Abstract
Enhancing the resilience of corals to rising temperatures is now a matter of urgency, leading to growing efforts to explore the use of heat tolerant symbiont species to improve their thermal resilience. The notion that adaptive traits can be retained by transferring the symbionts alone, however, challenges the holobiont concept, a fundamental paradigm in coral research. Holobiont traits are products of a specific community (holobiont) and all its co‐evolutionary and local adaptations, which might limit the retention or transference of holobiont traits by exchanging only one partner. Here we evaluate how interchanging partners affect the short‐ and long‐term performance of holobionts under heat stress using clonal lineages of the cnidarian model system Aiptasia (host and Symbiodiniaceae strains) originating from distinct thermal environments. Our results show that holobionts from more thermally variable environments have higher plasticity to heat stress, but this resilience could not be transferred to other host genotypes through the exchange of symbionts. Importantly, our findings highlight the role of the host in determining holobiont productivity in response to thermal stress and indicate that local adaptations of holobionts will likely limit the efficacy of interchanging unfamiliar compartments to enhance thermal tolerance.
Keywords: adaptation, climate change, coral reefs, Exaiptasia pallida, heat stress, photosynthesis, respiration, Symbiodiniaceae, SymPortal
The cnidarian holobiont is a distinct, interactive biological entity comprised of three main components: the animal host, algal symbionts of the family Symbiodiniaceae and an extensive microbiota. Holobiont traits, such as thermal resilience, are thus a product of a specific species assemblage and therefore not necessarily retained when only one partner is exchanged (holobionts iii and iv). Consequently, non‐native associations tend to perform at a suboptimal level (here shown as puzzle pieces that do not fit together).
1. INTRODUCTION
The interaction between animals and dinoflagellates of the family Symbiodiniaceae is one of the most abundant, widespread, and ecologically successful symbioses found in nature (Kirk & Weis, 2016). Numerous taxa, ranging from protists and sponges to cnidarians, flatworms, and mollusks (Kirk & Weis, 2016; Decelle et al., 2018) benefit from this symbiotic association. For shallow‐water corals—the foundation species of coral reefs—this relationship can be highly obligate such that its breakdown (i.e., bleaching) often results in death (Hoegh‐Guldberg, 1999). Thus, the maintenance of this partnership is vital for the continual growth and survival of entire ecosystems.
Warming sea surface temperatures, as a consequence of climate change, are causing more frequent and severe mass bleaching events worldwide that precipitate the global decline of coral reefs (Hoegh‐Guldberg, Poloczanska, Skirving, & Dove, 2017). Predictions reveal that current trends of global warming will result in a further loss of up to 90% compared to today (Masson‐Delmotte et al., 2018; van Hooidonk et al., 2016). In light of this, efforts to enhance corals’ resilience have accelerated (van Oppen et al., 2017; van Oppen, Oliver, Putnam, & Gates, 2015), including microbiome engineering (Damjanovic, Blackall, Webster, & van Oppen, 2017; Epstein, Smith, Torda, & van Oppen, 2019; Rosado et al., 2019). Indeed, there is ample evidence for the importance of microbes in the adaptive responses of corals to changes in the environment. Composition of their Symbiodiniaceae communities may vary before, during, and after stress exposure (i.e., symbiont shuffling and/or switching; Baker, 2003; Boulotte et al., 2016).
Certain symbiont species have been shown to increase heat tolerance of corals by up to 1.5°C (Berkelmans & van Oppen, 2006). Thus, it has been proposed that interchanging symbionts (and also hosts) with more resistant types might be a promising way to increase the thermal resilience of cnidarian holobionts (Chakravarti, Beltran, & van Oppen, 2017; Coles & Riegl, 2013; Cunning, Silverstein, & Baker, 2018; McIlroy et al., 2016; Morikawa & Palumbi, 2019; Palumbi, Barshis, Traylor‐Knowles, & Bay, 2014; Thomas et al., 2018; Figure 1). These approaches, however, conflict with the growing body of work that points toward the emergent properties of a holobiont system being “larger than the sum of its parts” (Bordenstein & Theis, 2015; Dittami et al., 2019; Rosenberg & Zilber‐Rosenberg, 2018). The host with its microbiota (Figure 1)—the holobiont—is without doubt a distinct, interactive biological entity, and ultimately can be, in many instances, a unit of selection (Rosenberg, Sharon, Atad, & Zilber‐Rosenberg, 2010; Rosenberg, Sharon, & Zilber‐Rosenberg, 2009; Rosenberg & Zilber‐Rosenberg, 2018; but see Douglas & Werren, 2016; Moran & Sloan, 2015). As holobionts constitute functional aggregates that are “interactors,” “reproducers,” and “manifestors of adaptation” (Roughgarden, Gilbert, Rosenberg, Zilber‐Rosenberg, & Lloyd, 2018), studying their closely intertwined ecology and evolution is uniquely challenging, especially in the context of environmental change. Present‐day corals have already, and will continue to experience, rapid changes in their thermal environment and must undergo short‐term adaptation, such as changes in their microbial communities toward more resilient types, to ensure their persistence. Novel (more resistant) symbioses are temporally limited and rarely persist over ecological timescales (LaJeunesse, Smith, Finney, & Oxenford, 2009; Thornhill, LaJeunesse, Kemp, Fitt, & Schmidt, 2006), thus highlighting the complex nature (metabolically, immunologically, developmental, etc.) of symbiotic associations. The viability of transferring more resilient symbiont types as a means of improving holobiont resilience is likely contingent on the long‐term evolution of the holobiont compartments (Figure 1) and yet, the level of importance given to this consideration is debatable.
FIGURE 1.
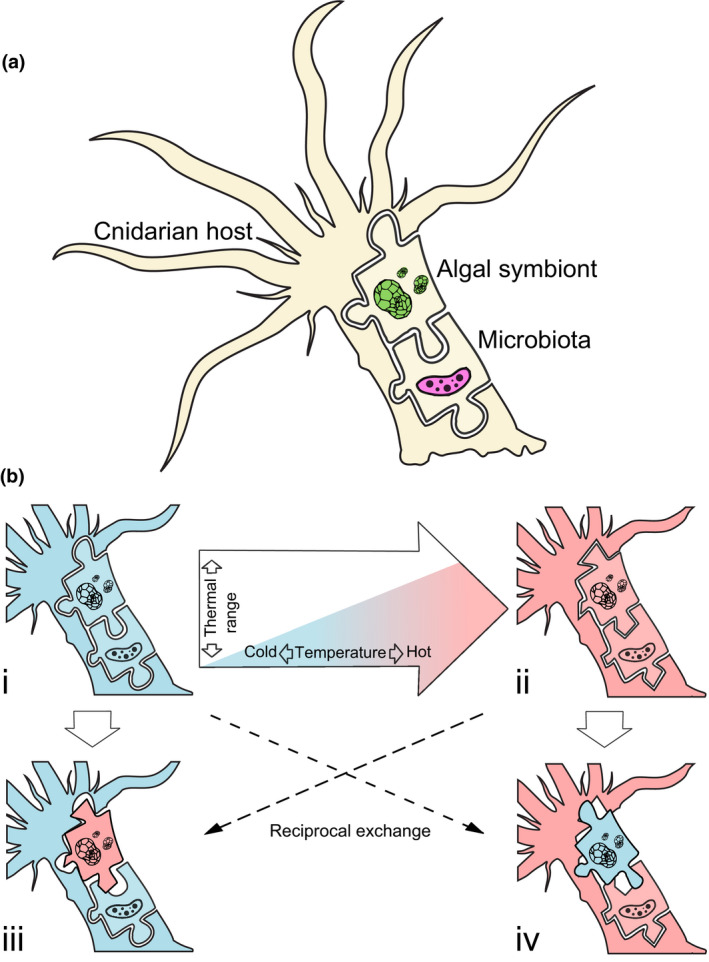
(a) The cnidarian holobiont is a distinct, interactive biological entity comprised by three main components: the animal host, algal symbionts of the family Symbiodiniaceae, and an extensive microbiota including bacteria, viruses, archaea, fungi, endolithic algae, and protists. (b) Holobiont traits (i.e., thermal resilience of holobionts i and ii) are not necessarily retained when translocating only one partner (holobionts iii and iv). Instead, non‐native associations tend to perform at a suboptimal level (here shown as puzzle pieces that do not fit together)
However, we still do not understand to what extent holobiont traits are products of co‐evolutionary processes and local adaptions that also underlie host–symbiont fidelity and performance, making them specific to a given host–symbiont combination and environment. Most studies focusing on understanding the cellular and physiological drivers that limit the capacity for novel symbioses to arise have, however, used foreign, non‐native photosymbiont species (Gabay, Parkinson, Wilkinson, Weis, & Davy, 2019; Gabay, Weis, & Davy, 2018; Goulet, Cook, & Goulet, 2005; Starzak, Quinnell, Nitschke, & Davy, 2014; Ye, Bhattacharjee, & Siemann, 2019). Here we built on these studies by using symbiont strains that are naturally found in the same host, and from different climatic regions of the host's distribution range. This approach allows us to test if it is possible to retain adaptations underlying host–symbiont specificity while increasing thermal resilience of hosts from colder regions. Furthermore, by testing the thermal performance of the different host–symbiont combinations, we can ascertain to what extent the partners contribute to thermal resilience and, therefore, their role in determining holobiont performance.
We examined the holobiont concept and the underlying complexity of host–microbe associations through time by actively manipulating the symbiotic communities of the sea anemone Exaiptasia pallida (Grajales & Rodríguez, 2014; hereafter referred to as “Aiptasia”). Aiptasia has been a model system for coral research for over 30 years (Baumgarten et al., 2015; Weis, Davy, Hoegh‐Guldberg, Rodriguez‐Lanetty, & Pringle, 2008) as it allows overcoming many of the limitations associated with corals. Aiptasia is a globally distributed species that can be bleached and colonized by different Symbiodiniaceae strains, facilitating the disentanglement of host and symbiont type effects (Gabay et al., 2018, 2019; Hambleton, Guse, & Pringle, 2014; Rädecker et al., 2018; Starzak et al., 2014; Wolfowicz et al., 2016). We used clonal lines from three different geographic locations with distinct temperature regimes: H2 from Hawaii (Xiang, Hambleton, DeNofrio, Pringle, & Grossman, 2013), CC7 from North Carolina (Sunagawa et al., 2009), and RS from the central Red Sea (Cziesielski et al., 2018). These lineages have almost certainly acquired specific genotypic adaptations to their local environment and differ in their natively associated symbionts (Breviolum minutum: Xiang et al., 2013; Symbiodinium linucheae: Bieri, Onishi, Xiang, Grossman, & Pringle, 2016; Symbiodinium microadriaticum: Cziesielski et al., 2018), thus resulting in different responses of both partners (Cziesielski et al., 2018). Red Sea Aiptasia may provide critical insight into mechanisms promoting the acclimatory capacity of symbiotic cnidarians due to their high thermal tolerance and ability to thrive in one of the warmest (as high as 35°C during summer; Ngugi, Antunes, Brune, & Stingl, 2012) seas on Earth (Berumen et al., 2019). Based on the premise that host and symbiont species from the Red Sea are more heat tolerant, we studied their contribution to the thermal resilience of Aiptasia holobionts that originate from lower temperature environments. Furthermore, we tested whether specific adaptations of the different Aiptasia‐Symbiodiniaceae combinations might limit the exchangeability of partners in novel symbioses. For this, we inoculated symbiont‐free polyps of all host lines with each pairwise combination of symbiont taxa and examined their functional variability under short‐ and long‐term thermal stress.
2. MATERIALS AND METHODS
2.1. Experimental set‐up: Aiptasia rearing, bleaching, and inoculations
Aiptasia from three different clonal laboratory strains were used in this study: H2 from Hawaii, CC7 from North Carolina and a Red Sea (RS) line obtained from the central coast of Saudi Arabia. H2’s homologous symbionts are B. minutum (Xiang et al., 2013; referred to as SSB01), whereas CC7 naturally associates with S. linucheae (Bieri et al., 2016; referred to as SSA01) but can also form stable symbioses with other species (Thornhill, Xiang, Pettay, Zhong, & Santos, 2013). Red Sea Aiptasia, on the other hand, occurs with both S. microadriaticum and Breviolum taxa (Cziesielski et al., 2018; Thornhill et al., 2013; here referred to as RS‐A and RS‐B, respectively; see Table 1). It is worth clarifying that all the symbiont strains we tested here are native to Aiptasia, yet, they are specific to each genotype so that homologous symbionts are those that regularly associate with a particular host and heterologous symbionts are those that typically do not associate with a given host. For example, while B. minutum is the homologous symbiont of H2‐Hawaii, it is a heterologous symbiont in CC7‐North Carolina.
TABLE 1.
Details of the Symbiodiniaceae cultures used to perform inoculations. Name of the culture followed by the host source, original geographic location, Symbiodiniaceae species, and majority ITS2 sequence as determined by SymPortal
| Symbiont culture | Aiptasia host source | Original geographic location | Symbiodiniaceae species | Majority ITS2 sequence |
|---|---|---|---|---|
| SSB01 | H2 | Hawaii, USA | Breviolum minutum | B1 |
| SSA01 | CC7 | North Carolina, USA | Symbiodinium linucheae | A4 |
| RS‐A | RS | Al Lith, Saudi Arabia (central Red Sea) | Symbiodinium microadriaticum | A1 |
| RS‐B | RS | Al Lith, Saudi Arabia (central Red Sea) | Breviolum sp. | RS‐B1 a |
SymPortal revealed different ITS2 type profiles for the Breviolum strains from Hawaii and the Red Sea, thus these were classified as B1 and RS‐B1, respectively.
Anemones were reared in autoclaved natural seawater (~39 psu and pH ~ 8) at 25°C under ~40 μmol photons m−2 s−1 white light on a 12:12 hr light:dark cycle (daytime of 06:00–18:00). These light levels, which are similar to those reported by other studies working with Aiptasia (Cui et al., 2019; Gegner et al., 2017; Lehnert et al., 2014; Röthig et al., 2016), were chosen to support optimal growth of the animals but also because all symbiont strains used here perform well under this irradiance. Individuals were fed twice per week with freshly hatched Artemia brine shrimp. All populations were kept in Percival incubators (Model I‐22LLVL, Percival Scientific) under identical conditions. Menthol‐induced bleaching (Matthews et al., 2016) was used to generate aposymbiotic individuals of each clonal line. Animals were incubated in autoclaved seawater with 0.19 mmol/L menthol during daytime, followed by a 5 μmol/L 3‐(3,4‐dichlorophenyl)‐1,1‐dimethylurea seawater incubation overnight. This treatment was repeated until complete bleaching was observed and confirmed via fluorescence microscopy. Anemones were then kept in a dark incubator for at least 2 months and further maintained for at least another month on a diurnal 12:12 hr light:dark cycle to ensure there was no re‐establishment of symbiosis.
Cultures of the strains SSA01 and SSB01 (courtesy of the John Pringle Lab) were used to perform inoculations with Symbiodinium and Breviolum taxa, respectively. Furthermore, Symbiodiniaceae from the Red Sea line were isolated. Briefly, one anemone was washed with 500 µl of f/2 medium + K/A/S (Kanamycin/Ampicillin/Streptomycin: 50/100/50 µg/ml) and then crushed in 500 µl of fresh f/2 medium + K/A/S using a glass tissue grinder (Duran Wheaton Kimble) to keep the algal cells intact. This was filtered through a 40 µm nylon mesh sterile cell strainer (Fisherbrand, Fisher Scientific), diluted in 15 ml of f/2 medium + K/A/S and subsequently transferred to liquid culture flasks (225 cm2 Nunc Cell Culture Treated EasYFlask, Thermo Scientific) with 5 ml of the diluted fresh extract and 150 ml of f/2 medium + K/A/S + GeO2 (4.47 µg/ml) saturated solution was added to prevent diatom and bacteria growth. Presence of algal cells was checked under a fluorescence microscope (Leica DMI3000 B inverted phase contrast microscope, Leica Microsystems GmbH). Moreover, solid agar f/2 + K/A/S plates were inoculated with the same fresh extract (normal concentration, 5‐fold and 25‐fold dilution) so that single colonies could be grown and isolated for further experiments. After 10 days, 15–20 single colonies were picked from each plate, grown in 1.5 ml Eppendorf tubes with 300 µl of f/2 medium + K/A/S and transferred to culture flasks (75 cm2 Nunc Cell Culture Treated EasYFlask, Thermo Scientific), as previously described. The K/A/S treatment was performed for every 10 subcultures to clear the bacterial load, and verified by growth on marine broth and inspected under the microscope. The above was performed under a flow hood (NuAire) to avoid environmental contamination. All liquid and solid cultures were kept in an incubator (Model I‐22LLVL, Percival Scientific) at 29°C on a 12:12 hr light:dark cycle (80–100 µmol photons m−2 s−1 of photosynthetically active radiation).
Inoculations were performed so that six heterologous host–symbiont combinations were obtained: H2‐Hawaii + SSA01, H2‐Hawaii + RS, CC7‐North Carolina + SSB01, CC7‐North Carolina + RS, RS‐Red Sea + SSA01, and RS‐Red Sea + SSB01. Algal strains were grown in cultures for more than 1 year before conducting experiments. Cell density of each culture was assessed 1 day prior by flow cytometry (BD LSRFortessa, BD Biosciences) to accurately calculate the desired amount of 105 cells/ml to perform the inoculations. In a flow hood, each culture flask was mixed well, and an aliquot was transferred to a 50 ml Falcon tube. The liquid was centrifuged for 5 min at 3,000 rcf, f/2 medium was removed and cells were resuspended in 30 ml of autoclaved seawater. The cell suspension was then poured in each 250 ml tank containing the aposymbiotic anemones, followed by immediate feeding with Artemia to facilitate the dinoflagellate uptake. Inoculations were verified by fluorescence microscopy. Individual anemones were inspected every day for the first 2 weeks until color pigmentation was visible to the naked eye. Colonization patterns (i.e., cell densities) in Aiptasia are generally stable after 12 weeks (Matthews et al., 2016); yet, inoculated animals were maintained (under the same conditions as described above) for at least 6 months before performing the following experiments.
2.2. Physiological performance under acute thermal stress
Fifteen individuals from each of the nine different Aiptasia host–symbiont combinations were subjected to acute heat stress as follows. Temperature was slowly ramped up starting at 25°C to 32.5°C with increments of 2.5°C every 1.5 hr, starting from 08:00 and reaching the target temperature by the afternoon (14:00). Anemones were transferred into 5 ml custom glass chambers fitted with an internal stir bar (chamber design by Dr. Julia Strahl, University of Oldenburg) and a FireSting O2 fiber‐optical oxygen sensor (PyroScience). Chambers were filled with autoclaved seawater and submerged in a 25°C water bath and maintained in darkness for 30 min to acclimate with the magnetic stirrers switched on to prevent stratification of the water column. O2 fluxes (μmol/L) were then recorded once every 15 s over the course of a 30 min incubation period in light (~40 μmol photons m−2 s−1), followed by 10 min acclimation in darkness (<1 μmol photons m−2 s−1) and then a 30 min incubation in dark. Temperature was increased by heating up the water tub and constantly monitoring it with a temperature probe. A 10 min acclimation time was allowed between each temperature increment to ensure the chamber had reached the target temperature. The above was repeated until a final temperature of 32.5°C was reached. Individuals were tested in different batches across several days so natural biological variation could be taken into account.
2.3. Gross photosynthesis, respiration, and P:R ratios
Net photosynthesis (Pnet) and respiration (R) rates were calculated from the slope of the linear increase and decrease in dissolved oxygen concentration during light and dark incubations, respectively, with each temperature increment. For every respirometry assay performed, values of Pnet and R were corrected for background microbial oxygen consumption (i.e., seawater controls) and transformed into their carbon equivalents by assuming quotients of 1.1 and 0.9 (over 24 hr), respectively (Muscatine, McCloskey, & Marian, 1981). These were then normalized to symbiont counts (see below) in order to generate values of gross photosynthesis (Pgross), expressed as pmol C cell−1 hr−1. Photosynthesis to respiration ratios (P:R) were calculated (Pgross/R, where Pgross = Pnet + |R|), as an indicator of the autotrophic capacity.
2.4. Thermal response under long‐term stress
This experiment was conducted ~6 months after performing the respirometry assays. Ten Aiptasia from each of the host–symbiont combinations above were subjected to long‐term heat stress as follows. In brief, temperature was gradually ramped up from 25°C to 32°C over the course of 8 hr at increments of 1°C per hour as previously described (Gegner et al., 2017). A subbleaching temperature of 32°C was chosen for this experiment (Aiptasia at 39 psu salinity can resist up to 34°C without bleaching according to Gegner et al., 2017) in order to investigate the thermal response (see below) among the different host–symbiont combinations. Anemones remained at this temperature for 28 days, during which daily maximum photochemical efficiency of PSII (Fv/Fm) was recorded with a Pulse Amplitude Modulated fluorometer (Mini‐PAM, Walz) to assess the photophysiological status of each individual. Polyps were dark acclimated for 30 min prior to measurements. Of note, all remaining individuals of the H2‐Hawaii + RS combination died within a few weeks following the previous experiment. Furthermore, homologous RS‐Red Sea Aiptasia was not available at the time we performed this test.
2.5. Activation energy effect sizes
The thermal dependence of the processes measured above was parameterized as the activation energy (E a), expressed in electronvolts (eV). This was calculated in an equivalent manner to an effect size per unit temperature so it could be used to compare the magnitude of responses across multiple biological traits (Marbà, Jorda, Agusti, Girard, & Duarte, 2015):
where V 0 is the value of the response variable observed previous to a thermal anomaly (V i) measured for temperatures (in Kelvin) T 0 and T i, respectively, multiplied by the Boltzmann constant (k = 8.617734 × 10–5 eV; Regaudie‐de‐Gioux & Duarte, 2012), following the Arrhenius model (Dell, Pawar, & Savage, 2011). Here, activation energies were calculated as the ratio of the response variable at 25°C (V 0) and 32.5°C (V i). E a values <0 (i.e., negative values) indicate a decrease in the trait response with increasing temperature. E a values ≈0 indicate no change in the trait response with increasing temperature, whereas E a values >0 (i.e., positive values) indicate an increase in the trait response with warming.
2.6. DNA extraction, protein content, and symbiont cell counts
Genomic DNA for Symbiodiniaceae typing was isolated from 10 ml of liquid culture that were centrifuged at 1,500 rcf for 10 min, where the supernatant was removed and the pellet was resuspended in 0.5 ml of cell lysis buffer from the DNeasy Plant Mini Kit (Qiagen) in a 2 ml screw top Eppendorf tube. The cells were then homogenized using glass beads in a TissueLyser II (Qiagen) set at 30 Hz for 30 s. The homogenate was spun down at 16,000 rcf for 10 s and the supernatant was taken for all downstream processing steps, according to the manufacturer's instruction manual for the DNeasy Plant Mini Kit. This was done for each strain, from which three technical replicates were independently DNA‐extracted. Thus, each Symbiodiniaceae strain in this study is represented by only one culture line.
For Aiptasia samples, a tentacle from each individual was plucked to extract DNA with the Chelex 100® (Bio‐Rad) resin method (Walsh, Metzger, & Higuchi, 1991). The tissue was vortexed with Chelex slurry for 20 s and briefly spun down in a picofuge. Samples were incubated for 20 min at 99°C, vortexed again for 20 s and spun down at 16,000 rcf for 2 min. Supernatant was later used as the template for PCR. Each anemone was then crushed in 500 µl of cell lysis buffer (200 mM TRIS pH 7.5, 2 M NaCl, 0.1% Triton 20%) and two aliquots of 100 and 400 µl were immediately snap‐frozen in liquid nitrogen and stored in −20°C for further protein content and symbiont concentration analysis, respectively. Total host protein content was quantified with a Micro BCA Protein Assay Kit (Thermo Scientific) using triplicates of 150 µl of 15×‐diluted tissue slurry as per manufacturer instructions. Protein concentrations were measured at 562 nm absorbance using a SpectraMax Paradigm Multi‐Mode Detection Platform (Molecular Devices). Symbiont cell counts were done by flow cytometry (BD LSRFortessa, BD Biosciences). To do this, tissue homogenate was spun down at 14,000 rcf for 5 min, supernatant was removed, and the pellet was resuspended in phosphate buffered saline solution. The latter was performed twice before filtering it through a 40 µm nylon mesh sterile cell strainer (Fisherbrand, Fisher Scientific). Cells were excited at a wavelength of 488 nm and fluorescence emission was recorded at 695/40 nm. Symbiont densities were quantified in triplicate measurements (20 μl each) based on forward‐scattered light and chlorophyll autofluorescence signals of recorded events.
2.7. ITS2 amplicon sequencing
Illumina sequencing of the Internal Transcriber Space 2 (ITS2) region was used to examine the Symbiodiniaceae community composition of all host–symbiont combinations. PCRs were performed in triplicates using the primers (Illumina adapters underlined below) SYM_VAR_5.82S2 (5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG‐GAATTGCAGAACTCCGTGAACC3′) and SYM_VAR_REV (5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG‐CGGGTTCWCTTGTYTGACTTCATGC3′; Hume et al., 2018). Each PCR reaction was run with a Qiagen Multiplex PCR Kit (Qiagen) and 10 μM primers in a final reaction volume of 15 μl. Thermal cycling conditions of 15 min at 95°C, followed by 30 cycles of 30 s at 95°C, 90 s at 56°C, and 30 s at 72°C, with a final extension step of 10 min at 72°C were used for amplification. For each sample, PCR products were run on a 1% agarose electrophoresis gel, pooled and cleaned using ExoProStar 1‐step (GE Healthcare). Indexing was then performed using the Nextera XT Index Kit (Illumina) according to the manufacturer's instructions followed by sample normalization and final library pooling. A SequalPrep Normalization Plate Kit (Invitrogen, Thermo Fisher Scientific) was used to do the normalization, avoiding the more labor‐intensive process of quantifying and aliquoting each individual sample. The final pooled library was quantified on a BioAnalyzer (Agilent Technologies) and sequenced at 7 pM with 20% phiX on the Illumina MiSeq, 2 × 300 bp end version 3 chemistry according to the manufacturer's specifications at the Bioscience Core Lab at KAUST, Saudi Arabia.
2.8. Identification of Symbiodiniaceae taxa
Sequencing data were analyzed with the SymPortal engine (Hume et al., 2019), a platform for phylogenetically resolving Symbiodiniaceae taxa using ITS2 amplicon data. SymPortal was run locally and all samples, including samples from the isolate symbiont cultures used to perform the inoculations, were analyzed together. Briefly, this software identifies sets of specific ITS2 sequences that reoccur in sufficient numbers of samples and considers them as “defining intra‐genomic variants” which in turn are then used to characterize an “ITS2 type profile” representative of putative Symbiodiniaceae taxa. Different terms have been used over time, often in an interchangeable manner, to describe taxonomic units of resolution within this group (e.g., “ITS2 type,” “ITS2 profile,” “type,” “subtype,” “clade,” “subclade”). Thus, for the purposes of this study we restricted our use to “majority ITS2 sequence” and “ITS2 type profile.” As defined by SymPortal, “majority ITS2 sequence”’ refers to the most abundant sequence(s) in each of the samples that have a “type profile,” which defines a putative taxon. For example, S. linucheae was characterized by the specific A4‐A4m ITS2 type profile, with A4 as the most abundant ITS2 sequence. Additional details on SymPortal are provided in Methods S1.
2.9. Statistical analyses
The response variables were analyzed using both analyses of variance (ANOVAs) and linear mixed models (LMMs). All data were first checked for normality and homoscedasticity using standardized residual plots and Q–Q plots and, if required, ln or ln (x + 1) transformations were applied. Symbiont cell densities were analyzed with a one‐way ANOVA using host‐symbiont combination as a fixed explanatory variable in R version 3.5.1 (R Core Team, 2018). If significant terms were detected, Tukey pairwise comparisons were conducted post hoc to determine where significant differences occurred.
Gross photosynthesis and respiration (pmol C cell−1 hr−1), and P:R ratios were analyzed using repeated‐measures LMMs in SPSS (Released 2013) as described previously (Klein et al., 2017). For each dependent variable, the fixed factors were host genotype, symbiont type (based on the majority ITS2 sequence), and temperature, which was the repeated measure. In all cases, several repeated covariance types (e.g., AR(1), AR(1) heterogeneous, CS) were investigated to assess the model‐of‐best fit by comparing numerous goodness‐of‐fit statistics (e.g., −2 restricted log likelihood, Akaike's information criterion and Bayesian information criterion). Preliminary analyses of the response variables included random factors (or blocks) to test for potential bias associated with the oxygen sensor ID fitted to each individual chamber, water tub ID in which chambers were submerged, and day on which the assay was performed. We used estimates of covariance parameters and the Wald Z test of simultaneous coefficients to assess the potential redundancy of these terms in each analysis. If any of these were revealed to significantly affect the fit of the model, they were retained to account for associated variance but were otherwise removed and the analyses rerun. Photochemical efficiency values (Fv/Fm) for each host–symbiont combination subjected to long‐term heat stress were analyzed using LMMs with day as the repeated measure. Likewise, AEs for each response variable measured were also analyzed with LMMs but without repeated covariance structure. For all LMM analyses, estimated marginal means (least‐squares means) were used to determine which means differed for the significant, highest‐order terms.
3. RESULTS
3.1. Colonization success
We identified three main putative symbiont taxa based on the majority ITS2 sequence: B1, A4, and A1 corresponding to B. minutum (SSB01), S. linucheae (SSA01), and S. microadriaticum (RS‐A), respectively (Table 1). The B1 taxon was particularly interesting as it exhibited distinct type profiles (i.e., different genotype representatives) for the Hawaii and Red Sea strains, which were designated as B1 and RS‐B1, respectively (Figure S1). In some cases, the presence of both A4 and A1 taxa also resulted in the recovery of the artefactual A4/A1 genotype (see Methods S1). Sequences belonging to other taxa (i.e., Cladocopium and Durusdinium sp.) with relative abundances below 1%, on the other hand, were classified as “others.”
Certainly, functional significance of rare and low abundant background Symbiodiniaceae taxa cannot be underestimated (Ziegler, Eguíluz, Duarte, & Voolstra, 2018). However, for the purpose of this study, we only considered individuals with majority ITS2 sequences corresponding to at least 80% of the targeted symbiont taxon. Thus, based on this, colonization success (i.e., proportion of individuals colonized by the desired symbiont) of each host–symbiont combination was examined and, on a case‐by‐case basis individuals were grouped according to the main symbiont they harbored (Table S1). It is noteworthy that the original isolate used to perform Red Sea inoculations was ~70% type A1 (Figure S1), yet the symbiont composition of homologous RS‐Red Sea Aiptasia was mainly characterized by the A4/A1 type (Figure S2; Discussion S1). Even though we only tested Symbiodiniaceae that are native to Aiptasia, we still observed limited interpartner compatibility in some cases. Specifically, holobionts with heterologous Red Sea taxa did not maintain a stable symbiosis (i.e., they died or reverted to their original symbiont composition) beyond 1 year after initial inoculations (Table S1; Figure S3).
3.2. Photosynthesis and respiration
Thermal performance varied greatly among host–symbiont combinations (Table S2). Overall, rates of gross photosynthesis differed among symbiont types, resulting in a significant main effect. However, the response of gross photosynthesis to thermal stress depended only on host identity, resulting in a significant host × temperature interaction. Surprisingly, rates of gross photosynthesis did not significantly vary between B1 and A4 symbionts but showed differences within Symbiodinium taxa (A4 and A1), and intraspecifically between B1 and RS‐B1 (Figure 2a). Overall, rates of productivity in the H2‐Hawaii holobionts were consistently higher across all temperatures, regardless of symbiont composition (Figure 2b). Respiration, on the other hand, depended on symbiont type, thus resulting in a significant symbiont × temperature interaction (Figure 2c). Interestingly, RS‐Red Sea strains exhibited lower respiration compared to others, yet, when in heterologous combinations (i.e., H2‐Hawaii and CC7‐North Carolina hosts harboring RS taxa), these had the highest respiration rates (Figure S6a). Increased respiratory rates also corresponded to the highest cell densities (Figure S5) which in turn explained the high productivity (Figure S6b; see Discussion S3). Photosynthesis to respiration ratio (P:R) responses to temperature differed according to host genotype and depended upon symbiont identity, resulting in a significant three‐way host × symbiont × temperature interaction, further highlighting the interplay between both partners (see below). Respiration rates increased much faster than photosynthesis with warming, leading to declining P:R ratios (Figure 2d) over time with accumulated stress. In agreement with our observations of gross photosynthesis, H2‐Hawaii Aiptasia also displayed the highest P:R ratios under the highest temperature of 32.5°C (Figure 2d).
FIGURE 2.
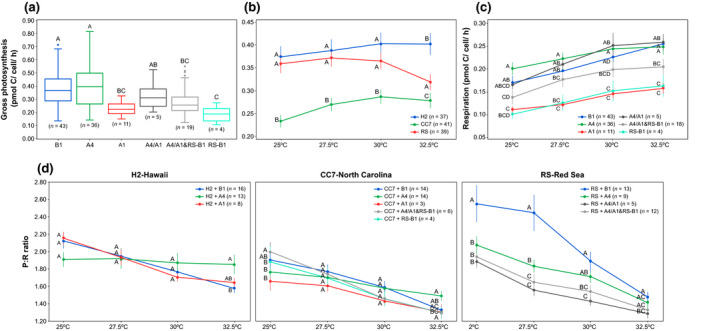
(a) Symbiont identity significantly affected gross photosynthesis rates yet (b) host genotype moderated the response to temperature. (c) Respiration rates varied with symbiont identity in response to temperature. (d) Photosynthesis to respiration (P:R) ratios across temperature increments for H2‐Hawaii, CC7‐North Carolina, and RS‐Red Sea Aiptasia harboring different Symbiodiniaceae taxa. All response variables are shown as mean values ±1 SE. Pairwise comparisons were carried out within each temperature increment. Letters next to data points indicate similarities (e.g., AA) or differences (e.g., AB) between host and/or symbiont types, as determined by estimated marginal means. Sample size (n) for each case is indicated in the legend
3.3. Photochemical efficiency (Fv/Fm)
Here, we characterized the thermal acclimation potential of the various host–symbiont combinations (noteworthy Red Sea hosts nor symbionts could be tested in this experiment; see Table S1) during long‐term heat stress by monitoring daily changes in symbiont photochemical efficiency (Fv/Fm). After 28 days at 32°C, Fv/Fm dropped from ~0.9 (on day 0 at 25°C) to ~0.6. Symbionts’ Fv/Fm depended on host and time (Table S3). CC7‐North Carolina and RS‐Red Sea genotypes exhibited higher heat tolerance than H2‐Hawaii (Figure 3a) as was also the case for A4 taxa compared to B1 (Figure 3b). Overall differences between the various host–symbiont combinations were also detected (Figure 3c), with those harboring B1 displaying lower yields. Although a significant host × symbiont × day interaction was not detected, we could nonetheless observe variability among host–symbiont combinations over time (Figure S7). We observed lower Fv/Fm for H2‐Hawaii and CC7‐North Carolina with B1 compared to the same genotypes harboring A4 taxa, which was further associated with a greater loss of symbionts (Figure S8).
FIGURE 3.
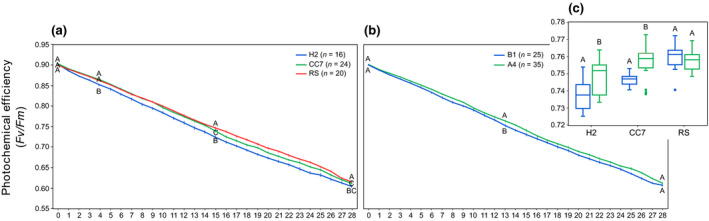
Mean (±1 SE) photochemical efficiencies of different (a) host genotypes, (b) symbiont types, and (c) host–symbiont combination. Letters indicate similarities (e.g., AA) or differences (e.g., AB), as determined by estimated marginal means. Sample sizes (n) for host genotypes (a) and symbiont types (b) are indicated in the legends
3.4. Thermal sensitivity of metabolic rates
We characterized the thermal dependence of metabolic processes from the associated activation energy (E a), calculated following the Boltzmann–Arrhenius model (Brown, Gillooly, Allen, Savage, & West, 2004; Dell et al., 2011); which is mathematically equivalent to an effect size per unit warming (Marbà et al., 2015), thereby facilitating comparisons of the magnitude of response across biological traits (Figure S9). E a estimates reported here were calculated in a manner where a value of zero indicates no (or limited) thermal dependence of the response, whereas greater deviations from zero (positive or negative) dictate increasing thermal dependence (Brown et al., 2004; Dell et al., 2011; see Figure 4a). Models comparing E a of photosynthesis, respiration, and P:R ratios revealed significant host × symbiont interactions in all cases (see Table S4). Notably, H2‐Hawaii holobionts differed from the rest as they exhibited smaller E a (Figure S9a). E a associated with photoinhibition of PS II depended on host genotype and not symbiont taxa (Table S4; Figure S9b), and followed the order H2‐Hawaii > CC7‐North Carolina > RS‐Red Sea. Overall, the magnitude of the response to temperature of the holobiont appeared to be strongly related with its location of origin. As H2‐Hawaii originates from a lower temperature and less variable environment, mean E a for P:R ratios was much lower relative to CC7‐North Carolina and RS‐Red Sea Aiptasia, where local environment is warmer and more variable (Figure 4b,c). In turn, this also corresponded with the degree of deterioration (Fv/Fm) suffered (Figure 4d). Surprisingly, we observed the opposite pattern for symbionts (Figure 4e). A4 taxa (S. linucheae, homologous to CC7‐North Carolina) seemed to be more affected by temperature than B1 from Hawaii despite originating from significantly more fluctuating thermal settings. Heat sensitivity (i.e., Fv/Fm) of B1 was not different from A4 either (Figure 4f).
FIGURE 4.
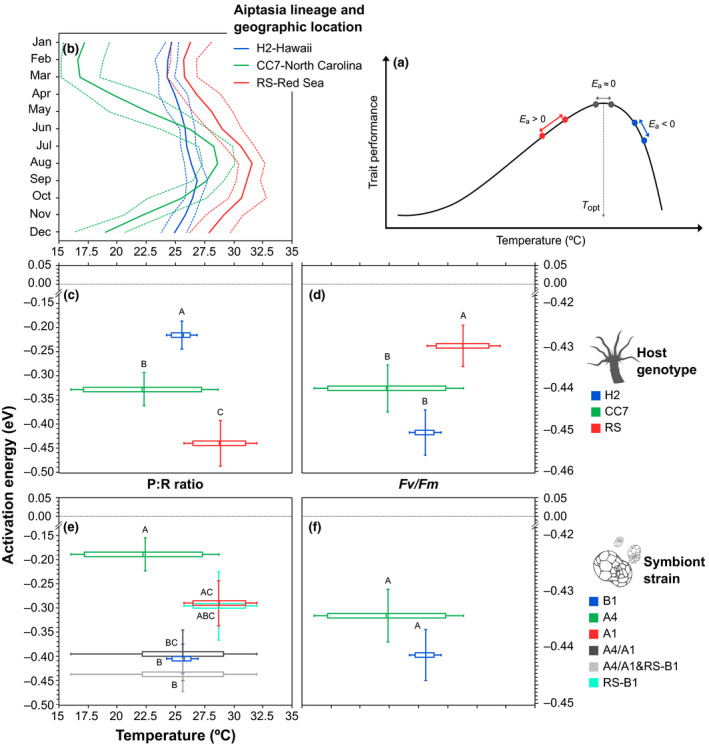
(a) Thermal response of physiological and metabolic processes has at least two different regimes: one above their optimal performance temperature (T opt), rise, and one below their optimum, fall. The Boltzmann–Arrhenius model involves calculating activation energies (E a) from the temperature dependence of reaction rates. Here E a < 0 (i.e., negative values) indicate a decrease in the trait response with increasing temperature, E a ≈ 0 indicate no change in the trait response with increasing temperature and E a > 0 (i.e., positive values) indicate an increase in the trait response with warming. The range of the rise regime is where organisms normally operate (highlighted by the green area), whereas the fall component (blue) is usually steeper and typically indicates biological collapse. (b) Thermal regimes of geographical locations of origin for the three Aiptasia lineages used here: Hawaii (Kāne'ohe Bay), North Carolina (Wilmington), and Red Sea (Al Lith, Saudi Arabia). Solid line denotes average, and dashed lines represent maximum and minimum sea surface temperatures (data taken from www.seatemperature.org; figure modified from Cziesielski et al., 2018). Box and whisker plots show the median and range of temperature variability where host (c, d) and symbiont taxa (e, f) originate versus the mean (±1 SE) activation energy (eV) associated to P:R ratios and photochemical efficiency (Fv/Fm). Thermal variation for symbiont taxa containing the artefactual “A4/A1” genotype was calculated based on the two corresponding locations (North Carolina and Red Sea). Letters next to data points indicate similarities (e.g., AA) or differences (e.g., AB) as determined by estimated marginal means
4. DISCUSSION
4.1. Symbiosis specificity in a cnidarian holobiont
Even though all symbiont strains initially colonized Aiptasia, long‐term interpartner compatibility was still limited in some cases (see Discussion S2), especially for hosts harboring heterologous RS taxa. Death of the H2‐Hawaii + RS combination, for example, was not surprising as other studies (Starzak et al., 2014; Tortorelli, Belderok, Davy, McFadden, & van Oppen, 2020) have already shown high mortality due to colonization of incompatible symbionts. For CC7‐North Carolina + RS, it is possible that if anemones were not completely bleached before inoculating them, communities could have shuffled to favor the homologous symbiont. It is important to take the latter into account as here we detected the presence of symbiont types (i.e., “others”) that were not included in the inoculation mix. Some individuals might not have been fully aposymbiotic at the time we performed the inoculations, thus contributing to the proliferation of “unwanted” symbionts. Similarly, we cannot exclude potential contamination while maintaining the animals, feeding (i.e., food was taken from a common batch) and/or subsequently during PCR amplification.
4.2. Photophysiological performance is driven by partner specificity
Consistent with previous reports showing the impacts of symbiont diversity on the physiology (Gabay et al., 2018; Goulet et al., 2005; Hawkins, Hagemeyer, Hoadley, Marsh, & Warner, 2016; Hoadley et al., 2019; Rädecker et al., 2018; Starzak et al., 2014), metabolic fluxes (Matthews et al., 2017, 2018), and protein expression levels (Medrano, Merselis, Bellantuono, & Rodriguez‐Lanetty, 2019; Sproles et al., 2019), we also observed differences in thermal performance among host–symbiont combinations. Not only are metabolic capabilities of Symbiodiniaceae are highly variable, but host identity can also play a fundamental role in determining the physiological response of the symbiont. For example, B. minutum is known to be thermally sensitive (Cziesielski et al., 2018; Grégoire, Schmacka, Coffroth, & Karsten, 2017; Rädecker et al., 2018; Robison & Warner, 2006; Swain, Chandler, Backman, & Marcelino, 2017) yet a more beneficial symbiont, at least in Aiptasia (Gabay et al., 2018; Rädecker et al., 2018; Starzak et al., 2014). Taxa from the genus Symbiodinium, on the other hand, can fix carbon at higher rates than other symbionts but tend to translocate less photosynthates to the host (Rädecker et al., 2018). Furthermore, the thermal response of Durusdinium trenchii can vary greatly depending on the coral species (Hoadley et al., 2019; Rädecker et al., 2018). Accordingly, our study demonstrates that different symbiont strains have a substantial impact on the stability and functionality of the symbiosis but that it is the host that largely determines holobiont productivity in response to heat stress.
Noteworthy, we also found that RS‐B1 (i.e., specific ITS2 type profiles found only in RS‐Red Sea Aiptasia) is distinctly different from B1 from Hawaii; and while questions remain regarding possible genotypic differences between these two, our data suggest that RS‐B1 might be indeed a different species or at least a different “eco‐type” worthy of further study. These observations highlight the importance of considering fine‐scale differences when comparing performance of different symbiont strains, as Symbiodiniaceae is already known to be greatly diverse at inter‐ and intraspecific levels (LaJeunesse et al., 2018).
4.3. Extent of thermal resilience depends on host–symbiont partnership
Most studies have suggested that it is the genetic identity and eco‐physiological attributes of Symbiodiniaceae that ultimately determine thermal acclimation of corals (Berkelmans & van Oppen, 2006; Cziesielski et al., 2018; Howells et al., 2011; Sampayo, Ridgway, Bongaerts, & Hoegh‐Guldberg, 2008). This is despite the increasing evidence that the host also exhibits signs of stress and may even be impaired before the symbiont (reviewed in Oakley & Davy, 2018). For example, it has recently been shown that host‐derived production of reactive oxygen species increases days before detecting bleaching or photoinhibition (Krueger et al., 2015; Oakley et al., 2017). Thus, even with the symbiont's own coping mechanisms, its performance still relies on (phenotypic and genotypic) plasticity of the host to respond under stress (Bellantuono, Hoegh‐Guldberg, & Rodriguez‐Lanetty, 2012; Kenkel & Matz, 2016). Certainly, the ability of the host to adapt to alternative environments plays a strong role (perhaps more than what has been acknowledged) in predicting thermal tolerance (Hoadley et al., 2019; Howells, Abrego, Meyer, Kirk, & Burt, 2016; Kenkel & Matz, 2016; Morikawa & Palumbi, 2019; Palumbi et al., 2014).
Here, and in line with Hoadley et al. (2019), we show that even within heat tolerant taxa like S. microadriaticum (Cziesielski et al., 2018; Díaz‐Almeyda et al., 2017; Swain et al., 2017; and potentially Breviolum types isolated from the RS lineage), host‐dependent (physiological) differences can strongly affect the overall thermal sensitivity of the holobiont. We see that even if the thermal response (measured as Fv/Fm) of H2‐Hawaii is improved when harboring heterologous, compatible symbionts (A4), it is still below CC7‐North Carolina and RS‐Red Sea holobionts. Indeed, strain‐specific responses have been identified for Aiptasia; particularly, RS‐Red Sea stands out as a more heat tolerant genotype (Cziesielski et al., 2018).
4.4. Local adaptation accounts for species‐specific responses to thermal stress
One species can be composed of different, locally adapted populations that differ in their ability to physiologically respond to changes in their environment (Bennett, Duarte, Marbà, & Wernberg, 2019), such that adaptation to a specific set of conditions can constrain and/or cause distribution shifts (Valladares et al., 2014). Populations are expected to adapt so that local genotypes have higher fitness in their native habitat than those from more distant populations (Kawecki & Ebert, 2004; Sanford & Kelly, 2010). Thus, local adaptation not only determines the spatial and temporal patterns of distribution, abundance, and ecological niches of populations of a given species (Valladares et al., 2014) but might well predict its response to environmental disturbances (Bennett et al., 2019; Kawecki & Ebert, 2004; Sanford & Kelly, 2010).
We tested the effect of temperature on the physiological performance of various Aiptasia lineages (host and symbionts) that originate from different geographic regions, and consequently have adapted to distinct thermal regimes. The locations of origin of the Aiptasia lineages tested exhibit different thermal regimes (Figure 4b), North Carolina has large seasonal fluctuations (almost 15°C), whereas temperatures in Hawaii are comparatively stable (temperature range <5°C). Organisms exhibit different levels of physiological compensation (i.e., phenotypic plasticity in the expression of metabolism) when coping with alternative conditions so that individuals from more variable environments show higher plasticity that might increase their ability to respond to extremes (Chevin & Hoffmann, 2017). For instance, a recent global analysis indicated a potentially fundamental role of high frequency temperature variability in reducing the severity of coral bleaching (Safaie et al., 2018). Thus, adding to the body of work shows that previous exposure to a thermally variable environment contributes substantially to the holobiont thermal tolerance beyond that provided by heat‐resistant symbionts alone (Carilli, Donner, & Hartmann, 2012; Howells, Berkelmans, van Oppen, Willis, & Bay, 2013; Oliver & Palumbi, 2011).
We hypothesized that temperature dependence of metabolic rates, characterized here as P:R ratio, reflects plasticity in response to heat stress. We therefore expected that higher plasticity would result in increased holobiont thermotolerance. Our data suggest that hosts from locations with lower and less variable temperatures show significantly lower plasticity (i.e., the ability to adjust their physiology in response to warming); that is, H2‐Hawaii is less plastic than CC7‐North Carolina and RS‐Red Sea Aiptasia. Indeed, H2‐Hawaii was the most susceptible to thermal stress (as reflected by lower photochemical yields and greater loss of symbionts) compared to the others. Furthermore, differences in the plastic responses vary among host and symbiont partners but specifically, that the former determines, in great part, acclimation to stress (as it has higher physiological plasticity). One possible explanation could be the dependence of the symbiont on the host (i.e., the symbiont relies on the host to provide a suitable environment that supports its functioning). Indeed, it has been shown that in hospite nutrient availability for the symbiont differs depending on the associated host (Rädecker et al., 2018), so that performance of the symbiont (e.g., gross productivity and carbon translocation) may be largely attributed to variations in host metabolism. Moreover, our results point toward a remarkable effect of local adaptation on holobiont metabolic plasticity.
5. CONCLUSIONS
This study provides valuable insight into the mechanisms (phenotypic plasticity and local adaptation) underlying the heat stress response of a cnidarian model system and highlights the role of the host and its natural local environment in this process. Coles and Jokiel (1977) first showed differences in the photosynthetic and respiratory capacities of corals in relation to their habitat and thermal histories. Here we build upon this knowledge, and further demonstrate that acclimation mechanisms are consistent with local adaptation to specific conditions; particularly, that individuals from a more thermally variable environment have the ability to dynamically regulate their response to temperature stress. We show that local adaptation may be a strong determinant of symbiosis specificity and as such, inoculation with more heat tolerant partners, even if naturally found in Aiptasia, does not automatically improve thermal resilience of the holobiont.
As global warming intensifies, novel communities and ecological networks are expected to emerge through species turnover and shifts in their distribution (Lurgi, López, & Montoya, 2012; Valladares et al., 2014) via phenotypic plasticity and/or rapid evolution (Hoffmann & Sgrò, 2011; Reusch, 2014; Torda et al., 2017). Thus, understanding the evolutionary constraints and trade‐offs of symbiosis is necessary to develop more realistic models of species survival (of symbiotic cnidarians) and its ecological consequences for future oceans. Particularly, quantifying the amount of plasticity and adaptive potential in metabolic rates may be important to forecast how organisms will cope with, in this case, warmer oceans (Bennett et al., 2019).
We are, nevertheless, cautious in drawing conclusions about thermal adaptation of symbiotic cnidarians, given the potentially oversimplified nature of our experimental approach. Conditions in laboratory settings are far more simplistic than the complex heterogeneity innate to natural environments, and so rates of adaptation here might not be consistent with those in nature where other important environmental traits vary with warming. Furthermore, all host and symbiont strains we tested here are not wild populations but have been cultured under the same, stable thermal regime for a considerable time (at least 2 years). Therefore, we cannot exclude a certain degree of acclimation or even adaptation to the culture conditions, which could account for some of the weak effects we observed here; especially with regard to the symbionts, which have a much shorter life cycle and, hence, higher evolutionary rates than the host (Chakravarti et al., 2017; Chakravarti & van Oppen, 2018; Pandolfi, Connolly, Marshall, & Cohen, 2011). More importantly, it should be noted that only one biological replicate (i.e., genotype) was examined for each geographic location.
Our results indicate that local adaptation is a holobiont trait (defined by the host and the symbionts alike) that cannot be retained when symbionts are transferred to new and unfamiliar hosts. These findings suggest that we may be limited in our capacity to manipulate cnidarian symbioses in light of climate change and that experimental acclimation (i.e., hardening) of the different, coevolved partners (Buerger et al., 2020; Chakravarti et al., 2017; Chakravarti & van Oppen, 2018; Cziesielski, Schmidt‐Roach, & Aranda, 2019) may be a more promising method to increase the persistence of corals in the Anthropocene.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
We thank the KAUST Bioscience Core Laboratory for library sequencing and Luke Esau for assisting with flow cytometry.
Herrera M, Klein SG, Schmidt-Roach S, et al. Unfamiliar partnerships limit cnidarian holobiont acclimation to warming. Glob Change Biol. 2020;26:5539–5553. 10.1111/gcb.15263
DATA AVAILABILITY STATEMENT
Sequencing data are available at NCBI under project number PRJNA577376.
REFERENCES
- Baker, A. C. (2003). Flexibility and specificity in coral‐algal symbiosis: Diversity, ecology, and biogeography of Symbiodinium. Annual Review of Ecology, Evolution, and Systematics, 34(1), 661–689. 10.1146/annurev.ecolsys.34.011802.132417 [DOI] [Google Scholar]
- Baumgarten, S. , Simakov, O. , Esherick, L. Y. , Liew, Y. J. , Lehnert, E. M. , Michell, C. T. , … Voolstra, C. R. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proceedings of the National Academy of Sciences of the United States of America, 112(38), 11893 10.1073/pnas.1513318112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellantuono, A. J. , Hoegh‐Guldberg, O. , & Rodriguez‐Lanetty, M. (2012). Resistance to thermal stress in corals without changes in symbiont composition. Proceedings of the Royal Society B: Biological Sciences, 279(1731), 1100–1107. 10.1098/rspb.2011.1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, S. , Duarte, C. M. , Marbà, N. , & Wernberg, T. (2019). Integrating within‐species variation in thermal physiology into climate change ecology. Philosophical Transactions of the Royal Society B: Biological Sciences, 374(1778), 20180550 10.1098/rstb.2018.0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans, R. , & van Oppen, M. J. H. (2006). The role of zooxanthellae in the thermal tolerance of corals: A “nugget of hope” for coral reefs in an era of climate change. Proceedings of the Royal Society B: Biological Sciences, 273(1599), 2305–2312. 10.1098/rspb.2006.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berumen, M. L. , Voolstra, C. R. , Daffonchio, D. , Agusti, S. , Aranda, M. , Irigoien, X. , … Duarte, C. M. (2019). The Red Sea: Environmental gradients shape a natural laboratory in a nascent ocean In Voolstra C. R. & Berumen M. L. (Eds.), Coral reefs of the Red Sea (pp. 1–10). Springer International Publishing; 10.1007/978-3-030-05802-9_1 [DOI] [Google Scholar]
- Bieri, T. , Onishi, M. , Xiang, T. , Grossman, A. R. , & Pringle, J. R. (2016). Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PLoS One, 11(4), e0152693 10.1371/journal.pone.0152693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein, S. R. , & Theis, K. R. (2015). Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLOS Biology, 13(8), e1002226 10.1371/journal.pbio.1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulotte, N. M. , Dalton, S. J. , Carroll, A. G. , Harrison, P. L. , Putnam, H. M. , Peplow, L. M. , & van Oppen, M. J. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef‐building corals. The ISME Journal, 10, 2693–2701. 10.1038/ismej.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. H. , Gillooly, J. F. , Allen, A. P. , Savage, V. M. , & West, G. B. (2004). Toward a metabolic theory of ecology. Ecology, 85(7), 1771–1789. 10.1890/03-9000 [DOI] [Google Scholar]
- Buerger, P. , Alvarez‐Roa, C. , Coppin, C. W. , Pearce, S. L. , Chakravarti, L. J. , Oakeshott, J. G. , … van Oppen, M. J. H. (2020). Heat‐evolved microalgal symbionts increase coral bleaching tolerance. Science Advances, 6(20), eaba2498. 10.1126/sciadv.aba2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carilli, J. , Donner, S. D. , & Hartmann, A. C. (2012). Historical temperature variability affects coral response to heat stress. PLoS One, 7(3). 10.1371/journal.pone.0034418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti, L. J. , Beltran, V. H. , & van Oppen, M. J. H. (2017). Rapid thermal adaptation in photosymbionts of reef‐building corals. Global Change Biology, 23(11), 4675–4688. 10.1111/gcb.13702 [DOI] [PubMed] [Google Scholar]
- Chakravarti, L. J. , & van Oppen, M. J. H. (2018). Experimental evolution in coral photosymbionts as a tool to increase thermal tolerance. Frontiers in Marine Science, 5, 227 10.3389/fmars.2018.00227 [DOI] [Google Scholar]
- Chevin, L. M. , & Hoffmann, A. A. (2017). Evolution of phenotypic plasticity in extreme environments. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1723), 20160138 10.1098/rstb.2016.0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles, S. L. , & Jokiel, P. L. (1977). Effects of temperature on photosynthesis and respiration in hermatypic corals. Marine Biology, 43(3), 209–216. 10.1007/BF00402313 [DOI] [Google Scholar]
- Coles, S. L. , & Riegl, B. M. (2013). Thermal tolerances of reef corals in the Gulf: A review of the potential for increasing coral survival and adaptation to climate change through assisted translocation. Marine Pollution Bulletin, 72(2), 323–332. 10.1016/j.marpolbul.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Cui, G. , Liew, Y. J. , Li, Y. , Kharbatia, N. , Zahran, N. I. , Emwas, A.‐H. , … Aranda, M. (2019). Host‐dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLOS Genetics, 15(6), e1008189 10.1371/journal.pgen.1008189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunning, R. , Silverstein, R. N. , & Baker, A. C. (2018). Symbiont shuffling linked to differential photochemical dynamics of Symbiodinium in three Caribbean reef corals. Coral Reefs, 37(1), 145–152. 10.1007/s00338-017-1640-3 [DOI] [Google Scholar]
- Cziesielski, M. J. , Liew, Y. J. , Cui, G. , Schmidt‐Roach, S. , Campana, S. , Marondedze, C. , & Aranda, M. (2018). Multi‐omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. Proceedings of the Royal Society B: Biological Sciences, 285(1877), 20172654 10.1098/rspb.2017.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziesielski, M. J. , Schmidt‐Roach, S. , & Aranda, M. (2019). The past, present, and future of coral heat stress studies. Ecology and Evolution, 9(17), 10055–10066. 10.1002/ece3.5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanovic, K. , Blackall, L. L. , Webster, N. S. , & van Oppen, M. J. H. (2017). The contribution of microbial biotechnology to mitigating coral reef degradation. Microbial Biotechnology, 10(5), 1236–1243. 10.1111/1751-7915.12769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decelle, J. , Carradec, Q. , Pochon, X. , Henry, N. , Romac, S. , Mahé, F. , … de Vargas, C. (2018). Worldwide occurrence and activity of the reef‐building coral symbiont Symbiodinium in the open ocean. Current Biology, 28(22), 3625–3633.e3. 10.1016/j.cub.2018.09.024 [DOI] [PubMed] [Google Scholar]
- Dell, A. I. , Pawar, S. , & Savage, V. M. (2011). Systematic variation in the temperature dependence of physiological and ecological traits. Proceedings of the National Academy of Sciences of the United States of America, 108(26), 10591–10596. 10.1073/pnas.1015178108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz‐Almeyda, E. M. , Prada, C. , Ohdera, A. H. , Moran, H. , Civitello, D. J. , Iglesias‐Prieto, R. , … Medina, M. (2017). Intraspecific and interspecific variation in thermotolerance and photoacclimation in Symbiodinium dinoflagellates. Proceedings of the Royal Society B: Biological Sciences, 284(1868), 20171767 10.1098/rspb.2017.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittami, S. M. , Arboleda, E. , Auguet, J.‐C. , Bigalke, A. , Briand, E. , Cardenas, P. , … Not, F. (2019). A community perspective on the concept of marine holobionts: Current status, challenges, and future directions. PeerJ Preprints, 7, e27519v3 10.7287/peerj.preprints.27519v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, A. E. , & Werren, J. H. (2016). Holes in the hologenome: Why host‐microbe symbioses are not holobionts. mBio, 7(2), e02099‐15 10.1128/mBio.02099-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, H. E. , Smith, H. A. , Torda, G. , & van Oppen, M. J. (2019). Microbiome engineering: Enhancing climate resilience in corals. Frontiers in Ecology and the Environment, 17(2), 100–108. 10.1002/fee.2001 [DOI] [Google Scholar]
- Gabay, Y. , Parkinson, J. E. , Wilkinson, S. P. , Weis, V. M. , & Davy, S. K. (2019). Inter‐partner specificity limits the acquisition of thermotolerant symbionts in a model cnidarian‐dinoflagellate symbiosis. The ISME Journal, 13(10), 2489–2499. 10.1038/s41396-019-0429-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay, Y. , Weis, V. M. , & Davy, S. K. (2018). Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model cnidarian‐dinoflagellate symbiosis. The Biological Bulletin, 234(1), 1–10. 10.1086/696365 [DOI] [PubMed] [Google Scholar]
- Gegner, H. M. , Ziegler, M. , Rädecker, N. , Buitrago‐López, C. , Aranda, M. , & Voolstra, C. R. (2017). High salinity conveys thermotolerance in the coral model Aiptasia. Biology Open, 6(12), 1943–1948. 10.1242/bio.028878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet, T. L. , Cook, C. B. , & Goulet, D. (2005). Effect of short‐term exposure to elevated temperatures and light levels on photosynthesis of different host‐symbiont combinations in the Aiptasia pallida/ Symbiodinium symbiosis. Limnology and Oceanography, 50(5), 1490–1498. 10.4319/lo.2005.50.5.1490 [DOI] [Google Scholar]
- Grajales, A. , & Rodríguez, E. (2014). Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actinaria, Metridioidea). Zootaxa, 3826, 55–100. [DOI] [PubMed] [Google Scholar]
- Grégoire, V. , Schmacka, F. , Coffroth, M. A. , & Karsten, U. (2017). Photophysiological and thermal tolerance of various genotypes of the coral endosymbiont Symbiodinium sp. (Dinophyceae). Journal of Applied Phycology, 29(4), 1893–1905. 10.1007/s10811-017-1127-1 [DOI] [Google Scholar]
- Hambleton, E. A. , Guse, A. , & Pringle, J. R. (2014). Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. The Journal of Experimental Biology, 217(9), 1613–1619. 10.1242/jeb.095679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, T. D. , Hagemeyer, J. C. G. , Hoadley, K. D. , Marsh, A. G. , & Warner, M. E. (2016). Partitioning of respiration in an animal‐algal symbiosis: Implications for different aerobic capacity between Symbiodinium spp. Frontiers in Physiology, 7, 128 10.3389/fphys.2016.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley, K. D. , Lewis, A. M. , Wham, D. C. , Pettay, D. T. , Grasso, C. , Smith, R. , … Warner, M. E. (2019). Host–symbiont combinations dictate the photo‐physiological response of reef‐building corals to thermal stress. Scientific Reports, 9(1), 9985 10.1038/s41598-019-46412-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh‐Guldberg, O. (1999). Climate change, coral bleaching and the future of the world’s coral reefs. Marine and Freshwater Research, 50(8), 839–866. 10.1071/MF99078 [DOI] [Google Scholar]
- Hoegh‐Guldberg, O. , Poloczanska, E. S. , Skirving, W. , & Dove, S. (2017). Coral reef ecosystems under climate change and ocean acidification. Frontiers in Marine Science, 4, 158 10.3389/fmars.2017.00158 [DOI] [Google Scholar]
- Hoffmann, A. A. , & Sgrò, C. M. (2011). Climate change and evolutionary adaptation. Nature, 470, 479–485. 10.1038/nature09670 [DOI] [PubMed] [Google Scholar]
- Howells, E. J. , Abrego, D. , Meyer, E. , Kirk, N. L. , & Burt, J. A. (2016). Host adaptation and unexpected symbiont partners enable reef‐building corals to tolerate extreme temperatures. Global Change Biology, 22(8), 2702–2714. 10.1111/gcb.13250 [DOI] [PubMed] [Google Scholar]
- Howells, E. J. , Beltran, V. H. , Larsen, N. W. , Bay, L. K. , Willis, B. L. , & van Oppen, M. J. H. (2011). Coral thermal tolerance shaped by local adaptation of photosymbionts. Nature Climate Change, 2, 116. [Google Scholar]
- Howells, E. J. , Berkelmans, R. , van Oppen, M. J. , Willis, B. L. , & Bay, L. K. (2013). Historical thermal regimes define limits to coral acclimatization. Ecology, 94(5), 1078–1088. 10.1890/12-1257.1 [DOI] [PubMed] [Google Scholar]
- Hume, B. C. C. , Smith, E. G. , Ziegler, M. , Warrington, H. J. M. , Burt, J. A. , LaJeunesse, T. C. , … Voolstra, C. R. (2019). SymPortal: A novel analytical framework and platform for coral algal symbiont next‐generation sequencing ITS2 profiling. Molecular Ecology Resources, 19(4), 1063–1080. 10.1111/1755-0998.13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume, B. C. C. , Ziegler, M. , Poulain, J. , Pochon, X. , Romac, S. , Boissin, E. , … Voolstra, C. R. (2018). An improved primer set and amplification protocol with increased specificity and sensitivity targeting the Symbiodinium ITS2 region. PeerJ, 6, e4816 10.7717/peerj.4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki, T. J. , & Ebert, D. (2004). Conceptual issues in local adaptation. Ecology Letters, 7(12), 1225–1241. 10.1111/j.1461-0248.2004.00684.x [DOI] [Google Scholar]
- Kenkel, C. D. , & Matz, M. V. (2016). Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nature Ecology & Evolution, 1, 0014. [DOI] [PubMed] [Google Scholar]
- Kirk, N. L. , & Weis, V. M. (2016). Animal‐Symbiodinium symbioses: Foundations of coral reef ecosystems In Hurst C. J. (Ed.), The mechanistic benefits of microbial symbionts (pp. 269–294). Springer International Publishing; 10.1007/978-3-319-28068-4_10 [DOI] [Google Scholar]
- Klein, S. G. , Pitt, K. A. , Nitschke, M. R. , Goyen, S. , Welsh, D. T. , Suggett, D. J. , & Carroll, A. R. (2017). Symbiodinium mitigate the combined effects of hypoxia and acidification on a noncalcifying cnidarian. Global Change Biology, 23(9), 3690–3703. 10.1111/gcb.13718 [DOI] [PubMed] [Google Scholar]
- Krueger, T. , Hawkins, T. D. , Becker, S. , Pontasch, S. , Dove, S. , Hoegh‐Guldberg, O. , … Davy, S. K. (2015). Differential coral bleaching – Contrasting the activity and response of enzymatic antioxidants in symbiotic partners under thermal stress. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 190, 15–25. 10.1016/j.cbpa.2015.08.012 [DOI] [PubMed] [Google Scholar]
- LaJeunesse, T. C. , Parkinson, J. E. , Gabrielson, P. W. , Jeong, H. J. , Reimer, J. D. , Voolstra, C. R. , & Santos, S. R. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Current Biology, 28(16), 2570–2580.e6. 10.1016/j.cub.2018.07.008 [DOI] [PubMed] [Google Scholar]
- LaJeunesse, T. C. , Smith, R. T. , Finney, J. , & Oxenford, H. (2009). Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proceedings of the Royal Society B: Biological Sciences, 276(1676), 4139–4148. 10.1098/rspb.2009.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert, E. M. , Mouchka, M. E. , Burriesci, M. S. , Gallo, N. D. , Schwarz, J. A. , & Pringle, J. R. (2014). Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3‐Genes|Genomes|Genetics, 4, 277–295. 10.1534/g3.113.009084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurgi, M. , López, B. C. , & Montoya, J. M. (2012). Novel communities from climate change. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 367(1605), 2913–2922. 10.1098/rstb.2012.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbà, N. , Jorda, G. , Agusti, S. , Girard, C. , & Duarte, C. M. (2015). Footprints of climate change on Mediterranean Sea biota. Frontiers in Marine Science, 2, 56 10.3389/fmars.2015.00056 [DOI] [Google Scholar]
- Masson‐Delmotte, V. , Zhai, P. , Pörtner, H. O. , Roberts, D. , Skea, J. , Shukla, P. R. , … Waterfield, T. (2018). IPCC, 2018: Global warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre‐industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty.
- Matthews, J. L. , Crowder, C. M. , Oakley, C. A. , Lutz, A. , Roessner, U. , Meyer, E. , … Davy, S. K. (2017). Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian‐dinoflagellate symbiosis. Proceedings of the National Academy of Sciences of the United States of America, 114(50), 13194–13199. 10.1073/pnas.1710733114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, J. L. , Oakley, C. A. , Lutz, A. , Hillyer, K. E. , Roessner, U. , Grossman, A. R. , … Davy, S. K. (2018). Partner switching and metabolic flux in a model cnidarian–dinoflagellate symbiosis. Proceedings of the Royal Society B: Biological Sciences, 285(1892), 20182336 10.1098/rspb.2018.2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, J. L. , Sproles, A. E. , Oakley, C. A. , Grossman, A. R. , Weis, V. M. , & Davy, S. K. (2016). Menthol‐induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. The Journal of Experimental Biology, 219(3), 306 10.1242/jeb.128934 [DOI] [PubMed] [Google Scholar]
- McIlroy, S. E. , Gillette, P. , Cunning, R. , Klueter, A. , Capo, T. , Baker, A. C. , & Coffroth, M. A. (2016). The effects of Symbiodinium (Pyrrhophyta) identity on growth, survivorship, and thermal tolerance of newly settled coral recruits. Journal of Phycology, 52(6), 1114–1124. 10.1111/jpy.12471 [DOI] [PubMed] [Google Scholar]
- Medrano, E. , Merselis, D. G. , Bellantuono, A. J. , & Rodriguez‐Lanetty, M. (2019). Proteomic basis of symbiosis: A heterologous partner fails to duplicate homologous colonization in a novel cnidarian–Symbiodiniaceae mutualism. Frontiers in Microbiology, 10, 1153 10.3389/fmicb.2019.01153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N. A. , & Sloan, D. B. (2015). The hologenome concept: Helpful or hollow? PLOS Biology, 13(12), e1002311 10.1371/journal.pbio.1002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa, M. K. , & Palumbi, S. R. (2019). Using naturally occurring climate resilient corals to construct bleaching‐resistant nurseries. Proceedings of the National Academy of Sciences of the United States of America, 116(21), 10586–10591. 10.1073/pnas.1721415116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatine, L. , McCloskey, L. R. , & Marian, R. E. (1981). Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnology and Oceanography, 26(4), 601–611. 10.4319/lo.1981.26.4.0601 [DOI] [Google Scholar]
- Ngugi, D. K. , Antunes, A. , Brune, A. , & Stingl, U. (2012). Biogeography of pelagic bacterioplankton across an antagonistic temperature–salinity gradient in the Red Sea. Molecular Ecology, 21(2), 388–405. 10.1111/j.1365-294X.2011.05378.x [DOI] [PubMed] [Google Scholar]
- Oakley, C. A. , & Davy, S. K. (2018). Cell biology of coral bleaching In van Oppen M. J. H. & Lough J. M. (Eds.), Coral bleaching: Patterns, processes, causes and consequences (pp. 189–211). Springer International Publishing; 10.1007/978-3-319-75393-5_8 [DOI] [Google Scholar]
- Oakley, C. A. , Durand, E. , Wilkinson, S. P. , Peng, L. , Weis, V. M. , Grossman, A. R. , & Davy, S. K. (2017). Thermal shock induces host proteostasis disruption and endoplasmic reticulum stress in the model symbiotic cnidarian Aiptasia. Journal of Proteome Research, 16(6), 2121–2134. 10.1021/acs.jproteome.6b00797 [DOI] [PubMed] [Google Scholar]
- Oliver, T. , & Palumbi, S. (2011). Do fluctuating temperature environments elevate coral thermal tolerance? Coral Reefs, 30(2), 429–440. 10.1007/s00338-011-0721-y [DOI] [Google Scholar]
- Palumbi, S. R. , Barshis, D. J. , Traylor‐Knowles, N. , & Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science, 344(6186), 895–898. 10.1126/science.1251336 [DOI] [PubMed] [Google Scholar]
- Pandolfi, J. M. , Connolly, S. R. , Marshall, D. J. , & Cohen, A. L. (2011). Projecting coral reef futures under global warming and ocean acidification. Science, 333(6041), 418–422. 10.1126/science.1204794 [DOI] [PubMed] [Google Scholar]
- R Core Team . (2018). R: A language and environment for statistical computing. Retrieved from https://www.R‐project.org/
- Rädecker, N. , Raina, J.‐B. , Pernice, M. , Perna, G. , Guagliardo, P. , Kilburn, M. R. , … Voolstra, C. R. (2018). Using Aiptasia as a model to study metabolic interactions in cnidarian‐Symbiodinium symbioses. Frontiers in Physiology, 9, 214 10.3389/fphys.2018.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regaudie‐de‐Gioux, A. , & Duarte, C. M. (2012). Temperature dependence of planktonic metabolism in the ocean. Global Biogeochemical Cycles, 26(1). 10.1029/2010GB003907 [DOI] [Google Scholar]
- Reusch, T. B. H. (2014). Climate change in the oceans: Evolutionary versus phenotypically plastic responses of marine animals and plants. Evolutionary Applications, 7(1), 104–122. 10.1111/eva.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison, J. D. , & Warner, M. E. (2006). Differential impacts of photoacclimation and thermal stress on the photobiology of four different phylotypes of Symbiodinium (Pyrrhophyta). Journal of Phycology, 42(3), 568–579. 10.1111/j.1529-8817.2006.00232.x [DOI] [Google Scholar]
- Rosado, P. M. , Leite, D. C. A. , Duarte, G. A. S. , Chaloub, R. M. , Jospin, G. , Nunes da Rocha, U. , … Peixoto, R. S. (2019). Marine probiotics: Increasing coral resistance to bleaching through microbiome manipulation. The ISME Journal, 13(4), 921–936. 10.1038/s41396-018-0323-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, E. , Sharon, G. , Atad, I. , & Zilber‐Rosenberg, I. (2010). The evolution of animals and plants via symbiosis with microorganisms. Environmental Microbiology Reports, 2(4), 500–506. 10.1111/j.1758-2229.2010.00177.x [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. , Sharon, G. , & Zilber‐Rosenberg, I. (2009). The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environmental Microbiology, 11(12), 2959–2962. 10.1111/j.1462-2920.2009.01995.x [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. , & Zilber‐Rosenberg, I. (2018). The hologenome concept of evolution after 10 years. Microbiome, 6(1), 78 10.1186/s40168-018-0457-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röthig, T. , Costa, R. M. , Simona, F. , Baumgarten, S. , Torres, A. F. , Radhakrishnan, A. , … Voolstra, C. R. (2016). Distinct bacterial communities associated with the coral model Aiptasia in aposymbiotic and symbiotic states with Symbiodinium. Frontiers in Marine Science, 3, 234 10.3389/fmars.2016.00234 [DOI] [Google Scholar]
- Roughgarden, J. , Gilbert, S. F. , Rosenberg, E. , Zilber‐Rosenberg, I. , & Lloyd, E. A. (2018). Holobionts as units of selection and a model of their population dynamics and evolution. Biological Theory, 13(1), 44–65. 10.1007/s13752-017-0287-1 [DOI] [Google Scholar]
- Safaie, A. , Silbiger, N. J. , McClanahan, T. R. , Pawlak, G. , Barshis, D. J. , Hench, J. L. , … Davis, K. A. (2018). High frequency temperature variability reduces the risk of coral bleaching. Nature Communications, 9(1), 1671 10.1038/s41467-018-04074-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampayo, E. M. , Ridgway, T. , Bongaerts, P. , & Hoegh‐Guldberg, O. (2008). Bleaching susceptibility and mortality of corals are determined by fine‐scale differences in symbiont type. Proceedings of the National Academy of Sciences of the United States of America, 105(30), 10444–10449. 10.1073/pnas.0708049105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, E. , & Kelly, M. W. (2010). Local adaptation in marine invertebrates. Annual Review of Marine Science, 3(1), 509–535. 10.1146/annurev-marine-120709-142756 [DOI] [PubMed] [Google Scholar]
- Sproles, A. E. , Oakley, C. A. , Matthews, J. L. , Peng, L. , Owen, J. G. , Grossman, A. R. , … Davy, S. K. (2019). Proteomics quantifies protein expression changes in a model cnidarian colonised by a thermally tolerant but suboptimal symbiont. The ISME Journal, 13(9), 2334–2345. 10.1038/s41396-019-0437-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPSS . (Released 2013). IBM SPSS statistics for windows (version 22.0). (n.d.) [Computer software].
- Starzak, D. E. , Quinnell, R. G. , Nitschke, M. R. , & Davy, S. K. (2014). The influence of symbiont type on photosynthetic carbon flux in a model cnidarian–dinoflagellate symbiosis. Marine Biology, 161(3), 711–724. 10.1007/s00227-013-2372-8 [DOI] [Google Scholar]
- Sunagawa, S. , Wilson, E. C. , Thaler, M. , Smith, M. L. , Caruso, C. , Pringle, J. R. , … Schwarz, J. A. (2009). Generation and analysis of transcriptomic resources for a model system on the rise: The sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics, 10, 258 10.1186/1471-2164-10-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, T. D. , Chandler, J. , Backman, V. , & Marcelino, L. (2017). Consensus thermotolerance ranking for 110 Symbiodinium phylotypes: An exemplar utilization of a novel iterative partial‐rank aggregation tool with broad application potential. Functional Ecology, 31(1), 172–183. 10.1111/1365-2435.12694 [DOI] [Google Scholar]
- Thomas, L. , Rose, N. H. , Bay, R. A. , López, E. H. , Morikawa, M. K. , Ruiz‐Jones, L. , & Palumbi, S. R. (2018). Mechanisms of thermal tolerance in reef‐building corals across a fine‐grained environmental mosaic: Lessons from ofu, American samoa. Frontiers in Marine Science, 4, 434 10.3389/fmars.2017.00434 [DOI] [Google Scholar]
- Thornhill, D. J. , LaJeunesse, T. C. , Kemp, D. W. , Fitt, W. K. , & Schmidt, G. W. (2006). Multi‐year, seasonal genotypic surveys of coral‐algal symbioses reveal prevalent stability or post‐bleaching reversion. Marine Biology, 148(4), 711–722. 10.1007/s00227-005-0114-2 [DOI] [Google Scholar]
- Thornhill, D. J. , Xiang, Y. , Pettay, D. T. , Zhong, M. , & Santos, S. R. (2013). Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Molecular Ecology, 22(17), 4499–4515. 10.1111/mec.12416 [DOI] [PubMed] [Google Scholar]
- Torda, G. , Donelson, J. M. , Aranda, M. , Barshis, D. J. , Bay, L. , Berumen, M. L. , … Munday, P. L. (2017). Rapid adaptive responses to climate change in corals. Nature Climate Change, 7, 627–636. 10.1038/nclimate3374 [DOI] [Google Scholar]
- Tortorelli, G. , Belderok, R. , Davy, S. K. , McFadden, G. I. , & van Oppen, M. J. H. (2020). Host genotypic effect on algal symbiosis establishment in the coral model, the anemone Exaiptasia diaphana, from the Great Barrier Reef. Frontiers in Marine Science, 6, 833 10.3389/fmars.2019.00833 [DOI] [Google Scholar]
- Valladares, F. , Matesanz, S. , Guilhaumon, F. , Araújo, M. B. , Balaguer, L. , Benito‐Garzón, M. , … Zavala, M. A. (2014). The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecology Letters, 17(11), 1351–1364. 10.1111/ele.12348 [DOI] [PubMed] [Google Scholar]
- van Hooidonk, R. , Maynard, J. , Tamelander, J. , Gove, J. , Ahmadia, G. , Raymundo, L. , … Planes, S. (2016). Local‐scale projections of coral reef futures and implications of the Paris Agreement. Scientific Reports, 6, 39666 10.1038/srep39666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oppen, M. J. H. , Gates, R. D. , Blackall, L. L. , Cantin, N. , Chakravarti, L. J. , Chan, W. Y. , … Putnam, H. M. (2017). Shifting paradigms in restoration of the world’s coral reefs. Global Change Biology, 23(9), 3437–3448. 10.1111/gcb.13647 [DOI] [PubMed] [Google Scholar]
- van Oppen, M. J. H. , Oliver, J. K. , Putnam, H. M. , & Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proceedings of the National Academy of Sciences of the United States of America, 112(8), 2307–2313. 10.1073/pnas.1422301112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, P. S. , Metzger, D. A. , & Higuchi, R. (1991). Chelex® 100 as a medium for simple extraction of DNA for PCR‐based typing from forensic material. BioTechniques, 10(4), 506–513. [PubMed] [Google Scholar]
- Weis, V. M. , Davy, S. K. , Hoegh‐Guldberg, O. , Rodriguez‐Lanetty, M. , & Pringle, J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends in Ecology & Evolution, 23(7), 369–376. 10.1016/j.tree.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Wolfowicz, I. , Baumgarten, S. , Voss, P. A. , Hambleton, E. A. , Voolstra, C. R. , Hatta, M. , & Guse, A. (2016). Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Scientific Reports, 6, 32366 10.1038/srep32366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, T. , Hambleton, E. A. , DeNofrio, J. C. , Pringle, J. R. , & Grossman, A. R. (2013). Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. Journal of Phycology, 49, 447–458. 10.1111/jpy.12055 [DOI] [PubMed] [Google Scholar]
- Ye, S. , Bhattacharjee, M. , & Siemann, E. (2019). Thermal tolerance in green hydra: Identifying the roles of algal endosymbionts and hosts in a freshwater holobiont under stress. Microbial Ecology, 77(2), 537–545. 10.1007/s00248-018-01315-1 [DOI] [PubMed] [Google Scholar]
- Ziegler, M. , Eguíluz, V. M. , Duarte, C. M. , & Voolstra, C. R. (2018). Rare symbionts may contribute to the resilience of coral–algal assemblages. The ISME Journal, 12(1), 161–172. 10.1038/ismej.2017.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Sequencing data are available at NCBI under project number PRJNA577376.


