Summary
Rice tiller angle determines plant growth density and further contributes grain production. Although a few genes have been characterized to regulate tiller angle in rice, the molecular mechanism underlying the control of tiller angle via microRNA is poorly understood. Here, we report that rice tiller angle is controlled by OsmiR167a‐targeted auxin response factors OsARF12, OsARF17 and OsARF25. In the overexpression of OsMIR167a plants, the expression of OsARF12, OsARF17 and OsARF25 was severely repressed and displayed larger tiller angle as well as the osarf12/osarf17 and osarf12/ osarf25 plants. In addition, those plants showed compromised abnormal auxin distribution and less sensitive to gravity. We also demonstrate that OsARF12, OsARF17 and OsARF25 function redundantly and might be involved in HSFA2D and LAZY1‐dependent asymmetric auxin distribution pathway to control rice tiller angle. Our results reveal that OsmiR167a represses its targets, OsARF12, OsARF17 and OsARF25, to control rice tiller angle by fine‐tuning auxin asymmetric distribution in shoots.
Keywords: OsmiR167a, auxin response factors, auxin asymmetric distribution, tiller angle, rice
Introduction
Rice plant architecture is regarded as one of the main agronomical traits that influence planting density in the field and thus contribute grain yield (Khush, 2003). Rice is a multi‐tiller crop, and its plant architecture is determined by tiller angle, tiller number, plant height, leaf inclination and panicle architecture as well (Wang and Li, 2008). Tiller angle is defined as the angle between the main culm and its side tillers, and it is one of the main target traits being selected in breeding for achieving ideal plant architecture to improve rice yield. Therefore, understanding the mechanism controlling till angle will not only elucidate the process of varieties artificial selection but also provide a way of improving grain production in rice.
With the implementation of rice functional genomics project, quite a few genes have been characterized involving the control of tiller angle. LAZY1 (LA1) is the first identified gene that controls rice tiller angle. Mutation in LA1 alters the endogenous IAA distribution in shoots, leading to the tiller‐spreading phenotype in rice (Li et al., 2007; Yoshihara and Iino, 2007). HSFA2D acts an upstream regulator of LA1 to initiate the asymmetric auxin distribution and further influences asymmetric expression of WOX6 and WOX11 to specify tiller angle (Zhang et al., 2018). Recent investigation showed that OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization (Li et al., 2019). It was reported that strigolactones (SLs) biosynthetic or signalling mutants could rescue the spreading phenotype of la1, indicating that SLs also participate in the tiller angle regulation by inhibiting auxin biosynthesis (Sang et al., 2014). The dominant PAY1 could optimize tiller angle via affecting auxin transport activity and distribution in rice (Zhao et al., 2015). OsLIC functions as a negative regulator for optimal tiller angle in rice through mediating the brassinosteroids (BRs) response (Wang et al., 2008). LPA1, encoding a plant‐specific INDETERMINATE DOMAIN protein, controls tiller angle by regulating the sedimentation rate of amyloplasts (Wu et al., 2013). Additionally, several quantitative trait loci (QTLs) affecting tiller angle have been characterized (Dong et al., 2016; He et al., 2017; Qian et al., 2001; Yu et al., 2007). Mutation in the major QTL Tiller Angle Control1 (TAC1) leads to a compact tiller in japonica rice compared with relatively wider tiller angle in indica rice (Yu et al., 2007). Another QTL TAC3, together with TAC1 and D2, greatly controls tiller angle in rice cultivars (Dong et al., 2016). Besides, tiller angle is also associated with rice domestication. The artificial selection of an amino acid substitution in the PROG1 protein during domestication resulted in the transition from the prostrate tillers of wild rice to the erect tillers of cultivated rice (Jin et al., 2008; Tan et al., 2008). TIG1 encoding a TCP transcription factor also contribute to the transition from inclined tiller growth in wild rice to erect tiller growth during rice domestication (Zhang et al., 2019).
MicroRNAs (miRNAs), a type of small (~21 nucleotides) non‐coding RNAs, play a crucial role in negative regulation of gene expression by binding to target mRNAs for cleavage and (or) translation repression at the post‐transcriptional level in both plants and animals (Bartel, 2009; Carrington and Ambros, 2003). Auxin response factors (ARFs) are transcription factors that bind specifically to TGTCTC‐containing auxin‐responsive elements (AuxREs) found in the promoters of early auxin response genes and mediate the signal transduction of auxin, and finally regulate plant growth and development (Chandler, 2016; Li et al., 2016; Wang et al., 2007). In plants, several ARF genes have been identified as the cleavage targets of miR390, miR160 and miR167. In Arabidopsis, AtARF2, AtARF3 and AtARF4 are the targets of miR390‐derived trans‐acting small interfering RNAs (ta‐siRNA) (Fahlgren et al., 2006; Williams et al., 2005). In Arabidopsis, miR160 and its targets ARF10, ARF16 and ARF17 have been suggested to function in growth and developmental processes. miR160 regulates the expression of ARF10 and ARF16 and determines the root cap formation (Wang et al., 2005). miR160‐directed ARF17 regulation is essential for proper development via modulating expression of early auxin response genes (Mallory et al., 2005). Repression of ARF10 by miR160 plays a critical role in seed germination and post‐embryonic developmental programmes (Liu et al., 2007). Furthermore, miR160‐controlled ARF10 is essential for seed germination through stimulation of both auxin and ABA signalling (Liu et al., 2013). ARF6 and ARF8 were identified as the cleavage targets of miR167 in Arabidopsis (Ru et al., 2006; Wu et al., 2006; Yao et al., 2019; Zheng et al., 2019). Previous investigation showed that miR167‐directed ARF6/ARF8 expression is essential for reproductive development in Arabidopsis (Ru et al., 2006; Wu et al., 2006). Recent studies suggested that miR167‐mediated negative regulation of ARF6 and ARF8 is necessary for anther dehiscence and ovule development (Zheng et al., 2019). Yao et al. (2019) reported that MIR167a acts as a maternal gene and functions largely through ARF6 and ARF8 in controlling of embryonic and seed development. Blue light could increase miR167 expression and then resulted in the enhanced transcription of ARF4 and ARF8 (Pashkovskiy et al., 2016). Interestingly, ARF17 (a target of miR160) is a negative regulator, and ARF6 and ARF8 (targets of miR167) are positive regulators of adventitious rooting, suggesting that miR160 and miR167 commonly control phenotypic plasticity of adventitious root initiation in Arabidopsis (Gutierrez et al., 2009).
MiR167 targeted the transcription factor ARF genes had been reported to be conserved among other plant species, including rice (Liu et al., 2012a), soybean (Wang et al., 2015), tomato (Liu et al., 2014), Ipomoea nil (Glazinska et al., 2014), Camelina sativa (Na et al., 2019) and so on. A total of 25 putative OsARFs have been identified in of rice (Guilfoyle and Hagen, 2007; Li et al., 2016; Wang et al., 2007). Four OsARFs (OsARF6, OsARF12, OsARF17 and OsARF25) were predicted to be the target of OsmiR167 (Li et al., 2010; Liu et al., 2012a). Yang et al. (2006) reported that OsmiR167‐OsARF8‐GH3 was response to exogenous auxin in cultured rice cells. OsmiR167‐directed OsARF12 expression regulates root elongation and affects iron accumulation in rice (Qi et al., 2012). In order to determine the biological function of OsmiR167, we generated OsMIR167a‐overexpressing lines in rice. OsARF12, OsARF17 and OsARF25 were examined as the cleavage targets of OsmiR167a. Both overexpressing OsMIR167a lines and osarf12/osarf17 and osarf12/osarf25 mutants showed larger tiller angle phenotype and disrupted auxin distribution in axillary buds. Our results indicate that OsmiR167a regulates the pattern of OsARF12, OsARF17 and OsARF25 expression, which is vital for modulation of tiller angle in rice.
Results
Overexpressing OsMIR167a resulted in larger tiller angle in rice
According to the annotation of miRNA database (miRBase: http://www.mirbase.org/index.shtml) (Kozomara and Griffiths‐Jones, 2011, 2014), there are ten OsMIR167 (OsMIR167a to OsMIR167j) genes in rice genome. Although OsMIR167 genes harbour different stem‐loop precursors, they generate the similar mature sequences of OsmiR167. There is a single nucleotide difference in the 3′ end between OsmiR167a‐c and OsmiR167d‐j, and OsmiR167a‐c show the same sequence with Arabidopsis AtmiR167a‐b (Table S1). In order to elucidate the function of miR167 in rice, we took OsMIR167a as the representative to generate its overexpressing plants (designated as MIR167a‐OE).
We first examined the expression pattern of OsMIR167a in different organs at vegetative stage. Quantitative real‐time RT‐PCR (qRT‐PCR) showed that pre‐OsmiR167a was detected in the examined organs including culm, leaf sheath, root and axillary bud (Figure 1a). The expression pattern of OsmiR167a was further confirmed by RNA gel blot analysis (Figure 1b). In the MIR167a‐OE lines, pre‐OsmiR167a and OsmiR167a had higher expression levels in those organs compared to that in wild‐type plants (WT) (Figure 1a,b). Then, we characterized the morphological differences between MIR167a‐OE and WT. At tillering stage, MIR167a‐OE plants showed clearly larger tiller angle compared with WT (Figure 1c). After heading, tiller angle was increased dramatically in MIR167a‐OE plants (Figure 1d,e). Compared to WT, MIR167a‐OE plants showed obvious semi‐dwarf and low fertility (Figure 1c–g). The maximum tiller angle of among MIR167a‐OE plants was 110°, approximately 40° wider than that in WT (Figure 1e,f). The plant height of MIR167a‐OE was reduced about 40% of the WT (Figure 1g). Our results suggest that overexpression of MIR167a results in larger tiller angle in rice.
Figure 1.
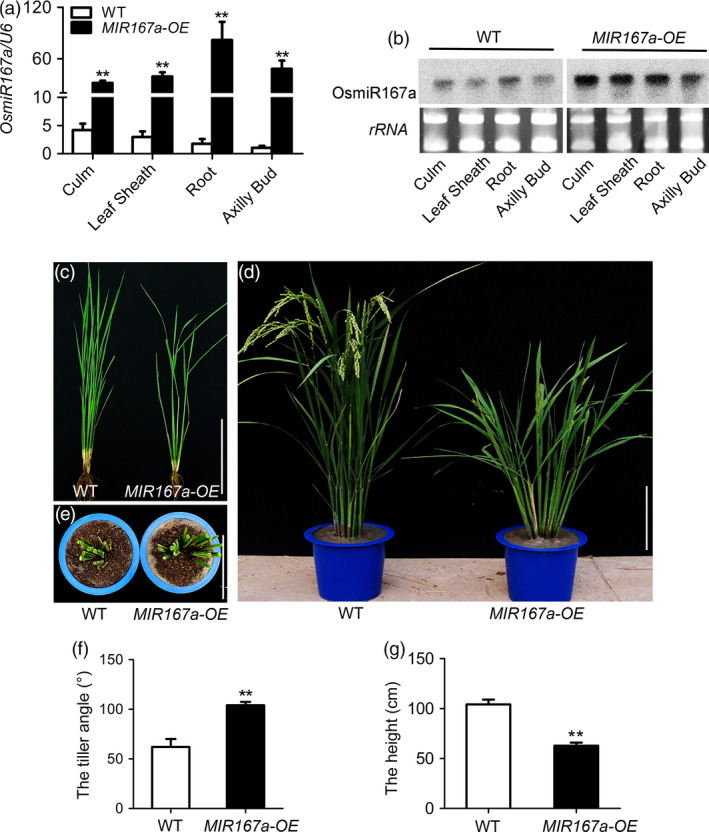
Phenotypic analysis of MIR167a‐OE lines. (a, b) Expression analysis of miR167a in the culm, leaf sheath, root and axillary bud by quantitative real‐time RT‐PCR (qRT‐PCR) (a) and small RNA gel blot (b). U6 was used as a control. (c, d) Plant morphology of WT (ZH11) and MIR167a‐OE lines at tillering (c) and heading (d) stages. (e) Vertical observation of WT and MIR167a‐OE transgenic lines. (f, g) Comparison of the tiller angle (f) and plant height (g) between WT and MIR167a‐OE lines. Data are shown as means ± SE (n = 10). Bars = 20 cm. Significant at **P < 0.01.
OsARF12, OsARF17 and OsARF25 are the promising target genes of OsmiR167a
There are 25 OsARFs in rice. Previous investigations predicted that OsARF6, OsARF12, OsARF17 and OsARF25 were the putative target genes of OsmiR167a (Liu et al., 2012a; Zhang, 2005). We further verified the predicted miRNA targets by CSRDB (Cereal small RNAs Database; http://sundarlab.ucdavis.edu/smrnas/) (Table S2). RT‐PCR analysis of OsARF genes expressed in axillary buds suggested that the expression levels of OsARF12, OsARF17 and OsARF25 were significantly reduced in MIR167a‐OE plants, whereas OsARF6 and other OsARFs (OsARF1, OsARF8, OsARF10‐11, OsARF13‐14, OsARF18, OsARF22) performed no significantly decreased expression (Figure 2a). We further examined the OsmiR167a‐directed cleavage sites of OsARFs using a 5′ RNA ligase‐mediated (RLM) rapid amplification of cDNA ends (RACE) assay. The cleavage products of OsARF12, OsARF17 and OsARF25 mRNA fragments were successfully detected in MIR167a‐OE plants, except for that of OsARF6 (Figure 2b). We randomly chose 12 clones from the cleavage products for sequencing, and more than 8 clones showed that the transcripts of OsARF12, OsARF17 or OsARF25 were cleaved at the OsmiR167a target site (Figure 2c). Thus, OsARF12, OsARF17 and OsARF25 might be the target genes of OsmiR167a in rice.
Figure 2.
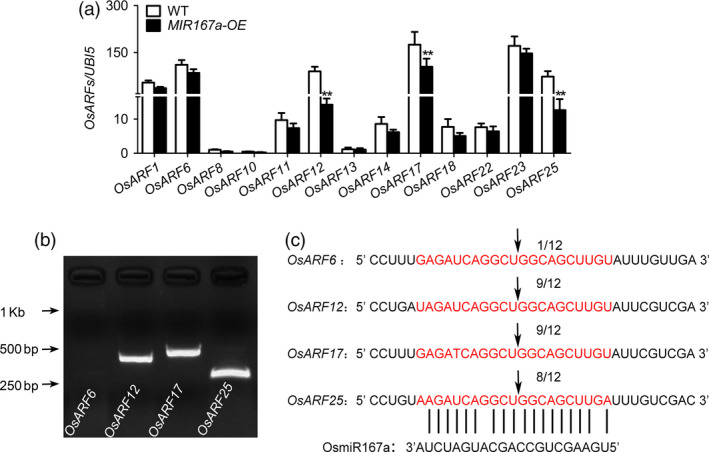
Mapping of miR167 target cleavage sites in OsARF12, OsARF17 and OsARF25. (a) Expression pattern of predicted target genes in WT and MIR167a‐OE plants. Ubiquitin 5 was used as a control. Data are presented as means ± SE (n = 3). Significant at **P < 0.01. (b) PCR products of OsARF6, OsARF12, OsARF17 and OsARF25 by RLM‐5'RACE was showed by gel electrophoresis. (c) The sequencing results of 12 clones from the 5′ RACE cleavage products of OsARF6, OsARF12, OsARF17 and OsARF25. The vertical lines represent the nucleotides that base‐pair with miR167a. The arrow points to the OsmiR167‐directed cleavage site at the OsARFs transcript. The numbers above the sequence indicate frequency of 5′RACE clones corresponding to each site.
OsmiR167a‐OsARFs modules regulate tiller angle in rice
To confirm that OsARF12, OsARF17 and OsARF25 are the target genes of OsmiR167a, we generated a series of transgenic plants overexpressing OsmiR167a‐resistant version of individual OsARFs (resulting plants named Ubi::mARF12, Ubi::mARF17 and Ubi::mARF25) in Zhonghua 11 (WT) and MIR167a‐OE background, respectively. Based on the conserved carboxy‐terminal dimerization domain (CTD) of OsARFs, which contained the OsmiR167a‐directed cleavage site, five synonymous mutations were introduced into the transgenic constructs (Figure 3a,b) without any amino acid changes. qRT‐PCR analysis confirmed the positive transgenic lines had overexpression of un‐cleaved version of OsARF12, OsARF17 and OsARF25 in WT (Figure 3c) and MIR167a‐OE (Figure 3d) background, respectively. Next, we characterized the morphological changes of the Ubi::mARF12, Ubi::mARF17 and Ubi::mARF25 transgenic plants compared to WT or MIR167a‐OE (Figure 3e–n). In WT background, transgenic plants with Ubi::mARF12, Ubi::mARF17 and Ubi::mARF25 showed slight reduced tiller angle compared with WT (Figure 3e,g–j). No remarkable plant height changes were observed between OsARFs‐resistant plants and WT (Figure 3f,g–j). However, in MIR167a‐OE background, overexpressing OsmiR167a‐resistant version of individual OsARF12, OsARF17 or OsARF25 resulted in obviously decreased tiller angle compared with MIR167a‐OE plants (Figure 3e,k–n). Meanwhile, the semi‐dwarf of MIR167a‐OE plants was partially restored in OsARFs‐resistant plants (Figure 3f,k–n). These observations suggest expression of mOsARF12, mOsARF17 or mOsARF25 could partially rescue the abnormal plant architecture when OsMIR167a overexpressed.
Figure 3.
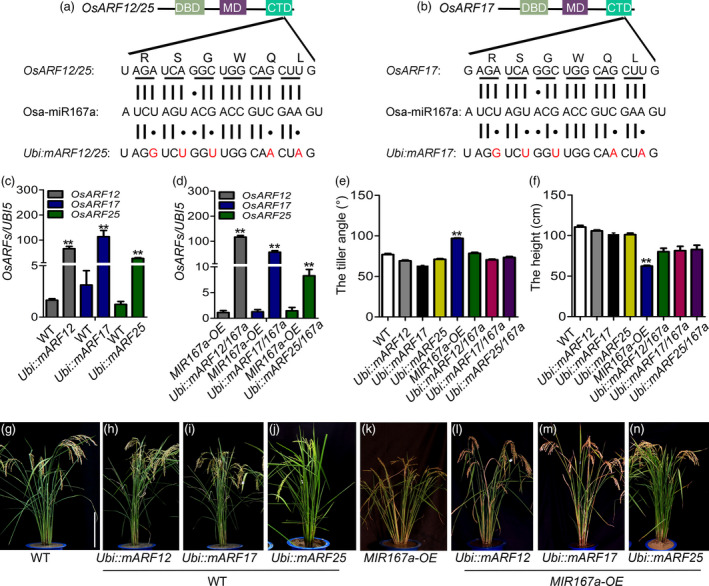
Generation of transgenic rice plants expressing an OsmiR167‐resistant version of OsARFs (mOsARFs). (a, b) Domain structure of the OsARF12, OsARF17 and OsARF25 protein. The B3 DNA binding domain (DBD), middle region (MR) and the C‐terminal dimerization domain (CTD) are indicated. The OsmiR167 complementary (or target) sequence in the OsARFs mRNA and the corresponding region of amino acid sequence (RSGWQL) are shown. The silent mutations were created in mOsARFs by introducing silent mutations. The vertical lines represent the nucleotides that base pair with miR167a and black dots show the mismatched nucleotide. (c, d) Expression levels of uncleaved OsARF12, OsARF17 and OsARF25 by qRT‐PCR in transgenic lines. (e, f) Comparison of the tiller angle (e) and plant height (f) between WT and transgenic lines. (g–n) Phenotype of Ubi::mOsARFs and Ubi::mOsARFs/167a plants. Bar = 20 cm. Significant at **P < 0.01.
Target MIMICs are known to inhibit miRNA activity (Franco‐Zorrilla et al., 2007; Todesco et al., 2010). Thus, we further verified the OsmiR167a‐OsARFs involved in tiller angle regulation by generation the MIM167a transgenic plants. Three‐nucleotide bulge (CAA) were introduced into the mature OsmiR167a, which replaced the OsmiR399 complementary motif in OsIPS1 (Franco‐Zorrilla et al., 2007) to engineer an artificial target mimicry construct of MIM167a (Figure S1a). In the MIM167a transgenic plants, the abundance of OsARF12, OsARF17 or OsARF25 were significantly increased relative to that in MIR167a‐OE plants, but not in WT background (Figure S1b). In WT background, overexpression of MIM167a has no effect on the tiller angle and plant height (Figure S1c,e,f). However, in MIR167a‐OE plants, the morphologies of larger tiller angle and semi‐dwarf were partially restored when overexpressing MIM167a (Figure S1d–f). Collectively, these results suggest that OsmiR167a modulates rice tiller angle by fine‐tuning the accumulation of OsARFs, mainly including OsARF12, OsARF17 and OsARF25.
OsARF12, OsARF17 and OsARF25 function redundantly to modulate tiller angle
To further confirm the role of OsARF12, OsARF17 and OsARF25 in tiller angle control, we collected their mutants and developed the double mutants to characterize the plant architecture. We obtained osarf12 (03Z11JV18) and osarf17 (03Z11BY76) mutants from our T‐DNA insertional mutant library in variety Zhonghua 11 (Wu et al., 2003). Analysis of T‐DNA insertion sites in the mutants shows that it is located in the fourteenth intron of OsARF12 (Figure 4a) and seventh intron of OsARF17 (Figure 4b), respectively. For osarf25 (PFG_2D‐11520) in variety Dongjing, T‐DNA insertion site in the fifth exon was characterized previously (Figure 4c; Qi et al., 2012). PCR analysis confirmed that the T‐DNA inserted into individual OsARF genes, and their homozygous were obtained (Figure 4d). qRT‐PCR results indicated the transcripts of OsARF12, OsARF17 and OsARF25 were suppressed in its corresponding mutants (Figure 4e), suggesting that these genes were knockout in mutants. We further characterized the tiller angle and plant height of osarf12, osarf17 and osarf25 compare with WT (Figure 4f–m). At the heading stage, osarf12 and osarf17 showed slight loosen plant types (Figure 4f,h–j) with tiny increase tiller angle compared to WT, and no obvious changes on the plant height were observed (Figure 4g–j). However, osarf25 showed obviously increased tiller angle and reduced plant height compared to WT (Figure 4f–k). The double mutants of osarf12/osarf17 and osarf12/osarf25 displayed remarkable phenotype defects with larger tiller angle and semi‐dwarf (Figure 4f–g,l–m). Thus, our results suggest that OsARF12, OsARF17 and OsARF25 function redundantly to modulate tiller angle.
Figure 4.
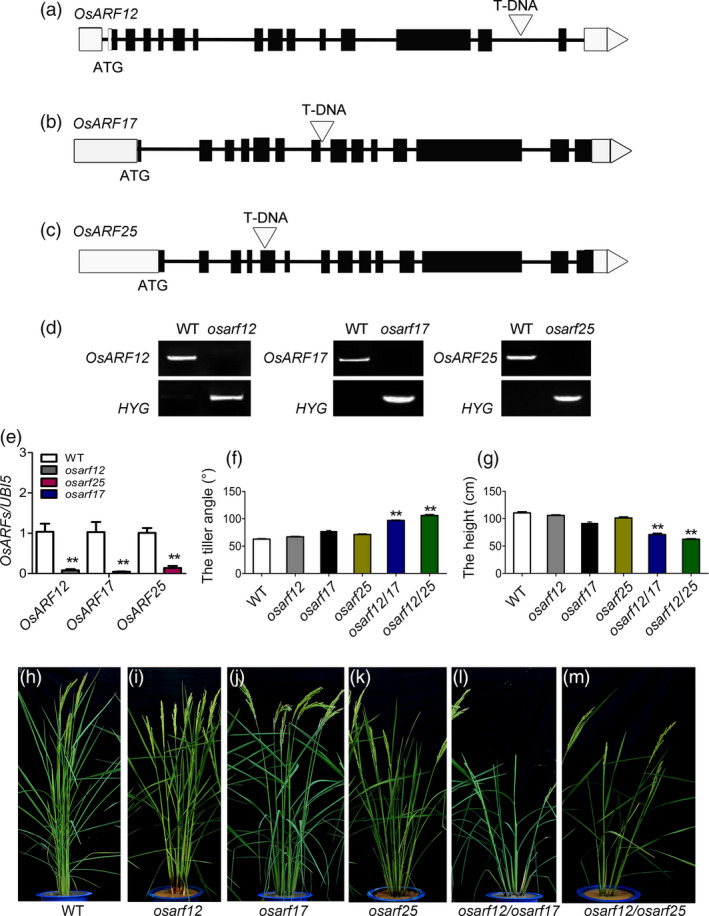
Identification of OsARFs mutants. (a–c) T‐DNA insertion sites in osarf12, osarf17 and osarf25. Exons (filled boxes) and introns (lines between the filled boxes) are shown. The open triangle indicates T‐DNA insertion site. (d) PCR analysis to confirm the integration of T‐DNA in OsARF mutants; the upper band indicates the OsARF gene fragment and the lower band indicates the T‐DNA insertion fragment. (e) Expression analysis the level of OsARF12, OsARF17 and OsARF25 in WT and osarf mutants by qRT‐PCR. (f–g) Comparison of the tiller angle (f) and plant height (g) of WT and mutants. (h–m) Morphologies of WT and mutants. Bar = 20 cm. Significant at **P < 0.01.
Auxin up‐regulated expression of OsARF12, OsARF17 and OsARF25
To test whether the expression of OsARF12, OsARF17 and OsARF25 was affected by phytohormones, we treated 7‐day seedlings with various hormones for 2 h. qRT‐PCR results showed that the expression of OsARF12, OsARF17 and OsARF25 were significantly induced by IAA treatment, but not by the other hormones (Figure S2a–c). Moreover, two other auxin compounds, IBA (indole‐3‐butyric acid) and NAA (naphthylacetic acid) also can induce the expression of OsARF12, OsARF17 and OsARF25, but with a less extent (Figure S2a–c). We further used tryptophan (a structurally similar molecule to IAA) to treat the seedlings, the expression of OsARF12, OsARF17 and OsARF25 remained unchanged, indicating tryptophan has no hormonal activity (Figure S2a–c). Next, we examined the time‐course response of OsARF12, OsARF17 and OsARF25 to 10 μM IAA treatment. The transcripts of OsARF12, OsARF17 and OsARF25 showed remarkable increase up to 60 min of IAA application (Figure S2d). However, the transcript levels of OsARF12, OsARF17 and OsARF25 maintained almost the same under different concentration of auxin (Figure S2e). Taken together, our results suggest that OsARF12, OsARF17 and OsARF25 might be involved in auxin response to modulate tiller angle in rice.
OsmiR167a‐OsARFs modules affect polar auxin transport
Considering OsARF12, OsARF17 and OsARF25 may control rice tiller angle via auxin signalling, we carried out to examine whether OsmiR167a‐OsARF module is involved in sensing gravistimulation and polar auxin transport (PAT). We first examined the free IAA concentration in axillary buds and roots among MIR167a‐OE, osarf12/osarf17, osarf12/osarf25 and WT plants. Compared to WT, the content of free IAA is almost consistent in MIR167a‐OE, osarf12/osarf17 or osarf12/osarf25 (Figure 5a), suggesting that OsmiR167a‐OsARFs may not affect free IAA accumulation in rice. However, gravitropism analysis of the WT and mutant seedlings showed that the coleoptiles of MIR167a‐OE, osarf12/osarf17 and osarf12/osarf25 could not grow upright completely. Correspondingly, MIM167a, MIM167a/167a and WT showed the normal gravity response, with upright coleoptiles (Figure 5b). When NPA (N‐1‐naphthylphthalamic acid, an auxin transport inhibitor) was added to the medium, the curved angle of coleoptiles of MIR167a‐OE, osarf12/osarf17 and osarf12/osarf25 were recurred, almost similar to that in WT (Figure 5b,c). However, this phenomenon of recurred curved angle was not observed when using IAA instead of NAA (Figure 5b,c). Thus, the gravitropism response requires the asymmetric distribution of auxin. Our result suggests that OsmiR167a‐OsARFs might affect gravity response by participating in auxin transport in rice.
Figure 5.
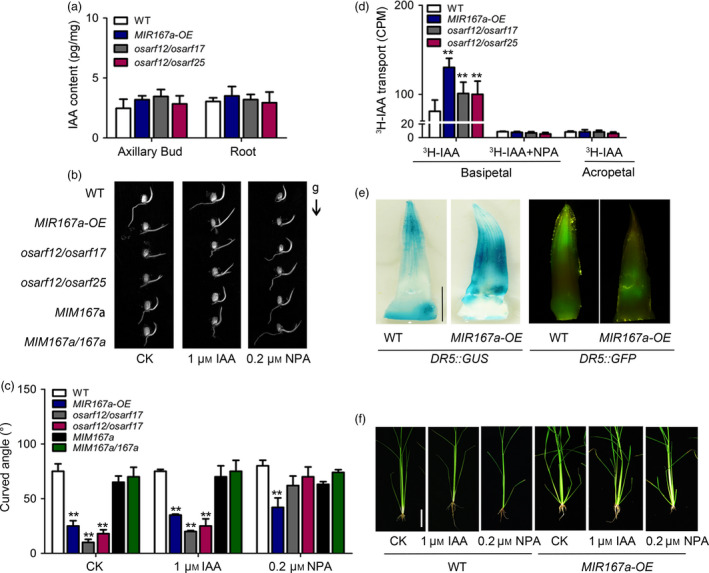
Altered auxin distribution and transport in MIR167a transgenic plant. (a) Quantification of auxin content in WT, MIR167a‐OE and OsARF mutant plants. (b) Three‐day‐old seedlings of the WT, MIR167a‐OE, ARF double mutants and MIM167a transgenic plants grown in dark after a 24‐h gravistimulation. The arrow indicates the direction of gravity (g). (c) Statistical data of ultimate curved angle of WT, MIR167a‐OE, OsARF double mutants and MIM167a transgenic lines under IAA or NPA treatment. Data are shown as means ± SE (n = 10). (d) Measurement of auxin transport ability in WT, MIR167a‐OE and OsARF mutant plants using coleoptile as materials. Significant at **P < 0.01. (e) Comparison of DR5 promoter activity in axillary bud of WT and MIR167a‐OE plants. Bar = 0.5 cm. (f) Morphology of MIR167a‐OE plants under treatment of 1 μM IAA and 0.2 μM NPA treatments for 40 days. Bar = 5 cm.
Next, we compared the auxin transport in etiolated coleoptiles between WT and mutant lines. Compared to WT, the basipetal transport of 3H‐IAA was significantly increased in MIR167a‐OE, osarf12/osarf17 and osarf12/osarf25, whereas the acropetal PAT of 3H‐IAA displayed no differences between WT and mutant lines (Figure 5d). When treated with NPA (N‐1‐naphthylphthalamic acid, an auxin transport inhibitor), the difference of basipetal PAT between WT and mutant lines was disappeared (Figure 5d). We further examined endogenous IAA distribution in the axillary buds by introducing the auxin reporter DR5::GUS and DR5::GFP into MIR167a‐OE background, respectively. In the axillary buds, the signal of GUS/GFP was mainly detected at the apex area in WT, whereas in MIR167a‐OE lines, the GUS/GFP signal moved to the middle part of axillary buds and expanded to a much broader area (Figure 5e), indicating that the enhanced basipetal PAT affected endogenous IAA distribution in axillary bud of MIR167a‐OE lines. Furthermore, the tiller angle was decreased in MIR167a‐OE when treating its seedlings with NPA (Figure 5f). Our results suggest that OsmiR167a‐OsARFs modules control rice tiller angle by affecting polar auxin transport in axillary buds.
OsmiR167a‐OsARFs modules regulate tiller angle through mediating asymmetric auxin distribution in shoots
Previous studies described a regulatory pathway of rice tiller angle by HSFA2D‐LAZY1 (LA1)–mediated auxin asymmetric distribution (Zhang et al., 2018). A knockout mutant hsfa2d (03Z11CE69) showing remarkable larger tiller angle has been identified from our T‐DNA insertional mutant library (http://rmd.ncpgr.cn/; Zhang et al., 2018). In order to investigate whether OsmiR167a‐OsARF module is involved in the LAZY1‐mediated asymmetric auxin distribution pathway, we obtained lazy1 mutant (03Z11UL88) from our T‐DNA insertional mutant library by mapping‐based cloning approach (Figure S3). We identified a 69 bp deletion in the 2nd exon of LAZY1 (LA1), which resulted in large tiller angle in la1 (Figure S3; Table S3). Both hsfa2d and la1 displayed an obvious larger tiller angle and semi‐dwarf phenotypes (Figure 6a,b). RT‐PCR results showed that the transcripts of HSFA2D and LA1 were significantly suppressed (Figure 6c), indicating that both HSFA2D and LA1 are loss of function in their corresponding mutants. Compared to that of WT, the expression levels of OsARF12, OsARF17 and OsARF25 were significantly reduced in hsfa2d or la1 in axillary shoots using qRT‐PCR analysis (Figure 6d). However, in the MIR167a‐OE, osarf12/osarf17 or osarf12/osarf25 backgrounds, the expression of HSFA2D and LA1 were obviously up‐regulated (Figure 6e), indicating that a feedback regulation network might exist between HSFA2D‐LA1 and OsmiR167a‐OsARFs modules. Of course, further analysis of the expression levels of OsMIR167a and OsARF12/1/7/25 in the overexpression of HSFA2D or LA1 plants will be needed to confirm this speculation. Compared to WT, the la1 showed more larger tiller angle than that of osarf12/osarf17 double mutant (Figure 6f–h). The la1/osarf12/osarf17 triple mutant exhibited an intermediate larger tiller angle between that of la1 and osarf12/osarf17 (Figure 6g–i), suggesting that OsmiR167a‐OsARFs modules might also be involved in the asymmetric auxin distribution in controlling tiller angle.
Figure 6.
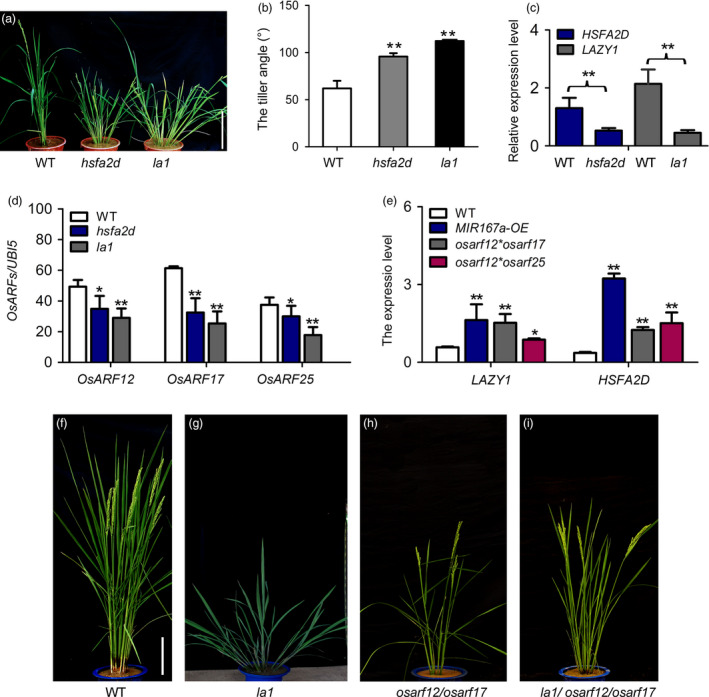
OsARF12/17/25 and LA1 affect auxin distribution in controlling tiller angle. (a, b) Morphology of WT, hsfa2d and la1 mutants. Bar = 15 cm. (c) Transcript levels of HSFA2D and LAZY1 in WT and mutants determined by qRT‐PCR. (d) Expression level of OsARF12, OsARF17 and OsARF25 in WT, hsfa2d and la1 mutants. (e) Expression level of HSFA2D and LAZY1 in WT and OsARF double mutants. Significant at *P < 0.05, **P < 0.01. (f–i) Phenotypes of WT, la1, osarf12/osarf17 and la1/osarf12/osarf17 plants. Bars = 20 cm.
Zhang et al. (2018) has reported that HSFA2D‐LA1 induces the asymmetric distribution of auxin, and then auxin triggers the asymmetric expression of WOX6 and WOX11 in shoots mediating the control of rice tiller angle. Thus, we further examined the expression patterns of WOX6 and WOX11 in seedling shoots upon gravistimulation under light for 6h in MIR167a‐OE, osarf12/osarf17 and osarf12/osarf25 plants. Similar to previous investigation (Zhang et al., 2018), the expression of WOX6 and WOX11 indeed performed asymmetric expression patterns with highly expressed in the lower side of shoot bases (Figure S4). However, in the MIR167a‐OE, osarf12/osarf17 or osarf12/osarf25 plants, the extent of the induction of WOX6 and WOX11 expression in the lower side of shoot bases was significantly reduced, compared to that in WT (Figure S4). Considering OsmiR167a‐OsARFs modules affect polar auxin transport in axillary buds (Figure 5d,e), these results suggest that OsmiR167a‐OsARFs modules might be involved in the asymmetric distribution of auxin in rice shoots and then resulted in the asymmetric expression of WOX6 and WOX11 to modulate tiller angle in rice.
Discussion
OsARF12, OsARF17 and OsARF25 are required to regulate tiller angle
There are 25 OsARF genes in rice genome (Guilfoyle and Hagen, 2007; Wang et al., 2007). All of the OsARF proteins contain a highly conserved N‐terminal domain for DNA binding, a central region, and a C‐terminal domain for protein‐protein interaction (Shen et al., 2010; Wang et al., 2007). Although a few OsARF genes have been functional characterized, the functions for most of them remain unknown. In this study, we identified OsmiR167a‐OsARF12/17/25 as a key regulatory module of tiller angle control, which affects plant architecture in rice.
OsARF1 is the first ARF gene that regards as an early auxin‐responsive gene in rice (Waller et al., 2002). The morphological defects of antisense‐OsARF1 plants suggested that OsARF1 is essential for vegetative growth and seed development (Attia et al., 2009). OsARF3 mediates the auxin‐cytokinin cross‐talk involving de novo shoot regeneration (Cheng et al., 2013). OsARF12 functions in phosphate homeostasis and required for primary root elongation (Qi et al., 2012; Wang et al., 2014). OsARF16 is required for the auxin and cytokinin response and is involved in phosphate transport and signalling (Shen et al., 2014). OsARF17 and OsARF19 control rice leaf inclination through regulating both auxin and BR signalling or cross‐talk between them (Chen et al., 2018; Zhang et al., 2015). OsARF23 and OsARF24 control cell growth and morphogenesis through heterodimerization to reduce the expression of RMD in rice (Li et al., 2014). In rice, 25 ARF protein sequences were classified into three clusters according to the phylogenetic tree (Wang et al., 2007). OsARF12, OsARF17 and OsARF25 belonged to the same subgroup and were phylogenetically closest to each other, suggesting that they might perform the similar biological function. In this study, we obtained the T‐DNA insertion mutant lines for OsARF12, OsARF17 and OsARF25 (Figure 4). Their single mutant plant showed tiny increase tiller angle compared to WT, but the double mutants of osarf12/osarf17 and osarf12/osarf25 displayed remarkable larger tiller angle (Figure 4h–m). Thus, our studies demonstrate that OsARF12, OsARF17 and OsARF25 function redundantly to modulate tiller angle.
miR167 target ARF genes are conserved in plants
Sequence similarities of mature miRNAs from various species indicated that miR167 is conserved throughout the plant kingdom (Li et al., 2010; Luo et al., 2013). Comparison of mature miR167s between Arabidopsis and rice showed the identical sequence (Table S1). Although some non‐conserved transcripts besides ARF gene family were predicted as targets for miR167 (Li et al., 2010), many ARF genes were experimentally validated as the miR167 targets among different species. In Arabidopsis, miR167 targets AtARF6 and AtARF8, and regulates some reproductive development process, such as anther dehiscence, ovule development, embryonic and seed development (Ru et al., 2006; Wu et al., 2006; Yao et al., 2019; Zheng et al., 2019). In rice, four OsARFs (OsARF6, OsARF12, OsARF17 and OsARF25) were predicted as the target of OsmiR167 (Liu et al., 2012a), and they belong to the same subgroup with AtARF6 and AtARF8 in Arabidopsis by phylogenetic analysis (Wang et al., 2007). In this study, we characterized the function of OsARF12/17/25, orthologues of Arabidopsis AtARF6/8. Overexpression of OsmiR167a or knock out of its targets, the OsARF genes (OsARF12, OsARF17 and OsARF25), has led to larger tiller angle in rice (Figures 1d, 4). Therefore, our data demonstrate that rice OsmiR167a target OsARF12/17/25 control plant architecture via affecting tiller angle. In soybean, GmARF8a and GmARF8b are the homologous genes of Arabidopsis AtARF8. The miR167‐GmARF8 module is required for soybean nodulation and lateral root development (Wang et al., 2015). The tomato SlARF6 and SlARF6 genes, as the orthologous AtARF6 and AtARF8 genes in Arabidopsis, are also the targets of miR167 (Liu et al., 2014). SlARF6 and SlARF6 have conserved roles in controlling growth and development of vegetative and flower organs in tomato (Liu et al., 2014). Recently, Na et al. (2019) reported that overexpression of miR167 targeted the CsARF8 in Camelina causes decreased content of α‐linolenic acid and increased seed size. Collectively, miR167 target ARF genes are conserved, but play diverse roles in plant growth and development.
Besides OsARF12, OsARF17 and OsARF25, OsARF6 also reported as the target of OsmiR167 (Liu et al., 2012a). However, the OsARF6 mRNA level was not reduced significantly in MIR167a‐OE plants in this study (Figure 2a). On the other hand, the cleavage products of OsARF6 mRNA fragments were rarely detected in MIR167a‐OE plants by RLM‐5'RACE (Figure 2b,c). It is possible that miR167‐mediated repression of OsARF6 function is at the level of translation. Whether OsARF6 also functions in rice tiller angle control, we need to develop its mutant for in‐depth dissection of OsARF6 functions.
Asymmetric auxin distribution affects tiller angle in rice
Rice tiller angle is a complex trait, which is affected by multiple factors and displays a strong phenotypic plasticity. Previous investigations suggested that rice tiller angle is strongly related to plant gravitropic responses. Zhang et al. (2018) described a core regulatory pathway controlling rice tiller angle: HSFA2D regulates auxin asymmetric distribution by regulating expression level of LA1, which in turn induce the asymmetric expression of WOX6 and WOX11 and thus modulates rice shoot gravitropism and tiller angle. Our study shows that the OsmiR167a‐OsARF12/17/25 modules also regulate tiller angle via the auxin‐mediated asymmetric distribution of WOX6 and WOX11 (Figure S4). Considering the expression of OsARF12/17/25 were induced by exogenous IAA treatment (Figure S2), we further examined the expression levels of OsARF12/17/25 at the lower and upper sides of the shoot based upon gravistimulation under light (Figure S5). It seems that the asymmetric distribution of auxin by gravistimulation have a feedback regulation on the asymmetric expression of OsARF12/17/25 in the shoot base. Since the expression of OsARF12/17/25 were suppressed in hsfa2d or la1, while the expression of HSFA2D and LA1 were increased in osarf12/osarf17 or osarf12/osarf25 plants, we speculate that HSFA2D‐LA1 can induce the asymmetric distribution of auxin as they response to gravistimulation, while OsARF12/OsARF17/OsARF25 is required for the asymmetric distribution of auxin, possibly by maintaining the location auxin to a suitable level to response to HSFA2D‐LA.
Based on our results, we propose that both OsmiR167a‐OsARF12/17/25 and HSFA2D‐LA1 could regulate the asymmetric distribution of auxin and subsequently induce the asymmetric expression of WOX6/11 and specify tiller angle (Figure 7). It was well known that auxin regulates cell division and elongation to drive plant growth and development (Woodward and Bartel, 2005). ARF transcription factor family regulates gene expression depend on the distribution of auxin in cells (Chandler, 2016; Li et al., 2016). Our investigation demonstrates that OsmiR167a‐OsARF12/17/25 regulates the asymmetric distribution of auxin in axillary buds (Figure 5e) and the auxin distribution also resulted in asymmetric expression of OsARF12/17/25 (Figure S5). Future studies should focus on examining the feedback loop regulation between OsmiR167a‐OsARF12/17/25 and dose‐sensitive auxin signalling, which might be essential for fine‐tuning tiller angle in rice.
Figure 7.
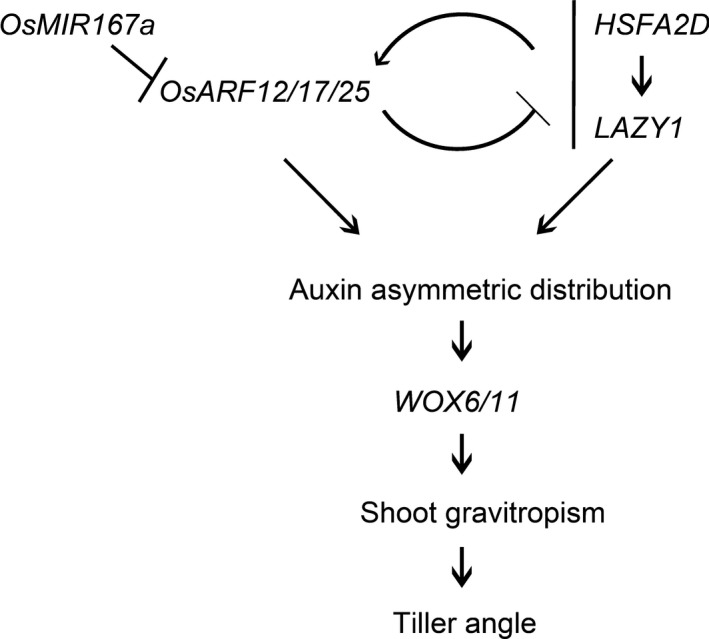
The integrated model controlling tiller angle mediated by OsmiR167a‐OsARFs and HSFA2D‐LA1 (Zhang et al., 2018) in rice.
Methods
Plant material and growth conditions
Rice plants used in this study were japonica (O. sativa ssp. geng) variety Zhonghua 11 (ZH11), except osarf25 in Dongjing background. Rice plants were cultivated in the experimental field of Huazhong Agriculture University in the normal growing season in Wuhan, China (latitude 30.5°N, 15 m above sea level; average daily temperature approximately 28°C).
Plasmid construction and rice transformation
The genomic sequence containing OsmiRNA167a precursor (http://www.mirbase.org/search.shtml) was cloned into the pU1301 vector, then electroporated into the Agrobacterium tumefaciens strain EHA105, and finally transformed into rice callus to generate miR167a overexpression transgenic plants. The target mimicry strategy was generated as described (Franco‐Zorrilla et al., 2007; Todesco et al., 2010). Three‐nucleotide bulge (CAA) was introduced between the position 10 and position 11 of mature miR167a, and then replaced the 24nt OsmiR399 complementary motif in OsIPS1 with the modified 24nt of miR167a. The generated artificial target mimics of MIM167a were under the control of a maize ubiquitin promoter. To construct the OsARF overexpression plasmids, the full‐length coding sequence (CDS) of OsARF12, OsARF17 and OsARF25 were amplified from the cDNA prepared from ZH11 seedlings and cloned into the pU2301 vector, then electroporated into EHA105 and transformed into ZH11 or MIR167a‐OE rice callus. Primers for plasmid construction are listed in Table S4.
RNA extraction and expression analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, http://www.invitrogen.com/). First‐strand cDNA synthesis was carried out using a reverse transcription kit (Invitrogen). Quantitative RT‐PCR was performed with the first‐strand cDNA as the template. Endogenous ubiquitin transcripts were used to normalize the expression levels. Quantitative RT‐PCR was performed on an ABI 7500 Sequence detection system using SYBR Green. Each set of experiments was repeated three times. The methods for miR167a reverse transcript detection were reported by Varkonyi‐Gasic et al. (2007). For the miRNA gel blot analysis, 40 μg total RNA was resolved on a urea‐containing 15% polyacrylamide gel (PAGE), transferred to a Hybond‐N + membrane (Amersham, Arlington Heights, IL) and hybridized with an antisense oligonucleotide probe with a 32Pi label as the miR167 probe. All primers used for expression analysis are listed in Table S5.
RLM‐5′RACE
RNA ligase‐mediated 5′RACE was conducted with the GeneRacer Kit according to the manufacturer's instructions (Invitrogen). Total RNA for RACE was obtained from the young tissues in rice, gene‐specific primers were designed according to each target ARF genes and used for each RACE PCR. The gene‐specific primers of OsARF6, ARF12, OsARF17 and OsARF25 for the first and second PCR products are OsARFs‐RACE and OsARFs‐RACE‐Nested (Table S6), respectively. The second PCR products were gel purified and subcloned into the pGEM‐T Easy Vector (Promega Corporation MD, USA) for sequencing.
Collection of mutants and DR5::GUS/GFP transgenetic plants
The osarf12, osarf17, hsfa2d and lazy1 mutants were identified in our T‐DNA insertion mutant library (http://rmd.ncpgr.cn/; Wu et al., 2003). The osarf25 mutant was previously identified (Qi et al., 2012). Homozygous plants for T‐DNA insertions were identified by PCR‐based genotyping. The primer sequences used for genotyping are listed in Table S7. The DR5::GUS and DR5::GFP transgenic lines were kindly provided by He Yubing from Huazhong Agricultural University.
Phytohormone treatments
For exogenous hormone treatment, 7‐day‐old ZH11 seedlings were treated in liquid 1/2 MS medium containing different kinds of phytohormones for 2 h, including ABA (abscisic acid, 10 μM), JA (jasmonic acid, 10 μM), GA3 (gibberellin, 10 μM), BR (brassinolide, 10 μM), IAA (auxin, 10 μM), 6‐BA (6‐benzylaminopurine, 10 μM), NAA (1‐naphthylacetic acid, 10 μM) and Trp (tryptophan, 10 μM), and DMSO as the control. A 1.5‐cm‐long piece of the rice seedling shoot base was harvested for RNA extraction after treatment. For IAA and 1‐naphthylphthalamic acid (NPA) treatment, 7‐day‐old ZH11 seedlings were transferred to hydroponic solution containing 1 μM IAA or 0.2 μM of NPA for 40 days.
IAA measurement and auxin transport assay
The young axillary buds and roots with about 5‐cm length were collected for IAA content analysis. IAA extraction and measurement were performed according to previous reports (Liu et al., 2012b). The polar auxin transport assays were performed using 7‐day‐old dark‐grown coleoptiles, as described previously with minor modifications (Li et al., 2007). The radioactivity was counted by a liquid scintillation counter (1450 MicroBetaTriLux, PerkinElmer, Boston, MA, USA).
Shoot gravitropism assay
The gravity response was measured according to the method described previously (Zhang et al., 2018). Three‐day‐old rice seedlings were grown in 1/2 MS medium after being dehusked and surface sterilized. The seeds were then grown at 28°C for 2 days (16 h light and 8 h dark) and were subsequently reoriented by 90° from the vertical for 1 day (gravistimulation treatment). The seedling shoots were analysed, and the shoot curvature was recorded.
Accession numbers
All related sequence and accession numbers are listed in Table S8.
Conflict of interest
The authors declare no competing financial interests.
Author contributions
C.W conceived and supervised this research. Y.L. and J.L. performed most of the experiments. Z.C. and Y.W. generated some transgenetic plants. Y.Q. characterized some mutant lines. C.W. and Y.L. wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Figure S1 Inactivation of miR167a activity by target mimicry.
Figure S2 OsARF12, OsARF17 and OsARF25 response to auxin treatment.
Figure S3 Map‐based cloning of LAZY1.
Figure S4 miR167 and OsARF12/17/25 regulate WOX6 and WOX11 asymmetric expression upon gravistimulation under light.
Figure S5 The expression levels of OsARF12/17/25 at the lower and upper sides of the shoot base upon gravistimulation under light.
Table S1 Bioinformatics analysis of MIR167 gene family in Oryza sativa and Arabidopsis thaliana based on miRU prediction.
Table S2 Putative target genes of miR167 in rice based on CSRDB (Cereal small RNAs Database) (http://sundarlab.ucdavis.edu/smrnas/) prediction.
Table S3 Primers of PCR‐based molecular markers for LAZY1.
Table S4 Primer sequences for plasmid construction.
Table S5 Primer sequences used for miR167 and gene expressions analysis.
Table S6 Primer sequences for RLM‐RACE.
Table S7 Primer sequences used for genotyping of T‐DNA mutants.
Table S8 Accession numbers of all related genes in this study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31630054, 31425018, 31821005), and Program for Chinese Outstanding Talents in Agricultural Scientific Research.
Li, Y. , Li, J. , Chen, Z. , Wei, Y. , Qi, Y. and Wu, C. (2020) OsmiR167a‐targeted auxin response factors modulate tiller angle via fine‐tuning auxin distribution in rice. Plant Biotechnol. J., 10.1111/pbi.13360
References
- Attia, K.A. , Abdelkhalik, A.F. , Ammar, M.H. , Wei, C. , Yang, J. , Lightfoot, D.A. , El‐Sayed, W.M. et al.(2009) Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr. Issues Mol. Biol. 11, 29–34. [PubMed] [Google Scholar]
- Bartel, D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. (2003) Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Chandler, J.W. (2016) Auxin response factors. Plant Cell. Environ. 39, 1014–1028. [DOI] [PubMed] [Google Scholar]
- Chen, S.H. , Zhou, L.J. , Xu, P. and Xue, H.W. (2018) SPOC domain‐containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet. 14, e1007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.J. , Wang, L. , Sun, W. , Zhang, Y. , Zhou, C. , Su, Y.H. , Li, W. et al. (2013) Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Zhao, H. , Xie, W. , Han, Z. , Li, G. , Yao, W. , Bai, X. et al. (2016) A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 12, e1006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. , Montgomery, T.A. , Howell, M.D. , Allen, E. , Dvorak, S.K. , Alexander, A.L. and Carrington, J.C. (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta‐siRNA affects developmental timing and patterning in Arabidopsis . Curr. Biol. 16, 939–944. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J.M. , Valli, A. , Todesco, M. , Mateos, I. , Puga, M.I. , Rubio‐Somoza, I. , Leyva, A. et al. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Glazinska, P. , Wojciechowski, W. , Wilmowicz, E. , Zienkiewicz, A. , Frankowski, K. and Kopcewicz, J. (2014) The involvement of InMIR167 in the regulation of expression of its target gene InARF8, and their participation in the vegetative and generative development of Ipomoea nil plants. J. Plant Physiol. 171, 225–234. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J. and Hagen, G. (2007) Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. [DOI] [PubMed] [Google Scholar]
- Gutierrez, L. , Bussell, J.D. , Pacurar, D.I. , Schwambach, J. , Pacurar, M. and Bellini, C. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J. , Shao, G. , Wei, X. , Huang, F. , Sheng, Z. , Tang, S. and Hu, P. (2017) Fine mapping and candidate gene analysis of qTAC8, a major quantitative trait locus controlling tiller angle in rice (Oryza sativa L.). PLoS ONE 12, e0178177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J. , Huang, W. , Gao, J.P. , Yang, J. , Shi, M. , Zhu, M.Z. , Luo, D. et al.(2008) Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369. [DOI] [PubMed] [Google Scholar]
- Khush, G. (2003) Productivity improvements in rice. Nutr. Rev. 61, S114–S116. [DOI] [PubMed] [Google Scholar]
- Kozomara, A. and Griffiths‐Jones, S. (2011) miRBase: integrating microRNA annotation and deep‐sequencing data. Nucleic Acids Res. 39, D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara, A. and Griffiths‐Jones, S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Wang, Y. , Qian, Q. , Fu, Z. , Wang, M. , Zeng, D. , Li, B. et al. (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. [DOI] [PubMed] [Google Scholar]
- Li, Y.F. , Zheng, Y. , Addo‐Quaye, C. , Zhang, L. , Saini, A. , Jagadeeswaran, G. , Axtell, M.J. et al. (2010) Transcriptome‐wide identification of microRNA targets in rice. Plant J. 62, 742–759. [DOI] [PubMed] [Google Scholar]
- Li, G. , Liang, W. , Zhang, X. , Ren, H. , Hu, J. , Bennett, M.J. and Zhang, D. (2014) Rice actin‐binding protein RMD is a key link in the auxin‐actin regulatory loop that controls cell growth. Proc. Natl Acad. Sci. USA 111, 10377–10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.B. , Xie, Z.Z. , Hu, C.G. and Zhang, J.Z. (2016) A review of auxin response factors (ARFs) in Plants. Front. Plant Sci. 7, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Liang, Y. , Yuan, Y. , Wang, L. , Meng, X. , Xiong, G. , Zhou, J. et al. (2019) OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization. Mol. Plant 12, 1143–1156. [DOI] [PubMed] [Google Scholar]
- Liu, P.P. , Montgomery, T.A. , Fahlgren, N. , Kasschau, K.D. , Nonogaki, H. and Carrington, J.C. (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post‐germination stages. Plant J. 52, 133–146. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Jia, S. , Shen, D. , Liu, J. , Li, J. , Zhao, H. , Han, S. et al.(2012a) Four AUXIN RESPONSE FACTOR genes downregulated by microRNA167 are associated with growth and development in Oryza sativa . Funct. Plant Biol. 39, 736. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012b) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice‐bacterium interaction. Plant Meth. 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhang, H. , Zhao, Y. , Feng, Z. , Li, Q. , Yang, H.Q. , Luan, S. et al. (2013) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF‐mediated ABI3 activation in Arabidopsis . Proc. Natl Acad. Sci. USA 110, 15485–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. , Wu, S. , Van Houten, J. , Wang, Y. , Ding, B. , Fei, Z. , Clarke, T.H. et al. (2014) Down‐regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 65, 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Guo, Z. and Li, L. (2013) Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 380, 133–144. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C. , Bartel, D.P. and Bartel, B. (2005) MicroRNA‐directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17, 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, G. , Mu, X. , Grabowski, P. , Schmutz, J. and Lu, C. (2019) Enhancing microRNA167A expression in seed decreases the alpha‐linolenic acid content and increases seed size in Camelina sativa . Plant J. 98, 346–358. [DOI] [PubMed] [Google Scholar]
- Pashkovskiy, P.P. , Kartashov, A.V. , Zlobin, I.E. , Pogosyan, S.I. and Kuznetsov, V.V. (2016) Blue light alters miR167 expression and microRNA‐targeted auxin response factor genes in Arabidopsis thaliana plants. Plant Physiol. Biochem. 104, 146–154. [DOI] [PubMed] [Google Scholar]
- Qi, Y. , Wang, S. , Shen, C. , Zhang, S. , Chen, Y. , Xu, Y. , Liu, Y. et al. (2012) OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 193, 109–120. [DOI] [PubMed] [Google Scholar]
- Qian, Q. , He, P. , Teng, S. , Zeng, D.L. and Zhu, L.H. (2001) QTLs analysis of tiller angle in rice (Oryza sativa L.). Acta Genet Sinica 28, 29–32. [PubMed] [Google Scholar]
- Ru, P. , Xu, L. , Ma, H. and Huang, H. (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 16, 457–465. [DOI] [PubMed] [Google Scholar]
- Sang, D. , Chen, D. , Liu, G. , Liang, Y. , Huang, L. , Meng, X. , Chu, J. et al. (2014) Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl Acad. Sci. USA 111, 11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. , Wang, S. , Bai, Y. , Wu, Y. , Zhang, S. , Chen, M. , Guilfoyle, T.J. et al. (2010) Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J. Exp. Bot. 61, 3971–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. , Yue, R. , Yang, Y. , Zhang, L. , Sun, T. , Tie, S. and Wang, H. .(2014) OsARF16 is involved in cytokinin‐mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.). PLoS ONE 9, e112906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L. , Li, X. , Liu, F. , Sun, X. , Li, C. , Zhu, Z. , Fu, Y. et al. (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. [DOI] [PubMed] [Google Scholar]
- Todesco, M. , Rubio‐Somoza, I. , Paz‐Ares, J. and Weigel, D. (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana . PLoS Genet. 6, e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi‐Gasic, E. , Wu, R. , Wood, M. , Walton, E.F. and Hellens, R.P. (2007) Protocol: a highly sensitive RT‐PCR method for detection and quantification of microRNAs. Plant Meth. 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller, F. , Furuya, M. and Nick, P. (2002) OsARF1, an auxin response factor from rice, is auxin‐regulated and classifies as a primary auxin responsive gene. Plant Mol. Biol. 50, 415–425. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Li, J. (2008) Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. [DOI] [PubMed] [Google Scholar]
- Wang, J.W. , Wang, L.J. , Mao, Y.B. , Cai, W.J. , Xue, H.W. and Chen, X.Y. (2005) Control of root cap formation by MicroRNA‐targeted auxin response factors in Arabidopsis . Plant Cell 17, 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Pei, K. , Fu, Y. , Sun, Z. , Li, S. , Liu, H. , Tang, K. et al. (2007) Genome‐wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene, 394, 13–24. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Xu, Y. , Zhang, C. , Ma, Q. , Joo, S.H. , Kim, S.K. , Xu, Z. et al. (2008) OsLIC, a novel CCCH‐type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3, e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Zhang, S. , Sun, C. , Xu, Y. , Chen, Y. , Yu, C. , Qian, Q. et al. (2014) Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 201, 91–103. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Li, K. , Chen, L. , Zou, Y. , Liu, H. , Tian, Y. , Li, D. et al. (2015) MicroRNA167‐directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 168, 984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L. , Carles, C.C. , Osmont, K.S. and Fletcher, J.C. (2005) A database analysis method identifies an endogenous trans‐acting short‐interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl Acad. Sci. USA 102, 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A.W. and Bartel, B. (2005) Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Li, X. , Yuan, W. , Chen, G. , Kilian, A. , Li, J. , Xu, C. et al. (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35, 418–427. [DOI] [PubMed] [Google Scholar]
- Wu, M.F. , Tian, Q. and Reed, J.W. (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development, 133, 4211–4218. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Tang, D. , Li, M. , Wang, K. and Cheng, Z. (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 161, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.H. , Han, S.J. , Yoon, E.K. and Lee, W.S. (2006) Evidence of an auxin signal pathway, microRNA167‐ARF8‐GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 34, 1892–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X. , Chen, J. , Zhou, J. , Yu, H. , Ge, C. , Zhang, M. , Gao, X. et al. (2019) An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 180, 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara, T. and Iino, M. (2007) Identification of the gravitropism‐related rice gene LAZY1 and elucidation of LAZY1‐dependent and ‐independent gravity signaling pathways. Plant Cell Physiol. 48, 678–688. [DOI] [PubMed] [Google Scholar]
- Yu, B. , Lin, Z. , Li, H. , Li, X. , Li, J. , Wang, Y. , Zhang, X. et al. (2007) TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 33, W701–W704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Wang, S. , Xu, Y. , Yu, C. , Shen, C. , Qian, Q. , Geisler, M. et al. (2015) The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3‐5 and OsBRI1 . Plant Cell Environ. 38, 638–654. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Yu, H. , Yu, H. , Cai, Y. , Huang, L. , Xu, C. , Xiong, G. et al. (2018) A core regulatory pathway controlling rice tiller angle mediated by the LAZY1‐dependent asymmetric distribution of auxin. Plant Cell 30, 1461–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Tan, L. , Sun, H. , Zhao, X. , Liu, F. , Cai, H. , Fu, Y. et al. (2019) Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol. Plant 12, 1075–1089. [DOI] [PubMed] [Google Scholar]
- Zhao, L. , Tan, L. , Zhu, Z. , Xiao, L. , Xie, D. and Sun, C. (2015) PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 83, 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L. , Nagpal, P. , Villarino, G. , Trinidad, B. , Bird, L. , Huang, Y. and Reed, J.W. (2019) miR167 limits anther growth to potentiate anther dehiscence. Development 146, dev174375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Inactivation of miR167a activity by target mimicry.
Figure S2 OsARF12, OsARF17 and OsARF25 response to auxin treatment.
Figure S3 Map‐based cloning of LAZY1.
Figure S4 miR167 and OsARF12/17/25 regulate WOX6 and WOX11 asymmetric expression upon gravistimulation under light.
Figure S5 The expression levels of OsARF12/17/25 at the lower and upper sides of the shoot base upon gravistimulation under light.
Table S1 Bioinformatics analysis of MIR167 gene family in Oryza sativa and Arabidopsis thaliana based on miRU prediction.
Table S2 Putative target genes of miR167 in rice based on CSRDB (Cereal small RNAs Database) (http://sundarlab.ucdavis.edu/smrnas/) prediction.
Table S3 Primers of PCR‐based molecular markers for LAZY1.
Table S4 Primer sequences for plasmid construction.
Table S5 Primer sequences used for miR167 and gene expressions analysis.
Table S6 Primer sequences for RLM‐RACE.
Table S7 Primer sequences used for genotyping of T‐DNA mutants.
Table S8 Accession numbers of all related genes in this study.


