2015). Many such events might remain undetected, either because of the lack of a sufficient phylogenetic signal or because they have been lost in the sampled extant lineages, but it is safe to assume that HGT was as important in past evolutionary transitions as it apparently is for short‐term adaptations in recent times.
(4). From genomes to pangenomes
The concept of pangenome arises from the realization that gene content differs among strains or isolates within a defined taxonomic unit, usually a species (Tetz, 2005). The pangenome refers to the complete gene pool of a single species, including copy number variations, horizontally acquired genes and mobile genetic elements, such as plasmids. For example, the different strains of Escherichia coli have genomes typically containing 3500–4000 protein‐coding genes, but with more than 2000 sequenced strains, the known pangenome of the species contains over 18000 genes (Jang et al., 2017). The pangenome can be divided into two distinct subsets: the core genome refers to the set of genes that are universally present in all analysed genomes of a given species, while the variable (also known as accessory or flexible) genome comprises genes which may be absent or present, depending on the analysed strain. Obviously, the composition of the pangenome and the core and accessory subsets depends on the number of analysed strains. Similar to HGT, this concept has been widely studied in Bacteria, while its impact is less recognized in eukaryotes. However, the concept can be applied to eukaryotes, including plants (Springer et al., 2009; Golicz, Batley & Edwards, 2016a; Golicz et al., 2016b), animals (Gerdol et al., 2019) and fungi.
Pangenome studies require the sequencing and comparison of multiple strains of the same species, and no other fungus has been studied in more detail than S. cerevisiae (Engel & Cherry, 2013; Strope et al., 2015; Gallone et al., 2016; Legras et al., 2018). Compared with other Fungi, particularly with moulds, the secretome and secondary metabolism of yeast is very limited. Even so, a significant number of genes are not fixed in the global yeast population. Comparative analyses of 100 strains characterized the patterns of presence/absence of several genes associated with highly variable phenotypic traits, such as resistance to copper, sulfite or cycloheximide (Strope et al., 2015). Some of these genes seem to have been acquired by introgression from other Saccharomyces species, mainly S. paradoxus, and quite often show copy number variation across isolates. A recent comparative analysis of over 1000 S. cerevisiae strains from diverse environments provided a reliable pangenome estimate of almost 7800 genes, of which approximately 4940 are core and 2860 are accessory (Peter et al., 2018). Accessory genes tend to be concentrated in subtelomeric regions, and are enriched in functions related to cell–cell interactions, secondary metabolism and stress responses. Furthermore, these genes have a higher tendency to show copy number variation and hemizygosity. Wine yeasts have a high tendency to form hybrids, and some HGT associated with adaptations to this niche have been described (Galeote et al., 2010; Borneman et al., 2011; Marsit et al., 2015; Legras et al., 2018). The same can be said for the sake strain K7. This strain is very similar to the reference S288c except for the presence of certain subtelomeric regions that include up to 48 open reading frames (ORFs) that are absent in the reference; 49 ORFs that are found in the reference are absent in K7 (Akao et al., 2011). Sequencing of the laboratory strain CEN.PK113‐7D revealed the absence of 83 genes that are present in S288c, and a small number of genes that appeared only in CEN.PK113‐7D (Nijkamp et al., 2012). Among the latter was the surprising finding of a functional biotin biosynthetic pathway (Nijkamp et al., 2012). All these studies illustrate that S. cerevisiae has a relatively small pangenome that, nonetheless, has a profound effect in shaping the phenotypic diversity of this species. It should be noted that most of these studies treat yeast hybridization as an anomaly, when instead we should consider the scope of yeast hybrid diversity as part of its total genetic pool. S. cerevisiae has some peculiar feaures compared to other yeast species, particularly regarding the frequency of sexual mating (Zeyl, 2009; Ni et al., 2011; Hittinger, 2013; Dujon & Louis, 2017), which probably makes it a poor model for Saccharomycotina as a whole with regard to pangenome dynamics.
Pangenomes have been studied for a handful of filamentous Ascomycota. Beauveria bassiana is an entomopathogenic member of Hypocreales (Sordariomycetes) with a wide host range. Valero‐Jiménez et al. (2016) compared five isolates with varying virulence phenotypes against mosquitoes. For B. bassiana, the core genome comprised around 7300 genes, with the pangenome size estimated at around 13000 genes. In the most virulent strain, 163 genes were strain specific, mostly containing secondary metabolic clusters located near telomeric regions. It is noteworthy that some of these strain‐specific genes show homology to other entomopathogenic Hypocreales or bacteria, suggesting an HGT origin. Aspergillus and Penicillium are two widely studied genera of Eurotiales (Eurotiomycetes) that produce a broad diversity of secondary metabolites and exhibit a wide range of genome and proteome sizes (de Vries et al., 2017; Nielsen et al., 2017). Several comparative genomics studies have shown that each strain carries a significant number of specific genes (Ropars et al., 2015; Julca et al., 2016; Gilbert et al., 2018). Pangenomic studies are of great relevance to understanding the emergence of virulent traits in plant pathogens. In this regard, accessory chromosomes in Fusarium spp. and Zymoseptoria tritici are a well‐known part of their pangenome. Fusarium in particular possesses a mitochondrial pangenome (Brankovics et al., 2018), with evidence of mitochondrial recombination between strains. Comparison of subtelomeric regions among six strains of Fusarium fujikuroi revealed the presence of lineage‐specific secondary metabolic pathways, some of which have emerged from HGT events (Chiara et al., 2015). Analysis of 60 F. graminearum isolates form North America identified an accessory genome close to 1700 genes that seems to concentrate in AT‐rich regions, probably centromeres or telomeres (Kelly & Ward, 2018). Similar results have been obtained for F. fujikuroi (Chiara et al., 2015; Niehaus et al., 2017), and F. oxysporum (Armitage et al., 2018). Together, these studies have focused either on geographically delimited isolates or have compared a handful of isolates. The pangenome of the wheat pathogen Z. tritici has recently been extensively investigated (Plissonneau et al., 2016, 2018) and it is probably the only filamentous fungus for which a reasonably complete pangenome has been described to date. Z. tritici contains a pangenome comprising more than 17400 protein‐coding genes. All analysed isolates possessed around 12000 genes, while the core genome for all analysed strains was around 9100 genes. This fungus has large effective population sizes and shows frequent genetic exchange between populations (Zhan, Pettway & McDonald, 2003). Finally, McCarthy & Fitzpatrick (2019) analysed the pangenome of four model fungal species: S. cerevisiae, C. albicans, Cr. neoformans and Aspergillus nidulans. Their study recovered an accessory genome ranging from almost 10% in C. albicans to almost 20% in Cr. neoformans. This probably represents a reasonable range for most fungi, although it is likely that particular species might have much greater or smaller pangenomes.
The development of a proper pangenomic paradigm for fungal genomics has been severely limited by the use of resequencing technologies in eukaryotes, where a single reference genome is used to map sequencing libraries from other strains. Applying de novo sequencing techniques to all strains in a given study is much more expensive, but should still be a feasible goal. However, a de novo assembly approach can still be limited by the fact that accessory genes tend to concentrate in AT‐rich regions such as telomeres, centromeres, genomic islands and transposable element‐rich regions, which in turn tend to be very difficult to assemble, at least without the use of long read‐sequencing technologies. As an alternative, RNA sequencing (RNAseq) techniques might be used to identify genes missing in specific strains, as well as novel transcripts without homology in the reference sequence. Accessory genes, however, tend to evolve at a faster rate than the rest of the genome (Dong, Raffaele & Kamoun, 2015), which of course further limits homology‐based annotation approaches. In any case, as more fungal genomes accumulate, the concept of the pangenome is starting to become more widely adopted, and the field is likely to expand in future years. The examples given here show that fungal pangenomes vary greatly in size and functional relevance across different lineages. The existence of an extensive pangenome requires efficient mechanisms for genetic exchange, which may or may not be sexual in nature. Our current knowledge in this area is severely lacking. Sex is only indirectly inferred from genomic data for many groups and mechanisms of HGT in Fungi are still unknown. Regarding the later, the pangenome concept forces us to reevaluate the hypothesis that HGT drives gene clustering in Fungi, as HGT among closely related Fungi might be much more prevalent than previously thought. It is important to point out the possible relationship between mesosynteny and pangenomes (Hane et al., 2011). Mesosynteny seems to be particularly prominent among Dothideomycetes (Hane et al., 2011), the group to which Z. tritici belongs.
The pangenome of the model arbuscular mycorrhizal fungus Rhizophagus irregularis has been studied only recently, and the results are controversial (Mathieu et al., 2018). After comparing just six laboratory strains, the results show that only 50% of the genes are shared among all of them, and the remaining, accessory genome contains over 150 000 genes (Chen et al., 2018). It remains to be seen whether these results can be interpreted in the context of the heterokaryotic nature of these fungi. First, the existence of such a large pangenome can only be explained in the light of rampant gene flow, either by sexual or sexual‐like interchange among strains or, alternatively, by high levels of HGT. Second, if these fungi are heterozygous, what are the implications for their intracytoplasmic pangenomes? How are these accessory genes distributed across the nuclear population within a certain mycelium? If distinct nuclei possess accessory genes that are not present in the entire cytoplasmic population, this would greatly affect genome assembly and annotation, as sequencing would be highly variable in these regions and reads spanning these regions would be incongruent. The large size of this pangenome might well be related to the fact that arbuscular mycorrhizal fungi have a species diversity and endemicity that is orders of magnitude lower than that of land plants, their obligate hosts. R. irregularis is currently the only member of Glomeromycota that can be cultured in laboratory. Assuming that the findings in this species can be extrapolated other members of the phylum, the genetic pool accessible to these fungi may be truly enormous (Mathieu et al., 2018).
V. CONCLUDING REMARKS
Despite the relevance of several species of fungi as model organisms, traditional genetics approaches during the last century have provided limited information about the peculiarities of fungal cell biology. Fungi have served well as model organisms for biochemistry and eukaryotic cell biology, but comparatively fewer studies have focused on understanding fungi themselves. Fungi exist in all shapes and sizes, contributing to their astounding evolutionary success. Unicellular fungi thrive in the environment mostly as saprobes and parasites, where their small size contributes to their successful dispersal and their ability to colonize specialized niches. The main multicellular organization in fungi, the mycelium, has a highly versatile cylindrical reticulated organization. The hyphal tip is able to exert physical force and can thus grow through solid substrates, while most microbes can access only the surface of such resources. Their network organization allows fungi to colonize their environment in a very flexible way. For example, they can simultaneously exploit spatially separated resources, using their network structure to transport nutrients and cell components across the whole colony. Efficient nutrient exploitation and secondary metabolite production generates a ‘territory’ for the fungus in which it exerts ecological control over other microorganisms. All of these traits require the fungus to regulate its gene expression in time and space, a feat that necessitates a complex network of sensory and signalling pathways, which in turn is a prerequisite for the emergence of complex multicellularity.
Genomics provides a powerful comparative and functional framework from where to start unravelling the intricacies of fungal biology. Comparative genomics is beginning to identify universal features over long evolutionary timescales. For example, the prevalence, evolution and global epigenetic regulation of gene clusters by the Velvet complex across the fungal kingdom was completely unknown three decades ago. The dynamic nature of fungal genomes over short evolutionary timescales is also receiving considerable attention due to its relevance to fungal diseases and industrial processes. Hybrids, polyploids, aneuploids and heterokaryons are an important and often overlooked dimension of fungal genetics whose importance to the kingdom can only be hypothesized at present. Heterokaryosis in particular is a relatively unexplored phenomenon with direct relevance to fungal physiology, potentially allowing new layers of phenotypic complexity without the need for complex regulatory pathways. As such, the effects of heterokaryosis should be considered for many aspects of the biology of fungi, as well as other syncytial eukaryotes.
Despite major advances in our understanding of fungi brought about by comparative genomics, sequencing approaches alone are not sufficient to solve most biological problems. The power of a comparative approach depends on previous functional knowledge, which can then be extrapolated to other organisms. Most functional knowledge in fungi derives from a few model species, and it is very limited outside Ascomycota. It is necessary to obtain direct empirical information from a wider diversity of species. Not only will this increase our ability to infer function based on homology, but it also will allow us to explore the realm of lineage‐specific genes. This is particularly relevant when dealing with secondary metabolism. Inferring the function and chemical nature of these products from exclusively in silico approaches is virtually impossible. Indeed, the extremely dynamic evolution of these genes imposes particular challenges for homology‐based methodologies. The recent technical revolution represented by novel genome editing technologies, which allow manipulation in non‐model species, promises further advances in our understanding of fungal biology (Ohm et al., 2010a; Nødvig et al., 2015; Deng et al., 2017) for both ancestral and taxonomically restricted traits.
VI. CONCLUSIONS
(1) Hyphal growth is the most common form of cellular organization found in the kingdom Fungi. Fungi were ancestrally unicellular and flagellated, and hyphal organization has been lost secondarily in several lineages independently. Mycelial growth requires the establishment of polarized cell extension, branching and anastomosis. While the cellular machinery required for cell polarization seems to be well preserved across the whole kingdom, little is known regarding the regulation and establishment of the latter two processes. Additionally, certain lineages in the Taphrinomycotina, Pezizomycotina and Agaricomycotina have developed complex structures consisting of different types of hyphae. The genetic mechanisms underlying complex multicellularity in fungi are starting to be revealed by comparative genomic approaches, uncovering many peculiarities and convergences among the different lineages.
(2) The mycelial network is able to exert considerable control over a territory. To achieve this, the mycelium must be able to coordinate global reactions to local stimuli. Most known sensory systems in fungi seem to be functionally similar to those found in plants, although the majority have, when known, an independent evolutionary origin.
(3) Most filamentous fungi can compartmentalize their cytoplasm by means of transverse perforated sections of their cell wall, known as septa. Septated hyphae should, in theory, be able spatially to restrict the biosynthetic machinery of secondary metabolites, which are often toxic or selective only under specific conditions. Non‐septated lineages tend to encode fewer secondary metabolism pathways.
(4) Genomic plasticity in Fungi is an essential aspect of adaptation to ever‐changing environments and it is one of the bases for long‐term evolutionary change. Chromosome aberrations act as unstable intermediates during adaptation to new conditions, and their deleterious consequences can be buffered in heterogeneous multinucleated mycelia. Genetic exchange between distantly related strains has been shown to be important in ecological adaptation for many species. However, studying these phenomena is challenging, due to the presence of artifacts in many standard genomic analyses.
(5) The metabolic diversity of fungi, particularly with regards to their secondary metabolism, is key to explaining their evolutionary success. This biochemical toolbox can evolve through gene loss, gene duplication and horizontal gene transfer. Many of these secondary metabolic pathways appear physically clustered in fungal genomes, a situation that appears to be much more common than in any other eukaryotic group. The selective pressures that cause gene clustering are not fully understood, and it has been proposed that it is caused by the need to coordinate the expression of their components or by frequent lateral gene transfer. Finally, genomic analyses of intraspecific variability has proved the value to fungal biology of the prokaryotic concept of a pangenome.
ACKNOWLEDGEMENTS
T.G. acknowledges support from the Spanish Ministry of Science and Innovation for grants ‘Centro de Excelencia Severo Ochoa’ and PGC2018‐099921‐B‐I00, cofunded by the European Regional Development Fund (ERDF); from the CERCA Programme/Generalitat de Catalunya; from the Catalan Research Agency (AGAUR) SGR423, and grants from the European Union's Horizon 2020 research and innovation programme under the grant agreement ERC‐2016‐724173, and the Marie Sklodowska‐Curie grant agreement No H2020‐MSCA‐IF‐2017‐793699. T.G.'s research group also receives support from a INB Grant (PT17/0009/0023 ‐ ISCIII‐SGEFI/ERDF).
REFERENCES
- Aanen, D. K. , Debets, A. J. M. , de Visser, J. A. G. M. & Hoekstra, R. F. (2008). The social evolution of somatic fusion. BioEssays 30, 1193–1203. [DOI] [PubMed] [Google Scholar]
- Adam, A. , Deimel, S. , Pardo‐Medina, J. , García‐Martínez, J. , Konte, T. , Limón, M. C. , Avalos, J. & Terpitz, U. (2018). Protein activity of the Fusarium fujikuroi rhodopsins CarO and OpsA and their relation to fungus‐plant interaction. International Journal of Molecular Sciences 19, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamatzky, A. (2018). On spiking behaviour of oyster fungi Pleurotus djamor . Scientific Reports 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerer, R. & Iosifidou, P. (2004). Rhizomorph structures of Hymenomycetes: a possibility to test DNA‐based phylogenetic hypothesis? In Frontiers in Basidiomycote Mycology (eds P. Agerer, M. Piepenbring, P. Blanz.), pp. 249–302. IHW Verlag, Eching. [Google Scholar]
- Ahrendt, S. R. , Quandt, C. A. , Ciobanu, D. , Clum, A. , Salamov, A. , Andreopoulos, B. , Cheng, J.‐F. , Woyke, T. , Pelin, A. , Henrissat, B. , Reynolds, N. K. , Benny, G. L. , Smith, M. E. , James, T. Y. , Grigoriev, I. V. , et al. (2018). Leveraging single‐cell genomics to expand the fungal tree of life. Nature Microbiology 3, 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aime, C. M. & Brearley, F. Q. (2012). Tropical fungal diversity: closing the gap between species estimates and species discovery. Biodiversity and Conservation 21, 2177–2180. [Google Scholar]
- Akagi, Y. , Akamatsu, H. , Otani, H. & Kodama, M. (2009). Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant‐pathogenic fungus. Eukaryotic Cell 8, 1732–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao, T. , Yashiro, I. , Hosoyama, A. , Kitagaki, H. , Horikawa, H. , Watanabe, D. , Akada, R. , Ando, Y. , Harashima, S. , Inoue, T. , Inoue, Y. , Kajiwara, S. , Kitamoto, K. , Kitamoto, N. , Kobayashi, O. , et al. (2011). Whole‐genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Research 18, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhberdi, O. , Zhang, Q. , Wang, D. , Wang, H. , Hao, X. , Liu, Y. , Wei, D. & Zhu, X. (2018). Distinct roles of velvet complex in the development, stress tolerance, and secondary metabolism in Pestalotiopsis microspora, a taxol producer. Genes 9, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin, W. & Marullo, P. (2012). Polyploidy in fungi: evolution after whole‐genome duplication. Proceedings of the Royal Society of London, Series B: Biological Sciences 279, 2497–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldabbous, M. S. , Roca, M. G. , Stout, A. , Huang, I.‐C. , Read, N. D. & Free, S. J. (2010). The ham‐5, rcm‐1 and rco‐1 genes regulate hyphal fusion in Neurospora crassa . Microbiology 156, 2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminnejad, M. , Diaz, M. , Arabatzis, M. , Castañeda, E. , Lazera, M. , Velegraki, A. , Marriott, D. , Sorrell, T. C. & Meyer, W. (2012). Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173, 337–346. [DOI] [PubMed] [Google Scholar]
- Anderson, C. A. , Eser, U. , Korndorf, T. , Borsuk, M. E. , Skotheim, J. M. & Gladfelter, A. S. (2013). Nuclear repulsion enables division autonomy in a single cytoplasm. Current Biology 23, 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C. A. , Roberts, S. , Zhang, H. , Kelly, C. M. , Kendall, A. , Lee, C. , Gerstenberger, J. , Koenig, A. B. , Kabeche, R. & Gladfelter, A. S. (2015). Ploidy variation in multinucleate cells changes under stress. Molecular Biology of the Cell 26, 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, I. C. & Cairney, J. W. G. (2004). Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environmental Microbiology 6, 769–779. [DOI] [PubMed] [Google Scholar]
- Anderson, M. Z. , Saha, A. , Haseeb, A. & Bennett, R. J. (2017). A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinical isolate of Candida albicans . Microbiology 163, 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J. O. (2005). Lateral gene transfer in eukaryotes. Cellular and Molecular Life Sciences 62, 1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J. O. (2009). Gene transfer and diversification of microbial eukaryotes. Annual Review of Microbiology 63, 177–193. [DOI] [PubMed] [Google Scholar]
- Andreou, A. , Brodhun, F. & Feussner, I. (2009). Biosynthesis of oxylipins in non‐mammals. Progress in Lipid Research 48, 148–170. [DOI] [PubMed] [Google Scholar]
- Andrés, F. & Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Angelard, C. , Tanner, C. J. , Fontanillas, P. , Niculita‐Hirzel, H. , Masclaux, F. & Sanders, I. R. (2014). Rapid genotypic change and plasticity in arbuscular mycorrhizal fungi is caused by a host shift and enhanced by segregation. ISME Journal 8, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz, R. A. & Bassilana, M. (2011). Polarized growth in fungi: symmetry breaking and hyphal formation. Seminars in Cell & Developmental Biology 22, 806–815. [DOI] [PubMed] [Google Scholar]
- Armijos‐Jaramillo, V. D. , Sukno, S. A. & Thon, M. R. (2014). Identification of horizontally transferred genes in the genus Colletotrichum reveals a steady tempo of bacterial to fungal gene transfer. BMC Genomics 16, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage, A. D. , Taylor, A. , Sobczyk, M. K. , Baxter, L. , Greenfield, B. P. J. , Bates, H. J. , Wilson, F. , Jackson, A. C. , Ott, S. , Harrison, R. & Clarkson, J. P. (2018). Pangenomic analysis reveals pathogen‐specific regions and novel effector candidates in Fusarium oxysporum f.sp. cepae. Scientific reports 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam, C. & Doohan, F. M. (2013). Trichothecene toxicity in eukaryotes: cellular and molecular mechanisms in plants and animals. Toxicology Letters 217, 149–158. [DOI] [PubMed] [Google Scholar]
- Asenova, E. , Lin, H.‐Y. , Fu, E. , Nicolau, D. V. & Nicolau, D. (2016). Optimal fungal space searching algorithms. IEEE Transactions on NanoBioscience 15, 613–618. [DOI] [PubMed] [Google Scholar]
- Atriztán‐Hernández, K. , Moreno‐Pedraza, A. , Winkler, R. , Markow, T. & Herrera‐Estrella, A. (2018). Trichoderma atroviride from predator to prey: role of the MAPK Tmk3 in fungal chemical defense against fungivory by Drosophila melanogaster larvae. Applied and Environmental Microbiology 85, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, M. , Cibrario, A. , Peltier, E. , Coton, M. , Coton, E. , Schacherer, J. , Spano, G. , Capozzi, V. , Blaiotta, G. , Salin, F. , Dols‐Lafargue, M. , Grbin, P. , Curtin, C. , Albertin, W. & Masneuf‐Pomarede, I. (2018). Brettanomyces bruxellensis population survey reveals a diploid‐triploid complex structured according to substrate of isolation and geographical distribution. Scientific Reports 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia, E. , Benoit, I. , van den Brink, J. , Wiebenga, A. , Coutinho, P. M. , Henrissat, B. & de Vries, R. P. (2011). Carbohydrate‐active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genomics 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram, Ö. & Braus, G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiology Reviews 36, 1–24. [DOI] [PubMed] [Google Scholar]
- Beadle, G. W. & Coonradt, V. L. (1944). Heterocaryosis in Neurospora crassa . Genetics 29, 291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebber, D. P. , Hynes, J. , Darrah, P. R. , Boddy, L. & Fricker, M. D. (2007). Biological solutions to transport network design. Proceedings of the Royal Society of London, Series B: Biological Sciences 274, 2307–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begerow, D. , Schäfer, A. M. , Kellner, R. , Yurkov, A. , Kemler, M. , Oberwinkler, F. & Bauer, R. (2014). 11 Ustilaginomycotina In The Mycota VII: Systematics and Evolution: Part A (ed K. Esser), pp. 295–329. Springer, Berlin Heidelberg. [Google Scholar]
- Benhamou, N. & Thériault, G. (1992). Treatment with chitosan enhances resistance of tomato plants to the crown and root pathogen Fusarium oxysporium f. sp. radicis‐lycopersici . Physiological and Molecular Plant Pathology 41, 33–52. [Google Scholar]
- Bennett, R. J. , Forche, A. & Berman, J. (2014). Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harbor Perspectives in Medicine 4, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benny, G. L. , Humber, R. A. & Voigt, K. (2014). Zygomycetous fungi: phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina In The Mycota VII: Systematics and Evolution: Part A, Second Edition (ed K. Esser), pp. 209–250. Springer, Berlin Heidelberg. [Google Scholar]
- Berbee, M. L. , James, T. Y. & Strullu‐Derrien, C. (2017). Early diverging Fungi: diversity and impact at the dawn of terrestrial life. Annual Review of Microbiology 71, 41–60. [DOI] [PubMed] [Google Scholar]
- Berman, J. , Wertheimer, N. B. & Stone, N. (2016). Ploidy dynamics and evolvability in fungi. Philosophical Transactions of the Royal Society, B: Biological Sciences 371, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini, P. P. , Frascella, A. , Kolarik, M. , Comparini, C. , Pepori, A. L. , Santini, A. , Scala, F. & Scala, A. (2014). Widespread horizontal transfer of the cerato‐ulmin gene between Ophiostoma novo‐ulmi and Geosmithia species. Fungal Biology 8, 663–674. [DOI] [PubMed] [Google Scholar]
- Bever, J. D. & Wang, M. (2005). Arbuscular mycorrhizal fungi: hyphal fusion and multigenomic structure. Nature 433, E3–E4. [DOI] [PubMed] [Google Scholar]
- Bills, G. F. & Gloer, J. B. (2016). Biologically active secondary metabolites from the Fungi In The Fungal Kingdom (Volume 4) (eds J. Heitman, B. J. Howlett, P. W. Crous, E. H. Stukenbrock, T. Y. James, and N. A. R. Gow.), pp. 1087–1119. American Society of Microbiology, Washington DC. [DOI] [PubMed] [Google Scholar]
- Blackwell, M. (2011). The Fungi: 1, 2, 3 … 5.1 million species? American Journal of Botany 98, 426–438. [DOI] [PubMed] [Google Scholar]
- Bleichrodt, R.‐J. , van Veluw, G. J. , Recter, B. , Maruyama, J. , Kitamoto, K. & Wösten, H. A. B. (2012). Hyphal heterogeneity in Aspergillus oryzae is the result of dynamic closure of septa by Woronin bodies. Molecular Microbiology 86, 1334–1344. [DOI] [PubMed] [Google Scholar]
- Boberg, J. B. , Finlay, R. D. , Stenlid, J. , Ekblad, A. & Lindahl, B. D. (2014). Nitrogen and carbon reallocation in fungal mycelia during decomposition of boreal forest litter. PLoS One 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, L. & Hiscox, J. (2016). Fungal ecology: principles and mechanisms of colonization and competition by saprotrophic Fungi In The Fungal Kingdom (eds J. Heitman, B. J. Howlett, P. W. Crous, E. H. Stukenbrock, T. Y. James, and N. A. R. Gow.), pp. 293–308. American Society of Microbiology, Washington DC. [DOI] [PubMed] [Google Scholar]
- de Boer, W. , Folman, L. B. , Summerbell, R. C. & Boddy, L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiology Reviews 29, 795–811. [DOI] [PubMed] [Google Scholar]
- Boersma, F. G. H. , Otten, R. , Warmink, J. A. , Nazir, R. & van Elsas, J. D. (2010). Selection of Variovorax paradoxus‐like bacteria in the mycosphere and the role of fungal‐released compounds. Soil Biology and Biochemistry 42, 2137–2145. [Google Scholar]
- Bok, J. W. & Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp . Eukaryotic Cell 3, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömke, C. & Tudzynski, B. (2009). Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry 70, 1876–1893. [DOI] [PubMed] [Google Scholar]
- Bonney, M. E. , Moriya, H. & Amon, A. (2015). Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage‐sensitive genes. Genes & Development 29, 898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon, E. , Halary, S. , Bapteste, E. & Hijri, M. (2015). Studying genome heterogeneity within the arbuscular mycorrhizal fungal cytoplasm. Genome Biology and Evolution 7, 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman, A. R. , Desany, B. A. , Riches, D. , Affourtit, J. P. , Forgan, A. H. , Pretorius, I. S. , Egholm, M. & Chambers, P. J. (2011). Whole‐genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae . PLoS Genetics 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman, A. R. , Zeppel, R. , Chambers, P. J. & Curtin, C. D. (2014). Insights into the Dekkera bruxellensis genomic landscape: comparative genomics reveals variations in ploidy and nutrient utilisation potential amongst wine isolates. PLoS Genetics 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boustie, J. & Grube, M. (2005). Lichens—a promising source of bioactive secondary metabolites. Plant Genetic Resources: Characterization and Utilization 3, 273–287. [Google Scholar]
- Boyce, G. , Gluck‐Thaler, E. , Slot, J. C. , Stajich, J. E. , Davis, W. J. , James, T. Y. , Cooley, J. R. , Panaccione, D. G. , Eilenberg, J. , Licht, H. H. D. F. , Macias, A. M. , Berger, M. C. , Wickert, K. L. , Stauder, C. M. , Spahr, E. J. , et al. (2019). Psychoactive plant‐ and mushroom‐associated alkaloids from two behavior modifying cicada pathogens. Fungal Ecology 41, 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva, S. , Daviet, L. , Wolfender, J.‐L. & Fitzpatrick, T. B. (2014). The rise of operon‐like gene clusters in plants. Trends in Plant Science 19, 447–459. [DOI] [PubMed] [Google Scholar]
- Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nature Reviews Microbiology 11, 21–32. [DOI] [PubMed] [Google Scholar]
- Brakhage, A. A. & Schroeckh, V. (2011). Fungal secondary metabolites – strategies to activate silent gene clusters. Fungal Genetics and Biology 48, 15–22. [DOI] [PubMed] [Google Scholar]
- Brankovics, B. , Kulik, T. , Sawicki, J. , Bilska, K. , Zhang, H. , de Hoog, G. S. , van der Lee, T. A. , Waalwijk, C. & van Diepeningen, A. D. (2018). First steps towards mitochondrial pan‐genomics: detailed analysis of Fusarium graminearum mitogenomes. PeerJ 6, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge, P. & Spooner, B. (2001). Soil fungi: diversity and detection. Plant and Soil 232, 147–154. [Google Scholar]
- Brodhun, F. & Feussner, I. (2011). Oxylipins in fungi. FEBS Journal 278, 1047–1063. [DOI] [PubMed] [Google Scholar]
- Brown, A. J. P. , Budge, S. , Kaloriti, D. , Tillmann, A. , Jacobsen, M. D. , Yin, Z. , Ene, I. V. , Bohovych, I. , Sandai, D. , Kastora, S. , Potrykus, J. , Ballou, E. R. , Childers, D. S. , Shahana, S. & Leach, M. D. (2014). Stress adaptation in a pathogenic fungus. The Journal of Experimental Biology 217, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J. J. , Huelsenbeck, J. P. , Cunningham, C. W. , Swofford, D. L. & Waddell, P. J. (1993). Partitioning and combining data in phylogenetic analysis. Systematic Biology 42, 384. [Google Scholar]
- Caballero Ortiz, S. , Trienens, M. & Rohlfs, M. (2013). Induced fungal resistance to insect grazing: reciprocal fitness consequences and fungal gene expression in the Drosophila–Aspergillus model system. PLoS One 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, M. A. , Ganley, A. R. D. , Gabaldón, T. & Cox, M. P. (2016). The case of the missing ancient fungal polyploids. The American Naturalist 188, 602–614. [DOI] [PubMed] [Google Scholar]
- Carretero‐Paulet, L. , Chang, T. H. , Librado, P. , Ibarra‐Laclette, E. , Herrera‐Estrella, L. , Rozas, J. & Albert, V. A. (2015). Genome‐wide analysis of adaptive molecular evolution in the carnivorous plant Utricularia gibba . Genome Biology and Evolution 7, 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana, J. (2007). Topological variation in single‐gene phylogenetic trees. Genome Biology 8, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnon, P.‐L. (2014). Ecological and evolutionary implications of hyphal anastomosis in arbuscular mycorrhizal fungi. FEMS Microbiology Ecology 88, 437–444. [DOI] [PubMed] [Google Scholar]
- Chanda, A. , Roze, L. V. , Kang, S. , Artymovich, K. A. , Hicks, G. R. , Raikhel, N. V. , Calvo, A. M. & Linz, J. E. (2009). A key role for vesicles in fungal secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America 106, 19533–19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E. C. H. , Morin, E. , Beaudet, D. , Noel, J. , Yildirir, G. , Ndikumana, S. , Charron, P. , St‐Onge, C. , Giorgi, J. , Krüger, M. , Marton, T. , Ropars, J. , Grigoriev, I. V. , Hainaut, M. , Henrissat, B. , et al. (2018). High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis . New Phytologist 220, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Hui, J. H. L. , Lee, Y. Y. , Wan Law, P. T. & Kwan, H. S. (2015). A “developmental hourglass” in fungi. Molecular Biology and Evolution 32, 1556–1566. [DOI] [PubMed] [Google Scholar]
- Chiang, Y.‐M. , Lee, K.‐H. , Sanchez, J. F. , Keller, N. P. & Wang, C. C. C. (2009). Unlocking fungal cryptic natural products. Natural Product Communications 4, 1505–1510. [PMC free article] [PubMed] [Google Scholar]
- Chiara, M. , Fanelli, F. , Mulè, G. , Logrieco, A. F. , Pesole, G. , Leslie, J. F. , Horner, D. S. & Toomajian, C. (2015). Genome sequencing of multiple isolates highlights subtelomeric genomic diversity within Fusarium fujikuroi . Genome Biology and Evolution 7, 3062–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S. A. & Kolomiets, M. V. (2011). The lipid language of plant–fungal interactions. Fungal Genetics and Biology 48, 4–14. [DOI] [PubMed] [Google Scholar]
- Cichewicz, R. H. (2010). Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Natural Product Reports 27, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claydon, N. (1978). Insecticidal secondary metabolites from entomogenous fungi: Entomophthora virulenta . Journal of Invertebrate Pathology 32, 319–324. [Google Scholar]
- Cogliati, M. , Barchiesi, F. , Spreghini, E. & Tortorano, A. M. (2012). Heterozygosis and pathogenicity of Cryptococcus neoformans AD‐hybrid isolates. Mycopathologia 173, 347–357. [DOI] [PubMed] [Google Scholar]
- Coleman, J. J. , Rounsley, S. D. , Rodriguez‐Carres, M. , Kuo, A. , Wasmann, C. C. , Grimwood, J. , Schmutz, J. , Taga, M. , White, G. J. , Zhou, S. , Schwartz, D. C. , Freitag, M. , Ma, L.‐J. , Danchin, E. G. J. , Henrissat, B. , et al. (2009). The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genetics 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant, G. C. & Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics 9, 938–950. [DOI] [PubMed] [Google Scholar]
- Corrochano, L. M. & Garre, V. (2010). Photobiology in the Zygomycota: multiple photoreceptor genes for complex responses to light. Fungal Genetics and Biology 47, 893–899. [DOI] [PubMed] [Google Scholar]
- Corrochano, L. M. , Kuo, A. , Marcet‐Houben, M. , Polaino, S. , Salamov, A. , Villalobos‐Escobedo, J. M. , Grimwood, J. , Álvarez, M. I. , Avalos, J. , Bauer, D. , Benito, E. P. , Benoit, I. , Burger, G. , Camino, L. P. , Cánovas, D. , et al. (2016). Expansion of signal transduction pathways in fungi by extensive genome duplication. Current Biology 26, 1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov, A. , Heller, J. , Herzog, S. , Fleißner, A. & Glass, N. L. (2017). Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous Fungi. Microbiology Spectrum 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Dee, J. M. , Mollicone, M. , Longcore, J. E. , Roberson, R. W. & Berbee, M. L. (2015). Cytology and molecular phylogenetics of Monoblepharidomycetes provide evidence for multiple independent origins of the hyphal habit in the Fungi. Mycologia 107, 710–728. [DOI] [PubMed] [Google Scholar]
- Dehal, P. & Boore, J. L. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biology 3, 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Guardia, M. D. , Aragón, C. M. , Murillo, F. J. & Cerdá‐Olmedo, E. (1971). A carotenogenic enzyme aggregate in Phycomyces: evidence from quantitive complementation. Proceedings of the National Academy of Sciences of the United States of America 68, 2012–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo, J. C. & Ramirez‐Parra, E. (2015). Whole genome duplications in plants: an overview from Arabidopsis. Journal of Experimental Botany 66, 6991–7003. [DOI] [PubMed] [Google Scholar]
- de Mattos‐Shipley, K. M. J. , Ford, K. L. , Alberti, F. , Banks, A. M. , Bailey, A. M. & Foster, G. D. (2016). The good, the bad and the tasty: the many roles of mushrooms. Studies in Mycology 85, 125–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, H. , Gao, R. , Liao, X. & Cai, Y. (2017). CRISPR system in filamentous fungi: current achievements and future directions. Gene 627, 212–221. [DOI] [PubMed] [Google Scholar]
- Depotter, J. R. , Seidl, M. F. , Wood, T. A. & Thomma, B. P. (2016). Interspecific hybridization impacts host range and pathogenicity of filamentous microbes. Current Opinion in Microbiology 32, 7–13. [DOI] [PubMed] [Google Scholar]
- de Vries, R. P. , Riley, R. , Wiebenga, A. , Aguilar‐Osorio, G. , Amillis, S. , Akemi Uchima, C. , Anderluh, G. , Asadollahi, M. , Askin, M. , Barry, K. , Battaglia, E. , Bayram, Ö. , Benocci, T. , Braus‐Stromeyer, S. A. , Caldana, C. , et al. (2017). Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus . Genome Biology 10, 1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepeveen, E. T. , Gehrmann, T. , Pourquié, V. , Abeel, T. & Laan, L. (2018). Patterns of conservation and diversification in the fungal polarization network. Genome Biology and Evolution 10, 1765–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirschnabel, D. E. , Nowrousian, M. , Cano‐Domínguez, N. , Aguirre, J. , Teichert, I. & Kück, U. (2014). New insights into the roles of NADPH oxidases in sexual development and ascospore germination in Sordaria macrospora . Genetics 196, 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson, S. E. , Kim, S. , Costanzo, M. , Baryshnikova, A. , Morse, D. L. , Kaiser, C. A. , Boone, C. & Amon, A. (2016). Chromosome‐specific and global effects of aneuploidy in Saccharomyces cerevisiae . Genetics 202, 1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döll, K. , Chatterjee, S. , Scheu, S. , Karlovsky, P. & Rohlfs, M. (2013). Fungal metabolic plasticity and sexual development mediate induced resistance to arthropod fungivory. Proceedings of the Royal Society of London, Series B: Biological Sciences 280, 20131219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Raffaele, S. & Kamoun, S. (2015). The two‐speed genomes of filamentous pathogens: waltz with plants. Current Opinion in Genetics & Development 35, 57–65. [DOI] [PubMed] [Google Scholar]
- Doolittle, F. W. (1998). You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends in Genetics 14, 307–311. [DOI] [PubMed] [Google Scholar]
- Dujon, B. (2010). Yeast evolutionary genomics. Nature Reviews. Genetics 11, 512–524. [DOI] [PubMed] [Google Scholar]
- Dujon, B. (2015). Basic principles of yeast genomics, a personal recollection. FEMS Yeast Research 15, 1–11. [DOI] [PubMed] [Google Scholar]
- Dujon, B. A. & Louis, E. J. (2017). Genome diversity and evolution in the budding yeasts (Saccharomycotina). Genetics 206, 717–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundon, S. E. R. , Chang, S.‐S. , Kumar, A. , Occhipinti, P. , Shroff, H. , Roper, M. & Gladfelter, A. S. (2016). Clustered nuclei maintain autonomy and nucleocytoplasmic ratio control in a syncytium. Molecular Biology of the Cell 27, 2000–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C. & Loros, J. J. (2004). The Neurospora circadian system. Journal of Biological Rhythms 19, 414–424. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C. & Loros, J. J. (2006). How fungi keep time: circadian system in Neurospora and other fungi. Current Opinion in Microbiology 9, 579–587. [DOI] [PubMed] [Google Scholar]
- Dupont, P.‐Y. & Cox, M. P. (2017). Genomic data quality impacts automated detection of lateral gene transfer in fungi. G3 7, 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger, I. , de Matos Simoes, R. , Kupczok, A. , Gube, M. , Kothe, E. , Voigt, K. & von Haeseler, A. (2012). A consistent phylogenetic backbone for the Fungi. Molecular Biology and Evolution 29, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibel, P. , Schimek, C. , Fries, V. , Grolig, F. , Schapat, T. , Schmidt, W. , Schneckenburger, H. , Ootaki, T. & Galland, P. (2000). Statoliths in Phycomyces: characterization of octahedral protein crystals. Fungal Genetics and Biology 29, 211–220. [DOI] [PubMed] [Google Scholar]
- Engel, S. R. & Cherry, J. M. (2013). The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database 2013, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh, I. & Nowrousian, M. (2010). Sordaria macrospora, a model organism to study fungal cellular development. European Journal of Cell Biology 89, 864–872. [DOI] [PubMed] [Google Scholar]
- Estiarte, N. , Lawrence, C. B. , Sanchis, V. , Ramos, A. J. & Crespo‐Sempere, A. (2016). LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. International Journal of Food Microbiology 238, 153–164. [DOI] [PubMed] [Google Scholar]
- Farrer, R. A. , Henk, D. A. , Garner, T. W. J. , Balloux, F. , Woodhams, D. C. & Fisher, M. C. (2013). Chromosomal copy number variation, selection and uneven rates of recombination reveal cryptic genome diversity linked to pathogenicity. PLoS Genetics 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtey, A. & Stukenbrock, E. H. (2018). Interspecific gene exchange as a driver of adaptive evolution in Fungi. Annual Review of Microbiology 72, 1–14. [DOI] [PubMed] [Google Scholar]
- Fischbach, M. A. , Walsh, C. T. & Clardy, J. (2008). The evolution of gene collectives: how natural selection drives chemical innovation. Proceedings of the National Academy of Sciences of the United States of America 105, 4601–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, G. J. & Keller, N. P. (2016). Production of cross‐kingdom oxylipins by pathogenic fungi: an update on their role in development and pathogenicity. Journal of Microbiology 54, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, R. , Aguirre, J. , Herrera‐estrella, A. & Corrochano, L. M. (2016). The complexity of fungal vision. Microbiology Spectrum 4, 1–22. [DOI] [PubMed] [Google Scholar]
- Fisher, K. E. & Roberson, R. W. (2016). Hyphal tip cytoplasmic organization in four zygomycetous fungi. Mycologia 108, 533–542. [DOI] [PubMed] [Google Scholar]
- Fisher, K. E. , Romberger, I. , Lowry, D. , Shange, P. & Roberson, R. W. (2018). Hyphal tip growth and cytoplasmic characters of Conidiobolus coronatus (Zoopagomycota, Entomophthoromycotina). Mycologia 110, 31–38. [DOI] [PubMed] [Google Scholar]
- Fisher, M. C. , Henk, D. A. , Briggs, C. J. , Brownstein, J. S. , Madoff, L. C. , McCraw, S. L. & Gurr, S. J. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, D. A. (2012). Horizontal gene transfer in fungi. FEMS Microbiology Letters 329, 1–8. [DOI] [PubMed] [Google Scholar]
- Fleißner, A. & Herzog, S. (2016). Signal exchange and integration during self‐fusion in filamentous fungi. Seminars in Cell & Developmental Biology 57, 76–83. [DOI] [PubMed] [Google Scholar]
- Forche, A. (2012). Large‐scale chromosomal changes and associated fitness consequences in pathogenic fungi. Current Fungal Infection Reports 29, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A. , Alby, K. , Schaefer, D. , Johnson, A. D. , Berman, J. & Bennett, R. J. (2008). The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biology 6, 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker, M. D. , Heaton, L. L. M. , Jones, N. S. & Boddy, L. (2017). The mycelium as a network In The Fungal Kingdom (eds J. Heitman, B. J. Howlett, P. W. Crous, E. H. Stukenbrock, T. Y. James, and N. A. R. Gow.) pp. 335–367. American Society of Microbiology, Washington DC. [Google Scholar]
- Fricker, M. D. , Lee, J. A. , Bebber, D. P. , Tlalka, M. , Hynes, J. , Darrah, P. R. , Watkinson, S. C. & Boddy, L. (2008). Imaging complex nutrient dynamics in mycelial networks. Journal of Microscopy 231, 317–331. [DOI] [PubMed] [Google Scholar]
- Friesen, T. L. , Stukenbrock, E. H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J. D. , Rasmussen, J. B. , Solomon, P. S. , McDonald, B. A. & Oliver, R. P. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nature Genetics 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Fromm, J. & Lautner, S. (2007). Electrical signals and their physiological significance in plants. Plant, Cell & Environment 30, 249–257. [DOI] [PubMed] [Google Scholar]
- Fu, C. , Iyer, P. , Herkal, A. , Abdullah, J. , Stout, A. & Free, S. J. (2011). Identification and characterization of genes required for cell‐to‐cell fusion in Neurospora crassa . Eukaryotic Cell 10, 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, K. K. , Loros, J. J. & Dunlap, J. C. (2015). Fungal photobiology: visible light as a signal for stress, space and time. Current Genetics 61, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón, T. , Naranjo‐Ortíz, M. A. & Marcet‐Houben, M. (2016). Evolutionary genomics of yeast pathogens in the Saccharomycotina. FEMS Yeast Research 16, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek, A. & Strauss, J. (2012). The chromatin code of fungal secondary metabolite gene clusters. Applied Microbiology and Biotechnology 95, 1389–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote, V. , Novo, M. , Salema‐Oom, M. , Brion, C. , Valerio, E. , Goncalves, P. & Dequin, S. (2010). FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high‐affinity fructose/H+ symporter. Microbiology 156, 3754–3761. [DOI] [PubMed] [Google Scholar]
- Gallone, B. , Steensels, J. , Prahl, T. , Soriaga, L. , Saels, V. , Herrera‐Malaver, B. , Merlevede, A. , Roncoroni, M. , Voordeckers, K. , Miraglia, L. , Teiling, C. , Steffy, B. , Taylor, M. , Schwartz, A. , Richardson, T. , et al. (2016). Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier, N. & Daubin, V. (2008). Dealing with incongruence in phylogenomic analyses. Philosophical Transactions of the Royal Society, B: Biological Sciences 363, 4023–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q. , Jin, K. , Ying, S.‐H. , Zhang, Y. , Xiao, G. , Shang, Y. , Duan, Z. , Hu, X. , Xie, X.‐Q. , Zhou, G. , Peng, G. , Luo, Z. , Huang, W. , Wang, B. , Fang, W. , et al. (2011). Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum . PLoS Genetics 7, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbelotto, M. & Gonthier, P. (2013). Biology, epidemiology, and control of Heterobasidion species worldwide. Annual Review of Phytopathology 51, 39–59. [DOI] [PubMed] [Google Scholar]
- Garbelotto, M. , Gonthier, P. , Linzer, R. , Nicolotti, G. & Otrosina, W. (2004). A shift in nuclear state as the result of natural interspecific hybridization between two North American taxa of the basidiomycete complex Heterobasidion . Fungal Genetics and Biology 41, 1046–1051. [DOI] [PubMed] [Google Scholar]
- García‐Martínez, J. , Brunk, M. , Avalos, J. & Terpitz, U. (2015). The CarO rhodopsin of the fungus Fusarium fujikuroi is a light‐driven proton pump that retards spore germination. Scientific Reports 5, 7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Vallvé, S. , Romeu, A. & Palau, J. (2000). Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Molecular Biology and Evolution 17, 352–361. [DOI] [PubMed] [Google Scholar]
- Gerdol, M. , Moreira, R. , Cruz, F. , Gomez‐Garrido, J. , Vlasova, A. , Rosani, U. , Venier, P. , Naranjo‐Ortiz, M. A. , Murgarella, M. , Balseiro, P. , Corvelo, A. , Frias, L. , Gut, M. , Gabaldon, T. , Pallavicini, A. , et al. (2019). Massive gene presence/absence variation in the mussel genome as an adaptive strategy: first evidence of a pan‐genome in Metazoa. bioRxiv, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhards, N. , Matuschek, M. , Wallwey, C. & Li, S.‐M. (2015). Genome mining of ascomycetous fungi reveals their genetic potential for ergot alkaloid production. Archives of Microbiology 197, 701–713. [DOI] [PubMed] [Google Scholar]
- Gerke, J. & Braus, G. H. (2014). Manipulation of fungal development as source of novel secondary metabolites for biotechnology. Applied Microbiology and Biotechnology 98, 8443–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, A. C. & Berman, J. (2015). Shift and adapt: the costs and benefits of karyotype variations. Current Opinion in Microbiology 26, 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, A. C. , Fu, M. S. , Mukaremera, L. , Li, Z. , Ormerod, K. L. , Fraser, J. A. , Berman, J. & Nielsen, K. (2015). Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. MBio 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaux, R. , Politi, A. Z. , Philippsen, P. & Nédélec, F. (2017). Mechanism of nuclear movements in a multinucleated cell. Molecular Biology of the Cell 28, 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, M. K. , Mack, B. M. , Moore, G. G. , Downey, D. L. , Lebar, M. D. , Joardar, V. , Losada, L. , Yu, J. , Nierman, W. C. & Bhatnagar, D. (2018). Whole genome comparison of Aspergillus flavus L‐morphotype strain NRRL 3357 (type) and S‐morphotype strain AF70. PLoS One 13, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano, L. , Sillo, F. , Garbelotto, M. & Gonthier, P. (2018). Mitonuclear interactions may contribute to fitness of fungal hybrids. Scientific Reports 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A. S. (2006). Nuclear anarchy: asynchronous mitosis in multinucleated fungal hyphae. Current Opinion in Microbiology 9, 547–552. [DOI] [PubMed] [Google Scholar]
- Glass, N. L. & Kaneko, I. (2003). Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryotic Cell 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck‐Thaler, E. , Slot, J. C. , Thomas, C. , Nielsen, K. , Baquero, F. , Smillie, C. , Smith, M. , Friedman, J. , Cordero, O. , David, L. , Barlow, M. , Kroken, S. , Glass, N. , Taylor, J. , Yoder, O. , et al. (2015). Dimensions of horizontal gene transfer in eukaryotic microbial pathogens. PLoS Pathogens 11, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojkovic, Z. , Knecht, W. , Zameitat, E. , Warneboldt, J. , Coutelis, J.‐B. , Pynyaha, Y. , Neuveglise, C. , Moller, K. , Loffer, M. & Piskur, J. (2004). Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Molecular Genetics and Genomics 271, 387–393. [DOI] [PubMed] [Google Scholar]
- Golicz, A. A. , Batley, J. & Edwards, D. (2016a). Towards plant pangenomics. Plant Biotechnology Journal 14, 1099–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz, A. A. , Bayer, P. E. , Barker, G. C. , Edger, P. P. , Kim, H. , Martinez, P. A. , Chan, C. K. K. , Severn‐Ellis, A. , McCombie, W. R. , Parkin, I. A. P. , Paterson, A. H. , Pires, J. C. , Sharpe, A. G. , Tang, H. , Teakle, G. R. , et al. (2016b). The pangenome of an agronomically important crop plant Brassica oleracea . Nature Communications 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonthier, P. , Warner, R. , Nicolotti, G. , Mazzaglia, A. & Garbelotto, M. M. (2004). Pathogen introduction as a collateral effect of military activity. Mycological Research 108, 468–470. [DOI] [PubMed] [Google Scholar]
- Gostinčar, C. , Stajich, J. E. , Zupančič, J. , Zalar, P. & Gunde‐Cimerman, N. (2018). Genomic evidence for intraspecific hybridization in a clonal and extremely halotolerant yeast. BMC Genomics 19, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttig, M. & Galland, P. (2014). Gravitropism in Phycomyces: violation of the so‐called resultant law ‐ evidence for two response components. Plant Biology 16, 158–166. [DOI] [PubMed] [Google Scholar]
- Gow, N. A. R. (1984). Transhyphal electrical currents in Fungi. Microbiology 130, 3313–3318. [DOI] [PubMed] [Google Scholar]
- Grant, J. R. & Katz, L. A. (2014). Phylogenomic study indicates widespread lateral gene transfer in Entamoeba and suggests a past intimate relationship with parabasalids. Genome Biology and Evolution 6, 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolig, F. , Döring, M. & Galland, P. (2006). Gravisusception by buoyancy: a mechanism ubiquitous among fungi? Protoplasma 229, 117–123. [DOI] [PubMed] [Google Scholar]
- Gromov, B. V. (2000). Algal parasites of the genera Aphelidium, Amoeboaphelidium, and Pseudaphelidium from the Cienkovski's ‘“monadinea”’ group as representatives of a new class. Zoologichesky Zhurnal 79, 517–525. [Google Scholar]
- Grube, M. & Hawksworth, D. L. (2007). Trouble with lichen: the re‐evaluation and re‐interpretation of thallus form and fruit body types in the molecular era. Mycological Research 111, 1116–1132. [DOI] [PubMed] [Google Scholar]
- Haegeman, A. , Jones, J. T. , Danchin, E. G. J. , Hall, C. , Brachat, S. , Dietrich, F. S. , Huang, J. , Mullapudi, N. , Lancto, C. A. , Scott, M. , Abrahamsen, M. S. , Kissinger, J. C. , Schönknecht, G. , Weber, A. P. M. , Lercher, M. J. , et al. (2014). Horizontal gene transfer in plants. BMC Evolutionary Biology 4, 1–8. [Google Scholar]
- Hall, C. , Welch, J. , Kowbel, D. J. & Glass, N. L. (2010). Evolution and diversity of a fungal self/nonself recognition locus. PLoS One 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek, O. & Nelson, D. R. (2008). Gene surfing in expanding populations. Theoretical Population Biology 73, 158–170. [DOI] [PubMed] [Google Scholar]
- Hamann, A. , Brust, D. & Osiewacz, H. D. (2008). Apoptosis pathways in fungal growth, development and ageing. Trends in Microbiology 16, 276–283. [DOI] [PubMed] [Google Scholar]
- Hamilton, C. E. , Faeth, S. H. & Dowling, T. E. (2009). Distribution of hybrid fungal symbionts and environmental stress. Microbial Ecology 58, 408–413. [DOI] [PubMed] [Google Scholar]
- Hane, J. K. , Rouxel, T. , Howlett, B. J. , Kema, G. H. , Goodwin, S. B. & Oliver, R. P. (2011). A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biology 12, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq, I. U. , Zhang, M. , Yang, P. & van Elsas, J. D. (2014). The interactions of bacteria with fungi in soil. Advances in Applied Microbiology 89, 185–215. [DOI] [PubMed] [Google Scholar]
- Harris, S. D. (2001). Septum formation in Aspergillus nidulans . Current Opinion in Microbiology 4, 736–739. [DOI] [PubMed] [Google Scholar]
- Harris, S. D. (2008). Branching of fungal hyphae: regulation, mechanisms and comparison with other branching systems. Mycologia 100, 823–832. [DOI] [PubMed] [Google Scholar]
- Harris, S. D. (2011). Cdc42/Rho GTPases in fungi: variations on a common theme. Molecular Microbiology 79, 1123–1127. [DOI] [PubMed] [Google Scholar]
- Harrison, B. D. , Hashemi, J. , Bibi, M. , Pulver, R. , Bavli, D. , Nahmias, Y. , Wellington, M. , Sapiro, G. & Berman, J. (2014). A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biology 12, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth, D. L. (2001). The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycological Research 105, 1422–1432. [Google Scholar]
- Hawksworth, D. L. & Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiology Spectrum 5, 1–17. [DOI] [PubMed] [Google Scholar]
- He, Q. , Cheng, P. , Yang, Y. , Wang, L. , Gardner, K. H. & Liu, Y. (2002). White collar‐1, a DNA binding transcription factor and a light sensor. Science 297, 840–843. [DOI] [PubMed] [Google Scholar]
- Healy, R. A. , Kumar, T. K. A. , Hewitt, D. A. & McLaughlin, D. J. (2013). Functional and phylogenetic implications of septal pore ultrastructure in the ascoma of Neolecta vitellina . Mycologia 105, 802–813. [DOI] [PubMed] [Google Scholar]
- Heinemann, J. A. & Sprague, G. F. (1989). Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340, 205–209. [DOI] [PubMed] [Google Scholar]
- Heinemann, J. A. & Sprague, G. F. (1991). Transmission of plasmid DNA to yeast by conjugation with bacteria. Methods in Enzymology 194, 187–195. [DOI] [PubMed] [Google Scholar]
- Heitman, J. , Sun, S. & James, T. Y. (2013). Evolution of fungal sexual reproduction. Mycologia 105, 1–27. [DOI] [PubMed] [Google Scholar]
- Hellborg, L. & Piskur, J. (2009). Complex nature of the genome in a wine spoilage yeast, Dekkera bruxellensis . Eukaryotic Cell 8, 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henk, D. A. & Fisher, M. C. (2012). The gut fungus Basidiobolus ranarum has a large genome and different copy numbers of putatively functionally redundant elongation factor genes. PLoS One 7, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Oñate, M. A. , Esquivel‐Naranjo, E. U. , Mendoza‐Mendoza, A. , Stewart, A. & Herrera‐Estrella, A. H. (2012). An injury‐response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proceedings of the National Academy of Sciences of the United States of America 109, 14918–14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Oñate, M. A. & Herrera‐Estrella, A. (2015). Damage response involves mechanisms conserved across plants, animals and fungi. Current Genetics 61, 359–372. [DOI] [PubMed] [Google Scholar]
- Herzog, S. , Schumann, M. R. & Fleißner, A. (2015). Cell fusion in Neurospora crassa . Current Opinion in Microbiology 28, 53–59. [DOI] [PubMed] [Google Scholar]
- Hibbett, D. S. (2006). A phylogenetic overview of the Agaricomycotina. Mycologia 98, 917–925. [DOI] [PubMed] [Google Scholar]
- Hibbett, D. S. , Bauer, R. , Binder, M. , Giachini, A. J. , Hosaka, K. , Justo, A. , Larsson, E. , Larsson, K. H. , Lawrey, J. D. , Miettinen, O. , Nagy, L. G. , Nilsson, R. H. , Weiss, M. & Thorn, R. G. (2014). Agaricomycetes In The mycota VII: Systematics and Evolution: Part A, Second Edition (ed K. Esser.). pp. 373–412. Springer, Berlin Heidelberg. [Google Scholar]
- Hickman, M. A. , Paulson, C. , Dudley, A. & Berman, J. (2015). Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans . Genetics 200, 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, M. P. , Martinez, D. A. , Sakthikumar, S. , Anderson, M. Z. , Berlin, A. , Gujja, S. , Zeng, Q. , Zisson, E. , Wang, J. M. , Greenberg, J. M. , Berman, J. , Bennett, R. J. & Cuomo, C. A. (2015). Genetic and phenotypic intra‐species variation in Candida albicans . Genome Research 25, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger, C. T. (2013). Saccharomyces diversity and evolution: a budding model genus. Trends in Genetics 29, 309–317. [DOI] [PubMed] [Google Scholar]
- Honegger, R. , Edwards, D. , Axe, L. & Strullu‐Derrien, C. (2018). Fertile Prototaxites taiti: a basal ascomycete with inoperculate, polysporous asci lacking croziers. Philosophical Transactions of the Royal Society, B: Biological Sciences 373, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken, J. , Frisvad, J. C. & Samson, R. A. (2010). Sex in Penicillium series Roqueforti. IMA Fungus 1, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Kang, M. & Xu, A. (2017). HaploMerger2: rebuilding both haploid sub‐assemblies from high‐heterozygosity diploid genome assembly. Bioinformatics 490, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber, F. M. (2001). Rotted wood–alga–fungus: the history and life of Prototaxites Dawson 1859. Review of Palaeobotany and Palynology 116, 123–158. [Google Scholar]
- Hur, G. H. , Vickery, C. R. & Burkart, M. D. (2012). Explorations of catalytic domains in non‐ribosomal peptide synthetase enzymology. Natural Product Reports 29, 1074–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik, F. & McCutcheon, J. P. (2017). Functional horizontal gene transfer from bacteria to eukaryotes. Nature Reviews Microbiology 16, 67–79. [DOI] [PubMed] [Google Scholar]
- Ibarra, R. U. , Edwards, J. S. & Palsson, B. O. (2002). Escherichia coli K‐12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189. [DOI] [PubMed] [Google Scholar]
- Idnurm, A. (2013). Light sensing in Aspergillus fumigatus highlights the case for establishing new models for fungal photobiology. MBio 4, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis, D. O. , Binkley, J. , Skrzypek, M. S. , Arnaud, M. B. , Cerqueira, G. C. , Shah, P. , Wymore, F. , Wortman, J. R. & Sherlock, G. (2013). Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae . BMC Microbiology 13, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata, K. , Nishikawa, M. & Yoshida, K. (1994). The yeast Saccharomyces kluyveri as a recipient eukaryote in transkingdom conjugation: behavior of transmitted plasmids in transconjugants. Journal of Bacteriology 176, 4770–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, F. H. , Souza, E. A. , Shoji, J.‐Y. , Connolly, L. , Freitag, M. , Read, N. D. & Roca, M. G. (2012). Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PLoS One 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa, M. , Tanabe, S. & Kamada, T. (1998). The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genetics and Biology 23, 110–116. [DOI] [PubMed] [Google Scholar]
- Jain, R. , Rivera, M. C. & Lake, J. A. (1999). Horizontal gene transfer among genomes: the complexity hypothesis. Proceedings of the National Academy of Sciences of the United States of America 96, 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S. & Keller, N. (2013). Insights to fungal biology through LaeA sleuthing. Fungal Biology Reviews 27, 51–59. [Google Scholar]
- James, S. a. , Bond, C. J. , Stratford, M. & Roberts, I. N. (2005). Molecular evidence for the existence of natural hybrids in the genus Zygosaccharomyces . FEMS Yeast Research 5, 747–755. [DOI] [PubMed] [Google Scholar]
- James, T. Y. , Johansson, S. B. K. & Johannesson, H. (2009). Trikaryon formation and nuclear selection in pairings between heterokaryons and homokaryons of the root rot pathogen Heterobasidion parviporum . Mycological Research 113, 583–590. [DOI] [PubMed] [Google Scholar]
- James, T. Y. , Porter, T. M. & Martin, W. W. (2014). Blastocladiomycota In The mycota VII: Systematics and Evolution: Part A, Second Edition (ed K. Esser.), pp. 177–207. Springer, Berlin Heidelberg. [Google Scholar]
- James, T. Y. , Stenlid, J. , Olson, Å. & Johannesson, H. (2008). Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum . Evolution 62, 2279–2296. [DOI] [PubMed] [Google Scholar]
- Jang, J. , Hur, H.‐G. , Sadowsky, M. J. , Byappanahalli, M. N. , Yan, T. & Ishii, S. (2017). Environmental Escherichia coli: ecology and public health implications‐a review. Journal of Applied Microbiology 123, 570–581. [DOI] [PubMed] [Google Scholar]
- Jany, J. & Pawlowska, T. E. E. (2010). Multinucleate spores contribute to evolutionary longevity of asexual glomeromycota. The American Naturalist 175, 424–435. [DOI] [PubMed] [Google Scholar]
- Jedd, G. & Pieuchot, L. (2012). Multiple modes for gatekeeping at fungal cell‐to‐cell channels. Molecular Microbiology 86, 1291–1294. [DOI] [PubMed] [Google Scholar]
- Jia, Q. , Chen, X. , Köllner, T. G. , Rinkel, J. , Fu, J. , Labbé, J. , Xiong, W. , Dickschat, J. S. , Gershenzon, J. & Chen, F. (2019). Terpene synthase genes originated from Bacteria through horizontal gene transfer contribute to terpenoid diversity in Fungi. Scientific Reports 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, D. , Zhu, W. , Wang, Y. , Sun, C. , Zhang, K.‐Q. & Yang, J. (2013). Molecular tools for functional genomics in filamentous fungi: recent advances and new strategies. Biotechnology Advances 31, 1562–1574. [DOI] [PubMed] [Google Scholar]
- Jiao, Y. , Wickett, N. J. , Ayyampalayam, S. , Chanderbali, A. S. , Landherr, L. , Ralph, P. E. , Tomsho, L. P. , Hu, Y. , Liang, H. , Soltis, P. S. , Soltis, D. E. , Clifton, S. W. , Schlarbaum, S. E. , Schuster, S. C. , Ma, H. , et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. [DOI] [PubMed] [Google Scholar]
- Jinks, J. L. (1952). Heterocaryosis in wild Penicillium . Heredity 6, 77–87. [Google Scholar]
- Johannesson, H. & Samils, N. (2014). Nuclear interactions in a heterokaryon: insight from the model Neurospora tetrasperma. Proceedings of the Royal Society of London, Series B: Biological Sciences 281, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M. & Staiger, D. (2015). Time to flower: interplay between photoperiod and the circadian clock. Journal of Experimental Botany 66, 719–730. [DOI] [PubMed] [Google Scholar]
- Jones, M. D. M. , Forn, I. , Gadelha, C. , Egan, M. J. , Bass, D. , Massana, R. & Richards, T. A. (2011). Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203. [DOI] [PubMed] [Google Scholar]
- Joneson, S. , Stajich, J. E. , Shiu, S.‐H. & Rosenblum, E. B. (2011). Genomic transition to pathogenicity in chytrid fungi. PLoS Pathogens 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers, W. , Leeder, A. C. , Ansong, C. , Wang, Y. , Yang, F. , Starr, T. L. , Camp, D. G. , Smith, R. D. , Glass, N. L. & Glass, N. L. (2014). HAM‐5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa . PLoS Genetics 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julca, I. , Droby, S. , Sela, N. , Marcet‐Houben, M. & Gabaldón, T. (2016). Contrasting genomic diversity in two closely related postharvest pathogens: Penicillium digitatum and Penicillium expansum . Genome Biology and Evolution 8, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani, R. , Toshimoto, K. , Noguchi, H. , Toyoda, A. , Ogura, Y. , Okuno, M. , Yabana, M. , Harada, M. , Nagayasu, E. , Maruyama, H. , Kohara, Y. , Fujiyama, A. , Hayashi, T. & Itoh, T. (2014). Efficient de novo assembly of highly heterozygous genomes from whole‐genome shotgun short reads. Genome Research 24, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, T. , Sano, H. , Nakazawa, T. & Nakahori, K. (2010). Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea . Fungal Genetics and Biology 47, 917–921. [DOI] [PubMed] [Google Scholar]
- Kämper, J. , Kahmann, R. , Bölker, M. , Ma, L.‐J. , Brefort, T. , Saville, B. J. , Banuett, F. , Kronstad, J. W. , Gold, S. E. , Müller, O. , Perlin, M. H. , Wösten, H. A. B. , de Vries, R. , Ruiz‐Herrera, J. , Reynaga‐Peña, C. G. , et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature 444, 97–101. [DOI] [PubMed] [Google Scholar]
- Karpov, S. A. , López‐García, P. , Mamkaeva, M. A. , Klimov, V. I. , Vishnyakov, A. E. , Tcvetkova, V. S. & Moreira, D. (2018). The Chytrid‐like parasites of algae Amoeboradix gromovi gen. et sp. nov. and Sanchytrium tribonematis belong to a new fungal lineage. Protist 169, 122–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov, S. A. , Mamkaeva, M. A. , Aleoshin, V. V. , Nassonova, E. , Lilje, O. & Gleason, F. H. (2014a). Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Frontiers in Microbiology 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov, S. A. , Mamkaeva, M. A. , Benzerara, K. , Moreira, D. & López‐García, P. (2014b). Molecular phylogeny and ultrastructure of Aphelidium aff. melosirae (Aphelida, Opisthosporidia). Protist 165, 512–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katicheva, L. , Sukhov, V. , Akinchits, E. & Vodeneev, V. (2014). Ionic nature of burn‐induced variation potential in wheat leaves. Plant and Cell Physiology 55, 1511–1519. [DOI] [PubMed] [Google Scholar]
- Katz, L. A. (2015). Recent events dominate interdomain lateral gene transfers between prokaryotes and eukaryotes and, with the exception of endosymbiotic gene transfers, few ancient transfer events persist. Philosophical Transactions of the Royal Society of London 370, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh, K. & Whittaker, P. A. (1996). Application of protoplast fusion to the nonconventional yeast. Enzyme and Microbial Technology 18, 45–51. [Google Scholar]
- Keller, N. P. (2015). Translating biosynthetic gene clusters into fungal armor and weaponry. Nature Chemical Biology 11, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, N. P. (2018). Fungal secondary metabolism: regulation, function and drug discovery. Nature Reviews Microbiology 17, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A. C. & Ward, T. J. (2018). Population genomics of Fusarium graminearum reveals signatures of divergent evolution within a major cereal pathogen. PLoS One 13, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, L. L. & Larcade, R. J. (1971). Basidiocarp development in Polyporus adustus . Mycologia 63, 69–78. [Google Scholar]
- Kenny, N. J. , Chan, K. W. , Nong, W. , Qu, Z. , Maeso, I. , Yip, H. Y. , Chan, T. F. , Kwan, H. S. , Holland, P. W. H. , Chu, K. H. & Hui, J. H. L. (2016). Ancestral whole‐genome duplication in the marine chelicerate horseshoe crabs. Heredity 116, 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, V. D. , Mendgen, K. & Hock, B. (1997). Flammulina as a model system for fungal graviresponses. Planta 203, 23–32. [DOI] [PubMed] [Google Scholar]
- Khaldi, N. , Collemare, J. , Lebrun, M.‐H. & Wolfe, K. H. (2008). Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biology 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey, J. A. (1990). Tad, a LINE‐like transposable element of Neurospora, can transpose between nuclei in heterokaryons. Genetics 126, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler, H. C. & Broz, K. (2015). Cellular compartmentalization of secondary metabolism. Frontiers in Microbiology 6, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul, M. & Singh, S. (2017). Penicillium spp.: prolific producer for harnessing cytotoxic secondary metabolites. Anti‐Cancer Drugs 28, 11–30. [DOI] [PubMed] [Google Scholar]
- Krause, D. J. , Kominek, J. , Opulente, D. A. , Shen, X.‐X. , Zhou, X. , Langdon, Q. K. , DeVirgilio, J. , Hulfachor, A. B. , Kurtzman, C. P. , Rokas, A. & Hittinger, C. T. (2018). Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proceedings of the National Academy of Sciences of the United States of America 115, 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravets, A. , Yang, F. , Bethlendy, G. , Sherman, F. & Rustchenko, E. (2014). Adaptation of Candida albicans to growth on sorbose via monosomy of chromosome 5 accompanied by duplication of another chromosome carrying a gene responsible for sorbose utilization. FEMS Yeast Research 14, 708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer, M. , Wang, J. & Kronstad, J. W. (2012). Peroxisomal and mitochondrial β‐oxidation pathways influence the virulence of the pathogenic fungus Cryptococcus neoformans . Eukaryotic Cell 11, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, P. , Ma, X. , McDonald, B. A. & Brunner, P. C. (2018). Widespread signatures of selection for secreted peptidases in a fungal plant pathogen. BMC Evolutionary Biology 18, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizsan, K. , Almasi, E. , Merenyi, Z. , Sahu, N. , Viragh, M. , Koszo, T. , Mondo, S. , Kiss, B. , Balint, B. , Kues, U. , Barry, K. , Cseklye, J. , Hegedus, B. , Henrissat, B. , Johnson, J. , et al. (2019). Transcriptomic atlas of mushroom development highlights an independent origin of complex multicellularity. Proceedings of the National Academy of Sciences of the United States of America 116, 7409–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiezopolska, E. & Gabaldón, T. (2018). Evolutionary emergence of drug resistance in Candida opportunistic pathogens. Genes 9, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües, U. (2000). Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiology and Molecular Biology Reviews 64, 316–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües, U. , Khonsuntia, W. & Subba, S. (2018). Complex fungi. Fungal Biology Reviews 32, 205–218. [Google Scholar]
- Kües, U. & Liu, Y. (2000). Fruiting body production in Basidiomycetes. Applied Microbiology and Biotechnology 54, 141–152. [DOI] [PubMed] [Google Scholar]
- Kües, U. & Navarro‐González, M. (2015). How do Agaricomycetes shape their fruiting bodies? 1. Morphological aspects of development. Fungal Biology Reviews 29, 63–97. [Google Scholar]
- Kumaran, R. , Yang, S. Y. & Leu, J. Y. (2013). Characterization of chromosome stability in diploid, polyploid and hybrid yeast cells. PLoS One 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler, M. (2018). How fungi defend themselves against microbial competitors and animal predators. PLoS Pathogens 14, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, A. , Kohler, A. , Martin, F. M. & Grigoriev, I. V. (2014). Expanding genomics of mycorrhizal symbiosis. Frontiers in Microbiology 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman, C. P. & Sugiyama, J. (2015). Saccharomycotina and Taphrinomycotina: the Yeasts and Yeastlike Fungi of the Ascomycota In The Mycota VII: Systematics and Evolution Part B, Second Edition (eds. D. McLaughlin. and J. Spatafora.) pp. 3–33. Springer, Berlin Heidelberg. [Google Scholar]
- Lacroix, B. & Citovsky, V. (2016). Transfer of DNA from bacteria to eukaryotes. MBio 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix, B. , Tzfira, T. , Vainstein, A. & Citovsky, V. (2006). A case of promiscuity: Agrobacterium's endless hunt for new partners. Trends in Genetics 22, 29–37. [DOI] [PubMed] [Google Scholar]
- Lan, N. , Zhang, H. , Hu, C. , Wang, W. , Calvo, A. M. , Harris, S. D. , Chen, S. & Li, S. (2014). Coordinated and distinct functions of velvet proteins in Fusarium verticillioides . Eukaryotic Cell 13, 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S. , Linton, L. M. , Birren, B. , Nusbaum, C. , Zody, M. C. , Baldwin, J. , Devon, K. , Dewar, K. , Doyle, M. , FitzHugh, W. , Funke, R. , Gage, D. , Harris, K. , Heaford, A. , Howland, J. , et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Landvik, S. , Schumacher, T. K. , Eriksson, O. E. & Moss, S. T. (2003). Morphology and ultrastructure of Neolecta species. Mycological Research 107, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Lara, E. , Moreira, D. & López‐García, P. (2010). The environmental clade LKM11 and Rozella form the deepest branching clade of Fungi. Protist 161, 116–121. [DOI] [PubMed] [Google Scholar]
- Lawrence, J. (1999). Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Current Opinion in Genetics & Development 9, 642–648. [DOI] [PubMed] [Google Scholar]
- Lebreton, A. , Meslet‐Cladière, L. , Morin‐Sardin, S. , Coton, E. , Jany, J.‐L. , Barbier, G. & Corre, E. (2018). Comparative analysis of five Mucor species transcriptomes. Genomics 111, 1306–1314. [DOI] [PubMed] [Google Scholar]
- Leducq, J.‐B. , Nielly‐Thibault, L. , Charron, G. , Eberlein, C. , Verta, J.‐P. , Samani, P. , Sylvester, K. , Hittinger, C. T. , Bell, G. & Landry, C. R. (2016). Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nature Microbiology 1, 1–10. [DOI] [PubMed] [Google Scholar]
- Lee, B. N. & Adams, T. H. (1994). The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes & Development 8, 641–651. [DOI] [PubMed] [Google Scholar]
- Lee, B. N. & Adams, T. H. (1995). FluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA beta activation. EMBO Journal 15, 299–309. [PMC free article] [PubMed] [Google Scholar]
- Lee, S. C. & Heitman, J. (2014). Sex in the mucoralean fungi. Mycoses 57, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. C. & Idnurm, A. (2017). Fungal sex: the mucoromycota In The Fungal Kingdom (eds J. Heitman, B. J. Howlett, P. W. Crous, E. H. Stukenbrock, T. Y. James, and N. A. R. Gow.), pp. 177–191. American Society for Microbiology, Washington DC. [Google Scholar]
- Lee, S. C. , Ni, M. , Li, W. , Shertz, C. & Heitman, J. (2010). The evolution of sex: a perspective from the fungal kingdom. Microbiology and Molecular Biology Reviews 74, 298–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras, J.‐L. , Galeote, V. , Bigey, F. , Camarasa, C. , Marsit, S. , Nidelet, T. , Sanchez, I. , Couloux, A. , Guy, J. , Franco‐Duarte, R. , Marcet‐Houben, M. , Gabaldon, T. , Schuller, D. , Sampaio, J. P. & Dequin, S. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Molecular Biology and Evolution 35, 1712–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, J. W. , Lapointe, F.‐J. , Lopez, P. & Bapteste, E. (2011). Evaluating phylogenetic congruence in the post‐genomic era. Genome Biology and Evolution 3, 571–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, A. R. & Leitch, I. J. (2012). Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytologist 194, 629–646. [DOI] [PubMed] [Google Scholar]
- Lenassi, M. , Gostinčar, C. , Jackman, S. , Turk, M. , Sadowski, I. , Nislow, C. , Jones, S. , Birol, I. , Cimerman, N. G. & Plemenitaš, A. (2013). Whole genome duplication and enrichment of metal cation transporters revealed by de novo genome sequencing of extremely halotolerant black yeast Hortaea werneckii . PLoS One 8, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher, P. M. , Powell, M. J. , Lee, P. A. , Lopez, S. & Burnett, M. (2017). Molecular phylogeny and ultrastructure of Aphelidium desmodesmi, a new species in Aphelida (Opisthosporidia). Journal of Eukaryotic Microbiology 64, 655–667. [DOI] [PubMed] [Google Scholar]
- Lever, M. C. , Robertson, B. E. M. , Buchan, A. D. B. , Miller, P. F. P. , Gooday, G. W. & Gow, N. A. R. (1994). pH and Ca2+ dependent galvanotropism of filamentous fungi: implications and mechanisms. Mycological Research 98, 301–306. [Google Scholar]
- Lew, R. R. (2011). How does a hypha grow? The biophysics of pressurized growth in fungi. Nature Reviews Microbiology 9, 509–518. [DOI] [PubMed] [Google Scholar]
- Li, W. , Averette, A. F. , Desnos‐Ollivier, M. , Ni, M. , Dromer, F. & Heitman, J. (2012). Genetic diversity and genomic plasticity of Cryptococcus neoformans AD Hybrid Strains. G3 2, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Min, M. , Du, N. , Gu, Y. , Hode, T. , Naylor, M. , Chen, D. , Nordquist, R. E. & Chen, W. R. (2013). Chitin, chitosan, and glycated chitosan regulate immune responses: the novel adjuvants for cancer vaccine. Clinical & Developmental Immunology 2013, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Tiley, G. P. , Galuska, S. R. , Reardon, C. R. , Kidder, T. I. , Rundell, R. J. & Barker, M. S. (2018). Multiple large‐scale gene and genome duplications during the evolution of hexapods. Proceedings of the National Academy of Sciences of the United States of America 115, 4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichius, A. , Berepiki, A. & Read, N. D. (2011). Form follows function – the versatile fungal cytoskeleton. Fungal Biology 115, 518–540. [DOI] [PubMed] [Google Scholar]
- Lin, H.‐C. , Hewage, R. T. , Lu, Y.‐C. & Chooi, Y.‐H. (2019). Biosynthesis of bioactive natural products from Basidiomycota. Organic & Biomolecular Chemistry 17, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Lin, K. , Limpens, E. , Zhang, Z. , Ivanov, S. , Saunders, D. G. O. O. , Mu, D. , Pang, E. , Cao, H. , Cha, H. , Lin, T. , Zhou, Q. , Shang, Y. , Li, Y. , Sharma, T. , van Velzen, R. , et al. (2014a). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genetics 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. , Alspaugh, J. A. , Liu, H. & Harris, S. (2014b). Fungal morphogenesis. Cold Spring Harbor Perspectives in Medicine 5, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. & Bell‐Pedersen, D. (2006). Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryotic Cell 5, 1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. J. & Hall, B. D. (2004). Body plan evolution of ascomycetes, as inferred from an RNA polymerase II phylogeny. Proceedings of the National Academy of Sciences of the United States of America 101, 4507–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. J. , Hodson, M. C. & Hall, B. D. (2006). Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes. BMC Evolutionary Biology 6, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, H.‐C. , Entwistle, R. , Guo, C.‐J. , Ahuja, M. , Szewczyk, E. , Hung, J.‐H. , Chiang, Y.‐M. , Oakley, B. R. & Wang, C. C. C. (2012). Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans . Journal of the American Chemical Society 134, 4709–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Berges, M. S. , Hera, C. , Sulyok, M. , Schäfer, K. , Capilla, J. , Guarro, J. & Di Pietro, A. (2013). The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Molecular Microbiology 87, 49–65. [DOI] [PubMed] [Google Scholar]
- Lopez‐Moya, F. & Lopez‐Llorca, L. V. (2016). Omics for Investigating chitosan as an antifungal and gene modulator. Journal of Fungi 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, K. M. & Read, N. D. (2011). Perithecium morphogenesis in Sordaria macrospora . Fungal Genetics and Biology 48, 388–399. [DOI] [PubMed] [Google Scholar]
- Ma, L.‐J. , Ibrahim, A. S. , Skory, C. , Grabherr, M. G. , Burger, G. , Butler, M. , Elias, M. , Idnurm, A. , Lang, B. F. , Sone, T. , Abe, A. , Calvo, S. E. , Corrochano, L. M. , Engels, R. , Fu, J. , et al. (2009). Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole‐genome duplication. PLoS Genetics 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadum, S. , Banerjee, U. , Murugan, P. , Gangapur, D. & Ravikesavan, R. (2013). Gene duplication as a major force in evolution. Journal of Genetics 92, 155–161. [DOI] [PubMed] [Google Scholar]
- Maheshwari, R. (2005). Nuclear behavior in fungal hyphae. FEMS Microbiology Letters 249, 7–14. [DOI] [PubMed] [Google Scholar]
- Malerba, M. & Cerana, R. (2016). Chitosan effects on plant systems. International Journal of Molecular Sciences 17, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm, J. , Christophersen, C. & Frisvad, J. C. (2000). Secondary metabolites characteristic of Penicillium citrinum, Penicillium steckii and related species. Phytochemistry 54, 301–309. [DOI] [PubMed] [Google Scholar]
- Manocha, M. S. (1965). Fine structure of the Agaricus carpophore . Canadian Journal of Botany 43, 1329–1333. [Google Scholar]
- Marahiel, M. A. (2009). Working outside the protein‐synthesis rules: insights into non‐ribosomal peptide synthesis. Journal of Peptide Science 15, 799–807. [DOI] [PubMed] [Google Scholar]
- Marcet‐Houben, M. , Ballester, A.‐R. , de la Fuente, B. , Harries, E. , Marcos, J. F. , González‐Candelas, L. & Gabaldón, T. (2012). Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics 13, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet‐Houben, M. & Gabaldón, T. (2010). Acquisition of prokaryotic genes by fungal genomes. Trends in Genetics 26, 5–8. [DOI] [PubMed] [Google Scholar]
- Marcet‐Houben, M. & Gabaldón, T. (2015). Beyond the whole‐genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker's yeast lineage. PLoS Biology 13, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet‐Houben, M. & Gabaldón, T. (2016). Horizontal acquisition of toxic alkaloid synthesis in a clade of plant associated fungi. Fungal Genetics and Biology 86, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet‐Houben, M. & Gabaldón, T. (2019). Evolutionary and functional patterns of shared gene neighbourhood in fungi. Nature Microbiology 4, 2383–2392. [DOI] [PubMed] [Google Scholar]
- Marsit, S. , Mena, A. , Bigey, F. , Sauvage, F.‐X. , Couloux, A. , Guy, J. , Legras, J.‐L. , Barrio, E. , Dequin, S. & Galeote, V. (2015). Evolutionary advantage conferred by an eukaryote‐to‐eukaryote gene transfer event in wine yeasts. Molecular Biology and Evolution 32, 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F. , Kohler, A. , Murat, C. , Balestrini, R. , Coutinho, P. M. , Jaillon, O. , Montanini, B. , Morin, E. , Noel, B. , Percudani, R. , Porcel, B. , Rubini, A. , Amicucci, A. , Amselem, J. , Anthouard, V. , et al. (2010). Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464, 1033–1038. [DOI] [PubMed] [Google Scholar]
- Martín, J. F. (2017). Key role of LaeA and velvet complex proteins on expression of β‐lactam and PR‐toxin genes in Penicillium chrysogenum: cross‐talk regulation of secondary metabolite pathways. Journal of Industrial Microbiology & Biotechnology 44, 525–535. [DOI] [PubMed] [Google Scholar]
- Martín, J. F. & Liras, P. (2016). Evolutionary formation of gene clusters by reorganization: the meleagrin/roquefortine paradigm in different fungi. Applied Microbiology and Biotechnology 100, 1579–1587. [DOI] [PubMed] [Google Scholar]
- Martín, J. F. , Ullán, R. V. & García‐Estrada, C. (2010). Regulation and compartmentalization of β‐lactam biosynthesis. Microbial Biotechnology 3, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. G. & Arkowitz, R. A. (2014). Cell polarization in budding and fission yeasts. FEMS Microbiology Reviews 38, 228–253. [DOI] [PubMed] [Google Scholar]
- Maruyama, J. , Escaño, C. S. & Kitamoto, K. (2010). AoSO protein accumulates at the septal pore in response to various stresses in the filamentous fungus Aspergillus oryzae . Biochemical and Biophysical Research Communications 391, 868–873. [DOI] [PubMed] [Google Scholar]
- Mathieu, S. , Cusant, L. , Roux, C. & Corradi, N. (2018). Arbuscular mycorrhizal fungi: intraspecific diversity and pangenomes. New Phytologist 220, 1129–1134. [DOI] [PubMed] [Google Scholar]
- McCarthy, C. G. P. & Fitzpatrick, D. A. (2019). Pan‐genome analyses of model fungal species. Microbial Genomics 5, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel, D. P. & Roberson, R. W. (1998). γ‐Tubulin is a component of the Spitzenkorper and centrosomes in hyphal‐tip cells of Allomyces macrogynus . Protoplasma 203, 118–123. [Google Scholar]
- McDonald, T. R. , Mueller, O. , Dietrich, F. S. & Lutzoni, F. (2013). High‐throughput genome sequencing of lichenizing fungi to assess gene loss in the ammonium transporter/ammonia permease gene family. BMC Genomics 14, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary, K. L. , Slot, J. C. & Rokas, A. (2013). Physical linkage of metabolic genes in fungi is an adaptation against the accumulation of toxic intermediate compounds. Proceedings of the National Academy of Sciences of the United States of America 110, 11481–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, D. J. , Kumar, T. K. A. , Blackwell, M. , Letcher, P. M. & Roberson, R. W. (2015). Subcellular structure and biochemical characters in fungal phylogeny In The Mycota VII: Systematics and Evolution: Part B, Second Edition (eds D. J. McLaughlin. and J. W. Spatafora.), pp. 229–258. Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- McLaughlin, D. J. , Kumar, T. K. A. , Padamsee, M. , Toome‐Heller, M. , Frieders, E. M. & Aime, M. C. (2017). Structural character evolution in Pucciniomycotina: mitosis, septa, and hyphal branch initiation in two Helicogloea species. Mycologia 109, 162–181. [DOI] [PubMed] [Google Scholar]
- Medina‐Castellanos, E. , Esquivel‐Naranjo, E. U. , Heil, M. & Herrera‐Estrella, A. (2014). Extracellular ATP activates MAPK and ROS signaling during injury response in the fungus Trichoderma atroviride . Frontiers in Plant Science 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina‐Castellanos, E. , Villalobos‐Escobedo, J. M. , Riquelme, M. , Read, N. D. , Abreu‐Goodger, C. & Herrera‐Estrella, A. (2018). Danger signals activate a putative innate immune system during regeneration in a filamentous fungus. PLoS Genetics 14, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. , Bahkali, A. H. , Abd‐Elsalam, K. A. , Moslem, M. , Ben M'Barek, S. , Gohari, A. M. , Jashni, M. K. , Stergiopoulos, I. , Kema, G. H. J. & de Wit, P. J. G. M. (2011). Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiology Reviews 35, 542–554. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Mirzadi Gohari, A. & Kema, G. H. J. (2017). Karyotype variability in plant‐pathogenic fungi. Annual Review of Phytopathology 55, 483–503. [DOI] [PubMed] [Google Scholar]
- Meiser, A. , Otte, J. , Schmitt, I. & Grande, F. D. (2017). Sequencing genomes from mixed DNA samples – evaluating the metagenome skimming approach in lichenized fungi. Scientific Reports 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, R. J. & Fuller, M. S. (1985). Structure and development of hyphal septa in Allomyces . American Journal of Botany 72, 1458–1465. [Google Scholar]
- Mhurchu, C. N. , Dunshea‐Mooij, C. , Bennett, D. & Rodgers, A. (2005). Effect of chitosan on weight loss in overweight and obese individuals: a systematic review of randomized controlled trials. Obesity Reviews 6, 35–42. [DOI] [PubMed] [Google Scholar]
- Micali, C. O. & Smith, M. L. (2006). A nonself recognition gene complex in Neurospora crassa . Genetics 173, 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millanes, A. M. , Diederich, P. , Ekman, S. & Wedin, M. (2011). Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 61, 12–28. [DOI] [PubMed] [Google Scholar]
- Mixão, V. & Gabaldón, T. (2018). Yeast Interspecies Hybrids Hybridization and emergence of virulence in opportunistic human yeast pathogens. Yeast 35, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol, P. C. , Vermeulen, C. A. & Wessels, J. G. H. (1990). Diffuse extension of hyphae in stipes of Agaricus bisporus may be based on a unique wall structure. Mycological Research 94, 480–488. [Google Scholar]
- Möller, M. & Stukenbrock, E. H. (2017). Evolution and genome architecture in fungal plant pathogens. Nature Reviews Microbiology 15, 756–771. [DOI] [PubMed] [Google Scholar]
- Molnár, K. & Farkas, E. (2010). Current results on biological activities of lichen secondary metabolites: a review. Journal of Biosciences 65, 157–173. [DOI] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance and new axes. Current Opinion in Microbiology 5, 580–585. [DOI] [PubMed] [Google Scholar]
- Mondo, S. J. , Dannebaum, R. O. , Kuo, R. C. , Louie, K. B. , Bewick, A. J. , LaButti, K. , Haridas, S. , Kuo, A. , Salamov, A. , Ahrendt, S. R. , Lau, R. , Bowen, B. P. , Lipzen, A. , Sullivan, W. , Andreopoulos, B. B. , et al. (2017). Widespread adenine N6‐methylation of active genes in fungi. Nature Genetics 49, 964–968. [DOI] [PubMed] [Google Scholar]
- Monzer, J. (1995). Actin filaments are involved in cellular graviperception of the basidiomycete Flammulina velutipes . European Journal of Cell Biology 66, 151–156. [PubMed] [Google Scholar]
- Monzer, J. (1996). Cellular graviperception in the basidiomycete Flammulina velutipes – Can the nuclei serve as fungal statoliths? European Journal of Cell Biology 71, 216–220. [PubMed] [Google Scholar]
- Moore, D. , Hock, B. , Greening, J. P. , Kern, V. D. , Frazer, L. N. & Monzer, J. (1996). Gravimorphogenesis in agarics. Mycological Research 100, 257–273. [DOI] [PubMed] [Google Scholar]
- Morales, L. & Dujon, B. (2012). Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiology and Molecular Biology Reviews 76, 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi, K. , Yamamoto, S. , Tanaka, K. , Kurata, N. & Suzuki, K. (2013). Trans‐kingdom horizontal DNA transfer from bacteria to yeast is highly plastic due to natural polymorphisms in auxiliary nonessential recipient genes. PLoS One 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, E. , Miyauchi, S. , San Clemente, H. , Chen, E. C. , Pelin, A. , de la Providencia, I. , Ndikumana, S. , Beaudet, D. , Hainaut, M. , Drula, E. , Kuo, A. , Tang, N. , Roy, S. , Viala, J. , Henrissat, B. , et al. (2019). Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. The New Phytologist 222, 1584–1598. [DOI] [PubMed] [Google Scholar]
- Morris, J. J. (2015). Black Queen evolution: the role of leakiness in structuring microbial communities. Trends in Genetics 31, 475–482. [DOI] [PubMed] [Google Scholar]
- Morrison, D. J. (2004). Rhizomorph growth habit, saprophytic ability and virulence of 15 Armillaria species. Forest Pathology 34, 15–26. [Google Scholar]
- Morrow, C. a. & Fraser, J. a. (2013). Ploidy variation as an adaptive mechanism in human pathogenic fungi. Seminars in Cell and Developmental Biology 24, 339–346. [DOI] [PubMed] [Google Scholar]
- Mosblech, A. , Feussner, I. & Heilmann, I. (2009). Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiology and Biochemistry 47, 511–517. [DOI] [PubMed] [Google Scholar]
- Motta, J. J. (1969). Cytology and morphogenesis in the rhizomorph of Armillaria mellea . American Journal of Botany 56, 610–619. [Google Scholar]
- Muller, W. H. , Montijn, R. C. , Humbel, B. M. , van Aelst, A. C. , Boon, E. J. M. C. , van der Krift, T. P. & Boekhout, T. (1998). Structural differences between two types of basidiomycete septal pore caps. Microbiology 144, 1721–1730. [DOI] [PubMed] [Google Scholar]
- Murat, C. , Payen, T. , Noel, B. , Kuo, A. , Morin, E. , Chen, J. , Kohler, A. , Krizsán, K. , Balestrini, R. , Da Silva, C. , Montanini, B. , Hainaut, M. , Levati, E. , Barry, K. W. , Belfiori, B. , et al. (2018). Pezizomycetes genomes reveal the molecular basis of ectomycorrhizal truffle lifestyle. Nature Ecology & Evolution 2, 1956–1965. [DOI] [PubMed] [Google Scholar]
- Murphy, C. , Youssef, N. , Hanafy, R. A. , Couger, M. , Stajich, J. E. , Wang, Y. , Baker, K. , Dagar, S. , Griffith, G. , Farag, I. , Callaghan, T. & Elshahed, M. S. (2019). Horizontal gene transfer as an indispensible driver for Neocallimastigomycota evolution into a distinct gut‐dwelling fungal lineage. Applied and environmental microbiology, 85 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, L. G. , Kovács, G. M. & Krizsán, K. (2018). Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biological Reviews 93, 1778–1794. [DOI] [PubMed] [Google Scholar]
- Nair, D. R. , Occhipinti, P. , Borsuk, M. E. & Gladfelter, A. S. (2010). A conserved G 1 regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm. Cell Cycle 9, 3771–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagiri, A. & Ito, T. (1991). Basidiocarp development of the cyphelloid gasteroid aquatic basidiomycetes Halocyphina villosa and Limnoperdon incarnatum . Canadian Journal of Botany 69, 2320–2327. [Google Scholar]
- Naranjo‐Ortíz, M. A. , Brock, M. , Brunke, S. , Hube, B. , Marcet‐Houben, M. & Gabaldón, T. (2016). Widespread inter‐ and intra‐domain horizontal gene transfer of d‐amino acid metabolism enzymes in eukaryotes. Frontiers in Microbiology 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo‐Ortiz, M. A. & Gabaldón, T. (2019a). Fungal evolution: diversity, taxonomy and phylogeny of the Fungi. Biological Reviews 94, 2101–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo‐Ortiz, M. A. & Gabaldón, T. (2019b). Fungal evolution: major ecological adaptations and evolutionary transitions. Biological Reviews 94, 1443–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir, R. , Warmink, J. A. , Boersma, H. & van Elsas, J. D. (2010). Mechanisms that promote bacterial fitness in fungal‐affected soil microhabitats. FEMS Microbiology Ecology 71, 169–185. [DOI] [PubMed] [Google Scholar]
- Neafsey, D. E. , Barker, B. M. , Sharpton, T. J. , Stajich, J. E. , Park, D. J. , Whiston, E. , Hung, C.‐Y. , Mcmahan, C. , White, J. , Sykes, S. , Heiman, D. , Young, S. , Zeng, Q. , Abouelleil, A. , Aftuck, L. , et al. (2010). Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Research 20, 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M. , Ekstrom, A. , Li, X. & Yin, Y. (2015). HGT‐Finder: a new tool for horizontal gene transfer finding and application to Aspergillus genomes. Toxins 7, 4035–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. A. , Cissé, O. H. , Yun Wong, J. , Zheng, P. , Hewitt, D. , Nowrousian, M. , Stajich, J. E. & Jedd, G. (2017). Innovation and constraint leading to complex multicellularity in the Ascomycota. Nature Communications 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. A. , Greig, J. , Khan, A. , Goh, C. & Jedd, G. (2018). Evolutionary novelty in gravity sensing through horizontal gene transfer and high‐order protein assembly. PLoS Biology 16, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M. , Feretzaki, M. , Sun, S. , Wang, X. & Heitman, J. (2011). Sex in Fungi. Annual Review of Genetics 45, 405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus, E.‐M. , Kim, H.‐K. , Münsterkötter, M. , Janevska, S. , Arndt, B. , Kalinina, S. A. , Houterman, P. M. , Ahn, I.‐P. , Alberti, I. , Tonti, S. , Kim, D.‐W. , Sieber, C. M. K. , Humpf, H.‐U. , Yun, S.‐H. , Güldener, U. , et al. (2017). Comparative genomics of geographically distant Fusarium fujikuroi isolates revealed two distinct pathotypes correlating with secondary metabolite profiles. PLoS Pathogens 13, 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J. C. , Grijseels, S. , Prigent, S. , Ji, B. , Dainat, J. , Nielsen, K. F. , Frisvad, J. C. , Workman, M. & Nielsen, J. (2017). Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nature Microbiology 2, 1–9. [DOI] [PubMed] [Google Scholar]
- Nijkamp, J. F. , van den Broek, M. , Datema, E. , de Kok, S. , Bosman, L. , Luttik, M. A. , Daran‐Lapujade, P. , Vongsangnak, W. , Nielsen, J. , Heijne, W. H. M. , Klaassen, P. , Paddon, C. J. , Platt, D. , Kötter, P. , van Ham, R. C. , et al. (2012). De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113‐7D, a model for modern industrial biotechnology. Microbial Cell Factories 11, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas, K. J. (2014). The evolutionary‐developmental origins of multicellularity. American Journal of Botany 101, 6–25. [DOI] [PubMed] [Google Scholar]
- Nobre, T. , Koopmanschap, B. , Baars, J. J. P. , Sonnenberg, A. S. M. & Aanen, D. K. (2014). The scope for nuclear selection within Termitomyces fungi associated with fungus‐growing termites is limited. BMC Evolutionary Biology 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nødvig, C. S. , Nielsen, J. B. , Kogle, M. E. & Mortensen, U. H. (2015). A CRISPR‐Cas9 system for genetic engineering of filamentous fungi. PLoS One 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr, M. C. , Erb‐Downward, J. R. & Huffnagle, G. B. (2003). Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clinical Microbiology Reviews 16, 517–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo, M. , Bigey, F. , Beyne, E. , Galeote, V. , Gavory, F. , Mallet, S. , Cambon, B. , Legras, J.‐L. , Wincker, P. , Casaregola, S. & Dequin, S. (2009). Eukaryote‐to‐eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proceedings of the National Academy of Sciences of the United States of America 106, 16333–16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian, M. (2018). Genomics and transcriptomics to study fruiting body development: an update. Fungal Biology Reviews 32, 231–235. [Google Scholar]
- Nowrousian, M. , Frank, S. , Koers, S. , Strauch, P. , Weitner, T. , Ringelberg, C. , Dunlap, J. C. , Loros, J. J. & Kück, U. (2007). The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordaria macrospora . Molecular Microbiology 64, 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann, H.‐W. , Huang, A. & Osbourn, A. (2016). Plant metabolic clusters ‐ from genetics to genomics. The New Phytologist 211, 771–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman, C. M. , Fuller, H. T. & Dyer, P. S. (2009). Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus . Nature 457, 471–474. [DOI] [PubMed] [Google Scholar]
- Oberwinkler, F. (2014). Dacrymycetes In The Mycota VII: Systematics and Evolution: Part A, Second Edition (ed K. Esser.), pp. 357–370. Springer, Berlin Heidelberg. [Google Scholar]
- Oberwinkler, F. (2017). Yeasts in Pucciniomycotina. Mycological Progress 16, 831–856. [Google Scholar]
- Ohm, R. A. , de Jong, J. F. , Berends, E. , Wang, F. , Wösten, H. A. B. & Lugones, L. G. (2010a). An efficient gene deletion procedure for the mushroom‐forming basidiomycete Schizophyllum commune . World Journal of Microbiology and Biotechnology 26, 1919–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm, R. A. , de Jong, J. F. , Lugones, L. G. , Aerts, A. , Kothe, E. , Stajich, J. E. , de Vries, R. P. , Record, E. , Levasseur, A. , Baker, S. E. , Bartholomew, K. A. , Coutinho, P. M. , Erdmann, S. , Fowler, T. J. , Gathman, A. C. , et al. (2010b). Genome sequence of the model mushroom Schizophyllum commune . Nature Biotechnology 28, 957–963. [DOI] [PubMed] [Google Scholar]
- Okagaki, L. H. & Nielsen, K. (2012). Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryotic Cell 11, 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte, R. A. , Menke, J. , Zhang, Y. , Sullivan, S. , Slot, J. C. , Huang, Y. , Badalamenti, J. P. , Quandt, A. C. , Spatafora, J. W. & Bushley, K. E. (2019). Chromosome rearrangements shape the diversification of secondary metabolism in the cyclosporin producing fungus Tolypocladium inflatum . BMC Genomics 20, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, R. P. & Solomon, P. S. (2008). Recent fungal diseases of crop plants: is lateral gene transfer a common theme? Molecular Plant‐Microbe Interactions 21, 287–293. [DOI] [PubMed] [Google Scholar]
- Olmedo, M. , Ruger‐Herreros, C. , Luque, E. M. & Corrochano, L. M. (2013). Regulation of transcription by light in Neurospora crassa: a model for fungal photobiology? Fungal Biology Reviews 27, 10–18. [Google Scholar]
- Orlovich, D. A. & Ashford, A. E. (1994). Structure and development of the dolipore septum in Pisolithus tinctorius . Protoplasma 178, 66–80. [Google Scholar]
- Oromendia, A. B. , Dodgson, S. E. & Amon, A. (2012). Aneuploidy causes proteotoxic stress in yeast. Genes & Development 26, 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrosky‐zeichner, L. (2012). Invasive mycoses: diagnostic challenges. American Journal of Medicine 125, 14–24. [DOI] [PubMed] [Google Scholar]
- Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. [DOI] [PubMed] [Google Scholar]
- Pacheco‐Arjona, J. R. & Ramirez‐Prado, J. H. (2014). Large‐scale phylogenetic classification of fungal chitin synthases and identification of a putative cell‐wall metabolism gene cluster in Aspergillus genomes. PLoS One 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma‐Guerrero, J. , Huang, I.‐C. , Jansson, H.‐B. , Salinas, J. , Lopez‐Llorca, L. V. & Read, N. D. (2009). Chitosan permeabilizes the plasma membrane and kills cells of Neurospora crassa in an energy dependent manner. Fungal Genetics and Biology 46, 585–594. [DOI] [PubMed] [Google Scholar]
- Palma‐Guerrero, J. , Jansson, H.‐B. , Salinas, J. & Lopez‐Llorca, L. V. (2008). Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. Journal of Applied Microbiology 104, 541–553. [DOI] [PubMed] [Google Scholar]
- Palmer, G. E. & Horton, J. S. (2006). Mushrooms by magic: making connections between signal transduction and fruiting body development in the basidiomycete fungus Schizophyllum commune . FEMS Microbiology Letters 262, 1–8. [DOI] [PubMed] [Google Scholar]
- Park, R. F. & Wellings, C. R. (2012). Somatic hybridization in the Uredinales. Annual Review of Phytopathology 50, 219–239. [DOI] [PubMed] [Google Scholar]
- Partida‐Martinez, L. P. , De Looß, C. F. , Ishida, K. , Ishida, M. , Roth, M. , Buder, K. & Hertweck, C. (2007). Rhizonin, the first mycotoxin isolated from the zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Applied and Environmental Microbiology 73, 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peberdy, J. F. (1979). Fungal protoplasts: isolation, reversion, and fusion. Annual Review of Microbiology 33, 21–39. [DOI] [PubMed] [Google Scholar]
- Peberdy, J. F. (1989). Presidential address: fungi without coats — protoplasts as tools for mycological research. Mycological Research 93, 11–20. [Google Scholar]
- Pelin, A. , Selman, M. , Aris‐Brosou, S. , Farinelli, L. & Corradi, N. (2015). Genome analyses suggest the presence of polyploidy and recent human‐driven expansions in eight global populations of the honeybee pathogen Nosema ceranae . Environmental Microbiology 17, 4443–4458. [DOI] [PubMed] [Google Scholar]
- Perlin, M. H. , Amselem, J. , Fontanillas, E. , Toh, S. S. , Chen, Z. , Goldberg, J. , Duplessis, S. , Henrissat, B. , Young, S. , Zeng, Q. , Aguileta, G. , Petit, E. , Badouin, H. , Andrews, J. , Razeeq, D. , et al. (2015). Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis‐dioicae, the biotrophic and plant‐castrating anther smut fungus. BMC Genomics 16, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, J. , De Chiara, M. , Friedrich, A. , Yue, J.‐X. , Pflieger, D. , Bergström, A. , Sigwalt, A. , Barre, B. , Freel, K. , Llored, A. , Cruaud, C. , Labadie, K. , Aury, J.‐M. , Istace, B. , Lebrigand, K. , et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza, D. , Lin, C.‐W. , van der Velden, N. S. , Aebi, M. & Künzler, M. (2014). Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue‐specific armories and a conserved circuitry for sexual development. BMC Genomics 15, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plissonneau, C. , Hartmann, F. E. & Croll, D. (2018). Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biology 16, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plissonneau, C. , Stürchler, A. , Croll, D. & Taylor, J. W. (2016). The evolution of orphan regions in genomes of a fungal pathogen of wheat. MBio 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler, S. , Nowrousian, M. & Kück, U. (2006). Fruiting‐body development in Ascomycetes In The mycota I: Growth, Differentiation and Sexuality (eds U. Kües, and R. Fischer.), pp. 325–355. Springer, Berlin Heidelberg. [Google Scholar]
- Pontecorvo, G. (1956). The parasexual cycle in fungi. Annual Review of Microbiology 10, 393–400. [DOI] [PubMed] [Google Scholar]
- Ponting, C. P. (2017). Biological function in the twilight zone of sequence conservation. BMC Biology 15, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe, S. , Dorsheimer, L. , Happel, P. & Stukenbrock, E. H. (2015). Rapidly evolving genes are key players in host specialization and virulence of the fungal wheat pathogen Zymoseptoria tritici (Mycosphaerella graminicola). PLoS Pathogens 11, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, M. J. & Letcher, P. M. (2014). Chytridiomycota, Monoblepharidomycota, and Neocallimastigomycota In The Mycota VII: Systematics and Evolution: Part A, Second Edition (ed. K. Esser.), pp. 141–175. Springer, Berlin Heidelberg. [Google Scholar]
- Prieto, M. , Wedin, M. , Ho, S. , Yang, Y. , Zhang, X. & Xiang, M. (2013). Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS One 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz, L. P. & Gabaldón, T. (2016). Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Research 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz, L. P. , Nemeth, T. , Gacser, A. & Gabaldon, T. (2014). Genome comparison of Candida orthopsilosis clinical strains reveals the existence of hybrids between two distinct subspecies. Genome Biology and Evolution 6, 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz, L. P. , Németh, T. , Saus, E. , Ksiezopolska, E. , Hegedűsová, E. , Nosek, J. , Wolfe, K. H. , Gacser, A. & Gabaldón, T. (2015). The genomic aftermath of hybridization in the opportunistic pathogen Candida metapsilosis . PLoS Genetics 11, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, H. , Cai, G. , Luo, J. , Bhattacharya, D. & Zhang, N. (2016). Extensive horizontal gene transfers between plant pathogenic fungi. BMC Biology 14, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quin, M. B. , Flynn, C. M. & Schmidt‐Dannert, C. (2014). Traversing the fungal terpenome. Natural Product Reports 31, 1449–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudaskoski, M. & Kothe, E. (2010). Basidiomycete mating type genes and pheromone signaling. Eukaryotic Cell 9, 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redecker, D. & Schüβler, A. (2014). Glomeromycota In The Mycota VII: Systematics and Evolution, Part A, Second Edition (ed K. Esser.), pp. 251–269. Springer, Berlin Heidelberg. [Google Scholar]
- Ren, R. , Sun, Y. , Zhao, Y. , Geiser, D. , Ma, H. & Zhou, X. (2016). Phylogenetic resolution of deep eukaryotic and fungal relationships using highly conserved low‐copy nuclear genes. Genome Biology and Evolution 8, 2683–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, H. T. , Vijayakumar, V. , Gluck‐Thaler, E. , Korotkin, H. B. , Matheny, P. B. & Slot, J. C. (2018). Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evolution Letters 2, 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, T. A. , Dacks, J. B. , Jenkinson, J. M. , Thornton, C. R. & Talbot, N. J. (2006). Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Current Biology 16, 1857–1864. [DOI] [PubMed] [Google Scholar]
- Richards, T. A. , Leonard, G. , Soanes, D. M. & Talbot, N. J. (2011). Gene transfer into the fungi. Fungal Biology Reviews 25, 98–110. [Google Scholar]
- Riquelme, M. (2013). Tip growth in filamentous fungi: a road trip to the apex. Annual Review of Microbiology 67, 587–609. [DOI] [PubMed] [Google Scholar]
- Riquelme, M. , Aguirre, J. , Bartnicki‐García, S. , Braus, G. H. , Feldbrügge, M. , Fleig, U. , Hansberg, W. , Herrera‐Estrella, A. , Kämper, J. , Kück, U. , Mouriño‐Pérez, R. R. , Takeshita, N. & Fischer, R. (2018). Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiology and Molecular Biology Reviews 82, 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme, M. & Bartnicki‐Garcia, S. (2004). Key differences between lateral and apical branching in hyphae of Neurospora crassa . Fungal Genetics and Biology 41, 842–851. [DOI] [PubMed] [Google Scholar]
- Riquelme, M. & Sánchez‐León, E. (2014). The Spitzenkörper: a choreographer of fungal growth and morphogenesis. Current Opinion in Microbiology 20, 27–33. [DOI] [PubMed] [Google Scholar]
- Riquelme, M. , Yarden, O. , Bartnicki‐Garcia, S. , Bowman, B. , Castro‐Longoria, E. , Free, S. J. , Fleißner, A. , Freitag, M. , Lew, R. R. , Mouriño‐Pérez, R. , Plamann, M. , Rasmussen, C. , Richthammer, C. , Roberson, R. W. , Sanchez‐Leon, E. , et al. (2011). Architecture and development of the Neurospora crassa hypha – a model cell for polarized growth. Fungal Biology 115, 446–474. [DOI] [PubMed] [Google Scholar]
- Rittenour, W. R. , Si, H. & Harris, S. D. (2009). Hyphal morphogenesis in Aspergillus nidulans . Fungal Biology Reviews 23, 20–29. [Google Scholar]
- Roberson, R. W. , Saucedo, E. , Maclean, D. , Propster, J. , Unger, B. , Oneil, T. A. , Parvanehgohar, K. , Cavanaugh, C. & Lowry, D. (2011). The hyphal tip structure of Basidiobolus sp.: a zygomycete fungus of uncertain phylogeny. Fungal Biology 115, 485–492. [DOI] [PubMed] [Google Scholar]
- Roberts, S. E. & Gladfelter, A. S. (2015). Nuclear autonomy in multinucleate fungi. Current Opinion in Microbiology 28, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs, M. (2014). Fungal secondary metabolite dynamics in fungus‐grazer interactions: novel insights and unanswered questions. Frontiers in Microbiology 5, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs, M. , Albert, M. , Keller, N. P. & Kempken, F. (2007). Secondary chemicals protect mould from fungivory. Biology Letters 3, 523–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas‐Jimenez, K. , Wurzbacher, C. , Bourne, E. C. , Chiuchiolo, A. , Priscu, J. C. & Grossart, H.‐P. (2017). Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities in ice‐covered lakes of the McMurdo Dry Valleys, Antarctica. Scientific Reports 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas, A. , Wisecaver, J. H. & Lind, A. L. (2018). The birth, evolution and death of metabolic gene clusters in fungi. Nature Reviews Microbiology 16, 731–744. [DOI] [PubMed] [Google Scholar]
- Ropars, J. & Corradi, N. (2015). Homokaryotic vs heterokaryotic mycelium in arbuscular mycorrhizal fungi: different techniques, different results? New Phytologist 208, 638–641. [DOI] [PubMed] [Google Scholar]
- Ropars, J. , Rodríguez de la Vega, R. C. , López‐Villavicencio, M. , Gouzy, J. , Sallet, E. , Dumas, É. , Lacoste, S. , Debuchy, R. , Dupont, J. , Branca, A. & Giraud, T. (2015). Adaptive horizontal gene transfers between multiple cheese‐associated fungi. Current Biology 25, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M. , Ellison, C. , Taylor, J. W. & Glass, N. L. (2012). Nuclear and genome dynamics in multinucleate ascomycete fungi. Current Biology 21, 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M. , Lee, C. , Hickey, P. C. & Gladfelter, A. S. (2015). Life as a moving fluid: fate of cytoplasmic macromolecules in dynamic fungal syncytia. Current Opinion in Microbiology 26, 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M. , Simonin, A. , Hickey, P. C. , Leeder, A. & Glass, N. L. (2013). Nuclear dynamics in a fungal chimera. Proceedings of the National Academy of Sciences of the United States of America 110, 12875–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum, E. B. , James, T. Y. , Zamudio, K. R. , Poorten, T. J. , Ilut, D. , Rodriguez, D. , Eastman, J. M. , Richards‐Hrdlicka, K. , Joneson, S. , Jenkinson, T. S. , Longcore, J. E. , Parra Olea, G. , Toledo, L. F. , Arellano, M. L. , Medina, E. M. , et al. (2013). Complex history of the amphibian‐killing chytrid fungus revealed with genome resequencing data. Proceedings of the National Academy of Sciences of the United States of America 110, 9385–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig, R. F. , Sharp, R. R. , Treves, D. S. & Adams, J. (1994). Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli . Genetics 137, 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewich, U. L. & Kistler, H. C. (2000). Role of horizontal gene transfer in the evolution of Fungi. Annual Review of Phytopathology 38, 325–363. [DOI] [PubMed] [Google Scholar]
- Roze, L. V. , Chanda, A. & Linz, J. E. (2011). Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genetics and Biology 48, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, F. J. , Beadle, G. W. & Tatum, E. L. (1943). The tube nethod of measuring the growth rate of Neurospora . American Journal of Botany 30, 784–799. [Google Scholar]
- Ryan, F. J. & Lederberg, J. (1946). Reverse‐mutation and adaptation in leucineless Neurospora . Proceedings of the National Academy of Sciences of the United States of America 32, 163–173. [PubMed] [Google Scholar]
- Saari, S. & Faeth, S. H. (2012). Hybridization of Neotyphodium endophytes enhances competitive ability of the host grass. New Phytologist 195, 231–236. [DOI] [PubMed] [Google Scholar]
- Safonova, Y. , Bankevich, A. & Pevzner, P. A. (2015). dipSPAdes: assembler for Highly polymorphic diploid genomes. Journal of Computational Biology 22, 528–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, Y. (2018). Influences of environmental factors on fruiting body induction, development and maturation in mushroom‐forming fungi. Fungal Biology Reviews 32, 236–248. [Google Scholar]
- Salichos, L. & Rokas, A. (2010). The diversity and evolution of circadian clock proteins in fungi. Mycologia 102, 269–278. [DOI] [PubMed] [Google Scholar]
- Salzberg, S. L. (2017). Horizontal gene transfer is not a hallmark of the human genome. Genome Biology 18, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, W. B. & de los Rios, A. (2012). Development of thallus axes in Usnea longissima (Parmeliaceae, Ascomycota), a fruticose lichen showing diffuse growth. American Journal of Botany 99, 998–1009. [DOI] [PubMed] [Google Scholar]
- Sanders, W. B. & de los Rios, A. (2017). Parenchymatous cell division characterizes the fungal cortex of some common foliose lichens. American Journal of Botany 104, 207–217. [DOI] [PubMed] [Google Scholar]
- Sanz, C. , Alvarez, M. I. , Orejas, M. , Velayos, A. , Eslava, A. P. & Benito, E. P. (2002). Interallelic complementation provides genetic evidence for the multimeric organization of the Phycomyces blakesleeanus phytoene dehydrogenase. European Journal of Biochemistry 269, 902–908. [DOI] [PubMed] [Google Scholar]
- Saper, R. B. , Eisenberg, D. M. & Phillips, R. S. (2004). Common dietary supplements for weight loss. American Family Physician 70, 1731–1738. [PubMed] [Google Scholar]
- Sarikaya Bayram, O. , Bayram, O. , Valerius, O. , Park, H. S. , Irniger, S. , Gerke, J. , Ni, M. , Han, K.‐H. , Yu, J.‐H. & Braus, G. H. (2010). LaeA control of velvet family regulatory proteins for light‐dependent development and fungal cell‐type specificity. PLoS Genetics 6, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathiyabama, M. , Akila, G. & Charles, R. E. (2014). Chitosan‐induced defence responses in tomato plants against early blight disease caused by Alternaria solani (Ellis and Martin) Sorauer. Archives of Phytopathology and Plant Protection 47, 1963–1973. [Google Scholar]
- Saupe, S. J. (2000). Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiology and Molecular Biology Reviews 64, 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasaki, Y. , Inomata, K. & Yoshida, K. (1996). Trans‐kingdom conjugation between Agrobacterium tumefaciens and Saccharomyces cerevisiae, a bacterium and a yeast. Plant & Cell Physiology 37, 103–106. [DOI] [PubMed] [Google Scholar]
- Scazzocchio, C. (2014). Fungal biology in the post‐genomic era. Fungal Biology and Biotechnology 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheib, U. , Stehfest, K. , Gee, C. E. , Körschen, H. G. , Fudim, R. , Oertner, T. G. & Hegemann, P. (2015). The rhodopsin–guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Science Signaling 8, 1–9. [DOI] [PubMed] [Google Scholar]
- Schimek, C. , Eibel, P. , Grolig, F. , Horie, T. , Ootaki, T. & Galland, P. (1999). Gravitropism in Phycomyces: a role for sedimenting protein crystals and floating lipid globules. Planta 210, 132–142. [DOI] [PubMed] [Google Scholar]
- Schmitt, I. (2011). Fruiting Body Evolution in the Ascomycota: a molecular perspective integrating lichenized and non‐lichenized groups In The Mycota XIV: Evolution of Fungi and Fungal‐like Organisms (ed S. Pöggeler, and J. Wöstemeyer.), pp. 187–204. Springer, Berlin Heidelberg. [Google Scholar]
- Schmitt, I. & Lumbsch, H. T. (2009). Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch, C. L. , Sung, G.‐H. , López‐Giráldez, F. , Townsend, J. P. , Miadlikowska, J. , Hofstetter, V. , Robbertse, B. , Matheny, P. B. , Kauff, F. , Wang, Z. , Gueidan, C. , Andrie, R. M. , Trippe, K. , Ciufetti, L. M. , Wynns, A. , et al. (2009). The Ascomycota tree of life: a phylum‐wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Systematic Biology 58, 224–239. [DOI] [PubMed] [Google Scholar]
- Schoenfelder, K. P. & Fox, D. T. (2015). The expanding implications of polyploidy. The Journal of Cell Biology 209, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüffler, A. & Anke, T. (2009). Secondary metabolites of basidiomycetes In The Mycota: Physiology and Genetics (eds T. Anke, and A. Schüffler.), pp. 209–231. Springer, Berlin Heidelberg. [Google Scholar]
- Schumacher, J. (2017). How light affects the life of Botrytis . Fungal Genetics and Biology 106, 26–41. [DOI] [PubMed] [Google Scholar]
- Schumacher, J. , Simon, A. , Cohrs, K. C. , Traeger, S. , Porquier, A. , Dalmais, B. , Viaud, M. & Tudzynski, B. (2015). The VELVET complex in the gray mold fungus Botrytis cinerea: impact of BcLAE1 on differentiation, secondary metabolism, and virulence. Molecular Plant‐Microbe Interactions 28, 659–674. [DOI] [PubMed] [Google Scholar]
- Schwager, E. E. , Sharma, P. P. , Clarke, T. , Leite, D. J. , Wierschin, T. , Pechmann, M. , Akiyama‐Oda, Y. , Esposito, L. , Bechsgaard, J. , Bilde, T. , Buffry, A. D. , Chao, H. , Dinh, H. , Doddapaneni, H. , Dugan, S. , et al. (2017). The house spider genome reveals an ancient whole‐genome duplication during arachnid evolution. BMC Biology 15, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A. L. , Richmond, P. A. , Dowell, R. D. & Selmecki, A. M. (2017). The influence of polyploidy on the evolution of yeast grown in a sub‐optimal carbon source. Molecular Biology and Evolution 34, 2690–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, I. & Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. The EMBO Journal 23, 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto, S. , Rochon, D. , Long, J. E. , Dee, J. M. & Berbee, M. L. (2011). A multigene phylogeny of Olpidium and its implications for early fungal evolution. BMC Evolutionary Biology 11, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse, M.‐A. (2002). Prototaxites: a 400 MYR old giant fossil, a saprophytic holobasidiomycete, or a lichen? Mycological Research 106, 642–644. [Google Scholar]
- Sharon, A. , Finkelstein, A. , Shlezinger, N. & Hatam, I. (2009). Fungal apoptosis: function, genes and gene function. FEMS Microbiology Reviews 33, 833–854. [DOI] [PubMed] [Google Scholar]
- Shepherd, V. A. , Orlovich, D. A. & Ashford, A. E. (1993). Cell‐to‐cell transport via motile tubules in growing hyphae of a fungus. Journal of Cell Science 105, 1173–1178. [DOI] [PubMed] [Google Scholar]
- Shirouzu, T. , Hirose, D. , Oberwinkler, F. , Shimomura, N. , Maekawa, N. & Tokumasu, S. (2013). Combined molecular and morphological data for improving phylogenetic hypothesis in Dacrymycetes. Mycologia 105, 1110–1125. [DOI] [PubMed] [Google Scholar]
- Shlezinger, N. , Doron, A. & Sharon, A. (2011). Apoptosis‐like programmed cell death in the grey mould fungus Botrytis cinerea: genes and their role in pathogenicity. Biochemical Society Transactions 39, 1493–1498. [DOI] [PubMed] [Google Scholar]
- Shlezinger, N. , Goldfinger, N. & Sharon, A. (2012). Apoptotic‐like programed cell death in fungi: the benefits in filamentous species. Frontiers in Oncology 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, J.‐Y. , Charlton, N. D. , Yi, M. , Young, C. A. & Craven, K. D. (2015). Vegetative hyphal fusion and subsequent nuclear behavior in Epichloë grass endophytes. PLoS One 10, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin, A. , Palma‐Guerrero, J. , Fricker, M. & Glass, N. L. (2012). Physiological significance of network organization in fungi. Eukaryotic Cell 11, 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. , Flibotte, S. , Niera, M. , Formby, S. , Plemenitaš, A. , Gunde Cimerman, N. , Lenassi, M. , Gostinčar, C. , Stajich, J. E. & Nislow, C. (2017). Insight into the recent genome duplication of the halophilic yeast Hortaea werneckii: combining an improved genome with gene expression and chromatin structure. G3 7, 2015–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov, E. , Lee, H. , Chang, Y. C. & Kwon‐Chung, K. J. (2010). Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathogens 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos, G. , Anderson, J. B. & Nagy, L. G. (2018). Armillaria . Current Biology 28, 297–298. [DOI] [PubMed] [Google Scholar]
- Sipos, G. , Prasanna, A. N. , Walter, M. C. , O'Connor, E. , Bálint, B. , Krizsán, K. , Kiss, B. , Hess, J. , Varga, T. , Slot, J. , Riley, R. , Bóka, B. , Rigling, D. , Barry, K. , Lee, J. , et al. (2017). Genome expansion and lineage‐specific genetic innovations in the forest pathogenic fungi Armillaria . Nature Ecology & Evolution 1, 1931–1941. [DOI] [PubMed] [Google Scholar]
- Slot, J. C. (2017). Fungal gene cluster diversity and evolution In Advances in Genetics (eds J. P. Townsend, and Z. Wang.), pp. 141–178. Academic Press Inc, Cambridge MA. [DOI] [PubMed] [Google Scholar]
- Slot, J. C. & Hibbett, D. S. (2007). Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS One 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot, J. C. & Rokas, A. (2011). Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Current Biology 21, 134–139. [DOI] [PubMed] [Google Scholar]
- Soanes, D. & Richards, T. A. (2014). Horizontal gene transfer in eukaryotic plant pathogens. Annual Review of Phytopathology 52, 583–614. [DOI] [PubMed] [Google Scholar]
- Soid‐Raggi, G. , Sánchez, O. , Ramos‐Balderas, J. L. & Aguirre, J. (2016). The adenylate‐forming enzymes AfeA and TmpB are involved in Aspergillus nidulans self‐communication during asexual development. Frontiers in Microbiology 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis, P. S. , Marchant, D. B. , Van de Peer, Y. & Soltis, D. E. (2015). Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development 35, 119–125. [DOI] [PubMed] [Google Scholar]
- Soucy, S. M. , Huang, J. & Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nature Reviews Genetics 16, 472–482. [DOI] [PubMed] [Google Scholar]
- Spatafora, J. W. & Bushley, K. E. (2015). Phylogenomics and evolution of secondary metabolism in plant‐associated fungi. Current Opinion in Plant Biology 26, 37–44. [DOI] [PubMed] [Google Scholar]
- Spatafora, J. W. , Chang, Y. , Benny, G. L. , Lazarus, K. , Smith, M. E. , Berbee, M. L. , Bonito, G. , Corradi, N. , Grigoriev, I. , Gryganskyi, A. , James, T. Y. , O'Donnell, K. , Roberson, R. W. , Taylor, T. N. , Uehling, J. , et al. (2016). A phylum‐level phylogenetic classification of zygomycete fungi based on genome‐scale data. Mycologia 108, 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora, J. W. , Sung, G.‐H. , Johnson, D. , Hesse, C. , O'Rourke, B. , Serdani, M. , Spotts, R. , Lutzoni, F. , Hofstetter, V. , Miadlikowska, J. , Reeb, V. , Gueidan, C. , Fraker, E. , Lumbsch, T. , Lücking, R. , et al. (2006). A five‐gene phylogeny of Pezizomycotina. Mycologia 98, 1018–1028. [DOI] [PubMed] [Google Scholar]
- Spribille, T. , Tuovinen, V. , Resl, P. , Vanderpool, D. , Wolinski, H. , Aime, M. C. , Schneider, K. , Stabentheiner, E. , Toome‐Heller, M. , Thor, G. , Mayrhofer, H. , Johannesson, H. & McCutcheon, J. P. (2016). Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 353, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, N. M. , Ying, K. , Fu, Y. , Ji, T. , Yeh, C.‐T. , Jia, Y. , Wu, W. , Richmond, T. , Kitzman, J. , Rosenbaum, H. , Iniguez, A. L. , Barbazuk, W. B. , Jeddeloh, J. A. , Nettleton, D. & Schnable, P. S. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genetics 5, e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S. , Vargas, M. M. & Roberson, R. W. (1996). Functional, organizational, and biochemical analysis of actin in hyphal tip cells of Allomyces macrogynus . Mycologia 88, 57–70. [Google Scholar]
- Sriswasdi, S. , Takashima, M. , Manabe, R.‐I. , Ohkuma, M. , Sugita, T. & Iwasaki, W. (2016). Global deceleration of gene evolution following recent genome hybridizations in fungi. Genome Research 26, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, M. & Hoffmeister, D. (2015). Fungal natural products ‐ the mushroom perspective. Frontiers in Microbiology 6, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich, J. E. , Wilke, S. K. , Ahrén, D. , Au, C. H. , Birren, B. W. , Borodovsky, M. , Burns, C. , Canbäck, B. , Casselton, L. A. , Cheng, C. K. , Deng, J. , Dietrich, F. S. , Fargo, D. C. , Farman, M. L. , Gathman, A. C. , et al. (2010). Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proceedings of the National Academy of Sciences of the United States of America 107, 11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J. T. & Konopka, A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annual Review of Microbiology 39, 321–346. [DOI] [PubMed] [Google Scholar]
- Stanhope, M. J. , Lupas, A. , Italia, M. J. , Koretke, K. K. , Volker, C. & Brown, J. R. (2001). Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature 411, 940–944. [DOI] [PubMed] [Google Scholar]
- States, J. S. (1975). Normal basidiocarp development of Gloeophyllum (Lenzites) saepiarium in culture. Mycologia 67, 1166–1175. [Google Scholar]
- Steinberg, G. (2007). Hyphal growth: a tale of motors, lipids, and the spitzenkörper. Eukaryotic Cell 6, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, G. , Harmer, N. J. , Schuster, M. & Kilaru, S. (2017a). Woronin body‐based sealing of septal pores. Fungal Genetics and Biology 109, 53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, G. , Peñalva, M. A. , Riquelme, M. , Wösten, H. A. & Harris, S. D. (2017b). Cell biology of hyphal growth In The Fungal Kingdom (eds J. Heitman, B. J. Howlett, P. W. Crous, E. H. Stukenbrock, T. Y. James, and N. A. R. Gow.) pp. 231–265. American Society of Microbiology, Washington DC. [DOI] [PubMed] [Google Scholar]
- Steinberg, G. , Schuster, M. , Hacker, C. , Kilaru, S. & Correia, A. (2017c). ATP prevents Woronin bodies from sealing septal pores in unwounded cells of the fungus Zymoseptoria tritici . Cellular Microbiology 19, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker‐Wörgötter, E. (2008). Metabolic diversity of lichen‐forming ascomycetous fungi: culturing, polyketide and shikimate metabolite production, and PKS genes. Natural Product Reports 25, 188–200. [DOI] [PubMed] [Google Scholar]
- Strack, D. , Fester, T. , Hause, B. , Schliemann, W. & Walter, M. H. (2003). Arbuscular mycorrhiza: biological, chemical, and molecular aspects. Journal of Chemical Ecology 29, 1955–1979. [DOI] [PubMed] [Google Scholar]
- Strom, N. B. & Bushley, K. E. (2016). Two genomes are better than one: history, genetics, and biotechnological applications of fungal heterokaryons. Fungal Biology and Biotechnology 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope, P. K. , Skelly, D. A. , Kozmin, S. G. , Mahadevan, G. , Stone, E. A. , Magwene, P. M. , Dietrich, F. S. & McCusker, J. H. (2015). The 100‐genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Research 25, 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E. H. (2016). The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology 106, 104–112. [DOI] [PubMed] [Google Scholar]
- Stukenbrock, E. H. , Bataillon, T. , Dutheil, J. Y. , Hansen, T. T. , Li, R. , Zala, M. , Mcdonald, B. A. , Wang, J. & Schierup, M. H. (2011). The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Research 21, 2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E. H. , Christiansen, F. B. , Hansen, T. T. , Dutheil, J. Y. & Schierup, M. H. (2012). Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proceedings of the National Academy of Sciences of the United States of America 109, 10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. , Billmyre, R. B. , Mieczkowski, P. a. & Heitman, J. (2014). Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans . PLoS Genetics 10, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K. , Moriguchi, K. & Yamamoto, S. (2015). Horizontal DNA transfer from bacteria to eukaryotes and a lesson from experimental transfers. Research in Microbiology 166, 753–763. [DOI] [PubMed] [Google Scholar]
- Swilaiman, S. S. , O'Gorman, C. M. , Balajee, S. A. & Dyer, P. S. (2013). Discovery of a sexual cycle in Aspergillus lentulus, a close relative of A. fumigatus. Eukaryotic Cell 12, 962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syme, R. A. , Tan, K.‐C. , Rybak, K. , Friesen, T. L. , McDonald, B. A. , Oliver, R. P. & Hane, J. K. (2018). Pan‐Parastagonospora comparative genome analysis – effector prediction and genome evolution. Genome Biology and Evolution 10, 2443–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllősi, G. J. , Davín, A. A. , Tannier, E. , Daubin, V. & Boussau, B. (2015). Genome‐scale phylogenetic analysis finds extensive gene transfer among fungi. Philosophical Transactions of the Royal Society, B: Biological Sciences 370, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, N. (2016). Coordinated process of polarized growth in filamentous fungi. Bioscience, Biotechnology, and Biochemistry 80, 1693–1699. [DOI] [PubMed] [Google Scholar]
- Tanabe, Y. , Watanabe, M. M. & Sugiyama, J. (2005). Evolutionary relationships among basal fungi (Chytridiomycota and Zygomycota): insights from molecular phylogenetics. Journal of General and Applied Microbiology 51, 267–276. [DOI] [PubMed] [Google Scholar]
- Tang, N. , San Clemente, H. , Roy, S. , Bécard, G. , Zhao, B. & Roux, C. (2016). A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Frontiers in Microbiology 7, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo, L. , Bahram, M. , Puusepp, R. , Nilsson, R. H. & James, T. Y. (2017). Novel soil‐inhabiting clades fill gaps in the fungal tree of life. Microbiome 45, 5–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelaar, M. & Wösten, H. A. B. (2017). Functional distinction of hyphal compartments. Scientific Reports 7, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert, I. , Lutomski, M. , Märker, R. , Nowrousian, M. & Kück, U. (2017). New insights from an old mutant: SPADIX4 governs fruiting body development but not hyphal fusion in Sordaria macrospora . Molecular Genetics and Genomics 292, 93–104. [DOI] [PubMed] [Google Scholar]
- Teixeira, M. M. , Moreno, L. F. , Stielow, B. J. , Muszewska, A. , Hainaut, M. , Gonzaga, L. , Abouelleil, A. , Patané, J. S. L. , Priest, M. , Souza, R. , Young, S. , Ferreira, K. S. , Zeng, Q. , da Cunha, M. M. L. , Gladki, A. , et al. (2017). Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Studies in Mycology 86, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetz, V. V. (2005). The pangenome concept: a unifying view of genetic information. Medical Science Monitor 11, 24–29. [PubMed] [Google Scholar]
- Tlalka, M. , Bebber, D. P. , Darrah, P. R. , Watkinson, S. C. & Fricker, M. D. (2007). Emergence of self‐organised oscillatory domains in fungal mycelia. Fungal Genetics and Biology 44, 1085–1095. [DOI] [PubMed] [Google Scholar]
- Todd, R. B. , Zhou, M. , Ohm, R. A. , Leeggangers, H. A. C. F. , Visser, L. & de Vries, R. P. (2014). Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics 15, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, R. T. , Forche, A. & Selmecki, A. (2017). Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution In The Fungal Kingdom (Volume 5) (eds J. Heitman, B. J. Howlett, P. W. Crous, E. H. Stukenbrock, T. Y. James, and N. A. R. Gow.), pp. 599–618. American Society of microbiology, Washington DC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo‐Hernández, C. , Gulis, V. , Ruiz‐Diaz, C. P. , Sabat, A. M. & Bayman, P. (2013). When aspergillosis hits the fan: disease transmission and fungal biomass in diseased versus healthy sea fans (Gorgonia ventalina). Fungal Ecology 6, 161–167. [Google Scholar]
- Torres, E. M. , Williams, B. R. & Amon, A. (2008). Aneuploidy: cells losing their balance. Genetics 179, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trail, F. , Wang, Z. , Stefanko, K. , Cubba, C. & Townsend, J. P. (2017). The ancestral levels of transcription and the evolution of sexual phenotypes in filamentous fungi. PLoS Genetics 13, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter, E. D. , Johnson, E. M. , Benny, G. L. , Lichtwardt, R. W. , Wang, Y. , Kandel, P. , Novak, S. J. , Smith, J. F. & White, M. M. (2014). An eight‐gene molecular phylogeny of the Kickxellomycotina, including the first phylogenetic placement of Asellariales. Mycologia 106, 912–935. [DOI] [PubMed] [Google Scholar]
- Tretter, E. D. , Johnson, E. M. , Wang, Y. , Kandel, P. & White, M. M. (2013). Examining new phylogenetic markers to uncover the evolutionary history of early‐diverging fungi: comparing MCM7, TSR1 and rRNA genes for single‐ and multi‐gene analyses of the Kickxellomycotina. Persoonia 30, 106–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis, D. I. & Keller, N. P. (2006). Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans . Molecular Microbiology 59, 882–892. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis, D. I. & Keller, N. P. (2007). Oxylipins as developmental and host–fungal communication signals. Trends in Microbiology 15, 109–118. [DOI] [PubMed] [Google Scholar]
- Valero‐Jiménez, C. A. , Faino, L. , Spring in't Veld, D. , Smit, S. , Zwaan, B. J. & van Kan, J. A. L. (2016). Comparative genomics of Beauveria bassiana: uncovering signatures of virulence against mosquitoes. BMC Genomics 17, 1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Does, H. C. & Rep, M. (2012). Horizontal transfer of supernumerary chromosomes in fungi. Methods in Molecular Biology 835, 427–437. [DOI] [PubMed] [Google Scholar]
- Van der Nest, M. A. , Olson, Å. , Lind, M. , Vélëz, H. , Dalman, K. , Durling, M. B. , Karlsson, M. & Stenlid, J. (2014). Distribution and evolution of het gene homologs in the basidiomycota. Fungal Genetics and Biology 64, 45–57. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y. (2004). Tetraodon genome confirms Takifugu findings: most fish are ancient polyploids. Genome Biology 5, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y. , Taylor, J. S. & Meyer, A. (2003). Are all fishes ancient polyploids? Journal of Structural and Functional Genomics 3, 65–73. [PubMed] [Google Scholar]
- van Driel, K. G. A. , Humbel, B. M. , Verkleij, A. J. , Stalpers, J. , Müller, W. H. & Boekhout, T. (2009). Septal pore complex morphology in the Agaricomycotina (Basidiomycota) with emphasis on the Cantharellales and Hymenochaetales. Mycological Research 113, 559–576. [DOI] [PubMed] [Google Scholar]
- Vargas, M. M. , Aronson, J. M. & Roberson, R. W. (1993). The cytoplasmic organization of hyphal tip cells in the fungus Allomyces macrogynus . Protoplasma 176, 43–52. [Google Scholar]
- Vlaardingerbroek, I. , Beerens, B. , Rose, L. , Fokkens, L. , Cornelissen, B. J. C. & Rep, M. (2016). Exchange of core chromosomes and horizontal transfer of lineage‐specific chromosomes in Fusarium oxysporum . Environmental Microbiology 18, 3702–3713. [DOI] [PubMed] [Google Scholar]
- Vodeneev, V. , Akinchits, E. & Sukhov, V. (2015). Variation potential in higher plants: mechanisms of generation and propagation. Plant Signaling & Behavior 10, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz, P. A. & Niederpruem, D. J. (1969). Dikaryotic fruiting in Schizophyllum commune fr.: morphology of the developing basidiocarp. Archiv für Mikrobiologie 68, 246–258. [DOI] [PubMed] [Google Scholar]
- Walther, A. , Hesselbart, A. & Wendland, J. (2014). Genome sequence of Saccharomyces carlsbergensis, the world's first pure culture lager yeast. G3 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. M. , Begerow, D. , Groenewald, M. , Liu, X. Z. , Theelen, B. , Bai, F. Y. & Boekhout, T. (2015a). Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology 81, 55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q.‐M. , Yurkov, A. M. , Göker, M. , Lumbsch, H. T. , Leavitt, S. D. , Groenewald, M. , Theelen, B. , Liu, X.‐Z. , Boekhout, T. & Bai, F.‐Y. (2015b). Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Studies in Mycology 81, 149–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Wang, J. , Li, N. , Li, J. , Trail, F. , Dunlap, J. C. & Townsend, J. P. (2018). Light sensing by opsins and fungal ecology: NOP‐1 modulates entry into sexual reproduction in response to environmental cues. Molecular Ecology 27, 216–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmink, J. A. & van Elsas, J. D. (2008). Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved? The ISME Journal 2, 887–900. [DOI] [PubMed] [Google Scholar]
- Wasternack, C. & Feussner, I. (2018). The oxylipin pathways: biochemistry and function. Annual Review of Plant Biology 69, 363–386. [DOI] [PubMed] [Google Scholar]
- Weber, T. & Marahiel, M. A. (2001). Exploring the domain structure of modular nonribosomal peptide synthetases. Structure 9, 3–9. [DOI] [PubMed] [Google Scholar]
- Weiss, M. , Bauer, R. , Sampaio, J. P. & Oberwinkler, F. (2014). Tremellomycetes and related groups In The Mycota VII: Systematics and Evolution (ed K. Esser.), pp. 331–355. Springer, Berlin Heidelberg. [Google Scholar]
- Whelan, W. L. , Markie, D. M. , Simpkin, K. G. & Poulter, R. M. (1985). Instability of Candida albicans . Journal of Bacteriology 161, 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann, P. , Albermann, S. , Niehaus, E.‐M. , Studt, L. , von Bargen, K. W. , Brock, N. L. , Humpf, H.‐U. , Dickschat, J. S. & Tudzynski, B. (2012). The Sfp‐type 4'‐phosphopantetheinyl transferase Ppt1 of Fusarium fujikuroi controls development, secondary metabolism and pathogenicity. PLoS One 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.‐J. , Britt, R. & Dickman, M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathogens 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. A. , Calvo, A. M. , Chang, P.‐K. & Keller, N. P. (2004). Characterization of the Aspergillus parasiticus delta12‐desaturase gene: a role for lipid metabolism in the Aspergillus‐seed interaction. Microbiology 150, 2881–2888. [DOI] [PubMed] [Google Scholar]
- Wisecaver, J. H. , Alexander, W. G. , King, S. B. , Hittinger, C. T. , Rokas, A. , Barlow, M. , Todd Hittinger, C. & Rokas, A. (2016). Dynamic evolution of nitric oxide detoxifying flavohemoglobins, a family of single‐protein metabolic modules in bacteria and eukaryotes. Molecular Biology and Evolution 33, 1979–1987. [DOI] [PubMed] [Google Scholar]
- Wisecaver, J. H. , Borowsky, A. T. , Tzin, V. , Jander, G. , Kliebenstein, D. J. & Rokas, A. (2017). A global coexpression network approach for connecting genes to specialized metabolic pathways in plants. Plant Cell 29, 944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver, J. H. & Hackett, J. D. (2014). The impact of automated filtering of BLAST‐determined homologs in the phylogenetic detection of horizontal gene transfer from a transcriptome assembly. Molecular Phylogenetics and Evolution 71, 184–192. [DOI] [PubMed] [Google Scholar]
- Wisecaver, J. H. & Rokas, A. (2015). Fungal metabolic gene clusters: caravans traveling across genomes and environments. Frontiers in Microbiology 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver, J. H. , Slot, J. C. & Rokas, A. (2014). The evolution of fungal metabolic pathways. PLoS Genetics 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H. & Shields, D. C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- Wortel, M. T. , Bosdriesz, E. , Teusink, B. & Bruggeman, F. J. (2016). Evolutionary pressures on microbial metabolic strategies in the chemostat. Scientific Reports 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrońska, A. K. , Boguś, M. I. , Kaczmarek, A. & Kazek, M. (2018). Harman and norharman, metabolites of entomopathogenic fungus Conidiobolus coronatus (Entomopthorales), disorganize development of Galleria mellonella (Lepidoptera) and affect serotonin‐regulating enzymes. PLoS One 13, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Gao, B. & Zhu, S. (2014). The fungal defensin family enlarged. Pharmaceuticals (Basel, Switzerland) 7, 866–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , Cheng, J. , Fu, Y. , Chen, T. , Jiang, D. , Ghabrial, S. A. & Xie, J. (2017). Virus‐mediated suppression of host non‐self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathogens 13, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss, T. , Masclaux, F. G. , Rosikiewicz, P. , Pagni, M. & Sanders, I. R. (2016). Population genomics reveals that within‐fungus polymorphism is common and maintained in populations of the mycorrhizal fungus Rhizophagus irregularis . The ISME Journal 10, 2514–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W.‐B. , Amaike, S. , Wohlbach, D. J. , Gasch, A. P. , Chiang, Y.‐M. , Wang, C. C. C. , Bok, J. W. , Rohlfs, M. & Keller, N. P. (2012). An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Molecular Microbiology 83, 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W. & Keller, N. P. (2011). Transcriptional regulatory elements in fungal secondary metabolism. Journal of Microbiology 49, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z. , Zhu, B. , Feng, H. & Huang, L. (2016). Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali . Scientific Reports 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharoff, D. A. , Rogers, C. J. , Hance, K. W. , Schlom, J. & Greiner, J. W. (2007). Chitosan solution enhances both humoral and cell‐mediated immune responses to subcutaneous vaccination. Vaccine 25, 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak, J. C. & Visser, S. (1996). An appraisal of soil fungal biodiversity: the crossroads between taxonomic and functional biodiversity. Biodiversity and Conservation 5, 169–183. [Google Scholar]
- Zeyl, C. (2009). The role of sex in fungal evolution. Current Opinion in Microbiology 12, 592–598. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Pettway, R. E. & McDonald, B. A. (2003). The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genetics and Biology 38, 286–297. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Chen, X. , Xu, C. , Zhao, H. , Zhang, X. , Zeng, G. , Qian, Y. , Liu, R. , Guo, N. , Mi, W. , Meng, Y. , St. Leger, R. J. & Fang, W. (2019). Horizontal gene transfer allowed the emergence of broad host range entomopathogens. Proceedings of the National Academy of Sciences of the United States of America 116, 7982–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.‐X. , Spiering, M. J. , Dawe, A. L. & Nuss, D. L. (2014). Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 197, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, J.‐J. & Xiao, J.‐H. (2009). Secondary metabolites from higher fungi: discovery, bioactivity, and bioproduction In Biotechnology in China I (eds J. Zhong, B. Feng‐Wu, and Z. Wei.), pp. 79–150. Springer, Berlin Heidelberg. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Xu, J. , Sun, C. , Zhou, S. , Xu, H. , Nelson, D. R. , Qian, J. , Song, J. , Luo, H. , Xiang, L. , Li, Y. , Xu, Z. , Ji, A. , Wang, L. , Lu, S. , et al. (2015). Chromosome‐level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense . Scientific Reports 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. O. , Sherlock, G. & Petrov, D. A. (2016). Whole genome analysis of 132 clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. G3 6, 2421–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Živanović, B. D. (2005). Ca2+ and H+ ion fluxes near the surface of gravitropically stimulated Phycomyces sporangiophore. Annals of the New York Academy of Sciences 1048, 487–490. [DOI] [PubMed] [Google Scholar]
- Živanović, B. D. (2012). Surface tip‐to‐base Ca2+ and H+ ionic fluxes are involved in apical growth and graviperception of the Phycomyces stage I sporangiophore. Planta 236, 1817–1829. [DOI] [PubMed] [Google Scholar]
- Živanović, B. D. (2013). Intracellular reorganization and ionic signaling of the Phycomyces stage I sporangiophore in response to gravity and touch. Communicative & Integrative Biology 6, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]


