Abstract
Objective: Ovarian cancer is one of the most common gynecological malignancies worldwide, and its mortality rate ranks first among gynecologic cancers. Ceramide synthases are closely related to cancer development. In this study, we investigated the role of ceramide synthase 6 (CerS6) in the development of serous ovarian cancer. Methods: Expression of CerS6 in cancerous and healthy ovarian tissue was assessed by database analysis and immunohistochemistry. The biological role of CerS6 in serous ovarian cancer cells was assessed by CerS6 knockdown followed by cell counting, colony formation, transwell migration, wound healing, and flow cytometry assays and measurement of tumor proliferation in nude mice. Signaling pathway components were analyzed by Western blotting. Gene enrichment was analyzed by GSEA and R, and RNA sequencing was used to compare the transcriptomes of serous ovarian cancer cells with and without CerS6 knockdown. Results: High CerS6 expression in ovarian cancer tissues was closely related to poor prognosis. Knockdown of CerS6 inhibited serous ovarian cancer cell proliferation, invasion, and metastasis and promoted their apoptosis. In addition, CerS6 knockdown increased the proportion of serous ovarian cancer cells in G2/M phase. CerS6 regulates cell cycle through the AKT/mTOR/4EBP1 signaling pathway, which affects cell proliferation and metastasis. The GSEA, R, and RNA sequencing analyses showed that knocking down CerS6 significantly affects cell cycle in serous ovarian cancer cells. Conclusions: CerS6 may have an oncogenic role in ovarian cancer and may represent a new prognostic marker and therapeutic target for serous ovarian cancer.
Keywords: High-grade serous ovarian cancer, CerS6, cell cycle, AKT/mTOR/4EBP1 signaling pathway
Introduction
Ovarian cancer is one of the three malignant gynecological tumors. According to the latest global cancer statistics, ovarian cancer has the highest mortality rate among gynecological tumors worldwide [1]. Ovarian cancer cases have increased worldwide in recent years [2] and currently account for 2.5% of all malignant tumors in women each year [3]. In the United States, approximately 22,530 new cases of ovarian cancer and 13,980 ovarian cancer-related deaths occurred in 2019 [1]. In China, 52,100 new cases of ovarian cancer and 22,500 ovarian cancer-related deaths occurred in 2015 [4]. A recent study reported that the 5-year survival rate of patients with stage I ovarian cancer is 90%, whereas the 5-year survival rate of patients with stage III and IV ovarian cancer is less than 10% [3]. Although the pathogenesis of ovarian cancer is complex and unclear, epidemiological and genetic analyses have linked ovarian cancer occurrence with specific genetic mutations [5]. The most common mutations associated with ovarian cancer, such as those in BRCA1, BRCA2, and p53, are related to defective homologous recombinant DNA repair [6-9] and are accompanied by changes in the PI3K/AKT/mTOR and RAS/RAF/MEK signaling pathways [10].
One of the six identifying hallmarks of tumor cells is abnormal metabolism [11]. Tumor tissues meet the biological energy, biosynthesis, and redox requirements for rapid growth through metabolic reprogramming [12]. Ceramide synthases, key enzymes in eukaryotic lipid metabolism that produce ceramide [13], have recently attracted increasing attention within the tumor field. The ceramide synthase family includes six genes (CerS1-6) [14] that are encoded on different chromosomes and synthesize fatty acyl-CoAs of different chain lengths [13]. Among them, CerS6, also known as LASS6, mainly synthesizes C16: 0-ceramide [15], which promotes cell migration and metastasis and is an important mediator of apoptosis [16,17]. Many recent studies have shown that CerS6 is involved in the occurrence and progression of a variety of tumors. For example, CerS6 is highly expressed in breast cancer [18] and non-small-cell lung cancer [17] cells. Knockdown of CerS6 in non-small-cell lung cancer cells attenuates lung metastasis [17], whereas overexpression of CerS6 in human head and neck squamous cell carcinoma xenografts leads to their rapid growth [19]. However, few studies have investigated the mechanistic role of CerS6 in the development of ovarian cancer.
In the early stage of this study, we found that CerS6 is highly expressed in serous ovarian cancer cells and is associated with poor prognosis. Immunohistochemistry results confirmed that high CerS6 expression is closely related to high-grade and advanced-stage serous ovarian cancer. Therefore, in the later stage of this study, we investigated the effect of CerS6 on the biological functions of serous ovarian cancer cells in vivo and in vitro and explored the possible mechanism for its role in ovarian cancer. Our results indicate that CerS6 may be a new molecular biomarker for the prognosis of serous ovarian cancer and may represent a new therapeutic target for the treatment of this common malignancy.
Materials and methods
Cell culture and reagents
The human epithelial ovarian cancer cell lines HO8910PM, OVCAR5, OVCAR8, ES-2, SKOV-3, OV15, IGROV1, and CAOV-3 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and the cell lines OVCAR3, OVCAR8, and HEK293T were obtained as gifts from Shanghai Cancer Institute (Shanghai, China). All cells were cultured according to American Type Culture Collection (ATCC) instructions. An anti-CerS6 antibody (ab115539, Abcam, Cambridge, UK) was used for Western blotting and immunohistochemistry. Additional Western blotting antibodies recognizing AKT (#4685), p-AKT (#4060), mTOR (#2972), p-mTOR (#2971), 4EBP1 (#9644), and p-4EBP1 (#9451) were obtained from Cell Signaling Technology (Danvers, MA, USA), and a primary anti-GAPDH antibody (M1210-2) was obtained from Sigma (St. Louis, MO, USA). Secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA).
Clinical samples and database
Human ovarian cancer tissue and normal ovarian tissue used in this study were obtained from the Department of Obstetrics and Gynecology, Fengxian District Center Hospital Graduate Student Training Base, Jinzhou Medical University and the Department of Obstetrics and Gynecology, The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University. None of the patients received radiotherapy, chemotherapy, or other related antitumor therapies before surgery. All human tissues were obtained with informed consent, and the study was approved by the Research Ethics Committee of Fengxian District Center Hospital Graduate Student Training Base, Jinzhou Medical University. We downloaded and analyzed ovarian cancer cohort data from the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) and the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/).
Immunohistochemical staining
Immunohistochemistry was performed as previously described in [20] with an anti-CerS6 antibody (1:200, ab115539, Abcam). The intensity of CerS6 staining was scored using the following criteria: 0-5% staining was scored as 0; 6-35% staining as 1; 36-70% staining as 2; and > 70% staining as 3. A total score < 2 was considered to represent negative expression, and a score ≥ 2 was considered to represent positive expression. The scoring was performed in a blinded manner by two senior pathologists.
Cell transfection and RNA interference
Cells were plated at 70% confluence and transfected with CerS6-specific small interfering RNA (siRNA) or with nontargeted siRNA (GenePharma, Shanghai, China) using Lipofectamine® RNAiMAX (13778150, Invitrogen, Waltham, MA, USA) following the manufacturer’s instructions. The CerS6 siRNA sequences were as follows: siRNA1 sense, 5’-GCAGGCUGAGGACCUCUAUTT-3’, anti-sense, 5’-AUAGAGGUCCUCAGCCUGCTT-3’; siRNA2 sense, 5’-GCGCCAUAGCCCUCAACAUTT-3’, anti-sense, 5’-AUGUUGAGGGCUAUGGCGCTT-3’; siRNA3 sense, 5’-GCCACUCACAACUGACCUUTT-3’, anti-sense, 5’-ACGUGACACGUUCGGAGAATT-3’. For stable CerS6 knockdown, short hairpin RNA (shRNA) or negative control (shNC) was cloned into the pLKO.1 plasmid (Sigma). Lentivirus packaging was performed in 293T cells according to standard protocols [20]. Puromycin (A1113802, Gibco, Waltham, MA, USA) was applied to virally infected cells to obtain stable knockdown cell lines. The CerS6 shRNA sequence was 5’-TTCTCCGAACGTGTCACGT-3’, and the negative control sequence was 5’-TTCTCCGAACGTGTCACGT-3’.
RNA sequencing and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells using TRIzol reagent (9109, Takara, Japan), and reverse transcription was performed using the PrimeScript RT-PCR kit (Takara) according to the manufacturer’s protocol. CerS6 mRNA expression was detected by qRT-PCR using SYBR Premix Ex Taq (Takara) on a 7500 real-time PCR system (Applied Biosystems, Waltham, MA, USA) with the following cycling settings: one initial cycle at 95°C for 10 s followed by 40 cycles of 95°C for 5 s and 60°C for 31 s. Data were normalized to 18S rRNA expression and represent the averages of three repeated experiments. The qRT-PCR primers for CerS6 were as follows: forward, 5’-TTCATGGTGCGGCTCATCTT-3’; reverse, 5’-GCTTGGAGAGGCCTTCCAAT-3’. The qRT-PCR primers for 18S were as follows: forward, 5’-TGCGAGTACTCAACACCAACA-3’; reverse, 5’-GCATATCTTCGGCCCACARNA-3’.
Western blotting
Cells were lysed and cell proteins extracted with protein extraction buffer (Sangon, Shanghai, China). Western blotting analysis was performed as described previously [20].
Cell proliferation assay (CCK-8 assay)
Cell viability was measured using the Cell Counting Kit-8 (CCK-8, Dojindo, Japan) according to the manufacturer’s instructions. Cells with the indicated treatments were grown in 96-well plates at 4,000 cells per well. CCK-8 was added to each well and incubated for 1 h at 37°C. Absorbance at 450 nm was measured with a microplate reader (M1000 PRO, Tecan, Zurich, Switzerland).
Colony formation assay
A total of 1000 cells with the indicated treatments were seeded per well into 6-well plates and cultured for 10 days. The culture medium was replaced every three days. Individual colonies (> 50 cells per colony) were fixed, stained with a solution of 1% crystal violet in methanol, and counted.
In vivo tumor xenograft model
Five- to six-week-old female BALB/c athymic nude mice were randomly divided into two groups and inoculated subcutaneously under the left front flank with 100 μl serum-free Dulbecco’s Modified Eagle’s Medium (DMEM) containing 5 × 106 cells (shCerS6 or shNC). Tumor volumes and body weights of the mice were monitored at intervals of 3 or 4 days. Tumor volume was calculated using the following formula: volume = (a × b2)/2, where “a” is the longer diameter and “b” is the shorter diameter. After the mice were sacrificed, tumors were surgically removed and weighed. Mice were manipulated and housed according to protocols approved by the East China Normal University Animal Care Commission.
Cell migration and invasion assays
For the transwell migration assay, cells with the indicated treatments were suspended in 200 µl medium without fetal bovine serum (FBS) in the upper chamber (Corning, NY, USA) and 600 µl medium with 10% FBS in the lower chamber. The invasion assay was performed with Matrigel-coated filters (BD Biosciences, San Jose, CA, USA). After incubation at 37°C, cells were allowed to migrate for 24 h or invade through Matrigel for 48 h. The migrated and invaded cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells were counted under a microscope in six random fields.
Wound healing assay
Cells with the indicated treatments were seeded into a 6-well plate and grown to a nearly confluent monolayer, and the wounds were carefully created by scraping with a pipette tip. The cultures were photographed with a DM IRB microscope (Leica, Wetzlar, Germany), and the number of migrated cells was quantified using the ImageJ software (National Institutes of Health, version 1.52u). The percentage of cell migration was calculated relative to the number of migrated wild-type cells, which was defined as 100%.
Cell apoptosis assay
Adherent cells with the indicated treatments were detached with 0.25% trypsin without EDTA in 1 × phosphate-buffered saline (PBS). Cells were harvested in complete RPMI 1640 medium or DMEM and centrifuged at 1000 rpm for 5 min. The collected cells were washed with 1 × PBS and stained with 50 μg/ml propidium iodide (PI) and annexin V-FITC (BD PharMingen, San Jose, CA, USA) following the manufacturer’s instructions. The percentages of annexin V (+) and PI (-) cells were analyzed by flow cytometry using a FACSCalibur system (BD Biosciences).
Cell cycle assay
Cells with the indicated treatments were plated in 6-well plates and grown for 24 h, washed twice with cold PBS, and fixed in cold 70% ethanol at -20°C overnight. The cells were then washed twice with PBS and incubated in PBS containing 10 mg/ml RNase A and 400 mg/ml PI at room temperature for 30 min. The cell cycle phase distribution was subsequently analyzed by flow cytometry using a FACSCalibur system (BD Biosciences).
Bioinformatics analyses with TCGA data
Gene Set Enrichment Analysis (GSEA) was performed using the Broad Institute web platform, and statistical significance (false discovery rate, FDR) was set at 0.25. Enrichment analyses of genes were performed with the Kyoto Encyclopedia of Genes and Genomes (KEGG) or Gene Ontology (GO) canonical pathways using clusterProfiler in the R package. P-values were calculated based on hypergeometric distribution with FDR correction using the Benjamini method.
Statistical analysis
SPSS 20.0 for Windows (IBM Corp., Armonk, NY, USA), Microsoft Excel (Microsoft Corp., version 2016) and GraphPad Prism (GraphPad Software Inc., version 7.00) software were used for statistical analysis. Two-tailed Student’s t test was used for statistical analysis unless otherwise indicated. The chi-square test or Student’s t-test was used for comparisons between groups. A P-value < 0.05 was considered statistically significant. Data are presented as the mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
Database analysis showed that CerS6 was highly expressed in epithelial ovarian cancer tissues and was closely related to poor prognosis
We first determined the expression of CerS6 in epithelial ovarian cancer tissue through database analysis. Our analysis of CerS6 mRNA expression data obtained from the Bonome Ovarian and Hendrix Ovarian databases on the Oncomine website (https://www.oncomine.org/resource/main.html) indicated that CerS6 mRNA expression was significantly higher in primary ovarian tumors and serous ovarian tumors than in the normal ovarian surface epithelium (P < 0.001, Figure 1A and 1B). Our subsequent analysis of independent ovarian cancer microarray data (GDS3592) from the GEO database (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS3592) also revealed significantly higher expression of CerS6 mRNA in ovarian cancer tissue compared to the normal ovarian surface epithelium (P = 0.0331, Figure 1C). Further, we analyzed sequencing data from the ovarian cancer U133A chip in the TCGA database and found significantly higher CerS6 mRNA expression in serous ovary cancer tissue compared to normal ovarian tissue (P < 0.001, Figure 1D).
Figure 1.
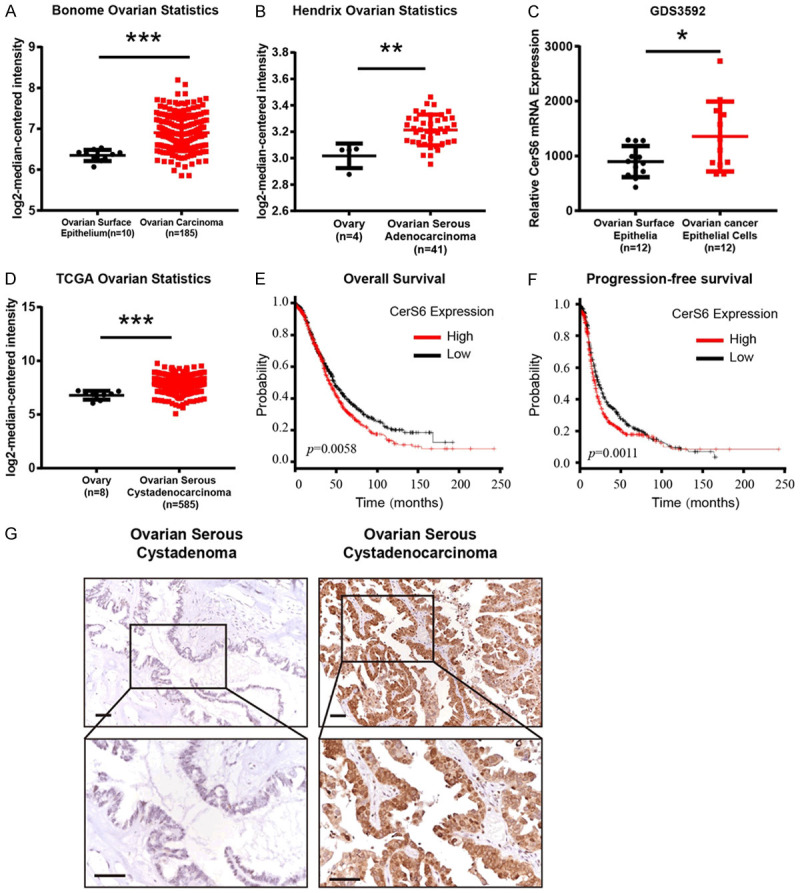
CerS6 was highly expressed in epithelial ovarian cancer tissues and closely related to poor prognosis. A. Analysis of CerS6 mRNA expression data from the Bonome Ovarian database on the Oncomine website. B. Analysis of CerS6 mRNA expression data from the Hendrix Ovarian database on the Oncomine website. C. Analyses of CerS6 mRNA levels in normal ovarian epithelial cells (n = 12) and ovarian cancer epithelial cells (n = 12) from the GEO database (GDS3592). D. The mRNA expression of CerS6 is upregulated in tumor tissues compared to normal tissues from the TCGA database. E. Analysis of overall survival period according to CerS6 expression in 1435 patients with epithelial ovarian cancer from the Kaplan-Meier plotter database. F. Analysis of progression-free survival according to CerS6 expression in 1657 patients with epithelial ovarian cancer from the Kaplan-Meier plotter database. G. Representative immunohistochemical staining of CerS6 expression in benign ovarian serous cystadenoma samples and serous ovarian cystadenocarcinoma samples. Scale bar: 50 μm. (The values represent mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. Experiments were statistically analyzed using two-tailed Student’s t-test).
To determine the relationship between CerS6 expression and patient prognosis, we divided 1657 cases of epithelial ovarian cancer from the Kaplan-Meier plotter database (http://kmplot.com/analysis/index.php?p=service&cancer=ovar) into groups with high (n = 826) and low (n = 830) CerS6 expression based on the median expression level of CerS6 mRNA. Univariate analysis of the overall survival rate revealed that patients in the high CerS6 expression group had significantly shorter overall survival times than those in the low CerS6 expression group (P = 0.0058, Figure 1E). We then divided 1435 cases of epithelial ovarian cancer in the Kaplan-Meier plotter database into groups with high (n = 717) and low (n = 718) CerS6 expression based on the median expression level of CerS6 mRNA. Univariate analysis of progression-free survival showed that patients with high CerS6 expression had a significantly shorter period of progression-free survival than patients with low CerS6 expression (P = 0.0011, Figure 1F). The source of the 1657 cases and 1435 cases were from Kalan-Meier Plotter database using the seleted parameters. These data suggest that high expression of CerS6 is closely related to poor prognosis in ovarian cancer patients.
Immunohistochemical results showed that CerS6 was closely related to advanced serous ovarian cancer
We sought to verify the database analysis results described above by using immunohistochemistry to detect CerS6 expression in tissues from 110 cases of serous ovarian cancer and 35 cases of benign serous cystadenoma. Expression of CerS6 was significantly higher in serous ovarian cancer tissue than in benign ovarian serous cystadenoma (P < 0.001, Figure 1G). Further analysis of the relationship between CerS6 and the clinicopathological characteristics of serous ovarian cancer in this group of cases (Table 1) revealed high CerS6 expression in 21% (3/11) of low-grade serous ovarian cancer cases and in 66% (63/96) of high-grade serous ovarian cancer cases, and the difference between the two groups was statistically significant (P = 0.003). Moreover, high CerS6 expression was detected in 48% (23/48) of early-stage (1+2) serous ovarian cancer cases and in 69% (43/62) of late-stage (3+4) serous ovarian cancer cases, and the difference between these groups was also statistically significant (P = 0.031). These results suggest that high expression of CerS6 is closely related to advanced high-grade serous ovarian cancer.
Table 1.
Correlation of CerS6 expression with patient clinical characteristics
| Variable | Expression of CerS6 | Total | Χ2 | P | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Group | |||||
| Serous Cystadenoma | 31 (88.57) | 4 (11.43) | 35 | ||
| Serous Ovarian cancer | 44 (40) | 66 (60) | 110 | 25.086 | 0.000 |
| Total | 75 (51.72) | 70 (48.28) | 145 | ||
| Age | |||||
| ≤ 50 | 15 (41.67) | 21 (58.33) | 36 | ||
| > 50 | 29 (39.19) | 45 (60.81) | 74 | 0.062 | 0.803 |
| Total | 44 (40) | 66 (60) | 110 | ||
| Lymph node metastasis | |||||
| Present | 8 (33.33) | 16 (66.67) | 24 | ||
| Absent | 36 (41.86) | 50 (58.14) | 86 | 0.568 | 0.451 |
| Total | 44 (40) | 66 (60) | 110 | ||
| Grade of Serous | |||||
| High | 33 (34.38) | 63 (65.63) | 96 | ||
| Low | 11 (78.57) | 3 (21.43) | 14 | 9.944 | 0.003 |
| Total | 44 (40) | 66 (60) | 110 | ||
| FIGO Stage | |||||
| I+II | 25 (52.1) | 23 (47.9) | 48 | ||
| III+IV | 19 (30.65) | 43 (69.35) | 62 | 5.181 | 0.023 |
| Total | 44 (40) | 66 (60) | 110 | ||
FIGO: The International Federation of Gynecology and Obstetrics.
Knockdown of CerS6 inhibited the proliferation of serous ovarian cancer cells in vivo and in vitro
Next, we investigated the effect of CerS6 knockdown in vitro and in vivo. First, we determined the expression levels of CerS6 in 10 ovarian cancer cell lines (OVCAR5, SNU840, OVCAR3, IGROV1, OV15, HO8910PM, SKOV-3, OVCAR8, ES-2, and CAOV-3) by qRT-PCR and Western blotting (Figure 2A and 2B). We then developed three anti-CerS6 siRNAs and tested them in the serous ovarian cancer cell lines OVCAR3 and OVCAR8, which have high CerS6 expression (Figure 2C and 2D). We selected siRNA1 and siRNA2 for use in our in vitro knockdown experiments. For in vivo knockdown experiments, we established stable CerS6 knockdown serous ovarian cancer cell lines by transducing OVCAR8 cells with lentivirus carrying CerS6 shRNA (shCerS6) or negative control (shNC) and verified the transduction efficiency by Western blotting (Figure 2E).
Figure 2.
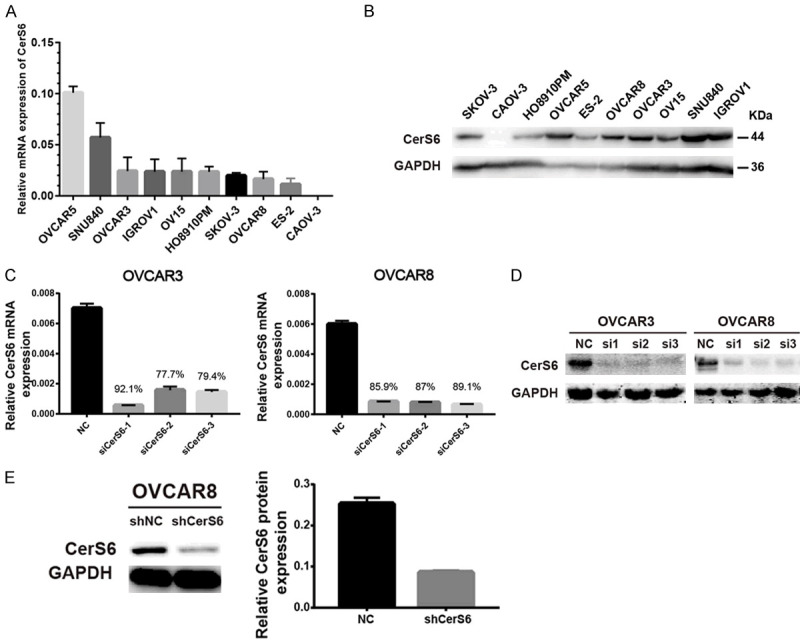
CerS6 expression in ovarian cancer cell lines. (A and B) Expression of CerS6 in 10 different ovarian cancer cell lines was measured by (A) qRT-PCR and (B) Western blotting. (C and D) Expression of CerS6 after siRNA knockdown in OVCAR3 and OVCAR8 cells was detected by (C) qRT-PCR and (D) Western blotting. (E) Expression of CerS6 in OVCAR8 cell lines carrying shRNA (shCerS6 or shNC) was detected by Western blotting. (The values represent mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. Experiments were statistically analyzed using two-tailed Student’s t-test).
We next determined the effect of CerS6 knockdown on serous ovarian cancer cell proliferation in vitro. After siRNA knockdown of CerS6 in OVCAR3 and OVCAR8 cells, the cells were subjected to CCK-8 and colony formation assays. Knockdown of CerS6 significantly inhibited the proliferation and clone formation abilities of serous ovarian cancer cells compared to the control group (P < 0.01, Figure 3A-C).
Figure 3.
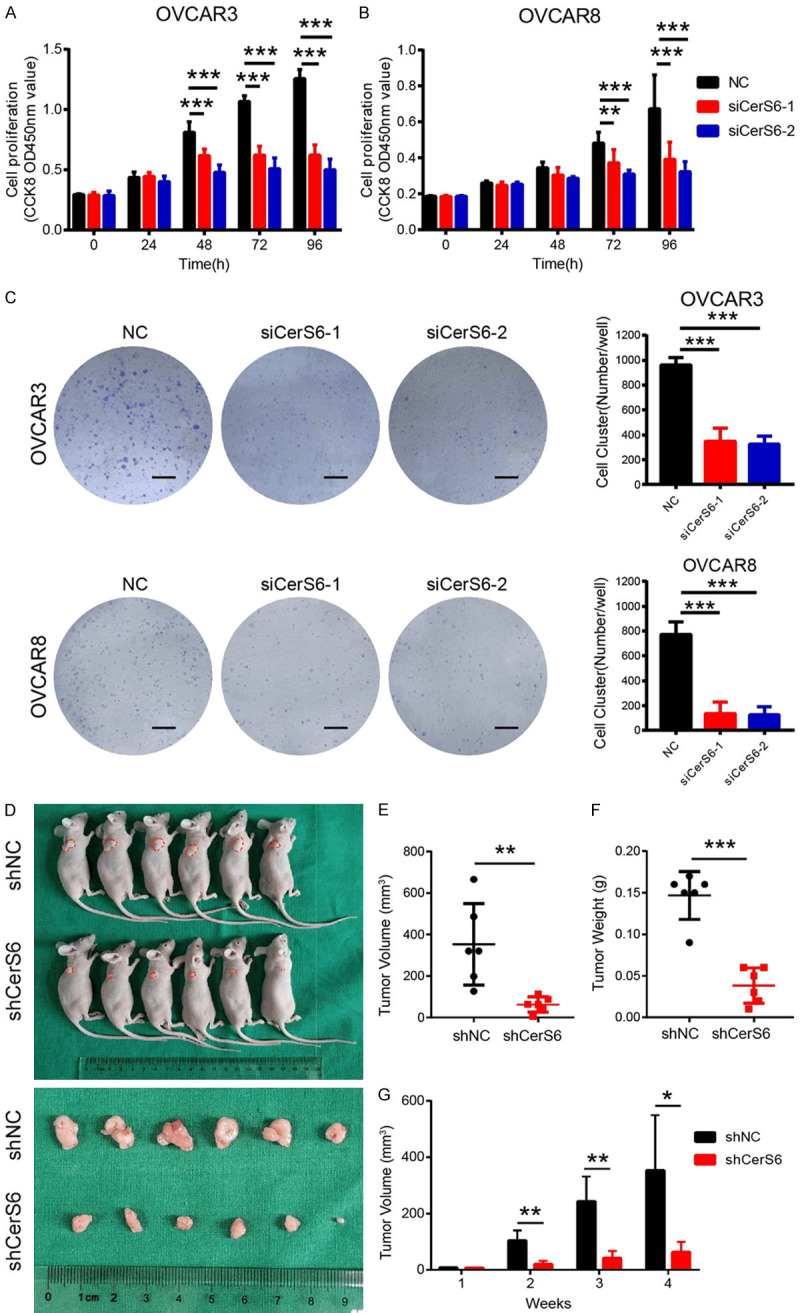
CerS6 knockdown inhibited the proliferation of serous ovarian cancer cells in vitro and in vivo. (A and B) Cell proliferation at 0, 24, 48, 72, and 96 h after CerS6 knockdown in (A) OVCAR3 and (B) OVCAR8 cells was determined by CCK8 assay. (C) Effect of CerS6 knockdown on the colony formation ability of OVCAR3 and OVCAR8 cells in vitro. Scale bar: 5 mm. (D) Morphological characteristics of tumors from mice inoculated with OVCAR8 cells carrying shNC or shCerS6. (E) Weights and (F) Volumes of tumors from the shNC and shCerS6 groups (n = 6). (G) Time-course of xenograft growth. The tumor volumes of mice described in (D) were measured every week. (The values represent mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. Experiments were statistically analyzed using two-tailed Student’s t-test).
To verify the proliferation results in vivo, we inoculated nude mice with stable shCerS6 knockdown and shNC control cells and evaluated the resulting tumors. Tumors from the shNC group had a larger mean volume and weight than those of the shCerS6 group, and CerS6 knockdown significantly decreased the size and inhibited the growth of ovarian tumors (Figure 3D-G). Collectively, these results show that CerS6 knockdown inhibits the proliferation of serous ovarian cancer cells.
Knockdown of CerS6 inhibited the invasion and migration of serous ovarian cancer cells and promoted their apoptosis
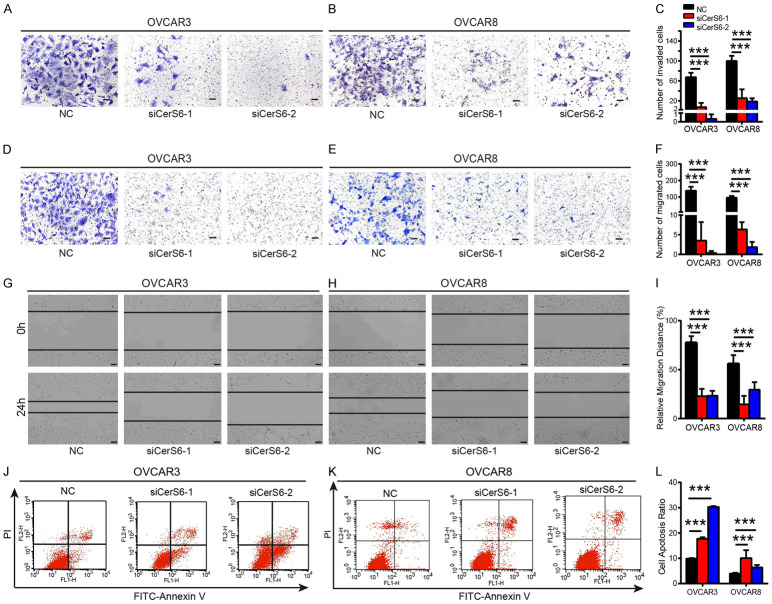
We next investigated the effect of CerS6 knockdown on the invasion and migration abilities of serous ovarian cancer cells using transwell invasion and migration assays. We found that siRNA knockdown of CerS6 in OVCAR3 and OVCAR8 cells significantly decreased their invasive and migratory abilities compared to the control group (P < 0.001, Figure 4A-F). We also used a wound healing assay to evaluate the effect of CerS6 knockdown on the migration of OVCAR3 and OVCAR8 cells and found that cell migration ability was significantly decreased in the CerS6 knockdown group compared to the control group (P < 0.001, Figure 4G-I). Thus, knockdown of CerS6 inhibited the invasion and migration of OVCAR3 and OVCAR8 serous ovarian cancer cells.
Figure 4.
CerS6 knockdown inhibits invasion and migration and promotes the apoptosis of serous ovarian cancer cells. (A and B) The level of invasion was probed in the CerS6 knockdown and negative control (NC) groups derived from (A) OVCAR3 and (B) OVCAR8 cells. (C) Quantification of invasion rates in OVCAR3 and OVCAR8 cells. (D and E) The level of migration was probed in the CerS6 knockdown and NC groups derived from (D) OVCAR3 and (E) OVCAR8 cells. (F) Quantification of migration rates in OVCAR3 and OVCAR8 cells. (G and H) Representative wound healing images of CerS6 knockdown (G) OVCAR3 and (H) OVCAR8 cells at 0 h and 24 h post-wounding. The cell boundary is indicated by the black outline. (I) Quantification of wound healing rates in OVCAR3 and OVCAR8 cells. Data are means ± SD of the wound area relative to the NC group. (J and K) CerS6 knockdown promotes (J) OVCAR3 and (K) OVCAR8 cell apoptosis, as detected by cell apoptosis assays. (L) Quantification of apoptosis rates in OVCAR3 and OVCAR8 cells. Scale bar: 100 μm. (The values represent mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. Experiments were statistically analyzed using two-tailed Student’s t-test).
Given its effect on cell proliferation, we next evaluated the effect of CerS6 knockdown on serous ovarian cancer cell apoptosis using flow cytometry. The apoptosis rates of OVCAR3 cells treated with siCerS6-1 and siCerS6-2 were 17.72 ± 0.3358% and 30.28 ± 0.2139%, respectively, whereas the apoptosis rate of the control group was 9.887% ± 0.1203%. The difference between the two groups was statistically significant (P < 0.001, Figure 4J). Knockdown of CerS6 also increased apoptosis in the OVCAR8 cell line (Figure 4K). Thus, CerS6 knockdown significantly promoted the apoptosis of OVCAR3 and OVCAR8 serous ovarian cancer cells.
CerS6 regulated the cell cycle by arresting serous ovarian cancer cells in G2/M phase
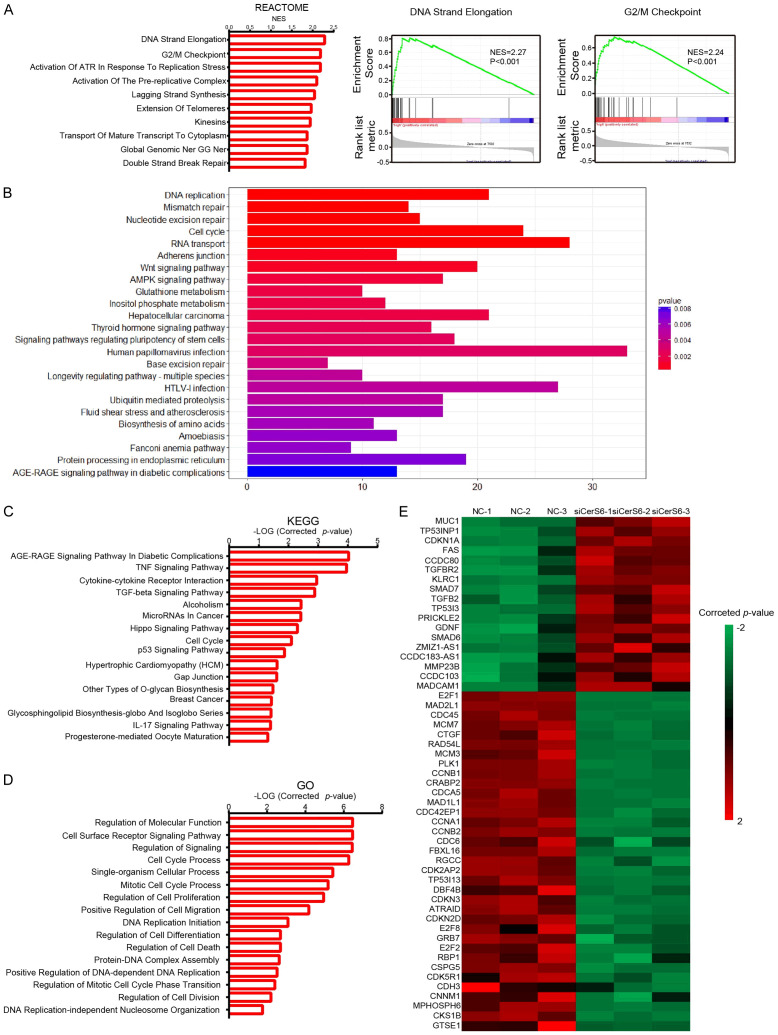
To further explore the possible mechanistic role of CerS6 in ovarian cancer, we first used GSEA and R to analyze differences in gene expression in the high and low CerS6 expression groups from the TCGA ovarian cancer database. In the Reactome gene set analyzed by GSEA, differentially expressed genes were mainly related to processes involved in DNA replication, including strand elongation, G2/M checkpoints, activation of ATR in response to replication stress, activation of the prereplicative complex, and lagging strand synthesis (Figure 5A). These results suggest that CerS6 may regulate ovarian cancer cell cycle function by affecting processes such as DNA replication or cell cycle checkpoints.
Figure 5.
The database and RNA-seq analysis of CerS6. A. GSEA using Reactome gene sets was performed to compare the data from the TCGA ovarian cancer database. NES, normalized enrichment score. B. R analysis using KEGG pathway gene sets was performed to compare the data from the TCGA ovarian cancer database. C. Representative KEGG categories affected by CerS6 expression in OVCAR8 cells. D. Representative GO categories affected by CerS6 expression in OVCAR8 cells. E. Heat map of cell cycle molecules (GO: 0022402) from gene expression profiling results based on GO analysis (control groups vs siCerS6 groups, P = 5.47E-7).
The R analysis results indicated that the differentially expressed genes in the KEGG pathway were also mostly related to cell cycle processes, including DNA replication, mismatch repair, nucleotide excision repair, and RNA transport (Figure 5B). These results also suggest a role for CerS6 in the cell cycle of serous ovarian cancer cells.
To verify the above database analysis results, we then compared the transcriptomes of OVCAR8 serous ovarian cancer cells with or without CerS6 knockdown using high-throughput transcriptome sequencing (RNA-seq) followed by GO analysis and KEGG enrichment analysis of differentially expressed genes. Knocking down CerS6 significantly affected the cell cycle of ovarian cancer cells (P < 0.05, Figure 5C and 5D). In addition, the cluster analysis heat map showed that CerS6 significantly affected cell cycle-related genes such as CDKN1A, CDK5R1, CDK2AP2, CDC45, and CDCA5 (Figure 5E).
Finally, we used flow cytometry and Western blotting to detect the effect of CerS6 knockdown on the cell cycle. The flow cytometry results showed that the proportion of OVCAR3 and OVCAR8 cells in G2/M phase of ovarian cancer increased after knocking down CerS6, but there was no significant effect on the proportion of cells in G1 phase (Figure 6A). Western blotting indicated that 48 h after CerS6 knockdown, the expression of G2/M phase-related proteins (CDK1 and CCNB1) in OVCAR3 and OVCAR8 ovarian cancer cells was significantly reduced (Figure 6B). Collectively, these results suggest that CerS6 knockdown causes serous ovarian cancer cells to arrest at G2/M and consequently affects their biological function.
Figure 6.
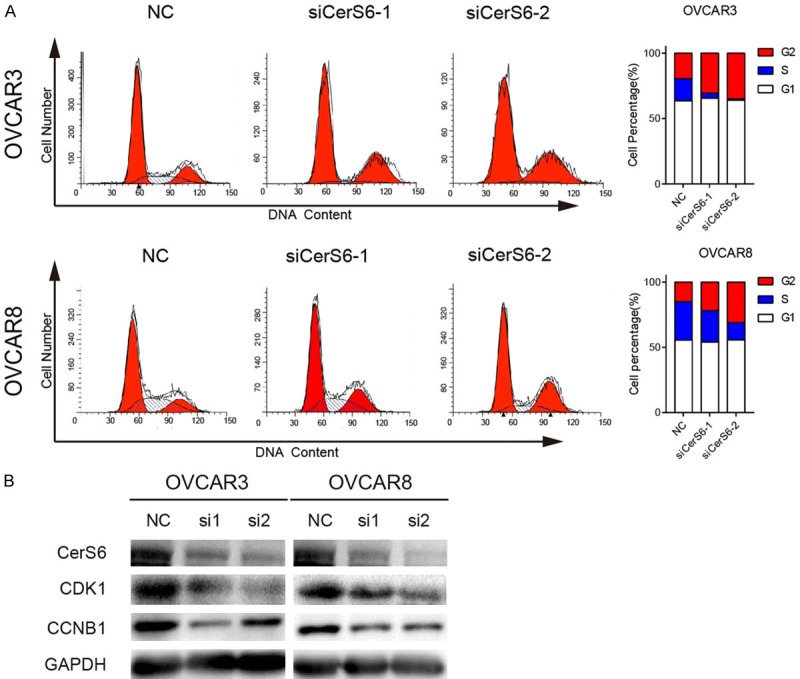
CerS6 regulates the cell cycle by arresting serous ovarian cancer cells in G2/M phase. A. CerS6 knockdown arrested ovarian cancer cells at G2/M. Cell cycle distribution of OVCAR3 and OVCAR8 cells was detected by flow cytometry. B. CerS6 knockdown inhibits activation of CDK1 and CCNB1. Expression of CDK1 and CCNB1 in CerS6 knockdown cell lines (OVCAR3 and OVCAR8) and NC cells was detected by Western blotting.
CerS6 promoted the proliferation and invasion of ovarian cancer cells by activating the AKT/mTOR/4EBP1 pathway
To investigate the possible mechanism by which CerS6 affects ovarian tumorigenesis, we used Western blotting to detect the activation of the AKT/mTOR signaling pathway in OVCAR3 and OVCAR8 serous ovarian cancer cells after CerS6 knockdown. The levels of p-AKT, p-mTOR, and p-4EBP1 detected by Western blotting were significantly reduced in CerS6 knockdown cells compared to the control cells, whereas expression of AKT, mTOR, and 4EBP1 was not significantly changed by CerS6 knockdown (Figure 7). These results indicate that knockdown of CerS6 can significantly inhibit the phosphorylation of AKT, mTOR, and the downstream molecule 4EBP1 in the AKT/mTOR pathway. Further, we speculate that CerS6 may promote the proliferation, invasion, and metastasis of ovarian cancer cells by activating the AKT/mTOR/4EBP1 pathway.
Figure 7.
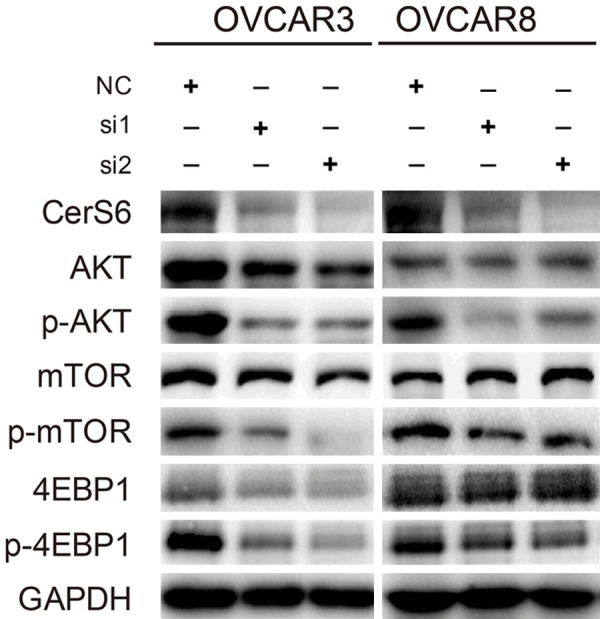
CerS6 knockdown inhibits AKT activation. Analysis of AKT, mTOR, and 4EBP1 activation in CerS6 knockdown OVCAR3 and OVCAR8 cell lines using GAPDH as a loading control.
Discussion
The main function of ceramide synthases (CerS) is to synthesize ceramide, which is an important component of eukaryotic cell membranes [21]. An increasing number of studies have shown that ceramide synthases play a significant role in the pathogenesis of cancer. For example, overexpression of CerS1 and CerS2 in breast cancer promotes tumor cell proliferation and is associated with poor prognosis [13,22]. The mRNA and protein levels of CerS4 are upregulated in human liver cancer tissues and promote the proliferation of liver cancer cells in vitro and in vivo [23]. The mRNA expression of CerS5 is elevated in colorectal cancer tissues [24-26], and CerS5 has been identified as a tumor marker [26] and a prognostic indicator [27] of colorectal cancer. Studies have also found that CerS6 is highly expressed in breast cancer, non-small-cell lung cancer, gastric cancer, and head and neck cancer [17-19,21]. However, no published studies have investigated the role of CerS6 in ovarian cancer.
In our study, database analysis and immunohistochemical assays showed that CerS6 is highly expressed in serous ovarian cancer tissues and that high CerS6 expression is closely related to a poor prognosis in patients with ovarian cancer, which is consistent with data previously reported in the literature [17-19,21]. Studies have also reported that CerS6 can be used as a prognostic marker of tumors in gastric cancer and non-small-cell lung cancer [17,21]. Our immunohistochemistry experiments showed that CerS6 expression level was closely related to the grade and stage of ovarian cancer and, further, that CerS6 expression was significantly higher in advanced high-grade serous ovarian cancer than in early stage cancer. Both clinical stage and grade have been reported to be important factors affecting the prognosis of ovarian cancer, and the prognosis of early-stage low-grade ovarian cancer is significantly different from that of late-stage high-grade ovarian cancer [28-31]. Therefore, we speculate that CerS6 may be a good prognostic marker of high-grade serous ovarian cancer.
Research has shown that CerS6 knockdown reduces gastric cancer cell proliferation, migration, and invasion [21]. It has also been reported that interference with CerS6 in non-small-cell lung cancer tissues reduces lung cancer cell invasion and metastasis and promotes lung cancer cell apoptosis [17]. In this study, we found that knockdown of CerS6 inhibits the proliferation, invasion, and metastasis of serous ovarian cancer cells and promotes their apoptosis, indicating that CerS6 affects ovarian cancer cell proliferation, invasion, and apoptosis.
Increased tumor self-proliferation ability is the most basic characteristic of cancer cells and is often associated with cell cycle dysregulation [11]. We used RNA sequencing and enrichment analysis of data from the TCGA ovarian cancer database to determine that changes in CerS6 expression affect the expression of genes closely related to the cell cycle. Further, our cell cycle and Western blotting experiments revealed that CerS6 knockdown significantly increased the proportion of ovarian cancer cells in the G2/M phase compared to the control cells.
The AKT/mTOR/4EBP1 pathway is an intracellular signaling pathway that is crucial for regulating not only cell cycle and tumorigenesis but also for cell nutrient absorption, cell proliferation, invasion, and survival [32,33]. The AKT/mTOR/4EBP1 pathway is activated in 70% of ovarian cancers, which leads to constitutive activation of signaling cascades associated with cell growth, proliferation, survival, metabolism, and angiogenesis [34]. Our study revealed that phosphorylation of AKT, mTOR, and 4EBP1 was significantly decreased after CerS6 knockdown, whereas the levels of non-phosphorylated AKT, mTOR, and 4EBP1 were not significantly changed. Therefore, we speculate that high expression of CerS6 may significantly promote the phosphorylation of the key AKT/mTOR signaling molecules AKT, mTOR, and 4EBP1, thus promoting the growth and metastasis of ovarian cancer cells.
In summary, CerS6 is highly expressed in high-grade advanced serous ovarian cancer and is closely related to poor prognosis. CerS6 promotes ovarian cancer cell proliferation, invasion, and metastasis and inhibits their apoptosis. After knockdown of CerS6, the serous ovarian cancer cell cycle was arrested in the G2/M phase. High expression of CerS6 may promote the proliferation, invasion, and metastasis of ovarian cancer cells by activating the AKT/mTOR/4EBP1 pathway. These results suggest that CerS6 may be a new prognostic marker and therapeutic target for serous ovarian cancer.
Acknowledgements
The authors would like to acknowledge Dr. Zhigang Zhang (State Key Laboratory for Oncogenes and Related Genes, Shanghai, China) for his valuable suggestions and technical assistance and Experimental Animal Center of East China Normal University for animal care. The study was supported by the National Natural Science Foundation of China (NO: 81672587, 81974407).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Bras J, Singleton A, Cookson MR, Hardy J. Emerging pathways in genetic Parkinson’s disease: potential role of ceramide metabolism in Lewy body disease. FEBS J. 2008;275:5767–5773. doi: 10.1111/j.1742-4658.2008.06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, Stewart C, Fereday S, Caldas C, DeFazio A, Bowtell D, Brenton JD. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, Soucy P, Fredericksen Z, Barrowdale D, Dennis J, Gaudet MM, Dicks E, Kosel M, Healey S, Sinilnikova OM, Lee A, Bacot F, Vincent D, Hogervorst FB, Peock S, Stoppa-Lyonnet D, Jakubowska A kConFab Investigators; Radice P, Schmutzler RK SWE-BRCA; Domchek SM, Piedmonte M, Singer CF, Friedman E, Thomassen M Ontario Cancer Genetics Network; Hansen TV, Neuhausen SL, Szabo CI, Blanco I, Greene MH, Karlan BY, Garber J, Phelan CM, Weitzel JN, Montagna M, Olah E, Andrulis IL, Godwin AK, Yannoukakos D, Goldgar DE, Caldes T, Nevanlinna H, Osorio A, Terry MB, Daly MB, van Rensburg EJ, Hamann U, Ramus SJ, Toland AE, Caligo MA, Olopade OI, Tung N, Claes K, Beattie MS, Southey MC, Imyanitov EN, Tischkowitz M, Janavicius R, John EM, Kwong A, Diez O, Balmaña J, Barkardottir RB, Arun BK, Rennert G, Teo SH, Ganz PA, Campbell I, van der Hout AH, van Deurzen CH, Seynaeve C, Gómez Garcia EB, van Leeuwen FE, Meijers-Heijboer HE, Gille JJ, Ausems MG, Blok MJ, Ligtenberg MJ, Rookus MA, Devilee P, Verhoef S, van Os TA, Wijnen JT HEBON; EMBRACE; Frost D, Ellis S, Fineberg E, Platte R, Evans DG, Izatt L, Eeles RA, Adlard J, Eccles DM, Cook J, Brewer C, Douglas F, Hodgson S, Morrison PJ, Side LE, Donaldson A, Houghton C, Rogers MT, Dorkins H, Eason J, Gregory H, McCann E, Murray A, Calender A, Hardouin A, Berthet P, Delnatte C, Nogues C, Lasset C, Houdayer C, Leroux D, Rouleau E, Prieur F, Damiola F, Sobol H, Coupier I, Venat-Bouvet L, Castera L, Gauthier-Villars M, Léoné M, Pujol P, Mazoyer S, Bignon YJ GEMO Study Collaborators; Złowocka-Perłowska E, Gronwald J, Lubinski J, Durda K, Jaworska K, Huzarski T, Spurdle AB, Viel A, Peissel B, Bonanni B, Melloni G, Ottini L, Papi L, Varesco L, Tibiletti MG, Peterlongo P, Volorio S, Manoukian S, Pensotti V, Arnold N, Engel C, Deissler H, Gadzicki D, Gehrig A, Kast K, Rhiem K, Meindl A, Niederacher D, Ditsch N, Plendl H, Preisler-Adams S, Engert S, Sutter C, Varon-Mateeva R, Wappenschmidt B, Weber BH, Arver B, Stenmark-Askmalm M, Loman N, Rosenquist R, Einbeigi Z, Nathanson KL, Rebbeck TR, Blank SV, Cohn DE, Rodriguez GC, Small L, Friedlander M, Bae-Jump VL, Fink-Retter A, Rappaport C, Gschwantler-Kaulich D, Pfeiler G, Tea MK, Lindor NM, Kaufman B, Shimon Paluch S, Laitman Y, Skytte AB, Gerdes AM, Pedersen IS, Moeller ST, Kruse TA, Jensen UB, Vijai J, Sarrel K, Robson M, Kauff N, Mulligan AM, Glendon G, Ozcelik H, Ejlertsen B, Nielsen FC, Jønson L, Andersen MK, Ding YC, Steele L, Foretova L, Teulé A, Lazaro C, Brunet J, Pujana MA, Mai PL, Loud JT, Walsh C, Lester J, Orsulic S, Narod SA, Herzog J, Sand SR, Tognazzo S, Agata S, Vaszko T, Weaver J, Stavropoulou AV, Buys SS, Romero A, de la Hoya M, Aittomäki K, Muranen TA, Duran M, Chung WK, Lasa A, Dorfling CM, Miron A BCFR; Benitez J, Senter L, Huo D, Chan SB, Sokolenko AP, Chiquette J, Tihomirova L, Friebel TM, Agnarsson BA, Lu KH, Lejbkowicz F, James PA, Hall P, Dunning AM, Tessier D, Cunningham J, Slager SL, Wang C, Hart S, Stevens K, Simard J, Pastinen T, Pankratz VS, Offit K, Easton DF, Chenevix-Trench G, Antoniou AC CIMBA. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köbel M, Reuss A, Du Bois A, Kommoss S, Kommoss F, Gao D, Kalloger SE, Huntsman DG, Gilks CB. The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol. 2010;222:191–198. doi: 10.1002/path.2744. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee S, Kaye S. New strategies in the treatment of ovarian cancer-current clinical perspectives and future potential. Clin Cancer Res. 2013;19:961–968. doi: 10.1158/1078-0432.CCR-12-2243. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Hiller K, Metallo CM. Profiling metabolic networks to study cancer metabolism. Curr Opin Biotechnol. 2013;24:60–68. doi: 10.1016/j.copbio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Park JW, Park WJ, Futerman AH. Ceramide synthases as potential targets for therapeutic intervention in human diseases. Biochim Biophys Acta. 2014;1841:671–681. doi: 10.1016/j.bbalip.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J. 2012;441:789–802. doi: 10.1042/BJ20111626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wegner MS, Schiffmann S, Parnham MJ, Geisslinger G, Grösch S. The enigma of ceramide synthase regulation in mammalian cells. Prog Lipid Res. 2016;63:93–119. doi: 10.1016/j.plipres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhäberle E, Kaufmann M, Ackermann H, Lötsch J, Schmidt H, Geisslinger G, Grösch S. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis. 2009;30:745–752. doi: 10.1093/carcin/bgp061. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki M, Cao K, Kato S, Komizu Y, Mizutani N, Tanaka K, Arima C, Chee Tai M, Yanagisawa K, Togawa N, Shiraishi T, Usami N, Taniguchi T, Fukui T, Yokoi K, Wakahara K, Hasegawa Y, Mizutani Y, Igarashi Y, Inokuchi JI, Iwaki S, Fujii S, Satou A, Matsumoto Y, Ueoka R, Tamiya-Koizumi K, Murate T, Nakamura M, Kyogashima M, Takahashi T. Targeting ceramide synthase 6-dependent metastasisprone phenotype in lung cancer cells. J Clin Invest. 2016;126:254–265. doi: 10.1172/JCI79775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Erez-Roman R, Pienik R, Futerman AH. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem Biophys Res Commun. 2010;391:219–223. doi: 10.1016/j.bbrc.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 19.Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2010;24:296–308. doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang SH, Li J, Dong FY, Yang JY, Liu DJ, Yang XM, Wang YH, Yang MW, Fu XL, Zhang XX, Li Q, Pang XF, Huo YM, Li J, Zhang JF, Lee HY, Lee SJ, Qin WX, Gu JR, Sun YW, Zhang ZG. Increased serotonin signaling contributes to the Warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;153:277–291. e19. doi: 10.1053/j.gastro.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Uen YH, Fang CL, Lin CC, Hseu YC, Hung ST, Sun DP, Lin KY. Ceramide synthase 6 predicts the prognosis of human gastric cancer: it functions as an oncoprotein by dysregulating the SOCS2/JAK2/STAT3 pathway. Mol Carcinog. 2018;57:1675–1689. doi: 10.1002/mc.22888. [DOI] [PubMed] [Google Scholar]
- 22.Moro K, Kawaguchi T, Tsuchida J, Gabriel E, Qi Q, Yan L, Wakai T, Takabe K, Nagahashi M. Ceramide species are elevated in human breast cancer and are associated with less aggressiveness. Oncotarget. 2018;9:19874–19890. doi: 10.18632/oncotarget.24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Li X, Ma D, Liu T, Tian P, Wu C. Ceramide synthase-4 orchestrates the cell proliferation and tumor growth of liver cancer in vitro and in vivo through the nuclear factor-κB signaling pathway. Oncol Lett. 2017;14:1477–1483. doi: 10.3892/ol.2017.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Chen H, Li Y, Li L, Qiu Y, Ren J. Endocannabinoid and ceramide levels are altered in patients with colorectal cancer. Oncol Rep. 2015;34:447–454. doi: 10.3892/or.2015.3973. [DOI] [PubMed] [Google Scholar]
- 25.Jang SW, Park WJ, Min H, Kwon TK, Baek SK, Hwang I, Kim S, Park JW. Altered mRNA expression levels of the major components of sphingolipid metabolism, ceramide synthases and their clinical implication in colorectal cancer. Oncol Rep. 2018;40:3489–3500. doi: 10.3892/or.2018.6712. [DOI] [PubMed] [Google Scholar]
- 26.Kijanka G, Hector S, Kay EW, Murray F, Cummins R, Murphy D, MacCraith BD, Prehn JH, Kenny D. Human IgG antibody profiles differentiate between symptomatic patients with and without colorectal cancer. Gut. 2010;59:69–78. doi: 10.1136/gut.2009.178574. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald S, Sheehan KM, Espina V, O’Grady A, Cummins R, Kenny D, Liotta L, O’Kennedy R, Kay EW, Kijanka GS. High CerS5 expression levels associate with reduced patient survival and transition from apoptotic to autophagy signalling pathways in colorectal cancer. J Pathol Clin Res. 2014;1:54–65. doi: 10.1002/cjp2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gockley A, Melamed A, Bregar AJ, Clemmer JT, Birrer M, Schorge JO, Del Carmen MG, Rauh-Hain JA. Outcomes of women with high-grade and low-grade advanced-stage serous epithelial ovarian cancer. Obstet Gynecol. 2017;129:439–447. doi: 10.1097/AOG.0000000000001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He W, Zhang P, Ye M, Chen Z, Wang Y, Chen J, Yao F. Polymorphisms of the ras-association domain family 1 isoform A (RASSF1A) gene are associated with ovarian cancer, and with the prognostic factors of grade and stage, in women in Southern China. Med Sci Monit. 2018;24:2360–2367. doi: 10.12659/MSM.910058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peres LC, Cushing-Haugen KL, Köbel M, Harris HR, Berchuck A, Rossing MA, Schildkraut JM, Doherty JA. Invasive epithelial ovarian cancer survival by histotype and disease stage. J Natl Cancer Inst. 2019;111:60–68. doi: 10.1093/jnci/djy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terzi A, Aktaş IY, Dolgun A, Ayhan A, Küçükali T, Usubütün A. Early stage epithelial ovarian cancers: a study of morphologic prognostic factors. Pathol Res Pract. 2013;209:359–364. doi: 10.1016/j.prp.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal J. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Kim C, Um JY, Sethi G, Ahn K. Casticin-induced inhibition of cell growth and survival are mediated through the dual modulation of Akt/mTOR signaling cascade. Cancers (Basel) 2019;11:254. doi: 10.3390/cancers11020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li HX, Zeng JF, Shen K. PI3K/AKT/mTOR signaling pathway as a therapeutic target for ovarian cancer. Arch Gynecol Obstet. 2014;290:1067–1078. doi: 10.1007/s00404-014-3377-3. [DOI] [PubMed] [Google Scholar]