Abstract
Although many long non-coding RNAs (lncRNAs) are modulators of biological events in hepatocellular carcinoma (HCC), the potential significance of most lncRNAs in HCC remains to be fully understood. The role of lncRNA ADAMTS9-AS1 in HCC was therefore determined. ADAMTS9-AS1 expression was higher in HCC cell lines compared to normal cells as determined by qPCR analyses. Furthermore, CCK-8, scratch wound healing, transwell migration, and invasion assays suggested that ADAMTS9-AS1 overexpression promoted the proliferation, migration, and invasion in MHCC97-H and HepG2 cells; ADAMTS9-AS1 knockdown showed the opposite results. Based on the results from GEO, the correlation between ADAMTS9-AS1 and PI3K/AKT/mTOR was identified. Thus, an association between ADAMTS9-AS1 and the PI3K/AKT/mTOR signaling pathway was further observed. To confirm the pathway protein levels, p-AKT, PIK3CB, and p-mTOR were selected. Western blot assays suggested that ADAMTS9-AS1 enhanced the expression levels of the three proteins. Because of their close relationship with PI3K/AKT/mTOR, apoptosis- or autophagy-related proteins were further investigated. ADAMTS9-AS1 expression was negatively related with LC3-II, BECN1, and pro-apoptotic Bax, whereas it was positively correlated SQSTM1 and anti-apoptotic Bcl-2 expression. Western blot results suggested that ADAMTS9-AS1 decreased ADAMTS9 expression. Our data revealed that ADAMTS9-AS1 contributed to proliferation, migration, and invasion in HCC cells, likely due to the activation of the PI3K/AKT/mTOR signaling pathway, to influence autophagy and apoptosis. These findings suggest that ADAMTS9-AS1 could serve as a molecular target in HCC treatment.
Keywords: Long non-coding RNA, ADAMTS9-AS1, PI3K/AKT/mTOR, hepatocellular carcinoma (HCC)
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer [1], with a rapid increase in prevalence and poor survival worldwide [2,3]. Surgical resection remains the most common choice for patients with HCC, but the long-term prognosis is unsatisfactory due to tumor metastasis and relapse [4]. In particular, patients with advanced HCC often have limited options and a low 5-year survival [5]. There have been studies reporting molecular alterations in HCC [6], and many molecular biomarkers have displayed significant values in the diagnosis and treatment of HCC [7-9]. However, it is still not possible to definitively identify the mechanisms underlying recurrence and metastasis of HCC. For a better understanding of the precise causes of HCC, as well as improving treatments, it is important to first identify more potential biomarkers that might be involved in HCC carcinogenesis.
Long non-coding RNAs (lncRNAs) belong to a group of non-protein-coding RNAs that are > 200 nucleotides in length [10] and possess limited or no protein coding potential [11]. A large class of lncRNAs is differentially expressed in various tumors, including colorectal cancer [12], bladder cancer [13], and lung adenocarcinoma [14]. In addition, the functions of abnormal lncRNAs are diverse and complex because of their own attributes. Currently, highly abundant lncRNAs serve as oncogenes or tumor suppressor genes to influence biological functions. For example, LCAT1 is an oncogene, whose knockdown causes growth arrest and cell invasion in lung cancer cells in vitro [15]; LINC02273 drives proliferation, migration, invasion, and metastasis in breast cancer [16]; FOXD3-AS1 inhibits neuroblastoma progression [17]; and TUSC7 inhibits proliferation of colorectal cancer [18]. Although studies have also reported that lncRNAs such as MAGI2-AS3, TPTEP1, and PVT1 play roles in HCC [19-21], the precise molecular mechanisms remain largely unknown. Therefore, it is important to identify additional candidate biomarkers associated with HCC that may offer a better understanding of HCC initiation, progression, and prognosis.
The lncRNA ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9) antisense RNA 1 (ADAMTS9-AS1) was of special interest given extensive research. ADAMTS9-AS1 is located on chromosome 3p14.1, ranging from 64, 536, 387-64, 592, and 757 bp (https://www.genecards.org/cgi-bin/carddisp.pl?gene=ADAMTS9-AS1&keywords=ADAMTS9-AS1) and is an antisense transcript of ADAMTS9 [22]. Previous studies have reported that ADAMTS9-AS1 is associated with certain types of tumors [22-25]. Nevertheless, the functional roles and potential mechanism of ADAMTS9-AS1 in HCC have not yet been fully investigated. The intracellular signaling pathway phosphatidylinositol 3-kinases (PI3K)/AKT/mammalian target of rapamycin (mTOR) (PI3K/AKT/mTOR) [26] plays an important role in many types of tumors and participates in biological processes such as cell cycle progression, autophagy, and apoptosis [27,28]. Notably, lncRNAs, such as OR3A4 [29], SNHG16 [30], and meg3 [31], have been reported to play roles in HCC via regulating the PI3K/AKT/mTOR pathway. Additionally, ADAMTS9 can function as a tumor suppressor by inhibiting the AKT/mTOR signaling pathway in gastric cancer [32]. These observations suggest that ADAMTS9-AS1 may be involved in the PI3K/AKT/mTOR signaling pathway.
The aim of the present study was, therefore, to evaluate the functional roles and potential mechanisms of ADAMTS9-AS1 in HCC cells. Based on a group of in vitro assays, we determined the effects of ADAMTS9-AS1 on HCC cell proliferation, migration, and invasion. Furthermore, the underlying mechanisms of ADAMTS9-AS1 on HCC cells were investigated. Our findings suggest possible roles of ADAMTS9-AS1 in HCC, as well as its possible clinical significance in HCC-targeted therapy.
Materials and methods
Cells culture
Four human HCC cell lines (HepG2, MHCC97-H, Hep3B, and SMCC-7721) and the LO2 normal liver cell line were obtained from the American Type Culture Collection (Manassas, VA, USA). The cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere of 5% CO2 at 37°C.
Cell transfection
The pcDNA3.1 vector and short interfering RNAs (siRNAs) were obtained from Invitrogen and Ribobio (Guangzhou, China), respectively. The pcDNA3.1-ADAMTS9-AS1 (pcDNA3.1-AS1) was constructed by cloning a fragment of ADAMTS9-AS1 into a pcDNA3.1 vector at the NheI/XhoI sites. The si1-ADAMTS9-AS1 (si1-AS1), si2-ADAMTS9-AS1 (si2-AS1), and the negative control siRNA (si-NC) were obtained by Ribobio (Guangzhou, China). Cells were cultured at a density of 5 × 105 cells/well in 12-well plates at least 24 h prior to transfection. The cells were transfected with pcDNA3.1 or pcDNA3.1-AS1, and si-NC, si1-AS1 or si2-AS1 using Lipofectamine™ 2000 (Invitrogen) following the manufacturer’s instructions. At 48-h post-transfection, the cells were collected for further assays. The sequences were as follows: pcDNA3.1-AS1, forward 5’-CTAGCTAGCCTAGCTAGCCCAGACTTGGAACA-3’ and reverse 5’-CCGCTCGAGTGCATGCTCCATTTATTGAATT-3’; si1-AS1 5’-CCAUACUGAUACAGCCAAATT-3’ (sense), and si2-AS1 5’-CCUAACGACAAGGUCCUAUTT-3’ (sense).
RNA extraction and quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted from the HCC cells using the TRIzol total RNA reagent (Invitrogen) based on the manufacturer’s instructions. cDNA synthesis was performed with 2 μg of total RNA using a PrimeScript RT Reagent Kit with a cDNA Eraser (Takara Biotech, Dalian, China). The following sequences were used, ADAMTS9-AS1, forward 5’-CCATCACTAATCGCCAGGAT-3’ and reverse 5’-CTGTTGTGGAGTTGCCCTTC-3’; GAPDH, forward 5’-ACGGATTTGGTCGTATTGGGCG-3’ and reverse 5’-GCTCCTGGAAGATGGTGATGGG-3’. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control.
qPCR was conducted using SYBR Premix Ex Taq (Takara Biotech, Shiga, Japan) on an ABI 7900 system (Applied Biosystems, Foster City, CA, USA). The reaction mixture started at 95°C for 30 s, followed by 40 amplification cycles of 95°C for 5 s and 60°C for 34 s. The quantification of gene expression was performed using a 2-ΔΔCT calculation with CT as the threshold cycle.
Cell proliferation assays
Cell Counting Kit-8 (CCK-8) assays (Dojindo Molecular Technologies, Kumamoto, Japan) were performed to determine cell proliferation. Cells were seeded at a density of 4 × 104 cells/well in 96-well culture plates. After culturing for 18 h, cells were transfected with the pcDNA3.1 vector or pcDNA3.1-AS1, si-NC, si1-AS1, or si2-AS1. The culture medium was replaced with DMEM after incubation for 4 h. At 0-3 days post-transfection, a new medium containing 10% CCK-8 solution was used. Absorbance was measured at 450 nm using an ELISA reader (PerkinElmer, Waltham, MA, USA). The growth curve was produced based on absorbance values at 0-3 days. Each experiment was independently repeated three times.
Clone formation assay
A total of 1 × 106 cells were cultured in 6-well plates. After transfection, the cells were transferred to DMEM medium with 10% FBS and cultured for 24 h. Afterwards, cells were added to a 1.5 mL centrifuge tube and then inoculated into new 6-well plates (2 × 103 cells/well) at 37°C with 5% CO2 for 2-3 weeks until colonies were visible. After the supernatant solution was removed, cells were washed twice with phosphate-buffered saline (PBS). Next, colonies were fixed with 4% paraformaldehyde solution for 15 min and stained with 0.2% crystal violet for 15 min. Images of each well were captured using a camera, and the number of clones in each well were determined using Image J software (National Institutes of Health, Bethesda, MD, USA). Clone formation rates of each well were then calculated.
The scratch wound healing assay
The cells were seeded in 24-well culture plates at a density of 2 × 105 cells/well and cultured for 18 h prior to transfection. Cells were scraped to form wounds using a sterile pipette tip after transfection with pcDNA3.1 vector or pcDNA3.1-AS1, and si-NC, si1-AS1, or si2-AS1, separately. Wells were washed three times with PBS to remove the exfoliated cells, medium containing 2% fetal bovine serum was added, and the cells were further incubated at 37°C in a 5% CO2 incubator. Photographs at a magnification of 40 × were taken at 0, 24, 48, and 72 h. All experiments were performed in triplicate.
Transwell migration and invasion assays
Cell migration and invasion assays were performed using transwell chambers uncoated or coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) in 24-well plates according to the manufacturer’s instruction. For migration assays, the cells were cultured in 100 μL serum-free DMEM after transfection for 48 h. Cells were seeded onto a fibronectin-coated polycarbonate membrane inserts in a Transwell apparatus. Complete medium was added to the lower chamber as a chemoattractant. After incubation for 24 h, the cells remaining on the upper membrane were removed with cotton swabs, and cells adhering to the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 20 min. For invasion assays, the procedure was similar to the cell migration assays, except that the transwell membranes were precoated with 24 mg/mL Matrigel. All experiments were performed in triplicate.
Western blot assay
Total cell lysates were collected after washing with precooled PBS. Lysates were incubated in boiling water for 5 min. Proteins samples were resolved using 10% polyacrylamide gel electrophoresis (SDS-PAGE) for 3 h, and then the samples were transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked for 80 min with 5% skimmed milk at room temperature and incubated with primary antibodies (1:1,000) for ADAMTS9, AKT, p-AKT, PIK3CB, mTOR, p-mTOR, LC3, BECN1, SQSTM1, Bax, or Bcl-2 at 4°C overnight. Peroxidase-conjugated goat anti-mouse antisera (1:10,000) was used as secondary antibodies for GAPDH and BECN1 at 37°C for 1 h. Antibodies were purchased from ABclonal (Wuhan, China). PVDF membranes were developed using ECL chemiluminescent reagent (Millipore, Billerica, MA, USA). GAPDH was used as an internal control. Images were obtained using a Bio-Rad Gel Doc XR+ system (Bio-Rad, Hercules, CA, USA). Each sample was examined in triplicate.
Gene Expression Omnibus (GEO) data extraction
Using GEO database (https://www.ncbi.nlm.nih.gov/geo/), gene expression data sets were obtained based on the clinical samples data of HCC. The heat map of gene expressions and correlation coefficient map were generated by R language. The data sets (GSE121711; GPL17586) including 18 samples were selected using Cor function in the R-language.
Statistical analyses
All in vitro experiments were performed in triplicate. All statistical calculations and analyses were performed using GraphPad Prism 7.0 software (GraphPad Software, San Diego, CA, USA). Student’s t-test was used to analyze differences between two groups. Data are presented as the mean ± standard deviation (SD). P < 0.05 was considered statistically significant.
Results
ADAMTS9-AS1 increased the proliferation of HCC cells in vitro
ADAMTS9-AS1 expression was observed in four HCC cell lines (HepG2, MHCC97-H, Hep3B, and SMCC-7721) and the LO2 normal liver cell line using qPCR. Except for SMCC-7721 cells, ADAMTS9-AS1 expression was higher in the remaining HCC cell lines, particularly in HepG2 and MHCC97-H cells when compared with the LO2 normal cell line (Figure S1). In addition, the transfection efficacy of ADAMTS9-AS1 overexpression or knockdown was examined in HepG2 and MHCC97-H cells, which suggested that compared to the pcDNA3.1 vector group, there was increased expression of ADAMTS9-AS1 in cells transfected with pcDNA3.1-AS1 (Figure S2A, S2B). However, in cells transfected with si1-AS1 or si2-AS1, ADAMTS9-AS1 expression decreased when compared to the si-NC groups (Figure S2C, S2D). The pcDNA3.1 vector, pcDNA3.1-AS1, si-NC, si1-AS1, and si2-AS1 were, therefore, selected for the following in vitro assays in the HepG2 and MHCC97-H cell lines.
CCK-8 and clone formation assays were then conducted in HepG2 and MHCC97-H cell lines to determine the influence of ADAMTS9-AS1 on proliferation. The cells were transfected with the pcDNA3.1 vector or pcDNA3.1-AS1 for ADAMTS9-AS1 overexpression experiments. CCK8 and clone formation assays showed that compared to the pcDNA3.1 vector group, proliferation was increased in cells transfected with pcDNA3.1-AS1 (Figure 1A, 1C). The cells were transfected with si-NC, si1-AS1, or si2-AS1 for ADAMTS9-AS1 knockdown experiments. CCK8 and clone formation assays showed that compared to the si-NC control group, proliferation was decreased in cells of the si1-AS1 and si2-AS1 transfected groups (Figure 1B, 1D). Together, these results suggested that ADAMTS9-AS1 expression increased the proliferation of HCC cells in vitro.
Figure 1.
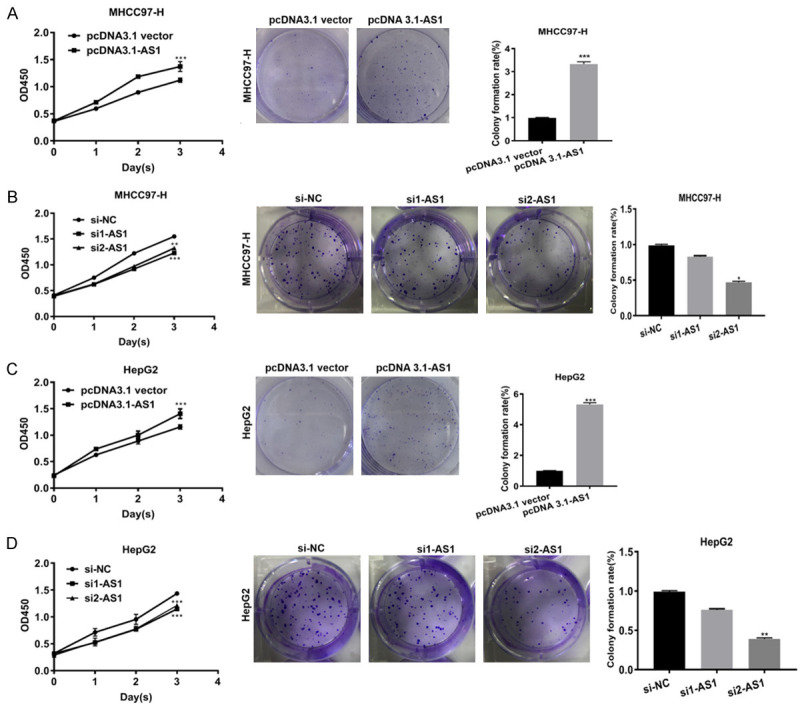
ADAMTS9-AS1 increased the proliferation of HCC cells in vitro. A, C. CCK-8 and clonogenic assays suggested that in HepG2 and MHCC97-H cells transfected with pcDNA3.1-AS1 groups, upregulated ADAMTS9-AS1 promoted proliferation of HCC cells. B, D. In cells transfected with si1-AS1, and si2-AS1 groups, downregulated ADAMTS9-AS1 inhibited proliferation of HCC cells. Error bars represented SD obtained from three independent experiments and all the data were shown as mean ± SD. CCK-8, Cell Counting Kit-8; ADAMTS9-AS1, ADAMTS9 antisense RNA 1; NC, negative control; si, short interfering RNA; *, P < 0.05; **, P < 0.01, ***, P < 0.001.
ADAMTS9-AS1 facilitated the migration of HCC cells in vitro
To further characterize the effect of ADAMTS9-AS1 on migration in HepG2 and MHCC97-H cell lines, scratch wound healing and transwell migration assays were separately conducted. The migration showed a difference in ADAMTS9-AS1 overexpression or knockdown experiments, suggesting that the migration was increased in cells transfected with pcDNA3.1-AS1 when compared with the corresponding pcDNA3.1 vector groups (Figure 2A, 2B, 2E, 2F). In contrast, the migration was suppressed in cells transfected with si1-AS1 or si2-AS1 when compared with the corresponding si-NC groups (Figure 2C, 2D, 2G, 2H). Together, the results showed that ADAMTS9-AS1 increased the migration of HCC cells in vitro.
Figure 2.
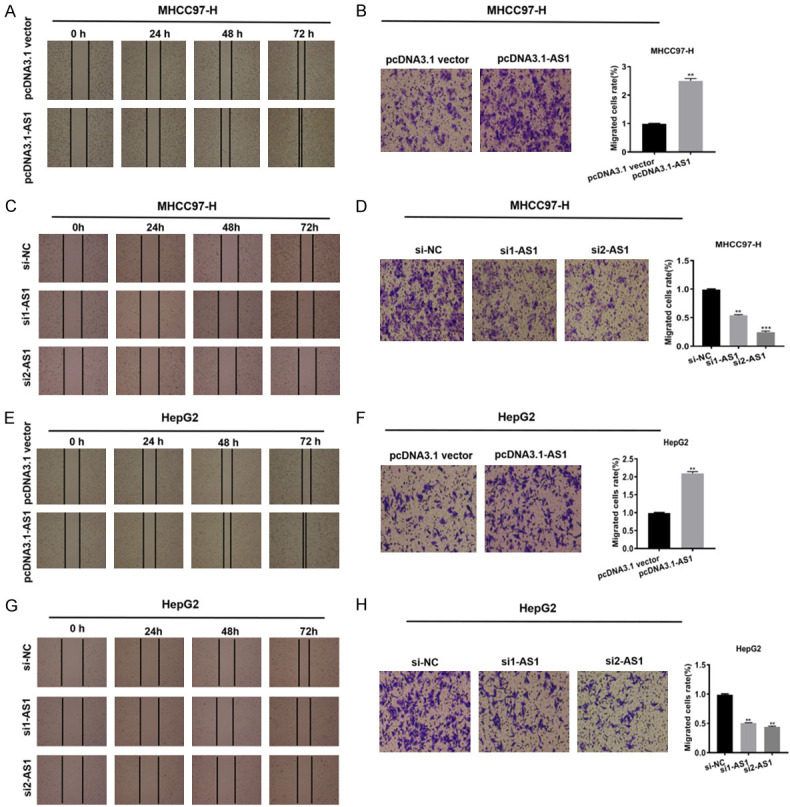
ADAMTS9-AS1 facilitated the migration of HCC cells in vitro. A, B, E, F. Scratch wound healing and transwell migration assays indicated that the migration ability was enhanced in HepG2 and MHCC97-H cells transfected with pcDNA3.1-AS1 compared with corresponding pcDNA3.1 vector groups. C, D, G, H. The migration ability was decreased in cells transfected with si1-AS1 compared with corresponding si-NC group, respectively. ADAMTS9-AS1, ADAMTS9 antisense RNA 1; NC, negative control; si, short interfering RNA; h, hour; **, P < 0.01, ***, P < 0.001.
ADAMTS9-AS1 increased the invasion in HCC cells in vitro
To test the effect of ADAMTS9-AS1 on invasion, transwell invasion assays using the HepG2 and MHCC97-H cell lines were conducted. In cells transfected with pcDNA3.1-AS1, ADAMTS9-AS1 overexpression promoted invasion (Figure 3A, 3C), while in cells transfected with si1-AS1 or si2-AS1, knockdown decreased invasion compared to the corresponding control groups (Figure 3B, 3D). Together, these results showed that ADAMTS9-AS1 increased the invasion capability of HCC cells.
Figure 3.
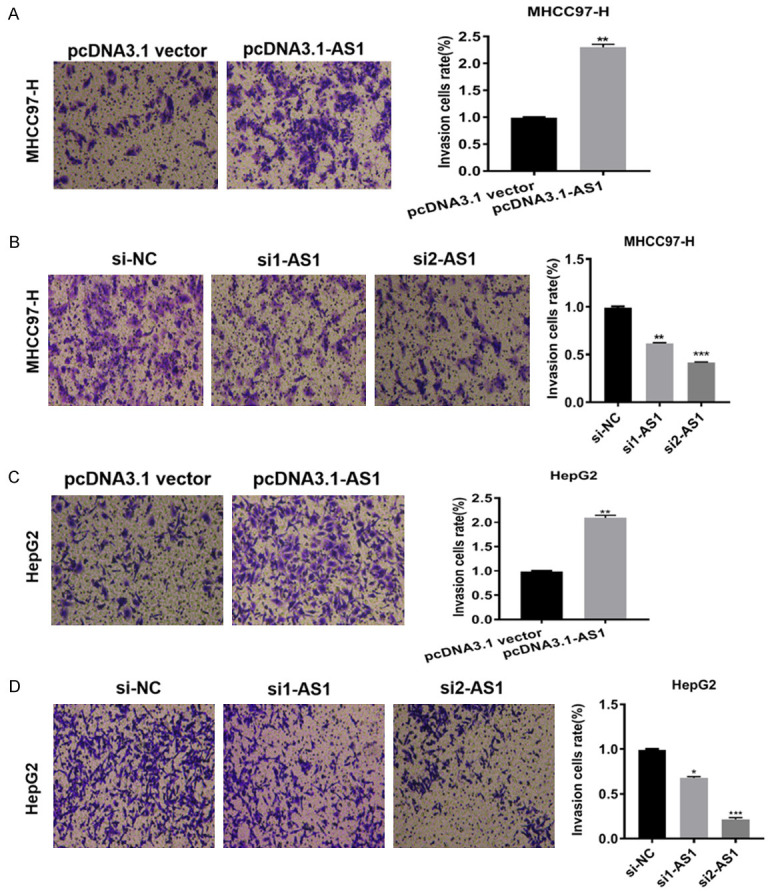
ADAMTS9-AS1 increased the invasion in HCC cells in vitro. A, C. Transwell invasion assays suggested that in HepG2 and MHCC97-H cells transfected with pcDNA3.1-AS1 groups, ADAMTS9-AS1 overexpression promoted the invasion ability. B, D. In cells transfected with si1-AS1 or si2-AS1 groups, ADAMTS9-AS1 knockdown curbed invasion ability compared with corresponding control groups. ADAMTS9-AS1, ADAMTS9-AS1 antisense RNA 1; NC, negative control; si, short interfering RNA; h, hour; *, P < 0.05; **, P < 0.01, ***, P < 0.001.
ADAMTS9-AS1 increased the PI3K/AKT/mTOR signaling pathway proteins in HCC cells in vitro
To increase our understanding of the functional effects of ADAMTS9-AS1 on cell proliferation, migration, and invasion, the potential mechanism of ADAMTS9-AS1 in HCC cells was determined. For starters, the data from GEO database suggested that ADAMTS9-AS1 has a weak correlation with PI3 and MTOR. And it was positively correlated with AKT (Figure S3). To further explore the potential relationship among them, the expression levels of PI3K/AKT/mTOR signaling pathway-related proteins (AKT, p-AKT, PIK3CB, mTOR, and p-mTOR) were determined using western blotting. ADAMTS9-AS1 overexpression or knockdown suggested that except for AKT and mTOR, expression levels of p-AKT, PIK3CB, and p-mTOR were increased in cells transfected with the pcDNA3.1-AS1 (Figure 4A, 4C), while there was decreased expression in cells transfected with the si1-AS1 or si2-AS1, when compared with their corresponding control cells (Figure 4B, 4D). Together, the results suggest that ADAMTS9-AS1 increased the PI3K/AKT/mTOR signaling pathway in HCC cells in vitro.
Figure 4.
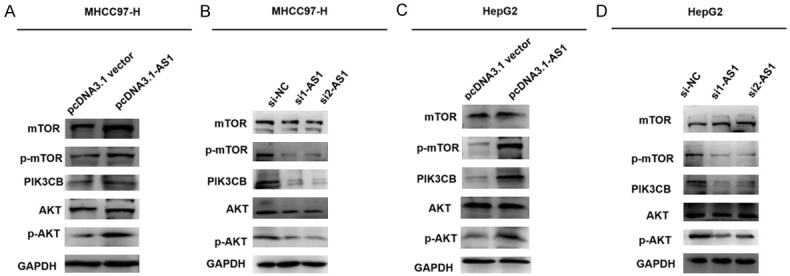
ADAMTS9-AS1 increased the PI3K/AKT/mTOR signaling pathway proteins in HCC cells in vitro. A, C. In HepG2 and MHCC97-H cells transfected with pcDNA3.1-AS1 groups, it suggested that overexpression of ADAMTS9-AS1 increased the expression levels of p-AKT, PIK3CB, and p-mTOR compared to their corresponding pcDNA3.1 vector groups. B, D. In HepG2 and MHCC97-H cells transfected with si1-AS1 or si2-AS1 groups, knockdown of ADAMTS9-AS1 decreased p-AKT, PIK3CB, and p-mTOR expression levels compared to corresponding their pcDNA3.1 vector groups. ADAMTS9-AS1, ADAMTS9 antisense RNA 1; p-AKT, phosphorylated AKT; p-mTOR, phosphorylated mTOR; NC, negative control; si, short interfering RNA.
ADAMTS9-AS1 decreased autophagy and apoptosis in HCC cells in vitro
As previously mentioned, ADAMTS9-AS1 activated the PI3K/AKT/mTOR signaling pathway. Previous studies have also reported that aberrant PI3K/AKT/mTOR signaling is closely related to apoptosis and autophagy [33,34]. Based on this information, we speculated that ADAMTS9-AS1 might decrease autophagy and apoptosis in HCC cells. To explore the possibility that ADAMTS9-AS1 interfered with autophagy and apoptosis, levels of key autophagy-related proteins (LC3-I, LC3-II, BECN1, and SQSTM1) and apoptotic proteins (pro-apoptotic Bax and anti-apoptotic Bcl-2) were determined using western blotting. Based on the results from the ADAMTS9-AS1 overexpression and knockdown experiments, protein levels of LC3-II, BECN1, and pro-apoptotic Bax were decreased in the pcDNA3.1-AS1 groups (Figure 5A, 5C), but were increased in the si1-AS1 or si2-AS1 groups, when compared with the corresponding control cell groups (Figure 5B, 5D). The results also showed that ADAMTS9-AS1 expression was negatively related with ADAMTS9. Overall, the results showed that ADAMTS9-AS1 expression decreased autophagy and apoptosis in HCC cells in vitro.
Figure 5.
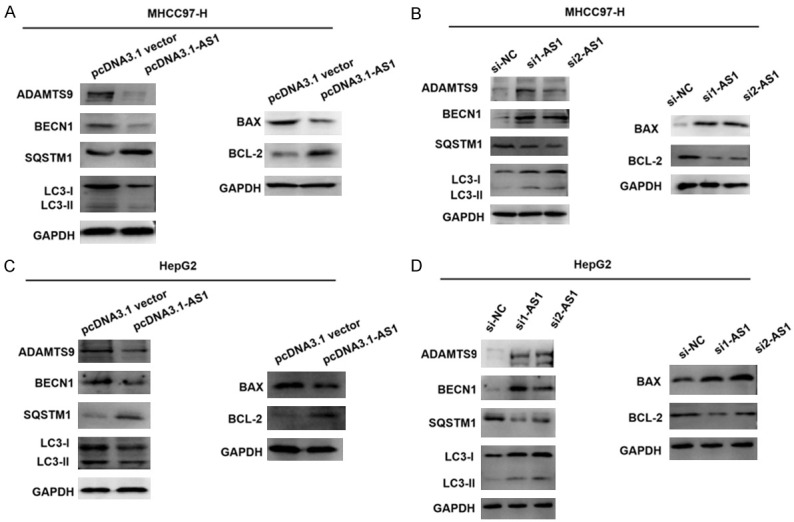
ADAMTS9-AS1 decreased autophagy and apoptosis in HCC cells in vitro. A, C. Western blot assays was conducted to examine the protein levels of autophagy and apoptosis-related proteins in HCC cells. Compared with corresponding control groups, the expression of LC3-II, BECN1, and pro-apoptotic Bax decreased in pcDNA ADAMTS9-AS1 groups compared to corresponding control groups, and no significant change was found in LC3-I expression. Besides, decrease of ADAMTS9 was also found in pcDNA ADAMTS9-AS1 groups. B, D. Increase of ADAMTS9, LC3-II, BECN1, and pro-apoptotic Bax was identified in si1-AS1 or si2-AS1 groups as compared with corresponding control groups. ADAMTS9-AS1, ADAMTS9 antisense RNA 1; NC, negative control; si, short interfering RNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
HCC is a common cause of cancer-related deaths, and patients with advanced or metastatic HCC have been reported in past studies [35,36]. Despite various therapies such as surgery, chemotherapy, and radiotherapy, patients rarely have positive prognoses [37-40]. An increasing number of lncRNAs have exhibited effects on tumorigenesis in many types of tumors [41-43]. Studies have also identified diverse roles of lncRNAs in HCC. F11-AS1 inhibits HBV-related HCC [44], but AURKAPS1 and ANCR potentiate HCC progression [45,46]. However, the exact mechanisms responsible for HCC pathogenesis remain to be fully understood.
ADAMTS9-AS1 has recently been identified as a cancer-related lncRNA and is the antisense of ADAMTS9 [22]. The functions of ADAMTS9 in HCC are widely known and some reports suggest that ADAMTS9-AS1 is associated with several tumors [22-25]; therefore, we investigated its possible role and potential mechanism in HCC. The qPCR assays showed that compared to the LO2 normal cell line, ADAMTS9-AS1 expression was higher in three HCC cell lines, including MHCC97-H and HepG2 cells. Therefore, functional assays were conducted in MHCC97-H and HepG2 cells. Based on the CCK-8 assay, clone formation assay, scratch wound healing, transwell migration, and invasion assays, ADAMTS9-AS1 enhanced the proliferation, migration, and invasion in MHCC97-H and HepG2 cells.
It is important to note that a few lncRNAs, such as CDKN2B-AS1 [47], DUXAP10 [48], and DCST1-AS1 [49], regulate HCC progression by regulating the PI3K/AKT/mTOR signaling pathway. However, one study reported that ADAMTS9 was considered a tumor suppressor because it regulated the AKT/mTOR signaling pathway in gastric cancer [32]. Our findings suggest that ADAMTS9-AS1 achieved its functional roles by interacting with the PI3K/AKT/mTOR signaling pathway in HCC. ADAMTS9-AS1 was negatively related with ADAMTS9. Based on the GEO analysis, we found that ADAMTS9-AS1 has a weak correlation with PI3 and mTOR and was positively correlated with AKT. The expression levels of proteins related to the PI3K/AKT/mTOR signaling pathway were therefore determined using western blot assays in HCC cells. We showed that ADAMTS9-AS1 promoted the expression of p-AKT, PIK3CB, and p-mTOR in HCC cells, which suggested that ADAMTS9-AS1 facilitated the proliferation, migration, and invasion via the PI3K/AKT/mTOR signaling pathway.
Typically, activation of the PI3K/AKT/mTOR signaling pathway is associated with apoptosis and autophagy [27,28]. To further characterize the potential mechanism, the effects of ADAMTS9-AS1 on apoptosis and autophagy were evaluated. Western blot analysis results suggested that ADAMTS9-AS1 reduced the expressions of autophagy proteins (LC3-II and BECN1) and the pro-apoptotic protein Bax, but increased the expression of SQSTM1 and the anti-apoptotic protein Bcl-2 in HCC cells. Our findings suggested that ADAMTS9-AS1 enhanced cell proliferation, migration, and invasion in HCC by promoting the PI3K/AKT/mTOR pathway to affect autophagy and apoptosis at the protein expression level.
Although our results highlight the biological functions of ADAMTS9-AS1 in HCC, study limitations did exist. The current research was only performed in vitro. ADAMTS9-AS1 was adversely related to ADAMTS9 and participated in the PI3K/AKT/mTOR signaling pathway, whereas the specified mechanism of regulation in HCC requires further studies including more clinical samples. It is possible that there may be other participants that affect the behavior of ADAMTS9-AS1 in HCC that will be investigated in future studies to further understand the roles of ADAMTS9-AS1 in HCC.
Conclusion
The biological role of ADAMTS9-AS1 in HCC cells was investigated. The findings showed that ADAMTS9-AS1 may facilitate cell proliferation, migration, and invasion ability by triggering the PI3K/AKT/mTOR signaling pathway against autophagy and apoptosis. Overall, our study provides novel insights into ADAMTS9-AS1, which is considered a potential therapeutic candidate in the treatment of HCC.
Acknowledgements
Youth Program of National Natural Science Foundation of China (Grant No. 81703875).
Disclosure of conflict of interest
None.
Abbreviations
- lncRNA
long non-coding RNA
- ADAMTS9
ADAM metallopeptidase with thrombospondin type 1 motif, 9
- ADAMTS9-AS1
ADAMTS9 antisense RNA 1
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- siRNA
short interfering RNA
- qPCR
quantitative real-time polymerase chain reaction
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- OD
optical density
- mTOR
kinase mammalian target of rapamycin
Supporting Information
References
- 1.Berndt N, Eckstein J, Heucke N, Gajowski R, Stockmann M, Meierhofer D, Holzhütter HG. Characterization of lipid and lipid droplet metabolism in human hCC. Cells. 2019;8:512. doi: 10.3390/cells8050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon J, Ouro A, Ala-Ibanibo L, Presa N, Delgado TC, Martínez-Chantar ML. Sphingolipids in non-alcoholic fatty liver disease and hepatocellular carcinoma: ceramide turnover. Int J Mol Sci. 2019;21:40. doi: 10.3390/ijms21010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayoub WS, Steggerda J, Yang JD, Kuo A, Sundaram V, Lu SC. Current status of hepatocellular carcinoma detection: screening strategies and novel biomarkers. Ther Adv Med Oncol. 2019;11:1758835919869120. doi: 10.1177/1758835919869120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin S, Hoffmann K, Schemmer P. Treatment of hepatocellular carcinoma: a systematic review. Liver Cancer. 2012;1:144–158. doi: 10.1159/000343828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng T, Xie F, Geng L, Sui CJ, Dai DH, Shen RX, Yan L, Yang JM. Safety and long-term outcomes of anatomic left hepatic trisectionectomy for intermediate and advanced hepatocellular carcinoma. Gastroenterol Hepatol. 2015;30:1015–23. doi: 10.1111/jgh.12887. [DOI] [PubMed] [Google Scholar]
- 6.Tian M, Zhao B, Martin FL, Morais CLM, Liu L, Huang Q, Zhang J, Shen H. Gene-environment interactions between GSTs polymorphisms and targeted epigenetic alterations in hepatocellular carcinoma following organochlorine pesticides (OCPs) exposure. Environ Int. 2020;134:105313. doi: 10.1016/j.envint.2019.105313. [DOI] [PubMed] [Google Scholar]
- 7.Li WT, Zou AE, Honda CO, Zheng H, Wang XQ, Kisseleva T, Chang EY, Ongkeko WM. Etiology-specific analysis of hepatocellular carcinoma transcriptome reveals genetic dysregulation in pathways implicated in immunotherapy efficacy. Cancers. 2019;11:1273. doi: 10.3390/cancers11091273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and regulation of long noncoding rnas in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi: 10.1158/0008-5472.CAN-19-0255. [DOI] [PubMed] [Google Scholar]
- 9.Harris WP, Wong KM, Saha S, Dika IE, Abou-Alfa GK. Biomarker-driven and molecular targeted therapies for hepatobiliary cancers. Semin Oncol. 2018;45:116–123. doi: 10.1053/j.seminoncol.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y, Li C, Chen J, Zhang K, Chu X, Wang R, Chen L. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem. 2016;40:219–229. doi: 10.1159/000452539. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Peng D, Sood AK, Dang CV, Zhong X. Shedding light on the dark cancer genomes: long noncoding RNAs as novel biomarkers and potential therapeutic targets for cancer. Mol Cancer Ther. 2018;17:1816–1823. doi: 10.1158/1535-7163.MCT-18-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi L, Hong X, Ba L, He X, Xiong Y, Ding Q, Yang S, Peng G. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10:150. doi: 10.1038/s41419-019-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, He W, Huang J, Wang B, Li H, Cai Q, Su F, Bi J, Liu H, Zhang B, Jiang N, Zhong G, Zhao Y, Dong W, Lin T. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9:3826. doi: 10.1038/s41467-018-06152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao M, Xin XF, Zhang JY, Dai W, Lv TF, Song Y. LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. Cancer Med. 2020;9:1196–1208. doi: 10.1002/cam4.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M, Li X, Li J, Mi C, Zhang J, Lu B, Chen E, Liu P, Lu Y. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18:171. doi: 10.1186/s12943-019-1107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, Guo R, Si J, Li L, Xue J, Shao ZM, Wu ZH, Huang S, Wu J. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18:187. doi: 10.1186/s12943-019-1115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Li D, Huang D, Song H, Mei H, Fang E, Wang X, Yang F, Zheng L, Huang K, Tong Q. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther. 2018;26:755–773. doi: 10.1016/j.ymthe.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Xu J, Zhang R, Zhao J. The novel long noncoding RNA TUSC7 inhibits proliferation by sponging miR-211 in colorectal cancer. Cell Physiol Biochem. 2017;41:635–644. doi: 10.1159/000457938. [DOI] [PubMed] [Google Scholar]
- 19.Pu J, Wang J, Wei H, Lu T, Wu X, Wu Y, Shao Z, Luo C, Lu Y. lncRNA MAGI2-AS3 prevents the development of HCC via recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 promoter. Mol Ther Nucleic Acids. 2019;18:351–362. doi: 10.1016/j.omtn.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Liu J, Zou R, Cheng P, Su Y. Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. J Exp Clin Cancer Res. 2019;38:189. doi: 10.1186/s13046-019-1193-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Xu Y, Luo X, He W, Chen G, Li Y, Li W, Wang X, Lai Y, Ye Y. Long non-coding RNA PVT1/miR-150/HIG2 axis regulates the proliferation, invasion and the balance of iron metabolism of hepatocellular carcinoma. Cell Physiol Biochem. 2018;49:1403–1419. doi: 10.1159/000493445. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Zhang C, Wu Y, He W, Gou X. Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in bladder urothelial carcinoma. Cancer Cell Int. 2019;19:327. doi: 10.1186/s12935-019-1052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JJ, Huang YQ, Song W, Li YF, Wang H, Wang WJ, Huang M. Comprehensive analysis of the lncRNA-associated competing endogenous RNA network in breast cancer. Oncol Rep. 2019;42:2572–2582. doi: 10.3892/or.2019.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li QW, Ma L, Qiu B, Yang H, Zhu YJ, Qiang MY, Liu SR, Chen NB, Guo JY, Cai LZ. Differential expression profiles of long noncoding RNAs in synchronous multiple and solitary primary esophageal squamous cell carcinomas: a microarray analysis. J Cell Biochem. 2019;120:2439–2453. doi: 10.1002/jcb.27536. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J, Lv M, Gu Y, Zhang J, Hua X. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci Rep. 2016;6:38983. doi: 10.1038/srep38983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan KH, Yap TA, Yan L, Cunningham D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J Cancer. 2013;32:253–65. doi: 10.5732/cjc.013.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owusu-Brackett N, Shariati M, Meric-Bernstam F. Role of PI3K/AKT/mTOR in cancer signaling. Predictive Biomarkers in Oncology. Springer; 2019. pp. 263–270. [Google Scholar]
- 28.Yang J, Pi C, Wang G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed Pharmacother. 2018;103:699–707. doi: 10.1016/j.biopha.2018.04.072. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Fu Q, Man W, Guo H, Yang P. LncRNA OR3A4 participates in the angiogenesis of hepatocellular carcinoma through modulating AGGF1/akt/mTOR pathway. Eur J Pharmacol. 2019;849:106–114. doi: 10.1016/j.ejphar.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 30.Zhong JH, Xiang X, Wang YY, Liu X, Qi LN, Luo CP, Wei WE, You XM, Ma L, Xiang BD. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2020;235:1090–1102. doi: 10.1002/jcp.29023. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu J, Lv Y, Zhang C, Guo S. LncRNA meg3 suppresses hepatocellular carcinoma in vitro and vivo studies. Am J Transl Res. 2019;11:4089–4099. [PMC free article] [PubMed] [Google Scholar]
- 32.Du W, Wang S, Zhou Q, Li X, Chu J, Chang Z, Tao Q, Ng EK, Fang J, Sung JJ, Yu J. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32:3319–3328. doi: 10.1038/onc.2012.359. [DOI] [PubMed] [Google Scholar]
- 33.Zhang DM, Liu JS, Deng LJ, Chen MF, Yiu A, Cao HH, Tian HY, Fung KP, Kurihara H, Pan JX. Arenobufagin, a natural bufadienolide from toad venom, induces apoptosis and autophagy in human hepatocellular carcinoma cells through inhibition of PI3K/Akt/mTOR pathway. Carcinogenesis. 2013;34:1331–1342. doi: 10.1093/carcin/bgt060. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Dong H, Li M, Wu Y, Liu Y, Zhao Y, Chen X, Ma M. Honokiol induces autophagy and apoptosis of osteosarcoma through PI3K/Akt/mTOR signaling pathway. Mol Med Rep. 2018;17:2719–2723. doi: 10.3892/mmr.2017.8123. [DOI] [PubMed] [Google Scholar]
- 35.Simonetti R, Liberati A, Angiolini C, Pagliaro L. Treatment of hepatocellular carcinoma: a systematic review of randomized controlled trials. Ann Oncol. 1997;8:117–136. doi: 10.1023/a:1008285123736. [DOI] [PubMed] [Google Scholar]
- 36.Uka K, Aikata H, Takaki S, Shirakawa H, Jeong SC, Yamashina K, Hiramatsu A, Kodama H, Takahashi S, Chayama K. Clinical features and prognosis of patients with extrahepatic metastases from hepatocellular carcinoma. World J Gastroenterol. 2007;13:414–20. doi: 10.3748/wjg.v13.i3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farmer DG, Rosove MH, Shaked A, Busuttil RW. Current treatment modalities for hepatocellular carcinoma. Ann Surg. 1994;219:236–47. doi: 10.1097/00000658-199403000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, He P, Xie X, Sun C. Knockdown of SUMO1P3 represses tumor growth and invasion and enhances radiosensitivity in hepatocellular carcinoma. Mol Cell Biochem. 2019;450:125–134. doi: 10.1007/s11010-018-3379-8. [DOI] [PubMed] [Google Scholar]
- 39.Tang S, Tan G, Jiang X, Han P, Zhai B, Dong X, Qiao H, Jiang H, Sun X. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget. 2016;7:73257. doi: 10.18632/oncotarget.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Bisceglie AM, Rustgi VK, Hoofnagle JH, Dusheiko GM, Lotze MT. Hepatocellular carcinoma. Ann Intern Med. 1988;108:390–401. doi: 10.7326/0003-4819-108-3-390. [DOI] [PubMed] [Google Scholar]
- 41.Li N, Yang G, Luo L, Ling L, Wang X, Shi L, Lan J, Jia X, Zhang Q, Long Z, Liu J, Hu W, He Z, Liu H, Liu W, Zheng G. LncRNA THAP9-AS1 promotes pancreatic ductal adenocarcinoma growth and leads to a poor clinical outcome via sponging miR-484 and interacting with YAP. Clin Cancer Res. 2020;26:1736–1748. doi: 10.1158/1078-0432.CCR-19-0674. [DOI] [PubMed] [Google Scholar]
- 42.Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14:309–328. doi: 10.1002/1878-0261.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan S, Shen M, Zhou M, Shi X, He R, Yin T, Wang M, Guo X, Qin R. Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis. 2019;10:883. doi: 10.1038/s41419-019-2123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng Y, Wei Z, Huang M, Xu G, Wei W, Peng B, Nong S, Qin H. Long non-coding RNA F11-AS1 inhibits HBV-related hepatocellular carcinoma progression by regulating NR1I3 via binding to microRNA-211-5p. J Cell Mol Med. 2020;24:1848–1865. doi: 10.1111/jcmm.14881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Guo W, Xue W, Xu P, Deng Z, Zhang D, Zheng S, Qiu X. Long noncoding RNA AURKAPS1 potentiates malignant hepatocellular carcinoma progression by regulating miR-142, miR-155 and miR-182. Sci Rep. 2019;9:19645. doi: 10.1038/s41598-019-56036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen Z, Lian L, Ding H, Hu Y, Xiao Z, Xiong K, Yang Q. LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 2020;17:381–394. doi: 10.1080/15476286.2019.1708547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. doi: 10.1016/j.canlet.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 48.Sun L, Wang L, Chen T, Yao B, Wang Y, Li Q, Yang W, Liu Z. microRNA-1914, which is regulated by lncRNA DUXAP10, inhibits cell proliferation by targeting the GPR39-mediated PI3K/AKT/mTOR pathway in HCC. J Cell Mol Med. 2019;23:8292–8304. doi: 10.1111/jcmm.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Zhai D, Huang Q, Chen H, Zhang Z, Tan Q. LncRNA DCST1-AS1 accelerates the proliferation, metastasis and autophagy of hepatocellular carcinoma cell by AKT/mTOR signaling pathways. Eur Rev Med Pharmacol Sci. 2019;23:6091–6104. doi: 10.26355/eurrev_201907_18423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


