Abstract
Newborn screening (NBS) programmes utilise information on a variety of clinical variables such as gestational age, sex, and birth weight to reduce false‐positive screens for inborn metabolic disorders. Here we study the influence of ethnicity on metabolic marker levels in a diverse newborn population. NBS data from screen‐negative singleton babies (n = 100 000) were analysed, which included blood metabolic markers measured by tandem mass spectrometry and ethnicity status reported by the parents. Metabolic marker levels were compared between major ethnic groups (Asian, Black, Hispanic, White) using effect size analysis, which controlled for group size differences and influence from clinical variables. Marker level differences found between ethnic groups were correlated to NBS data from 2532 false‐positive cases for four metabolic diseases: glutaric acidemia type 1 (GA‐1), methylmalonic acidemia (MMA), ornithine transcarbamylase deficiency (OTCD), and very long‐chain acyl‐CoA dehydrogenase deficiency (VLCADD). In the result, 79% of the metabolic markers (34 of 43) had ethnicity‐related differences. Compared to the other groups, Black infants had elevated GA‐1 markers (C5DC, Cohen's d = .37, P < .001), Hispanics had elevated MMA markers (C3, Cohen's d = .13, P < .001, and C3/C2, Cohen's d = .27, P < .001); and Whites had elevated VLCADD markers (C14, Cohen's d = .28, P < .001, and C14:1, Cohen's d = .22, P < .001) and decreased OTCD markers (citrulline, Cohen's d = −.26, P < .001). These findings correlated with the higher false‐positive rates in Black infants for GA‐1, in Hispanics for MMA, and in Whites for OTCD and for VLCADD. Web‐based tools are available to analyse ethnicity‐related changes in newborn metabolism and to support developing methods to identify false‐positives in metabolic screening.
Keywords: inborn metabolic disorders, informatics and statistics, newborn screening, paediatric clinical chemistry, racial/ethnic‐disparities
1. INTRODUCTION
Newborn screening (NBS) is a public health programme that identifies infants with heritable disorders before the onset of clinical signs, allowing for early and life‐saving intervention. Screening for metabolic disorders using tandem mass spectrometry (MS/MS) identifies most affected newborns, along with a large number of false‐positive infants. 1 , 2 In some states, initial screen‐positive results are followed by second‐tier MS/MS‐based testing at higher specificity. 3 Diagnostic biochemical and/or DNA testing is performed to confirm (true positive) or reject (false positive) the screening result, to establish the final diagnosis and guide patient treatment. 4 , 5 , 6 , 7 False‐positive screens for metabolic disorders can also be identified using post‐analytical interpretive tools in Clinical Laboratory Integrated Reports (CLIR, formerly R4S). 8 , 9 , 10 Using these algorithms, NBS data can be adjusted based on a number of clinical variables associated with false‐positive screens such as gestational age (GA), sex, birth weight (BW), age at blood collection, season of birth, and nutritional therapy. 11 , 12 , 13 , 14 , 15
We recently reported elevated screening marker levels for methylmalonic acidemia (MMA) in healthy Hispanic infants, which correlated with a higher MMA false positive rate for this group. 16 Ethnicity‐related differences in NBS marker levels have also been reported for cystic fibrosis (CF), congenital hypothyroidism (CH), and for total galactose (TGAL). Isolated TGAL elevations have been found in certain ethnic groups, like the Hmong, which resulted in a disproportionate number of false‐positive cases in this population and implementation of Hmong‐specific TGAL cutoffs in the Minnesota NBS programme (A. Gaviglio, personal communication). For CF, higher levels of immunoreactive trypsinogen (IRT) were found in African‐American infants in the United States, 17 in infants of North African descent in the Rhone‐Alpes region of France, 18 and in Roma infants in Slovakia. 19 For CH, elevated mean concentrations of thyroid‐stimulating hormone (TSH) were found for Pakistani, Bangladeshi, and Chinese infants, suggesting that ethnic diversity in populations should be considered when establishing screening TSH cutoffs. 20
In this study, we investigated whether ethnicity status could be associated with differences in the blood levels of NBS markers for inborn metabolic disorders on the Recommended Universal Screening Panel (RUSP). 21 To identify the effects of ethnicity on metabolic marker levels, we analysed a large and ethnically diverse population of screen‐negative infants reported by the California NBS programme. Importantly, GA and BW are highly correlated with race/ethnicity, 22 , 23 and they are known to influence metabolic marker levels. 13 , 24 To account for this confounding, we first studied the effects of these clinical variables on marker levels, and then controlled for them in the analysis of marker levels between ethnicity groups. Additionally, we explored the effect of ethnicity on false‐positive newborn screens. The identified ethnicity‐related differences in marker levels were correlated to false‐positive cases for four inborn errors of metabolism. Based on these findings, web‐based software was established to aid the interpretation of screening data (https://RUSPtools.shinyapps.io/MetaDB), and to support development of algorithms that incorporate information on ethnicity and clinical variables in disease screening.
2. MATERIAL AND METHODS
2.1. Data summary
NBS data from 100 000 screen‐negative singleton babies born between 2013 and 2015 were analysed. The cohort was selected at random by the California NBS programme from over half a million newborns screened each year. Babies that are screen‐negative in first‐tier NBS are reported as negative with no additional testing. The data included 41 metabolic analytes measured by MS/MS, 1 two analyte ratios (C3/C2, C0/(C16+C18)), and clinical variables of BW, GA, sex, race/ethnicity, age at blood collection (AaC), and total parenteral nutrition (TPN) status. In addition, we analysed data from 2767 screen‐positive infants for four inborn metabolic disorders reported by the California NBS programme between 2005 and 2015. This cohort consisted of confirmed true‐positive (TP) cases and of first‐tier false‐positive (FP) cases for glutaric acidemia type 1 (GA‐1, TP = 48, FP = 1344), methylmalonic acidemia (MMA, TP = 103, FP = 502), ornithine transcarbamylase deficiency (OTCD, TP = 24, FP = 496), and very long‐chain acyl‐CoA dehydrogenase deficiency (VLCADD, TP = 60, FP = 200). Of the 2532 false‐positive cases, 10 infants were false‐positive for two disorders. Table 1 and S1 show the cohort characteristics.
TABLE 1.
Participant and sub‐group demographics
| Screen‐negative infants (%) | Screen‐positive infants (%) | |
|---|---|---|
| Variable | (n = 100 000) | (n = 2767) |
| Gestational age (wk) | ||
| >42 a | 25 (0.03) | 40 (1.4) |
| 42 | 472 (0.5) | 50 (1.8) |
| 41 | 7649 (7.6) | 161 (5.8) |
| 39‐40 | 62 440 (62.4) | 890 (32.1) |
| 37‐38 | 23 813 (23.8) | 890 (32.1) |
| 28‐36 | 5553 (5.6) | 648 (23.4) |
| <28 a | 73 (0.07) | 88 (3.2) |
| Birth weight (g) | ||
| >5000 a | 124 (0.1) | 6 (0.2) |
| 4001‐5000 | 8246 (8.2) | 179 (6.5) |
| 3501‐4000 | 28 406 (28.4) | 507 (18.3) |
| 3001‐3500 | 41 497 (41.5) | 795 (28.7) |
| 2500‐3000 | 17 537 (17.5) | 673 (24.3) |
| 1000‐2499 | 4101 (4.1) | 507 (18.3) |
| <1000 a | 89 (0.09) | 100 (3.6) |
| Sex | ||
| Male | 51 625 (51.6) | 1651 (59.7) |
| Female | 48 071 (48.1) | 1105 (39.9) |
| Unknown | 304 (0.3) | 11 (0.4) |
| Race/ethnicity | ||
| Asian | 14 320 (14.3) | 272 (9.8) |
| Black | 6668 (6.7) | 302 (10.9) |
| Hispanic | 49 627 (49.6) | 1164 (42.1) |
| White | 26 481 (26.5) | 941 (34.0) |
| Other/unknown | 2904 (2.9) | 88 (3.2) |
| Age at collection (h) | ||
| <12 a | 48 (0.05) | 13 (0.5) |
| 12‐24 | 21 598 (21.6) | 466 (16.8) |
| 24‐48 | 71 562 (71.6) | 1631 (58.9) |
| 49‐168 | 6625 (6.6) | 615 (22.2) |
| >168 a | 167 (0.2) | 42 (1.5) |
| TPN | ||
| No | 97 646 (97.6) | 2263 (81.8) |
| Yes | 1068 (1.1) | 392 (14.2) |
| Unknown | 1286 (1.3) | 112 (4.0) |
Abbreviation: TPN, total parenteral nutrition.
Screen‐negative (n = 463) and screen‐positive (n = 188) infants recorded outside of the indicated ranges were removed from analysis.
2.2. Analysis of clinical variables
The analysis of clinical variables included 99 537 screen‐negative infants with BW range of 1000 to 5000 g, GA range of 28 to 42 weeks, and AaC from 12 hours to 7 days after birth (Table 1, Figure 1A). Infants recorded outside these ranges were removed from analysis (n = 463). The five GA categories included preterm (<37 weeks), early‐term (37‐38 weeks), full‐term (39‐40 weeks), late‐term (41 weeks), and post‐term (42 weeks) birth. 25 The five BW categories included low BW (<2500 g), high BW (>4000 g), and normal BW, 26 which was divided into the normal BW categories of 2500 to 3000, 3001 to 3500, and 3501 to 4000 g.
FIGURE 1.
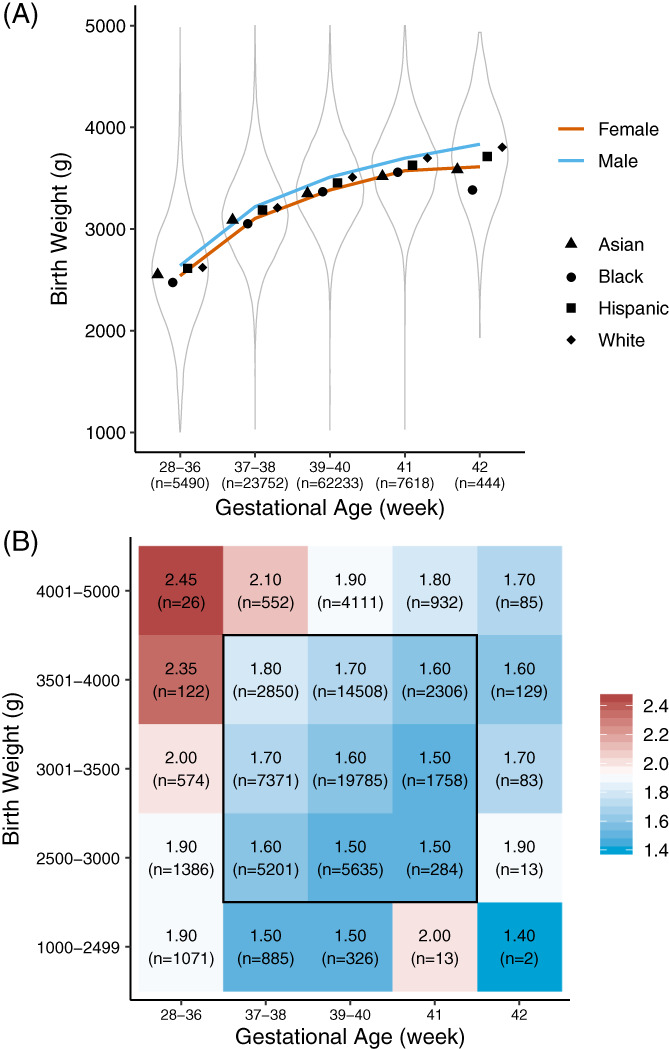
Newborn birth weight (BW), gestational age (GA), and race/ethnicity. A, Distribution of BW for all infants (n = 99 537) and sub‐groups defined by GA. Symbols indicate the mean of BW in each of four race/ethnicity groups. Male‐to‐female BW differences remained significant as BW increased with GA in the growth curve (Table S3). B, The study cohort (n = 70 008) was divided into 25 groups defined by BW and GA. Only infants without total parenteral nutrition and age at blood collection AaC between 24 and 48 hours were included. The colour code indicates median of marker level for each group (C3 in this example) with group size in parenthesis. The central 3 × 3 area indicates the study cohort selected to identify ethnicity‐related marker level differences while controlling for the influence of GA and BW on marker levels. Race/ethnicity information was available for 58 056 infants born at term with a normal BW. Marker plots are available at https://RUSPtools.shinyapps.io/MetaDB
2.3. Analysis of race/ethnicity
The race/ethnicity status of the newborn was self‐reported by the parents. Of the 99 537 screen‐negative infants studied, 79% (n = 78 671) were reported as being of Asian, Black, Hispanic, or White origin. Newborns recorded with more than one race/ethnicity (18%, n = 17 975) were classified according to NBS programme guidelines 27 as follows: (a) Hispanic, if reported Hispanic and any other race/ethnicity; (b) Black, if reported Black and any other race/ethnicity except Hispanic; (c) Asian, if reported Asian and any other race/ethnicity except Hispanic and Black; (d) White, if reported White only. All other ethnicities and unknown race/ethnicity were recorded as Other/Unknown (2.9%, n = 2891). The relative percentage of each race/ethnicity group in our study cohort closely matched those of the larger California NBS population of 5 624 000 newborns tested between 2005 and 2015.
2.4. Analysis of metabolic screening markers
To identify differences in marker levels between the four major race/ethnicity groups, 29 529 infants were removed from the analysis, which included 2268 with positive or unknown TPN status and 28 141 with AaC outside the recommended blood collection time of 24 to 48 hours. 28 Eight hundred and eighty infants overlapped between these categories. The remaining 70 008 infants were divided into 25 groups, each defined by a specific BW and GA range (Figure 1B). For each of the 25 groups, the median for each marker was calculated (https://RUSPtools.shinyapps.io/MetaDB). We first analysed the effect of race/ethnicity on metabolic markers in 19 247 full‐term (39‐40 weeks) infants with normal BW (3001‐3500 g) and AaC between 24 and 48 hours in order to control for the confounding effects of GA, BW, and of AaC on marker levels. To identify sex‐related effects on marker levels in this cohort, an analysis was performed between full‐term females (n = 10 054) and full‐term males (n = 9152) with normal BW (3001‐3500 g) in each race/ethnicity group. Finally, the effect of race/ethnicity on marker levels was studied in a larger cohort of 58 056 term (37‐41 weeks), normal BW infants (2500‐4000 g).
2.5. Statistical analysis
Statistical analyses were performed in R 3.5.3. 29 One‐way analysis of variance and Tukey's honest significance test were used to compare BW differences between race/ethnicity groups (Figure S1). Cohen's d 30 and t test were used to evaluate BW differences between males and females (Table S3). Fisher's exact test 31 and binomial test were used to compare differences between screen‐negative and false‐positive infants (Table 2). Difference in difference (DID) was calculated using the formula: (BWmale − BWfemale)White − (BWmale − BWfemale)Other. The analysis of metabolic differences between race/ethnicity groups was based on 41 markers and 2 ratios, which required multiple test correction. We performed effect size analysis using Cohen's d 30 to compare marker levels between groups, which were not affected by differences in sample size (Figure 2, Figures S4 and S5). Given the large sample size, the smallest group analysed was the group of Black female infants (n = 616). In this study Cohen's d of .2, which is considered a small effect size, was selected as the cutoff (medium effect 0.5, large effect 0.8). 30 If Cohen's d is larger than .2, the P value is less than .001 for a two‐sample t test with 600 samples per group, which is significant after a Bonferroni correction (0.001 × 43 markers = 0.043). Some of the metabolic marker values follow a log‐normal distribution instead of a Gaussian distribution. However, some marker values are very close to 0, which would generate large negative values after log‐transformation. Here we choose to use raw data marker values. Cohen's d effect size values might be an underestimate for some markers, particularly those that follow log‐normal distribution and show a large Cohen's d value.
TABLE 2.
Correlation of marker levels between screen‐negatives and false‐positives
| Disease | NBS marker | Race/ethnicity a | Gestational age b | Birth weight c | Sex d | ||||
|---|---|---|---|---|---|---|---|---|---|
| (FP) | SN | FP | SN | FP | SN | FP | SN | FP | |
| n = 58 056 | No. (%) | n = 70 008 | No. (%) | n = 44 365 | (g) | n = 44 245 | No. (%) | ||
|
GA‐1 n = 1344 |
↑C5DC | ↑Black |
B: 100 (19.9) (P = .005) |
— |
PT: 139 (17.9) (P < .001) |
— |
FP: 3363 (n = 299) SN: 3443 (P = .005) |
— |
M: 179 (59.9) (P = .002) |
|
MMA n = 502 |
↑C3 | ↑Hispanic |
H: 69 (71.9) (P < .001) |
↑PT |
PT: 29 (19.3) (P < .001) |
↑BW |
FP: 3539 (n = 54) SN: 3443 (P = .14) |
— |
M: 23 (42.6) (P = .22) |
| ↑C3/C2 | ↑Hispanic | ↑PT | ↑BW | — | |||||
|
OTCD n = 496 |
↓CIT | ↓White |
W: 46 (37.1) (P = .005) |
↓PT |
PT: 58 (27.2) (P < .001) |
— |
FP: 3398 (n = 59) SN: 3443 (P = .49) |
— |
M: 39 (66.1) (P = .026) |
|
VLCADD n = 200 |
↑C14 | ↑White |
W: 40 (43.0) (P < .001) |
↑PT |
PT: 36 (19.7) (P < .001) |
↓BW |
FP: 3209 (n = 61) SN: 3443 (P < .001) |
↑Male |
M: 37 (60.7) (P = .16) |
| ↑C14:1 | ↑White | ↑PT | — | — | |||||
Abbreviations: B, Black; BW, birth weight; FP, false‐positive infants; GA, gestational age; H, Hispanic; M, male; PT, preterm birth; SN, screen‐negative controls; TPN, total parenteral nutrition; W, white. P values less than .05 are shaded grey. Only data without TPN and AaC of 24 to 48 hours were analysed.
Percentage of race/ethnicity groups in SN controls used in binomial testing was based on Table 1. GA was controlled from 37 to 41 weeks. BW was controlled from 2500 to 4000 g.
Elevated marker levels were associated with preterm birth (↑PT). Preterm birth rate of 8.3% in the general California population in 2014 42 was used in binomial testing.
Elevated marker levels were associated with high (↑BW) and with low (↓BW) birth weight. BW between FP and SN infants was compared using a t test. GA was controlled to 39 to 40 weeks.
SN male percentage of 50.9% was used in binomial testing. GA was controlled to 39 to 40 weeks.
FIGURE 2.
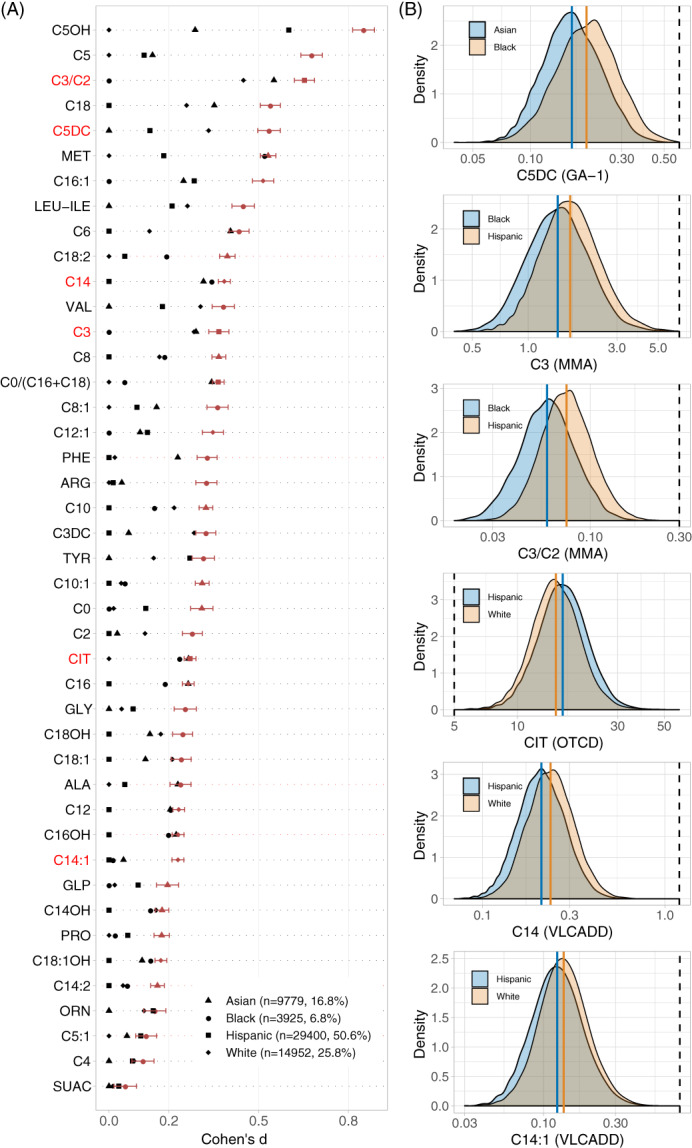
Newborn metabolic differences in race/ethnicity groups. A, To evaluate marker level differences between race/ethnicity groups, the group with the lowest mean marker level was defined as the reference group for each screening marker. Effect size differences for all markers between the four groups was ranked from top to bottom. 79% of the markers showed significant differences between race/ethnicity groups (Cohen's d ≥ .2), including 28 markers for 38 metabolic diseases on the RUSP 21 (Table S5). Six markers (red label) are used to detect four metabolic disorders through elevated (GA‐1, MMA, VLCADD), or decreased (OTCD) marker levels. B, For each of the six markers, the distribution of marker levels (Log scale X‐axis) is shown for the two race/ethnicity groups with the largest Cohen's d difference. The coloured vertical lines correspond to the mean marker levels in the respective race/ethnicity group. The black dashed line indicates the screening cutoff value in the California NBS programme. Black infants had elevated C5DC, Hispanics had elevated C3 and C3/C2, and Whites had elevated C14 and C14:1, and decreased citrulline
3. RESULTS
3.1. Analysis of clinical variables and race/ethnicity
The relation between BW, GA, sex, and race/ethnicity was analysed in our screen‐negative cohort. The mean infant BW was higher in Whites (3429 g) than Hispanics (3350 g), Blacks (3258 g), and Asians (3247 g; Table S3). For term infants (94% of total cohort), BW and GA showed a positive linear relationship (Pearson Cor = .5, Spearman Cor = .41, P < .01) with no significant changes in the ethnic‐related BW differences (Figure 1A). Males had a higher mean BW than females by 111 g (P < .001). This overall effect of sex on BW varied significantly between racial/ethnic groups, ranging from a difference of 60 g (P < .001) between White (n = 6782) and Hispanic (n = 23 898) females, to 200 g (P < .001) between White (n = 13 635) and Asian (n = 7448) males (Figure S1). The BW difference between White males and males in other race/ethnicity groups was larger than that between White females to females in other groups. The findings correlated with male‐to‐female BW difference within race/ethnicity groups, which was largest for Whites (137 g, P < .001). The difference in male‐to‐female BW difference (difference in difference, DID) between Whites and other race/ethnicity groups was 35 g (P < .001). An analysis of GA and race/ethnicity showed that preterm birth was higher in Blacks (n = 427, 6.4%, P < .001) and Hispanics (n = 2863, 5.8%, P < .001), while late‐ and post‐term births were higher in Blacks (n = 590, 8.9%, P = .008) and Whites (n = 3028, 11.5%, P < .001; Figure S2). Asians and Hispanics had a high percentage of males in preterm and early‐term births (Figure S3).
3.2. Analysis of race/ethnicity and metabolic markers
Effect‐size analysis between infants in the four race/ethnicity groups identified significant differences for 79.1% of the markers (34/43 markers, Cohen's d > .2). This analysis was performed in 19 247 full‐term infants in the BW category of 3001 to 3500 g in order to control for GA and BW effects on marker levels (Figure 1B). Infants with positive/unknown TPN status and AaC outside the 24 to 48 hours window were excluded from this analysis. Significant findings included elevated C5, C5OH, and C5DC in Blacks, elevated C3 and C3/C2 in Hispanics, and elevated C14, C14:1, as well as decreased citrulline in Whites (Figure S4). A separate analysis in this cohort between females (n = 9152) and males (n = 10 054) within each race/ethnicity group showed that marker differences between females and males were similar between race/ethnicity groups. An analysis in a larger cohort of 58 056 term infants with a normal BW confirmed the effect of ethnicity on markers levels (Figure 2, Table S4). Finally, similar ethnicity‐related marker level differences were found for 47 800 infants recorded with only a single race/ethnicity category (Figure S5), which are a sub‐group (82%) of the 58 056 infants.
3.3. Correlation of marker differences to NBS false‐positive results
The four metabolic diseases are detected in NBS by elevated (GA‐1, MMA, VLCADD), or decreased (OTCD) marker levels (Table S5). Ethnic differences in NBS false‐positive rates were found for each disease. These findings correlated with the physiologic differences in marker levels in the respective race/ethnicity groups (Table 2). For example, elevated C5DC found in screen‐negative Blacks correlated with the higher percentage of Blacks among GA‐1 false‐positives (19.9% vs 6.7% in Table 1). Elevated C3 and C3/C2 in screen‐negative Hispanics correlated with the high percentage of Hispanics in MMA false‐positives. Decreased citrulline in screen‐negative Whites correlated with the high percentage of Whites in OTCD false‐positives. Elevated C14 and C14:1 in screen‐negative Whites correlated with the high percentage of Whites in VLCADD false‐positives. False‐positives were also associated with preterm birth (GA‐1, MMA, OTCD, VLCADD), BW (GA‐1, VLCADD), and male sex (GA‐1, OTCD).
4. DISCUSSION
NBS programmes consider information on a variety of clinical variables such as GA, sex, and BW that can lead to false‐positive screens. These important clinical variables are known to influence metabolic marker levels, 13 , 16 , 24 while they themselves may be modulated by ethnic differences. 22 , 23 Here we investigate whether ethnicity status could be associated with differences in metabolic marker levels for inborn metabolic disorders on the RUSP. 21 To explore this question, we first studied the relationship between ethnicity and clinical variables, which was analysed in a large and ethnically diverse population of screen‐negative infants (n = 96 646) reported by the California NBS programme. We found the mean BW lower in Asian and Black infants, highest in Whites, and second‐highest in Hispanics, which was consistent across different GAs at delivery (Figure 1A). Compared with same‐sex infants in other race/ethnicity groups, BW differences for White males were larger than for White females (Figure S1). These results correlated with the larger male‐to‐female BW difference in White infants (137 g, P < .001) compared to other race/ethnicity groups (100‐104 g, P < .001). Notably, the racial/ethnic differences in BW correlated with differences in GA (Figure S2). Black and Hispanic infants were more likely to be born premature, White and Black infants had significantly more late‐term and post‐term births, and Asian and Hispanic infants had the highest male percentage among preterm and early‐term births. These findings confirmed the complex relationship between clinical variables (BW, GA, sex), and race/ethnicity. 32 , 33 , 34 , 35 , 36
In order to account for this confounding, we followed a stringent study design by controlling for the influence of important clinical variables in the analysis of marker levels between ethnicity groups (Figure 1B). In a cohort of 58 056 infants born at term with normal BW and age of blood collection between 24 and 48 hours after birth, ethnicity‐related differences were found for 79.1% of the NBS metabolic analytes (34 of 43, Cohen's d > .2) (Figure 2). These analytes included 28 primary markers for 38 metabolic disorders on the RUSP. 21 Highly similar and robust metabolic patterns in relation to race/ethnicity were identified in a smaller cohort of full‐term infants with a BW of 3001 to 3500 g (n = 19 247, Figure S4). A separate analysis between males and females in each race/ethnicity group confirmed that sex had no significant effect on the identified marker level differences.
We reasoned that these marker differences could lead to false‐positive newborn screens. To test this hypothesis, we selected four diseases with frequent false‐positive results that included GA‐1 with a ratio of 29 infants without the condition to 1 infant with the condition (PPV = 0.03). Analysis of false‐positive cases for these diseases revealed racial/ethnic disparities, which correlated with the differences in marker levels discovered in the respective race/ethnicity groups. For example, Black infants were more likely false‐positive in GA‐1 screening, which correlated with elevated C5DC in screen‐negative Black infants (Table 2). In addition to race/ethnicity‐associated marker differences identified for all four diseases, NBS false‐positives were also associated with pre term birth (GA‐1, MMA, OTCD, VLCADD), BW (GA‐1, VLCADD), and sex (GA‐1, OTCD). These findings show that newborn physiological metabolism is variably confounded by GA, sex, BW, and by ethnic differences.
A limiting factor in our study was confounding, which could distort the association between clinical variables and race/ethnicity. Studying a large newborn population (n = 100 000) allowed us to select sub‐populations to reduce the influence of confounding factors. To stratify BW and GA by sex and race/ethnicity, analysis was performed in normal weight, full‐term infants. Such stratification, however, was limited in false‐positive cases due to the much smaller sample size. We noted that GA‐1 false‐positives had lower GA and BW compared to screen‐negatives, which could be related to the larger number of Black infants among false‐positives (Table 2). For OTCD, false‐positives had a higher percentage of males compared to screen‐negatives, which could be related to higher BW in males and in particular White males. For VLCADD, while full‐term screen‐negative males had elevated C14, the male percentage in full‐term false‐positive males was not significantly increased. Notably, expanding the analysis to the larger cohort of false‐positive males born at term (37‐41 weeks) revealed an association with elevated C14 (n = 96, 62.7%, P = .006), which indicated that statistical significance for this association was lost due to sample size.
Here we uncovered an association between MS/MS disease markers and race/ethnicity, and show that these differences could lead to false‐positive newborn screens for metabolic disorders. However, we do not suggest to select different NBS marker cutoffs solely based on race/ethnicity information. While such ethnicity‐adjusted marker levels could reduce false‐positives, it could also lower screening sensitivity and increase false‐negatives results, which is the primary concern of NBS. Nevertheless, results from this study could inform the second‐tier interpretive analysis of screen‐positive cases. For example, CLIR tools simultaneously correct analyte levels and ratios for covariates such as preterm birth, sex, and BW in order to reduce false‐positives. 10 Accordingly, race/ethnicity information could be used in a regression model together with other covariates (eg, BW, GA, AaC, sex) to adjust metabolic marker levels, or by building a data mining model that incorporates all covariates to predict metabolic disease status. Each method requires a large amount of self‐reported race/ethnicity data that may not be recorded by every NBS programme. 27 Overcoming these challenges could improve our ability to screen and diagnose metabolic diseases in diverse and admixed populations.
The cause of the identified marker level differences in race/ethnicity groups is unknown. It is possible that systematic disparities based on socioeconomic status and ethnicity could affect BW, GA, 32 , 33 , 34 , 36 and maternal nutrition and access to prenatal vitamins could affect B12 levels in MMA screen‐positive infants. 37 , 38 It is also possible that genetic differences associated with ethnicity or ancestry could contribute to these variable infant metabolic phenotypes.39, 40, 41
5. CONCLUSION
This study provides evidence for association between race/ethnicity status and the levels of NBS markers for GA‐1, MMA, OTC, and VLCADD, which could lead to false‐positive screening results for these disorders. While maintaining high sensitivity is the primary goal of NBS, ethnic diversity in populations should be considered together with the clinical variables of GA, sex, and BW in the second‐tier analysis of screening data for inborn metabolic disorders.
CONFLICT OF INTEREST
Gang Peng, Yishuo Tang, Neeru Gandotra, Gregory M. Enns, Tina M. Cowan, Hongyu Zhao, and Curt Scharfe declare that they have no conflict of interest.
ETHICS STATEMENT
This study was overseen by the institutional review boards at Yale University, Stanford University, and the State of California Committee for the Protection of Human Subjects.
Supporting information
Figure S1. Birth weight differences between race/ethnicity groups.
Figure S2. Newborn race/ethnicity groups in different gestational age categories.
Figure S3. Percentage of male infants in different gestational age categories.
Figure S4. Metabolic differences between race/ethnicity groups.
Figure S5. Metabolic differences between infants with a single race/ethnicity category.
Table S1. Demographics of false‐positive newborns in four metabolic diseases.
Table S2. Sample size of screen‐negatives in GA, sex, and race/ethnicity groupings.
Table S3. Difference of birth weight between male and female infants.
Table S4. Comparison of NBS disease markers between race/ethnicity groups.
Table S5. Newborn screening markers for inborn metabolic disorders.
ACKNOWLEDGMENTS
We thank Robin Cooley, Steve Graham, Hao Tang, Stanley Sciortino, Lisa Feuchtbaum, and Robert Currier at the Genetic Disease Screening Program (GDSP) for advising on data analysis. The data used in this study were obtained from the California Biobank Program (SIS request 886). The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication.
Peng G, Tang Y, Gandotra N, et al. Ethnic variability in newborn metabolic screening markers associated with false‐positive outcomes. J Inherit Metab Dis. 2020;43:934–943. 10.1002/jimd.12236
Funding information National Institute of Child Health and Human Development, Grant/Award Number: R01HD081355; Yale University
REFERENCES
- 1. McHugh D, Cameron CA, Abdenur JE, et al. Clinical validation of cutoff target ranges in newborn screening of metabolic disorders by tandem mass spectrometry: a worldwide collaborative project. Genet Med. 2011;13(3):230‐254. [DOI] [PubMed] [Google Scholar]
- 2. Waisbren SE, Albers S, Amato S, et al. Effect of expanded newborn screening for biochemical genetic disorders on child outcomes and parental stress. JAMA. 2003;290(19):2564‐2572. [DOI] [PubMed] [Google Scholar]
- 3. Matern D, Tortorelli S, Oglesbee D, Gavrilov D, Rinaldo P. Reduction of the false‐positive rate in newborn screening by implementation of MS/MS‐based second‐tier tests: the Mayo Clinic experience (2004‐2007). J Inherit Metab Dis. 2007;30(4):585‐592. [DOI] [PubMed] [Google Scholar]
- 4. Strauss KA, Puffenberger EG, Robinson DL, Morton DH. Type I glutaric aciduria, part 1: natural history of 77 patients. Am J Med Genet C Semin Med Genet. 2003;121C(1):38‐52. [DOI] [PubMed] [Google Scholar]
- 5. Manoli I, Sloan JL, Venditti CP. Isolated methylmalonic acidemia. In: Adam MP, Ardinger HH, Pagon RA, eds. GeneReviews. Seattle, WA: University of Washington, Seattle; 1993. [Google Scholar]
- 6. Pena LD, van Calcar SC, Hansen J, et al. Outcomes and genotype‐phenotype correlations in 52 individuals with VLCAD deficiency diagnosed by NBS and enrolled in the IBEM‐IS database. Mol Genet Metab. 2016;118(4):272‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Posset R, Garbade SF, Boy N, et al. Transatlantic combined and comparative data analysis of 1095 patients with urea cycle disorders—a successful strategy for clinical research of rare diseases. J Inherit Metab Dis. 2019;42(1):93‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marquardt G, Currier R, McHugh DM, et al. Enhanced interpretation of newborn screening results without analyte cutoff values. Genet Med. 2012;14(7):648‐655. [DOI] [PubMed] [Google Scholar]
- 9. Tortorelli S, Eckerman JS, Orsini JJ, et al. Moonlighting newborn screening markers: the incidental discovery of a second‐tier test for Pompe disease. Genet Med. 2018;20(8):840‐846. [DOI] [PubMed] [Google Scholar]
- 10. Minter Baerg MM, Stoway SD, Hart J, et al. Precision newborn screening for lysosomal disorders. Genet Med. 2018;20(8):847‐854. [DOI] [PubMed] [Google Scholar]
- 11. Blanco CL, Gong AK, Green BK, Falck A, Schoolfield J, Liechty EA. Early changes in plasma amino acid concentrations during aggressive nutritional therapy in extremely low birth weight infants. J Pediatr. 2011;158(4):543‐548. e541. [DOI] [PubMed] [Google Scholar]
- 12. Sarafoglou K, Banks K, Gaviglio A, Hietala A, McCann M, Thomas W. Comparison of one‐tier and two‐tier newborn screening metrics for congenital adrenal hyperplasia. Pediatrics. 2012;130(5):e1261‐e1268. [DOI] [PubMed] [Google Scholar]
- 13. Ryckman KK, Berberich SL, Shchelochkov OA, Cook DE, Murray JC. Clinical and environmental influences on metabolic biomarkers collected for newborn screening. Clin Biochem. 2013;46(1–2):133‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hall PL, Marquardt G, McHugh DM, et al. Postanalytical tools improve performance of newborn screening by tandem mass spectrometry. Genet Med. 2014;16(12):889‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark RH, Kelleher AS, Chace DH, Spitzer AR. Gestational age and age at sampling influence metabolic profiles in premature infants. Pediatrics. 2014;134(1):e37‐e46. [DOI] [PubMed] [Google Scholar]
- 16. Peng G, de Fontnouvelle CA, Enns GM, Cowan TM, Zhao H, Scharfe C. Elevated methylmalonic acidemia (MMA) screening markers in Hispanic and preterm newborns. Mol Genet Metab. 2019;126(1):39‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giusti R, New York State Cystic Fibrosis Newborn Screening Consortium . Elevated IRT levels in African‐American infants: implications for newborn screening in an ethnically diverse population. Pediatr Pulmonol. 2008;43(7):638‐641. [DOI] [PubMed] [Google Scholar]
- 18. Cheillan D, Vercherat M, Chevalier‐Porst F, Charcosset M, Rolland MO, Dorche C. False‐positive results in neonatal screening for cystic fibrosis based on a three‐stage protocol (IRT/DNA/IRT): should we adjust IRT cut‐off to ethnic origin? J Inherit Metab Dis. 2005;28(6):813‐818. [DOI] [PubMed] [Google Scholar]
- 19. Dluholucky S, Knapkova M. The first results of extended newborn screening in Slovakia—differences between the majority and the Roma ethnic group. Int J Neonatal Screen. 2017;3:25. [Google Scholar]
- 20. Peters C, Brooke I, Heales S, et al. Defining the newborn blood spot screening reference interval for TSH: impact of ethnicity. J Clin Endocrinol Metab. 2016;101(9):3445‐3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. American College of Medical Genetics Newborn Screening Expert Group . Newborn screening: toward a uniform screening panel and system—executive summary. Pediatrics. 2006;117(5 Pt 2):S296‐S307. [DOI] [PubMed] [Google Scholar]
- 22. Patel RR, Steer P, Doyle P, Little MP, Elliott P. Does gestation vary by ethnic group? A London‐based study of over 122,000 pregnancies with spontaneous onset of labour. Int J Epidemiol. 2004;33(1):107‐113. [DOI] [PubMed] [Google Scholar]
- 23. Fulda KG, Kurian AK, Balyakina E, Moerbe MM. Paternal race/ethnicity and very low birth weight. BMC Pregnancy Childbirth. 2014;14:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Slaughter JL, Meinzen‐Derr J, Rose SR, et al. The effects of gestational age and birth weight on false‐positive newborn‐screening rates. Pediatrics. 2010;126(5):910‐916. [DOI] [PubMed] [Google Scholar]
- 25. Spong CY. Defining "term" pregnancy: recommendations from the defining “term” pregnancy workgroup. JAMA. 2013;309(23):2445‐2446. [DOI] [PubMed] [Google Scholar]
- 26. Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A. Birth weight and subsequent risk of type 2 diabetes: a meta‐analysis. Am J Epidemiol. 2007;165(8):849‐857. [DOI] [PubMed] [Google Scholar]
- 27. Feuchtbaum L, Carter J, Dowray S, Currier RJ, Lorey F. Birth prevalence of disorders detectable through newborn screening by race/ethnicity. Genet Med. 2012;14(11):937‐945. [DOI] [PubMed] [Google Scholar]
- 28. Newborn Screening Process . Health Resources & Service Administration. https://mchb.hrsa.gov/maternal-child-health-initiatives/newborn-screening/process. Accessed December 1, 2019.
- 29. R‐Core‐Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Core Team; 2019. https://www.R-project.org/. Accessed November 3, 2019. [Google Scholar]
- 30. Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. Hillsdale, NJ: Erlbaum; 1988. ISBN 0‐8058‐0283‐5. [Google Scholar]
- 31. Fisher RA. Mathematics of a lady tasting tea. World Mat. 1956;3:1512‐1521. [Google Scholar]
- 32. Shiono PH, Klebanoff MA, Graubard BI, Berendes HW, Rhoads GG. Birth weight among women of different ethnic groups. JAMA. 1986;255(1):48‐52. [PubMed] [Google Scholar]
- 33. Madan A, Holland S, Humbert JE, Benitz WE. Racial differences in birth weight of term infants in a northern California population. J Perinatol. 2002;22(3):230‐235. [DOI] [PubMed] [Google Scholar]
- 34. Morisaki N, Kawachi I, Oken E, Fujiwara T. Social and anthropometric factors explaining racial/ethnical differences in birth weight in the United States. Sci Rep. 2017;7:46657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin JA, Hamilton BE, Osterman MJK, Driscoll AK, Drake P. Births: final data for 2017. Natl Vital Stat Rep. 2018;67(8):1‐50. [PubMed] [Google Scholar]
- 36. Brown CC, Moore JE, Felix HC, et al. Association of State Medicaid Expansion Status with low birth weight and preterm birth. JAMA. 2019;321(16):1598‐1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bjorke Monsen AL, Ueland PM, Vollset SE, et al. Determinants of cobalamin status in newborns. Pediatrics. 2001;108(3):624‐630. [DOI] [PubMed] [Google Scholar]
- 38. Bryant AS, Worjoloh A, Caughey AB, Washington AE. Racial/ethnic disparities in obstetric outcomes and care: prevalence and determinants. Am J Obstet Gynecol. 2010;202(4):335‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Worgan LC, Niles K, Tirone JC, et al. Spectrum of mutations in mut methylmalonic acidemia and identification of a common Hispanic mutation and haplotype. Hum Mutat. 2006;27(1):31‐43. [DOI] [PubMed] [Google Scholar]
- 40. Morel CF, Lerner‐Ellis JP, Rosenblatt DS. Combined methylmalonic aciduria and homocystinuria (cblC): phenotype‐genotype correlations and ethnic‐specific observations. Mol Genet Metab. 2006;88(4):315‐321. [DOI] [PubMed] [Google Scholar]
- 41. Almannai M, Marom R, Divin K, et al. Milder clinical and biochemical phenotypes associated with the c.482G>A (p.Arg161Gln) pathogenic variant in cobalamin C disease: implications for management and screening. Mol Genet Metab. 2017;122(1–2):60‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Center for Health Statistics; 2014. https://www.cdc.gov/nchs/pressroom/states/california/california.htm. Accessed November 28, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Birth weight differences between race/ethnicity groups.
Figure S2. Newborn race/ethnicity groups in different gestational age categories.
Figure S3. Percentage of male infants in different gestational age categories.
Figure S4. Metabolic differences between race/ethnicity groups.
Figure S5. Metabolic differences between infants with a single race/ethnicity category.
Table S1. Demographics of false‐positive newborns in four metabolic diseases.
Table S2. Sample size of screen‐negatives in GA, sex, and race/ethnicity groupings.
Table S3. Difference of birth weight between male and female infants.
Table S4. Comparison of NBS disease markers between race/ethnicity groups.
Table S5. Newborn screening markers for inborn metabolic disorders.


