Abstract
Dysregulation of Hippo signaling by long non-coding RNA (lncRNA) contributes to colon adenocarcinoma (COAD) progression, while the underlying mechanisms remain elusive. Our study shows that lncRNA USP2-AS1 is a Yes-associated protein 1 (YAP1) binding lncRNA, and inactivates Hippo signaling in COAD cells. Moreover, our data indicated that USP2-AS1 lowered the phosph-YAP (S127), elevated the total level of YAP1, and triggered the expression of downstream target genes in COAD cells. The loss- and gain-of function assays demonstrated that USP2-AS1 promotes cellular proliferation and metastasis of COAD cells. Clinically, the USP2-AS1 levels were significantly elevated in COAD tissues and were positively correlated with tumor grade, size, and TNM stage. Collectively, these findings demonstrated that USP2-AS1 modulates and regulates Hippo signaling in COAD and could be a valuable therapeutic target.
Keywords: lncRNA, USP2-AS1, cell proliferation, metastasis, Hippo signaling
Introduction
Colon adenocarcinoma (COAD) is the third leading cause of cancer-related mortalities in the world. Cancer mortality rates have been increasing gradually. Diagnostic and treatment advancements have only been of benefit to a few patients [1]. There is an urgent need to understand the molecular characteristics of this tumor and uncover new therapeutic targets. Hippo signaling is a highly conserved regulator of organ size, stem cell proliferation and maintenance. Its a modulator of several pathways in cancers, including COAD [2-4]. Core Hippo components include the kinases MST1/2 and LATS1/2. Activated LATS1/2 kinases phosphorylate YAP (Yes-associated protein) thereby inactivating it through cytoplasmic sequestration and degradation. Phosphorylation of YAP and a transcriptional co-activator with a PDZ-binding motif (TAZ; also known as WWTR1) inhibits its activity by creating 14-3-3 binding sites that allow their accumulation in the cytoplasm. This stimulates their ubiquitin-mediated proteolysis [5]. Contrarily, dephosphorylated YAP promotes cellular proliferation and organ growth that is primarily mediated by the TEAD (TEA domain) family transcription factors [6]. The YAP and TAZ are closely related paracyclic homologous substances that mediate the downstream effects of Hippo signal transductions. They are transcriptional co-activators that shuttle between the cytoplasm and the nucleus. They induce cellular proliferation and expression of anti-apoptotic genes. This is accomplished through their interactions with transcription factors, particularly members of the TEAD family. When the inhibitory Hippo kinase module “turns on”, LATS1 and LATS2 phosphorylate and inactivate YAP and TAZ, thereby blocking the production of output genes [7].
Long non-coding RNA (lncRNA) are long RNA transcripts that are more than 200 nucleotides in length without any coding potential [8]. Studies have demonstrated that lncRNAs serve important roles in carcinogenesis and cancer metastasis. Uncharacteristic expression of lncRNAs have been implicated in COAD progression [9-11]. For instance, lncRNA H19 modulates the expression of multiple genes involved in EMT (epithelial-mesenchymal transition) by acting as a competing endogenous RNA in colorectal cancer [12]. The human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC loci [13]. lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance through miR-181a-5p-mediated regulation of the Wnt/β-catenin signaling pathway [14]. In colorectal cancer, the lncRNA GAS5 interacts with the WW domain of YAP to facilitate the translocation of endogenous YAP from the nucleus to the cytoplasm. Furthermore, GAS5 also promotes the phosphorylation of YAP that leads to ubiquitin-mediated YAP degradation and ultimately inhibits colorectal cancer progression [16]. Equally, lncRNA KCNA3 inhibits colorectal cancer progression by down-regulating the level of YAP [16]. By interacting with LATS1 or recruiting YAP to the nucleus to modulate the Hippo signaling pathway, lncRNAs can serve as oncogenes in other types of cancers [17,18].
In the current study, we report that a previously unrecognized lncRNA USP2-AS1 plays an oncogenic role in COAD progression. We demonstrate that USP2-AS1 binds to YAP1 and inactivates Hippo signaling by downregulating phosphorylated YAP in COAD cells.
Materials and methods
Clinical specimens
Forty three samples of fresh paired colon cancer tissues were collected from surgical resections. This was done at the Department of Digestive Surgery at the First Affiliated Hospital of Zhengzhou University. Healthy tissues were also collected at a distance of 2 cm from the malignant tissues. The inclusion criteria for the study participants were: (i) aged 30-70; (ii) diagnostically free from other types of tumors or diseases; and (iii) without preoperative chemotherapy or radiation therapy. All tissues were obtained under sterile conditions, fast-frozen in liquid nitrogen, and stored at -80°C. Patients were provided with an informed consent before the samples could be obtained. Ethical approvals were obtained from the ethical review committee of Zhengzhou University.
Cell cultures
Normal human colon epithelial cells (NCM460), and colon cancer cell lines (SW480, SW620, and Lovo) were purchased from ATCC (American Type Culture Collection, Manassas, VA). They were cultured in ATCC recommended media, supplemented with 10% FBS (fetal bovine serum, sigma aldrich), 100 units/mL penicillin, and 100 µg/ml streptomycin (GIBCO).
Lentivirus infection and establishment of stable cell lines
The USP2-AS1 (pLVX-USP2-AS1) or vector (pLVX) were purchased from GeneCopoeia (Guangzhou, China), while shRNA targeting USP2-AS1 (sh#1, sh#2) or the negative control (NC) were obtained from RiboBio (RiboBio, China). Lentivirus were cultured in HEK-293T cells and collected from the supernatant at 24 and 48 hours post-transfection. These lentiviruses were then introduced into the SW480, SW620, or Lovo cells before selction using 2 μg/mL of puromycin (Millipore, USA) after 48 hours.
Quantitative real-time PCR (qRT-PCR)
Total RNAs were isolated from tissues and cells using TRIzol reagent (Invitrogen, USA) following the manufacturer’s instructions. 1 µg of total RNA was subjected to reverse transcription with the MMLV system (Promega, USA). Quantitative real-time PCR (qRT-PCR) assays were then performed using an ABI 7500 real-time RT-PCR system with SYBR® Green Real-time PCR Master Mix (ABI, USA). The 18S RNA was used as the reference. Primer sequences used are shown in Table 1.
Table 1.
Primer Sequenes and shRNAs
| Name | Sequence (5’-3’) |
|---|---|
| USP2-AS1 primer | F: GGGAGAGGCAGAGAGATCCA |
| R: CAAGGGAGCAGGACAGTGAG | |
| GAPDH primer | F: GTAACCCGTTGAACCCCATT |
| R: CCATCCAATCGGTAGTAGCG | |
| U1 primer | F: TCCTCTCCAAAATGCCAGAG |
| R: GGCGGATTGGAAATGCTT | |
| CTGF primer | F: CAGCATGGACGTTCGTCTG |
| R: AACCACGGTTTGGTCCTTGG | |
| CYR61 | F: CTCGCCTTAGTCGTCACCC |
| R: CGCCGAAGTTGCATTCCAG | |
| SOX9 | F: AGCGAACGCACATCAAGAC |
| R: CTGTAGGCGATCTGTTGGGG | |
| Sh-#1 | CCGGTTCAGCGATATGAAGTTAAATCTCGAGATTTAACTTCATATCGCTGAATTTTTG |
| Sh-#2 | CCGGCTCAGCGAGATGAAGAGTTTACTCGAGTAAACTCTTCATCTCGCTGAGTTTTTG |
Western blot analysis
Marked cells were seeded into a 6 cm plate at a concentration of 5 × 105 cells per plate for 48 hours. Cell lysates were subjected to an SDS-PAGE electrophoretic system. They were then transferred onto a nitrocellulose membrane and blocked with 5% skim milk at room temperature for 1 hour. The resulting mixture was incubated with primary antibodies overnight at 4°C. The membranes were then washed with PBS buffer and incubated with anti-mouse or rabbit IgG-HRP. The p-LAST1 (CST) and LAST1 (CST), LAST2 (Abcam), p-YAP (Abcam), YAP1 (Abcam), and GAPDH (Proteintech) antibodies served as loading controls.
Cellular colony formation assays
The indicated cells (1000 cells/3 ml) were seeded into 6-well plates and incubated for 14 days at 37°C and 5% CO2. The culture medium was changed after every 2 days. On the 14th day, colonies were washed once using phosphate-buffered saline and fixed with 4% paraformaldehyde for 20 min. Thereafter, the cells were stained with crystal violet, and the number of clones per well counted. All assays were conducted in triplicate, and their mean values calculated.
Transwell invasion assay
Marked cells were seeded into 6-well plates at a density of 5 × 103 cells per well. After 24 hours, shRNA, and pLVX or pLVX-USP2-AS1 were transferred and incubated for 48 hours. Cells were then harvested and transferred into a serum-free media with cell densities adjusted to 4 × 105 cells/ml. Approximately 100 µL of cells were transferred to the upper chamber. The bottom of the upper chamber was coated with 1 mg/ml fibronectin (Millipore; Bedford, CA, USA), while the lower chamber was covered with a medium containing 20% FBS. After incubation at 37°C for 48 hours, cells were gently removed from the upper chamber using cotton swabs. The chamber was fixed using methanol for 15 min, dried, and stained with 0.1% crystal violet. Five fields of vision were randomly selected, and the number of cells in each field was counted under an inverted microscope. All experiments were done in triplicates.
Cytoplasm and nuclear isolation
The SW620 cells (1 × 107) were used for isolation using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo) as per the manufacturer’s instructions. The cytoplasmic lysate and nuclear pellet were added to 1 ml Trizol for RNA extraction. Fractionated RNAs were used for cDNA synthesis and qRT-PCR.
The RNA-FISH assays
Subcellular distribution of USP2-AS1 was detected using RNA FISH. Briefly, cells were fixed with 4% PFA for 15 min, followed by permeabilization with 0.5% Triton X-100 for 5 min on ice. They were then incubated with RNA-FISH probes in a hybridization buffer at 37°C overnight. The nuclei were counterstained with DAPI. FISH probes were purchased from RiboBio (RiboBio, China).
RNA pull-down assay
Both USP2-AS1 and USP2-AS1-AS were in vitro transcribed with Biotin-RNA Transcription Kit (Roche), and purified with TRIzol reagent (Invitrogen). In vitro transcripted RNAs were incubated with RNA structure buffer (10 mM Tris-HCl, pH = 7, 0.1 M KCl, and 10 mM MgCl2), and heated at 72°C for 2 min to yield suitable secondary structures. The resulting RNAs were incubated with SW480 cell lysates at 4°C for 4 hours, followed by incubation with Streptavidin beads (Thermo) at room temperature for 1 hour. They were then washed 5 times, afterwhich the pull-down complexes were analyzed using Western blot.
The RNA immunoprecipitation (RIP) assay
Briefly, SW620 cells (2 × 107) were crosslinked with 0.3% formaldehyde medium for 10 min at room temperature followed by neutralization with 250 mM glycine for 5 min. Cells were washed twice using cold PBS, lysed in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% NP-40 and 0.5% sodium deoxycholate, 0.5 mM dithiothreitol, RNase inhibitor, and protease inhibitor cocktail), followed by sonication on ice and DNase treatment. Resulting lysates were pre-cleared with protein A/G beads for 30 min and incubated with Flag antibody (BD bioscience) or equivalent mouse IgG (GeneTex) at 4°C overnight. The antibodies were then precipitated with protein A/G beads by incubation. Cells were then washed for 5 × 10 min, and the RNA extracted using Trizol reagent (Invitrogen) and quantified using Qrt-PCR. The obtained protein samples were subjected to Western blots analysis.
Subcutaneous tumor formation assay
Marked stable cells (2 × 106) were subcutaneously injected into at least 5 parts of two flanks of 6 weeks old female BALB/c nude mice (Linchang biotech). After 24 days, the mice were sacrificed and the tumors were photographed. The use of mice in the experiments was approved by the Institutional Animal Care and Use Committee.
Tail vein injection
The Lovo-shNC/sh#2-Luc cells (2 × 106) in 100 ml PBS were injected into the tail veins of 6 weeks old female nu/nu mice. After the initial injection, mice were monitored weekly for lung metastases using an IVIS Lumina Kinetic optical imaging system with an EMCCD camera (PerkinElmer, Waltham, MA, USA). At autopsy, the lungs were collected, fixed in 10% formalin, embedded in paraffin (FFPE), sectioned into 5 mm slices at 100 mm intervals and finally stained with H&E.
Immunohistochemistry (IHC) staining
Tumor slides were stained based on standard Immunohistochemistry (IHC) protocols (Reference). The slides were blocked with 10% bovine serum albumin (BSA, Sangon) for 1 hour and incubated with the PCNA antibody (Abcam) at 4°C overnight. They were then incubated with a HRP-labeled secondary antibody (Dako) for 1 hour at room temperature. The diaminobenzidine substrate chromogen (DAB) was used for detection. All slides were counterstained with hematoxylin before dehydration and mounting.
Luciferase reporter assay
Cells (10 × 104) were seeded in triplicates in 12-well plates and cultured for 24 hours. The luciferase reporter assay was performed as described [19]. Cells were then transfected with 100 ng HOP-Flash (83467, Addgene) or HIP-Flash luciferase reporter plasmid (83466, Addgene), plus 5 ng pRL-TK Renilla plasmid (Promega) and Lipofectamine 2000 (Invitrogen) as per the manufacturer’s recommendations. Luciferase and Renilla signals were measured at 48 hours post-transfection using a Dual-Luciferase Reporter Assay Kit (Promega), based on the manufacturer’s protocols.
Statistical analysis
Data are presented as mean ± SD. Statistical analyses were performed with Graphpad Prism software (version 8.0). Statistical significance was compared via unpaired Student’s t test or one-way analysis of variance (ANOVA) followed with Bonferroni’s multiple as appropriate. Differences were considered statistically significant at the level of P < 0.05. Kaplan-Meier survival analysis were determined by log rank test.
Results
USP2-AS1 is highly expressed in colon cancer and closely correlated with COAD progression
Wen et al. performed RIP-seq assay and revealed that lncRNA GAS5 binds to YAP1. The binding promoted YAP1 degradation, thereby inhibiting colorectal cancer progression [15]. We analyzed their RIP-seq dataset and found that there are mounts of natural antisense lncRNAs that are enriched by YAP1 (Table S1). Among these natural antisense lncRNAs, 17 of them (Supplementary Table number?) could be enriched by YAP1 with two or more transcripts. These lncRNAs exhibited affinity to YAP1, and regulated the Hippo pathway. From the TCGA database http://gepia.cancer-pku.cn [20], we noted that USP2-AS1 was significantly elevated in COAD. This indicated poor prognosis (Table S2; Figure 1A and 1B). Moreover, higher levels of USP2-AS1 reflect poor prognosis in liver cancer (LIHC), head and neck squamous cell carcinoma (HNSC), brain cancer (GBM and LGG), and Acute Myeloid Leukemia (LAML) (Figure S1). These results suggest that USP2-AS1 might be an oncogenic lncRNA in COAD. To confirm these findings, we examined USP2-AS1 expression in 43 paired fresh COAD tissues. Fold changes were defined as (FC, Tumor vs Adjacent) ≥ 1.5 as a higher expression, 0.66 < FC < 1.5 as unchanged, and FC < 0.66 as down-regulated (Figure 1C and 1D). The expression of USP2-AS1 was higher in COAD tissues. Clinicopathological analysis revealed that USP2-AS1 was not correlated with age and gender. However, it was significantly correlated with tumor grade, size, and TNM stage (Table 2). These findings suggest that USP2-AS1 might be positively correlated with COAD progression. USP2-AS1 was detected in the normal human colon mucosal epithelial cell line NCM460, and COAD cell lines (SW480, SW620, and LoVo). The USP2-AS1 was found to be elevated in cancer cell lines and was positively correlated with the degree of malignancy (Figure 1E).
Figure 1.
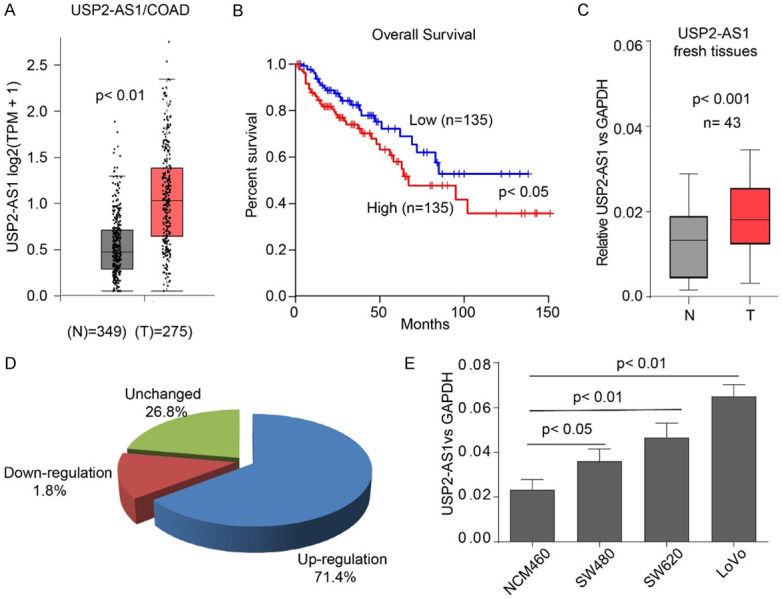
The USP2-AS1 is highly expressed in COAD and predicts poor prognosis. A. The expression of USP2-AS1 in COAD (n = 275) and normal tissues (n = 349), data from Gepia analysis. B. Data from TCGA showed that a higher expression of Linc00337 in COAD is a marker for poor prognostic outcomes. C. The qRT-PCR analysis of USP2-AS1 expression in 43 paired fresh COAD patients. D. Pie chart illustration of the proportions of different USP2-AS1 levels in 43 patients. E. The expression levels of USP2-AS1 in indicated cell lines. C-E. Data is represented as mean ± SD for n = 3.
Table 2.
Correlations between USP2-AS1 and key clinicopathological parameters
| Variable | USP2-AS1 (n = 42) | |||
|---|---|---|---|---|
|
| ||||
| Low | High | P value | ||
| Age | ≤ 50 years | 6 | 13 | 0.205 |
| > 50 years | 6 | 17 | ||
| Gender | Female | 1 | 7 | 0.402 |
| Male | 11 | 23 | ||
| Grade | I | 8 | 8 | 0.031* |
| II and III | 4 | 22 | ||
| Tumor size | ≤ 2 cm | 7 | 6 | 0.015* |
| > 2 cm | 5 | 24 | ||
| TNM stage | T1 | 10 | 9 | 0.016* |
| T2 | 2 | 12 | ||
| T3-T4 | 0 | 9 | ||
P < 0.05.
USP2-AS1 binds to YAP1 in SW620 cells
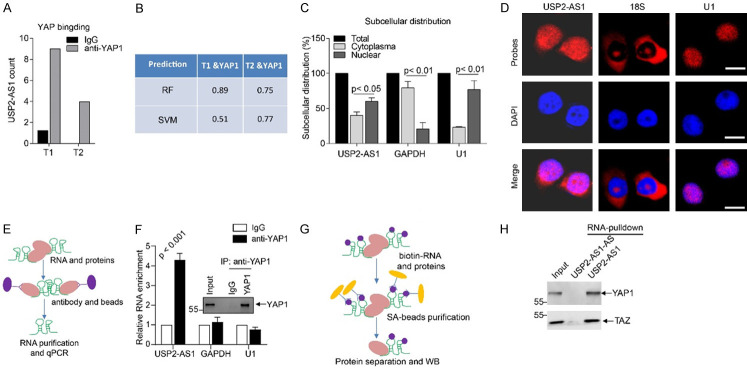
From the Wen et al. results [15], we observed that the two transcripts (ENST00000500970, T1; ENST00000577297, T2) of USP2-AS1 bind to YAP1 (Figure 2A). To confirm the interactions between USP2-AS1 and YAP1, we evaluated the binding potential between USP2-AS1 and YAP1 using RNA-Protein Interaction Prediction (RPISeq), an online prediction software. The RF and SVM classifiers of the two isoforms of USP2-AS1 and YAP1 were higher than 0.5 (Figure 2B). These two isoforms of USP2-AS1 were noted to be capable of binding to YAP1. Upon inactivation of the Hippo signaling, YAP1 was transported from the cytoplasm to the nucleus. The qRT-PCR results revealed that USP2-AS1 was located in the nucleus and cytoplasm, but primarily in the nucleus (Figure 2C). Its subcellular distribution was confirmed using RNA-FISH assays (Figure 2D). Localization of USP2-AS1 is, therefore, similar to that of YAP1. RIP assays in SW620 cells verified that YAP1 binds to USP2-AS1, but not to GAPDH and U1 (Figure 2E and 2F). These findings were further confirmed using RNA-pulldown assays (Figure 2G, 2H). Notably, USP2-AS1 was found to bind YAP1 in COAD cells.
Figure 2.
The USP2-AS1 binds to YAP1 in SW620 cells. A. Two transcripts of USP2-AS1 (T1, T2) are enriched by YAP1, data from Wu et al. [15]. B. USP2-AS1 binding to YAP1. C. The qRT-PCR analysis showed that USP2-AS1 is mainly located in the nucleus in SW620 cells. The GAPDH was used as a cytoplasm marker while U1 as a nuclear marker. D. The RNA-FISH results revealed that USP2-AS1 is mainly located in the nucleus in SW620 cells. The 18S was used as a cytoplasm marker whereas U1 was the nuclear marker. Scale bar, 10 μm. E. Flow chart of RIP assays. F. The RIP-WB results indicated that YAP1 binds to itself, while RIP-qPCR analysis showed that YAP1 binds to USP2-AS1, but not to GAPDH or U1, the negative controls. G. Flow chart of biotin-labeled RNA pulldown assays. H. The RNA-pulldown assays showed that USP2-AS1 could pulldown YAP1 and TAZ. Each bar represents the mean ± SD for n = 3.
USP2-AS1 promotes COAD cell proliferation and invasion in vitro
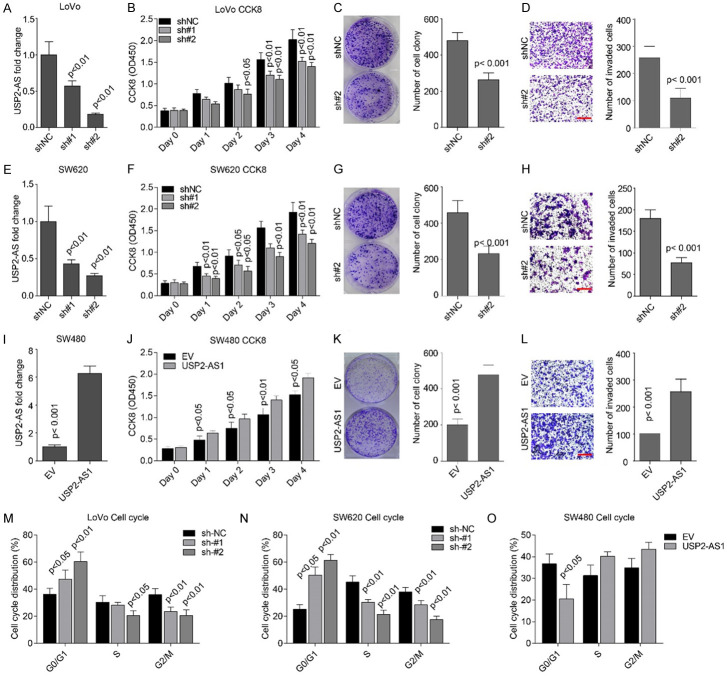
To investigate the biological functions of USP2-AS1 in COAD, we silenced its expression in LoVo and SW620 cell lines using shRNAs. The levels of USP2-AS1 in these two cell lines was significantly inhibited by shRNAs, while sh#2 was more effective in both cell lines (Figure 3A and 3E). The CCK8 cell viability and colony formation assays showed that USP2-AS1 knockdown significantly inhibited cellular proliferation, growth (Figure 3B, 3C, 3F and 3G), and the invasive ability of cells (Figure 3D and 3H) in the same cell lines. A higher expression of USP2-AS1 in SW480 cells (Figure 3I) promoted cellular proliferation, growth, and invasion (Figure 3J-L). Cell cycle assays showed that USP2-AS1 depletion inhibited the transition from the G0/G1 stage to the S stage, while its higher expression enhanced transition through the same stages (Figure 3M-O). These findings indicate that highly expressed USP2-AS1 promotes COAD cell proliferation, growth, and invasion.
Figure 3.
The USP2-AS1 promotes cellular proliferation and invasion in COAD cells. (A, E) The qRT-PCR assays showed that the level of USP2-AS1 is significantly inhibited by two shRNAs in LoVo (A) and SW620 (E) cells. (B, F) The CCK8 cell viability assays showed that silencing USP2-AS1 using two shRNAs significantly inhibited cellular proliferation in LoVo (B) and SW620 (F) cells. (C, G) Cellular colony formation assays revealed that USP2-AS1 knockdown inhibited LoVo (C) and SW620 (G) cell growths. (D, H) Matrigel-coated transwell invasion assays showed that USP2-AS1 knockdown significantly inhibited cellular invasion in LoVo (D) and SW620 (H) cells. Scale bar, 50 μm. (I) Overexpression of USP2-AS1 considerably increased the levels of USP2-AS1 in SW480 cells. (J) The CCK8 cell viability assays showed that higher expressions of USP2-AS1 significantly promoted cellular proliferation in SW480 cells. (K) Cellular colony formation assays showed that a higher expression of USP2-AS1 significantly promoted cellular proliferation and growth. (L) Matrigel-coated transwell invasion assays showed that USP2-AS1 overexpression significantly elevated the ability of cellular invasion in SW480 cells. Scale bar, 50 μm. (M, N) Cell cycle assays showed that depletion of USP2-AS1 triggered cell cycle arrest at the G0/G1 stage in LoVo cells (M) and SW620 cells (N). (O) Overexpression of USP2-AS1 accelerated cell cycle progression in SW480 cells. Each bar represents the mean ± SD for n = 3.
USP2-AS1 promotes COAD cell proliferation and metastasis in vivo
The in vitro effects of USP2-AS1 on cellular proliferation and invasion were validated in vivo. Subcutaneous tumor formation assays revealed that tumor volumes and weight in the mice injected with SW620-sh#2 cells were considerably smaller and lighter than in the SW620-shNC group (Figure 4A-C). Additionally, Ki67 staining revealed a low number of positive cells in the sh#2 group (Figure 4D). Caudal vein lung metastasis assays indicated that metastasis ability was significantly impaired in SW620-sh#2 cells compared to SW620-shNC cells (Figure 4E-G). Therefore, USP2-AS1 promotes COAD cell proliferation and metastasis in vivo.
Figure 4.
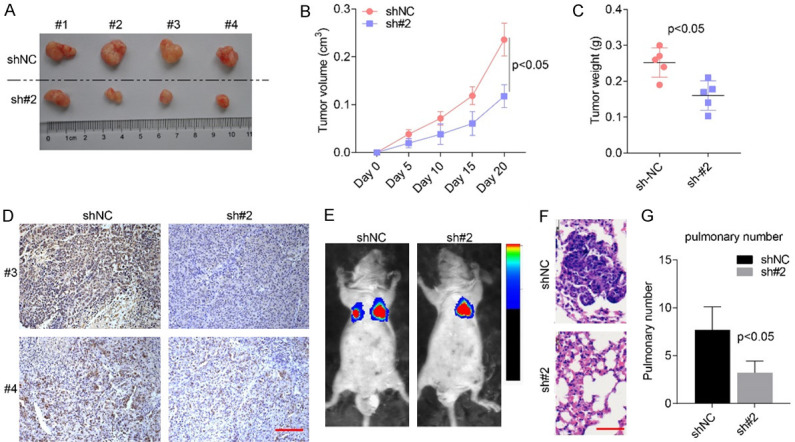
USP2-AS1 knockdown significantly inhibited COAD cellular proliferation and metastasis in vivo. (A) Xenograft Tumor formed by SW620 stable cell lines. (B, C) Tumor growth curve (B) and weight (C) showed that USP2-AS1 knockdown significantly inhibited tumor formation in SW620 in vivo. (D) The IHC staining of Ki67 in indicated tumors. Scale bar, 50 μm. (E-G) Caudal vein lung metastasis assays showed that USP2-AS1 silencing was significantly reduced in SW620 cells. (E) The fluorescence intensity, while (F and G) is the number of intrapulmonary metastasis nodes. Scale bar, 50 μm.
USP2-AS1 modulates Hippo signaling through YAP1
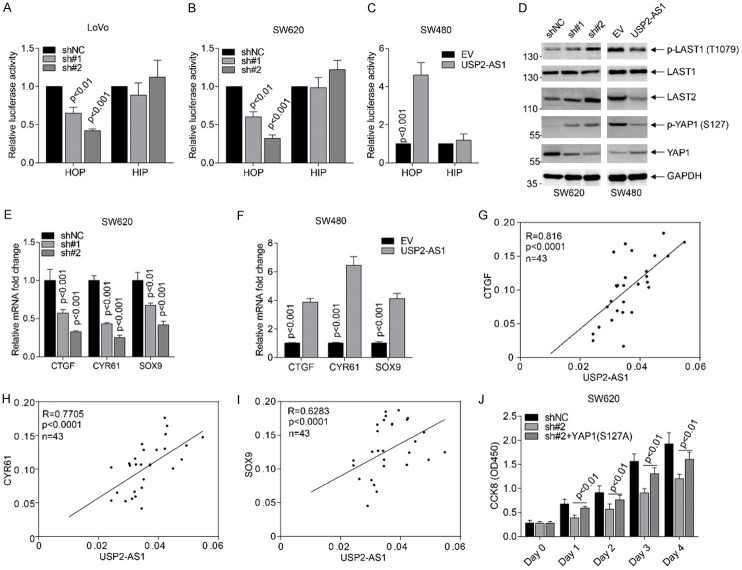
Based on the above findings, we demonstrated that USP2-AS1 binds to YAP1 and promotes COAD progression. The HOP-flash/HIP-flash luciferase reporter assays reflected the downstream YAP1 signaling activity in LoVo, SW620, and SW480 transfected cell lines (Figure 5A-C). Silencing USP2-AS1 in LoVo and SW620 significantly inhibited luciferase activity (Figure 5A, 5B), and vice versa (Figure 5C). Knockdown of USP2-AS increased the levels of p-LAST1 (T1079), LAST1, LAST2, phospho-YAP1 (S127) and inhibited the total YAP1 in LoVo and SW620 cells (Figure 5D). Conversely, overexpression of USP2-AS1 down-regulated the levels of p-LAST1 (T1079), LAST1, LAST2, phospho-YAP1 (S127), and elevated total YAP1 (Figure 5D). Furthermore, silencing of USP2-AS1 inhibited the expression of Hippo signaling target genes in SW620 cells (Figure 5E), whereas a higher expression enhanced the expression of these target genes (Figure 5F). In fresh COAD tissues the expression of these three targets was positively correlated with USP2-AS1 (Figure 5G-I). The CCK8 cell viability rescue assays showed that YAP1 (S127A) greatly improved the inhibitory effects of sh#2 in SW620 cells (Figure 5G). This demonstrated that USP2-AS1 inactivates Hippo signaling and promotes COAD progression.
Figure 5.
The USP2-AS1 regulates Hippo-YAP1 signaling in COAD cells. (A-C) Luciferase reporter assays showed that USP2-AS1 silencing significantly inhibited TEAD transcriptional activity in LoVo (A) and SW620 (B), while USP2-AS1 overexpression enhanced TEAD transcriptional activity (C). (D) Western blot analysis showed that USP2-AS1 regulated the level of p-LAST1 (T1079), LAST1, LAST2, p-YAP1 (S127), and the total YAP1 in indicated cell lines. (E, F) The silencing of USP2-AS1 inhibited the expression of indicated Hippo signaling targets in SW620 cells (E), while its overexpression elevated the levels of these genes (F). (G-I) The expression of USP2-AS1 was positively correlated with CTGF (G), CYR61 (H) and SOX9 (I) in fresh COAD tissues. (J) Constitutively activated YAP1 (S127A) partly improved the proliferation of inhibitory effects through USP2-AS1 knockdown. Each bar represents the mean ± SD for n = 3.
Discussion
In this study, we uncovered a previously unrecognized lncRNA USP2-AS1 that binds to YAP1. Besides, high USP2-AS1 levels protected YAP1 from phosphorylation by upstream kinases and degradation through the ubiquitin-proteasome system. We reveal that USP2-AS1 biologically functions in promoting COAD cell proliferation, growth, and metastasis. Clinicopathological analysis showed that USP2-AS1 is positively correlated with tumor progression and can be used as a marker for poor prognosis.
The COAD-related lncRNAs have previously been found to regulate COAD formation and development by either stimulating or inhibiting different cellular processes such as tumor cell proliferation, apoptosis, differentiation, invasion, and metastasis. For instance, lncRNA GAS5 inhibits colorectal cancer cell proliferation through the miR-182-5p/FOXO3a axis [15,21], whereas lncRNA-APC1 inhibits colorectal cancer tumorigenesis through reducing exosome production [22]. Some lncRNAs such as H19, CRNDE, NEAT1, and DANCR promote COAD progression in various ways [12,23-25]. Our findings suggest that USP2-AS1 exerts oncogenic functions in COAD tumorigenesis. However, its role in other cancer types remain elusive.
YAP1/TAZ plays a central role in the Hippo pathway [26]. Studies have shown that YAP is a highly expressed oncogenic transcription co-activator in a variety of tumors, which can regulate their occurrence and development (References). However, multiple studies have also shown that YAP inhibits tumorigenesis in colorectal cancer by affecting cell growth, apoptosis, and maintenance of dryness and inflammation [27]. When the Hippo pathway is disabled or inactivated, YAP is dephosphorylated and transferred to the nucleus, thereby functioning as a transcriptional co-activator [28]. The nuclear YAP acts as an oncogene, enhancing invasion and proliferation. Although YAP functions as an oncogene, many reports are supporting the idea that YAP functions as a tumor suppressor in colorectal cancer [27,29]. We have shown that USP2-AS1 can decrease the levels of p-YAP (S127), enhance the levels of YAP1 and activate downstream signaling of?. We also report that USP2-AS1 protects YAP1 from phosphorylation and degradation, thereby triggering downstream signaling of the YAP/TEAD axis. Mechanisms by which USP2-AS1 inhibits YAP phosphorylation in our study is unknown. It has been postulated that USP2-AS1 decreases the expression of upstream kinase LAST thereby inhibiting YAP1 phosphorylation.
In conclusion, we provide a first-time report revealing that natural antisense lncRNA USP2-AS1 plays an oncogenic role in COAD by promoting cellular proliferation and metastasis. Besides, USP2-AS1 binds to YAP1 and modulates Hippo signaling that promotes COAD progression. In addition, USP2-AS1 positively correlates with tumor progression and could be used as a marker for poor prognosis.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huh HD, Kim DH, Jeong HS, Park HW. Regulation of TEAD transcription factors in cancer biology. Cells. 2019;8:600. doi: 10.3390/cells8060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HB, Myung SJ. Clinical implications of the Hippo-YAP pathway in multiple cancer contexts. BMB Rep. 2018;51:119–125. doi: 10.5483/BMBRep.2018.51.3.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misra JR, Irvine KD. The hippo signaling network and its biological functions. Annu Rev Genet. 2018;52:65–87. doi: 10.1146/annurev-genet-120417-031621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 7.Moroishi T, Hansen CG, Guan KJ. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 9.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. Regulatory roles of non-coding RNAs in colorectal cancer. Int J Mol Sci. 2015;16:19886–19919. doi: 10.3390/ijms160819886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong X, Lü M, Wan J, Zhou T, Qin B. Long noncoding RNA kcna3 inhibits the progression of colorectal carcinoma through down-regulating YAP1 expression. Biomed Pharmacother. 2018;107:382–389. doi: 10.1016/j.biopha.2018.07.118. [DOI] [PubMed] [Google Scholar]
- 17.Guan ZB, Cao YS, Li Y, Tong WN, Zhuo AS. Knockdown of lncRNA GHET1 suppresses cell proliferation, invasion and LATS1/YAP pathway in non small cell lung cancer. Cancer Biomark. 2018;21:557–563. doi: 10.3233/CBM-170431. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Fang Z, Guo X, Dong H, Zhou K, Huang Z, Xiao Z. lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity. J Cell Physiol. 2019;234:18524–18534. doi: 10.1002/jcp.28489. [DOI] [PubMed] [Google Scholar]
- 19.Ren D, Dai Y, Yang Q, Zhang X, Guo W, Ye L, Huang S, Chen X, Lai Y, Du HJ. Wnt5a induces and maintains prostate cancer cells dormancy in bone. J Exp Med. 2019;216:428. doi: 10.1084/jem.20180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. J Exp Med. 2019;216:428–449. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng K, Zhao Z, Wang G, Wang J, Zhu W. lncRNA GAS5 inhibits colorectal cancer cell proliferation via the miR-182-5p/FOXO3a axis. Oncol Rep. 2018;40:2371–2380. doi: 10.3892/or.2018.6584. [DOI] [PubMed] [Google Scholar]
- 22.Wang FW, Cao CH, Han K, Zhao YX, Cai MY, Xiang ZC, Zhang JX, Chen JW, Zhong LP, Huang Y, Zhou SF, Jin XH, Guan XY, Xu RH, Xie D. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129:727–743. doi: 10.1172/JCI122478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Lu Z, Wang N, Feng J, Zhang J, Luan L, Zhao W, Zeng X. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50:1–17. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding J, Li J, Wang H, Tian Y, Xie M, He X, Ji H, Ma Z, Hui B, Wang K. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, Xu M, Sun H, Liu C, Wei P. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11:113. doi: 10.1186/s13045-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 27.Ou C, Sun Z, Li S, Li G, Li X, Ma J. Dual roles of yes-associated protein (YAP) in colorectal cancer. Oncotarget. 2017;8:75727–75741. doi: 10.18632/oncotarget.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson RL, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehsanian R, Brown MA, Lu H, Yang X, Pattatheyil A, Yan B, Duggal P, Chuang R, Doondeea J, Feller SM. YAP dysregulation by phosphorylation or ΔNp63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene. 2010;29:6160–6171. doi: 10.1038/onc.2010.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.