Abstract
Persistent atrial fibrillation (PeAF) is a progressive cardiovascular disease with a high risk for most patients after diagnosis. Poor molecular description of PeAF has led to unsatisfactory interpretation of the pathogenesis of it, resulting in the lack of effective treatments. The aim of the present study was to find several new potential biomarkers for early prevention, diagnosis and treatment of this disease and explore the underlying molecular mechanisms. An absolute quantitation Tandem Mass Tag (TMT)-liquid chromatography-tandem mass spectrometry (LC-MS/MS) approach was applied to identify differentially expressed proteins (DEPs) in left atrial appendage. Totally, 4682 proteins were identified and 4159 proteins were quantified. Compared with control subjects, 118 DEPs (85 upregulated proteins and 33 downregulated proteins) were identified in the atrial tissues of PeAF patients. Using String software, a regulatory network containing 87 nodes and 244 edges was built, and the functional enrichment showed that DEPs were predominantly involved in protein digestion and absorption, regulation of metabolism and focal adhesion. Four proteins, collagen 1 (COL-I), collagen 2 (COL-II), ras-related protein 1 (RAP1) and leucine-rich alpha-2-glycoprotein 1 (LRG1) were selected for validation using Western blot analysis to distinguish PeAF patients and control subjects. The present results provide a comprehensive understanding of the pathophysiological mechanisms of PeAF and the validated biomarkers for the diagnosis of PeAF, which facilitate the development of therapeutic targets.
Keywords: Persistent atrial fibrillation, cardiovascular disease, proteome, parallel reaction monitoring
Introduction
Atrial fibrillation (AF) is the most common arrhythmia in the clinic and it mainly occurs in the elderly [1]. It is well recognized that AF leads to adverse consequences such as stroke, thromboembolism and heart failure, resulting in increased mortality and disability in patients [2,3]. According to statistics, the risk of stroke in patients with AF is 5 times higher than that in patients without AF, and about 4-5% of patients with AF are suffering from cerebral apoplexy every year [4]. Pulmonary vein isolation (PVI), a standard ablative strategy for paroxysmal AF, is unideal in patients with persistent AF (PeAF), thus the mortality of AF still maintains an upward trend. In recent years, people have also deeply studied the pathogenesis of AF, mainly including inflammatory response, atrial remodeling, oxidative stress and genetic inheritance. However, the differentially expressed proteins (DEPs) in atrial tissue of PeAF patients and control subjects have not been investigated. Identifying DEPs in atrial tissue of PeAF patients and control subjects is beneficial to the better understanding of the molecular mechanisms of AF, which may be of great importance for the early prevention, diagnosis and treatment of PeAF.
The emergence of proteomics has provided a new research direction for current biology. Compared with the conventional biochemical approaches, which analyze only one or several specific proteins at a time, proteomics conducts a large-scale analysis of protein expression, forming the more accurate molecular mechanisms related to the development of diseases [5]. At present, protein biomarkers are gradually becoming the important biomarkers for multiple diseases in clinical practice [6,7]. Zou et al [8] found that three verified proteins biomarkers could diagnose the myocardial infarction (MI) and monitor the disease state and the therapeutic effects of MI. Sun et al [9] utilized iTRAQ coupled with 2D LC-MS/MS to find the differential urinary proteins between patients with coronary artery disease and healthy subjects. Ilias P. Doulamis et al [10] indicated that PeAF was related to changes in the proteome of patients undergoing cardiac surgery and the differential protein biomarkers could stratify risk for PeAF. Importantly, the application of proteomics in the diagnosis of heart diseases is gradually prevalent.
The TMT coupled with LC-MS/MS-based approach is a powerful sensitive proteomic tool, which can simultaneously quantify proteins in 2-plex, 6-plex and 10-plex samples to discover protein biomarkers of multiple diseases [11-15]. The TMT technique has been widely used in the proteomic research due to its advantages such as high sensitivity, labeling efficiency and resolution, good sample compatibility and ideal repeatability. Bioinformatics analysis based on TMT-coupled proteomics was also performed to find DEPs in pathophysiological mechanisms of several cardiovascular diseases [13,16,17]. However, as far as we know, no study has identified protein biomarkers in atrial tissue from AF patients and control subjects with TMT-based approaches.
In this study, we used TMT with nano-LC-MS/MS to find the DEPs in atrial tissue from PeAF patients and control subjects to better understand the molecular mechanism in signaling pathway and biological processes, which could provide important theoretical basis for the early prevention, diagnosis and treatment of PeAF in clinical practice.
Materials and methods
Materials and experimental design
Human left atrial appendage tissue samples were obtained from PAF patients (n = 6; mean age ± SD: 56.3 ± 8.2 years; 2 female, 4 males) or PeAF patients (n = 6; mean age ± SD: 56.3 ± 8.3 years; 2 female, 4 males) who had received atrial transplantation surgery at the cardiothoracic surgery department of Nanjing Drum Tower Hospital (Nanjing, PR China). Histological examination of the explanted atrial confirmed the diagnosis of PAF or PeAF conducted by pathologists. All patients met the diagnostic criteria for PAF or PeAF defined by an official ATS/ERS/JRS/ALAT statement: PeAF: evidence-based guidelines for diagnosis and management. However, PeAF-2 sample was eliminated due to the great individual differences. Control atrial tissue (Control subjects; mean age ± SD: 56.7 ± 8.6 years; 2 females, 4 males) was collected from 6 patients having cardiac surgery at thoracic surgery of Nanjing Drum Tower Hospital. 5 mL peripheral venous blood of patients and control subjects was collected into a test tube without anticoagulant reagent. After centrifugation at 3,000 r/min for 5 min, the supernatant was obtained and placed in an EP tube. The samples of tissues and serum were stored in an ultra-low temperature freezer at -80°C for subsequent experiments. The study protocol was approved by the Ethics Committee of Nanjing Drum Tower Hospital of Medical School of Nanjing University. Informed consent was obtained in writing from each subject for the study protocol. Demographic and clinical data (atrial function test parameters) on control subjects or patients receiving transplantation are summarized in Supporting Information.
Protein extraction
The atrial tissue sample was ground into cell powder with liquid nitrogen and transferred to a 5-mL centrifuge tube. Subsequently, four volumes of lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail) was added to the cell powder and sonicated on ice using a high-intensity ultrasonic processor (Scientz) for three times. (Note: For PTM experiments, inhibitors were also added to the lysis buffer, e.g. 3 µM TSA and 50 mM NAM for acetylation). The remaining debris was removed after cell powder centrifugation at 12,000 g and 4°C for 10 min. Finally, the supernatant was collected, and the protein concentration was determined by BCA kit according to the manufacturer’s instructions.
Trypsin digestion
For digestion, the protein solution was volatilized with 5 mM dithiothreitol for 30 min at 56°C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was diluted by adding 100 mM TEAB when the urea concentration was less than 2 M. Finally, the protein sample was digested with trypsin (trypsin/protein = 1:50) overnight and then digested with trypsin (trypsin/protein = 1:100) for 4 h.
TMT labeling
Peptide was reconstituted in 0.5 M TEAB and processed in accordance with the manufacturer’s protocols for TMT kit/TMT kit. Briefly, one unit of TMT/TMT reagent was thawed which was reconstituted in acetonitrile. The peptide mixtures were then incubated for 2 h at room temperature and then pooled, desalted and dried by vacuum centrifugation.
HPLC fractionation
The tryptic peptides were separated into fractions by high pH reverse-phase HPLC using Thermo Betasil C18 column (5 µm particles, 10 mm ID, 250 mm length). Concisely, peptides were separated into 60 fractions with a gradient of 8% to 32% acetonitrile (pH 9.0) over 60 min. Then, the peptides were synthesized into 18 fractions and dried by vacuum centrifugation.
LC-MS/MS analysis
The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and directly loaded onto a home-made reversed-phase analytical column (15-cm length, 75 µm i.d.). The gradient was changed as follows: solvent B (0.1% formic acid in 98% acetonitrile) increased from 6% to 23% in 26 min, increased from 23% to 35% in 8 min, reached 80% in 3 min and maintained at 80% for the last 3 min. The constant flow rate of mobile phase was 400 nL/min in an EASY-nLC 1000 UPLC system. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC with 2.0 kV electrospray voltage. The m/z scan ranged from 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Further, the peptides were selected for MS/MS using NCE setting as 28 and the fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure alternated between one MS scan and another followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5E4 and fixed first mass was set as 100 m/z.
Database search and bioinformatics methods
The MS/MS data were processed using Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against Uniprot database concatenated with reverse decoy database. Trypsin/P was defined as a cleavage enzyme that tolerated up to 4 missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in First search and 5 ppm in Main search, and that for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification and oxidation on Met were defined as variable modifications. FDR was adjusted to < 1% and the minimum score of modified peptides was set > 40. The classification of DEPs was performed based on the annotations obtained from the UniProt knowledge base (http://www.uniprot.org/). In addition, the terms of Gene Ontology (GO) (http://geneontology.org/) were utilized to elaborate cellular components (CC), biological process (BP), and molecular function (MF). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (http://www.genome.ad.jp/kegg/pathway.html) was used to elucidate the knowledge of biochemical pathways and other types of molecular interactions.
Masson staining
Atrial tissue sections (3 mm thick) were obtained and treated with masson compound staining solution for 5 minutes. Then they were slightly washed with 0.2% acetic acid, treated with 5% phosphatotungstic acid, soaked and washed with 0.2% acetic acid three times. After washing with acetic acid water three times and dewaxing with anhydrous ethanol, the sections were transparently treated with dimethylbenzene and neutral gum to seal the slides. Finally, the slides were observed and photographed by an optical microscope (CKX41, Olympus, Tokyo, Japan).
HL-1 cell pacing model
Mouse atrial cardiomyocyte cell line HL-1 (SCC065; Sigma-Aldrich) was used for cell pacing model. HL-1 cells were cultured and maintained in supplemented Claycomb Medium (Sigma-Aldrich) [18]. The cells were serum-starved for 24 h on the third day after passaging and then depolarized spontaneously at a rate of 0.5-1 Hz. Pacing was administered using a C-Pace100TM-culture pacer and CDish100TM culture dishes (IonOptix Corporation, Netherlands) with a 1 V/cm, 5-ms pulse width and 5-Hz frequency, according to previous studies [19,20]. HL-1 cells in control group without stimulation were also performed in parallel.
Western blot analysis
For atrial tissue, proteins were extracted from approximately 100 mg of atrial tissue and quantified with a BCA Protein Concentration Determination Kit (Beyotime Biotech). The separation of proteins (40 µg) was performed by electrophoresis in 10%, 12% or 15% SDS-PAGE gels. The proteins were then transferred onto polyvinylidene difluoride (PVDF) membranes, and blocked with 5% skimmed milk in TBST under 25-30°C for 1 h. The membranes were then incubated with the corresponding primary antibodies at 4°C overnight. The primary antibodies of COL1A1 (COL-I), COL3A1 (COL-III) and GAPDH were purchased from Bioworld Technology (St.Louis Park, MN, USA) and LRG1 and Rap-1a/b were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Subsequently, the blots were incubated with peroxidase-conjugated secondary antibodies at 25-30°C for 1 h and visualized with enhanced chemiluminescence (ECL) detection reagent (Sigma Chemical Co). For HL-1 cell, HL-1 cells were washed with PBS for 3 times and treated with RIPA lysate on ice for 30 min, which was then transferred to a new EP tube for centrifugation for 10 min. The supernatant was quantified with a BCA Protein Concentration Determination Kit (Beyotime Biotech). The subsequent procedure was similar to above.
Immunofluorescence
Atrial tissue sections (3 mm thick) were obtained, blocked with 10% serum for 1 h and incubated against RAP1 and LRG1 overnight at 4°C. After washing for 3 times, the sections were incubated with a secondary antibody at 20-37°C for 1 h. Finally, the sections were stained with DAPI staining for 15 min, which were observed by an optical microscope (CKX41, Olympus, Tokyo, Japan).
ELISA assay
The expression level of LRG1 in serum of PeAF patients and control subjects was detected by RayBio® Human LRG1 ELISA Kit (RayBiotech. Inc) according to instructions. Absorbance at 450 nm was recorded, and concentration (μg/ml) was calculated from a standard curve.
Statistical analysis
Data are presented as mean ± standard deviation (SD). Unpaired and two-tailed Student’s t test was used to compare the results of Western blot analysis and ELISA assay of two groups. All data were statistically analyzed using SPSS 21.0. Differences were considered statistically significant at P < 0.05.
Results
Cohort characteristics
To quantitatively describe the proteomic variation in human left atrial appendage tissues, we specially selected the most differentiated tissue extremes represented by clinical indexes, such as BMI, LAD, LVEF, BNP and CRP. Table 1 summarizes the characteristics of patients with associated heart functions. All subjects were between 55 and 60 years old. The mean value of FEV1/FVC was 0.34 measured by spirometry tests in PeAF patients, which was obviously below the COPD diagnostic criterion of FEV1/FVC < 0.70.
Table 1.
Clinic characters of left atrium appendage samples
| PAF (n = 6) | SR (n = 6) | PeAF (n = 6) | |
|---|---|---|---|
| Age (y) | 56.3 ± 8.2 | 56.7 ± 8.6 | 56.3 ± 8.3 |
| Gender (Male/Female) | 4/2 | 4/2 | 4/2 |
| BMI (kg/m2) | 25.5 ± 3.6 | 22.9 ± 2.9 | 23.7 ± 4.6 |
| Hypertension (n) | 3 | 3 | 1 |
| Diabetes mellitus (n) | 0 | 0 | 1 |
| CKD (n) | 0 | 0 | 0 |
| Smoke abuse | 3 | 1 | 1 |
| Alcohol abuse | 0 | 1 | 1 |
| Coronary artery disease (n) | 1 | 3 | 0 |
| Stroke (n) | 1 | 0 | 1 |
| LAD (cm) | 5.3 ± 0.9 | 4.8 ± 0.9 | 5.8 ± 0.6 |
| LVEF (%) | 50.3 ± 10.1 | 54.5 ± 3.5 | 45.0 ± 11.2 |
| BNP (pg/ml) | 165.1 ± 96.6 | 122.6 ± 94.7 | 219.3 ± 91.0 |
| CRP (mg/dl) | 3.1 ± 1.4 | 5.4 ± 4.2 | 5.6 ± 4.3 |
| eGFR (mL/(min·1.73 m2)) | 97.3 ± 18.1 | 88.8 ± 20.9 | 99.2 ± 16.6 |
| Beta blockers | 3 | 4 | 5 |
| Anticoagulants | 4 | 3 | 3 |
| Digoxin | 0 | 0 | 0 |
| PGI2 | 1 | 1 | 2 |
| Heart rate (bpm) | 70.8 ± 5.4 | 85.6 ± 15.6 | 80.5 ± 37.4 |
BMI = Body mass index, CKD = Chronic kidney disease, LAD = Left atrium diameter, LVEF = Left ventricular ejection fraction, BNP = Brain natriuretic peptide, CRP = C-reactive protein, GFR = Glomerular filtration rate, PGI2 = Prostacyclin.
Masson staining of left atrial appendage
The histomorphology changes of left atrial appendage in control subjects (Figure 1A), PAF patients (Figure 1B) and PeAF patients (Figure 1C) were observed by masson staining. There were some collagen fibers appearing in left atrial appendage in PAF patients and many collagen fibers appearing in left atrial appendage in PeAF patients, compared with the control subjects (CTRL).
Figure 1.
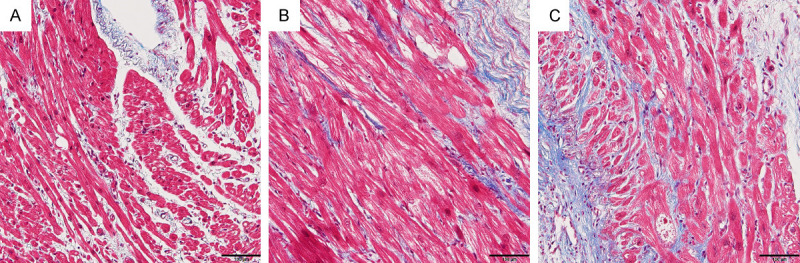
The fibrosis in left atrial appendage tissue samples of control subjects (A), PAF patients (B) and PeAF patients (C) was observed by Masson staining.
Bioinformatics analysis
The samples of left atrial appendage from patients and control subjects were randomly analyzed by TMT-based proteomics method to determine the DEPs which may be related to the molecular mechanism of PAF or PeAF. Our experimental design was shown in Figure 2. Figure 3 is a volcano plot of the log2 fold-change (x-axis) versus -log10 p value (the y-axis represents the probability that the protein could be differentially abundant). The red points in the upper right (ratio > 1.3) and the green points in the upper left (ratio < 0.77) sections with P < 0.05 represent significantly dysregulated proteins in PAF (left) and PeAF (right) patients. In total, 4682 proteins were identified, of which 4159 proteins were quantified. 118 DEPs (85 upregulated proteins and 33 downregulated proteins) (fold change = 1.3) were identified in atrial tissue from PeAF patients and control subjects.
Figure 2.
Workflow of global proteome analyses (SR: sinus rhythm).
Figure 3.
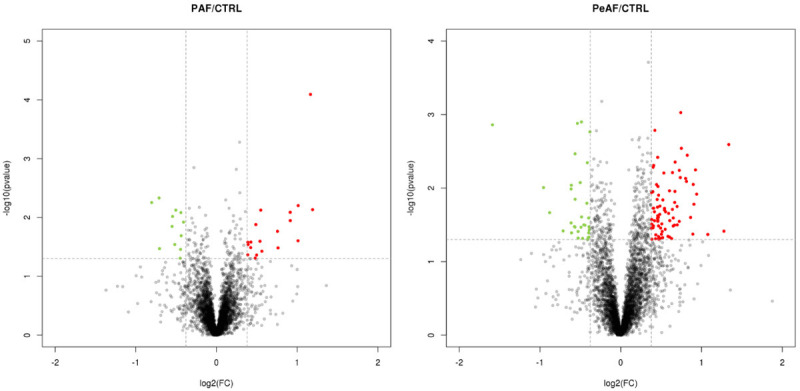
Differentially expressed proteins in left atrial appendage tissue of PAF and PeAF patients compared with control subjects (CTRL). PAF/CTRL: Volcano plot of protein changes in left atrial appendage tissue of PAF and CTRL groups. There were 30 proteins with fold change of > 1.5 or < 0.67 and p-values < 0.05 were identified as significantly dysregulated. PeAF/CTRL: There were 108 proteins with fold change of > 1.5 or < 0.67 and p-values < 0.05 were identified as significantly dysregulated.
According to Venn Diagram analysis, there are only 8 altered proteins in common between PAF and PeAF, such as COL1A1, COL1A2, COL3A1 and LRG1 (Figure 4). However, the difference of protein abundance between PeAF patients and control subjects was more obvious than that between PAF patients and control subjects, and patients with PeAF are at a higher risk than those with PAF. Therefore, we decided to focus our research on PeAF analysis.
Figure 4.
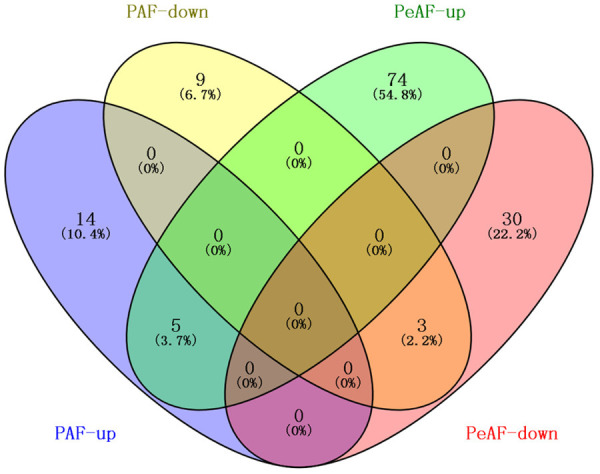
Venn diagram showing the overlapping differentially expressed proteins in left atrial appendage tissue from PAF and PeAF patients.
To investigate the functional classification of all DEPs, we performed GO analysis and categorized these proteins according to their molecular function (MF), biological process (BP), and cellular component (CC) by employing the Blast2 GO software. The DEPs derived from PeAF patients and control subjects (controls) were classified into biological process, cellular component and molecular function by the GO analysis tool (Figure 5A).
Figure 5.
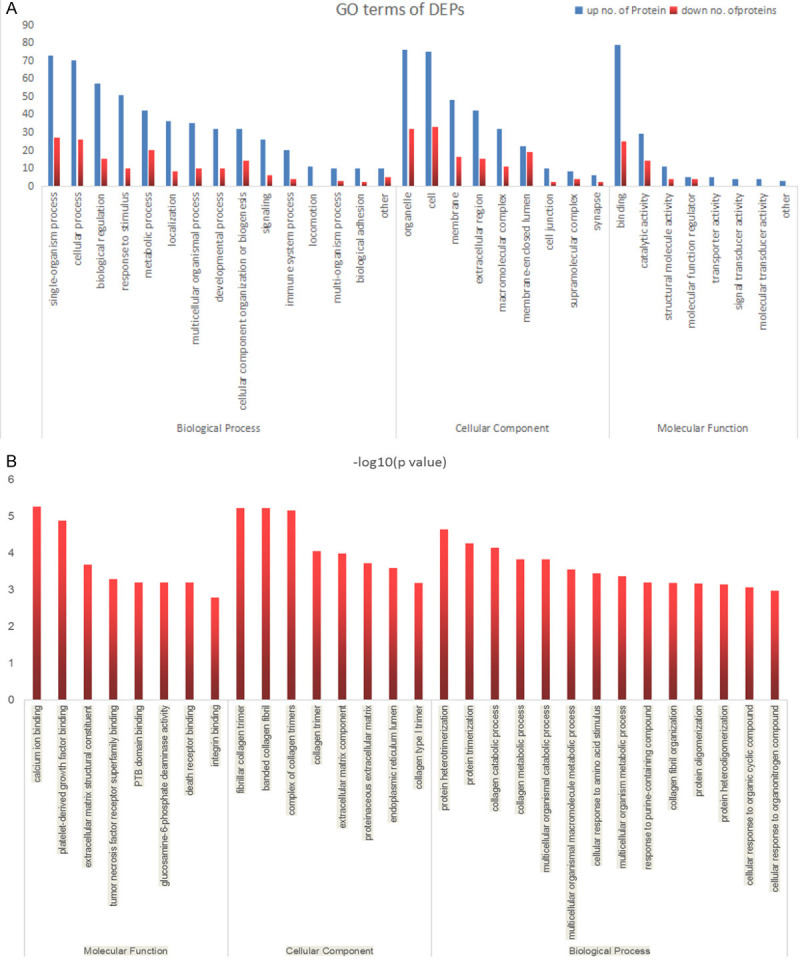
Go terms of differentially expressed proteins in left atrial appendage tissue from PeAF patients and control subjects. The identified proteins were classified into biological process, cellular components and molecular function (A). Go enrichment of differentially expressed proteins in left atrial appendage tissue from PeAF patients and control subjects (B).
To elucidate the functional differences between down-regulated and up-regulated proteins, three types of enrichment-based clustering analyses were conducted for the quantified proteins: Gene Ontology (GO) enrichment-based clustering analysis, KEGG pathway enrichment-based clustering analysis and protein complex enrichment-based clustering analysis. Figure 5B demonstrated that DEPs between PeAF patients and control subjects (controls) show a significant enrichment tendency in Calcium ion binding and other functional types. Negative logarithm (-log10) transformation is carried out for the p value obtained from the enrichment test (Fisher’s exact test is used here). The larger the value is obtained after conversion, the more significant the enrichment of this functional type will be.
Overexpression of DEPs on a given pathway refers to the pathway enrichment analysis of DEPs. Figure 6 exhibited the results of KEGG pathway enrichment analysis of DEPs in atrial tissue from PeAF patients and control subjects (controls). Each point in the graph represents a KEGG pathway, the name of which can be clearly seen on the left coordinate axis. The x-axis represents enrichment factor. The larger the enrichment factor is, the more reliable the enrichment significance of the differential protein in this pathway will be. KEGG pathway enrichment analysis showed that the altered proteins in atrial tissue were predominantly involved in protein digestion and absorption, ECM-receptor interaction, focal adhesion and PI3K-AKT signaling pathway. All of these pathways have reported to be related to the myocardial fibrosis, especially focal adhesion [21].
Figure 6.

KEGG pathway enrichment analysis of differentially expressed proteins in left atrial appendage tissue from PeAF patients and control subjects.
Figure 7 is a heat map of the most significant DEPs in atrial tissue from PeAF patients and control subjects (controls). The x-axis represents different atrial tissue samples and the DEPs in atrial tissue from PeAF patients and control subjects (controls) are shown on the right coordinate axis. The color of each square represents different expression levels of proteins. The protein abundance showed by heat map was different in atrial tissue between PeAF patients and control subjects (controls).
Figure 7.
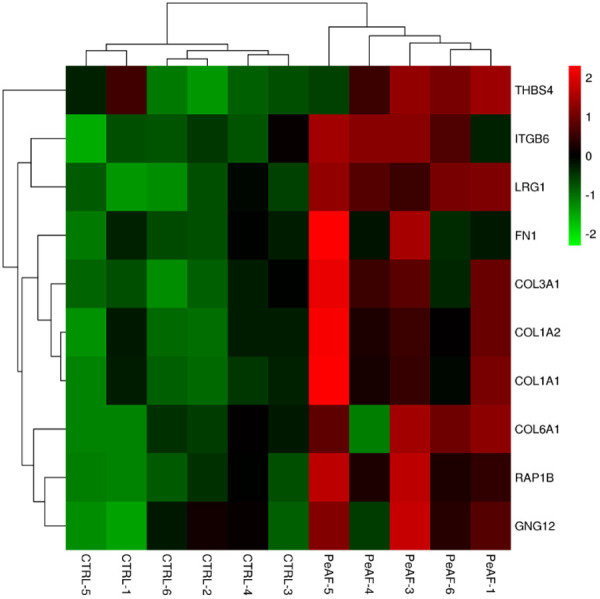
Heat map of the most significant differentially expressed proteins in left atrial appendage tissue from PeAF patients and control subjects (CTRL). The colors represent the up-regulation (red) or down-regulation (green) of the gene for each type.
Proteins often cannot act alone, but they can interact with other proteins to perform multiple functions in different pathways. To understand the molecular mechanism between PeAF patients and control subjects, the identified DEPs were connected by string to form the protein-protein interaction (PPI) regulatory network [22,23] (Figure 8). The network nodes mean proteins. Moreover, the different types of lines connecting the proteins represent various protein-protein interactions. COL1A1, COL1A2, COL3A1, COL5A2, COL6A1 and COL6A2 were collagens that participated in the composition of myocardial fibers. These collagens interacted with each other and connected with FN1, RAP1B and LRG1.
Figure 8.
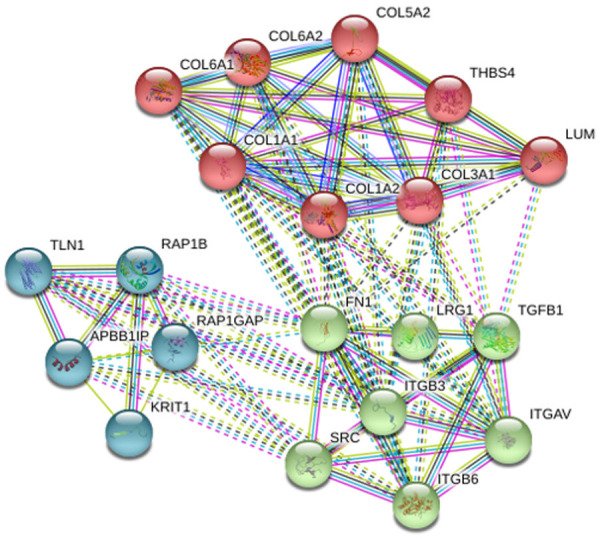
Protein interaction network generated using STRING software. Protein-protein interaction regulatory network of differentially expressed proteins between PeAF patients and control subjects.
Verification of the differently expressed proteins
In this experiment, 4682 proteins were identified in total atrial tissue samples, of which 4159 proteins were quantified. A total of 118 DEPs were identified in atrial tissue from PeAF patients and control subjects (controls). Four DEPs, which were COL-I, COL-III, RAP1 and LRG1, were validated using western blot to verify the proteomics results. As shown in Figure 9A, the expression of COL-I, COL-III, Rap-1a/b and LRG1 was increased in atrial tissue in PeAF patients than in control subjects (Ctrl). The results of Western blot analysis were consistent with the results of TMT-based quantitative proteomics. Immunofluorescence experiment was conducted to determine the expression of Rap1 and leucine-rich alpha-2-glycoprotein 1 (LRG1) in atrial tissue from PeAF patients (Figure 9B). RAP1 (orange) was prominent in the atrial myocardial cells. The expression of LRG1 (red) was found both in myocardial cells and extracellular matrix. The expression level of LRG1 in serum of PeAF patients was decreased compared with that in control subjects (Ctrl). As shown in Figure 9C, the expression level of LRG1 in serum of PeAF patients was lower than that in control subjects (Ctrl). In pacing model, the expression of Rap-1a/b and LRG1 in pacing HL-1 cells was increased compared with control subjects (Ctrl) (Figure 9D).
Figure 9.
Several altered proteins in atrial tissue, human serum and pacing HL-1 cells were selected to verify the results in proteomes analysis. A. The expression of COL-I, COL-III, Rap-1a/b and LRG1 in atrial tissue were detected by Western Blot. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. Ctrl group. B. The expression of Rap1 and LRG1 in left atrial appendage tissue from PeAF patients and control subjects was showed by Immunofluorescence experiment. C. The expression level of LRG1 in human serum was detected by ELISA assay. **P < 0.01 vs. Ctrl group. D. The expression of Rap-1a/b and LRG1 in pacing HL-1 cells were detected by Western Blot. Data are shown as the mean ± SEM. ***P < 0.001 vs. Ctrl group.
Discussion
AF refers to the loss of regular atrial electrical activity, replaced by irregular atrial fibrillation waves, which is one of the most common clinical arrhythmias [1]. In the process of AF, effective contraction of atria disappears, and cardiac output decreases by 25% or less in sinus rhythm. AF-induced hemodynamic instability and high blood coagulation lead to cerebral apoplexy and other thromboembolism complications, resulting in high disability and mortality rates, which seriously harm the human health [24]. Stroke is one of the biggest hazards of PeAF and the stroke caused by atrial fibrillation can bring more serious consequences [25].
The diagnosis of AF relies on clinical symptoms and signs and can be confirmed by an electrocardiogram [26,27]. In previous studies, a set of AF biomarkers were chosen for AF patients with hypertension or metabolic syndrome [28,29]. Recently, Akshay Goel et al [30] have found that L-arginine derivatives, ADMA and ADMA, appear to be promising biomarkers for the prediction of prognosis and risk stratification of AF patients. However, there is little research on biomarkers for early diagnosis of AF. Therefore, differentiating the proteins in atrial tissue from PeAF patients and control subjects could provide an effective way to identify diagnostic biomarkers. In the present study, we used TMT-based proteomics coupled with nano-LC-MS/MS to screen the DEPs in atrial tissue from PAF or PeAF patients and control subjects. Because the difference of protein abundance between PeAF patients and control subjects is more obvious than that between PAF patients and control subjects, and PeAF patients have a five-year survival rate only, DEPs in atrial tissue from PeAF patients and control subjects are explored in this study.
Among the differentially expressed extracellular matrix (ECM) proteins, COL-I, COL-III, RAP1 and LRG1 were selected for their biological functions in the process of myocardial fibrosis in PeAF. One kind of DEPs was collagens, including COL-I and COL-III. In the heart, collagens are the main ECM proteins with unique physiological properties encoded by different genes. ECM is the general term of structural glycoproteins, protein polysaccharide and glycosaminoglycan. With the in-depth research on the pathophysiology of heart disease, ECM is found to be changed in the heart disease. At present, collagens in the heart mainly include COL-I, COL-III, COL-IV, COL-V and COL-VI, among which COL-I content is the highest followed by COL-III. COL-I is a main component of large collagenous fibers parallel to the myocardial bundles and its increase may enhance the myocardial tensile ability. In addition, COL-III mainly surrounds cardiomyocytes with good extensibility and resilience. Myocardial collagen network is an important factor in determining cardiac muscle extension and resilience. COL-III constitutes a lateral connection between cardiomyocytes and muscle bundles with spring-like spiral structure [31]. COL-I and COL-III also play important roles in cardiac remodeling. A study has shown that at the early stage of remodeling, collagen degradation is enhanced while COL-III is often significantly increased, and COL-I is mainly increased at the advanced stage of remodeling [32]. In this study, the expression of COL-I and COL-III is increased in atrial tissue from PeAF patients compared with control subjects (controls), indicating the decreased cardiac muscle stretching and myocardial resilience.
RAP1, a member of the Ras superfamily, is a GTP binding protein involved in the cell-matrix interactions and it mediates the cell adhesion and apoptosis by binding to GTP (active form) or GDP (inactive form) signaling pathway [33,34]. In this study, it can be seen from the PPI regulatory network that RAP1 can interact with LRG1, and it is increased in atrial tissue from PeAF patients compared with control subjects (controls). In addition, the expression of Rap-1a/b in pacing HL-1 cells was also increased compared with control subjects (Ctrl). LRG1, a newly discovered angiogenic factor in recent years, is a secretory glycoprotein mainly involved in cell differentiation, cell adhesion, cell migration, apoptosis and angiogenesis [35,36]. A study indicated that RAP1, which is located in cytoplasm, is expressed mainly in Golgi apparatus in normal rat spermatogenic cells [37]. After RAP1 activation, the signal is transmitted to the nucleus through a series of downstream effectors, regulating the biological functions of the cell [38-40]. The results of immunofluorescence experiment showed that RAP1 mainly appeared in atrial myocardial cells and LRG1 appeared both in myocardial cells and extracellular matrix. Thus, we speculated that RAP1 may regulate the secretion process of LRG1 in myocardial cells. Further studies are required to explore the effect of RAP1 on the expression of LRG1 in PeAF and to investigate the possibility of preventing or reversing PeAF by up-regulating the RAP1 expression.
LRG1 is a member of leucine-rich repeat (LRR) protein family, many of which participate in protein-protein interactions, signaling and cell adhesion [41]. AF often results from cardiomyopathy, hypertension, coronary heart disease, valvular disease, heart failure and congenital heart disease. Studies have shown that different LRG1 expressions are associated with various heart diseases [42,43]. Gopalakrishnan et al [44] demonstrated that LRG1 expression was obviously decreased in the left ventricle tissue of hypertensive rat with concentric cardiac hypertrophy. LRG1 expression was decreased at the later period of heart failure due to dilated cardiomyopathy [45]. Transitory activation of AKT1 caused reversible cardiac hypertrophy, which was related to the decrease of LRG1 [46]. Similarly, PI3K down-regulation obviously alleviated cardiac hypertrophy in transgenic mice, which was also related to the simultaneous LRG1 overexpression [47]. LRG1 expression was decreased in mouse models of pathological cardiac hypertrophy. LRG1 expression was obviously down-regulated in the heart of mice with compensated pressure overload hypertrophy [48,49]. Moreover, the LRG1 expression was obviously decreased in the ventricle of α-TM E180G transgenic mice, while LRG1 overexpression could reverse cardiac hypertrophy in α-TM E180G transgenic mice [50]. LRG1 was also expressed in cardiac fibroblasts and cardiomyocytes [51,52]. These findings indicated that LRG1 expression is related to the fibrosis, abnormal vascular properties and heart failure induced by genetic modification, pressure overload, or hypertension. The results of the present study indicated that LRG1 expression is increased in atrial tissue while decreased in serum of PeAF patients compared with control subjects (controls). We speculated that the increase of LRG1 in tissues may lead to the decrease of LRG1 in serum. Because LRG1 can compete with TGF-β1 for TGF-β receptor binding to inhibit the proliferation of fibroblasts induced by TGF-β1 [53], compensatory increase of LRG1 expression in AF process was to inhibit tissue fibrosis.
In conclusion, the DEPs between PeAF patients and control subjects were discovered by a TMT-based quantitative proteomics approach through analyzing the left atrial appendage. Four DEPs (COL1A1, COL3A1, RAP1 and LRG1) were validated by Western blot analysis in samples of left atrial appendage of PeAF patients and control subjects. Consistent with the previous reports, our results confirmed fibrosis-related proteins COL1A1 and COL3A1, and novel biomarkers LRG1 and RAP1B as specific PeAF biomarkers. The present results provide a novel understanding of the pathophysiological mechanisms of PeAF and validated biomarkers for the diagnosis of PeAF, which may facilitate the development of therapeutic targets. Combining TMT and LC-MS/MS coupled with subsequent investigation may provide further insights into the molecular mechanisms in pathogenesis of PeAF.
Acknowledgements
This work was supported in part by Fundamental Research Funds for the Central Universities [YG1903002], Jiangsu Provincial Medical Youth Talent [QNRC2016034], Jiangsu Province Health Department Program Grant [Z201411], Key Project supported by Medical Science and technology development Foundation, Nanjing Department of Health [YKK17066].
Disclosure of conflict of interest
None.
References
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O’Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics-2018 update: a report from the American heart association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Menezes AR, Lavie CJ, Dinicolantonio JJ, O’Keefe J, Morin DP, Khatib S, Milani RV. Atrial fibrillation in the 21st century: a current understanding of risk factors and primary prevention strategies. Mayo Clin Proc. 2013;88:394–409. doi: 10.1016/j.mayocp.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Deplanque D, Corea F, Arquizan C, Parnetti L, Mas JL, Gallai V, Leys D. Stroke and atrial fibrillation: is stroke prevention treatment appropriate beforehand? SAFE I Study Investigators. Heart. 1999;82:563–569. doi: 10.1136/hrt.82.5.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma M, Cornelius VR, Patel JP, Davies JG, Molokhia M. Efficacy and harms of direct oral anticoagulants in the elderly for stroke prevention in atrial fibrillation and secondary prevention of venous thromboembolism: systematic review and meta-analysis. Circulation. 2015;132:194–204. doi: 10.1161/CIRCULATIONAHA.114.013267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson NL, Anderson NG. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson CL. Bacterial proteomics and vaccine development. Am J Pharmacogenomics. 2002;2:59–65. doi: 10.2165/00129785-200202010-00005. [DOI] [PubMed] [Google Scholar]
- 7.Lahner E, Bernardini G, Possenti S, Renzone G, Scaloni A, Santucci A, Annibale B. Immunoproteomics of helicobacter pylori infection in patients with atrophic body gastritis, a predisposing condition for gastric cancer. Int J Med Microbiol. 2011;301:125–132. doi: 10.1016/j.ijmm.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Zou L, Wang X, Guo Z, Sun H, Shao C, Yang Y, Sun W. Differential urinary proteomics analysis of myocardial infarction using iTRAQ quantification. Mol Med Rep. 2019;19:3972–3988. doi: 10.3892/mmr.2019.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, Wang D, Liu D, Guo Z, Shao C, Sun W, Zeng Y. Differential urinary proteins to diagnose coronary heart disease based on iTRAQ quantitative proteomics. Anal Bioanal Chem. 2019;411:2273–2282. doi: 10.1007/s00216-019-01668-7. [DOI] [PubMed] [Google Scholar]
- 10.Doulamis IP, Samanidis G, Tzani A, Antoranz A, Gkogkos A, Konstantopoulos P, Pliaka V, Minia A, Alexopoulos LG, Perrea DN, Perreas K. Proteomic profile of patients with atrial fibrillation undergoing cardiac surgery. Interact Cardiovasc Thorac Surg. 2019;28:94–101. doi: 10.1093/icvts/ivy210. [DOI] [PubMed] [Google Scholar]
- 11.Stryinski R, Mateos J, Pascual S, Gonzalez AF, Gallardo JM, Lopienska-Biernat E, Medina I, Carrera M. Proteome profiling of L3 and L4 anisakis simplex development stages by TMT-based quantitative proteomics. J Proteomics. 2019;201:1–11. doi: 10.1016/j.jprot.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Chen YH, Huang AQ, Ao W, Wang ZW, Yuan JJ, Song Q, Wei DH, Ye HH. Proteomic analysis of serum proteins from HIV/AIDS patients with talaromyces marneffei infection by TMT labeling-based quantitative proteomics. Clin Proteomics. 2018;15:1–12. doi: 10.1186/s12014-018-9219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilic P, Guillemin N, Kovacevic A, Ljubic BB, Jovic I, Galan A, Eckersall PD, Burchmore R, Mrljak V. Serum proteome profiling in canine idiopathic dilated cardiomyopathy using TMT-based quantitative proteomics approach. J Proteomics. 2018;179:110–121. doi: 10.1016/j.jprot.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Shi XL, Liang BC, Shi ZY, Wang B, Wu P, Kong LC, Yao JL, Li CW. Discovery and identification of serum biomarkers for postmenopausal osteoporosis based on TMT labeling and HPLC-MS/MS technology. Int J Clin Exp Med. 2017;10:334–346. [Google Scholar]
- 15.Zhou Q, Xie F, Zhou B, Wang J, Wu B, Li L, Kang Y, Dai R, Jiang Y. Differentially expressed proteins identified by TMT proteomics analysis in bone marrow microenvironment of osteoporotic patients. Osteoporos Int. 2019;30:1089–1098. doi: 10.1007/s00198-019-04884-0. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki-Hatano S, Saha M, Soustek MS, Kang PB, Byrne BJ, Cade WT, Pacak CA. AAV9-TAZ gene replacement ameliorates cardiac TMT proteomic profiles in a mouse model of barth syndrome. Mol Ther Methods Clin Dev. 2019;13:167–179. doi: 10.1016/j.omtm.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romanick SS, Ulrich C, Schlauch K, Hostler A, Payne J, Woolsey R, Quilici D, Feng Y, Ferguson BS. Obesity-mediated regulation of cardiac protein acetylation: parallel analysis of total and acetylated protein via TMT-tagged mass spectrometry. Biosci Rep. 2018;38:1–18. doi: 10.1042/BSR20180721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claycomb W, Lanson N, Stallworth B, Egeland D, Delcarpio J, Bahinski A, Izzo N. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Shen W, Rottman JN, Wikswo JP, Murray KT. Rapid stimulation causes electrical remodeling in cultured atrial myocytes. J Mol Cell Cardiol. 2005;38:299–308. doi: 10.1016/j.yjmcc.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Brundel B, Kampinga H, Henning R. Calpain inhibition prevents pacing-induced cellular remodeling in a HL-1 myocyte model for atrial fibrillation. Cardiovasc Res. 2004;62:521–528. doi: 10.1016/j.cardiores.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Fan G, Zhao H, Wang Z, Li F, Zhang P, Zhang J, Wang X, Wang W. Targeted inhibition of focal adhesion kinase attenuates cardiac fibrosis and preserves heart function in adverse cardiac remodeling. Sci Rep. 2017;7:43146. doi: 10.1038/srep43146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing T, Wang C, Zhao X, Dai C, Xu X. Proteome analysis using iTRAQ reveals the alterations in stress-induced dysfunctional chicken muscle. J Agric Food Chem. 2017;65:2913–2922. doi: 10.1021/acs.jafc.6b05835. [DOI] [PubMed] [Google Scholar]
- 23.Yu Q, Wu W, Tian X, Hou M, Dai R, Li X. Unraveling proteome changes of Holstein beef M. semitendinosus and its relationship to meat discoloration during post-mortem storage analyzed by label-free mass spectrometry. J Proteomics. 2016;154:85. doi: 10.1016/j.jprot.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 24.January CT, Wann LS, Alpert JS, Calkins H, Cigarroa JE, Cleveland JC, Conti JB, Ellinor PT, Ezekowitz MD, Field ME, Murray KT, Sacco RL, Stevenson WG, Tchou PJ, Tracy CM, Yancy CW. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the american college of cardiology/American heart association task force on practice guidelines and the heart rhythm society. J Am Coll Cardiol. 2014;64:e1–e76. doi: 10.1016/j.jacc.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Li YG, Lip GYH. Stroke prevention in atrial fibrillation: state of the art. Int J Cardiol. 2019;287:201–209. doi: 10.1016/j.ijcard.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 26.Farris GR, Smith BG, Oates ET, Colon C, Doppalapudi H. New atrial fibrillation diagnosed by 30-day rhythm monitoring. Am Heart J. 2019;209:29–35. doi: 10.1016/j.ahj.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Narui R, Yamane T, Tokuda M, Ikewaki H, Okajima E, Sato H, Oseto H, Isogai R, Tokutake K, Yokoyama K, Kato M, Ito K, Tanigawa S, Yamashita S, Inada K, Matsuo S, Miyanaga S, Sugimoto K, Yoshimura M. Atrial fibrillation diagnosed by a medical checkup is associated with a poor outcome of catheter ablation. Heart Vessels. 2018;33:770–776. doi: 10.1007/s00380-017-1115-z. [DOI] [PubMed] [Google Scholar]
- 28.Tsioufis C, Konstantinidis D, Nikolakopoulos E, Vemou E, Kalos T, Georgiopoulos G, Vogiatzakis N, Ifantis A, Konstantinou K, Gennimata V. Biomarkers of atrial fibrillation in hypertension. Curr Med Chem. 2019;26:888–897. doi: 10.2174/0929867324666171006155516. [DOI] [PubMed] [Google Scholar]
- 29.Onuchina EL, Solov’Ev OV, Onuchin SG, Mochalova OV, Kononov SK. Assessment of risk factors of atrial fibrillation in patients with metabolic syndrome. Klin Med. 2012;90:72–6. [PubMed] [Google Scholar]
- 30.Goel A, Agnihotri K, Ashish K, Subramany S, Paydak H. Novel biomarkers for atrial fibrillation: a recent update. Int J Cardiol. 2019;277:248. doi: 10.1016/j.ijcard.2018.08.077. [DOI] [PubMed] [Google Scholar]
- 31.Weber K, Sun Y. Remodeling of the cardiac interstitium in ischemic cardiomyopathy. Congestive Heart Failure. 2000:117–136. [Google Scholar]
- 32.Weber KT. Cardiac interstitium in health and disease: the fibrillar collagen network. J Am Coll Cardiol. 1989;13:1637–1652. doi: 10.1016/0735-1097(89)90360-4. [DOI] [PubMed] [Google Scholar]
- 33.Frische EW, Zwartkruis FJ. Rap1, a mercenary among the Ras-like GTPases. Dev Biol. 2010;340:1–9. doi: 10.1016/j.ydbio.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 34.Kooistra MR, Dubé N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 35.Serada S, Fujimoto M, Terabe F, Iijima H, Shinzaki S, Matsuzaki S, Ohkawara T, Nezu R, Nakajima S, Kobayashi T, Plevy SE, Takehara T, Naka T. Serum leucine-rich alpha-2 glycoprotein is a disease activity biomarker in ulcerative colitis. Inflamm Bowel Dis. 2012;18:2169–2179. doi: 10.1002/ibd.22936. [DOI] [PubMed] [Google Scholar]
- 36.Druhan LJ, Lance A, Li S, Price AE, Emerson JT, Baxter SA, Gerber JM, Avalos BR. Leucine rich α-2 glycoprotein: a novel neutrophil granule protein and modulator of myelopoiesis. PLoS One. 2017;12:e0170261. doi: 10.1371/journal.pone.0170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berruti G. Signaling events during male germ cell differentiation: bases and perspectives. Front Biosci. 1998;3:D1097–1108. doi: 10.2741/a347. [DOI] [PubMed] [Google Scholar]
- 38.Stork PJ. Does Rap1 deserve a bad Rap? Trends Biochem Sci. 2003;28:267–75. doi: 10.1016/S0968-0004(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 39.Su L, Hattori M, Moriyama M, Murata N, Harazaki M, Kaibuchi K, Minato N. AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem. 2003;278:15232–8. doi: 10.1074/jbc.M211888200. [DOI] [PubMed] [Google Scholar]
- 40.Boettner B, Harjes P, Ishimaru S, Heke M, Qing Fan H, Qin Y, Van Aelst L, Gaul U. The AF-6 homolog canoe acts as a rap1 effector during dorsal closure of the drosophila embryo. Genetics. 2003;165:159–69. doi: 10.1093/genetics/165.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng AC, Eisenberg JM, Heath RJ, Huett A, Robinson CM, Nau GJ, Xavier RJ. Colloquium paper: human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4631–8. doi: 10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson CJ, Ledwidge MT, Phelan D, Collier P, Byrne JC, Dunn MJ, McDonald KM, Baugh JA. Proteomic analysis of coronary sinus serum reveals leucine-rich alpha 2-glycoprotein as a novel biomarker of ventricular dysfunction and heart failure. Circ Heart Fail. 2011;4:188–97. doi: 10.1161/CIRCHEARTFAILURE.110.952200. [DOI] [PubMed] [Google Scholar]
- 43.Chia RN, Song WH, Wang XM. Role of leucine-rich alpha-2-glycoprotein 1 (LRG1) in diabetes-related ischaemic heart diseases. J Vasc Res. 2019;56:21–21. [Google Scholar]
- 44.Gopalakrishnan K, Morgan E, Yerga-Woolwine S, Farms P, Kumarasamy S, Kalinoski A, Liu X, Wu J, Liu L, Joe B. Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-QT interval and hypertension. Hypertension. 2011;57:764–71. doi: 10.1161/HYPERTENSIONAHA.110.165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blaxall BC, Spang R, Rockman HA, Koch WJ. Differential myocardial gene expression in the development and rescue of murine heart failure. Physiol Genomics. 2003;17:105–14. doi: 10.1152/physiolgenomics.00087.2003. [DOI] [PubMed] [Google Scholar]
- 46.Schiekofer S, Shiojima I, Sato K, Galasso G, Oshima Y, Walsh K. Microarray analysis of Akt1 activation in transgenic mouse hearts reveals transcript expression profiles associated with compensatory hypertrophy and failure. Physiol Genomics. 2006;27:156–170. doi: 10.1152/physiolgenomics.00234.2005. [DOI] [PubMed] [Google Scholar]
- 47.McMullen J, Shioi T, Zhang L, Tarnavski O, Sherwood MC, Dorfman A, Longnus S, Pende M, Martin K, Blenis J, Thomas G, Izumo S. Deletion of ribosomal S6 kinases does not attenuate pathological, physiological, or insulin-like growth factor 1 receptor-phosphoinositide 3-kinase-induced cardiac hypertrophy. Mol Cell Biol. 2004;24:6231–6240. doi: 10.1128/MCB.24.14.6231-6240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao MM, Amy C, Jennifer P, Giovanni F, Daniel B. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol Genomics. 2004;19:93–105. doi: 10.1152/physiolgenomics.00040.2004. [DOI] [PubMed] [Google Scholar]
- 49.Smeets PJ, de Vogel-van den Bosch HM, Willemsen PH, Stassen AP, Ayoubi T, van der Vusse GJ, van Bilsen M. Transcriptomic analysis of PPARalpha-dependent alterations during cardiac hypertrophy. Physiol Genomics. 2008;36:15–23. doi: 10.1152/physiolgenomics.90296.2008. [DOI] [PubMed] [Google Scholar]
- 50.Rajan S, Williams SS, Jagatheesan G, Ahmed RP, Fuller-Bicer G, Schwartz A, Aronow BJ, Wieczorek DF. Microarray analysis of gene expression during early stages of mild and severe cardiac hypertrophy. Physiol Genomics. 2006;27:309–317. doi: 10.1152/physiolgenomics.00072.2006. [DOI] [PubMed] [Google Scholar]
- 51.Ifkovits JL, Addis RC, Epstein JA, Gearhart JD. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PloS one. 2014;9:e89678. doi: 10.1371/journal.pone.0089678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, Huang XN, Stewart AF, Sepulveda JL. Gene expression changes associated with fibronectin-induced cardiac myocyte hypertrophy. Physiol Genomics. 2004;18:273–283. doi: 10.1152/physiolgenomics.00104.2004. [DOI] [PubMed] [Google Scholar]
- 53.Yu RB, Li K, Wang G, Gao GM, Du JX. MiR-23 enhances cardiac fibroblast proliferation and suppresses fibroblast apoptosis via targeting TGF-β1 in atrial fibrillation. Eur Rev Med Pharmacol Sci. 2019;23:4419–4424. doi: 10.26355/eurrev_201905_17950. [DOI] [PubMed] [Google Scholar]




