Abstract
Bladder cancer (BLCA) is a common malignancy arising from the urinary bladder and therapeutic options are limited. However, the mechanisms underlying BLCA development are poorly understood. In this study, robust rank aggregation was used to integrate five GEO BLCA microarray datasets for identifying differentially expressed genes (DEGs) between non-muscular invasive BLCA and muscular invasive BLCA. One-hundred fifty-four DEGs related to the degree of BLCA infiltration, including 24 immune-related genes (IRGs), were identified. Missense mutations were the most common type in IRGs. Ten hub IRGs were identified by protein-protein interaction network analysis. Gene set enrichment analysis and gene set variation analysis of two novel BLCA-related genes (TYROBP and FCER1G) revealed that they were related to immunity. Nine survival-related IRGs were identified, and their potential regulation by transcription factors was analyzed. An immune-related gene-based prognostic index (IRGPI) comprising CTSE, CXCL10, FAM3B, MMP9, OLR1, and S100P was constructed using multivariate analysis. The reliability of the IRGPI was evaluated using independent datasets, and correlations between the IRGPI and clinicopathological characteristics, as well as the immune microenvironment, were evaluated. Finally, a nomogram was established to evaluate the prognosis of patients with BLCA. Our data provide new insights into the pathogenesis of BLCA and target genes for immunotherapy. The application of molecular markers for hierarchical prediction paves the way for precision medicine.
Keywords: Immune-related genes, invasive bladder cancer, non-invasive bladder cancer, prognostic index, immune cell infiltration
Introduction
In the 1940s, Juett and Strong conducted a detailed study of bladder cancer (BLCA) autopsy samples [1]. They found that the incidence of extravesical spread and metastasis was higher in patients with tumors invading the muscular layer than in patients with tumors confined to the submucosa. Over the next few decades, the hypothesis that muscle-invading primary bladder tumors reflect a unique and fatal disease biology was confirmed [2]. It has been estimated that there will be 81,400 new BLCA cases and approximately 17,980 deaths in the USA in 2020 [3]. Among the BLCA patients, 70% have non-muscular invasive BLCA (NMIBC), characterized by a high recurrence rate and low mortality, and 30% have muscular invasive BLCA (MIBC), which is prone to early metastasis and is fatal in half of the cases [4,5], indicating that the molecular characteristics of NMIBC and MIBC are very different.
The state of myometrial invasion can be judged from basal tumor tissue obtained by transurethral resection of the bladder tumor. It is worth noting that it is difficult to precisely determine the tumor stage based on the resected transurethral bladder, as this requires expertise. Additionally, tissue destruction often occurs because of the use of a high-energy laser. Studies have shown that the tumor stage may be underestimated in 25% of cases [6,7]. A lack of expertise on the part of the surgeon can result in incomplete resection and residual lesions, which can lead to recurrence and progression and, eventually, death [8]. Therefore, it is very important to develop a more accurate diagnostic method for predicting whether NMIBC would progress to MIBC.
Cisplatin-based combination chemotherapy and surgery are currently the main treatment approaches for MIBC. In patients undergoing radical cystectomy, the 5-year disease-free survival (DFS) and overall survival (OS) are 68% and 60%, respectively, and the 10-year DFS and OS are 66% and 43%, respectively [9]. Patients with metastatic urothelial cancers generally receive chemotherapy combined with cisplatin, but the prognosis is poor, with a 5-year OS of only 15% [5,10]. Despite the time, effort, and funds devoted to BLCA-related research, the overall morbidity and mortality have barely changed in the past 20 years, and no new drugs have been approved [11].
The 2018 Nobel Prize in Physiology or Medicine was awarded to the American immunologist James Allison and the Japanese biologist Tasuku Honjo for their contribution to the discovery of immunomodulatory therapy for cancer, which represents a milestone in the fight against cancer. Immune checkpoint inhibitors have revolutionized cancer treatment [12]. As BLCA has a very high tumor mutation burden [13,14], immunotherapy has become a key treatment for advanced BLCA. While immunotherapy is approved for the treatment of MIBC, only a small subset of patients are responsive to the treatment [15]. Studies have shown that in order for these drugs to achieve clinical efficacy, the tumor must be in a pre-activated immune state [16]. Thus, there is an urgent clinical need to identify molecular biomarkers that can more accurately judge the degree of tumor invasion and identify the immune state of the tumor. Therefore, this study aimed to evaluate the potential functions of DEGs and immune-related genes (IRGs) in the prognosis of BLCA and their potential significance as biomarkers for targeted therapy.
Materials and methods
Data collection
BLCA RNA-sequencing data, data on copy number variation, and clinical data were downloaded from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). In addition, the GSE13507, GSE31684, GSE32548, GSE32894, and GSE48075 BLCA microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and used for further bioinformatics analysis. An updated list of IRGs actively involved in immune activity and associated with cancer was downloaded from the Immunology Database and Analysis Portal (ImmPort).
Identification of differentially expressed genes (DEGs)
The ‘limma’ package in R was used to normalize the data and identify DEGs between NMIBC and MIBC [17]. Robust rank aggregation (RRA) was used to integrate the five datasets before DEG identification. Genes with P < 0.05 were identified as DEGs in RRA analysis. Differentially expressed IRGs were screened from the IRG list from ImmPort (https://www.immport.org/shared/home).
Functional enrichment analyses
The ‘clusterprofiler’ package in R was used to analyze Gene Ontology (GO) term (biological processes, molecular function, and cellular component) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Significance was defined as P < 0.05, and data were graphically visualized.
Survival analysis and molecular characteristics of survival-related immune-related genes (IRGs)
IRGs showing a significant correlation with OS in the TCGA BLCA data were identified by univariate Cox proportional hazard regression analysis. P < 0.05 was used as a threshold. Transcription factors (TFs) among DEGs were identified using the Cistrome cancer database (http://cistrome.org/), and a regulatory network of IRGs and TFs was constructed. Finally, gene mutations in 24 IRGs were identified based on TCGA copy number variation data. The GenVisR package was used to generate a waterfall map of gene mutations.
Protein-protein interaction (PPI) network analysis and analysis of the molecular characteristics of hub IRGs
The STRING database (https://string-db.org/) was employed to analyze relationships among proteins. The 24 IRGs were imported into the STRING database, and the combined score was set to ≥ 0.4 [18]. The Molecular Complex Detection plugin in Cytoscape was utilized for network analysis. Gene set variation analysis (GSVA) provides an overall pathway or gene set activity score for each sample, and the ‘GSVA’ R package was used to identify pathways that were most relevant to the hub genes. TCGA samples were divided into two groups based on the median expression of hub genes, using a p-value cut-off of 0.05. In addition, hub genes were analyzed by Gene set enrichment analysis (GSEA) software v. 4.0, and a GSEA map was generated.
Construction of an IRG-based prognostic index (IRGPI) and a prognostic signature
The survival-related IRGs obtained by univariate Cox analysis were treated as candidate prognostic biomarkers. Multivariate Cox regression analysis was then used to build an optimal prognostic signature. The threshold used was P < 0.05. The final prognostic signature consisted of six genes. An IRGPI risk score was calculated for each tumor type based on the relative expression of each IRG and its correlation coefficient, using the following formula: risk score = Symbol (coef β × Expr β), where coef β is the multivariate Cox coefficient of gene β and Expr β is the relative expression of the gene in the IRGPI. The six IRGs in the model were subjected to GO functional enrichment analysis and KEGG pathway analysis.
Evaluation of the prognostic value of the IRGPI
Based on the risk score for the IRG signature in the TCGA training set, the median value was set as a threshold, and patients were divided into low- and high-risk groups. Kaplan-Meier (KM) survival analysis was carried out, and the prognostic value of the prediction model was evaluated based on the area under the time-dependent receiver operating characteristic (ROC) curve (AUC). The survivalROC software package in R was used to test the prediction accuracy of the model.
Validation of the prognostic value of the IRGPI in an independent dataset
OS in GSE13507 and GSE48075 and DFS in GSE13507, GSE48075, GSE31684, and GSE32894 were used as external verification sets to verify the clinical prognostic value of the IRGPI. KM survival and time-dependent ROC analyses were used to evaluate the survival prediction accuracy of the model. A p-value < 0.05 was considered statistically significant.
Correlation between the IRGPI and clinicopathological characteristics
The relationships between age, sex, tumor grade and cancer stage, pathological TNM stage, and the prognostic index (PI) were analyzed. The relationships between six genes in the IRGPI and clinical pathology were also analyzed. Wilcox test was used to analyze statistical significance. A p-value < 0.05 was considered statistically significant.
Generation and validation of a predictive nomogram
Univariate and multivariate Cox regression analyses were used to determine independent prognostic factors for constructing a nomogram. All independent prognostic factors obtained by multivariate Cox regression analysis were selected to construct a combined prognostic model to evaluate the 1-, 3-, and 5-year OS in patients with BLCA. A calibration curve was used to evaluate the consistency between the actual and the predicted survival rate; the 45° line represents the best prediction, and the higher the conformation with this line, the better the prediction value of the model.
Validation of immune correlation
CIBERSORT is a gene expression-based algorithm used to infer the relative proportions of 22 types of immune cells based on bulk gene expression profiles of nearly 11,000 tumors. Pearson correlation analysis was used to determine the linear relationship between 22 immune cell types and the IRGPI. The correlation index r and the corresponding p-value were plotted.
Results
DEG identification by the RRA method
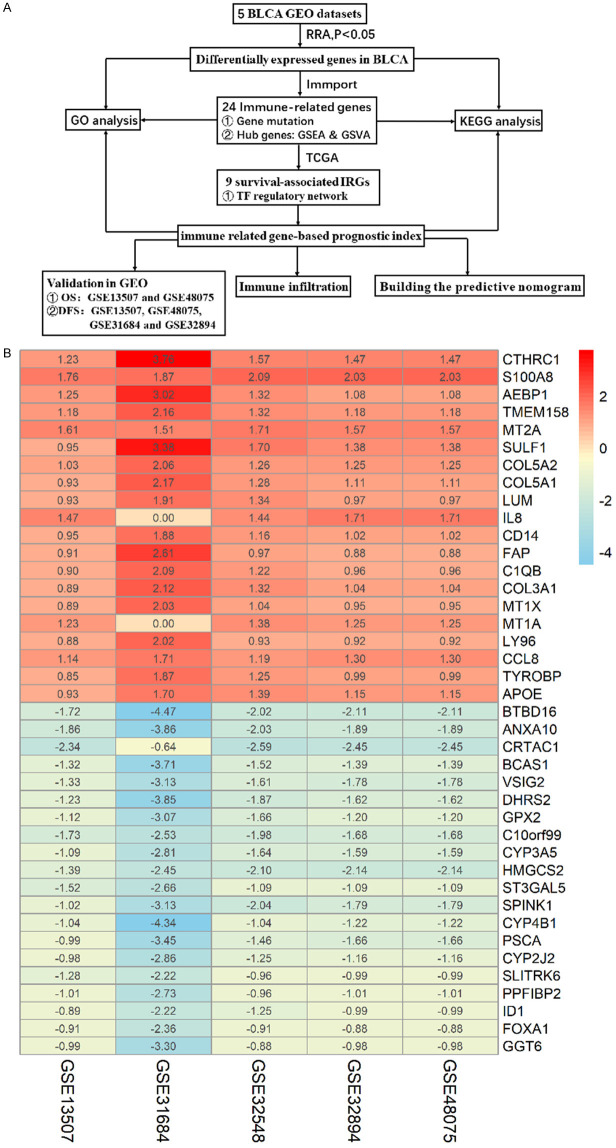
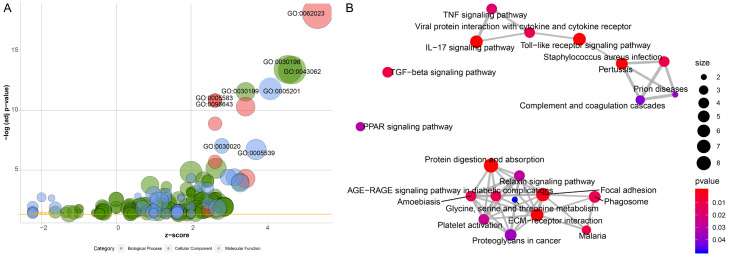
Five BLCA-related GEO microarray datasets were used to identify DEGs between NMIBC and MIBC based on the RRA method. We thus identified 154 DEGs, including 82 genes that were upregulated and 72 that were downregulated in MIBC as compared to NMIBC. Figure 1A shows the workflow used for DEG identification, verification, and functional analysis. The 20 most upregulated and downregulated DEGs are shown in a heatmap in Figure 1B. GO analysis revealed that extracellular matrix organization, collagen-containing extracellular matrix, and extracellular matrix structural constituent were the most frequent biological terms in the biological process, cellular component, and molecular function categories, respectively (Figure 2A). According to the results of the KEGG pathway analysis, DEGs were enriched for ECM-receptor interaction and the IL-17 signaling pathway (Figure 2B).
Figure 1.
A. Study workflow; B. Heatmap of DEGs identified by RRA analysis. The 20 most up- and downregulated genes are displayed according to the p-value. Each column represents a dataset, and each row represents a gene. Red indicates upregulation, and blue indicates downregulation. The numbers in the heat map represent logarithmic fold changes in each dataset calculated by the ‘limma’ R package.
Figure 2.
A. Bubble plot of enriched GO terms. The x-axis represents the z fraction, and the y-axis represents the negative logarithm of the p-value. The higher the location of the bubble in the graph, the more significant the difference. The circle size represents the number of genes assigned to the corresponding term. Green circles correspond to biological processes, red circles to cellular components, and blue circles to molecular function categories; B. Pathway analysis of differentially expressed DEGs. Red circles indicate the number of differentially expressed IRGs in each pathway. Lines between the two red circles represent the ratio of differentially expressed IRGs to the commonly expressed genes in the two pathways; the thicker the line, higher the ratio of the differentially expressed IRGs.
Identification and characterization of differentially expressed IRGs
We identified 24 IRGs among the DEGs (Table S1). The IRGs were mostly enriched for GO terms related to receptor-ligand activity and biological behavior of neutrophils and leukocytes (Figure S1A). IL-17 signaling was the most frequently identified KEGG pathway (Figure S1B). Missense mutation was the most common type of mutation among the differentially expressed IRGs (Figure S2A).
Identification of hub IRGs
PPI network analysis based on the gene expression data revealed that CXCL10, CCL2, MMP9, CXCL9, CCL8, S100A8, TYROBP, FCER1G, S100A9, and CD14 are the IRGs that actively participate in BLCA development. Among the 10 hub genes identified, we selected two genes (TYROBP and FCER1G) that have not been previously reported in BLCA to explore pathways in which they are involved in BLCA. According to the median expression of the hub genes in the TCGA data set, BLCA samples were divided into high- and low-expression group. Notably, GSEA and GSVA revealed that the gene sets with high scores were significantly enriched in immune-related pathways (Figure 3A-D).
Figure 3.
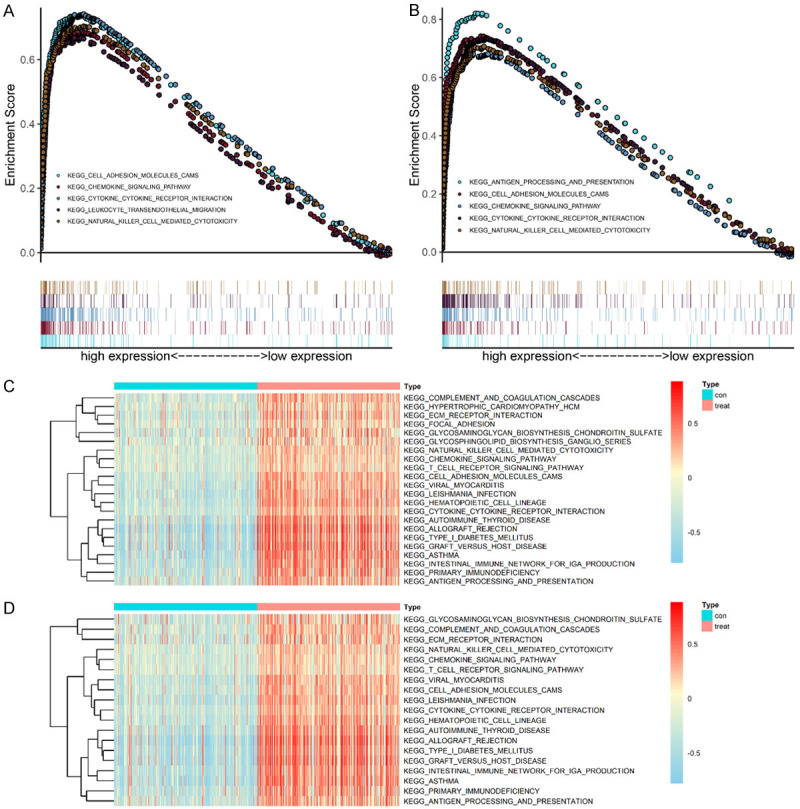
GSEA and GSVA of hub genes in the TCGA-BLCA dataset. A, B. TYROBP and FCER1G were enriched in the high hub gene-expression group (according to the GSEA score). A. TYROBP; B. FCER1G; C, D. GSVA-derived clustering heatmaps of differentially expressed pathways for individual hub genes. C. TYROBP; D. FCER1G. Only signaling pathways with log(fold change) >0.2 are shown.
Identification of survival-associated IRGs and TF regulatory network establishment
Based on the expression levels of 24 IRGs from the TCGA dataset, we identified nine genes that were significantly related to survival, based on univariate Cox regression analysis. To explore the clinical significance of the survival-related IRGs and the potential underlying molecular mechanism, we studied the regulatory network of these genes. Two TFs were identified from the DEGs. The regulatory network in Figure S2B shows the regulatory relationships between the two TFs and the nine survival-related IRGs.
Establishment of an IRGPI
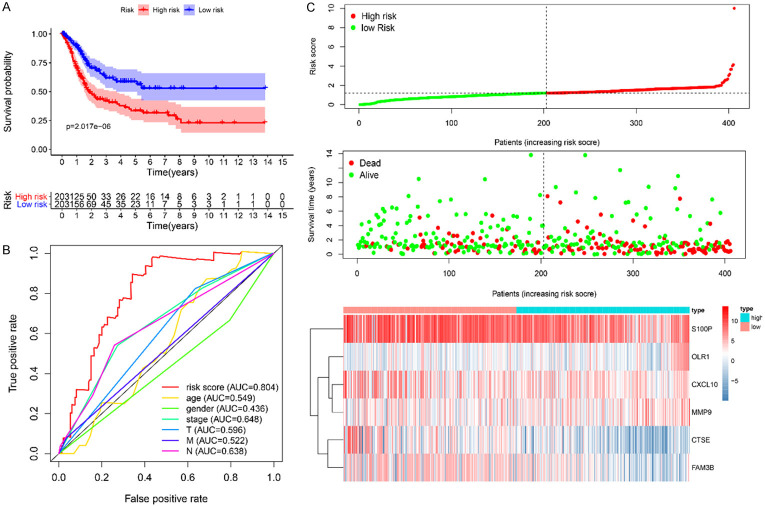
The nine survival-related IRGs were then subjected to multivariate Cox regression analysis to construct an IRGPI. We calculated the biomarker index as the model’s prediction probability multiplied by 100, using the following formula: biomarker index = [CTSE expression * (-0.5426) + CXCL10 expression * (-0.3986) + FAM3B expression * (-0.9789) + MMP9 expression * 0.0203 + OLR1 expression * 0.7072 + S100P expression * (-0.0305)]. According to this model, the patients in the TCGA training set were divided into a high- and low-risk group according to the median risk score. A KM survival curve showed that the prognosis was significantly better in the low-risk group than in the high-risk group (P < 0.0001). Time-dependent ROC analysis revealed that the AUC for the immune-related gene signature was 0.804, which was significantly higher than that for other predictive factors (AUC for age = 0.549, AUC for sex = 0.436, AUC for stage = 0.648, AUC for T = 0.596, AUC for M = 0.522, and AUC for N = 0.638), indicating that it has good prediction potential. Figure 4A-C shows the IRGPI distribution for patients in the training set, the number of patients in the different risk groups, and expression heatmaps for the six genes included in the model.
Figure 4.
A. KM survival analysis of high- and low-risk groups in the TCGA-BLCA data set; B. Time-dependent ROC of the indicated predictive factors in the TCGA-BLCA training set; C. Rank of prognostic index and distribution of groups, survival status of patients in different groups, and expression heatmap of the genes included.
Validation of the IRGPI
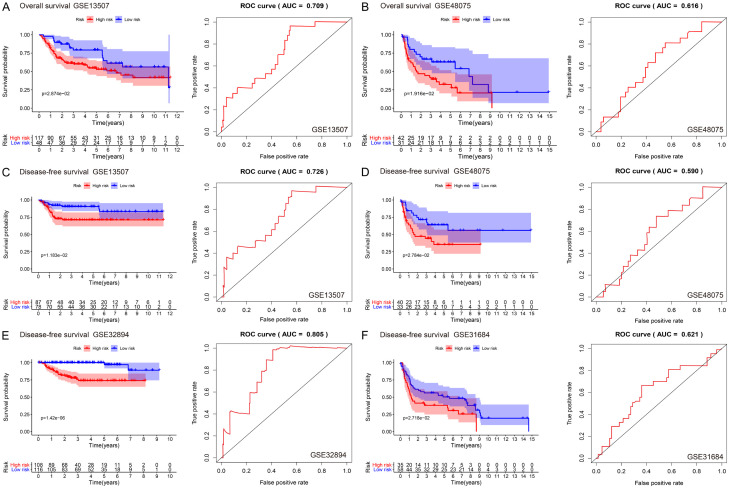
Next, we evaluated the predictive ability of the prognostic marker genes in different BLCA cohorts in the GEO database. Two external datasets with OS as a prognostic indicator (GSE13507 and GSE48075) and four external datasets with DFS as a prognostic indicator (GSE13507, GSE48075, GSE31684, and GSE32894) were used to verify the predictive value of the IRGPI.
In the external verification set with OS as the prognostic indicator, the survival rate in the low-risk group was significantly higher than that in the high-risk group (P < 0.05), and the IRGPI had significant prognostic value, with AUCs of 0.709 and 0.616 in GSE13507 and GSE48075, respectively (Figure 5A, 5B). The DFS trends were the same in the four datasets used: the prognosis of the low-risk group was significantly better than that of the high-risk group (P < 0.05, P < 0.05, P < 0.05, and P < 0.001 for GSE13507, GSE48075, GSE31684, and GSE32894, respectively). The AUC values were 0.726, 0.590, 0.621, and 0.805, respectively (Figure 5C-F). Thus, the IRGPI could accurately predict patient prognosis in internal and external datasets.
Figure 5.
A, B. KM survival (overall survival) and time-dependent ROC analysis of NMIBC and MIBC patients in the GSE13507 and GSE48075 datasets; C-F. KM survival (disease-free survival) and time-dependent ROC analysis of NMIBC and MIBC patients in the GSE13507, GSE48075, GSE32894, and GSE31684 datasets.
Clinical utility of the IRGPI
We analyzed the relationship between the IRGPI and age, sex, tumor grade, cancer stage, tumor T stage, tumor N stage, and tumor M stage. The results showed that the IRGPI increased significantly with late cancer stage, high grade, and late T and N stages (Figure 6A-D). Table S2 shows the relationships between clinicopathological factors and the IRGPI as well as the six genes in the IRGPI.
Figure 6.
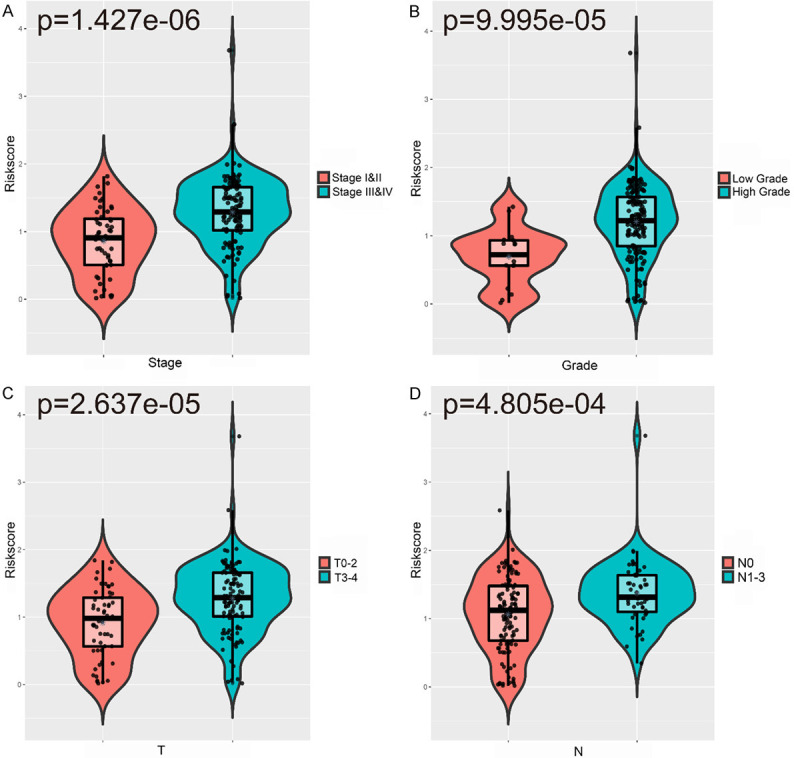
Relationships between the IRGPI and (A) cancer stage; (B) grade; (C) stage T; (D) stage N.
Clinical application of a nomogram incorporating the IRGPI
A nomogram is a powerful tool to quantitatively assess the individual risk in a clinical environment by integrating multiple risk factors. Through univariate and multivariate Cox regression analyses, IRGPI and age were identified as potential independent predictors, based on which a nomogram was constructed (Figure 7A). The calibration map shows that the actual survival rate was in good agreement with the predicted survival rate (Figure 7B), indicating that the nomogram has high clinical application potential.
Figure 7.
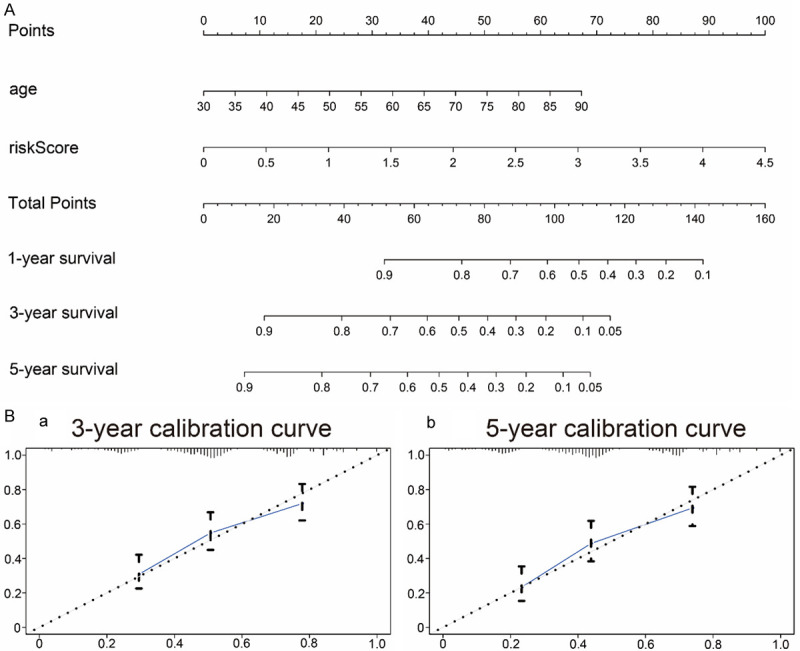
Nomogram and calibration curves. A. Nomogram for the prediction of 1-, 3-, and 5-year OS in the TCGA-BLCA training set; B. Calibration curves for the prediction of 3- and 5-year OS.
Validation of the immune correlation
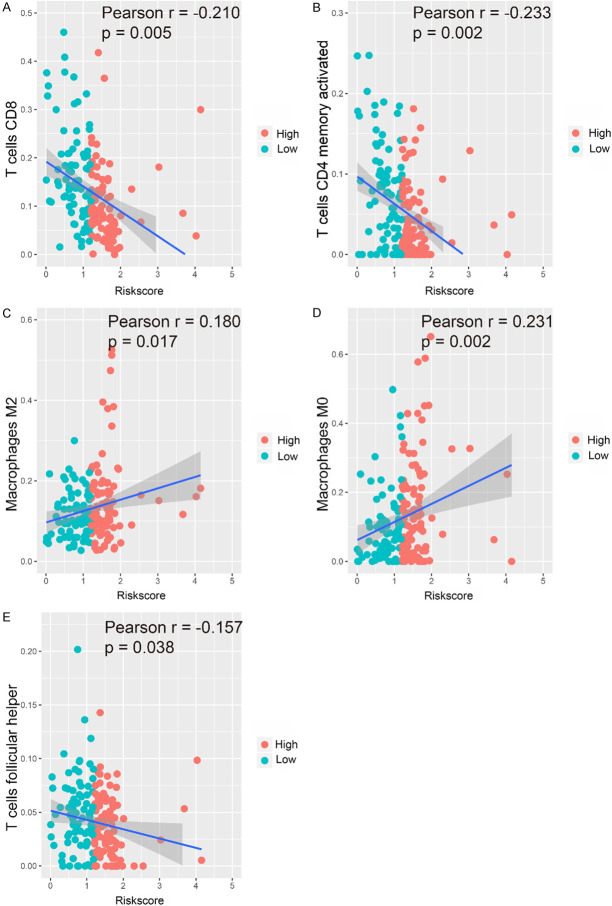
We analyzed the relationship between the IRGPI and immune cell infiltration to determine whether it can accurately reflect the immune microenvironment of BLCA. The IRGPI was positively correlated with M0 and M2 macrophages, whereas the abundance of CD4+ memory activated T cells, T follicular helper cells, and CD8+ T cells decreased with increasing of IRGPI (Figure 8A-E).
Figure 8.
Relationships between the IRGPI and immune cell infiltration. Correlations were determined by Pearson correlation analysis. A. CD8+ T cells; B. CD4+ memory activated T cells; C. M2 macrophages; D. M0 macrophages; E. T follicular helper cells.
Discussion
Using five BLCA-related GEO datasets that were integrated with the RRA method, we identified 154 DEGs in MIBC vs. NMIBC. In agreement with previously published data [19], these DEGs were mainly enriched for ECM organization, which indicated that they are indeed involved in the development of BLCA. Among these genes, we identified 24 IRGs and subjected them to GO and KEGG analyses, which revealed that they were mainly enriched for neutrophil and leukocyte-related biological behavior and the IL-17 signaling pathway. Further, we found that missense mutation was the most common type of mutation in these IRGs. In accordance with Le et al.’s report of a strong link between the response to immune checkpoint inhibitors and defects in the mismatch repair pathway in tumors [20]. To explore the molecular mechanisms associated with biomarkers of potential clinical value, we constructed a TF-IRG network to identify TFs that regulate prognosis-related genes. This network also provides useful information and guidance for future analyses. Among the hub genes, we selected two novel BLCA-related genes, and GSEA and GSVA revealed that they were closely related to immunity.
We developed a prognostic prediction model for targeted therapy based on six IRGs. Blaveri et al. found that CTSE is overexpressed in NMIBC when compared with MIBC [21]. During a long-term follow-up study of 693 cases of NMIBC, low CTSE expression was confirmed to be significantly related to the progression of NMIBC to MIBC [22]. It has also been confirmed that MMP9 can lead to tumor recurrence, high invasiveness, and poor prognosis in BLCA [23,24]. This may be related to the important roles of MMP9 in promoting angiogenesis, maintaining the tumor microenvironment, and promoting the proliferation of malignant tumor cells [25,26]. S100P is a member of the S100 protein family. The positive expression rate of S100P in high-grade urothelial carcinoma is 88% [27], and this protein is used as a marker to support urothelial cell differentiation [28]. While a role for CXCL10 in MIBC has not been reported, its immune effect in NMIBC has been confirmed in previous studies. It is suggested that CXCL10 may be involved in mediating the recruitment of effector cells to the tumor site and that it can attract granulocytes and effector leukocytes into the bladder for tumor clearance, which is conducive to a long-term tumor immune response. Therefore, CXCL10, together with other pro-inflammatory cytokines, may be a biomarker of a good anti-tumor immune response in BLCA treated with BCG [29]. Relationships between the other two genes (FAM3B and OLR1) and BLCA have not been thoroughly investigated to date, and only a few studies have shown that these genes are related to the occurrence of some tumors. High FAM3B expression has been confirmed to be associated with the specific prolongation of survival in papillary thyroid carcinoma [30]. In addition, FAM3B expression is increased in esophageal cancer, and the expression level is positively correlated with the T/TNM stage [31]. Further, FAM3B can inhibit esophageal cancer cell death, increase tumor growth in vivo, and promote esophageal cancer cell migration and invasion [31]. OLR1 was found to be overexpressed in pancreatic cancer tissues, and it was confirmed that it promotes the transcription of HMGA2 through c-Myc, thus promoting the metastasis of pancreatic cancer cells [32].
KEGG analysis revealed that all the DEGs identified were associated with IL-17 signaling. Under the action of IL-17, IL-17 signal pathway components bind with a receptor complex to form a signal complex composed of IL17 receptor, Actl, TRAF6, and other factors. Wang et al. found that IL-17 mRNA expression was positively correlated with the expression of the immunosuppressants CD274, CTLA4, and LAG3, and patients with high IL-17 mRNA expression were more likely to benefit from immunosuppressants [33]. However, the role of IL-17 in cancer remains controversial. IL-17 can promote tumor development by directly stimulating cancer cells and indirectly inducing an immunosuppressive tumor environment. IL-17 also indirectly shapes the immune cell microenvironment through chemokines and cytokines, supports cancer cell proliferation, stimulates early tumor growth, and suppresses the immune system [34]. However, IL-17 expression has also been shown to be associated with a better prognosis of cancer patients; in colorectal and lung cancer, IL-17 recruits antineoplastic neutrophils to the tumor environment, thus stimulating T cell responses and improving overall survival [35,36]. The regulatory potential of IL-17 in the immune microenvironment and immune axis renders it an attractive target for cancer immunotherapy.
The performance of the IRGPI in different datasets was evaluated using GEO data as external data and OS as a prognosis indicator. The results showed that the model is highly robust. BLCA characteristically has a high recurrence rate compared to other tumors, and recurrence can lead to a worse prognosis. Based on the results of four independent GEO data sets, we found that the IRGPI also has a high predictive ability for DFS. Interestingly, our data showed that the IRGPI is associated with tumor stage, T stage, and N stage. A nomogram was used as a predictive tool to evaluate the OS in BLCA. The actual survival was closely related to the predicted survival, indicating that the nomogram has good predictive potential.
There is evidence that immune cells in the tumor microenvironment can affect tumor occurrence and development and patient prognosis [37,38]. Importantly, the pretreatment tumor immune status can be used as a predictor of the treatment response and survival [39,40]. In this study, high-throughput data were used to explore the relationship between the IRGPI and immune cell infiltration as a reflection of the immune microenvironment of BLCA. Our analysis showed that the infiltration of M0 and M2 macrophages among 22 types of tumor-infiltrating immune cells was positively correlated with the IRGPI, whereas the infiltration of CD8+ T cells, CD4+ memory activated T cells, and T follicular helper cells were negatively correlated with the IRGPI. CD8+ T cells have been a major focus in cancer research, and it has been confirmed that they mediate anti-tumor functions. Patients with higher CD8+ T cell infiltration in metastatic urothelial carcinoma exhibited better DFS and OS [41], which is in line with our findings. Pre-existing CD8+ T cells are located at the edge and in the center of invasive tumors, and CD8+ T cells are the main cellular source of PD-1 expression [42]. CD8+ T cells are also associated with the expression of the PD-1/PD-L1 immune inhibitory axis [14,43]. The induction of memory T cells is a key focus in the development of tumor vaccines and immunotherapy. CD4+ memory T cells ‘remember’ their previous effector lineage after antigen clearance and can regain lineage-specific effector function when they re-encounter antigens [44]. Vahidi et al. indicated that CD4+ memory T cells might play a role in preventing lymph node metastasis and tumor progression [45]. The role of T follicular helper cells in immune responses against tumors may be dual: (i) they may help to develop or support ectopic lymphoid structures that aggregate CD8+ T cells, NK cells, and macrophages involved in anti-tumor immunity; (ii) they may support the anti-tumor antibody response of B cells [46]. Macrophages are multifunctional antigen-presenting cells that play a central role in cancer. Increasing evidence suggests that M2 macrophages have an immunosuppressive function and promote tumor progression and metastasis [47,48]. Xue et al. used ssGSEA and CIBERSORT algorithms to explore the infiltration of various immune cells based on BLCA TCGA data, and found that M2 macrophages were the main tumor-infiltrating immune cells in the BLCA microenvironment and that they were related to histopathological grade and stage as well as patient prognosis [49]. Studies have shown that M2 macrophages can accumulate in hypoxic regions, and hypoxia-induced transcription factors of M2 macrophages can induce the production of various proangiogenic genes, which leads to tumor progression [50,51]. Therefore, M2 macrophage infiltration may be a potential target for immunotherapy for BLCA [47].
While we used bioinformatics tools to identify immune-related candidate genes that affect the prognosis of BLCA based on a large dataset, findings based on bioinformatics analysis alone are insufficient and require experimental verification in vitro or in vivo; the lack thereof is a clear limitation of our study.
The main purpose of this study was to gain an in-depth understanding of the potential functions of IRGs in the prognosis of BLCA and their potential significance as biomarkers for targeted therapy. More importantly, by applying the CIBERSORT algorithm to large-scale data to explore the infiltration of various immune cells.
Conclusion
Based on bioinformatics analysis of large-scale data, we identified DEGs and IRGs in MIBC, and we developed an IRGPI and a predictive nomogram. The results of this study provide a basis for more in-depth immune-related studies of BLCA and are of great significance to provide predictive and prognostic biomarkers and for guiding effective immunotherapy.
Acknowledgements
The results published here are based on data generated by the TCGA Research Network (http://cancergenome.nih.gov/) and GEO Research Network (http://www.ncbi.nlm.nih.gov/geo/). We would like to thank Editage (www.editage.cn) for English language editing. This study was financially supported by the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (grant number: 2018-I2M-1-002) and the Beijing Hospital Clinical Research 121 Project (BJ-2018-090). The funders had no roles in study design, data collection, data analysis, and interpretation, or writing the manuscript.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Jewett HJ, Strong GH. Infiltrating carcinoma of the bladder; relation of depth of penetration of the bladder wall to incidence of local extension and metastases. J Urol. 1946;55:366–372. doi: 10.1016/S0022-5347(17)69924-5. [DOI] [PubMed] [Google Scholar]
- 2.Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W, Guo CC, Lotan Y, Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 5.Babjuk M, Bohle A, Burger M, Capoun O, Cohen D, Comperat EM, Hernandez V, Kaasinen E, Palou J, Roupret M, van Rhijn BW, Shariat SF, Soukup V, Sylvester RJ, Zigeuner R. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 6.Ark JT, Keegan KA, Barocas DA, Morgan TM, Resnick MJ, You C, Cookson MS, Penson DF, Davis R, Clark PE, Smith JA Jr, Chang SS. Incidence and predictors of understaging in patients with clinical T1 urothelial carcinoma undergoing radical cystectomy. BJU Int. 2014;113:894–899. doi: 10.1111/bju.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkarni GS, Hakenberg OW, Gschwend JE, Thalmann G, Kassouf W, Kamat A, Zlotta A. An updated critical analysis of the treatment strategy for newly diagnosed high-grade T1 (previously T1G3) bladder cancer. Eur Urol. 2010;57:60–70. doi: 10.1016/j.eururo.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Mariappan P, Zachou A, Grigor KM Edinburgh Uro-Oncology Group. Detrusor muscle in the first, apparently complete transurethral resection of bladder tumour specimen is a surrogate marker of resection quality, predicts risk of early recurrence, and is dependent on operator experience. Eur Urol. 2010;57:843–849. doi: 10.1016/j.eururo.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 9.Stein JP, Lieskovsky G, Cote R, Groshen S, Feng AC, Boyd S, Skinner E, Bochner B, Thangathurai D, Mikhail M, Raghavan D, Skinner DG. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J. Clin. Oncol. 2001;19:666–675. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- 10.Alfred Witjes J, Lebret T, Comperat EM, Cowan NC, De Santis M, Bruins HM, Hernandez V, Espinos EL, Dunn J, Rouanne M, Neuzillet Y, Veskimae E, van der Heijden AG, Gakis G, Ribal MJ. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71:462–475. doi: 10.1016/j.eururo.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser J, Couzin-Frankel J. Cancer immunotherapy sweeps Nobel for medicine. Science. 2018;362:13. doi: 10.1126/science.362.6410.13. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, Mega AE, Britten CD, Ravaud A, Mita AC, Safran H, Stinchcombe TE, Srdanov M, Gelb AB, Schlichting M, Chin K, Gulley JL. Avelumab, an anti-programmed death-ligand 1 antibody, in patients with refractory metastatic urothelial carcinoma: results from a multicenter, phase Ib study. J. Clin. Oncol. 2017;35:2117–2124. doi: 10.1200/JCO.2016.71.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, Zhang C, Lunceford JK, Joe A, Cheng J, Webber AL, Ibrahim N, Plimack ER, Ott PA, Seiwert TY, Ribas A, McClanahan TK, Tomassini JE, Loboda A, Kaufman D. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deb B, Patel K, Sathe G, Kumar P. N-glycoproteomic profiling reveals alteration in extracellular matrix organization in non-type bladder carcinoma. J Clin Med. 2019;8:1303. doi: 10.3390/jcm8091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, Waldman FM. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 22.Fristrup N, Ulhoi BP, Birkenkamp-Demtroder K, Mansilla F, Sanchez-Carbayo M, Segersten U, Malmstrom PU, Hartmann A, Palou J, Alvarez-Mugica M, Zieger K, Borre M, Orntoft TF, Dyrskjot L. Cathepsin E, maspin, Plk1, and survivin are promising prognostic protein markers for progression in non-muscle invasive bladder cancer. Am J Pathol. 2012;180:1824–1834. doi: 10.1016/j.ajpath.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Zhao Z, Sun Z, Liu C, Cheng X, Ruge F, Yang Y, Jiang WG, Ye L. Increased expression of Psoriasin is correlated with poor prognosis of bladder transitional cell carcinoma by promoting invasion and proliferation. Oncol Rep. 2020;43:562–570. doi: 10.3892/or.2019.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reis ST, Leite KR, Piovesan LF, Pontes-Junior J, Viana NI, Abe DK, Crippa A, Moura CM, Adonias SP, Srougi M, Dall’Oglio MF. Increased expression of MMP-9 and IL-8 are correlated with poor prognosis of Bladder Cancer. BMC Urol. 2012;12:18. doi: 10.1186/1471-2490-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fouad H, Salem H, Ellakwa DE, Abdel-Hamid M. MMP-2 and MMP-9 as prognostic markers for the early detection of urinary bladder cancer. J Biochem Mol Toxicol. 2019;33:e22275. doi: 10.1002/jbt.22275. [DOI] [PubMed] [Google Scholar]
- 26.Khatibi AS, Roodbari NH, Majidzade AK, Yaghmaei P, Farahmand L. In vivo tumor-suppressing and anti-angiogenic activities of a recombinant anti-CD3epsilon nanobody in breast cancer mice model. Immunotherapy. 2019;11:1555–1567. doi: 10.2217/imt-2019-0068. [DOI] [PubMed] [Google Scholar]
- 27.Mohanty SK, Smith SC, Chang E, Luthringer DJ, Gown AM, Aron M, Amin MB. Evaluation of contemporary prostate and urothelial lineage biomarkers in a consecutive cohort of poorly differentiated bladder neck carcinomas. Am J Clin Pathol. 2014;142:173–183. doi: 10.1309/AJCPK1OV6IMNPFGL. [DOI] [PubMed] [Google Scholar]
- 28.Suryavanshi M, Sanz-Ortega J, Sirohi D, Divatia MK, Ohe C, Zampini C, Luthringer D, Smith SC, Amin MB. S100P as a marker for urothelial histogenesis: a critical review and comparison with novel and traditional urothelial immunohistochemical markers. Adv Anat Pathol. 2017;24:151–160. doi: 10.1097/PAP.0000000000000150. [DOI] [PubMed] [Google Scholar]
- 29.Ashiru O, Esteso G, Garcia-Cuesta EM, Castellano E, Samba C, Escudero-Lopez E, Lopez-Cobo S, Alvarez-Maestro M, Linares A, Ho MM, Leibar A, Martinez-Pineiro L, Vales-Gomez M. BCG therapy of bladder cancer stimulates a prolonged release of the chemoattractant CXCL10 (IP10) in patient urine. Cancers (Basel) 2019:11. doi: 10.3390/cancers11070940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin P, Guo YN, Shi L, Li XJ, Yang H, He Y, Li Q, Dang YW, Wei KL, Chen G. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY) 2019;11:480–500. doi: 10.18632/aging.101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He SL, Wang WP, Yang YS, Li EM, Xu LY, Chen LQ. FAM3B promotes progression of oesophageal carcinoma via regulating the AKT-MDM2-p53 signalling axis and the epithelial-mesenchymal transition. J Cell Mol Med. 2019;23:1375–1385. doi: 10.1111/jcmm.14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang G, Xiong G, Feng M, Zhao F, Qiu J, Liu Y, Cao Z, Wang H, Yang J, You L, Zheng L, Zhang T, Zhao Y. OLR1 promotes pancreatic cancer metastasis via increased c-Myc expression and transcription of HMGA2. Mol Cancer Res. 2020;18:685–697. doi: 10.1158/1541-7786.MCR-19-0718. [DOI] [PubMed] [Google Scholar]
- 33.Wang JT, Li H, Zhang H, Chen YF, Cao YF, Li RC, Lin C, Wei YC, Xiang XN, Fang HJ, Zhang HY, Gu Y, Liu X, Zhou RJ, Liu H, He HY, Zhang WJ, Shen ZB, Qin J, Xu JJ. Intratumoral IL17-producing cells infiltration correlate with anti-tumor immune contexture and improved response to adjuvant chemotherapy in gastric cancer. Ann Oncol. 2019;30:266–273. doi: 10.1093/annonc/mdy505. [DOI] [PubMed] [Google Scholar]
- 34.Vitiello GA, Miller G. Targeting the interleukin-17 immune axis for cancer immunotherapy. J Exp Med. 2020;217:e20190456. doi: 10.1084/jem.20190456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, Stephen TL, Ranganathan A, Deshpande C, Akimova T, Vachani A, Litzky L, Hancock WW, Conejo-Garcia JR, Feldman M, Albelda SM, Singhal S. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest. 2014;124:5466–5480. doi: 10.1172/JCI77053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y, Xu J, Su H, Zhong W, Yuan Y, Yu Z, Fang Y, Zhou H, Li C, Huang K. Interleukin-17 is a favorable prognostic marker for colorectal cancer. Clin Transl Oncol. 2015;17:50–56. doi: 10.1007/s12094-014-1197-3. [DOI] [PubMed] [Google Scholar]
- 37.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 38.Van den Eynde M, Mlecnik B, Bindea G, Fredriksen T, Church SE, Lafontaine L, Haicheur N, Marliot F, Angelova M, Vasaturo A, Bruni D, Jouret-Mourin A, Baldin P, Huyghe N, Haustermans K, Debucquoy A, Van Cutsem E, Gigot JF, Hubert C, Kartheuser A, Remue C, Leonard D, Valge-Archer V, Pages F, Machiels JP, Galon J. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell. 2018;34:1012–1026. e1013. doi: 10.1016/j.ccell.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Tabernero J, Hozak RR, Yoshino T, Cohn AL, Obermannova R, Bodoky G, Garcia-Carbonero R, Ciuleanu TE, Portnoy DC, Prausova J, Muro K, Siegel RW, Konrad RJ, Ouyang H, Melemed SA, Ferry D, Nasroulah F, Van Cutsem E. Analysis of angiogenesis biomarkers for ramucirumab efficacy in patients with metastatic colorectal cancer from RAISE, a global, randomized, double-blind, phase III study. Ann Oncol. 2018;29:602–609. doi: 10.1093/annonc/mdx767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilky BA, Trucco MM, Subhawong TK, Florou V, Park W, Kwon D, Wieder ED, Kolonias D, Rosenberg AE, Kerr DA, Sfakianaki E, Foley M, Merchan JR, Komanduri KV, Trent JC. Axitinib plus pembrolizumab in patients with advanced sarcomas including alveolar soft-part sarcoma: a single-centre, single-arm, phase 2 trial. Lancet Oncol. 2019;20:837–848. doi: 10.1016/S1470-2045(19)30153-6. [DOI] [PubMed] [Google Scholar]
- 41.Sharma P, Callahan MK, Bono P, Kim J, Spiliopoulou P, Calvo E, Pillai RN, Ott PA, de Braud F, Morse M, Le DT, Jaeger D, Chan E, Harbison C, Lin CS, Tschaika M, Azrilevich A, Rosenberg JE. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. 2016;17:1590–1598. doi: 10.1016/S1470-2045(16)30496-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J, Plimack ER, Vaena D, Grimm MO, Bracarda S, Arranz JA, Pal S, Ohyama C, Saci A, Qu X, Lambert A, Krishnan S, Azrilevich A, Galsky MD. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18:312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 44.Bethune MT, Joglekar AV. Personalized T cell-mediated cancer immunotherapy: progress and challenges. Curr Opin Biotechnol. 2017;48:142–152. doi: 10.1016/j.copbio.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 45.Vahidi Y, Faghih Z, Talei AR, Doroudchi M, Ghaderi A. Memory CD4(+) T cell subsets in tumor draining lymph nodes of breast cancer patients: a focus on T stem cell memory cells. Cell Oncol (Dordr) 2018;41:1–11. doi: 10.1007/s13402-017-0352-6. [DOI] [PubMed] [Google Scholar]
- 46.Garaud S, Zayakin P, Buisseret L, Rulle U, Silina K, de Wind A, Van den Eyden G, Larsimont D, Willard-Gallo K, Line A. Antigen specificity and clinical significance of IgG and IgA autoantibodies produced in situ by tumor-infiltrating b cells in breast cancer. Front Immunol. 2018;9:2660. doi: 10.3389/fimmu.2018.02660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99:180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Xue Y, Tong L, LiuAnwei Liu F, Liu A, Zeng S, Xiong Q, Yang Z, He X, Sun Y, Xu C. Tumorinfiltrating M2 macrophages driven by specific genomic alterations are associated with prognosis in bladder cancer. Oncol Rep. 2019;42:581–594. doi: 10.3892/or.2019.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronte V, Murray PJ. Understanding local macrophage phenotypes in disease: modulating macrophage function to treat cancer. Nat Med. 2015;21:117–119. doi: 10.1038/nm.3794. [DOI] [PubMed] [Google Scholar]
- 51.Jiang J, Wang GZ, Wang Y, Huang HZ, Li WT, Qu XD. Hypoxia-induced HMGB1 expression of HCC promotes tumor invasiveness and metastasis via regulating macrophage-derived IL-6. Exp Cell Res. 2018;367:81–88. doi: 10.1016/j.yexcr.2018.03.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.