Abstract
Clear cell renal cell carcinoma (ccRCC) is an aggressive tumor with frequent metastatic rate. In order to explore the mechanisms, we determined the roles of miR-125b-2-3p in metastatic ccRCC progression. In the study, both clinical and experimental evidences supported the critical role of miR-125b-2-3p in accelerating ccRCC metastasis. Elevated miR-125b-2-3p expression correlated with lymphatic invasion, distant metastasis and poor survival. Functional study showed that high miR-125b-2-3p expression significantly increased ccRCC cell migration in vitro and lung metastasis in vivo. Furthermore, we demonstrated that miR-125b-2-3p directly targeted EGR1, and miR-125b-2-3p accelerated ccRCC cell migration through down-regulating EGR1. Taken together, this study demonstrated that miR-125b-2-3p associates with ccRCC prognosis via promoting tumor metastasis through targeting EGR1.
Keywords: miR-125b-2-3p, ccRCC, metastasis, prognosis, EGR1
Introduction
Renal cell carcinoma (RCC) is one of the most prevalent cancers and accounts for 2-3% in adults worldwide [1-3]. Clear cell RCC (ccRCC) is the most common subtypes, which comprises nearly 85% of all RCCs [4,5]. ccRCC is an aggressive tumor with frequent metastatic rate [6,7], and about one third patients have metastasized at the time of diagnosis. Another third may develop metastases eventually [8,9]. Since metastatic ccRCCs are insensitive to radiotherapy and chemotherapy [10,11] and novel targeted agents fail to work on a large amount of metastatic ccRCC patients [12,13], the 5-yr survival rate of metastatic ccRCCs is < 10%, compared to 75% for organ-confined disease [14]. The most recent immune checkpoint therapy had been shown to be efficacious to RCCs, but unfortunately for only a minority of individuals [15-17]. Thus, metastatic ccRCC is the focus of RCC management and prevention. However, the mechanism of ccRCC metastasis is not completely understood.
MicroRNAs (miRNAs), a class of natural small noncoding RNAs, play important roles in gene expression regulation by binding to the 3’-UTR of their targets, resulting in down-regulation of target genes at post-transcriptional or transcriptional levels [18-21]. Emerging evidences have shown that miRNAs can function as potential tumor oncogenes or suppressors during the initiation and progression of cancers [22-24].
MiR-125b-2-3p, located in 21q21.1, is cleaved from stem-loop pre-miRNA miR-125b-2. Murray et al. found that the serum level of miR-125b-2-3p was elevated in pleuropulmonary blastoma (PPB). Its level showed a reduction following treatment [25]. Up-regulated miR-125b-2-3p served as a characteristic of ERG-related B cell precursor acute lymphoblastic leukemia [26]. Furthermore, miR-125b-2-3p was up-regulated in colorectal cancer with liver metastasis compared with the non-metastatic counterparts. Bioinformatics analysis with KEGG and gene ontology showed that miR-125b-2-3p was involved in liver metastasis during colorectum carcinogenesis [27]. Therefore, it seemed that miR-125b-2-3p involved in tumorigenesis and metastasis, but its role in ccRCC is unknown.
In this study, miR-125b-2-3p is up-regulated in metastatic ccRCCs and its high level associates with distant metastasis and worse prognosis. Exogenous miR-125b-2-3p expression in ccRCC cells significantly promotes cell migration in vitro and lung metastasis in vivo via targeting EGR1. These findings provide strong evidence that miR-125b-2-3p impacts prognosis of ccRCCs through promoting tumor metastasis.
Materials and methods
Tissue samples
Biopsies were obtained from patients diagnosed with ccRCC and underwent surgery at Department of Urology, Ningbo First Hospital, Ningbo, China. The study protocol was approved by the ethics committee of Ningbo First Hospital. Written consent was obtained from all subjects in the study.
Cell culture
Three cancer cells (Caki-1, 786-O and ACHN), proximal tubular epithelial cell HK-2, and 293T cell were purchased from ATCC (USA). OS-RC-2 cell was provided by Stem Cell Bank, Chinese Academy of Sciences. RPMI, McCoy’s 5A and DMEM medium (Gibco, USA) were used to culture these cells with 10% fetal bovine serum (ExCell Bio, China) added. All cells were cultured in chamber at 37°C with 5% CO2.
Transfection
MiRNA mimic, inhibitor and the corresponding negative control of miR-125b-2-3p, siRNA targeted EGR1 and negative control siRNA were provided by GenePharma (China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, USA) following the instructions.
RT-qPCR
Total RNAs were extracted from tumor tissues and cells using TRIzol Reagent (Invitrogen, USA). RNA concentrations were determined using NANODROP 2000 Spectrophotometer (Thermo Fisher SCIENTIFIC, USA).
For the detection of miR-125b-2-3p, 2 µg total RNA was used to synthesize cDNA using miScript® II RT Kit (QIAGEN, Germany). miScript SYBR® Green PCR Kit (QIAGEN, Germany) was used for quantitative detection of miR-125b-2-3p, and snRNA U6 was used as the control.
For the detection of EGR1, 2 µg total RNA was used to synthesize cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher SCIENTIFIC, USA). SYBR Green PCR Master Mix (Roche, US) was used for quantitative detection of EGR1 on a LightCycler480 system (Roche, US), and GAPDH was used as the control.
The primer sequences were as follows: miR-125b-2-3p 5’-TCACAAGTCAGGCTCTTGGGA-3’, U6 5’-ACGCAAATTCGTGAAGCGTTC-3’, Forward primer: 5’-ACCCACTCCTCCACCTTTGAC-3’ and Reverse primer: 5’-TGTTGCTGTAGCCAAATTCGTT-3’ for GAPDH, Forward primer: 5’-ACTTAAAGGACAGGAGGAGGAGATGG-3’ and Reverse primer: 5’-AGGGAGGACTTGGCTCTGAGAAC-3’ for EGR1.
Cell migration assay
Scratch wound healing assay and transwell assay were used to detected cell migration with 8 μm filters (Costar, Corning). For wound healing assay, when seeded cells reached a confluence > 90% in 24-well plate, a linear scratch was made across the well. PBS was used to rinse the dislodged cells, and cells were cultured with fresh medium without serum. The wound width was then monitored. For transwell assay, a suspension of cells (5 × 104 for both Caki-1 and 786-O) in basal medium was added to the upper well. After 24 h, removed non-migratory cells and the migratory cells were fixed, stained with crystal violet, and then imaged.
Plasmid construction, lentivirus package and cell infection
To construct EGR1 expression vector, EGR1 was cloned into pcDNA3.1-3xFlag. We cloned miR-125b-2-3p into pCDH-CMV-EF1-Puro to construct over-expression vector pCDH/miR-125b-2-3p (pCDH-CMV-EF1-Puro/miR-125b-2-3p). The pCDH-CMV-EF1-Puro vector was used as control (pCDH/Control). Three plasmids pCMV-dR8.2 dvpr, pCMV-VSVG and pCDH/miR-125b-2-3p or pCDH/Control were co-transfected into 293T cells and cultured for at least 72 h. Lentivirus-containing supernatants were then harvested using 0.45 μm sterilizing filter. Lentivirus was then used to infect 786-O cell to acquire steadily up-regulated cells of miR-125b-2-3p (786-O/miR-125b-2-3p) and control (786-O/Control).
Western blotting analysis
RIPA buffer (Solarbio, China) was used to lyse tumor cells and proteins were quantified by BCA analysis (Beyotime, China). After the proteins were transferred onto PVDF membranes (BIO-RAD, USA) and blocked with 5% skim milk, the specific primary antibodies were used to incubate the membranes overnight at 4°C. The following primary antibodies were used: anti-E-Cadherin, anti-N-Cadherin, anti-Vimentin, anti-Fibronectin, anti-EGR1 and anti-GAPDH (Cell Signaling Technology, USA). HRP-labeled secondary antibody (Boster, China) was then used to incubate the membranes. The protein bands were visualized using enhanced chemiluminescence reagent.
Luciferase reporter assay
We constructed the sensor vector by joining the regions with the possible binding sites from EGR1 3’-UTR to a luciferase reporter pmirGLO Vector (Promega, USA) to examine the target sequence recognized by miR-125b-2-3p. The full-length EGR1 3’-UTR and the shorting EGR1 3’-UTR without putative miR-125b-2-3p-binding site were cloned into pmirGLO downstream of the firefly luciferase gene. The primary pmirGLO was used as control.
786-O cells were co-transfected with miR-125b-2-3p mimic and pmirGLO/full-length EGR1 3’-UTR, pmirGLO/shorting EGR1 3’-UTR or empty pmirGLO Vector. The luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega, USA) according to the protocols.
Tail injection
In tail injection, 786-O/miR-125b-2-3p and 786-O/Control cells were harvested and concentration was adjusted with 1 × 107 cells/ml. 100 μl cell suspension was then injected into the nude mice tails (5 per group). Fifty days after tail injection, the mice were sacrificed and the numbers of metastatic nodules on lung surface were counted. H&E staining was used to detect the pulmonary metastases.
Statistical analysis
The SPSS 18.0 software (SPSS Inc., USA) was used for statistical analyses. The difference of the mean value between two groups was analyzed with t-test. Survival analysis was estimated using Kaplan-Meier and log-rank test. The correlation between miR-125b-2-3p expression and the clinicopathologic features was analyzed with chi-square test. The data were showed as mean ± SD. P < 0.05 was considered statistically significant.
Results
MiR-125b-2-3p expression associated with ccRCC prognosis and metastasis
We defined miR-125b-2-3p expression in 66 ccRCC samples by RT-qPCR and the results demonstrated that miR-125b-2-3p was up-regulated in metastatic ccRCCs (n=14) compared with non-metastatic tissues (n=52) (P < 0.001) (Figure 1A). Survival analysis showed that the overall survival of ccRCCs with high miR-125b-2-3p level was worse than those with low miR-125b-2-3p expression (The median is used as the cutoff value; P=0.0039; HR, 2.471; 95% CI, 1.337 to 4.566) (Figure 1B).
Figure 1.
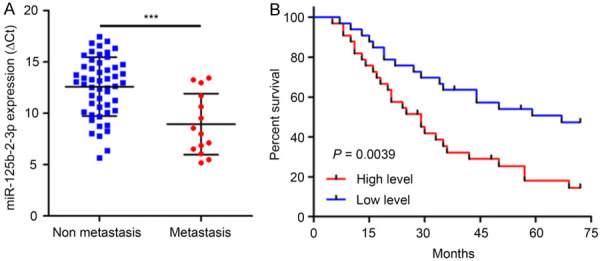
MiR-125b-2-3p associates with ccRCC prognosis. A. MiR-125b-2-3p was up-regulated in metastasis ccRCC tissues (n=14) compared to non-metastasis tissues (n=52) detected by RT-qPCR (P < 0.001). Data are presented as ∆Ct value and are shown as the mean ± SD. B. The overall survival was significantly poorer in patients with high level of miR-125b-2-3p (P=0.0039; HR, 2.471; 95% CI, 1.337 to 4.566). The median is used as the cutoff value.
Next, we analyzed the relationship between miR-125b-2-3p expression and clinicopathologic parameters. As shown in Table 1, higher-expression of miR-125b-2-3p contained more cases with high-grade tumor stage (P < 0.05), low differentiation (P < 0.05), lymphatic invasion (P < 0.05) and distant metastasis (P < 0.05). These data strongly indicated that high miR-125b-2-3p level may represent worse prognosis and tumor metastasis of ccRCCs.
Table 1.
Correlation between miR-125b-2-3p expression and clinicopathologic characteristics of ccRCC patients
| Variables | No. of patients | miR-125b-2-3p expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age | ||||
| ≤ 60 | 30 | 18 | 12 | 0.138 |
| > 60 | 36 | 15 | 21 | |
| Gender | ||||
| Male | 40 | 23 | 17 | 0.131 |
| Female | 26 | 10 | 16 | |
| Tumor size (cm) | ||||
| ≤ 7 | 43 | 24 | 19 | 0.196 |
| > 7 | 23 | 9 | 14 | |
| Tumor stage | ||||
| I+II | 49 | 29 | 20 | 0.011 |
| III+IV | 17 | 4 | 13 | |
| Differentiation | ||||
| Well and moderate | 51 | 29 | 22 | 0.04 |
| Low | 15 | 4 | 11 | |
| N status | ||||
| No | 46 | 27 | 19 | 0.032 |
| Yes | 20 | 6 | 14 | |
| Distant metastasis | ||||
| No | 52 | 30 | 22 | 0.016 |
| Yes | 14 | 3 | 11 | |
No significant effects on ccRCC cell proliferation of miR-125b-2-3p
While analyzing miR-125b-2-3p expression in renal cancer cell lines, miR-125b-2-3p was increased significantly in high metastatic tumor cells Caki-1 and ACHN, however, a marked decrease was demonstrated in 786-O cell with low metastatic capability (Figure 2A). These results are consistent with the fact that high miR-125b-2-3p level associates with tumor metastasis (Table 1).
Figure 2.
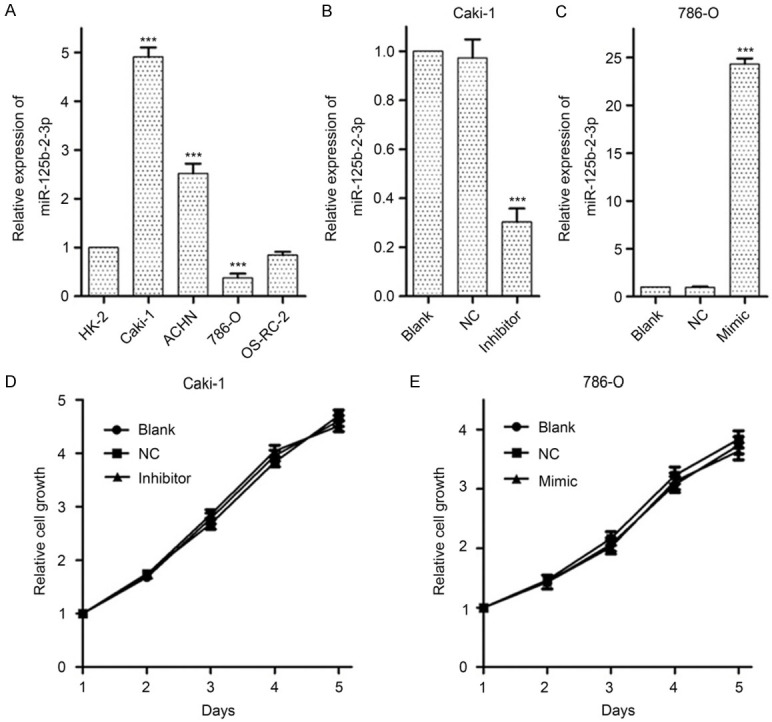
MiR-125b-2-3p shows no significant effects on ccRCC cell growth. A. MiR-125b-2-3p expression in renal cancer cells detected by RT-qPCR. A significant increase of miR-125b-2-3p is shown in Caki-1 and ACHN cells, and a significant reduction is shown in 786-O cells. Data are normalized to normal cell HK-2. B, C. The expression of miR-125b-2-3p in Caki-1 and 786-O cells when transfected miR-125b-2-3p inhibitor and mimic, respectively. Treatment with inhibitor and mimic of miR-125b-2-3p could specifically reverse its levels in Caki-1 and 786-O cells, respectively. D, E. The MTS assay showed miR-125b-2-3p had no significant effect on cell proliferation. Data of the relative expression of miR-125b-2-3p are presented as 2-∆∆Ct value, and are shown as the mean ± SD. ***P < 0.001
As Caki-1 and 786-0 are all ccRCC cell lines, we treated them with miR-125b-2-3p inhibitor and mimic, respectively (Figure 2B, 2C), and then detected its role on cell growth in vitro. The MTS assay suggested that miR-125b-2-3p expression shows no significant change on cell growth (Figure 2D, 2E).
Over-expression of miR-125b-2-3p stimulated ccRCC cell migration in vitro
As miR-125b-2-3p showed no significant change on ccRCC cell proliferation, we further studied whether miR-125b-2-3p affects the cell motility by scratch wound healing assay and transwell assay. The results showed that down-regulation of miR-125b-2-3p reduced the migration distances (Figure 3A) and restrained cell crossing the wells (Figure 3B) in Caki-1 cells. Meanwhile, the contrary effect was demonstrated in 786-O cells with increased miR-125b-2-3p levels (Figure 3A, 3B).
Figure 3.
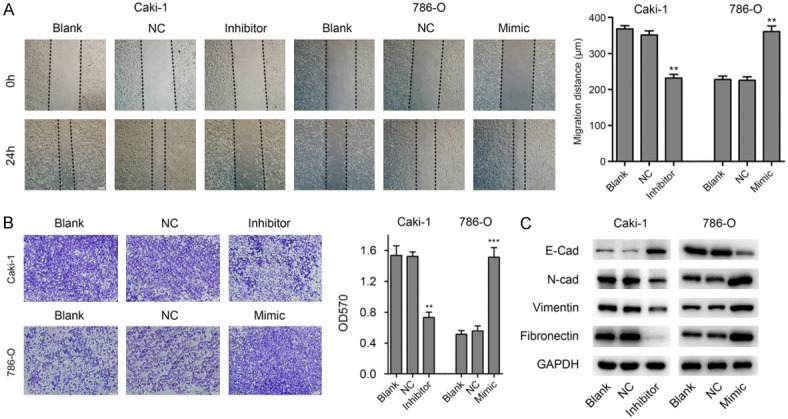
MiR-125b-2-3p stimulates migration of ccRCC cells in vitro. A. MiR-125b-2-3p over-expression promotes cell migration by wound healing assay. The wound healing assay showed that down-regulation of miR-125b-2-3p in Caki-1 cells inhibits migration, however, the contrary effect was demonstrated in 786-O cells with the increased levels of miR-125b-2-3p. **P < 0.01. B. MiR-125b-2-3p over-expression promotes cell migration by Transwell assay. The scale bar in the image is 200 μm. **P < 0.01, ***P < 0.001. C. The expression of EMT markers were detected by western blot in ccRCC cells after transfected miR-125b-2-3p inhibitor or mimic.
Furthermore, we found that knockdown of miR-125b-2-3p in Caki-1 cells inhibited mesenchymal cell proteins expression, including N-Cad, Vimentin and Fibronectin, and up-regulated the expression of epithelial cell marker E-Cad (Figure 3C). However, contrary results were demonstrated in 786-O cells with miR-125b-2-3p exogenous expression (Figure 3C). Collectively, these results indicated that miR-125b-2-3p over-expression promotes ccRCC cell migration in vitro.
EGR1 is a direct target of miR-125b-2-3p
To illuminate the mechanisms mediating the function of miR-125b-2-3p, online computational algorithms miRDB, TargetMiner and miRanda were used to predict the targets of miR-125b-2-3p, and EGR1 was predicted as a candidate gene. A miR-125b-2-3p complementary binding site (1113 site) was found in the 3’-UTR of EGR1 mRNA predicted by all three algorithms and the perfect base pairing was observed (Figure 4A). Furthermore, TargetMiner also predicted another two candidate miR-125b-2-3p binding sites (780 and 1089 site) (Figure 4A).
Figure 4.
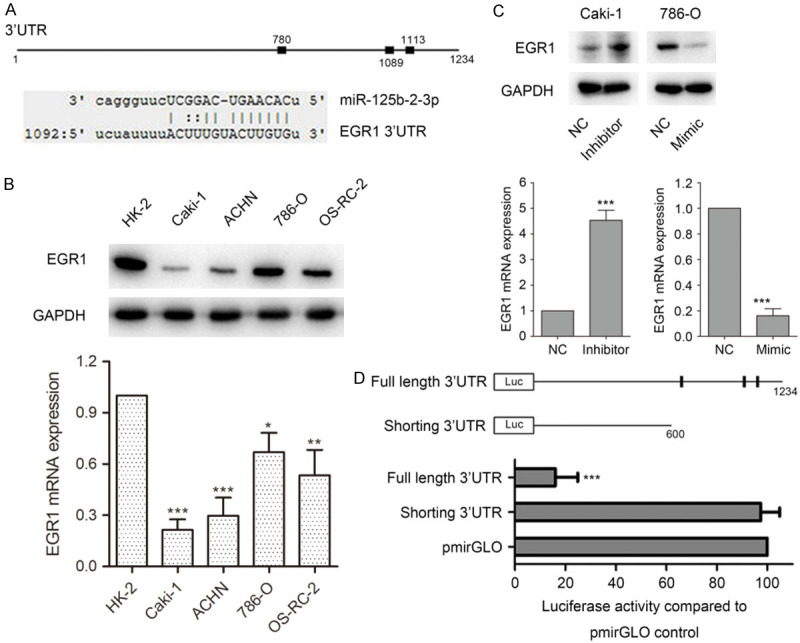
MiR-125b-2-3p directly targets EGR1. A. Schematic diagram of interaction between miR-125b-2-3p and EGR1 3’-UTR. The predicted binding sites of miR-125b-2-3p on EGR1 3’-UTR by online computational algorithms miRDB, TargetMiner and miRanda (Up); the interaction between miR-125b-2-3p and EGR1 3’-UTR at 1113 site predicted by all three algorithms (Down). B. EGR1 was down-regulated in cancer cells compare to HK-2 in both protein and mRNA levels. C. MiR-125b-2-3p negatively regulated EGR1 expression in both protein and mRNA levels. EGR1 expression was detected in both protein and mRNA levels after ccRCC cells transfected miR-125b-2-3p inhibitor or mimic. D. MiR-125b-2-3p directly targets EGR1. Schematic diagram of structures for luciferase assay (Up). Luciferase assay was determined on 786-O cells transfected luciferase vectors together with miR-125b-2-3p mimic (Down).
To identify whether EGR1 is a candidate target of miR-125b-2-3p, we detected EGR1 expression in renal cancer cells. As shown in Figure 4B, EGR1 was down-regulated in cancer cells, compared to HK-2 in both protein and mRNA level. Furthermore, western blotting and qPCR analysis revealed that a reduction of EGR1 was found in 786-O cells when transfected with miR-125b-2-3p mimic. However, Caki-1 cells transfected with miR-125b-2-3p inhibitor showed increased EGR1 expression (Figure 4C).
In addition, luciferase reporter assay was used to verify whether EGR1 is a direct target of miR-125b-2-3p. We subcloned the full-length (with miR-125b-2-3p binding site) and the shorting (without miR-125b-2-3p binding site) EGR1 3’-UTR into pmirGLO vector to structure the luciferase reporter gene vectors (Figure 4D). When 786-O cells were co-transfected with reporter gene vectors (full-length 3’-UTR, shorting 3’-UTR or pmirGLO vector) and miR-125b-2-3p mimic, we found that miR-125b-2-3p dramatically suppressed the luciferase activity of full-length EGR1 3’-UTR group. As shorting 3’-UTR luciferase vector does not contain miR-125b-2-3p binding sites, the luciferase activity was not influenced (Figure 4D). These results indicated that miR-125b-2-3p could directly target EGR1 and negatively regulate the expression of the EGR1.
MiR-125b-2-3p accelerates cell migration via negatively regulating EGR1
In light of the findings described above, we hypothesized that EGR1 might contribute to miR-125b-2-3p mediated ccRCC migration. To confirm this hypothesis, we transfected miR-125b-2-3p inhibitor together with siRNA targeted EGR1 into Caki-1 cells followed by wound healing assay. The results showed that depletion of EGR1 was found to rescue Caki-1 cells from migration inhibition caused by miR-125b-2-3p inhibitor (Figure 5A). However, over-expression of EGR1 rescued the effects of up-regulated miR-125b-2-3p on cell migration in 786-O (Figure 5A). In addition, transwell assay got a consistent result with wound healing assay in both Caki-1 and 786-O cells (Figure 5B).
Figure 5.
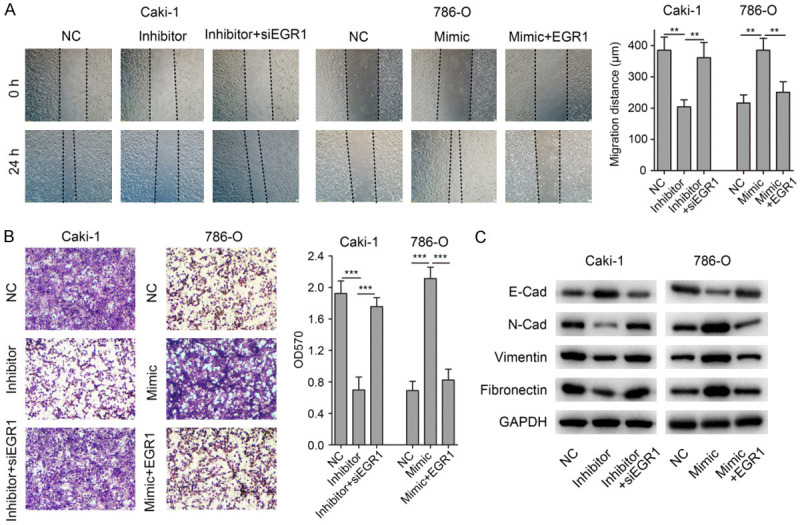
MiR-125b-2-3p accelerates cell migration via negatively regulating EGR1. (A) Depletion of EGR1 was found to rescue Caki-1 cells from migration inhibition caused by miR-125b-2-3p inhibitor. Meanwhile, over-expression of EGR1 rescued the effects of miR-125b-2-3p up-regulation on cell migration in 786-O. (B) Transwell assay got a consistent result with wound healing assay in (A). (C) The expression of EMT markers was detected by western blot in ccRCC cells after co-transfecting miR-125b-2-3p inhibitor and EGR1 siRNA, or miR-125b-2-3p mimic and EGR1 expression vector.
Furthermore, we determined the levels of EMT markers in ccRCC cells. As showed in Figure 5C, when Caki-1 cells co-transfected miR-125b-2-3p inhibitor and EGR1 siRNA, the EMT markers’ expression were rescued in the co-transfected group. Consistent results were found in 786-O cells co-transfected miR-125b-2-3p mimic and EGR1 expression vector (Figure 5C). Taken together, these results showed that miR-125b-2-3p regulates ccRCC cell migration via EGR1.
MiR-125b-2-3p facilitates ccRCC cell migration in vivo
When testing the effect of miR-125b-2-3p on ccRCC cell pulmonary metastasis, we performed an experimental study on nude mice by injecting 786-O/miR-125b-2-3p or 786-O/Control cells through tail vein. The number of metastatic nodules in 786-O/miR-125b-2-3p group was significantly more than the 786-O/Control group (Figure 6A, 6B), which exhibited a good consistency with wound healing and transwell assay. To further confirm the presence of tumor cells in the lesions, we sectioned the tissues following H&E staining and Western blotting. Microscopic examination revealed the presence of tumor cell nests as depicted in representative photomicrographs (Figure 6C), and EGR1 was down-regulated in the lesions of 786-O/miR-125b-2-3p group (Figure 6D).
Figure 6.
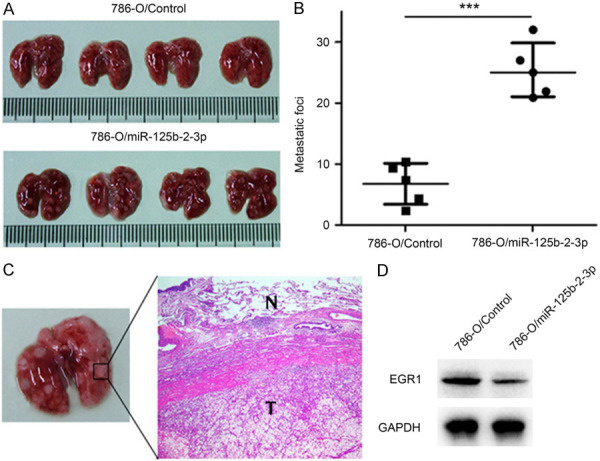
MiR-125b-2-3p promotes migration of ccRCC cells in vivo. A, B. Injecting 786-O/miR-125b-2-3p and 786-O/Control cells into nude mice through tail vein. Increased lung nodule number is shown in miR-125b-2-3p over-expression group compared to the control group. C. H&E staining of lung nodules. D. EGR1 expression in lung nodules between 786-O/miR-125b-2-3p and 786-O/Control group.
Discussion
Previous studies showed that miR-125b-2-3p served as a characteristic of DICER1-mutated pleuropulmonary blastoma and ERG-related B cell precursor acute lymphoblastic leukemia [25,26]. Furthermore, miR-125b-2-3p involved in colorectum cancer liver metastasis [27]. Therefore, miR-125b-2-3p played an important role in cancer.
In this study, we identified that miR-125b-2-3p was up-regulated in metastasis ccRCCs (Figure 1A), which suggested that its expression associates with ccRCC distant metastasis. In the present study, both clinical and experimental evidences supported the critical role of miR-125b-2-3p on accelerating ccRCC metastasis. Elevated miR-125b-2-3p expression is correlated with lymphatic invasion and distant metastasis (Table 1). Furthermore, functional study showed that high miR-125b-2-3p expression significantly increased ccRCC cell migration in vitro (Figure 3) and lung metastasis in vivo (Figure 6).
Early growth response 1 (EGR1) is a zinc-finger transcription factor and has been shown to play roles in multiple processes [28-30]. EGR1 is also crucial in tumorigenesis and progression. Studies showed that EGR1 was undetectable in the great mass of breast cancers and deleted in 50% of AML [31,32]. In addition, it could inhibit cell growth, metastasis and induce cell apoptosis via up-regulating tumor suppressors, including PTEN, TP53, BCL-2, and TGFb1 [33-37]. We demonstrated that miR-125b-2-3p directly targeted EGR1 (Figure 4), and miR-125b-2-3p accelerated ccRCC cell migration through down-regulating EGR1 (Figure 5).
ccRCC is an aggressive tumor with frequent metastatic rate [6,7]. One third patients have metastasized at the time of diagnosis, and another third may develop metastases eventually [8,9]. Because metastasis is the leading cause of worse prognosis [38-42] and miR-125b-2-3p increased ccRCC metastasis, we are interested in finding possible correlations between miR-125b-2-3p expression and the clinical outcome of ccRCCs. We found that elevated miR-125b-2-3p expression represented worse clinical outcomes (Figure 1B). Combined with the result that high miR-125b-2-3p level stimulated ccRCC migration, we suggested that up-regulated miR-125b-2-3p associated with poor prognosis of ccRCC through accelerating migration.
In summary, this study identified miR-125b-2-3p as a significant promoter of metastasis in ccRCC via targeting EGR1. Over-expressed miR-125b-2-3p associates with worse prognosis of ccRCC via stimulating tumor migration.
Acknowledgements
This study was funded by National Natural Science Foundation of China (81272828 and 81372209), Ningbo Natural Science Foundation (2017A610185) and the Medical Technology Project of Ningbo (2018A01).
Disclosure of conflict of interest
None.
References
- 1.Guo S, He X, Chen Q, Yang G, Yao K, Dong P, Ye Y, Chen D, Zhang Z, Qin Z, Liu Z, Xue Y, Zhang M, Liu R, Zhou F, Han H. The C-reactive protein/albumin ratio, a validated prognostic score, predicts outcome of surgical renal cell carcinoma patients. BMC Cancer. 2017;17:171. doi: 10.1186/s12885-017-3119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta S, Kang HC, Ganeshan DM, Bathala TK, Kundra V. Diagnostic approach to hereditary renal cell carcinoma. AJR Am J Roentgenol. 2015;204:1031–1041. doi: 10.2214/AJR.14.13514. [DOI] [PubMed] [Google Scholar]
- 3.Jiao D, Huan Y, Zheng J, Wei M, Zheng G, Han D, Wu J, Xi W, Wei F, Yang AG, Qin W, Wang H, Wen W. UHRF1 promotes renal cell carcinoma progression through epigenetic regulation of TXNIP. Oncogene. 2019;38:5686–5699. doi: 10.1038/s41388-019-0822-6. [DOI] [PubMed] [Google Scholar]
- 4.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turajlic S, Larkin J, Swanton C. SnapShot: renal cell carcinoma. Cell. 2015;163:1556–1556. e1. doi: 10.1016/j.cell.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Xie Y, Chen L, Ma X, Li H, Gu L, Gao Y, Fan Y, Zhang Y, Zhang X. Prognostic and clinicopathological role of high Ki-67 expression in patients with renal cell carcinoma: a systematic review and meta-analysis. Sci Rep. 2017;7:44281. doi: 10.1038/srep44281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girgis H, Masui O, White NM, Scorilas A, Rotondo F, Seivwright A, Gabril M, Filter ER, Girgis AH, Bjarnason GA, Jewett MA, Evans A, Al-Haddad S, Siu KM, Yousef GM. Lactate dehydrogenase A is a potential prognostic marker in clear cell renal cell carcinoma. Mol Cancer. 2014;13:101. doi: 10.1186/1476-4598-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He H, Wang L, Zhou W, Zhang Z, Wang L, Xu S, Wang D, Dong J, Tang C, Tang H, Yi X, Ge J. MicroRNA expression profiling in clear cell renal cell carcinoma: identification and functional validation of key miRNAs. PLoS One. 2015;10:e0125672. doi: 10.1371/journal.pone.0125672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia Y, Zhang QF, Zhen Q, Zhao Y, Liu NJ, Li T, Hao YN, Zhang Y, Luo CL, Wu XH. Negatively regulation of tumor-infiltrating NK cell in clear cell renal cell carcinoma patients through the exosomal pathway. Oncotarget. 2017;8:37783–37795. doi: 10.18632/oncotarget.16354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji SQ, Su XL, Cheng WL, Zhang HJ, Zhao YQ, Han ZX. Down-regulation of CD74 inhibits growth and invasion in clear cell renal cell carcinoma through HIF-1alpha pathway. Urol Oncol. 2014;32:153–161. doi: 10.1016/j.urolonc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Vaziri SA, Grabowski DR, Hill J, Rybicki LR, Burk R, Bukowski RM, Ganapathi MK, Ganapathi R. Inhibition of proteasome activity by bortezomib in renal cancer cells is p53 dependent and VHL independent. Anticancer Res. 2009;29:2961–2969. [PMC free article] [PubMed] [Google Scholar]
- 12.Rini BI, Rathmell WK, Godley P. Renal cell carcinoma. Curr Opin Oncol. 2008;20:300–306. doi: 10.1097/CCO.0b013e3282f9782b. [DOI] [PubMed] [Google Scholar]
- 13.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 14.Heinzelmann J, Unrein A, Wickmann U, Baumgart S, Stapf M, Szendroi A, Grimm MO, Gajda MR, Wunderlich H, Junker K. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: a comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21:1046–1054. doi: 10.1245/s10434-013-3361-3. [DOI] [PubMed] [Google Scholar]
- 15.Ghatalia P, Zibelman M, Geynisman DM, Plimack ER. Checkpoint inhibitors for the treatment of renal cell carcinoma. Curr Treat Options Oncol. 2017;18:7. doi: 10.1007/s11864-017-0458-0. [DOI] [PubMed] [Google Scholar]
- 16.Lee CH, Motzer RJ. Immune checkpoint therapy in renal cell carcinoma. Cancer J. 2016;22:92–95. doi: 10.1097/PPO.0000000000000177. [DOI] [PubMed] [Google Scholar]
- 17.Massari F, Santoni M, Ciccarese C, Santini D, Alfieri S, Martignoni G, Brunelli M, Piva F, Berardi R, Montironi R, Porta C, Cascinu S, Tortora G. PD-1 blockade therapy in renal cell carcinoma: current studies and future promises. Cancer Treat Rev. 2015;41:114–121. doi: 10.1016/j.ctrv.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7:6476–6505. doi: 10.18632/oncotarget.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet. 2014;4:295. doi: 10.3389/fgene.2013.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song L, Li D, Zhao Y, Gu Y, Zhao D, Li X, Bai X, Sun Y, Zhang X, Sun H, Wang Y, Peng L. miR-218 suppressed the growth of lung carcinoma by reducing MEF2D expression. Tumour Biol. 2016;37:2891–2900. doi: 10.1007/s13277-015-4038-2. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Zou L, Zhu L, Zhang H, Du C, Li Z, Gao C, Zhao X, Bao S, Zheng H. miRNA mediated up-regulation of cochaperone p23 acts as an anti-apoptotic factor in childhood acute lymphoblastic leukemia. Leuk Res. 2012;36:1098–1104. doi: 10.1016/j.leukres.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Murray MJ, Bailey S, Raby KL, Saini HK, de Kock L, Burke GA, Foulkes WD, Enright AJ, Coleman N, Tischkowitz M. Serum levels of mature microRNAs in DICER1-mutated pleuropulmonary blastoma. Oncogenesis. 2014;3:e87. doi: 10.1038/oncsis.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vendramini E, Giordan M, Giarin E, Michielotto B, Fazio G, Cazzaniga G, Biondi A, Silvestri D, Valsecchi MG, Muckenthaler MU, Kulozik AE, Gattei V, Izraeli S, Basso G, Te Kronnie G. High expression of miR-125b-2 and SNORD116 noncoding RNA clusters characterize ERG-related B cell precursor acute lymphoblastic leukemia. Oncotarget. 2017;8:42398–42413. doi: 10.18632/oncotarget.16392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin M, Chen W, Huang J, Gao H, Ye Y, Song Z, Shen X. MicroRNA expression profiles in human colorectal cancers with liver metastases. Oncol Rep. 2011;25:739–747. doi: 10.3892/or.2010.1112. [DOI] [PubMed] [Google Scholar]
- 28.Ngiam N, Post M, Kavanagh BP. Early growth response factor-1 in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1089–1091. doi: 10.1152/ajplung.00265.2007. [DOI] [PubMed] [Google Scholar]
- 29.Zhu QB, Unmehopa U, Bossers K, Hu YT, Verwer R, Balesar R, Zhao J, Bao AM, Swaab D. MicroRNA-132 and early growth response-1 in nucleus basalis of Meynert during the course of Alzheimer’s disease. Brain. 2016;139:908–921. doi: 10.1093/brain/awv383. [DOI] [PubMed] [Google Scholar]
- 30.Duclot F, Kabbaj M. The estrous cycle surpasses sex differences in regulating the transcriptome in the rat medial prefrontal cortex and reveals an underlying role of early growth response 1. Genome Biol. 2015;16:256. doi: 10.1186/s13059-015-0815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Liu YG, Huang R, Yao C, Li S, Yang W, Yang D, Huang RP. Concurrent down-regulation of Egr-1 and gelsolin in the majority of human breast cancer cells. Cancer Genomics Proteomics. 2007;4:377–385. [PubMed] [Google Scholar]
- 32.Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin SY, Bahk YY, Ko J, Chung IY, Lee YS, Downward J, Eibel H, Sharma PM, Olefsky JM, Kim YH, Lee B, Lee YH. Suppression of Egr-1 transcription through targeting of the serum response factor by oncogenic H-Ras. EMBO J. 2006;25:1093–1103. doi: 10.1038/sj.emboj.7600987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci U S A. 2011;108:632–637. doi: 10.1073/pnas.1008848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Kang HS, Lee YJ, Lee HJ, Yun J, Shin JH, Lee CW, Kwon BM, Hong SH. EGR1-dependent PTEN upregulation by 2-benzoyloxycinnamaldehyde attenuates cell invasion and EMT in colon cancer. Cancer Lett. 2014;349:35–44. doi: 10.1016/j.canlet.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Chen X, Wang J, Guang W, Han W, Zhang H, Tan X, Gu Y. EGR1 decreases the malignancy of human non-small cell lung carcinoma by regulating KRT18 expression. Sci Rep. 2014;4:5416. doi: 10.1038/srep05416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin SJ, Jeon YK, Cho YM, Lee JL, Chung DH, Park JY, Go H. The association between PD-L1 expression and the clinical outcomes to vascular endothelial growth factor-targeted therapy in patients with metastatic clear cell renal cell carcinoma. Oncologist. 2015;20:1253–1260. doi: 10.1634/theoncologist.2015-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krabbe LM, Westerman ME, Bagrodia A, Gayed BA, Darwish OM, Haddad AQ, Khalil D, Kapur P, Sagalowsky AI, Lotan Y, Margulis V. Dysregulation of beta-catenin is an independent predictor of oncologic outcomes in patients with clear cell renal cell carcinoma. J Urol. 2014;191:1671–1677. doi: 10.1016/j.juro.2013.11.052. [DOI] [PubMed] [Google Scholar]
- 40.Na N, Li H, Xu C, Miao B, Hong L, Huang Z, Jiang Q. High expression of Aldolase A predicts poor survival in patients with clear-cell renal cell carcinoma. Ther Clin Risk Manag. 2017;13:279–285. doi: 10.2147/TCRM.S123199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinmei S, Sakamoto N, Goto K, Sentani K, Anami K, Hayashi T, Teishima J, Matsubara A, Oue N, Kitadai Y, Yasui W. MicroRNA-155 is a predictive marker for survival in patients with clear cell renal cell carcinoma. Int J Urol. 2013;20:468–477. doi: 10.1111/j.1442-2042.2012.03182.x. [DOI] [PubMed] [Google Scholar]
- 42.Kapoor S. Re: microRNA-155 is a predictive marker for survival in patients with clear cell renal cell carcinoma. Int J Urol. 2013;20:739. doi: 10.1111/iju.12025. [DOI] [PubMed] [Google Scholar]


