Abstract
Aims: This study was to investigate the effect of TUG1 on apoptosis and ECM degradation of human degenerative intervertebral disc nucleus pulposus cells (NPCs) and its mechanism. Methods: Human degenerative intervertebral disc NP tissues were obtained from 10 patients with lumbar disc herniation (LDH) who underwent lumbar spine surgery (IDD group), normal intervertebral disc NP tissues were obtained from 10 patients with lumbar vertebrae fractures (LVF group). Results: The expression of TUG1 and HMGB1 protein in human degenerative disc NP tissues and NPCs was significantly increased, while the level of miR-26a was significantly decreased. Overexpression of TUG1 inhibited the proliferation while promoted apoptosis and ECM degradation of human degenerative intervertebral disc NPCs. Simultaneously, the effect of TUG1 knockdown on NPCs was opposite. Interestingly, TUG1 acted as an endogenous sponge to down-regulate the expression of miR-26a in NPCs by direct binding to miR-26a. Overexpression of miR-26a reversed the effects of TUG1 overexpression on apoptosis and ECM degradation. Additionally, HMGB1 was a target gene of miR-26a. The increased expression of HMGB1 induced by TUG1 overexpression could be reversed by the introduction of miR-26a mimic. Overexpression of TUG1 significantly upregulated the expression of p65 in the nucleus, while overexpression of TUG1 partially abolished the inhibition of NF-κB by QNZ pretreatment. Conclusion: TUG1 could promote the apoptosis and ECM degradation of degenerated intervertebral disc NPCs by regulating the miR-26a/HMGB1, which may be involved in the activation of NF-κB pathway.
Keywords: Intervertebral disc degeneration, nucleus pulposus cells, long-chain non-coding RNAs taurine upregulated gene 1, apoptosis, extracellular matrix degradation
Introduction
Intervertebral disc (IVD) degeneration (IDD) is a typical common and frequently-occurring disease in clinical practice that can cause a series of clinical syndromes, including cervical spondylosis, disc herniation, etc. [1]. Current research suggests that IDD occurs under a variety of physiological and pathological conditions that can be affected by a variety of factors, such as genetics, cellular senescence, mechanical load, matrix degradation, inflammation and apoptosis. The decrease in the number of nucleus pulposus cells (NPCs) and the loss of extracellular matrix (ECM) are the core characteristics of IDD [2]. However, there is no unified theory on the exact mechanism of IDD. Once IDD is degenerated, it is difficult to reverse, and there is still no effective means to resist degeneration of the IVD [3]. Therefore, it is necessary to carry out basic research in depth to clarify the mechanism of IDD.
Long noncoding RNA (lncRNA) is a group of endogenous, non-coding transcripts of more than 200 nucleotides in length. Increasing evidences suggest that lncRNA acts as a key regulator in biological processes. Abnormal expression of lncRNA has been shown to be involved in a variety of human diseases, including IDD and osteoarthritis [4]. However, lncRNA is still in its infancy in the study of the regulation mechanism of IDD.
Taurine upregulated gene 1 (TUG1) is a lncRNA located on chromosome 22q12.2 with a length of 7598 nt. As an oncogene, TUG1 has been shown to be involved in cell proliferation, apoptosis, differentiation and metastasis [5]. Recently, Chen et al. [6] found that TUG1 is up-regulated in NP samples from patients with lumbar disc herniation, and knocking out TUG1 can inhibit TNF-α-induced apoptosis and senescence of NPCs. However, the mechanism of action of TUG1 in the pathogenesis of IDD remains to be further explored. lncRNA can competitively adsorb certain miRNAs through endogenous RNA pathways to regulate gene expression to participate in the development of IDD [7]. Xi et al. [8] confirmed that lncRNA HCG18 acts as an endogenous sponge to inhibit the proliferation and induce apoptosis of NPCs by directly binding to miR-146a-5p to promote IDD development. Recent studies have shown that there may be a targeted regulatory relationship between TUG1 and miR-26a [9-11]. We also confirmed that miR-26a is one of the targets of TUG1 through bioinformatics software. We speculated that TUG1 may participate in the promotion of IDD by targeting miR-26a. As such, we collected human degenerative disc NP tissue samples and explored the role of TUG1 in IDD and related mechanisms by in vitro culture of human degenerated intervertebral disc NPCs.
Methods
Patients and samples
All specimens were taken from March 2017 to March 2019 in the Orthopedic Department of Chinese Academy of Medical Sciences Peking Union Medical College Hospital for lumbar spine surgery. Human degenerative intervertebral disc NP tissues were obtained from 10 patients with lumbar disc herniation (LDH) who underwent lumbar spine surgery (IDD group), including 5 males and 5 females (age 45-65 years, mean age 52 ± 10 years). Preoperative symptoms, signs and imaging were clearly diagnosed as LDH, which was ineffective by conservative treatment and had surgical indications. Postoperative pathological findings confirmed degenerative disc tissues. The grade was III-IV according to the Pfirrmann degenerative intervertebral disc sagittal MRI grading standard [12]. Normal intervertebral disc NP tissues were obtained from 10 patients with lumbar vertebrae fractures (LVF group), including 5 males and 5 females (age 45-65 years, mean age 53 ± 11 years). Intraoperative human NP tissues were placed in DMEM/F12 medium (HyClone, USA) containing 10% fetal bovine serum (Gibco, USA), stored in an ice box and transferred to a clean bench for subsequent processing. This study was approved by the Ethics Committee of Chinese Academy of Medical Sciences Peking Union Medical College Hospital.
Quantitative real-time (qRT-PCR)
Total RNA in NP tissues and NPCs was extracted using the Total RNA Rapid Extraction Kit (Sigma-Aldrich, USA). 500 ng total RNA was used to analyze the relative expression of TUG1 and miR-26a by using SYBR Green PCR kit (Takara, Japan) on a 7500 fast realtime PCR system (Applied Biosystems, USA). GAPDH and U6 were used as internal parameters. The relative expression of the gene of interest was calculated by the 2-ΔΔt method. The primers are as follows: TUG1, 5’-ACGACTGAGCAAGCACTACC-3’(F) and 5’-CTCAGCAATCAGGAGGCACA-3’(R); miR-26a, 5’-CTGTCAACGATACGCTAC-3’(F) and 5’-GTAATCCAGGATAGGCTG-3’(R); GAPDH, 5’-ACAACTTTGGTATCGTGGAAGG-3’(F) and 5’-GCCATCACGCCACAGTTTC-3’(R); U6, 5’-GCCAGCACCATGCTCTTCTA-3’(F) and 5’-GGTTCCACAGATGCTCAGGTC-3’(R).
Immunohistochemistry
Paraffin sections (4 μm) were prepared after NP tissue was fixed in 10% formalin for 24 h. Paraffin sections were dewaxed and hydrated and placed in citrate buffer (pH 6.0) for antigen retrieval. Paraffin sections were dewaxed and hydrated and placed in citrate buffer (pH 6.0) for antigen retrieval. The primary antibody anti-HMGB1 (Abcam, Cambridge, MA, USA) was added dropwise to the sections and placed at 4°C overnight. After washing with PBS, a multi-polymer secondary antibody was added dropwise to the sections. After washing, the sections were developed with DAB. The sections were counterstained with hematoxylin and returned to blue with an aqueous solution of lithium carbonate, followed by dehydration, transparency and sealing. The expression of HMGB1 in the NP tissue of the intervertebral disc was observed under an optical microscope.
Western blot
Cells were completely lysed in lysis buffer (Takara, Japan) and extracted nuclear proteins and cytoplasmic proteins. Protein concentration was determined using the BCA Protein Assay Kit (Pierce). 50 μg of the sample was taken for SDS-PAGE electrophoresis, and transfered to the PVDF membrane. Then, membranes were blocked with 5% skim milk for 1 h at room temperature. Membranes were then incubated with the corresponding primary antibody, including anti-HMGB1 (Abcam, USA), anti-Bcl-2 (Abcam, USA), anti-Bax (Abcam, USA), anti-cleaved caspase-3 (Abcam, USA), anti-collagen II (Abcam, USA), anti-aggrecan (Abcam, USA), anti-MMP-3 (Abcam, USA), anti-MMP-13 (Abcam, USA), anti-p65 (Abcam, USA), anti-Histone (Abcam, USA) and anti-GAPDH (Abcam, USA) overnight at 4°C. GAPDH and Histone were used as internal controls. After washing with TBST, membranes were incubated with horseradish peroxidase-labeled secondary antibody for 1 h at room temperature. After washing with TBST, specific protein bands were visualized using an enhanced chemiluminescent (ECL; Millipore, Shanghai, Chian).
Isolation, culture and identification of NPCs
The NP tissues were cut into pieces of 1 mm3 size with an ophthalmic scissors. Then, samples were incubated with 0.25% trypsin for 30 min, and centrifuged at 1000 r/min for 10 min. The supernatant was discarded and type II collagenase was added and digested at 37°C for 4 h. After filtration through a 200 mesh filter and centrifugation, the cells were collected. The NPCs were resuspended in DMEM/F12 medium (ThermoFisher, USA) containing 20% fetal bovine serum (FBS; Biosera, USA) and cultured in a 37°C, 5% CO2 incubator. After the cells were attached, the cells were observed under an inverted microscope. The cells were cultured to 70% to 80% confluence, trypsinized, and passaged at 1:2. The passage 2 of NPCs were seeded in 6-well plates to make cell slides, and the NPCs were identified by immunocytochemical staining of collagen II [13] and Safranin O staining [14] as described previously.
Cell transfection
The TUG1 sequence was synthesized from Sangon Biotech (Shanghai, China) and cloned into the pcDNA3.1 plasmid (Thermo Fisher Scientific, USA) to construct the TUG1 overexpression vector (TUG1 group). The pcDNA3.1 empty vector was used as a control (NC group). The pcDNA-HMGB1 and the corresponding pcDNA-Ctrl were also synthesized and constructed by Sangon Biotech. All siRNAs (si-TUG1 and si-NC, si-HMGB1 and si-Ctrl), miRNAs mimics (miR-26a mimic, miR-NC) and miRNAs inhibitors (miR-26a inhibitor, inhibitor-NC) were purchased from RiboBio (Guangzhou, China). Plasmids or oligonucleotides were transfected into passage 3 degenerative intervertebral disc NPCs using a Lipofectamine 2000 Transfection kit (Life Technologies, USA) according to the manufacturer’s instructions. For rescue experiments, human degenerative intervertebral disc NPCs were pretreated with NF-κB specific inhibitor QNZ (6-amino-4-quinazoline, 40 nmol/L) before transfection with pcDNA-TUG1 or/and si-HMGB1.
5-ethynyl-20-deoxyuridine (EdU) assay
To measure the proliferation of cells, the Cell Light EdU DNA imaging kit (Ribobio, Guangzhou, China) was used for EdU incorporation experiments. All procedures were performed in strict accordance with the kit instructions. The ratio of EdU-stained cells (with red fluorescence) to Hoechst-stained cells (with blue fluorescence) was used to evaluate the cell proliferation activity.
Flow cytometry
Human degenerated intervertebral disc NPCs were transfected for 48 h and cells were harvested. After washing with PBS, the cells were uniformly mixed with 500 μL of pre-cooled 1× binding buffer. Cells were incubated with 5 μL of Annexin V-FITC (Bioscience, USA) for 15 min at room temperature in the dark, and then incubated with 2.5 μL of PI for 5 min at room temperature in the dark. Apoptosis rate was measured within 1 h using a FACS Calibur flow cytometer (FACSCalibur, Becton-Dickinson, Franklin Lakes, USA).
Luciferase reporter assay
The partial sequences of TUG1 or 3’-UTR of HMGB1 mRNA containing the predicted miR-26a binding site were amplified by PCR, and then cloned into a luciferase vector (Promega, USA) to construct luciferase reporter vectors, which were named as TUG1-wild-type (TUG1-WT) or HMGB1-wild-type (HMGB1-WT), respectively. The counterparts with mutated miR-26a binding sequences were replaced as indicated and named as TUG1-mutated-type (TUG1-MUT) or HMGB1-mutated-type (HMGB1-MUT). Then, the TUG1-WT (or TUG1-MUT) or HMGB1-WT (or HMGB1-MUT) plasmid vectors were co-transfected with miR-26a mimic or miR-NC into HEK293T cells. The luciferase activity was analyzed with the dual luciferase reporter assay system (Promega, USA) 48 h post-transfection according to the manufacturer’s instructions.
Immunofluorescence staining
NP cells were seeded onto a cover slip of a 24-well plate. After the corresponding treatment, the slides with NP cells were washed with pre-cooled PBS. NP cells were fixed for 10 min by adding 200 μL of 4% paraformaldehyde per well. After washing the cells, they were incubated with 0.1% Triton X-100 for 10 min at room temperature, followed by 5% goat serum for 30 min. 200 μL of anti-NF-κB p65 primary antibody (Abcam) was added to each well and incubated for 1 h at room temperature. After washing with PBST, 200 μL of IgG secondary antibody (Abcam) was added to each well and incubated at room temperature for 30 min in the dark. After washing, the nuclei were stained by adding 200 μL of 0.1% DAPI to each well. After washing, the slides were removed from the 24-well plates and a small amount of anti-fluorescence quenching was added. The images were observed and acquired under a laser confocal microscope.
Statistical analysis
Statistical analyses were performed by Student’s t-test or one-way ANOVA using software SPSS 15.0 (SPSS Inc., Chicago, IL, USA). All data were presented as the mean ± standard deviation (mean ± SD). Pearson’s Correlation Coefficient was applied to explore the correlation between TUG1 and miR-26a, or TUG1 and HMGB1, or miR-26a and HMGB1 in the IDD tissues. A p-value less than 0.05 was considered statistically significant.
Results
Up-regulation of LncRNA TUG1 expression in degenerated intervertebral disc NP tissues
The expression of TUG1 in the NP tissues of the IDD group was significantly up-regulated compared to the LVF group (P<0.05, Figure 1A). And the expression of miR-26a in the IDD group was significantly down-regulated compared to the LVF group (P<0.05, Figure 1B). Correlation analysis found that miR-26a was significantly negatively correlated with TUG1 in IDD tissues (P<0.05, Figure 1C). Immunohistochemical analysis (Figure 1D) and western blot assay also revealed a significant up-regulation of HMGB1 expression in IDD tissues, which was positively correlated with TUG1 while negatively correlated with miR-26a (Figure 1E, 1F). Additionally, we also analyzed the relationship between TUG1, miR-26a and HMGB1 in NP tissues of LVF group. The results showed that HMGB1 and miR-26a showed a significant negative correlation in LVF group (P<0.05), while TUG1 and miR-26a, as well as TUG1 and HMGB1 were not significantly correlated (P>0.05, Figure S1), which may be due to our small sample size.
Figure 1.
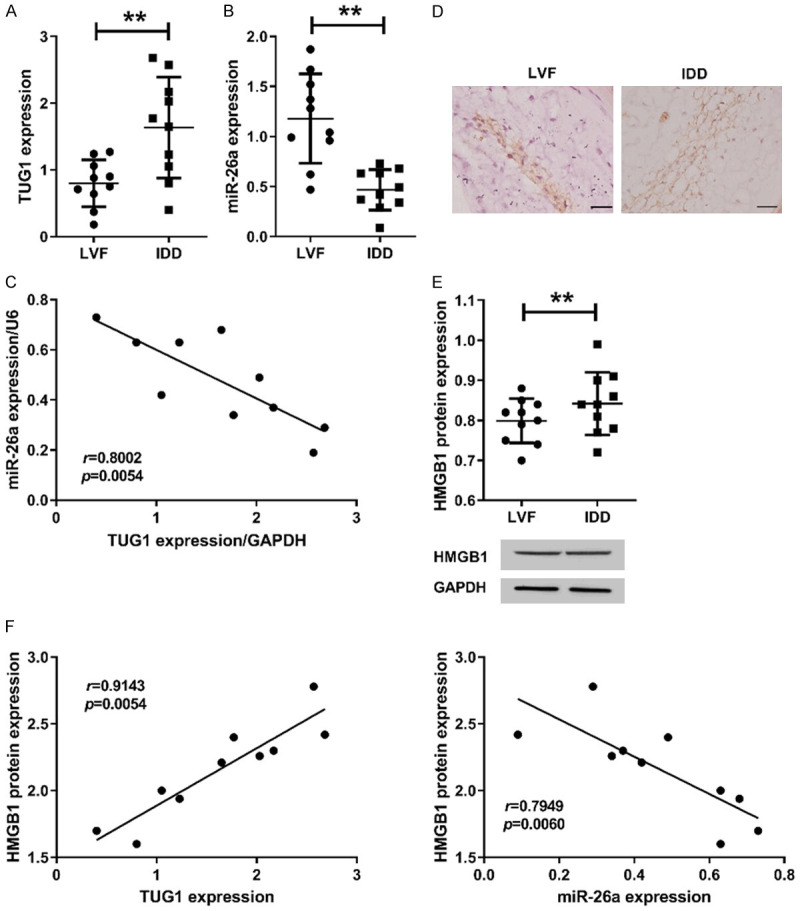
LncRNA TUG1 was significantly up-regulated in degenerated intervertebral disc NP tissues. The expression of TUG1 (A) and miR-26a (B) were assessed by qRT-PCR in human disc NP tissues of LVF group (n=10) and IDD group (n=10). (C) The correlation between TUG1 and miR-26a expression in human disc NP tissues of IDD group by qRT-PCR. GAPDH and U6 were used as internal reference. (D) Immunohistochemistry was used to analyze the localization of HMGB1 in NP tissues. (E) Western blot assay was used to analyze the expression of HMGB1 protein in NP tissues. Bar =100 μm. The original images are available in Figure S2. (F) The relationship between HMGB1 and TUG1 or miR-26a in NP tissues of IDD group. Data are presented as mean ± standard deviation. *P<0.05, **P<0.01.
Morphology and identification of NPCs
The morphology of primary NPCs was observed under an inverted microscope. The results showed that the morphology of normal NPCs (LVF group) was mostly fusiform and polygonal, while the degenerative NPCs (IDD group) were mostly spindle-shaped, fusiform and irregular (Figure 2A). The NPCs in the IDD group were sparse and disorderly arranged, and some cells were necrotic. Moreover, the number of NPCs was reduced, which may be related to cell senescence and apoptosis [15]. The second generation of NPCs were scrambled and identified by collagen II immunocytochemical staining and Safranin O staining. Collagen II immunocytochemical staining showed a strong positive reaction in the cytoplasm of cells, while the nucleus was not stained, suggesting that the cultured cells were intervertebral disc NPCs. Additionally, the degree of coloration of the LVF group was higher than that of the IDD group (Figure 2B). Safranin O staining showed that the NPCs of normal intervertebral disc stained deeper and the cytoplasm showed uniform pink coloration. And the nuclei of degenerated NPCs were swollen and stained lightly, and the cytoplasm was stained unevenly with vacuoles (Figure 2C). These results confirmed that the NPCs of the IDD group do exhibit degenerative properties.
Figure 2.
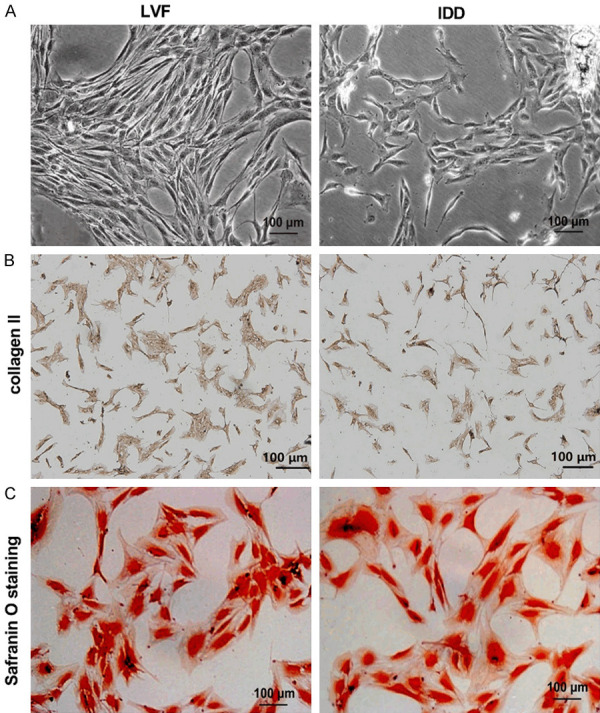
Morphological and phenotypic identification of NPCs. (A) Morphological changes of NPCs in the LVF group and the IDD group were observed under a phase contrast microscope. The NPCs were identified by collagen II immunohistochemical staining (B) and Safranin O staining (C). Scale bar =100 μm.
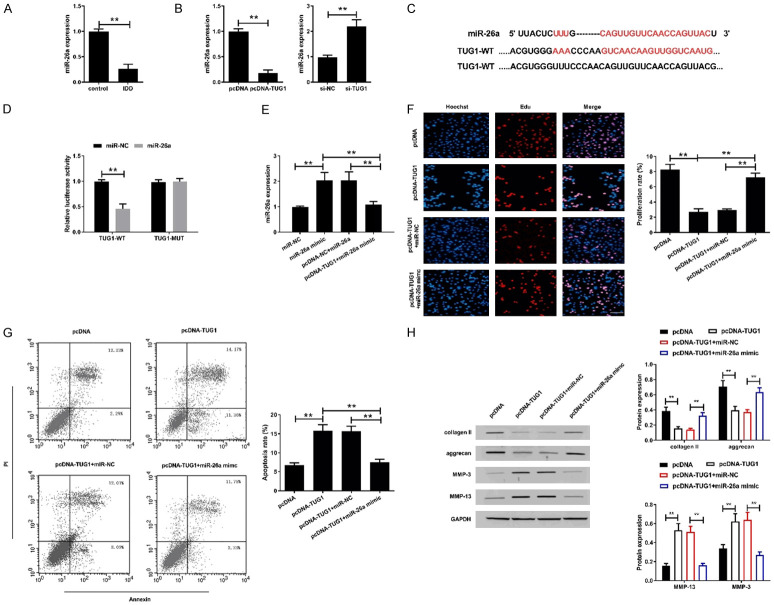
TUG1 inhibited the proliferation of human degenerative disc nucleus cells and promoted apoptosis and ECM degradation
qRT-PCR showed that the expression of TUG1 in the NPCs of the IDD group was significantly higher than that of the LVF group (P<0.05, Figure 3A). In view of the abnormal expression of TUG1 in degenerated intervertebral disc NP tissues and cells, we investigated the role of TUG1 in IDD by transfecting human degenerative disc NPCs with pcDNA-TUG1 or si-TUG1. qRT-PCR showed that transfection of TUG1 pcDNA3.1 significantly increased the expression of TUG1 in the cells compared with the pcDNA group, while transfection of TUG1 siRNA significantly reduced the expression of TUG1 in the cells compared with the si-NC group (P<0.05, Figure 3B).
Figure 3.
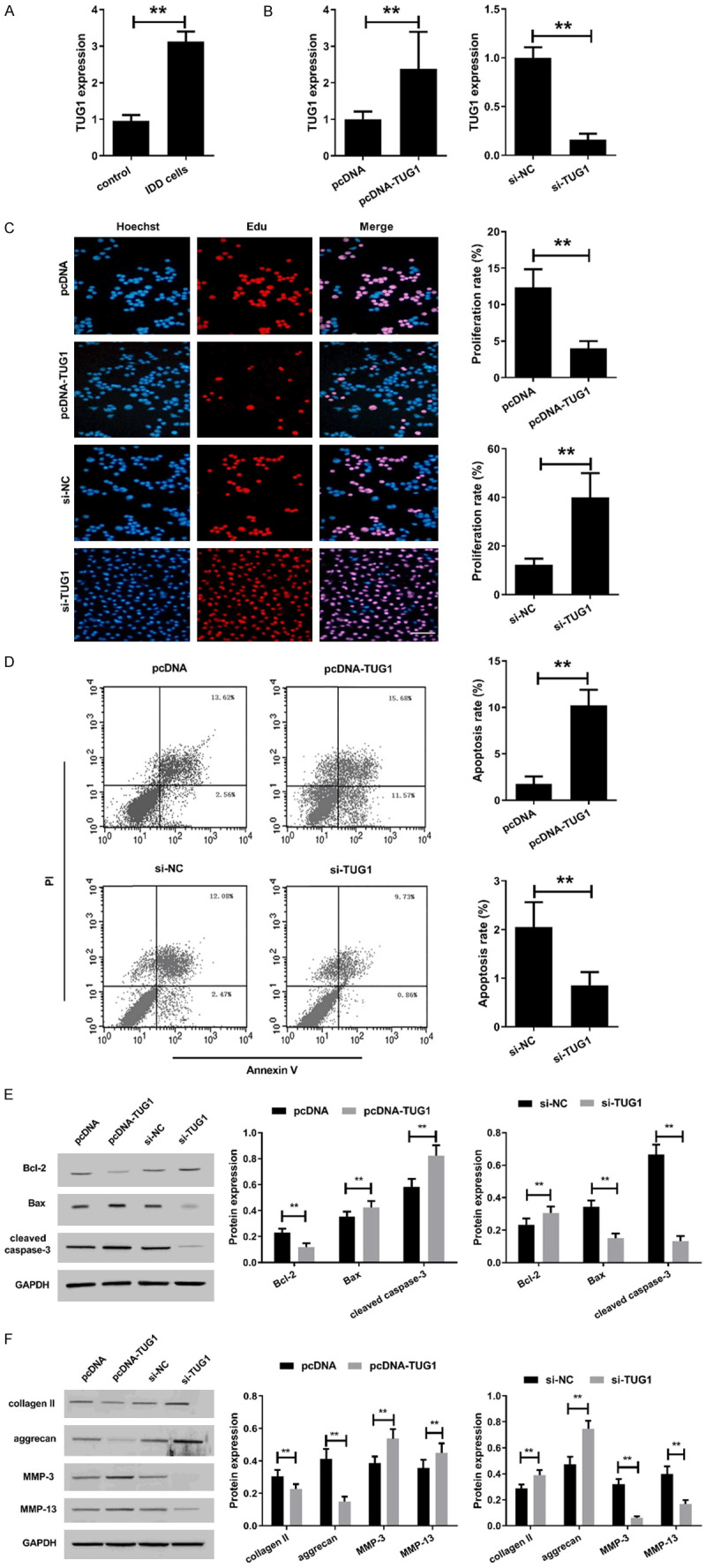
Effect of TUG1 on proliferation, apoptosis and ECM degradation of human degenerated intervertebral disc NPCs. A. The expression of TUG1 was assessed by qRT-PCR in NPCs of LVF group and IDD group. *P<0.05, **P<0.01. After isolated from human degenerative disc nucleus pulposus, the NPCs were transfected with pcDNA, pcDNA-TUG1, si-NC or si-TUG1. B. After transfection for 48 h, TUG1 expression was detected by qRT-PCR. C. Cell viability of human degenerated intervertebral disc NPCs transfected for 48 h was detected by EdU assay. Bar =100 μm. D. Cell apoptosis of human degenerated intervertebral disc NPCs transfected for 48 h was analyzed by flow cytometry using Annexin V/PI staining assay kit. E. Apoptosis-related proteins Bcl-2, Bax and cleaved caspase-3 expression were detected by western blot assay in human degenerated intervertebral disc NPCs transfected for 48 h. The original images are available in Figure S3. F. ECM degradation-related proteins collagen II, aggrecan, MMP-3 and MMP-13 expression were detected by western blot assay in human degenerated intervertebral disc NPCs transfected for 48 h. The original images are available in Figure S4. *P<0.05, **P<0.01.
The effect of TUG1 overexpression or knockdown on proliferation of human degenerative intervertebral disc NPCs was analyzed using EdU assay (Figure 3C). The results showed that overexpression of TUG1 significantly inhibited the proliferation of human degenerated intervertebral disc NPCs, while si-TUG1 significantly promoted cell proliferation (Figure 3C). Additionally, we evaluated the apoptosis of degenerated intervertebral disc NPCs after transfection by flow cytometry. The apoptotic rate of the pcDNA-TUG1 group was significantly lower than that of the pcDNA group, while it was higher in the si-TUG1 group than that of the si-NC group (Figure 3D). Simultaneously, the expression of apoptosis-related proteins was determined by western blot. Compared with the pcDNA group, overexpression of TUG1 significantly down-regulated the expression of Bcl-2 while up-regulated the expression of Bax and cleaved caspase-3. Conversely, TUG1 knockdown significantly up-regulated Bcl-2 expression while down-regulated Bax and cleaved caspase-3 expression (Figure 3E).
Next, we analyzed the effect of TUG1 on ECM degradation in human degenerative intervertebral disc NPCs. We found that overexpression of TUG1 reduced the expression of collagen II and aggrecan in human degenerative NPCs, while significantly increased the expression of MMP-3 and MMP-13 (Figure 3F), suggesting that TUG1 promoted ECM degradation of human degenerative disc NPCs. Conversely, TUG1 knockdown improved ECM degradation by up-regulating the expression of collagen II and aggrecan, whereas down-regulating the expression of MMP-3 and MMP-13. These results suggested that TUG1 may participate in the promotion of IDD by inhibiting the proliferation while promoting apoptosis and ECM degradation of human degenerative disc NPCs.
TUG1 targeted miR-26a to promote IDD
Compared with normal intervertebral disc NPCs (control group), the expression of miR-26a in human degenerative intervertebral disc NPCs (IDD group) was significantly down-regulated (Figure 4A). Moreover, overexpression of TUG1 down-regulated the expression of miR-26a in human degenerative disc NPCs, while TUG1 knockdown up-regulated miR-26a expression (Figure 4B).
Figure 4.
TUG1 negatively regulated the expression of miR-26a in degenerated intervertebral disc nucleus cells. A. The expression of miR-26a was assessed by qRT-PCR in NPCs of control group and IDD group. B. The expression of miR-26a was examined in human degenerative disc NPCs transfected with pcDNA-TUG1 or si-TUG1. C. Sequence alignment of miR-26a and the putative binding sites within the wild-type TUG1, and mutation in the TUG1. D. The luciferase activity was detected in HEK293T cells transfected with TUG1-WT or TUG1-MUT reporter vector together with miR-26a mimics or miR-NC. E. The expression of miR-26a was examined in human degenerated intervertebral disc NPCs transfected with miR-NC, miR-26a mimic, pcDNA-NC+miR-26a or pcDNA-TUG1+miR-26a mimic. F. Proliferation of treated human degenerated intervertebral disc NPCs was detected by EdU assay. Scale bar =100 μm. G. Apoptosis of treated human degenerated intervertebral disc NPCs was detected by flow cytometry. H. The expression of collagenII, aggrecan, MMP-3 and MMP-13 were determined in treated human degenerated intervertebral disc NPCs by western blot assay. The original images are available in Figure S5. *P<0.05, **P<0.01.
To analyze the potential molecular mechanisms of TUG1 in human degenerative NPCs, bioinformatics analysis was performed using starBase v2.0 (http://starbase.sysu.edu.cn/) and Miranda (http://www.microrna.org). The results revealed that miR-26a contained a sequence complementary to TUG1 (Figure 4C). To confirm the interaction between TUG1 and miR-26a, we performed a luciferase reporter assay. The results showed that miR-26a significantly reduced the luciferase activity of TUG1-WT without affecting the luciferase activity of TUG1-MUT (Figure 4D). To investigate whether TUG1 exerts its biological effects by targeting miR-26a, we co-transfected miR-26a mimic into human degenerated intervertebral disc NPCs were co-transfected with miR-26a mimic and pcDNA-TUG1. As expected, miR-26a expression was increased after miR-26a mimic transfection (Figure 4E). Meanwhile, overexpression of TUG1 attenuated the expression of miR-26a that induced by mimics. These results indicated that miR-26a was a direct target gene of TUG1.
The EdU assay showed that miR-26a overexpression abolished the inhibition of TUG1 overexpression on proliferation of human degenerative disc NPCs (Figure 4F). Similarly, miR-26a mimic transfection reversed the promotion of apoptosis by TUG1 (Figure 4G). Furthermore, TUG1-induced matrix degradation was inhibited due to overexpression of miR-26a (Figure 4H). These data indicated that TUG1 could exert its function of promoting IDD through inhibiting miR-26a.
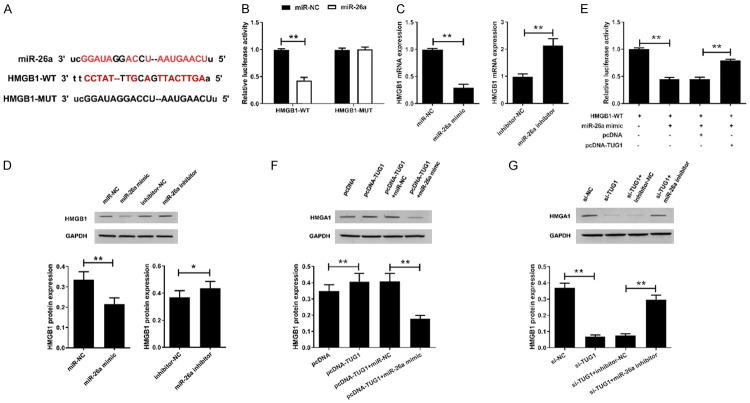
TUG1 regulated miR-26a/HMGB1 axis in human degenerated intervertebral disc NPCs
We found through the online software TargetScan that miR-26a has some complementary bases to the 3’-UTR of HMGB1 (Figure 5A), suggesting that HMGB1 may be a target gene for miR-26a. We further constructed HMGB1-WT and HMGB1-MUT luciferase vectors and co-transfected with miR-26a mimic or miR-NC into HEK 293T cells to verify the interaction between miR-26a and HMGB1. The results showed that miR-26a significantly reduced the luciferase activity of the luciferase reporter vectors containing HMGB1-WT, but did not substantially affect the luciferase activity of the reporter gene of HMGB1-MUT (Figure 5B). Next, we overexpress or inhibit miR-26a in human degenerative NPCs. The results revealed that miR-26a mimic significantly down-regulated the expression of HMGB1 mRNA and protein in human degenerative NPCs, whereas miR-26a inhibitor up-regulated the expression of HMGB1 (Figure 5C and 5D).
Figure 5.
TUG1 regulates miR-26a/HMGB1 axis in human degenerated intervertebral disc NPCs. (A) Sequence alignment of miR-26a and the putative binding sites within HMGB1-WT and HMGB1-MUT. (B) The luciferase activity was detected in HEK293T cells transfected with HMGB1-WT or HMGB1-MUT reporter vector together with miR-26a mimics or miR-NC. The expression of HMGB1 mRNA (C) and protein (D) was examined in human degenerated intervertebral disc NPCs transfected with miR-26a mimic or miR-26a inhibitor. The original images are available in Figure S6. (E) The luciferase activity was detected in HEK293T cells co-transfected HMGB1-WT with miR-26a mimic, miR-26a mimic+pcDNA or miR-26a mimic+pcDNA-TUG1. (F) The expression of HMGB1 protein in NPCs transfected with pcDNA, pcDNA-TUG1, pcDNA-TUG1+miR-NC and pcDNA-TUG1+miR-26a mimic. The original images are available in Figure S7. (G) The expression of HMGB1 protein in NPCs transfected with si-NC, si-TUG1, si-TUG1+inhibitor-NC, si-TUG1+miR-26a inhibitor. The original images are available in Figure S8. *P<0.05, **P<0.01.
To investigate whether TUG1 was involved in the regulation of the miR-26a/HMGB1 axis, we transfected the HMGB1-WT vector with miR-26a mimic, miR-26a mimic+pcDNA and miR-26a mimic+pcDNA-TUG1 into HEK 293T cells, then the dual luciferase reporter assay was performed. Co-transfection of miR-26a with HMGB1-WT significantly reduced luciferase activity, whereas overexpression of TUG1 significantly reversed the effect of miR-26a overexpression on luciferase activity (Figure 5E). These results suggested that HMGB1 was the target of miR-26a.
Furthermore, in human degenerative NPCs, an increase in TUGB1 expression induced by TUG1 overexpression could be reversed by the introduction of miR-26a mimic (Figure 5F). Moreover, overexpression of miR-26a inhibited the upregulation of HMGB1 expression induced by pcDNA-TUG1, and miR-26a inhibitor reversed the inhibitory effect of si-TUG1 on HMGB1 expression (Figure 5G). These results confirmed that TUG1 could participate in degenerative disc degeneration by modulating HMGB1 by antagonizing miR-26a.
TUG1 activited the NF-κB pathway by affecting HMGB1 expression
HMGB1 participates in IDD by activating NF-κB to promote the release of inflammatory factors [16]. Based on this, we further analyzed the relationship between TUG1 and NF-κB pathway. We found that overexpression of TUG1 significantly upregulated the expression of p65 in the nucleus (Figure 6A). Immunofluorescence staining also showed that overexpression of TUG1 induced nuclear transfer of p65 (Figure 6B). Simultaneously, si-HMGB1 inhibited nuclear transfer of p65. Additionally, transfection of pcDNA-TUG1 reversed the inhibitory effect of si-HMGB1 on NF-κB activation. More importantly, overexpression of TUG1 partially abolished the inhibition of NF-κB by NF-κB specific inhibitor QNZ pretreatment. These results suggested that TUG1 participated in IDD by activating the NF-κB signaling pathway.
Figure 6.
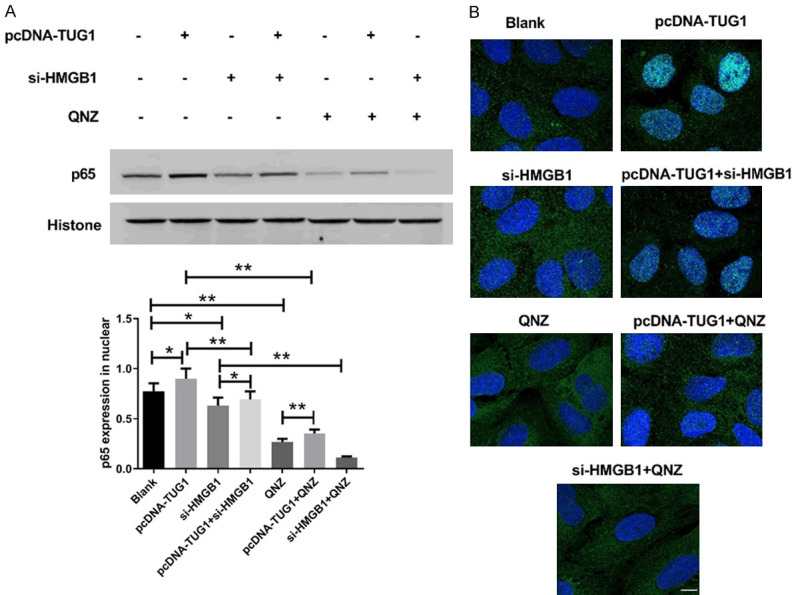
TUG1 activited the NF-κB pathway by affecting HMGB1 expression. A. The expression of NF-κB p65 in nuclear was analyzed by western blot. Histone were used as internal references. The original images are available in Figure S9. B. Immunofluorescence staining for nuclear translocation of p65 in human intervertebral disc degeneration NPCs. Scale bar =25 μm. *P<0.05, **P<0.01.
Discussion
Degeneration of the intervertebral disc is accompanied by a decrease in the number of cells and synthesis of extracellular matrix [17]. The role of lncRNA in IDD has received extensive attention recently. We found that TUG1 was significantly up-regulated in human degenerative disc NP tissue. Additionally, TUG1 participated in the promotion of IDD by inhibiting the proliferation while promoting apoptosis and ECM degradation of human degenerative disc NPCs in vitro. More importantly, TUG1 promoted IDD by targeting miR-26a/HMGB1 axis, which may also involve activation of the NF-κB signaling pathway.
Recently, some studies have reported that the expression of many lncRNAs in IDD has changed a lot [4]. Chen et al. [7] reported that 8 lncRNAs were differentially expressed in human normal NP and degenerative NP. Wan et al. [18] also found that 116 lncRNAs were differentially expressed in IDD, and overexpression of lncRNA RP11 296A18.3 induced up-regulation of Fas-associated protein factor 1 (FAF1) and ultimately enhanced abnormal apoptosis of intervertebral disc NPCs. Xi et al. [8] confirmed that lncRNA HCG18 promoted IDD development by inhibiting proliferation and inducing apoptosis of NPCs. Exploring the mechanism by which lncRNA regulates the development of IDD may have important implications for the discovery of new therapeutic targets. TUG1, a type of lncRNAs, has been shown to be highly expressed in a variety of malignancies and is closely related to the development of tumors [19,20]. Recent studies have found that TUG1 is up-regulated in NP samples from patients with lumbar disc herniation that is involved in the regulation of NPCs apoptosis [6]. The present study also found that TUG1 expression was significantly up-regulated in degenerated intervertebral disc NP tissues and cells, suggesting that TUG1 may be involved in the occurrence and development of IDD.
Apoptosis of the NPCs of the intervertebral disc plays an important role in the IDD process. Mitochondrial apoptosis pathway is an important pathway for apoptosis during IDD [21]. Mitochondria regulate endogenous and exogenous apoptotic pathways through caspase-dependent and non-caspase-dependent apoptotic pathways. Bcl-2 and caspase are two evolutionarily conserved protein families in eukaryotes. Bcl-2 can prevent the release of Cyt-c from mitochondria to the cytoplasm to inhibit the apoptosis of NPCs, while caspase mediates the initiation and execution of apoptosis. Caspase-3 is the main performer of apoptosis, and silencing caspase-3 in vitro can reduce the apoptosis of NPCs induced by mechanical overload to inhibit IDD [22]. We found that the expression of this two key protein family members on the mitochondrial apoptotic pathway in NPCs after TUG1 overexpression or knockdown has changed, suggesting that TUG1 could activate mitochondrial apoptosis pathway in NPCs by down-regulating Bcl-2 and up-regulating caspase-3. It is worth noting that recent studies have also found evidence that TUG1 is involved in the regulation of mitochondrial apoptosis. Li et al. [9] demonstrated that TUG1 promoted apoptosis in glioma cells by mediating caspase-3 and -9-mediated intrinsic pathways and inhibiting Bcl-2-mediated anti-apoptotic pathways. Chen et al. [23] found that TUG1 could directly target and adsorb miR-9 to indirectly inhibit the expression of Bcl-2 to promote apoptosis. These studies suggested that TUG1 regulated the balance between pro-apoptotic and apoptotic molecules in the mitochondrial apoptotic pathway, and the expression of TUG1 determined the sensitivity of NPCs to apoptotic stimuli.
The NPCs in the IDD mainly produce two kinds of extracellular matrix (ECM) components, aggrecan and collagen II, which play an important role in maintaining the integrity of the IDD [24]. IDD is often accompanied by degradation of ECM, which is caused by the imbalance of ECM synthesis and catabolism. Catabolism is primarily controlled by proteolytic enzymes, in which MMPs play a key role. MMP-3 and -13 are considered to be the most important MMPs in IDD. MMP-13 is the major rate-limiting factor in the degradation of collagen II, and MMP-3 is an important aggrecan-degrading enzyme that also degrades other extracellular matrix components [25]. We found that overexpression of TUG1 increased the expression of MMP-13 and MMP-3 while decreased the expression of collagen II and aggrecan in NPCs, whereas TUG1 exerted the opposite effect when silenced. These results suggested that TUG1 could promote the degradation of ECM in NPCs by regulating the expression of MMPs. However, the mechanism of action of TUG1 in ECM degradation remains to be explored.
lncRNA can act as a “sponge” to play a competitive endogenous RNA (ceRNA) role by targeting downstream miRNAs to participate in the regulation of pathophysiological processes. As an important “sponge”, TUG1 competitively binds to multiple miRNAs. TUG1 can be used as a miR-335-5p ceRNA to promote the migration and invasion of myeloma cells [26], and can also promote the tumorigenesis of myeloma by regulating the expression of miR-144-3p [27]. Yu et al. [28] showed TUG1 could sponge miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. As a miRNA, miR-26a has a complex role in carcinogenesis, in the sense that both oncogenic and tumor suppressive effects were reported in cancers [29,30]. Furthermore, miR-26a is one of the essential molecules in skeletal muscle cell differentiation and regeneration after skeletal muscle injury [31]. Recently, Shen et al. [32] reported that miR-26a was up-regulated in osteoarthritis, a degenerative disease, and lncRNA SNG5 could competitively absorb miR-26a as a ceRNA to promote chondrocyte proliferation and migration. Li et al. [9] demonstrated that TUG1 could target miR-26a to regulate the direct and functional downstream target PTEN of miR-26a to exert tumor suppressive effects in glioma development. However, the role of miR-26a in IDD is unclear. The present study also found a continuous miR-26a binding site on the TUG1 sequence by bioinformatics prediction. Moreover, luciferase reporter assay also showed a targeted regulatory relationship between TUG1 and miR-26a. Furthermore, TUG1 silencing could significantly promote the expression of miR-26a in degenerated intervertebral disc NPCs, while overexpression of TUG1 significantly inhibited the expression of miR-26a, and attenuated miR-26a mimic-induced miR-26a expression. Additionally, we also found that overexpression of miR-26a reversed the effect of TUG1 on apoptosis and ECM degradation of human degenerative disc NPCs. In the present study, we also confirmed that miR-26a directly targeted HMGB1 mRNA and inhibited the expression of HMGB1 protein. Therefore, we speculated that TUG1 may play its role in the development of intervertebral disc degeneration by regulating the miR-26a/HMGB1 axis.
HMGB1 is a member of the HMG family that is involved in a variety of pathophysiological processes including inflammation, tumors, autoimmune diseases, osteoarthritis and intervertebral disc degeneration [33,34]. Gruber et al. [35] found that HMGB1 was up-regulated 8-fold in human degenerative disc tissue compared to healthy intervertebral discs, and in vitro studies had shown that HMGB1 was up-regulated 24-fold in NPCs exposed to TNF-α. HMGB1 promotes the release of inflammatory cytokines and the expression of MMPs in human IVD cells, which aggravates the progression of IDD [16]. Additionally, HMGB1 phosphorylates p65 in the NF-κB p65/p50 heterodimer to inducing nuclear translocation to activating NF-κB, and then induces transcription of downstream genes [36]. Chronic activation of the NF-κB pathway has been shown to be the leading cause of some degenerative diseases, including osteoarthritis [37] and IDD [38]. Inhibition of NF-κB signaling pathway activation can significantly delay degeneration of the IVD [39,40]. Wang et al. [41] confirmed that NF-κB p65 protein was localized in the nucleus of human degenerative lumbar disc tissue, but was located in the cytoplasm in normal lumbar disc tissue. Subsequently, they also found that the NF-κB specific blocker BY11-7082 significantly down-regulated the expression of ADAMTS and MMPs in the IVD tissues, while up-regulated the expression of collagen II and aggrecan, suggesting that NF-κB played an important role in the degradation of ECM. Studies have also shown that inhibition of NF-κB signaling pathway could inhibit apoptosis of human NPCs [42]. To elucidate the molecular mechanism of TUG1/miR-26a/HMGB1 axis in IDD progression, we validated the NF-κB signaling pathway. We found that overexpression of TUG1 significantly promoted nuclear translocation of NF-κB p65, whereas HMGB1 silencing inhibited the induction of NF-κB activation by overexpression of TUG1 in human degenerative intervertebral disc NPCs. More importantly, overexpression of TUG1 could eliminated the inhibitory effect of NF-κB specific inhibitor QNZ pretreatment on NF-κB pathway in human IDD NPCs. Therefore, we hypothesized that TUG1 could promote apoptosis and ECM degradation in human IDD NPCs by targeting miR-26a/HMGB1 axis, a process involved in the activation of the NF-κB signaling pathway.
Conclusion
TUG1 could participate in the promotion of apoptosis and ECM degradation of degenerated intervertebral disc NPCs by targeting miR-26a/HMGB1 axis, which may be related to the activation of NF-κB pathway. Our study further elucidated the mechanism by which TUG1 promotes the progression of IDD, which may provide new ideas for the treatment of degenerative diseases.
Acknowledgements
The present study was approved by the Ethic Committee of Chinese Academy of Medical Sciences Peking Union Medical College Hospital (Beijing, China). All subjects provided written informed consent to participate in the present study.
Disclosure of conflict of interest
None.
Abbreviations
- IDD
intervertebral disc degeneration
- NPCs
nucleus pulposus cells
- ECM
extracellular matrix
- lncRNAs
long-chain non-coding RNAs
- TUG1
taurine upregulated gene 1
- LDH
lumbar disc herniation
- LVF
lumbar vertebrae fractures
- IVD
intervertebral disc
Supporting Information
References
- 1.van Uden S, Silva-Correia J, Oliveira JM, Reis RL. Current strategies for treatment of intervertebral disc degeneration: substitution and regeneration possibilities. Biomater Res. 2017;21:22. doi: 10.1186/s40824-017-0106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cappello R, Bird JL, Pfeiffer D, Bayliss MT, Dudhia J. Notochordal cell produce and assemble extracellular matrix in a distinct manner, which may be responsible for the maintenance of healthy nucleus pulposus. Spine (Phila Pa 1976) 2006;31:873–882. doi: 10.1097/01.brs.0000209302.00820.fd. [DOI] [PubMed] [Google Scholar]
- 3.Sakai D, Schol J. Cell therapy for intervertebral disc repair: clinical perspective. J Orthop Translat. 2017;9:8–18. doi: 10.1016/j.jot.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50:e12313. doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang WY, Wang YF, Ma P, Xu TP, Shu YQ. Taurineupregulated gene 1: a vital long noncoding RNA associated with cancer in humans (Review) Mol Med Rep. 2017;16:6467–6471. doi: 10.3892/mmr.2017.7472. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma S, Wang YJ. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulposus cells via regulating Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2017;491:668–674. doi: 10.1016/j.bbrc.2017.07.146. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Ni H, Zhao Y, Chen K, Li M, Li C, Zhu X, Fu Q. Potential role of lncRNAs in contributing to pathogenesis of intervertebral disc degeneration based on microarray data. Med Sci Monit. 2015;21:3449–3458. doi: 10.12659/MSM.894638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X, He Y. Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep. 2017;7:13234. doi: 10.1038/s41598-017-13364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, An G, Zhang M, Ma Q. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem Biophys Res Commun. 2016;477:743–748. doi: 10.1016/j.bbrc.2016.06.129. [DOI] [PubMed] [Google Scholar]
- 10.Yang B, Tang X, Wang Z, Sun D, Wei X, Ding Y. TUG1 promotes prostate cancer progression by acting as a ceRNA of miR-26a. Biosci Rep. 2018;38:BSR20180677. doi: 10.1042/BSR20180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Cheng G, Yang X, Li C, Shi R, Zhao N. Tanshinol suppresses endothelial cells apoptosis in mice with atherosclerosis via lncRNA TUG1 up-regulating the expression of miR-26a. Am J Transl Res. 2016;8:2981–2991. [PMC free article] [PubMed] [Google Scholar]
- 12.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001;26:1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 13.Song X, Peng S. Transplantation of transforming growth factor beta3 gene-modified nucleus pulposus cells for intervertebral disc degeneration in rabbits. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2012;26:790–5. [PubMed] [Google Scholar]
- 14.Wang D, Vo NV, Sowa GA, Hartman RA, Ngo K, Choe SR, Witt WT, Dong Q, Lee JY, Niedernhofer LJ, Kang JD. Bupivacaine decreases cell viability and matrix protein synthesis in an intervertebral disc organ model system. Spine J. 2011;11:139–146. doi: 10.1016/j.spinee.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horner HA, Roberts S, Bielby RJ, Evans H, Urban JP. Cells from different regions of the intervertebral disc: effect of culture system on matrix expression and cell phenotype. Spine (Phila Pa 1976) 2002;27:1018–1028. doi: 10.1097/00007632-200205150-00004. [DOI] [PubMed] [Google Scholar]
- 16.Fang F, Jiang D. IL-1β/HMGB1 signalling promotes the inflammatory cytokines release via TLR signalling in human intervertebral disc cells. Biosci Rep. 2016;36:e00379. doi: 10.1042/BSR20160118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 2006;31:2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 18.Wan ZY, Song F, Sun Z, Chen YF, Zhang WL, Samartzis D, Ma CJ, Che L, Liu X, Ali MA, Wang HQ, Luo ZJ. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther. 2014;16:456. doi: 10.1186/s13075-014-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Tian G, Tian F, Shao L. Long non-coding RNA TUG1 promotes osteosarcoma cell proliferation and invasion through inhibition of microRNA-212-3p expression. Exp Ther Med. 2018;16:779–787. doi: 10.3892/etm.2018.6216. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Sun J, Hu J, Wang G, Yang Z, Zhao C, Zhang X, Wang J. LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res. 2018;37:106. doi: 10.1186/s13046-018-0771-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Shi L, Teng H, Zhu M, Li C, Huang K, Chen BI, Dai Y, Wang J. Paeoniflorin inhibits nucleus pulposus cell apoptosis by regulating the expression of Bcl-2 family proteins and caspase-9 in a rabbit model of intervertebral disc degeneration. Exp Ther Med. 2015;10:257–262. doi: 10.3892/etm.2015.2501. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Yamada K, Sudo H, Iwasaki K, Sasaki N, Higashi H, Kameda Y, Ito M, Takahata M, Abumi K, Minami A, Iwasaki N. Caspase 3 silencing inhibits biomechanical overload-induced intervertebral disk degeneration. Am J Pathol. 2014;184:753–764. doi: 10.1016/j.ajpath.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Wang M, Yang H, Mao L, He Q, Jin H, Ye Z, Luo X, Xia Y, Hu B. LncRNA TUG1 sponges microRNA-9 to promote neurons apoptosis by up-regulated Bcl2l11 under ischemia. Biochem Biophys Res Commun. 2017;485:167. doi: 10.1016/j.bbrc.2017.02.043. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Song Y, Liu W, Wang K, Gao Y, Li S, Duan Z, Shao Z, Yang S, Yang C. IAPP modulates cellular autophagy, apoptosis, and extracellular matrix metabolism in human intervertebral disc cells. Cell Death Discovery. 2017;3:16107. doi: 10.1038/cddiscovery.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Z, Yang H, Bau B, Soder S, Aigner T. Role of mitogen-activated protein kinases and NFkappaB on IL-1beta-induced effects on collagen type II, MMP-1 and 13 mRNA expression in normal articular human chondrocytes. Rheumatol Int. 2006;26:900–903. doi: 10.1007/s00296-006-0114-7. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao J, Han X, Qi X, Jin X, Li X. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol. 2017;51:1115–1123. doi: 10.3892/ijo.2017.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu C, Li L, Xie F, Guo S, Liu F, Dong N, Wang Y. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res. 2018;114:168–179. doi: 10.1093/cvr/cvx180. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, Li MF, Chen GQ, Zhao Q. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011;32:2–9. doi: 10.1093/carcin/bgq209. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q, Xu K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta. 2012;1822:1692–1704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen H, Wang Y, Shi W, Sun G, Hong L, Zhang Y. LncRNA SNHG5/miR-26a/SOX2 signal axis enhances proliferation of chondrocyte in osteoarthritis. Acta Biochim Biophys Sin (Shanghai) 2018;50:191–198. doi: 10.1093/abbs/gmx141. [DOI] [PubMed] [Google Scholar]
- 33.Ugrinova I, Pasheva E. HMGB1 protein: a therapeutic target inside and outside the cell. Adv Protein Chem. 2017;107:37. doi: 10.1016/bs.apcsb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, Huang J, Yu Y, Fan XG, Yan Z. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. doi: 10.1016/j.mam.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruber HE, Hoelscher GL, Synthia B, Jane I, Michael C, Hanley EN. High-mobility group box-1 gene, a potent proinflammatory mediators, is upregulated in more degenerated human discs in vivo and its receptor upregulated by TNF-α exposure in vitro. Exp Mol Pathol. 2015;98:427–430. doi: 10.1016/j.yexmp.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Afrin R, Arumugam S, Rahman A, Wahed MI, Karuppagounder V, Harima M, Suzuki H, Miyashita S, Suzuki K, Yoneyama H, Ueno K, Watanabe K. Curcumin ameliorates liver damage and progression of NASH in NASH-HCC mouse model possibly by modulating HMGB1-NF-κB translocation. Int Immunopharmacol. 2017;44:174–182. doi: 10.1016/j.intimp.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Roman-Blas JA, Jimenez SA. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthritis Cartilage. 2006;14:839–848. doi: 10.1016/j.joca.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Ma T, Guo CJ, Zhao X, Wu L, Sun SX, Jin QH. The effect of curcumin on NF-κB expression in rat with lumbar intervertebral disc degeneration. Eur Rev Med Pharmacol Sci. 2015;19:1305–1314. [PubMed] [Google Scholar]
- 39.Zhongyi S, Sai Z, Chao L, Jiwei T. Effects of nuclear factor kappa B signaling pathway in human intervertebral disc degeneration. Spine (Phila Pa 1976) 2015;40:224–232. doi: 10.1097/BRS.0000000000000733. [DOI] [PubMed] [Google Scholar]
- 40.Nasto LA, Seo HY, Robinson AR, Tilstra JS, Clauson CL, Sowa GA, Ngo K, Dong Q, Pola E, Lee JY, Niedernhofer LJ, Kang JD, Robbins PD, Vo NV. ISSLS prize winner: inhibition of NF-kappaB activity ameliorates age-associated disc degeneration in a mouse model of accelerated aging. Spine (Phila Pa 1976) 2012;37:1819–1825. doi: 10.1097/BRS.0b013e31824ee8f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang XF, Zhang AP, Sun ZY, Liu C, Kuang LH, Tian JW. Expression of NF-κB in a degenerative human intervertebral disc model. Zhonghua Yi Xue Za Zhi. 2017;97:1324–1329. doi: 10.3760/cma.j.issn.0376-2491.2017.17.011. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Ma C, Shen J, Wang D, Hao J, Hu Z. SDF1/CXCR4 axis induces apoptosis of human degenerative nucleus pulposus cells via the NFkappaB pathway. Mol Med Rep. 2016;14:783–789. doi: 10.3892/mmr.2016.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.